Abstract
Background
This study analyzed the effect of silicon (Si) application on the occurrence of ginseng black spot caused by Alternaria panax. We explored the differences in soil physical and chemical factors and microbial community structure following Si application as well as the key factors that affected the occurrence of ginseng black spot in soil. Potted Panax ginseng plants were used to assess the effect of Si treatment on ginseng black spot. Soil physical and chemical properties were comprehensively analyzed. Bacterial communities were analyzed using Illumina HiSeq sequencing targeting the 16S rRNA gene.
Results
After inoculation with A. panax, the morbidity (and morbidity index) of ginseng with and without Si was 52% (46) and 83% (77), respectively. Soil physical and chemical analysis showed that under the ginseng black spot inoculation, bacterial communities were mainly affected by pH and available potassium, followed by ammonium nitrogen and available Si. NMDS and PLS-DA analyses and the heat maps of relative abundance revealed that Si application elevated the resistance of ginseng black spot as regulated by the abundance and diversity of bacterial flora in rhizosphere soils. Heatmap analysis at the genus level revealed that A. panax + Si inoculations significantly increased the soil community abundance of Sandaracinus, Polycyclovorans, Hirschia, Haliangium, Nitrospira, Saccharothrix, Aeromicrobium, Luteimonas, and Rubellimicrobium and led to a bacterial community structure with relative abundances that were significantly similar to that of untreated soil.
Conclusions
Short-term Si application also significantly regulated the structural impact on soil microorganisms caused by ginseng black spot. Our findings indicated that Si applications may possibly be used in the prevention and treatment of ginseng black spot.
Keywords: Alternaria panax, Silicon, Ginseng black spot, Soil bacterial community, Panax ginseng, Illumina HiSeq sequencing
Background
Ginseng black spot, caused by Alternaria panax Whetz, is a common soil-borne disease and one of the most serious diseases affecting the above-ground parts, especially the leaves, of Panax ginseng. This pathogen is distributed widely in the Changbai Mountains of China and other ginseng production regions, and accounts for more than 20—30% of the annual incidence, which is very common in cultivated and wild ginseng. Alternaria panax infestation may lead to 10—20% yield loss of the total crop. Infection first appears as elongated reddish to dark brown crevices in the infected areas. In seedlings, the stems are gradually girdled and thus collapse, resulted in damping-off [1]. In older plants, foliar infections appear later in summer, characterized by rapidly enlarging dark brown necrotic spots (circular, ellipsoid, or wedge-shaped) surrounded by chlorotic margins.
Silicon (Si) has been demonstrated to play an important role in enhancing plant resistance to disease. Si deposition has been suggested to create a physical barrier along cell walls and prevent fungal penetration into the plant [2]. Additional studies have indicated that Si is related to plant-pathogen interactions for the control in diseases in different plant species [3], and aids in the enhancement of plant resistance against disease caused by viruses, fungi, bacteria, and nematodes. Recently, it was suggested that the deposition of Si in the apoplast may prevent fungal effectors from entering the target cells, thus altering the development of the pathogens [4]. Another recently study showed that Si treatment conferred an effective protection of soybean plants against Phytophthora sojae in a hydroponic experiment [5]. Agricultural soil productivity largely depends on microbial diversity and community composition, which significantly affects plant growth and crop quality [6]. The homeostasis of the soil microbial community can suppress pathogens and promote plant growth [7]. Plant—microbe interactions remodel the complex biological and ecological processes in soil, where roots are influenced by the rhizosphere [8]. Many studies have assessed the effect of Si on plant-microbe interactions and have demonstrated that Si enhances plant resistance to pathogens by activating defense reactions [9, 10]. Recently, a pot experiment demonstrated that Si addition decreased the concentrations of water-soluble and exchangeable arsenic in soil and, therefore, decreased the bioavailability of red soil arsenic in Panax notoginseng [11].
The present study, therefore, aimed to investigate if Si treatment would enhance the resistance of ginseng against A. panax. The study objectives were to evaluate the effect of Si on the prevention and treatment of ginseng black spot and to analyze the interaction between soil properties and plant growth responses. Further objectives were to determine the changes in the dynamics, i.e., the structure, composition, and abundance, of the soil microbial community in response to infection with A. panax and treatment with Si to determine the underlying factors that may influence the quantity and composition of soil bacteria.
Results
Disease index and incidence and plant weights
Figure 1 shows the phenotypic differences in leaves of P. ginseng in 9 dpi among treatment groups: Control, A, AS, and S. Significant differences were observed in the severity of A. panax infections under Si treatment (Fig. 1). As shown in Fig. 1, no effect of Si on biomass was observed compared to the Control group. The differences between Control plants and group S plants were not obvious, however, group AS plants were obviously healthier than group A plants (Fig. 1). The first symptom of leaf spots appeared soon (3 days) after post inoculation (dpi), followed by stunting and blight within a few days. As shown in Table 1, Si treatment significantly reduced the disease incidence and disease index of ginseng black spot.
Fig. 1.

The effect in different treatments of soil. Abbreviations: CK, ginseng control plants; A, plants only inoculated with A. panax; AS, plants inoculated with A. panax + Si; S, plants only inoculated with Si
Table 1.
Effect of silicon application on the disease incidence and disease index of ginseng black spot
| Treatment | Disease incidence (%) | Disease index |
|---|---|---|
| A | 83.5 ± 6.5a | 77.8 ± 7.5a |
| AS | 52.6 ± 9.7b | 46 ± 5.6b |
Abbreviations: A, A.panax inoculated ginseng; AS, Silicon inoculated in soil with A. panax infection
There was no significant difference in dry weight among non-inoculated (pathogen) plants: Control plants (1.12 ± 0.81 g) and group S plants (1.23 ± 0.59 g). However, the plant dry weight was significantly reduced in group A plants. Apparently, Si treatment resulted in significantly heavier plants (Table 2). After 9 days post-treatment, the fresh weight of group AS plants was 15% higher than that of group A plants (Table 3).
Table 2.
The fresh weight and dry weight of the ginseng after different treatments
| Treatment | fresh weight | dry weitht |
|---|---|---|
| CK | 5.53 ± 1.25a | 1.12 ± 0.81a |
| A | 4.329 ± 1.35b | 0.56 ± 0.12b |
| AS | 5.12 ± 1.36a | 0.97 ± 0.24b |
| S | 5.67 ± 1.28a | 1.23 ± 0.59a |
Table 3.
The fresh weight and dry weight of ginseng shoots and ginseng roots after different treatments
| ginseng shoots | ginseng roots | |||
|---|---|---|---|---|
| Treatment | fresh weight | dry weitht | fresh weight | root dry weight |
| CK | 2.51 ± 0.21a | 0.38 ± 0.11a | 3.02 ± 1.21b | 0.75 ± 0.12b |
| A | 1.56 ± 0.12d | 0.20 ± 0.02d | 2.57 ± 0.89d | 0.52 ± 0.11c |
| AS | 2.23 ± 0.25c | 0.26 ± 0.13c | 2.89 ± 0.98c | 0.61 ± 0.12b |
| S | 2.39 ± 0.31b | 0.35 ± 0.13b | 3.28 ± 1.02a | 0.79 ± 0.19a |
Different letters within a column indicates significant difference among treatments p < 0.05
Soil properties and plant growth responses
Soil properties are presented in Table 4. A one-way ANOVA showed that the treatments significantly recovered the soil property parameters from disease treatment (P < 0.05). (Table 4). The pH value of the Control soil samples was ~ 7.39. Compared with the Control group, the soil pH, NO3−-N, and NH4+-N were significantly reduced in Group A (P < 0.05). In contrast, the ratio of available P and available K (P < 0.05) were significantly increased. Furthermore, AS significantly increased the soil pH, and NO3−-N and NH4+-N contents (P < 0.05), and significantly reduced the ratio of available P and available K (P < 0.05) compared to the A treatment, i.e., without Si (P < 0.05). No significant differences in the above-mentioned nutrients, except available Si, was detected between group S and Control.
Table 4.
Characteristics of soils after different treatments
| Indexes | CK | A | AS | S |
|---|---|---|---|---|
| pH | 7.39 ± 0.12ab | 7.05 ± 0.24c | 7.39 ± 0.26a | 7.38 ± 0.36b |
| Available phosphorus AP (mg/kg) | 13.15 ± 2.61ab | 14.95 ± 3.25a | 12.25 ± 2.68b | 12.73 ± 2.62b |
| Available potassium AK (mg/kg) | 194.48 ± 3.26b | 217.15 ± 2.35a | 187.64 ± 3.69b | 199.06 ± 2.65b |
| Ammonium nitrogen NH4(g/kg) | 16.42 ± 2.35a | 4.58 ± 1.02c | 9.38 ± 2.36b | 14.38 ± 1.25a |
| Nitrate nitrogen NO3(g/kg) | 1.31 ± 0.25 | 1.15 ± 0.36 | 1.89 ± 0.28 | 1.97 ± 0.69 |
| Available silicon ASi (mg/kg) | 457.99 ± 20.35b | 451.78 ± 25.69b | 457.02 ± 19.89b | 492.06 ± 26.98a |
Different letters within a column indicates significant difference among treatments p < 0.05
Analysis of bacterial composition and diversity of soil bacterial community structure based on 16S rRNA gene sequencing
Bacteria-targeted regions were completely amplified by PCR and fully sequenced for all soil samples. The raw sequence libraries were screened to remove reads that originated from sequencing noise or putative chimeric sequences. Using the 12 soil samples from the different treatments, a total of 815,609 valid 16S rDNA sequences were obtained by filtering and processing according to a 97% similarity. Variation of a single soil sample ranged from 56,510 to 76,384 sequences, and the above sequences were retained for further analysis.
The effective sequence number and OTU number of each group of samples did not significantly differ between the treatment groups and the Control group, as shown in Table 5. The sequencing coverage of samples ranged from 98.5 to 98.6%. After sample diversity (alpha diversity) analysis, the indexes reflecting the abundance and diversity of microbial communities were calculated, and the results of all treatments were analyzed using a one-way ANOVA (Table 5). The coverage index of the sample library was more than 98.5%, which indicated that the sequencing results represented the real situation of the bacterial population in the sample. The microflora richness index (Chao1, ACE) and biodiversity index (Shannon, Simpson) of the samples revealed that the diversity of the bacterial populations in the soil samples was relatively high (Table 5). Further analysis revealed that at a 97% similarity level, the Shannon index and Simpson index of soil bacteria of each treatment group were not significantly different from those in the Control group.
Table 5.
The bacterial diversity indices of ginseng rhizosphere soil samples with different treatment
| Sequence number | OTU | Coverage % |
Shannon | Simpson | ACE | Chao1 | |
|---|---|---|---|---|---|---|---|
| CK | 207,437 | 7381 | 98.6 | 11.2 ± 0.2 | 0.9989 ± 0.01 | 5250.6 ± 23.6a | 5149.3 ± 123.3a |
| A | 209,093 | 7210 | 98.6 | 11.1 ± 0.3 | 0.9989 ± 0.02 | 5048.9 ± 69.7b | 5007.6 ± 369.9b |
| AS | 206,227 | 6974 | 98.5 | 11.1 ± 0.2 | 0.9988 ± 0.01 | 5138.2 ± 102.3a | 5063.6 ± 325.3b |
| S | 192,852 | 7063 | 98.5 | 11.1 ± 0.1 | 0.9988 ± 0.02 | 5141.5 ± 69.8a | 5163.2 ± 345.6a |
Different letters within a column indicates significant difference among treatments p < 0.05
Analysis of soil bacterial community structure
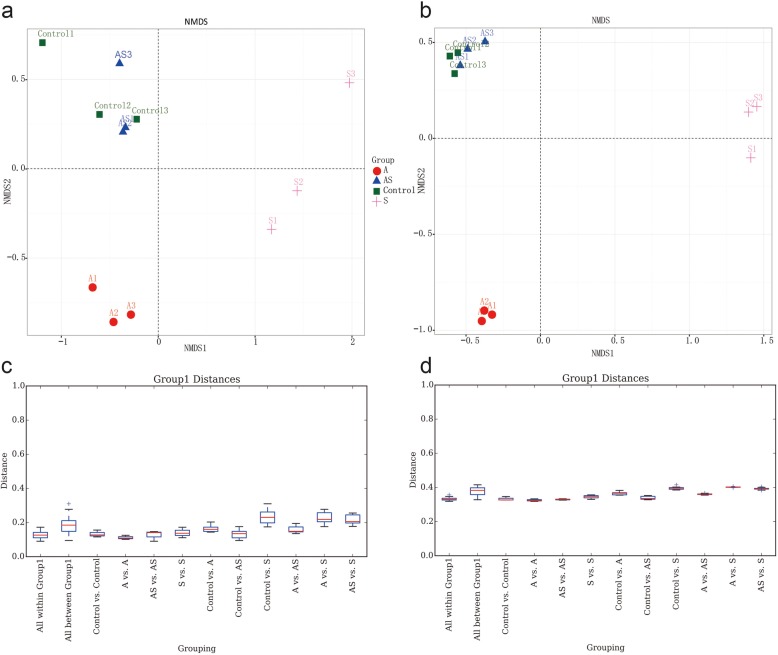
According to the abundance of bacterial OTU types in the 12 soil samples, a non-metric multidimensional scale (NMDS) diversity analysis was conducted to determine the differences in the bacterial compositions of the different samples and treatments (Fig. 2). The NMDS results were evaluated using the UniFrac distances to estimate the phylogenetic relatedness among the bacterial communities (Fig. 3a, c). The soil bacterial communities were found to be totally distinct between groups A and S, i.e., when treated with A. panax or Si (NMDS). Among treatment groups, the soil bacterial composition of group Control was most similar to that of group AS, i.e., had the highest phylogenetic relatedness, and the group AS bacterial flora could be independently distinguished from that in the infected soil (group A). However, the composition of bacterial flora in group S differed from that of the other treatments. In summary, Si application significantly regulated the changes in bacterial flora (back to the composition of Control) that were induced by inoculation of ginseng black spot (group AS).
Fig. 2.
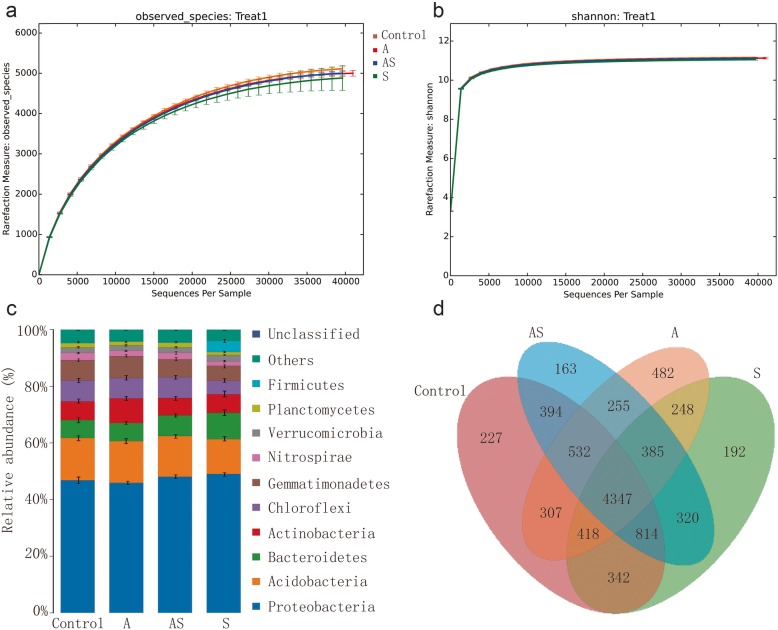
Overall analysis of bacterial communities in different treatments of soil. a The bacterial composition of in different treatments of soil at the phylum taxonomic level. b The Venn map of bacterial communities in different treatments of soil. c The bacterial composition of in different treatments of soil at the phylum taxonomic level. d The Venn map of bacterial communities in different treatments of soil
Fig. 3.
NMDS analysis of bacterial community diversity in different samples (a) Non-metric multidimensional scaling (NMDS) plots of operational taxonomic unit tables from all substrates based on abundances of bacterial community similarities using unweighted unifrac-distance of matrix (b) Unweighted weighted unifrac-distance box-line graph. c NMDS plots of operational taxonomic unit tables from all substrates based on abundances of bacterial community similarities using weighted unifrac-distance of matrix (d) Weighted unifrac-distance box-line graph
Cluster analysis of soil bacterial community structure
Based on a Beta diversity analysis, a distance matrix was obtained for the 12 soil samples, and a hierarchical clustering analysis was conducted using the unweighted group average method (UPGMA) (Fig. 3b, d). The soil samples of groups S and AS were classified as one branch, and those of groups Control and AS group were classified as one branch. The results were consistent with those of the NMDS analysis, which fully demonstrated that the soils inoculated with Si + ginseng black spot (AS group) were significantly recovered compared with the soils inoculated with only ginseng black spot (S group). PLS-DA analysis showed that the microbial composition of soil in the AS group was significantly altered following Si treatment. The results suggested similarities between group Control and AS, but not with nor among the other two groups. In summary, Si was again shown to have alleviated the changes in soil bacteria caused by ginseng black spot (Fig. 4).
Fig. 4.
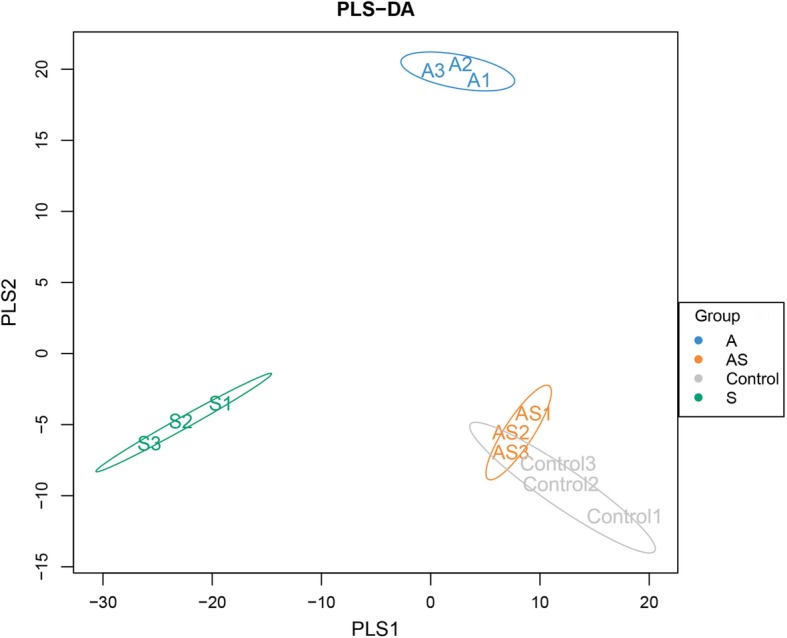
PLS-DA analysis of different soil samples
Heat map analysis of the soil bacterial community structure
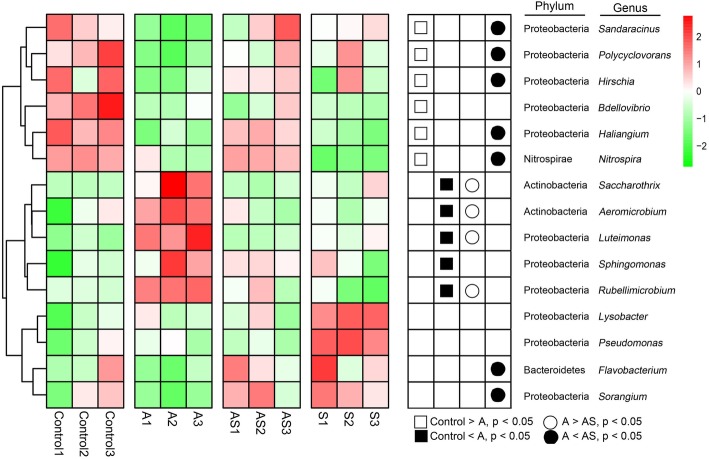
A heat map of the bacterial community structure among different samples (Fig. 5) revealed the relative abundances of the various bacterial groups (at phylum and genus levels) and that significant differences were observed among different groups of samples. The results showed that, at the phylum level, Proteobacteria, Nitrospirae, Actinobacteria, and Bacteroidetes were the four main groups (Fig. 5). The relative abundances (represented by the color depth in Fig. 5) of Sandaracinus, Polycyclovorans, Hirschia, Bdellovibrio, Haliangium, and Nitrospira were significantly higher in group Control than those of group A (P < 0.05). In addition, the relative abundances of Sandaracinus, Polycyclovorans, Hirschia, Haliangium, and Nitrospira in group AS were significantly higher than those in A (P < 0.05). The results showed that Si application significantly regulated the structural impact of soil microorganisms caused by ginseng black spot inoculation.
Fig. 5.
Heat map comparison of the dominant bacterial with average relative abundance from blue to red means relative abundance from low to high
Factors influencing the quantity and composition of soil bacteria
Correlation analysis showed (Table 6) that most of the other dominant bacterial groups had significant correlations with soil chemical properties, except Arenimonas, H16, and RB41, which showed no correlations with all chemical indicators. Haliangium and available K were significantly negatively correlated; Phenylobacterium (phenyl coli) was very significantly negatively correlated with pH and Gemmatimonas (bacillus); Nitrospira (nitrification spirillum) was negatively correlated with NO3−-N; Mesorhizobium (rhizobia) was very significantly positively correlated with NO3−-N; Gemmatimonas (bacillus), Nitrospira (nitrobacteria), and available Si were significantly negatively correlated; Lactobacillus (lactobacillus), Mesorhizobium (rhizobium), and available Si were significantly positively correlated. Haliangium was significantly positively correlated with pH.
Table 6.
Pearson’s correlation coefficients between various physicochemical variables and the relative abundances of main genera (> 1%) across all samples
| pH | AP | AK | NH4+-N | NO3−-N | ASi | |
|---|---|---|---|---|---|---|
| Arenimonas | 0.21 | −0.33 | − 0.21 | − 0.03 | 0.57 | 0.50 |
| Gemmatimonas | − 0.40 | 0.27 | 0.41 | 0.05 | −0.66* | −0.62* |
| H16 (Proteobacteria) | 0.3 | 0.01 | −0.19 | 0.50 | −0.38 | −0.23 |
| Haliangium | 0.65* | −0.21 | −0.67* | 0.50 | −0.45 | − 0.27 |
| Lactobacillus | 0.13 | −0.37 | −0.01 | 0.09 | 0.96** | 0.92** |
| Mesorhizobium | −0.01 | −0.36 | 0.01 | 0.05 | 0.80** | 0.84** |
| Nitrospira | 0.48 | −0.02 | 0.51 | 0.26 | −0.69* | −0.61* |
| Phenylobacterium | −0.72** | 0.42 | 0.72** | −0.07 | 0.04 | 0.03 |
| RB41 (Acidobacteria) | 0.04 | 0.10 | 0.11 | 0.06 | NO3−-N | −0.29 |
Values in bold indicate significant correlations at **p < 0.01 and *p < 0.05
Discussion
Silicon reduced disease incidence and disease index of ginseng black spot
Si has been shown to effectively improve the mechanical and physiological capacities of plants and enhance plant resistance to overcome various biotic and abiotic stresses [12, 13]. To examine the effect of Si application on ginseng black spot-infected plants, a pot experiment was performed with pretreatment of Si for 2 weeks, following 9 dpi with A. panax. Si application significantly reduced the disease incidence and index of ginseng black spot (Table 1) and clearly alleviated the incidence of leaf blight caused by A. panax (Fig. 1). Similarly, Si has been reported to enhance plant resistance to diseases, potentially through interacts with several key factors of the stress signaling pathway [14]. In comparison to group A plants, Si application was shown to increase the accumulation of shoot and root biomass in group AS P. ginseng plants. These findings suggest that Si triggered plant-microbial response mechanisms that directly limited ginseng black spot index and incidence in the leaves and thus enhanced P. ginseng biomass accumulation. However, the root and shoot biomass of group S plants was not significantly different compared with Control, which opposes the notion that Si promotes plant biomass accumulation [15–17]. In the present study, a short-term pot experiment was used to determine the effects of Si application on ginseng black spot and the soil bacterial community, however, future research is needed to clarify the effects of long-term applications on Si-P. ginseng-soil interactions.
Soil properties
Compared with the Control, inoculation of ginseng black spot (in group A) significantly reduced the soil pH and NH4+-N content, and significantly increased the ratio of available K. After Si application, the reduced soil pH and NH4+-N content were significantly recovered to the levels of group Control. Compared with group A, the ratio of available P and available K was reduced in group AS, which resulted in similar soil physical and chemical indexes to that of group CK. In the present study, Si application led to the amendment of the soil pH changes caused by the inoculation of ginseng black spot. However, the soil pH of group Control and group S were not significantly different, and thus Si application alone did not alter the pH. A similar result was found in a study with P. notoginseng, whereby Si increased the soil pH when in the presence of arsenic, which may have been because the Si treatment decreased the bioavailability of arsenic [11]. Although our study does rule out the possibility of other chemical differences among treatments, our data doe does suggest that nutrient availabilities were not the driving differences in soil properties without pathogen inoculation. It is important to consider plant root exudates and their great impact on the population and community structure of soil microbes [18]. In our study, Si application may have affected the root exudates, and other root-derived molecules; as was observed in another study when plants were infected by a fungus [19]. Further research is needed to elucidate the root exudates-Si, plants-Si, and root exudates-plants interactions in the soil-P. ginseng system. However, besides available Si, there were no significant differences in the above-mentioned nutrients between groups S and Control. Therefore, it is likely that Si altered the root exudes rather than physicochemical soil characteristics, which caused the recovery of the bacterial community from A to AS, which was similar to Control.
Soil microbial community composition and diversity
Microbial community diversity is an important component of soil health [20]. The impact of Si on bacterial richness and diversity was analyzed by high-throughput sequencing. The soil bacterial richness (Shannon index and Simpson index) was not significantly different under the different treatments (Fig. 2), which indicated that Si and A. panax treatments did not alter the number of bacterial species in the short-term. However, genes-level differences were found in the relative abundances of bacterial species. Interestingly, the relative abundances of Saccharothrix, Aeromicrobium, Luteimonas, and Rubellimicrobium recovered (P < 0.05) from lower levels in group A to higher levels (similar to Control) following Si application (Fig. 5). The results showed that Si application significantly regulated the structural impact of soil microorganisms caused by ginseng black spot. Aeromicrobium as a potential disease suppression indicator [21, 22] and a member of phylum Actinobacteria. Moreover, the antibiotics produced by Actinobacteria are able to suppress various plant diseases [23, 24]. Disease-suppressive natural soils, with reference to a variety of agricultural crop diseases, have been reported for wheat Take-all and Rhizoctonia bare patch diseases [25], Fusarium wilt on strawberry and vanilla [26], and Rhizoctonia solani on sugar beet [27]. This characteristic relates to the abundance of certain beneficial soil microbes [26, 27], which produce antimicrobial compounds that directly inhibit pathogens. In addition, indirect pathogen inhibition via induced systemic resistance (ISR) may occur, via the triggering of plant immune responses [28]. However, in the event of a severe disease outbreak, consecutive cropping cycles of the same species are stipulated for disease-suppressive microbes to flourish. The proposed hypothesis suggests that, when invaded, certain favorable microbes are amplified and sustained in plants [25, 29, 30]. The bacterial composition of the group Control soil was similar to that of group AS (Si-treated), i.e., their compositions had the highest phylogenetic relatedness. The group AS bacterial flora differed from that of the group A, and the composition of bacterial flora in group S differed from that of the other treatments. The results showed that Si application significantly regulated the changes in bacterial flora caused by ginseng black spot inoculation, and increased the levels in group AS to almost the same as group Control. Recent reports additionally revealed that Arabidopsis plants can stimulate specific favorable microbes in the rhizosphere, even in natural soils [31]. Another validating instance was seen when a Xanthomonas sp., Stenotrophomonas sp., and Microbacterium sp. beneficial consortium was activated in the rhizosphere as part of the downy mildew Hyaloperonospora arabidopsidis induced foliar defense [19]. Furthermore, these strains, when isolated, collectively induced downy mildew resistance, when inoculated back into Arabidopsis; and interestingly, the resistance of a second plant population grown in the same soil was considerably amplified as a result of the downy mildew infection in the first population. These outcomes collectively suggested that beneficial microbes ensue from plant invasions, which in turn prompt a memory or “soil-borne legacy” that amplifies the next plant generation defenses against harmful pathogens [31–34]. The implication here is that Si triggered the soil bacterial community response, which might have directly regulated plant growth. In the present study, we observed that the bacterial community differed in group AS compared with group A, i.e., between Si and no Si treatments. Overall, the increase in the soil bacterial diversity after Si application may contribute to the suppression of ginseng black spot disease. The functionality of root exudates and other root-derived molecules, is indicated in this process [31, 32, 35–37]; albeit this hypothesis requires validation. The most recent research, however, also reports that plants secure favorable rhizosphere communities via the modification of plant exudation patterns, induced by exposure to aboveground pathogens, which subsequently benefits future plant generations [19].
In summary, Si can alter the structure and diversity of the soil microbial community by directly and indirectly affecting the growth of plants, and the altered soil microbial community can, in turn, affect the plants [38–40].
Conclusion
This study provided a detailed outline of the bacterial community compositions in Si applications inoculated with ginseng black spot using Illumina HiSeq sequencing. Si application of ginseng black spot-inoculated plants significantly optimized soil bacterial population structure, improved soil bacterial activity and diversity, and thus effectively prevented and controlled the occurrence of ginseng black spot. In addition, we speculated that Si indirectly altered the structure, composition, and abundance of the soil microbial community by directly altering the root exudates or inducing plant systemic resistance. In conclusion, the present study demonstrated the good application prospect of Si and that it is recommended for use as ginseng fertilizer for the prevention and treatment of ginseng black spot.
Methods
Experimental design
Two-year-old fresh ginseng roots (Panax ginseng Meyer) were provided by dongdu ginseng technology development co., LTD in April 2017 and placed in sand at 23 °C. After 6 days, the roots sprouted and were then washed with deionized water, and transplanted into PVC pots (120 × 180 mm, diameter × height) containing turfy soil (6 seedlings per pot). The ginseng seedlings were grown under greenhouse conditions: temperatures of 17–28 °C, a relative humidity of 70%~ 80%, and a 14 h photoperiod. Before A. panax inoculation, half of the plants were pretreated for 2 weeks with potassium silicate (pH = 7.0) as the Si source. After Si pretreatment, the plants were inoculated with conidia of the appropriate A. panax pathogen. The conidia of A. panax infecting P. ginseng were identified by PCR of the internal transcribed spacer (ITS) region generating 553~554 bp fragments and the glyceraldehyde 3-phosphate dehydrogenase (gpd) for 565~566 bp fragments, respectively. Sequence showed 100% identical to that of A. panax (JF417572 of ITS, JF417653 of gpd). The A. panax strain was deposited in the Culture Collection Center of Yangtze University in Jingzhou, China. Spores were flushed from colonies and then resuspended in sterile distilled water at 1 × 105 spores/mL. The sterilized surfaces of detached spring ginseng leaves were inoculated with 20 μL conidial suspension and incubated under the same greenhouse conditions for 9 days, when black spot symptoms became visible on the leaves.
Plants were grown under four kinds of treatment: ginseng control plants (Control), plants only inoculated with A. panax (A), plants inoculated with A. panax + Si (AS), and plants only inoculated with Si (S), with 18 plants (3 pots) per treatment. To test the prophylactic role of Si, the Si concentration was set at 1.7 mM, i.e., the highest possible concentration of silica acid in solution [4].
Six seedlings of ginseng were randomly selected from each treatment group, and the soils were mixed to form a single representative sample. After inoculation with A. panax for 9 days, plants were removed from the soil and the excess soil was carefully shaken off. The rhizosphere soil (i.e., adhering to the roots) was collected as previously described by Bulgarelli et al. [41], with some modifications. Three replicate rhizosphere soil samples were obtained per treatment. Soil samples (n = 12) were air-dried for 2 weeks, passed through a 2 mm sieve, and stored at − 80 °C.
Plant dry weights and analysis of disease index and incidence
For A. panax infected plants, ginseng black spot incidence was recorded from 9 days after A. panax inoculation. The 18 plants (3 pots) per treatment were collected to calculate the percentage of diseased plants and count disease index, using the following equations [42]:
where A is the disease class (0, 1, 2, 3, 4) and B is the number of plants in the corresponding disease class.
For each plant, the shoots and roots were separated and weighed after air drying (dry weight, g) for 2 weeks at 30 °C.
Sampling and chemical analysis
Air-dried plants and soil samples were used in the nutrient analysis. About 50 mg oven-dried plant tissue was digested with a mixture of 8 mL HNO3 and 2 mL HClO4 at 200 °C for 120 min in a semi-closed system. The digestates were cooled down to 25 °C and made up to 50 mL with 4% (v/v) HNO3 solution. The soil pH (1:5, soil: water) was measured using a glass electrode (SK220, Switzerland). Soil nitrate nitrogen (NO3−-N) was assayed using a continuous flow analytical system (SJAKAR SAN++, The Netherlands). Ammonium nitrogen (NH4+-N) in the soil was extracted with 0.01 M CaCl2, and the concentration was measured by an Auto Analyzer (Auto Analyzer 3, Germany). Potassium (K) in the soil was dissolved with ammonium acetate and calculated by flame photometry. Soluble phosphorus (P) was dissolved with sodium bicarbonate and its concentration measured using the molybdenum blue method [43].
High-throughput sequencing
The total DNA was extracted from 0.5 g of each soil sample using a bacterial DNA Isolation Kit (Omega Bio-tek, Norcross, GA, USA) following the manufacturer’s instructions [44]. To assess the bacterial community composition, Illumina HiSeq platform (Illumina, San Diego, California, USA) was used in present study. The quantity and quality of extracted DNAs were measured using a Nanodrop 1000 (Thermo Fisher Scientific, Wilmington, DE, USA) and agarose gel electrophoresis, respectively. Primers for amplification and preamplification sequence: bacterial 16S rRNA gene V3-V4 region primers: 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). DNA was amplified by PCR under conditions of 95 °C 2 min, followed by 27 cycles of 95 °C extends 30 s, 55 °C for 30s and 72 °C for 45 s; and a final extension at 72 °C for10 min, then maintained at 10 °C until halted. The PCR reactions were performed triplicate in a 20 μL mixture contained 4 μL 5 Mix × FastPfu Buffffer, 2 μL of 2.5 mM dNTPs, 0.4 μL of each primer (5 μM), 0.4 μL of TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China), and 10 ng of template DNA [45]. PCR amplicons were purified with A gencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2 × 300 bp sequencing was performed using the Illumina HiSeq platform (Illumina, San Diego, California, USA) at Biomarker Technologies, Beijing, China.
The Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) pipeline was employed to process the sequencing data [46]. The low-quality sequences were filtered through the criteria [47, 48]. Paired-end reads were assembled using FLASH [49]. After chimera detection, the remaining high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity by UCLUST [50]. A representative sequence was selected from each OTU using default parameters. OTU taxonomic classification was conducted by BLAST searching the representative sequences set against the Greengenes Database [51]. Each OUT in each sample and the taxonomy was recorded in an OTU table, and OTUs containing less than 0.001% of total sequences across all samples were discarded. Sequences were deposited at the NCBI Short Read Archive and accession numbers are SRR9822023-SRR9822034.
Sequence data analyses were mainly performed using QIIME and R packages (v3.2.0). OTU-level alpha diversity indices, were calculated in QIIME. Beta diversity analysis was performed to investigate the structural variation of microbial communities across samples using UniFrac distance metrics [52, 53] and nonmetric multidimensional scaling (NMDS) [54]. Venn diagram was generated to visualize the shared and unique OTUs among groups using R package [55]. Taxa abundances at the phylum, class, order, family, genus and species levels were statistically compared among groups by Metastats [56]. PLS-DA (Partial least squares discriminant analysis) was also introduced as a supervised model to reveal the microbiota variation among groups, using the “plsda” function in R package “mixOmics” [48].
Data analysis
One-way analysis of variance (ANOVA) was used to calculate the difference between treatments with variable soil pathogen abundance. The significance threshold was set at 0.05. The statistical analyses was performed using SAS 9.1 software (SAS Institute Inc., Cary, NC).
Acknowledgements
We thank Jianxin Deng (Yangtze University) for constructive input and discussions. We also acknowledge Hai Sun for Biotechnology, for providing the necessary facilities. YZ acknowledges Cai Shao and Yiming Guan for assistance in seedling and sample collection.
Abbreviations
- ANOVA
Analysis of variance
- dpi
Days after post inoculation
- gpd
Glyceraldehyde 3-phosphate dehydrogenase
- ITS
Internal transcribed spacer
- NMDS
Non-metric multidimensional scale
- OTU
Operational taxonomic unit
- PLS-DA
Partial least squares discriminant analysis
- Si
Silicon
- UPGMA
Unweighted group average method
Authors’ contributions
ML carried out the experimental plan, collected samples. QW, ZL and XP extracted DNA and conducted data analysis. YZ participated in the experimental design, and drafted the manuscript. All authors read and approved the final manuscript.
Funding
YZ acknowledges funding from China Agriculture Research System (CARS-21), the Agricultural Science and Technology Innovation Program (CAAS-XTCX2016012), National Key R&D Program of China (2018YFD0201100), Collaborative innovation project of science and technology innovation project of Chinese academy of agricultural sciences (2018XTCX01), the National Natural Science Foundation of China (31501828), the National Natural Science Foundation of China (81903755), and the Central Public-interest Scientific Institution Basal Research Foundation of CAAS (No. 1610342017017, No. 1610342018011, No. 1610342018020) on high-throughput sequencing. The funding body had no role in the design of the study, analysis and interpretation of data and in writing the manuscript.
Availability of data and materials
The dataset(s) supporting the conclusions of this article are available in the following repositories: The raw read sequences were deposited at the NCBI Short Read Archive and accession numbers are SRR9822023-SRR9822034.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Putnam M. L., du Toit L. J. First report of alternaria blight caused by Alternaria panax on ginseng (Panax quinquefolius) in Oregon and Washington, USA. Plant Pathology. 2003;52(3):406–406. doi: 10.1046/j.1365-3059.2003.00828.x. [DOI] [Google Scholar]
- 2.Epstein E. Silicon. Ann. Rev. Plant Physiol Plant Mol Biol. 1999;50:641–664. doi: 10.1146/annurev.arplant.50.1.641. [DOI] [PubMed] [Google Scholar]
- 3.Fauteux F, Remus-Borel W, Menzies JG, Belanger RR. Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol Lett. 2005;249:1–6. doi: 10.1016/j.femsle.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 4.Vivancos J, Labbé C, Menzies JG, Bélanger RR. Silicon-mediated resistance of Arabidopsis against powdery mildew involves mechanisms other than the salicylic acid (SA)-dependent defence pathway. Mol Plant Pathol. 2015;16:572–582. doi: 10.1111/mpp.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aliyeh R, Caroline L, Humira S, et al. Silicon protects soybean plants against Phytophthora sojae by interfering with effector-receptor expression. BMC Plant Biol. 2018;18(1):97. doi: 10.1186/s12870-018-1312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayyar A, Hamel C, Lafond G, Gossen BD, Hanson K, Germida J. Soil microbial quality associated with yield reduction in continuous-pea. Appl Soil Ecol. 2009;43(1):115–121. doi: 10.1016/j.apsoil.2009.06.008. [DOI] [Google Scholar]
- 7.Liu X, Zhang S, Jiang Q, Bai Y, Shen G, Li S, Ding W. Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci Rep 2016;6:36773. https://doi.org/10.1038/srep36773. [DOI] [PMC free article] [PubMed]
- 8.Fang S, Liu D, Ye T, Deng S, Shang X. Tree species composition influences enzyme activities and microbial biomass in the Rhizosphere: a Rhizobox approach. PLoS One. 2013;8(4):e61461. doi: 10.1371/journal.pone.0061461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai Kunzheng, Gao Dan, Luo Shiming, Zeng Rensen, Yang Jianyuan, Zhu Xiaoyuan. Physiological and cytological mechanisms of silicon-induced resistance in rice against blast disease. Physiologia Plantarum. 2008;134(2):324–333. doi: 10.1111/j.1399-3054.2008.01140.x. [DOI] [PubMed] [Google Scholar]
- 10.Ghareeb H, Bozsó Z, Ott PG, Repenning C, Stahl F, Wydra K. Transcriptome of silicon-induced resistance against Ralstonia solanacearum in the silicon non-accumulator tomato implicates priming effect. Physiol Mol Plant Pathol. 2011;75(3):83–89. doi: 10.1016/j.pmpp.2010.11.004. [DOI] [Google Scholar]
- 11.Yue Y, Aichen Z, Yanjiao C, et al. Impacts of silicon addition on arsenic fractionation in soils and arsenic speciation in Panax notoginseng planted in soils contaminated with high levels of arsenic. Ecotox Environ Safe. 2018;162:400–407. doi: 10.1016/j.ecoenv.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Richmond Kathryn E, Sussman Michael. Got silicon? The non-essential beneficial plant nutrient. Current Opinion in Plant Biology. 2003;6(3):268–272. doi: 10.1016/S1369-5266(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 13.Ma JF, Yamaji N. Silicon uptake and accumulation in higher plants. Trends Plant Sci. 2006;11:392–397. doi: 10.1016/j.tplants.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Fawe A, Abou-Zaid M, Menzies JG, Bélanger RR. Silicon-mediated accumulation of flavonoid phytoalexins in cucumber. Phytopathology. 1998;88:396–401. doi: 10.1094/PHYTO.1998.88.5.396. [DOI] [PubMed] [Google Scholar]
- 15.Fleck AT, Mattusch J, Schenk MK. Silicon decreases the arsenic level in rice grain by limiting arsenite transport. J Plant Nutr Soil Sci. 2013;176:785–794. [Google Scholar]
- 16.Li RY, Stroud JL, Ma JF, Mcgrath SP, Zhao FJ. Mitigation of arsenic accumulation in rice with water management and silicon fertilization. Environ Sci Technol. 2009;43:3778–3783. doi: 10.1021/es803643v. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Zou Q, Xue S, Pan W, Yue X, Hartley W, Huang L, Mo J. Effect of silicate on arsenic fractionation in soils and its accumulation in rice plants. Chemosphere. 2016;165:478–486. doi: 10.1016/j.chemosphere.2016.09.061. [DOI] [PubMed] [Google Scholar]
- 18.Kong HG, Kim BK, Song GC, Lee S, Ryu C-M. Aboveground whitefly infestation-mediated reshaping of the root microbiota. Front Microbiol. 2016;7:1314. doi: 10.3389/fmicb.2016.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun Y, Jun Z, Tao W, et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome. 2018;6(1):156. doi: 10.1186/s40168-018-0537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garbeva P, Van Veen JA, Van Elsas JD. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressivenss. Annu Rev Phytopathol. 2004;42:243–270. doi: 10.1146/annurev.phyto.42.012604.135455. [DOI] [PubMed] [Google Scholar]
- 21.Palaniyandi SA, Yang SH, Zhang L, Suh JW. Effects of actinobacteria on plant disease suppression and growth promotion. Appl Microbiol Biotechnol. 2013;97:9621–9636. doi: 10.1007/s00253-013-5206-1. [DOI] [PubMed] [Google Scholar]
- 22.Shen G, Zhang S, Liu X, Jiang Q, Ding W. Soil acidification amendments change the rhizosphere bacterial community of tobacco in a bacterial wilt affected field. Appl Microbiol Biotech. 2018;102(22):9781–9791. doi: 10.1007/s00253-018-9347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YS, Kim HM, Chang C, Hwang IC, Oh H, Ahn JS, Kim KD, Hwang BK, Kim BS. Biological evaluation of neopeptins isolated from a Streptomyces strain. Pest Manag Sci. 2007;63:1208–1214. doi: 10.1002/ps.1450. [DOI] [PubMed] [Google Scholar]
- 24.Lee SY, Tindwa H, Lee YS, Naing KW, Hong SH, Nam Y, Kim KY. Biocontrol of anthracnose in pepper using chitinase, β-1,3 glucanase, and 2-furancarboxaldehyde produced by Streptomyces cavourensis SY224. J Microbiol Biotechnol. 2012;22:1359–1366. doi: 10.4014/jmb.1203.02056. [DOI] [PubMed] [Google Scholar]
- 25.Weller DM, Raaijmakers JM, Gardener BB, Thomashow LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol. 2002;40:309–348. doi: 10.1146/annurev.phyto.40.030402.110010. [DOI] [PubMed] [Google Scholar]
- 26.Cha JY, Han S, Hong HJ, Cho H, Kim D, Kwon Y, Kwon SK, Crüsemann M, Bok Lee Y, Kim JF, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016;10:119–129. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendes R, Kruijt K, De Bruijn I, Dekkers E, Van Der Voort M, Schneider JH, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 28.Pieterse CM, Van Der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 29.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T. Disease suppressive soils: new insights from the soil microbiome. Phytopathology. 2017;107:1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- 31.Berendsen RL, Vismans G, Yu K, Song Y, de Jonge R, Burgman WP, Burmølle M, Herschend J, Bakker PAHM, Pieterse CMJ. Disease-induced assemblage of a plant-benefificial bacterial consortium. ISME J. 2018;12:1496–1507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudrappa T, Czymmek KJ, Pare PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raaijmakers JM, Mazzola M. Soil immune responses. Science. 2016;352:1392–1393. doi: 10.1126/science.aaf3252. [DOI] [PubMed] [Google Scholar]
- 34.Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL. The soil-borne legacy. Cell. 2018;172:1178–1180. doi: 10.1016/j.cell.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 35.Badri DV, Chaparro JM, Zhang RF, Shen QR, Vivanco JM. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 2013;288:4502–4512. 10.1074/jbc. M112.433300. [DOI] [PMC free article] [PubMed]
- 36.Gu Y, Wei Z, Wang X, Friman V-P, Huang J, Wang X, Mei X, Xu Y, Shen Q, Jousset A. Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol Fertil Soils. 2016;52:997–1005. doi: 10.1007/s00374-016-1136-2. [DOI] [Google Scholar]
- 37.Sasse J, Martinoia E, Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2018;23:25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Kardol Paul, Martijn Bezemer T., van der Putten Wim H. Temporal variation in plant-soil feedback controls succession. Ecology Letters. 2006;9(9):1080–1088. doi: 10.1111/j.1461-0248.2006.00953.x. [DOI] [PubMed] [Google Scholar]
- 39.Reinhart KO, Callaway RM. Soil biota and invasive plants. New Phytol. 2006;170(3):445–457. doi: 10.1111/j.1469-8137.2006.01715.x. [DOI] [PubMed] [Google Scholar]
- 40.Bever James D., Dickie Ian A., Facelli Evelina, Facelli Jose M., Klironomos John, Moora Mari, Rillig Matthias C., Stock William D., Tibbett Mark, Zobel Martin. Rooting theories of plant community ecology in microbial interactions. Trends in Ecology & Evolution. 2010;25(8):468–478. doi: 10.1016/j.tree.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulgarelli Davide, Rott Matthias, Schlaeppi Klaus, Ver Loren van Themaat Emiel, Ahmadinejad Nahal, Assenza Federica, Rauf Philipp, Huettel Bruno, Reinhardt Richard, Schmelzer Elmon, Peplies Joerg, Gloeckner Frank Oliver, Amann Rudolf, Eickhorst Thilo, Schulze-Lefert Paul. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488(7409):91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 42.Faheem M, Raza W, Zhong W, Nan Z, Shen Q, Xu Y. Evaluation of the biocontrol potential of streptomyces goshikiensis YCXU against Fusarium oxysporum f. sp. niveum: theory and applications in pest management. Biol Control. 2015;81(10):101–110. doi: 10.1016/j.biocontrol.2014.11.012. [DOI] [Google Scholar]
- 43.Sun H, Wang Q, Liu N, Zhang C, Liu Z, Zhang Y. Effects of different leaf litters on the physicochemical properties and bacterial communities in Panax ginseng-growing soil. Appl Soil Ecol. 2016;111:17–24. doi: 10.1016/j.apsoil.2016.11.008. [DOI] [Google Scholar]
- 44.Zhang H, Feng J, Chen S, et al. Geographical patterns of nirS gene abundance and nirS-type denitrifying bacterial community associated with activated sludge from different wastewater treatment plants. Microb Ecol. 2019;77(2):304–316. doi: 10.1007/s00248-018-1236-7. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q, Sun H, Xu C, et al. Analysis of rhizosphere bacterial and fungal communities associated with rusty root disease of Panax ginseng. Appli Soil Ecol. 2019;138:245–252. doi: 10.1016/j.apsoil.2019.03.012. [DOI] [Google Scholar]
- 46.Caporaso JG, Kuczynski J, Stombaugh K, Bittinger FD, Bushman EK, Costello N, Fierer AG, Pena JK, Goodrich JI, Gordon GA, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen YF, Yang FL, Lu HF, Wang BH, Chen YB, Lei DJ, Wang YZ, Zhu BL, Li LJ. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54(2):562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 49.Magoc T., Salzberg S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microb. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbio Ecol. 2007;62(2):142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaura E, Keijser BJF, Huse SM, Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009;9:12. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White James Robert, Nagarajan Niranjan, Pop Mihai. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Computational Biology. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset(s) supporting the conclusions of this article are available in the following repositories: The raw read sequences were deposited at the NCBI Short Read Archive and accession numbers are SRR9822023-SRR9822034.