Abstract
Identification and characterization of small molecule–protein interactions is critical to understanding the mechanism of action of bioactive small molecules. Photo-affinity labeling (PAL) enables the capture of non-covalent interactions for enrichment and unbiased analysis by mass spectrometry (MS). Quantitative proteomics of the enriched proteome reveals potential interactions, and MS characterization of binding sites provides validation and structural insight into the interactions. Here, we describe the identification of the protein targets and binding sites of a small molecule using Small molecule Interactome Mapping by PAL (SIM-PAL). Cells are exposed to a diazirine-alkyne-functionalized small molecule and binding interactions are covalently captured upon UV irradiation. An isotopically-coded, acid-cleavable biotin azide handle is attached to the conjugated proteins using copper-catalyzed azide-alkyne cycloaddition. Biotin-labeled proteins are enriched for on-bead digestion and quantitative proteomics. Acid cleavage of the handle releases the bead-bound conjugated peptides for MS analysis and isotope-directed assignment of the binding site.
Keywords: photo-affinity labeling, binding site mapping, chemical proteomics, small molecule target identification, structural proteomics
INTRODUCTION:
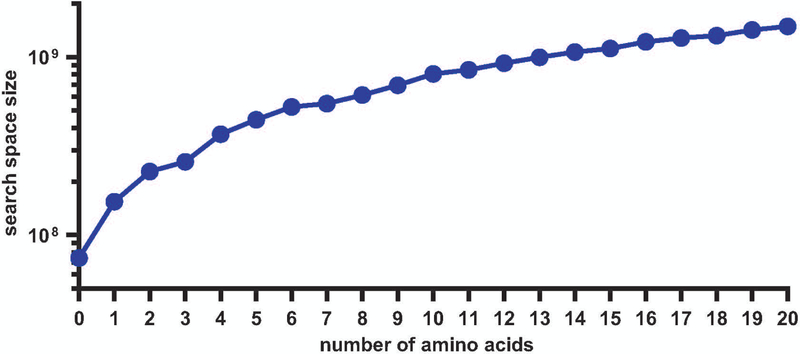
Photo-affinity labeling (PAL) combined with mass spectrometry (MS) analysis is a powerful technique for identifying non-covalent small molecule–protein interactions in cells. In a typical PAL experiment, a small molecule of interest functionalized with a photo-activatable group is introduced into a biological system and the molecule is conjugated to interacting proteins upon photo-irradiation. A specific advantage of PAL chemistry is the UV-induced production of reactive carbene intermediates that are relatively agnostic to the amino acids available within a given binding site for covalent conjugation. In general, methods for target identification by PAL use one of several photo-activatable groups, such as the azide, benzophenone, or diazirine (Das, 2011; Fleming, 1995). Recently, the small size of diazirines and their ease of incorporation into functionalized tags for further applications through copper-catalyzed azide-alkyne cycloaddition (CuAAC) have increased their use in the field (Chang, Mfuh, Gao, Wu, & Woo, 2018; Hill & Robertson, 2018; Li et al., 2013). With a diazirine-alkyne functionalized tag embedded in the small molecule of interest, several detection methods are available to visualize or enrich the captured proteins, including Western blotting and MS analysis. For identification in an unbiased manner by MS, one strategy is to append a biotin-containing handle by CuAAC for enrichment on streptavidin–agarose beads. On-bead digestion with trypsin or other proteases results in the release of non-conjugated peptides, which upon LC-MS/MS analysis yields identification of interacting proteins. Comparison between samples can be achieved using quantification methods (e.g., label-free, SILAC, TMT). These approaches have been widely used to characterize the protein interactomes of metabolites, fragment libraries, bioactive small molecules, and drugs (Das, 2011; Flaxman & Woo, 2018; Gao, Mfuh, Amako, & Woo, 2018; Gertsik et al., 2017; MacKinnon, Garrison, Hegde, & Taunton, 2007; Parker et al., 2017). Cleavage of the enrichment handle by chemical or enzymatic means then allows for recovery of the small molecule-conjugated peptide for analysis of the binding site itself (Speers & Cravatt, 2005; Szychowski et al., 2010). Identification of directly-conjugated peptides can aid in proposal of a structure for the identified small molecule–protein interaction and provides additional evidence that the enriched proteins interact directly with the small molecule. However, while characterization of the PAL-enriched proteome is straightforward, characterization of the directly interacting peptides is significantly more challenging due to the large number of potential products generated by PAL (Figure 1).
Figure 1:
Increase in size of the search space with the number of modified amino acid possibilities in standard database search of the tryptic human proteome (3 missed cleavages).
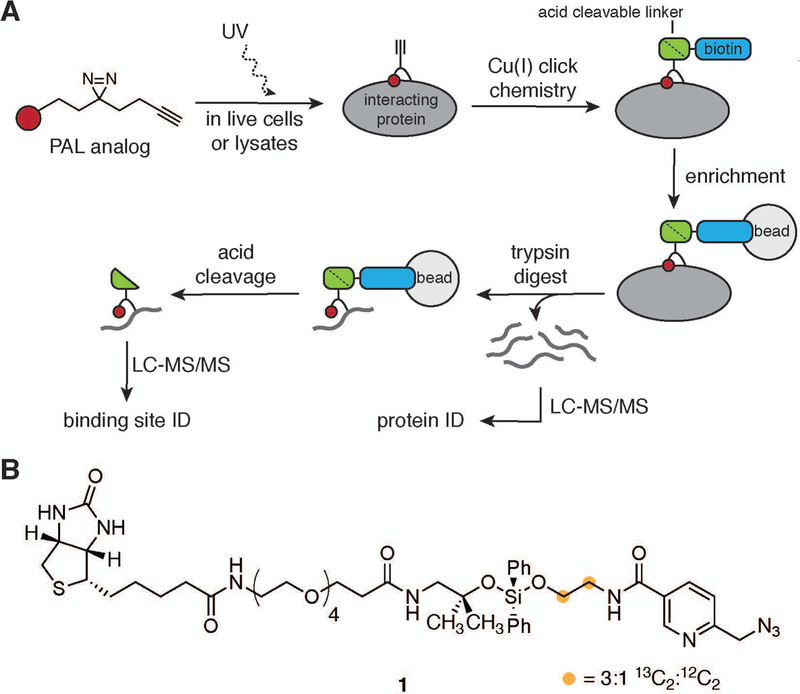
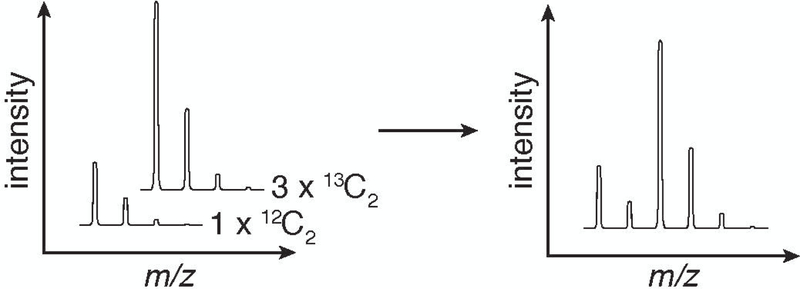
To ameliorate the complexity challenge in binding site assignment, we developed a method termed Small molecule Interactome Mapping by Photo-Affinity Labeling (SIM-PAL), summarized in Figure 2A. The SIM-PAL workflow maps both the interacting proteins and the underlying binding site, thus providing direct validation of and structural insight into small molecule–protein interaction (Flaxman, Chang, Wu, Nakamoto, & Woo, 2019; Flaxman & Woo, 2018; Gao et al., 2018). Key to this method is the use of a multi-functional acid-cleavable, isotopically-coded biotin picolyl azide, shown in Figure 2B, which utilizes a picolyl group to chelate Cu(I) near the reaction site, thus increasing the rate of the CuAAC reaction (Miyamoto, Flaxman, Wu, Gao, & Woo; Uttamapinant et al., 2012). The acid-cleavable diphenyl silane enables easy recovery of conjugated peptides from beads under MS-compatible conditions. The isotopic code, with two carbon atoms installed such that the ratio of 13C2:12C2 is 3:1, produces a distinct pattern in the full-scan MS (MS1) as shown in Figure 3, providing additional validation for small molecule-conjugated peptide spectral matches (PSMs).
Figure 2:
Summary of SIM-PAL method. A. Workflow used to identify interacting proteins and directly conjugated peptides. B. Structure of acid-cleavable picolyl biotin azide handle.
Figure 3:
Generation of the characteristic isotopic code from a handle with a 3:1 ratio of 13C2:12C2 stable isotopes incorporated.
The SIM-PAL protocol takes a small molecule functionalized with a diazirine and an alkyne through cellular treatment, photo-conjugation, enrichment, on-bead digestion, and recovery of directly conjugated peptides prior to analysis by LC-MS/MS. The major steps of the procedure are: (1) generation of small molecule-conjugated proteins, either in live cells or cell lysate, (2) attachment of an acid cleavable, isotopically-coded biotin picolyl azide via CuAAC and enrichment with streptavidin–agarose beads, followed by collection of trypsin and acid cleavage fractions for protein and conjugation site identification, respectively, and (3) LC-MS/MS analysis to identify the interacting proteins and conjugated peptides. The example data shown was generated by implementing the SIM-PAL procedure following treatment of A549 cells with photo-celecoxib (Gao et al., 2018; Miyamoto et al.).
STRATEGIC PLANNING
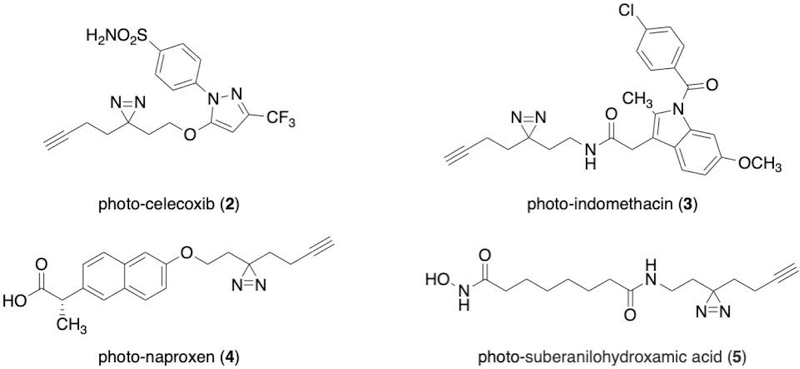
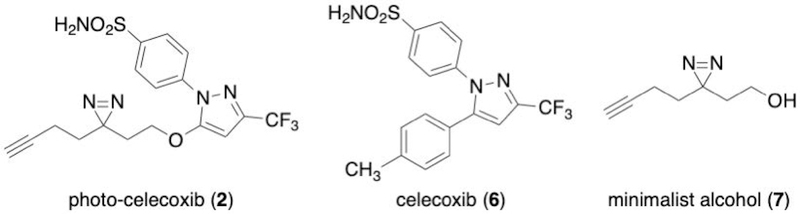
Prior to beginning a SIM-PAL experiment, a suitably functionalized derivative of the small molecule of interest needs to be developed and validated. Examples of PAL analogs previously used in SIM-PAL experiments are shown in Figure 4, including photo-celecoxib, which was used to generate the data presented in this protocol. The SIM-PAL protocol is broadly applicable to many of the PAL analogs and libraries that have been previously synthesized. To use the protocol as described, the molecule must contain a diazirine for PAL and an alkyne for CuAAC. Though these functional groups can be incorporated independently, tags such as the “minimalist tag” may be used to simultaneously incorporate both functional groups. The flexibility of the minimalist tag to be functionalized as an alcohol, amine, iodide, or triflate provides synthetic options for tagging of many small molecules (Flaxman et al., 2019; Li et al., 2013). Importantly, the biological effect of the PAL analog should be benchmarked against the parent molecule using appropriate in vivo and/or in vitro assays. For some small molecules, several PAL analogs may need to be assayed to identify an analog with activity that mimics the parent molecule.
Figure 4:
Selected examples of PAL analogs used in SIM-PAL.
BASIC PROTOCOL 1
Generation of a small molecule-conjugated protein sample following treatment of live cells
This protocol details the preparation of small molecule-conjugated cell lysates by PAL for subsequent enrichment. A cell line of interest is treated with the PAL analog and separately with the minimalist alcohol as a negative control for non-specific labeling due to the PAL functional group and the PAL analog with an excess of the parent molecule for competitive displacement. After incubation, cells are photo-irradiated and lysed. Protein concentrations in the lysates are normalized. This protocol uses A549 cells treated with photo-celecoxib to generate the example data. The compounds used are shown in Figure 5. The efficiency of labeling in the protocol will depend on factors including the cell line (permeability, presence of target proteins), the PAL analog (solubility, cell permeability, chemical complexity), and experimental variables (incubation time, cell number, vehicle, handle concentrations used). The lamp used for UV irradiation is particularly critical. The power of the lamp will determine the irradiation time; the emission profile may affect the generation of the reactive intermediates in PAL and thus the product distribution. A high-power lamp with broad emission, for example the lamp listed in the Materials section, will reduce UV irradiation time. If needed, optimization of labeling can be performed by Western blotting of enrichment fractions as described in Support Protocol 2 or by visualization of in-gel fluorescence following attachment of a fluorophore by CuAAC. SIM-PAL is a versatile method that can be applied to many small molecules, cell types, and lysates. In this this protocol, we focus on the use of SIM-PAL to identify the interactions of celecoxib in A549 cells. Alternate procedures have been provided for suspension cells and for labeling in cell lysates.
Figure 5:
Compounds used in SIM-PAL evaluation of celecoxib binding in A549 cells.
Materials:
Cell line of interest (e.g., A549)
Complete media (See Reagents and Solutions)
Serum-free media (See Reagents and Solutions)
150 mm diameter cell culture dishes (Corning # 353025)
Cell culture flasks
12 well plates (Greinger Bio-One #665–180)
Benchtop ultracentrifuge
PBS (sterile)
1 mM PAL analog (e.g., photo-celecoxib 2)/DMSO
10 mM PAL analog (e.g., photo-celecoxib 2)/DMSO
11.1 mM parent compound (e.g., celecoxib 6)/DMSO
1 mM minimalist alcohol (7)/DMSO (See Reagents and Solutions)
1 mM PAL analog (e.g., photo-celecoxib 2), 10 mM parent compound (e.g., celecoxib 6)/DMSO
Cell culture incubator
0.25% trypsin-EDTA
Cell scrapers
Light microscope
UV lamp (cooled to 4 °C) (ECE 5000 UV Light-Curing Flood Lamp system, Dymax #41060)
-
1% RapiGest/PBS (See Reagents and Solutions)
RapiGest is a MS-compatible denaturing detergent. Other detergents such as sodium dodecyl sulfate (SDS) may also be used, but extra care should be taken in washing the bead-bound proteins to ensure that such detergents do not reach the LC-MS/MS instrument.
25x Protease inhibitor stock (cOmplete, EDTA-free Protease Inhibitor Cocktail Tablets, Sigma #11873580001)
1.7-mL microcentrifuge tubes
Refrigerated microcentrifuge
Sonicator (Branson SFX250 with microtip)
BCA assay kit (G Biosciences #786–570)
96 well plate, non-TC treated (Research Products International, #141380)
Plate reader capable of reading absorbance at 590 nm in 96 well plate
Treatment of cells with PAL analog and UV irradiation - adherent cells (e.g., A549 cells)
Note: If you are using suspension cells skip steps 1–10 and start with step 11.
-
1
Grow cells to 90% confluence in a 150-mm diameter plate in complete media. One confluent plate per treatment condition should be prepared; three plates are required for a complete set of minimalist alcohol, PAL analog, and parent competition samples.
This protocol is written using A549 cells as an example. A confluent 150-mm plate of A549 cells yields approximately 1 × 107 cells per plate, corresponding to 1–2.5 mg protein in upon cell lysis. For other cell lines, adjustment of the cellular input may be necessary. At least 750 µg of protein per condition is required for the protocol as described.
This experiment can be performed with each condition in replicate to increase statistical power if desired.
-
2
Remove media by aspiration. Carefully wash adhered cells twice with 10 mL of PBS to remove residual serum-containing media and remove washes by aspiration.
-
3
For minimalist alcohol (7): In a 15-mL centrifuge tube, prepare a 10 µM solution of minimalist alcohol by adding 100 µL of 1 mM minimalist alcohol/DMSO to 10 mL of serum-free media. Add solution to a washed plate of cells.
For the PAL analog (2): In a 15-mL centrifuge tube, prepare a 10 µM solution of PAL analog by adding 100 µL of 1 mM PAL analog/DMSO to 10 mL of serum-free media. Add solution to a washed plate of cells.
For competition (2 + excess 6): In a 15-mL centrifuge tube, prepare a solution of 100 µM parent compound, 10 µM PAL analog by adding 10 µL of 10 mM PAL analog/DMSO and 90 µL of 11.1 mM parent compound/DMSO to 10 mL of adherent cell labeling media. Add solution to a washed plate of cells.
For some PAL analogs, concentrations of 10 µM–100 µM produce sufficient labeling without excessive background from the labeling. A 10x competition with the parent generally shows reduction in labeling of known target. The concentration and vehicle (e.g., DMSO, EtOH, water) used may be an important factor for each small molecule. The compound can alternately be added directly to the serum-free media dispensed in the culture dishes.
-
4
Incubate the treated cells for 60 min at 37 °C, 5% CO2. Meanwhile, turn on the UV lamp to warm up the bulb for at least 30 min.
The incubation time should be optimized for each PAL analog.
-
5
Remove the lid of the cell culture dish and UV irradiate for 60 s at 4 °C.
The UV irradiation time may need to be optimized if a different light source is used.
-
6
Remove media by aspiration. Carefully wash the cells twice with 10 mL PBS to remove residual serum. Discard each PBS wash by aspiration.
-
7
Add 10 mL of 0.25% trypsin-EDTA solution to each plate. Incubate at 37 °C, 5% CO2 until cells detach, approximately 5 min.
Cells may be more difficult to detach after photo-irradiation. Check that cells have completely detached by microscopy. Some cells may require tapping or scaping to completely detach from the plate.
-
8
Collect detached cells in 15 mL tubes. Pellet cells by centrifugation (3 min, 500 rcf, 24 °C), and remove supernatant using a serological pipette.
Cells may form loose pellets after photo-irradiation; pipetting rather than aspirating prevents loss of material.
-
9
Add 10 mL of PBS to each sample to wash cells. Pellet cells by centrifugation (3 min, 500 rcf, 24 °C), and remove supernatant by pipetting.
-
10
Add 300 µL of 1% RapiGest/PBS with 1x protease inhibitor to each pellet, and pipette to resuspend the cells. Transfer to a 1.7 mL microcentrifuge tube. Proceed to step 20.
Treatment of cells with PAL analog and UV irradiation - suspension cells
-
11
Grow cells to saturation in 45 mL of complete media (approximately 1 × 106 cells/mL for Jurkat or K562 cells). This is enough cells for a complete set of minimalist alcohol, PAL analog, and parent competition samples.
The values in this protocol are based on experiments with Jurkat and K562 cells.
This experiment can be performed with each condition in replicate to increase statistical power if desired. Depending the protein yield of the cell line of interest, adjustment of the cellular inputs may be necessary. At least 750 µg of protein per condition is required for the protocol as described.
-
12
Transfer 45 mL of culture to a 50 mL centrifuge tube. Pellet cells by centrifugation (3 min, 300 rcf, 24 °C), and remove supernatant by aspiration. Wash twice with 25 mL of PBS, repeating centrifugation and aspiration after each wash.
-
13
Resuspend cells in 1.5 mL of serum-free media. Transfer 0.5 mL of resuspended cells to 3 wells of a 12 well plate.
-
14
For minimalist alcohol (7) : Add 5 µL of 1 mM minimalist alcohol in DMSO to cells. Gently mix the sample after addition.
For the PAL analog (2): Add 5 µL of 1 mM PAL analog in DMSO to cells. Gently mix the sample after addition.
For competition (2 + excess 6): Add 5 µL of 1 mM PAL analog, 10 mM parent in DMSO to cells. Gently mix the sample after addition.
For some PAL analogs, concentrations of 10 µM–100 µM produce sufficient labeling without excessive background from the labeling. A 10x competition with the parent generally shows reduction in labeling of known target. The concentration and vehicle (ex. DMSO, EtOH, water) used may be an important factor for each small molecule. The compound can alternately be added directly to the serum-free media dispensed in the culture dishes.
-
15
Incubate the treated cells for 30–60 min at 37 °C, 5% CO2. Meanwhile, turn on the UV lamp to warm up bulb for at least 30 min.
The incubation time should be optimized for each PAL analog.
-
16
Remove the lid of the 12-well plate and UV irradiate for 60 s.
The UV irradiation time may need to be optimized if a different light source is used.
-
17
Transfer samples to 1.7 mL microcentrifuge tubes. Pellet cells by centrifugation (1 min, 500 rcf, 24 °C). Wash with 1 mL of PBS per sample, repeating centrifugation and aspiration after the wash.
Some suspension cells adhere upon serum starvation, in which case a cell scraper may be used to gently detach the cells.
-
18
Add 300 µL of 1% RapiGest, 1x protease inhibitor/PBS to each pellet, and pipette to resuspend the cells.
-
19
Proceed to Step 20.
Cell lysis
-
20
Sonicate each sample (2 s on, 5 s off, 10% amplitude, 10 s total).
Sonication can be performed on ice if desired.
-
21
Clarify lysates by centrifugation (10 min, 10,000 rcf, 4 °C). Collect supernatants.
If supernatant is viscous and difficult to remove without damaging the pellet, repeat sonication.
-
22
Measure protein concentration by BCA assay according to kit instructions. Normalize protein concentration to at most 2.5 mg/mL with 1% RapiGest/PBS such that each 250 µL sample has equal protein concentration.
The lysates can be taken forward to Basic Protocol 2 immediately, or stored at –20 °C and thawed prior to proceeding to Basic Protocol 2.
ALTERNATE PROTOCOL 1
Generation of small molecule-conjugated protein sample following treatment of cell lysate
Treatment of cell lysates rather than live cells may be performed for small molecules with limited cell permeability and may allow for harsher treatment conditions. The protocol described is analogous to Basic Protocol 1 with minor reordering of the steps. The tubes used for irradiation are particularly important; polypropylethylene (PPE) PCR tubes made of thin plastic are ideal for this application.
Materials:
-
Cell line of interest
The protocol for labeling lysates may be useful for small molecules with limited cellular permeability. Any cell lysate that is generated in an EDTA-free non-denaturing lysis buffer may be used.
Complete media (See Reagents and Solutions)
150 mm diameter cell culture dishes (Corning # 353025)
Cell culture flasks
PBS (sterile) (See Reagents and Solutions)
0.25% trypsin-EDTA
Cell culture incubator
Benchtop ultracentrifuge
0.3% Triton-X 100/PBS
25x protease inhibitor stock (cOmplete, EDTA-free Protease Inhibitor Cocktail Tablets, Sigma #11873580001)
Sonicator (Branson SFX250 with microtip)
Refrigerated microcentrifuge
BCA assay kit (G Biosciences #786–570)
0.5 mL PPE PCR tubes (VWR #490004–478)
1 mM PAL analog (e.g., photo-celecoxib 2)/DMSO
1 mM PAL analog (e.g., photo-celecoxib 2),10 mM parent compound (e.g., celecoxib 6)/DMSO
1 mM minimalist alcohol (7)/DMSO (See Reagents and Solutions)
Tube rotator
UV lamp (ECE 5000 UV Light-Curing Flood Lamp system, Dymax #41060)
10% RapiGest/PBS (See Reagents and Solutions)
1.7 mL microcentrifuge tubes
Cell lysis
-
1
For adherent cells: Grow cells to confluence in (3) 150 mm diameter plates (approximately 1 × 107 cells per plate for A549 cells). Remove media by aspiration. Carefully wash adhered cells with 10 mL of PBS to remove residual serum, removing PBS by aspiration. Add 5 mL of trypsin solution to each plate. Incubate at 37 °C, 5% CO2 until cells detach, approximately 5 min. Combine detached cells in a 50 mL centrifuge tube. Pellet cells by centrifugation (3 min, 300 rcf, 24 °C), and remove supernatant by aspiration. Wash twice with 25 mL of PBS, repeating centrifugation and aspiration for each wash.
For suspension cells: Transfer 45 mL of saturated culture to a 50 mL centrifuge tube (approximately 1 × 106 cells/mL for Jurkat or K562 cells). Pellet cells by centrifugation (10 min, 300 rcf, 24 °C), and remove supernatant by aspiration. Wash twice with 25 mL of PBS, repeating centrifugation and aspiration for each wash.
Cell pellets can be flash-frozen in liquid nitrogen and stored at –80 °C until use.
-
2
Resuspend cell pellets in 1 mL of 0.3% Triton-X 100, 1x protease inhibitor/PBS.
Other non-denaturing, non-chelating buffers may also be used.
-
3
Sonicate the cell lysate (2 s on, 5 s off, 10% amplitude, 10 s total).
Sonication can be performed on ice if desired.
-
4
Clarify lysate by centrifugation (10 min, 10,000 rcf, 4 °C). Collect supernatant.
If supernatant is viscous and difficult to remove without damaging the pellet, repeat sonication.
-
5
Measure protein concentration by BCA assay according to kit instructions. Normalize protein concentration to at most 2.5 mg/mL with 0.3% Triton-X 100/PBS such that each 250 µL sample has equal protein concentration.
Treatment of lysate with PAL analog
-
6
For minimalist alcohol (7): In a 0.5 mL PPE PCR tube, add 2.5 µL of 1 mM minimalist alcohol/DMSO to 250 µL of cell lysate.
For the PAL analog (2): In a 0.5 mL PPE PCR tube, add 2.5 µL of 1 mM PAL analog/DMSO to 250 µL of cell lysate.
For competition (2 + excess 6): In a 0.5 mL PPE PCR tube, add 2.5 µL of 10 mM parent PAL analog, 1 mM PAL analog/DMSO to 250 µL of cell lysate.
For some PAL analogs, concentrations of 10 µM–100 µM produce sufficient labeling without excessive background from the labeling. A 10x competition with the parent generally shows reduction in labeling of known target. The concentration and vehicle (ex. DMSO, EtOH, water) used may be an important factor for each small molecule. The compound can alternately be added directly to the serum-free media dispensed in the culture dishes.
-
7
Incubate for 30 min at 24 °C with inversion.
The incubation time should be optimized for each PAL analog.
UV irradiation
-
8
Irradiate with UV lamp for 60 s.
Tubes should be placed on their sides to expose the maximum surface area to the light source. The UV irradiation time may need to be optimized if a different light source is used.
Protein denaturation
-
9
Add 25 µL of 10% RapiGest/PBS to each sample and transfer samples to 1.7 mL microcentrifuge tubes.
The lysates can be taken forward to Basic Protocol 2 immediately, or stored at –20 °C and thawed prior to proceeding to Basic Protocol 2.
BASIC PROTOCOL 2
Copper-catalyzed azide-alkyne cycloaddition (CuAAC) functionalization and enrichment of labeled peptides
Following the preparation of PAL analog-conjugated peptides in Basic Protocol 1 or Alternate Protocol 1, the acid-cleavable, isotopically-coded biotin azide handle is attached to small molecule-conjugated proteins using CuAAC. This enables enrichment of the interacting proteins with streptavidin agarose, on-bead digest, and recovery of the conjugated peptides upon acid cleavage. Tandem mass tag (TMT) labeling of the peptides released by on-bead digest allows for quantification of protein enrichment ratios from multiple samples in one LC-MS/MS run.
Materials:
Treated lysates (Basic Protocol 1 or Alternate Protocol 1)
10 mM cleavable picolyl biotin azide (1, synthesized according to Support Protocol 2)/DMSO
50 mM copper sulfate/water (ultra-pure)
10 mM Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA, Sigma #762342)/water (ultra-pure)
50 mM sodium ascorbate/water(ultra-pure), prepared fresh
Methanol (ACS solvent grade)
–80 °C freezer
Refrigerated microcentrifuge
1% RapiGest/PBS (See Reagents and Solutions)
Sonicator (Branson SFX250 with microtip)
50% streptavidin agarose/PBS (Pierce Streptavidin Agarose, Thermo Scientific #20353)
Mini bio-spin columns (Bio-Rad #7326207)
Vacuum manifold (for example, Promega #A7231)
Vacuum source
8 M urea/PBS
PBS
100 mM dithiothreitol (DTT)/PBS, prepared fresh
Tube rotator
500 mM iodoacetamide/PBS, prepared fresh
50 mM HEPES pH 8.5
0.5 M guanidine-HCl/50 mM HEPES pH 8.5
Sequencing grade trypsin (20 µg resuspended in 40 µL of resuspension buffer, Promega, #V5111)
Heated incubator with tube rotator
2 mL microcentrifuge tubes
Ultra-pure water
50% acetonitrile (HPLC-grade)/water (ultra-pure)
1.7 mL low protein binding microcentrifuge tubes (Sorenson BioScience, # 39640T)
Speed-vac (for example, Eppendorf #022920109)
2% formic acid/water (ultra-pure)
TMT10plex isobaric label reagent set (Thermo Scientific # 90110)
5% hydroxylamine/water (ultra-pure)
C18 P10 Ziptip (Millipore #ZTC18M096)
1% formic acid/water (ultra-pure)
0.1% formic acid/water (ultra-pure)
0.1% formic acid/50% acetonitrile (HPLC-grade)/water (ultra-pure)
Autosampler vials with septa caps
Install enrichment handle via CuAAC
-
1
Prepare a CuAAC master mix based on the total volume of labeled lysate to be treated. We have found that 250 µL lysate per sample at a protein concentration of 2.5 mg/mL is generally sufficient. Use the amounts given in Table 1 below to calculate components to be used. Mix components 1–3 in sequence, followed by addition of sodium ascorbate immediately before adding the master mix to cell lysates.
-
2
Add the calculated volume of master mix to each sample.
-
3
Incubate at 24 °C with inversion for 90 min.
-
4
Add 4 volumes of methanol to each sample to precipitate protein. Incubate at –80 °C until protein forms a visible, flocculent precipitate, at least 1 h. The sample may alternatively be left to precipitate overnight (16–18 h) without ill effects.
-
5
Pellet the precipitated protein by centrifugation (4 °C, 21,130 rcf, 10 min).
-
6
Carefully remove the supernatant by decanting. Invert tubes on a clean paper towel and allow pellets to dry for 5 min.
-
7
Add 500 µL of 1% RapiGest/PBS to each sample to resuspend protein. To ensure complete resuspension, sonicate (2 s on, 5 s off, 15% amplitude, 10 s total).
Sonication can be performed on ice if desired.
Table 1:
Calculation of reagent amounts for CuAAC reaction master mix. Volume per sample (µL) = v; number of samples = n. Volume of master mix to add to each sample = 0.1 * v (i. e. for a 250 µL sample add 25 µL of master mix)
| Component | Stock concentration | Final concentration | Dilution | Amount to add (µL) |
|---|---|---|---|---|
| 1. Cleavable picolyl biotin azide | 10 mM | 100 µM | 100 | n * v * 1.2 /100 |
| 2. Copper sulfate | 50 mM | 250 µM | 200 | n * v * 1.2 /200 |
| 3. THPTA | 10 mM | 250 µM | 40 | n * v * 1.2 /40 |
| 4. Sodium ascorbate | 50 mM | 2.5 mM | 20 | n * v * 1.2 /20 |
Enrichment of the small molecule-conjugated proteins
-
8
Wash 50% streptavidin–agarose slurry twice with PBS prior to use. Add 100 µL of the washed 50% streptavidin–agarose bead slurry to each sample.
More or less streptavidin agarose may be required depending on the amount of labeling in the sample. This can be optimized by blotting samples taken throughout the enrichment to observe transfer of biotin signal using the procedure described in Support Protocol 2.
If performing Western blotting as described in Support Protocol 2, collect 20 µL this solution as the enrichment “load.”
-
9
Incubate samples 12–18 h at 24 °C with inversion.
Samples can be incubated for a shorter time and/or at 4 °C if protein degradation is a concern.
-
10
Transfer each sample to a mini bio-spin column, with the bottom stopper removed, in a 2 mL microcentrifuge tube. Collect the supernatant by centrifugation (1 min, 1000 rcf). After collecting the supernatants, remove the column lids and place the columns on a vacuum manifold.
Save the stoppers that are removed when opening the bottom of the columns, as they will be used to re-seal the columns.
If performing Western blotting as described in Support Protocol 2, collect 20 µL of the flow through as the enrichment “supernatant.”
-
11
Wash each tube with 500 µL of 1% RapiGest/PBS and transfer wash into the column corresponding to the sample. Remove wash by applying vacuum.
-
12
Continuing to use the vacuum manifold, wash beads with 5 × 1 mL 8 M urea/PBS and 5 × 1 mL PBS per sample.
-
13
Remove the columns from vacuum manifold and seal the bottoms of the columns with the stoppers.
Reduction and alkylation of enriched proteins
-
14
Add 175 µL of PBS and 25 µL of 100 mM DTT/PBS to each sample to reduce cysteines. Cap the columns and incubate at 24 °C with inversion for 30 min.
If performing Western blotting as described in Support Protocol 2, collect 20 µL the resuspended beads as the enrichment “capture” prior to adding the DTT solution.
As mini bio-spin columns are not securely held by standard rotators, it may be useful to encase the columns in foil such that they can be securely attached to a rotator with tape.
-
15
Add 10 µL of 500 mM iodoacetamide/PBS to each column to alkylate reduced cysteines. Cap the columns and incubate at 24 °C with inversion for 30 min.
-
16
After removing the caps and stoppers, return the columns to the vacuum manifold and wash the beads with 1 mL PBS and 1 mL 50 mM HEPES pH 8.5 per sample.
On-bead trypsin digest of enriched proteins
-
17
Remove columns from the vacuum manifold and seal the bottoms of the columns with the stoppers. Add 250 µL of 0.5 M GdnHCl/50 mM HEPES pH 8.5 to each sample. Add 1 µg of trypsin (2 µL) to each sample. Cap the columns and incubate at 37 °C with inversion for 12–18 h.
-
18
Carefully remove the stoppers from the column and place each column in a 2 mL microcentrifuge tube for collection of the tryptic digestion.
-
19
Centrifuge (24 °C, 1000 rcf, 1 min) the columns to collect the tryptic digest flow through in the collection tubes. Keep the cap on the columns during this centrifugation to ensure no sample is stuck in the lid.
-
20
Remove lids and add 200 µL of ultra-pure water to each column. Centrifuge (24 °C, 1000 rcf, 1 min), collecting flow-through in tube with elution.
-
21
Add 200 µL of 50% acetonitrile/water to each column. Centrifuge (24 °C, 1000 rcf, 1 min), collecting flow-through in tube with elution and the first wash.
-
22
Remove each column from the collection tube and insert the stopper. Transfer the collected digests and washes from each sample (“trypsin fraction”) to a 1.7 mL low protein binding tube. Dry digests using speed-vac.
Acid cleavage to release enriched conjugated peptides (for binding site analysis)
Performing binding site analysis on the PAL analog-treated samples allows for identification of interaction sites. Additionally, binding site analysis of the minimalist alcohol and parent competition samples may be helpful in differentiating direct versus indirect binding partners or nonspecific enrichment.
-
23
Add 200 µL of 2% formic acid/water to each column with the stopper inserted. Cap the columns and incubate at 24 °C for 30 min with inversion.
-
24
Remove the stoppers from the columns and collect the flow through from each sample in a 2 mL tube by centrifugation (24 °C, 1000 rcf, 1 min). Keep the cap on the column during this centrifugation to ensure no sample is stuck in the lid.
-
25
Add 200 µL of 50% acetonitrile/water to each column to elute hydrophobic conjugated peptides. Centrifuge (24 °C, 1000 rcf, 1 min), collect the flow-through with the elution from step 24.
-
26
Transfer the collected acid cleavage flow through and wash to a 1.7 mL low protein binding tube. Dry the collected elutions (“acid cleavage”) using speed-vac.
TMT labeling of the trypsin fraction
The TMT 10plex kit contains 10 isobaric reagents that produce characteristic fragment masses upon LC-MS/MS analysis (126, 127N, 127C, 128N, 128C, 129N, 129C, 130N, 130C, 131). Each trypsin fraction is labeled with a different TMT reagent so that the relative amount of different proteins in each trypsin fractions can be quantified.
-
27
Resuspend each trypsin fraction in 25 µL of water.
-
28
Add 10 µL of TMT reagent to each sample. Incubate at 24 °C for 1 h.
Record which TMT reagent is added to each sample.
-
29
Add 6 µL of 5% hydroxylamine/water to each sample to quench TMT reagent. Incubate at 24 °C for 15 min.
-
30
Combine all of the TMT-labeled trypsin digests in a new 1.7 mL protein low binding microcentrifuge tube and dry using the speed-vac.
Sample desalting
If additional depth of coverage is desired, high-pH fractionation (e. g. with the Pierce High pH Reversed-Phase Peptide Fractionation Kit, Thermo #84868) may be performed prior to LC-MS/MS analysis.
-
31
Resuspend the dried, TMT-labeled, combined trypsin fraction in 500 µL of 5% formic acid/water.
-
32
Wet a C18 Ziptip with MeOH by pipetting 10 µL of MeOH three times, disposing of the eluent.
-
33
Equilibrate the Ziptip in 0.1% formic acid/water by pipetting 10 µL three times, disposing of the eluent.
-
34
Load the sample onto the Ziptip, transferring solution to a clean tube after it has passed through the Ziptip to ensure that all sample is desalted.
If the pipetting resistance increases to the point that it is difficult to load additional sample onto the Ziptip, a large amount of protein (> 5 µg) is present in the sample. If this occurs, stop loading the sample and proceed to washing the Ziptip and eluting the peptides (Step 35). If desired, Step 34 may be repeated with an additional Ziptip, although it is not critical. Five µg of desalted peptides is sufficient for LC-MS/MS analysis. Larger capacity desalting cartridges (ex. Waters #WAT054945)) may also be used.
-
35
Wash the loaded Ziptip with 50 µL of 0.1% formic acid/water by pipetting 10 µL five times, disposing of the eluent.
-
36
Elute the desalted proteins into a MS-compatible autosampler vial with 20 µL of 0.1% formic acid/50% acetonitrile/water by pipetting up and down five times.
-
37
Resuspend each acid cleavage fraction in 100 µL of 1% formic acid/water. Repeat steps 32–36 to desalt.
For particularly hydrophobic PAL analogs, the elution solution may be modified to 0.1% formic acid/75% acetonitrile/water to ensure complete elution of conjugated peptides.
-
38
Dry desalted samples in speed-vac.
At the end of this protocol, the user should have one sample vial containing the combined, desalted, TMT-labeled “trypsin fraction” for identification of interacting proteins and one sample vial per condition containing the desalted “acid cleavage” fraction for identification of interaction sites. The samples are ready to be resuspended and subjected to LC-MS/MS analysis. The dried samples can be stored at –20 °C or –80 °C until analysis.
SUPPORT PROTOCOL 2
Synthesis of acid-cleavable, isotopically-coded biotin picolyl azide handle
The acid-cleavable, isotopically-coded biotin picolyl azide handle is essential to the SIM-PAL protocol, as it enables the identification of both the interacting proteins and the conjugated peptides. Preparation of the isotopically-coded handle requires synthesis of a 13C2 version and a 12C2 version of an intermediate, then incorporation of the two intermediates into the final compound at the desired 3:1 isotopic ratio. Characterization of the intermediates and handle have been reported previously (Miyamoto et al.).
Materials:
Picolyl pyridine, synthesized using the procedure of Ting and co-workers (Uttamapinant et al., 2012)
Dimethylformamide (DMF), dried
N-(3-Dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC)
N-hydroxysuccinimide (NHS)
Methanol (MeOH), solvent grade
Dichloromethane (DCM), solvent grade
Dichloromethane (DCM), dried
Triethylamine (TEA), dried over calcium hydride
Ethanolamine, dried over calcium hydride
Isopropanol, solvent grade
Ethylacetate, solvent grade
Ethanolamine-13C2 hydrochloride (Cambridge Isotopic Laboratories # CLM-274–0.1)
Biotin-CA(PEG)4-alcohol, synthesized using the procedure of Tirrell and co-workers (Szychowski et al., 2010)
Dichlorodiphenylsilane (Gelest #SID4510.1)
General synthetic chemistry outfit, including:
Chemical fume hood with vacuum, nitrogen gas lines
Scale
Standard synthetic glassware
Airtight glass syringes
Needles for chemical transfer
Sodium sulfate
Filter paper
Saturated sodium chloride in water
Saturated sodium bicarbonate in water
Filter paper
Rotary evaporator
NMR
Silica column chromatography equipment
Thin layer chromatography equipment
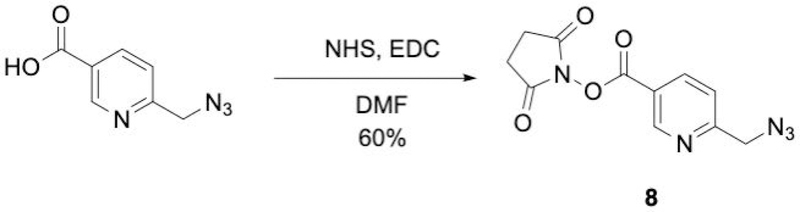
Synthesis of 2,5-dioxopyrrolidin-1-yl 6-(azidomethyl)nicotinate (8, Figure 6)
Figure 6:
Synthesis of 2,5-dioxopyrrolidin-1-yl 6-(azidomethyl)nicotinate.
-
1
In an oven-dried round bottom flask equipped with a stir bar prepare a 0.2 M solution of picolyl pyridine in DMF by adding 819 mg (4.6 mmol, 1 equiv) of the picolyl pyridine to 23 mL DMF at 24 °C.
-
2
To the stirred solution, add EDC (1.32 g, 6.89 mmol, 1.50 equiv) at 24 °C. Continue to stir for 5 min.
-
3
To the stirred solution, add NHS (793 mg, 6.89 mmol, 1.50 equiv) at 24 °C.
-
4
Continue to stir at 24 °C for 18 h.
-
5
Remove the solvent by rotary evaporation.
-
6
Take a small portion of the crude product to record an NMR spectrum.
-
7
Purify the crude product by silica gel chromatography with a 0–5% gradient of MeOH in DCM with 3 steps. Monitor the purification by silica thin-layer chromatography with 5% MeOH–DCM as the mobile phase and UV detection (Rf = 0.52).
-
8
Combine fractions that contain the purified product and dry by rotary evaporation.
-
9
Take a small portion of the product to record an NMR spectrum.
The purified product is a light orange solid. Typical yield is approximately 60%. The product 8 is highly water soluble. Additional extractions from the aqueous layer may increase the yield.
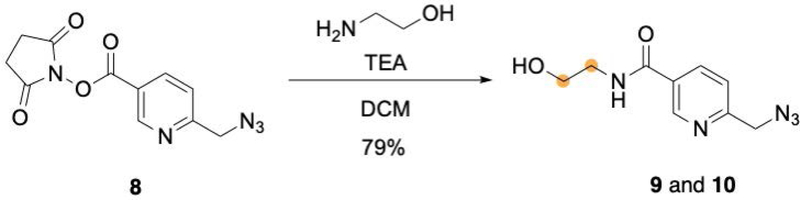
Synthesis of 6-(azidomethyl)-N-(2-hydroxyethyl)nicotinamide (9, Figure 7)
Figure 7:
Synthesis of 6-(azidomethyl)-N-(2-hydroxyethyl)nicotinamide.
-
10
In an oven-dried round bottom flask equipped with a stir bar prepare a 0.1 M solution of NHS ester 8 in DCM by adding 191 mg (0.7 mmol, 1 eq) of the NHS ester to 7 mL DCM at 24 °C.
-
11
While stirring under nitrogen, add 0.2 mL of TEA (0.770 mmol, 1.10 equiv) to the round bottom flask by syringe.
-
12
While stirring under nitrogen, add 60 µL of ethanolamine (1.04 mmol, 1.50 equiv) to the round bottom flask by syringe.
-
13
Continue to stir the solution at 24 °C for 18 h.
-
14
Dilute the solution with 15 mL of saturated aqueous sodium chloride solution and transfer to a separatory funnel. Shake the solution and allow the layers to separate. Collect the organic layer.
-
15
Extract the saturated aqueous sodium chloride solution three times with 15 mL of 10% isopropanol/ethyl acetate. Combine the organic layers.
-
16
Dry the organic layers over sodium sulfate and filter the dried solution into a collection flask.
-
17
Concentrate the filtered organic layers by rotary evaporation to yield the crude product.
-
18
Take a small portion of the crude product to record an NMR spectrum.
-
19
Purify the crude product by silica gel chromatography with a 0–10% gradient of MeOH in DCM with 5 steps. Monitor the purification by silica thin-layer chromatography with 5% MeOH–DCM as the mobile phase and UV detection (Rf = 0.32).
-
20
Combine the purification fractions that contain product and dry by rotary evaporation.
-
21
Take a small portion of the product to record an NMR spectrum.
The product is an off-white solid. Typical yield is approximately 79%.
Synthesis of 6-(azidomethyl)-N-(2-hydroxyethyl-1,2-13C2)nicotinamide (10, Figure 7)
Repeat steps 10–21, using ethanolamine-13C2 hydrochloride in place of ethanolamine. The final product is an off-white solid. Typical yield is approximately 66%.
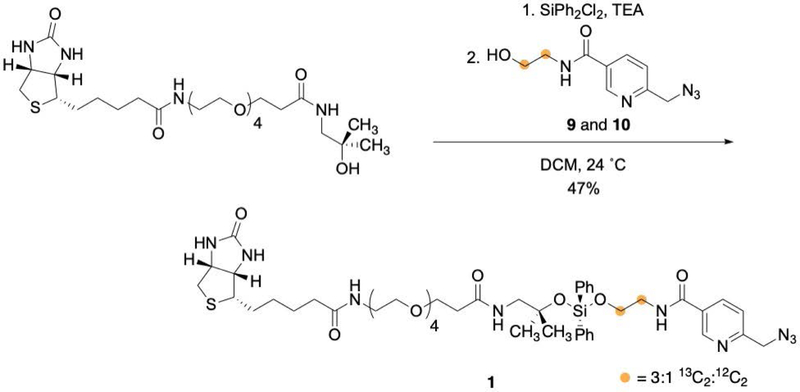
Synthesis of the cleavable biotin picolyl azide handle (1, Figure 8)
Figure 8:
Synthesis of the cleavable biotin picolyl azide handle.
-
22
In an oven-dried round bottom flask equipped with a stir bar prepare a 0.2 M solution of biotin-CA(PEG)4-alcohol in DCM by adding 9.5 mg (17 µmol, 1 equiv) of biotin-CA(PEG)4-alcohol to 85 µL DCM at 24 °C.
-
23
While stirring under nitrogen, add 47.4 µL of TEA (339 µmol, 20 equiv) to the solution by syringe.
-
24
While stirring under nitrogen, add 10.7 µL of dichlorodiphenylsilane (51.0 µmol, 3.0 equiv) to the solution by syringe.
-
25
Continue to stir at 24 °C for 2 h.
-
26
To the stirring solution, add 1:3 mixture of 12C2 amide (9) and 13C2 amide (10) (22.7 mg, 12.0 µmol, 6.0 equiv) in DCM (300 µL) by syringe.
In order to add the correct isotope ratio, weigh a 3:1 ratio of compound 9: compound 10, mix the solids together, and dissolve in methanol. Dry by rotary evaporation and take a small portion to check the ratio by 1H NMR. Adjust as necessary.
-
27
Continue to stir at 24 °C for 18 h.
-
28
Dilute the reaction mixture with 1 mL of saturated sodium bicarbonate and transfer to a separatory funnel.
-
29
Collect the organic layer.
-
30
Extract the aqueous layer twice with 1 mL of DCM, combining the organic layers.
-
31
Dry the organic layers over sodium sulfate and filter into a collection flask.
-
32
Concentrate the dried organic layers by rotary evaporation to yield the crude product.
-
33
Take a small portion of the crude product to record an NMR spectrum.
-
34
Purify the crude product by silica gel chromatography with a 0–11% gradient of MeOH in DCM with 11 steps. Monitor the purification by silica thin-layer chromatography with 10% MeOH–DCM as the mobile phase and UV detection (Rf = 0.30).
-
35
Combine purification fractions that contain product and dry by rotary evaporation.
-
36
Take a small portion of the product to record an NMR spectrum.
The final product is a yellow oil. Typical yield is approximately 40%. The reaction is water sensitive and hydrolysis of the diphenyldichlorosilane may reduce the overall yield. In order to maximize the yield, extra care should be taken to ensure that all glassware and reagents are dry and that reagents are fresh.
SUPPORT PROTOCOL 2
Monitoring enrichment by Western blotting
Analyzing samples taken throughout the enrichment process by Western blotting provides a means to monitor the overall success of the enrichment or the enrichment of a specific protein of interest. In order to test the overall success of the enrichment, streptavidin-HRP is used to visualize the transfer of biotin signal from the lysate to the beads, which can be particularly useful when troubleshooting the enrichment procedure. If there is a particular protein of interest, one can use Western blotting with a protein-specific antibody to determine the relative enrichment of the protein under different conditions. Blotting can accelerate the screening of conditions for the use a particular PAL analog.
Materials:
5x SDS-PAGE sample loading buffer (See Reagents and Solutions)
Heat block
Tabletop centrifuge
Polyacrylamide gel(s)
SDS-PAGE system
Gel transfer system and membranes, with filter paper, buffers, etc. as required by manufacturer
Ponceau stain (See Reagents and Solutions)
Scanner or camera
Blotting boxes
TBST (See Reagents and Solutions)
5% BSA/TBST (See Reagents and Solutions)
Orbital shaker
Streptavidin-HRP (Pierce High Sensitivity Streptavidin-HRP, Thermo Scientific #21130)
Chemiluminescence reagent (Azure Radiance ECL #AC2204)
Chemiluminescence imaging system (i.e. CCD camera-equipped imager or film and developer)
Other antibodies as desired (e.g., to blot for PTGES, anti-PGES (Invitrogen #PA5–51036) and anti-rabbit HRP secondary antibody (Rockland Immunochemical #611–1302))
-
1
Collect 20 µL of the enrichment “load” in step 8 of Basic Protocol 2.
-
2
Collect 20 µL of the enrichment “supernatant” from the solution collected by centrifugation in step 10 of Basic Protocol 2.
-
3
After washing and resuspending the beads in step 11 of Basic Protocol 2, collect 20 µL of the resulting slurry as the “capture” fraction.
-
4
Add 5 µL of 5x SDS-PAGE gel loading buffer to each sample.
-
5
Heat samples to 95 °C for 5 min.
-
6
Briefly centrifuge samples to collect any evaporation that may have collected in the lids.
-
7
Run the samples on an SDS-PAGE gel.
When loading the “capture” fraction, use a P20 pipette tip as the beads may clog P10 pipette tips. Pipette capture samples up and down several times before loading to make sure the beads are evenly distributed.
-
8
Transfer the proteins to a nitrocellulose membrane using a Western blotting transfer system according to manufacturer instructions.
-
9
In a Western blotting box or a tray, add enough Ponceau solution to cover the membrane. Swirl for approximately 30 s, then decant the Ponceau. Wash the membrane with deionized water, avoiding pouring water directly onto the membrane, until there is no decrease in the pink background signal. Image the total protein by scanner or camera.
If using streptavidin-HRP to visualize biotin on the membrane, proceed to step 10. If interested in blotting for a specific protein, proceed to step 14.
Blotting for biotinylated proteins
-
10
Block the membrane in 5% BSA/TBST for 1 h at 24 °C on an orbital shaker.
-
11
Incubate the membrane in streptavidin-HRP diluted 1:50,000 in 5% BSA/TBST for 1 h at 24 °C on an orbital shaker.
-
12
Wash membrane three times with TBST, allowing the membrane to incubate for 5 min on an orbital shaker between washes.
-
13
Visualize chemiluminescence using chemiluminescent reagents and imager according to manufacturer instructions. An example blot (with contrast adjusted to clearly visualize signal) is shown in Figure 9A.
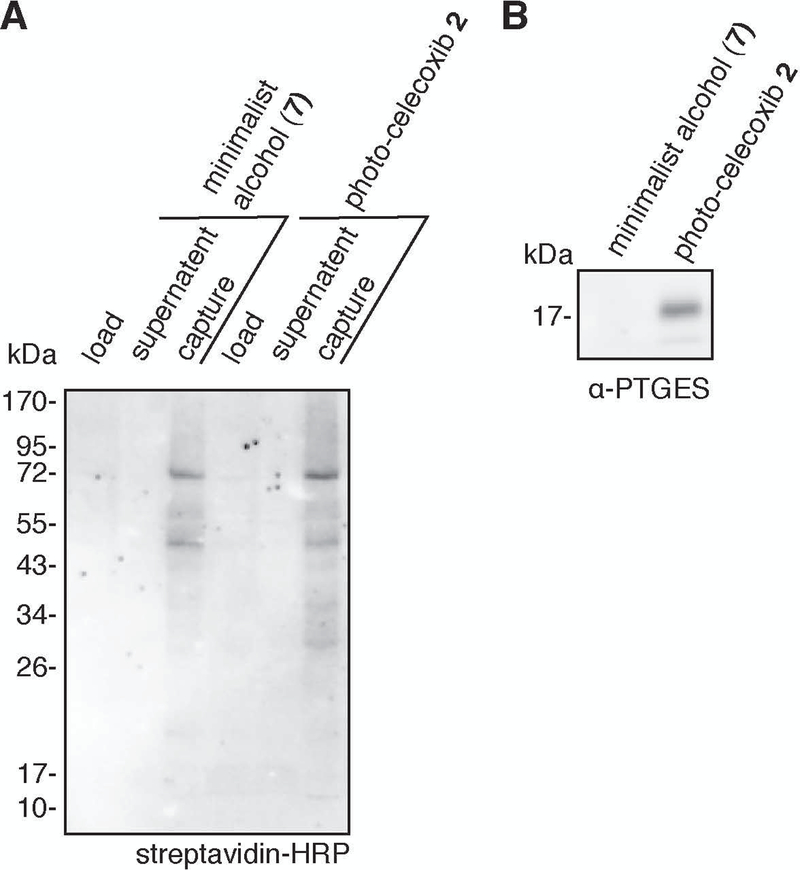
If enrichment was successful, a small amount of biotin signal will be observed in the “load” lanes, very little biotin signal will be observed in “supernatant” lanes, and strong biotin signal will be observed in “capture” lanes.
Figure 9:
Examples of Western blotting results. A. Western blotting for biotinylation using streptavidin-HRP. B. Western blotting for PTGES.
Blotting for specific protein
When blotting for a specific protein of interest, it may be preferable to analyze only the “capture” lanes.
The procedure given is optimized for blotting for PTGES. Blotting for other proteins may require optimization of the blotting protocol.
-
14
Block the membrane in 5% BSA/TBST for 1 h at 24 °C on an orbital shaker.
-
15
Incubate the membrane in primary antibody diluted 1:1,000 in 5% BSA/TBST for 12–18 h at 4 °C on an orbital shaker.
-
16
Wash membrane three times with TBST, allowing the membrane to incubate for 5 min on an orbital shaker between washes.
-
17
Incubate the membrane in secondary antibody diluted 1:10,000 in 5% BSA/TBST for 1 h at 24 °C on an orbital shaker.
-
18
Wash membrane three times with TBST, allowing the membrane to incubate for 5 min on an orbital shaker between washes.
-
19
Visualize chemiluminescence using chemiluminescent reagents and imager according to manufacturer instructions. An example blot is shown in Figure 9B.
BASIC PROTOCOL 3
Mass spectrometry analysis to identify interacting proteins and conjugation sites
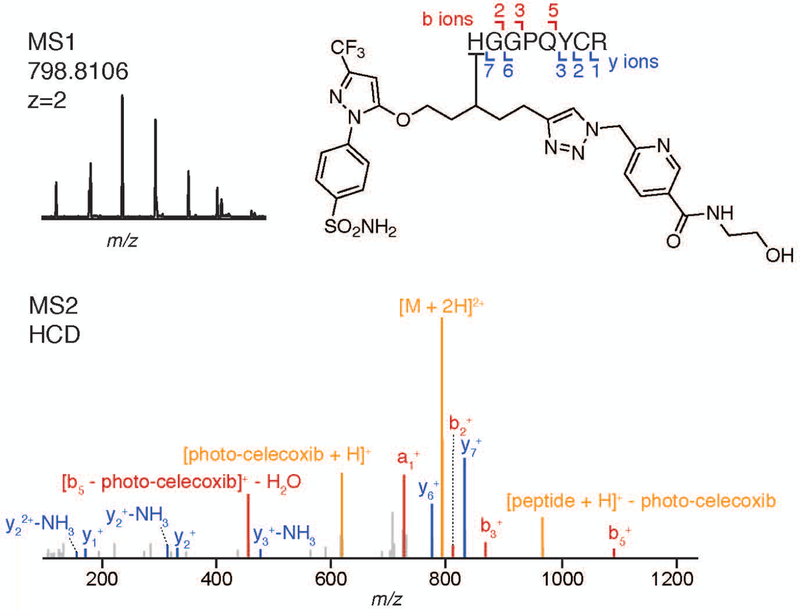
Both the trypsin fractions and the acid cleavage fractions are analyzed by LC-MS/MS; the former yielding identification of the interacting proteins and the latter yielding identification of the small molecule-conjugated peptides. Analysis of the trypsin fraction can be performed using standard methods for TMT-labeled samples. Due the unique isotopic coding of the acid cleavage fractions, both LC-MS/MS parameters and the analysis workflow have been optimized to enable identification of conjugated peptides. Example data showing the identification of a photo-celecoxib-conjugated peptide on prostaglandin E synthase (PTGES) is presented in Figures 8.
Materials:
TMT-labeled, pooled, desalted trypsin fractions (Basic Protocol 2)
Desalted acid cleavage sample(s) (Basic Protocol 2)
nano-LC system with C18 column
Buffer A (0.1% formic acid/water)
Buffer B (0.1% formic acid/acetonitrile)
PC computer
Thermo Orbitrap Lumos (or other LC-MS/MS instrument capable of CID and HCD fragmentation, such as Thermo Orbitrap Elite)
Proteome Discoverer software (Thermo, version 2.3)
MSConvert software (ProteoWizard)
Isostamp software (http://mass-spec-169.herokuapp.com/; for a more recent version developed in-house, contact the corresponding author)
LC-MS/MS analysis of trypsin fraction
-
1
Resuspend TMT-labeled, pooled, desalted trypsin fractions in 20 µL of buffer A and cap the autosampler vial.
-
2
Analyze the sample by injecting 2 µL of sample on a nano-LC system equipped with a C18 column, connected in line to a Thermo Orbitrap Lumos or Fusion.
Identification of interacting proteins
-
3
Import data into a project file in Thermo Proteome Discoverer software with TMT10- or 11-plex as the quantification method.
-
4
Perform a search of the data file (e.g., with the SEQUEST HT search engine) against the human proteome and contaminant proteins (FASTA files containing the desired protein database may be obtained from www.Uniprot.org), normalizing quantification values to the total amount of protein per quantification channel.
-
5
Analyze the results of the search to identify proteins that were enriched in PAL analog-treated cells relative to minimalist alcohol-treated cells and to parent competition treated cells. If multiple replicates were performed, p-value analysis can be performed.
For a guide to setting up quantitative proteomics searches in Thermo Proteome Discoverer, see the Proteome Discoverer 2.2 Quick Start document (Proteome Discoverer 2.2 Quick Start, 2017).
-
6
Optional: Export a list of the proteins identified with high confidence from the trypsin fraction to a FASTA file for use as a protein database in the acid cleavage fraction search.
Searching the acid cleavage fraction against the proteins identified in the trypsin fraction can improve automated assignment of binding sites and reduce the number of false positives, but may result in missed assignments of conjugated peptides from proteins not identified in the trypsin fraction.
LC-MS/MS analysis of acid-cleavage fraction
-
7
Resuspend desalted acid cleavage fraction(s) in 20 µL of buffer A and cap the autosampler vial.
-
8
Analyze the samples by injecting 8 µL of sample on a nano-LC system equipped with a C18 column, connected in line to a Thermo Orbitrap Lumos.
The isotopic isolation window should be increased to 3 m/z to account for isotopic coding. Depending on the PAL analog used, the LC gradient may need to be modified to ensure the elution of hydrophobic conjugated peptides. Further, using multiple activation modes (CID, HCD, EThcD) may increase the likelihood of conclusive assignment of the conjugated peptides.
Identification of conjugated peptides via manual or automated analysis (Figure 10)
Figure 10:
Identification of conjugated peptide from PTGES, with MS1 spectra showing isotopic coding and MS2 assignment showing peptide sequence match. Fragments resulting from modification loss were manually annotated (tolerance = 0.03 Da).
-
9
Import data into a project file in Thermo Proteome Discoverer software.
-
10
Perform a search of the data file (e.g., with the SEQUEST HT search engine) against the human proteome, or the FASTA file generated from the trypsin fraction in step 6, and contaminant proteins with a modification mass corresponding the PAL analog (including the remaining portion of the CuAAC handle) with modification at any amino acid.
-
11
For manual analysis: For each PAL analog-conjugated peptide identified with medium or high confidence (<5% FDR), examine the MS1 spectra and determine whether the peptide has a normal isotopic distribution (which can be calculated using software at chemcalc.org) or is isotopically recoded. The presence of an isotopic peak at [M-2] is a key signature in determining whether a feature is isotopically recoded.
For automated analysis:- Export the modified peptide list as a text file.
- Use MS Convert to convert the data file into an mzML file in both profile and vendor formats.
- Run the IsoStamp program (Woo et al., 2017), with the peptide list and the mzML file as input, to identify isotopically coded peaks. A file with IsoStamp probability scores between 0 and 1.0 for each peptide will be produced.
- Determine Isostamp probability cutoff score for filtering data by analysis of data collected using a DMSO-treated sample or by visual inspection of the species at intermediate IsoStamp scores.
The analysis workflow and software described work in conjunction with Thermo Proteome Discoverer 2.3 software. Using other versions or other vendor software may require modification of the analysis methods.
REAGENTS AND SOLUTIONS:
Complete media – base media as required for cell line (e.g. DMEM or RPMI) + 10% fetal bovine serum + 100 units/mL penicillin + 0.1 mg/mL streptomycin
Serum-free media – base media as required for cell line (e.g. DMEM or RPMI) + 100 units/mL penicillin + 0.1 mg/mL streptomycin
PBS – 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mL KCl, pH 7.4
TBST – 19 mM Tris, 137 mM NaCl, 2.7 mM KCl, 0.1% v/v Tween-20, pH 7.4
5x SDS-PAGE gel loading buffer – 5% v/v β-mercaptoethanol, 0.02% w/v bromophenol blue, 30% v/v glycerol, 10% w/v SDS/ 250 mM Tris, pH 6.8 (from Cold Springs Harbor Protocols)
Ponceau stain – 0.002% w/v Ponceau S, 3% v/v acetic acid/water
RapiGest was prepared by the method of Bouvier and co-workers (Bouvier, 2013)
Minimalist alcohol was prepared by the method of Yao and co-workers (Li et al., 2013)
COMMENTARY
BACKGROUND INFORMATION:
PAL was first implemented by Frank Westheimer, who used diazoacetic acid to label proteins in a rapid and unbiased manner to elucidate protein structure and function (Browne, Hixson, & Westheimer, 1971; Westheimer, 1980). Several PAL functional groups have been used over the years, including aryl azides, benzophenones, and aryl and alkyl diazirines. In the context of PAL-functionalization of a small molecule, alkyl diazirines are currently most commonly used due to their small, minimally perturbative size and ease of incorporation into functionalized tags that can be appended to molecules of interest (Chang et al., 2018; Li et al., 2013).
Many robust chemical proteomics methods have been developed to identify the targets of covalent small molecules on a proteome-wide scale using MS have recently been extended to PAL chemistry, which enables the conjugation of non-covalent small molecules to their binding sites (Flaxman & Woo, 2018). Enrichment and recovery of the conjugated peptides allows for structural characterization of the profiled interactions. Methods for conjugated peptide recovery include the use of biotin analogs with weaker binding, enzymatically cleavable handles, and the acid cleavable handle used in this method (Lee et al., 2017; Lemeer, Zörgiebel, Ruprecht, Kohl, & Kuster, 2013; Speers & Cravatt, 2005; Szychowski et al., 2010). The acid cleavable handle is readily cleaved upon exposure to low pH, conditions that are highly MS-compatible. Further enabling identification of the small molecule-conjugated peptide, SIM-PAL uses a set of isotopes embedded into the CuAAC handle that are transferred to the small molecule-conjugated peptides, which provides independent verification of small molecule-conjugated peptides by MS. The use of isotope coding for confident assignment of modified peptides has been used to accelerate identification of a variety complex modifications, including PAL analogs, glycans, and chemical crosslinkers (Tomohiro, Morimoto, Shima, Chiba, & Hatanaka, 2014; Wacker, Kashyap, Li, & Kapoor, 2011; Weerapana et al., 2010; Woo, Iavarone, Spiciarich, Palaniappan, & Bertozzi, 2015).
CRITICAL PARAMETERS:
Successful identification of small molecule binding sites depends both on the system being analyzed and experimental design factors. Design of the PAL analog is critical; it must recapitulate the activity of the parent molecule after incorporation of a photo-activatable group and an alkyne. Additionally, the cell type selected must allow for the PAL analog to interact with and conjugate to target protein(s) in amounts that can be enriched. In cases where multiple cell lines are sensitive to a small molecule, it may be advantageous to perform SIM-PAL in several cell lines to identify the ideal system. Optimal treatment conditions will vary by molecule and cell line. Photo-irradiation conditions are particularly sensitive to the UV light source used. The irradiation conditions given are for a high-power photo-curing lamp. Using a different UV light source will require optimization of irradiation times.
During enrichment, the streptavidin-agarose beads used must be compatible with MS analysis downstream; we have found the listed product to work well. Additionally, one should be careful to close the bottom stopper and top lid on the mini bio-spin columns securely during the enrichment and subsequent steps to prevent sample from leaking during incubations. If enrichment problems are encountered, it may be useful to troubleshoot by blotting the load, supernatant, and capture fractions with streptavidin-HRP to check that the biotin signal is being transferred from the load to the beads as described in Supplemental Protocol 2.
LC-MS/MS analysis protocols will vary depending on the instrument used. For particularly complex trypsin fractions, fractionation prior to analysis may provide deeper coverage. When analyzing the data for conjugated peptides, the IsoStamp cut-off score may need to be modified; this can be done by comparison to a DMSO-treated sample and/or manually inspecting spectra of interest and comparing the observed isotopic distribution to models of the isotopic distribution with and without recoding.
TROUBLESHOOTING:
| problem | possible cause | solutions |
|---|---|---|
| Few/no enriched or competed proteins in trypsin fraction | PAL analog was not cell-permeable | • Try alternate PAL analogs • Treat cell lysate rather than live cells • Increase PAL analog concentration |
| PAL analog did not effectively conjugate known interacting proteins | • Try alternate PAL analogs | |
| PAL analog did not conjugate enough target protein for identification | • Increase PAL analog concentration • Check protein expression in cell line |
|
| Irradiation was not sufficient | • Increase irradiation time • Use alternate irradiation light source • Check that there is minimal interference between the light source and sample (plate lids, non-PPE tubes, etc.) |
|
| CuAAC reaction failed | • Check that lysis buffer did not contain reducing agents or metal chelators (e.g., EDTA) | |
| Precipitation following CuAAC reaction failed | • Increase incubation time at –80 °C • Precipitate with acetone rather than methanol • Use a spin concentrator |
|
| Resuspension following precipitation was incomplete | • Increase sonication time or amplitude • Increase amount of RapiGest in buffer to 2% |
|
| Enrichment failed | • Use fresh streptavidin-agarose beads • Try shorter or longer incubation times |
|
| Trypsin cleavage failed | • Use fresh, sequencing-grade trypsin | |
| No isotopically-coded assigned species in acid-cleavage fraction | Early cleavage of tag | • Make sure that sample is not prematurely exposed to acid • Make sure to use fresh 8 M urea for washes |
| Tag not cleaved | • Increase cleavage time • Make fresh 2% formic acid/water |
|
| Desired binding site not observed | Binding site not on detectible tryptic peptide | • Try other proteases (chymotrypsin, etc.) |
| Binding site occurs at low frequency | • Immunoprecipitate the target protein instead of enriching for biotinylated proteins • Use a cell line that overexpresses the target protein |
STATISTICAL ANALYSIS:
If the trypsin fraction includes replicates, the standard statistical treatment incorporated into Proteome Discoverer can be used to calculate p-values for labeling with the PAL analog relative to minimalist alcohol and parent competition.
UNDERSTANDING RESULTS:
Successful implementation of the SIM-PAL protocol will yield identification of both interacting proteins and sites of interaction. In the example data, we present the identification of a specific conjugated peptide on prostaglandin E synthase-1 (PTGES). This peptide was validated upon examination of its isotopic coding. These data point to the identification of a new target and binding site for celecoxib and highlight the strength of MS-based target ID to profile interactomes in an unbiased manner. However, a binding site on each target protein should not be expected due to the potential occurrence of binding events in regions of the proteome that are difficult to visualize by trypsin proteolysis.
TIME CONSIDERATIONS:
Steps requiring overnight (i.e. 16–18 h) incubations are underlined.
Basic Protocol 1 (0.5 day)
sample set up – 30–60 min
incubation – 30–60 min
irradiation – 5 min
cell lysis – 30 min
BCA assay – 45 min
lysate dilution – 15 min
Basic Protocol 2 (2.5 days)
preparation of CuAAC reagents – 15 min
CuAAC reaction – 90 min
precipitation – 1–18 h
set up enrichment – 30 min
enrichment – 16–18 h
washing beads – 30 min
reduction – 30 min
alkylation – 30 min
set up trypsin digest – 15 min
trypsin digest – 12–18 h
collect trypsin digest – 15 min
dry trypsin digest – 4 h
TMT labeling – 1.5 h
dry TMT-labeled sample – 4 h
acid cleavage – 1.5 h
dry acid cleavage – 4 h
desalt – 30 min
Basic Protocol 3 (1 day)
run trypsin sample – 1.5 h
run acid cleavage sample – 1.5 h
search trypsin data – variable
search acid cleavage data – variable
run isostamp – variable
SIGNIFICANCE STATEMENT:
Photo-affinity labeling (PAL) enables the chemical capture of non-covalent small molecule–protein interactions for enrichment and identification of both protein targets and binding sites by mass spectrometry (MS), an unbiased analytical method for profiling interactions across the proteome. PAL is widely used for chemical capture due to its ability to react with a range of amino acids and binding sites, but the heterogeneity of the potential conjugation sites poses challenges for routine data analysis pipelines. This protocol describes an optimized method for the enrichment, isotopic coding, recovery, and analysis of the small molecule-conjugated peptides for routine, high-confidence assignment of binding sites and access to the structural information encoded in the observed binding interactions.
ACKNOWLEDGEMENTS:
Support from the Burroughs Wellcome Fund (C.M.W.), Ono Pharma Foundation (C.M.W.), Sloan Research Foundation (C.M.W.), the National Institutes of Health (DP1DA046586-02, C.M.W.), National Science Foundation (H.A.F.), and Harvard University are gratefully acknowledged.
Contributor Information
Hope A. Flaxman, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St., Cambridge, MA 02138
David K. Miyamoto, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St., Cambridge, MA 02138
Christina M. Woo, Department of Chemistry and Chemical Biology, Harvard University, 12 Oxford St., Cambridge, MA 02138
LITERATURE CITED:
- Bouvier ESPS, MA, US), Compton Bruce J. (Lexington, MA, US), Gebler John C. (Hopkinton, MA, US), Gilar Martin (Franklin, MA, US), Yu Ying-qing (Milford, MA, US), Lee Peter Jeng-jong (Westborough, MA, US), Brown Elizabeth K. (Sutton, MA, US). (2013). United States Patent No
- Browne DT, Hixson SS, & Westheimer FH (1971). A diazo compound for the photochemical labeling of yeast alcohol dehydrogenase. Journal of Biological Chemistry, 246(14), 4477–4484. Retrieved from http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=4328443&retmode=ref&cmd=prlinks. [PubMed] [Google Scholar]
- Chang C-F, Mfuh A, Gao J, Wu H-Y, & Woo CM (2018). Synthesis of an electronically-tuned minimally interfering alkynyl photo-affinity label to measure small molecule-protein interactions. Tetrahedron, 74(26), 3273–3277. Retrieved from 10.1016/j.tet.2018.03.024. doi: 10.1016/j.tet.2018.03.024 [DOI] [Google Scholar]
- Das J (2011). Aliphatic Diazirines as Photoaffinity Probes for Proteins: Recent Developments. Chemical reviews, 111(8), 4405–4417. Retrieved from http://pubs.acs.org/doi/abs/10.1021/cr1002722. doi: 10.1021/cr1002722 [DOI] [PubMed] [Google Scholar]
- Flaxman HA, Chang C-F, Wu H-Y, Nakamoto CH, & Woo CM (2019). A Binding Site Hotspot Map of the FKBP12-Rapamycin-FRB Ternary Complex by Photoaffinity Labeling and Mass Spectrometry-Based Proteomics. J Am Chem Soc, 141(30), 11759–11764. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/31309829. doi: 10.1021/jacs.9b03764 [DOI] [PubMed] [Google Scholar]
- Flaxman HA, & Woo CM (2018). Mapping the Small Molecule Interactome by Mass Spectrometry. Biochemistry, 57(2), 186–193. Retrieved from http://pubs.acs.org/doi/10.1021/acs.biochem.7b01038. doi: 10.1021/acs.biochem.7b01038 [DOI] [PubMed] [Google Scholar]
- Fleming SA (1995). Chemical reagents in photoaffinity labeling. Tetrahedron, 51(46), 12479–12520. Retrieved from http://linkinghub.elsevier.com/retrieve/pii/0040402095005983. doi: 10.1016/0040-4020(95)00598-3 [DOI] [Google Scholar]
- Gao J, Mfuh A, Amako Y, & Woo CM (2018). Small Molecule Interactome Mapping by Photoaffinity Labeling Reveals Binding Site Hotspots for the NSAIDs. J Am Chem Soc, 140(12), 4259–4268. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/29543447. doi: 10.1021/jacs.7b11639 [DOI] [PubMed] [Google Scholar]
- Gertsik N, am Ende CW, Geoghegan KF, Nguyen C, Mukherjee P, Mente S, … Li Y-M (2017). Mapping the Binding Site of BMS-708163 on γ-Secretase with Cleavable Photoprobes. Cell Chemical Biology, 24(1), 3–8. Retrieved from http://linkinghub.elsevier.com/retrieve/pii/S2451945616304457. doi: 10.1016/j.chembiol.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JR, & Robertson AAB (2018). Fishing for Drug Targets: A Focus on Diazirine Photoaffinity Probe Synthesis. J Med Chem, 61(16), 6945–6963. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/29683660. doi: 10.1021/acs.jmedchem.7b01561 [DOI] [PubMed] [Google Scholar]
- Lee S-Y, Kang M-G, Shin S, Kwak C, Kwon T, Seo JK, … Rhee H-W (2017). Architecture Mapping of the Inner Mitochondrial Membrane Proteome by Chemical Tools in Live Cells. Journal of the American Chemical Society, 139(10), 3651–3662. Retrieved from http://pubs.acs.org/doi/abs/10.1021/jacs.6b10418. doi: 10.1021/jacs.6b10418 [DOI] [PubMed] [Google Scholar]
- Lemeer S, Zörgiebel C, Ruprecht B, Kohl K, & Kuster B (2013). Comparing Immobilized Kinase Inhibitors and Covalent ATP Probes for Proteomic Profiling of Kinase Expression and Drug Selectivity. Journal of Proteome Research, 12(4), 1723–1731. Retrieved from http://pubs.acs.org/doi/abs/10.1021/pr301073j. doi: 10.1021/pr301073j [DOI] [PubMed] [Google Scholar]
- Li Z, Hao P, Li L, Tan CYJ, Cheng X, Chen GYJ, … Yao SQ (2013). Design and Synthesis of Minimalist Terminal Alkyne-Containing Diazirine Photo-Crosslinkers and Their Incorporation into Kinase Inhibitors for Cell- and Tissue-Based Proteome Profiling. Angewandte Chemie International Edition, 52(33), 8551–8556. Retrieved from http://doi.wiley.com/10.1002/anie.201300683. doi: 10.1002/anie.201300683 [DOI] [PubMed] [Google Scholar]
- MacKinnon AL, Garrison JL, Hegde RS, & Taunton J (2007). Photo-Leucine Incorporation Reveals the Target of a Cyclodepsipeptide Inhibitor of Cotranslational Translocation. Journal of the American Chemical Society, 129(47), 14560–14561. Retrieved from http://pubs.acs.org/doi/abs/10.1021/ja076250y. doi: 10.1021/ja076250y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DK, Flaxman HA, Wu H-Y, Gao J, & Woo CM A cleavable chelation-assisted biotin probe enables the discovery of a celecoxib binding site on prostaglandin E synthase. ACS Chem Biol(Submitted). [DOI] [PubMed]
- Parker CG, Galmozzi A, Wang Y, Correia BE, Sasaki K, Joslyn CM, … Cravatt BF (2017). Ligand and Target Discovery by Fragment-Based Screening in Human Cells. Cell, 168(3), 527–541.e529 Retrieved from http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=28111073&retmode=ref&cmd=prlinks. doi: 10.1016/j.cell.2016.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proteome Discoverer 2.2 Quick Start. (2017). Retrieved from https://assets.thermofisher.com/TFS-Assets/CMD/manuals/QS-XCALI-97809-Proteome-Discoverer-QSXCALI97809-EN.pdf
- Speers AE, & Cravatt BF (2005). A Tandem Orthogonal Proteolysis Strategy for High-Content Chemical Proteomics. Journal of the American Chemical Society, 127(28), 10018–10019. Retrieved from http://pubs.acs.org/doi/abs/10.1021/ja0532842. doi: 10.1021/ja0532842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szychowski J, Mahdavi A, Hodas JJL, Bagert JD, Ngo JT, Landgraf P, … Tirrell DA (2010). Cleavable Biotin Probes for Labeling of Biomolecules via Azide–Alkyne Cycloaddition. Journal of the American Chemical Society, 132(51), 18351–18360. Retrieved from http://pubs.acs.org/doi/abs/10.1021/ja1083909. doi: 10.1021/ja1083909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomohiro T, Morimoto S, Shima T, Chiba J, & Hatanaka Y (2014). An Isotope-Coded Fluorogenic Cross-Linker for High-Performance Target Identification Based on Photoaffinity Labeling. Angewandte Chemie International Edition, 53(49), 13502–13505. Retrieved from http://doi.wiley.com/10.1002/anie.201408580. doi: 10.1002/anie.201408580 [DOI] [PubMed] [Google Scholar]
- Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, … Ting AY (2012). Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew Chem Int Ed Engl, 51(24), 5852–5856. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/22555882. doi: 10.1002/anie.201108181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker SA, Kashyap S, Li X, & Kapoor TM (2011). Examining the Mechanism of Action of a Kinesin Inhibitor Using Stable Isotope Labeled Inhibitors for Cross-Linking (SILIC). Journal of the American Chemical Society, 133(32), 12386–12389. Retrieved from http://pubs.acs.org/doi/abs/10.1021/ja204561q. doi: 10.1021/ja204561q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, … Cravatt BF (2010). Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature, 468(7325), 790–795. Retrieved from 10.1038/nature09472. doi: 10.1038/nature09472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westheimer FH (1980). Photoaffinity Labeling-Retrospect and Prospect. Annals of the New York Academy of Sciences, 346(1 Applications), 134–143. Retrieved from message:%3C6B563075-6F23-43FD-892D-30E43D6C6044@chemistry.harvard.edu%3E. doi: 10.1111/j.1749-6632.1980.tb22097.x [DOI] [Google Scholar]
- Woo CM, Felix A, Byrd WE, Zuegel DK, Ishihara M, Azadi P, … Bertozzi CR (2017). Development of IsoTaG, a Chemical Glycoproteomics Technique for Profiling Intact N- and O-Glycopeptides from Whole Cell Proteomes. Journal of Proteome Research, 16(4), 1706–1718. Retrieved from http://pubs.acs.org/doi/abs/10.1021/acs.jproteome.6b01053. doi: 10.1021/acs.jproteome.6b01053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, & Bertozzi CR (2015). Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nature methods, 12(6), 561–567. Retrieved from http://www.nature.com/articles/nmeth.3366. doi: 10.1038/nmeth.3366 [DOI] [PMC free article] [PubMed] [Google Scholar]