Abstract
Background
The development of vaccines and therapeutics has relied on healthy volunteer influenza challenge studies. A validated human infection model with wild-type A(H1N1)pdm09 was reported previously. Our objective was to characterize a wild-type influenza A/Bethesda/MM1/H3N2 challenge virus in healthy volunteers.
Methods
Participants received a single dose of a cell-based, reverse-genetics, Good Manufacturing Practices–produced wild-type influenza A(H3N2)2011 virus intranasally and were isolated at the National Institutes of Health Clinical Center for ≥9 days. Dose escalation was performed from 104 to 107 TCID50 (50% tissue culture infectious dose). Viral shedding and clinical disease were evaluated daily.
Results
Of 37 participants challenged, 16 (43%) had viral shedding and 27 (73%) developed symptoms, with 12 (32%) participants experiencing mild to moderate influenza disease (MMID), defined as shedding and symptoms. Only participants receiving 106 and 107 TCID50 experienced MMID at 44% and 40%, respectively. Symptom severity peaked on day 3, whereas most viral shedding occurred 1–2 days after challenge. Only 10 (29%) participants had a ≥4-fold rise in hemagglutination inhibition antibody titer after challenge.
Conclusions
The A/Bethesda/MM1/H3N2 challenge virus safely induced MMID in healthy volunteers, but caused less MMID than the A(H1N1)pdm09 challenge virus even at the highest dose. There was less detection of shedding though the incidence of symptoms was similar to A(H1N1)pdm09. Fewer serum anti-hemagglutinin (HA) antibody responses with less MMID indicate that preexisting immunity factors other than anti-HA antibody may limit shedding in healthy volunteers. This A/Bethesda/MM1/H3N2 challenge virus can be utilized in future studies to further explore pathogenesis and immunity and to evaluate vaccine candidates.
Clinical Trials Registration
Keywords: influenza A, H3N2, healthy volunteer, challenge
We successfully characterized an influenza A/Bethesda/MM1/H3N2 challenge virus and show that it can be administered safely to induce mild to moderate influenza disease.
Seasonal influenza causes significant morbidity and mortality each year and pandemic influenza continues to pose a worldwide threat. Government health agencies [1, 2] and private organizations [3, 4] have made developing an improved and more broadly protective universal influenza vaccine a high priority. Healthy volunteer human challenge models of wild-type influenza will play a major role in this development process [1, 2]. Improvement of these models and development of new models are needed so that studies evaluating novel vaccines can be more efficient but can also inform novel vaccine design through comprehensive evaluation of influenza pathogenesis and immunity in humans. For example, validation of a human infection model with a 2009 (H1N1)pdm virus has helped to characterize influenza A disease and identify correlates of protection demonstrating the differential predictive value of antibodies to the hemagglutinin (HA) head and stem regions, as well as the neuraminidase (NA) [5–7]. Data such as these are valuable to further understand immune responses to influenza and for future development of improved influenza vaccines.
Expansion of human influenza challenge models through the development of new challenge strains is critical to advancing this work as influenza viruses are a complex set of types, subtypes, and strains that will ultimately require broadly protective vaccines to prevent and mitigate disease. The rapid evolution of influenza viruses and the unpredictable emergence of novel pandemics require careful consideration when evaluating broadly protective vaccines. In addition, the complex and ever-changing immunity and prior exposure history of people in different age groups, along with the still poorly understood correlates of immunity, make comprehensive study in human models essential. Ultimately, breadth of protection of novel vaccines will need to be evaluated, and the availability of a diverse set of challenge models may be the only way to do this before another novel seasonal or pandemic strain emerges.
This report describes the first seasonal influenza A(H3N2) virus challenge model using a mammalian cell–grown, reverse genetics–produced human challenge virus administered under a US Food and Drug Administration (FDA) Investigational New Drug (IND) application in the United States. Previous H3N2 challenge studies have been performed with egg-grown viruses. Many of these studies were completed decades ago and with limited characterization of the challenge virus in the setting of evaluating vaccines and therapeutics showing highly variable results [8–20]. More recent dose-finding approaches to challenge studies have been taken with more consistent results. In a previous study we demonstrated, with a consistent approach to virus production, dosing, administration, and clearly defined endpoints, that a careful dose-finding study of an (H1N1)pdm challenge virus could be performed to produce a validated model [5]. Using this experience, we sought to characterize a seasonal wild-type H3N2 challenge virus in a dose escalation study. We also sought to understand some of the factors, in particular preexisting immune factors that may affect viral performance and disease in healthy volunteers, which are key factors to consider when designing challenge trials to study human pathogenesis or evaluate novel vaccines.
MATERIALS AND METHODS
Clinical Study
A dose escalation trial was performed at the National Institutes of Health (NIH) Clinical Center. The primary objective was to determine the dose of influenza A/Bethesda/MM1/H3N2 challenge virus that induced mild to moderate influenza disease (MMID) in ≥60% of healthy volunteers. MMID was defined as at least 1 day of viral shedding by clinical molecular testing and at least 1 symptom of influenza disease [5]. Participants were inoculated intranasally using the MAD Nasal, an intranasal mucosal atomization device (Teleflex, Morrisville, North Carolina) attached to a 1-mL syringe at doses ranging from 104 to 107 TCID50 (50% tissue culture infectious dose). These doses were administered to cohorts in a stepwise fashion based on the outcome of the previous dose until ≥60% MMID was achieved or the maximum dose of 107 TCID50 (Supplementary Figure 1).
Isolation, evaluation, and testing were performed as previously described [5]. Clinical outcomes were measured through the use of clinician assessments and the Influenza Patient-Reported Outcome (FLU-PRO) tool, a standardized and validated questionnaire for evaluating influenza severity [21–24].
The study (ClinicalTrials.gov identifier NCT02594189) was performed under FDA IND number 16586, approved by the National Institute of Allergy and Infectious Diseases Institutional Review Board, and conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines. All participants gave written informed consent prior to enrollment.
Study Participants
Healthy volunteers between the ages of 18 and 50 were recruited and enrolled between December 2015 and July 2017. Volunteers were nonsmokers, healthy, had not received seasonal influenza vaccine within 1 year of challenge, and were screened for a serum hemagglutination inhibition (HAI) antibody titer of <1:40 within 60 days of enrollment.
Challenge Virus
An investigational influenza A challenge virus (A/Bethesda/MM1/H3N2) was manufactured under Good Manufacturing Practices (GMP) from a seed stock made by reverse genetics using plasmids encoding the gene segments of a wild-type seasonal H3N2 strain A/Bethesda/NIH12/2011 (GenBank accession numbers KR137643–K137650) using methods previously described [5]. This wild-type strain was collected from a naturally infected patient in Bethesda, Maryland, and is representative of the H3N2 viruses circulating during the 2011–2012 influenza season. It is most antigenically similar to the A/Victoria/361/2011 vaccine strain included in the 2012–2013 influenza vaccine. After manufacturing, the challenge virus was sequenced identifying 3 nucleic acid changes, 1 each in PB1, PB2, and NA. All 3 were nonsynonymous changes leading to amino acid change T566I in PB1, R144K in PB2, and I148T in NA (Table 1). The GMP-produced challenge virus was characterized in both Balb/C mice and ferrets demonstrating no significant phenotypic differences from A/Bethesda/NIH12/2011 (Supplementary Figures 2–6).
Table 1.
Genomic Changes Found in the Influenza A/Bethesda/MM1/H3N2 Human Challenge Virus (Lot Number 001A) Compared to Wild-type
| Gene Segment | No. of Nucleic Acid Changes | No. of Amino Acid Changes | Nucleic Acid Change Position | Corresponding Amino Acid Change Position |
|---|---|---|---|---|
| Segment 1: PB2 | 1 | 1 | G431A | R144K |
| Segment 2: PB1 | 1 | 1 | C1701T | T566I |
| Segment 3: PA | 0 | 0 | N/A | N/A |
| Segment 4: HA | 0 | 0 | N/A | N/A |
| Segment 5: NP | 0 | 0 | N/A | N/A |
| Segment 6: NA | 1 | 1 | C443T | I148T |
| Segment 7: M | 0 | 0 | N/A | N/A |
| Segment 8: NS | 0 | 0 | N/A | N/A |
Abbreviations: HA, hemagglutinin; M, matrix; N/A, not applicable; NA, neuraminidase; NP, nucleocapsid protein; NS, nonstructural; PA, polymerase acid; PB1, polymerase PB1; PB2, polymerase PBS.
Immunologic and Virologic Assays
Serum HAI and neuraminidase inhibition (NAI) antibody titers were measured against the challenge virus using standard methods [25–27]. HAI assays were performed with fresh turkey red blood cells. Nasal washes were analyzed for the presence of respiratory pathogens daily using the FilmArray Respiratory Panel (BioFire Diagnostics, Salt Lake City, Utah) [28].
Statistical Methods
Geometric mean titers with 95% confidence intervals were calculated to compare HAI and NAI titers. Fisher’s exact test was used to compare MMID among the dosing groups, with P < .05 considered to be significant. Statistical analyses were performed using GraphPad Prism 7 software (GraphPad, La Jolla, California).
RESULTS
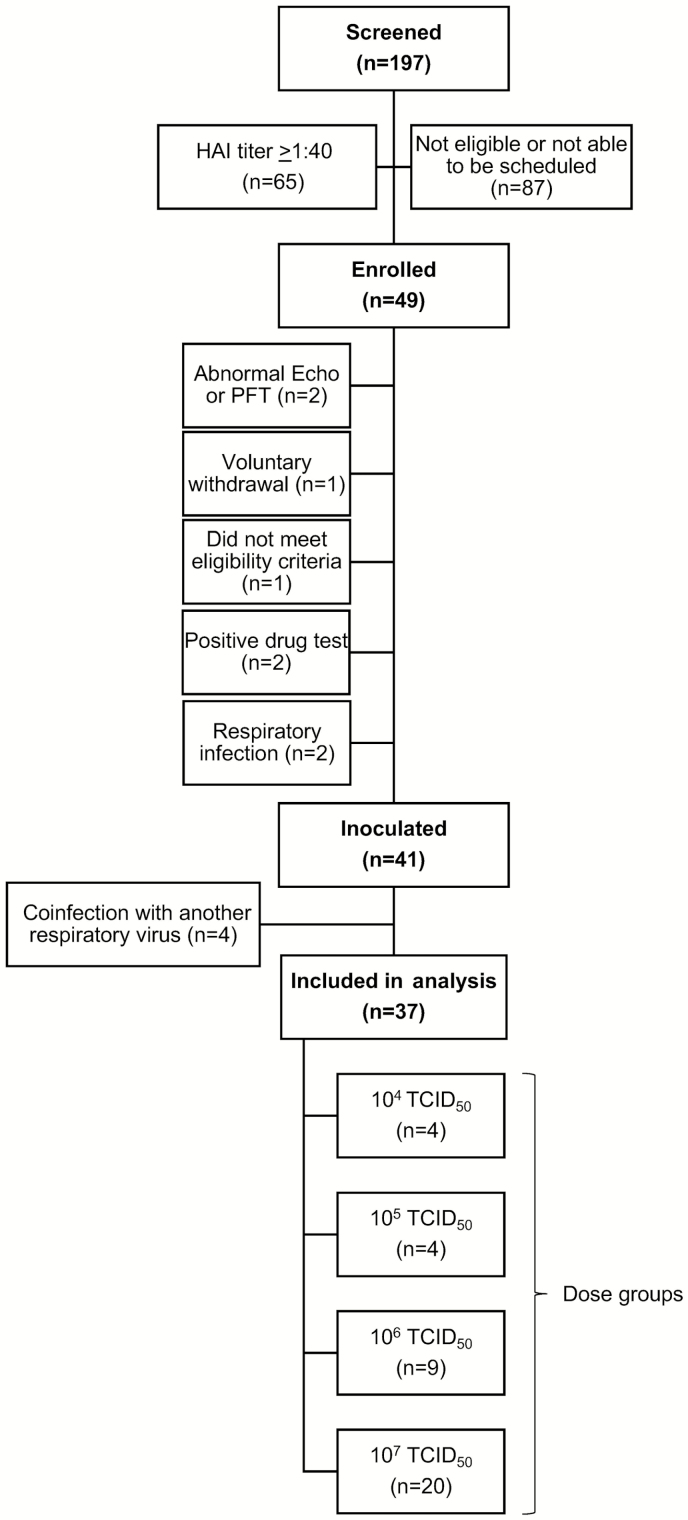
A total of 49 participants were enrolled. Of these, 41 were challenged with influenza, then 4 were excluded for a positive respiratory viral panel for a pathogen other than influenza A(H3N2) after inoculation (Figure 1). Therefore, 37 participants were included in the analysis.
Figure 1.
Study enrollment. One hundred ninety-seven participants were screened, with 65 (33%) having hemagglutination inhibition titers ≥1:40, making them ineligible for the study. A total of 49 participants were enrolled. Eight were excluded prior to challenge. Forty-one participants were inoculated with 1 of 4 doses of influenza A(H3N2). Four were excluded due to infection with another respiratory virus, resulting in a total of 37 participants included in the analysis. Abbreviations: echo, echocardiogram; HAI, hemagglutination inhibition; PFT, pulmonary function test; TCID50, 50% tissue culture infectious dose.
The median participant age was 31 years (range, 23–47 years); 51% of participants were female. Fifteen (41%) participants were white, 14 (38%) black, and 4 (11%) multiple races; 4 (11%) did not report race. Eight (22%) of the participants were Hispanic or Latino.
After challenge, 16 (43%) participants demonstrated viral shedding and 27 (73%) developed influenza symptoms, with 12 (32%) experiencing MMID (Table 2). Only participants receiving 106 or 107 TCID50 experienced viral shedding and MMID. These higher dose groups demonstrated a similar MMID of 44% and 40%, respectively (Table 2). Neither was statistically different from the lowest doses given (P = .262 and P = .228, respectively).
Table 2.
Clinical Outcomes of Participants Receiving the Influenza A(H3N2) Challenge Virus
| Dose | Total | Symptoms | Shedding | MMID |
|---|---|---|---|---|
| 104 TCID50 | 4 | 4 (100) | 0 | 0 |
| 105 TCID50 | 4 | 2 (50) | 0 | 0 |
| 106 TCID50 | 9 | 6 (67) | 5 (56) | 4 (44) |
| 107 TCID50 | 20 | 15 (75) | 11 (55) | 8 (40) |
| Total | 37 | 27 (73) | 16 (43) | 12 (32) |
Data are presented as No. (%). MMID is defined as positive clinical molecular testing AND at least 1 symptom.
Abbreviations: MMID, mild to moderate influenza disease; TCID50, 50% tissue culture infectious dose.
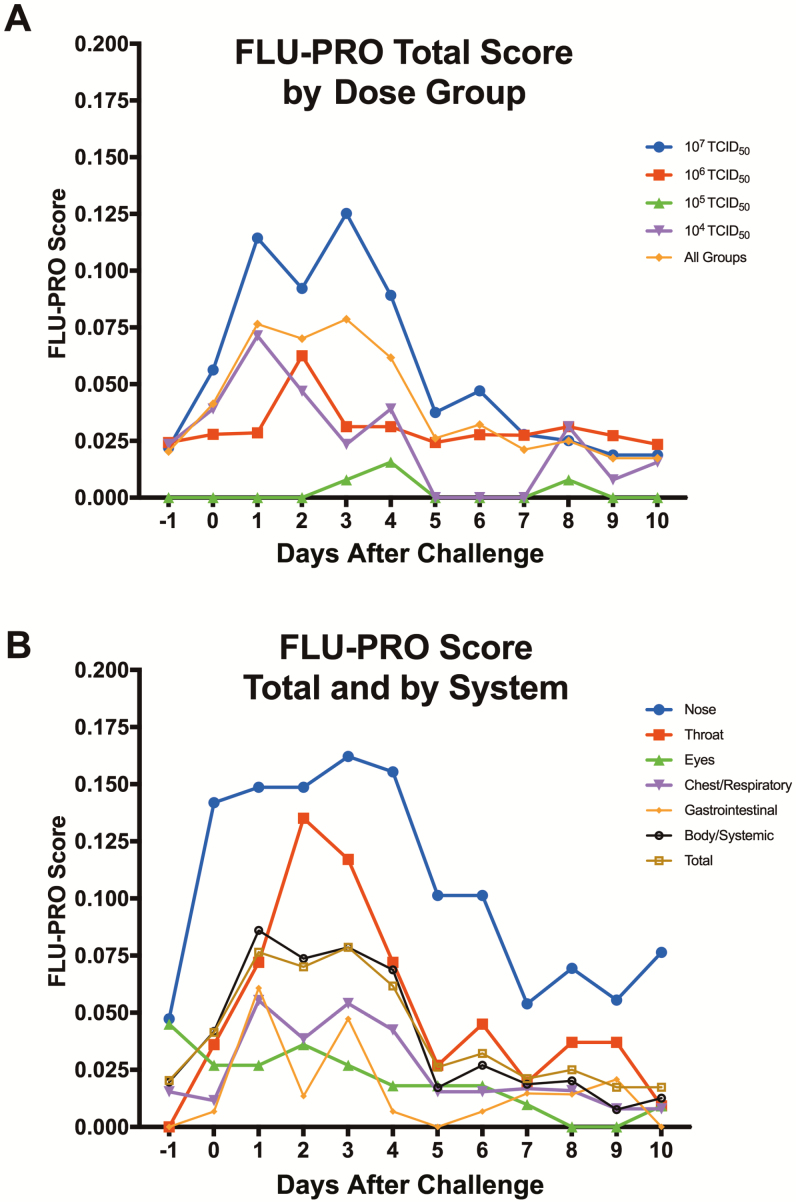
Participants in every dose group reported symptoms with a range of severity on the FLU-PRO questionnaire. The 107 TCID50 group demonstrated the highest severity, while all other dose groups demonstrated variable severity (Figure 2A). The total severity score peaked day 3 postchallenge (Figure 2B). The nose, throat, and systemic domains were the primary components of the FLU-PRO scores (Figure 2B), consistent with the clinician assessments that identified nasal/sinus congestion, rhinorrhea, sore throat, headache, and fatigue as the 5 most common symptoms, similar to the (H1N1)pdm challenge virus (Table 3).
Figure 2.
A, Average Influenza Patient-Reported Outcome (FLU-PRO) total scores reported by each dose group. Participants completed FLU-PRO questionnaires daily starting on day –1 through day 14 (only day –1 through day 10 are shown). Symptoms were reported by all dose groups, although the highest severity was reported by the highest dose group of 107 50% tissue culture infectious dose. B, Types of symptoms reported by participants using FLU-PRO. For each question, participants reported 0 (none) to 4 (most severe), with a maximum daily total score and a maximum daily domain score of 4. On average, the nose scores were most severe overall, with the peak of throat and nose scores occurring on days 2 and 3, respectively. Point estimates represent means. Abbreviations: FLU-PRO, Influenza Patient-Reported Outcome; TCID50, 50% tissue culture infectious dose.
Table 3.
Percentage of Participants With Influenza Symptoms for Both This H3N2 Dose-finding Study and for a Previously Reported H1N1 Dose-finding Challenge Study by Viral Shedding Status
| H3N2 | H1N1 [5] | |||||
|---|---|---|---|---|---|---|
| Symptoma | Viral Shedding (n = 16) | No Viral Shedding (n = 21) | All (n = 37) | Viral Shedding (n = 20) | No Viral Shedding (n = 26) | All (n = 46) |
| Arthralgia | 1 (6.3) | 0 | 1 (2.7) | 2 (10.0) | 2 (7.7) | 4 (8.7) |
| Chills | 1 (6.3) | 0 | 1 (2.7) | 6 (30.0) | 1 (3.8) | 7 (15.2) |
| Red/watery eyes | 1 (6.3) | 0 | 1 (2.7) | 5 (25.0) | 2 (7.7) | 7 (15.2) |
| Nasal/sinus congestion | 7 (43.8) | 11 (52.4) | 18 (48.6) | 15 (75.0) | 9 (34.6) | 24 (52.2) |
| Diarrhea | 3 (18.8) | 1 (4.8) | 4 (10.8) | 1 (5.0) | 1 (3.8) | 2 (4.3) |
| Dry cough | 2 (12.5) | 4 (19.0) | 6 (16.2) | 7 (35.0) | 6 (23.1) | 13 (28.3) |
| Dyspnea on exertion | 0 | 0 | 0 | 6 (30.0) | 4 (15.4) | 10 (21.7) |
| Fatigue | 7 (43.8) | 7 (33.3) | 14 (37.8) | 12 (60.0) | 7 (26.9) | 19 (41.3) |
| Fever (>38°C) | 0 | 1 (4.8) | 1 (2.7) | 2 (10.0) | 1 (3.8) | 3 (6.5) |
| Headache | 8 (50) | 6 (28.6) | 14 (37.8) | 12 (60.0) | 8 (30.8) | 20 (43.5) |
| Myalgia | 4 (25) | 0 | 4 (10.8) | 9 (45.0) | 3 (11.5) | 12 (26.1) |
| Nausea | 3 (18.8) | 1 (4.8) | 4 (10.8) | 3 (15.0) | 2 (7.7) | 5 (10.9) |
| Productive cough | 1 (6.3) | 1 (4.8) | 2 (5.4) | 3 (15.0) | 1 (3.8) | 4 (8.7) |
| Rhinorrhea | 5 (31.3) | 4 (19.0) | 9 (24.3) | 15 (75.0) | 5 (19.2) | 20 (43.5) |
| Sore throat | 8 (50) | 4 (19.0) | 12 (32.4) | 13 (65.0) | 7 (26.9) | 20 (43.5) |
| Sweats | 2 (12.5) | 0 | 2 (5.4) | 6 (30.0) | 2 (7.7) | 8 (17.4) |
Data are presented as No. (%).
aThe 5 most common symptoms for both challenge studies were nasal/sinus congestion, rhinorrhea, sore throat, headache, and fatigue.
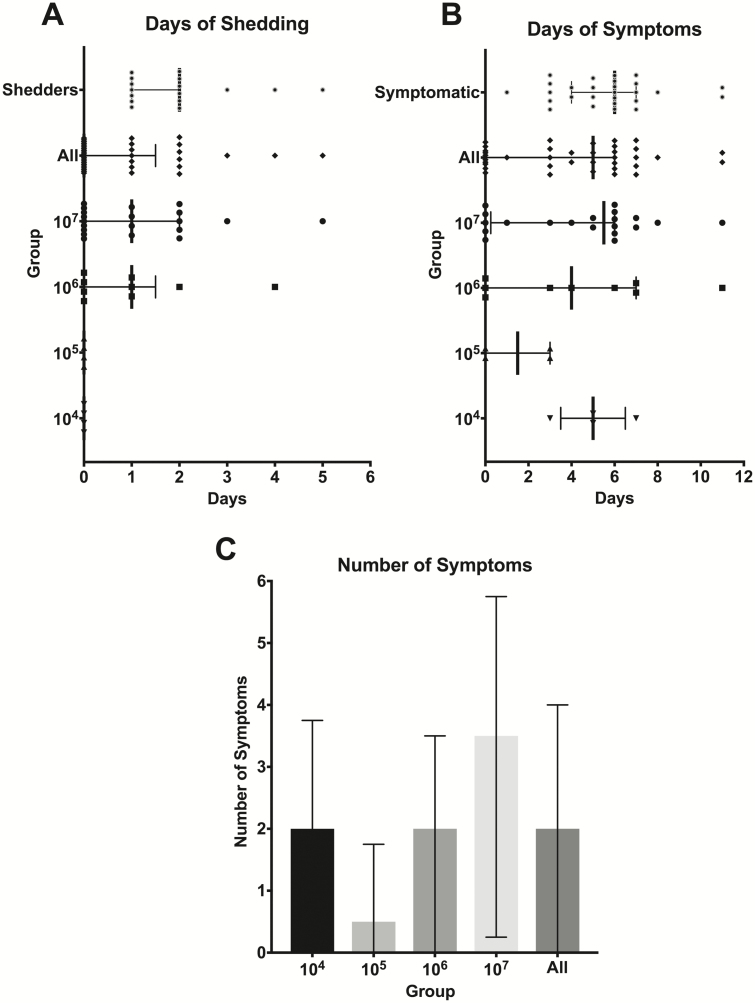
The median number of symptoms experienced by all participants was 2, with the highest median number of symptoms (3.5 symptoms) in the highest dose group, 107 TCID50 (Figure 3C). On average, participants reached their peak number of symptoms on day 3 (Figure 4C and 4D).
Figure 3.
Disease severity measures, by days of shedding and symptoms and by number of symptoms. A, Participants underwent daily nasal washes after challenge to identify the presence of influenza A(H3N2) after challenge. There was no viral shedding in the lowest 50% tissue culture infectious dose groups. The median duration overall for all shedders was 2 days and the median duration of viral shedding in the 2 highest TCID50 dose groups was 1 day. B, Study clinicians assessed clinical symptoms daily. Participants in all TCID50 dose groups experienced influenza symptoms with a median duration of 5 days for all participants. C, The highest number of symptoms occurred among the highest TCID50 dose group with a median of 3.5 symptoms over the study period. Lines represent medians with error bars representing the interquartile range. Abbreviations: TCID50, 50% tissue culture infectious dose.
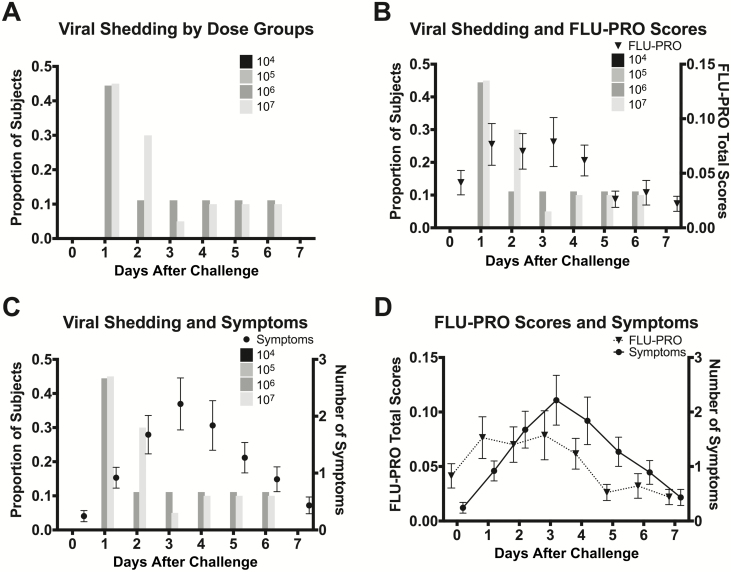
Figure 4.
Proportion of participants with viral shedding by dose group and symptom severity. A, Participants receiving 104 or 105 TCID50 did not have any viral shedding. Participants receiving 106 and 107 TCID50 had the highest proportion of shedding 1 day after challenge and no shedding by day 7. B, Participants completed daily FLU-PRO questionnaires indicating type and severity of symptoms. The maximum daily score (most severe) is 4. The peak of average symptom severity by FLU-PRO occurred on day 3, with the most severe symptoms being reported on days 1–3. C, Study clinicians evaluated symptoms daily during the inpatient stay. The number of symptoms by day ranged from 0 to 12, with the average number of symptoms peaking on day 3. D, FLU-PRO total scores and number of symptoms had similar trajectories. Circles and triangles represent means and the error bars represent standard errors of the mean. Abbreviation: FLU-PRO, Influenza Patient-Reported Outcome; TCID50, 50% tissue culture infectious dose.
The duration of symptoms varied by dose group and the overall median duration of symptomatic illness in all participants was 5 days (Figure 3B). The 2 highest dose groups experienced a median of 4 and 5.5 days of symptoms, respectively. Among only those individuals who developed clinical disease, the median duration of illness was 6 days (Figure 3B).
In participants who developed viral shedding, the viral shedding preceded significant symptoms in most cases and the proportion of those shedding was highest on days 1 and 2 postchallenge (Figure 4A). This was prior to peak number and severity of symptoms by approximately 1–2 days (Figure 4B and 4C). No viral shedding was observed in participants receiving the 2 lowest doses of 104 and 105 TCID50. Similar proportions of participants shed in the 2 highest dosing groups, 56% in the 106 TCID50 group and 55% in the 107 TCID50 group. Both dosing groups included individuals with asymptomatic shedding (Table 2). The median duration of shedding in the 2 highest dose groups was 1 day and the median duration of shedding in those who had any shedding was 2 days (Figure 3A).
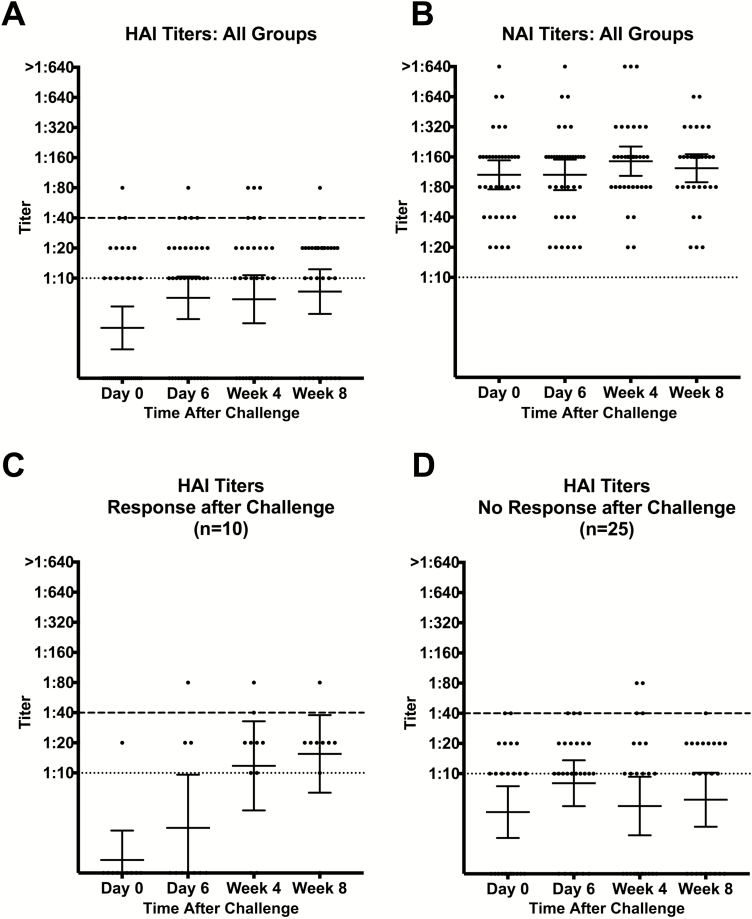
At the time of challenge, all but 3 participants had HAI titers <1:40, 2 participants had titers of 1:40, and 1 had a titer of 1:80. Two (5%) participants were lost to follow-up, so week 4 and week 8 HAI and NAI titers could not be obtained. After challenge, only 10 (29%) of the remaining participants had a ≥4-fold increase in HAI titer while 25 (71%) did not. Of those with an HAI titer response, none occurred in the lowest 104 TCID50 group; higher dose groups had a 25%–37.5% response. There was a statistically significant rise in geometric mean HAI titer from day 0 to week 8, but overall titers were quite low (Figure 5A). When broken down into who did and did not respond with an increase in HAI titer, there was a statistically significant rise in HAI titer in the responder participants by week 8, while the other 71% of participants demonstrated no significant increase in HAI titer (Figure 5C and 5D). There was no significant difference in symptoms or shedding between the HAI responder and nonresponder participants.
Figure 5.
Hemagglutination inhibition (HAI) and neuraminidase inhibition (NAI) titers. HAI and NAI titers were evaluated pre- and postchallenge and in follow-up. A, There was an overall increase in HAI titers among all participants by week 8 compared to day 0 (prechallenge). B, NAI titers were elevated even prior to challenge and remained elevated throughout the study period. C, Participants were grouped by response after challenge. Those participants with a response after challenge had a ≥4-fold increase in HAI titer after challenge and showed a significant rise by week 8. D, Many participants did not have a rise in HAI titer after challenge, and HAI titers remained low throughout the study period. Dotted lines indicate the lowest level of detection. Dashed lines indicate the level of protection (≥1:40). Lines represent geometric mean titers and error bars represent 95% confidence intervals. Abbreviations: HAI, hemagglutination inhibition; NAI, neuraminidase inhibition.
Baseline NAI titers were much higher as participants were not screened by NAI titer prior to enrollment. Three (9%) participants had a ≥4-fold or higher increase in NAI titer; however, the remaining participants maintained similarly high NAI titers from baseline through the convalescent timepoints (Figure 5B).
DISCUSSION
The A/Bethesda/MM1/H3N2 challenge virus at 107 TCID50 was effective in inducing MMID in 40% of healthy volunteers screened for a titer of <1:40 in a human challenge study. The study was performed safely and influenza illness was induced, demonstrating that this mammalian cell–grown, reverse genetics–produced virus at 107 TCID50 can be used to model seasonal H3N2 influenza infection. However, this model was not as effective at inducing MMID as our previously published A(H1N1)pdm model that induced MMID in >70% of participants with an HAI titer of <1:40 at the same 107 TCID50 [5]. The difference in these results was due to less detectable shedding and more asymptomatic shedding in this H3N2 model.
Clinical symptoms occurred in the majority of participants given the highest dose of the H3N2 challenge virus (75%) (Table 2), slightly less than what was observed in the (H1N1)pdm model (85%) [5]. The duration of clinical disease was also shorter in this H3N2 model. Symptoms lasted for a median of 5 days (Figure 3B) and the median duration of detectable shedding in those who shed virus was only 2 days (Figure 3A) as compared to 8 days of symptoms and 4 days of shedding in the (H1N1)pdm model [5].
Despite these differences, there were similarities between the models. A much larger number of individuals developed symptoms than shedding, and the primary symptoms observed were very similar to the primary influenza symptoms observed in the (H1N1)pdm model (Table 3). These symptoms were typical influenza symptoms of headache, fatigue, nasal and sinus congestion, rhinorrhea, and sore throat. Also of note was the timing of illness. Similar to the (H1N1)pdm model, more shedding occurred before the peak of symptom severity, suggesting that these individuals would likely be contagious before they were clinically symptomatic (Figure 4B and 4C). In addition, these data, in conjunction with the data from the (H1N1)pdm model, demonstrate that influenza infections are likely underrecognized as shedding is often undetectable once individuals are suffering from significant symptoms that would lead them to seek medical attention.
This range of symptom severity and duration of illness noted in the H3N2 and (H1N1)pdm models was accompanied by variability in the immune response. The greatest variability was seen in antibody responses against the HA head as measured by HAI assay. A clear delineation of 2 groups of individuals, those who responded after challenge with an increase in HAI and those who did not, was again observed in this H3N2 model, similar to the (H1N1)pdm model [6]. More than half of the participants demonstrated no increase in titer, suggesting that by screening for individuals with low titers against seasonal H3N2 influenza, we may be selecting for individuals who do not develop antibody titers against HA despite years of exposure to seasonal H3N2 and vaccines. This indicates that a percentage of individuals with low HAI titers who are considered unprotected may not develop antibody responses to current or future HA-based vaccines and could be a factor in vaccine failures. This study, along with our previous challenge studies with (H1N1)pdm, suggests that this phenomenon is prevalent in both H1N1 and H3N2 infections. In addition, these individuals may have other prechallenge protective immune correlates that were not assessed.
Antibodies against the NA were less variable, and overall NA titers were much higher at baseline. Unlike the (H1N1)pdm model, no participants had undetectable titers against H3N2 NA at baseline. Similar to the (H1N1)pdm model, some individuals demonstrated an increase in titer, but there may be a threshold limiting the overall rise in titer that could be achieved after infection if at baseline the levels are already elevated. Most participants, including those with and without an HAI response after challenge, had an elevated NAI titer at baseline, indicating that even those without a serum HAI response can develop a strong serum antibody response to the NA. This experience, now demonstrated in both H1N1 and H3N2 models, is important to note as influenza surveillance is based on screening for anti-HA antibodies. This suggests that serosurveys are likely lacking and measurements of NA immunity should be considered in the future to obtain a broader understanding of serologic influenza immune correlates and exposure in the population.
Although we demonstrated that the A/Bethesda/MM1/H3N2 challenge virus can be used to model seasonal influenza infection, there are some significant limitations as compared to the A(H1N1)pdm model. The biggest limitation was that even at the highest dose of 107 TCID50, only 40% MMID was achieved and the clinical disease was more limited by all severity measures than in the (H1N1)pdm model. While this does not preclude using the H3N2 challenge virus in future phase 2 efficacy trials, this will need to be considered in the trial design and would likely require larger trials. However, smaller studies using this model can be instructive in understanding influenza immunity and correlates of protection.
Although the challenge virus stock was determined to have 3 nonsynonymous mutations as compared to the wild-type virus (Table 1), we do not believe these mutations played a significant role in influencing any of the outcomes. The PB2 R114K and PB1 T566I mutations were both mapped to experimentally determined T-cell epitopes [29] and have been observed in other human H3N2 sequences. An analysis of 2029 unique N2 protein sequences in the National Center for Biotechnology Information Influenza Virus Resource [30] demonstrated that 94% of N2 protein sequences from 1968 to 2014 encode T148, so the I148T NA mutation observed in this challenge virus appears to be a reversion to the predominant amino acid encoded at this site that has also been mapped to an antigenic epitope [31]. These observations, in conjunction with the preclinical animal data demonstrating no difference in phenotype, give us confidence that the virus was representative of a wild-type 2011 seasonal H3N2 virus.
These data suggest that preexisting immune factors other than anti-HA antibody likely played a role in protection against challenge with this H3N2 influenza A virus. Almost all of the participants in this study had what would be considered low, nonprotective HAI titers at baseline, and yet >60% did not demonstrate MMID, and both viral shedding and clinical symptoms/severity were short-lived in many. These important observations indicate that there are correlates of protection other than anti-HA antibody titer acting to greatly minimize disease in many of these individuals. These participants likely had previous experience with antigenically related strains of this seasonal virus as H3N2 has been the predominant virus identified in most years since the 2009 pandemic [32–36]; however, these individuals had low HAI titers. Further, the high baseline NAI titers may be very important indicators of other immune factors present offering protection. Significant questions remain that should be addressed in future challenge studies with this virus and other influenza viruses regarding what aspects of immunity are the most protective, including antibodies against the NA, T-cell immunity, mucosal immunity, and other factors that are potentially playing a very important role in protection.
CONCLUSIONS
This is the first mammalian cell–grown, reverse genetics–produced wild-type seasonal H3N2 challenge virus model in humans. The results reported here demonstrate that this model is safe and can induce MMID in healthy volunteers, and the challenge model can be used successfully to study pathogenesis and immunity. The model was not as effective as the (H1N1)pdm model in that it induced less severe disease. This limited disease in those with low HAI titer suggests that other aspects of immunity beyond serum anti-HA antibody titer are likely playing important roles in protection, and these immune factors and correlates of protection should be explored further. This further exploration will ultimately be the key to reaching our goals of developing universal, broadly protective influenza vaccines.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors acknowledge Dr H. Clifford Lane, Dr Richard T. Davey, the National Institutes of Health (NIH) Clinical Center Special Clinical Studies Unit staff, and the Department of Laboratory Medicine for their support of the clinical protocol.
Financial support. This research was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Erbelding EJ, Post DJ, Stemmy EJ, et al. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 2018; 218:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Paules CI, Marston HD, Eisinger RW, Baltimore D, Fauci AS. The pathway to a universal influenza vaccine. Immunity 2017; 47:599–603. [DOI] [PubMed] [Google Scholar]
- 3. McCoy D, Kembhavi G, Patel J, Luintel A. The Bill & Melinda Gates Foundation’s grant-making programme for global health. Lancet 2009; 373:1645–53. [DOI] [PubMed] [Google Scholar]
- 4. Gates B. The next epidemic is coming. Here’s how we can make sure we’re ready. Boston, MA: Shattuck Lecture, 2018. [Google Scholar]
- 5. Memoli MJ, Czajkowski L, Reed S, et al. Validation of the wild-type influenza A human challenge model H1N1pdMIST: an A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis 2015; 60:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Memoli MJ, Shaw PA, Han A, et al. Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio 2016; 7:e00417–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park JK, Han A, Czajkowski L, et al. Evaluation of preexisting anti-hemagglutinin stalk antibody as a correlate of protection in a healthy volunteer challenge with influenza A/H1N1pdm virus. mBio 2018; 9. doi:10.1128/mBio.02284-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Nakib W, Higgins PG, Willman J, et al. Prevention and treatment of experimental influenza A virus infection in volunteers with a new antiviral ICI 130,685. J Antimicrob Chemother 1986; 18:119–29. [DOI] [PubMed] [Google Scholar]
- 9. Cate TR, Couch RB. Live influenza A/Victoria/75 (H3N2) virus vaccines: reactogenicity, immunogenicity, and protection against wild-type virus challenge. Infect Immun 1982; 38:141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clements ML, Betts RF, Murphy BR. Advantage of live attenuated cold-adapted influenza A virus over inactivated vaccine for A/Washington/80 (H3N2) wild-type virus infection. Lancet 1984; 1:705–8. [DOI] [PubMed] [Google Scholar]
- 11. Clements ML, Sears SD, Christina K, Murphy BR, Snyder MH. Comparison of the virologic and immunologic responses of volunteers to live avian-human influenza A H3N2 reassortant virus vaccines derived from two different avian influenza virus donors. J Clin Microbiol 1989; 27:219–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clements ML, Subbarao EK, Fries LF, Karron RA, London WT, Murphy BR. Use of single-gene reassortant viruses to study the role of avian influenza A virus genes in attenuation of wild-type human influenza A virus for squirrel monkeys and adult human volunteers. J Clin Microbiol 1992; 30:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murphy BR, Chanock RM, Clements ML, et al. Evaluation of A/Alaska/6/77 (H3N2) cold-adapted recombinant viruses derived from A/Ann Arbor/6/60 cold-adapted donor virus in adult seronegative volunteers. Infect Immun 1981; 32:693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reuman PD, Bernstein DI, Keefer MC, Young EC, Sherwood JR, Schiff GM. Efficacy and safety of low dosage amantadine hydrochloride as prophylaxis for influenza A. Antiviral Res 1989; 11:27–40. [DOI] [PubMed] [Google Scholar]
- 15. Reuman PD, Bernstein DI, Keely SP, Young EC, Sherwood JR, Schiff GM. Differential effect of amantadine hydrochloride on the systemic and local immune response to influenza A. J Med Virol 1989; 27:137–41. [DOI] [PubMed] [Google Scholar]
- 16. Sears SD, Clements ML, Betts RF, Maassab HF, Murphy BR, Snyder MH. Comparison of live, attenuated H1N1 and H3N2 cold-adapted and avian-human influenza A reassortant viruses and inactivated virus vaccine in adults. J Infect Dis 1988; 158:1209–19. [DOI] [PubMed] [Google Scholar]
- 17. Snyder MH, Betts RF, DeBorde D, et al. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol 1988; 62:488–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snyder MH, Clements ML, Betts RF, et al. Evaluation of live avian-human reassortant influenza A H3N2 and H1N1 virus vaccines in seronegative adult volunteers. J Clin Microbiol 1986; 23:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Treanor JJ, Kotloff K, Betts RF, et al. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine 1999; 18:899–906. [DOI] [PubMed] [Google Scholar]
- 20. Treanor JJ, Roth FK, Betts RF. Use of live cold-adapted influenza A H1N1 and H3N2 virus vaccines in seropositive adults. J Clin Microbiol 1990; 28: 596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han A, Poon JL, Powers JH 3rd, Leidy NK, Yu R, Memoli MJ. Using the Influenza Patient-Reported Outcome (FLU-PRO) diary to evaluate symptoms of influenza viral infection in a healthy human challenge model. BMC Infect Dis 2018; 18:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Powers JH 3rd, Bacci ED, Guerrero ML, et al. Reliability, validity, and responsiveness of Influenza Patient-Reported Outcome (FLU-PRO) scores in influenza-positive patients. Value Health 2018; 21:210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Powers JH 3rd, Bacci ED, Leidy NK, et al. Performance of the Influenza Patient-Reported Outcome (FLU-PRO) diary in patients with influenza-like illness (ILI). PLoS One 2018; 13:e0194180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Powers JH, Guerrero ML, Leidy NK, et al. Development of the FLU-PRO: a patient-reported outcome (PRO) instrument to evaluate symptoms of influenza. BMC Infect Dis 2016; 16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull 1979; 35:69–75. [DOI] [PubMed] [Google Scholar]
- 26. Wan H, Gao J, Xu K, et al. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 2013; 87:9290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cottey R, Rowe CA, Bender BS. Influenza virus. Curr Protoc Immunol 2001; Chapter 19: Unit 19.11. [DOI] [PubMed] [Google Scholar]
- 28. Poritz MA, Blaschke AJ, Byington CL, et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 2011; 6:e26047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan PT, Heiny AT, Miotto O, et al. Conservation and diversity of influenza A H1N1 HLA-restricted T cell epitope candidates for epitope-based vaccines. PLoS One 2010; 5:e8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bao Y, Bolotov P, Dernovoy D, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol 2008; 82:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Venkatramani L, Bochkareva E, Lee JT, et al. An epidemiologically significant epitope of a 1998 human influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. J Mol Biol 2006; 356:651–63. [DOI] [PubMed] [Google Scholar]
- 32. Appiah GD, Blanton L, D’Mello T, et al. ; Centers for Disease Control and Prevention (CDC) Influenza activity—United States, 2014-15 season and composition of the 2015-16 influenza vaccine. MMWR Morb Mortal Wkly Rep 2015; 64:583–90. [PMC free article] [PubMed] [Google Scholar]
- 33. Blanton L, Alabi N, Mustaquim D, et al. Update: influenza activity in the United States during the 2016-17 season and composition of the 2017-18 influenza vaccine. MMWR Morb Mortal Wkly Rep 2017; 66:668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davlin SL, Blanton L, Kniss K, et al. Influenza activity—United States, 2015-16 season and composition of the 2016-17 influenza vaccine. MMWR Morb Mortal Wkly Rep 2016; 65:567–75. [DOI] [PubMed] [Google Scholar]
- 35. Epperson S, Blanton L, Kniss K, et al. ; Influenza Division, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention Influenza activity—United States, 2013-14 season and composition of the 2014-15 influenza vaccines. MMWR Morb Mortal Wkly Rep 2014; 63:483–90. [PMC free article] [PubMed] [Google Scholar]
- 36. Garten R, Blanton L, Elal AIA, et al. Update: influenza activity in the United States during the 2017-18 season and composition of the 2018-19 influenza vaccine. MMWR Morb Mortal Wkly Rep 2018; 67:634–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.