Abstract
Background
As all we know, gastric cancer (GC) is a highly aggressive disease. Recently, circular RNA (circRNA) was found to play a vital role in regulation of GC. Some circRNAs could regulate messenger RNA (mRNA) expression by functioning as a microRNA (miRNA) sponge. Nevertheless, the circRNA-miRNA-mRNA regulatory network involved GC rarely has been explored and researched.
Material/Methods
All the differentially expressed circRNAs, miRNAs, and mRNA were derived from Gene Expression Omnibus (GEO) microarray data (GSE78092, GSE89143, GSE93415, and GSE54129). GC level 3 miRNA-sequencing data and clinical information were downloaded from The Cancer Genome Atlas (TCGA) database. Furthermore, a circRNA-miRNA-mRNA regulatory network was constructed by Cytoscape (version 3.6.1). Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway revealed the functions and signaling pathways associated with these target genes. Hub genes of protein-protein interaction (PPI) network were identified by STRING database and cytoHubba.
Results
The regulatory network consists of 3 circRNAs, 22 miRNAs, and 128 mRNAs. Only 3 miRNAs of the network were consistent with the expression of TCGA and were associated with some clinical features. The results of the functional analysis of 128 mRNAs showed that GO analysis and KEGG pathways of inclusion criteria were 49 and 24, respectively. PPI network and Cytoscape showed that the top 10 hub genes were MYC, CTGF, TGFBR2, TGFBR1, SERPINE1, KRAS, ZEB1, THBS1, CDK6, and TNS1; 4 of which were verified by GEPIA based on TCGA. Highly expressed SERPINE1 had a poor OS (over survival) and DFS (disease-free survival), and TGFBR1 expression increased along with the increase of clinical stages.
Conclusions
This study looked at a circRNA-miRNA-mRNA regulatory network associated with GC and explored the potential functions of mRNA in the network, then identified a new molecular marker for prediction, prognosis, and therapeutic targets for clinical patients.
MeSH Keywords: Computational Biology, Biological Markers, Stomach Neoplasms, Gene Expression Profiling
Background
Gastric cancer (GC) is a disease with very high morbidity and mortality worldwide; it was responsible for over 1 000 000 new cases and 783 000 deaths in 2018, which makes it the fifth most frequently diagnosed cancer and the third leading cause of cancer death [1]. Although there has been significant progress in personalizing treatment for GC, it is still a clinically challenging disease characterized by a lack of effective treatment options and scarcely reliable molecular tools to predict patient outcomes. Compared with other cancer types, the clinical management of GC has not yet achieved the expected benefits from the era of personalized medicine [2]. Almost one third of patients experience recurrence and distant metastasis after undergoing GC surgery [3]. Therefore, the detection of molecular markers for early diagnosis, prognosis, and therapeutic targets of GC has become very urgent.
Most of the human genome is transcribed into non-protein-coding RNA, while protein-coding genome is less than 2% [4]. Circular RNA (circRNA), as one of noncoding RNAs, are derived from the precursor messenger RNA (mRNA) of RNA transcriptase II, consisting of continuous covalently closed loop without the 5′-cap structure and the 3′-poly A tail. Due to this structure, circRNAs are not easily degraded by exonuclease RNase R [5]. In particular, circRNAs are reported to play crucial roles in cancer occurrence, metastasis, and therapy resistance owning to its abundant biological effect on tumor cells including proliferation, apoptosis, and invasion [6]. CircRNAs function primarily as transcriptional and post-transcriptional regulators through various functional mechanisms, such as RNA binding protein (RBP) sponges and protein scaffolds [7], translate proteins [8], RNAP II elongation [9], RNA-RNA interactions [10], and RNA maturation [11]. At present, circRNAs function mainly by adsorbing microRNAs (miRNAs) as miRNA response elements (MRE) based on competing endogenous RNA (ceRNA) hypothesis in GC [12,13]. For instance, augmented expression of circNF1 obviously promotes cell proliferation by sponging miR-16 in GC [14]. Another study reported that has_circ_0001461 (termed circFAT1) low expression inhibited GC cell line proliferation by regulating the miR-548g/RUNX1 axis in the cytoplasm and targeting YBX1 in the nucleus, meanwhile, it was correlated with overall survival (OS) of GC patients [15]. In addition, circRNA circPDSS1 has been shown to promote GC progression by sponging miR-186-5p to modulate NEK2 [16].
Although several circRNAs have been identified as participating in the pathogenesis of GC, it is still necessary to conduct a circRNA-miRNA-mRNA regulatory network in GC, to help to advance our understanding of the molecular mechanism of GC. In the present study, we constructed a regulatory network consisting of 3 circRNAs, 22 miRNAs, and 128 mRNAs through multiple sets of the Gene Expression Omnibus (GEO) database and some online prediction websites, and analyzed the miRNAs and mRNAs in the network using The Cancer Genome Atlas (TCGA) database, the Gene Ontology (GO) analysis, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and the protein-protein interaction (PPI) network to indirectly understand the potential mechanism of circRNAs in the occurrence and development of GC
Material and Methods
Data collection
The microarray data used in the current study were acquired from the GEO database (http://www.ncbi.nlm.nih.gov/gds/). The circRNAs expression data were obtained from GSE78092 (3 pairs of primary GC tissue and normal gastric mucosa) and GSE89143 (3 pairs of GC tissue and matched paracancerous tissue). The miRNA and mRNA expression data were respectively derived from GSE93415 (20 pairs of gastric tumor and adjacent healthy gastric mucosa) and GSE54129 (111 tumor samples and 21 normal gastric mucosa). GC level 3 miRNA-sequencing data and clinical information were downloaded from TCGA database (https://cancergenome.nih.gov/) on January 07, 2019. A total of 491 samples were included in this study, containing 446 GC samples and 45 matched normal samples. The detailed clinical information included sex, age at diagnosis, grade, T stage, N stage, M stage, and clinical stage, which is shown in Supplementary Table 1.
Differential expression analysis of circRNAs, miRNAs, and mRNAs
The downloaded platform file(s) and series of matrix file(s) were converted through using the R language software and annotation package. The ID of the corresponding probe name was converted into an international standard name (circRNA symbol). The analysis of differentially expressed RNAs was performed using the limma package based on the Bioconductor package. The criteria for selection of differentially expressed circRNAs (DEcircRNAs) were P-value <0.05 and |log2FC| >1. Differentially expressed miRNAs (DEmiRNAs) were identified by using GEO2R in dataset GSE93415, as the |log2FC| of most miRNAs is less than 1, we set the criterion that FDR values <0.05 and |log2FC| >0.5 were considered significantly. Differentially expressed mRNAs (DEmRNAs) were also identified by using GEO2R in dataset GSE54129, with the criterion that FDR values <0.05 and |log2FC| >1 were considered significantly. In addition, miRNA-sequencing data derived from TCGA were processed with edgeR [17], a Bioconductor package based on the R language, to screen differentially expressed miRNAs (TDEmiRNAs) between GC tissue and adjacent normal tissue. Differentially expressed miRNAs with FDR values <0.05 and |log2FC| >1 were considered significantly.
Construction of the circRNA-miRNA-mRNA regulatory network
According to the results of the differential expression analysis, we first get the intersection of DEcircRNAs between GSE78092 and GSE89143, called IDEcircRNAs. Targeting miRNAs of IDEcircRNAs were predicted via Cancer-Specific CircRNA Database (CSCD) http://gb.whu.edu.cn/CSCD/ (we called it CPmiRNAs), then we took the intersection of it and DEmiRNAs, which we called ICPDEmiRNAs. Furthermore, we used Perl language to predict respectively their target genes according to downloaded miRNA databases on the 3 target gene prediction website including miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), targetScan (http://www.targetscan.org) and miRDB (http://www.mirdb.org/). Not all target genes were included, the miRNA target genes that could be found in all 3 databases were used as the final target genes, which were named TmRNA. We took the intersection of it and the DEmRNAs in the same way, and got the final functional genes, which were denominated FmRNAs, therefore, through IDEcircRNAs, ICPDEmiRNAs, and FmRNAs, we constructed the circRNA-miRNA-mRNA regulatory network using Cytoscape.
Functional enrichment analysis
We firstly converted the gene symbol of FmRNAs in the network into entrezIDs, meanwhile, installing R language packages including “colorspace”, “stringi”, “DOSE”, “clusterProfiler”, and “pathview”. Furthermore, we obtained the outcomes of the GO analysis and the KEGG pathway through R studio and R scripting language, however, not all results were included. We set a criterion that the P value of the GO analysis was less than 0.05, besides the P value and q value of the KEGG analysis were both less than 0.05, and the network diagram of the KEGG pathway containing mRNA was constructed by Cytoscape.
The analysis of miRNAs and mRNA in the network
We compared ICPDEmiRNAs and TDEmiRNAs by Venn diagram to find the sharing expressed miRNAs, whose expression in tumor tissues and normal tissues was plotted by GraphPad Prism 7, then we analyzed the clinical relevance via SPSS21.0. The hub genes of FmRNAs were screened by PPI and cytoHubba plugin. The medium confidence in the network was 0.400. In addition, their expression, survival prognosis and correlation with clinical stage were identified in GEPIA.
Statistical analysis
Statistical analysis was performed using SPSS 21.0 (Chicago, IL, USA). Significant differential expression levels of circRNAs were analyzed by R language limma packages and FDR filtering was used for comparative analysis. The P-value <0.05 and absolute fold change ≥2.0 were considered statistically significant. The correlation between miRNA expression and clinical characteristics was tested by chi-square test.
Results
Identification of DEcircRNA, DEmiRNAs, DEmRNAs
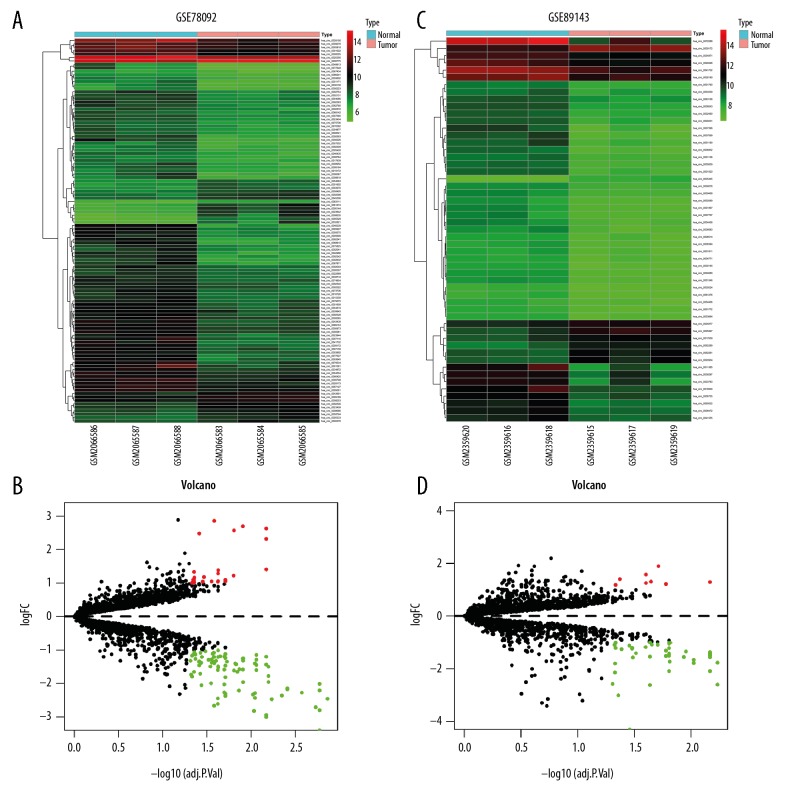
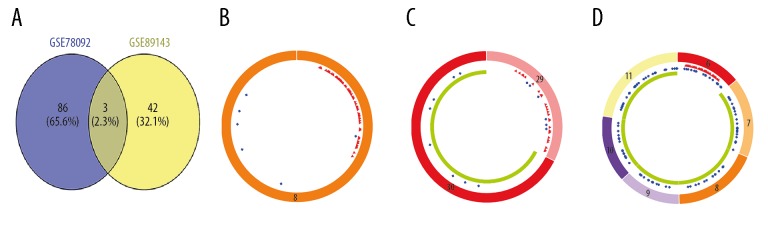
The integrated analysis of GSE78092 and GSE89143 dataset respectively identified 112 and 54 differentially expressed circRNAs (DEcircRNAs) by R language limma packages, the former included 23 upregulated and 89 downregulated circRNAs, the latter had 8 and 54 (Figure 1). Then, we took the intersection of DEcircRNAs of the 2 datasets, the outcome showed that they didn’t have common upregulated circRNAs, while having 3 sharing downregulated circRNAs (IDEcircRNAs) (Figure 2A), which were known as hsa_circ_004173, hsa_circ_0009076, hsa_circ_0028190 (Figure 2B–2D). Besides, we also used GEO2R online analysis on GSE93415 to obtain differentially expressed miRNAs. A total of 344 differentially expressed miRNAs were obtained, 149 of which were downregulated and 195 of which were upregulated (Supplementary Table 2). Similarly, we also did the same analysis on GSE54129, and found 3916 differentially expressed mRNAs, including 1896 upregulated mRNAs and 2020 downregulated mRNAs (Supplementary Table 3).
Figure 1.
Differentially expressed circular RNAs (DEcircRNAs). (A) Heatmap of GSE78092. (B) Volcano plot of GSE78092. (C) Heatmap of GSE89143. (D) Volcano plot of GSE89143.
Figure 2.
The intersection of circular RNAs (circRNAs). (A) Three sharing downregulated circRNAs. (B) Structure diagram of hsa_circ_0041732. (C) Structure diagram of hsa_circ_0009076. (D) Structure diagram of hsa_circ_0028190. The red, blue, and green regions inside the circular RNA molecule respectively represent MRE (microRNA response element), RBP (RNA binding protein), ORF (open reading frame).
Searching for the relationship among circRNA, miRNA, and mRNA
The results of predicted targeting miRNAs on 3 circRNAs shown that hsa_circ_0009076 had 48 targeting miRNAs, hsa_circ_0028190 had 69, and hsa_circ_0041732 had 108. A total of 202 miRNAs were obtained after the removal of the repeatedly targeted miRNAs (Supplementary Table 4). We got 22 common miRNAs (ICPDEmiRNAs) (Supplementary Figure 1A) by taking the intersection of 202 targeting miRNAs and the precious 344 DEmiRNAs. In terms of target gene prediction, we obtained 431 target genes (TmRNAs) of 22 miRNAs (Supplementary Table 5). Finally, we obtained 128 common mRNAs (FmRNAs) (Supplementary Figure 1B) by intersecting the TmRNAs with the previously obtained 3916 DEmRNAs.
Constructing of circRNA-miRNA-mRNA network
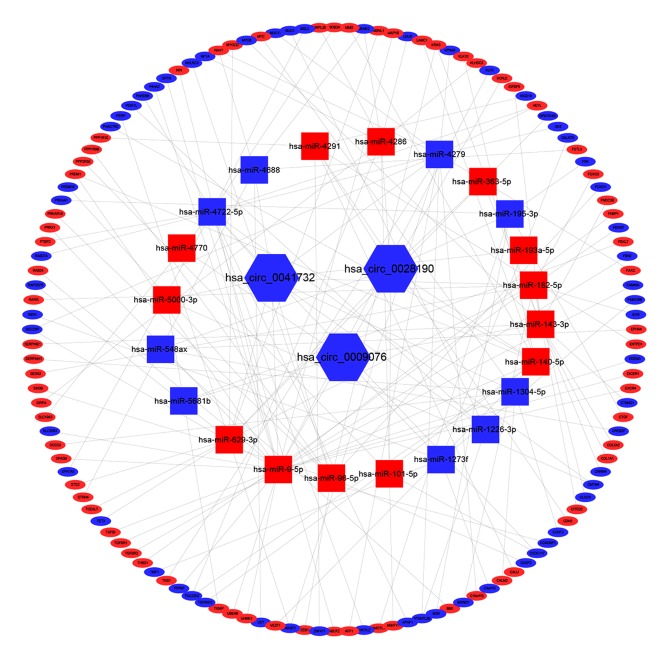
From the previous data analysis, we got IDEcircRNAs including 3 downregulated circRNAs, 22 ICRDEmiRNAs including 13 upregulated, 9 downregulated, 128 FmRNAs including 69 upregulated and 59 downregulated. Then, we use Cytoscape Version3.6.1 soft to describe their relationships in the network, 3 circRNAs had 24 targeted relationship with 22 miRNAs, 22 miRNAs have 139 targeted relationship with 128 DEmiRNA-mRNAs (Figure 3).
Figure 3.
The circRNA-miRNA-mRNA regulatory network. The hexagon, ellipse, rectangle respectively circRNAs, mRNAs, and miRNAs. Red represents upregulated RNAs, Blue represents downregulated RNAs. circRNA – circular RNA; miRNA – microRNA; mRNA – messenger RNA.
The correlation of clinical characteristics and miRNAs
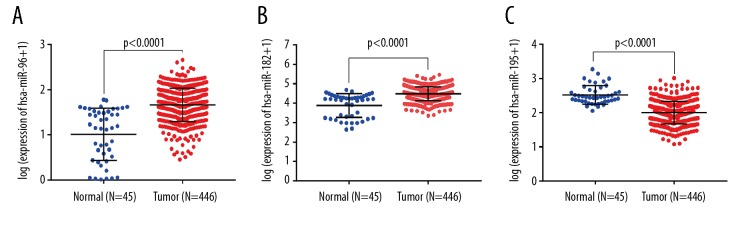
In order to further explore clinical correlation of miRNAs in the network, we first got 242 TDEmiRNAs including 178 upregulated and 69 downregulated (Supplementary Table 6, Supplementary Figure 2) in the stomach TCGA database. Compared with the precious 22 ICPDE miRNAs, we got the 3 same expression mode miRNAs. The situation of 3 miRNAs was displayed in Figure 4, then we analyzed their correlation with the clinical characteristics. The results were shown in Table 1: hsa-mir-182 was associated with T stage (P=0.006) and N stage (P=0.013), hsa-mir-96 was associated with age (P=0.025), T stage (P=0.003) and N stage (P=0.042), and hsa-mir-195 was associated with N stage (P=0.029).
Figure 4.
Expression of 3 microRNAs in gastric cancer and normal tissues: (A) hsa-miR-96; (B) hsa-miR-182; (C) hsa-miR-195.
Table 1.
Clinical correlation of 3 miRNAS.
| Variables | Numbers | hsa-miR-182 | χ2 test P value | hsa-miR-96 | χ2 test P value | hsa-miR-195 | χ2 test P value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low expression | High expression | Low expression | High expression | Low expression | High expression | |||||
| Gender | ||||||||||
| Female | 138 | 66 | 72 | 0.495 | 68 | 70 | 0.799 | 74 | 64 | 0.134 |
| Male | 235 | 121 | 114 | 119 | 116 | 113 | 122 | |||
| Age at diagnosis | ||||||||||
| >50 | 341 | 176 | 165 | 0.062 | 177 | 164 | 0.025 | 20 | 12 | 0.134 |
| ≤50 | 32 | 11 | 21 | 10 | 22 | 167 | 174 | |||
| Grade | ||||||||||
| G1 | 6 | 4 | 2 | 0.414 | 2 | 4 | 0.407 | 4 | 2 | 0.414 |
| G2+G3 | 367 | 183 | 184 | 185 | 182 | 183 | 184 | |||
| T stage | ||||||||||
| T1+2 | 97 | 37 | 60 | 0.006 | 36 | 61 | 0.003 | 56 | 41 | 0.405 |
| T3+4 | 276 | 150 | 126 | 151 | 125 | 131 | 145 | |||
| Metastasis | ||||||||||
| M0 | 345 | 168 | 177 | 0.051 | 170 | 175 | 0.244 | 174 | 171 | 0.683 |
| M1 | 28 | 199 | 9 | 17 | 11 | 13 | 15 | |||
| Lymph node status | ||||||||||
| N0 | 120 | 49 | 71 | 0.013 | 51 | 69 | 0.042 | 70 | 50 | 0.029 |
| N1–2 | 253 | 138 | 115 | 136 | 117 | 117 | 136 | |||
| Stage | ||||||||||
| I+II | 169 | 81 | 88 | 0.438 | 79 | 90 | 0.234 | 89 | 80 | 0.374 |
| III+IV | 204 | 106 | 98 | 108 | 96 | 98 | 106 | |||
GO and KEGG analysis of FmRNAs
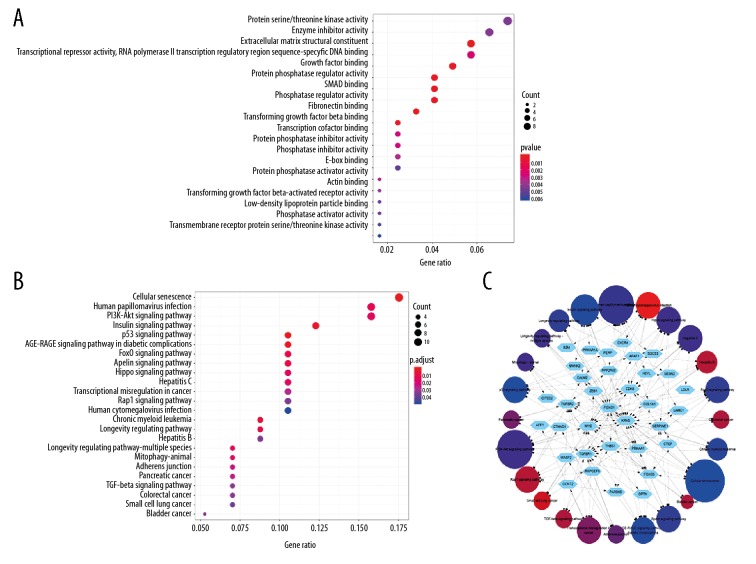
128 FmRNAs were used to detect the potential functions of circRNA, including GO enrichment analysis and KEGG pathway analysis. In the GO analysis, including biological process, cellular component and molecular function, we obtained 49 results, which were shown in Table 2. In KEGG pathway analysis, we got 24 results, which were shown in Table 3. The dotplot showed the results of the top 20 GO analysis results with P value from small to large, as well as the results of KEGG pathway, most of which were related to the density of tumor (Figure 5A, 5B). In addition, we also constructed the network diagram of the relationship between KEGG pathway and each mRNA (Figure 5C). In the tumor-associated signaling pathway, KEGG analysis showed that the PI3K-Akt signaling pathway contained the most genes (PRKAA1/MYC/CDK6/KRAS/LAMC1/THBS1/COL1A1/PPP2R5E/FOXO3) (Supplementary Figure 3A), and the p53 signaling pathway including PERP/APAF1/CDK6/SESN2/THBS1/SERPINE1 had the smallest P-value (Supplementary Figure 3B), which showed the most significance. The 2 signaling pathways became the focus of our observation.
Table 2.
The results of GO enrichment analysis.
| ID | Description | Gene ratio |
|---|---|---|
| GO: 0001968 | Fibronectin binding | 4/122 |
| GO: 0005201 | Extracellular matrix structural constituent | 7/122 |
| GO: 0019888 | Protein phosphatase regulator activity | 5/122 |
| GO: 0046332 | SMAD binding | 5/122 |
| GO: 0019838 | Growth factor binding | 6/122 |
| GO: 0050431 | Transforming growth factor beta binding | 3/122 |
| GO: 0019208 | Phosphatase regulator activity | 5/122 |
| GO: 0001227 | Transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding | 7/122 |
| GO: 0001221 | Transcription cofactor binding | 3/122 |
| GO: 0004864 | Protein phosphatase inhibitor activity | 3/122 |
| GO: 0019212 | Phosphatase inhibitor activity | 3/122 |
| GO: 0072542 | Protein phosphatase activator activity | 2/122 |
| GO: 0048185 | Activin binding | 2/122 |
| GO: 0004857 | Enzyme inhibitor activity | 8/122 |
| GO: 0004674 | Protein serine/threonine kinase activity | 9/122 |
| GO: 0005024 | Transforming growth factor beta-activated receptor activity | 2/122 |
| GO: 0030169 | Low-density lipoprotein particle binding | 2/122 |
| GO: 0070888 | E-box binding | 3/122 |
| GO: 0019211 | Phosphatase activator activity | 2/122 |
| GO: 0004675 | Transmembrane receptor protein serine/threonine kinase activity | 2/122 |
| GO: 0003779 | Actin binding | 8/122 |
| GO: 0001223 | Transcription coactivator binding | 2/122 |
| GO: 0030742 | GTP-dependent protein binding | 2/122 |
| GO: 0019955 | Cytokine binding | 4/122 |
| GO: 0005518 | Collagen binding | 3/122 |
| GO: 0017022 | Myosin binding | 3/122 |
| GO: 0019903 | Protein phosphatase binding | 4/122 |
| GO: 0005520 | Insulin-like growth factor binding | 2/122 |
| GO: 0071813 | Lipoprotein particle binding | 2/122 |
| GO: 0071814 | Protein-lipid complex binding | 2/122 |
| GO: 0030332 | Cyclin binding | 2/122 |
| GO: 0035035 | Histone acetyltransferase binding | 2/122 |
| GO: 0051721 | Protein phosphatase 2A binding | 2/122 |
| GO: 0019199 | Transmembrane receptor protein kinase activity | 3/122 |
| GO: 0003713 | Transcription coactivator activity | 6/122 |
| GO: 1901682 | Sulfur compound transmembrane transporter activity | 2/122 |
| GO: 0030020 | Extracellular matrix structural constituent conferring tensile strength | 2/122 |
| GO: 0043531 | ADP binding | 2/122 |
| GO: 0001078 | Transcriptional repressor activity, RNA polymerase II proximal promoter sequence-specific DNA binding | 4/122 |
| GO: 0019902 | Phosphatase binding | 4/122 |
| GO: 0000982 | Transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding | 7/122 |
| GO: 0019887 | l kinase regulator activity | 4/122 |
| GO: 0001228 | Transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding | 7/122 |
| GO: 0016706 | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 2/122 |
| GO: 0051018 | Protein kinase A binding | 2/122 |
| GO: 0045309 | Protein phosphorylated amino acid binding | 2/122 |
| GO: 0005160 | Transforming growth factor beta receptor binding | 2/122 |
| GO: 0031625 | Ubiquitin protein ligase binding | 5/122 |
| GO: 0031490 | Chromatin DNA binding | 3/122 |
| ID | Bg ratio | p Value | p Adjust | q Value | Gene ID | Count |
|---|---|---|---|---|---|---|
| GO: 0001968 | 27/17632 | 3.39E-05 | 0.012118429 | 0.011010275 | CTGF/FSTL3/THBS1/IGFBP5 | 4 |
| GO: 0005201 | 155/17632 | 0.00010192 | 0.018243653 | 0.016575386 | FBN2/MUC17/LAMC1/THBS1/TGFBI/COL1A1/COL5A2 | 7 |
| GO: 0019888 | 80/17632 | 0.000232027 | 0.020766389 | 0.018867433 | PHACTR2/PPP2R5E/PPP1R9B/PPP1R10/CALM2 | 5 |
| GO: 0046332 | 80/17632 | 0.000232027 | 0.020766389 | 0.018867433 | PRDM16/MYOCD/TGFBR1/COL5A2/TGFBR2 | 5 |
| GO: 0019838 | 137/17632 | 0.000381094 | 0.025089597 | 0.022795311 | CTGF/THBS1/IGFBP5/TGFBR1/COL1A1/TGFBR2 | 6 |
| GO: 0050431 | 22/17632 | 0.000452113 | 0.025089597 | 0.022795311 | THBS1/TGFBR1/TGFBR2 | 3 |
| GO: 0019208 | 94/17632 | 0.000490579 | 0.025089597 | 0.022795311 | PHACTR2/PPP2R5E/PPP1R9B/PPP1R10/CALM2 | 5 |
| GO: 0001227 | 267/17632 | 0.002578004 | 0.098990696 | 0.089938622 | FOXO1/MYC/ZEB1/HEYL/PRDM1/PRRX1/FOXO3 | 7 |
| GO: 0001221 | 41/17632 | 0.002844169 | 0.098990696 | 0.089938622 | CCNT2/FOXO1/FOXO3 | 3 |
| GO: 0004864 | 41/17632 | 0.002844169 | 0.098990696 | 0.089938622 | PHACTR2/PPP1R9B/PPP1R10 | 3 |
| GO: 0019212 | 44/17632 | 0.003480829 | 0.098990696 | 0.089938622 | PHACTR2/PPP1R9B/PPP1R10 | 3 |
| GO: 0072542 | 13/17632 | 0.003523642 | 0.098990696 | 0.089938622 | PPP2R5E/CALM2 | 2 |
| GO: 0048185 | 14/17632 | 0.004092371 | 0.098990696 | 0.089938622 | FSTL3/TGFBR1 | 2 |
| GO: 0004857 | 371/17632 | 0.004286672 | 0.098990696 | 0.089938622 | PHACTR2/TXNIP/SOCS3/PRKAR1A/PPP1R9B/PPP1R10/SERPINH1/SERPINE1 | 8 |
| GO: 0004674 | 455/17632 | 0.004378555 | 0.098990696 | 0.089938622 | TRPM7/CCNT2/MKNK2/PDIK1L/PRKAA1/CDK6/UHMK1/TGFBR1/TGFBR2 | 9 |
| GO: 0005024 | 15/17632 | 0.004700676 | 0.098990696 | 0.089938622 | TGFBR1/TGFBR2 | 2 |
| GO: 0030169 | 15/17632 | 0.004700676 | 0.098990696 | 0.089938622 | LDLR/THBS1 | 2 |
| GO: 0070888 | 50/17632 | 0.004998838 | 0.099421342 | 0.090329887 | CLOCK/MYC/ZEB1 | 3 |
| GO: 0019211 | 16/17632 | 0.005347991 | 0.100767416 | 0.091552872 | PPP2R5E/CALM2 | 2 |
| GO: 0004675 | 17/17632 | 0.006033758 | 0.108004271 | 0.098127961 | TGFBR1/TGFBR2 | 2 |
| GO: 0003779 | 421/17632 | 0.00895487 | 0.148900927 | 0.135284877 | MYO6/WASF2/TRPM7/PHACTR2/TNS1/MAP1B/CXCR4/PPP1R9B | 8 |
| GO: 0001223 | 21/17632 | 0.009150336 | 0.148900927 | 0.135284877 | CCNT2/FOXO1 | 2 |
| GO: 0030742 | 22/17632 | 0.010020159 | 0.155965948 | 0.141703845 | RAPGEF6/RAB34 | 2 |
| GO: 0019955 | 125/17632 | 0.011123838 | 0.160730102 | 0.146032348 | THBS1/CXCR4/TGFBR1/TGFBR2 | 4 |
| GO: 0005518 | 67/17632 | 0.011224169 | 0.160730102 | 0.146032348 | THBS1/TGFBI/SERPINH1 | 3 |
| GO: 0017022 | 68/17632 | 0.011683934 | 0.160878783 | 0.146167433 | TRPM7/RAB27A/CXCR4 | 3 |
| GO: 0019903 | 133/17632 | 0.013712012 | 0.181811121 | 0.16518564 | FOXO1/SIRPA/STRN4/PPP1R9B | 4 |
| GO: 0005520 | 28/17632 | 0.015960482 | 0.185140606 | 0.168210665 | CTGF/IGFBP5 | 2 |
| GO: 0071813 | 28/17632 | 0.015960482 | 0.185140606 | 0.168210665 | LDLR/THBS1 | 2 |
| GO: 0071814 | 28/17632 | 0.015960482 | 0.185140606 | 0.168210665 | LDLR/THBS1 | 2 |
| GO: 0030332 | 29/17632 | 0.017066033 | 0.185140606 | 0.168210665 | FBXW7/CDK6 | 2 |
| GO: 0035035 | 29/17632 | 0.017066033 | 0.185140606 | 0.168210665 | MYOCD/CITED2 | 2 |
| GO: 0051721 | 29/17632 | 0.017066033 | 0.185140606 | 0.168210665 | FOXO1/STRN4 | 2 |
| GO: 0019199 | 81/17632 | 0.018648344 | 0.196356092 | 0.178400566 | EPHA4/TGFBR1/TGFBR2 | 3 |
| GO: 0003713 | 321/17632 | 0.024230576 | 0.247844173 | 0.225180387 | PRDM16/RARG/ZEB1/PRRX1/MYOCD/CITED2 | 6 |
| GO: 1901682 | 36/17632 | 0.025665148 | 0.255225634 | 0.231886859 | SLC35B3/SLC19A3 | 2 |
| GO: 0030020 | 37/17632 | 0.027010862 | 0.261348336 | 0.237449679 | COL1A1/COL5A2 | 2 |
| GO: 0043531 | 39/17632 | 0.02978569 | 0.274678286 | 0.249560689 | MYO6/APAF1 | 2 |
| GO: 0001078 | 169/17632 | 0.029923053 | 0.274678286 | 0.249560689 | FOXO1/HEYL/PRDM1/PRRX1 | 4 |
| GO: 0019902 | 178/17632 | 0.035210199 | 0.304397033 | 0.276561844 | FOXO1/SIRPA/STRN4/PPP1R9B | 4 |
| GO: 0000982 | 447/17632 | 0.03582466 | 0.304397033 | 0.276561844 | KLF5/FOXO1/MYC/HEYL/PRDM1/PRRX1/MYOCD | 7 |
| GO: 0019887 | 180/17632 | 0.036454143 | 0.304397033 | 0.276561844 | CCNT2/SOCS3/PRKAR1A/CALM2 | 4 |
| GO: 0001228 | 449/17632 | 0.036561655 | 0.304397033 | 0.276561844 | KLF5/CLOCK/MYC/HEYL/VEZF1/MYOCD/FOXO3 | 7 |
| GO: 0016706 | 47/17632 | 0.041933365 | 0.341185106 | 0.30998588 | TET3/P4HA2 | 2 |
| GO: 0051018 | 48/17632 | 0.043562833 | 0.346566539 | 0.314875215 | WASF2/PRKAR1A | 2 |
| GO: 0045309 | 49/17632 | 0.045215614 | 0.351895431 | 0.319716813 | FBXW7/SOCS3 | 2 |
| GO: 0005160 | 51/17632 | 0.04858954 | 0.352879061 | 0.320610497 | TGFBR1/TGFBR2 | 2 |
| GO: 0031625 | 286/17632 | 0.048696742 | 0.352879061 | 0.320610497 | FOXO1/FBXW7/TXNIP/CXCR4/PRKAR1A | 5 |
| GO: 0031490 | 119/17632 | 0.049642361 | 0.352879061 | 0.320610497 | CLOCK/PRDM1/FOXO3 | 3 |
Table 3.
The results of KEGG pathway analysis.
| ID | Description | Gene ratio | Bg ratio | p Value |
|---|---|---|---|---|
| hsa04218 | Cellular senescence | 10/57 | 160/7466 | 2.80E-07 |
| hsa04115 | p53 signaling pathway | 6/57 | 72/7466 | 1.60E-05 |
| hsa04910 | Insulin signaling pathway | 7/57 | 137/7466 | 7.39E-05 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 6/57 | 99/7466 | 9.81E-05 |
| hsa05220 | Chronic myeloid leukemia | 5/57 | 76/7466 | 0.000265202 |
| hsa04068 | FoxO signaling pathway | 6/57 | 132/7466 | 0.000472257 |
| hsa04211 | Longevity regulating pathway | 6/57 | 89/7466 | 0.000552582 |
| hsa04371 | Apelin signaling pathway | 6/57 | 137/7466 | 0.000575675 |
| hsa05165 | Human papillomavirus infection | 9/57 | 330/7466 | 0.000794412 |
| hsa04390 | Hippo signaling pathway | 6/57 | 154/7466 | 0.001064365 |
| hsa05160 | Hepatitis C | 6/57 | 155/7466 | 0.001100757 |
| hsa04213 | Longevity regulating pathway - multiple species | 6/57 | 62/7466 | 0.00122559 |
| hsa04151 | PI3K-Akt signaling pathway | 9/57 | 354/7466 | 0.0013066 |
| hsa04137 | Mitophagy – animal | 4/57 | 65/7466 | 0.001462455 |
| hsa04520 | Adherens junction | 4/57 | 72/7466 | 0.002136126 |
| hsa05212 | Pancreatic cancer | 4/57 | 75/7466 | 0.002481334 |
| hsa05202 | Transcriptional misregulation in cancer | 6/57 | 186/7466 | 0.002786785 |
| hsa05219 | Bladder cancer | 3/57 | 41/7466 | 0.003662102 |
| hsa04350 | TGF-beta signaling pathway | 4/57 | 85/7466 | 0.003907022 |
| hsa05210 | Colorectal cancer | 4/57 | 86/7466 | 0.004074595 |
| hsa04015 | Rap1 signaling pathway | 6/57 | 206/7466 | 0.004611998 |
| hsa05161 | Hepatitis B | 5/57 | 144/7466 | 0.004659537 |
| hsa05222 | Small cell lung cancer | 4/57 | 93/7466 | 0.005385847 |
| hsa05163 | Human cytomegalovirus infection | 6/57 | 225/7466 | 0.007051458 |
| ID | p. Adjust | q Value | Gene ID | Count |
|---|---|---|---|---|
| hsa04218 | 4.68E-05 | 3.51E-05 | TRPM7/FOXO1/MYC/CDK6/KRAS/TGFBR1/TGFBR2/SERPINE1/CALM2/FOXO3 | 10 |
| hsa04115 | 0.001338182 | 0.001003742 | PERP/APAF1/CDK6/SESN2/THBS1/SERPINE1 | 6 |
| hsa04910 | 0.004095217 | 0.003071736 | FOXO1/MKNK2/PRKAA1/KRAS/SOCS3/PRKAR1A/CALM2 | 7 |
| hsa04933 | 0.004095217 | 0.003071736 | FOXO1/KRAS/TGFBR1/COL1A1/TGFBR2/SERPINE1 | 6 |
| hsa05220 | 0.008857732 | 0.006643997 | MYC/CDK6/KRAS/TGFBR1/TGFBR2 | 5 |
| hsa04068 | 0.012017218 | 0.00901386 | FOXO1/PRKAA1/KRAS/TGFBR1/TGFBR2/FOXO3 | 6 |
| hsa04211 | 0.012017218 | 0.00901386 | FOXO1/PRKAA1/KRAS/SESN2/FOXO3 | 5 |
| hsa04371 | 0.012017218 | 0.00901386 | PRKAA1/KRAS/CTGF/TGFBR1/SERPINE1/CALM2 | 6 |
| hsa05165 | 0.014740748 | 0.011056723 | FOXO1/PARD6B/CDK6/KRAS/HEYL/LAMC1/THBS1/COL1A1/PPP2R5E | 9 |
| hsa04390 | 0.016711491 | 0.012534935 | PARD6B/MYC/CTGF/TGFBR1/TGFBR2/SERPINE1 | 6 |
| hsa05160 | 0.016711491 | 0.012534935 | LDLR/APAF1/MYC/CDK6/KRAS/SOCS3 | 6 |
| hsa04213 | 0.016784782 | 0.012589909 | FOXO1/PRKAA1/KRAS/FOXO3 | 4 |
| hsa04151 | 0.016784782 | 0.012589909 | PRKAA1/MYC/CDK6/KRAS/LAMC1/THBS1/COL1A1/PPP2R5E/FOXO3 | 9 |
| hsa04137 | 0.017444997 | 0.013085122 | OPTN/KRAS/CITED2/FOXO3 | 4 |
| hsa04520 | 0.023782202 | 0.017838526 | WASF2/CTNND1/TGFBR1/TGFBR2 | 4 |
| hsa05212 | 0.025898925 | 0.019426234 | CDK6/KRAS/TGFBR1/TGFBR2 | 4 |
| hsa05202 | 0.027376065 | 0.020534206 | CCNT2/FOXO1/MYC/ZEB1/AFF1/TGFBR2 | 6 |
| hsa05219 | 0.033976167 | 0.025484802 | MYC/KRAS/THBS1 | 3 |
| hsa04350 | 0.03402287 | 0.025519833 | MYC/THBS1/TGFBR1/TGFBR2 | 4 |
| hsa05210 | 0.03402287 | 0.025519833 | MYC/KRAS/TGFBR1/TGFBR2 | 4 |
| hsa04015 | 0.03537012 | 0.026530377 | PARD6B/RAPGEF6/CTNND1/KRAS/THBS1/CALM2 | 6 |
| hsa05161 | 0.03537012 | 0.026530377 | APAF1/MYC/CDK6/KRAS/TGFBR1 | 5 |
| hsa05222 | 0.03910593 | 0.029332528 | APAF1/MYC/CDK6/LAMC1 | 4 |
| hsa05163 | 0.049066392 | 0.03680366 | B2M/MYC/CDK6/KRAS/CXCR4/CALM2 | 6 |
Figure 5.
Functional analysis of 128 DEmi-mRNAs. (A) The dotplot of top 20 GO analysis. (B) The dotplot of KEGG pathway. (C) The network diagram between KEGG pathways and mRNA. The larger the circle, the more genes it contained; conversely, the smaller the circle, the fewer genes it contained. The color of the circle is correlated with the P-value. The smaller the P-value is, the closer it is to the red value. The larger the P-value is, the closer it is to the blue value. DE – differentially expressed; mi – micro; m – messenger; GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
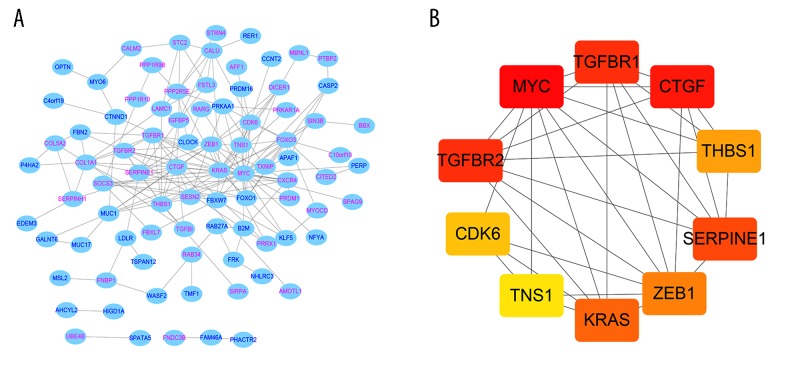
Screening of hub genes
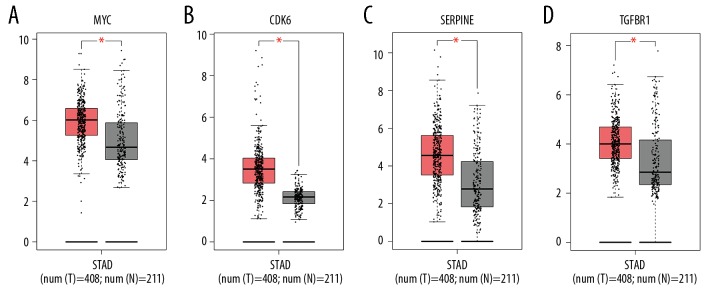
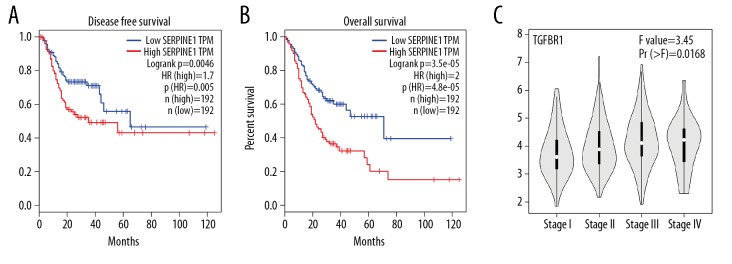
In order to further find the hub genes of 128 FmRNAs in the network, STRING database (http://string-db.org), Cytoscape and its plugin (cytoHubba) were applied, and the results showed that 86 genes were related to each other (Figure 6A). According to cytoHubba plugin’s MCC ranking, the top 10 hub genes were MYC (v-myc avian myelocytomatosis viral oncogene homolog), CTGF (connective tissue growth factor), TGFBR2 (TGF-beta receptor type-2), TGFBR1 (TGF-beta receptor type-1), SERPINE1 (plasminogen activator inhibitor 1), KRAS (Kirsten rat sarcoma viral oncogene homolog), ZEB1 (zinc finger E-box-binding homeobox 1), THBS 1 (thrombospondin-1), CDK6 (cyclin-dependent kinase 6), TNS1 (tensin-1) (Figure 6B, Table 4). The expression of all the 10 genes was verified on the GEPIA, and it was found that MYC, TGFBR1, SERPINE1, and CDK6 showed statistically significant differences in expression (Figure 7). In addition, we also found that highly expressed SERPINE1 had a poor OS and disease-free survival (DFS), and TGFBR1 expression increased along with the increase of clinical stages (Figure 8) [18].
Figure 6.
(A) PPI network diagram of 86 DEmi-mRNAs. (B) The network diagram of top 10 hub genes. PPI – protein-protein interaction; DE – differentially expressed; miRNA – microRNA; mRNA – messenger RNA.
Table 4.
The rank of hub genes via various of situations.
| Node name | MCC | DMNC | MNC | Degree | EPC | Bottle neck | Ec centricity | Closeness | Radiality | Betweenness | Stress | Clustering coefficient |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYC | 609 | 0.24794 | 25 | 30 | 36.302 | 49 | 0.22965 | 50.91667 | 5.72361 | 2930.436 | 6766 | 0.13563 |
| CTGF | 401 | 0.37042 | 13 | 14 | 35.37 | 8 | 0.18372 | 40.61667 | 5.33497 | 676.2413 | 2134 | 0.31868 |
| TGFBR2 | 396 | 0.49882 | 10 | 10 | 34.088 | 3 | 0.18372 | 38.03333 | 5.25253 | 183.2476 | 1158 | 0.55556 |
| TGFBR1 | 396 | 0.49882 | 10 | 10 | 33.892 | 1 | 0.18372 | 38.03333 | 5.25253 | 183.2476 | 1158 | 0.55556 |
| SERPINE1 | 370 | 0.5012 | 9 | 11 | 34.56 | 8 | 0.18372 | 39.45 | 5.34675 | 868.6996 | 2280 | 0.38182 |
| KRAS | 322 | 0.36053 | 15 | 17 | 36.072 | 16 | 0.18372 | 42.76667 | 5.39386 | 828.0731 | 2704 | 0.26471 |
| ZEB1 | 318 | 0.45368 | 12 | 12 | 35.42 | 8 | 0.18372 | 39.2 | 5.28787 | 194.055 | 870 | 0.4697 |
| THBS1 | 259 | 0.41901 | 10 | 11 | 33.93 | 6 | 0.18372 | 38.61667 | 5.26431 | 402.2999 | 1202 | 0.38182 |
| CDK6 | 250 | 0.47733 | 9 | 11 | 33.692 | 3 | 0.18372 | 38.26667 | 5.20543 | 428.6176 | 1188 | 0.36364 |
| TNS1 | 240 | 0.62199 | 7 | 7 | 32.135 | 1 | 0.18372 | 35.18333 | 5.07588 | 8.55088 | 56 | 0.80952 |
| COL1A1 | 142 | 0.32239 | 11 | 11 | 33.115 | 4 | 0.1531 | 34.35 | 4.85212 | 350.8601 | 1722 | 0.34545 |
| FOXO1 | 134 | 0.31924 | 10 | 10 | 33.841 | 4 | 0.18372 | 37.95 | 5.24076 | 203.7761 | 800 | 0.35556 |
| FOXO3 | 131 | 0.4082 | 8 | 11 | 32.054 | 2 | 0.18372 | 36.85 | 5.09943 | 398.0874 | 1268 | 0.25455 |
| DICER1 | 121 | 0.64826 | 5 | 6 | 29.184 | 2 | 0.18372 | 34.18333 | 5.02877 | 157.1734 | 434 | 0.66667 |
| CXCR4 | 58 | 0.37904 | 8 | 8 | 32.284 | 11 | 0.18372 | 36.86667 | 5.2172 | 178.6964 | 600 | 0.46429 |
| MUC1 | 52 | 0.47549 | 6 | 8 | 30.378 | 5 | 0.18372 | 36.43333 | 5.1701 | 366.1695 | 864 | 0.39286 |
| IGFBP5 | 32 | 0.36588 | 7 | 7 | 28.18 | 1 | 0.1531 | 32.31667 | 4.82856 | 193.0638 | 622 | 0.47619 |
| LAMC1 | 28 | 0.38039 | 6 | 6 | 26.728 | 1 | 0.1531 | 30.46667 | 4.67546 | 88.60129 | 278 | 0.53333 |
| CALU | 27 | 0.56839 | 4 | 7 | 27.046 | 7 | 0.1531 | 32.88333 | 4.87567 | 368.5913 | 1006 | 0.33333 |
| STC2 | 25 | 0.56839 | 4 | 5 | 21.431 | 2 | 0.13123 | 27.22619 | 4.34571 | 112.2128 | 258 | 0.6 |
| FSTL3 | 24 | 0.56839 | 4 | 4 | 21.367 | 1 | 0.13123 | 26.34286 | 4.28682 | 0 | 0 | 1 |
| SOCS3 | 21 | 0.38039 | 6 | 7 | 29.643 | 3 | 0.18372 | 34.51667 | 5.02877 | 250.4977 | 792 | 0.38095 |
| FBXW7 | 20 | 0.38039 | 6 | 6 | 29.119 | 1 | 0.18372 | 34.18333 | 5.02877 | 77.23846 | 304 | 0.53333 |
| FBN2 | 8 | 0.37893 | 4 | 4 | 23.641 | 2 | 0.1531 | 27.75 | 4.48703 | 26.03518 | 280 | 0.66667 |
| KLF5 | 7 | 0.46346 | 3 | 4 | 24.062 | 2 | 0.18372 | 32.68333 | 4.96989 | 59.30048 | 238 | 0.5 |
| PRKAA1 | 7 | 0.30898 | 3 | 6 | 25.204 | 1 | 0.1531 | 30.03333 | 4.6048 | 61.26671 | 174 | 0.13333 |
| PRRX1 | 6 | 0.46346 | 3 | 3 | 23.368 | 1 | 0.1531 | 27.36667 | 4.48703 | 0 | 0 | 1 |
| TGFBI | 6 | 0.46346 | 3 | 3 | 23.103 | 1 | 0.1531 | 28.15 | 4.53414 | 0 | 0 | 1 |
| COL5A2 | 6 | 0.2842 | 4 | 4 | 19.627 | 1 | 0.13123 | 24.70952 | 3.9924 | 5 | 10 | 0.5 |
| PPP2R5E | 5 | 0 | 1 | 5 | 19.69 | 3 | 0.1531 | 30.61667 | 4.69902 | 302.8195 | 790 | 0 |
| CASP2 | 5 | 0.30898 | 3 | 4 | 20.746 | 2 | 0.18372 | 31.93333 | 4.88745 | 143.4932 | 330 | 0.33333 |
| SERPINH1 | 5 | 0.30898 | 3 | 4 | 20.532 | 2 | 0.1531 | 28.4 | 4.52236 | 171.9268 | 490 | 0.33333 |
| TXNIP | 4 | 0.30898 | 3 | 3 | 20.773 | 2 | 0.18372 | 32.1 | 4.93456 | 12.23252 | 76 | 0.66667 |
| PRDM1 | 4 | 0.30898 | 3 | 3 | 22.727 | 1 | 0.18372 | 31.1 | 4.85212 | 0.68182 | 4 | 0.66667 |
| B2M | 4 | 0.30779 | 2 | 4 | 18.355 | 7 | 0.18372 | 32.53333 | 4.96989 | 716.2094 | 1514 | 0.33333 |
| CTNND1 | 4 | 0.30779 | 2 | 4 | 18.921 | 5 | 0.18372 | 32.68333 | 4.95811 | 480.2316 | 832 | 0.16667 |
| APAF1 | 4 | 0.30898 | 3 | 3 | 20.821 | 1 | 0.18372 | 31.76667 | 4.89922 | 4.66667 | 16 | 0.66667 |
| SIN3B | 3 | 0.30779 | 2 | 3 | 18.56 | 2 | 0.18372 | 32.26667 | 4.95811 | 154 | 238 | 0.33333 |
| FNBP1 | 3 | 0 | 1 | 3 | 4.766 | 3 | 0.1531 | 22.36667 | 3.81574 | 277.7906 | 550 | 0 |
| LDLR | 3 | 0 | 1 | 3 | 11.833 | 5 | 0.18372 | 27.81667 | 4.58125 | 537.7906 | 1026 | 0 |
| FRK | 3 | 0.30779 | 2 | 3 | 7.811 | 2 | 0.18372 | 24.4 | 4.15727 | 154 | 298 | 0.33333 |
| MYO6 | 3 | 0 | 1 | 3 | 8.446 | 3 | 0.1531 | 24.88333 | 4.19261 | 218.1825 | 368 | 0 |
| SESN2 | 3 | 0.30779 | 2 | 3 | 21.783 | 1 | 0.18372 | 32.85 | 5.01699 | 34.11001 | 178 | 0.33333 |
| RAB27A | 3 | 0.30779 | 2 | 3 | 8.435 | 4 | 0.18372 | 24.86667 | 4.20438 | 326.2094 | 692 | 0.33333 |
| RAB34 | 3 | 0 | 1 | 3 | 4.339 | 3 | 0.1531 | 20.68333 | 3.43888 | 210.7381 | 444 | 0 |
| CLOCK | 3 | 0 | 1 | 3 | 16.66 | 1 | 0.1531 | 28.1 | 4.58125 | 65.63025 | 158 | 0 |
| PERP | 2 | 0 | 1 | 2 | 12.381 | 2 | 0.18372 | 30.6 | 4.84034 | 43.91725 | 162 | 0 |
| CALM2 | 2 | 0 | 1 | 2 | 8.433 | 1 | 0.13123 | 22.04286 | 3.76863 | 29.95094 | 44 | 0 |
| RARG | 2 | 0 | 1 | 2 | 14.151 | 2 | 0.18372 | 30.93333 | 4.87567 | 28.30952 | 88 | 0 |
| WASF2 | 2 | 0 | 1 | 2 | 3.067 | 1 | 0.13123 | 18.80952 | 3.06202 | 38.52872 | 68 | 0 |
| MBNL1 | 2 | 0 | 1 | 2 | 10.944 | 1 | 0.1531 | 24.3 | 4.15727 | 10.72727 | 40 | 0 |
| MYOCD | 2 | 0 | 1 | 2 | 14.687 | 1 | 0.1531 | 25.3 | 4.23971 | 2.05556 | 8 | 0 |
| FAM46A | 2 | 0 | 1 | 2 | 1.568 | 3 | 0.03488 | 2 | 0.12209 | 2 | 2 | 0 |
| PRKAR1A | 2 | 0 | 1 | 2 | 16.389 | 1 | 0.1531 | 27.86667 | 4.53414 | 4.43252 | 46 | 0 |
| CITED2 | 2 | 0 | 1 | 2 | 12.229 | 1 | 0.1531 | 25.05 | 4.21616 | 3.06667 | 8 | 0 |
| PPP1R10 | 2 | 0 | 1 | 2 | 11.151 | 1 | 0.1531 | 24.55 | 4.18083 | 27.70586 | 98 | 0 |
| PPP1R9B | 2 | 0 | 1 | 2 | 8.77 | 1 | 0.13123 | 22.55952 | 3.8393 | 7 | 8 | 0 |
| PTBP2 | 2 | 0 | 1 | 2 | 8.932 | 1 | 0.1531 | 23.3 | 4.01595 | 3.55385 | 8 | 0 |
| P4HA2 | 2 | 0.30779 | 2 | 2 | 13.847 | 1 | 0.13123 | 23.54286 | 3.95707 | 0 | 0 | 1 |
| GALNT6 | 2 | 0.30779 | 2 | 2 | 11.642 | 1 | 0.1531 | 24.96667 | 4.27504 | 0 | 0 | 1 |
| FBXL7 | 2 | 0.30779 | 2 | 2 | 15.652 | 1 | 0.1531 | 24.63333 | 4.19261 | 0 | 0 | 1 |
| MUC17 | 2 | 0.30779 | 2 | 2 | 11.691 | 1 | 0.1531 | 24.96667 | 4.27504 | 0 | 0 | 1 |
| RER1 | 1 | 0 | 1 | 1 | 8.885 | 1 | 0.13123 | 22.69286 | 3.96884 | 0 | 0 | 0 |
| AMOTL1 | 1 | 0 | 1 | 1 | 12.027 | 1 | 0.1531 | 26.06667 | 4.42815 | 0 | 0 | 0 |
| SPAG9 | 1 | 0 | 1 | 1 | 10.867 | 1 | 0.18372 | 29.93333 | 4.81679 | 0 | 0 | 0 |
| SIRPA | 1 | 0 | 1 | 1 | 10.606 | 1 | 0.1531 | 25.31667 | 4.35748 | 0 | 0 | 0 |
| AHCYL2 | 1 | 0 | 1 | 1 | 1.322 | 1 | 0.02326 | 1 | 0.06977 | 0 | 0 | 0 |
| HIGD1A | 1 | 0 | 1 | 1 | 1.322 | 1 | 0.02326 | 1 | 0.06977 | 0 | 0 | 0 |
| PHACTR2 | 1 | 0 | 1 | 1 | 1.374 | 1 | 0.01744 | 1.5 | 0.10465 | 0 | 0 | 0 |
| MSL2 | 1 | 0 | 1 | 1 | 2.175 | 1 | 0.13123 | 17.24286 | 2.90891 | 0 | 0 | 0 |
| AFF1 | 1 | 0 | 1 | 1 | 10.166 | 1 | 0.1531 | 25.1 | 4.2986 | 0 | 0 | 0 |
| TMF1 | 1 | 0 | 1 | 1 | 2.402 | 1 | 0.13123 | 16.14524 | 2.53205 | 0 | 0 | 0 |
| C4orf19 | 1 | 0 | 1 | 1 | 6.234 | 1 | 0.1531 | 22.86667 | 4.05128 | 0 | 0 | 0 |
| STRN4 | 1 | 0 | 1 | 1 | 6.291 | 1 | 0.13123 | 21.64286 | 3.79219 | 0 | 0 | 0 |
| BBX | 1 | 0 | 1 | 1 | 6.08 | 1 | 0.1531 | 22.75 | 4.05128 | 0 | 0 | 0 |
| SPATA5 | 1 | 0 | 1 | 1 | 1.289 | 1 | 0.02326 | 1 | 0.06977 | 0 | 0 | 0 |
| UBE4B | 1 | 0 | 1 | 1 | 1.289 | 1 | 0.02326 | 1 | 0.06977 | 0 | 0 | 0 |
| PRDM16 | 1 | 0 | 1 | 1 | 11.749 | 1 | 0.18372 | 29.93333 | 4.81679 | 0 | 0 | 0 |
| C10orf10 | 1 | 0 | 1 | 1 | 9.952 | 1 | 0.1531 | 24.38333 | 4.19261 | 0 | 0 | 0 |
| OPTN | 1 | 0 | 1 | 1 | 3.585 | 1 | 0.13123 | 18.7619 | 3.28578 | 0 | 0 | 0 |
| TSPAN12 | 1 | 0 | 1 | 1 | 4.562 | 1 | 0.1531 | 20.43333 | 3.67442 | 0 | 0 | 0 |
| FNDC3B | 1 | 0 | 1 | 1 | 1.358 | 1 | 0.01744 | 1.5 | 0.10465 | 0 | 0 | 0 |
| EDEM3 | 1 | 0 | 1 | 1 | 7.224 | 1 | 0.13123 | 20.55952 | 3.61553 | 0 | 0 | 0 |
| CCNT2 | 1 | 0 | 1 | 1 | 10.511 | 1 | 0.1531 | 25.1 | 4.2986 | 0 | 0 | 0 |
| NHLRC3 | 1 | 0 | 1 | 1 | 3.536 | 1 | 0.1531 | 18.5 | 3.25045 | 0 | 0 | 0 |
| NFYA | 1 | 0 | 1 | 1 | 11.172 | 1 | 0.18372 | 29.93333 | 4.81679 | 0 | 0 | 0 |
Figure 7.
Four hub genes expression in GEPIA; (A) MYC, (B) CDK6, (C) SERPINE, (D) TGFBR1.
Figure 8.
(A) Disease-free survival of SERPINE; (B) overall survival of SERPINE. (C) Relationship between clinical stage and TGFBR1 expression.
Discussion
Although the levels of diagnosis and treatment of GC is constantly improving, GC is still a high-risk disease, and a large part of its potential occurrence and development mechanism is still unclear. Prior to this, many studies on the pathogenesis of GC have been presented, but mainly focusing on genes encoding proteins. Since non-coding protein RNAs have appeared in everyone’s field of vision and have been found to have specific regulatory functions, researchers have been investigating more and more non-coding RNAs, especially circRNAs. In this study, we discovered a few new circRNAs that target the regulation of downstream genes through sponge-adsorbed miRNAs. We also performed functional analysis of these target genes to understand the potential functions of these circRNAs.
In our study, we obtained a network of 3 circRNAs, 22 miRNAs, and 128 mRNAs using the GEO datasets. In addition, we found that on the CSCD website that hsa_circ_0009076 was composed of the 29th and 30th exons of the reverse transcript of the gene NRDC (location: 1p32.3, exon count: 35), hsa_circ_0028190 was composed of the 6th–11th exons of the reverse transcript of the gene ANAPC7 (location: 12q24.11, exon count: 13), and hsa_circ_0041732 was composed of the 8th exons of the forward transcript of the gene FAM64A (location: 17p13.2, exon count: 16). The 3 circRNAs have not been reported previous in the literature. We named hsa_circ_0009076, hsa_circ_0028190, and hsa_circ_0041732 respectively as circNRDC, circANAPC7, and circFAM64A. In the future, more experiments will be needed to verify the expression of these circRNAs and their effects on the proliferation, apoptosis, invasion, and metastasis of GC cells.
In order to explore the indirect mechanism of circRNAs on GC, miRNAs and mRNAs of circRNAs downstream in the regulatory network were further analyzed. We validated the 22 differentially expressed miRNAs in TCGA database and found that the 3 miRNAs had the same expression (has-miR-96, has-miR-182, and has-miR-195). The correlation with clinical features by chi-square test demonstrated that hsa-mir-182 was associated with T stage and N stage, hsa-mir-96 was associated with age, T stage, and N stage, and hsa-mir-195 was associated with N stage, although only 3 of the 22 miRNAs have been confirmed, which may be due to different sample and statistical methods; more experiments are needed to verify our results. Hsa-miR-96-5p has been reported in many studies, for example, miR-96 was successfully shown to increase expression in GC compared to normal or adenoma samples and was further validated with real-time quantitative polymerase chain reaction (RT-qPCR) in 77 samples [19]. Hsa-miR-96 was shown to be upregulated in tumor tissues and HepG2 cells, and to promote tumorigenesis and progression by inhibiting FOXO1 and activating of AKT/GSK-3β/β-catenin signaling pathway in hepatocellular carcinoma [20]. In addition, hsa-miR-96 accelerates invasion and migration of bladder cancer through epithelial-mesenchymal transition in response to transforming growth factor-β1 [21]. One circRNA can adsorb multiple miRNAs, meanwhile, one miRNA can also be adsorbed by multiple circRNAs. In addition, we found that mir-182-5p in our network could be adsorbed by hsa_circ_0041732. Sun et al. showed that high expression of circ-SFMBT2 was related to the advanced progression of GC. The functional mechanism experiment showed that circ-SFMBT2 could regulate CREB1 by sponging mir-182-5p to promote the progression of GC [22]. Li et al. showed that miR-182-5p had a higher expressed level through comparison between GC tissue and normal tissue, and improved the viability, mitosis, migration, and invasion ability of human GC cells by downregulating RAB27A [23]. Wang et al. reported that expression levels of miR-195-5p and bFGF showed negative correlation in human GC tissues and miRNA-195 suppressed human GC by binding basic fibroblast growth factor [24]. Ye et al. demonstrated that miR-195 overexpression restrained invasion, migration, and proliferation of GC cells in vitro and enhanced the chemotherapy sensitivity of cisplatin in GC cells, furthermore, benefiting survival prognosis of GC patients [25]. These studies confirmed that the expression of the 3 miRNAs in our network was indeed involved in the occurrence and progression of GC.
Among KEGG tumor-related signaling pathways, PI3K-Akt signaling pathway contains the largest number of genes (PRKAA1/MYC/CDK6/KRAS/LAMC1/THBS1/COL1A1/PPP2R5E/FOXO3), while p53 signaling pathway (PERP/APAF1/CDK6/SESN2/THBS1/SERPINE1) has the smallest P value. We focused on these 2 signaling pathways. The PI3K/Akt signaling pathway functions as a vital intermediary to facilitate various cellular and physiological processes, including the cell cycle, cellular growth, differentiation, survival, apoptosis, metabolism, angiogenesis, and migration [26]. Accumulating evidence has displayed that crucial epigenetic modifiers are directly or indirectly regulated by PI3K/AKT signaling and involved in oncogenesis of PI3K cascade in cancers [27]. The core of p53 signaling pathway is p53, which functions as safeguarding the integrity of the genome. Activation of p53 affects apoptosis, cell cycle arrest, angiogenesis, DNA repair, and metastasis by mainly regulating downstream targeting genes [28]. Wei et al. showed that overexpression of lncRNA MEG3 could increase the expression of p53, then its conclusion demonstrated that lncRNA MEG3 restrain proliferation and metastasis of GC via p53 signaling pathway [29]. In our regulatory network, both THBS1 and CDK6 are involved in the regulation of PI3K-Akt and p53 signaling pathways. THBS1 was the first member of extracellular matrix (ECM) proteins family, which acts as an angiogenesis inhibitor and regulates tumor cell adhesion, invasion, migration, proliferation, apoptosis, and tumor immunity [30,31]. CDK6 function cell-cycle progression, transcription, tissue homeostasis and differentiation as one member of cyclin-dependent kinases [32]. A recent study found that the inhibition of hsa_circ_0081143 reduced GC cells invasion ability, viability, and increased the sensitivity of GC cells to cisplatin (DDP) in vitro act as an endogenous sponge adsorption by directly binding to miR-646 and efficiently inversing the inhibition of CDK6 [33].
The 4 of top 10 hub genes (THBS1, CDK6, SERPINE1, TGFBR1) in the PPI network were consistent with the expression of GEPIA. Their survival analysis and clinical stage correlated with the expression were further explored. It was found that highly expressed SERPINE1 had a poor prognosis, and the expression of TGFBR1 was positively correlated with clinical stage. SERPINE1 encodes a member of the serine proteinase inhibitor (serpin) superfamily, which is the principal inhibitor of tissue plasminogen activator (tPA) and urokinase (uPA). Overexpression of uPA/uPAR and SERPINE1 promotes tumor cell invasion and migration, playing an important role in metastasis development, conferring poor prognosis. In addition, both uPA/uPAR and SERPINE1 are directly associated with the induction of the acquisition of stem cell properties, epithelial-to-mesenchymal transition and resistance to antitumor agents [34]. The protein encoded by TGFBR1, as a serine/threonine protein kinase, forms a heteromeric complex with type II TGF-beta receptors when bound to TGF-beta, transducing the TGF-beta signal from the cell surface to the cytoplasm, which participated in the regulation of various cell physiology and pathological processes, including adhesion, motility, differentiation, division and apoptosis, and plays a vital role in tumor invasion and metastasis by mediating epithelial-to-mesenchymal transition (EMT) [35]. So in our regulatory network, we found 4 important circRNA-miRNA-mRNA axis including hsa_circ_0041732/hsa-mir-182-5p/THBS1, hsa_circ_0028190/hsa-mir-4291/CDK6, hsa_circ_0009076/hsa-mir-143-3p/SERPINE1, and hsa_circ_0028190/hsa-mir-140-5p/TGFBR1 about the 3 circRNAs. In future studies, more experiments are expected to verify this possible ceRNA mechanism in GC.
Conclusions
Although there are some studies on circRNA in GC, the number and methods of research are different. In this study, we constructed a circRNA-miRNA-mRNA regulatory network including 3 important circRNAs (hsa_circ_0009076, hsa_circ_0028190, and hsa_circ_0041732) through multiple GEO databases and TCGA database and performed functional enrichment analysis on these final target genes for understanding the potential functional mechanisms of circRNA. Of course, it is more important to combine basic experiments with clinical data to further explore the feasibility of these circRNAs, so as to provide new molecular marker of prediction, prognosis, and therapeutic targets for clinical patients.
Supplementary Data
Supplementary Tables 1–6 available from the corresponding author on request.
(A) The intersection of targeting mRNAs and DEmRNAs. (B) The intersection of targeting miRNAs and DEmiRNAs. mRNA – messenger RNA; DE – differentially expressed; miRNA – microRNA.
Heatmap of differentially expressed miRNAs in gastric cancer on TCGA. miRNA – microRNA; TCGA – The Caner Genome Atlas
(A) PI3K-Akt signaling pathway diagram; (B) p53 signaling pathway diagram. Red represents upregulated mRNAs, blue represents downregulated mRNAs. mRNA –messenger RNA; miRNA – mircoRNA.
Acknowledgements
We thank Zhaowu Ma for the technical advice.
Footnotes
Source of support: Departmental sources
Conflict of interests
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer Jo Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Duarte HO, Gomes J, Machado JC, Reis CA. Gastric cancer: Basic aspects. Helicobacter. 2018;23(Suppl 1):e12523. doi: 10.1111/hel.12523. [DOI] [PubMed] [Google Scholar]
- 3.Spolverato G, Ejaz A, Kim Y, et al. Rates and patterns of recurrence after curative intent resection for gastric cancer: A United States multi-institutional analysis. J Am Coll Surg. 2014;219(4):664–75. doi: 10.1016/j.jamcollsurg.2014.03.062. [DOI] [PubMed] [Google Scholar]
- 4.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–88. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Huang C, Bao C, et al. Corrigendum: Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2017;24(2):194. doi: 10.1038/nsmb0217-194a. [DOI] [PubMed] [Google Scholar]
- 11.Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–58. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Ma K, Pitts S, et al. Novel circular RNA NF1 acts as a molecular sponge, promoting gastric cancer by absorbing miR-16. Endocr Relat Cancer. 2018 doi: 10.1530/ERC-18-0478. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang J, Hong H, Xue X, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–32. doi: 10.1016/j.canlet.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang Y, Li Y, Huang Y, et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234(7):10458–69. doi: 10.1002/jcp.27714. [DOI] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang J, Min BH, Jang J, et al. MicroRNA expression profiles in gastric carcinogenesis. Sci Rep. 2018;8(1):14393. doi: 10.1038/s41598-018-32782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang N, Zhou J, Li Q, et al. MiR-96 exerts carcinogenic effect by activating AKT/GSK-3beta/beta-catenin signaling pathway through targeting inhibition of FOXO1 in hepatocellular carcinoma. Cancer Cell Int. 2019;19:38. doi: 10.1186/s12935-019-0756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C, Zhang Q, Gu R, et al. MiR-96 regulates migration and invasion of bladder cancer through epithelial-mesenchymal transition in response to transforming growth factor-beta1. J Cell Biochem. 2018;119(9):7807–17. doi: 10.1002/jcb.27172. [DOI] [PubMed] [Google Scholar]
- 22.Sun H, Xi P, Sun Z, et al. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res. 2018;10:5725–34. doi: 10.2147/CMAR.S172592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Chen S, Shan Z, et al. miR-182-5p improves the viability, mitosis, migration, and invasion ability of human gastric cancer cells by down-regulating RAB27A. Biosci Rep. 2017;37(3) doi: 10.1042/BSR20170136. pii: BSR20170136. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Wang J, Li L, Jiang M, Li Y. MicroRNA-195 inhibits human gastric cancer by directly targeting basic fibroblast growth factor. Clin Translat Oncol. 2017;19(11):1320–28. doi: 10.1007/s12094-017-1668-4. [DOI] [PubMed] [Google Scholar]
- 25.Ye R, Wei B, Li S, et al. Expression of miR-195 is associated with chemotherapy sensitivity of cisplatin and clinical prognosis in gastric cancer. Oncotarget. 2017;8(57):97260–72. doi: 10.18632/oncotarget.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye B, Jiang LL, Xu HT, et al. Expression of PI3K/AKT pathway in gastric cancer and its blockade suppresses tumor growth and metastasis. Int J Immunopathol Pharmacol. 2012;25(3):627–36. doi: 10.1177/039463201202500309. [DOI] [PubMed] [Google Scholar]
- 27.Yang Q, Jiang W, Hou P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.04.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Harris SL, Levine AJ. The p53 pathway: Positive and negative feedback loops. Oncogene. 2005;24(17):2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 29.Wei GH, Wang X. LncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(17):3850–56. [PubMed] [Google Scholar]
- 30.Naumov GN, Bender E, Zurakowski D, et al. A model of human tumor dormancy: An angiogenic switch from the nonangiogenic phenotype. J Natl Cancer Inst. 2006;98(5):316–25. doi: 10.1093/jnci/djj068. [DOI] [PubMed] [Google Scholar]
- 31.Jeanne A, Schneider C, Martiny L, Dedieu S. Original insights on thrombospondin-1-related antireceptor strategies in cancer. Front Pharmacol. 2015;6:252. doi: 10.3389/fphar.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tigan AS, Bellutti F, Kollmann K, et al. CDK6-a review of the past and a glimpse into the future: From cell-cycle control to transcriptional regulation. Oncogene. 2016;35(24):3083–91. doi: 10.1038/onc.2015.407. [DOI] [PubMed] [Google Scholar]
- 33.Xue M, Li G, Fang X, et al. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int. 2019;19:25. doi: 10.1186/s12935-019-0737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavon MA, Arroyo-Solera I, Cespedes MV, et al. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget. 2016;7(35):57351–66. doi: 10.18632/oncotarget.10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vander Ark A, Cao J, Li X. TGF-beta receptors: In and beyond TGF-beta signaling. Cell Signal. 2018;52:112–20. doi: 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The intersection of targeting mRNAs and DEmRNAs. (B) The intersection of targeting miRNAs and DEmiRNAs. mRNA – messenger RNA; DE – differentially expressed; miRNA – microRNA.
Heatmap of differentially expressed miRNAs in gastric cancer on TCGA. miRNA – microRNA; TCGA – The Caner Genome Atlas
(A) PI3K-Akt signaling pathway diagram; (B) p53 signaling pathway diagram. Red represents upregulated mRNAs, blue represents downregulated mRNAs. mRNA –messenger RNA; miRNA – mircoRNA.