Abstract
Wine production is a complex process from the vineyard to the winery. On this journey, microbes play a decisive role. From the environment where the vines grow, encompassing soil, topography, weather and climate through to management practices in vineyards, the microbes present can potentially change the composition of wine. Introduction of grapes into the winery and the start of winemaking processes modify microbial communities further. Recent advances in next-generation sequencing (NGS) technology have progressed our understanding of microbial communities associated with grapes and fermentations. We now have a finer appreciation of microbial diversity across wine producing regions to begin to understand how diversity can contribute to wine quality and style characteristics. In this review, we highlight literature surrounding wine-related microorganisms and how these affect factors interact with and shape microbial communities and contribute to wine quality. By discussing the geography, climate and soil of environments and viticulture and winemaking practices, we claim microbial biogeography as a new perspective to impact wine quality and regionality. Depending on geospatial scales, habitats, and taxa, the microbial community respond to local conditions. We discuss the effect of a changing climate on local conditions and how this may alter microbial diversity and thus wine style. With increasing understanding of microbial diversity and their effects on wine fermentation, wine production can be optimised with enhancing the expression of regional characteristics by understanding and managing the microbes present.
Keywords: wine quality, microbial biogeography, climate, soil, bacteria, fungi
Introduction
Wine production is a global billion-dollar industry for which regionally distinct wine characteristics, collectively known as “terroir,” are an important collection of traits that wine industry would like to define. Wines made from the same grape cultivar but grown in different regions are appreciated for their distinctive features (Van Leeuwen and Seguin, 2006). Regionality can be identified with chemical compositions and sensory properties (Pereira et al., 2005; Gonzálvez et al., 2009; Robinson et al., 2012), and are likely to be ascribed to local environmental parameters that influence grapevine growth and development, such as soil types, climate, topography and human management, but the mechanisms by which these factors affect wine composition remain tenuous (Van Leeuwen and Seguin, 2006; Gladstones, 2011; Vaudour et al., 2015).
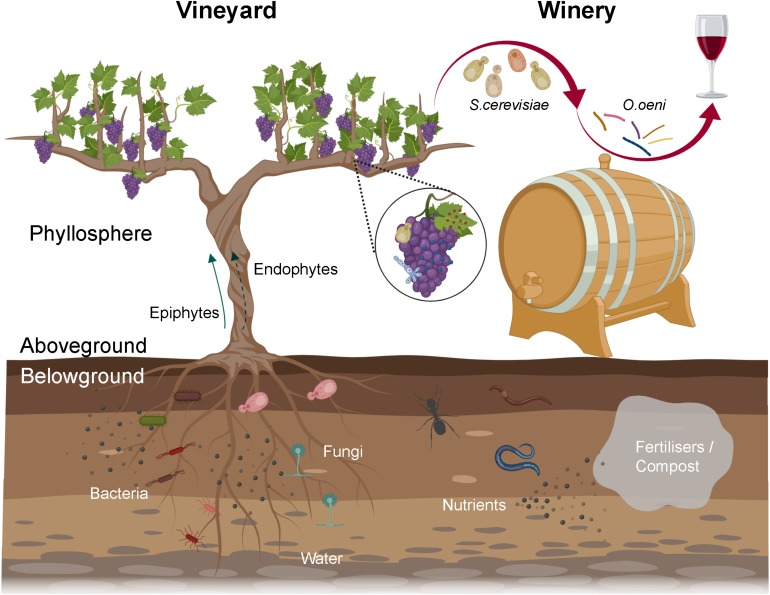
From the vineyard to the winery, microorganisms play key roles in wine production and quality. The grapevine (Vitis vinifera) phyllosphere harbours diverse microbes including yeasts, filamentous fungi and bacteria that substantially modulate grapevine health, growth, and grape and wine production (Figure 1; Barata et al., 2012; Gilbert et al., 2014). Microbes could originate from the vineyard soil (Zarraonaindia et al., 2015; Morrison-Whittle and Goddard, 2018), air, precipitation (rainfall, hail, snow), be transported by animal vectors (bees, insects, and birds) (Goddard et al., 2010; Francesca et al., 2012; Stefanini et al., 2012; Lam and Howell, 2015), and be resident in nearby native forests (Figures 1, 2; Knight and Goddard, 2015; Morrison-Whittle and Goddard, 2018). Microbes that are grapevine-associated and are transferred to the must have a profound influence on wine composition, flavour and quality (Figure 1; Barata et al., 2012). Fermentative yeasts (primarily Saccharomyces cerevisiae) and lactic acid bacteria (LAB, predominantly Oenococcus oeni) in the must modulate the flavour and aroma of wine (Swiegers et al., 2005). Beyond these species, many microorganisms in the must could release metabolites changing the chemical environment and/or fermentation processes and thus affect wine compositions and characteristics, for example, release of inhibitory molecules altering Saccharomyces metabolism (Swiegers et al., 2005; Bokulich et al., 2016).
FIGURE 1.
Overview of the wine-related microbiota from the vineyard to the winery. Microbiota associated with grapevine phyllosphere, especially grapes, can enter musts and constitute wine microbial consortium, in which fermentative yeasts and LAB conduct alcoholic and malolactic fermentation, respectively. The rhizosphere harbours diverse microbes that can benefit plants by enabling nutrient uptake and tolerance to (a)biotic stress. Soil borne microorganisms might translocate to the phyllosphere internally (endophytes) or externally (epiphytes), thereby entering wine fermentation. Viticulture practices, for example fertilisers/compost addition, can modify soil microbiota via shifting nutrient pools or adding manure borne microorganisms (created with BioRender, https://app.biorender.com/).
Increasing evidence supports a microbial aspect to wine regionality may be due, in part, to regionally structured microbial communities, or microbial biogeography. Advanced genetic-based methodologies, in particular next-generation sequencing (NGS), have allowed researchers to sample microbial diversity more deeply and widely and encouraged more comprehensive biogeographical surveys. This has marked the beginning of a new era of information on the grapevine-associated microbiome across multiple scales (region, vineyard, and vine) to elucidate wine quality and regional variation (Bokulich et al., 2014; Taylor et al., 2014; Burns et al., 2015; Morrison-Whittle and Goddard, 2018). This review aims to disentangle the role of microbial biogeography on wine production, by considering how microbes interact with environmental conditions and thus drive wine quality and style.
We commence our discussion with a review of current knowledge on microbial geographic patterns and how they differ from the vineyard to the winery, and then describe how climate, soil, and anthropogenic practices can affect microbial communities through the winemaking process to the finished wine. The concept of scale is crucial to define microbial biogeography, as large-scale geographic and climatic features significantly affect microbial communities, but at smaller scales these differences may not be apparent. We propose that microbial biogeography gives a theoretical basis to wine terroir, which further provides information to the industry to produce distinctive and quality wines through microbial manipulation. This is particularly important as wine styles are altered by weather patterns and we conclude by considering how microbial biogeography and activity may respond to changing climates.
Microbial Biogeography: A Microbial Insight to Understand Variation in Wine Quality
Global studies of microbial biogeography have shown that distinct microbial populations are present in soil, bodies of water and ocean biomes and associated with plants (Martiny et al., 2006). Soil fungi and bacteria show global niche differentiation that is associated with contrasting diversity responses to environmental filtering (for example by precipitation, soil pH) and biotic interactions (Bahram et al., 2018). Culture-based microbiological methods revealed only a small portion of the diversity of environments (Martiny et al., 2006), and so previous studies on the vineyard and wine microbiome do not reveal regional boundary restrained patterns (Barata et al., 2012). New technologies such as NGS benefit from being culture-independent and can therefore reveal a distinct scenario of microbial biogeography. We propose that geographic patterns favour fungi- or bacteria-driven metabolites and thus contribute to wine composition and quality. Some key wine microbial biogeography studies are listed in Table 1, where we highlight the influence of climate, soil and anthropogenic practices (a comparative list of these studies is given in Supplementary Table S1).
TABLE 1.
Recent findings on wine microbial ecology from the vineyard to the winery.
| Microorganisms | Scale | Habitat | Methodology | Major Conclusions | References |
| Yeasts | Three regions | Grape juice | Culture-dependent method, ITS-RFLP and D1/D2 26S sequencing | (i) Regional delineations were found on yeast communities and S. cerevisiae populations | Gayevskiy and Goddard, 2012 |
| (ii) Reasonable levels of gene flow were found in S. cerevisiae populations among regions | |||||
| Bacteria, fungi | Four regions | Grape must | Culture-independent method, 16S rRNA and ITS amplicon sequencing | (i) Regional origin defined grape must microbial patterns, with some influences by the cultivar | Bokulich et al., 2014 |
| (ii) Weather and climate were responsible for driving biogeographical diversity | |||||
| (iii) Vintage exerted seasonal shifts in grape microbiota within single vineyards, especially bacteria | |||||
| S. cerevisiae | Six regions | Vineyard soil, grape juice, native forest soil and fruits | Culture-dependent method, microsatellite loci amplification and genotyping | (i) Regionally genetically differentiated S. cerevisiae populations drove different wine phenotype | Knight and Goddard, 2015; Knight et al., 2015 |
| (ii) Genetic similarity of S. cerevisiae populations was found between vineyards and forests within regions | |||||
| Fungi | Within region, three vineyards | Grapes | Culture-dependent method, ITS-ARISA fingerprinting | (i) Intravineyard variations were greater than intervineyard variations, possibly due to microclimate’s influences on grape microbiota | Setati et al., 2012 |
| (ii) The least treated vineyard (biodynamic and integrated) displayed significantly higher fungal species richness | |||||
| Bacteria | Within region, seven vineyards | Grape must, end malolactic ferments | Culture-independent method, 16S rRNA amplicon sequencing | (i) Bacterial community heterogeneities were influenced by the cultivar and geographic orientation | Portillo et al., 2016 |
| (ii) Intervineyard variations were greater than intravineyard variations | |||||
| Bacteria | Within region, 19 vineyards | Soil | Culture-independent method, 16S rRNA amplicon sequencing | (i) Soil bacterial communities were structured with respect to soil properties, location, geographic features, and management practices, e.g., conventional/organic/biodynamic systems | Burns et al., 2015; Burns et al., 2016 |
| (ii) High relative abundances of the majority of dominant taxa were found in soils with lower carbon or nitrogen contents | |||||
| Bacteria | Within region, five vineyards | Soil, roots, leaves, flowers/grapes | Culture-independent method, 16S rRNA amplicon sequencing | (i) Most grapevine OTUs originated in the soil | Zarraonaindia et al., 2015 |
| (ii) Soil-borne bacteria were selected by plants | |||||
| (iii) Microbial structure was influenced by edaphic factors, i.e., pH, C:N ration, soil carbon, etc. | |||||
| Fungi | Six regions | Vineyard soil, bark, juice and ferments, native forest soil and fruits | Culture-independent method, 26S rRNA amplicon sequencing | (i) Vineyard fungi accounted for ∼40% of the diversity in juice and ferments | Morrison-Whittle and Goddard, 2018 |
| (ii) The geographical diversification of must microbiome weakened during fermentation | |||||
| Fungi | Within region, 12 vineyards | Soil, bark, grapes, juice and ferments | Culture-independent method, 26S rRNA amplicon sequencing | Biodynamic practices significantly affected soil and grapevine-associated microbiome but not the harvest juice communities, nor on final wine quality | Morrison-Whittle and Goddard, 2017 |
| Fungi | Within vineyard | Grapes, must and ferments | Culture-independent method for fungi with 18S rRNA amplicon sequencing, culture-dependent method for yeasts | (i) Lower biodiversity of yeasts and fungal populations was measured in organically- than conventionally-farmed grapes and ferments | Grangeteau et al., 2017 |
| (ii) SO2 addition favoured the domination of S. cerevisiae during fermentation | |||||
| Yeasts | Within vineyard | Must and ferments | Culture-dependent method, D1/D2 26S sequencing | Prefermentative cold soak modified yeast dynamics in a temperature-dependent manner | Maturano et al., 2015 |
| Bacteria, fungi | Within vineyard | Must and ferments | Culture-independent method, 16S rRNA and ITS amplicon sequencing | SO2 treatment altered wine microbial diversity in a dose-dependent manner | Bokulich et al., 2015 |
Microbiota-Metabolome Geographic Patterns to Elucidate Wine Regionality
Geographic delineations of S. cerevisiae populations and cultivable yeasts were first reported in New Zealand vineyards, providing evidence for regional distribution of yeast populations (Table 1; Gayevskiy and Goddard, 2012). In the United States, Bokulich et al. (2014) used NGS of 16S rRNA and internal transcribed spacer (ITS) ribosomal sequence to demonstrate regionally structured bacterial and fungal consortia in grape musts, with some influences from cultivar and vintage (Table 1). These studies posit that microbial biogeography is a contributor to wine regionality expression. Further biogeographical correlations between wine-related microbiota and regional origins have been reported, holding for newly planted or older vineyards and are representative from vineyards planted around the world (Tofalo et al., 2014; Pinto et al., 2015; Wang et al., 2015; Bokulich et al., 2016; Capece et al., 2016; Garofalo et al., 2016; Marzano et al., 2016; Portillo et al., 2016; El Khoury et al., 2017; Mezzasalma et al., 2017, 2018; Singh et al., 2018; Vitulo et al., 2019). Notably, the microbial geographic diversification in the must weakens as fermentation processes, due to S. cerevisiae yeasts dominance breaks the community diversity (Morrison-Whittle and Goddard, 2018). In addition, S. cerevisiae can persist perennially in a particular vineyard or winery within one single region, thereby enabling wine style consistency between vintages (Börlin et al., 2016; Guzzon et al., 2018).
The geographic diversification observed in microbiota has been verified in wine chemical attributes (Knight et al., 2015; Bokulich et al., 2016). Knight et al. (2015) empirically showed that regionally differentiated S. cerevisiae populations drove different metabolites in the resultant wines (Table 1; Knight et al., 2015). Further, microbial patterns correlating with regional metabolite profiles were reported by Bokulich et al. (2016), and showed the importance of the fermentative yeasts (for example, S. cerevisiae, Hanseniaspora, Pichia) and LAB (Leuconostocaceae). These correlations between microbiome and metabolome can be used to predict wine compositions and styles with their microbial patterns and deserve future study. For example, exploration of non-Saccharomyces yeasts that enhance wine aroma complexity, the importance of microbial diversity in early fermentation, and the likelihood of significant interactions between yeasts and bacteria will held unpick mechanisms to explain the correlations described in the studies above.
Geospatial Scales Shape Geographic Diversification of Microbes
The biogeographic patterns of microbes in the wine environment are generally based on regional scales which are interpreted differently within each winegrowing country. This means that the notion of a “region” is not strictly defined and varies considerably. For example, a wine region can describe an association of vineyards spanning hundreds or even thousands of kilometres (Gayevskiy and Goddard, 2012; Bokulich et al., 2014; Taylor et al., 2014; Knight and Goddard, 2015) or be quite a small geographic area (Pinto et al., 2015). When comparing microbial communities in smaller geographical scales (at the scale of individual vineyards), the geographical patterns among populations are more evident for fungi than for bacteria (Bokulich et al., 2014; Miura et al., 2017). The leaf and grape fungal community dissimilarities between sampling sites increase as geographic distance increases (Miura et al., 2017).
Non-mobile yeasts require animals (insects and birds) to be transferred across regions (Figure 2; Francesca et al., 2012; Stefanini et al., 2012; Lam and Howell, 2015), animal vectors are one potential biotic factor shaping the yeast and fungal community dissimilarities. When the studied scale is a small area, more factors maybe involved in the uniqueness of a site. Grape varieties and clones exert marked impact on grape surface bacteria within vineyard. For example, Bacteroidetes, Chloroflexi, Acidobacteria, and Planctomycetes are clone- specific phyla alongside the prevalent phylum Cyanobacteria, Proteobacteria, and Firmicutes (Zhang et al., 2019). This is in contrast with findings by Bokulich et al. (2014) who showed that the cultivar-specific influence on microbial diversity is weak in larger scales. While Portillo et al. (2016) reported higher variability of bacteria between vineyards compared to intra-vineyard, Setati et al. (2012) demonstrated that greater intra-vineyard variation was evident than inter-vineyard based on fungal populations, including yeasts (Table 1). In these cases, microclimate may play a more important role in structuring fungal communities (details in the section “Microclimate”), but this supposition requires further empirical studies for confirmation.
FIGURE 2.
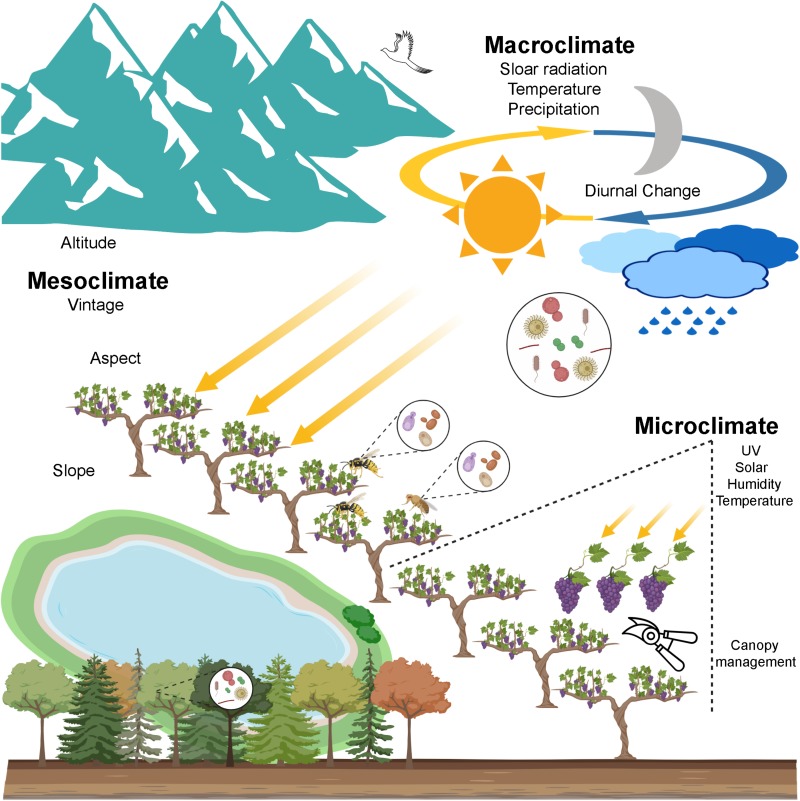
A scenario of wine microbial biogeography. Grapevine-associated microbiota originates from the local ecosystem encompassing soil, air, precipitation, native forests, etc. Genetic isolation is one driver of the geographic pattern that long distance decreases the gene flow that depends on physical forces and animal vectors (e.g., insects and birds). Climate is a profound environmental element shaping the microbial geographic pattern and thus affects wine quality. Macroclimate exerts influences on the regional pattern of bacteria and fungi. Mesoclimate at the vineyard scale shows weaker influences on the microbial distribution, especially for bacteria. Microclimate within the grapevine, modified by canopy management, may influence associated microbiota, this still remains to be shown (created with BioRender, https://app.biorender.com/).
Microbial biogeography is indelibly shaped by genetic isolation, as gene flow decreases with longer distances and depends on vectors (Martiny et al., 2006; Kuehne et al., 2007). Likewise, wine-related microbiota from relatively small scales are more similar to one another than those from a larger geographic areas (Martínez et al., 2007; Miura et al., 2017), although whether microbial patterns present a distance-dependant model is not clear. At the same time, adaptation to local environments influences how microbial ecology develops and diversifies (Martiny et al., 2006). For example, S. cerevisiae shows remarkable ability to adapt and thrive in human-associated food fermentative ecosystems (Legras et al., 2018). S. cerevisiae could be dispersed within, and successfully co-exist with other yeasts over a very small scale (10–200 m from the winery) (Valero et al., 2005). How physical environments modulate wine-related microbiota are affected by climate and weather, soil, and anthropogenic practices is considered in the following sections.
Sampling and Methodologies
It is noteworthy that a lack of standardised sampling strategies and analytical framework impedes comparisons among studies, hindering the global insight of microbial ecology in wine production. For example, the depth of soil, the volume size of soil and plant materials (roots, bark, leaves, flowers, grapes), as well as must conditions (grapes crushed under aseptic conditions or collected directly from commercial wineries), vary among studies and generate different datasets (Supplementary Table S1) (reviewed in Morgan et al., 2017). Standardised sampling procedures are indispensable to obtain sound data for microbial biogeography studies. Metagenomic methodologies, covering from DNA extraction, target genes for sequencing (e.g., 16S V4 region, ITS region) to bioinformatics pipelines (e.g., QIIME), can generate technical variation among individual studies [reviewed in Morgan et al. (2017) and Stefanini and Cavalieri (2018)]. The example set by the Earth Microbiome Project1 to standardise sampling and analysis could be a good solution to reduce the technical variation and help understand the contribution of biological variation, weather, climate and other general trends.
Climate Affects Wine Quality Via Microbiota
Climate, the long-term weather pattern of an area, is a profound element determining wine styles and regional characteristics and thus wine quality. Cooler climates are better suited to producing light and delicate wines, while warmer climates tend to shape heavy and rich flavour profiles. The climate impacts viticulture and wine quality through temperature, precipitation, and solar radiation (Van Leeuwen and Seguin, 2006). In viticulture, climatic influences are recognised at multiple geospatial scales. Macroclimate, or regional climate, is largely determined by latitude and altitude but also modified by moderating influences from water such as seas or large lakes (Figure 2; Van Leeuwen, 2010; White, 2015). The local climate of a particular vineyard is given as the mesoclimate, which is determined primarily by topography including altitude, aspect, and slope to impact upon wine quality and style (Figure 2; Gladstones, 1992; Van Leeuwen et al., 2004; Bramley et al., 2011). For example, vineyard orientation can affect the warmth and sunlight interception of grapevines, and steeper slopes benefit even more from this influence. At the smallest scale, microclimate, the temperature, humidity and solar variations within the canopy and between vines, may be affected in part by soil conditions and leaf shading and manipulated by canopy management (Figure 2; Smart and Robinson, 1991; Van Leeuwen and Seguin, 2006). Here, we highlight the microbial presence at these different scales to disentangle the intersection of microbes and climate and how this influences wine composition and style.
Macroclimate
Microorganisms are mainly distributed by physical forces, such as air and wind (Zhu et al., 2017). Incorporation into clouds and precipitation and into nearby ecosystems increases their long-distance dispersal (Hamilton and Lenton, 1998; Figure 2). These forces shape microbial biogeographic patterns in a similar way to those of plants and animals (Hamilton and Lenton, 1998; Zhu et al., 2017). Climate, as the most important environmental factor of grapegrowing regions, exerts influences on local microbial incidence and persistence in both space and time (described as the vintage effect). At the macroclimate scale, grape must microbiota present regional distribution patterns which are significantly conditioned by local environments (e.g., temperatures and rainfall) and weakly affected by the vintage, for example maximum temperature and average low temperature are negatively associated with Penicillium, Pseudomonas, Enterobacteriaceae, and Leuconostocaceae (O. oeni) (Bokulich et al., 2014). Rainfall and humidity positively correlate with yeast Hanseniaspora and Metschnikowia, and negatively correlate with Torulaspora, Saccharomyces and Meyerozyma (Jara et al., 2016). As many of these species (in particular Saccharomyces and O. oeni) are the most abundant species present in a wine fermentation, is can be said that local climatic conditions can shape wine compositions by affecting their presence as seen in these association studies.
Mesoclimate
At the mesoclimate level, or vineyard scale, consistency between vintages is one dimension of wine quality that is targeted for the terroir expression (Van Leeuwen et al., 2004; Beverland, 2006). Vintage variations substantially influence microbial communities in small geospatial scales (individual vineyards) rather than large scales (regions), with more stable fungal patterns observed between vintages (Bokulich et al., 2014). Vintage can significantly affect the biodiversity of yeast populations in the grape and must (Sabate et al., 2002; Vigentini et al., 2015). As a driver to shape mesoclimate, topographical features also exert influence on grape microbiota (Figure 2; Portillo et al., 2016). For example, Oxalobacteraceae, Haemophilus, Sphingomonas, and Pseudomonas were identified in grapes from east-facing vineyards, while Streptococcus, Micrococcaceae, Staphylococcus, Enhydrobacter, and Aeromonadaceae were shown as typical taxa of flat vineyards (Portillo et al., 2016).
Microclimate
It could be argued that microclimate is the environment most likely to affect the presence, growth and activity of microbes. Modifying the grapevine leaf area through training, pruning, trellising, and defoliation alters canopy microclimate, in particular the solar radiation onto grapes and leaves, and to a lesser extent, by air movement in the leaf canopy (strongly affecting humidity) and temperature (Figure 2; Reynolds and Heuvel, 2009). The effect of light in the microclimate environment have been reported to affect a wide range of aspects of berry composition, and can strongly affect the colour and flavours present in the final wine (Haselgrove et al., 2000; Liu et al., 2015). However, studies on the effects of microclimate variability on microbial communities are rare. Fungal communities can be discriminated within vineyards and this effect was ascribed to microclimate variability, but no experimentally test has validated this correlation (Setati et al., 2012). From other studies it is clear that sunlight interception could affect the grapevine microbiome. For example, an aquatic yeast study suggested that Cryptococcus spp. and pigmented yeasts Rhodotorula spp. predominate in certain environments as they can produce photoprotective compounds (carotenoid pigments and mycosporines) to adapt to pelagic sites with high UV radiation (Brandão et al., 2011). These yeasts are also associated with grapevines (Sabate et al., 2002; Setati et al., 2012), and so a similar response is possible in the fruit zone affect the composition of fungal communities. Whether microclimate can influence the wine-related microbiome and influence resulting wine compositions remains speculative and deserves further study.
The Influence of the Soil Borne Microbiome on Wine Composition and Quality
The soil substrate provides the grapevine with water and nutrients and soil type, composition and structure profoundly affects vine growth and development. Soil composition can affect the composition of wine, as wine can be related to its origin by tracking multiple major and trace elements from the soil to the wine (Almeida and Vasconcelos, 2003; Kment et al., 2005). Significant correlations were attributed to the movement of elements from the soil to grapes and resultant wines (Almeida and Vasconcelos, 2003). This contributes to our understanding of how soil geochemistry affects wine composition but has not been established mechanistically. An appreciation of the soil microbiota to vine health is relatively recent, and we review the knowledge in this area below.
Soil Borne Microorganisms in Association With Grapevines
Grapevines live in biogeochemically diverse soils harbouring diverse microbiota that affect plant health and growth in beneficial, commensal, or pathogenic ways (Müller et al., 2016). Soil-borne microbes can affect crop yields and metabolite synthesis in other agricultural systems, and thus may shape wine colour, aroma, flavour, and quality. Serving as a reservoir for plant-associated bacteria (Zarraonaindia et al., 2015), soil borne bacteria can colonise plant organs by physical contact or travel from the rhizosphere toward the phyllosphere on the surface (epiphytes) or within plants (endophytes) (Figure 1; Chi et al., 2005; Compant et al., 2005; Compant et al., 2011; Martins et al., 2013; Ruiz-Pérez et al., 2016). For example, plant-growth-promoting (PGP) bacteria Burkholderia sp. strain PsJN can endophytically colonise Chardonnay plantlets and translocate onto the root surfaces, root internal tissues, and finally the internode and leaf (Compant et al., 2005). Some dominant taxa in the soil, such as Gammaproteobacteria (including Pseudomonas spp.) and Firmicutes (including Bacillus spp.) have been visualised by fluorescence in situ hybridisation as endophytes inside the flower ovules, and berries and seeds of Zweigelt grapevines (Vitis vinifera) (Compant et al., 2011).
As well as bacteria, the vineyard soil is one of the natural sources of fungi in musts (Morrison-Whittle and Goddard, 2018). Notably, identical genotypes of S. cerevisiae are found shared between soil and fruit niches within regions (Knight and Goddard, 2015). A vineyard field experiment suggested that S. cerevisiae yeasts can be adsorbed from the soil by roots and transported via vine to stems and surface of grapes, and finally entered fermentation musts (Mandl et al., 2015). This translocation process can enable soil borne microorganisms be a part of grapevine-associated microbiome that influence resultant wine quality and characteristics directly; these microbes can survive fermentation and release secondary metabolites affecting wine profiles or interact with fermentative yeasts or bacteria within the community. Of particular note for this discussion is the root associated rhizobia which has not been extensively studied in grapevines. This community is likely to have a profound affect as it was observed in rice plants (Oryza sativa L.) that an ascending endophytic migration of rhizobia starting on the rhizoplane surface, within root tissues, to reach the stem base, leaf sheath, and leaves, and this process benefits rice growth physiology afterward (Chi et al., 2005).
Soil Properties and Interactions With Plants Shape Vineyard Microbiota
Biogeographic patterns have been observed in the microbial communities of vineyard soils (Table 1). Soil bacterial diversity and composition associate with the vineyard geographical location, with significant influences from soil physicochemical properties, such as soil texture, soil pH, temperature, moisture, carbon and nitrogen pools (Burns et al., 2015; Zarraonaindia et al., 2015). The majority of dominant taxa such as Proteobacteria (especially Beta- and Gamma-proteobacteria), Bacteroidetes, Gemmatimonadetes, and Firmicutes show higher relative abundance in soils with lower carbon or nitrogen contents, while abundance of Actinobacteria shows a negative trend (Burns et al., 2015). In addition, local climate (average annual precipitation) and topography (altitude, aspect, and slope) could indirectly influence soil microbial communities through their impacts on soil properties (Burns et al., 2015). Noticeably, fermentation-related bacteria in the musts are found in the vineyard soil as dominant taxa, such as Firmicutes (encompassing LAB) and spoilage bacteria Acidobacteria and Proteobacteria (Burns et al., 2015). As a reservoir of grapevine-associated bacteria, soil-borne microbes can shape phyllosphere bacterial assemblages, in particular grapes (Zarraonaindia et al., 2015). When the grapes are transferred into the winery and the microbiota persist in alcoholic fermentation, soil microbiota directly correlate with resulting wine metabolites.
Differentiation of vineyard soil bacterial community structure is reflected in the roots microbiome, with the relative abundance of several taxa occurring in a vineyard-depending manner. These taxa outcompete other bacteria for colonisation and/or are selected by grapevines. For example, PGP bacteria Rhizobiales, especially Bradyrhizobium spp., can benefit nitrogen fixation and antibiotic production that would promote plant growth, disease suppression and grape composition, thereby indirectly influencing wine quality and characteristics (Zarraonaindia et al., 2015). Within a vineyard, interactions between the root compartments (rhizosphere and root endosphere) and the rootstock can also exert a unique selective pressure enhancing niche differentiation of bacteria, in particular on the taxa with PGP potential, for example by producing indole acetic acid which affect plant hormone balances (Marasco et al., 2018).
Vineyard niches (soil, bark, fruit) account for approximately 40% of fungal populations in the musts and ferments. The clear similarity of fungi is between musts and grapes, that communities are dominated by fermenting yeasts (e.g., S. cerevisiae and Hanseniaspora uvarum) and filamentous fungi (Aureobasidium pullulans, Cladosporium spp.) (Morrison-Whittle and Goddard, 2018). While Saccharomyces spp. and wine spoilage species are mostly present in low numbers and in low frequencies in the soil, the most abundant genera (Amniculicola, Doratomyces, Endocarpon, and Tricellulortus) do not appear to be directly involved in wine production (Barata et al., 2012). In addition, regional delineations of fungal communities were observed in musts but not in the vineyard habitats at the regional scale (Morrison-Whittle and Goddard, 2018). Interestingly, significant differentiation of small-scale fungal communities was observed in a vineyard system (distance < 2 km), although with no vineyard demarcation between fermenting populations of S. cerevisiae (Knight et al., 2019). In-depth insight in fungal ecology in vineyards will inform how grapevines recruit fungal communities during the annual plant cycle and how this affects the communities present on the grape. For example, incidence of fermenting yeasts on grapes changes during ripening (Barata et al., 2012), and can thus contribute to wine metabolite profiles.
Anthropogenic Practices Affect Quality Wine Production by Microbial Modulation
The natural environment of the vineyard determines grape production and composition. Based on the local conditions, thoughtful human management can optimise wine quality and style across vintages (White, 2015). Wine producers select vineyard sites with favourable environmental conditions (especially climate and soil) for quality wine production, as well as grapevine cultivars that adapt to the local environment (Van Leeuwen and Seguin, 2006). The grapegrower manages the vine to achieve balance between vine vigour and yield and may use different training, pruning, trellising and canopy methodologies, to harvest a healthy crop, with a desired fruit composition and to optimise wine quality (Jackson and Lombard, 1993; Reynolds and Heuvel, 2009). Pesticides (including fungicides against downy mildew, powdery mildew and grey rot) and fertiliser application are common interventions in the vineyard, which are now embedded within viticulture management practices of conventional, organic and biodynamic systems. Some evidence exists to differentiate the superiority of microbial diversity of organic or biodynamic wines with respect to specific management practice (Ross et al., 2009; Pagliarini et al., 2013), yet spontaneous fermentation and related winemaking techniques (for example cold soak, limited SO2 usage) are thought to encourage showing the indigenous microbial diversity, and thus enhance wine terroir expression (Capozzi et al., 2015). Recent studies focusing on the wine-related microbiome offer new information to this area and provide more information to allow expression of local individuality for viticultural production.
Viticultural Practices
Specific human interventions including pesticides, fungicides, and herbicides usage can affect microbial diversity in specific habitats in vineyards (Čadež et al., 2010; Fierer et al., 2012a; Perazzolli et al., 2014; Pinto et al., 2014; Chou et al., 2018). Here we highlight the effect of viticulture practices on the vineyard microbiology, and whether these microbes and their effects can persist into wine production to ultimately influence wine quality.
Viticulture practices can modify the belowground microbiome. Soil borne bacterial communities are structured with respect to commercial/organic/biodynamic systems, as mediated by shifts in soil resource pools, particularly carbon and nitrogen (Burns et al., 2016). Compost addition in organic and biodynamic vineyards increases overall bacterial diversity and alter the community composition, and can the effect can be enhanced by tillage and cover crop management. Similarly, soil fungal diversity and community composition of biodynamic vineyards differ from those which are conventionally managed (Morrison-Whittle and Goddard, 2017; Table 1). Biodynamic management enhance fungal diversity in the grapevines niches (bark, fruits) but this effect did not persist into the harvested juice and fermentation (Morrison-Whittle and Goddard, 2017).
The phyllosphere microbiota can be shaped by agricultural management, in particular fungicide usage (Gilbert et al., 2014). Conventional vineyards are usually treated with several agricultural chemicals, while organic/biodynamic vineyards only recieve sulphur- and/or copper-based formulations. Several studies have shown a higher microbial diversity in grapes present in organic and biodynamic vineyards for both yeasts (Saccharomyces and non-Saccharomyces) and total fungi (yeasts and filamentous fungi) (Setati et al., 2012; Martins et al., 2014; Setati et al., 2015), and this could be because chemical treatments reduce microbial richness and diversity associated with grapevines and wine (Pinto et al., 2014; Escribano-Viana et al., 2018). This effect can maintain in spontaneous fermentations from organic/biodynamic musts where higher yeast species richness and diversity is observed compared to musts from conventionally managed vineyards, and this is particularly true for fermentative yeasts species such as H. uvarum, H. vineae, H. guilliermondii, Streptomyces bacillaris, Lachancea thermotolerans, and S. cerevisiae (Cordero-Bueso et al., 2011; Bagheri et al., 2015). Regarding grapevine-associated bacteria, responses to viticulture practices are weak compared to fungi, especially grape surface bacteria showing more resilience than that of leaves (Schmid et al., 2011; Miura et al., 2017). Biodynamic berries are found rich in Bacillales including genera of Lysinibacillus, Bacillus, and Sporosarcin, which are typical microbes in manure (Mezzasalma et al., 2017), but their influences on wine composition are not clear. Heavy use of sulphur and copper fungicides decrease the biodiversity of yeasts and fungi in resultant fermentations (Milanoviæ et al., 2013; Grangeteau et al., 2017). Particular fungi are associated with differently managed vineyards, where Basidiomycota (especially Cryptococcus) is mainly associated with organic vineyards, while fermentative yeasts Saccharomyces, Metschnikowia, and Hanseniaspora were mainly found in conventionally managed vineyards (Grangeteau et al., 2017; Table 1). The yeast-like fungus Aureobasidium pullulans dominate phyllosphere fungi in organic/biodynamic vineyards (Schmid et al., 2011; Pancher et al., 2012; Setati et al., 2012; Martins et al., 2014; Setati et al., 2015), but tends to be present only in the early stages of fermentation. Some debate exists, as Kecskeméti et al. (2016) claimed conventional/organic/biodynamic practices in the same vineyard do not significantly influence grape microbial diversity. Inconsistency is not surprising, as local vineyard conditions, specific management practices and sampling methods differ in the surveys published.
Management of Microbes During Winemaking
The wine industry has developed a series of methods to promote wine fermentations, of which the most effective methods are the inoculation of cultured S. cerevisiae strains and use of sulphur dioxide (SO2). Commercial fermentations are widely used to reduce the risk of spoilage and unpredictable wine composition, and to ensure a stable wine flavour. However, inoculated fermentations reduce the potential of microbiota to contribute to regional characteristics. Spontaneous fermentations, comprising a diversity of yeast species and S. cerevisiae strains originating from vineyard and winery, enhance wine regional expression (Capozzi et al., 2015). The diversity of yeast species present can have profound impacts on the flavour of the resultant wine. For example, non-Saccharomyces yeasts can produce and secrete several enzymes (esterases, β-glucosidases, proteases), to synthesise volatile compounds and playing a role in varietal aroma (Romano et al., 2003). H. uvarum can positively interact with S. cerevisiae to enhance fermentation (Romano et al., 2003). Interactions within S. cerevisiae populations provide regional microbial signatures positively correlating with wine aroma profiles (Knight et al., 2015). Pre-fermentative cold soak, a technique widely used in red wine production to favour wine colour, taste and mouthfeel attributes, can influence yeast population dynamics depending the temperature. For example, a cold soak at 14 ± 1°C can increases total yeast populations and favour growth of H. uvarum and Candida zemplinina, whereas cold soak at 8 ± 1°C favours growth of S. cerevisiae (Maturano et al., 2015; Table 1). SO2 treatment favours the early implantation and domination of S. cerevisiae and alters wine microbial diversity and fermentation progression in a dose-dependent manner (Bokulich et al., 2015; Grangeteau et al., 2017; Table 1). A concentration of 25 mg/L SO2 is ideal to stabilise the microbial communities, which inhibits the growth of LAB and Gluconobacter but not other bacteria and fungi at early fermentation, thereby maximising microbial diversity to benefit wine regionality expression. Much less “manipulating the terroir,” production of distinctive and quality wine is achievable through microbial manipulation in the winery.
How Will Climate Change Affect the Incidence and Activity of Microbes Involved in Winemaking?
Human activity has dramatically affected the global climate and the weather patterns that grapevines experience (Olesen et al., 2011). Wine production is particularly sensitive to climate change as there is an inherent link between climate and wine quality and style. Matching grape varieties to a unique combination of climate and soil enables production of distinctive wines worldwide (Van Leeuwen and Seguin, 2006), but this is likely to change as the temperature and water profiles of specific areas change. Trends in grapevine phenology associated with global warming are widely reported, and report earlier maturation with undesirable influences on grape and wine aroma and flavour (Palliotti et al., 2014). For example, wine grapes have been ripening earlier in Australia in recent decades, driven by warming and declines in soil moisture (Webb et al., 2012). A deficit of water will affect production of colourful and flavoursome wines rich in phenolic substances in a warmer and drier future (Bonada et al., 2015). In the long term, climate change will affect the geographical distribution of viticulture and will require changing varieties grown to adapt to warming or providing artificial shading to reduce temperatures (Jones et al., 2005; Webb et al., 2012), whereas in the short term, management techniques may be able to mitigate negative impacts and include managing soil water content (irrigation), crop yield (pruning regime), and vine response (rootstock selection, leaf removal) (Webb et al., 2012).
Microorganisms involved in winemaking are not discussed in the context of climate change. Increasing temperature and drought would strongly affect the ability to grow grapes and wine production, but in largely unknown ways. One area where these effects will be felt is in the soil and its microbial composition and activity. Studies outside of agriculture show that soil pH, moisture, temperature and nutrient availability are the main drivers of microbial community assembly (Fierer et al., 2012b) and that soil fungi and bacteria occupy specific niches and respond differently to precipitation and soil pH, and therefore would respond differently to climate change on their diversity, abundance, and function potential (Bahram et al., 2018). For example, fungal networks are more stable under drought conditions than bacteria in a grassland ecosystem (de Vries et al., 2018). Warming increases soil bacterial populations but decreases diversity and changes the composition (Sheik et al., 2011). Warming also increases soil respiration (Luo et al., 2001), which can in turn reduce the abundance of Actinobateria as sensitive to high carbon dioxide production (Sheik et al., 2011). Soil water saturation affects microbial composition by changing oxygen availability and thus microbial respiration (Carbone et al., 2011). Drought can also increase the resistance of soil bacteria (Bouskill et al., 2013), for example Actinobateria, which is tolerant of low moisture conditions (Fierer et al., 2012b). While these studies have been performed outside vineyards, it is likely that the same overall principals are at play. We suggest that superimposing warming and drying mechanisms will affect vineyard soil microbiota abundance, diversity and functions, and thus change their capacity to support plant growth. Further studies are needed, but it is clear that understanding wine microbial biogeography under the changing climate will help wine industry to adapt to climate change and enable quality wine production in the future.
Conclusion and Future Perspectives
Fundamental questions about how wine quality and distinctiveness can be derived from local environments have been very difficult to answer. Here, we present microbial biogeography as core to understanding wine regionality. As the driver of wine fermentation, microorganisms inhabit and adapt to local geography, climate, soil and anthropogenic practices. The climate shapes microbial geographic diversification at multiple scales thereby affecting wine compositions. Soil-borne microbiota shapes the grapevine-associated microbiota and physiology and ultimately the flavour of resulting wines, but the mechanism by which this occurs remains to be elucidated. Human management practices modify wine-related microbiota to improve quality wine production. However, our knowledge of how geographically diverse microbiota shape wine chemical and organoleptic characteristics is limited. In-depth insights have emerged from studies of environment-plant-microbe interactions and will inform us about how grapevines recruit their microbiome to maximise both nutrition and microbial diversity under local conditions. The grapevine microbiota can then be sensibly exploited in wine production by introducing and/or managing specific microbes in combination with optimised wine metabolites to sculpt fermentation consortia for quality and distinctive wine production. Anthropogenic climate change will have profound consequences on wine-associated microbiota and thus affect wine quality and style. We believe that carefully designed empirical experiments to unpick microbial ecology and wine metabolome, surveys, and international and interdisciplinary collaborations will be indispensable to obtain a comprehensive understanding of climate change and the future of wine industry.
Author Contributions
DL wrote the first draft of the review. KH revised and added to the draft. All authors approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. DL acknowledges support from a Ph.D. scholarship and funding from Wine Australia (AGW Ph1602) and a Melbourne Research Scholarship from the University of Melbourne.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02679/full#supplementary-material
References
- Almeida C. M. R., Vasconcelos M. T. S. (2003). Multielement composition of wines and their precursors including provenance soil and their potentialities as fingerprints of wine origin. J. Agric. Food Chem. 51 4788–4798. 10.1021/jf034145b [DOI] [PubMed] [Google Scholar]
- Bagheri B., Bauer F., Setati M. (2015). The diversity and dynamics of indigenous yeast communities in grape must from vineyards employing different agronomic practices and their influence on wine fermentation. South Afr. J. Enol. Vitic. 36 243–251. [Google Scholar]
- Bahram M., Hildebrand F., Forslund S. K., Anderson J. L., Soudzilovskaia N. A., Bodegom P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560 233–237. 10.1038/s41586-018-0386-6 [DOI] [PubMed] [Google Scholar]
- Barata A., Malfeito-Ferreira M., Loureiro V. (2012). The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153 243–259. 10.1016/j.ijfoodmicro.2011.11.025 [DOI] [PubMed] [Google Scholar]
- Beverland M. (2006). The ‘real thing’: branding authenticity in the luxury wine trade. J. Bus. Res. 59 251–258. 10.1016/j.jbusres.2005.04.007 [DOI] [Google Scholar]
- Bokulich N. A., Collins T. S., Masarweh C., Allen G., Heymann H., Ebeler S. E., et al. (2016). Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7:e00631-16. 10.1128/mBio.00631-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Swadener M., Sakamoto K., Mills D. A., Bisson L. F. (2015). Sulfur dioxide treatment alters wine microbial diversity and fermentation progression in a dose-dependent fashion. Am. J. Enol. Vitic. 66 73–79. 10.5344/ajev.2014.14096 [DOI] [Google Scholar]
- Bokulich N. A., Thorngate J. H., Richardson P. M., Mills D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111 E139–E148. 10.1073/pnas.1317377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonada M., Jeffery D., Petrie P. R., Moran M. A., Sadras V. O. (2015). Impact of elevated temperature and water deficit on the chemical and sensory profiles of B arossa S hiraz grapes and wines. Aust. J. Grape Wine Res. 21 240–253. 10.1111/ajgw.12142 [DOI] [Google Scholar]
- Börlin M., Venet P., Claisse O., Salin F., Legras J.-L., Masneuf-Pomarede I. (2016). Cellar-associated Saccharomyces cerevisiae population structure revealed high diversity and perennial persistence in Sauternes wine estates. Appl. Environ. Microbiol. 82 2909–2918. 10.1128/AEM.03627-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouskill N. J., Lim H. C., Borglin S., Salve R., Wood T. E., Silver W. L., et al. (2013). Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J. 7 384–394. 10.1038/ismej.2012.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley R., Ouzman J., Boss P. K. (2011). Variation in vine vigour, grape yield and vineyard soils and topography as indicators of variation in the chemical composition of grapes, wine and wine sensory attributes. Aust. J. Grape Wine Res. 17 217–229. 10.1111/j.1755-0238.2011.00136.x [DOI] [Google Scholar]
- Brandão L. R., Vaz A. B. M., Rosa C. A., Espírito Santo L. C., Van Broock M., Moliné M., et al. (2011). Yeasts from an oligotrophic lake in Patagonia (Argentina): diversity, distribution and synthesis of photoprotective compounds and extracellular enzymes. FEMS Microbiol. Ecol. 76 1–13. 10.1111/j.1574-6941.2010.01030.x [DOI] [PubMed] [Google Scholar]
- Burns K. N., Bokulich N. A., Cantu D., Greenhut R. F., Kluepfel D. A., O’geen A. T., et al. (2016). Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by vineyard management. Soil Biol. Biochem. 103 337–348. 10.1016/j.soilbio.2016.09.007 [DOI] [Google Scholar]
- Burns K. N., Kluepfel D. A., Strauss S. L., Bokulich N. A., Cantu D., Steenwerth K. L. (2015). Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by geographic features. Soil Biol. Biochem. 91 232–247. 10.1016/j.soilbio.2015.09.002 [DOI] [Google Scholar]
- Čadež N., Zupan J., Raspor P. (2010). The effect of fungicides on yeast communities associated with grape berries. FEMS Yeast Res. 10 619–630. 10.1111/j.1567-1364.2010.00635.x [DOI] [PubMed] [Google Scholar]
- Capece A., Granchi L., Guerrini S., Mangani S., Romaniello R., Vincenzini M., et al. (2016). Diversity of Saccharomyces cerevisiae strains isolated from two Italian wine-producing regions. Front. Microbiol. 7:1018. 10.3389/fmicb.2016.01018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi V., Garofalo C., Chiriatti M. A., Grieco F., Spano G. (2015). Microbial terroir and food innovation: the case of yeast biodiversity in wine. Microbiol. Res. 181 75–83. 10.1016/j.micres.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Carbone M. S., Still C. J., Ambrose A. R., Dawson T. E., Williams A. P., Boot C. M., et al. (2011). Seasonal and episodic moisture controls on plant and microbial contributions to soil respiration. Oecologia 167 265–278. 10.1007/s00442-011-1975-3 [DOI] [PubMed] [Google Scholar]
- Chi F., Shen S.-H., Cheng H.-P., Jing Y.-X., Yanni Y. G., Dazzo F. B. (2005). Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 71 7271–7278. 10.1128/aem.71.11.7271-7278.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.-Y., Heuvel J. V., Bell T. H., Panke-Buisse K., Kao-Kniffin J. (2018). Vineyard under-vine floor management alters soil microbial composition, while the fruit microbiome shows no corresponding shifts. Sci. Rep. 8:11039. 10.1038/s41598-018-29346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Mitter B., Colli-Mull J. G., Gangl H., Sessitsch A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62 188–197. 10.1007/s00248-011-9883-y [DOI] [PubMed] [Google Scholar]
- Compant S., Reiter B., Sessitsch A., Nowak J., Clément C., Barka E. A. (2005). Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71 1685–1693. 10.1128/aem.71.4.1685-1693.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Bueso G., Arroyo T., Serrano A., Tello J., Aporta I., Vélez M. D., et al. (2011). Influence of the farming system and vine variety on yeast communities associated with grape berries. Int. J. Food Microbiol. 145 132–139. 10.1016/j.ijfoodmicro.2010.11.040 [DOI] [PubMed] [Google Scholar]
- de Vries F. T., Griffiths R. I., Bailey M., Craig H., Girlanda M., Gweon H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:3033. 10.1038/s41467-018-05516-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury M., Campbell-Sills H., Salin F., Guichoux E., Claisse O., Lucas P. M. (2017). Biogeography of oenococcus oeni reveals distinctive but nonspecific populations in wine-producing regions. Appl. Environ. Microbiol. 83:e02322-16. 10.1128/AEM.02322-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Viana R., López-Alfaro I., López R., Santamaría P., Gutiérrez A. R., González-Arenzana L. (2018). Impact of chemical and biological fungicides applied to grapevine on grape biofilm, must, and wine microbial diversity. Front. Microbiol. 9:59. 10.3389/fmicb.2018.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Lauber C. L., Ramirez K. S., Zaneveld J., Bradford M. A., Knight R. (2012a). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6 1007–1017. 10.1038/ismej.2011.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Leff J. W., Adams B. J., Nielsen U. N., Bates S. T., Lauber C. L., et al. (2012b). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. 109 21390–21395. 10.1073/pnas.1215210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesca N., Canale D. E., Settanni L., Moschetti G. (2012). Dissemination of wine-related yeasts by migratory birds. Environ. Microbiol. Rep. 4 105–112. 10.1111/j.1758-2229.2011.00310.x [DOI] [PubMed] [Google Scholar]
- Garofalo C., Russo P., Beneduce L., Massa S., Spano G., Capozzi V. (2016). Non-Saccharomyces biodiversity in wine and the ‘microbial terroir’: a survey on Nero di Troia wine from the Apulian region Italy. Ann. Microbiol. 66 143–150. 10.1007/s13213-015-1090-5 [DOI] [Google Scholar]
- Gayevskiy V., Goddard M. R. (2012). Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 6 1281–1290. 10.1038/ismej.2011.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Van Der Lelie D., Zarraonaindia I. (2014). Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. U.S.A. 111 5–6. 10.1073/pnas.1320471110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstones J. (1992). Viticulture and Environment. Broadview, SA: Winetitles. [Google Scholar]
- Gladstones J. (2011). Wine, Terroir and Climate Change. Adelaide, SA: Wakefield Press. [Google Scholar]
- Goddard M. R., Anfang N., Tang R., Gardner R. C., Jun C. (2010). A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ. Microbiol. 12 63–73. 10.1111/j.1462-2920.2009.02035.x [DOI] [PubMed] [Google Scholar]
- Gonzálvez A., Llorens A., Cervera M., Armenta S., De La Guardia M. (2009). Elemental fingerprint of wines from the protected designation of origin Valencia. Food Chem. 112 26–34. 10.1016/j.foodchem.2008.05.043 [DOI] [Google Scholar]
- Grangeteau C., Roullier-Gall C., Rousseaux S., Gougeon R. D., Schmitt-Kopplin P., Alexandre H., et al. (2017). Wine microbiology is driven by vineyard and winery anthropogenic factors. Microb. Biotechnol. 10 354–370. 10.1111/1751-7915.12428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzon R., Labagnara T., Toffanin A., Villegas T. R. (2018). Oenological characterisation of indigenous strains of S. cerevisiae isolated in a biodynamic winery in the Cortona DOC area. Ann. Microbiol. 68 963–967. 10.1007/s13213-018-1405-4 [DOI] [Google Scholar]
- Hamilton W. D., Lenton T. (1998). Spora and Gaia: how microbes fly with their clouds. Ethol. Ecol. Evol. 10 1–16. 10.1080/08927014.1998.9522867 [DOI] [Google Scholar]
- Haselgrove L., Botting D., Van Heeswijck R., Høj P., Dry P., Ford C., et al. (2000). Canopy microclimate and berry composition: the effect of bunch exposure on the phenolic composition of Vitis vinifera L cv. Shiraz grape berries. Aust. J. Grape Wine Res. 6 141–149. 10.1111/j.1755-0238.2000.tb00173.x [DOI] [Google Scholar]
- Jackson D., Lombard P. (1993). Environmental and management practices affecting grape composition and wine quality-a review. Am. J. Enol. Vitic. 44 409–430. [Google Scholar]
- Jara C., Laurie V. F., Mas A., Romero J. (2016). Microbial terroir in chilean valleys: diversity of non-conventional yeast. Front. Microbiol. 7:663. 10.3389/fmicb.2016.00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. V., White M. A., Cooper O. R., Storchmann K. (2005). Climate change and global wine quality. Clim. Change 73 319–343. 10.1007/s10584-005-4704-2 [DOI] [Google Scholar]
- Kecskeméti E., Berkelmann-Löhnertz B., Reineke A. (2016). Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS One 11:e0160852. 10.1371/journal.pone [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kment P., Mihaljeviè M., Ettler V., Šebek O., Strnad L., Rohlová L. (2005). Differentiation of Czech wines using multielement composition–A comparison with vineyard soil. Food Chem. 91 157–165. 10.1016/j.foodchem.2004.06.010 [DOI] [Google Scholar]
- Knight S., Goddard M. (2015). Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J. 9 361–370. 10.1038/ismej.2014.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S. J., Karon O., Goddard M. R. (2019). Small scale fungal community differentiation in a vineyard system. Food Microbiol. 10.1016/j.fm.2019.103358 [DOI] [PubMed] [Google Scholar]
- Knight S., Klaere S., Fedrizzi B., Goddard M. R. (2015). Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci. Rep. 5:14233. 10.1038/srep14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne H. A., Murphy H. A., Francis C. A., Sniegowski P. D. (2007). Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Curr. Biol. 17 407–411. 10.1016/j.cub.2006.12.047 [DOI] [PubMed] [Google Scholar]
- Lam S. S., Howell K. S. (2015). Drosophila-associated yeast species in vineyard ecosystems. FEMS Microbiol. Lett. 362:fnv170. 10.1093/femsle/fnv170 [DOI] [PubMed] [Google Scholar]
- Legras J.-L., Galeote V., Bigey F., Camarasa C., Marsit S., Nidelet T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35 1712–1727. 10.1093/molbev/msy066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Gao Y., Li X.-X., Li Z., Pan Q.-H. (2015). Attenuated UV radiation alters volatile profile in cabernet sauvignon grapes under field conditions. Molecules 20 16946–16969. 10.3390/molecules200916946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Wan S., Hui D., Wallace L. L. (2001). Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413 622–625. 10.1038/35098065 [DOI] [PubMed] [Google Scholar]
- Mandl K., Schieck J., Silhavy-Richter K., Alexander P., Schneider V., Schmidt H.-P. (2015). Vines take up yeasts from soil and transport them through the vine to the stem and skins of grapes. Ithaka J. 349–355. [Google Scholar]
- Marasco R., Rolli E., Fusi M., Michoud G., Daffonchio D. (2018). Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6:3. 10.1186/s40168-017-0391-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C., Cosgaya P., Vásquez C., Gac S., Ganga A. (2007). High degree of correlation between molecular polymorphism and geographic origin of wine yeast strains. J. Appl. Microbiol. 103 2185–2195. 10.1111/j.1365-2672.2007.03493.x [DOI] [PubMed] [Google Scholar]
- Martins G., Lauga B., Miot-Sertier C., Mercier A., Lonvaud A., Soulas M.-L., et al. (2013). Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLoS One 8:e73013. 10.1371/journal.pone.0073013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G., Vallance J., Mercier A., Albertin W., Stamatopoulos P., Rey P., et al. (2014). Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int. J. Food Microbiol. 177 21–28. 10.1016/j.ijfoodmicro.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Martiny J. B. H., Bohannan B. J., Brown J. H., Colwell R. K., Fuhrman J. A., Green J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4 102–112. 10.1038/nrmicro1341 [DOI] [PubMed] [Google Scholar]
- Marzano M., Fosso B., Manzari C., Grieco F., Intranuovo M., Cozzi G., et al. (2016). Complexity and dynamics of the winemaking bacterial communities in berries, musts, and wines from apulian grape cultivars through time and space. PLoS One 11:e0157383. 10.1371/journal.pone.0157383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturano Y. P., Mestre M. V., Esteve-Zarzoso B., Nally M. C., Lerena M. C., Toro M. E., et al. (2015). Yeast population dynamics during prefermentative cold soak of cabernet sauvignon and malbec wines. Int. J. Food Microbiol. 199 23–32. 10.1016/j.ijfoodmicro.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Mezzasalma V., Sandionigi A., Bruni I., Bruno A., Lovicu G., Casiraghi M., et al. (2017). Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS One 12:e0184615. 10.1371/journal.pone.0184615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzasalma V., Sandionigi A., Guzzetti L., Galimberti A., Grando M. S., Tardaguila J., et al. (2018). Geographical and cultivar features differentiate grape microbiota in Northern Italy and Spain vineyards. Front. Microbiol. 9:946. 10.3389/fmicb.2018.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanoviæ V., Comitini F., Ciani M. (2013). Grape berry yeast communities: influence of fungicide treatments. Int. J. Food Microbiol. 161 240–246. 10.1016/j.ijfoodmicro.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Miura T., Sánchez R., Castañeda L. E., Godoy K., Barbosa O. (2017). Is microbial terroir related to geographic distance between vineyards? Environ. Microbiol. Rep. 9 742–749. 10.1111/1758-2229.12589 [DOI] [PubMed] [Google Scholar]
- Morgan H. H., Du Toit M., Setati M. E. (2017). The grapevine and wine microbiome: insights from high-throughput amplicon sequencing. Front. Microbiol. 8:820. 10.3389/fmicb.2017.00820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison-Whittle P., Goddard M. (2017). Fungal communities are differentially affected by conventional and biodynamic agricultural management approaches in vineyard ecosystems. Agric. Ecosyst. Environ. 246 306–313. 10.1016/j.agee.2017.05.022 [DOI] [Google Scholar]
- Morrison-Whittle P., Goddard M. R. (2018). From vineyard to winery: a source map of microbial diversity driving wine fermentation. Environ. Microbiol. 20 75–84. 10.1111/1462-2920.13960 [DOI] [PubMed] [Google Scholar]
- Müller D. B., Vogel C., Bai Y., Vorholt J. A. (2016). The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50 211–234. 10.1146/annurev-genet-120215-034952 [DOI] [PubMed] [Google Scholar]
- Olesen J. E., Trnka M., Kersebaum K. C., Skjelvåg A., Seguin B., Peltonen-Sainio P., et al. (2011). Impacts and adaptation of European crop production systems to climate change. Eur. J. Agron. 34 96–112. [Google Scholar]
- Pagliarini E., Laureati M., Gaeta D. (2013). Sensory descriptors, hedonic perception and consumer’s attitudes to Sangiovese red wine deriving from organically and conventionally grown grapes. Front. Psychol. 4:896. 10.3389/fpsyg.2013.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palliotti A., Tombesi S., Silvestroni O., Lanari V., Gatti M., Poni S. (2014). Changes in vineyard establishment and canopy management urged by earlier climate-related grape ripening: a review. Sci. Hortic. 178 43–54. 10.1016/j.scienta.2014.07.039 [DOI] [Google Scholar]
- Pancher M., Ceol M., Corneo P. E., Longa C. M. O., Yousaf S., Pertot I., et al. (2012). Fungal endophytic communities in grapevines (Vitis vinifera L.) respond to crop management. Appl. Environ. Microbiol. 78 4308–4317. 10.1128/AEM.07655-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzolli M., Antonielli L., Storari M., Puopolo G., Pancher M., Giovannini O., et al. (2014). Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides. Appl. Environ. Microbiol. 80 3585–3596. 10.1128/aem.00415-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G. E., Gaudillere J.-P., Van Leeuwen C., Hilbert G., Lavialle O., Maucourt M., et al. (2005). 1H NMR and chemometrics to characterize mature grape berries in four wine-growing areas in Bordeaux, France. J. Agric. Food Chem. 53 6382–6389. 10.1021/jf058058q [DOI] [PubMed] [Google Scholar]
- Pinto C., Pinho D., Cardoso R., Custódio V., Fernandes J., Sousa S., et al. (2015). Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front. Microbiol. 6:905. 10.3389/fmicb.2015.00905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto C., Pinho D., Sousa S., Pinheiro M., Egas C., Gomes A. C. (2014). Unravelling the diversity of grapevine microbiome. PLoS One 9:e85622. 10.1371/journal.pone.0085622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo M. D. C., Franquès J., Araque I., Reguant C., Bordons A. (2016). Bacterial diversity of grenache and carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 219 56–63. 10.1016/j.ijfoodmicro.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Reynolds A. G., Heuvel J. E. V. (2009). Influence of grapevine training systems on vine growth and fruit composition: a review. Am. J. Enol. Vitic. 60 251–268. [Google Scholar]
- Robinson A. L., Adams D. O., Boss P. K., Heymann H., Solomon P. S., Trengove R. D. (2012). Influence of geographic origin on the sensory characteristics and wine composition of Vitis vinifera cv. Cabernet Sauvignon wines from Australia. Am. J. Enol. Vitic. 63 467–476. 10.5344/ajev.2012.12023 [DOI] [Google Scholar]
- Romano P., Fiore C., Paraggio M., Caruso M., Capece A. (2003). Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 86 169–180. 10.1016/s0168-1605(03)00290-3 [DOI] [PubMed] [Google Scholar]
- Ross C. F., Weller K. M., Blue R. B., Reganold J. P. (2009). Difference testing of merlot produced from biodynamically and organically grown wine grapes. J. Wine Res. 20 85–94. 10.1080/09571260903169423 [DOI] [Google Scholar]
- Ruiz-Pérez C. A., Restrepo S., Zambrano M. M. (2016). Microbial and functional diversity within the phyllosphere of Espeletia species in an Andean high-mountain ecosystem. Appl. Environ. Microbiol. 82 1807–1817. 10.1128/AEM.02781-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabate J., Cano J., Esteve-Zarzoso B., Guillamón J. M. (2002). Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol. Res. 157 267–274. 10.1078/0944-5013-00163 [DOI] [PubMed] [Google Scholar]
- Schmid F., Moser G., Müller H., Berg G. (2011). Functional and structural microbial diversity in organic and conventional viticulture: organic farming benefits natural biocontrol agents. Appl. Environ. Microbiol. 77 2188–2191. 10.1128/AEM.02187-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setati M. E., Jacobson D., Andong U.-C., Bauer F. (2012). The vineyard yeast microbiome, a mixed model microbial map. PLoS One 7:e52609. 10.1371/journal.pone.0052609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setati M. E., Jacobson D., Bauer F. F. (2015). Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Microbiol. 6:1358. 10.3389/fmicb.2015.01358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik C. S., Beasley W. H., Elshahed M. S., Zhou X., Luo Y., Krumholz L. R. (2011). Effect of warming and drought on grassland microbial communities. ISME J. 5 1692–1700. 10.1038/ismej.2011.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Santoni S., This P., Péros J.-P. (2018). Genotype-environment interaction shapes the microbial assemblage in grapevine’s phyllosphere and carposphere: an NGS approach. Microorganisms 6:E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R., Robinson M. (1991). Sunlight Into Wine: a Handbook for Winegrape Canopy Management. Broadview, SA: Winetitles. [Google Scholar]
- Stefanini I., Cavalieri D. (2018). Metagenomic approaches to investigate the contribution of the vineyard environment to the quality of wine fermentation: potentials and difficulties. Front. Microbiol. 9:991. 10.3389/fmicb.2018.00991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini I., Dapporto L., Legras J.-L., Calabretta A., Di Paola M., De Filippo C., et al. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U.S.A. 109 13398–13403. 10.1073/pnas.1208362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiegers J., Bartowsky E., Henschke P., Pretorius I. (2005). Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11 139–173. 10.1111/j.1755-0238.2005.tb00285.x [DOI] [Google Scholar]
- Taylor M. W., Tsai P., Anfang N., Ross H. A., Goddard M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16 2848–2858. 10.1111/1462-2920.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofalo R., Perpetuini G., Fasoli G., Schirone M., Corsetti A., Suzzi G. (2014). Biodiversity study of wine yeasts belonging to the “terroir” of Montepulciano d’Abruzzo “Colline Teramane” revealed Saccharomyces cerevisiae strains exhibiting atypical and unique 5.8 S-ITS restriction patterns. Food Microbiol. 39 7–12. 10.1016/j.fm.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Valero E., Schuller D., Cambon B., Casal M., Dequin S. (2005). Dissemination and survival of commercial wine yeast in the vineyard: a large-scale, three-years study. FEMS Yeast Res. 5 959–969. 10.1016/j.femsyr.2005.04.007 [DOI] [PubMed] [Google Scholar]
- Van Leeuwen C. (2010). Terroir: the effect of the physical environment on vine growth, grape ripening and wine sensory attributes. Manag. Wine Qual. 1 273–315. 10.1533/9781845699284.3.273 [DOI] [Google Scholar]
- Van Leeuwen C., Friant P., Chone X., Tregoat O., Koundouras S., Dubourdieu D. (2004). Influence of climate, soil, and cultivar on terroir. Am. J. Enol. Vitic. 55 207–217. [Google Scholar]
- Van Leeuwen C., Seguin G. (2006). The concept of terroir in viticulture. J. Wine Res. 17 1–10. 10.1080/09571260600633135 [DOI] [Google Scholar]
- Vaudour E., Costantini E., Jones G., Mocali S. (2015). An overview of the recent approaches to terroir functional modelling, footprinting and zoning. Soil 1 287–312. 10.5194/soil-1-287-2015 [DOI] [Google Scholar]
- Vigentini I., De Lorenzis G., Fabrizio V., Valdetara F., Faccincani M., Panont C. A., et al. (2015). The vintage effect overcomes the terroir effect: a three year survey on the wine yeast biodiversity in Franciacorta and Oltrepò Pavese, two northern Italian vine-growing areas. Microbiology 161 362–373. 10.1099/mic.0.000004 [DOI] [PubMed] [Google Scholar]
- Vitulo N., Lemos W. J. F., Jr., Calgaro M., Confalone M., Felis G. E., Zapparoli G., et al. (2019). Bark and grape microbiome of Vitis vinifera: influence of geographic patterns and agronomic management on bacterial diversity. Front. Microbiol. 9:3203. 10.3389/fmicb.2018.03203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., García-Fernández D., Mas A., Esteve-Zarzoso B. (2015). Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE. Front. Microbiol. 6:1156. 10.3389/fmicb.2015.01156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb L., Whetton P., Bhend J., Darbyshire R., Briggs P., Barlow E. (2012). Earlier wine-grape ripening driven by climatic warming and drying and management practices. Nat. Clim. Change 2 259–264. 10.1038/nclimate1417 [DOI] [Google Scholar]
- White R. (2015). Understanding Vineyard Soils. Oxford: Oxford University Press. [Google Scholar]
- Zarraonaindia I., Owens S. M., Weisenhorn P., West K., Hampton-Marcell J., Lax S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. mBio 6:e02527-14. 10.1128/mBio.02527-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang E. T., Singh R. P., Guo C., Shang Y., Chen J., et al. (2019). Grape berry surface bacterial microbiome: impact from the varieties and clones in the same vineyard from central China. J. Appl. Microbiol. 126 204–214. 10.1111/jam.14124 [DOI] [PubMed] [Google Scholar]
- Zhu Y.-G., Gillings M., Simonet P., Stekel D., Banwart S., Penuelas J. (2017). Microbial mass movements. Science 357 1099–1100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.