Abstract
Background
Recently, some studies have showed that miR‐200 families act as novel biomarkers for the prediction of cancer outcomes.
Aims
This meta‐analysis was designed to investigate the associations between miR‐200 families and the prognosis of patients with various cancers.
Materials & Methods
Eligible published databases including PubMed, Embase and Chinese National Knowledge Infrastructure (CNKI) databases were searched for articles until October 18, 2016. We performed a meta‐analysis by calculating pooled hazard ratios (HR) and 95% confidence intervals (CI). Data were extracted from studies comparing overall survival (OS), progression‐free survival (PFS) or recurrence‐free survival (RFS).
Results
For OS, the pooled HR was 1.54 (95% CI: 1.01‐2.33), showing that high miR‐200 family was clearly related to poor survival in various carcinomas, but no significantly association was found in PFS or RFS. Subgroup analysis indicated that upregulated miR‐200 family was linked to poor OS in Asians (HR = 2.19, 95% CI: 1.27‐3.78) but not in Caucasians (HR = 0.94, 95% CI: 0.46‐1.91). Similarly, high miR‐200 expression could not clearly predict the relationship with PFS and RFS. For cancer type, high miR‐200 also predicted poor OS among lung cancer patients (HR = 3.09, 95% CI: 1.75‐5.46). Besides, only elevated miR‐200c of the miR‐200 family indicated a significantly poor OS (HR = 2.25, 95% CI: 1.39‐3.64).
Discussion
Aberrant expression of miRNAs played a crucial role in the area of human carcinomas. Many studies have indicated that miRNAs are considered promising tumor biomarkers for prognosis and potential targets for clinical treatment. We have testified that high levels of miR‐200 family expression (predominantly miR‐200c) are significantly associated with poor survival and prognostic outcomes of patients with cancers, especially in lung cancer. However, no statistically significant results were calculated for miR‐200a/b and miR‐429, and this might result from a relatively small number of articles about them. In other tumor models except lung cancer, our results indicated that high miR‐200 family was not obviously associated with OS (Gastric or Colorectal cancer; Ovarian cancer; Others). In addition, some other records showed the opposite results, for they exhibited that upregulated miR‐200 family level was linked to longer survival. For ethnic group, our stratified analyses showed that the Asian population predicted poor OS. While the Caucasian population did not exhibit an significant association with OS. This discrepancy might result from different hereditary backgrounds and environment exposure. Although these results have indicated that miR‐200 families were promising biomarkers to predict prognosis for patients with cancers, there were several limitations in this analysis that would impact its quality. Generally, further studies should be warranted to clarify this question and to provide a new novel idea for routine clinical application.
Conclusion
Our findings suggest that miR‐200 family might be a potentially useful biomarker for predicting cancer prognosis, especially for lung cancer in Asians.
Keywords: biomarker, carcinoma, miR‐200 family, prognosis
1. INTRODUCTION
MicroRNAs (miRNAs) are a new class of small non‐coding, single‐stranded RNA molecules with approximately 23 nucleotides in length, and they could pair to complementary sequences in the 3′ untranslated region (3′ UTR) of target mRNAs.1 Functionally, the miRNAs can not only regulate gene expression but also involve in diverse cellular biological processing, including differentiation, proliferation, growth, apoptosis, migration, and survival.1 Each miRNA can affect a variety of mRNAs, depending on its targets, and can play different roles in biological or pathological processes.2 The miR‐200 family of miRNAs is composed of miR‐200a, miR‐200b, miR‐200c, miR‐141, and miR‐429, which are located in two different clusters: miR‐200a, miR‐200b, and miR‐429 on chromosome 1, miR‐200c and miR‐141 on chromosome 12.3 Recent studies have demonstrated that miR‐200 families play significant roles in human various cancers, such as lung cancer, gastric cancer, colorectal cancer, ovarian cancer, breast cancer, and glioma.4, 5, 6, 7, 8, 9, 10, 11, 12, 13
To date, some studies have investigated that downregulation of miR‐200 family correlated with progressive pathological feature and poor prognosis in cancer patients.2, 4, 5, 6 However, some others showed insignificant or opposite results.1, 7, 8, 9, 10, 11, 12, 13 According to this, it remains unclear whether miRNA‐200 families act as tumor suppressors or oncogenes in human malignancies and the prognostic significance.14 Thus, the prognostic value of miR‐200 family in patients with various carcinomas remains controversial. In terms of the limits of the single study, we performed this meta‐analysis of all eligible studies to discuss the relationship between miR‐200 family and prognosis in human malignant neoplasms.
2. MATERIALS AND METHODS
2.1. Search strategy
We searched online in PubMed, EMBASE, and CNKI databases up to October 18, 2016, to identify relevant studies, with a combination of the following keywords used simultaneously, namely “cancer,” “carcinoma,” “tumor,” “micro‐RNA‐200 family,” “miRNA‐200,” “miR‐200,” “hazard ratio (HR),” “follow‐up,” “survival,” and “prognosis.” We evaluated potentially relevant studies by examining their titles, abstracts, and full texts matching the eligible criteria retrieved.
Studies were considered eligible if they met the following criteria: (i) studies focused on patients with any type of cancers; (ii) studies measuring the expression of miR‐200 family in tissue or serum/plasma; (iii) studies investigating the association between miR‐200 family expression and prognosis outcomes. Articles were excluded if they were review articles or letters, not focusing on human carcinomas. Other exclusion criteria included studies lacking key information such as HR, 95% confidence intervals (CIs), and P value, without survival curves.
2.2. Data extraction
We extracted relevant data from included studies and recorded these data based on a standardized form. Extracted data elements included: (i) the name of first author and publication year; (ii) characteristics of the studied population, including total number of patients, country, ethnicity, cancer type, and sample category; (iii) test method; (iv) study design; (v) cutoff value; (vi) follow‐up time; (vii) HRs of elevated miR‐200 family expression for OS, PFS, and RFS, along with their 95% CIs and P values. If HRs and 95% CIs were not directly reported, data would be extracted from Kaplan‐Meier curves of graphical survival plots to calculate HRs and 95% CIs.15 The extracted information was summarized in a consistent manner to prevent bias.
2.3. Statistical methods
The aggregation of HRs and 95% CIs was calculated in the following Tierney method.16 Forest plots were used to estimate the effect of miR‐200 family expression on patients’ survival. A test of heterogeneity among included studies was carried out using Cochran Q test and Higgins I‐squared statistic. If P < .1 and the percentage of I 2 > 50%, then random‐effects model (Der Simonian and Laird method) was adopted, otherwise, the fixed‐effects model (Mantel‐Haenszel test) was used.17 Subgroup analysis was performed by the ethnicity, cancer type, and miRNA type method. Publication bias was assessed by Egger's linear regression test with a funnel plot.18 For all analyses, a P value <.05 was considered to be statistically significant, and all the P values were two‐sided. All analyses were performed with the STATA 11.0 software (Stata Corp LP, College Station, TX, USA).
3. RESULTS
A total of 274 potentially relevant studies were identified from literature search in PubMed, EMBASE and CNKI databases; 257 papers were excluded after manual screening of titles, abstracts and key words, because they were not related to the current study. After reading the full texts of the remaining 17 studies, 12 eligible studies were included for the final analysis. A flow diagram of the study selection process is shown in Figure 1.
Figure 1.
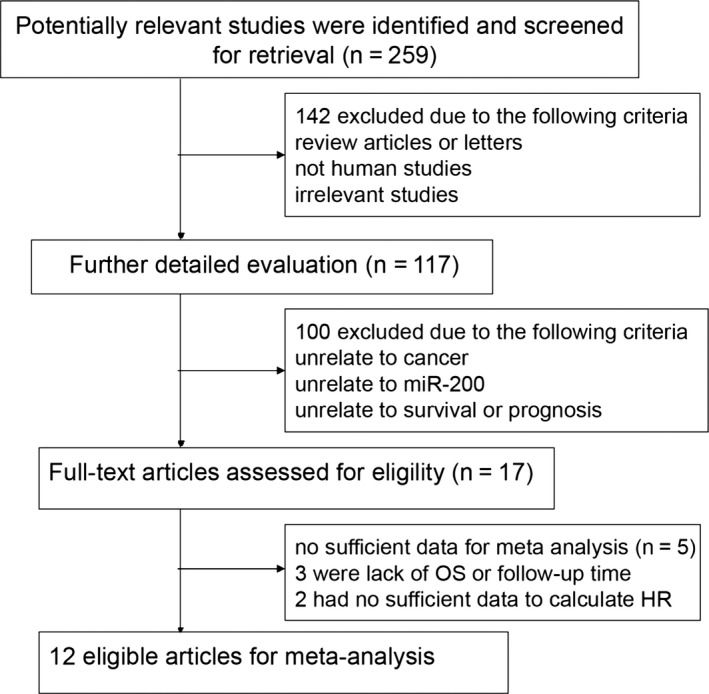
Flow diagram of the study selection process
We collected data from enrolled 12 retrospective studies. Ten of these studies examined cancerous tissue, and only two did with serum or plasma. The expression level of miR‐200 was widely detected by quantitative real‐time polymerase chain reaction (qRT‐PCR) assay. The cutoff values of miR‐200 family were different in each study, such as median, mean, quintile, or twofold. The main characteristics of analyzed studies are systematically listed in Table 1. For prognosis, all the studies analyzed OS and significant heterogeneity between studies was shown (P < .001, I 2 = 79%); hence, a random‐effects model was applied to estimate a pooled HR (HR = 1.54) along with its 95% CI (1.01‐2.33). We found that higher expression of miR‐200 family significantly associated with poorer OS. Three studies evaluated PFS or RFS, and the results indicated that there was no statistical association (Table 2; Figure 2 ).
Table 1.
Main characteristics of all studies included in the meta‐analysis
| Author & Year | Country | Ethnicity | N | Study design | Cancer type | Cutoff | Method | Sample | Follow‐up (mo) | miR type | Survival |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tejero 2014 | Spain | Caucasian | 155 | R | Lung | NG | qRT‐PCR | Tissue | 43 (2‐160) | miR‐200c | OS |
| Cao 2014 | China | Asian | 100 | R | Ovarian | Median | qRT‐PCR | Tissue | 36.8 (6‐56) | miR‐200a | OS |
| Cao 2014 | China | Asian | 100 | R | Ovarian | Median | qRT‐PCR | Tissue | 36.8 (6‐56) | miR‐200b | OS |
| Cao 2014 | China | Asian | 100 | R | Ovarian | Median | qRT‐PCR | Tissue | 36.8 (6‐56) | miR‐200c | OS |
| Men 2014 | China | Asian | 168 | R | Glioma | Median | qRT‐PCR | Tissue | 60 | miR‐200b | OS |
| DIAZ 2014 | Spain | Caucasian | 127 | R | Colorectal | NG | qRT‐PCR | Tissue | 120 | miR‐200a | OS |
| DIAZ 2014 | Spain | Caucasian | 127 | R | Colorectal | NG | qRT‐PCR | Tissue | 120 | miR‐200c | OS |
| DIAZ 2014 | Spain | Caucasian | 127 | R | Colorectal | NG | qRT‐PCR | Tissue | 120 | miR‐429 | OS |
| Toiyama 2013 | Japan | Asian | 182 | R | Colorectal | NG | qRT‐PCR | Serum | 60 | miR‐200c | OS |
| Leskela 2011 | Spain | Caucasian | 72 | R | Ovarian | NG | qRT‐PCR | Tissue | 128 | miR‐429 | OS |
| Ayerbes 2012 | Spain | Caucasian | 52 | R | Gastric | Mean | qRT‐PCR | Serum | 26.3 (6‐53) | miR‐200c | OS |
| Hu 2009 | China | Asian | 55 | R | Ovarian | NG | qRT‐PCR | Tissue | 50 | miR‐200a | OS |
| Song 2013 | China | Asian | 180 | R | Gastric | Quintile | Microarray | Tissue | 35 (1‐112) | miR‐200a | OS |
| Song 2013 | China | Asian | 180 | R | Gastric | Quintile | Microarray | Tissue | 35 (1‐112) | miR‐200b | OS |
| Song 2013 | China | Asian | 180 | R | Gastric | Quintile | Microarray | Tissue | 35 (1‐112) | miR‐200c | OS |
| Liu 2012 | China | Asian | 70 | R | Lung | Twofold | qRT‐PCR | Tissue | 24 | miR‐141 | OS |
| Liu 2012 | China | Asian | 70 | R | Lung | Twofold | qRT‐PCR | Tissue | 24 | miR‐200c | OS |
| Tuomarila 2014 | Finland | Caucasian | 172 | R | Breast | Median | qRT‐PCR | Tissue | 240 | miR‐200c | OS |
| Li 2012 | China | Asian | 107 | R | Colorectal | NG | qRT‐PCR | Tissue | 65 (11‐82) | miR‐429 | OS |
| Men 2014 | China | Asian | 168 | R | Glioma | Median | qRT‐PCR | Tissue | 60 | miR‐200b | PFS |
| Ayerbes 2012 | Spain | Caucasian | 52 | R | Gastric | Mean | qRT‐PCR | Serum | 26.3 (6‐53) | miR‐200c | PFS |
| Song 2013 | China | Asian | 180 | R | Gastric | Quintile | Microarray | Tissue | 35 (1‐112) | miR‐200a | PFS |
| Song 2013 | China | Asian | 180 | R | Gastric | Quintile | Microarray | Tissue | 35 (1‐112) | miR‐200b | PFS |
| Song 2013 | China | Asian | 180 | R | Gastric | Quintile | Microarray | Tissue | 35 (1‐112) | miR‐200c | PFS |
| Leskela 2011 | Spain | Caucasian | 72 | R | Ovarian | NG | qRT‐PCR | Tissue | 128 | miR‐429 | RFS |
| Leskela 2011 | Spain | Caucasian | 72 | R | Ovarian | NG | qRT‐PCR | Tissue | 128 | miR‐200c | RFS |
| Leskela 2011 | Spain | Caucasian | 72 | R | Ovarian | NG | qRT‐PCR | Tissue | 128 | miR‐141 | RFS |
| Hu 2009 | China | Asian | 55 | R | Ovarian | NG | qRT‐PCR | Tissue | 50 | miR‐200a | RFS |
| Tuomarila 2014 | Finland | Caucasian | 172 | R | Breast | Median | qRT‐PCR | Tissue | 240 | miR‐200c | RFS |
N, number of patients; R, retrospective; qRT‐PCR, quantitative real‐time PCR; OS, overall survival; PFS, progression‐free survival; RFS, recurrence‐free survival; CI, confidence interval; NG, not given.
Table 2.
Patient survival by total and stratified analyses
| Subgroup | OS | PFS | RFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | HR (95% CI) | P | n | HR (95% CI) | P | n | HR (95% CI) | P | |
| Overall | 19 | 1.54 (1.01‐2.33)a | .044 | 5 | 0.92 (0.67‐1.26)a | .582 | 5 | 0.65 (0.29‐1.48)a | .307 |
| Ethnicity | |||||||||
| Caucasian | 7 | 0.94 (0.46‐1.91)a | .862 | 1 | – | – | 4 | 0.74 (0.28‐2.01)a | .559 |
| Asian | 12 | 2.19 (1.27‐3.78)a | .005 | 4 | 0.82 (0.65‐1.02)a | .080 | 1 | – | – |
| Cancer type | |||||||||
| Lung | 3 | 3.09 (1.75‐5.46)b | <.001 | – | – | – | – | – | – |
| Gastric or Colorectal | 9 | 1.00 (0.67‐1.50)a | .992 | 4 | 0.95 (0.67‐1.33)a | .745 | – | – | – |
| Ovarian | 5 | 3.49 (0.47‐25.62)a | .220 | – | – | – | 4 | 0.43 (0.29‐0.66)a | <.001 |
| Others | 2 | 1.63 (0.40‐6.69)a | .498 | 1 | – | – | 1 | – | – |
| miRtype | |||||||||
| miR‐200a | 4 | 1.03 (0.28‐3.82)a | .961 | 1 | – | – | 1 | – | – |
| miR‐200b | 3 | 1.83 (0.44‐7.65)a | .406 | 2 | 0.79 (0.55‐1.14)a | .213 | – | – | – |
| miR‐200c | 8 | 2.25 (1.39‐3.64)a | .001 | 2 | 1.28 (0.79‐2.09)a | .314 | 2 | 1.20 (0.18‐8.09)a | .854 |
| miR‐429 | 3 | 0.69 (0.22‐2.17)a | .522 | – | – | – | 1 | – | – |
| miR‐141 | 1 | – | – | – | – | – | 1 | – | – |
OS, overall survival; PFS, progression‐free survival; RFS, recurrence‐free survival; HR, hazard ratio; CI, confidence interval.
The HRs and 95% CIs of analyzed studies were pooled by the random‐effects model if the P value for heterogeneity was <.10 or I 2 was >50%.
The HRs and 95% CIs of analyzed studies were pooled by the fixed‐effects model if the P value for heterogeneity was more than .10 or I 2 was <50%.
Figure 2.
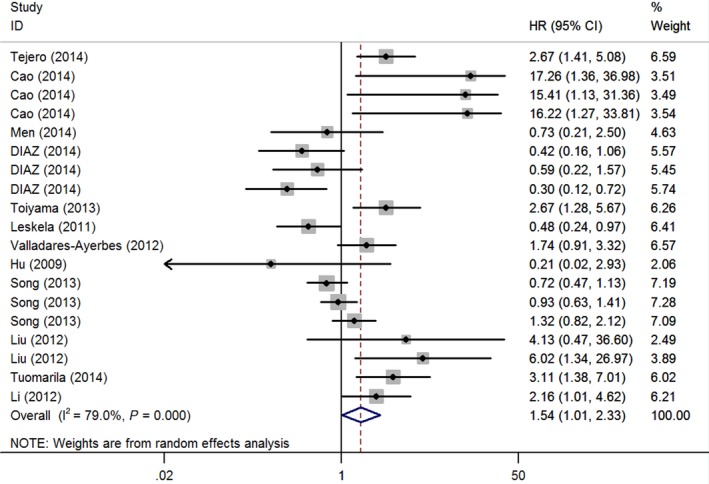
Forest plots of total analyses for patient survival associated with miR‐200 family expression. The squares and horizontal lines correspond to study‐specific hazard ratio (HR) and 95% confidence interval (CI). The area of the squares reflects the weight. The diamond represents HR and 95% CI
Furthermore, stratified analyses were performed, such as ethnicity, cancer type, and miRNA type. Firstly, for ethnicity, seven studies in Asian subgroup investigated that elevated miR‐200 family significantly predicted poor survival (HR = 2.19, 95% CI 1.27‐3.78) by a random‐effects model (P < .001, I 2 = 78%), and the other five studies in Caucasian subgroup could not show a significant association between miR‐200 family and Caucasian cases (HR = 0.94, 95% CI 0.46‐1.91). Secondly, for cancer type, two studies in lung carcinoma subgroup displayed a worse survival outcome linked to the elevated miR‐200 family (HR = 3.09, 95% CI 1.75‐5.46) using a fixed‐effects model (P = .60, I 2 < 0). And in other subgroups of gastric or colorectal cancer, ovarian cancer, breast cancer, and glioma, there was no obviously significant association with poor survival (HR = 1.00, 95% CI 0.67‐1.50; HR = 3.49, 95% CI 0.47‐25.62; HR = 1.63, 95% CI 0.40‐6.69, respectively). In addition, for miRNA type, eight studies also indicated that high miR‐200c expression was a significant predictor for worse outcome (HR = 2.25, 95% CI 1.39‐3.64). No significant association was found in miR‐200a, miR‐200b, and miR‐429 subgroups (Table 2).
Finally, the funnel plot and Egger tests were used to detect publication bias of the included studies. As shown in Figure 3, the funnel plot was almost symmetric. In OS meta‐analysis, the P value of the Egger test was .120; therefore, there was no evidence for significant publication bias in the meta‐analysis.
Figure 3.
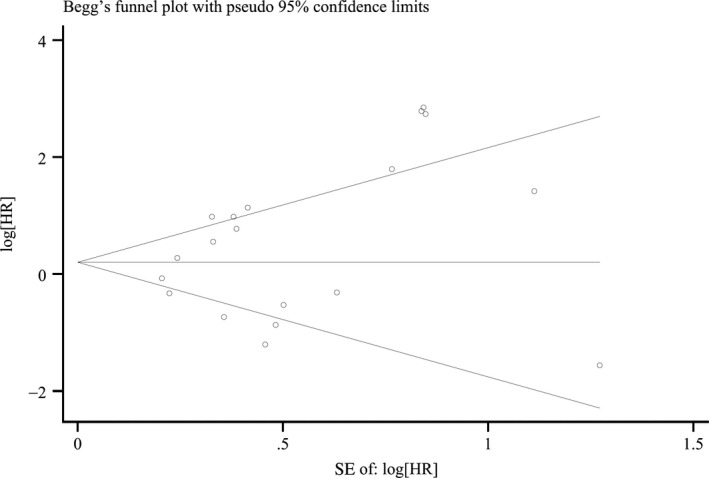
Funnel plot of publication bias test for overall analysis of overall survival. Each point represents a separate study
4. DISCUSSION
Aberrant expression of miRNAs played a crucial role in the area of human carcinomas.19, 20, 21 Many studies have indicated that miRNAs are considered promising tumor biomarkers for prognosis and potential targets for clinical treatment.22 As described above, we have testified that high levels of miR‐200 family expression (predominantly miR‐200c, HR = 2.25, 95% CI 1.39‐3.64) are significantly associated with poor survival and prognostic outcomes of patients with cancers, especially in lung cancer (HR = 3.09 95% CI 1.75‐5.46). Liu et al11 reported that expression of miR‐200c was upregulated in non‐small cell lung cancer (NSCLC) tissues, and the patients with high miR‐200c expression had a short survival time in a cohort of 70 NSCLC patients. This might indicate that high miR‐200c plays a potential role in promoting progression of NSCLC. Some previous literatures have mentioned that miR‐200c has a binding site in the 3′ UTR of β‐tubulin III and elevated miR‐200c in cell lines decreased β‐tubulin III protein. And high tumoral β‐tubulin III was related to worse survival in NSCLC, breast and ovarian cancer.23 In addition, it was also shown to play a role in the regulation of mesenchymal‐epithelial transition (MET), induction of MET is important at a later point in the metastasis process and MET enables primary tumor to colonize and produce metastases in distant organs.3 Tejero et al7 recommended that only miR‐200c influenced MET/EMT in the H23, A‐549, and HCC‐44 cell lines, and Liu et al11 also found that overexpression of miR‐200c was associated with advanced clinical stage and lymph node metastasis. Thus, miR‐200c could be used as a potential biomarker in the prognosis of cancer patients, mainly in lung cancer. However, no statistically significant results were calculated for miR‐200a/b and miR‐429, and this might result from a relatively small number of articles about them.
In other tumor models except lung cancer, our results indicated that high miR‐200 family was not obviously associated with OS (gastric or colorectal cancer: HR = 1.00, 95% CI 0.67‐1.50; ovarian cancer: HR = 3.49, 95% CI 0.47‐25.62; others: HR = 1.63, 95% CI 0.40‐6.69). We speculated that the discordancy might result from different extent of tumor growth in cancer cells.24 Pacurari et al25 indicated that expression of miR‐200 family was rather complex during tumorigenesis, which elevated in primary tumors and downregulated in metastatic cells. Thus, it can be seen that the role of miR‐200 may differ depending on cancer type and stage. In addition, some other records showed the opposite results,2, 5, 6 for they exhibited that upregulated miR‐200 family level was linked to longer survival.1, 4, 7, 8, 9, 10, 11, 12, 13 MiR‐200 family was previously showed to inhibit the epithelial‐mesenchymal transition (EMT) and cell migration. EMT plays a key role in invasion and metastasis during carcinogenesis. Herein, these patients with high miR‐200 family expression levels might have longer survival by inhibiting EMT.
In this meta‐analysis, our stratified analyses showed that the combined HR of Asian population was 2.19 (95% CI 1.37‐3.78) and predicted poor OS. While the HR of Caucasian population was 0.94 (95% CI 0.46‐1.91), it did not exhibit an significant association with OS. This discrepancy might result from different hereditary backgrounds and environment exposure.26
Although these results have indicated that miR‐200 families were promising biomarkers to predict prognosis for patients with cancers, there were several limitations in this analysis that would impact its quality. First, the number of records was only 12 with a small sample size. In particular, all studies were Asians or Caucasians, and no African population were included in the analysis. Therefore, we need more proper studies included in our meta‐analysis. Second, the relatively large heterogeneity in the present study must make caution, although it signified that the variation was not due to chance. This may arises from many factors, including tumor types, different ethnicities, or selection of subjection. The expression of miR‐200 family was detected in tumor tissues and blood samples such as plasma and serum may partly contribute to the heterogeneity. Therefore, we strongly recommend future investigations should be performed with matched cancer type, specimen, and ethnicity to confirm the conclusion. Third, the cutoff definition of miR‐200 family appeared to be different in each study, while we could not provide a clear interpretation. And then, estimates derived from different tumors are not comparable, and then inconsistent cutoff value may be the reason that provided contradictory results. Finally, as a cancer biomarker, the prognostic role was proved, besides, it remained unknown whether miR‐200 family should be used as an independent biomarker or used with several other biomarkers for predicting outcomes. Generally, further studies should be warranted to clarify this question and to provide a new novel idea for routine clinical application.
5. CONCLUSIONS
In summary, our meta‐analysis represents that high level of miR‐200 family expression is significantly associated with poor survival in patients with a variety of cancers. However, the evidence was not so convincing due to the limitations mentioned above, and further studies and more clinical investigations are required to predict the clinical outcome of patients with various carcinomas.
CONFLICT OF INTEREST
Financial & competing interests disclosure: Support was provided by Jiangsu Provincial Key Discipline of Medicine (ZDXKA2016003), the Priority Academic Program Development of Jiangsu Higher Education Institutions (Jiangsu, China), and was also supported by grants from the International Science and Technology Cooperation Program of China (no. 2014DFA31940), the National Natural Science Foundation of China (Beijing, China; nos. 81302014 and 81572259), the Six Talent Peaks Project (Jiangsu, China; no. 2015‐WSN‐038) and the top talent project of “six one engineering” (Jiangsu, China; no. LGY2017071). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Yin Y, Song WW, Wang Y, Zhao W, Wu J, Xu W. MicroRNA‐200 families and prognostic value in various carcinomas: A systematic review and meta‐analysis. Aging Med. 2018;1:39–45. 10.1002/agm2.12005
REFERENCES
- 1. Toiyama Y, Hur K, Tanaka K, et al. Serum miR‐200c is a novel prognostic and metastasis‐predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;00:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Men DH, Liang YS, Chen LY. Decreased expression of miR‐200b is an independent unfavorable prognostic factor for glioma patients. Cancer Epidemiol. 2014;38:152‐156. [DOI] [PubMed] [Google Scholar]
- 3. Gregory PA, Bert AG, Paterson EL, et al. The miR‐200 family and miR‐205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593‐601. [DOI] [PubMed] [Google Scholar]
- 4. Leskela S, Garcia LJ, Mendiola M, et al. The miR‐200 family controls β‐tubulin III expression and is associated with paclitaxel‐based treatment response and progression‐free survival in ovarian cancer patients. Endocr Relat Cancer. 2011;18:85‐95. [DOI] [PubMed] [Google Scholar]
- 5. Hu XX, Macdonald DM, Huettner PC, et al. A miR‐200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457‐464. [DOI] [PubMed] [Google Scholar]
- 6. Diaz T, Tejero R, Moreno I, et al. Role of miR‐200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J Surg Oncol. 2014;109:676‐683. [DOI] [PubMed] [Google Scholar]
- 7. Tejero R, Navarro A, Campayo M, et al. MiR‐141 and miR‐200c as markers of overall survival in early stage non‐small cell lung cancer adenocarcinoma. PLoS ONE. 2014;9:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valladares‐Ayerbes M, Reboredo M, Villaamil VM, et al. Circulating miR‐200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2014;10:186‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao Q, Lu KL, Dai S, Hu Y, Fan W. Clinicopathological and prognostic implications of the miR‐200 family in patients with epithelial ovarian cancer. Int J Clin Exp Pathol. 2014;7:2392‐2401. [PMC free article] [PubMed] [Google Scholar]
- 10. Song FJ, Yang D, Liu B, et al. Integrated miRNA network analyses identify a poor‐prognosis subtype of gastric cancer characterized by the miR‐200 family. Clin Cancer Res. 2014;20:878‐889. [DOI] [PubMed] [Google Scholar]
- 11. Liu XG, Zhu WY, Huang YY, et al. High expression of serum miR‐21 and tumor miR‐200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618‐626. [DOI] [PubMed] [Google Scholar]
- 12. Tuomarila M, Luostari K, Soini Y, Kataja V, Kosma VM, Mannermaa A. Overexpression of miR‐200c predicts poor outcome in patients with PR‐negative breast cancer. PLoS ONE. 2014;9:e109508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J, Du LT, Yang YM, et al. MiR‐429 is an independent prognostic factor in colorectal cancer and exerts its anti‐apoptotic function by targeting SOX2. Cancer Lett. 2013;329:84‐90. [DOI] [PubMed] [Google Scholar]
- 14. Esquela‐Kerscher A, Slack FJ. Oncomirs‐microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259‐269. [DOI] [PubMed] [Google Scholar]
- 15. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta‐analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815‐2834. [DOI] [PubMed] [Google Scholar]
- 16. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time‐to‐event data into meta analysis. Trials. 2007;8:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DerSimonian R, Laird N. Meta‐analysis in clinical trials. Control Clin Trials. 1986;7:177‐188. [DOI] [PubMed] [Google Scholar]
- 18. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta‐analysis detected by a simple, graphical test. BMJ. 1997;315:629‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297‐308. [DOI] [PubMed] [Google Scholar]
- 20. Landi MT, Zhao Y, Routunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16:430‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48‐57. [DOI] [PubMed] [Google Scholar]
- 22. Windeler J. Prognosis‐what does the clinician associate with this notion? Stat Med. 2000;19:425‐430. [DOI] [PubMed] [Google Scholar]
- 23. Rosell R, Scagliotti G, Cooc J, et al. Transcripts in pretreatment biopsies from a three‐arm randomized trial in metastatic non‐small‐cell lung cancer. Oncogene. 2003;22:3548‐3553. [DOI] [PubMed] [Google Scholar]
- 24. Artells R, Navarro A, Diaz T, Monzó M. Ultrastructural an immunohistochemical analysis of intestinal myofibroblasts during the early organogenesis of the human small intestine. Anat Rec. 2011;294:462‐471. [DOI] [PubMed] [Google Scholar]
- 25. Pacurari M, Addison JB, Bondalapati N, et al. The microRNA‐200 family targets multiple non‐small cell lung cancer prognostic markers in H1299 cells and BEAS‐2B cells. Int J Oncol. 2013;43:548‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang RS, Gamazon ER, Ziliak D, et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8:692‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]


