Abstract
Obesity is associated with adipose inflammation, defined by macrophages encircling dead adipocytes, as well as extracellular matrix (ECM) remodeling and increased risk of breast cancer. Whether ECM affects macrophage phenotype in obesity is uncertain. A better understanding of this relationship could be strategically important to reduce cancer risk or improve outcomes in the obese. Using clinical samples, computational approaches, and in vitro decellularized ECM models, this study quantified the relative abundance of pro-inflammatory (M1) and anti-inflammatory (M2) macrophages in human breast adipose tissue, determined molecular similarities between obesity and tumor-associated macrophages, and assessed the regulatory effect of obese versus lean ECM on macrophage phenotype. Our results suggest that breast adipose tissue contains more M2- than M1-biased macrophages across all body mass index categories. Obesity further increased M2-biased macrophages but did not affect M1-biased macrophage density. Gene Set Enrichment Analysis suggested that breast tissue macrophages from obese versus lean women are more similar to tumor-associated macrophages. These changes positively correlated with adipose tissue interstitial fibrosis, and in vitro experiments indicated that obese ECM directly stimulates M2-biased macrophage functions. However, mammographic density cannot be used as a clinical indicator of these changes. Collectively, these data suggest that obesity-associated interstitial fibrosis promotes a macrophage phenotype similar to tumor-associated macrophages, which may contribute to the link between obesity and breast cancer.
Excess body weight has been associated with both an increased risk of cancer and worse prognosis for some tumor types.1 In particular, obesity has been linked with the development of both hormone-receptor positive, postmenopausal,2, 3, 4 and triple-negative breast cancer.5 Furthermore, obesity contributes to worse clinical outcome in both premenopausal and postmenopausal breast cancer patients.6, 7 Historically, the link between obesity and breast cancer has been attributed to metabolic dysregulation, altered secretion of adipokines, elevated levels of estrogen, and inflammation.8, 9, 10, 11 Yet, experimental evidence suggests that obesity-dependent interstitial fibrosis of breast adipose tissue may also be important.12
Fibrosis is a hallmark of obese white adipose tissue that is characterized by excess amounts of interstitial extracellular matrix (ECM) of varying composition, structure, and mechanical properties.12, 13, 14 Given that myofibroblasts are considered key players in mediating fibrosis, it is not surprising that these cells are also increased in white adipose tissue. More specifically, adipose stromal cells (ASCs) isolated from the adipose tissue interstitium of obese versus lean mice are enriched in myofibroblasts that lay down abundant ECM. The resulting ECM contains increased amounts of aligned collagen type I and fibronectin (Fn) fibers and is mechanically stiffer, ultimately promoting malignant tumor cell behavior due to increased mechanosignaling.12 This finding is directly relevant to humans, as demonstrated by analysis of human breast adipose tissue and cancer samples12 and because ECM remodeling genes are highly enriched in the transcriptomic signature of s.c. white adipose tissue in obese individuals.15 Although the pathophysiology underlying obesity-associated interstitial fibrosis is complex, it is commonly linked to white adipose tissue inflammation (WATi), another hallmark of obesity.15, 16, 17
WATi has been identified in breast tissue from the obese and is associated with both an increased risk of breast cancer and shorter relapse-free survival in women with recurrent metastatic breast cancer.18, 19, 20 Histologically, WATi appears as a grouping of macrophages surrounding dead or dying adipocytes, termed crown-like structures (CLSs). CLSs have been described in s.c.,16 visceral,21 and breast18 adipose tissue and are considered to consist of pro-inflammatory (M1) macrophages as their presence correlates with increased circulating levels of pro-inflammatory cytokines and positive acute-phase proteins.22, 23 Nevertheless, recent experimental evidence in mice and humans indicates that anti-inflammatory (M2) macrophages may also be increased with obesity.24, 25, 26 Whether similar findings apply to human breast adipose tissue remains unclear. Gaining an improved understanding of how obesity modulates M1 and M2 macrophage density and function in the human breast is critical because M2 macrophages share similarities with tumor-associated macrophages (TAMs)27 and, thus, could play an important role in tumorigenesis28 by suppressing antitumor immune responses,29 promoting angiogenesis,30 and assisting with cancer cell intravasation and metastasis.31 We recognize that macrophages are phenotypically diverse and plastic and that their phenotypes overlap on both an individual32 and cell population33, 34 level. Nevertheless, the simplified classification of macrophages as M1 or M2 biased that is used herein provides a framework to gain valuable new insights.
Although most previous work studying the functional link between inflammation and the ECM has focused on how macrophages mediate fibrotic ECM remodeling,35, 36, 37 the opposite may be equally relevant (ie, how the ECM influences macrophages). Indeed, TAMs often reside along the tumor-stroma border, an area that is characterized by increased fibrotic ECM remodeling.38, 39 Although TAMs have been shown to enhance collagenous deposition within tumors,40 experimental evidence suggests that the converse, tumor-associated ECM remodeling directly modulating macrophage functions,41 may also be true. Within adipose tissue, M2 macrophages also interact with ECM as they exist within the interstitial space between adipocytes24 in which fibrotic remodeling is abundant in obesity. Nevertheless, it remains unknown whether obesity-associated differences in interstitial ECM remodeling modulate macrophage function toward an anti-inflammatory and possibly protumorigenic phenotype. Yet, this possibility is conceivable as macrophages can transition to an M2 phenotype when interacting with substrates mimicking biochemical and biophysical changes representative of fibrotic ECM remodeling.42, 43, 44
Herein, we investigated the overall hypothesis that obesity-associated changes to adipose tissue interstitial ECM promote an anti-inflammatory M2-biased macrophage phenotype similar to TAMs. To test this hypothesis, the spatial distribution of pro-inflammatory and anti-inflammatory macrophages in tumor-free regions of breast adipose tissue was evaluated via immunohistochemistry (IHC). Macrophages were compared in both lean and obese breast adipose tissue using gene expression profiling, and the detected signatures were related to those of tumor-associated macrophages. Subsequently, the influence of obesity-associated ECM remodeling on macrophage phenotype was explored with an in vitro model of lean and obese ECM. Finally, it was determined whether obese ECM-mediated regulation of macrophage function could impact endothelial cell behavior, a hallmark of protumorigenic macrophages.
Materials and Methods
Clinical Data, Biospecimen Collection, and Breast Tissue Assessment
Clinical data and archived formalin-fixed, paraffin-embedded samples were obtained from a cohort of women who underwent mastectomy for breast cancer risk reduction or treatment between January 2011 and August 2013 at Memorial Sloan Kettering Cancer Center (New York, NY). Due to known racial and ethnic disparities in breast cancer prognosis and survival, race and ethnicity was queried on the Memorial Sloan Kettering oncology intake questionnaire and self-reported. For race, patients could choose from the following options: American Indian, Asian, Native Hawaiian, Black or African American, White, or unknown. For ethnicity, patients could choose from the following-options: Hispanic or Latino, Not Hispanic or Latino, or unknown. In this cohort, non–tumor-containing breast white adipose tissue was prospectively collected at the time of surgery. CLS presence in breast adipose tissue had been previously classified in this cohort.19 Samples were randomly selected on the basis of a power analysis estimating 80% power to detect a correlation of 0.4 using a two-sided hypothesis test with a significance level of 0.05. Macrophage polarization states in breast white adipose tissue were evaluated via IHC on archived formalin-fixed, paraffin-embedded breast tissue. Sections (5 μm thick) were probed with rabbit polyclonal anti-mannose receptor (CD206; Abcam, Cambridge, MA; ab64693) or rabbit monoclonal anti-CD11c (Abcam; ab52623). The signal was amplified using Vectastain ABC universal kit (Vector Laboratories, Burlingame, CA) and detected with peroxidase substrate and 3,3'-diaminobenzadine chromogen (ThermoFisher, Waltham, MA). Tissue sections were also stained with hematoxylin and eosin, picrosirius red, and IHC for CD31 (Abcam; ab28364). Samples with repeatable severe sectioning artifact were excluded from analysis. Stained slides were digitally archived with an Aperio CS2 microscope (Leica Biosystems, Buffalo Grove, IL) and were analyzed with the included ImageScope software positive pixel count algorithm version 9 (Leica Biosystems) (Supplemental Figure S1). Picrosirius red–stained sections were imaged under crossed polarized light45 with a Nikon (Melville, NY) TE2000-S microscope and an RTKE (Spot Imaging Solutions, Inc., Sterling Heights, MI) color camera, and images were analyzed with ImageJ software version 1.48 (NIH, Bethesda, MD; https://imagej.nih.gov/ij) using an algorithm for positive pixel counts. Mammographic data were retrieved from the patient medical record, when available (n = 26), and anonymized before use. Mammographic density pattern was determined by a radiologist (M.S.J.).
Breast Tissue Transcriptomic and Computational Analysis
At the time of the collection, fresh tissue specimens were snap frozen and stored in RNAlater (Ambion, Foster City, CA), and total RNA was extracted for RNA sequencing using the RNeasy Mini Kit (Qiagen, Valencia, CA). Polyadenylated RNA sequencing was performed using the standard Illumina (San Diego, CA) Truseq kits, and samples were sequenced using the HiSeq2000 platform. All reads were independently aligned with STAR_2.4.0f1 (http://code.google.com/p/rna-star)46 for sequence alignment against the human genome build hg19 and downloaded via the University of California, Santa Cruz, genome browser, and SAMTOOLS version 0.1.19 (http://samtools.sourceforge.net)47was used for sorting and indexing reads. Cufflinks version 2.0.2 (http://cufflinks.cbcb.umd.edu)48 was used to get the expression values (fragments per kilobase per million), and Gencode version 19 (https://www.gencodegenes.org)49 gene transfer format (GTF) file was used for annotation. The gene counts from htseq-count50 and DESeq2 Bioconductor package51 were used to identify differentially expressed genes. The hypergeometric test and Gene Set Enrichment Analysis52 were used to identify enriched signatures using the different pathway collection in the MSigDB database.53 Gene name–based enrichment analysis was also performed using the webtool ENRICHR.54, 55
The resultant transcripts were analyzed with the LM22 CIBERSORT signature (http://cibersort.stanford.edu) 56 to estimate macrophage polarization subsets, on the basis of a signature matrix from 547 genes that effectively differentiate 22 distinct human hematopoietic cell populations, in lean versus obese breast tissue. RNA transcripts were additionally evaluated using Gene Set Enrichment Analysis52 to determine whether gene expression from our data set is concordant with a previously published TAM gene signature.57
Animal Use
The Cornell University and Kansas State University Institutional Animal Care and Use Committee approved mouse use for both ASC and bone marrow–derived macrophage (BMDM) isolation under protocol numbers 2009-0117 and 4094, respectively.
Preparation of the Decellularized ECM Model System
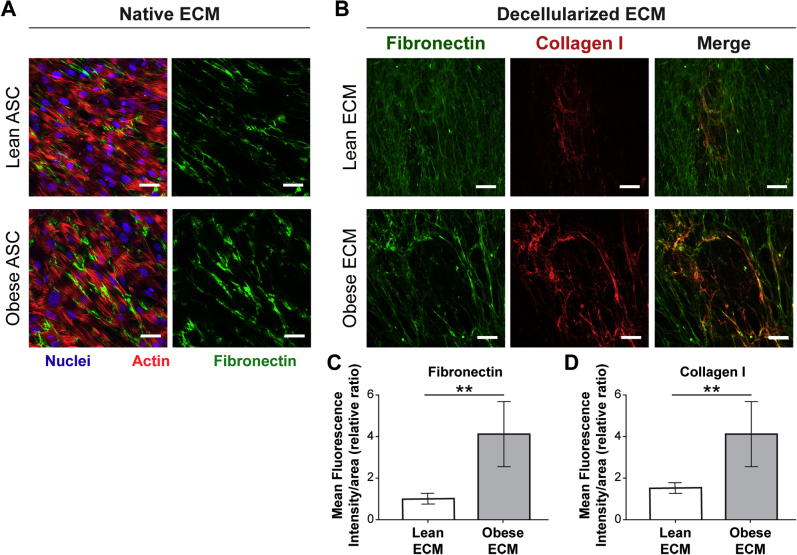
ASCs were prepared using an aseptic technique from the inguinal fat pad of age-matched 10-week–old female wild-type C57Bl6/J mice and genetically obese B6.Cg-Lepop/J (ob/ob) mice (both strains from The Jackson Laboratory, Bar Harbor, ME) after CO2 euthanasia. The fat pad was maintained under sterile conditions, minced, and digested in collagenase at 37°C for 1 hour. After digestion, the stromal-vascular fraction of fat was separated from adipocytes by density centrifugation and subsequent filtration and then expanded in 1:1 Dulbecco's modified Eagle's medium (DMEM) and Ham's F12 Nutrient Mixture (both obtained from Gibco, Waltham, MA) enriched with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA) and 1% penicillin/streptomycin (Gibco) to select for ASCs. Expanded ASCs were cultured on plastic coverslips (Nunc Thermanox, Rochester, NY) for 10 days before detergent-based decellularization. Decellularization was achieved as previously described.12, 58 Briefly, ASC cultures were washed with phosphate-buffered saline and subsequently incubated in 0.5% Triton-X + 200 mmol/L NH4OH,12, 59, 60 followed by incubation in 1 U/mL DNase (VWR Amresco, Radnor, PA) at 37°C to remove cellular membranes, organelles, and nucleic acids. After decellularization, maintenance of native matrix structure and key compositional components was confirmed by immunofluorescence analysis of Fn (Millipore-Sigma, St. Louis, MO; F7387) and collagen type I (Abcam; ab34710) via confocal microscopy (Zeiss, Pleasanton, CA; 710) and image analysis.
In Vitro Macrophage Experiments
BMDM Isolation
Bone marrow was isolated from the femurs and tibiae of 10- to 16-week–old wild-type C57Bl6/J mice of mixed sex. Briefly, after bone extraction, remaining musculature was stripped and the bones were cut via sterile scissors at the proximal and distal physis. Using a syringe and 25-gauge needle, the bone marrow was flushed from the metaphyseal regions and diaphysis with DMEM cell culture media into a sterile tissue culture plate. Bone marrow was suspended and plated in 9-cm dishes and cultured at 37°C. Progenitor cells were differentiated into BMDM in DMEM + 10% FBS + 1% penicillin/streptomycin + 10 ng/mL recombinant mouse macrophage colony stimulating factor (CSF; R&D Systems, Minneapolis, MN) and were used for experiments between days 7 and 10 after collection.
Assessment of Macrophage Morphology and Activation
BMDMs were seeded on lean and obese ECM at a concentration of 10,000 cells/coverslip and cultured in DMEM + 10% FBS + 1% penicillin/streptomycin and 10 ng/mL recombinant mouse macrophage CSF. After 72 hours of culture, BMDMs and ECM were fixed with 4% paraformaldehyde. Cells were labeled with DAPI and phalloidin (AlexaFluor; Molecular Probes, Waltham, MA), whereas decellularized ECM was visualized by immunostaining with rabbit anti-Fn antibody (Millipore-Sigma; F3648). Macrophage polarization state was evaluated by quantifying the murine M1 marker inducible nitric oxide synthase (Abcam; ab3523) and M2 marker arginase-134 protein levels (Santa Cruz Biotechnology, Dallas, TX; sc-18351) via immunofluorescence image analysis. Macrophage proliferation was assessed via bromodeoxyuridine (Invitrogen, Waltham, MA) incorporation and immunostaining with antibromodeoxyuridine antibody (Invitrogen) after 10 hours of culture. For select experiments, BMDMs were stimulated to undergo M1 polarization with 100 ng/mL lipopolysaccharide (Millipore-Sigma) or M2 polarization with 20 ng/mL IL-4 and IL-13 (R&D Systems), as indicated in figure legends.
Assessment of Macrophage-Secreted Factors on Endothelial Cell Behavior
A transwell assay was used to assess the effect of macrophage-secreted factors on endothelial cell migration. BMDMs were seeded onto lean and obese ECM, as described above, at a density of 50,000 cells/coverslip in the presence of 10% serum and 10 ng/mL recombinant mouse macrophage CSF. After overnight adhesion, BMDM coverslips were transferred to a new 24-well plate and medium was changed to DMEM + 5% FBS + 20 ng/mL IL-4 and 20 ng/mL IL-13. An 8.0-μm pore polycarbonate membrane tissue culture insert (Corning Falcon, Corning, NY) that had been previously coated with 70 μg/mL rat-tail collagen type I was placed over each coverslip. After 6 hours of culture time, human umbilical vein endothelial cells (HUVECs; passages 2 to 3; Lonza, Allendale, NJ) cultured in M199 medium (BioWhittaker, Walkersville, MD) + 20% FBS + 1% penicillin/streptomycin + 30 μg/mL endothelial cell growth supplement (Millipore-Sigma), 2 mmol/L GlutaMAX (Gibco), and 2500 U heparin salt (Millipore-Sigma) were seeded on top of the transwell insert at 50,000 cells/insert. After a subsequent 12 hours of culture at 37°C and 5% CO2, the transwell inserts were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and the distal side of the membrane was stained with DAPI. Nuclei of cells that had migrated through the membrane were manually counted (ImageJ) on four 10× objective fluorescent microscopy images (12-, 3-, 6-, and 9-o'clock positions) per membrane with three membranes per condition per experiment.
Endothelial cell tubulogenesis was assessed via a Matrigel (Corning Falcon) tube formation assay. BMDMs were cultured on lean or obese ECM in medium containing 5% serum and no macrophage CSF. After 24 hours of culture, conditioned medium was collected, concentrated 10× in a 3-kDa molecular weight centrifugation filter unit (Millipore-Sigma), and diluted back to a 1× concentration using EBM2 (Lonza, Basel, Switzerland) without growth factors. Using the reconstituted, conditioned media, HUVECs were seeded onto growth factor–reduced, Matrigel-coated, 96-well plates at a density of 10,000 cells/well and cultured for 12 hours. Subsequently, the medium was carefully removed from the wells and HUVECs were incubated in Calcein AM (Invitrogen) to stain viable cells. After 15 minutes of incubation at 37°C, the wells were rinsed with phosphate-buffered saline. A single 2× objective representative fluorescent image (covering the entire monolayer of each well) was captured and analyzed for HUVEC tube branch points via manual counting in ImageJ. Three wells were analyzed per condition per experiment.
Statistical Analysis
All statistical analysis was performed using Prism version 7.0 (GraphPad Software, La Jolla, CA) and R version 3.4.3. Differences in patient characteristics across body mass index (BMI) categories were examined using analysis of variance for continuous variables and Fisher exact test for categorical variables. Normality assumption of the parametric method was assessed by the Shapiro-Wilk test. Continuous data are represented as the means ± SD or the median and interquartile range. Categorical data were summarized in terms of counts and percentages. Correlations between two continuous variables were assessed using Spearman's method. Specific statistical tests used and number of independent samples and replicates per experiment are listed in the figure legends. A Tukey's post-hoc test was used when needed to account for multiple comparisons. Significance was set at P < 0.05.
Results
Study Population
Demographic data for the 50 women selected for this study are presented in Table 1. On the basis of BMI, women were stratified into lean (BMI < 25 kg/m2), overweight (BMI 25-29.9 kg/m2), and obese (BMI ≥ 30 kg/m2) groups. No differences were found between groups with respect to age, race, breast cancer 1, early-onset gene mutation, or subtype of breast cancer (eg, hormone-receptor positive versus triple-negative breast cancer). Menopausal status and breast inflammation in the form of histologically identified CLSs were significantly different between groups. Obese women (66.7%) were more likely to be postmenopausal than lean women (17.4%). The incidence of CLSs increased from lean (34.8%) to overweight (50%) to obese (100%).
Table 1.
Demographic and Clinicopathologic Features of a Cohort of 50 Women Undergoing Mastectomy for Preventative or Therapeutic Reasons
| Features | All (n = 50) | Lean (n = 23) | Overweight (n = 18) | Obese (n = 9) | P value |
|---|---|---|---|---|---|
| Age in years, mean ± SD | 47.1 ± 8.6 | 45.0 ± 8.7 | 49.5 ± 8.2 | 47.6 ± 8.8 | 0.243 |
| Race, n (%) | |||||
| Asian | 5 (10.2) | 2 (8.7) | 3 (17.7) | 0 (0) | |
| Black | 5 (10.2) | 1 (4.4) | 2 (11.8) | 2 (22.2) | |
| White | 39 (79.6) | 20 (86.9) | 12 (70.6) | 7 (77.8) | 0.379 |
| Missing | 1 (2) | 0 (0) | 1 (5.6) | 0 (0) | 0.54 |
| BMI, mean ± SD, kg/m2 | 26.3 ± 5.5 | 22.1 ± 1.8 | 27.5 ± 1.6 | 34.6 ± 6.1 | <0.001 |
| Menopausal, n (%) | |||||
| Premenopausal | 33 (66) | 19 (82.6) | 11 (61.1) | 3 (33.3) | |
| Postmenopausal | 17 (34) | 4 (17.4) | 7 (38.9) | 6 (66.7) | 0.024 |
| CLSs, n (%) | |||||
| No | 24 (48) | 15 (65.2) | 9 (50) | 0 (0) | |
| Yes | 26 (52) | 8 (34.8) | 9 (50) | 9 (100) | 0.002 |
| BRCA mutation, n (%) | |||||
| BRCA1 | 4 (8) | 1 (4.4) | 2 (11.1) | 1 (11.1) | |
| BRCA2 | 1 (2) | 0 (0) | 0 (0) | 1 (11.1) | |
| Negative | 45 (90) | 22 (95.7) | 16 (88.9) | 7 (77.8) | 0.248 |
| Invasive, n (%) | |||||
| No | 11 (22) | 5 (21.7) | 5 (27.8) | 1 (11.1) | |
| Yes | 39 (78) | 18 (78.3) | 13 (72.2) | 8 (88.9) | 0.674 |
| ER, n (%) | |||||
| Negative | 9 (22) | 4 (20) | 3 (23.1) | 2 (25) | |
| Positive | 32 (78.1) | 16 (80) | 10 (76.9) | 6 (75) | 1 |
| Missing | 9 (18) | 3 (13.0) | 5 (27.8) | 1 (11.1) | 0.493 |
| PR, n (%) | |||||
| Negative | 12 (30.8) | 5 (27.8) | 4 (30.8) | 3 (37.5) | |
| Positive | 27 (69.2) | 13 (72.2) | 9 (69.2) | 5 (62.5) | 0.905 |
| Missing | 11 (22) | 5 (21.7) | 5 (27.8) | 1 (11.1) | 0.674 |
| HER2, n (%) | |||||
| Negative | 32 (82.1) | 13 (72.2) | 11 (84.6) | 8 (100) | |
| Positive | 7 (18) | 5 (27.8) | 2 (15.4) | 0 (0) | 0.315 |
| Missing | 11 (22) | 5 (21.7) | 5 (27.8) | 1 (11.1) | 0.674 |
| Triple negative, n (%) | 8 (20.5) | 3 (16.7) | 3 (23.1) | 2 (25) | 0.639 |
Statistically significant P values, as determined by either analysis of variance for continuous variables or Fisher exact test for categorical variables, are in bold.
BMI, body mass index; BRCA, breast cancer 1, early-onset gene; CLS, crown-like structure; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
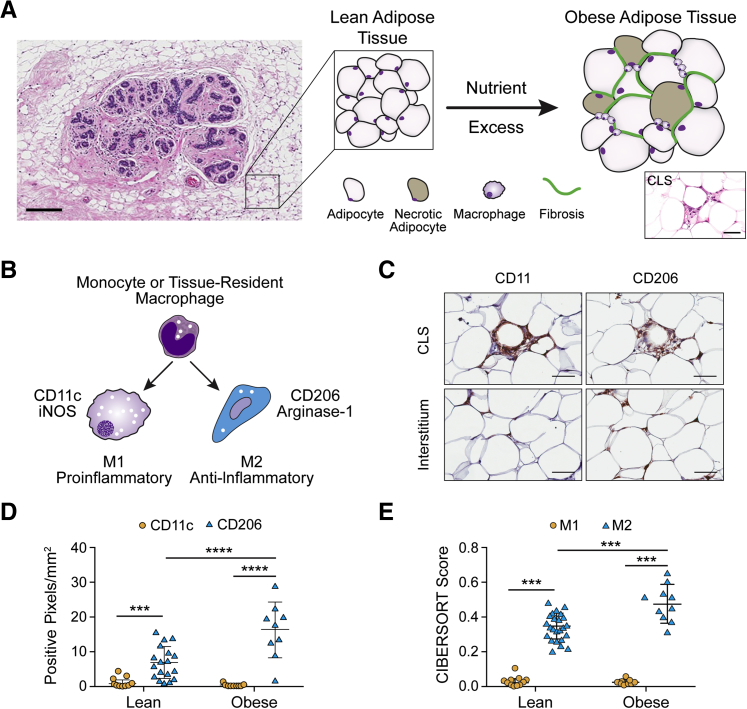
Macrophage Polarization States Are Spatially Regulated in Breast Adipose Tissue
Breast tissue is a composite of fibroglandular tissue surrounded by abundant adipose tissue (Figure 1A). Therefore, obesity-dependent structural changes to adipose tissue, including fibrosis and inflammation (Figure 1A), alter the breast microenvironment and, thus, potentially tumorigenesis. For the purposes of these studies, only the adipose tissue, and not the fibroglandular compartment of breast tissue, is being modeled. Recent discoveries from diabetes research showed that local proliferation of CD206+ (typically considered M2-biased) macrophages within interstitial regions of s.c. and epididymal white adipose tissue contributes to the macrophage burden in mice.24 In addition, obesity in humans increases the number of CLSs22 (Figure 1A). However, a direct comparison of macrophage phenotype in human breast adipose tissue CLSs versus interstitial regions is missing. By immunostaining for the M1 marker CD11c and the M2 marker CD206, the density of pro-inflammatory and anti-inflammatory macrophages was examined in both interstitial regions and CLSs of tumor-free regions of human breast tissue (Figure 1, B and C).19 Although both CD11c+ and CD206+ macrophages were present in CLSs (Figure 1C), quantitative image analysis revealed that CD206+ macrophages were the dominant phenotype in the adipose tissue interstitium in both lean and obese individuals. Although obesity increased the density of CD206+ macrophages, no difference was detected for CD11c+ macrophages (Figure 1D).
Figure 1.
CD206+ macrophages dominate in breast adipose tissue and increase with obesity. A: Fibroglandular breast tissue exists in close proximity to adipose tissue that undergoes structural alterations during obesity, including increased fibrosis and infiltrates of macrophages, visualized as crown-like structures (CLSs). B: Schematic representation of macrophage polarization states and associated membrane molecule and enzymatic markers. C: Representative photomicrographs illustrating two spatially distinct compartments of macrophages in breast adipose tissue, CLSs that are composed predominantly of CD11c+ M1-biased macrophages, and interstitial macrophages, composed predominantly of CD206+ M2-biased macrophages, as determined by immunohistochemistry (IHC). D: The interstitium of breast adipose tissue contains more CD206+ versus CD11c+ macrophages regardless of obesity; however, CD206+ macrophage density is increased in obese versus lean women. E: Computational analysis of macrophage phenotype via CIBERSORT analysis of RNA transcripts corroborates IHC data with significant enrichment of M2-biased macrophages in obese versus lean breast tissue. D and E: Two-way analysis of variance was performed. Data are expressed as means ± SD (D and E). n = 26 sections, with 5 representative images per section (D); n = 26 (E). ***P < 0.001, ****P < 0.0001. Scale bars: 300 μm (A, main image); 100 μm (A, inset, and C). iNOS, inducible nitric oxide synthase.
To corroborate the IHC data with a more comprehensive molecular data set, CIBERSORT analysis was performed to estimate the proportions of M2- versus M1-biased macrophages in lean and obese human breast adipose tissue56 from the same cohort of women used for IHC studies. CIBERSORT is a computational approach that allowed us to infer the representation of different macrophage phenotypes on the basis of the transcriptome of bulk breast tissue. Similar to IHC results, CIBERSORT analysis predicted more M2-biased macrophages in breast adipose tissue regardless of BMI (Figure 1E) and that the burden of M2 macrophages was higher in obese versus lean adipose tissue, whereas no difference was detected for RNA transcript patterns indicative of M1-biased macrophages (Figure 1E).
Collectively, these results suggest that although obesity is associated with a pro-inflammatory M1 macrophage phenotype both histologically and biochemically, M2-biased macrophages are the predominant macrophage population in breast adipose tissue interstitium and increase with obesity.
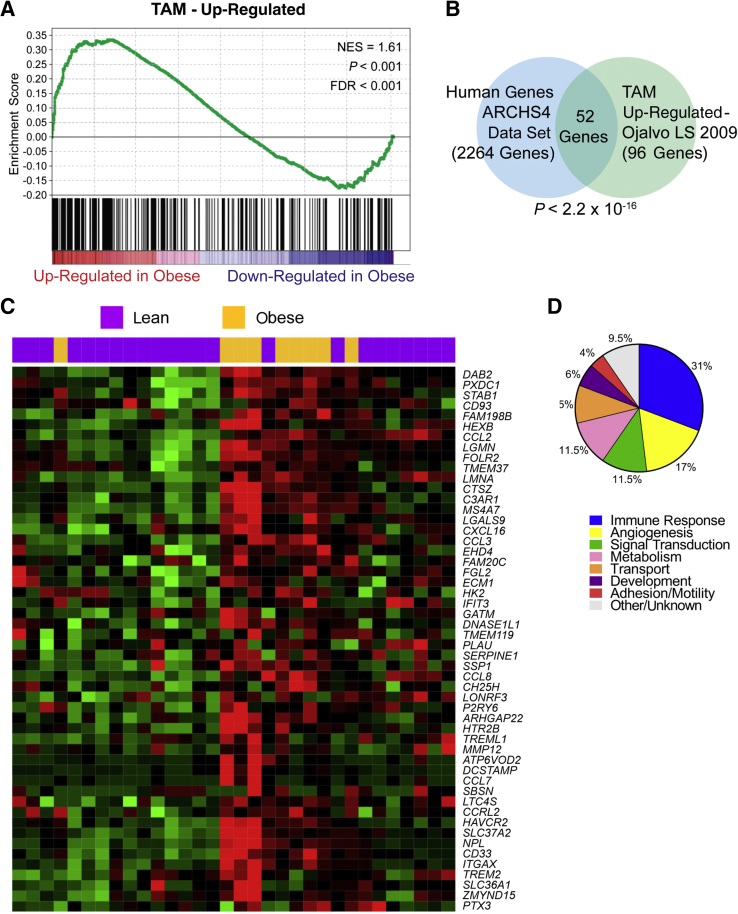
Macrophages from Obese versus Lean Tumor-Free Breast Tissue More Closely Resemble Tumor-Associated Macrophages
Given that M2 macrophage functions, such as promoting fibrosis and angiogenesis and immune suppression, are commonly associated with increased tumorigenesis, it was studied whether the transcriptomic signature of obesity-associated M2-like macrophages in cancer-free breast tissue resembles that of TAMs. Accordingly, a published gene expression signature of mammary tumor TAMs57 was compared with the RNA transcripts of obesity-associated M2 macrophages via Gene Set Enrichment Analysis. This analysis revealed 96 genes that are significantly up-regulated in both the TAM gene signature and tumor-free breast tissue in obese versus lean women (Figure 2A). As the TAM data set was derived from the mouse mammary tumor virus-polyoma virus middle T antigen (MMTV-PyMT) mouse model of breast tumorigenesis, it was evaluated whether the identified gene set would have human relevance by querying the 96 up-regulated TAM genes against the ARCHS4 human gene database (https://amp.pharm.mssm.edu/archs4/data.html, last accessed November 27, 2018). The human macrophage signature was significantly enriched (52/96 genes; P < 2.2 × 10−6), indicating the human relevance of macrophage enrichment in mouse-derived TAMs (Figure 2B). More important, these 52 human-relevant macrophage genes were capable of differentiating most obese versus lean individuals on the basis of differential expression levels in breast tissue (Figure 2C). Interestingly, two lean women clustered with the obese rather than lean individuals, but histologic analysis revealed that their breast adipose tissue had comparable levels of breast inflammation as the obese samples, as determined by quantification of CLS number. Both women also had CIBERSORT M2 scores in the top 50% of our cohort. These findings could be compatible with recent evidence suggesting that a subset of women with normal BMI but histologic evidence of breast inflammation23 or increased adiposity, when measured via dual-energy X-ray absorptiometry,61 is at increased risk for breast cancer development, similar to obese individuals. When the 52 genes were divided into categories based on cellular function (Supplemental Table S1), differences in immune response– and angiogenesis-related genes were the most enriched subsets (Figure 2D), compatible with known functional properties of TAMs.30, 62, 63, 64
Figure 2.
Obesity increases the expression of genes associated with a tumor-associated macrophage (TAM)–like phenotype in human, tumor-free breast tissue. A: Gene Set Enrichment Analysis (GSEA)53 illustrates that anti-inflammatory macrophage–related genes in obese breast tissue samples are concordant to a published gene profile of TAM.57B: Comparison of TAM gene profile57 to ARCHS4 human tissue data set (https://amp.pharm.mssm.edu/archs4/data.html, last accessed November 27, 2018) indicates significant overlap between the MMTV-PyMT–derived TAM genes and human macrophage genes via ENRICHR54, 55 analysis tool. C: The 52 genes identified via GSEA and ENRICHR analysis are capable of delineating most lean versus obese women on the basis of differential gene expression. D: Differential gene expression analysis of macrophage-associated genes suggests varied physiological functions of macrophages in obese breast tissue. n = 26 (A); n = 52 (B and D). FDR, false discovery rate; NES, normalized enrichment score.
Collectively, these results suggest that obesity is associated with macrophage phenotypic changes that lead to an increase in M2-biased macrophages, as determined by IHC of CD206 and computational analysis of a comprehensive panel of macrophage-associated genes. In addition, these differences are associated with transcript levels in tumor-free breast adipose tissue that resemble those of tumor-associated macrophages.
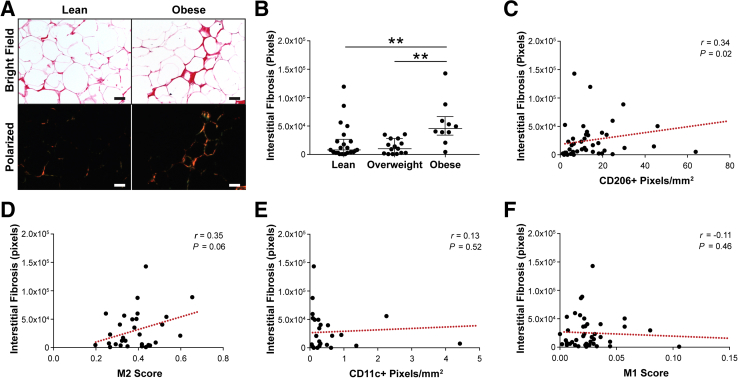
Interstitial Fibrosis of Human Samples Correlates to Increased M2 Macrophage-Biased State
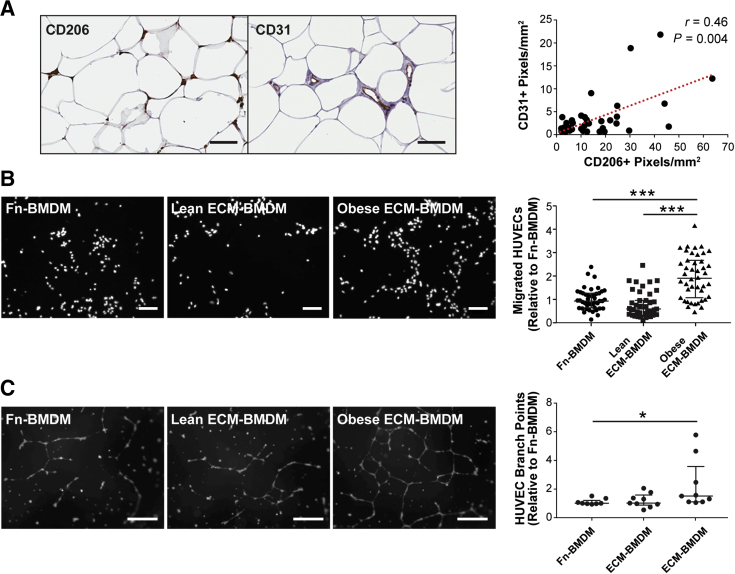
Prior work suggested that obesity causes interstitial fibrosis,14, 65 increased fibrosis correlates to increased macrophage infiltration,66 and an anti-inflammatory macrophage phenotype can be induced by compositional, mechanical, and topographical cues associated with fibrotic ECM remodeling.42, 43, 67, 68 Therefore, it was next tested whether fibrosis in obese breast adipose tissue might mediate the detected differences in macrophage populations. First, by image analysis of picrosirius red–stained cross-sections, it was confirmed that obesity is associated with increased interstitial collagen, a hallmark of fibrosis, in our patient cohort.14, 69 Indeed, interstitial collagen deposition between adipocytes was significantly increased in samples from obese individuals (Figure 3, A and B) and positively correlated with BMI (Supplemental Figure S2). Image analysis of consecutive cross-sections revealed that increased interstitial fibrosis correlated with greater numbers of CD206+ macrophages (Figure 3C). This finding was supported by analysis of paired histologic and RNA samples, which suggested that interstitial fibrosis and CIBERSORT M2 score were similarly interrelated (Figure 3D). More important, the same trends were not evident for CD11c+ macrophages within the interstitium or CIBERSORT M1 score (Figure 3, E and F). Similarly, no correlation existed between the density of interstitial CD11c+ macrophages and BMI (Supplemental Figure S3). These results suggest that obesity-associated interstitial fibrosis may have a direct effect on macrophage polarization toward an M2- but not an M1-biased phenotype in breast adipose tissue.
Figure 3.
Macrophage phenotype is associated with obesity-associated interstitial fibrosis. A and B: Interstitial extracellular matrix is more fibrotic in obese versus lean or overweight breast adipose tissue, as determined by image analysis and pixel quantification of picrosirius red–stained sections under polarized light. Kruskal-Wallis analysis was used. C–F: Interstitial fibrosis positively correlates to CD206+ immunohistochemistry (IHC) and CIBERSORT anti-inflammatory (M2) score (C and D) but not CD11c IHC (E) or CIBERSORT pro-inflammatory (M1) (F) score. C–F: Red dotted lines illustrate the best fit line, as determined by linear regression. Data are expressed as median and interquartile range (B). n = 37 with 5 representative images per section (A and B); n = 37 (C, D, and F); n = 26 (E). **P < 0.01. Scale bars = 50 μm (A).
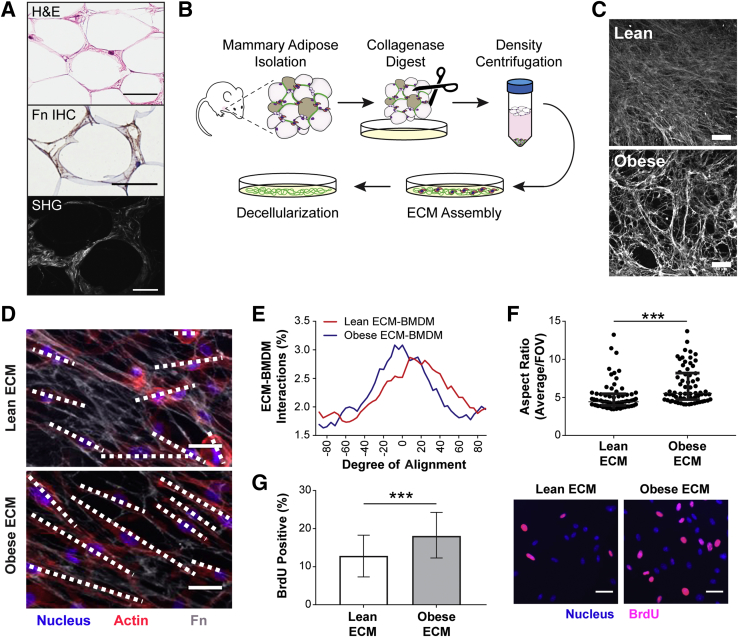
ECM Assembled by Obese ASCs Promotes an M2 Phenotype of Macrophages in Vitro
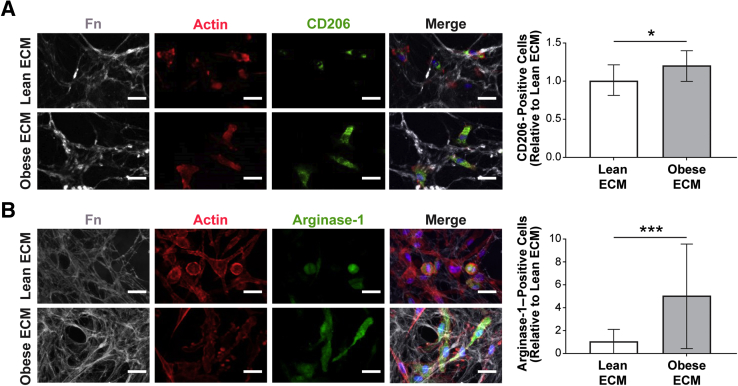
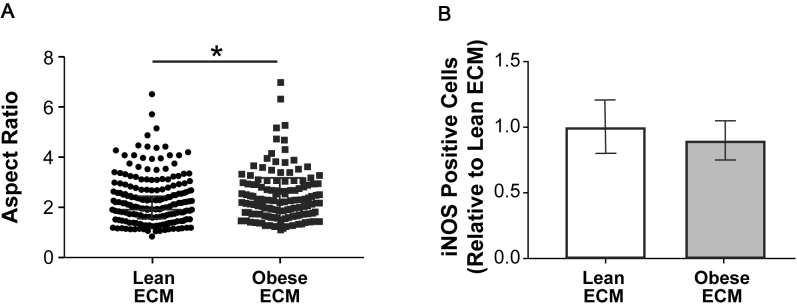
To assess whether the in vivo correlation between adipose tissue interstitial fibrosis and M2-biased macrophage phenotype has a causal relationship, an in vitro model system was used that allowed direct interrogation of macrophage phenotypic changes in response to obesity-specific adipose tissue interstitial ECM remodeling. As mentioned above, obesity-associated, fibrotic ECM remodeling is characterized by increased concentrations of fibrillar ECM components, including Fn and collagen type I, that can be visualized by IHC and second harmonic generation imaging, respectively (Figure 4A). More important, detergent-based decellularization of ECMs, deposited by lean and obese ASCs isolated from wild-type and genetically obese (ob/ob) mice (Figure 4B), can be used to generate cell culture substrates with similar compositional and structural properties as the interstitial ECM identified in lean versus obese breast adipose tissue that is evident in both mice and humans (Figure 4C and Supplemental Figure S4).12 These substrates were used to evaluate the behavior of syngeneic, primary BMDMs in response to lean and obese ECMs. Interestingly, BMDMs cultured on obese ECM exhibited several characteristics that were previously associated with the M2 phenotype, independent of prototypical stimuli for differentiation.43, 70, 71 First, BMDMs cultured on obese ECM aligned more significantly in the direction of ECM fibers than their counterparts cultured on lean ECMs (Figure 4, D and E). Accordingly, analysis of the cells' aspect ratio revealed that macrophages were more elongated when they were cultured on obese versus lean ECM, regardless if the macrophages were stimulated to polarize toward an M2 phenotype (Figure 4F) or an M1 phenotype (Supplemental Figure S5A). This is an important finding as elongated versus rounded morphology has been described for M2 versus M1 macrophage polarization states in vitro.43, 70 In addition, interactions with ECM, resulting in increased alignment and elongation, have been shown to up-regulate phenotypic markers of M2 macrophages, even in the absence of cytokine stimulation.43 Previous reports have also indicated that proliferation of adipose tissue macrophages occurs exclusively in CD206+ M2 macrophages.24 Accordingly, BMDMs cultured on obese versus lean ECMs were also more proliferative, as detected by bromodeoxyuridine staining (Figure 4G). To test whether the different ECMs also affected the expression of the murine M2 macrophage markers CD206 and arginase-1 (a marker not validated for human macrophages34) under the influence of appropriate cytokine stimulation, IL-4 and IL-13, its expression was assessed via immunofluorescence. Immunofluorescence image analysis indicated increased numbers of both CD206+ (Figure 5A) and arginase-1+ (Figure 5B) BMDMs cultured on obese ECM versus lean ECM. Although neither IHC nor CIBERSORT data suggested obesity affected M1-biased macrophage polarization in breast adipose interstitium, for completeness, the expression of M1 marker inducible nitric oxide synthase was evaluated. When BMDMs cultured on lean and obese ECM were treated with lipopolysaccharide to induce M1-biased polarization, there was a trend for reduced inducible nitric oxide synthase expression in BMDMs cultured on obese versus lean ECM that neared significance (P = 0.06) (Supplemental Figure S5B). Taken together, the described characteristic morphologic changes in conjunction with increased proliferation, increased CD206 and arginase-1 expression, and a trend toward reduced inducible nitric oxide synthase expression suggest that macrophage interactions with obese ECM enhance an M2-biased polarization state in comparison to lean ECM.
Figure 4.
Obesity-associated extracellular matrix (ECM) regulates macrophage morphology and proliferation in vitro. A: Representative photomicrographs illustrating the distribution of human adipose tissue interstitial ECM in between adipocytes via hematoxylin and eosin (H&E; top panel) and visualizing fibronectin (Fn) and collagen, two key components of interstitial fibrosis, by immunohistochemistry (IHC; middle panel) and second harmonic generation (SHG) imaging (bottom panel), respectively. B: Schematic illustrating protocol for adipose stromal cell (ASC) isolation from the inguinal fat pad of lean wild-type and obese (ob/ob) age-matched mice and their use for preparation of decellularized ECMs. C: Representative confocal microscopy images of decellularized ECM assembled by lean and obese ASCs after immunostaining for Fn. D–F: Bone marrow–derived macrophages (BMDMs) cultured on lean and obese decellularized matrices (dashed lines highlight the long axis of the macrophages; D) are more aligned with the matrix (0 on x axis represents perfect alignment; E) and exhibit increased elongation (F), as determined by confocal microscopy and image analysis (top 25% of cells plotted) after stimulation with IL-4 and IL-13. F:U-test was performed. G: BMDMs cultured on obese decellularized ECM are more proliferative, as determined by bromodeoxyuridine (BrdU) incorporation with immunofluorescence image analysis. A t-test analysis was performed. Data are expressed as median and interquartile range (F) or means ± SD (G). n = 3 independent experiments with 10 images per condition per experiment (D and E); n = 3 independent experiments with 30 representative images per condition per experiment (G). ***P < 0.001. Scale bars: 50 μm (A and C); 25 μm (D and G).
Figure 5.
Obesity-associated extracellular matrix (ECM) impacts macrophage polarization in vitro. A and B: A greater proportion of bone marrow–derived macrophages cultured on obese decellularized ECM expresses the anti-inflammatory macrophage markers CD206 (A) and arginase-1 (B), when exposed to IL-4 and IL-13, as determined by immunofluorescence confocal image analysis. A t-test analysis was used. Data are expressed as means ± SD (A and B). n = 3 independent experiments, with 15 images per condition per experiment (A and B). *P < 0.05, ***P < 0.001. Scale bars = 25 μm (A and B). Fn, fibronectin.
Obesity-Associated ECM Promotes Proangiogenic Functions of Macrophages in Vitro
Next, it was validated whether the detected ECM-dependent changes in macrophage phenotype may have functional consequences. Obesity is associated with increased vascular density in adipose tissue,72 a finding that is typically attributed to an adipocyte hypertrophy-mediated increase in hypoxia and consequential up-regulation of proangiogenic factor secretion.73 However, it remains unclear whether obesity-associated, fibrotic ECM remodeling may independently activate angiogenesis by stimulating macrophage transition into a proangiogenic M2 phenotype. As increased angiogenesis is a necessary checkpoint for tumor growth and metastasis74, 75 and because macrophages in obese adipose tissue up-regulated angiogenesis-related genes (Figure 2D), it was evaluated whether vascular density and presence of M2-biased macrophages correlated in histologic samples from our patient cohort. Indeed, this analysis revealed that the presence of CD206+ macrophages in the adipose tissue interstitium correlated with positive pixel counts for the endothelial cell marker CD31 (Figure 6A). Given that many other mechanisms are active in vivo and could be responsible for these observations, it was studied whether BMDMs cultured on obese ECM would possess increased proangiogenic capability compared with BMDMs cultured on lean ECM. Using a transwell migration setup and subsequent image analysis, it was found that BMDMs cultured on obese ECM increased HUVEC migration relative to BMDMs cultured on either lean ECM or Fn-coated coverslips (Figure 6B). In addition, conditioned medium collected from BMDM cultures on obese matrices stimulated increased tubulogenesis of HUVECs on growth factor–reduced Matrigel relative to BMDM-conditioned medium from lean ECM or Fn-coated coverslips (Figure 6C). These results suggest that macrophages in contact with obesity-associated ECM could promote vascular remodeling by releasing proangiogenic factors, possibly playing a role in the recognized link between obesity and tumor development and progression.
Figure 6.
Interactions with obese extracellular matrix (ECM) promote macrophage proangiogenic function. A: CD206+ macrophages and CD31+ vascular profiles are positively correlated on the basis of immunohistochemistry image analysis of clinical samples. Red dotted line illustrates the best fit line, as determined by linear regression. B: Endothelial cell transwell migration toward bone marrow–derived macrophages (BMDMs) is significantly increased when IL-4– and IL-13–stimulated BMDMs are in contact with obese versus lean or fibronectin (Fn) control ECMs; Kruskal-Wallis analysis was performed. C: Endothelial cell tubulogenesis is increased in the presence of conditioned medium collected from unstimulated BMDMs cultured on obese versus lean ECM or Fn control substrates. Kruskal-Wallis analysis was performed. Data are expressed as median and interquartile range (B and C). n = 43 (A); n = 3 independent experiments with 3 transwells per condition per experiment and 4 images per transwell (B); n = 3 independent experiments with 3 wells per condition and 1 representative image per well (C). *P < 0.05, ***P < 0.001. Scale bars: 100 μm (A); 50 μm (B and C). HUVEC, human umbilical vein endothelial cell.
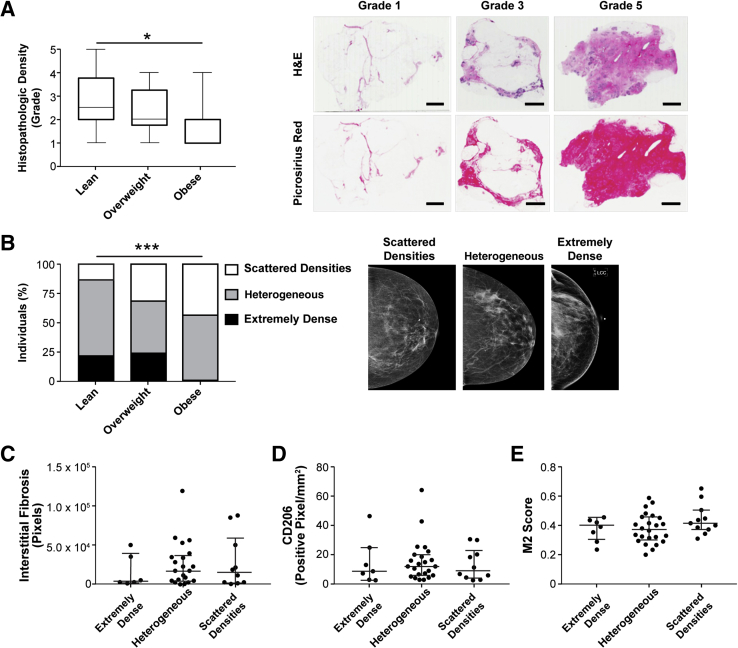
Mammographic and Histologic Breast Densities Are Not Predictive of Interstitial Fibrosis or Macrophage Content
Given the observations that obesity causes an increase in interstitial fibrosis, but that obesity is typically associated with decreased breast tissue density due to accumulation of fatty tissue, which appears radiolucent on mammography,76 it was next evaluated whether bulk breast fibrosis, as determined via both mammographic and histologic77 findings, would correlate to M2 macrophage phenotype. Twenty-six women in our cohort had available mammographic data. As expected, histologic density grade was inversely related to BMI (Figure 7A) and there was a significant difference in the proportions of mammographic patterns between lean, overweight, and obese women (Figure 7B). The differences between mammographic density pattern and histopathologic density score between lean, overweight, and obese women (Figure 3, A and B) did not predict interstitial fibrosis (Figure 7C) or M2-biased macrophage burden, as detected by CD206 IHC and CIBERSORT M2 score (Figure 7, D and E). Collectively, these results indicate that obesity-associated interstitial fibrosis might play a role in modulating macrophage phenotype and function in obese breast adipose tissue, but that these changes cannot be diagnosed with conventional clinical imaging techniques that focus on detection of gross breast tissue density.
Figure 7.
Global breast density is not predictive of either fibrosis or macrophage burden within adipose tissue interstitium. A: Histologic breast density, assessed by the ratio of fibrous tissue/adipose tissue,77 is inversely correlated with body mass index. Kruskal-Wallis analysis was performed. B and C: The distribution of mammographic density patterns is significantly different between body condition (χ2 analysis) and is not predictive of interstitial fibrosis (Kruskal-Wallis analysis). D and E: Macrophage burden, as determined by both CD206 immunohistochemistry and CIBERSORT analysis of RNA transcripts, cannot be predicted by mammographic density pattern; Kruskal-Wallis analysis was performed. Data are expressed as median, interquartile range, and minimum-maximum (A) or median and interquartile range (C–E). n = 50 sections (A); n = 26 (B–E). *P < 0.05, ***P < 0.001. Scale bars = 4 mm (A). H&E, hematoxylin and eosin; M2, anti-inflammatory.
Discussion
Obesity has been linked to an increase in breast cancer risk in postmenopausal women and increased risk of distant metastasis and worse clinical outcome independent of menopause status, although the underlying mechanisms are poorly understood. Using a combination of clinical samples, conventional and computational pathology, and in vitro model systems, this study suggests that obesity-associated adipose tissue interstitial fibrosis could contribute to a tumor-permissive microenvironment by modulating macrophage function to a TAM-like state.
The link between chronic inflammation and cancer development is well established78, 79, 80; however, in the case of obesity, much of the focus has been on WATi in the form of CLSs. WATi has been associated with both an increased risk of breast cancer20 and worse prognosis.18, 19 WATi has been shown to be reversible with weight loss in mouse models of obesity.81 However, a recent secondary analysis of a large randomized clinical trial indicated that weight loss in postmenopausal women did not significantly reduce risk for invasive breast cancer development.82 This suggests that, although WATi in the form of CLSs appears to be reversible with weight loss, other changes to adipose tissue during obesity, such as interstitial fibrosis or expanded M2-biased macrophage populations, may be playing a role.
The recognition of anti-inflammatory macrophages and up-regulated cancer-related pathways in obese adipose tissue macrophages spans the past decade.25, 26 Indeed, similar to our study, M2-biased macrophages have been previously identified to reside within the interstitium of obese adipose tissue.24, 83 However, our hypothesis that the interactions between obesity-associated interstitial fibrosis and macrophages might promote M2 bias or TAM-like polarization is a unique inquiry brought about by the recent characterization of interstitial fibrosis within breast adipose tissue12 and the fact that macrophages are sensitive to both biochemical and biophysical changes of the ECM that occur with fibrosis.42, 43, 71 Because of substantial plasticity of macrophage function and overlapping phenotypes of macrophage subsets, therapeutically targeting tumor-promoting macrophages has yet to yield positive results in clinical trials.84 Therefore, targeting the underlying interstitial fibrosis might be a novel approach to modulate the microenvironment and diminish TAM-like functions of adipose tissue macrophages in obesity.
Macrophages are a heterogeneous population of cells in which the M1 and M2 nomenclature used herein represents extremes. Indeed, recent studies have illustrated that adipose tissue macrophages have activation states that do not conform to the classic definitions of M1 or M2 macrophages.32, 85, 86 We acknowledge that the characterization of macrophages as either M1 or M2 biased in this study is oversimplified; however, approaches that afford detailed characterization of macrophage phenotype, such as flow cytometry, gene expression profiling, and computational methods, sacrifice the spatial organization and relationships that were essential findings in this study. The corroboration of both markers of M2 macrophages, CD206 in human and murine samples and arginase-1 for murine cells, to CIBERSORT analysis of macrophage RNA transcripts and our IHC data are compelling evidence supporting our hypothesis. Unlike IHC, which is binary or, at best, semiquantitative on the basis of labeling intensity, CIBERSORT analysis provides a score on a continuous scale that is more likely to recapitulate the spectrum of macrophage activation states. Furthermore, CIBERSORT uses a large data set of 547 genes to predict macrophage activation; thus, it is potentially more comprehensive in determining macrophage polarization than any feasible number of IHC markers. Nevertheless, future studies could be performed using additional IHC markers to increase characterization of macrophage populations localized within the breast adipose interstitium.
A limitation of the study is the use of a single in vitro model system using hematogenously derived macrophages. Many tissue macrophages are not hematogenously derived but rather are a self-renewing population originating from the embryonic yolk sac.87, 88 One could argue that bone marrow–derived macrophages are not an ideal choice to model adipose tissue macrophages; however, CSF-1 cultured macrophages from murine bone marrow are recognized as a reproducible in vitro experimental standard34 and, thus, were an appropriate choice for these experiments. In addition, experimental evidence from breast cancer research indicates increased recruitment of hematogenously derived macrophages into adipose tissue during the process of obesity,89 further supporting the relevance of the BMDM model system within this context. However, future experiments using alternative in vitro and ex vivo macrophage model systems would be worthwhile.
The tumor-promoting functions of macrophages are manifold and—in addition to modulating angiogenesis—include direct effects on tumor growth, invasion, metastasis, and stroma remodeling.28, 40, 90, 91 The effect of obesity-associated ECM on transition to M2-like macrophages and possible functional implications on endothelial cells were studied to demonstrate the broad significance of obesity-associated ECM in regulating stromal cell functions contributing to tumorigenesis. Although future studies will be necessary to further validate the molecular mechanisms underlying these results, these findings are complementary to previous work. For example, others have shown that peritumoral adipose tissue not only contains increased collagen content, but also represents a rich depot of proangiogenic macrophages.92 In addition, obese (and, therefore, inflamed) adipose tissue has been shown to promote stromal vascularization and angiogenesis before tumor development, with implications for tumorigenesis.89 On the basis of these data, it is plausible that interstitial fibrosis in adipose tissue can contribute to increased angiogenesis via modulation of macrophage phenotype. Indeed, the field of regenerative medicine and biomaterials has been using alterations in matrix composition to modulate macrophage response and guide proper wound healing for surgical implant devices for some time.67, 93, 94, 95, 96 Future studies using engineered co-culture models that integrate ASCs, macrophages, endothelial cells, and varying ECM composition and mechanics to recapitulate the complex obese adipose tissue microenvironment are necessary to identify the impact of obese adipose tissue on macrophages, angiogenesis, and, ultimately, tumor cell behavior.97, 98, 99
Several angiogenesis-related genes were up-regulated in the obese macrophage gene signature, including secreted phosphoprotein 1 (SPP1; alias and referred to herein as osteopontin or OPN), serpine 1, and stabilin 1. Of these three, OPN is the best characterized regarding cancer and TAMs and, thus, is an enticing target for future studies. OPN is a multifunctional cytokine, expressed by many cell types with both paracrine and autocrine activities, that plays a role in multiple hallmarks of cancer.100 Although cancer cells are a large source of OPN in the tumor microenvironment, TAMs are capable of supplying sufficient OPN to rescue cancer cell motility and invasion in OPN knockdown models,101 suggesting that a source of OPN within the obese breast adipose tissue microenvironment might accelerate tumorigenesis, through paracrine interactions, during the initiating stages of neoplastic transformation. TAM expression of OPN stimulates cyclooxygenase-2 production and subsequent prostaglandin E2 production, both known players in tumor growth and angiogenesis.102 Interestingly, cyclooxygenase-2–expressing TAM numbers have recently been correlated to collagen deposition and density within tumor stroma but not epithelial nests,103 suggesting that ECM remodeling might play a regulatory role in the OPN–cyclooxygenase-2 signaling axis contributing to breast cancer.
Mammographic density has consistently been identified as an independent risk factor for breast cancer.76, 104 Recent evidence indicates that an increase in macrophage numbers within the breast fibroglandular region positively correlates with the amount of stromal collagen,77 cancer invasion, and aggression.66 However, neither of these studies assessed BMI within its cohort, macrophage populations residing adjacent to the fibroglandular compartment within the adipose tissue, or distinct subsets of macrophage populations. Interestingly, this study found that mammography is insensitive to detect interstitial fibrosis within breast adipose tissue. This finding alters the paradigm of the definition of breast density, suggesting that microscale fibrosis might be similarly important to gross fibrosis. Knowing that the interstitial adipose tissue M2-biased macrophages are migratory,24 future studies are needed to evaluate whether this population is a source for recruitment of TAMs into the fibroglandular compartment during tumor development.
In conclusion, this study identifies an association between interstitial fibrosis in obese adipose tissue and TAM-like macrophage functions. Others had previously demonstrated that TAMs promote fibrotic ECM remodeling within the tumor microenvironment,40 whereas our data suggest that profibrotic ECM remodeling itself can promote a TAM-like phenotype of macrophages. Hence, these processes are not mutually exclusive, but rather occur simultaneously and via a positive feedback mechanism. Additional studies are necessary to identify the underlying mechanisms; however, these findings support a model of obese adipose tissue priming the microenvironment for future cancer development. More important, our findings highlight that microscale changes in breast adipose tissue structure could have pathophysiological implications for tumorigenesis, but these changes are undetectable by standard diagnostic imaging methods.
Acknowledgments
We thank Mal Hoover, certified medical illustrator (Kansas State University College of Veterinary Medicine), for assistance in figure preparation; the Cornell Biotechnology Resource Center Imaging Facility; the Confocal Microscopy and Microfluorometry Core (Kansas State University); and both the Cornell Center for Animal Resources and Education and the Kansas State University Comparative Medicine Group for animal care and veterinary support.
Footnotes
Supported by the NIH National Cancer Institute through grants R01CA185239 (C.F., A.J.D.), R01CA215797 (A.J.D.), the Center on the Physics of Cancer Metabolism grant U54CA21084 (C.F., A.J.D., O.E.), and the Office of the Director grant T32OD0011000 (N.L.S.); Breast Cancer Research Foundation grant BCR-18-034 (A.J.D.); and the Botwinick-Wolfensohn Foundation (in memory of Mr.and Mrs. Benjamin Botwinick) (A.J.D.).
Disclosures: N.M.I. is a consultant for Novartis and Puma Biotechnology; M.S.J. is a speaker for GE Healthcare.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2019.06.005.
Supplemental Data
Supplemental Figure S1.
Methods for the ImageScope positive pixel count algorithm. The negative selection tool is used to exclude cross-reactive or non-specific staining (illustrating CD206 immunoreactive vasculature; arrows). Pixels are categorized as negative, weak, moderate, or strong. For the purposes of this study, only strong positive pixels were included in the analysis. Scale bars = 200 μm.
Supplemental Figure S2.
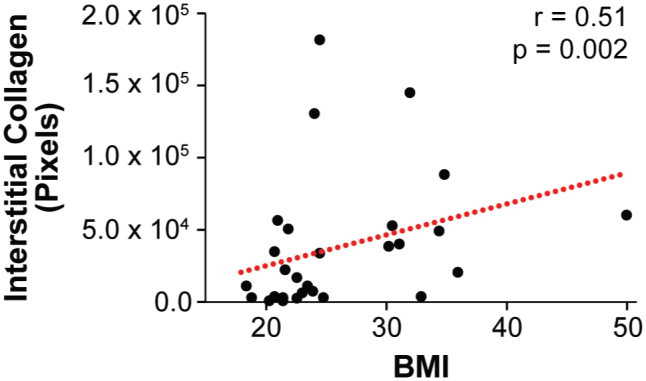
Interstitial fibrosis is positively correlated to body mass index (BMI). Spearman rank correlation between BMI and interstitial fibrosis, as determined by image analysis and positive pixel quantification of picrosirius red–stained sections from lean and obese women imaged under polarized light. Red dotted line illustrates the best fit line, as determined by linear regression. n = 26.
Supplemental Figure S3.
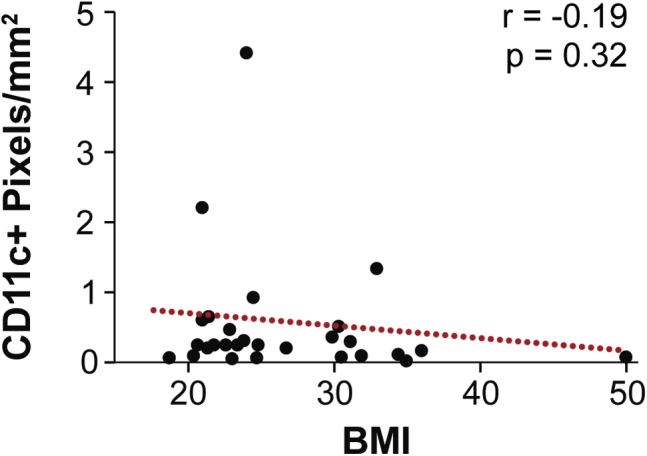
Interstitial adipose tissue CD11c macrophage density does not correlate to body mass index (BMI). Spearman rank correlation between BMI and CD11c labeling, as determined by image analysis and positive pixel quantification of immunohistochemistry sections from lean and obese women. Red dotted line illustrates the best fit line, as determined by linear regression. n = 28.
Supplemental Figure S4.
The in vitro decellularized extracellular matrix (ECM) model mimics key features of fibrotic ECM in obesity. A: Composite and single-channel confocal microscopy images of ECM, assembled by lean and obese adipose stromal cells (ASCs). B–D: Decellularization does not affect the intrinsic fibrillar structure of ECMs regardless of ASC source (B) but recapitulates increased fibronectin and collagen I content in ECMs assembled by obese versus lean ASCs (C and D), as determined by immunofluorescence image analysis. Data are expressed as means ± SD (C and D). n = 1 representative experiment (C and D). **P < 0.01 (t-test analysis). Scale bars = 25 μm (A and B).
Supplemental Figure S5.
Obesity-associated differences in extracellular matrix (ECM) impact macrophage polarization in vitro. A: When stimulated with lipopolysaccharide (LPS), bone marrow–derived macrophages (BMDMs) cultured on decellularized obese ECM are more elongated relative to BMDMs cultured on lean ECM, as determined by confocal image analysis of F-actin–stained cells. U-test was performed. B: Culture of LPS-treated BMDMs on obese versus lean decellularized ECMs suggests a trend—albeit no statistically significant difference (P = 0.06)—for reduced inducible nitric oxide synthase (iNOS) expression, as determined by immunofluorescence image analysis. A t-test analysis was performed. Data are expressed as median and interquartile range (A) or means ± SD (B). n = 1 representative experiment (A). *P < 0.05 (t-test analysis).
References
- 1.Arnold M., Pandeya N., Byrnes G., Renehan A.G., Stevens G.A., Ezzati M., Ferlay J., Miranda J.J., Romieu I., Dikshit R., Forman D., Soerjomataram I. Global burden of cancer attributable to high body-mass index in 2012: a population-based study. Lancet Oncol. 2015;16:36–46. doi: 10.1016/S1470-2045(14)71123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Brandt P.A., Spiegelman D., Yaun S.S., Adami H.O., Beeson L., Folsom A.R., Fraser G., Goldbohm R.A., Graham S., Kushi L., Marshall J.R., Miller A.B., Rohan T., Smith-Warner S.A., Speizer F.E., Willett W.C., Wolk A., Hunter D.J. Pooled analysis of prospective cohort studies on height, weight, and breast cancer risk. Am J Epidemiol. 2000;152:514–527. doi: 10.1093/aje/152.6.514. [DOI] [PubMed] [Google Scholar]
- 3.Trentham-Dietz A., Newcomb P.A., Storer B.E., Longnecker M.P., Baron J., Greenberg E.R., Willett W.C. Body size and risk of breast cancer. Am J Epidemiol. 1997;145:1011–1019. doi: 10.1093/oxfordjournals.aje.a009057. [DOI] [PubMed] [Google Scholar]
- 4.Endogenous Hormones Breast Cancer Collaborative Group Body mass index, serum sex hormones, and breast cancer risk in postmenopausal women. J Natl Cancer Inst. 2003;95:1218–1226. doi: 10.1093/jnci/djg022. [DOI] [PubMed] [Google Scholar]
- 5.Pierobon M., Frankenfeld C.L. Obesity as a risk factor for triple-negative breast cancers: a systematic review and meta-analysis. Breast Cancer Res Treat. 2013;137:307–314. doi: 10.1007/s10549-012-2339-3. [DOI] [PubMed] [Google Scholar]
- 6.Protani M., Coory M., Martin J.H. Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res Treat. 2010;123:627–635. doi: 10.1007/s10549-010-0990-0. [DOI] [PubMed] [Google Scholar]
- 7.Chan D.S.M., Vieira A.R., Aune D., Bandera E.V., Greenwood D.C., McTiernan A., Navarro Rosenblatt D., Thune I., Vieira R., Norat T. Body mass index and survival in women with breast cancer: systematic literature review and meta-analysis of 82 follow-up studies. Ann Oncol. 2014;25:1901–1914. doi: 10.1093/annonc/mdu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown K.A. Impact of obesity on mammary gland inflammation and local estrogen production. J Mammary Gland Biol Neoplasia. 2014;19:183–189. doi: 10.1007/s10911-014-9321-0. [DOI] [PubMed] [Google Scholar]
- 9.Zahid H., Simpson E.R., Brown K.A. Inflammation, dysregulated metabolism and aromatase in obesity and breast cancer. Curr Opin Pharmacol. 2016;31:90–96. doi: 10.1016/j.coph.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Cleary M.P., Grossmann M.E. Minireview: obesity and breast cancer: the estrogen connection. Endocrinology. 2009;150:2537–2542. doi: 10.1210/en.2009-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleary M.P., Grossmann M.E., Ray A. Effect of obesity on breast cancer development. Vet Pathol. 2010;47:202–213. doi: 10.1177/0300985809357753. [DOI] [PubMed] [Google Scholar]
- 12.Seo B.R., Bhardwaj P., Choi S., Gonzalez J., Eguiluz R.C.A., Wang K., Mohanan S., Morris P.G., Du B., Zhou X.K., Vahdat L.T., Verma A., Elemento O., Hudis C.A., Williams R.M., Gourdon D., Dannenberg A.J., Fischbach C. Obesity-dependent changes in interstitial ECM mechanics promote breast tumorigenesis. Sci Transl Med. 2015;7:1–12. doi: 10.1126/scitranslmed.3010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun K., Tordjman J., Clément K., Scherer P.E. Fibrosis and adipose tissue dysfunction. Cell Metab. 2013;18:470–477. doi: 10.1016/j.cmet.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divoux A., Tordjman J., Lacasa D., Veyrie N., Hugol D., Aissat A., Basdevant A., Guerre-Millo M., Poitou C., Zucker J., Bedossa P., Clément K. Fibrosis in human adipose tissue: composition, distribution, and link with lipid metabolism and fat. Diabetes. 2010;59:2817–2825. doi: 10.2337/db10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henegar C., Tordjman J., Achard V., Lacasa D., Cremer I., Guerre-Millo M., Poitou C., Basdevant A., Stich V., Viguerie N., Langin D., Bedossa P., Zucker J.D., Clement K. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 2008;9:1–32. doi: 10.1186/gb-2008-9-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H., Barnes G.T., Yang Q., Tan G., Yang D., Chou C.J., Sole J., Nichols A., Ross J.S., Tartaglia L.A., Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris P.G., Hudis C.A., Giri D., Morrow M., Falcone D.J., Zhou X.K., Du B., Brogi E., Crawford C.B., Kopelovich L., Subbaramaiah K., Dannenberg A.J. Inflammation and increased aromatase expression occur in the breast tissue of obese women with breast cancer. Cancer Prev Res. 2011;4:1021–1029. doi: 10.1158/1940-6207.CAPR-11-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyengar N.M., Zhou X.K., Gucalp A., Morris P.G., Howe L.R., Giri D.D., Morrow M., Wang H., Pollak M., Jones L.W., Hudis C.A., Dannenberg A.J. Systemic correlates of white adipose tissue inflammation in early-stage breast cancer. Clin Cancer Res. 2016;22:2283–2289. doi: 10.1158/1078-0432.CCR-15-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter J.M., Hoskin T.L., Pena M.A., Brahmbhatt R., Winham S.J., Frost M.H., Stallings-Mann M., Radisky D.C., Knutson K.L., Visscher D.W., Degnim A.C. Macrophagic “crown-like structures” are associated with an increased risk of breast cancer in benign breast disease. Cancer Prev Res. 2018;11:113–119. doi: 10.1158/1940-6207.CAPR-17-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gucalp A., Iyengar N.M., Zhou X.K., Giri D.D., Falcone D.J., Wang H., Williams S., Krasne M.D., Yaghnam I., Kunzel B., Morris P.G., Jones L.W., Pollak M., Laudone V.P., Hudis C.A., Scher H.I., Scardino P.T., Eastham J.A., Dannenberg A.J. Periprostatic adipose inflammation is associated with high-grade prostate cancer. Prostate Cancer Prostatic Dis. 2017;20:418–423. doi: 10.1038/pcan.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wentworth J.M., Naselli G., Brown W.A., Doyle L. Pro-inflammatory CD11c+ CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes. 2010;59:1648–1656. doi: 10.2337/db09-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyengar N.M., Brown K.A., Zhou X.K., Gucalp A., Subbaramaiah K., Giri D.D., Zahid H., Bhardwaj P., Wendel N.K., Falcone D.J., Wang H., Williams S., Pollak M., Morrow M., Hudis C.A., Dannenberg A.J. Metabolic obesity, adipose inflammation and elevated breast aromatase in women with normal body mass index. Cancer Prev Res. 2017;10:235–243. doi: 10.1158/1940-6207.CAPR-16-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haase J., Weyer U., Immig K., Klöting N., Blüher M., Eilers J., Bechmann I., Gericke M. Local proliferation of macrophages in adipose tissue during obesity-induced inflammation. Diabetologia. 2014;57:562–571. doi: 10.1007/s00125-013-3139-y. [DOI] [PubMed] [Google Scholar]
- 25.Zeyda M., Farmer D., Todoric J., Aszmann O., Speiser M., Györi G., Zlabinger G.J., Stulnig T.M. Human adipose tissue macrophages are of an anti-inflammatory phenotype but capable of excessive pro-inflammatory mediator production. Int J Obes. 2007;31:1420–1428. doi: 10.1038/sj.ijo.0803632. [DOI] [PubMed] [Google Scholar]
- 26.Mayi T.H., Daoudi M., Derudas B., Gross B., Bories G., Wouters K., Brozek J., Caiazzo R., Raverdi V., Pigeyre M., Allavena P., Mantovani A., Pattou F., Staels B., Chinetti-Gbaguidi G. Human adipose tissue macrophages display activation of cancer-related pathways. J Biol Chem. 2012;287:21904–21913. doi: 10.1074/jbc.M111.315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medrek C., Pontén F., Jirström K., Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian B.-Z., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sousa S., Brion R., Lintunen M., Kronqvist P., Sandholm J., Mönkkönen J., Kellokumpu-Lehtinen P.-L., Lauttia S., Tynninen O., Joensuu H., Heymann D., Määttä J.A. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 2015;17:101. doi: 10.1186/s13058-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin L., Chen Y.-S., Yao Y.-D., Chen J.-Q., Chen J.-N., Huang S.-Y., Zeng Y.-J., Yao H.-R., Zeng S.-H., Fu Y.-S., Song E.-W. CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer. Oncotarget. 2015;6:34758–34773. doi: 10.18632/oncotarget.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyckoff J.B., Wang Y., Lin E.Y., Li J.F., Goswami S., Stanley E.R., Segall J.E., Pollard J.W., Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 32.Hill D.A., Lim H.-W., Kim Y.H., Ho W.Y., Foong Y.H., Nelson V.L., Nguyen H.C.B., Chegireddy K., Kim J., Habertheuer A., Vallabhajosyula P., Kambayashi T., Won K.-J., Lazar M.A. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci U S A. 2018;115:E5096–E5105. doi: 10.1073/pnas.1802611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Italiani P., Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol. 2014;5:1–22. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T., Locati M., Mantovani A., Martinez F.O., Mege J.L., Mosser D.M., Natoli G., Saeij J.P., Schultze J.L., Shirey K.A., Sica A., Suttles J., Udalova I., vanGinderachter J.A., Vogel S.N., Wynn T.A. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka M., Ikeda K., Suganami T., Komiya C., Ochi K., Shirakawa I., Hamaguchi M., Nishimura S., Manabe I., Matsuda T., Kimura K., Inoue H., Inagaki Y., Aoe S., Yamasaki S., Ogawa Y. Macrophage-inducible C-type lectin underlies obesity-induced adipose tissue fibrosis. Nat Commun. 2014;5:4982. doi: 10.1038/ncomms5982. [DOI] [PubMed] [Google Scholar]
- 36.Wynn T.A., Barron L. Macrophages: master regulators of inflammation and fibrosis. Semin Liver Dis. 2010;30:245–257. doi: 10.1055/s-0030-1255354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wynn T.A., Vannella K.M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laoui D., Movahedi K., Van Overmeire E., Van den Bossche J., Schouppe E., Mommer C., Nikolaou A., Morias Y., De Baetselier P., Van Ginderachter J.A. Tumor-associated macrophages in breast cancer: distinct subsets, distinct functions. Int J Dev Biol. 2011;55:719–729. doi: 10.1387/ijdb.113371dl. [DOI] [PubMed] [Google Scholar]
- 39.Provenzano P.P., Eliceiri K.W., Campbell J.M., Inman D.R., White J.G., Keely P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afik R., Zigmond E., Vugman M., Klepfish M., Shimshoni E., Pasmanik-Chor M., Shenoy A., Bassat E., Halpern Z., Geiger T., Sagi I., Varol C. Tumor macrophages are pivotal constructors of tumor collagenous matrix. J Exp Med. 2016;213:2315–2331. doi: 10.1084/jem.20151193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinto M.L., Rios E., Silva A.C., Neves S.C., Caires H.R., Pinto A.T., Durães C., Carvalho F.A., Cardoso A.P., Santos N.C., Barrias C.C., Nascimento D.S., Pinto-do-Ó P., Barbosa M.A., Carneiro F., Oliveira M.J. Decellularized human colorectal cancer matrices polarize macrophages towards an anti-inflammatory phenotype promoting cancer cell invasion via CCL18. Biomaterials. 2017;124:211–224. doi: 10.1016/j.biomaterials.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Patel N.R., Bole M., Chen C., Hardin C.C., Kho A.T., Mih J., Deng L., Butler J., Tschumperlin D., Fredberg J.J., Krishnan R., Koziel H. Cell elasticity determines macrophage function. PLoS One. 2012;7:e41024. doi: 10.1371/journal.pone.0041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McWhorter F.Y., Wang T., Nguyen P., Chung T., Liu W.F. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci U S A. 2013;110:17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruber E., Heyward C., Cameron J., Leifer C. Toll-like receptor signaling in macrophages is regulated by extracellular substrate stiffness and Rho-associated coiled-coil kinase (ROCK1/2) Int Immunol. 2018;30:267–278. doi: 10.1093/intimm/dxy027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Junqueira L.C., Bignolas G., Brentani R.R. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem J. 1979;11:447–455. doi: 10.1007/BF01002772. [DOI] [PubMed] [Google Scholar]
- 46.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks [Erratum appeared in Nat Protoc 2014, 9:2513] Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S., Pyl P.T., Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A., Tamayo P., Mootha V. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ojalvo L.S., King W., Cox D., Pollard J.W. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am J Pathol. 2009;174:1048–1064. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lourenço B.N., Springer N.L., Ferreira D., Oliveira C., Granja P.L., Fischbach C. CD44v6 increases gastric cancer malignant phenotype by modulating adipose stromal cell-mediated ECM remodeling. Integr Biol. 2018;10:145–158. doi: 10.1039/c7ib00179g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crapo P.M., Gilbert T.W., Badylak S.F. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Badylak S.F. Decellularized allogeneic and xenogeneic tissue as a bioscaffold for regenerative medicine: factors that influence the host response. Ann Biomed Eng. 2014;42:1517–1527. doi: 10.1007/s10439-013-0963-7. [DOI] [PubMed] [Google Scholar]
- 61.Iyengar N.M., Arthur R., Manson J.E., Chlebowski R.T., Kroenke C.H., Peterson L., Cheng T.-Y.D., Feliciano E.C., Lane D., Luo J., Nassir R., Pan K., Wassertheil-Smoller S., Kamensky V., Rohan T.E., Dannenberg A.J. Association of body fat and risk of breast cancer in postmenopausal women with normal body mass index. JAMA Oncol. 2018;10065:1–9. doi: 10.1001/jamaoncol.2018.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang Z., Wen C., Chen X., Yin R., Zhang C., Wang X., Huang Y. Myeloid-derived suppressor cell and macrophage exert distinct angiogenic and immunosuppressive effects in breast cancer. Oncotarget. 2017;8:54173–54186. doi: 10.18632/oncotarget.17013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koru-Sengul T., Santander A.M., Miao F., Sanchez L.G., Jorda M., Glück S., Ince T.A., Nadji M., Chen Z., Penichet M.L., Cleary M.P., Torroella-Kouri M. Breast cancers from black women exhibit higher numbers of immunosuppressive macrophages with proliferative activity and of crown-like structures associated with lower survival compared to non-black Latinas and Caucasians. Breast Cancer Res Treat. 2016;158:113–126. doi: 10.1007/s10549-016-3847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu H., Clauser K.R., Tam W.L., Fröse J., Ye X., Eaton E.N., Reinhardt F., Donnenberg V.S., Bhargava R., Carr S.A., Weinberg R.A. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun K., Kusminski C.C.M., Scherer P.E.P. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acerbi I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y.Y., Liphardt J., Hwang E.S., Weaver V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol. 2015;7:1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meng F.W., Slivka P.F., Dearth C.L., Badylak S.F. Solubilized extracellular matrix from brain and urinary bladder elicits distinct functional and phenotypic responses in macrophages. Biomaterials. 2015;46:131–140. doi: 10.1016/j.biomaterials.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 68.Franz S., Allenstein F., Kajahn J., Forstreuter I., Hintze V., Möller S., Simon J.C. Artificial extracellular matrices composed of collagen I and high-sulfated hyaluronan promote phenotypic and functional modulation of human pro-inflammatory M1 macrophages. Acta Biomater. 2013;9:5621–5629. doi: 10.1016/j.actbio.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 69.Chun T.-H. Peri-adipocyte ECM remodeling in obesity and adipose tissue fibrosis. Adipocyte. 2012;1:89–95. doi: 10.4161/adip.19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wheeler A.P. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J Cell Sci. 2006;119:2749–2757. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- 71.McWhorter F.Y., Davis C.T., Liu W.F. Physical and mechanical regulation of macrophage phenotype and function. Cell Mol Life Sci. 2015;72:1303–1316. doi: 10.1007/s00018-014-1796-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu P., Xu A. Interplay between adipose tissue and blood vessels in obesity and vascular dysfunction. Rev Endocr Metab Disord. 2013;14:49–58. doi: 10.1007/s11154-012-9230-8. [DOI] [PubMed] [Google Scholar]
- 73.Halberg N., Khan T., Trujillo M.E., Wernstedt-Asterholm I., Attie A.D., Sherwani S., Wang Z.V., Landskroner-Eiger S., Dineen S., Magalang U.J., Brekken R.A., Scherer P.E. Hypoxia-inducible factor 1alpha induces fibrosis and insulin resistance in white adipose tissue. Mol Cell Biol. 2009;29:4467–4483. doi: 10.1128/MCB.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carmeliet P., Jain R.K. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 75.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 76.Boyd N.F., Lockwood G.A., Byng J.W., Tritchler D.L., Yaffe M.J. Mammographic densities and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1998;7:1133–1144. [PubMed] [Google Scholar]
- 77.Huo C.W., Chew G., Hill P., Huang D., Ingman W., Hodson L., Brown K.A., Magenau A., Allam A.H., Mcghee E., Timpson P., Henderson M.A. High mammographic density is associated with an increase in stromal collagen and immune cells within the mammary epithelium. Breast Cancer Res. 2015;17:1–20. doi: 10.1186/s13058-015-0592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pesic M., Greten F.R. Inflammation and cancer: tissue regeneration gone awry. Curr Opin Cell Biol. 2016;43:55–61. doi: 10.1016/j.ceb.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 80.Wu Y., Antony S., Meitzler J.L., Doroshow J.H. Molecular mechanisms underlying chronic inflammation-associated cancers. Cancer Lett. 2014;345:164–173. doi: 10.1016/j.canlet.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhardwaj P., Du B., Zhou X.K., Sue E., Harbus M.D., Falcone D.J., Giri D., Hudis C.A., Kopelovich L., Subbaramaiah K., Dannenberg A.J. Caloric restriction reverses obesity-induced mammary gland inflammation in mice. Cancer Prev Res. 2013;6:282–289. doi: 10.1158/1940-6207.CAPR-12-0467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Neuhouser M.L., Aragaki A.K., Prentice R.L., Manson J.E., Chlebowski R., Carty C.L., Ochs-Balcom H.M., Thomson C.A., Caan B.J., Tinker L.F., Urrutia R.P., Knudtson J., Anderson G.L. Overweight, obesity, and postmenopausal invasive breast cancer risk. JAMA Oncol. 2015;1:611. doi: 10.1001/jamaoncol.2015.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lumeng C.N., Delproposto J.B., Westcott D.J., Saltiel A.R. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang L., Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol. 2017;10:58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu X., Grijalva A., Skowronski A., Van Eijk M., Serlie M.J., Ferrante A.W. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kratz M., Coats B.R., Hisert K.B., Hagman D., Mutskov V., Peris E., Schoenfelt K.Q., Kuzma J.N., Larson I., Billing P.S., Landerholm R.W., Crouthamel M., Gozal D., Hwang S., Singh P.K., Becker L. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., Greter M., Mortha A., Boyer S.W., Forsberg E.C., Tanaka M., van Rooijen N., García-Sastre A., Stanley E.R., Ginhoux F., Frenette P.S., Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., De Bruijn M.F., Geissmann F., Rodewald H.R. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arendt L.M., McCready J., Keller P.J., Baker D.D., Naber S.P., Seewaldt V., Kuperwasser C. Obesity promotes breast cancer by CCL2-mediated macrophage recruitment and angiogenesis. Cancer Res. 2013;73:6080–6093. doi: 10.1158/0008-5472.CAN-13-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lewis C.E., Pollard J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 91.Tariq M., Zhang J., Liang G., Ding L., He Q., Yang B. Macrophage polarization: anti-cancer strategies to target tumor-associated macrophage in breast cancer. J Cell Biochem. 2017;118:2484–2501. doi: 10.1002/jcb.25895. [DOI] [PubMed] [Google Scholar]
- 92.Wagner M., Bjerkvig R., Wiig H., Melero-Martin J.M., Lin R.Z., Klagsbrun M., Dudley A.C. Inflamed tumor-associated adipose tissue is a depot for macrophages that stimulate tumor growth and angiogenesis. Angiogenesis. 2012;15:481–495. doi: 10.1007/s10456-012-9276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spiller K.L., Anfang R.R., Spiller K.J., Ng J., Nakazawa K.R., Daulton J.W., Vunjak-Novakovic G. The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials. 2014;35:4477–4488. doi: 10.1016/j.biomaterials.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sridharan R., Cameron A.R., Kelly D.J., Kearney C.J., O'Brien F.J. Biomaterial based modulation of macrophage polarization: a review and suggested design principles. Mater Today. 2015;18:313–325. [Google Scholar]
- 95.Spiller K.L., Koh T.J. Macrophage-based therapeutic strategies in regenerative medicine. Adv Drug Deliv Rev. 2017;122:74–83. doi: 10.1016/j.addr.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Petrosyan A., Da Sacco S., Tripuraneni N., Kreuser U., Lavarreda-Pearce M., Tamburrini R., De Filippo R.E., Orlando G., Cravedi P., Perin L. A step towards clinical application of acellular matrix: a clue from macrophage polarization. Matrix Biol. 2017;57–58:334–346. doi: 10.1016/j.matbio.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Truong D., Puleo J., Llave A., Mouneimne G., Kamm R.D., Nikkhah M. Breast cancer cell invasion into a three dimensional tumor-stroma microenvironment. Sci Rep. 2016;6:1–18. doi: 10.1038/srep34094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han S., Shin Y., Jeong H.E., Jeon J.S., Kamm R.D., Huh D., Sohn L.L., Chung S. Constructive remodeling of a synthetic endothelial extracellular matrix. Sci Rep. 2015;5:18290. doi: 10.1038/srep18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li R., Hebert J.D., Lee T.A., Xing H., Boussommier-Calleja A., Hynes R.O., Lauffenburger D.A., Kamm R.D. Macrophage-secreted TNFα and TGFβ1 influence migration speed and persistence of cancer cells in 3D tissue culture via independent pathways. Cancer Res. 2017;77:279–290. doi: 10.1158/0008-5472.CAN-16-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shevde L.A., Samant R.S. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng J., Huo D.H., Kuang D.M., Yang J., Zheng L., Zhuang S.M. Human macrophages promote the motility and invasiveness of osteopontin-knockdown tumor cells. Cancer Res. 2007;67:5141–5147. doi: 10.1158/0008-5472.CAN-06-4763. [DOI] [PubMed] [Google Scholar]
- 102.Kale S., Thorat D., Soundararajan G., Patil T.V., Kundu G.C. Osteopontin signaling upregulates cyclooxygenase-2 expression in tumor-associated macrophages leading to enhanced angiogenesis and melanoma growth via α9β1 integrin. Oncogene. 2014;33:2295–2305. doi: 10.1038/onc.2013.184. [DOI] [PubMed] [Google Scholar]
- 103.Esbona K., Yi Y., Saha S., Yu M., Van Doorn R.R., Conklin M.W., Graham D.S., Wisinski K.B., Ponik S.M., Eliceiri K.W., Wilke L.G., Keely P.J. The presence of cyclooxygenase 2, tumor-associated macrophages, and collagen alignment as prognostic markers for invasive breast carcinoma patients. Am J Pathol. 2018;188:559–573. doi: 10.1016/j.ajpath.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boyd N.F., Guo H., Martin L.J., Sun L., Stone J., Fishell E., Jong R.A., Hislop G., Chiarelli A., Minkin S., Yaffe M.J. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356:227–236. doi: 10.1056/NEJMoa062790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.