Abstract
Background
Dysregulation of the microRNA (miRNA) network is a typical feature of many cancers. However, the key specific miRNAs involved in uveal melanoma carcinogenesis are largely unknown.
Material/Methods
RT-qPCR was used to detected miR-652 expression in uveal melanoma tissues and cell lines. miR-652 inhibitor was transfected into uveal melanoma cells to decrease miR-652 expression and determine the biological role of miR-652 by CCK-8 and wound healing assays. Bioinformatic analysis and dual luciferase reporter assay were used to predict and validate the target gene of miR-652. HOXA9 siRNA was transfected into cells to confirm that miR-652 relies on regulation of HOXA9 to regulate cell proliferation and migration.
Results
RT-qPCR showed that miR-652 was overexpressed in uveal melanoma cell lines (MUM-2B, MEL270) compared with melanocyte cells (ARPE-19). Overexpression of miR-652 was also observed in uveal melanoma compared to paired non-tumor tissues. Downregulation of miR-652 inhibited the cell proliferation ability and migration ability of uveal melanoma cells. Using bioinformatic analysis, HOXA9 was found to be a potential target gene of miR-652. The direct regulation of HOXA9 by miR-652 was experimentally validated in uveal melanoma cells by dual luciferase assay and Western blotting. We also observed that miR-652 promoted HIF-1α signaling via repression of HOXA9 in uveal melanoma cells. Silencing of HOXA9 attenuated the miR-652 inhibitor decreased cell growth rate and decreased migration ability in uveal melanoma cells.
Conclusions
Our data demonstrate an oncogenic role of miR-652 in uveal melanoma, showing that miR-652 may be a useful biomarker for prediction of prognosis for patients with uveal melanoma.
MeSH Keywords: Cell Migration Assays, Cell Proliferation, Melanoma, MicroRNAs
Background
Uveal melanoma (UM) is one of the most commonly diagnosed adult primary intraocular malignancies [1]. With strong invasive potential, UM cells are highly metastatic and show a propensity to metastasis to the liver [2]. Although great progress has been made in the diagnosis and treatment of UM, the prognosis of patients with UM remains dismal, with a median overall survival of only several months after diagnosis of metastasis [3]. Certain mutations, such as monosomy 3, has been identified as drivers of metastasis of patients with UM, and were proved to be prognostic predictors for patients [4,5]. The discovery of new molecular mechanisms underlying the proliferation and metastasis of UM is crucial for providing the basis for novel targeted therapies to improve the clinical outcome of patients with UM.
MicroRNAs (miRNAs) are small, non-coding, single-stranded RNA molecules in mammalian cells [6]. Through binding to 3′UTR of target gene mRNA, miRNAs destabilize the mRNA or inhibit the translation process, leading to altered expression of genes at the post-transcriptional level [7]. As the target genes can be oncogenes or tumor suppressors, dysregulation of miRNAs is critical for cancer progression and is observed in many cancer types [8,9]. Several studies have identified miRNAs that are pivotal for development of UM [10]. For example, miR-144 and miR-34a were experimentally confirmed as being downregulated miRNAs in UM cells, and these 2 miRNAs can target c-MET, a well-characterized oncogene in UM, to enhance the proliferation and metastasis ability of UM cells [11,12]. Additionally, microarray analysis has revealed several differentially expressed miRNAs between UM with low risk and high risk of metastasis [13] and between metastatic and non-metastatic UM [14]. A 6-miRNAs (let-7b, miR-143, miR-193b, miR-199a, miR-199a*, and miR-652) classifier was showed to accurately distinguish UM patients with high risk metastasis from those with low risk [13]. A later study indicated that monosomy 3 can promote miR-199-5p expression to facilitate metastasis in UM [15]. However, the underlying mechanism by which these miRNAs mediate cancer progression remains elusive.
HOX members are transcription factors that mediate embryonic development via regulation of gene expression [16]. Consistent with its other members, HOXA9 is also a regulator of embryonic development [17]. Recent studies discovered that HOXA9 is associated with tumorigenesis [18,19]. The role of HOXA9 in cancer is not unique. In non-small cell lung cancer, HOXA9 suppressed the invasiveness of cancer cells and its expression is an indicator of cancer occurrence [20]. In ovarian cancer, HOXA9 activated transcription of TGF-β2 to promote cancer progression [21]. Via inhibition of HIF-1α, HOXA9 repressed glucose metabolism in cutaneous squamous cell carcinoma to function as a tumor suppressor [22].
In the present study, we discovered that miR-652 was overexpressed in UM. The function assays indicated that miR-652 downregulation inhibited cell proliferation and migration of UM cells. It was predicted and confirmed that HOXA9 was a target gene of miR-652 in UM cells. Moreover, silencing of HOXA9 attenuated miR-652 inhibitor-induced inhibition of cell proliferation and migration in UM cells. Our study demonstrates an oncogenic role of miR-652 in UM, which provides a novel therapeutic target for patients with UM.
Material and Methods
Tissue collection
The present study was supervised and approved by the Ethic Committee of the Sixth Affiliated Hospital, Sun Yat-sen University (dated 2016.03.05). We collected 26 pairs of uveal melanoma and matched normal tissues during surgery from patients diagnosed with uveal melanoma. The tissues were immediately stored at −80°C before use in the following experiments. All participants provided written informed consent.
Cell culture
Human uveal melanoma cell lines MUM-2B and MEL270 and human immortalized retinal pigment epithelial cell line ARPE-19 were purchased from the Cell Bank of the Chinese Academy of Sciences (Beijing, China). All cell lines were cultured in DMEM (Gibco, Waltham, MA) containing 10% FBS (HyClone, Logan, UT) in a humidified incubator with 5% CO2 at 37°C.
RNA extraction and RT-qPCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The RNA was reverse transcribed to cDNA using a PrimeScript RT reagent kit (TaKaRa, Shiga, Japan). RT-qPCR was performed with SYBR Premix Ex Taq (TaKaRa) on a CFX96 Realtime System (Bio-Rad Laboratories, Inc., Hercules, CA). The qPCR conditions were predenatured at 95°C for 30 s, followed by denature at 95°C for 10 s, annealing and elongation at 60°C for 30 s for 35 cycles. The relative expressions of miRNA and mRNA were calculated using 2−ΔΔCt method [23]. β-actin and U6 were used as internal controls for mRNA and miRNA, respectively. The primer sequences are listed in Table 1.
Table 1.
Primer sequences for RT-qPCR.
| Primer | Sequences |
|---|---|
| Stem-loop | 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGAATG-3′ |
| miR-652-forward | 5′-GCCGAGCAACCCTAGGAGAG-3′ |
| miR-652-reverse | 5′-CTCAACTGGTGTCGTGGA-3′ |
| U6-forward | 5′-TGCGGGTGCTCGCTTCGCAGC-3′ |
| U6-reverse | 5′-CCAGTGCAGGGTCCGAGGT-3′ |
| HOXA9-forward | 5′-TACGTGGACTCGTTCCTGCT-3′ |
| HOXA9-reverse | 5′-CGTCGCCTTGGACTGGAAG-3′ |
| ACTB-forward | 5′-CATGTACGTTGCTATCCAGGC-3′ |
| ACTB-reverse | 5′-CTCCTTAATGTCACGCACGAT-3′ |
Downregulation and upregulation of miR-652
miR-NC mimic, miR-652 mimic, miR-NC inhibitor, and miR-652 inhibitor were synthesized and purchased from Ribo Bio (Guangzhou, China). We seeded 2×106 MUM-2B cells in each well of 6-well plates. On the next day, cells were transfected with 100 nM miR-NC inhibitor or miR-652 inhibitor using Lipofectamine RNAiMAX reagent (Invitrogen) following the manufacturer’s instructions. After 72 h, the cells were subjected to RNA or protein extraction.
Protein extraction and Western blotting
HOXA9 (sc-81291, 1: 1000), HIF-1α (sc-13515, 1: 1000), and HK2 (sc-130358, 1: 1000) antibodies were bought from Santa Cruz Biotechnology (Santa Cruz, CA). PDK1 (ab52893, 1: 1000), c-Myc (ab32027, 1: 1000), cyclinD1 (ab40754, 1: 1000), and β-actin (ab6276, 1: 10000) antibodies were purchased from Abcam (Cambridge, UK). Secondary HRP-conjugated antibodies against rabbit (#7074, 1: 100000) and mouse (#7076, 1: 100000) were products of Cell Signaling Technology (Beverly, MA). The lysates were prepared with RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. The concentration of protein lysates was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). We loaded 20 μg lysates on each well of the SDS-PAGE gel. The proteins were separated and transferred to a PVDF membrane. After that, membranes were blocked in 5% non-fat milk followed by incubation with primary antibody overnight at 4°C. On the next day, the membrane was incubated with secondary antibody for 1 h at room temperature. The blots were developed with an ECL Western Blotting Substrate Kit (Thermo Fisher Scientific).
Cell proliferation and migration assay
The proliferation ability of MUM-2B and MEL270 cells was detected with the Cell Counting Kit-8 (DoJinDo, Kumamoto, Japan) following the manufacturer’s protocol. Briefly, cells were seeded in 96-well plates. The cells were transfected with miR-NC inhibitor or miR-652 inhibitor and HOXA9 siRNA or control siRNA on the next day. At 0, 24, 48, and 72 h after transfection, 20 μL CCK-8 solution was added into each well and sustained for 2 h. The absorbance of each well was detected using a microplate reader (Bio-Rad Laboratories, Inc.).
The migration ability of MUM-2B and MEL270 cells was detected by the wound healing assay. Briefly, 1×107 cells were seeded in each well of 6-well plates and grown to 100% confluence. A wound area was made at the central area of each well by a 10-μL pipette tip. The medium was replaced with serum-free medium and cultured for another 30 h. At 0 and 30 h, images of the wound area were captured and the percentage of migrated area was calculated.
Bioinformatic analysis
To predict the potential target genes of miR-652, TargetScan software was used [24]. Seventeen potential target genes were predicted to contain putative binding sites for miR-652 on the 3′UTR of mRNAs. The predicted target genes are listed in Table 2.
Table 2.
List of predicted target genes of miR-652.
| Target gene | Representative transcript | Gene name |
|---|---|---|
| ISL1 | ENST00000230658.7 | ISL LIM homeobox 1 |
| TNRC6A | ENST00000395799.3 | Trinucleotide repeat containing 6A |
| NPTN | ENST00000345330.4 | Neuroplastin |
| CAPZB | ENST00000375142.1 | Capping protein (actin filament) muscle Z-line, beta |
| PRRX1 | ENST00000367760.3 | Paired related homeobox 1 |
| KPNA1 | ENST00000344337.6 | Karyopherin alpha 1 (importin alpha 5) |
| NXF1 | ENST00000531709.2 | Nuclear RNA export factor 1 |
| KCNN3 | ENST00000271915.4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| HSD3B7 | ENST00000353250.5 | Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| KCNB1 | ENST00000371741.4 | Potassium voltage-gated channel, Shab-related subfamily, member 1 |
| RPL28 | ENST00000560583.1 | Ribosomal protein L28 |
| TP53 | ENST00000420246.2 | Tumor protein p53 |
| UBE2I | ENST00000355803.4 | Ubiquitin-conjugating enzyme E2I |
| RP1-170O19.20 | ENST00000470747.4 | Uncharacterized protein |
| HOXA9 | ENST00000396345.1 | Homeobox A9 |
| GGCX | ENST00000233838.4 | gamma-Glutamyl carboxylase |
| SLC35C2 | ENST00000372227.1 | Solute carrier family 35 (GDP-fucose transporter), member C2 |
Plasmid construction and dual luciferase reporter assay
3′UTR of HOXA9 was amplified from cDNA of MUM-2B and ligated into pGL3-basic (Promega Corporation, Madison, WI). Site mutations were introduced into pGL3-HOXA9 3′UTR-WT to construct pGL3-HOXA9 3′UTR-Mut using the QuickChange Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). MUM-2B and MEL270 cells were transfected with HOXA9 3′UTR-WT or HOXA9 3′UTR-Mut and miR-NC mimic or miR-652 mimic. After 48 h, the relative luciferase activity of each well was detected with the Dual Luciferase Reporter Assay System (Promega Corporation).
Small interference RNA (siRNA) mediated gene silencing
HOXA9 siRNA and control siRNA were synthesized and purchased from GenePharma (Suzhou, China). We seeded 2×106 MUM-2B cells in each well of 6-well plates. On the next day, cells were transfected with 50 nM HOXA9 siRNA or control siRNA using Lipofectamine RNAiMAX reagent (Invitrogen) following the manufacturer’s instructions. After 72 h, the cells were subjected to RNA or protein extraction.
Statistical analysis
All experiments were performed at least 3 times. The data were analyzed by GraphPad Prism 6.0 and are presented as mean±SD. Two groups were compared using the t test, and 3 groups were compared first with one-way ANOVA followed by Newman Keuls analysis. A p value less than 0.05 was considered statistically significant.
Results
miR-652 was overexpressed in uveal melanoma
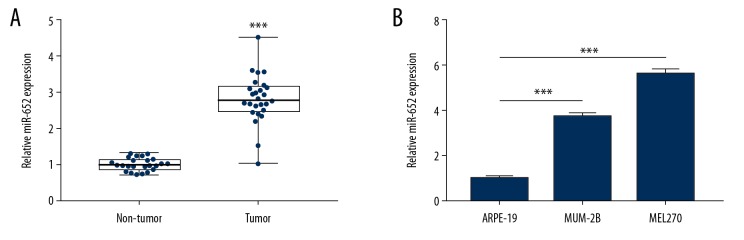
As one of 6 miRNAs prognostic biomarkers for patients with uveal melanoma, the role of miR-652 has not been studied before in uveal melanoma. We collected uveal melanoma and matched normal tissues from 26 patients with uveal melanoma. RT-qPCR showed that miR-652 was significantly overexpressed in uveal melanoma compared with normal tissues (Figure 1A). Consistently, it was observed that miR-652 was elevated in uveal melanoma cell lines (MUM-2B and MEL270) compared to the immortal retinal pigment epithelial cell line ARPE-19 (Figure 1B).
Figure 1.
Overexpression of miR-652 in uveal melanoma. (A) In 26 pairs of uveal melanoma and normal uveal tissues, RT-qPCR showed that miR-652 was overexpressed in uveal melanoma. (B) Compared with the immortalized retinal pigment epithelial cell line ARPE-19, RT-qPCR showed that miR-652 was overexpressed in uveal melanoma cell lines MUM-2B and MEL270 (*** P<0.001).
Downregulation of miR-652 inhibited cell proliferation and migration of uveal melanoma
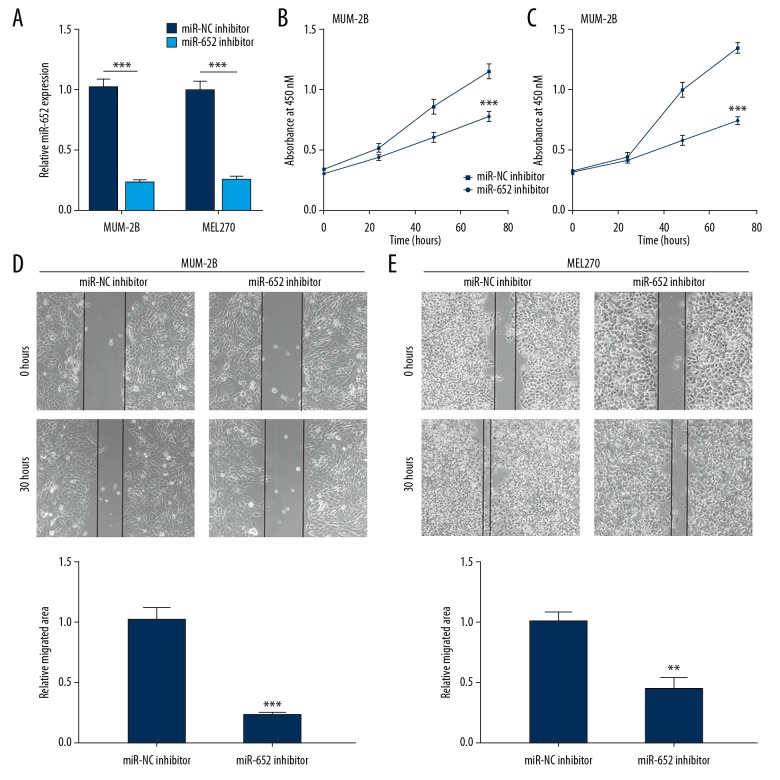
To investigate the function of miR-652, we downregulated miR-652 by transfection of miR-652 inhibitor into MUM-2B and MEL270 cells. Transfection of miR-652 inhibitor decreased miR-652 levels in both MUM-2B and MEL270 cells (Figure 2A). In the cell proliferation assay, it was observed that miR-652 inhibitor decreased the cell growth rate in MUM-2B and MEL270 cells (Figure 2B, 2C). In the cell migration assay, downregulation of miR-652-inhibited cells migrated towards the wound area in MUM-2B and MEL270, suggesting the cell migration ability was inhibited (Figure 2D, 2E). The data indicated that miR-652 exhibited a tumor suppressive function in uveal melanoma.
Figure 2.
Downregulation of miR-652 inhibited cell proliferation and migration in uveal melanoma cells. (A) Transfection of miR-652 inhibitor decreased miR-652 levels in MUM-2B and MEL270. (B) Downregulation of miR-652 inhibited cell proliferation in MUM-2B cells. (C) Downregulation of miR-652 inhibited cell proliferation in MEL270 cells. (D) Downregulation of miR-652 inhibited cell migration in MUM-2B cells. (E) Downregulation of miR-652 inhibited cell migration in MEL270 cells (** P<0.01, *** P<0.001).
miR-652 directly repressed HOXA9 expression in uveal melanoma
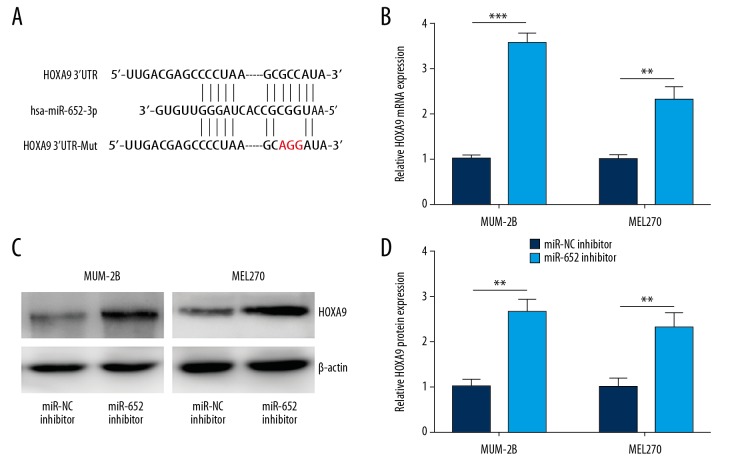
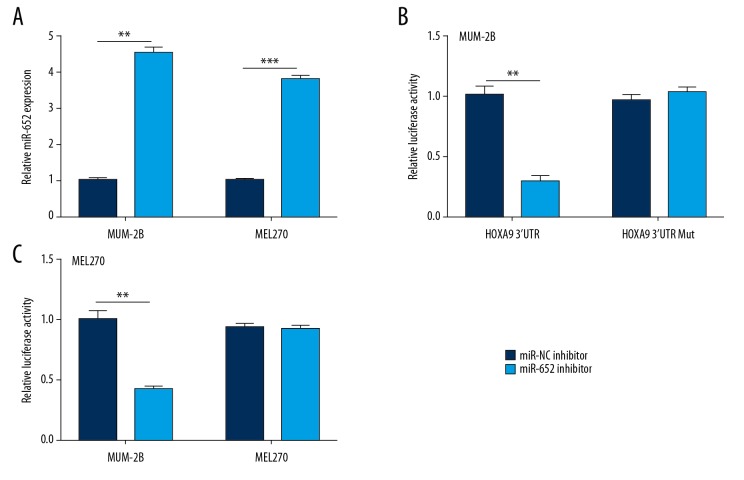
Using TargetScan, we predicted 17 potential target genes of miR-652. Among them, the 3′UTR of HOXA9 mRNA could complementary bind to miR-652 (Figure 3A) and was reported to be involved in melanoma progression [25]. RT-qPCR confirmed that miR-652 downregulation increased HOXA9 mRNA expression in MUM-2B and MEL270 cells (Figure 3B). In addition, Western blotting indicated that miR-652 downregulation elevated HOXA9 protein expression in MUM-2B and MEL270 cells (Figure 3C, 3D). To further evaluate their direct interaction, we transfected miR-652 mimic to overexpress miR-652 in MUM-2B and MEL270 cells (Figure 4A). Overexpression of miR-652 decreased relative luciferase activity in MUM-2B cells transfected with HOXA9 3′UTR; however, miR-652 mimic showed no effect on the relative luciferase content of HOXA9 3′UTR-Mut (Figure 4B). Similarly, miR-652 mimic also decreased relative luciferase activity in MEL270 cells transfected with HOXA9 3′UTR (Figure 4C).
Figure 3.
HOXA9 was negatively regulated by miR-652 in uveal melanoma. (A) The putative target genes and mutant complementary sequences of the HOXA9 mRNA 3′UTR were predicted by miR-652 sequences. (B) Downregulation of miR-652 increased HOXA9 mRNA levels in MUM-2B and MEL270. (C) Western blotting showed that the protein expression of HOXA9 was increased towards miR-652 downregulation in MUM-2B and MEL270. (D) Using Image J software, the HOXA9 protein levels in C were quantified and normalized to cells transfected with miR-NC inhibitor (** P<0.01, *** P<0.001).
Figure 4.
HOXA9 is a direct target gene of miR-652 in uveal melanoma. (A) Transfection of miR-652 mimic elevated miR-652 levels in MUM-2B and MEL270. (B) Dual luciferase reporter assay showed that miR-652 overexpression decreased the relative luciferase activity of MUM-2B cells transfected with HOXA9 3′UTR. (C) The dual luciferase reporter assay showed that miR-652 overexpression decreased the relative luciferase activity of MEL270 cells transfected with HOXA9 3′UTR (** P<0.01).
miR-652 activated the HIF-1 signaling pathway via repression of HOXA9
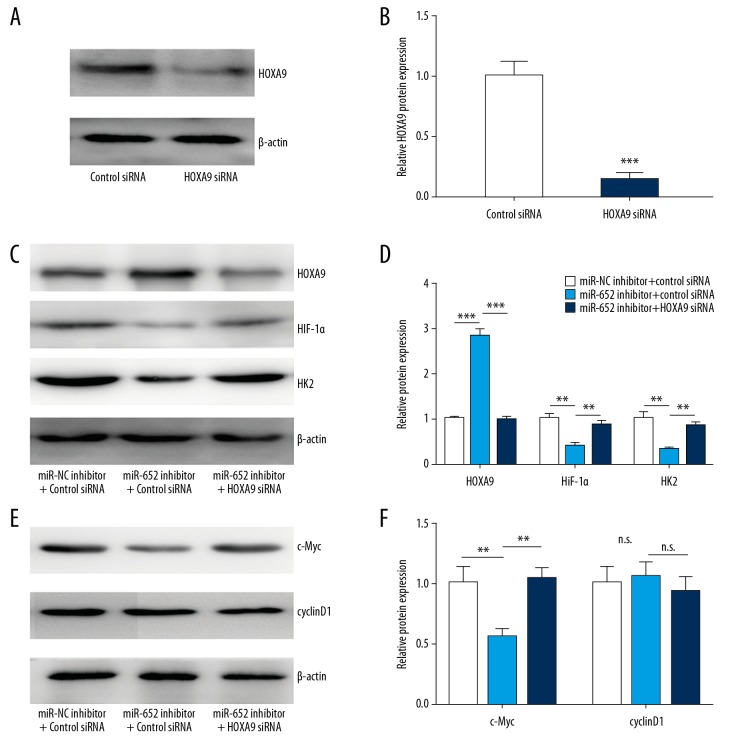
HOXA9 is known as a negative regulator of the HIF-1 signaling pathway in cancer cells [22]. To study whether miR-652 depends on regulation of HOXA9 to control activity of HIF-1 signaling, we transfected HOXA9 siRNA to silence HOXA9 expression in MUM-2B cells (Figure 5A, 5B). The Western blotting results showed that miR-652 downregulation increased HOXA9 and decreased HIF-1α and HK2 protein expression, and silencing of HOXA9 reversed downregulation of HIF-1α and HK2 in MUM-2B cells (Figure 5C, 5D). HOXA9 also regulates Wnt signaling in glioblastoma [26]. We also checked the expression of c-Myc and cyclinD1, the target genes of Wnt signaling. We observed decreased c-Myc after miR-652 downregulation, and silencing of HOXA9 reversed downregulation of c-Myc (Figure 5E, 5F). However, the expression of cyclinD1 was not changed (Figure 5E, 5F). These data suggest that miR-652 enhanced the HIF-1 signaling pathway via downregulation of HOXA9 in uveal melanoma.
Figure 5.
miR-652 activated HIF-1 signaling via HOXA9. (A) Western blotting showed that transfection of HOXA9 siRNA decreased HOXA9 protein expression in MUM-2B. (B) Using Image J software, the HOXA9 protein levels in A were quantified and normalized to cells transfected with control siRNA. (C) Western blotting showed that downregulation of miR-652 led to increase of HOXA9 and decrease of HIF-1α and HK2 protein expression, which was reversed after HOXA9 silencing. (D) Using Image J software, the HOXA9, HIF-1α and HK2 protein levels in C were quantified. (E) Western blotting showed that downregulation of miR-652 led to decrease of c-Myc protein expression, which was reversed after HOXA9 silencing. The protein level of cyclinD1 was not changed after miR-652 downregulation and HOXA9 silencing. (F) Using Image J software, the c-Myc and cyclinD1 protein levels in E were quantified (** P<0.01, *** P<0.001, n.s. – not significant).
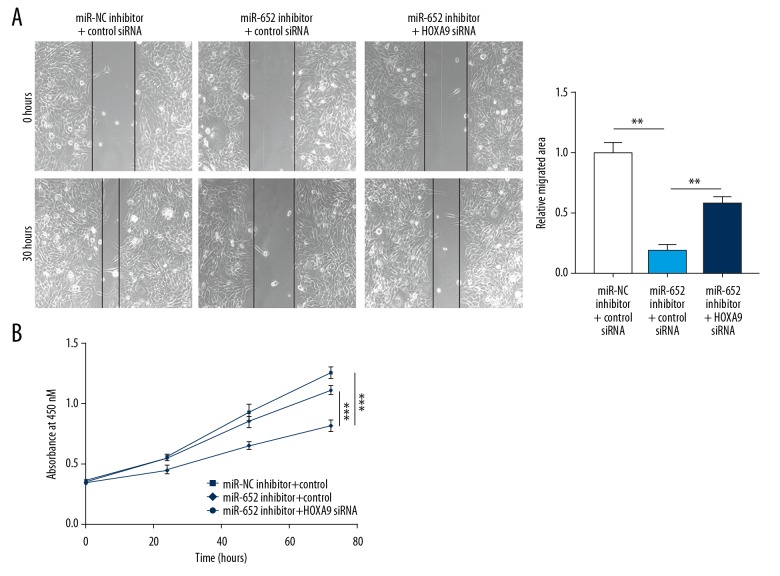
miR-652 promoted cell migration and proliferation via repression of HOXA9
In the function assays, we observed that downregulation of miR-652 inhibited MUM-2B cells from migrating towards the wound area, which was partially attenuated after HOXA9 silencing (Figure 6A). Additionally, the decreased MUM-2B cell growth rate induced by miR-652 inhibitor was also reversed after HOXA9 silencing (Figure 6B). The results indicated that HOXA9 is involved in the function of miR-652 in uveal melanoma.
Figure 6.
miR-652 promoted cell migration and proliferation via HOXA9. (A) Downregulation of miR-652 inhibited MUM-2B cell migration, which was partially reversed after HOXA9 silencing. (B) Downregulation of miR-652 inhibited MUM-2B cell proliferation, which was reversed after HOXA9 silencing. (** P<0.01, *** P<0.001)
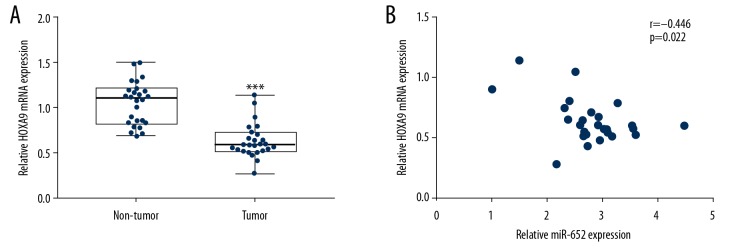
HOXA9 expression was associated with miR-652 levels in uveal melanoma
We assessed HOXA9 mRNA expression in 26 pairs of tumors and normal tissues from patients with uveal melanoma, showing that HOXA9 was significantly downregulated in tumors (Figure 7A). More importantly, Pearson correlation analysis indicated a strong negative correlation between miR-652 and HOXA9 mRNA levels in tumors (Figure 7B).
Figure 7.
The expression of HOXA9 was negatively associated with miR-652 expression in uveal melanoma. (A) RT-qPCR showed that HOXA9 mRNA expression was decreased in uveal melanoma compared with normal uveal tissues from 26 patients with uveal melanoma. (B) Pearson correlation analysis showed a significant negative correlation between HOXA9 mRNA levels and miR-652 levels in uveal melanoma (*** P<0.001).
Discussion
Numerous differentially expressed miRNAs between uveal melanoma and normal uveal tissues have been identified by miRNA profiling [14,27]. Although several of them have been experimentally validated as oncogenes or tumor suppressors [12,28,29], the function and molecular mechanism of many differentially expressed miRNAs are unknown. miR-652 is a recently discovered biomarker for patients with uveal melanoma [13]. In the present study, we for the first time confirmed a pro-proliferative and metastasis role of miR-652 and further showed that HOXA9 is a target gene of miR-652 in uveal melanoma cells.
miR-652 is associated with cell proliferation and metastasis of several types of cancers. The role of miR-652 in cell proliferation and metastasis is dependent on the cell background. For example, studies showed that miR-652 promotes cell proliferation and metastasis in non-small cell lung cancer and endometrial cancer [30,31]; in pancreatic cancer, however, miR-652 inhibits cell proliferation and metastasis of cancer cells via suppression of epithelial-mesenchymal transition [32]. Using RT-qPCR, we observed significant upregulation of miR-652 in uveal melanoma tissues and cells, which is consistent with a previous report [13]. Furthermore, in the function assays, we discovered that downregulation of miR-652 inhibited uveal melanoma cell proliferation and migration. These data collectively support a potential oncogenic role of miR-652 in uveal melanoma.
HOXA9 is involved in embryonic development and was recently identified as a cancer-related gene in several cancer types, either as an oncogene or a tumor suppressor. In acute myeloid leukemia, overexpression of HOXA9 cooperated with Meis1 to drive myeloid leukemogenesis [33]. Decreased expression of HOXA9 in non-small lung cancer promotes cancer cell migration [34]. The downregulation of HOXA9 was observed in our collected uveal melanoma tissues, suggesting a potential tumor suppressive role. The expression of HOXA9 was regulated by methylation [34] and miRNAs in different backgrounds [35,36]. For example, miR-182 directly inhibits HOXA9 expression to control cell proliferation and migration in gastric cancer cells [37]. The TargetScan prediction showed that HOXA9 might be a target gene of miR-652. In uveal melanoma cells, we observed that HOXA9 was upregulated after miR-652 downregulation. The dual luciferase reporter assay further confirmed HOXA9 as a target gene of miR-652 in uveal melanoma cells. Most recently, HOXA9 was discovered to inhibit cancer progression via inactivation of the HIF-1 signaling pathway [22]. Consistently, in uveal melanoma, we observed downregulation of HIF-1α and HK2, 2 key genes in HIF-1 signaling, in response to miR-652 downregulation. Moreover, the downregulation of HIF-1α and HK2 was recovered after HOXA9 silencing, indicating miR-652 controls HIF-1 signaling via HOXA9. HOXA9 was also confirmed as a negative regulator of Wnt signaling in glioblastoma [26]; however, it was not observed in uveal melanoma due to different cell backgrounds. Finally, the decreased cell growth rate and migration inhibition caused by miR-652 inhibitor was also partially reversed by HOXA9 silencing. miR-652 can repress HOXA9 expression to activate HIF-1 signaling, which leads to accelerated cell metabolism and proliferation.
Conclusions
The present study revealed the pivotal role of miR-652 in regulation of cell proliferation and migration in uveal melanoma. Mechanistically, overexpression of miR-652 directly repressed HOXA9 expression and led to activation of the HIF-1 signaling pathway. Our findings provide evidence suggesting the miR-652/HOXA9/HIF-1 axis is a potential new target for treating uveal melanoma.
Footnotes
Source of support: This study was supported by the Natural Science Foundation of Guangdong Province (No. 2017A030313770)
Availability of data and materials
Available by special request.
Conflict of interests
None.
References
- 1.Shields CL, Shields JA. Ocular melanoma: Relatively rare but requiring respect. Clin Dermatol. 2009;27:122–33. doi: 10.1016/j.clindermatol.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Kivela T, Eskelin S, Kujala E. Metastatic uveal melanoma. Int Ophthalmol Clin. 2006;46:133–49. doi: 10.1097/01.iio.0000195861.71558.13. [DOI] [PubMed] [Google Scholar]
- 3.Spagnolo F, Caltabiano G, Queirolo P. Uveal melanoma. Cancer Treat Rev. 2012;38:549–53. doi: 10.1016/j.ctrv.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Shields CL, Ganguly A, Bianciotto CG, et al. Prognosis of uveal melanoma in 500 cases using genetic testing of fine-needle aspiration biopsy specimens. Ophthalmology. 2011;118:396–401. doi: 10.1016/j.ophtha.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay C, Kim DW, Gombos DS, et al. Uveal melanoma: From diagnosis to treatment and the science in between. Cancer. 2016;122:2299–312. doi: 10.1002/cncr.29727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomase VS, Parundekar AN. microRNA: Human disease and development. Int J Bioinform Res Appl. 2009;5:479–500. doi: 10.1504/IJBRA.2009.028678. [DOI] [PubMed] [Google Scholar]
- 9.Giulietti M, Occhipinti G, Principato G, Piva F. Identification of candidate miRNA biomarkers for pancreatic ductal adenocarcinoma by weighted gene co-expression network analysis. Cell Oncol (Dordr) 2017;40:181–92. doi: 10.1007/s13402-017-0315-y. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Yu X, Shen J, Jiang Y. MicroRNA dysregulation in uveal melanoma: A new player enters the game. Oncotarget. 2015;6:4562–68. doi: 10.18632/oncotarget.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Bian G, Meng Z, et al. MiR-144 inhibits uveal melanoma cell proliferation and invasion by regulating c-Met expression. PLoS One. 2015;10:e0124428. doi: 10.1371/journal.pone.0124428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Yan D, Zhou X, Chen X, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci. 2009;50:1559–65. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 13.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18:184–90. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 14.Radhakrishnan A, Badhrinarayanan N, Biswas J, Krishnakumar S. Analysis of chromosomal aberration (1, 3, and 8) and association of microRNAs in uveal melanoma. Mol Vis. 2009;15:2146–54. [PMC free article] [PubMed] [Google Scholar]
- 15.Triozzi PL, Achberger S, Aldrich W, et al. Association of tumor and plasma microRNA expression with tumor monosomy-3 in patients with uveal melanoma. Clin Epigenetics. 2016;8:80. doi: 10.1186/s13148-016-0243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan R. Hox genes: A continuation of embryonic patterning? Trends Genet. 2006;22:67–69. doi: 10.1016/j.tig.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Domingo-Reines J, Lopez-Ornelas A, Montes R, et al. Hoxa9 and EGFP reporter expression in human Embryonic Stem Cells (hESC) as useful tools for studying human development. Stem Cell Res. 2017;25:286–90. doi: 10.1016/j.scr.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Jiang G, Zhang N, et al. HOXA9, PCDH17, POU4F2, and ONECUT2 as a urinary biomarker combination for the detection of bladder cancer in Chinese patients with hematuria. Eur Urol Focus. 2018 doi: 10.1016/j.euf.2018.09.016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Watanabe Y, Saito M, Saito K, et al. Upregulated HOXA9 expression is associated with lymph node metastasis in colorectal cancer. Oncol Lett. 2018;15:2756–62. doi: 10.3892/ol.2017.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son JW, Jeong KJ, Jean WS, et al. Genome-wide combination profiling of DNA copy number and methylation for deciphering biomarkers in non-small cell lung cancer patients. Cancer Lett. 2011;311:29–37. doi: 10.1016/j.canlet.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Ko SY, Barengo N, Ladanyi A, et al. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J Clin Invest. 2012;122:3603–17. doi: 10.1172/JCI62229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L, Wang Y, Zhou M, et al. HOXA9 inhibits HIF-1alpha-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nat Commun. 2018;9:1480. doi: 10.1038/s41467-018-03914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian m. RNAs Elife. 2015:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wouters J, Vizoso M, Martinez-Cardus A, et al. Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med. 2017;15:101. doi: 10.1186/s12916-017-0851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng DH, Wang X, Lu LN, et al. MiR-638 serves as a tumor suppressor by targeting HOXA9 in glioma. Eur Rev Med Pharmacol Sci. 2018;22:7798–806. doi: 10.26355/eurrev_201811_16404. [DOI] [PubMed] [Google Scholar]
- 27.Venza M, Dell’Aversana C, Visalli M, et al. Identification of microRNA expression patterns in cutaneous and uveal melanoma cell lines. Tumori. 2014;100:e4–7. doi: 10.1700/1430.15828. [DOI] [PubMed] [Google Scholar]
- 28.Yan D, Dong XD, Chen X, et al. Role of microRNA-182 in posterior uveal melanoma: Regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS One. 2012;7:e40967. doi: 10.1371/journal.pone.0040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Q, Cong R, Yan H, et al. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol Rep. 2009;22:563–67. doi: 10.3892/or_00000472. [DOI] [PubMed] [Google Scholar]
- 30.Yang W, Zhou C, Luo M, et al. MiR-652-3p is upregulated in non-small cell lung cancer and promotes proliferation and metastasis by directly targeting Lgl1. Oncotarget. 2016;7:16703–15. doi: 10.18632/oncotarget.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X, Dongol S, Qiu C, et al. miR-652 promotes tumor proliferation and metastasis by targeting RORA in endometrial cancer. Mol Cancer Res. 2018;16:1927–39. doi: 10.1158/1541-7786.MCR-18-0267. [DOI] [PubMed] [Google Scholar]
- 32.Deng S, Li X, Niu Y, et al. MiR-652 inhibits acidic microenvironment-induced epithelial-mesenchymal transition of pancreatic cancer cells by targeting ZEB1. Oncotarget. 2015;6:39661–75. doi: 10.18632/oncotarget.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohr S, Doebele C, Comoglio F, et al. Hoxa9 and Meis1 cooperatively induce addiction to Syk signaling by suppressing miR-146a in acute myeloid leukemia. Cancer Cell. 2017;31:549–62e11. doi: 10.1016/j.ccell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang JA, Lee BB, Kim Y, et al. HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E72–80. doi: 10.1002/mc.22180. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Bu J, Liu X, et al. miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer. Oncotarget. 2017;8:63935–48. doi: 10.18632/oncotarget.19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang ZF, Li GR, Cao CN, et al. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:8582–88. doi: 10.26355/eurrev_201812_16621. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, Tian X, Chang J, et al. RUNX3 inhibits the proliferation and metastasis of gastric cancer through regulating miR-182/HOXA9. Biomed Pharmacother. 2017;96:782–91. doi: 10.1016/j.biopha.2017.08.144. [DOI] [PubMed] [Google Scholar]