Abstract
One major challenge to delimiting species with genetic data is successfully differentiating population structure from species-level divergence, an issue exacerbated in taxa inhabiting naturally fragmented habitats. Many fields of science are now using machine learning, and in evolutionary biology supervised machine learning has recently been used to infer species boundaries. These supervised methods require training data with associated labels. Conversely, unsupervised machine learning (UML) uses inherent data structure and does not require user-specified training labels, potentially providing more objectivity in species delimitation. Here we demonstrate the utility of three UML approaches (random forests, variational autoencoders, t-distributed stochastic neighbor embedding) for species delimitation in an arachnid taxon with high population genetic structure (Opiliones, Laniatores, Metanonychus). We find that UML approaches successfully cluster samples according to species-level divergences and not high levels of population structure, while model-based validation methods severely over-split putative species. UML offers intuitive data visualization in two-dimensional space, the ability to accommodate various data types, and has potential in many areas of systematic and evolutionary biology. We argue that machine learning methods are ideally suited for species delimitation and may perform well in many natural systems and across taxa with diverse biological characteristics.
Keywords: Random Forest, t-SNE, Variational Autoencoders, ultraconserved elements, integrative taxonomy, Opiliones
1. INTRODUCTION
One of the fundamental issues in biology is how to recognize and delimit species in practice. Species are fundamental units in biology, and correctly identifying species level divergences is important for many reasons, including accurate estimates of biodiversity and conservation assessments. Species delimitation research is continually advancing, incorporating new theoretical frameworks, data types, and computational approaches. Modern species delimitation is increasingly quantitative and objective relying on, for example, statistical thresholds and/or clustering algorithms to identify species in multivariate morphological space (e.g., Ezard et al. 2010; Seifert et al. 2014), or using genetic data and a multispecies coalescent model to identify the boundary between population and species-level divergences (e.g., Yang and Rannala 2010). Similarly, species delimitation is becoming increasingly integrative, combining multiple data types in a reciprocally-illuminating framework providing more robust species hypotheses (Dayrat 2005; Schlick-Steiner et al. 2010).
Of utmost interest when delimiting species with genetic data is successfully distinguishing population structure from species-level divergences, an often blurry distinction considering that speciation is a continuous process. This issue is exacerbated in certain systems, for example, in taxa that are found in naturally fragmented habitats resulting in high levels of population genetic structure. The biological characteristics of organisms can also confound a clear distinction, for example, taxa with lower dispersal capabilities and higher ecological constraints might have inherently high population structure throughout the speciation process. Regardless of where taxa lie on the speciation continuum, these characteristics can lead to overestimation of the actual number of species as delimitation analyses may detect population structure (e.g., Sukumaran and Knowles 2017), an assertion previously demonstrated empirically (e.g., Niemiller et al. 2012; Hedin et al. 2015).
Machine learning approaches include algorithms that can be trained to make future decisions without user input. Recently, machine learning methods like Random Forest (RF; Breiman 2001) have been incorporated into evolutionary biology with applications in DNA barcoding (e.g., Austerlitz et al. 2010), environmental DNA metabarcoding (e.g., Cordier et al. 2018), predicting cryptic diversity (Espíndola et al. 2016), phylogeographic model selection (Pudlo et al. 2016; Smith et al. 2017) and speciation/species delimitation (Pei et al. 2018; Smith and Carstens 2018). Similarly, non-RF ML approaches have been used to model biogeographic processes (Sukumaran et al. 2015). In these examples supervised machine learning is used, where simulated datasets based on user-specified priors are used as training data, and a classifier is built to choose among different models given observed data. While supervised approaches are powerful and potentially transformative (Schrider and Kern 2018), unsupervised machine learning (UML) may also be useful in many areas including species delimitation, using only the inherent structure in the data to cluster samples. UML is conducted without a priori hypotheses regarding the underlying model, population parameters, species number, sample assignments, or levels of divergence needed to classify different species. For example, a UML approach was used to visualize and cluster barcode data via nonlinear dimension reduction and projection methods, showing successful clustering of named, unnamed, and potentially undescribed species (Olteanu et al. 2013).
Many machine learning algorithms are essentially dimensionality reduction in some form and can be used unsupervised. While dimensionality reduction methods like principal components analyses and clustering algorithms like k-means are widely considered machine learning, we focus on three less-familiar UML approaches representing diverse algorithm types (Table 1): RF (Breiman 2001), Variational Autoencoders (VAE; Kingma and Welling 2013), and t-Distributed Stochastic Neighbor Embedding (t-SNE; van der Maaten and Hinton 2008). RF is an ensemble learning method that relies on classification trees and tree bagging (Breiman 1996; 2001). In unsupervised RF, a synthetic training dataset based on observed data is used as a null hypothesis of no structure, and a classifier is built to distinguish synthetic data from observed, producing a pairwise “proximity matrix” (i.e., distance matrix) which can be used in multidimensional scaling (MDS) and clustering. A VAE is rooted in Bayesian statistics and learns a data distribution using latent variables in two stages approximated by neural networks and optimized simultaneously via unsupervised learning: 1) inference of the posterior distribution of latent variables and 2) generation of data sampled from a given set of latent values (Supplemental Fig. S1). Widely used in diverse biological fields (e.g., Bauer et al. 2015; Yoshida et al. 2016; Mallet et al. 2017), t-SNE is a nonlinear dimensionality reduction algorithm that attempts to preserve probability distributions of distances among samples within a cluster but repels samples that are in different clusters. UML algorithms, particularly neural network-based algorithms, are especially useful in determining the underlying structure in datasets for which prior information is uncertain or lacking. Applied to the issue of species delimitation, these approaches may provide an objective and assumption-free means to assess structure in biological datasets where species-level divergences are hypothesized.
Table 1.
Comparison of unsupervised machine learning methods used in this study.
| Method | Purpose | Approach used |
General algorithm | Relevant output |
|---|---|---|---|---|
| Random Forest (RF) | Classification and regression | Supervised Unsupervised | Ensemble method that grows many classification trees based on training data, runs input data down trees, and the classification with the most votes is chosen. | Proximity matrix |
| Variational Autoencoder (VAE) | Generative model | Unsupervised | Compresses data through multiple encoding layers into latent variables, then un-compresses latent variables through multiple decoder layers into reconstructed data. Learns the marginal likelihood distribution of the data using latent variables. | Latent variables (two-dimensional encoding) |
| t-Distributed Stochastic Neighbor Embedding (t-SNE) | Data embedding and visualization | Unsupervised | Constructs probability distribution of sample pairs, then minimizes divergence between high dimensional space and low dimension embedding, such that similar pairs are embedded nearby while dissimilar pairs are repelled. | Low dimensional embedding |
Here we demonstrate the utility of UML approaches in species delimitation through successful identification of clusters corresponding to species corroborated by multiple discovery and validation methods (Carstens et al. 2013). Using original data for an arachnid taxon with high population structure we first combine phylogenetic analysis of mitochondrial DNA sequences (cytochrome oxidase subunit I, COI) and examination of morphology to generate a priori species hypotheses. Then, using single nucleotide polymorphisms (SNPs) derived from sequence capture of ultraconserved elements (UCEs) we demonstrate the ability of three UML approaches to successfully cluster a priori species, including comparisons to commonly used methods. Using UCE data we validate species hypotheses using a standard method and a novel RF-based approach. We also demonstrate the utility of UML on previously published empirical and simulated datasets.
2. MATERIALS AND METHODS
2.1. Study System
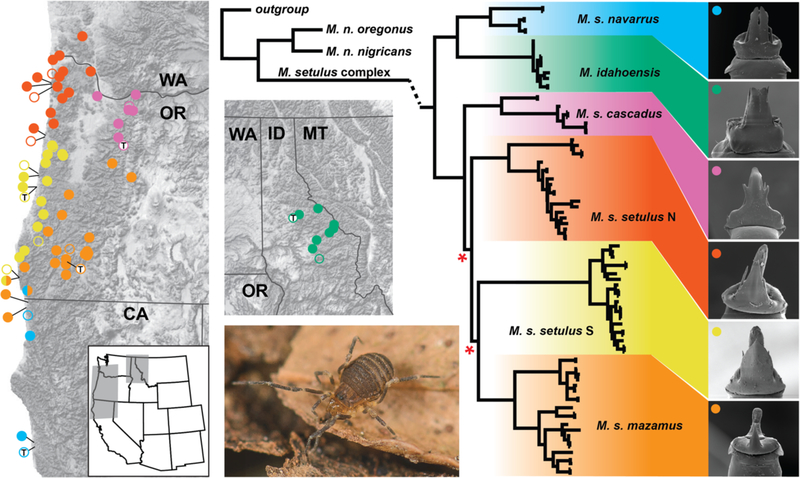
As a primary example we focus on the Pacific Northwest endemic genus Metanonychus Briggs, 1971, cryophilic harvestmen typically found beneath rotting logs/bark and in leaf litter in moist forests. Like many other harvestmen (Opiliones), these animals have low dispersal abilities and high ecological constraints leading to high population genetic structure and allopatric distributions, likely driven by niche conservatism (Wiens and Graham 2005). As currently described (Briggs 1971), Metanonychus includes three species: M. idahoensis, M. oregonus with two subspecies, and M. setulus with five subspecies (Fig. 1). Metanonychus is an ancient lineage; recent phylogenomic analyses show more genetic divergence between two samples of Metanonychus than divergences between the vast majority of sister genera pairs across Travunioidea (Derkarabetian et al. 2018). Despite the ancient origin of this group relatively few species were described, even though all subspecies were diagnosed based on apparently fixed differences in male genitalic morphology (Briggs 1971). Given geographically distinct subspecific lineages that show fixed morphological differences, the elevation of these taxa using modern systematic approaches is an expectation. Recent systematic studies on related taxa corroborate the conservative nature of subspecies in these harvestmen (Derkarabetian and Hedin 2014). We consider species as “separately evolving metapopulation lineages” (de Queiroz 2007), operationally identified as genetic clusters corresponding to monophyletic lineages with fixed morphological differences.
Figure 1.
(Left) Geographic distribution of the Metanonychus setulus complex. Filled circles are collecting locations sampled for this study. Open circles are published records; open circles with “T” indicate type localities. The single half-circle indicates sympatry. (Middle) RAxML COI phylogeny of Metanonychus, with detail for the M. setulus complex. Internal nodes with bootstrap support <90 are indicated with a red asterisk. Branch lengths for outgroups and M. nigricans not to scale. (Right) Representative scanning electron microscope images of male genitalia for each clade. Live photo: M. setulus navarrus.
2.2. Species Discovery Analyses
DNA was extracted using the Qiagen DNeasy kit (Qiagen, Valencia, CA) using 2–3 legs, PCR experiments followed a previous study (Derkarabetian and Hedin 2014) and amplified fragments were Sanger sequenced at Macrogen USA. COI was Sanger-sequenced for at least one sample from every collecting site, plus two outgroups. Sanger data were supplemented with COI derived as “UCE-bycatch” (e.g., Zarza et al. 2017; Hedin et al. 2018) for all UCE samples. COI sequence alignment was trivial (without any indels), with sequences added manually based on an initial COI alignment from a previous study (Derkarabetian and Hedin 2014). A phylogeny was reconstructed using RAxML v.8 (Stamatakis 2014) with 500 bootstrap replicates and the GTRGAMMA model with data partitioned by codon position as determined by PartitionFinder (Lanfear et al. 2012). COI divergence dating and phylogenetic estimation was conducted with BEAST 2.4.8 (Bouckaert et al. 2014) using two calibrations on the partitioned dataset: a strict clock calibrated with a substitution rate of 0.0178 based on well-calibrated insect taxa (Papadopolou et al. 2010), and a biogeographic calibration for the outgroups S. nondimorphicus (coastal Oregon/Washington) and S. idahoensis (Idaho), a well-known biogeographic break typically dated to 2–5 MY (Brunsfeld et al. 2001, and references therein), set to a uniform distribution of [2–5].
Male genitalia in Opiliones tend to be species-specific and have been used reliably in systematic studies across all taxonomic levels, especially the species level, since the mid-1900s (e.g., Forster 1954; Martens 1986; Pérez-González and Werneck 2018). We examined male genitalia for multiple samples of all described taxa using scanning electron microscopy. Images were taken using the FEI Quanta 450 FEG environmental SEM at the San Diego State University Electron Microscope Facility.
2.3. Sequence Capture and SNPs
DNA extractions were conducted as above, except in most cases whole bodies were used in digestion. UCE sequence capture followed protocols available on ultraconserved.org and previous studies (Starrett et al. 2017; Derkarabetian et al. 2018) using the Arachnida 1.1Kv1 myBaits kit (Arbor Biosciences) designed from an arachnid-specific probeset (Faircloth 2017). Illumina sequencing was done at the Brigham Young University DNA Sequencing Center on a HiSeq 2500 with 125 bp paired-end reads. Raw reads were processed using phyluce (Faircloth 2005), adapter removal and quality control were done with an illumiprocessor wrapper (Faircloth 2013), and contigs were assembled with Trinity version r2013–02-25 (Grabherr et al. 2011). When matching contigs to probes, conservative values of 82 and 80 were used for minimum coverage and minimum identity, respectively, to filter potential non-target contamination (Bossert and Danforth 2018). Loci were aligned using MAFFT (Katoh and Standley 2013) and trimmed using gblocks (Castresana 2000; Talavera and Castresana 2007) with settings --b1 0.5 --b2 0.5 --b3 10 --b4 4. All loci were manually inspected in Geneious (Kearse et al. 2012) to fix obvious alignment errors and filtered for obvious non-orthologs as evidenced by highly divergent sequences (Hedin et al. 2019). Mitochondrial contigs were identified by a local BLAST search in Geneious against available Metanonychus COI sequences. Although not used in species delimitation, a concatenated matrix of UCE loci with 70% taxon coverage was used to reconstruct a phylogeny using RAxML v. 8 (Stamatakis 2014) with 500 bootstraps and the GTRGAMMA model.
SNP datasets were assembled using published approaches for sequence capture data (e.g., Zarza et al. 2017). Due to relatively ancient divergence in Metanonychus, preliminary exploration of SNP data with all 38 samples produced datasets with too few loci or too sparse a matrix, with M. nigricans samples missing ~60% of SNPs (~11% average for M. setulus complex). As such, SNP datasets only included the 30 samples in the M. setulus complex. The sample with the highest number of recovered UCE loci was used as a reference genome (M. idahoensis, OP2432). After adapter removal and quality control, reads for all samples were aligned to the reference using bwa (Li and Durbin 2009), resulting SAM files were sorted using samtools (Li et al. 2009), PCR duplicates were identified and removed using picard (http://broadinstitute.github.io/picard), and all BAM files were merged. The Genome Analysis Toolkit 3.2 (McKenna et al. 2010) was used to realign reads, remove indels, and recalibrate SNPs using “best practices” (van der Auwera et al. 2013). SNPs were called and vcftools (Danecek et al. 2011) was used to create datasets which varied in taxon coverage percentage needed to include a SNP in the final matrix (50% and 70%). One random SNP from each locus was selected and the script adegenet_from_vcf.py (github.com/mgharvey/seqcap_pop) was used to create STRUCTURE-formatted (.str) files.
2.4. Standard Genetic Clustering
STRUCTURE version 2.3.4 (Pritchard et al. 2000) was run for 1 million generations and 100,000 burnin on K values ranging from 2–10, with five replicates each. Structure Harvester (Earl and vonHoldt 2012) was used to determine optimal K via calculation of ΔK (Evanno et al. 2005) and Clumpak (Kopelman et al. 2015) was used to visualize output (http://clumpak.tau.ac.il/). We used the adegenet R package (Jombart 2008; Jombart and Ahmed 2011) to conduct principal components analysis (PCA; dudi.pca function) and determine the optimal number of clusters and cluster assignment with discriminant analysis of principal components (DAPC) on scaled data.
2.5. UML Visualization
All UML analyses were run multiple times. RF was executed through the randomForest R package (Liaw and Wiener 2002), using scaled data from DAPC. There are two important parameters associated with RF: 1) ntree, the number of classification trees to create, was set to 5000; and 2) mtry, the number of splits in the classification tree, was left at default for classification analysis. The resulting proximity matrix was then used in cMDS using the MDSplot function in the randomForest R package and isoMDS using the isoMDS function in the MASS R package (Venables and Ripley 2002).
VAE was implemented utilizing the Keras python deep learning library (https://keras.io; Chollet 2015) and the TensorFlow machine learning framework (www.tensorflow.org; Abadi et al. 2015). Details of the VAE and training procedure are in Fig. S1. As input we used SNP matrices converted to “one-hot encoding” where nucleotides are transformed into four binary variables unique to each nucleotide (e.g., A = 1,0,0,0; C = 0,1,0,0; etc.) including ambiguities (e.g., M = 0.5,0.5,0,0) using a custom script. In the VAE, the encoder takes the one-hot encoded SNP data and infers the distribution of latent variables, given as a normal distribution with a mean (μ) and standard deviation (σ), then the decoder maps the latent distribution to a reconstruction of the one-hot encoded SNP data. Given two latent variables, SNP data is visualized as a two-dimensional representation.
t-SNE was executed using the R package tsne (Donaldson 2016). After preliminary testing, several parameters were specified: maximum iterations (max_iter=5000), perplexity=5, initial dimensions (initial_dims=5), and number of dimensions for the resulting embedding (k=2). The maximum iterations value is straightforward to determine as the KL divergence (a measure of the difference between high and low dimensional representations) should stabilize at a minimum. Perplexity measures the balance between the local and global elements of the data; essentially how many neighbors a particular sample can have. This parameter is somewhat subjective, where lower values produce tight well separated clusters, and higher values will produce more diffuse clusters. However, results and clusters are typically robust across a wide range of perplexity values (Pedregosa et al. 2011). Following recommendations for large datasets (Pedregosa et al. 2011), we performed t-SNE using the results of the initial PCA as input.
With RF and t-SNE, we also tested three different types of input format using the 70% SNP dataset: 1) SNPs represented as raw nucleotides with ambiguities in standard IUPAC coding extracted directly from .vcf files using the vcf2phylip script (github.com/edgardomortiz/vcf2phylip); 2) raw SNPs converted to haplotypes using the script SNPtoAFSready.py (github.com/jordansatler/SNPtoAFS); 3) raw unphased nucleotides converted into numerical format via one-hot encoding. For the first two datasets, missing data (N) were coded as blank, and PCA could not be conducted as the variables are categorical. As such, t-SNE was run using the cMDS output.
2.6. UML Clustering
Four sets of clustering analyses were conducted on UML outputs. 1) To confirm that UML cluster assignments are equivalent to DAPC and STRUCTURE, PAM clustering was conducted using the cluster R package (Maechler et al. 2018) with the optimal K taken from DAPC. To test whether the optimal K can be inferred correctly solely with UML, we conducted: 2) PAM clustering across K of 2–10 on all output, including the raw RF proximity matrix, with the optimal K having the highest average silhouette width (Rousseeuw 1987); 3) PAM clustering with optimal K determined via gap statistic using k-means clustering implemented in the factoextra R package (Kassambara and Mundt 2017); and 4) optimal K and clusters determined via hierarchical clustering with the mclust R package (Scrucca et al. 2017) using only components retained via broken stick implemented in the PCDimension R package (Coombes and Wang 2018).
2.7. Species validation
We implemented the commonly used Bayes Factor delimitation approach (*BFD; Leaché et al. 2014), a hypothesis testing framework for species delimitation, with SNAPP (Bryant et al. 2012) using a 70% UCE SNP matrix created by the phyluce script “phyluce_snp_convert_vcf_to_snapp”. Multiple species hypotheses were tested based on current taxonomy, a priori species, UML clustering, and analyses where each collecting site (n=29) and specimen (n=30) were treated as species. SNAPP was run with default settings for 100,000 generations, 10,000 burnin, and 48 steps, parameters used in previous studies with datasets of comparable size (e.g., Leaché et al. 2014). Each analysis was run twice to ensure consistency. Bayes Factors (Kass and Raftery 1995) were calculated as [2 * log likelihood difference] to determine relative support of species hypotheses.
We also used the recently developed supervised RF program CLADES (Pei et al. 2018), which treats delimitation as a classification issue. Here, a classifier is built from labeled data simulated on a two-species model with varying divergence times and population sizes. Several population genetic statistics are calculated and used as variables for both simulated training data and observed data with species defined a priori. The classifier is then used to infer whether the observed a priori species are equivalent to the same or different species. UCE loci were used as input in two analyses: 1) an analysis validating a priori species hypotheses (“spp” dataset); and 2) an analysis in which every individual was treated as a distinct species (“ind” dataset). CLADES requires that each locus have data for at least one sample within every a priori species. As a result, the “spp” dataset included 177 loci and the “ind” dataset included 12 loci.
2.8. Analysis of Published Datasets
We conducted UML on a published dataset for lizards of the Uma notata species complex (Gottscho et al. 2017), a group with a complicated history including gene flow and hybridization. Using multilocus Sanger-sequenced loci and ddRAD data, Gottscho et al. (2017) found significant levels of gene flow between multiple species and concluded that U. rufopunctata is a hybrid population. Several genetic clustering algorithms were used with differing results: DAPC favored an optimal K=5 (grouping the hybrid U. rufopunctata with U. cowlesi), while a model with admixture favored an optimal K=6 (splitting U. scoparia and showing varying levels of admixture for U. rufopunctata samples between U. cowlesi and U. notata). We reanalyzed their data with the intention of assessing UML clustering/visualization in the face of significant gene flow and possible hybrids. The published dataset of 597 unlinked SNPs was downloaded from Dryad (https://doi.org/10.5061/dryad.8br5c).
We additionally tested a number of simulated datasets with a varying range of scenarios (number of loci, sample size, population size [theta]) for four species in an asymmetric tree with no gene flow after fixed divergence times. These published datasets (https://doi.org/10.5061/dryad.r55fb) were previously used to demonstrate the suitability of *BFD (Leaché et al. 2014).
3. RESULTS
3.1. Metanonychus taxon sampling and phylogenetics
Metanonychus specimens were collected from 79 different localities. Specimens morphologically identifiable as M. setulus obrieni could not be collected. This subspecies is only known from five type specimens from the type locality (Fort Dick, CA), and the forests near this locality are now heavily disturbed. Multiple attempts to collect this taxon from near the type locality and localities within ~10 km produced samples that morphologically agree with several other Metanonychus taxa. A total of 117 sequences were included in COI analyses (alignment length of 1182 bp); all new COI sequences have been deposited to GenBank (Supplemental File 2; MN125375-MN125486). Seventy-seven sequences were acquired via Sanger sequencing and 38 were sequenced as UCE bycatch, with five samples being sequenced by both approaches, for a total of 110 Metanonychus specimens plus two outgroups (Supplemental File 2). COI sequences did not include stop codons, and for samples sequenced via Sanger and as UCE bycatch, sequences were identical. COI divergence dating supports the ancient origin of this genus dating to ~25 Ma (Fig. S2).
UCE analyses included 38 Metanonychus, 36 of which were newly sequenced (Supplemental File 2). Raw reads have been deposited to SRA (BioProject ID: PRJNA551762). The 70% UCE locus matrix included 185 loci (average of 158 per sample) with a mean locus length of 411 bp and a total length of 75,944 bp. Phylogenetic analysis of this matrix recovers all COI clades with high support (Fig. S3). The UCE phylogeny confirms deep divergence and reciprocal monophyly of M. nigricans and M. setulus complexes and COI clades are fully supported (Fig. S3). The setulus subspecies is monophyletic, albeit with reciprocally monophyletic northern and southern lineages. Analysis of a 50% UCE matrix with 278 loci (mean locus length of 384 bp, total length of 106,786 bp) returned results identical to the 70% matrix (not shown). The 70% UCE SNP dataset containing only M. setulus complex samples contained 316 SNPs (mean of 250), while a 50% dataset contained 1263 SNPs (mean of 774). All Metanonychus input matrices (COI, UCE SNPs, and .csv files), individual UCE alignments, and resulting trees are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.nj2mg77.
3.2. Species Discovery
COI analyses recover a deep and relatively ancient divergence between M. nigricans (with two subspecies) and the “M setulus complex” (M. idahoensis and M. setulus with five subspecies) (Fig. 1, Figs. S2 and S3). All named taxa are monophyletic with full support. The setulus subspecies is split into geographically cohesive northern and southern clades (setulus N and S), although support for relevant nodes are weak. Clear differences in male genitalic morphology are observed between all species/subspecies, including setulus N and S clades (Fig. 1, Fig. S4). Examination of the types of M. setulus obrieni (Fig. S4) indicates that it is likely conspecific with M. setulus navarrus (we examined all type specimens to assess distinctiveness). Overall, the species discovery phase identified eight a priori species corresponding to currently named species and subspecies.
3.3. Standard and UML Clustering
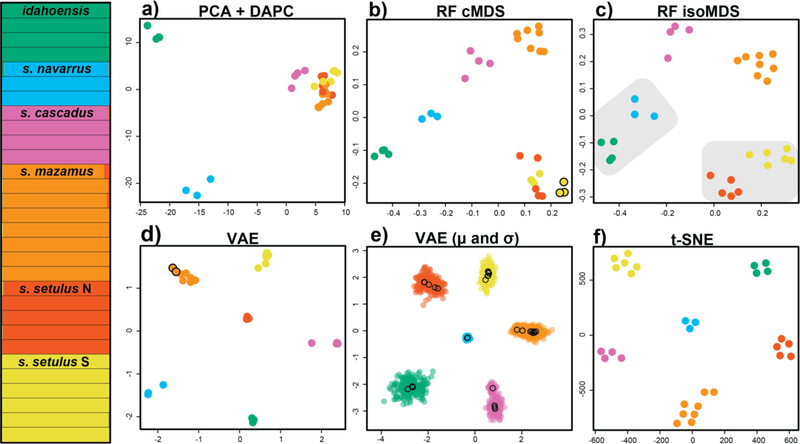
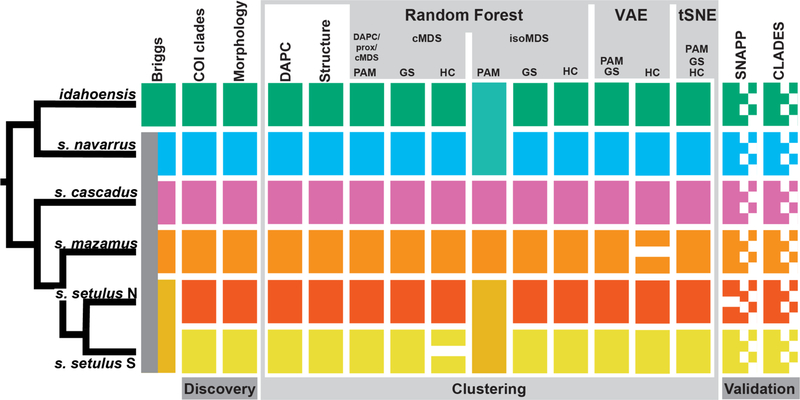
Two custom scripts were created to run ML analyses: an R script to run random forest, t-SNE, and all clustering algorithms (github.com/shahanderkarabetian/uml_species_delim), and a python script to run VAE (github.com/sokrypton/sp_deli). For the purposes of species delimitation, we focus only on the M. setulus complex (30 samples from 29 collecting sites), for which discovery analyses identified six a priori species. For 70% and 50% taxon coverage SNP datasets standard genetic clustering analyses favored an optimal K=6 (Figs. 2 and 3, Figs. S5 and S6), recovering all six a priori M. setulus complex lineages, including setulus N and S. For the 70% dataset all clustering approaches for all UML resulted in an optimum of K=6, except hierarchical clustering of cMDS (K=7, splitting setulus S) and VAE (K=7, splitting mazamus) (Figs. 2 and 3). Importantly, all K=6 UML clustering assignments were identical to standard analyses. For the 50% dataset an optimal of K=6 was found for the majority of analyses (Figs. S5 and S6). All VAE and t-SNE clusters were obvious. VAE clusters show clear and robust separation when σ (standard deviation) is visualized (Fig. 2e and Fig. S7). t-SNE clusters were robust to varying perplexity parameter values from 5–25 (Fig. S8). Similar plots for RF and t-SNE were obtained using input SNPs coded in multiple ways (Fig. S9). More consistent and accurate clustering results were obtained with the 70% taxon coverage dataset. Although we do not aim to test the relative performance of various clustering algorithms, we note that the gap statistic outperforms hierarchical clustering across multiple UML analyses. More consistent and accurate RF clustering was achieved with cMDS (classic MDS) relative to isoMDS (isotonic MDS) as multiple dimensions are used to inform the optimal clustering with cMDS, while isoMDS by default only outputs two dimensions.
Figure 2.
Clustering results for the Metanonychus setulus complex based on 70% SNP dataset. (Left) STRUCTURE plot. a) PCA plot with DAPC clusters. b) random forest cMDS plot, all clustering algorithms favored K=6, except hierarchical clustering with K=7 (seventh cluster indicated with black outline). c) random forest isoMDS plot, all clustering algorithms favored K=6, except PAM clustering of RF output with K=4 (lumped clusters indicated with grey shading). d) VAE plot, all clustering algorithms favored K=6, except hierarchical clustering with K=7 (seventh cluster indicated with black outline). e) VAE results with encoded mean (μ – open circles) and standard deviation (σ – closed circles) for each sample. f) t-SNE plot, all clustering algorithms favored K=6.
Figure 3.
Integrative species delimitation results for Metanonychus. Clustering and validation results based on 70% SNP dataset. Species tree at left adapted from RAxML analysis of 70% dataset of UCE loci. cMDS = classic multidimensional scaling, isoMDS = isotonic multidimensional scaling, GS = gap statistic, HC = hierarchical clustering.
3.4. Species Validation
To further test discovery-based species hypotheses (as recovered above) we used UCE data in two validation approaches. *BFD showed increasing likelihood with increasing number of species (Table 2), heavily favoring a model in which specimens from all collecting locations were treated as distinct species (K=29). Only considering hypotheses recovered in the discovery phase, the “7N” species hypothesis was favored, with all six a priori species plus a split in M. s. setulus N. Validation with the supervised RF program CLADES (Pei et al. 2018) supported species status of all six a priori species. However, species status was also supported when each specimen was treated as a species (Fig. S6).
Table 2.
Results of *BFD hypothesis testing. Multiple species hypotheses were tested, with hypotheses derived from analyses conducted in this study, or previous hypotheses. Each hypothesis was run twice, with Bayes Factors estimated from the average of the two runs.
| #Species | Justification | Run 1 | Run 2 | Bayes Factor |
|---|---|---|---|---|
| 2 | Briggs’ species | −3674.33 | −3885.74 | ~6927 |
| 4 | 70% isoMDS PAM, 50% isoMDS HC | −2917.94 | −2910.14 | ~5195 |
| 5 | Briggs’ species + subspecies | −2384.94 | −2386.83 | ~4139 |
| 6 | a priori species | −2210.48 | −2211.17 | ~3789 |
| 7 M | split s. mazamus: VAE HC | −2135.1 | −2136.23 | ~3638 |
| 7 N | split s. setulus N: 50% cMDS HC | −1797.95 | −1798.71 | ~2967 |
| 7 S | split s. setulus S: 70% cMDS HC | −2165.62 | −2166.25 | ~3699 |
| 29 | all collecting localities | −314.69 | −318.29 | 0 |
| 30 | all individuals | −320.3 | −316.24 | ~4 |
3.5. Taxonomy
As a result of integrative species delimitation, we elevate all subspecies of the setulus group to full species, now consisting of M. idahoensis, M. navarrus new comb., M. cascadus new comb., M. mazamus new comb., and M. setulus. In addition, all analyses supported the northern clade of the setulus subspecies as a distinct species, which we describe here as M. mechanicus n. sp. Derkarabetian and Hedin (Appendix A). Also, based on examination of all type specimens of M. obrieni deposited at the California Academy of Sciences, M. obrieni is synonymized with M. navarrus (see Appendix A). The nigricans group had too few samples for reliable clustering when analyzed alone. However, both the morphological divergence seen in male genitalia and nuclear divergence supports elevating the M. nigricans subspecies to full species: M. nigricans, and M. oregonus n. comb.
3.6. Analysis of Published Datasets
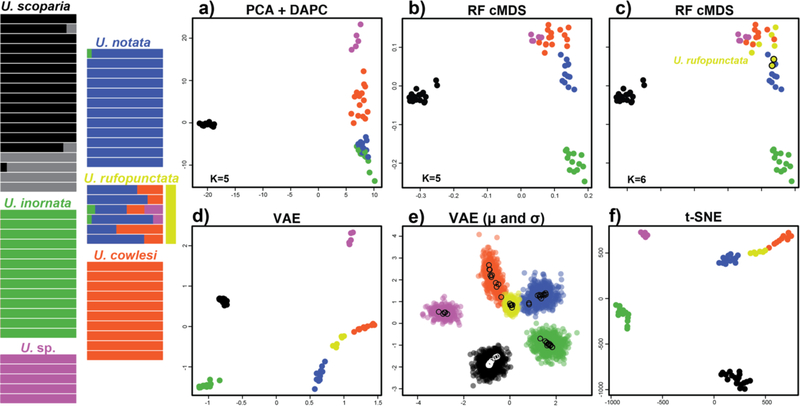
The analyses of Gottscho et al. (2017) support five species, four in the U. notata complex, plus an undescribed species. Model-based analyses support unidirectional gene flow from U. notata and U. cowlesi into U. rufopunctata which they consider a hybrid population of the two parental species. The t-SNE and VAE plots recover the hybrid species U. rufopunctata as a linear “grade” between the parental species U. cowlesi and U. notata, and the lack of complete distinctiveness of the hybrid samples from the parental species is seen in the VAE when σ is included (Fig. S7). Several genetic clustering algorithms were used in Gottscho et al. (2017) with differing results: DAPC favored an optimal K=5 (grouping the hybrid U. rufopunctata with U. cowlesi), while a model with admixture favored an optimal K=6 (splitting U. scoparia and showing varying levels of admixture for U. rufopunctata samples between U. cowlesi and U. notata). Here, both VAE and t-SNE plots place U. rufopunctata in closer proximity to U. cowelsi. Additionally, the undescribed species is clearly differentiated in all UML analyses. The optimal of K=6 recovered in the original study (Gottscho et al. 2017) does not differentiate U. rufopunctata, instead splitting U. scoparia. The cMDS plots do show two somewhat distinct samples of U. scoparia, which correspond to samples placed in the sixth cluster. All RF clustering with cMDS favored K=5, except hierarchical clustering favoring K=6 (Fig. 4). Here, a distinct cluster was identified for all U. rufopunctata and two samples of U. notata. Clustering results ranged from K=4 in PAM, lumping U. cowlesi, U. notata, and U. rufopunctata, to K=7 in some replicates of t-SNE clustered with gap statistic splitting U. scoparius. Simulated datasets show clear separation of each species and all clustering analyses favored K=4 except hierarchical clustering on three t-SNE plots (Fig. S10).
Figure 4.
Clustering results for Uma dataset. STRUCTURE plot at left adapted from Gottscho et al. (2017). a) PCA with DAPC clusters. b) random forest cMDS plot with clusters identified via DAPC, PAM, and gap statistic. c) random forest cMDS plot with clusters identified via hierarchical clustering. d) VAE plot with K=6 a priori species. e) VAE results with encoded mean (μ – open circles) and standard deviation (σ – closed circles) for each sample. f) t-SNE plot with K=6 a priori species. Species are color coded as in Gottscho et al. (2017). Note: algorithmic clustering was only conducted on random forest output.
5. DISCUSSION
5.1. Machine Learning in Species Delimitation
One goal of this study was to explore how well UML methods can successfully identify clusters equivalent to putative species and correctly infer the expected number of clusters. UML methods do not make assumptions about data type; data are merely treated as data. However, the underlying assumption here is that the analyses are operating at the species level. As with many dimensionality reduction techniques, UML methods will uncover any underlying structure regardless of the taxonomic level or data type. As such, integrative taxonomy with multiple data types and analytical approaches is ideal. Conversely, this insensitivity to taxonomic scale makes UML potentially relevant to population level analyses and phylogeography as well as species delimitation in taxa across varying divergence times, for example, ~20 Ma in the Metanonychus setulus group down to much more recent divergences of <1 Ma reported for Uma (Gottscho et al. 2017). While these UML approaches seemingly work well with relatively clear species showing congruence across data types, their ability to correctly cluster samples in more difficult speciation scenarios (e.g., rapid and recent divergence, divergence with gene flow, etc.) remains to be thoroughly tested, although results in Uma and simulated data are promising.
VAE and t-SNE clusters were clear regardless of input type, and robust across multiple iterations and varying parameters. t-SNE was designed purely for visualization of high dimensional data, although given a low dimensional embedding as output, clustering is an obvious application. It has been noted that t-SNE clusters, cluster size, and distances between clusters may lack biological meaning (Wattenberg et al. 2016) and clusters should be interpreted with caution. An alternative to t-SNE, the recently developed UMAP (McInnes et al. 2018; uniform manifold approximation and projection) produces clusters which are shown to have biological meaning (Becht et al. 2018). In the datasets used here, inferred clusters have obvious meaning corresponding to species corroborated by other analyses and data types. Regardless of whether downstream clustering is performed, UML methods offer excellent options for relatively quick and informative data visualization that can help uncover uncertainty in a priori groupings or detect misidentifications and paraphyly, both of which are problematic for species hypotheses if data are destined for downstream model-based analyses.
In cases where genetic data are perhaps the only source of information, supervised machine learning approaches can be used such that training data are made for specific studies and organismal types. For example, if cryptic species are suspected in taxa with high population structure, training data could consist of multiple “curated” datasets of biologically/ecologically similar taxa where species are known and well-supported, thus taking organismal biology more directly into account. While CLADES oversplits Metanonychus, we do not see this as a negative, but rather as imperative to create and use curated training datasets reflecting the biological characteristics of the study organism. More recently, another supervised approach was developed that treats species delimitation as a model selection problem, using the binned multidimensional Site Frequency Spectrum as the predictor variable to build an RF classifier that can distinguish among different speciation models (Smith and Carstens 2018). Training data is simulated based on specification of several priors (guide tree, population size, divergence time, migration) either known or estimated for the particular study system. This is a promising approach as the priors make the analysis more specific to the biology of focal taxa.
Speciation is a continuous process, and as such, species boundaries may be uncertain creating the need for assessments of probability associated with species distinctiveness and sample assignment. Here we show that VAEs, which leverage neural networks to learn a probability distribution of the data, can learn phylogenetic structure with the latent variables. In contrast to t-SNE, VAEs are derived from formal Bayesian probability theory, and can hence be used to score the probability that new data belongs to a trained set of data or is a new species. The standard deviation around samples/clusters is an inherent result of VAE and visualization makes the assessment of cluster distinctiveness or uncertainty relatively straightforward. Given results presented here, the robustness of output to parameter variation, and its Bayesian nature, VAEs are promising for future work. Recent applications of neural networks demonstrate the potentially transformative effect these approaches may have, for example in classification (Boer and Vos 2018), identification (Valan et al. 2019), citizen science (www.inaturalist.org), and population genetics (Schrider and Kern 2016; Flagel et al. 2019).
5.1. Rethinking Species Delimitation
Commonly used species delimitation approaches relying on genomic-scale data have the potential to identify population structure and over-split taxa (Sukumaran and Knowles 2017), a problem exacerbated when studying taxa with inherently high levels of population genetic structure. While the issue of population structure in species delimitation has recently come under focus from a methodological perspective, the potential misinterpretation of population structure as species-level divergences in empirical data has been a concern for taxonomists for a relatively long time (Hedin 1997; Harvey 2002), and continues to be so (e.g., Boyer et al. 2007; Bond and Stockman 2008; Niemiller et al. 2012; Keith and Hedin 2012; Barley et al. 2013; Satler et al. 2013; Fernández and Giribet 2014; Hedin 2015; Hedin et al. 2015; Chambers and Hillis 2019). While Metanonychus taxa show clear morphological divergence in genitalic morphology, many taxa with similar biological characteristics and deep genetic structuring are morphologically conserved (e.g., cryptic species), adding another level of difficulty where species delimitation relies primarily, if not entirely, on genetic data. In our dataset UML clustering provided reasonable species hypotheses that were largely identical to commonly used discovery-based analyses. Most importantly, the clusters identified in UML clearly correspond to species determined via integrative taxonomy with essentially no discordance between datasets, implying that cluster “learning” was dominated by species-level divergences and not population structure. Validation methods applied to the same data supported unrealistic results severely overestimating the number of species. Of course, our use of the multispecies coalescent model, which assumes panmictic species, in a taxon like Metanonychus is clearly a violation of the assumptions, encouraging the need for refinement of the multispecies coalescent model and new species delimitation approaches that are useful for all taxa including those with high population structure (Leaché et al. 2018).
Any given species delimitation approach may not be suitable for all taxa given the diversity of biological characteristics unique to particular groups or organismal types with differing degrees of population structure and isolation, etc. (Sukumaran and Knowles 2017). For example, assessments of reproductive isolation and/or the associated requirement of sympatry are not feasible for taxa found in highly fragmented landscapes or in short-range endemics showing high morphological and niche conservatism (e.g., Czekanski-Moir and Rundell 2019). In these taxa, allopatry between congeneric species is the rule and sympatry is extremely rare, if present (e.g., Bond and Stockman 2008; Leavitt et al. 2015; Wachter et al. 2015; Starrett et al. 2018). Despite overwhelming evidence for species status of six species in the M. setulus complex, only two instances of sympatry are recorded, both in extreme southwest Oregon. Similarly, in the related genus Sclerobunus, twelve species were described from western North America based on integrative taxonomy, but only a single instance of sympatry is known, albeit with very clear differences in microhabitat preference and morphology between the two sympatric species (Derkarabetian et al. 2010; Derkarabetian and Hedin 2014).
Taxa with different biological, ecological, and life history characteristics will have different underlying genetic patterns that are manifested at the population level, community level, during speciation, and through deep time (e.g., Massatti and Knowles 2016; Satler and Carstens 2016; Fang et al. 2017; Czekanski-Moir and Rundell 2019; Giribet and Baker 2019). As such, we argue that when testing new species delimitation methods/models, multiple empirical datasets should be included in which the taxa vary in their biological and ecological characteristics, and correspondingly, their underlying genetic patterns. The inclusion of multiple datasets covering a diversity of organismal and ecological types will ultimately lead to more robust methods that are more broadly applicable across the diversity of life, or conversely, will identify the biological limits of the methods. Here we show that UML approaches recover species level divergences across different empirical datasets derived from taxa with very different biological and ecological characteristics, and in simulated datasets with varying genetic parameters and dataset characteristics. Machine learning approaches, whether used alone or potentially combined with models like the multispecies coalescent, may possess the algorithmic flexibility needed to accommodate the inherent biological variability found across empirical datasets.
6. CONCLUSIONS
Machine learning algorithms, even those designed for image analysis or pattern and text recognition, all seek to identify and learn the underlying structure of input data via dimensionality reduction in some form. Considering this, the UML used here produced plots with easier interpretability relative to PCA, with species clusters showing obvious separation in two-dimensional space. In addition, many UML approaches offer the ability to accommodate various data types common to an integrative taxonomic framework (e.g., genetic, morphometric, continuous, categorical). Many machine learning algorithms are well-suited for species delimitation, providing promising avenues of incorporation into standard systematics protocols and excellent resources are available for implementation (e.g., Keras, TensorFlow). With a basic understanding of the types of algorithms, the applications to species delimitation and all aspects of evolutionary biology hold remarkable future promise.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Science Foundation [grant number DEB #1354558] to M.H. and a National Science Foundation Doctoral Dissertation Improvement Grant [grant number DEB #1601208] to S.D. Research reported in this publication was supported by Office of the Director of the National Institutes of Health under award number DP5OD026389. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. For assistance during fieldwork we thank Casey Richart, James Starrett, Alan Cabrero, Erik Ciaccio, and Morganne Sigismonti. Initial inspiration was provided by Ty Roach. Nick Vinciguerra helped during SNP processing and Morganne Sigismonti assisted with initial morphological examinations. Type specimen loans and permission to use a sample for SEM imaging were kindly provided by Darrell Ubick (California Academy of Sciences). The content of this manuscript was improved through discussion with Jeet Sukumaran. An earlier version of this manuscript was improved by comments from Damien de Vienne, Laura Kubatko, and three anonymous reviewers.
APPENDIX A
Taxonomy
Family Paranonychidae Briggs, 1971
Genus Metanonychus Briggs, 1971
Metanonychus mechanicus Derkarabetian and Hedin, new species
Zoobank LSID. urn:lsid:zoobank.org:act:94667695-BA2B-4069–9159-2FAD9DFC89FF
Metanonychus setulus Briggs, 1971 [in part]
Type material:
Holotype male and allotype female from Oregon, Tillamook County, Highway 101 at Neahkahnie Beach Road, 45.7243 −123.9288, collected 16 August 2018 by M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio. Paratypes: 1 female with identical collecting information (used for SEM). 1 male (used for SEM) and 2 females from Oregon, Lincoln County, along Highway 18, H.B. Van Duzer Forest Corridor Wayside, 44.0385 – 123.8088, collected 15 August 2014 by M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio. 1 female from Oregon, Clatsop County, Saddle Mountain State Natural Area, 45.9612 – 123.6877, collected 16 August 2014 by M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio. Type specimens deposited at the California Academy of Sciences.
Etymology:
The specific epithet is derived from the greek mēchanikós meaning “of machines”, and refers to the use of machine learning methods in identifying this species.
Diagnosis:
Differentiated from all other Metanonychus (except M. setulus) by the dorsal plate of the penis (DP): DP entire without medial separation, acute apex, and broader than and non-parallel to the ventral plate. Differentiated from M. setulus by a triangular DP (broadly ovoid with acute tip in M. setulus). Metanonychus mechanicus is the only setulus group species found in their distribution. Sympatric with M. nigricans but can be differentiated by their smaller body size: scute length 0.9–1.3 mm (1.4 −1.6 in M. nigricans).
The following subset of site changes are diagnostic for COI (positions correspond to the COI matrix available on Dryad): 334 – 335 GG (vs. CC or TC), 412 T (vs. A), 414 A (vs. C or T), 708 T (vs. G or A).
Description:
Male holotype: Scute length 1.03 mm (average of all males examines 1.1), width 0.94. Integument yellow, with patterned black pigmentation on scutum, lateral margins of scutum unpigmented. Palps with slight pigmentation distally, palpal femur length 0.32, depth 0.11, with 3–4 small setae bearing tubercles ventrally. Legs pigmented, leg II length 2.9. Tarsal formula: 3,5,4,4. Penis with DP entire and triangular, not parallel to the ventral plate.
Female allotype: Similar to male, except slightly larger, scute length 1.2 (average of all females examined 1.25). Margins of lateral lobes of ovipositor each with two spines ventrally and three spines dorsally.
Other Material Examined:
For a complete list of all specimens examined for all species, see Supplemental File 2. Here we list only those specimens examined for Metanonychus mechanicus.
Oregon: Lincoln Co.: Fogarty Creek State Park, South Creek Area, north of Depoe Bay, 44.8375 −124.0503, 15 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (1 male, 2 females, 1 juvenile). Tillamook Co.: Hwy 6, along Wilson River, near Kansas Creek, 45.4914 −123.6346, 16 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (1 male, 2 juveniles); road to Elk Creek Campground, off Hwy 6, along S Fork Wilson River, 45.6081 −123.4617, 15 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (2 males, 1 female); near Nehalem Falls Campground, along Nehalem River, Foss Road, E of Nehalem, 45.7299 −123.7687, 16 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (2 males, 1 female); Neahkahnie Mountain Trail, Oswald West State Park, 45.74057 −123.93447, 22 May 2011, S. Derkarabetian, S. Moore (1 male, 1 female). Clatsop Co.: Saddle Mountain State Natural Area, Saddle Mountain Road 0.4 mi N of US 26, 45.908 −123.7442, 3 April 2008, S. Derkarabetian, C. Richart (2 females); Saddle Mountain State Natural Area, along Saddle Mountain Trail, 45.9612 −123.6877, 16 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (1 male, 1 female, 1 juvenile); Fort Stevens State Park, Battery Russell Trail, 46.1893 −123.9704, 9 October 2010, S. Derkarabetian (2 females, 1 juvenile). Columbia Co.: Highway 47, 5 miles south of Clatskanie, 46.0636 −123.2678, 17 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (1 female).
Washington: Pacific Co.: Hwy 401, N of Knappton, 46.2906 −123.8114, 17 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (1 female). Lewis Co.: Rainbow Falls State Park, south side hiking trail, off Hwy 6, 46.6301 −123.2332, 17 August 2014, M. Hedin, J. Starrett, S. Derkarabetian, A. Cabrero, E. Ciaccio (1 male).
Distribution:
Known from the coastal ranges of northern Oregon north of the Yaquina River, to southwestern Washington around the Willapa Hills.
Metanonychus navarrus
Metanonychus navarrus Briggs, 1971
Metanonychus obrieni Briggs, 1971 (synonymy)
Comments:
Specimens morphologically agreeing with M. setulus obrieni could not be collected. This subspecies is only known from five type specimens from the type locality. The forests in the type locality of Fort Dick are now heavily disturbed. Multiple attempts to collect this taxon from near the type locality and localities within ~10 km produced samples that morphologically agree with several other Metanonychus subspecies. As such, we examined topotype specimens to assess distinctiveness. As originally described by Briggs (1971), the dorsal plate of obrieni is entire, while that of navarrus is cleft medially with an acute apex. All male obrieni type specimens possessed a cleft dorsal plate with a blunt apex (Fig. S4). While the blunt apex differs from the acute apex of navarrus, we do not consider this as warranting species status considering a sample collected north of the type locality of obrieni clusters with navarrus in all SNP analyses (Figs. 1 and 2).
Type material examined:
Metanonychus setulus obrieni holotype male and paratype female from California: Del Norte County, Fort Dick, Berlese – redwood litter, 2 July 1966,
C.W. Obrien (California Academy of Sciences); M. s. obrieni topotypes, two males (one used for SEM) and one female, with same collecting information as holotype.
APPENDIX B. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ympev.2019.106562.
Footnotes
Declarations of interest: none
REFERENCES
- Abadi M. et al. 2016. Tensorflow: a system for large-scale machine learning. 12th USENIX Symposium on Operating Systems Design and Implementation 16:265–283. www.tensorflow.org. [Google Scholar]
- Austerlitz F. et al. 2009. DNA barcode analysis: A comparison of phylogenetic and statistical classification methods. BMC Bioinformatics 10, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barley AJ, White J, Diesmos AC & Brown RM 2013. The challenge of species delimitation at the extremes: diversification without morphological change in Philippine sun skinks. Evolution 67, 3556–3572. [DOI] [PubMed] [Google Scholar]
- Bauer E, Laczny CC, Magnusdottir S, Wilmes P. & Thiele I. 2015. Phenotypic differentiation of gastrointestinal microbes is reflected in their encoded metabolic repertoires. Microbiome 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IW, Ng LG, Ginhoux F, Newell EW 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnol. 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Boer MJA, Vos RA 2018. Taxonomic classification of ants (Formicidae) from images using deep learning. Preprint at https://www.biorxiv.org/content/early/2018/09/04/407452.
- Bond JE, Stockman AK 2008. An integrative method for delimiting cohesion species: finding the population-species interface in a group of Californian trapdoor spiders with extreme genetic divergence and geographic structuring. Syst. Biol 57, 628–646. [DOI] [PubMed] [Google Scholar]
- Boyer SL, Baker JM, Giribet G. 2007. Deep genetic divergences in Aoraki denticulate (Arachnida, Opiliones, Cyphophthalmi): a widespread ‘mite harvestman’ defies DNA taxonomy. Mol. Ecol 16, 4999–5016. [DOI] [PubMed] [Google Scholar]
- Breiman L. 1996. Bagging predictors. Mach. Learn 24, 123–140. [Google Scholar]
- Breiman L. 2001. Random Forests. Mach. Learn 45, 5–32. [Google Scholar]
- Briggs TS 1971. The harvestmen of family Triaenonychidae in North America (Opiliones). Occas. Pap. Cal. Acad. Sci 90, 1–43. [Google Scholar]
- Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. 2012. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol 29, 1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna L, Benites P, Lougheed SC, Lijtmaer DA, Di Giacomo AS, Eaton MD, Tubaro PL 2011. Rapid phenotypic evolution during incipient speciation in a continental avian radiation. Proc. R. Soc. B. Biol. Sci 279, 1847–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens BC, Pelletier TA, Reid NM, Satler JD 2013. How to fail at species delimitation. Mol. Ecol 22, 4369–4383. [DOI] [PubMed] [Google Scholar]
- Chambers EA, Hillis DM 2019. The multispecies coalescent over-splits in the case of geographically widespread taxa. Syst. Biol. In Press [DOI] [PubMed] [Google Scholar]
- Chollet F. 2015. Keras. https://keras.io.
- Coombes KR, Wang M. 2018. PCDimension: finding the number of significant principal components. R package version 1.1.9. [Google Scholar]
- Cordier T, Forster D, Dufresne Y, Martins CIM, Stoeck T, Pawlowski J. 2018. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour 18, 1381–1391. [DOI] [PubMed] [Google Scholar]
- Czekanski-Moir JE, Rundell RJ 2019. The ecology of nonecological speciation and nonadaptive radiations. Trends Ecol. Evol 34, 400–415. [DOI] [PubMed] [Google Scholar]
- Dayrat B. 2005. Towards integrative taxonomy. Biol. J. Linn. Soc 85, 407–417. [Google Scholar]
- de Queiroz K. 2007. Species concepts and species delimitation. Syst. Biol 56, 879–86. [DOI] [PubMed] [Google Scholar]
- Derkarabetian S, Starrett J, Tsurusaki N, Ubick D, Castillo S, Hedin M. 2018. A stable phylogenomic classification of Travunioidea (Arachnida, Opiliones, Laniatores) based on sequence capture of ultraconserved elements. ZooKeys 760, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. 2016. tsne: T-Distributed Stochastic Neighbor Embedding for R (t-SNE). R package version 0.1–3. [Google Scholar]
- Earl DA, vonHoldt BM 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour 4, 359–361. [Google Scholar]
- Espíndola A, Ruffley M, Smith ML, Carstens BC, Tank DC, Sullivan J. 2016. Identifying cryptic diversity with predictive phylogeography. Proc. Rol. Soc. B 283, 20161529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Ezard THG, Pearson PN, Purvis A. 2010. Algorithmic approaches to aid species’ delimitation in multidimensional morphospace. BMC Evol. Biol 10, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faircloth BC 2013. Illumiprocessor: a trimmomatic wrapper for parallel adapter and quality trimming. Available from: 10.6079/J9ILL. [DOI]
- Faircloth BC 2017. Identifying conserved genomic elements and designing universal bait sets to enrich them. Meth. Ecol. Evol 8, 1103–1112. [Google Scholar]
- Faircloth BC 2015. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788. [DOI] [PubMed] [Google Scholar]
- Fang F, Chen J, Jiang LY, Chen R, Qiao GX 2017. Biological traits yield divergent phylogeographical patterns between two aphids living on the same host plants. J. Biogeogr 44, 348–360. [Google Scholar]
- Fernández R, Giribet G. 2014. Phylogeography and species delimitation in the New Zealand endemic, genetically hypervariable harvestman species, Aoraki denticulata (Arachnida, Opiliones, Cyphophthalmi). Invertebr. Syst 28, 401–414. [Google Scholar]
- Flagel L, Brandvain YJ, Schrider DR 2019. The unreasonable effectiveness of convolutional neural networks in population genetic inference. Mol. Biol. Evol 36, 220–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster RR 1954. The New Zealand harvestmen (sub-order Laniatores) (No. 2). Canterbury Museum Trust Board. [Google Scholar]
- Giribet G, Baker CM 2019. Further discussion on the Eocene drowning of New Caledonia: Discordances from the point of view of zoology. J. Biogeogr In press. 10.1111/jbi.13635 [DOI] [Google Scholar]
- Gottscho AD, Wood DA, Vandergast AG, Lemos-Espinal J, Gatesy J, Reeder TW 2017. Lineage diversification of fringe-toed lizards (Phrynosomatidae: Uma notata complex) in the Colorado Desert: Delimiting species in the presence of gene flow. Mol. Phylogenet. Evol 106, 103–117. [DOI] [PubMed] [Google Scholar]
- Grabherr MG et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey MS 2002. Short-range endemism amongst the Australian fauna: some examples from non-marine environments. Invertebr. Syst 16, 555–570. [Google Scholar]
- Hedin M. 2015. High-stakes species delimitation in eyeless cave spiders (Cicurina, Dictynidae, Araneae) from central Texas. Mol. Ecol 24, 346–361. [DOI] [PubMed] [Google Scholar]
- Hedin M, Carlson D, Coyle F. 2015. Sky island diversification meets the multispecies Coalescent–divergence in the spruce-fir moss spider (Microhexura montivaga, Araneae, Mygalomorphae) on the highest peaks of southern Appalachia. Mol. Ecol 24, 3467–3484. [DOI] [PubMed] [Google Scholar]
- Hedin M, Derkarabetian S, Blair J, Paquin P. 2018. Sequence capture phylogenomics of eyeless Cicurina spiders from Texas caves, with emphasis on US federally-endangered species from Bexar County (Araneae, Hahniidae). ZooKeys 769, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin M, Derkarabetian S, Alfaro A, Ramírez MJ, Bond JE 2019. Phylogenomic analysis and revised classification of atypoid mygalomorph spiders (Araneae, Mygalomorphae), with notes on arachnid ultraconserved element loci. PeerJ, 7, e6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin MC 1997. Molecular phylogenetics at the population/species interface in cave spiders of the Southern Appalachians (Araneae: Nesticidae: Nesticus). Mol. Biol. Evol 14, 309–324. [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. 2011. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. [DOI] [PubMed] [Google Scholar]
- Kass RE, Raftery AE 1995. Bayes factors. J. Am. Stat. Assoc 90, 773–795. [Google Scholar]
- Kassambara A, Mundt F. 2017. factoextra: extract and visualize the results of multivariate data analyses. R package version 1.0.5. [Google Scholar]
- Keith R, Hedin M. Extreme mitochondrial population subdivision in southern Appalachian paleoendemic spiders (Araneae: Hypochilidae: Hypochilus), with implications for species delimitation. J. Arachnol 40, 167–181. [Google Scholar]
- Kingma DP, Welling M. 2013. Auto-encoding variational Bayes. In: Proceedings of the International Conference on Learning Representations (ICLR) arXiv:1312.6114v10 [stat.ML]. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour 15, 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol 29, 1695–1701. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Fujita MK, Minin VN, Bouckaert RR 2014. Species delimitation using genome-wide SNP data. Syst. Biol 63, 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Zhu T, Rannala B, Yang Z. 2018. The spectre of too many species. Syst. Biol 68, 168–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt DH, Starrett J, Westphal MF, Hedin M. 2015. Multilocus sequence data reveal dozens of putative cryptic species in a radiation of endemic Californian mygalomorph spiders (Araneae, Mygalomorphae, Nemesiidae). Mol. Phylo. Evol 91, 56–67. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2, 18–22. [Google Scholar]
- Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. 2018. cluster: cluster analysis basics and extensions. R package version 2.0.7–1. [Google Scholar]
- Mallet L, Bitard-Feildel T, Cerutti F, Chiapello H. 2017. PhylOligo: a package to identify contaminant or untargeted organism sequences in genome assemblies. Bioinformatics 33, 3283–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. 1986. Die Grossgliederung der Opiliones und die evolution der ordnung (Arachnida) In Actas 10 Congreso Internacional de Aracnologia, Jaca/Espana (Barrienos JA, ed.). Instituto Pirenaico de Ecologia & Grupo de Aracnologia, Barcelona: (pp. 289–310). [Google Scholar]
- Massatti R, Knowles LL 2016. Contrasting support for alternative models of genomic variation based on microhabitat preference: Species-specific effects of climate change in alpine sedges. Mol. Ecol 25, 3974–3986. [DOI] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2018. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426v2 [stat.ML]. [Google Scholar]
- Niemiller ML, Near TJ, Fitzpatrick BM 2012. Delimiting species using multilocus data: diagnosing cryptic diversity in the southern cavefish, Typhlichthys subterraneus (Teleostei: Amblyopsidae). Evolution 66, 846–866. [DOI] [PubMed] [Google Scholar]
- Olteanu M, Nicolas V, Schaeffer B, Denys C, Missoup AD, Kennis J, Larédo C. 2013. Nonlinear projection methods for visualizing barcode data and application on two data sets. Mol. Ecol. Resour 13, 976–90. [DOI] [PubMed] [Google Scholar]
- Pedregosa F. Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. 2011. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res 12, 2825–2830. [Google Scholar]
- Pei J. Chong C, Xin L, Bin L, Yufeng W. 2018. CLADES: A classification-based machine learning method for species delimitation from population genetic data. Mol. Ecol. Resour 18, 1144–1156. [DOI] [PubMed] [Google Scholar]
- Pérez-González A, Werneck RM. 2018. A fresh look over the genital morphology of Triaenonychoides (Opiliones: Laniatores: Triaenonychidae) unravelling for the first time the functional morphology of male genitalia. Zool. Anz 272, 81–92. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudlo P. Marin JM, Estoup A, Cornuet JM, Gautier M, Robert CP 2016. Reliable ABC model choice via Random Forests. Bioinformatics 32, 859–66. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: https://www.R-project.org. [Google Scholar]
- Rousseeuw PJ 1987. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comp. Appl. Math 20, 53–65. [Google Scholar]
- Satler JD, Carstens BC, Hedin M. 2013. Multilocus species delimitation in a complex of morphologically conserved trapdoor spiders (Mygalomorphae, Antrodiaetidae, Aliatypus). Syst. Biol 62, 805–823. [DOI] [PubMed] [Google Scholar]
- Satler JD, Carstens BC 2016. Phylogeographic concordance factors quantify phylogeographic congruence among co-distributed species in the Sarracenia alata pitcher plant system. Evolution. 70, 1105–1119. [DOI] [PubMed] [Google Scholar]
- Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH 2010. Integrative taxonomy: a multisource approach to exploring biodiversity. Ann. Rev. Entomol 55, 421–438. [DOI] [PubMed] [Google Scholar]
- Schrider DR, Kern AD 2016. S/HIC: robust identification of soft and hard sweeps using machine learning. PLoS Genet 12, e1005928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD 2018. Supervised machine learning for population genetics: a new paradigm. Trends. Gene.t 34, 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Fop M, Murphy TB, Raftery AE 2017. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. The R Journal 8/1, 205–233. [PMC free article] [PubMed] [Google Scholar]
- Seifert B, Ritz M, Csősz S. 2014. Application of exploratory data analyses opens a new perspective in morphology-based alpha-taxonomy of eusocial organisms. Myrmecol. News 19, 1–15. [Google Scholar]
- Smith ML, Carstens BC 2018. Disentangling the process of speciation using machine learning. Preprint at https://www.biorxiv.org/content/early/2018/06/27/356345.
- Smith ML, Ruffley M, Espíndola A, Tank DC, Sullivan J, Carstens BC 2017. Demographic model selection using random forests and the site frequency spectrum. Mol. Ecol 26, 4562–73. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrett J, Derkarabetian S, Hedin M, Bryson RW Jr, McCormack JE, Faircloth BC 2017. High phylogenetic utility of an ultraconserved element probe set designed for Arachnida. Mol. Ecol. Resour 17, 812–823. [DOI] [PubMed] [Google Scholar]
- Starrett J, Hayashi CY, Derkarabetian S, Hedin M. 2018. Cryptic elevational zonation in trapdoor spiders (Araneae, Antrodiaetidae, Aliatypus janus complex) from the California southern Sierra Nevada. Mol. Phylo. Evol 118, 403–413. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Knowles LL 2017. Multispecies coalescent delimits structure, not species. Proc. Nat. Acad. Sci 114, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Economo EP, Knowles LL 2015. Machine learning biogeographic processes from biotic patterns: a new trait-dependent dispersal and diversification model with model choice by simulation-trained discriminant analysis. Syst. Biol 65, 525–545. [DOI] [PubMed] [Google Scholar]
- Valan M, Makonyi K, Maki A, Vondráček D, Ronquist F. 2019. Automated taxonomic identification of insects with expert-level accuracy using effective feature transfer from convolutional networks. Syst. Biol 10.1093/sysbio/syz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Maaten LVD, 2008. Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Venables WN, Ripley BD 2002. Statistics and Computing Modern Applied Statistics with S (Springer; ). [Google Scholar]
- Wächter GA, Muster C, Arthofer W, Raspotnig G, Föttinger P, Komposch C, Steiner FM, Schlick-Steiner BC 2015. Taking the discovery approach in integrative taxonomy: decrypting a complex of narrow-endemic Alpine harvestmen (Opiliones: Phalangiidae: Megabunus). Mol. Ecol 24(4), 863–889. [DOI] [PubMed] [Google Scholar]
- Wattenberg M, Viégas F, Johnson I. 2016. How to use t-SNE effectively, Distill. 10.23915/distill. [DOI]
- Wiens JJ, Graham CH 2005. Niche conservatism: integrating evolution, ecology, and conservation biology. Ann. Rev. Eco.l Evol. Syst 36, 519–539. [Google Scholar]
- Yang Z, Rannala B. 2010. Bayesian species delimitation using multilocus sequence data. Proc. Nat. Acad. Sci 107, 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Fukumizu K, Vogiatzis C. 2016. Multilocus phylogenetic analysis with gene tree clustering. Ann. Oper. Res 1–21. [Google Scholar]
- Zarza E, Connors EM, Maley JM, Tsai WL, Heimes P, Kaplan M, McCormack JE 2018. Bridging multilocus species delimitation and DNA barcoding through target enrichment of UCEs: A case study with Mexican highland frogs. PeerJ 6, e6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.