SUMMARY
Dysregulation of neuronal excitability underlies the pathogenesis of tauopathies, including frontotemporal dementia (FTD) with tau inclusions. A majority of FTD-causing tau mutations are located in the microtubule-binding domain, but how these mutations alter neuronal excitability is largely unknown. Here, using CRISPR/Cas9-based gene editing in human iPSC-derived neurons and isogenic controls, we show that the FTD-causing V337M tau mutation impairs activity-dependent plasticity of the cytoskeleton in the axon initial segment (AIS). Extracellular recordings by multi-electrode arrays (MEAs) revealed that the V337M tau mutation in human neurons leads to an abnormal increase in neuronal activity in response to chronic depolarization. Stochastic optical reconstruction microscopy of human neurons with this mutation showed that AIS plasticity is impaired by the abnormal accumulation of end-binding protein 3 (EB3) in the AIS submembrane region. These findings expand our understanding of how FTD-causing tau mutations dysregulate components of the neuronal cytoskeleton, leading to network dysfunction.
Keywords: Tau, FTD, Axon Initial Segment, EB3, Homeostasis, Neuronal activity, Cytoskeleton
INTRODUCTION
Dysregulation of neuronal excitability underlies the cognitive impairment of neurodegenerative diseases characterized by tau inclusions (Dickerson et al., 2005; Olazaran et al., 2010; Vossel et al., 2013). Excitability of neuronal networks is aberrantly increased in patients with frontotemporal dementia (FTD) (Beagle et al., 2017) and in mouse models with FTD-associated tau mutations (Crimins et al., 2012; Garcia-Cabrero et al., 2013). Tau has been implicated in the regulation of neuronal excitability. Genetic reduction of tau levels attenuates neuronal network hyperexcitability in flies (Holth et al., 2013) and mice (DeVos et al., 2013; Hall et al., 2015; Holth et al., 2013; Roberson et al., 2011). Transgenic mice with expression of a human FTD tau mutant have increased network excitability in the brain (Maeda et al., 2016). However, the mechanisms by which tau regulates neuronal excitability are not known.
Action potentials are initiated in the proximal region of the axon, the axon initial segment (AIS), a specialized compartment that has a high density of voltage-gated ion channels. The AIS has a unique cytoskeletal organization that consists of cytoskeletal submembrane networks (Jones et al., 2014; Leterrier et al., 2015; Rasband, 2010), which serve as scaffolds for ion channels on the membrane. Components of the AIS, including the cytoskeleton and ion channels, undergo activity-dependent structural changes to modulate neuronal excitability and maintain steady-state firing rates (Evans et al., 2015; Evans et al., 2013; Grubb and Burrone, 2010; Kuba et al., 2010; Yamada and Kuba, 2016).
Functional defects of the AIS due to cytoskeletal alterations have been reported in cellular models of tauopathies (Li et al., 2011; Sun et al., 2014; Tsushima et al., 2015; Zempel et al., 2017). Moreover, pathogenic tau with abnormal post-translational modifications destabilizes the cytoskeletal submembrane protein ankyrin G (AnkG) and microtubules in the AIS (Sohn et al., 2016), and changes the location of the AIS (Hatch et al., 2017). Thus, tau may have a key role in regulating the structural integrity of the AIS. Tau may affect the localization of structural components of the AIS such as end-binding proteins (EBs) (Ramirez-Rios et al., 2016; Sayas et al., 2015), which promote the stability of the AIS cytoskeletal networks by linking the cytoplasmic microtubules to AnkG in the AIS (Freal et al., 2016; Leterrier et al., 2011). However, it is unclear whether pathogenic tau affects activity-dependent AIS plasticity, leading to abnormal hyperexcitability of neurons.
In this study, we sought to determine whether the FTD-associated V337M mutation in MAPT (tauV337M) impairs activity-dependent AIS plasticity and disrupts neuronal function. We established a cellular model of FTD in cortical neurons differentiated from human induced pluripotent stem cells (iPSCs) derived from patients carrying tauV337M. Isogenic controls were established by genetically correcting the mutation with CRISPR/Cas9 genome editing system (Ran et al., 2013). We also introduced the V337M tau mutation into the genome of an iPSC line derived from a healthy subject. We used extracellular recordings by multi-electrode arrays (MEAs) to assess the function of human tauV337M neurons and super-resolution microscopy to assess the ultrastructure of the AIS cytoskeleton. Our findings revealed a novel pathway by which FTD mutant tau disrupts neuronal plasticity and function.
RESULTS
AIS Plasticity Is Impaired in Human iPSC-Derived Neurons with Pathogenic Tau Mutations
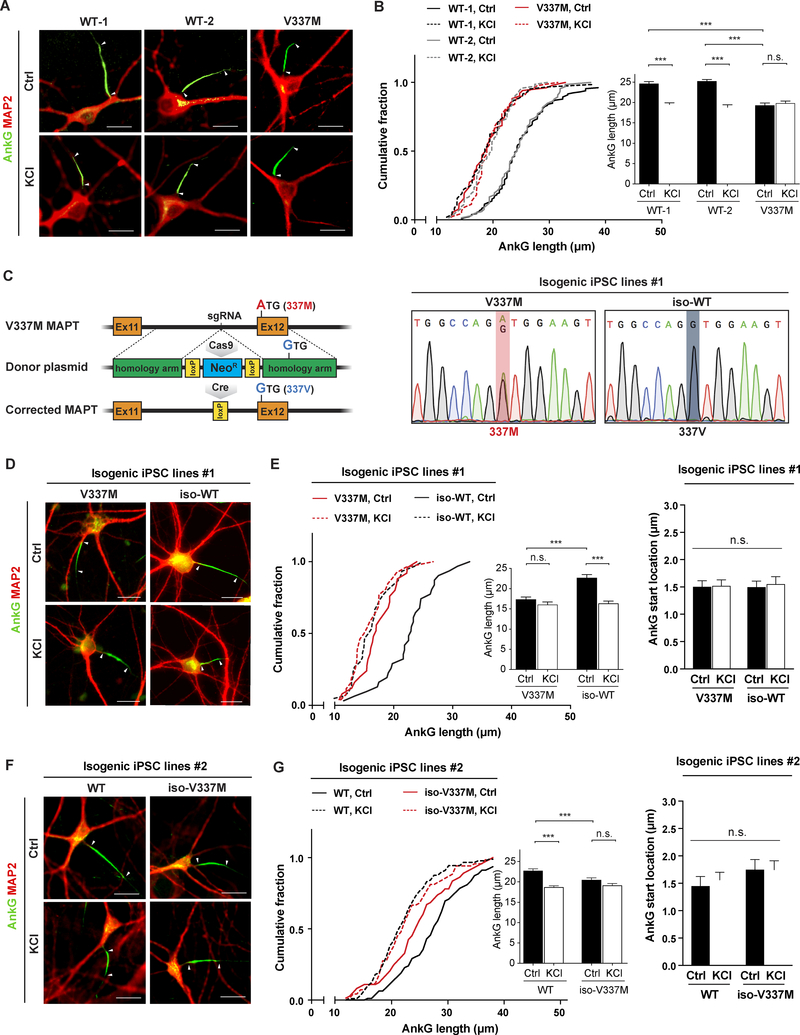
AIS plasticity in response to altered neuronal activity has been described as a homeostatic mechanism for modulating neuronal excitability in mouse, rat, chicken, and other vertebrates (Grubb and Burrone, 2010; Grubb et al., 2011; Kuba et al., 2010). To examine AIS plasticity in human neurons, we immunostained human iPSC-derived excitatory neurons for AnkG (Wang et al., 2017; Zhang et al., 2013). After chronic depolarization with KCl for 48 hr to increase neuronal activity in two independent lines of wild-type (WT) human neurons, AnkG was significantly shorter than in controls treated with NaCl (Figures 1A and 1B).
Figure 1. TauV337M Shortens the AIS and Impairs AIS Plasticity in Human iPSC-Derived Neurons.
(A) Representative images show immunostaining for AnkG in the AIS and MAP2 in the somatodendritic compartment in human iPSC-derived WT and tauV337M neurons treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS.
(B) Quantification of the length of AnkG shown by immunostaining in WT and tauV337M neurons treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 43–72 cells/group from three to five independent experiments. ***p < 0.001 by one-way ANOVA and Tukey’s post-hoc analysis.
(C) Schematic diagram of CRISPR gene editing at V337M MAPT and the DNA sequences before and after targeted gene editing.
(D) Representative images show immunostaining for AnkG in the AIS and MAP2 in the somatodendritic compartment in tauV337M and the corrected isogenic WT control (iso-WT) neurons treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS.
(E) Quantification of the length and start location of AnkG shown by immunostaining in tauV337M and iso-WT neurons treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 68–73 cells/group and n = 49–55 cells/group from three independent experiments for length and start location, respectively. ***p < 0.001 by one-way ANOVA and Tukey’s post-hoc analysis.
(F) Representative images show immunostaining for AnkG in the AIS and MAP2 in the somatodendritic compartment in WT and isogenic tauV337M (iso-V337M) neurons treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS.
(G) Quantification of the length and start location of AnkG shown by immunostaining in WT and iso-V337M neurons treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 54–56 cells/group and n = 45–49 cells/group from three independent for length and start location, respectively. ***p < 0.001 by one-way ANOVA and Tukey’s post-hoc analysis. Values are mean ± SEM. Scale bars, 15 μm.
To determine whether FTD mutant tau affects AIS plasticity in human neurons, we measured AnkG length in neurons derived from iPSCs from a patient with tauV337M after KCl treatment for 48 hr. AnkG was significantly shorter in unstimulated tauV337M neurons than in WT neurons, as were other AIS components βIV-spectrin and voltage-gated sodium channels (Figures 1A and 1B; Figures S1A and S1b). Notably, AnkG was not further decreased by KCl treatment (Figures 1A and 1B), suggesting that AIS plasticity is impaired in human tauV337M neurons. To exclude the possibility that the impairment reflected differences in the genetic background of WT and tauV337M neurons, we generated isogenic control iPSCs by using CRISPR/Cas9 genome editing to correct the mutation (Figure 1C). In the gene-corrected isogenic (iso-WT) neurons, AnkG, PIV- spectrin, and voltage-gated sodium channels were longer than in tauV337M neurons (Figures 1D and 1E; Figures S1A and S1B), and the gene correction restored AIS plasticity by reducing AnkG length after KCl treatment (Figures 1D and 1E). AnkG start location, measured by distance from the cell body, was not different between tauV337M and iso-WT neurons (Figures 1D and 1E). Moreover, KCl treatment did not alter the length of AnkG in tauV337M neurons differentiated from an independent iPSC line derived from a different patient carrying tauV337M (Figures S2A and S2B). In contrast, gene correction in an isogenic control elongated AnkG and restored AIS plasticity (Figures S2A and S2B), as expected.
To further dissect the significance of tauV337M in regulating AIS plasticity, we used CRISPR/Cas9 genome editing to introduce the mutation into WT human neurons (Figure S3A). Interestingly, gene-edited tauV337M neurons (iso-V337M) had the same phenotype as patient- derived tauV337M neurons; AnkG was significantly shorter than in WT controls, and chronic depolarization with KCl did not alter its length (Figures 1F and 1G), suggesting that tauV337M is sufficient to impair activity-dependent AIS plasticity. The AnkG start location was not different between WT and iso-V337M neurons (Figures 1F and 1G). We confirmed that the iPSC lines had a normal karyotype and were pluripotent, as shown by positivity for Oct4 and Sox2 (Figures S3B and S3C). These data from multiple independent isogenic iPSC lines suggest that tauV337M causes impaired AIS plasticity in human neurons.
Homeostatic Control of Neuronal Excitability Is Impaired in TauV337M Neurons
Since AIS plasticity underlies homeostatic control of neuronal excitability (Grubb and Burrone, 2010; Kuba et al., 2010), we next characterized the electrophysiological properties of tauV337M and isogenic control neurons. We recorded spontaneous neuronal activity of tauV337M and isogenic control neurons using two types of multi-electrode arrays (MEAs) with different layouts of electrodes (Figures 2A and 2J) (Tovar et al., 2017; Wainger et al., 2014). We first measured action potentials with a grid of 16 electrodes implanted in each well (Figure 2A) and analyzed parameters for individual spikes, single-channel bursts, and network bursts characterized by highly synchronized single-channel bursts over a period up to a few seconds. The firing and burst rates were not significantly different between tauV337M and isogenic control neurons (Figures 2B–2E). Interestingly, while tauV337M neurons showed significantly lower network burst rate than isogenic control neurons, they exhibited a significant increase in the duration of network burst and in the number of spikes per network burst (Figures 2F–2H), consistent with network hyper-synchrony. Indeed, tauV337M neurons exhibited more synchronized occurrence of spikes compared to isogenic control neurons (Figure 2I). Moreover, additional iPSC lines with gene-edited tauV337M mutation had the same abnormal activity profiles (Figures S4A–S4F) as patient-derived tauV337M neurons.
Figure 2. Homeostatic Control of Neuronal Excitability Is Impaired in TauV337M Neurons.
(A) MEA system #1. These MEAs have 16 electrodes per well arranged in a 4×4 grid and spaced at 350 μm.
(B) Representative raster plots of action potentials of tauV337M and iso-WT neurons detected from 16 electrodes in a single well of MEA system #1. Blue raster plots and boxes indicate action potentials involved in network bursts.
(C) Representative recordings from tauV337M and iso-WT neurons on MEA system #1.
(D-I) Spike, burst, and network burst parameters measured for tauV337M and iso-WT neurons on MEA system #1. n = 31–45 wells/group from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 by unpaired t-test.
(J) MEA system #2. These MEAs have 120 recording electrodes arranged in a 12×12 grid and spaced at 100 μm.
(K) Neuronal firing rate of tauV337M (n = 12) and iso-WT (n = 16) neurons on MEA system #2 from three independent recordings before and after chronic depolarization with KCl. **p < 0.01 by two-way ANOVA. Values are mean ± SEM.
We next measured the action potentials of tauV337M and isogenic control neurons before and after KCl-induced depolarization to determine whether excitability homeostasis is impaired in tauV337M neurons. The MEA layout with a grid of 120 electrodes with shorter inter-electrode distance allowed us to monitor the action potentials from the identical sets of neurons before and after KCl-induced depolarization (Figure 2J). Notably, the firing rate after depolarization was significantly increased in tauV337M neurons; however, the firing rate of isogenic control neurons before and after depolarization did not differ significantly (Figure 2K). These findings suggest that homeostatic control of spontaneous neuronal activity in response to depolarization is impaired in tauV337M neurons.
Increased EB3 Accumulation in the AIS in TauV337M Neurons
To examine mechanisms of impaired AIS plasticity, we assessed the possibility that the V337M mutation alters the binding affinity of tau with a component of AIS cytoskeleton. Since AnkG is stabilized by its interaction with EB3 in the AIS (Freal et al., 2016; Leterrier et al., 2011), and the accumulation of EB3 is regulated by tau (Sayas et al., 2015), we first examined whether tau binds directly to EB3 in vitro. Recombinant EB3 was titrated into 15N-K18 tau, a fragment that includes the microtubule-binding domains (MTBDs), and the putative interaction was measured by NMR spectroscopy. In this assay, potential binding sites were revealed by changes in peak signal intensity (I/I0) (Jinwal et al., 2013). We mapped three specific sites for EB3 in K18 tau (Figure 3A), including a site near V337, supporting a direct and potentially multivalent interaction between tau and EB3. We then directly compared binding of tau WT and V337M to EB3 by ELISA-based binding assay (Young et al., 2016). Notably, the V337M mutation led to tighter binding of tau to EB3 with a smaller dissociation constant (Kd = 5.6 nM) than WT tau (Kd = 22.0 nM) (Figure 3B).
Figure 3. EB3 Shows Increased Binding with Tau V337M and Is Accumulated in the AIS of Tau V337M Neurons.
(A) Profile of tau binding to EB3 as probed by 15N-HSQC of K18 tau containing the four microtubule-binding repeats. The NMR intensity ratio I/I0 is shown for individual peaks derived from HSQC spectra (I, chaperone present, I0, tau alone) where EB3 was present at a 2:1 molar ratio to tau. Red lines indicate potential EB3 binding sites of tau indicated by decreases in the NMR intensity ratio.
(B) Tau binding to EB3 measured by ELISA-based assay. Cumulative binding percentage of full-length WT tau and tauV337M to EB3 from three independent experiments. Dissociation constants (Kd) are indicated on top of the graph.
(C) Representative images show immunostaining of AnkG and EB3 in the AIS in WT, tauV337M, and iso-WT neurons. Arrowheads indicate the start and end of the AIS. Dashed lines indicate the boundary of the cell body. Scale bars, 15 μm.
(D, E) Quantification of the intensity of AnkG and EB3 immunostaining in WT, tauV337M, and iso-WT neurons. n = 55–59 cells/group from three independent experiments. ***p < 0.001 by one-way ANOVA and Tukey’s post-hoc analysis.
(F) Intensity profile of EB3 immunostaining along the axon in WT, tauV337M, and iso-WT neurons. n = 45–47 cells/group from three independent experiments. **p < 0.01 by multilevel mixed model analysis. Values are mean ± SEM.
Given this tighter binding of tauV337M to EB3 than WT tau and the effect of AnkG stabilization by EB3 (Freal et al., 2016; Leterrier et al., 2011), we hypothesized that tauV337M augments EB3 accumulation in the AIS, leading to increased AnkG stability and resistance of AnkG to rearrangement in response to chronic depolarization. Indeed, EB3 levels in the AIS were significantly higher in tauV337M neurons than in WT and isogenic controls, as shown by immunostaining (Figures 3C, 3E, and 3F), whereas AnkG levels were not different in the AIS of WT, tauV337M, and isogenic control neurons (Figures 3C and 3D).
EB3 Is Associated with AnkG and Tau in the AIS Submembrane Region of TauV337M Neurons
To examine the subcellular nanoscale distribution of EB3 and its spatial relationship with AnkG in the AIS, we immunostained neurons for EB3 and AnkG and analyzed cellular ultrastructure with super-resolution microscopy STORM (Huang et al., 2008; Rust et al., 2006). In WT and isogenic control neurons, EB3 was present throughout the cytoplasm, and its radial distribution was decreased in the vicinity of the AIS membrane; however, in tauV337M neurons, EB3 was concentrated in the submembrane region of the AIS (Figures 4A and 4B). STORM imaging after AnkG immunostaining showed no difference in the radial distribution of AnkG, but revealed AnkG enrichment in the AIS submembrane in WT, tauV337M, and isogenic control neurons (Figures 4C and 4D). Furthermore, based on the nanoscale distribution of tau in the AIS, the submembrane region was enriched in tauV337M (Figures 4E and 4F). We validated these results using cross-correlation analysis, and found that EB3 and AnkG co-localized more in the AIS submembrane region of tauV337M neurons than WT and isogenic controls (Figure 4G), and that tau also co-localized with EB3 (Figure 4H).
Figure 4. Increased EB3 Level Is Associated with AnkG and Tau in the AIS Submembrane Region of TauV337M Neurons.
(A) Representative 3D-STORM images of EB3 immunostaining in the AIS of WT, tauV337M, and iso-WT neurons. Color is used to indicate depth in z according to the color scale. Scale bar, 1 μm. Right panel: cross-sections of boxed regions in the left panel. Dashed circles represent the membrane of the axon; white dots represent the center of the cytoplasm. Scale bar, 200 nm.
(B) Radial distribution of EB3 in cross section of the AIS of WT (n = 8), tauV337M (n = 14), and iso-WT (n = 5) neurons. The distance from the center to the periphery of the cell was normalized to 1. ***p < 0.001 by multilevel mixed model analysis.
(C) Representative dual-color 3D-STORM images of EB3 (magenta) and AnkG (green) in the AIS of WT, tauV337M, and iso-WT neurons. Dashed circles represent the membrane of the axon; white dots represent the center of the cytoplasm. Scale bars, 1 pm. Right panel: cross-sections of boxed regions in the left panel. Scale bars, 200 nm.
(D) Radial distribution of AnkG in the AIS of WT (n = 7), tauV337M (n = 12), and iso-WT (n = 5) neurons. Values are mean ± SEM.
(E) Representative dual-color 3D-STORM images of tau (magenta) and EB3 (green) in the ATS of WT, tauV337M, and iso-WT neurons. Scale bars, 1 μm. Right panel: cross-sections of boxed regions in the left panel. Dashed circles represent the membrane of the axon; white dots represent the center of the cytoplasm. Scale bars, 200 nm.
(F) Radial distribution of tau in the AIS of WT (n = 8), tauV337M (n = 16), and iso-WT (n = 7) neurons.
(G, H) Cross-correlation analysis of EB3-AnkG and EB3-tau from dual-color 3D-STORM images. The pair cross-correlation function between the two channels was calculated from the distribution of intermolecular distances (x-axis) and normalized to that of randomly distributed sets of molecules with similar apparent sizes. Each curve represents the average of three independent neurons within each group. Cross-correlation indices for the first three distances (<30 nm) are taken as quantitation of co-localization, which is subject to the two-sample t test. *p < 0.05, ***p < 0.001. Values are mean ± SEM.
Critical Role of EB3 in AIS Plasticity and Activity Homeostasis
To determine whether EB3 accumulation changes the location and stability of AnkG, we compared the distribution and mobility of AnkG tagged with green fluorescent protein (GFP) in the absence and presence of overexpressed EB3 tagged with blue fluorescent protein (BFP). Consistent with previous studies on the interaction between AnkG and EB3 (Freal et al., 2016; Leterrier et al., 2011), overexpression of EB3 in Cos7 cells induced recruitment of AnkG-GFP to the lattice of microtubules and led to immobility of AnkG-GFP, whereas in the absence of EB3, AnkG-GFP decorated only the tips of microtubules and was highly mobile (Figures S5A and S5B). These findings suggest that upregulation of EB3 increases its interaction with AnkG, promoting the structural stability of AnkG via physical interaction with microtubules.
To assess the importance of EB3 in regulating AIS plasticity, we first lowered EB3 levels with siRNA. Immunostaining confirmed a significant reduction (~80%) of EB3 in the AIS after 1 week of EB3 knockdown in tauV337M neurons (Figures 5A and 5B). AnkG length in tauV337M neurons was significantly greater after EB3 knockdown and decreased in response to KCl (Figures 5C and 5D). In tauV337M neurons treated with control nontargeting siRNA, AnkG length did not change after KCl treatment (Figures 5C and 5D). EB3 knockdown in isogenic WT control neurons did not alter AnkG length and maintains AIS plasticity (Figures S6A and S6B). In addition, EB3 knockdown in an additional tauV337M line (iso-V337M) confirmed that reducing EB3 levels elongated AnkG length and restored AIS plasticity (Figures S6C and S6D). To further assess the functional consequence of EB3 knockdown in tauV337M neurons, we measured the firing rate of neurons before and after KCl-induced depolarization using the MEA system with 16 electrodes (Figure 5E). Consistent with results using 120 electrodes (Figures 2J and 2K), the firing rate was significantly increased after KCl treatment in tauV337M neurons, whereas KCl treatment did not significantly alter the firing rate of iso-WT neurons (Figure 5E). Importantly, the firing rate of tauV337M neurons with EB3 knockdown did not increase after KCl-induced depolarization (Figure 5F), indicating that EB3 knockdown in tauV337M neurons restores not only structural plasticity in the AIS but also functional homeostatic regulation of neuronal activity.
Figure 5. Critical Role of EB3 in Regulating AIS Plasticity and Activity Homeostasis.
(A) Representative images show immunostaining of EB3 in tauV337M neurons treated with control siRNA or EB3 siRNA. Arrowheads indicate the start and end of the AIS stained with EB3.
(B) Quantification of EB3 intensity in tauV337M neurons treated with control siRNA or EB3 siRNA. n = 12 images/group from three independent experiments. ***p < 0.001 by unpaired t test.
(C) Representative images show immunostaining for AnkG in the AIS and MAP2 in the somatodendritic compartment in tauV337M neurons treated with control siRNA or EB3 siRNA. The neurons were treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS.
(D) Quantification of the length of AnkG shown by immunostaining of tauV337M neurons treated with control siRNA or EB3 siRNA and then with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 43–52 cells/group from three independent experiments.
(E) Neuronal firing rate of tauV337M and iso-WT neurons before and after chronic depolarization with 10mM KCl for 48 hr on MEA system #1 with 16 recording electrodes spaced at 350 μm. n = 11–18 wells/group from two to three independent experiments. *p < 0.05 by paired t-test.
(F) Neuronal firing rate of tauV337M neurons treated with control siRNA or EB3 siRNA. Recordings were performed before and after chronic depolarization with 10mM KCl for 48 hr on MEA system #1. n = 9–13 wells/group from two independent experiments. *p < 0.05 by paired t-test.
(G) Representative images show immunostaining for GFP and AnkG in iso-WT neurons transfected with GFP or EB3-GFP and treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS. Dashed lines indicate the axon.
(H) Quantification of the length of AnkG shown by immunostaining in iso-WT neurons transfected with GFP or EB3-GFP and treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 37–98 cells/group from four independent experiments. ***p < 0.001 by one-way ANOVA and Tukey’s post-hoc analysis. Values are mean ± SEM. Scale bars, 15 μm.
To determine whether the increased stability of AnkG induced by EB3 overexpression impairs AIS plasticity in human neurons, we transfected gene-corrected neurons with EB3-GFP and measured AnkG length after KCl treatment. In these neurons with EB3 overexpression, AnkG was shorter than in GFP-transfected controls, and its length did not decrease in response to KCl, whereas it did in GFP-transfected neurons (Figures 5G and 5H). These findings strongly suggest that the abnormal increase in EB3 levels impairs AIS plasticity in tauV337M neurons.
Reducing Tau Restores AIS Plasticity by an EB3-Dependent Mechanism
To test whether tauV337M contributes to EB3 accumulation, we treated neurons with siRNA against tau, which lowered tau levels by ~40% in tauV337M neurons (Figure 6A). The knockdown significantly decreased EB3 levels in the AIS without altering EB3 length (Figure 6B). These findings attest to the critical role of tauV337M in EB3 accumulation in the AIS.
Figure 6. Tau Knockdown in TauV337M Neurons Elongates the AIS and Restores AIS Plasticity by an EB3-Dependent Mechanism.
(A) Representative images of the immunostaining for tau with HT7 antibody in tauV337V neurons treated with control siRNA or tau siRNA (left). Quantification of tau intensity in tauV337V neurons treated with control siRNA or tau siRNA (right). n = 12 images/group from three independent experiments. *** p < 0.001 by unpaired t test. Scale bars, 40 μm.
(B) Representative images show immunostaining for EB3 in tauV337V neurons treated with control siRNA or tau siRNA. Arrowheads indicate the start and end of the AIS stained for EB3. Dashed lines indicate the boundary of the cell bodies (left). Quantification of EB3 intensity and length in tauV337V neurons treated with control siRNA or tau siRNA (right). n = 43–48 cells/group from three independent experiments. *** p < 0.001 by unpaired t test. Scale bars, 15 μm.
(C) Representative images of the immunostaining of AnkG in the AIS and MAP2 in the somatodendritic compartment in tauV337M neurons treated with control siRNA or tau siRNA and treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS.
(D) Quantification of the length of AnkG shown by immunostaining in tauV337M neurons treated first with control siRNA or tau siRNA and then with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 57–65 cells/group from three independent experiments.
(E) Representative images show immunostaining for GFP and AnkG in tauV337M neurons treated with tau siRNA, transfected with GFP or EB3-GFP, and treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. Arrowheads indicate the start and end of the AIS. Dashed lines indicate the axon.
(F) Quantification of the length of AnkG shown by immunostaining in tauV337M neurons treated with tau siRNA, transfected with GFP or EB3-GFP, and treated with 10 mM NaCl (Ctrl) or KCl for 48 hr. n = 41–53 cells/group from three independent experiments. ***p < 0.001 by one-way ANOVA and Tukey’s post-hoc analysis. Values are mean ± SEM. Scale bars, 15 μm.
Next, we tested whether tau knockdown restores AIS plasticity in tauV337M neurons. After knockdown with siRNA, tauV337M neurons were treated with KCl and immunostained for AnkG. In these neurons, reducing tau elongated AnkG in the AIS, and its length was reduced in response to KCl, whereas in control neurons treated with nontargeting siRNA, AnkG length did not change after KCl treatment (Figures 6C and 6D). Furthermore, tau knockdown in an additional tauV337M line (iso-V337M) also elongated AnkG length and restored AIS plasticity (Figures S7A and S7B). To determine whether the restoration of AIS plasticity after tau lowering depends on downregulation of EB3, we overexpressed EB3-GFP in tauV337M neurons, knocked down tau, and measured AIS plasticity. In these neurons, AnkG was not shortened by KCl treatment (Figures 6E and 6F), suggesting that the reduction in EB3 levels was responsible for restoring AIS plasticity mediated by tau lowering. These results support a novel EB3-dependent mechanism by which a FTD-associated tau mutation reduces AIS length and impairs AIS plasticity, leading to neuronal dysfunction.
DISCUSSION
In this study, we showed that the FTD-causing tau V337M mutation in the microtubule-binding domain disrupts homeostatic rearrangement of the AIS cytoskeleton and leads to neuronal dysfunction in human neurons derived from FTD patient iPSCs. Gene editing by CRISPR/Cas9 in multiple independent iPSC lines showed that tauV337M impairs activity-dependent AIS plasticity by increasing EB3 levels in the AIS submembrane (Figure 7). TauV337M binds to EB3 tighter than tauWT and appeared to augment AIS localization of EB3, contributing to the impairment of AIS plasticity. Elevating EB3 levels shortened the AIS and blocked AIS plasticity, whereas reducing EB3 levels restored AIS plasticity and activity homeostasis in tauV337M neurons.
Figure 7. V337M Tau Reduces AIS Length and Impairs AIS Plasticity by an EB3-Dependent Mechanism.
Model of the cytoskeletal composition of the AIS and activity-dependent plasticity in WT and tauV337M neurons. In WT neurons (top), EB3 is evenly distributed throughout the cytoplasm of the AIS. A portion of AnkG remains unbound to EB3 and is susceptible to reorganization, resulting in depolarization-induced AIS shortening. In tauV337M neurons (bottom), EB3 accumulates abnormally in the AIS submembrane and binds to more AnkG and tau, making the AIS cytoskeleton shorter and resistant to reorganization. Consequently, the AIS remains unchanged in response to depolarization.
Mutations in MAPT, the gene encoding the microtubule-associated protein tau, cause FTD (Goedert et al., 1998; Hutton et al., 1998). Many FTD-associated tau mutations are located in the microtubule-binding domain (Rossi and Tagliavini, 2015; Wang and Mandelkow, 2016). Tau-mediated pathogenesis in FTD was previously thought to be mediated largely by the loss of microtubule stabilization as a result of reduced microtubule binding of tau caused by mutations in the microtubule-binding domain (Hong et al., 1998). However, the lack of microtubule perturbation and detrimental effects by tau reduction (Morris et al., 2011; Roberson et al., 2007) raised doubts about the essential role of microtubule destabilization in tau-dependent pathogenesis. Since cytoskeletal organization differs in distinct subcellular compartments in neurons (Conde and Caceres, 2009; Kevenaar and Hoogenraad, 2015), it is critical to determine which components of the neuronal cytoskeleton and their locations are perturbed by FTD-associated tau mutations. We found that tauV337M alters the structure of the cytoskeletal submembrane region of the AIS. It would be interesting to extend the investigation to examine whether other FTD-associated tau mutations also alter cytoskeletal structures in the AIS.
Our findings provide the first evidence that chronic depolarization shortens the AIS of human neurons. Activity-dependent AIS plasticity for homeostasis of neuronal excitability has been reported in nonhuman model systems, but the structural adaption of the AIS varies depending on the model system, method for activity perturbation, and cell type. The AIS is elongated in avian neurons deprived of synaptic inputs for several days (Kuba et al., 2010), but is shortened in murine neurons in response to short-term depolarization (Evans et al., 2015). In response to chronic depolarization, the AIS location shifts distally in murine excitatory neurons (Evans et al., 2013; Grubb and Burrone, 2010), but proximally in inhibitory interneurons (Chand et al., 2015). To maximize relevance to the human central nervous system and dissect AIS plasticity in a neuronal subtype, we engineered an inducible neurogenin-2 transgene into human iPSC lines to reliably convert them to homogenous cortical excitatory neurons with minimal variability (Wang et al., 2017). After chronic depolarization of these human cortical excitatory neurons with KCl, the proximal end of the AIS remained in the immediate vicinity of the cell body, but AIS length was shortened significantly. In contrast, when neurons are differentiated from human neuronal precursor line that gives rise to both excitatory and inhibitory neurons, excitatory neurons show a distal shift of the AIS in response to chronic depolarization (Horschitz et al., 2015). It is possible that the presence of inhibitory neurons in the heterogeneous culture can alter synaptic input to excitatory neurons, leading to differential AIS alterations in response to chronic depolarization.
At baseline, the AIS of the tauV337M neurons is shorter than that of WT neurons and has a concomitant increase in EB3 levels. Strikingly, gene correction of tauV337M decreased EB3 levels in the AIS and increased its length at the same time. Moreover, reducing EB3 levels lengthened the AIS in tauV337M neurons, whereas increasing EB3 levels shortened the AIS, suggesting a critical role for EB3 in regulating AIS length. Indeed, when AIS plasticity was restored, AIS length increased and EB3 levels fell. How elongation of the AIS contributes to AIS plasticity remains enigmatic. Possible scenarios could involve increased localization of Ca2+ channels in the elongated AIS as Ca2+ influx is required for AIS plasticity (Evans et al., 2015; Evans et al., 2013; Grubb and Burrone, 2010). Since the AIS is enriched in Ca2+ channels (Bender and Trussell, 2009), AIS elongation will likely result in greater abundance of Ca2+ channels and elevated local Ca2+ influx. Subsequently, Ca2+-dependent effectors such as the cysteine protease calpain could be activated; the resulting proteolysis of the AIS cytoskeleton (Czogalla and Sikorski, 2005; Schafer et al., 2009) could promote AIS plasticity.
Mechanistic dissection uncovered a novel role of EB3 in regulating AIS plasticity. We observed accumulation of EB3 in the AIS of tauV337M neurons. Super-resolution microscopy revealed enrichment of the submembrane region in EB3 associated with AnkG in tauV337M neurons. Unlike EB3 in other subcellular compartments in which EB3 only decorates the plus-end of microtubules (Galjart, 2010), EB3 in the AIS binds primarily to the microtubule lattice and links microtubules to the AIS submembrane cytoskeletal protein AnkG (Freal et al., 2016; Leterrier et al., 2011). This EB3-AnkG interaction may be essential for AIS stability (Freal et al., 2016). Consistent with this notion, we showed that in AIS of tauV337M neurons, the increased association between EB3 and AnkG remained static in response to rearrangement-inducing stimulation. In previous studies, activity-dependent AIS shortening appeared to require intracellular calcium and the calcium-dependent phosphatase calcineurin (Evans et al., 2015). This mechanism may not be mutually exclusive with the EB3-dependent mechanism that we propose, since EB3 regulates calcium signaling through its interaction with the inositol 1,4,5-trisphosphate receptor type 3 (Geyer et al., 2015). Excessive levels of EB3 in the AIS could perturb calcium signaling, preventing calcineurin from mediating AIS plasticity. On the other hand, calcineurin can regulate the stability of EB3 dimers by altering the phosphorylation status of EB3 (Komarova et al., 2012). Thus, EB3 might function downstream of calcineurin to relay to the AIS cytoskeleton signals required for plasticity.
How does tauV337M promote the accumulation of EB3 in the AIS submembrane? Our NMR findings suggest that tau binds to EB3 through tau’s microtubule-binding domains. The V337M mutation in this domain enhances tau’s binding affinity for EB3. Given the substantial enrichment of EB3 in the AIS compared to other compartments, it is likely that most significant effect of the enhanced tau-EB3 interaction may occur in the AIS. We demonstrated by STORM that tauV337M is abundant in the AIS submembrane and co-localized with EB3. Thus, the increased binding of tauV337M to EB3 may increase recruitment of EB3 to the AIS submembrane. It is possible that tauV337M neurons fail to recycle EB3 from the AnkG complex due to a tighter-than-expected affinity, leading to aberrant EB3 accumulation and AIS dysfunction. Indeed, knocking down tauV337M abolished EB3 accumulation in the AIS and restored AIS plasticity in an EB3-dependent manner. Reducing tau levels in pathological conditions attenuates network hyperexcitability (DeVos et al., 2013; Holth et al., 2013; Roberson et al., 2011) and thus restoration of AIS plasticity by tau lowering could contribute to homeostatic control of neuronal excitability and reduce excess activity in neural networks.
In conclusion, this study shows that tauV337M impairs the structural plasticity of the AIS by inducing abnormal accumulation of EB3 in the AIS, leading to loss of activity homeostasis—a novel pathway by which this FTD-causing tau mutation disrupts neuronal plasticity and function.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Li Gan (lig2033@med.cornell.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human neurons were differentiated as described (Wang et al., 2017). The Tet-ON 3G-controlled Neurogenin2 (Ngn2) transgene was integrated into the AAVS1 locus of human iPSC lines through a TALEN nuclease pair (Wang et al., 2017). Ngn2-integrated iPSC lines were generated from WT human iPSC line (male; WTC11) (Miyaoka et al., 2014; Wang et al., 2017) and a tauV337M-carrying human iPSC line (female; Bristol Myers Squibb, BMS). Human neurons were differentiated with a simplified two-step protocol (pre-differentiation and maturation). For pre-differentiation, iPSCs were incubated with doxycycline (2 μg/ml) for 3 days at a density of 2.0–2.5 × 106 cells/well in six-well plates coated with Matrigel in knockout Dulbecco’s modified Eagle’s medium/F12 medium containing N2 supplement, nonessential amino acids, mouse laminin (0.2 μg/ml), brain-derived neurotrophic factor (10 ng/ml), neurotrophin-3 (10 ng/ml; Peprotech), and rock inhibitor (Y-27632). For maturation, pre-differentiated precursor cells were dissociated and plated on Matrigel-coated coverslips with rat astrocytes in maturation medium containing 50% DMEM/F12, 50% Neurobasal-A medium, 0.5× B27 supplement, 0.5× N2 supplement, GlutaMax, nonessential amino acids, mouse laminin (1 μg/ml), brain-derived neurotrophic factor (10 ng/ml), and neurotrophin-3 (10 ng/ml). Half of the medium was replaced on days 3 and 7, and the medium volume was doubled on day 14. Thereafter, one-third of the medium was replaced weekly until the cells were used.
For siRNA-mediated knockdown of EB3 and tau, 4–5-week-old human neurons were treated with 1-μM nontargeting control siRNA (Dharmacon, D-001910–01), EB3 siRNA (Dharmacon, A-013109–13), or tau siRNA (Dharmacon, A-012488–13) for 6–8 days. For transfection, the neurons were treated overnight with 1 μg of EB3-GFP plasmid (The Nikon Imaging Center, UCSF) mixed with Lipofectamine 2000 (ThermoFisher Scientific) in OPTI-MEM (ThermoFisher Scientific). The transfection medium was then replaced with conditioned maturation medium.
For generation of iPSC lines without Ngn2 transgene integration (isogenic iPSC line 3 in Figure S2), dermal fibroblasts carrying the MAPT V337M mutation were transduced with nonintegrating Sendai virus carrying OCT3/4, SOX2, KLF4, and cMYC (Life Technologies) (Ban et al., 2011; Takahashi and Yamanaka, 2006). Cells that showed morphological evidence of reprogramming were selected manually. iPSC lines were analyzed for pluripotency markers by immunocytochemistry and for chromosomal abnormalities by karyotyping (Cell Line Genetics). Mutations were confirmed by Sanger sequencing.
Cos7 cells (ATCC) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum (FBS) in 35mm glass-bottom dishes (MatTek). 480AnkG-GFP (gift from Dr. Hoogenraad, Utrecht University, Netherlands) and EB3-BFP (The Nikon Imaging Center, UCSF) were transfected with Lipofectamine 2000 (ThermoFisher Scientific) in OPTI-MEM (ThermoFisher Scientific). Live-cell confocal imaging of Cos7 cells was performed with a Zeiss spinning disk confocal microscope (The Gladstone Histology and Light Microscopy Core) with 63× and 100× objectives. Images of AnkG-GFP and EB3-BFP were acquired at a rate of 1 frame / s for 1 min and analyzed with ImageJ software (NIH) for generating kymographs. All cell lines were cultured and imaged at 37°C in 5% CO2.
METHOD DETAILS
Immunofluorescence
Human iPSC-derived neurons differentiated for 4–6 weeks and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were then blocked with 5% normal goat serum and 0.3% Triton X-100 in PBS and incubated overnight at 4°C with mouse anti-AnkG (N106/36, NeuroMab), rabbit anti-MAP2 (AB5622, Millipore), mouse anti-Pan-Nav (S8809, Sigma-Aldrich), rabbit anti-βIV-spectrin (gift from Dr. Matthew N. Rasband, Baylor College of Medicine) (Yang et al., 2007), mouse anti-tau (HT7, ThermoFisher Scientific), rat anti-EB3 (KT36, Abcam), mouse anti-Oct4 (SC5279, Santa Cruz Biotechnology), rabbit anti-Sox2 (09–0024, Stemgent), and rabbit anti-GFP conjugated with Alexa Fluor 488 (ThermoFisher Scientific, A21311). The cells were then incubated with anti-mouse, anti-rabbit, anti-rat antibodies (ThermoFisher Scientific) conjugated with either Alexa Fluor 488, 555, or 647 for 1 h at room temperature. Images of the AIS were acquired with a Leica DM5000B fluorescence microscope with 20× and 40× objectives. ImageJ software (NIH) was used to analyze fluorescence intensity and length of the AIS manually in a blinded manner.
For immunofluorescence for STORM imaging, human iPSC-derived neurons differentiated for 4–5 weeks were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Cells were then blocked with 3% bovine serum albumin and 0.1% Triton X-100 in PBS and incubated with mouse anti-AnkG (N106/36, NeuroMab), rat anti-EB3 (KT36, Abcam), and mouse anti-tau (HT7, ThermoFisher Scientific) overnight at 4°C. The cells were then incubated with Alexa Fluor 647 goat anti-mouse (ThermoFisher Scientific, A21236), custom-labeled CF568 (Biotium, 92131), donkey anti-mouse (Jackson ImmunoResearch), CF568 donkey anti-rat (Jackson ImmunoResearch), and Alexa Fluor 647 donkey anti-rat (Life Technologies, A37573) for 1 h at room temperature.
Targeted Gene Editing
CRISPR/Cas9 gene editing by homologous recombination was used to create WT isogenic controls of iPSC lines carrying tauV337M (BMS) and to introduce the mutations into an iPSC line from a healthy donor (WTC11) (Miyaoka et al., 2014). iPSCs (1 × 106) were transfected with the Human Stem Cell Nucleofector Kit (Lonza) with 1.5 μg of sgRNA (5′-CTTGTGGGGTCA-TGGTTTACAGG-3’ for V337M) plasmid (Addgene, 68463) (Chen et al., 2015), 0.5 μg of Cas9 plasmid, and 2.5 μg of donor DNA plasmid containing a neomycin-resistance cassette (adapted from Addgene, PL552). Transfected cells were selected with neomycin for 1 week. Neomycin-resistant clones were selected and verified by genomic PCR and DNA sequencing. After sequence validation of the V337 site, 1 μM Cre recombinase (Excellgen) was added to remove the neomycin-resistance cassette from gene-targeted iPSCs. In addition, the top 10 potential off-target sites (http://crispr.mit.edu) were sequenced. For each genotype, the correct clones were karyotyped (Cell Line Genetics) and expanded for further experiments.
Multi-Electrode Array (MEA)
For the MEA system #1, we used Maestro Pro MEAs (Axion Biosystems). The MEA plate has 48 wells (Cytoview MEA48, Axion Biosystems), and each well contains 16 electrodes arranged in 4×4 grid and spaced at 350 μm. Each MEA well was coated with 1% polyethylenimine (Sigma-Aldrich) and seeded with 200,000 human iPSC-derived neurons that were co-cultured with 70,000 human glial cells. Recordings were done 4–5 weeks after the initiation of neuronal differentiation. Recordings were 10 minutes long and obtained with the AxIS Navigator software (Axion Biosystems), at a sampling rate of 12.5 kHz. Spikes were detected using a threshold set to 8 times the estimated standard deviation of the noise. Single-channel bursts were detected with minimum 5 spikes and maximum 100 ms inter-spike intervals. Network bursts were detected with minimum 12 spikes, maximum 100 ms inter-spike intervals, and minimum 25% participating electrodes. The Neural Metric Tool (Axion Biosysetms) was used to analyze parameters of individual spikes, bursts, and network bursts.
For the MEA system #2, we used MEAs from MultiChannel Systems (Reutlingen). The MEAs have 120 30-μm recording electrodes arranged in a 12×12 grid, and spaced at 100 μm. Each MEA was coated with 1% polyethylenimine (Sigma-Aldrich), and seeded with 120,000 human iPSC-derived neurons that were co-cultured with mouse glial cells. Cultures were maintained in BrainPhys Neuronal Medium (Stemcell Technologies) and fed weekly. Recordings were done 4 weeks after the initiation of neuronal differentiation. Each MEA was rested for 5 min on the acquisition stage before the recordings. Recordings were 3 minutes long and obtained with the MEA 2100 acquisition system (MultiChannel Systems), at a sampling rate of 20 kHz and using a band-pass filter with 200 and 4000 Hz cutoff frequencies. Spikes were detected using a threshold set to 6 times the estimated standard deviation of the noise. To study single neurons, we used custom software written in Python and Mathematica to identify propagation signals (Tovar et al., 2017). In brief, action potential propagation signals were detected with MEA Viewer (Bridges et al., 2018), a software program used to visualize MEA data. Subsequently, custom software was used to detect and export propagation signal traces. Propagation signals were detected by finding coincident spikes (spikes occurring frequently within a consistent interval) in groups of electrodes (Tovar et al., 2017). For chronic depolarization experiments, neuronal cultures in the MEAs were depolarized by increasing the KCl concentration in the medium from 5 mM to 15 mM. After 48 hr, the 15 mM KCl medium was removed and replaced with conditioned medium that was saved prior to KCl treatment. Recordings were obtained after 1 hr rest.
Protein Purification
For 0N4R WT, V337M tau, and K18 tau, protein was purified as previously described (Mok et al., 2018). Full-length EB3 was cloned into the pMCSG7 vector (Stols et al., 2002), which encodes an N-terminal 6XHis-tag linked to EB3 by a TEV cleavage site. The protein was expressed in Rosetta (DE3) competent cells with 600 μM IPTG for 16 hr at 18°C. 6XHis-tagged EB3 was affinity purified with Ni-NTA resin (Qiagen) in Tris buffer (50 mM Tris, 100 – 500 mM NaCl, 10–30 mM imidazole, pH 8.0), and eluted in Tris buffer (50mM Tris, 300mM NaCl, 300mM imidazole, pH 8.0). Aliquots of purified protein were flash frozen in liquid nitrogen and stored at −80°C.
NMR Spectroscopy
All proteins were dialyzed overnight in NMR buffer (25 mM HEPES pH 7.4, 10 mM KCl, 5 mM MgCl2, 1 mM TCEP, 10% (v/v) D2O). Samples were prepared at a final concentration of 50 μM 15N-labeled tau and 100 μM unlabeled EB3 in NMR buffer. Then, 2D 15N−1H heteronuclear single quantum coherence (HSQC) spectra were acquired at 10°C on a Bruker Avance AV800 spectrometer equipped with a cryoprobe. Sixteen scans were acquired per t1 value, and spectral widths of 2100 Hz and 10416 Hz were used in the 1H and 15N dimensions, respectively. Spectra were processed with rNMR and Sparky based on deposited tau assignments (Barre and Eliezer, 2013). Signal intensity ratios were calculated with GraphPad Prism.
Tau Binding Assay
This procedure was amended from a previous report (Young et al., 2016). Briefly, 0.005 mg mL−1 EB3 in Tris buffer (50mM Tris, 300mM NaCl, 300mM imidazole, pH 8.0) was immobilized overnight at 37°C in non-sterile 96 well plates (Fisher Scientific, clear, non-sterile, flat bottom) along with buffer matched negative controls. The wells were then washed with 100 μl of PBS-T (3×3 min, with rocking). To these wells, tau solution in binding buffer (25 mM HEPES, 40 mM KCl, 8 mM MgCl2, 100 mM NaCl, 0.01% Tween, 1mM DTT, pH 7.4) was added at indicated concentrations and incubated for 3 hours at RT with rocking. After blocking with 5% milk, the plates were quantified using rabbit anti-tau (H-150) primary antibody (Santa Cruz, sc5587) (1:2000 dilution, in TBS-T, 50 μl/well) and goat anti-rabbit HRP conjugated secondary antibody (Anaspec, 28177) (1:2,000 dilution in TBS-T, 50 μL/well), each for 1 hour. PBS-T washes (3×3 min, with rocking) were done after incubation of each antibody. TMB substrate (Cell Signaling Technology) was used to detect binding, and reactions were quenched using 1M HCl. The absorbance was measured using a SpectraMax M5 plate reader (OD450). Binding curves were fit using non-linear regression fits with a non-zero intercept and normalized to the maximum absorbance in GraphPad Prism version 8.0 (GraphPad Software).
STORM Imaging
For 3D-STORM imaging (Huang et al., 2008; Rust et al., 2006), we used a home-built setup (Wojcik et al., 2015) based on a Nikon Eclipse Ti-U inverted optical microscope with an oil immersion objective (Nikon CFI Plan Apochromat λ 100, NA 1.45). Lasers at 647 nm, 560 nm, and 405 nm were introduced into the sample from the back focal plane of the microscope. The laser beams were shifted toward the edge of the objective to illuminate a few micrometers into the sample. Continuous illumination of 647-nm laser (~2 kW cm−2) was used to excite Alexa Fluor 647 molecules and switch them into the dark state. Illumination of the 405-nm laser was added after ~2 min to reactivate the fluorophores to the emitting state. The power of the 405-nm laser (0–1 W cm−2) was tuned during image acquisition so that at any given instant, only a small fraction of the fluorophores in the sample were in the emitting state, which is optically resolvable. A 560-nm laser (~1.5 kW cm−2) was used to excite CF568 molecules after imaging of Alexa Fluor 647. For z position determination, a cylindrical lens was inserted between the electron- multiplying charge-coupled device (EMCCD) camera (iXon Ultra 897, Andor) and the microscope, so that images of single molecules were elongated differently for molecules at different z positions. The imaging buffer consisted of 100 mM Tris-Cl (pH 7.5) containing 140 mM cysteamine, 5% glucose, 0.8 mg mL−1 glucose oxidase, and 40 μg mL−1 catalase.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of 3D-STORM Data
3D-STORM raw data were reconstructed as described (Huang et al., 2008). Briefly, the centroid positions and ellipticity of single-molecule images determined in each frame were used to deduce the lateral and axial positions of each molecule. Single molecules from all frames were overlaid in one image, and sample drift was corrected by correlation across every 500 frames.
Radial distribution profiles (Figures 4B, 4D, and 4F) were generated with a custom MATLAB program. We used the relative distance to the center to account for variation in AIS width. An AIS segment of 1 μm long was toggled to the cross-sectional view, and an elliptical fit was computed to contain the outermost molecules. Twenty concentric ellipses with equally spaced radii within the outermost layer were drawn, and the number of contained molecules was counted for each ring. The radial histogram was scaled by the area of each ring. The border of the 12th ring was artificially assigned as the cell periphery (1.0 of the x-axis) since the normalized counts outside it decayed fast.
Two-dimensional cross-correlation analysis (Figures 4G–H) between two color channels (Stone and Veatch, 2015; Veatch et al., 2012) was done with a custom MATLAB program. Pairwise intermolecular distances between single molecules identified in the 647-nm and 560-nm channels were calculated. A histogram was generated for the counts of molecule pairs that fell into different ranges of distances, and normalized by using histograms generated from 10 sets of randomly distributed molecules in the same region. The resultant normalized cross-correlation values were expected to be >1 (for co-localizing patterns) or <1 (for excluding patterns) in the two channels. The x-axis indicates the distance of two-dimensional shift between two channels for the correlation analysis: Corr(Δr) = 〈Ired (r) ⋅ Igreen(r + Δr)〉/(〈Ired (r)〉 ⋅ 〈Igreen(r)〉). Cross-correlation indices with |Δr| ≤ 30 nm (first 3 data points) were used to quantify co-localization, as determined by the spatial resolution of STORM.
Statistical Analysis
Data were analyzed with GraphPad Prism and STATA. Differences between groups were assessed with the unpaired t test, one-way ANOVA with post-hoc test, and mixed-model linear regression analysis as indicated. Longitudinal data were fitted with mixed-model linear regression using the xtmixed command from STATA. All data were presented as the mean ± SEM. p < 0.05 was considered statistically significant.
DATA AND SOFTWARE AVAILABILITY
The custom MATLAB programs used in this study will be provided upon request to the corresponding author.
Supplementary Material
The FTD-causing V337M tau mutation impairs axon initial segment (AIS) plasticity.
The V337M tau mutation impairs activity homeostasis.
The V337M tau mutation leads to accumulation of EB3 in the AIS.
EB3 is critical for regulating AIS plasticity and activity homeostasis.
ACKNOWLEDGEMENTS
We thank Dr. Bruce Conklin (Gladstone Institutes) for the WTC11 iPSC line, Bristol-Myers Squibb (BMS) for the iPSC line with the tauV337M mutation, Dr. Lorena Saavedra and Dr. Jonathan Davila (Neucyte Inc.) for technical support for MEAs, and Erica Nguyen and Todd Paulsen for administrative assistance. This work was supported by NIH grants to L.G. (1R01AG054214-01A1, U54NS100717, R01AG051390), and K.S.K.; C.M.K.(AG046374) and J.E.G. (R01NS059690), and the Tau Consortium (to L.G., K.S.K, and J.E.G.). K.X. is a Chan Zuckerberg Biohub Investigator. C.M.C. is a CNPq fellow in Brazil. The Gladstone Institutes received support from National Center for Research Resources Grant RR18928.
Footnotes
DECLARATION OF INTERESTS
L.G is Founder of Aeton Therapeutics Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, et al. (2011). Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A 108, 14234–14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre P, and Eliezer D (2013). Structural transitions in tau k18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation. Protein Sci 22, 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagle AJ, Darwish SM, Ranasinghe KG, La AL, Karageorgiou E, and Vossel KA (2017). Relative Incidence of Seizures and Myoclonus in Alzheimer’s Disease, Dementia with Lewy Bodies, and Frontotemporal Dementia. J Alzheimers Dis 60, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender KJ, and Trussell LO (2009). Axon initial segment Ca2+ channels influence action potential generation and timing. Neuron 61, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges DC, Tovar KR, Wu B, Hansma PK, and Kosik KS (2018). MEA Viewer: A high- performance interactive application for visualizing electrophysiological data. PLoS One 13, e0192477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand AN, Galliano E, Chesters RA, and Grubb MS (2015). A distinct subtype of dopaminergic interneuron displays inverted structural plasticity at the axon initial segment. J Neurosci 35, 1573–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, Huang CT, Du Z, and Zhang SC (2015). Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell 17, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, and Caceres A (2009). Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci 10, 319–332. [DOI] [PubMed] [Google Scholar]
- Crimins JL, Rocher AB, and Luebke JI (2012). Electrophysiological changes precede morphological changes to frontal cortical pyramidal neurons in the rTg4510 mouse model of progressive tauopathy. Acta Neuropathol 124, 777–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czogalla A, and Sikorski AF (2005). Spectrin and calpain: a ‘target’ and a ‘sniper’ in the pathology of neuronal cells. Cell Mol Life Sci 62, 1913–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVos SL, Goncharoff DK, Chen G, Kebodeaux CS, Yamada K, Stewart FR, Schuler DR, Maloney SE, Wozniak DF, Rigo F, et al. (2013). Antisense reduction of tau in adult mice protects against seizures. J Neurosci 33, 12887–12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Greve DN, Chua EF, Rand-Giovannetti E, Rentz DM, Bertram L, Mullin K, Tanzi RE, Blacker D, et al. (2005). Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology 65, 404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MD, Dumitrescu AS, Kruijssen DLH, Taylor SE, and Grubb MS (2015). Rapid Modulation of Axon Initial Segment Length Influences Repetitive Spike Firing. Cell Rep 13, 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MD, Sammons RP, Lebron S, Dumitrescu AS, Watkins TB, Uebele VN, Renger JJ, and Grubb MS (2013). Calcineurin signaling mediates activity-dependent relocation of the axon initial segment. J Neurosci 33, 6950–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freal A, Fassier C, Le Bras B, Bullier E, De Gois S, Hazan J, Hoogenraad CC, and Couraud F (2016). Cooperative Interactions between 480 kDa Ankyrin-G and EB Proteins Assemble the Axon Initial Segment. J Neurosci 36, 4421–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N (2010). Plus-end-tracking proteins and their interactions at microtubule ends. Curr Biol 20, R528–537. [DOI] [PubMed] [Google Scholar]
- Garcia-Cabrero AM, Guerrero-Lopez R, Giraldez BG, Llorens-Martin M, Avila J, Serratosa JM, and Sanchez MP (2013). Hyperexcitability and epileptic seizures in a model of frontotemporal dementia. Neurobiol Dis 58, 200–208. [DOI] [PubMed] [Google Scholar]
- Geyer M, Huang F, Sun Y, Vogel SM, Malik AB, Taylor CW, and Komarova YA (2015). Microtubule-Associated Protein EB3 Regulates IP3 Receptor Clustering and Ca(2+) Signaling in Endothelial Cells. Cell Rep 12, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Crowther RA, and Spillantini MG (1998). Tau mutations cause frontotemporal dementias. Neuron 21, 955–958. [DOI] [PubMed] [Google Scholar]
- Grubb MS, and Burrone J (2010). Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature 465, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb MS, Shu Y, Kuba H, Rasband MN, Wimmer VC, and Bender KJ (2011). Short- and long-term plasticity at the axon initial segment. J Neurosci 31, 16049–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AM, Throesch BT, Buckingham SC, Markwardt SJ, Peng Y, Wang Q, Hoffman DA, and Roberson ED (2015). Tau-dependent Kv4.2 depletion and dendritic hyperexcitability in a mouse model of Alzheimer’s disease. J Neurosci 35, 6221–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch RJ, Wei Y, Xia D, and Gotz J (2017). Hyperphosphorylated tau causes reduced hippocampal CA1 excitability by relocating the axon initial segment. Acta Neuropathol 133, 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holth JK, Bomben VC, Reed JG, Inoue T, Younkin L, Younkin SG, Pautler RG, Botas J, and Noebels JL (2013). Tau loss attenuates neuronal network hyperexcitability in mouse and Drosophila genetic models of epilepsy. J Neurosci 33, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Zhukareva V, Vogelsberg-Ragaglia V, Wszolek Z, Reed L, Miller BI, Geschwind DH, Bird TD, McKeel D, Goate A, et al. (1998). Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 282, 1914–1917. [DOI] [PubMed] [Google Scholar]
- Horschitz S, Matthaus F, Gross A, Rosner J, Galach M, Greffrath W, Treede RD, Utikal J, Schloss P, and Meyer-Lindenberg A (2015). Impact of preconditioning with retinoic acid during early development on morphological and functional characteristics of human induced pluripotent stem cell-derived neurons. Stem Cell Res 15, 30–41. [DOI] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, and Zhuang X (2008). Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. (1998). Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705. [DOI] [PubMed] [Google Scholar]
- Jinwal UK, Akoury E, Abisambra JF, O’Leary JC 3rd, Thompson AD, Blair LJ, Jin Y, Bacon J, Nordhues BA, Cockman M, et al. (2013). Imbalance of Hsp70 family variants fosters tau accumulation. FASEB J 27, 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SL, Korobova F, and Svitkina T (2014). Axon initial segment cytoskeleton comprises a multiprotein submembranous coat containing sparse actin filaments. J Cell Biol 205, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevenaar JT, and Hoogenraad CC (2015). The axonal cytoskeleton: from organization to function. Front Mol Neurosci 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova YA, Huang F, Geyer M, Daneshjou N, Garcia A, Idalino L, Kreutz B, Mehta D, and Malik AB (2012). VE-cadherin signaling induces EB3 phosphorylation to suppress microtubule growth and assemble adherens junctions. Mol Cell 48, 914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba H, Oichi Y, and Ohmori H (2010). Presynaptic activity regulates Na(+) channel distribution at the axon initial segment. Nature 465, 1075–1078. [DOI] [PubMed] [Google Scholar]
- Leterrier C, Potier J, Caillol G, Debarnot C, Rueda Boroni F, and Dargent B (2015). Nanoscale Architecture of the Axon Initial Segment Reveals an Organized and Robust Scaffold. Cell Rep 13, 2781–2793. [DOI] [PubMed] [Google Scholar]
- Leterrier C, Vacher H, Fache MP, d’Ortoli SA, Castets F, Autillo-Touati A, and Dargent B (2011). End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc Natl Acad Sci U S A 108, 8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kumar Y, Zempel H, Mandelkow EM, Biernat J, and Mandelkow E (2011). Novel diffusion barrier for axonal retention of Tau in neurons and its failure in neurodegeneration. EMBO J 30, 4825–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Djukic B, Taneja P, Yu GQ, Lo I, Davis A, Craft R, Guo W, Wang X, Kim D, et al. (2016). Expression of A152T human tau causes age-dependent neuronal dysfunction and loss in transgenic mice. EMBO Rep 17, 530–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, Lizarraga PP, So PL, and Conklin BR (2014). Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods 11, 291–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok SA, Condello C, Freilich R, Gillies A, Arhar T, Oroz J, Kadavath H, Julien O, Assimon VA, Rauch JN, et al. (2018). Mapping interactions with the chaperone network reveals factors that protect against tau aggregation. Nat Struct Mol Biol 25, 384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, and Mucke L (2011). The many faces of tau. Neuron 70, 410–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazaran J, Prieto J, Cruz I, and Esteban A (2010). Cortical excitability in very mild Alzheimer’s disease: a long-term follow-up study. J Neurol 257, 2078–2085. [DOI] [PubMed] [Google Scholar]
- Pan L, Yan R, Li W, and Xu K (2018). Super-resolution microscopy reveals the native ultrastructure of the erythrocyte cytoskeleton. Cell Rep 22, 1151–1158. [DOI] [PubMed] [Google Scholar]
- Ramirez-Rios S, Denarier E, Prezel E, Vinit A, Stoppin-Mellet V, Devred F, Barbier P, Peyrot V, Sayas CL, Avila J, et al. (2016). Tau antagonizes end-binding protein tracking at microtubule ends through a phosphorylation-dependent mechanism. Mol Biol Cell 27, 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband MN (2010). The axon initial segment and the maintenance of neuronal polarity. Nat Rev Neurosci 11, 552–562. [DOI] [PubMed] [Google Scholar]
- Roberson ED, Halabisky B, Yoo JW, Yao J, Chin J, Yan F, Wu T, Hamto P, Devidze N, Yu GQ, et al. (2011). Amyloid-beta/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer’s disease. J Neurosci 31, 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, and Mucke L (2007). Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science 316, 750–754. [DOI] [PubMed] [Google Scholar]
- Rossi G, and Tagliavini F (2015). Frontotemporal lobar degeneration: old knowledge and new insight into the pathogenetic mechanisms of tau mutations. Front Aging Neurosci 7, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, and Zhuang XW (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayas CL, Tortosa E, Bollati F, Ramirez-Rios S, Arnal I, and Avila J (2015). Tau regulates the localization and function of End-binding proteins 1 and 3 in developing neuronal cells. J Neurochem 133, 653–667. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Jha S, Liu F, Akella T, McCullough LD, and Rasband MN (2009). Disruption of the axon initial segment cytoskeleton is a new mechanism for neuronal injury. J Neurosci 29, 13242–13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn PD, Tracy TE, Son HI, Zhou Y, Leite RE, Miller BL, Seeley WW, Grinberg LT, and Gan L (2016). Acetylated tau destabilizes the cytoskeleton in the axon initial segment and is mislocalized to the somatodendritic compartment. Molecular neurodegeneration 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stols L, Gu M, Dieckman L, Raffen R, Collart FR, and Donnelly MI (2002). A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif 25, 8–15. [DOI] [PubMed] [Google Scholar]
- Stone MB, and Veatch SL (2015). Steady-state cross-correlations for live two-colour superresolution localization data sets. Nat Commun 6, 7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wu Y, Gu M, Liu Z, Ma Y, Li J, and Zhang Y (2014). Selective filtering defect at the axon initial segment in Alzheimer’s disease mouse models. Proc Natl Acad Sci U S A 111, 14271–14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Bridges DC, Wu B, Randall C, Audouard M, Jang J, Hansma PK, and Kosik KS (2017). Recording action potential propagation in single axons using multi-electrode arrays. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Tsushima H, Emanuele M, Polenghi A, Esposito A, Vassalli M, Barberis A, Difato F, and Chieregatti E (2015). HDAC6 and RhoA are novel players in Abeta-driven disruption of neuronal polarity. Nature communications 6, 7781. [DOI] [PubMed] [Google Scholar]
- Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, and Baird BA (2012). Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS One 7, e31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossel KA, Beagle AJ, Rabinovici GD, Shu H, Lee SE, Naasan G, Hegde M, Cornes SB, Henry ML, Nelson AB, et al. (2013). Seizures and epileptiform activity in the early stages of Alzheimer disease. JAMA Neurol 70, 1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G et al. (2014). Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ward ME, Chen R, Liu K, Tracy TE, Chen X, Xie M, Sohn PD, Ludwig C, Meyer-Franke A, et al. (2017). Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem Cell Reports 9, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Mandelkow E (2016). Tau in physiology and pathology. Nat Rev Neurosci 17, 521. [DOI] [PubMed] [Google Scholar]
- Wojcik M, Hauser M, Li W, Moon S, and Xu K (2015). Graphene-enabled electron microscopy and correlated super-resolution microscopy of wet cells. Nature Commun 6, 7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, and Kuba H (2016). Structural and Functional Plasticity at the Axon Initial Segment. Front Cell Neurosci 10, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ogawa Y, Hedstrom KL, and Rasband MN (2007). betaIV spectrin is recruited to axon initial segments and nodes of Ranvier by ankyrinG. J Cell Biol 176, 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ZT, Rauch JN, Assimon VA, Jinwal UK, Ahn M, Li X, Dunyak BM, Ahmad A, Carlson GA, Srinivasan SR, et al. (2016). Stabilizing the Hsp70-Tau Complex Promotes Turnover in Models of Tauopathy. Cell Chem Biol 23, 992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempel H, Dennissen FJA, Kumar Y, Luedtke J, Biernat J, Mandelkow EM, and Mandelkow E (2017). Axodendritic sorting and pathological missorting of Tau are isoform-specific and determined by axon initial segment architecture. J Biol Chem 292, 12192–12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The custom MATLAB programs used in this study will be provided upon request to the corresponding author.