Abstract
Aging is progressive physiological degeneration and consequently declined function, which is linked to senescence on both cellular and organ levels. Accumulating studies indicate that long noncoding RNAs (lncRNAs) play important roles in cellular senescence at all levels—transcriptional, post‐transcriptional, translational, and post‐translational. Understanding the molecular mechanism of lncRNAs underlying senescence could facilitate interpretation and intervention of aging and age‐related diseases. In this review, we describe categories of known and novel lncRNAs that have been involved in the progression of senescence. We also identify the lncRNAs implicated in diseases arising from age‐driven degeneration or dysfunction in some representative organs and systems (brains, liver, muscle, cardiovascular system, bone pancreatic islets, and immune system). Improved comprehension of lncRNAs in the aging process on all levels, from cell to organismal, may provide new insights into the amelioration of age‐related pathologies and prolonged healthspan.
Keywords: age‐related diseases, long noncoding RNAs, senescence
1. INTRODUCTION
Aging is progressive physiological degeneration and consequently declined function, which is characterized by several tentative hallmarks at molecular and cellular levels.1 Apart from genomic instability and telomere attrition, advances in aging research exhibit a lot more determinants of aging, rendering this physiological process complex and complicated. Senescence on both cellular and organ levels gradually causes age‐related diseases, such as cardiovascular diseases, Alzheimer's disease (AD), cancer, and sarcopenia, most in forms of comorbidities. Meanwhile, consequent fragility and frailty result in high mortality. As world population above 60 is expected to double and reach 22% by 2050, the increases in morbidity and mortality are noted in elderly populations.2, 3 Therefore, the boosting aging global population becomes a critical healthcare issue, which demands further exploration through explicit mechanisms underlying the aging process.4, 5, 6
Age‐related changes in the cellular proteome and transcriptome levels are indispensable in physiological alterations in cells, tissues, and organ systems during aging. Recent advancement in microarrays and sequencing techniques has lead to a better understanding of various important mammalian genomes (eg, human, rat, and mouse) and their respective cellular, tissue, and organ‐specific transcriptomes. Series of multitude projects, including Functional Annotation of the Mammalian Genome and Encyclopedia of DNA elements, have revealed that only about 2% of transcripts are protein‐coding RNAs, and the reminders are pervasively transcribed into myriad multifunctional forms of RNA molecules known as noncoding RNAs (ncRNAs).7, 8 Based on the transcript length, these ncRNAs are divided into small (20‐30 nt) ncRNAs and long (>200 nt) ncRNAs (lncRNAs).8 lncRNAs are poorly conserved but abundant heterogeneous regulatory ncRNAs. Based on their genomic location, orientation, and mode of transcription, they are further classified into sense, antisense, bidirectional, promoter‐associated, enhancer‐associated, pseudogene‐associated, telomere‐associated, and circular lncRNAs in a broad but mutually nonexclusive manner.9, 10 They act as regulatory players with versatile roles in different modes. lncRNAs regulate gene expression virtually at all levels—transcriptional, RNA processing, translational, and post‐translational—by interacting with DNA, RNA, or proteins11 (Figure 1). The subcellular localization of lncRNAs may also bring additional complexity to their function.12
Figure 1.
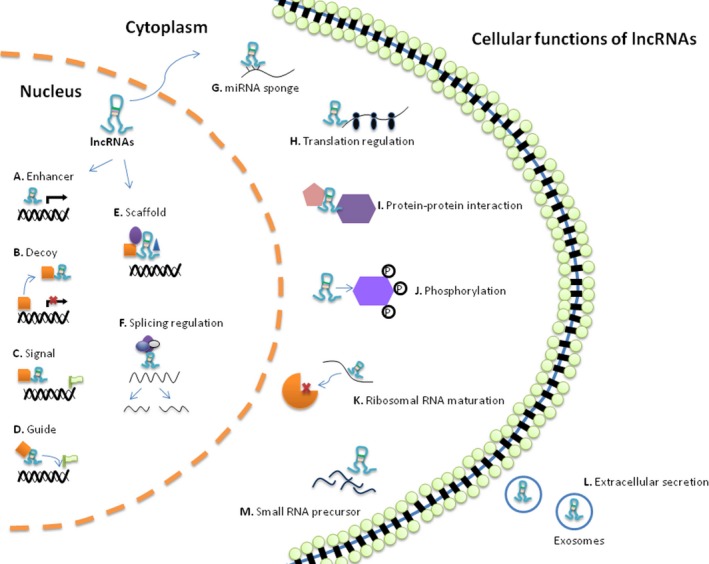
Cellular functions of long noncoding RNAs (lncRNAs). Genomic location relative to regulatory mechanisms of lncRNAs in the nucleus, cytoplasm, and extracellular compartments. Nuclear‐localized lncRNAs can act as (A) enhancers to induce transcription in cis or in trans; or (B) decoy to induce transcription factors and chromatin modifiers, blocking their binding to DNA; or (C) molecular signals to activate or silence gene expression through signaling to regulatory pathways; or (D) guide to instruct transcriptional elements (eg, chromatin modifiers) to specific target sites; or (E) scaffolds, binding proteins complexes to affect gene expression, and (F) then can modulate alternative splicing of pre‐mRNAs. In the cytoplasm, lncRNAs can serve as (G) microRNAs (miRNAs) sponge to block their effect and then can control (H) translational events, or (I) protein‐protein interaction, or (J) protein phosphorylation and activation of signaling pathways. K, They can regulate the maturation of ribosomal RNAs. Finally, some lncRNAs can be (L) released in the form of exosomes and transferred to other cells to (M) function as precursors of miRNAs and other regulatory small RNA
lncRNAs are increasingly recognized as essential in various cellular processes such as proliferation, apoptosis, differentiation, and senescence for the impact on gene expression.13, 14, 15, 16, 17, 18 lncRNAs also underly important pathologic processes in age‐mediated function, including metabolic imbalances, neurodegeneration, and cancer.19, 20 In this review, the emphasis is given to the association of lncRNAs with the aging process in cellular and organic levels with the forms of age‐related frequently occurring diseases.
2. lncRNAs IN CELLULAR AGING
Senescence is characterized as a stable form of growth arrest in untransformed cells, triggered by telomere attrition, chromosome destabilization, DNA damage, mitochondrial dysfunction, oncogene activation, and other cellular stress linked to cell cycle.21 Senescent cells are featured in morphological, secretory, and molecular aspects. Distinctive features include flattened, enlarged cell size, increased SA‐β‐galactosidase activity, production of senescence‐associated secretory phenotype (SASP), and differential expression of senescence‐associated pathways (eg, upregulated p53, p21, p27 and downregulated Sirt1).22, 23 Cellular senescence is implicated in normal aging. However, pathological effects of senescent cells could influence organisms wholly due to the accumulation of them during aging.24 These influences may possibly be on account of the following aspects: (a) impaired regeneration due to exhaustion of stem cells; (b) malfunction in tissues and organs caused by SASP; and (c) disturbed energy homeostasis resulted by various stress.12 On the contrary, cellular senescence plays a protective role against tumorigenesis, which is consistent with the counterplay of senescence pathway with tumor response pathway. Based on this counterinteraction theory, oncogene‐induced senescence (OIS) model is generally utilized. Recent studies have demonstrated that numerous lncRNAs mediate cellular senescence in different stages of the cell cycle by modulating senescence‐associated pathways, such as p53/p21, pRB/p16, and p14.25
2.1. Cell cycle–associated lncRNAs
Senescence represents a permanent withdrawal from the normal cell cycle progression in response to a diverse range of cellular stress, such as DNA damage, oxidative stress, telomere attrition, and environmental stress. Characterized cell cycle inhibitors include p16, p21, and p53, all of which are also senescence‐related tumor suppressors. lncRNAs involved in cell cycle could possibly influence senescence and organismal aging.
2.1.1. MALAT1
Transcript of metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) is a cell cycle regulator localized to the nuclear speckles.26 Abundantly expressed in several solid tumors, MALAT1 is involved in cancer metastasis and recurrence.27, 28, 29 Tripathi et al firstly declared the role of MALAT1 in cell cycle progression. He found that higher level of MALAT1 at G1/S phase and mitosis, but lower level at G1‐G2 phase.26 Several cell line studies have further confirmed that depletion of MALAT1 triggered G1 or G1/S arrest, thus repressing cell growth and proliferation but enhancing senescence phenotype.25, 26, 30, 31 However, MALAT1‐knockout mice showed no obvious phenotype of abnormalities.32, 33, 34 The overall studies have indicated MALAT1 is inessential for organismal development, but might be pivotal under specific pathological or environmental condition.
2.1.2. ANRIL
As an antisense to p15/CDKN2B/CDKN2A/ARF gene cluster, ANRIL is known to suppress the expression of CDKN2A (p16 INK4A, p14 ARF) and CDKN2B (p15 INK4B) genes in cis.35 This lncRNA plays an established role in cell proliferation, senescence, and aging. Depletion of ANRIL in WI‐38 and IMR‐90 cells results in upregulation of p15 INK4B with decreased cell growth and induced senescent phenotype.36 Recent studies have focused on its association with inflammation.37, 38, 39 According to the hypothesis of inflammaging, the positive link with ANRIL to TNF‐α and NF‐κB suggests the role of ANRIL in aging and age‐related diseases, such as certain cardiovascular diseases and AD.37, 38, 40, 41
2.1.3. 7SL
7SL has been identified in various cancers.42 As a highly conserved cytoplasmic lncRNA with six signal recognition proteins, 7SL forms a partial hybrid with the 3′‐untranslated region of p53 mRNA and competes with HuR protein for binding to p53 mRNA.43, 44 7SL silencing studies in HeLa and HCT116 cells displayed cell cycle arrest and senescence by increasing p53 translation through enhanced interaction between HuR and p53 mRNA.45
2.1.4. MEG3
Maternally expressed gene 3 (MEG3) is a maternally expressed and imprinted noncoding transcript.46 This lncRNA participates in biological processes including central nervous system development, angiogenesis, and liver metabolism.47, 48, 49 MEG3 is highly expressed in certain normal tissues but repressed in many tumors.48, 50, 51, 52 MEG3 affects the activities of multiple key cell cycle regulators, such as p53, MDM2, GDF15, and RB1.53, 54 Restoring the expression of MEG3 in HeLa, C‐33A, MCF‐7, and H4 cell lines rightly suppressed tumor cell growth via inducting G2/M cell cycle arrest and apoptosis, while downregulating level of MEG3 enhanced autophagy, cell proliferation, and inhibited cell death.55, 56, 57 As a tumor repressor, MEG3 could be a potential target for cancer diagnosis and prognosis and treatment.53 Decreased levels of MEG3 have also been observed in some age‐related neurodegenerative disorders including Huntington's disease (HD), whose mechanisms of epigenetic gene regulation in neurons may seem to contradict with those in cancer cells.58 Detailed mechanisms on its regulation of senescence and apoptosis need further elucidation to understand the role in brain aging.
2.1.5. H19
H19 is a highly conserved and maternally expressed lncRNA, whose location is near the paternally expressed insulin‐like growth factor 2 (IGF2) genes.59 As an epigenetic regulatory RNA, H19 positively affects cell growth and proliferation and delays senescence, thus promoting tumorigenesis.60, 61, 62 Due to the adjacent localization of H19 and IGF2 (H19‐IGF2) genes, expressions of both genes are always balanced, which is necessary in cell growth, proliferation, senescence, and apoptosis.63, 64 Loss of imprinting at H19‐IGF2 locus has been involved in the onset of cellular senescence. Interestingly, erasure (hypomethylation) of imprinting at this locus observed in aging is accompanied by enhanced expression of H19 but by reduced expression of IGF2, which indicates longevity and low incidence of tumor growth. Contrarily, imprinting loss (hypermethylation) in aging leads to overexpression of both genes, which may correspond to a higher incidence of cancer in advanced age.64, 65 Additionally, H19 is also associated with the development of other age‐related diseases, such as fat deposition and skeletal muscle regeneration.66, 67
2.1.6. UCA1
Firstly identified in bladder transitional cell carcinoma, urothelial cancer‐associated 1 (UCA1) has been demonstrated to promote cell proliferation and attenuate apoptosis as precursors to multiple miRNAs in malignant tumors.68, 69 As cellular senescence is considered as tumor suppression, UCA1 overexpression could induce cellular senescence.70 Relevant mechanism studies highlight the role of CAPERα/TBX3 repressor complex, which is required to prevent senescence in primary cells and mouse embryos. Certain stress induces separation of CAPERα and TBX3, thus activating production of UCA1 RNA, and causes senescence. Furthermore, CAPERα/TBX3 is known to regulate chromatin structure and to repress transcription of p16 INK4A and the RB pathway. In proliferating cells, hsRNPA1 binds and destabilizes p16 INK4A mRNA, whereas during senescence, UCA1 stabilizes p16 INK4A mRNA by sequestering hsRNPA1 from the binding with p16 INK4A.70, 71
2.1.7. FAL1
Focally amplified lncRNA on chromosome 1 (FAL1) was firstly identified among somatic copy number alterations of lncRNAs in 2394 tumor specimens from 12 cancer types through a genomewide survey. FAL1 displays striking oncogenic activity partly by suppressing p21 through association with BM1. On the contrary, FAL1 silencing or downregulation leads to G0/G1 arrest and cellular senescence.72, 73
2.1.8. Gadd7
Gadd7 was isolated from Chinese hamster ovary cells, whose levels were detected in response to DNA damage.74 Overexpression of gadd7 results in G1 arrest and promotes apoptosis by directly binding to TAR DNA‐binding protein (TDP‐43) and interfering with its interaction with Cdk6 mRNA.75 As consequent Cdk6 degradation induces cell cycle arrest and senescent phenotype, the possible impact of gadd7 on aging is expecting.76
2.1.9. MR31HG
MR31HG (MIR31 host gene/LOC554202) is located 400 kb upstream of the p16 INK4A locus in humans. MR31HG harbors miR‐31, which is upregulated in senescent human umbilical vein endothelial cells (ECs) but downregulated in various cancers.77, 78 Previous studies have shown that MR31HG could modulate cell growth and suppress tumorigenesis via miR‐31.79, 80, 82 Interestingly but intriguingly, a recent study reported that MR31HG was upregulated in OIS, whereas silencing of this lncRNA promoted p16 INK4A‐dependent senescence phenotype.83 MR31HG is present in both nucleus and cytoplasm in presenescent cells, but then located mainly in the cytoplasm after BRAF activation. MR31HG binds to both p16 INK4A and MR31HG genomic regions with polycomb group (PcG) proteins. During OIS, PcG proteins and enhanced MR31HG are required for PcG‐mediated repression of p16 INK4A locus.83
2.1.10. PANDA
p21‐associated ncRNA DNA damage activated (PANDA), a bidirectional transcript from the p21 promoter induced upon DNA damage via p53, modulates cell proliferation, apoptosis, and senescence in human fetal lung fibroblasts and neonatal foreskin, as a decoy for pro‐proliferative transcriptional factor, NF‐YA.84, 85, 86 Additionally, PANDA induced by p53 results in G1 cell cycle arrest in lymphoma through inactivation of MAPK/ERK pathway.87 Surprisingly, it has been demonstrated to determine entry and exit from senescence via dual regulation. PANDA at low level inhibits expressions of multiple prosenescence genes through the formation of PANDA‐SAFA‐PRC‐BMI complex in proliferative cells, whereas increased PANDA dissociated from this complex in senescent cells induces senescence arrest by repressing proliferation‐promoting genes and enforcing prosenescence genes.88 Consistently, depletion of PANDA by siRNA results in exit from senescence in senescent fibroblasts.88 The flexibility in switching between proliferation and senescence enables PANDA as a potential target for senescence and age‐related intervention.
2.1.11. lincRNA‐p21
P53‐mediated lincRNA‐p21 is firstly identified as a regulator of p21 by recruiting hnRNP‐K to the promoter region of p21, thus diminishing cell proliferation in mouse embryonic fibroblasts.89, 90 Meanwhile, lincRNA‐p21 is proved to provide positive feedback to p53 transcription via interacting with multiple factors, including MDM2 and Rck.89, 91 HuR/Ago2/let‐7 complex destabilizes lincRNA‐p21 and relieves its translational inhibition on target mRNAs.92 Further studies have found that lincRNA‐p21 impaired somatic cell reprogramming through cell senescence or apoptosis epigenetically.89, 93 This lncRNA participates in various cancers and age‐related coronary artery diseases, such as atherosclerosis and myocardial infarction.91, 94, 95, 96
2.1.12. PINT
p53‐induced noncoding transcript (PINT) is also controlled by p53 and in turn affects p53, MAPK, and TGF‐β signaling by PRC2‐mediated modulation on relevant gene promoter regions.97 PINT negatively associates with senescence and age‐related diseases.97
2.1.13. TUG1
Taurine upregulated gene 1 (TUG1) is primarily known as a growth regulator induced by p53 upon DNA damage.98, 99 Apart from p53‐mediated growth arrest and apoptosis, TUG1 disrupts the expressions of HOX genes family (eg, HOXB7), which results in aging.99 Moreover, TUG1 controls glycolysis in proliferation and metastasis of tumor cells through regulation of hexokinase 2 via miR‐455‐3p/AMPKβ2.100 As TUG1 is highly expressed in the human subependymal zone, it has been involved in age‐related neurodegenerative diseases, such as ischemic stroke and HD.101, 102, 103 TUG1 also has an impact on other tissue‐specific aging, such as intervertebral disk and age‐related cataract, through Wnt/β‐catenin or caspase pathways.104, 105 TUG1 is upregulated in the murine retina,106 but its influence in retinal degenerative diseases is not clear.
HEIH and HULC are both highly expressed in hepatitis B virus‐related hepatocellular carcinoma.107, 108, 109 They are involved in tumorigenesis by promoting hepatoma cell growth and proliferation. Suppression targets for HEIH are p15, p16, p21, and p57, while the target for HULC is p18.108, 109 BRAF‐activated noncoding RNA exerts oncogenic function in cancers via epigenetic regulation on various genes, such as p38 MAPK, MEK1/2, ERK1/2, JNK, NF‐κB, and p38.110 Abundant studies of BRAF in last 6 years have already revealed complex signaling pathways involved in tumor cell growth, proliferation, and apoptosis, yet findings on senescent phenotypes are seldom reported. As target genes for BRAF contain those involved in regulation of cell cycle and metabolism, its role in senescence calls for future exploration.
2.2. Telomere‐associated lncRNAs
Telomeres are the protective nucleoprotein caps at the end of chromosomes, which shorten with every cell division. Preservation of the telomere lengths requires telomerase reverse transcriptase combined with telomere RNA component (TERC).111 Telomere attrition is characterized as a key hallmark in cellular senescence and organismal aging.1, 22 lncRNAs play roles in the organization of telomere dynamics, indicating a possible correlation with telomere‐associated diseases.
2.2.1. TERC
TERC functions as a template for telomeric DNA synthesis by telomerase. Its involvement in senescence and aging is probably due to gradual loss of telomerase activity. TERC‐deficient mice displayed pulmonary premature aging and osteoporosis.112, 113 The pulmonary senescence‐associated inflammatory phenotype could partly be explained by telomerase‐mediated NF‐κB transcription.114 Introduction of TERC in telomerase‐deficient mice was confirmed to rescue premature aging phenotypes by restoring functional telomerase.115 Apart from that, TERC could affect angiogenesis and metastasis‐related genes’ expression without affecting telomere length.116
2.2.2. TERRA
Since the identification of telomeric repeat‐containing RNA (TERRA) in yeast, roles of this lncRNA have been highlighted in telomere functions throughout senescence and aging process.117, 118, 119 TERRA is transcribed by RNA polymerase II in a conserved manner.120 Altered expression of TERRA affects the formation of telomeric heterochromatin and the regulation of telomerase activity.121 However, the association between telomere length and TERRA expression is heterogeneous according to types of cells or species observed and methods or protocols applied.122 Therefore, conflicting results have been published on TERRA expression in cancers. TERRA levels were elevated in various cancers but decreased in advanced stages of them.118, 123, 124 Again, conflicting results have been uncovered on the relationship between TERRA and cellular senescence. Some studies revealed that overexpression of TERRA triggered premature senescence by the accumulation of itself and defective telomeric recombination.111, 119, 125 On the other hand, increased TERRA expression in telomerase‐negative cells was reported to delay the onset of senescence.126, 127 Another study even found no difference in TERRA expression between early and late passage human primary fibroblast, even in the state of repressed telomeric maintenance during senescence.128 The mystery of TERRA in senescence is expecting to be unveiled.
2.3. Chromatin‐modulating lncRNAs
Chromatin remodeling occurs within senescence and aging process. Alterations in chromatin features include epigenetic changes, heterochromatinization, histone modification, and DNA methylation. lncRNAs usually serve as modifiers, decoys, or guides, by recruiting various histone and DNA methyltransferase to the site of chromosome inactivation (eg, Xist, HOTAIR, and lncRNA‐p21) or by directing transcriptional factors to bind with regulatory DNA elements (eg, AIR). Several representative lncRNAs are mentioned in the previous parts, such as H19, ANRIL, and TERRA. In this part, we will focus on those unmentioned related lncRNAs.
2.3.1. Xist
Transcribed from the inactive X chromosome, Xist is responsible for gene imprinting and X chromosome inactivation in females by blocking the access of RNA polymerase II.129, 130 Level of Xist declines in senescent cells,131 yet its function in senescence is unclear.
2.3.2. Kcnq1ot1
KCNQ1‐overlapping transcript 1 (Kcnq1ot1) is a paternally expressed antisense lncRNA to Kcnq1ot1 gene.132 It exerts an impact on nearby imprinted genes, including CDKN1C and KCNQ1, by recruiting chromatin remodeling complexes to the paternal DMR‐LIT1 locus.133, 134 As the role of CDKN1C in cell cycle progression, Kcnq1ot1 affects cellular senescence and aging process. Moreover, the suppressed level of Kcnq1ot1 is relevant to age‐related diseases, such as type 2 diabetes, atherosclerosis, myocardial infarction, and various cancers.135, 136, 137, 138
2.3.3. ANRASSF1
As a member of poorly characterized RNAs, ANRASSF1 is an unspliced, nuclear‐localized, intronic antisense lncRNA targeting to the tumor suppressor gene, Ras‐associated domain‐containing protein 1A (RASSF1A), which is involved in G1/S cell cycle arrest and apoptosis upon DNA damage.139 Increasing DNA methylation of RASSF1A is observed in tumors, aging noncancerous liver, and chronic gastritis relevant to age.140, 141, 142 ANRASSF1 could reduce the transcription of RASSF1A by forming a DNA‐RNA hybrid and recruiting PRC2 to RASSF1A promoter region,139 indicating the role of ANRASSF1 in senescence and aging.
There are another couple of lncRNAs whose target genes have unambiguous roles in senescence and age‐related processes, yet the indirect involvement of these lncRNAs in the same field is not clear. Like Air, or antisense Igf2 receptor (Igf2r) RNA, is a paternally expressed and imprinted antisense lncRNA to maternally derived Igf2r promoter region.143 Air controls transcription of Igf2r in cis via allele‐specific methylation.144 Igf2 is directly linked to senescence and longevity.145, 146 Another example is ecCEBPA, or extra coding CEBPA, which recruits DNMT1 to silence C/EBP gene.147 The encoded C/EBP family proteins could promote growth arrest by inhibiting CDK2 and CDK4.148 C/EBP is dramatically decreased in aged tissues and causes age‐related liver injury and impaired adipogenesis and altered fat tissue function, whereas restoring aged‐like isoform of C/EBPα favors liver proliferation.149, 150 Heterodimerization of C/EBPβ and C/EBPγ promotes cell proliferation and suppress senescence.151 Similarly, pRNA serves to silence repeated nucleolar ribosomal RNA (rRNA) through the formation of DNA‐RNA triplex and subsequent repressive DNA methylation at the rRNA promoter.152 As levels of rRNA are tightly correlated with senescence, aging process and age‐related neurodegenerative diseases (eg, AD and Werner syndrome), and symptoms (eg, depression),153, 154, 155 the implication of pRNA in this field remains to be confirmed. PTENpg1 negatively regulates PTEN level, the latter of which is known suppressor of senescence, aging, and tumor.
2.4. SASP‐associated lncRNAs
SASP is a critical trait of senescent cells. Also, the accumulation of senescent cells during aging provokes production of SASP factors, facilitating low‐grade chronic inflammation and age‐related diseases. Regulation of lncRNAs contributes to innate immune responses, such as macrophage polarization and inflammatory factor secretion.
2.4.1. 17A
17A controls the alternate spicing of GABA receptor and subsequent downstream signaling.156, 157 It was reported to be triggered by inflammation in AD brains, leading to increase in Aβ accumulation.157
2.4.2. FIRRE
Functional intergenic repeating RNA element (FIRRE) is a newly discovered, conserved lncRNA, which has an impact on the nuclear architecture across chromosome through interacting with hnRNP‐U.158 Controlled by NF‐κB signaling in macrophages, FIRRE positively regulates several inflammatory genes following LPS stimulation by affecting the stability of relevant mRNAs.159
2.4.3. lnc‐IL7R
lnc‐IL7R is remarkably upregulated in THP‐1 cells with stimulation of LPS and then, in turn, diminishes LPS‐mediated proinflammatory cytokine secretion, characterized by reduced expression of E‐selectin, VCAM‐1, IL‐6, and IL‐8 through epigenetic regulation.160 This finding indicates contribution of lnc‐IL7R to SASP factor production.
2.4.4. lncRNA‐LET
lncRNA‐LET (low expression in tumors) is poorly expressed in multiple tumors. Further study has shown silencing this lncRNA allows accumulation of nuclear factor 90 (NF90), the latter of which suppresses the translation of MCP1, CXCL1, and IL‐6.161, 162 As downregulated NF90 is observed in senescent cells, lncRNA‐LET has a positive link to low levels of SASP through actions of NF90.162
2.4.5. lincRNA‐COX2
lincRNA‐COX2 is a broad‐acting regulatory component of the TLR/MyD88/NF‐κB pathway upon TLR activation. lincRNA‐COX2 represses transcription of a series of proinflammatory genes by interacting with hnRNP‐A/B and A2/B1.163 This lncRNA could form a complex with the switch/sucrose nonfermentable to modulate the assembly of NF‐κB and subsequently transactivate downstream inflammatory response genes.164 lincRNA‐COX2 enhances TLR‐induced IL‐6 and simultaneously suppressing chemokines CCL5, the latter of which is still controversial.163, 164
2.4.6. Lethe
The pseudogene, Lethe, is selectively induced by TNF‐α and IL‐1β upon NF‐κB activation. On the other hand, Lethe regulates NF‐κB pathway by interacting with the NF‐κB subunit p65 (RelA) to inhibit DNA binding to downstream cytokines genes.165 Age‐related reduction in Lethe could be explained by increased NF‐κB in aging tissues.166
2.4.7. NEAT1
Localized in nucleus’ interchromatin space, nuclear‐enriched abundant transcript 1 (NEAT1) is an essential component of nuclear paraspeckles.167 Paraspeckles can sequester many transcripts or multifunctional protein complex in the nucleus, and inhibit the translation or biological activity of these captives. NEAT1 serves as a novel inflammatory regulator by affecting the formation of paraspeckles.168 NEAT1 facilitates the expression of IL‐8 by relocating SFPQ, a repressor of IL8 transcription, to the paraspeckles.169 NEAT1 partly mediates LPS‐induced cytokine expressions via the NF‐κB pathway, as well as TLR4‐activated inflammatory process via MAPK pathway.170, 171 Recent studies have revealed the involvement of NEAT1 in osteoarthritis (OA) and formation and inflammation of foam cells,172, 173 suggesting its potential role in age‐related treatment.
2.4.8. PACER
p50‐associated COX‐2 extragenic RNA (PACER) is expressed in the upstream region of COX‐2 and regulates COX‐2 expression in monocyte‐derived cells upon LPS stimulation. PACER is modulated by CTCF/cohesion complex, which favors PACER transcription, and in turn, PACER functions to activate COX‐2 expression by directly sequestering the repressive NF‐κB p50 subunit from the COX‐2 promoter.174 PACER is reported to be induced in OA chondrocytes by multiple proinflammatory cytokines, suggesting its involvement in inflammation‐driven age‐related diseases.175
2.4.9. THRIL
Identified in human monocyte cell line THP1 macrophages, TNF and hnRNPL‐related immunoregulatory lincRNA (THRIL) promotes TNF transcription by forming THRIL‐hsRNPL complex through binding to TNF promoter.176
2.5. Other lncRNAs in cellular aging
2.5.1. HOTAIR
HOX transcript antisense RNA (HOTAIR) has been involved in senescence via multiple mechanisms. Transcribed from intergenic region between HOXC11 and HOXC12 within the homeobox (HOXC) gene cluster, HOTAIR regulates genes on HOXC foci epigenetically by acting as a scaffold and guide for various histone modification complexes.177, 178, 179 HOTAIR can activate senescence through NF‐κB pathway after DNA damage and even maintains the activation of this pathway in the presence of a positive feedback loop.180 HOTAIR can be suppressed by HuR in the way similar to lincRNA‐p21.181 As HOTAIR is upregulated in senescent cells, HuR deficiency in various cells leads to dramatically increased HOTAIR expression, characteristic senescent phenotypes, and HOTAIR‐mediated ubiquitination and proteolysis of ataxin‐1 and snurportin‐1.181 Yet, the role of protein ubiquitination and degradation in cellular senescence is still unknown.
2.5.2. ASncmtRNA‐2
Mitochondria play a significant role in the onset of senescence, as accumulated mitochondrial‐derived ROS induces senescence by adaptive modulation on the transcription of nuclear‐encoded factors.182 Antisense noncoding mitochondrial RNA‐2 (ASncmtRNA‐2) is exported from mitochondria to nucleus, whose flow direction is consistent with the mitochondria retrograde signaling. This lncRNA is involved in replicative senescence in ECs by maintaining the cell cycle arrest in G2/M phase through the production of has‐miR‐4485 and has‐miR‐1973. Meanwhile, p16 displayed similar ASncmtRNA‐2 pattern in the senescent cells, suggesting a possible coregulation of the two genes.183 Expression of ASncmtRNA‐2 was preponderant in aged murine aortas,183 indicating its impact on vascular aging.
3. SPECIFIC EXPRESSION Of lncRNAs IN DIFFERENT TISSUES/ORGANS DURING AGING
Changes in morphology and physiology determine specific age‐related diseases in different tissues and organs. We firstly summarized the various changes and characterized diseases found in the elderly. Then, we reviewed the reported specific expressed lncRNAs according to the localization or diseases (Table 1).
Table 1.
List of lncRNAs potentially implied in the aging process and age‐related diseases
| lncRNAs (References) | Samples studied | Processes | Effect during aging or other implications |
|---|---|---|---|
| Six3os, Dlx1as 258 | Adult mice brain | Neurogenesis | Upregulated in neuroblasts; downregulated in NSCs |
| Pnky 259 | Postnatal mice brain | Neurogenesis | Depletion of Pnky potentiates neuronal lineage commitment |
| MALAT1, GOMAFU, NEAT1, TUG1 101 | Human brain | Neurogenesis | Upregulated in the subependymal zone with age |
| RMST 260 | Human cell line | Neurogenesis | Required to promote neuronal differentiation |
| TERC 261 | Embryonic and postnatal mice brain | Neurogenesis | Balanced pattern with telomerase reverse transcriptase to determine NSC proliferation and survival |
| TERRA 123 | Postnatal mice brain | Neurogenesis | Upregulated in proliferating cerebellar neuronal progenitors |
| BC200 190, 262, 263, 264 | Rat/human brain, cell line | Cognitive decline | Act as a scaffold to bind with translational factors to repress neuronal protein synthesis; downregulated in the aged brain; upregulated in aging brain |
| BC1 262, 263, 264, 265 | Rat brain, human cell line | Cognitive decline | Act as a scaffold to bind with translational factors to repress neuronal protein synthesis; maintain neuronal excitability, mood, and exploratory behavior |
| BDNF‐AS, GDNF‐AS, EPHB2‐AS 266 | Mice brain, human brain neurons | Cognitive decline | Suppress protein synthesis (BDGF, GDNF, and EPHB2) involved in neurite elaboration |
| GOMAFU 267 | Human brain | Cognitive decline | Instruct alternate splicing in synaptic plasticity |
| NEAT1268 | Mice brain | Cognitive decline | Modulate ion channel components |
| BACE1‐AS 186 | Human and Mice brain | Neurodegeneration | Modulate BACE1 expression and Aβ aggregation |
| SORL1‐AS 187 | Human brain | Neurodegeneration | Direct alternate splicing of SORL1 and Aβ formation |
| UCHL1‐AS 188, 269, 270 | Human brain | Neurodegeneration | Regulate UCHL1 expression, which facilitates pathogenic protein aggregation in AD and PD |
| LRP1‐AS 185, 271 | Human brain | Neurodegeneration | Regulate LRP1 expression and Aβ metabolism in AD |
| 17A 157 | Human brain, cell line | Neurodegeneration |
Induce alternate splicing of GABA protein isoform Enhance Aβ secretion in AD |
| ANRIL 189 | Human brain | Neurodegeneration | Regulate CDKN2B expression, which is accumulated in neurofibrillary tangles and amyloid plaques in AD |
| SNHG1 191 | Mice brain, human cell line | Neurodegeneration | Promote α‐synuclein in PD by targeting miR‐15b‐5p |
| G069488 192 | Human cell line | Neurodegeneration | Regulate neurite regeneration and neural restoration by suppressing NEDD9 under α‐synuclein accumulation in AD |
| RP11‐142J21.2 192 | Human cell line | Neurodegeneration | Promote apoptosis by suppressing SEMA6D via MAPK under α‐synuclein accumulation in AD |
| NEAT1, MEG3, Rian, Mirg 198 | Mice liver | Liver aging | Upregulated in healthy aging liver |
| H19 60, 67 | Mice cell line | Myogenesis | Modulate myoblast differentiation and muscle regeneration |
| lncMD1 202, 203 | Mice cell line | Myogenesis | Modulate myoblast differentiation during aging |
| SIRT1‐AS 206 | Mice cell line | Myogenesis | Modulate myoblast differentiation |
| MALAT1 204, 205 | Mice muscle, Mice and human cell | Myogenesis | Promote myoblast proliferation and differentiation in aging muscle |
| YY1 209 | Mice cell line | Myogenesis |
Upregulated in myoblasts but downregulated during differentiation Regulate myogenesis at the transcriptional level |
| Glt2/Meg3 210 | Mice cell line | Myogenesis | Maintain muscle development |
| MAR1 208 | Mice cell line | Myogenesis | Attenuate muscle atrophy induced by aging |
| MALAT1 212, 213, 214, 215, 216 | Human and mice cell | Angiogenesis, vascular remodeling | Control EC proliferation and senescence; mediate angiogenesis and vascular inflammation |
| MEG3 217, 218 | Mice vessel, human cell | Angiogenesis | Upregulated in senescent ECs; depletion of MEG3 promotes sprouting and EC proliferation |
| ANRIL 219, 220, 221, 222, 223 | Human artery and cell | Atherosclerosis | Distinct modulation on VSMC proliferation and plaque formation according to different splicing variants |
| H19 272, 273, 274 | Rat artery, human cell | Atherosclerosis | Modulate EC and VSMC proliferation and homeostasis |
| ASncmtRNA‐2 183 | Mice cell | Vascular aging | Upregulated in aortas from aged mice and senescent ECs |
| HOTAIR 225 | Human artery, cell line | Atherosclerosis | Downregulated in ECs form atherosclerotic plaques; regulate EC proliferation and migration |
| MIAT 224 | Rat artery, human cell | Angiogenesis | Regulate EC function |
| TUG1 226, 227 | Rat and mice cell | Atherosclerosis | Regulate EC apoptosis and VSMC homeostasis |
| linc‐p21 91, 95, 228 | Mice cell | Atherosclerosis | Promote apoptosis and suppress proliferation in VSMCs and macrophages |
| Gas5 229, 230, 231 | Rat artery, human cell | Atherosclerosis, vascular remodeling | Promote VSMC proliferation and migration; guide macrophage polarization |
| HOXC‐AS 232 | Human artery | Atherosclerosis | Downregulated in atherosclerotic plaques through inflammatory responses |
| linc00305 233 | Human cell | Atherosclerosis | Promote monocyte activation and vascular inflammation |
| lncRNA OTTHUMT00000387022 234 | Human plasma and cell, | Atherosclerosis | Promote inflammation in macrophages |
| lncRNA RP5‐833A20.1 275 | Mice artery and cell | Atherosclerosis | Regulated cholesterol homeostasis and inflammatory responses in foam cells |
| H19 235 | Human cell | Osteogenesis | Promote osteoblast differentiation |
| MALAT1 236 | Human cell | Osteogenesis | Induce osteogenic differentiation |
| HOTAIR 237 | Human cell | Osteogenesis | Suppress osteogenic differentiation |
| DANCER 238 | Human cell | Osteogenesis | Suppress osteogenic differentiation |
| MEG3 239, 240 | Human cell | Osteogenesis | Suppress osteogenic differentiation |
| MIAT 241 | Human cell | Osteogenesis | Suppress osteogenic differentiation under inflammation |
| MIR31HG 242 | Human cell | Osteogenesis | Rescue osteogenic differentiation inhibited by inflammation |
| DANCER 238, 243 | Human bone and cell | Osteoporosis | Promote osteoblast differentiation; suppress osteogenic differentiation |
| H19 107 | Mice tissue | Lipid deposition | Imprint IGF2 and affect lipid deposition |
| PLUTO 251 | Human islets | T2DM | Regulate β‐cell function and pancreatic formation |
| βlinc1 252 | Mice islets | T2DM | Associated with β‐cell loss |
| HI‐LNC901 253 | Human islets | T2DM | Correlated with insulin exocytosis |
| Kcnq1ot1, HI‐LNC78, HI‐LNC80 254 | Human islets | T2DM |
Upregulated in T2DM Sense blood glucose level |
| HI‐LNC45 254 | Human islets | T2DM |
Downregulated in T2DM Sense blood glucose level |
AD, Alzheimer's disease; ECs, endothelial cells; NSCs, neural stem cells; PD, Parkinson's disease; VSMCs: vascular smooth muscle cells.
3.1. Brain
Brain aging is characterized by declined cognition, reduced neurogenesis, and neurodegeneration. Neurogenesis occurs even in adult life, but generally declines throughout aging. Current studies have revealed multiple functions of lncRNAs in embryonic and adult neurogenesis from different species (eg, MALAT1, TUG1, RMST, Dlx1as, Six3os, Pnky, TERC, and TERRA). Firstly, lncRNAs influence self‐renewal of neural stem cells (NSCs) and amplification of intermediated progenitors and neuroblasts. Secondly, lncRNAs determine the fate specification of NSCs, as this progenitor can generate astrocytes and oligodendrocytes, aside from neuroblasts. Lastly, lncRNAs are known key regulators of telomere dynamics in NSCs.
Impaired cognition is supposed to be a direct consequence of the alterations in synaptic connectivity.184 lncRNAs modulate pathological protein aggregation, and the subnuclear compartment‐specific lncRNAs regulate neuronal splicing, transcription, and sponging of ion channels in aging (detailed lncRNAs seen in Figure 1). Relative abundance of specific lncRNAs allows for beneficial functional processes. On the contrary, shifts in their abundance may trigger alterations in pretranscriptional and post‐transcriptional regulations of neuronal genes and consequent age‐related neurodegenerative diseases, including AD and Parkinson's disease (PD), which are featured by impaired cognitive and motor function. In AD, lncRNAs are known to contribute to Aβ aggregation and dysregulated synaptic plasticity. Certain differentially expressed antisense lncRNAs, including BACE1‐AS, SORL1‐AS, UCHL1‐AS, and LRP1‐AS, modulate expression or splicing of proteins involved in the generation and trafficking of Aβ.185, 186, 187, 188 On the other hand, 17A is involved in Aβ accumulation through local inflammatory responses.157 ANRIL regulates the expression of CDKN2B that accumulates in neurofibrillary tangles and amyloid plaques in AD brain.189 Expression of BC200 was decreased in the normal aging brain, but elevated in AD brain.190 The accumulated pathological protein in PD brain is α‐synuclein, contained in Lewy body. The identified genes involved in PD pathology include Parkin, PINK1, PARK‐7, and LRRK2. Therefore, further investigations regarding lncRNAs targeting these genes or linked to the pathogenesis of α‐synuclein would be a promising strategy in PD therapy.103, 188, 191, 192
3.2. Liver
Liver blood flow is estimated to be reduced by 20%‐40%, which seems to be consistent with the shrinkage of liver volume.193, 194 Accumulated lipofuscin in hepatocytes contributes to chronic oxidative stress, and vacuolation of hepatocyte nuclei is linked to diabetes and nonalcoholic fatty liver diseases (NAFLD), both of which are possible markers of hepatocyte senescence.195, 196 Age‐related decline in drug metabolism and regeneration capacity, and abnormal immune responses enhance vulnerability to acute liver injury, liver fibrosis, hepatitis C, NAFLD, alcoholic liver diseases, and liver tumor. Alterations in C/EBP family and telomere reverse transcriptase by repressive chromatin remodeling are observed in aged drug‐induced liver injury, resulting in impaired regenerative capacity and fibrosis.197 A group of differentially expressed lncRNAs in mouse have been identified in the above pathophysiologies, including NEAT1, MEG3, Rian, and Mirg.198 Rian and MEG3 could regulate proliferation by directly recruiting PRC2.199 Mirg could predict certain cell cycle factors, such as Myc and p53.200 Moreover, the involvement of ANRASSF1, ecCEBPA, and some other lncRNAs, whose target genes are involved in liver metabolism, cell cycle, or local inflammatory responses, remains to be elucidated.
3.3. Muscle
Muscle mass declines progressively during aging. Sarcopenia is a common age‐related skeletal muscle degeneration, characterized by reduced muscle mass and muscle fibers. The underlying mechanisms are multifaceted, including a sedentary lifestyle, reduced hormonal level, and increased inflammation, loss of proteostasis, and mitochondrial dysfunction.201 H19 is implicated in skeletal muscle differentiation by acting as a molecular sponge to bind the miRlet‐7.60 H19 is highly expressed in skeletal muscle, as well as H19‐encoded miRNAs, miRlet‐7, and miRlet‐7 during muscle regeneration, all of which are regulated by SMAD1/5.67 The muscle‐specific lncMD1 exerts as a decoy for miR‐133 and miR‐135, which is enhanced by HuR, to limit its impact on the expression of Elavl1 in muscle differentiation during muscle aging. HuR plays a direct role in muscle wasting and sarcopenia.202, 203 Stimulated by myostatin, MALAT1 regulates muscle cell proliferation and differentiation, thus influencing muscle aging.204, 205 SIRT1‐AS was recently reported to play a role in myogenesis, as its antisense target SIRT1 could prevent senescence and aging through myogenic program.206, 207 There are other lncRNAs involved in myogenesis, such as YY1, Glt2/Meg3, and MAR1, whose function in muscle aging needs further exploration.29, 208, 210
3.4. Cardiovascular system
Cardiovascular aging is generally accompanied by the occurrence of ischemic cardiovascular diseases (eg, hypertension, coronary artery disease [CAD], atherosclerosis, myocardial infarction, stroke).211 An expanding number of lncRNAs have been identified in the series of pathophysiologies by regulating EC and vascular smooth muscle cell (VSMC) proliferation, angiogenesis, vascular remodeling, macrophage polarization, and cholesterol metabolism.49 MALAT1 is significantly important in promoting EC proliferation, vessel outgrowth, and sprouting and in protecting ECs against apoptosis induced by oxygen‐glucose deprivation and ox‐LDL–related inflammation via various targets (eg, miR‐22‐3p and encoded genes CXCR2 and Akt, miR‐26a) through multiple signaling, including p21, p38, PI3K/Akt.212, 213, 214, 215, 216 MEG3 is upregulated during vascular aging. Silencing MEG3 could prevent aging‐mediated inhibition of sprouting activity and EC proliferation.217, 218 ANRIL is known as an independent risk factor for CAD. However, functional annotation of this lncRNA in atherosclerosis is controversial, as different splicing variants of ANRIL might play distinct roles.219, 220, 221, 222, 223 Also, H19, ASncmtRNA‐2, HOTAIR, MIAT, TUG1, linc‐p21, and Gas5 play similar roles in angiogenesis and atherosclerosis by regulating the function of ECs and VSMCs.91, 95, 183, 224, 225, 226, 227, 228, 229, 230, 231 On the other hand, lncRNAs including HOXC‐AS, Gas5, linc00305, lncRNA OTTHUMT00000387022, and lncRNA RP5‐833A20.1 could activate macrophages, mediate inflammatory responses, or regulate lipid metabolism, exerting impacts on atherosclerotic plaque formation.232, 233, 234 Additionally, other lncRNAs related to macrophage activation and polarization, which are mentioned in the previous part, such as PACER, THRIL, and lincRNA‐COX2, might be conducible to the progression of atherosclerosis.
3.5. Bone
The process of aging breaks the balance between bone formation and resorption. The changes in bone turnover cause osteoporosis, which can also be induced by endogenous estrogen deficiency or corticosteroid treatment. Plentiful lncRNAs have already been revealed to take part in osteogenesis ossification (eg, H19, MALAT1, HOTAIR, DANCR, MEG3, MIAT, and MIR31HG) and osteoclast differentiation (eg, DANCER) via specific target mRNAs or miRNAs.235, 236, 237, 238, 239, 240, 241, 242 DANCER is involved in the pathology of osteoporosis, as it promotes inflammation‐induced osteoclastogenesis and suppresses osteogenic differentiation, which implies a potential biomarker for osteoporosis.238, 243 As the half‐lives of lncRNAs are less than those of mRNAs, recent strategies have applied the systematic analysis of lncRNAs‐miRNAs‐mRNAs regulatory network as to search for more potential biomarkers for osteoporosis.244 Only a handful of lncRNAs have been screened out in these studies, including LOC105376834, LOC101929866, and mmu_12821_PI428960544, all of whose biological significances are required to be addressed in further studies.245, 246
3.6. Adipose tissue
Adipose tissue exerts immune and endocrine actions throughout life, besides being a major source of energy source. Compared to the subcutaneous distribution in adult years, visceral redistribution, and ectopic deposition in liver, bone marrow and muscle are adopted in the old age. lncRNAs is involved in this extensive remodeling process by controlling adipogenesis and lipid metabolism. H19 affects fat deposition and metabolism. In adult mice, low expressed IGF2 is associated with increased lipid deposition. Then during aging, the expression of H19‐IGF2 is enhanced due to loss of imprinting of this gene locus.66 linc‐DMRT2 and linc‐TP53I13 were reported to be downregulated by lipopolysaccharide in adipose tissue of obese humans, providing clues to age‐related diseases derived from interrupted homeostasis of adipose tissue.247 Sun et al firstly identified a group of lncRNAs, termed as lnc‐RAP‐n, which are specifically regulated during adipogenesis through PPARγ and CEBPα. However, the direct impacts of individual lnc‐RAP‐n on adipose tissue aging warrant further study.248
3.7. Pancreatic islets
Type 2 diabetes mellitus (T2DM) is considered as an age‐related disease, as it is well documented that aging is associated with declined insulin action and β‐cell secretory activity.249 Moreover, pancreatic islet cell senescence partly contributes to the rise of T2DM in the elderly.250 Growing evidence implicates lncRNAs in the etiology of T2DM. PLUTO is involved in pancreas development and β‐cell function, as it regulates PDX1 transcriptional activity.251 βlinc1, a β‐cell long intergenic noncoding RNA, could modulate β‐cell formation and function.252 HI‐LNC901 was reported to be directly correlated with insulin exocytosis.253 In addition, high levels of Kcnq1ot1, HI‐LNC78, and HI‐LNC80 and low level of HI‐LNC45 were observed in pancreatic islets from diabetic individuals or in the presence of high glucose, indicating the function of sensing blood glucose levels.254
3.8. Immune system
Immunosenescence refers to the acquisition of senescent features in the immune system, which result in increased susceptibility to infection and a higher incidence of age‐related diseases. Moreover, aging is considered as a low‐grade chronic inflammation state, termed inflammaging, where SASP plays an important role.255 SASP‐associated lncRNAs have been stated above in Section 2.1.8. Apart from that, loss of CD4+T cells partly leads to dysfunction of innate immunity. Only limited lncRNAs, such as linc‐MAF‐4 and rmrp, post‐transcriptionally regulated CD4+T‐cell subsets, but no direct or indirect evidences point to their involvement in aging.256, 257
4. CONCLUSION AND PERSPECTIVES
As concluding remarks, the emerging role of lncRNAs as regulators of cellular senescence and age‐related diseases is still in its infancy. Numerous diseases arise with advancing age, yet we just pick a couple of them to discuss in our review. Cancer is another kind of age‐related disease, in which the function of lncRNAs has been deeply investigated; thus, it is difficult for us to list all of them in limited words. At present, aging and age‐related diseases have become a heavy burden in society. Illustrating lncRNAs function in aging physiology and pathology is of great significance under this context. Samples from elderly populations and a few animal models are adopted to obtain the comprehensive spectrum of lncRNAs implicated in age‐associated diseases. On the other hand, applications of recent advanced technologies facilitate detailed elucidation of mechanisms on the regulation and function of lncRNAs systematically. Although the potential usefulness of lncRNAs in aging and age‐related diseases cannot be fully realized at present, we can expect fast progress in technologies will enable us to make good use of lncRNAs in aging.
CONFLICT OF INTEREST
The authors declared no conflict of interests regarding this manuscript.
ACKNOWLEDGMENTS
This work was financially supported by the Natural Science Foundation of Hunan Province, China (2018JJ3759 and 2018JJ3716).
He J, Tu C, Liu Y. Role of lncRNAs in aging and age‐related diseases. Aging Med. 2018;1:158–175. 10.1002/agm2.12030
REFERENCES
- 1. Lopez‐Otin C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell. 2013;153(6):1194‐1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Larson KJ, Hamlin RJ, Sprung J, et al. Associations between Charlson Comorbidity Index and surgical risk severity and the surgical outcomes in advanced‐age patients. Am Surg. 2014;80(6):555‐560. [PubMed] [Google Scholar]
- 3. Divo MJ, Celli BR, Poblador‐Plou B, et al. Chronic Obstructive Pulmonary Disease (COPD) as a disease of early aging: evidence from the EpiChron Cohort. PLoS ONE. 2018;13(2):e0193143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990‐2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1211‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dwolatzky T, Brodsky J, Azaiza F, et al. Coming of age: health‐care challenges of an ageing population in Israel. Lancet. 2017;389(10088):2542‐2550. [DOI] [PubMed] [Google Scholar]
- 6. Dieleman JL, Squires E, Bui AL, et al. Factors associated with increases in US health care spending, 1996–2013. JAMA. 2017;318(17):1668‐1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Consortium EP, Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799‐816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elling R, Chan J, Fitzgerald KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur J Immunol. 2016;46(3):504‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Degirmenci U, Lei S. Role of lncRNAs in cellular aging. Front Endocrinol (Lausanne). 2016;7:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loewer S, Cabili MN, Guttman M, et al. Large intergenic non‐coding RNA‐RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113‐1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McHugh CA, Chen CK, Chow A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232‐236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300‐307. [DOI] [PubMed] [Google Scholar]
- 17. Rossi MN, Antonangeli F. LncRNAs: new players in apoptosis control. Int J Cell Biol. 2014;2014:473857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lazorthes S, Vallot C, Briois S, et al. A vlincRNA participates in senescence maintenance by relieving H2AZ‐mediated repression at the INK4 locus. Nat Commun. 2015;6:5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grammatikakis I, Panda AC, Abdelmohsen K, et al. Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY). 2014;6(12):992‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greco S, Gorospe M, Martelli F. Noncoding RNA in age‐related cardiovascular diseases. J Mol Cell Cardiol. 2015;83:142‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ben‐Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37(5):961‐976. [DOI] [PubMed] [Google Scholar]
- 22. Hernandez‐Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28(6):436‐453. [DOI] [PubMed] [Google Scholar]
- 23. Frenk S, Houseley J. Gene expression hallmarks of cellular ageing. Biogerontology. 2018;(Suppl 1):1‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aravinthan A. Cellular senescence: a hitchhiker's guide. Hum Cell. 2015;28(2):51‐64. [DOI] [PubMed] [Google Scholar]
- 25. Abdelmohsen K, Gorospe M. Noncoding RNA control of cellular senescence. Wiley Interdiscip Rev RNA. 2015;6(6):615‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B‐MYB. PLoS Genet. 2013;9(3):e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong Y, Liang G, Yuan B, et al. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36(3):1477‐1486. [DOI] [PubMed] [Google Scholar]
- 28. Hu L, Wu Y, Tan D, et al. Up‐regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang MH, Hu ZY, Xu C, et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852(1):166‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo F, Li Y, Liu Y, et al. Inhibition of metastasis‐associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai). 2010;42(3):224‐229. [DOI] [PubMed] [Google Scholar]
- 31. Zhao Z, Chen C, Liu Y, et al. 17beta‐Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT‐1 RNA level. Biochem Biophys Res Commun. 2014;445(2):388‐393. [DOI] [PubMed] [Google Scholar]
- 32. Eissmann M, Gutschner T, Hammerle M, et al. Loss of the abundant nuclear non‐coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487‐1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis‐regulatory role in the adult. Cell Rep. 2012;2(1):111‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pasmant E, Laurendeau I, Heron D, et al. Characterization of a germ‐line deletion, including the entire INK4/ARF locus, in a melanoma‐neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67(8):3963‐3969. [DOI] [PubMed] [Google Scholar]
- 36. Kotake Y, Nakagawa T, Kitagawa K, et al. Long non‐coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956‐1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou X, Han X, Wittfeldt A, et al. Long non‐coding RNA ANRIL regulates inflammatory responses as a novel component of NF‐kappaB pathway. RNA Biol. 2016;13(1):98‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chi JS, Li JZ, Jia JJ, et al. Long non‐coding RNA ANRIL in gene regulation and its duality in atherosclerosis. J Huazhong Univ Sci Technolog Med Sci. 2017;37(6):816‐822. [DOI] [PubMed] [Google Scholar]
- 39. Song CL, Wang JP, Xue X, et al. Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell Physiol Biochem. 2017;42(3):1202‐1212. [DOI] [PubMed] [Google Scholar]
- 40. Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17(6):806‐814. [DOI] [PubMed] [Google Scholar]
- 41. Cunnington MS, Santibanez Koref M, Mayosi BM, et al. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6(4):e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen W, Bocker W, Brosius J, et al. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183(3):345‐351. [DOI] [PubMed] [Google Scholar]
- 43. Baryakin DN, Semenov DV, Savelyeva AV, et al. Alu‐ and 7SL RNA analogues suppress MCF‐7 cell viability through modulating the transcription of endoplasmic reticulum stress response genes. Acta Naturae. 2013;5(4):83‐93. [PMC free article] [PubMed] [Google Scholar]
- 44. Abdelmohsen K, Panda AC, Kang MJ, et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014;42(15):10099‐10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kour S, Rath PC. Long noncoding RNAs in aging and age‐related diseases. Ageing Res Rev. 2016;26:1‐21. [DOI] [PubMed] [Google Scholar]
- 46. Miyoshi N, Wagatsuma H, Wakana S, et al. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5(3):211‐220. [DOI] [PubMed] [Google Scholar]
- 47. McLaughlin D, Vidaki M, Renieri E, et al. Expression pattern of the maternally imprinted gene Gtl2 in the forebrain during embryonic development and adulthood. Gene Expr Patterns. 2006;6(4):394‐399. [DOI] [PubMed] [Google Scholar]
- 48. Zhao Y, Wu J, Liangpunsakul S, et al. Long non‐coding RNA in liver metabolism and disease: current status. Liver Res. 2017;1(3):163‐167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simion V, Haemmig S, Feinberg MW. LncRNAs in vascular biology and disease. Vascul Pharmacol. 2018. 10.1016/j.vph.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Braconi C, Kogure T, Valeri N, et al. microRNA‐29 can regulate expression of the long non‐coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750‐4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X, Zhou Y, Mehta KR, et al. A pituitary‐derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88(11):5119‐5126. [DOI] [PubMed] [Google Scholar]
- 52. Lu KH, Li W, Liu XH, et al. Long non‐coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. He Y, Luo Y, Liang B, et al. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget. 2017;8(42):73282‐73295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou Y, Zhong Y, Wang Y, et al. Activation of p53 by MEG3 non‐coding RNA. J Biol Chem. 2007;282(34):24731‐24742. [DOI] [PubMed] [Google Scholar]
- 55. Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol BioSyst. 2013;9(3):407‐411. [DOI] [PubMed] [Google Scholar]
- 56. Anwar SL, Krech T, Hasemeier B, et al. Loss of imprinting and allelic switching at the DLK1‐MEG3 locus in human hepatocellular carcinoma. PLoS ONE. 2012;7(11):e49462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Qin R, Chen Z, Ding Y, et al. Long non‐coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60(5):486‐492. [DOI] [PubMed] [Google Scholar]
- 58. Johnson R. Long non‐coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis. 2012;46(2):245‐254. [DOI] [PubMed] [Google Scholar]
- 59. Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12(23):3693‐3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let‐7 microRNAs. Mol Cell. 2013;52(1):101‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR‐675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281(16):3766‐3775. [DOI] [PubMed] [Google Scholar]
- 62. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318‐2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sun H, Wang G, Peng Y, et al. H19 lncRNA mediates 17beta‐estradiol‐induced cell proliferation in MCF‐7 breast cancer cells. Oncol Rep. 2015;33(6):3045‐3052. [DOI] [PubMed] [Google Scholar]
- 64. Ratajczak MZ. Igf2‐H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia Histochem Cytobiol. 2012;50(2):171‐179. [DOI] [PubMed] [Google Scholar]
- 65. Fu VX, Dobosy JR, Desotelle JA, et al. Aging and cancer‐related loss of insulin‐like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 2008;68(16):6797‐6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jones BK, Levorse J, Tilghman SM. Deletion of a nuclease‐sensitive region between the Igf2 and H19 genes leads to Igf2 misregulation and increased adiposity. Hum Mol Genet. 2001;10(8):807‐814. [DOI] [PubMed] [Google Scholar]
- 67. Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR‐675‐3p and miR‐675‐5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28(5):491‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang XS, Zhang Z, Wang HC, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12(16):4851‐4858. [DOI] [PubMed] [Google Scholar]
- 69. Wang H, Guan Z, He K, et al. LncRNA UCA1 in anti‐cancer drug resistance. Oncotarget. 2017;8(38):64638‐64650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar PP, Emechebe U, Smith R, et al. Coordinated control of senescence by lncRNA and a novel T‐box3 co‐repressor complex. Elife. 2014;3:e02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xue M, Chen W, Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J Cancer Res Clin Oncol. 2016;142(7):1407‐1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hu X, Feng Y, Zhang D, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26(3):344‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhong X, Hu X, Zhang L. Oncogenic long noncoding RNA FAL1 in human cancer. Mol Cell Oncol. 2015;2(2):e977154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hollander MC, Alamo I, Fornace AJ Jr. A novel DNA damage‐inducible transcript, gadd7, inhibits cell growth, but lacks a protein product. Nucleic Acids Res. 1996;24(9):1589‐1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu X, Li D, Zhang W, et al. Long non‐coding RNA gadd7 interacts with TDP‐43 and regulates Cdk6 mRNA decay. EMBO J. 2012;31(23):4415‐4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rader J, Russell MR, Hart LS, et al. Dual CDK4/CDK6 inhibition induces cell‐cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19(22):6173‐6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Augoff K, McCue B, Plow EF, et al. miR‐31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple‐negative breast cancer. Mol Cancer. 2012;11:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dellago H, Preschitz‐Kammerhofer B, Terlecki‐Zaniewicz L, et al. High levels of oncomiR‐21 contribute to the senescence‐induced growth arrest in normal human cells and its knock‐down increases the replicative lifespan. Aging Cell. 2013;12(3):446‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xi S, Yang M, Tao Y, et al. Cigarette smoke induces C/EBP‐beta‐mediated activation of miR‐31 in normal human respiratory epithelia and lung cancer cells. PLoS ONE. 2010;5(10):e13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shih JW, Chiang WF, Wu ATH, et al. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF‐1alpha co‐activator driving oral cancer progression. Nat Commun. 2017;8:15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu Z, Bai J, Zhang L, et al. Conditional knockout of microRNA‐31 promotes the development of colitis associated cancer. Biochem Biophys Res Commun. 2017;490(1):62‐68. [DOI] [PubMed] [Google Scholar]
- 82. Qin J, Ning H, Zhou Y, et al. LncRNA MIR31HG overexpression serves as poor prognostic biomarker and promotes cells proliferation in lung adenocarcinoma. Biomed Pharmacother. 2018;99:363‐368. [DOI] [PubMed] [Google Scholar]
- 83. Montes M, Nielsen MM, Maglieri G, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015;6:6967. [DOI] [PubMed] [Google Scholar]
- 84. Di Agostino S, Strano S, Emiliozzi V, et al. Gain of function of mutant p53: the mutant p53/NF‐Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10(3):191‐202. [DOI] [PubMed] [Google Scholar]
- 85. Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell‐cycle promoters. Nat Genet. 2011;43(7):621‐629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Matuoka K, Chen KY. Possible role of subunit A of nuclear factor Y (NF‐YA) in normal human diploid fibroblasts during senescence. Biogerontology. 2000;1(3):261‐271. [DOI] [PubMed] [Google Scholar]
- 87. Wang Y, Zhang M, Xu H, et al. Discovery and validation of the tumor‐suppressive function of long noncoding RNA PANDA in human diffuse large B‐cell lymphoma through the inactivation of MAPK/ERK signaling pathway. Oncotarget. 2017;8(42):72182‐72196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Puvvula PK, Desetty RD, Pineau P, et al. Long noncoding RNA PANDA and scaffold‐attachment‐factor SAFA control senescence entry and exit. Nat Commun. 2014;5:5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dimitrova N, Zamudio JR, Jong RM, et al. LincRNA‐p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wu G, Cai J, Han Y, et al. LincRNA‐p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA‐p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bao X, Wu H, Zhu X, et al. The p53‐induced lincRNA‐p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25(1):80‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cekin N, Ozcan A, Goksel S, et al. Decreased FENDRR and LincRNA‐p21 expression in atherosclerotic plaque. Anatol J Cardiol. 2018;19(2):131‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tang SS, Cheng J, Cai MY, et al. Association of lincRNA‐p21 haplotype with coronary artery disease in a Chinese Han population. Dis Markers. 2016;2016:9109743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tang SS, Zheng BY, Xiong XD. LincRNA‐p21: implications in human diseases. Int J Mol Sci. 2015;16(8):18732‐18740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Marin‐Bejar O, Marchese FP, Athie A, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li Z, Shen J, Chan MT, et al. TUG1: a pivotal oncogenic long non‐coding RNA of human cancers. Cell Prolif. 2016;49(4):471‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667‐11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lin YH, Wu MH, Huang YH, et al. Taurine up‐regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology. 2018;67(1):188‐203. [DOI] [PubMed] [Google Scholar]
- 101. Barry G, Guennewig B, Fung S, et al. Long non‐coding RNA expression during aging in the human subependymal zone. Front Neurol. 2015;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bao MH, Szeto V, Yang BB, et al. Long non‐coding RNAs in ischemic stroke. Cell Death Dis. 2018;9(3):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wu P, Zuo X, Deng H, et al. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69‐80. [DOI] [PubMed] [Google Scholar]
- 104. Chen J, Jia YS, Liu GZ, et al. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulposus cells via regulating Wnt/beta‐catenin signaling pathway. Biochem Biophys Res Commun. 2017;491(3):668‐674. [DOI] [PubMed] [Google Scholar]
- 105. Li G, Song H, Chen L, et al. TUG1 promotes lens epithelial cell apoptosis by regulating miR‐421/caspase‐3 axis in age‐related cataract. Exp Cell Res. 2017;356(1):20‐27. [DOI] [PubMed] [Google Scholar]
- 106. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501‐512. [DOI] [PubMed] [Google Scholar]
- 107. Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up‐regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330‐342. [DOI] [PubMed] [Google Scholar]
- 108. Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54(5):1679‐1689. [DOI] [PubMed] [Google Scholar]
- 109. Du Y, Kong G, You X, et al. Elevation of highly up‐regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down‐regulating p18. J Biol Chem. 2012;287(31):26302‐26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zou Y, Li J, Chen Y, et al. BANCR: a novel oncogenic long non‐coding RNA in human cancers. Oncotarget. 2017;8(55):94997‐95004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yu TY, Kao YW, Lin JJ. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci USA. 2014;111(9):3377‐3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Chen R, Zhang K, Chen H, et al. Telomerase deficiency causes alveolar stem cell senescence‐associated low‐grade inflammation in lungs. J Biol Chem. 2015;290(52):30813‐30829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Saeed H, Qiu W, Li C, et al. Telomerase activity promotes osteoblast differentiation by modulating IGF‐signaling pathway. Biogerontology. 2015;16(6):733‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ghosh A, Saginc G, Leow SC, et al. Telomerase directly regulates NF‐kappaB‐dependent transcription. Nat Cell Biol. 2012;14(12):1270‐1281. [DOI] [PubMed] [Google Scholar]
- 115. Samper E, Flores JM, Blasco MA. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc‐/‐ mice with short telomeres. EMBO Rep. 2001;2(9):800‐807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li S, Crothers J, Haqq CM, et al. Cellular and gene expression responses involved in the rapid growth inhibition of human cancer cells by RNA interference‐mediated depletion of telomerase RNA. J Biol Chem. 2005;280(25):23709‐23717. [DOI] [PubMed] [Google Scholar]
- 117. Luke B, Panza A, Redon S, et al. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat‐containing RNA and promotes telomere elongation in Saccharomyces cerevisiae . Mol Cell. 2008;32(4):465‐477. [DOI] [PubMed] [Google Scholar]
- 118. Misino S, Bonetti D, Luke‐Glaser S, et al. Increased TERRA levels and RNase H sensitivity are conserved hallmarks of post‐senescent survivors in budding yeast. Differentiation. 2018;100:37‐45. [DOI] [PubMed] [Google Scholar]
- 119. Sinha S, Shukla S, Khan S, et al. Telomeric Repeat Containing RNA (TERRA): aging and cancer. CNS Neurol Disord Drug Targets. 2015;14(7):936‐946. [DOI] [PubMed] [Google Scholar]
- 120. Redon S, Reichenbach P, Lingner J. The non‐coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38(17):5797‐5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Deng Z, Campbell AE, Lieberman PM. TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle. 2010;9(1):69‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cusanelli E, Chartrand P. Telomeric noncoding RNA: telomeric repeat‐containing RNA in telomere biology. Wiley Interdiscip Rev RNA. 2014;5(3):407‐419. [DOI] [PubMed] [Google Scholar]
- 123. Deng Z, Wang Z, Xiang C, et al. Formation of telomeric repeat‐containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J Cell Sci. 2012;125(Pt 18):4383‐4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ng LJ, Cropley JE, Pickett HA, et al. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37(4):1152‐1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Maicher A, Kastner L, Dees M, et al. Deregulated telomere transcription causes replication‐dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012;40(14):6649‐6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Balk B, Maicher A, Dees M, et al. Telomeric RNA‐DNA hybrids affect telomere‐length dynamics and senescence. Nat Struct Mol Biol. 2013;20(10):1199‐1205. [DOI] [PubMed] [Google Scholar]
- 127. Cusanelli E, Romero CA, Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell. 2013;51(6):780‐791. [DOI] [PubMed] [Google Scholar]
- 128. Thijssen PE, Tobi EW, Balog J, et al. Chromatin remodeling of human subtelomeres and TERRA promoters upon cellular senescence: commonalities and differences between chromosomes. Epigenetics. 2013;8(5):512‐521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Penny GD, Kay GF, Sheardown SA, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131‐137. [DOI] [PubMed] [Google Scholar]
- 130. Fukuda A, Tomikawa J, Miura T, et al. The role of maternal‐specific H3K9me3 modification in establishing imprinted X‐chromosome inactivation and embryogenesis in mice. Nat Commun. 2014;5:5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Abdelmohsen K, Panda A, Kang MJ, et al. Senescence‐associated lncRNAs: senescence‐associated long noncoding RNAs. Aging Cell. 2013;12(5):890‐900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Du M, Zhou W, Beatty LG, et al. The KCNQ1OT1 promoter, a key regulator of genomic imprinting in human chromosome 11p15.5. Genomics. 2004;84(2):288‐300. [DOI] [PubMed] [Google Scholar]
- 133. Kanduri C. Functional insights into long antisense noncoding RNA Kcnq1ot1 mediated bidirectional silencing. RNA Biol. 2008;5(4):208‐211. [DOI] [PubMed] [Google Scholar]
- 134. Mohammad F, Mondal T, Guseva N, et al. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137(15):2493‐2499. [DOI] [PubMed] [Google Scholar]
- 135. Travers ME, Mackay DJ, Dekker Nitert M, et al. Insights into the molecular mechanism for type 2 diabetes susceptibility at the KCNQ1 locus from temporal changes in imprinting status in human islets. Diabetes. 2013;62(3):987‐992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wan J, Huang M, Zhao H, et al. A novel tetranucleotide repeat polymorphism within KCNQ1OT1 confers risk for hepatocellular carcinoma. DNA Cell Biol. 2013;32(11):628‐634. [DOI] [PubMed] [Google Scholar]
- 137. Arslan S, Berkan O, Lalem T, et al. Long non‐coding RNAs in the atherosclerotic plaque. Atherosclerosis. 2017;266:176‐181. [DOI] [PubMed] [Google Scholar]
- 138. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115(7):668‐677. [DOI] [PubMed] [Google Scholar]
- 139. Beckedorff FC, Ayupe AC, Crocci‐Souza R, et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS Genet. 2013;9(8):e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Damaschke NA, Yang B, Bhusari S, et al. Epigenetic susceptibility factors for prostate cancer with aging. Prostate. 2013;73(16):1721‐1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Nishida N, Nagasaka T, Nishimura T, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47(3):908‐918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Strunnikova M, Schagdarsurengin U, Kehlen A, et al. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol. 2005;25(10):3923‐3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Nagano T, Mitchell JA, Sanz LA, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717‐1720. [DOI] [PubMed] [Google Scholar]
- 144. Santoro F, Mayer D, Klement RM, et al. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development. 2013;140(6):1184‐1195. [DOI] [PubMed] [Google Scholar]
- 145. Xu W, Zhuang ZX, Yang JP, et al. [The profile of IGF2R gene expression and H3 histone modifications in replicative cell senescence]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2014;45(1):6‐9. [PubMed] [Google Scholar]
- 146. Abdul Rahman A, Abdul Karim N, Abdul Hamid NA, et al. Senescence‐related changes in gene expression of peripheral blood mononuclear cells from octo/nonagenarians compared to their offspring. Oxid Med Cell Longev. 2013;2013:189129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Di Ruscio A, Ebralidze AK, Benoukraf T, et al. DNMT1‐interacting RNAs block gene‐specific DNA methylation. Nature. 2013;503(7476):371‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang H, Iakova P, Wilde M, et al. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8(4):817‐828. [DOI] [PubMed] [Google Scholar]
- 149. Hong IH, Lewis K, Iakova P, et al. Age‐associated change of C/EBP family proteins causes severe liver injury and acceleration of liver proliferation after CCl4 treatments. J Biol Chem. 2014;289(2):1106‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Karagiannides I, Tchkonia T, Dobson DE, et al. Altered expression of C/EBP family members results in decreased adipogenesis with aging. Am J Physiol Regul Integr Comp Physiol. 2001;280(6):R1772‐R1780. [DOI] [PubMed] [Google Scholar]
- 151. Huggins CJ, Malik R, Lee S, et al. C/EBPgamma suppresses senescence and inflammatory gene expression by heterodimerizing with C/EBPbeta. Mol Cell Biol. 2013;33(16):3242‐3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Schmitz KM, Mayer C, Postepska A, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24(20):2264‐2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Johnson R, Strehler BL. Loss of genes coding for ribosomal RNA in ageing brain cells. Nature. 1972;240(5381):412‐414. [DOI] [PubMed] [Google Scholar]
- 154. Pietrzak M, Rempala G, Nelson PT, et al. Epigenetic silencing of nucleolar rRNA genes in Alzheimer's disease. PLoS ONE. 2011;6(7):e22585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Machwe A, Orren DK, Bohr VA. Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. FASEB J. 2000;14(12):1715‐1724. [DOI] [PubMed] [Google Scholar]
- 156. Alemany R, Perona JS, Sanchez‐Dominguez JM, et al. G protein‐coupled receptor systems and their lipid environment in health disorders during aging. Biochim Biophys Acta. 2007;1768(4):964‐975. [DOI] [PubMed] [Google Scholar]
- 157. Massone S, Vassallo I, Fiorino G, et al. 17A, a novel non‐coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41(2):308‐317. [DOI] [PubMed] [Google Scholar]
- 158. Nakagawa S, Hirano T. Gathering around Firre. Nat Struct Mol Biol. 2014;21(3):207‐208. [DOI] [PubMed] [Google Scholar]
- 159. Lu Y, Liu X, Xie M, et al. The NF‐kappaB‐responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J Immunol. 2017;199(10):3571‐3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Cui H, Xie N, Tan Z, et al. The human long noncoding RNA lnc‐IL7R regulates the inflammatory response. Eur J Immunol. 2014;44(7):2085‐2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA‐LET by histone deacetylase 3 contributes to hypoxia‐mediated metastasis. Mol Cell. 2013;49(6):1083‐1096. [DOI] [PubMed] [Google Scholar]
- 162. Tominaga‐Yamanaka K, Abdelmohsen K, Martindale JL, et al. NF90 coordinately represses the senescence‐associated secretory phenotype. Aging (Albany NY). 2012;4(10):695‐708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789‐792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hu G, Gong AY, Wang Y, et al. LincRNA‐Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF‐mediated chromatin remodeling. J Immunol. 2016;196(6):2799‐2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Rapicavoli NA, Qu K, Zhang J, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. Elife. 2013;2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Adler AS, Sinha S, Kawahara TL, et al. Motif module map reveals enforcement of aging by continual NF‐kappaB activity. Genes Dev. 2007;21(24):3244‐3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717‐726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non‐coding RNAs. RNA Biol. 2013;10(3):456‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1‐dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393‐406. [DOI] [PubMed] [Google Scholar]
- 170. Chen Y, Qiu J, Chen B, et al. Long non‐coding RNA NEAT1 plays an important role in sepsis‐induced acute kidney injury by targeting miR‐204 and modulating the NF‐kappaB pathway. Int Immunopharmacol. 2018;59:252‐260. [DOI] [PubMed] [Google Scholar]
- 171. Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96‐104. [DOI] [PubMed] [Google Scholar]
- 172. Huang‐Fu N, Cheng JS, Wang Y, et al. Neat1 regulates oxidized low‐density lipoprotein‐induced inflammation and lipid uptake in macrophages via paraspeckle formation. Mol Med Rep. 2018;17(2):3092‐3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Wang Q, Wang W, Zhang F, et al. NEAT1/miR‐181c regulates Osteopontin (OPN)‐mediated synoviocyte proliferation in osteoarthritis. J Cell Biochem. 2017;118(11):3775‐3784. [DOI] [PubMed] [Google Scholar]
- 174. Krawczyk M, Emerson BM. p50‐associated COX‐2 extragenic RNA (PACER) activates COX‐2 gene expression by occluding repressive NF‐kappaB complexes. Elife. 2014;3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Pearson MJ, Philp AM, Heward JA, et al. Long intergenic noncoding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol. 2016;68(4):845‐856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. 2014;111(3):1002‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Somarowthu S, Legiewicz M, Chillon I, et al. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58(2):353‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Zhang K, Sun X, Zhou X, et al. Long non‐coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Ozes AR, Miller DF, Ozes ON, et al. NF‐kappaB‐HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35(41):5350‐5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Yoon JH, Abdelmohsen K, Kim J, et al. Scaffold function of long non‐coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Lauri A, Pompilio G, Capogrossi MC. The mitochondrial genome in aging and senescence. Ageing Res Rev. 2014;18:1‐15. [DOI] [PubMed] [Google Scholar]
- 183. Bianchessi V, Badi I, Bertolotti M, et al. The mitochondrial lncRNA ASncmtRNA‐2 is induced in aging and replicative senescence in endothelial cells. J Mol Cell Cardiol. 2015;81:62‐70. [DOI] [PubMed] [Google Scholar]
- 184. Morrison JH, Baxter MG. The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat Rev Neurosci. 2012;13(4):240‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yamanaka Y, Faghihi MA, Magistri M, et al. Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin‐associated protein, HMGB2. Cell Rep. 2015;11(6):967‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med. 2008;14(7):723‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ciarlo E, Massone S, Penna I, et al. An intronic ncRNA‐dependent regulation of SORL1 expression affecting Abeta formation is upregulated in post‐mortem Alzheimer's disease brain samples. Dis Model Mech. 2013;6(2):424‐433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Carrieri C, Cimatti L, Biagioli M, et al. Long non‐coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454‐457. [DOI] [PubMed] [Google Scholar]
- 189. Pereira Fernandes D, Bitar M, Jacobs FMJ, et al. Long non‐coding RNAs in neuronal aging. Noncoding RNA. 2018;4(2):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci USA. 2007;104(25):10679‐10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Chen Y, Lian YJ, Ma YQ, et al. LncRNA SNHG1 promotes alpha‐synuclein aggregation and toxicity by targeting miR‐15b‐5p to activate SIAH1 in human neuroblastoma SH‐SY5Y cells. Neurotoxicology. 2017;S0161‐813X(17)30235‐8. [DOI] [PubMed] [Google Scholar]
- 192. Lin D, Liang Y, Jing X, et al. Microarray analysis of an synthetic alpha‐synuclein induced cellular model reveals the expression profile of long non‐coding RNA in Parkinson's disease. Brain Res. 2018;1678:384‐396. [DOI] [PubMed] [Google Scholar]
- 193. Schmucker DL. Age‐related changes in liver structure and function: implications for disease ? Exp Gerontol. 2005;40(8–9):650‐659. [DOI] [PubMed] [Google Scholar]
- 194. Wynne HA, Cope LH, Mutch E, et al. The effect of age upon liver volume and apparent liver blood flow in healthy man. Hepatology. 1989;9(2):297‐301. [DOI] [PubMed] [Google Scholar]
- 195. Jung T, Bader N, Grune T. Lipofuscin: formation, distribution, and metabolic consequences. Ann N Y Acad Sci. 2007;1119:97‐111. [DOI] [PubMed] [Google Scholar]
- 196. Aravinthan A, Verma S, Coleman N, et al. Vacuolation in hepatocyte nuclei is a marker of senescence. J Clin Pathol. 2012;65(6):557‐560. [DOI] [PubMed] [Google Scholar]
- 197. Kim IH, Kisseleva T, Brenner DA. Aging and liver disease. Curr Opin Gastroenterol. 2015;31(3):184‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. White RR, Milholland B, MacRae SL, et al. Comprehensive transcriptional landscape of aging mouse liver. BMC Genom. 2015;16:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zhao J, Ohsumi TK, Kung JT, et al. Genome‐wide identification of polycomb‐associated RNAs by RIP‐seq. Mol Cell. 2010;40(6):939‐953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Hagan JP, O'Neill BL, Stewart CL, et al. At least ten genes define the imprinted Dlk1‐Dio3 cluster on mouse chromosome 12qF1. PLoS ONE. 2009;4(2):e4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Picca A, Calvani R, Bossola M, et al. Update on mitochondria and muscle aging: all wrong roads lead to sarcopenia. Biol Chem. 2018;399(5):421‐436. [DOI] [PubMed] [Google Scholar]
- 202. Legnini I, Morlando M, Mangiavacchi A, et al. A feedforward regulatory loop between HuR and the long noncoding RNA linc‐MD1 controls early phases of myogenesis. Mol Cell. 2014;53(3):506‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Watts R, Johnsen VL, Shearer J, et al. Myostatin‐induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol. 2013;304(10):C995‐C1001. [DOI] [PubMed] [Google Scholar]
- 205. Thomas M, Langley B, Berry C, et al. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J Biol Chem. 2000;275(51):40235‐40243. [DOI] [PubMed] [Google Scholar]
- 206. Wang Y, Pang WJ, Wei N, et al. Identification, stability and expression of Sirt1 antisense long non‐coding RNA. Gene. 2014;539(1):117‐124. [DOI] [PubMed] [Google Scholar]
- 207. Pardo PS, Boriek AM. The physiological roles of Sirt1 in skeletal muscle. Aging (Albany NY). 2011;3(4):430‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Zhang ZK, Li J, Guan D, et al. A newly identified lncRNA MAR1 acts as a miR‐487b sponge to promote skeletal muscle differentiation and regeneration. J Cachexia Sarcopenia Muscle. 2018;9(3):613‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Lu L, Sun K, Chen X, et al. Genome‐wide survey by ChIP‐seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013;32(19):2575‐2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Zhou Y, Cheunsuchon P, Nakayama Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137(16):2643‐2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Costantino S, Camici GG, Mohammed SA, et al. Epigenetics and cardiovascular regenerative medicine in the elderly. Int J Cardiol. 2018;250:207‐214. [DOI] [PubMed] [Google Scholar]
- 212. Li Z, Li J, Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen‐glucose deprivation/reoxygenation‐induced injury by sponging miR‐26b and upregulating ULK2 expression. Neuroscience. 2017;354:1‐10. [DOI] [PubMed] [Google Scholar]
- 213. Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389‐1397. [DOI] [PubMed] [Google Scholar]
- 214. Puthanveetil P, Chen S, Feng B, et al. Long non‐coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19(6):1418‐1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Tang Y, Jin X, Xiang Y, et al. The lncRNA MALAT1 protects the endothelium against ox‐LDL‐induced dysfunction via upregulating the expression of the miR‐22‐3p target genes CXCR2 and AKT. FEBS Lett. 2015;589(20 Pt B):3189‐3196. [DOI] [PubMed] [Google Scholar]
- 216. Xin JW, Jiang YG. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen‐glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp Ther Med. 2017;13(4):1225‐1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Boon RA, Hofmann P, Michalik KM, et al. Long noncoding RNA Meg3 controls endothelial cell aging and function: implications for regenerative angiogenesis. J Am Coll Cardiol. 2016;68(23):2589‐2591. [DOI] [PubMed] [Google Scholar]
- 218. He C, Yang W, Yang J, et al. Long noncoding RNA MEG3 negatively regulates proliferation and angiogenesis in vascular endothelial cells. DNA Cell Biol. 2017;36(6):475‐481. [DOI] [PubMed] [Google Scholar]
- 219. Holdt LM, Stahringer A, Sass K, et al. Circular non‐coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wan G, Mathur R, Hu X, et al. Long non‐coding RNA ANRIL (CDKN2B‐AS) is induced by the ATM‐E2F1 signaling pathway. Cell Signal. 2013;25(5):1086‐1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Congrains A, Kamide K, Katsuya T, et al. CVD‐associated non‐coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun. 2012;419(4):612‐616. [DOI] [PubMed] [Google Scholar]
- 222. Holdt LM, Beutner F, Scholz M, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30(3):620‐627. [DOI] [PubMed] [Google Scholar]
- 223. Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29(10):1671‐1677. [DOI] [PubMed] [Google Scholar]
- 224. Yan B, Yao J, Liu JY, et al. lncRNA‐MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143‐1156. [DOI] [PubMed] [Google Scholar]
- 225. Peng Y, Meng K, Jiang L, et al. Thymic stromal lymphopoietin‐induced HOTAIR activation promotes endothelial cell proliferation and migration in atherosclerosis. Biosci Rep. 2017;37(4):BSR20170351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Chen C, Cheng G, Yang X, et al. Tanshinol suppresses endothelial cells apoptosis in mice with atherosclerosis via lncRNA TUG1 up‐regulating the expression of miR‐26a. Am J Transl Res. 2016;8(7):2981‐2991. [PMC free article] [PubMed] [Google Scholar]
- 227. Chen R, Kong P, Zhang F, et al. EZH2‐mediated alpha‐actin methylation needs lncRNA TUG1, and promotes the cortex cytoskeleton formation in VSMCs. Gene. 2017;616:52‐57. [DOI] [PubMed] [Google Scholar]
- 228. He C, Ding JW, Li S, et al. The role of long intergenic noncoding RNA p21 in vascular endothelial cells. DNA Cell Biol. 2015;34(11):677‐683. [DOI] [PubMed] [Google Scholar]
- 229. Chen L, Yang W, Guo Y, et al. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE. 2017;12(9):e0185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Ito I, Asai A, Suzuki S, et al. M2b macrophage polarization accompanied with reduction of long noncoding RNA GAS5. Biochem Biophys Res Commun. 2017;493(1):170‐175. [DOI] [PubMed] [Google Scholar]
- 231. Wang YN, Shan K, Yao MD, et al. Long noncoding RNA‐GAS5: a novel regulator of hypertension‐induced vascular remodeling. Hypertension. 2016;68(3):736‐748. [DOI] [PubMed] [Google Scholar]
- 232. Huang C, Hu YW, Zhao JJ, et al. Long noncoding RNA HOXC‐AS1 suppresses Ox‐LDL‐induced cholesterol accumulation through promoting HOXC6 expression in THP‐1 macrophages. DNA Cell Biol. 2016;35(11):722‐729. [DOI] [PubMed] [Google Scholar]
- 233. Zhang DD, Wang WT, Xiong J, et al. Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR‐NF‐kappaB pathway in human monocytes. Sci Rep. 2017;7:46204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Cai Y, Yang Y, Chen X, et al. Circulating ‘lncRNA OTTHUMT00000387022’ from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res. 2016;112(3):714‐724. [DOI] [PubMed] [Google Scholar]
- 235. Liang WC, Fu WM, Wang YB, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Xiao X, Zhou T, Guo S, et al. LncRNA MALAT1 sponges miR‐204 to promote osteoblast differentiation of human aortic valve interstitial cells through up‐regulating Smad4. Int J Cardiol. 2017;243:404‐412. [DOI] [PubMed] [Google Scholar]
- 237. Wei B, Wei W, Zhao B, et al. Long non‐coding RNA HOTAIR inhibits miR‐17‐5p to regulate osteogenic differentiation and proliferation in non‐traumatic osteonecrosis of femoral head. PLoS ONE. 2017;12(2):e0169097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Jia Q, Jiang W, Ni L. Down‐regulated non‐coding RNA (lncRNA‐ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. 2015;60(2):234‐241. [DOI] [PubMed] [Google Scholar]
- 239. Li Z, Jin C, Chen S, et al. Long non‐coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose‐derived mesenchymal stem cells via miR‐140‐5p. Mol Cell Biochem. 2017;433(1–2):51‐60. [DOI] [PubMed] [Google Scholar]
- 240. Wang Q, Li Y, Zhang Y, et al. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR‐133a‐3p. Biomed Pharmacother. 2017;89:1178‐1186. [DOI] [PubMed] [Google Scholar]
- 241. Jin C, Zheng Y, Huang Y, et al. Long non‐coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose‐derived stem cells. Cell Biol Int. 2017;41(1):33‐41. [DOI] [PubMed] [Google Scholar]
- 242. Jin C, Jia L, Huang Y, et al. Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose‐derived stem cells. Stem Cells. 2016;34(11):2707‐2720. [DOI] [PubMed] [Google Scholar]
- 243. Tong X, Gu PC, Xu SZ, et al. Long non‐coding RNA‐DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem. 2015;79(5):732‐737. [DOI] [PubMed] [Google Scholar]
- 244. Clark MB, Johnston RL, Inostroza‐Ponta M, et al. Genome‐wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885‐898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Fei Q, Bai X, Lin J, et al. Identification of aberrantly expressed long non‐coding RNAs in postmenopausal osteoporosis. Int J Mol Med. 2018;41(6):3537‐3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Hao L, Fu J, Tian Y, et al. Systematic analysis of lncRNAs, miRNAs and mRNAs for the identification of biomarkers for osteoporosis in the mandible of ovariectomized mice. Int J Mol Med. 2017;40(3):689‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Liu Y, Ferguson JF, Xue C, et al. Tissue‐specific RNA‐Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler Thromb Vasc Biol. 2014;34(4):902‐912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Sun L, Goff LA, Trapnell C, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA. 2013;110(9):3387‐3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. De Tata V. Age‐related impairment of pancreatic Beta‐cell function: pathophysiological and cellular mechanisms. Front Endocrinol (Lausanne). 2014;5:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Ammon HP, Fahmy A, Mark M, et al. The effect of glucose on insulin release and ion movements in isolated pancreatic islets of rats in old age. J Physiol. 1987;384:347‐354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Akerman I, Tu Z, Beucher A, et al. Human pancreatic beta cell lncRNAs control cell‐specific regulatory networks. Cell Metab. 2017;25(2):400‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Arnes L, Akerman I, Balderes DA, et al. betalinc1 encodes a long noncoding RNA that regulates islet beta‐cell formation and function. Genes Dev. 2016;30(5):502‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Fadista J, Vikman P, Laakso EO, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci USA. 2014;111(38):13924‐13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Moran I, Akerman I, van de Bunt M, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue‐specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Castelo‐Branco C, Soveral I. The immune system and aging: a review. Gynecol Endocrinol. 2014;30(1):16‐22. [DOI] [PubMed] [Google Scholar]
- 256. Zhang F, Liu G, Wei C, et al. Linc‐MAF‐4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J. 2017;31(2):519‐525. [DOI] [PubMed] [Google Scholar]
- 257. Huang W, Thomas B, Flynn RA, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528(7583):517‐522. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 258. Ramos AD, Diaz A, Nellore A, et al. Integration of genome‐wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12(5):616‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Ramos AD, Andersen RE, Liu SJ, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16(4):439‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Ng SY, Bogu GK, Soh BS, et al. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51(3):349‐359. [DOI] [PubMed] [Google Scholar]
- 261. Klapper W, Shin T, Mattson MP. Differential regulation of telomerase activity and TERT expression during brain development in mice. J Neurosci Res. 2001;64(3):252‐260. [DOI] [PubMed] [Google Scholar]
- 262. Wang H, Iacoangeli A, Popp S, et al. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22(23):10232‐10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Lin D, Pestova TV, Hellen CU, et al. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28(9):3008‐3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Kondrashov AV, Kiefmann M, Ebnet K, et al. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)‐binding protein (PABP). J Mol Biol. 2005;353(1):88‐103. [DOI] [PubMed] [Google Scholar]
- 265. Lewejohann L, Skryabin BV, Sachser N, et al. Role of a neuronal small non‐messenger RNA: behavioural alterations in BC1 RNA‐deleted mice. Behav Brain Res. 2004;154(1):273‐289. [DOI] [PubMed] [Google Scholar]
- 266. Modarresi F, Faghihi MA, Lopez‐Toledano MA, et al. Inhibition of natural antisense transcripts in vivo results in gene‐specific transcriptional upregulation. Nat Biotechnol. 2012;30(5):453‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Tsuiji H, Yoshimoto R, Hasegawa Y, et al. Competition between a noncoding exon and introns: GOMAFU contains tandem UACUAAC repeats and associates with splicing factor‐1. Genes Cells. 2011;16(5):479‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Barry G, Briggs JA, Hwang DW, et al. The long non‐coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci Rep. 2017;7:40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Barrachina M, Dalfo E, Puig B, et al. Amyloid‐beta deposition in the cerebral cortex in Dementia with Lewy bodies is accompanied by a relative increase in AbetaPP mRNA isoforms containing the Kunitz protease inhibitor. Neurochem Int. 2005;46(3):253‐260. [DOI] [PubMed] [Google Scholar]
- 270. Choi J, Levey AI, Weintraub ST, et al. Oxidative modifications and down‐regulation of ubiquitin carboxyl‐terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279(13):13256‐13264. [DOI] [PubMed] [Google Scholar]
- 271. Holtzman DM, Herz J, Bu G. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(3):a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Pan JX. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF‐kB signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(2):322‐328. [PubMed] [Google Scholar]
- 273. Lv J, Wang L, Zhang J, et al. Long noncoding RNA H19‐derived miR‐675 aggravates restenosis by targeting PTEN. Biochem Biophys Res Commun. 2018;497(4):1154‐1161. [DOI] [PubMed] [Google Scholar]
- 274. Voellenkle C, Garcia‐Manteiga JM, Pedrotti S, et al. Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA‐sequencing. Sci Rep. 2016;6:24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Hu YW, Zhao JY, Li SF, et al. RP5‐833A20.1/miR‐382‐5p/NFIA‐dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. 2015;35(1):87‐101. [DOI] [PubMed] [Google Scholar]


