Abstract
In the past decade, advances in precision oncology have resulted in an increased demand for predictive assays that enable the selection and stratification of patients for treatment. The enormous divergence of signalling and transcriptional networks mediating the crosstalk between cancer, stromal and immune cells complicates the development of functionally relevant biomarkers based on a single gene or protein. However, the result of these complex processes can be uniquely captured in the morphometric features of stained tissue specimens. The possibility of digitizing whole-slide images of tissue has led to the advent of artificial intelligence (AI) and machine learning tools in digital pathology, which enable mining of subvisual morphometric phenotypes and might, ultimately, improve patient management. In this Perspective, we critically evaluate various Al-based computational approaches for digital pathology, focusing on deep neural networks and ‘hand-crafted’ feature-based methodologies. We aim to provide a broad framework for incorporating AI and machine learning tools into clinical oncology, with an emphasis on biomarker development. We discuss some of the challenges relating to the use of AI, including the need for well-curated validation datasets, regulatory approval and fair reimbursement strategies. Finally, we present potential future opportunities for precision oncology.
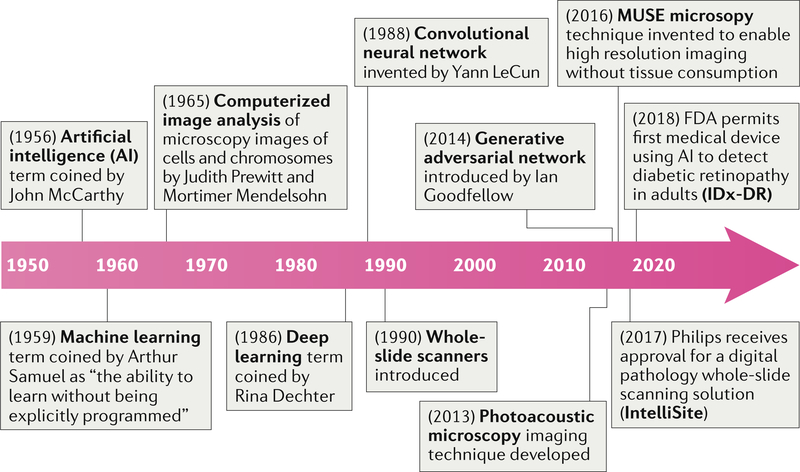
Digital pathology includes the process of digitizing histopathology slides using whole-slide scanners as well as the analysis of these digitized whole-slide images (WSI) using computational approaches. Such scanners were introduced two decades ago (ca. 1999), but the roots of digital pathology can be traced back to the 1960s, when Prewitt1–3 and Mendelsohn3 devised a method to scan simple images from a microscopic field of a common blood smear, converting the optical data into a matrix of optical density values while preserving spatial and grey-scale relationships, and then discerning the presence of different cell types based on the information in the scanned image (FIG. 1).
Fig. 1 |. Milestones in computational pathology.
Over the past two decades, technological advances have enabled efficient digitization of whole-slide images, subsequently helping to streamline pathology workflows across pathology labs worldwide. Slide digitization has enabled the creation of large-scale digital-slide libraries, the most popular of which is probably The Cancer Genome Atlas188, which has enabled researchers around the world to freely access a richly curated and annotated dataset of pathology images linked with clinical, outcome and genomic information, in turn spurring substantial research activity into artificial intelligence for digital pathology and oncology189.
Artificial intelligence (AI) is a broadly encompassing term, coined by McCarthy et al.4,5 in the 1950s, referring to the branch of computer science in which machine-based approaches are used to attempt to make a prediction — emulating what an intelligent human might do in the same situation. Machine learning (ML)-based approaches, which involve the machine ‘learning’ from data that are fed into it in order to make a prediction, fall under the broad umbrella of AI. Deep learning (DL) is a particular ML approach developed through the advancement of artificial neural networks6,7, which were devised in the 1980s as artificial representations of the human neural architecture. A DL network8,9 typically comprises multiple layers of artificial neural networks and tends to include an input layer, an output layer and multiple hidden layers. Interestingly, the hidden layers are also used to generate new representations of the image and, with a sufficient number of training instances, can be used to identify the representations that best distinguish categories of interest. In the past 15 years, the findings of many research groups (such as Hinton and others10–15) and a parallel increase in computational processing power have resulted in increasing interest in the use of DL for a number of different applications, including digital pathology.
The term digital pathology was initially coined to include the process of digitizing WSIs using advanced slide-scanning techniques, and now also refers to AI-based approaches for the detection, segmentation, diagnosis and analysis of digitized images (FIG. 2). To our knowledge, the first large-scale multicentre comparison of diagnostic performance between digital pathology and conventional microscopy was a comprehensive study by Mukhopadhyay et al.16 including specimens from 1,992 patients with different tumour types and involving 16 surgical pathologists. This study showed that primary diagnostic performance with digitized WSIs was non-inferior to that achieved with traditional microscopy-based approaches (with a major discordance rate from the reference standard of 4.9% for WSI and 4.6% for microscopy).
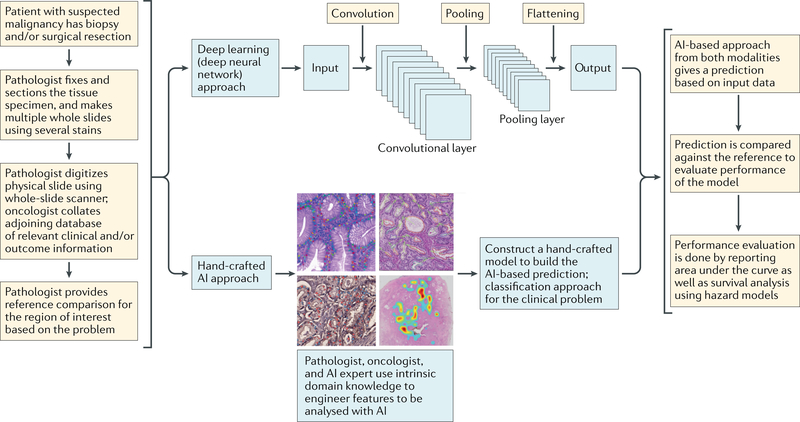
Fig. 2 |. Workflow and general framework for artificial intelligence (Ai) approaches in digital pathology.
Typical steps involved in the use of two popular categories of AI approaches: deep learning and hand-crafted feature engineering.
The development of new AI-based image analysis approaches in pathology and oncology is being led by computer engineers and data scientists, who are developing and applying AI tools for a variety of tasks, such as helping to improve diagnostic accuracy and to identify novel biomarker approaches for precision oncology. Pathologists and oncologists are the primary end users of these image analysis approaches. In routine clinical practice, pathologists (particularly anatomical pathologists) base their histological diagnosis in the visual recognition, semi-quantification and integration of multiple morphological features of the analysed sample and in the context of the underlying disease process. With extensive, postgraduate systematic training, pathologists are able to rapidly extract dominant morphological patterns that are associated with predefined criteria and established pre-existing clinical features in order to classify their observations. Most commonly, the result of this process is a histopathological diagnosis that is delivered in a written report to the treating physicians. While the systematic training and use of standardized guidelines can support harmonization of the analytical process and diagnostic accuracy, the histopathology analysis is inherently limited by its subjective nature and the natural differences in visual perception, data integration and judgement between independent observers17–20. Discrepancies in opinion can arise even between pathologists with the same training, leading to diagnostic inconsistencies21,22 and potential suboptimal patient care. The nature of this problem is, most likely, multifactorial and effective solutions remain elusive. In addition, the widespread use of non-invasive or minimally invasive procedures to acquire diagnostic samples has considerably reduced the size and quality of specimens obtained, making the work of pathologists more challenging. This difficulty is paradoxically accompanied by an increasing demand to perform refined diagnostics, including reporting of variables with prognostic or predictive value. Even with the use of molecular analysis strategies (in principle more objective), frequent limitations complicate definitive diagnosis or characterization of the biological drivers of the disease process to anticipate a clinical course. Patient preferences and reimbursement considerations can also considerably influence the diagnostic process and should be leveraged carefully by the clinical team23. These issues are further compounded by the variability in companion diagnostic assays and biomarkers24 for guiding treatment decisions, which is often the result of a lack of standardization but also of spatial and/or temporal biological heterogeneity in samples25,26.
In cancer, the complexity of genomic alterations that affect cell signalling and cellular interactions with their environment can influence the biological course of the disease and affect responses to therapeutic interventions. The assessment of such alterations requires the simultaneous interrogation of multiple features with highly sensitive and precise approaches. In addition, most biological features are continuous variables, and the reduction of these characteristics into categorical and/or discrete variables is necessary for their use in clinical decision-making. Nevertheless, biomarker development is often unidimensional, qualitative and does not account for the complex signalling and cellular network of tumour cells and/or tissues. Automated AI-based extraction of multiple subvisual morphometric features on routine haematoxylin and eosin (H&E)-stained preparations remains limited by sampling issues and tumour heterogeneity, but can help to overcome limitations of subjective visual assessment and to integrate multiple measurements in order to capture the complexity of tissue architecture. These histopathological features could potentially be used in conjunction with other radiological, genomic and proteomic measurements to provide a more objective, multi-dimensional and functionally relevant diagnostic output.
Thus, AI-based approaches, which are robust and reproducible, are a starting point in alleviating some of the challenges faced by oncologists and pathologists. This premise is supported by the results of several studies showing that AI-based approaches have a similar level of accuracy to that of expert pathologists27–29 and, more importantly, can improve the performance of the human reader when used in tandem with standard protocols in detection and diagnostic scenarios30,31. Herein, we present an overview of how AI-based approaches can be integrated into the workflow of pathologists and oncologists, and discuss the challenges associated with the implementation of such tools in the routine management of patients with cancer.
AI approaches in pathology
In digital pathology, AI approaches have been applied to a variety of image processing and classification tasks, including low-level tasks, focused around object recognition problems such as detection27,32–37 and segmentation38–41 as well as higher-level tasks such as predicting disease diagnosis and prognosis of treatment response on the basis of patterns in the image42–53. Independently of the final application, AI approaches are built to initially extract appropriate image representations, which can then be used to train a machine classifier for a particular segmentation, diagnostic or prognostic task. Several AI applications in digital pathology have been focused on the need to automate tasks that are time consuming for the pathologist27,32,36–38,54–57, subsequently enabling them to spend additional time on high-level decision-making tasks — especially those related to disease presentations with more confounding features31,58,59. In addition, AI approaches in digital pathology have been increasingly applied towards helping to address issues faced by oncologists, for example, through the development of prognostic assays to evaluate disease severity and outcome42,45,47,60,61 as well as assays to predict response to therapy62–64.
Hand-crafted feature-based approaches
Feature engineering is the process of delineating and developing the building blocks of ML algorithms, either by leveraging intrinsic domain knowledge (domain-inspired features) of pathologists and oncologists to create a particular hand-crafted feature-based ML approach, or without inherent domain knowledge (domain-agnostic features). In the hand-crafted ML approach, attempts are made to engineer new features that are anchored to the problem domain — that is, the algorithms are usually targeted towards a specific cancer or tissue type, focusing on particular features that might not be applicable for a broad use (FIG. 3). For example, these hand-crafted features could reflect the quantitative enumeration of mitotic figures36,65,66, which are currently qualitatively (and hence, subjectively) assessed by pathologists during the grading of breast cancers. Nevertheless, hand-crafted features can also encompass domain-agnostic features, such as subvisual textural heterogeneity measurements43,44,62,67 of the tissue or quantitative measurements of nuclear shape and size, which could be applied across disease and tissue types. Other domain-agnostic features that are broadly used include graph-based approaches, which enable quantification of the spatial distribution, arrangement and architecture of individual types of discrete tissue elements or primitives (for example, nuclei, lymphocytes or glandular structures) or between different tissue-specific primitives. Herein, we highlight a selection of numerous hand-crafted feature-based approaches that are being developed. These domain-agnostic and domain-specific hand-crafted feature-based approaches have been used for the diagnosis, grading, prognosis and prediction of response to therapy for cancer subtypes, including prostate68, breast69, oropharyngeal carcinomas60,70 and brain tumours71,72.
Fig. 3 |. Visual representations of hand-crafted features across cancer types.
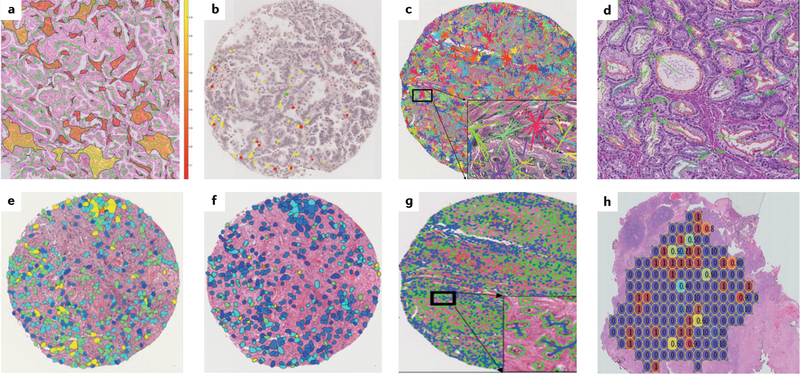
a | Spatial arrangement of clusters of tissue-infiltrating lymphocytes in a non-small-cell lung carcinoma (NSCLC) whole-slide image. b | Features developed using quantitative immunofluorescence of tissue-infiltrating lymphocyte subpopulations (including detection of CD4+ and CD8+ T cells and CD20+ B cells) in NSCLC samples. c | Features reflecting the distribution and entropy of global cell cluster graphs constructed using NSCLC specimens. d | Features computing the relative orientation of the glands present in prostate cancer tissue. e | Diversity of texture of cancer cell nuclei in an oral cavity squamous cell carcinoma. f | Nuclear shape feature computed on cancer cell nuclei in a human papillomavirus-positive oropharyngeal carcinoma. g | Graph feature showing the spatial relationships of different cancer cell nuclei in an oral cavity carcinoma. h | Hand-crafted feature capturing cellular heterogeneity in an oestrogen receptor-positive breast cancer.
Diagnostic applications.
In the diagnostic setting, Osareh et al.73 presented a supervised ML model focused on 10 cellular features identified by an expert breast pathologist in order to differentiate between malignant and benign breast tumours using images of samples obtained by fine-needle aspiration biopsy. Veta et al.74 showed that, in a tissue microarray (TMA) of male breast cancers, features such as nuclear shape or texture had prognostic value. In work from our group, Lee et al.42 presented a novel gland angularity feature associated with the level of disorganization of the glandular architecture in pathology images from prostate specimens; this feature occurred more frequently in advanced-stage prostate cancers compared with indolent prostate cancers. We have also presented novel hand-crafted features relating to disorders in nuclear shape, orientation and architecture in the tumour and tumour-associated benign regions43. These features were subsequently used in conjunction with an ML classifier developed using H&E-stained surgical prostate specimens to predict the likelihood of biochemical recurrence within 5 years of surgery. In a study of oral cavity squamous cell carcinoma, we used computerized features relating to the diversity of nuclear shape and texture to stratify patients into risk categories predictive of disease-free survival (DFS)75. The study44 also showed that a combination of nuclear shape and orientation features enables patients with oestrogen receptor (ER)-positive breast cancer with short-term survival (<10 years) to be distinguished from those with long-term survival (>10 years). Our team67 showed that quantitative features of nuclear shape, texture and architecture independently enable prediction risk of recurrence in patients with ER-positive breast tumours (on the basis of the 21-gene expression-based companion diagnostic test Oncotype DX).
Prognostic applications.
Graphical approaches are currently being used to evaluate the spatial arrangement and architecture of different types of tissue elements in order to predict clinical outcome. In an article published in 2018, Saltz et al.76 described the use of a convolutional neural network (CNN) in conjunction with feedback from pathologists to automatically detect the spatial organization of tumour-infiltrating lymphocytes (TILs) in images of tissue slides from The Cancer Genome Atlas, and found that this feature was prognostic of outcome for 13 different cancer subtypes. In a related study, we showed45 that the spatial arrangement of clusters of TILs in early stage non-small-cell lung cancer (NSCLC), which was calculated by computing the graph-derived relationships between TILs proximally located to each other and between TILs and cancer cell nuclei, was strongly prognostic of recurrence risk to a greater extent than TIL density alone. Yuan77 proposed a method to model and analyse the spatial distribution of lymphocytes among tumour cells on triple-negative breast cancer WSIs. Using this model, they identified three different categories of lymphocytes according to their spatial proximity to cancer cells. The ratio of intratumoural lymphocytes to cancer cells was found to be independently prognostic of survival and correlated with the levels of cytotoxic T lymphocyte protein 4 (CTLA-4) expression determined by TMA gene expression profiling. These investigators78 further expanded this method and found that the spatial distribution of immune cells was also associated with late recurrence in ER-positive breast cancer.
While many hand-crafted ML approaches have primarily been focused on analysing cells of epithelial origin within the tumour, increasing interest exists in trying to identify prognostic patterns within the tumour stroma. In a seminal study by Beck et al.79, 6,642 features relating to morphology as well as to spatial relationships and global image features of epithelial and stromal regions were extracted from digitized WSIs of specimens from patients with breast cancer. The features were used to train a prognostic model, and were found to be strongly associated with overall survival (OS) in cohorts of patients with breast cancer from two different institutions; features extracted from the stromal compartment had a stronger prognostic value (P = 0.004) than features extracted from the epithelial compartment (P = 0.02). In a study presented by our team, Ali et al.60 showed that combining nuclear features of the stromal and the epithelial compartments enabled prediction of the likelihood of progression of human papillomavirus-positive oropharyngeal cancer. Our team80 discovered statistically significant differences in nuclear features of the stromal compartment (23 discriminative features were selected with a P < 0.05) in images of surgically excised tissues from African American versus white American patients with prostate cancer, after correcting for disease stage and grade, indicating that population-specific features need to be taken into account in model design. Additionally, these investigators found that a prognostic model based on stromal features alone and trained in a cohort of African American patients with prostate cancer had a stronger prognostic accuracy for biochemical recurrence after surgery in an independent test cohort of African American patients than a model trained with both African American and white American patients with prostate cancer (average area under the curve (AUC) 0.82 versus 0.68).
Drug discovery and development applications.
Over the past few years, interest in the use of ML-based approaches for drug discovery and development has increased81. The fact that a substantial proportion of patients receiving certain therapeutic modalities, such as cytotoxic agents or immune-checkpoint inhibitors (ICIs), do not respond to treatment has resulted in increased interest in combining AI with digital pathology to identify the patients who are most likely to derive therapeutic benefit. Wang et al.64 developed an approach whereby nuclear and perinuclear features (shape, orientation and spatial arrangement) could be used to stratify patients with early stage NSCLC treated with surgery alone into two distinct groups according to the risk of disease recurrence; patients in the high-risk group were considered potential candidates who might benefit from adjuvant chemotherapy. Hand-crafted ML approaches have also been focused on probing therapeutic response to particular therapeutic agents, including targeted agents, chemotherapy drugs and ICIs. Wang et al. have developed an approach whereby the spatial arrangement of nuclei62 or TILs63 enables prediction of the responsiveness of patients with late-stage NSCLC to the anti-programmed cell death 1 (PD-1) antibody nivolumab.
Deep neural network-based approaches
DL approaches have been increasingly used and are being adapted in the context of digital pathology because they do not rely on engineered features and they can learn representations directly from the primary data82. DL approaches typically involve a learning set of images with associated class labels (for example, whether the tumour is benign or malignant83,84) and subsequent interrogation of new input data with no pre-existing assumptions. The process involves generating and subsequently learning the optimal image features to best separate the categories of interest. Other reasons for the wide acceptability of DL-based methods are their easier application (compared with hand-crafted feature engineering) and high accuracy. Consequently, several DL models or algorithms for analysing pathology images have been developed.
Convolutional neural networks.
CNNs are the most extensively used DL algorithms to date and have been applied to a variety of pathology image analysis applications83–86. A CNN is a type of deep, feedforward network that comprises multiple layers in order to infer an output (typically a fixed category) from an input (for example, an image), and is so named because of the presence of multiple convolutional sheets, which are the building blocks of a CNN in which the network learns and extracts feature maps from the image using filters between the input and output layers. The layers in a CNN are not fully connected: the neurons in one layer interact with only a fixed region of the previous layer and not with all neurons. In addition, CNNs also comprise pooling layers in between, whose primary function is to scale down or reduce the dimensionality of the features. The CNN thus works by hierarchically deconstructing the image into low-level cues, such as edges, curves or shapes, which are then aggregated to form high-order structural relationships in order to identify features of interest. CNN DL-based approaches have been used for image-based detection and segmentation tasks to identify and quantify cells32,33,82,87 (such as neutrophils, lymphocytes and blast cells), histological features (for example, nuclei34,35,88,89, mitotic figures, stroma90 and glandular structures38,39) or regions of interest (such as the tumoural54 or peritumoural areas). In addition, Senaras et al.91 have developed the CNN-based DeepFocus system to automatically detect and segment out-of-focus and blurry areas in digitized WSIs, with an average accuracy of 93.2% (±9.6%).
With regard to AI-based diagnostic approaches, Araujo et al.33 used a CNN to classify WSI images of suspected breast cancer specimens as containing non-malignant tissue, benign lesion, in situ carcinoma or invasive carcinoma. Esteva et al.92 used a deep CNN trained on images of skin lesions and compared its performance against that of 21 board-certified dermatologists in differentiating keratinocyte carcinoma from benign seborrheic keratosis and malignant melanoma from benign nevi. The CNN achieved results comparable to those of evaluations by expert dermatologists. Tschandl et al.93 compared the diagnostic accuracy of different CNN-based DL models against human readers in correctly classifying pigmented skin lesions into one of seven predefined disease categories using digital dermatoscopic images. The investigators found that the DL models consistently outperformed human physicians. In a comparison of 511 human readers against 139 DL algorithms, the algorithms achieved a mean of 2.01 more correct diagnoses (17.91 versus 19.92; P < 0.0001).
In terms of prognostic applications, Couture et al.94 applied a CNN to images of H&E-stained TMA in order to determine the histological and intrinsic molecular subtypes of the component breast cancers. Our group95 used a CNN combined with a fully connected network to automatically detect mitotic figures in WSIs of ER-positive breast cancer specimens, and found that the distribution of mitotic figures differed significantly between those with a high versus a low Oncotype DX-defined risk of disease recurrence (P = 0.00001). Nagpal et al.28 used a DL system to automatically assign Gleason scores following detection of cancerous regions in WSIs of radical prostatectomy specimens. In a validation set of 331 slides, with the reference diagnostic standard set by an expert genitourinary pathologist who also had access to initial diagnostic comments from prior reviews of at least three general pathologists, the DL approach had an accuracy of 0.70 in predicting Gleason scores, whereas 29 general pathologists had a mean accuracy of 0.61. Geessink et al.96 used CNN-based approaches with prognostic intent in patients with colorectal cancer. This CNN was leveraged to quantify the presence of the stromal component within the tumour using pathologist-identified ‘stromal hotspots’; the ratio of tumour-to-stroma was independently prognostic for DFS (HR 2.05, CI 1.11–3.78) in a multivariable analysis incorporating clinicopathological factors. Kather et al.97 used a CNN to generate a ‘deep stroma score’ and found it to be independently prognostic of recurrence-free survival (HR 1.92, CI 1.34–2.76) and OS (HR 1.63, CI 1.14–2.33) in an independent validation set of 409 patients.
Fully convolutional networks.
Another popular DL model is the fully convolutional network (FCN), which lacks fully connected layers and comprises only a hierarchy of convolutional layers. Unlike a CNN, which is used to aggregate information locally for a global prediction, FCNs can be used to learn representations from every pixel and, therefore, potentially enable detection of an element or feature that might occur sparsely in an entire pathology image. This attribute enables a FCN to make pixel-level predictions with a possible advantage over a CNN, which learns from repetitive features that occur throughout the entire image. In digital pathology, FCNs have been used by Rodner et al.98 to differentiate cancerous regions from non-malignant epithelium in histopathology images of head and neck cancer specimens. Using co-registered H&E images with multimodal microscopy techniques, the FCN was used to segment the WSIs into four classes: cancer, non-malignant epithelium, background and other tissue. Our group99 used a FCN, which was trained on 500 images from 349 patients, in 195 patients in order to detect invasive breast cancer regions on WSIs, resulting in a detection accuracy of 71% (Sørensen-Dice coefficient) when compared with the assessment of an expert breast pathologist.
Recurrent neural networks.
Unlike CNNs and FCNs, which are limited to the analysis of data from one individual time point, recurrent neural networks (RNN) can store inputs over different time intervals or time points in order to process them sequentially and learn from (often several million) discrete earlier steps100. The RNN takes into account the status of the input at different time points, thus exhibiting dynamic behaviour. Long short-term memory101 (LSTM) networks are a type of RNN augmented by the presence of recurrent gates, referred to as ‘forget’ gates, which enable the CNN to learn tasks by looking back at propagated errors. Bychkov et al.29 built a network combining a LSTM and a CNN to predict the risk of disease recurrence (high versus low, based on retrospective DFS data) using images of H&E-stained colorectal cancer TMA specimens. The investigators first deconstructed the entire TMA image into small patches, which were then fed into the CNN. Thus, the advantage of using the LSTM network was that the model was able to learn patterns from each patch, all of which were aggregated to generate a patient-level prediction. Accordingly, the predictive performance of this LSTM network (average AUC 0.69) was higher than that based on histological grade alone (average AUC 0.57) and a visual risk score agreed by three expert pathologists (average AUC 0.58). The analysis of tissue images obtained at different time points, for example, serial follow-up biopsy samples from patients with prostate cancer who are under active surveillance, is another potential use of the RNN-based AI approach.
Generative adversarial networks.
In addition to CNNs, generative adversarial network102 (GAN)-based approaches are showing increasing promise in DL-based digital pathology approaches, including feature segmentation103 and stain transfer (a term that refers to the correction of colour variations)104,105. A GAN works by implementing two simultaneous neural networks, which compete with each other. One network is the generator and produces synthetic data from training exemplars fed to the network, while the second network evaluates the agreement between the generated and the original data. The objective of a GAN is to decrease the degree of classification error of the second network, such that the generated images more closely resemble the original images. Gadermayr et al.106 used a GAN to segment out glomeruli from images of renal pathology specimens obtained from resected mouse kidneys (which show high similarity to human kidneys). A GAN-based approach107 has also been used for training a DL method to automatically score tumoural programmed cell death 1 ligand 1 (PD-L1) expression in images of NSCLC biopsy samples. This approach helped minimize the number of pathologist annotations necessary and thus compensate for the lack of tissue available in a biopsy specimen. Similarly, Xu et al.105 proposed a novel GAN-based approach to convert the H&E staining of WSIs to virtual immunohistochemistry staining based on cytokeratins 18 and 19, an approach that potentially obviates the need for destructive immunohistochemistry-based tissue testing (TABLE 1).
Table 1 |.
Comparison of hand-crafted feature engineering and DL-based approaches
| Aspect | Hand-crafted feature engineering | DL-based approaches |
|---|---|---|
| Development | Usually developed in close collaboration with expert pathologists and oncologists; these approaches are based on a given aspect of domain knowledge, and thus their development can be complicated and time consuming | Developed though unsupervised feature learning, dependent on the existence of learning sets and annotated exemplars from the categories of interest; network design usually involves a focus on fine-tuning the algorithm to maximize accuracy while minimizing processing time |
| Generalizability | Approaches usually tailored for a specific cancer subtype and/ or tissue of origin; for example, features relating to glandular morphology would only apply to diseases with an abundance of tubules and glands | Using approaches such as transfer learning, a network trained on a particular disease subtype could be applied to other subtypes as well |
| Training datasets | The particular set of features or markers of interest are known, and thus small-sized training datasets are needed for feature engineering | Large amounts of well-annotated data with several exemplars from the different target categories or classes of interest are often required |
| Interpretability | Feature engineering is typically associated with attributes of the disease domain, and thus the features tend to have greater interpretability and a stronger morphological and biological underpinning in comparison with DL-based approaches | Representations can often be difficult to interpret and, despite new emerging approaches aimed at providing clarity (such as visual attention maps), these approaches are still largely considered ‘black-box' methods |
| Clinical deployment | On account of being more interpretable than DL-based approaches, hand-crafted features might be more likely to be used for high-level decision-making, such as that relating to disease prognosis or prediction of benefit from therapy | Might be more appropriate in situations in which the need to ‘explain the decision' is reduced; such situations could include low-level tasks such as object detection or segmentation |
DL, deep learning.
Integration of pathology and oncology
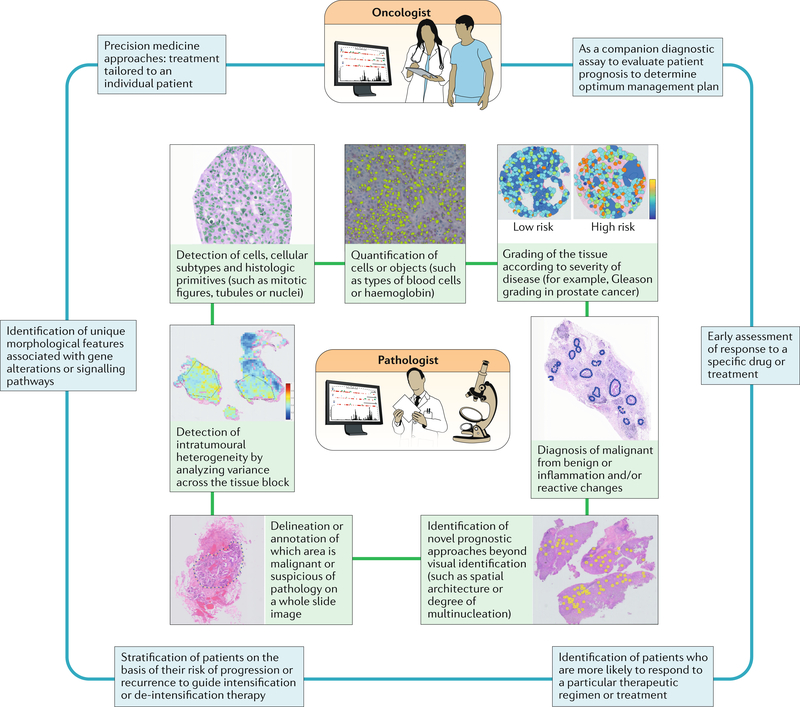
Successful Al-based approaches for digital pathology need to closely integrate the work of the pathologist with that of the oncologist (FIG. 4). For example, most supervised AI algorithms are crucially dependent on carefully annotated digital slide images for classifier training. The pathologist also has an essential role in providing domain-specific knowledge to the computational and data scientists involved in designing and developing the AI algorithms. This collaboration might entail the pathologist directing the developers towards the specific attributes of the pathology image that should be examined and helping in the training of the algorithms through the annotation and segmentation of cells, tissue types, biological structures or regions of interest. The pathologist also often provides the diagnostic reference or gold standard against which the ML algorithms will be compared.
Fig. 4 |. Artificial intelligence (Al) and machine learning approaches complement the expertise and support the pathologist and oncologist.
Some of the existing AI approaches currently used by pathologists to analyse images from tumours are depicted. For the practicing oncologist, AI approaches can be used to aid decision making for different aspects of the management of patients with cancer.
A major dilemma for oncologists is the lack of consensus when deciding whether to provide a particular treatment or not when tailoring therapy to an individual patient. In addition to their utility as decision support systems, AI approaches can also serve as companion tools for precision patient-centred approaches. For example, a thoracic oncologist might be initially undecided regarding offering chemotherapy to a patient with stage IA (early stage) NSCLC according to guidelines, despite a symptomatic decline following surgery. However, if an AI-based predictive approach assigns a high risk of recurrence to the patient on the basis of the histomorphometric analysis of a surgical specimen, this clinician would be more likely to recommend chemotherapy. In addition to enabling the standardization of management plans, AI could help oncologists to handle some of the limitations of current companion diagnostic assays based on genetic or tissue-based biomarkers108–110. Besides being tissue-destructive and requiring a large amount of viable tissue for analysis, one of the challenges associated with genomic-based tests is the increasingly well recognized intra-tumour and inter-tumour heterogeneity25,26,111,112 across cancer types, whereby diverse spatial locations can yield different prognostic information (for example, differences in the tumour genetic signatures according to the tissue sampling coordinates113). Thus, the final results of those tests are dependent on the specific area of the tumour from which the tissue is sampled. AI-based interrogation tools could help overcome this limitation by enabling the analysis of all the individual slides from the tumour to generate an integrated consistent signature representative of the entire lesion. Implementing ML methods into routine clinical practice is also potentially easier, less expensive and less disruptive than using genomic tests because routine standard-of-care images can be used. Genomic approaches have the advantage that they could provide considerable insight into the underlying molecular characteristics of the disease and, thus, several groups are exploring ways to combine molecular and morphological attributes of the tumour to improve the prognostic and predictive performance of ML approaches61,114,115. Indeed, future companion diagnostic tests for precision medicine applications will likely involve a combination of tumour morphological and molecular attributes58,116–119.
Oncologists also have to contend with rapidly changing treatment guidelines incorporating new therapeutic agents such as ICIs and targeted agents that require substantial out-of-pocket expenses and might not be covered by insurance. Optimized AI-based biomarker approaches could provide oncologists in this situation with an accurate and inexpensive tool to potentially pre-select patients for treatment with novel agents from which they would actually derive benefit, not only saving patients from incurring unnecessary expenses but also unwanted systemic toxicities without any substantial physiological improvement.
Challenges towards clinical adoption
Regulatory roadblocks
One of the key questions with regard to the clinical adoption of AI approaches for digital pathology in the clinic is the pathway for approval by regulatory agencies. The key principle guiding the approval process in most countries is the requirement of ‘an explanation of how the software works’120–122; this is obviously crucial for DL-based AI approaches, which are perceived as being a ‘black-box’ and lacking interpretability123. In the USA, the FDA uses a three-class system for the approval of medical devices on the basis of the risks that the device poses to the patient, as well as the intended use of the device, with Class I devices deemed to have the lowest risk and Class III devices to have the highest risk — AI-based devices tend to be assigned to Classes II or III. Typically, Class II devices are allowed to be marketed through the 510(k) approval pathway, whereas Class III devices require a more rigorous premarket approval.
In addition, under proposed changes published by the FDA in 2018124, the De Novo pathway can be used as an alternative approval mechanism for novel Class I or II devices. Applications for licensing of devices can be submitted directly through this pathway or after the device receives a ‘not substantially equivalent’ designation after submission for approval via the 510(k) pathway. In the European Union (EU), any software, including AI-based devices, “to be used for the purpose, among others, of diagnosis, prevention, monitoring, treatment, or alleviation of disease”125,126 is considered a medical device. The Medical Devices Regulation127 and the In Vitro Diagnostic Device Regulation128 will be effective from 26 May 2020 as a reform of the existing directives from the 1990s, bringing them into line to regulate AI-based health-care systems. Similar to the FDA’s system, the EU regulations that will be effective from May 2020 will use a four-tiered medical device risk classification, with Class A denoting the lowest risk and Class D implying the highest risk129.
In the USA, the FDA has started granting approval to DL-based approaches for clinical use. In 2018, the cloud-based Arterys130 imaging platform received approval via the 510(k) pathway to be used in radiology in order to help physicians track tumours in MRI and CT scans from patients with lung or liver cancer131. In 2017, Philips received De Novo pathway clearance to market the IntelliSite Pathology Solution as a comprehensive digital pathology system132. In 2019, the digital pathology solution PAIGE.AI133 was granted Breakthrough Device designation by the FDA134. Meanwhile, current guidelines in the EU require a Conformité Européenne (European Conformity) marking on devices and software before they can be applied to human tissue for diagnostic purposes129,135. Currently, no AI solutions with prognostic and/or predictive intent have a Conformité Européenne marking, but digital pathology solutions developed by Sectra, Philips and OptraSCAN have secured clearance to carry such a designation, thus paving the way for their deployment in the EU. In 2017, the FDA decided to downgrade radiology-based companion diagnostic assays from Class III to Class II medical devices136. With the new proposed changes to the De Novo pathway, as well as the intention to implement the Software Precertification programme under it, this pathway could become a viable mechanism for approving novel AI-based devices with low-to-moderate risk without predicates. MammaPrint137, an RNA-based genomic test to determine the likelihood of benefit from adjuvant chemotherapy in patients with early stage breast cancer, received 510(k) clearance138 from the FDA, suggesting that AI-based digital pathology companion diagnostic approaches might be able to leverage MammaPrint as a predicate device. Interestingly, FDA approval has not been sought for Oncotype DX139, another gene expression-based test for patients with breast cancer, owing to the status of this assay as a Clinical Laboratory Improvement Amendments (CLIA)-certified central test. Thus, developers of AI-based digital pathology tests could potentially take a similar approach and apply for CLIA certification as a Laboratory Developed Test. Actions taken in 2019 by the FDA, such as sending a warning letter140 to a lab illegally using CLIA-based genomic tests to predict response to specific drugs and publishing a discussion paper141 about the needs for future oversight of CLIA tests, suggest that this agency is reinforcing the concept that regulatory approval be obtained for predictive assays before they are used in the clinic. In addition, the College of American Pathologists, which is the organization that grants CLIA certification, has also called for the FDA to regulate high-risk prognostic and predictive tests currently considered to be Laboratory Developed Tests owing to the complexity and lack of transparency in how the test results are obtained with these tests142.
Quality of data
The performance of any AI-based approach is primarily dependent on both the quantity and quality of the input data. The data used to train an AI algorithm need to be clean, curated, with a maximal signal-to-noise ratio, and as accurate and comprehensive as possible in order to achieve the maximum predictive performance58,82,143,144. For example, if an AI approach is meant to segment a particular biological structure present in a WSI, the performance of the approach is primarily dependent on the fidelity of the reference annotations by expert pathologists in the learning set54,145. The importance of well-curated data is apparent in the work of Doyle et al.145, who have developed an ML-based AI approach to automatically detect regions of prostate cancer in WSIs. The investigators noted a decrease in the performance of the method when the magnification was increased, which they determined was due to the apparent loss in granularity and detail of the reference learning manual annotations at increased resolution. This situation also highlights the issue of the scanning resolution when digitizing WSIs. Most existing slide scanners have a maximum capability to scan at ×40. Higher resolution images (>×20) can be scaled down to be used by an algorithm trained at a resolution of ×20, but the use of an AI approach developed at ×40 when the maximum scanning resolution available is ×20 would likely result in a loss of data fidelity. Super-resolution microscopy techniques146 enable focus to be placed on specific biological elements (such as mitotic figures or nucleoli) at much higher resolution than can be obtained with standard optical microscopy techniques. An AI approach could be used to identify sites on the slides for subsequent imaging with super-resolution techniques, thus helping to scan certain key structures and locations in the image at substantially higher resolution while also reducing the total amount of digitally scanned data generated. Taking this approach, Kleppe et al.147 used an ML-based algorithm and determined that, across different solid tumour types, patients with chromatin defined as homogeneous had more favourable survival outcomes than those with heterogeneous chromatin.
Situations such as those discussed above warrant the need for the creation of accurate, manually annotated reference datasets by expert pathologists in order to standardize the evaluation of the performance of AI algorithms. In addition to efforts by the FDA to curate these datasets148, global digital pathology image analysis challenges at major AI and imaging conferences149 have led to the development of well-curated, accurate WSI reference datasets across cancer subtypes, with annotated cancerous regions as well as regions of interest, including mitotic figures, glandular structures and lymph node metastases, among others.
Interpretability
Despite their high accuracy and ease of application, criticism regarding the lack of interpretability and contrasting domain-inspired intuitiveness found in hand-crafted networks58,123,150 is a possible obstacle towards the clinical adoption of deep neural networks. A few studies31,55 have been aimed at providing biological interpretability to DL tools with current approaches, including post hoc methods or supervised ML models, to explain the output after the DL model has already made its prediction. Mobadersany et al.61 used a CNN trained using images of brain tumour biopsy specimens to predict OS. In one of the patients with high-grade tumours, the heat-map visualization of the AI prediction corresponded to pathologist-identified areas of early microvascular proliferation, which is a hallmark of malignant progression, thus providing unique interpretability to the analysis. These visual attention maps and post hoc analyses of DL methods have also been criticized123 on the basis that additional models should not be required to explain how a DL model works.
Hand-crafted AI approaches can provide greater interpretability because they are typically developed in conjunction with domain experts. Nevertheless, engineering hand-crafted features is often a challenging and non-trivial task owing to the substantial time investment of the pathologist or oncologist in the development of such approaches. In the past few years, fusion approaches involving integration of DL and hand-crafted strategies have started to gain attention. These strategies might involve the use of DL algorithms for the initial detection of cells or elements with subsequent reliance on hand-crafted ML approaches for prediction, thereby leveraging domain knowledge to ensure the biological interpretability of the approach. For example, our team47 used a DL approach to segment nuclei in images of TMAs comprising early stage NSCLC specimens before applying a hand-crafted approach involving the interrogation of nuclear shape and texture to predict which patients were more likely to have disease recurrence.
Algorithm validation
Prior to clinical adoption, AI-based and ML-based tools need to be sufficiently validated using multi-institutional data in order to ensure generalizability of the approaches. The available data for building an AI approach is often partitioned into training and validation sets. The initial dataset, often referred to as a training, learning or discovery set, is typically class balanced and exemplars from the categories of interest are equally represented. Once the model has been trained and locked down on the learning set (that is, no more alterations need to be made to the AI model), validation without further optimization is typically performed on a test set, which is either extracted from the original set of cases or obtained from a different institution.
One of the critical reasons for attempting to independently validate AI approaches using separate test sets is to ensure that these approaches are resilient to pre-analytical sources of variation, which can include variations in slide preparation, scanner models and/or protocols. Coudray et al.151 showed that a CNN trained on NSCLC images from The Cancer Genome Atlas could not only distinguish adenocarcinomas from squamous cell carcinomas with an average AUC of 0.97 but also, more interestingly, had consistent performance when validated on frozen tissue preparations, formalin-fixed paraffin-embedded tissues or biopsy-derived samples from a separate institutional cohort. By contrast, Zech et al.152 found that a CNN trained to detect pneumonia showed significantly poorer performance when it was trained using data from one institution and validated independently using data from two other institutions (P < 0.001) than when it was trained using data pooled from all three institutions. AI algorithms have been developed to standardize the data153, including stain154,155 and colour normalization40,156 techniques. In the past few years, research has also been aimed at building comprehensive quality control and standardization tools157,160. For example, we159,160 have developed novel stability measures in an attempt to instil AI algorithms with resilience to pre-analytic variations, and found that stable features (all of which were relating to gland shape) enabled better discrimination of Gleason grade and the presence of prostate cancer within surgical resection specimens.
Reimbursement and clinical adoption
Reimbursement of the costs of performing AI-based companion assays or decision-support systems is one of the major issues that needs to be overcome for assays to be implemented in routine clinical practice144. In the USA, insurance companies presently standardize expenses according to the current procedural terminology codes maintained by the American Medical Association and reported by medical professionals135,161–163. In the EU, procedure codes (analogous to current procedural terminology codes) differ among countries. In many other regions of the world, for example in India, no standardized system for medical billing exists and individual institutions typically implement their own procedures.
Currently, no dedicated procedure codes exist for the use of AI approaches in digital pathology with diagnostic or prognostic intent. New procedure codes might need to be established, but AI-based tools will probably first need to be approved by the FDA before they are reimbursable and thus implementable in the clinic — especially considering the possibility that the FDA will regulate complex CLIA-based tests in the near future. AI-based pathology companion diagnostic assays, however, could follow the model established by CLIA-based genomic tests, which again could be challenging owing to the apparent FDA intention to regulate CLIA-based tests more stringently. The needs of the end user might also dictate the way in which AI-based tools will be deployed.
Perspective of the pathologist.
For a pathologist, AI-based tools will be mainly needed to detect structures or specific regions of interest in digitized WSIs. Hence, the key element in the development of such applications is the digital slide scanner in order to enable quick turnaround times and the ability to control the clinical workflow. Moving forward, when slide scanners become more ubiquitous at hospitals and medical institutions, the deployment of AI tools might be app-based, with integration into the data cloud in order to enable pathologists to instantly share images and AI-based prediction with collaborators and patients around the world. Current challenges associated with cloud-based usage include the massive bandwidth required to transmit gigapixel-sized WSI images into data clouds as well as managing permanent and uninterrupted communication channels between end users and the cloud. Another consideration relating to the transition to digital pathology is the possibility of having automated approaches to assess the quality of slide digital images. Automated algorithms (for example, HistoQC157 and DeepFocus91) have been developed in the past few years to standardize the quality of WSIs; these tools can be used to automatically evaluate and detect optimum quality regions for analysis while eliminating out-of-focus regions or those with artefacts.
The performance thresholds that AI algorithms would have to achieve in order for pathologists to feel comfortable using them is an issue that has not been explicitly addressed. Nevertheless, Wang et al.30 demonstrated that the combination a DL-based model predictions with pathologist diagnoses of breast cancer metastases in WSIs of sentinel lymph-node biopsy specimens decreased the human error rate by almost 85%. In a study addressing a similar problem, Steiner et al.31 showed that algorithm-assisted pathologists were more accurate than the pathologist or the algorithm alone in the detection of micrometastases (sensitivity 91% versus 83%; P = 0.02).
Perspective of the oncologist.
For oncologists, digital pathology-based companion diagnostic tests could provide additional valuable information for disease risk stratification and patient selection for targeted therapies. Genomic companion diagnostic tests involve shipping tissue to a centralized location (usually one or two central labs worldwide), with a turnaround time of ~2 weeks. These AI-based tests could potentially be set up as CLIA-based tests, whereby tissue blocks are shipped and slides are centrally constructed and then digitized in order to maximize control over the process of creating the digital slides and to minimize pre-analytic variance. Obviously, this approach limits scalability, and hence a cloud-based approach using locally scanned WSIs stored in a Health Insurance Portability and Accountability Act-secure cloud environment might be another viable option for deploying AI-based companion diagnostic assays. These images would be accessible for interrogation and the generated risk score would be sent electronically to the ordering oncologist. Another important question for the oncologist is deciding the minimum performance and accuracy of companion diagnostic assays required for routine clinical use. In situations in which these tests are directly competing with existing tests, non-inferiority would be a minimum requirement but, ideally, superiority in direct comparisons is the level of performance that would convince the oncologist to adopt a new AI-based test.
The adoption of a prognostic or predictive biomarker is usually guided by the levels of evidence demonstrated in the validation of the test164,165. The highest level of evidence is level IA, generated by prospective validation of the assay in a large multi-centre prospective phase III clinical trial (possibly from cooperative oncology groups). Previously, prospective clinical trials were considered the highest level of evidence, and evidence from retrospective studies was relegated to level II or lower165, yet updated evidence guidelines indicate that retrospective studies using controlled archived samples from at least two independent prospective phase III trials can provide evidence of up to level IB164. Genomic assays currently in clinical use need at least level IB evidence of clinical validity before they can be incorporated into practice guidelines. For instance, the recent National Comprehensive Cancer Network guidelines for breast cancer166 included Oncotype DX and MammaPrint owing to level IA evidence following the completion of the landmark 10-year analyses of the phase III TAILORx167 (involving 9,719 patients) and MINDACT168 (in 6,693 patients) trials, respectively. Interestingly, Oncotype DX was CLIA certified without traditional prospective validation, following level IB evidence generated using archived samples of the already completed NSABP B-14169 (in 668 patients) and B-20170 (in 651 patients) studies. Interestingly, Decipher171, an assay for predicting the risk of metastasis in patients with prostate cancer treated with radical prostatectomy, currently only has level IB evidence172, but is CLIA certified and currently in the National Comprehensive Cancer Network guidelines173.
Likewise, AI-based prognostic and predictive assays will probably need to achieve at least level I evidence to support clinical deployment. Another consideration is whether AI assays are validated in a way that enables their use in clinically relevant applications. For example, the use of Oncotype DX in routine oncology practice was spurred by an evolution from being merely prognostic to also being predictive of benefit from adjuvant chemotherapy in patients with ER-positive, HER2-negative, lymph node-negative, early stage breast cancer. In 2018, our team114 presented the first findings of a digital pathology AI test to predict the risk of recurrence in patients with breast cancer on archived samples from a completed clinical trial (ECOG 2197). We showed that the AI-derived risk groups enabled prediction of breast cancer recurrence with a HR of 2.41 (CI 1.21–4.79) in 378 patients from the trial. In a subset of 116 patients with available Oncotype DX scores, patients in the low-risk group predicted with the AI tool had a 10-year recurrence rate of 17.2% as compared to 19.6% for those in the Oncotype DX-defined low-risk group.
Conclusions
The advent of whole-slide digital scanning and the concomitant rise of DL-based neural networks for interrogating digital images of slides has resulted in an explosion of interest in AI-based digital pathology technologies. Despite the ambiguity and challenges surrounding regulatory strategies, reimbursement and deployment, interest in the development and use of these technologies, both from the pathology and oncology communities, is increasing58,144,174–177. Startups like PAIGE.AI178,179, Proscia180, DeepLens181, PathAI182 and Inspirata183 are using DL-based AI tools for detection, diagnosis and prognosis of several cancer subtypes. Some of them, such as Inspirata and PAIGE.AI, are spending substantial time and resources on creating large libraries of digital WSI for use in training AI algorithms. Interestingly, the landscape of digital pathology is, in parallel, also undergoing important innovation and rapid changes. Breakthroughs from the past few years include open-top light sheet microscopy184, which generates 3D images of a tissue sample without the need for destructive sectioning or slide preparation, and MUSE microscopy185, which can be used to gather high-resolution images of tissue surfaces almost instantly through the use of ultraviolet illumination, in some cases without the need for tissue processing or staining. These non-destructive, slide-less techniques might provide a substantially greater degree of 3D spatial and volumetric information and might render conventional 2D whole-slide scanners redundant in the future. The AI approaches that have been developed to date using 2D-stained slides would have to be suitably adapted to leverage the novel techniques that generate 3D images of the entire tissue, keeping in mind that a substantially larger volume of data would need to be analysed. In addition, future companion diagnostic approaches might need to incorporate multimodal measurements, such as proteomics, genomics and measurements from multiplexed marker-staining platforms (for example, fluorescence in situ hybridization, immunofluorescence or digital spatial profiling), in order to provide a comprehensive and holistic patient-specific portrait of the tumour.
Despite these challenges and obstacles, the potential of AI approaches for digital pathology is promising. In the past few years, several institutions around the world have decided to digitize their entire pathology workflow177,186,187, and the FDA approval of the Philips whole-slide scanner in 2017 marked a major inflexion point in the path towards truly digital pathology laboratories. It is worth remembering that changes are hard to accept: the light microscope popularized by Anton van Leuwenhoek has been the mainstay for pathologists for the past 100 years, and the advent of digital mammography in radiology involved a sudden transition to a film-less workflow. Now, after radiology has transitioned from 2D X-ray images to 3D CT scans and MRIs, pathology is on the cusp of incorporating 3D tissue representations with more extensive sampling to improve diagnostic, prognostic and predictive decision-making. AI approaches will thus be key in analysing and interpreting these high-volume data, aiding pathologists and oncologists in the process.
Acknowledgements
Research discussed in this publication was supported by the Department of Defence and Department of Veterans, the National Cancer Institute of the National Institutes of Health, the National Centre for Research Resources, the Ohio Third Frontier Technology Validation Fund, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and the Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the institutions named.
Footnotes
Competing interests
K.A.S. has received fees as a speaker for Merck and Takeda, is a consultant for Celgene, Moderna and Shattuck Labs, and receives research funding from Genoptix (Novartis), Onkaido, Pierre Fabre, Surface Oncology, Takeda, Tesaro and Tioma. D.L.R. is a consultant and research adviser to Agendia, Agilent, AstraZeneca, Biocept, BMS, Cell Signalling Technology, Cepheid, Merck, PAIGE, Perkin Elmer and Ultivue, owns equity in AstraZeneca, Cepheid, Lilly, Navigate-Novartis, NextCure, PixelGear and Ultivue, and receives research funding from Perkin Elmer. A.M. is a consultant and scientific adviser to Elucid Bioimaging and Inspirata, owns stock options in Elucid Bioimaging and Inspirata, and receives research funding from Philips. K.B. and V.V. declare no competing interests.
Peer review information
Nature Reviews Clinical Oncology thanks J. Liu, A. Tsirigos and the other, anonymous, reviewer(s), for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Prewitt JMS Intelligent microscopes: recent and near-future advances. Proc. SPIE 10.1117/12.958214 (1979). [DOI] [Google Scholar]
- 2.Prewitt JMS Parametric and nonparametric recognition by computer: an application to leukocyte image processing. Adv. Comput. 12, 285–414 (1972). [Google Scholar]
- 3.Prewitt JMS & Mendelsohn ML The analysis of cell images. Ann. NY Acad. Sci. 128, 1035–1053 (1966). [DOI] [PubMed] [Google Scholar]
- 4.McCarthy J, Minsky ML, Rochester N & Shannon CE A proposal for the Dartmouth summer research project on artificial intelligence, August 31, 1955 AI Mag. 27, 12 (2006). [Google Scholar]
- 5.McCarthy JJ, Minsky ML & Rochester N Artificial intelligence. Research Laboratory of Electronics (RLE) at the Massachusetts Institute of Technology (MIT) https://dspace.mit.edu/handle/1721.1/52263 (1959). [Google Scholar]
- 6.Yao X Evolving artificial neural networks. Proc. IEEE 87 1423–1447 (1999). [Google Scholar]
- 7.Haykin S Neural Networks (Prentice Hall, 1994). [Google Scholar]
- 8.Deng L Deep learning: methods and applications. Found. Trends® Signal Process. 7, 197–387 (2014). [Google Scholar]
- 9.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Hinton GE & Salakhutdinov RR Reducing the dimensionality of data with neural networks. Science 313, 504–507 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Krizhevsky A, Sutskever I & Hinton GE ImageNet classification with deep convolutional neural networks. Nips.cc http://papers.nips.cc/paper/4824-imagenetclassification-with-deep-convolutional-neural-networks.pdf (2012). [Google Scholar]
- 12.LeCun Y, Huang FJ & Bottou L in Proc. 2004 IEEE Comput. Soc; Conf. Comput. Vis. Pattern Recognit. II-104 (IEEE, 2004). [Google Scholar]
- 13.LeCun Y & Bengio Y in The handbook of brain theory and neural networks (ed. Arbib MA) 255–258 (MIT Press, 1998). [Google Scholar]
- 14.LeCun Y, Bottou L, Bengio Y & Haffner P Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998). [Google Scholar]
- 15.Deng J, et al. in 2009 IEEE Conf. Comput. Vis. Pattern Recognit. 248–255 (IEEE, 2009). [Google Scholar]
- 16.Mukhopadhyay S et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: a multicenter blinded randomized noninferiority study of 1992 cases (Pivotal Study). Am. J. Surg. Pathol. 42, 39–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kujan O et al. Why oral histopathology suffers inter-observer variability on grading oral epithelial dysplasia: an attempt to understand the sources of variation. Oral Oncol. 43, 224–231 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Chi AC, Katabi N, Chen H-S & Cheng Y-SL Interobserver variation among pathologists in evaluating perineural invasion for oral squamous cell carcinoma. Head Neck Pathol. 10, 451–464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans AJ et al. Interobserver variability between expert urologic pathologists for extraprostatic extension and surgical margin status in radical prostatectomy specimens. Am. J. Surg. Pathol. 32, 1503–1512 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Shanes JG et al. Interobserver variability in the pathologic interpretation of endomyocardial biopsy results. Circulation 75, 401–405 (1987). [DOI] [PubMed] [Google Scholar]
- 21.Elmore JG et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313, 1122–1132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brimo F, Schultz L & Epstein JI The value of mandatory second opinion pathology review of prostate needle biopsy interpretation before radical prostatectomy. J. Urol. 184, 126–130 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Kilgore ML & Goldman DP Drug costs and out-of-pocket spending in cancer clinical trials. Contemp. Clin. Trials 29, 1–8 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Agarwal A, Ressler D & Snyder G The current and future state of companion diagnostics. Pharmacogenomics Pers. Med. 8, 99–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michor F & Polyak K The origins and implications of intratumor heterogeneity. Cancer Prev. Res. 3, 1361–1364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cyll K et al. Tumour heterogeneity poses a significant challenge to cancer biomarker research. Br. J. Cancer 117, 367–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bejnordi BE et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagpal K et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. npj Digital Med. 2, 48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bychkov D et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8, 3395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Khosla A, Gargeya R, Irshad H & Beck AH Deep learning for identifying metastatic breast cancer. ArXiv.org https://arxiv.org/abs/1606.05718 (2016). [Google Scholar]
- 31.Steiner D et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am. J. Surg. Pathol. 42, 1636–1646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J & Srinivas C Automatic lymphocyte detection in H&E images with deep neural networks. ArXiv.org https://arxiv.org/abs/1612.03217 (2016). [Google Scholar]
- 33.Garcia E, et al. in 2017 IEEE 30th Int. Symp. Comput.-Based Med. Sys. (CBMS). 200–204 (IEEE, 2017). [Google Scholar]
- 34.Lu C et al. Multi-pass adaptive voting for nuclei detection in histopathological images. Sci. Rep. 6, 33985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sornapudi S et al. Deep learning nuclei detection in digitized histology images by superpixels. J. Pathol. Inform. 10.4103/jpi.jpi_74_17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H et al. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J. Med. Imaging (Bellingham) 1, 034003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Kofahi Y, Lassoued W, Lee W & Roysam B Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans. Biomed. Eng. 57, 841–852 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Naik S, et al. in 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 284–287 (IEEE, 2008). [Google Scholar]
- 39.Nguyen K, Jain AK & Allen RL in 2010 20th Int. Conf. Pattern Recognit. 1497–1500 (IEEE, 2010). [Google Scholar]
- 40.Kothari S, et al. in 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 657–660 (2011, IEEE; ). [Google Scholar]
- 41.Sirinukunwattana K et al. Gland segmentation in colon histology images: the glas challenge contest. Med. Image Anal. 35, 489–502 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Lee G et al. Co-occurring gland angularity in localized subgraphs: predicting biochemical recurrence in intermediate-risk prostate cancer patients. PLOS ONE 9, e97954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee G et al. Nuclear shape and architecture in benign fields predict biochemical recurrence in prostate cancer patients following radical prostatectomy: preliminary findings. Eur. Urol. Focus 3, 457–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu C et al. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab. Investig. J. Tech. Methods Pathol. 98, 1438–1448 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corredor G et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin. Cancer Res. 25, 1526–1534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mungle T et al. MRF-ANN: a machine learning approach for automated ER scoring of breast cancer immunohistochemical images. J. Microsc. 267, 117–129 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Wang X et al. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci. Rep. 7, 13543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosado B et al. Accuracy of computer diagnosis of melanoma: a quantitative meta-analysis. Arch. Dermatol. 139, 361–367 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Rosenbaum BE et al. Computer-assisted measurement of primary tumor area is prognostic of recurrence-free survival in stage IB melanoma patients. Mod. Pathol. 30, 1402–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teramoto A, Tsukamoto T, Kiriyama Y & Fujita H Automated classification of lung cancer types from cytological images using deep convolutional neural networks. Biomed. Res. Int. 10.1155/2017/4067832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu M, Yan C, Liu H & Liu Q Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci. Rep. 10.1042/BSR20180289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ali S, Basavanhally A, Ganesan S & Madabhushi A Histogram of Hosoya indices for assessing similarity across subgraph populations: breast cancer prognosis prediction from digital pathology [abstract 118]. Lab. Invest. (supplement) 95, 32A (2015). [Google Scholar]
- 53.Yu K-H et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 7, 12474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cruz-Roa A et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci. Rep. 7, 46450 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y et al. Artificial intelligence-based breast cancer nodal metastasis detection. Arch. Pathol. Lab. Med. 143, 859–868 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Litjens G et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience 7, giy065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y et al. Detecting cancer metastases on gigapixel pathology images. ArXiv.org https://arxiv.org/abs/1703.02442 (2017). [Google Scholar]
- 58.Madabhushi A & Lee G Image analysis and machine learning in digital pathology: challenges and opportunities. Med. Image Anal. 33, 170–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansen I et al. Histopathology: ditch the slides, because digital and 3D are on show. World J. Urol. 36, 549–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ali S, Lewis J & Madabhushi A Spatially aware cell cluster(spACC1) graphs: predicting outcome in oropharyngeal pl6+ tumors. Med. Image Comput. Comput. Assist. Interv. 16, 412–419 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Mobadersany P et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X et al. Computer extracted features of cancer nuclei from H&E stained tissues of tumor predicts response to nivolumab in non-small cell lung cancer. J. Clin. Oncol. 36(15_suppl), 12061–12061 (2018). [Google Scholar]
- 63.Barrera C et al. Computer-extracted features relating to spatial arrangement of tumor infiltrating lymphocytes to predict response to nivolumab in non-small cell lung cancer (NSCLC). J. Clin. Oncol. 36, 12115–12115 (2018). [Google Scholar]
- 64.Wang X, et al. Computerized nuclear morphometric features from H&E slide images are prognostic of recurrence and predictive of added benefit of adjuvant chemotherapy in early stage non-small cell lung cancer. Presented at the United States and Canadian Academy of Pathology’s 108th Annual Meeting. (2019). [Google Scholar]
- 65.Gisselsson D et al. Abnormal nuclear shape in solid tumors reflects mitotic instability. Am. J. Pathol. 158, 199–206 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malon CD & Cosatto E Classification of mitotic figures with convolutional neural networks and seeded blob features. J. Pathol. Inform. 4, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitney J et al. Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer 18, 610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon I, Pound CR, Partin AW, Clemens JQ & Christens-Barry WA Automated image analysis system for detecting boundaries of live prostate cancer cells. Cytom. J. Int. Soc. Anal. Cytol. 31, 287–294 (1998). [DOI] [PubMed] [Google Scholar]
- 69.Basavanhally A et al. Multi-field-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Trans. Biomed. Eng. 60, 2089–2099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewis JS, Ali S, Luo J, Thorstad WL & Madabhushi A A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am. J. Surg. Pathol. 38, 128–137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barker J, Hoogi A, Depeursinge A & Rubin DL Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med. Image Anal. 30, 60–71 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Kong J et al. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLOS ONE 8, e81049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Osareh A & Shadgar B in 2010 5th Int. Symp. Health Informat. Bioinformat. 114–120 (2010, IEEE; ). [Google Scholar]
- 74.Veta M et al. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod. Pathol. 25, 1559 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Lu C et al. An oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier of nuclear morphology can risk stratify patients for disease-specific survival. Mod. Pathol. 30, 1655–1665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saltz J et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 23, 181–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan Y Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. J. R. Soc. Interface 10.1098/rsif.2014.1153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heindl A, et al. Relevance of spatial heterogeneity of immune infiltration for predicting risk of recurrence after endocrine therapy of ER+ breast cancer. J. Natl Cancer. Inst. 10.1093/jnci/djx137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beck AH et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci. Transl Med. 3, 108ra113 (2011). [DOI] [PubMed] [Google Scholar]
- 80.Bhargava HK et al. Computer-extracted stromal features of African-Americans versus Caucasians from H&E slides and impact on prognosis of biochemical recurrence. J. Clin. Oncol. 36(15_suppl), 12075–12075 (2018). [Google Scholar]
- 81.Vamathevan J et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov 18, 463–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janowczyk A & Madabhushi A Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 7, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Araujo T et al. Classification of breast cancer histology images using Convolutional Neural Networks. PLOS ONE 12, e0177544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ehteshami Bejnordi B et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 31, 1502–1512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haenssle HA et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 29, 1836–1842 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Sheikhzadeh F, Guillaud M & Ward RK Automatic labeling of molecular biomarkers of whole slide immunohistochemistry images using fully convolutional networks. PLOS ONE 13, e0190783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basavanhally AN et al. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Trans. Biomed. Eng. 57, 642–653 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Alzubaidi L et al. A robust deep learning approach to detect nuclei in histopathological images. Int. J. Innov. Res. Comp. Commun. Eng. 5, 4461–4467 (2007). [Google Scholar]
- 89.Hofener H et al. Deep learning nuclei detection: a simple approach can deliver state-of-the-art results. Comput. Med. Imaging Graph. 70, 43–52 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Xu J, Luo X, Wang G, Gilmore H & Madabhushi A A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 191, 214–223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Senaras C, Niazi MKK, Lozanski G & Gurcan MN DeepFocus: detection of out-of-focus regions in whole slide digital images using deep learning. PLOS ONE 13, e0205387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Esteva A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tschandl P et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. 20, 938–947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Couture HD et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. npj Breast Cancer 4, 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Romo-Bucheli D, Janowczyk A, Gilmore H, Romero E & Madabhushi A A deep learning based strategy for identifying and associating mitotic activity with gene expression derived risk categories in estrogen receptor positive breast cancers. Cytomtery A 91, 566–573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Geessink OGF, et al. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell Oncol. 42, 331–341 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Kather JN et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLOS Med. 16, e1002730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rodner E et al. Fully convolutional networks in multimodal nonlinear microscopy images for automated detection of head and neck carcinoma: pilot study. Head Neck 41, 116–121 (2019). [DOI] [PubMed] [Google Scholar]
- 99.Cruz-Roa A et al. High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: application to invasive breast cancer detection. PLOS ONE 13, e0196828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rumelhart DE, Hinton GE & Williams RJ Learning representations by back-propagating errors. Nature 323, 533–536 (1986). [Google Scholar]
- 101.Hochreiter S & Schmidhuber J Long short-term memory. Neural Comput. 9, 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 102.Li W, Gauci M & Gross R in Proc. 15th Ann. Conf. Genet. Evolut. Computat. 223–230. (ACM, 2013). [Google Scholar]
- 103.Hou L, et al. Unsupervised histopathology image synthesis. ArXiv.org https://arxiv.org/abs/1712.05021 (2017). [Google Scholar]
- 104.Shaban MT, Baur C, Navab N & Albarqouni S StainGAN: stain style transfer for digital histological images. IEEE 10.1109/ISBI.2019. 8759152 (2018). [DOI] [Google Scholar]
- 105.Xu Z, Moro CF, Bozoky B & Zhang Q GAN-based virtual re-staining: a promising solution for whole slide image analysis. ArXiv.org https://arxiv.org/abs/1901.04059 (2019). [Google Scholar]
- 106.Gadermayr M, Gupta L, Klinkhammer BM, Boor P & Merhof D Unsupervisedly training GANs for segmenting digital pathology with automatically generated annotations. Proc. Machine Learn. 102, 175–184 (2018). [Google Scholar]
- 107.Kapil A, et al. Deep semi supervised generative learning for automated PD-L1 tumor cell scoring on NSCLC tissue needle biopsies. Sci. Rep. 8,17343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boutros PC The path to routine use of genomic biomarkers in the cancer clinic. Genome Res. 25, 1508–1513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chatterjee SK & Zetter BR Cancer biomarkers: knowing the present and predicting the future. Future Oncol. 1, 37–50 (2005). [DOI] [PubMed] [Google Scholar]
- 110.Grigg C & Rizvi NA PD-L1 biomarker testing for non-small cell lung cancer: truth or fiction? J. Immunother. Cancer 4, 48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mroz EA & Rocco JW Intra-tumor heterogeneity in head and neck cancer and its clinical implications. World J. Otorhinolaryngol. Head Neck Surg. 2, 60–67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morris LGT et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7, 10051–10063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sottoriva A et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl Acad. Sci. USA 110, 4009–4014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Verma N et al. Image-based risk score to predict recurrence of ER+ breast cancer in ECOG-ACRIN Cancer Research Group E2197. J. Clin. Oncol. 36 (15_suppl), 540 (2018). [Google Scholar]
- 115.Ginsburg SB, Lee G, Ali S & Madabhushi A Feature importance in nonlinear embeddings (FINE): applications in digital pathology. IEEE Trans. Med. Imaging. 35, 76–88 (2016). [DOI] [PubMed] [Google Scholar]
- 116.Cooper LAD et al. Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab. Investig. J. Tech. Methods Pathol. 95, 366–376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lloyd MC, Johnson JO, Kasprzak A & Bui MM Image analysis of the tumor microenvironment. Adv. Exp. Med. Biol. 936, 1–10 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Hamilton PW et al. Digital pathology and image analysis in tissue biomarker research. Methods 70, 59–73 (2014). [DOI] [PubMed] [Google Scholar]
- 119.Barsoum I, Tawedrous E, Faragalla H & Yousef GM Histo-genomics: digital pathology at the forefront of precision medicine. Diagnosis 10.1515/dx-2018-0064 (2018). [DOI] [PubMed] [Google Scholar]
- 120.FDA. Developing a software precertification program: a working model. Fda.gov https://www.fda.gov/media/119722/download (2019).
- 121.Daniel G, Silcox C, Sharma I & Wright M Current state and near-term priorities for AI-enabled diagnostic support software in health care. Duke.edu https://healthpolicy.duke.edu/sites/default/files/atoms/files/dukemargolisaienableddxss.pdf (2019). [Google Scholar]
- 122.Pesapane F, Volonté C, Codari M & Sardanelli F Artificial intelligence as a medical device in radiology: ethical and regulatory issues in Europe and the United States. Insights Imaging 9, 745–753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rudin C Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1, 206–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Food and Drug Administration. FDA in brief: FDA proposes improvements to the de novo pathway for novel medical devices to advance safe, effective, and innovative treatments for patients. Fda.gov https://www.fda.gov/NewsEvents/Newsroom/FDAInBrief/ucm627522.htm (2018). [Google Scholar]
- 125.Commission European. MDCG 2018–2 Future EU medical device nomenclature - description of requirements. Europa.eu https://ec.europa.eu/docsroom/documents/28668 (2018). [Google Scholar]
- 126.European Economic Community. Council Directive 93/42/EEC of 14 June 1993 concerning medical devices. Europa.eu https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=C0NSLEG:1993L0042: 20071011:EN:PDF (1993). [Google Scholar]
- 127.Parliament European. Regulation (EU) 2017/745 of the European Parliament and of the Council of 5 April 2017 on medical devices, amending directive 2001/83/EC, regulation (EC) no 178/2002 and regulation (EC) no 1223/2009 and repealing council directives 90/385/EEC and 93/42/EEC (text with EEA relevance). Europa.eu http://data.europa.eu/eli/reg/2017/745/oj/eng (2017). [Google Scholar]
- 128.Parliament European. Regulation (EU) 2017/746 of the European Parliament and of the council of 5 April 2017 on in vitro diagnostic medical devices and repealing directive 98/79/EC and commission decision 2010/227/EU (Text with EEA relevance) Europa.eu http://data.europa.eu/eli/reg/2017/746/oj/eng (2017). [Google Scholar]
- 129.García-Rojo M, et al. New European Union regulations related to whole slide image scanners and image analysis software. J. Pathol. Inform. 10, 10.4103/jpi.jpi_33_18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Arterys. Medical imaging cloud AI. Arterys.com https://www.arterys.com/ (2019).
- 131.Arterys Inc. Arterys receives first FDA clearance for broad oncology imaging suite with deep learning. Prnewswire.com https://www.prnewswire.com/news-releases/arterys-receives-first-fda-clearance-for-broad-oncology-imaging-suite-with-deep-learning-300599275.html (2018). [Google Scholar]
- 132.Food and Drug Administration. IntelliSite Pathology Solution (PIPS, Philips Medical Systems). Fda.gov https://www.fda.gov/drugs/resources-information-approved-drugs/intellisite-pathology-solution-pips-philips-medical-systems (2017). [Google Scholar]
- 133.PAIGE. PAIGE. Paige.ai https://paige.ai/ (2019). [Google Scholar]
- 134.Wire Business. FDA grants breakthrough designation to Paige.AI. Businesswire.com https://www.businesswire.com/news/home/20190307005205/en/FDA-Grants-Breakthrough-Designation-Paige.AI (2019). [Google Scholar]
- 135.Bueno G, Fernández-Carrobles MM, Deniz O & García-Rojo M New trends of emerging technologies in digital pathology. Pathobiology 83, 61–69 (2016). [DOI] [PubMed] [Google Scholar]
- 136.Food and Drug Administration. Radiological computer-assisted diagnostic (CADx) software for lesions suspicious for cancer. Fda.gov. https://www.accessdata.fda.gov/cdrh_docs/pdf17/DEN170022.pdf (2017). [Google Scholar]
- 137.Agendia. MammaPrint Test. Agendia.com https://www.agendia.com/our-tests/mammaprint/(2019). [Google Scholar]
- 138.Food and Drug Administration. MammaPrint 510(k) premarket notification. Fda.gov https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=k070675 (2019). [Google Scholar]
- 139.OncotypeIQ. Smarter Cancer Care - Genomic Testing. Oncotypeiq.com https://www.oncotypeiq.com/en-US/ (2019). [Google Scholar]
- 140.Food and Drug Administration. FDA issues warning letter to genomics lab for illegally marketing genetic test that claims to predict patients’ responses to specific medications. Fda.gov https://www.fda.gov/news-events/press-announcements/fda-issues-warning-letter-genomics-lab-illegally-marketing-genetic-test-claims-predict-patients (2019). [Google Scholar]
- 141.Food and Drug Administration. Discussion paper on laboratory developed tests (LDTs) January 13, 2017 Fda.gov https://www.fda.gov/media/102367/download (2019). [Google Scholar]
- 142.Paxton A Laboratory-developed tests CAP suggests added oversight of homebrews. Captodayonline.com http://www.captodayonline.com/Archives/1009/1009d_laboratory_developed_tests.html (2009). [Google Scholar]
- 143.Higgins C Applications and challenges of digital pathology and whole slide imaging. Biotech. Histochem. 90, 341–347 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Tizhoosh HR & Pantanowitz L Artificial intelligence and digital pathology: challenges and opportunities. J. Pathol. Inform. 9, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Doyle S, Feldman M, Tomaszewski J & Madabhushi A A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans. Biomed. Eng. 59, 1205–1218 (2012). [DOI] [PubMed] [Google Scholar]
- 146.Huang B, Bates M & Zhuang X Super resolution fluorescence microscopy. Annu. Rev. Biochem. 78, 993–1016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kleppe A et al. Chromatin organisation and cancer prognosis: a pan-cancer study. Lancet Oncol. 19, 356–369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gallas BD NCIP Hub - Group: eeDAP studies ~ Wiki: High-throughput Truthing - Year 2. Ncihub.org https://nciphub.org/groups/eedapstudies/wiki/HighThroughputTruthing/HighThroughputTruthingYear2 (2018). [Google Scholar]
- 149.Challenge Grand. All Challenges. Grand-challenge.org https://grand-challenge.org/challenges/ (2019). [Google Scholar]
- 150.Ching P et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15, 20170387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Coudray N et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zech JR et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLOS Med. 15, e1002683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yagi Y & Gilbertson JR Digital Imaging in Pathology: The Case for Standardization. (SAGE Publications, 2005). [DOI] [PubMed] [Google Scholar]
- 154.Janowczyk A, Basavanhally A & Madabhushi A Stain Normalization using Sparse AutoEncoders (StaNoSA): application to digital pathology. Comput. Med. Imaging Graph. 57, 50–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Khan AM, Rajpoot N, Treanor D & Magee D A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans. Biomed. Eng. 61, 1729–1738 (2014). [DOI] [PubMed] [Google Scholar]
- 156.Vahadane A et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging. 35, 1962–1971 (2016). [DOI] [PubMed] [Google Scholar]
- 157.Janowczyk A, Zuo R, Gilmore H, Feldman M & Madabhushi A HistoQC: an open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inform. 3, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.choosehappy. HistoQC is an Open-source Quality Control Tool for Digital Pathology Slides: choosehappy/ HistoQC. Github.com https://github.com/choosehappy/HistoQC (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Leo P et al. Stable and discriminating features are predictive of cancer presence and Gleason grade in radical prostatectomy specimens: a multi-site study. Sci. Rep. 8, 14918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Leo P et al. Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J. Med. Imaging 3, 047502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lowe A et al. Validation of digital pathology in a healthcare environment. Digitalpathologyassociation. org https://digitalpathologyassociation.org/_data/files/DPA-Healthcare-White-Paper--FINAL_v1.0.pdf (2011). [Google Scholar]
- 162.Montalto MC An industry perspective: an update on the adoption of whole slide imaging. J. Pathol. Inform. 7, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Digital Pathology Association. Healthcare FAQs. Digitalpathologyassociation.org https://digitalpathologyassociation.org/healthcare-faqs (2019). [Google Scholar]
- 164.Simon RM, Paik S & Hayes DF Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 101, 1446–1452 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hayes DF et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J. Natl Cancer Inst. 88, 1456–1466 (1996). [DOI] [PubMed] [Google Scholar]
- 166.Goetz MP et al. NCCN Guidelines insights: breast cancer, version 3.2018. J. Natl Compr. Canc. Netw. 17, 118–126 (2019). [DOI] [PubMed] [Google Scholar]
- 167.Sparano JA et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 379, 111–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cardoso F et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 375, 717–729 (2016). [DOI] [PubMed] [Google Scholar]
- 169.Paik S et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004). [DOI] [PubMed] [Google Scholar]
- 170.Paik S et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 (2006). [DOI] [PubMed] [Google Scholar]
- 171.DecipherBio. GenomeDx Biosciences: Where to get decipher prostate cancer classifier. decipherbio.com https://genomedx.com/decipher-test/get-decipher/(2019). [Google Scholar]
- 172.Erho N et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLOS ONE. 8, e66855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Carroll PH & Mohler JL NCCN guidelines updates: prostate cancer and prostate cancer early detection. J. Natl Compr. Canc. Netw. 16(5S), 620–623 (2018). [DOI] [PubMed] [Google Scholar]
- 174.Bhargava R & Madabhushi A Emerging themes in image informatics and molecular analysis for digital pathology. Annu. Rev. Biomed. Eng. 18, 387–412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Madabhushi A Digital pathology image analysis: opportunities and challenges. Imaging Med. 1, 7–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Madabhushi A, Agner S, Basavanhally A, Doyle S &Lee G Computer-aided prognosis: predicting patient and disease outcome via quantitative fusion of multi-scale, multi-modal data. Comput. Med. Imaging Graph. 35, 506–514 (2011). [DOI] [PubMed] [Google Scholar]
- 177.Pantanowitz L et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch. Pathol. Lab. Med. 137, 1710–1722 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fuchs TJ, Wild PJ, Moch H & Buhmann JM Computational pathology analysis of tissue microarrays predicts survival of renal clear cell carcinoma patients. Med. Image Comput. Comput. Assist. Interv. 11, 1–8 (2008). [DOI] [PubMed] [Google Scholar]
- 179.Hayback J Mechanisms of Molecular Carcinogenesis - Volume 2. (Springer, 2017). [Google Scholar]
- 180.Proscia. Proscia digital pathology. Proscia.com https://proscia.com (2019). [Google Scholar]
- 181.Lens Deep. Digital pathology cloud platform. Deeplens.ai https://www.deeplens.ai (2019). [Google Scholar]
- 182.PathAI. PathAI. Pathai.com https://www.pathai.com/(2019). [Google Scholar]
- 183.Aifora. WebMicroscope. Big pictures. Deep Diagnosis. Aiforia.com https://www.aiforia.com/ (2019). [Google Scholar]
- 184.Glaser AK, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fereidouni F et al. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat. Biomed. Eng. 1, 957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fraggetta F, Garozzo S, Zannoni GF, Pantanowitz L & Rossi ED Routine digital pathology workflow: the Catania experience. J. Pathol. Inform. 8, 51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cheng CL et al. Enabling digital pathology in the diagnostic setting: navigating through the implementation journey in an academic medical centre. J. Clin. Pathol. 69, 784–792 (2016). [DOI] [PubMed] [Google Scholar]
- 188.Tomczak K, Czerwinska P & Wiznerowicz M The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 19, A68–77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gutman DA et al. Cancer Digital Slide Archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J. Am. Med. Inform. Assoc. 20, 1091–1098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]