Abstract
The protein composition of the plasma membrane is rapidly remodeled in response to changes in nutrient availability or cellular stress. This occurs, in part, through the selective ubiquitylation and endocytosis of plasma membrane proteins, which in the yeast Saccharomyces cerevisiae is mediated by the HECT E3 ubiquitin ligase Rsp5 and arrestin-related trafficking (ART) adaptors. Here, we provide evidence that the ART protein family members are composed of an arrestin fold with interspersed disordered loops. Using Art1 as a model, we show that these loop and tail regions, while not strictly required for function, regulate its activity through two separate mechanisms. Disruption of one loop mediates Art1 substrate specificity. Other loops are subjected to phosphorylation in a manner dependent on the Pho85 cyclins Clg1 and Pho80. Phosphorylation of the loops controls Art1’s localization to the plasma membrane, which promotes cargo ubiquitylation and endocytosis, demonstrating a mechanism through which Art1 activity is regulated.
INTRODUCTION
Cells interact with, and respond to, the extracellular environment through the plasma membrane (PM). The PM contains a complex collection of channels, receptors, and transporters, and it must be rapidly remodeled to respond to changes in the environment and maintain cellular homeostasis. This occurs through the opposing processes of protein trafficking to the PM and endocytosis. Selective endocytosis occurs via ubiquitin conjugation to a PM protein, which commonly serves as a signal for endocytosis, and lysosomal sorting. The budding yeast Saccharomyces cerevisiae has proved to be a powerful and useful model to understand selective ubiquitin-mediated endocytosis. Numerous nutrient permeases have been shown to be specifically endocytosed and down-regulated in response to changes in the extracellular concentration of each permease’s substrate (Haguenauer-Tsapis and André, 2004; Gournas et al., 2016).
In yeast, ubiquitylation of PM proteins is mainly mediated by the HECT-type E3 ubiquitin ligase Rsp5 (Dupré et al., 2004; Léon and Haguenauer-Tsapis, 2009; Lauwers et al., 2010; MacGurn et al., 2012). Rsp5 contains 3 WW domains that specifically recognize PPXY motifs that allow binding and ubiquitylation of substrates (Macias et al., 1996; Dunn and Hicke, 2001; Gajewska et al., 2001). Many Rsp5 substrates, including most PM proteins, do not contain their own PPXY motifs but instead rely on PPXY-containing adaptor proteins to facilitate Rsp5 recruitment and ubiquitylation (Léon and Haguenauer-Tsapis, 2009). Although yeast contain membrane bound Rsp5 adaptor proteins that function on internal membranes (Hettema et al., 2004; Léon et al., 2008; MacDonald et al., 2011; Sardana et al., 2018), most PM proteins are recognized by a family of soluble arrestin-related trafficking (ART) adaptor proteins (Lin et al., 2008; Nikko and Pelham, 2009).
ART proteins belong to the α-arrestin protein family, a conserved protein family that consists of 14 members in yeast and at least six in mammals (Alvarez, 2008; Aubry and Klein, 2013). Similar to visual and β-arrestins, α-arrestins contain conserved N- and C-terminal arrestin domains (Aubry et al., 2009). Previous sequence analysis of the ART proteins suggested that most of the ART family members contain an N-terminal arrestin fold and a long C-terminal tail, which contains PPXY motifs (Lin et al., 2008; Becuwe et al., 2012a; Aubry and Klein, 2013).
Each ART protein is able to recruit Rsp5 to its own specific subset of PM cargoes in response to specific stimuli, resulting in cargo ubiquitylation and endocytosis (reviewed in Lauwers et al., 2010; O’Donnell and Schmidt, 2019). For example, Art1/Ldb19 targets Rsp5 to the methionine permease Mup1 in response to increased extracellular methionine and the arginine permease Can1 in response to increased extracellular arginine (Lin et al., 2008; Ghaddar et al., 2014); whereas Art3/Aly2 mediates the endocytosis of the aspartic acid/glutamic acid transporter Dip5 in response to increased aspartic acid concentrations (Hatakeyama et al., 2010). Further, multiple ARTs can regulate a single permease, as is the case for the uracil transporter Fur4 (Nikko and Pelham, 2009). Finally, different stimuli can activate different ARTs, leading to the ubiquitylation of the same cargo; the lysine permease Lyp1 is endocytosed in response to increased lysine concentrations via Art1, and in response to cycloheximide via Art2 (Lin et al., 2008). Thus, the ART adaptors recruit Rsp5 to specific cargoes in response to specific stimuli, forming a complicated signaling network that specifically modulates the PM proteome. Currently, little is known regarding how ARTs are able to achieve this striking cargo specificity.
Substrate transport facilitates endocytosis both by inducing movement out of specific PM domains to membrane domains where endocytosis is more favored (Busto et al., 2018; Gournas et al., 2018) and by exposing an ART binding site on the permease to allow ubiquitylation to occur (Keener and Babst, 2013; Guiney et al., 2016; Fujita et al., 2018). In addition to regulatory mechanisms involving the permeases, the ARTs themselves are subject to regulation. Most ARTs are ubiquitylated by Rsp5 (Gupta et al., 2007), and for many ARTs this ubiquitylation promotes ART activity (Lin et al., 2008; Becuwe et al., 2012b; Hovsepian et al., 2017). Finally, many ARTs can be regulated by phosphorylation. As examples, Art1 is phosphorylated by Npr1 in response to TORC1 activity (MacGurn et al., 2011), Art4 by Snf1 (Becuwe et al., 2012b), Art8 by PKA, and Art6/Aly1 is dephosphorylated by calcineurin (O’Donnell et al., 2013). In most cases described so far, phosphorylation inactivates, while dephosphorylation activates, ARTs. However, the exact mechanism of how phosphorylation regulates the ARTs is currently unclear. Thus, much remains to be discovered regarding how ARTs are able to specifically and temporally direct PM cargo ubiquitylation and endocytosis.
Here, we show that Art1, as well as the other ART proteins in S. cerevisiae, likely form an arrestin fold using the full length of their primary sequence, with multiple large insertions between sequences that form the arrestin fold. These “loops” and “tails” are not strictly required for Art1 function, but regulate its activity. Loop 3 confers cargo specificity to Art1, where it is required for Can1 endocytosis but is antagonistic for Mup1 and Lyp1 endocytosis. Further, loop 1 and the C-terminal tail, as well as a poorly conserved region within the arrestin fold (termed a “mini-loop”), are phosphorylated in a manner dependent on the Pho85 cyclins Clg1 and Pho80. This phosphorylation inactivates Art1 and prevents cargo endocytosis by reducing the ability of Art1 to associate with the PM.
RESULTS
Art1 contains an arrestin domain disrupted by multiple insertions
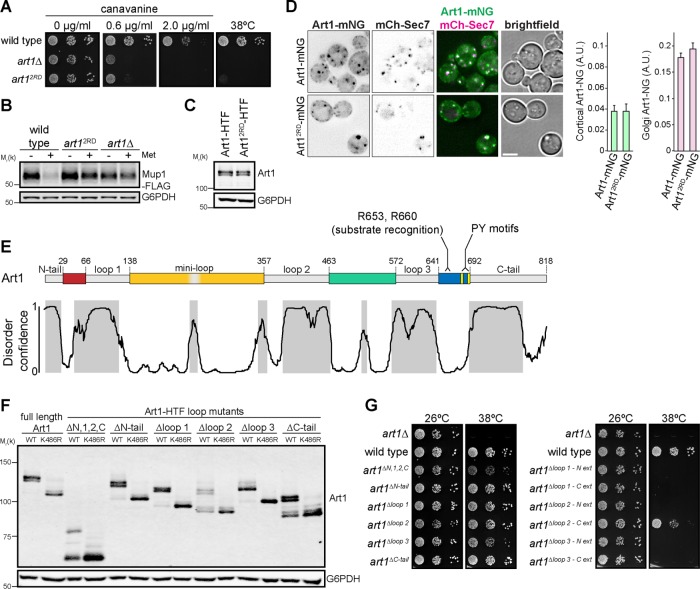
Sequence analysis and structural predictions suggest that the ART family members typically contain an N-terminal arrestin fold with a PPXY motif–containing C-terminal tail (Lin et al., 2008; Becuwe et al., 2012a; Aubry and Klein, 2013). However, we have previously provided evidence that Mup1 and Can1 interact with Art1 by binding to a motif in the Art1 C-terminal tail, rather than binding within the predicted N-terminal arrestin fold (Guiney et al., 2016). This was unexpected, because the arrestin domain directly binds substrate in a variety of arrestin domain–containing proteins (Aubry and Klein, 2013). Mutating the substrate binding motif (residues R653 and R660 mutated to Asp, termed “art12RD”) completely ablated Art1 function. Cells expressing art12RD were hypersensitive to the toxic arginine analogue canavanine (Figure 1A). Canavanine is transported into cells via Can1 (Grenson et al., 1966; Ahmad and Bussey, 1986), but toxicity can be mitigated by Can1 endocytosis and vacuolar degradation, and canavanine hypersensitivity occurs when Can1 cannot be endocytosed and degraded, providing a readout of Art1 function (Lin et al., 2008). Additionally, similar to art1Δ, cells expressing art12RD were unable to grow at 38°C (Figure 1A). At elevated temperatures, Art1 is required to endocytose misfolded PM proteins, including Lyp1, preventing membrane permeability and cell death (Zhao et al., 2013). Growth at high temperature is currently the most permissive assay for Art1 function. Thus, the inability of art12RD to grow at 38°C indicates a severe defect in Art1 function. Further, in cells expressing art12RD, methionine-induced Mup1-FLAG degradation was inhibited to a similar extent as in art1Δ (Figure 1B). Thus, R653 and R660 are required for Art1 function. Importantly, Art12RD is expressed similarly to wild-type (WT) Art1 (Figure 1C), and when fused to mNeonGreen (Art12RD-mNG) exhibited WT-like localization (Figure 1D), consistent with a disruption in substrate binding rather than a more general defect in folding or expression.
FIGURE 1:
Art1 contains an arrestin domain disrupted by multiple insertions. (A) Serial dilutions of the indicated yeast strains spotted on synthetic media containing canavanine. (B) Immunoblot of the yeast strains in A expressing Mup1-FLAG before and after treatment with 20 µg/ml Met for 60 min. (C) Immunoblot of yeast expressing Art1 or Art12RD with a C-terminal HTF (6xHis-TEV-3xFLAG) tag. Art1-HTF was detected with an α-FLAG antibody. (D) Localization of Art1-mNG and Art12RD-mNG in minimal media. Scale bar = 2 µm. Right, quantification of Art1 localization at the PM and Golgi; error bars are 95% CI, n > 300 cells for each condition. (E) Top, Art1 schematic. Nonconserved loop regions shown in gray. Conserved regions predicted to form an arrestin fold are colored. Bottom, disorder confidence predicted DISOPRED3. Gray shading indicates predicted disordered regions. (F) Immunoblot of Art1-HTF tail and loop mutants, with and without the K486R mutation, detected with an α-FLAG antibody. (G) Serial dilutions of Art1 tail and loop mutants spotted on synthetic media.
To further investigate this C-terminal substrate binding region, we analyzed the conservation of this region among all the fungal Art1 homologues. The region surrounding R653 and R660 is well conserved among the Art1 homologues examined, suggesting that they all bind their substrates using a similar motif. Strikingly, applying this analysis to the entire sequence revealed that Art1 (and its closely related homologues) contained multiple large, variable insertions (herein referred to as “loops” and “tails”) between regions that were well conserved. Shorter, more distantly related Art1 homologues retained only the well conserved regions, while the loops and tails were absent (Figure 1E and Supplemental Figure S1A). As an example, the distantly related Any1/Arn1 from Schizosaccharomyces pombe contains only a short N-terminal tail, and no loops. Structural modeling predicts that Any1 still forms an arrestin fold (Supplemental Figure S1B), suggesting that the inserted loops and tails in Art1 are not part of the arrestin domain. In the new structural model, loop 1 is inserted between the second and third β-strand in the N-lobe, loop 2 is positioned between the third and fourth β-strand of the C-lobe, and loop 3 occurs before the final β-strand in the C-lobe. There is also a shorter nonconserved region, termed a “mini-loop,” which also maps to a turn between two β-strands (Figure 1E and Supplemental Figure S1C). The regions where the loops are predicted to be inserted occur in turns between β-strands or coils, and therefore are not expected to disrupt the core arrestin fold. Thus, Art1 may form its arrestin fold using the conserved regions with interspersed loops, rather than forming an arrestin fold with its N-terminal half. Further supporting this model, the variable loops and tails are predicted to be disordered, unlike the conserved core regions, suggesting that these regions do not tightly fold into a structured domain (Figure 1E).
To test this hypothesis, the N- and C-terminal tails, and each loop, were removed individually and tested for function. Steady-state expression of each loop mutant was similar to WT Art1 (Figure 1F). Combining each loop mutant with the K486R mutation, which prevents the activating ubiquitylation of Art1 (Lin et al., 2008), shows that each mutant is ubiquitylated and that the multiple bands represent posttranslational modifications rather than degradation products. In contrast to art12RD, each loop/tail mutant could grow at an elevated temperature (Figure 1G), demonstrating that the loop and tail deletions did not completely ablate Art1 function. Further, after shifting to elevated temperature, most loop and tail deletions were as stable as WT Art1 (Supplemental Figure S1E), with the exception of art1Δloop 2. However, art1Δloop 2 can still support growth at 38°C, and its steady-state expression is similar to WT, suggesting a compensatory response, potentially via increased transcription/translation. When the N-terminus, loop 1, loop 2, and the C-terminus deletions were combined (termed “art1ΔN,1,2,C”), cells exhibited a reduced ability to grow at an elevated temperature, but not to the extent of art1Δ, indicating that many of the loops and tails contribute to, but are not strictly required for, Art1 function. Coimmunoprecipitation analysis showed that each loop and tail mutant was still able to interact with Rsp5, unlike the PPXY mutant (art12xPY; Supplemental Figure S1D).
To test the accuracy of our loop predictions, and to establish that Art1 does not tolerate arbitrary large deletions, we extended each loop deletion 10–15 additional amino acids into the predicted core arrestin fold. All of these loop extensions completely abolished Art1 activity (Figure 1G), except the loop 2 C-terminal extension, further supporting the identification of the predicted loops (with the exception of the precise loop 2 C-terminal boundary). Both loop 1 extension mutants and the loop 2 N-terminal extension mutant were not ubiquitylated (Supplemental Figure S1F), suggesting that protein folding was disrupted to the extent that Rsp5 binding was affected, despite retaining the PPXY motifs. However, both loop 3 extension mutants were ubiquitylated, signifying that Art1 function was abolished through a mechanism distinct from decreased Rsp5 binding. Additionally, most of the loop extension mutants were rapidly turned over compared with WT Art1 (Supplemental Figure S1E), consistent with protein misfolding. Taken together, these data support a model where the arrestin fold of Art1 consists of multiple conserved regions that are disrupted by long insertions.
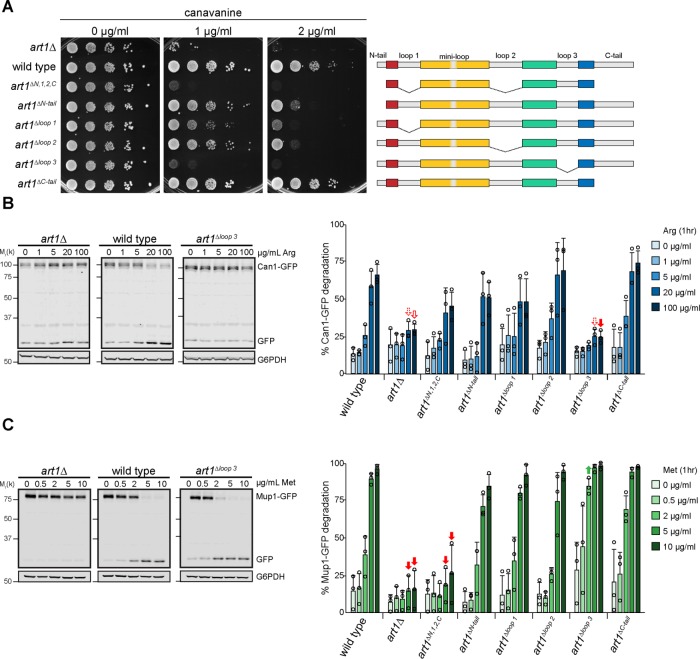
Art1 loops regulate cargo endocytosis
Because individually deleting the Art1 loops and tails did not result in a measurable defect in Art1-mediated PM quality control, but the combined art1ΔN,1,2,C mutant did show a decrease in function (Figure 1G), we hypothesized that the loops and tails could be involved in regulating Art1. We further examined the effect of the loop and tail mutants on two Art1 cargoes, Can1 and Mup1. When grown on canavanine, the strains expressing art1ΔN,1,2,C and art1Δloop 3 were canavanine hypersensitive (Figure 2A) implying a Can1 endocytosis defect, while the remaining mutants exhibited WT-like or slightly impaired canavanine growth. This canavanine sensitivity pattern was consistent with Can1-GFP degradation after addition of its substrate, Arg (Figure 2B and Supplemental Figure S3A). Can1-GFP degradation occurs after treatment with Arg in a dose-dependent manner; thus higher Arg concentrations applied for the same amount of time cause more Can1-GFP degradation in WT cells. Cells expressing art1ΔN,1,2,C exhibited less Can1-GFP degradation at higher Arg concentrations compared with WT ART1, and expression of art1Δloop 3 caused a nearly complete block in Can1-GFP. The remaining loop deletion mutants showed a degradation profile similar to that of WT ART1.
FIGURE 2:
Art1 loop 3 dictates substrate specificity. (A) Serial dilutions of the Art1 tail and loop mutant strains spotted on media containing canavanine. (B ) Left, immunoblot analysis of Can1-GFP endocytosis induced with the indicated concentration of Arg for 60 min. Right, band intensities were quantified and expressed as the mean % Can1-GFP degradation. Individual experiments are represented as open circles. Error bars indicate 95% CI. n = 3. Filled arrow, p < 0.001; outlined arrow, p < 0.01; dashed arrow, p < 0.05 vs. WT at matched treatment. (C) Immunoblot analysis of Mup1-GFP. As in B, except Met was used to induce endocytosis. n = 3.
Methionine-induced Mup1-GFP degradation in the loop and tail mutants was similar to that of Can1-GFP, with two exceptions: art1ΔN,1,2,C caused a stronger block in Mup1-GFP degradation, and art1Δloop3 unexpectedly showed enhanced Mup1-GFP degradation compared with WT ART1 (Figure 2C and Supplemental Figure S3C). In agreement with the differential effects on Can1 and Mup1 endocytosis, in art1Δloop3 Mup1 is ubiquitylated in response to its substrate, whereas Can1 is not (Supplemental Figure S3C).
Growth on thialysine, a toxic Lys analogue that is transported by the Art1 cargo Lyp1, revealed a pattern similar (although not identical) to Can1 (Supplemental Figure S3D). Cells expressing art1ΔN,1,2,C and art1Δloop 3 were the most thialysine hypersensitive, but unlike Can1, neither mutation causes a defect as severe as art1Δ.
Together, these data demonstrate that individually removing most of the loops results in defective substrate-induced cargo endocytosis, with stronger effects when multiple loop deletions are combined. Further, the defect in Can1 and Lyp1 endocytosis but enhanced Mup1 endocytosis in art1Δloop 3 suggest that loop 3 contributes to Art1 function by dictating substrate specificity.
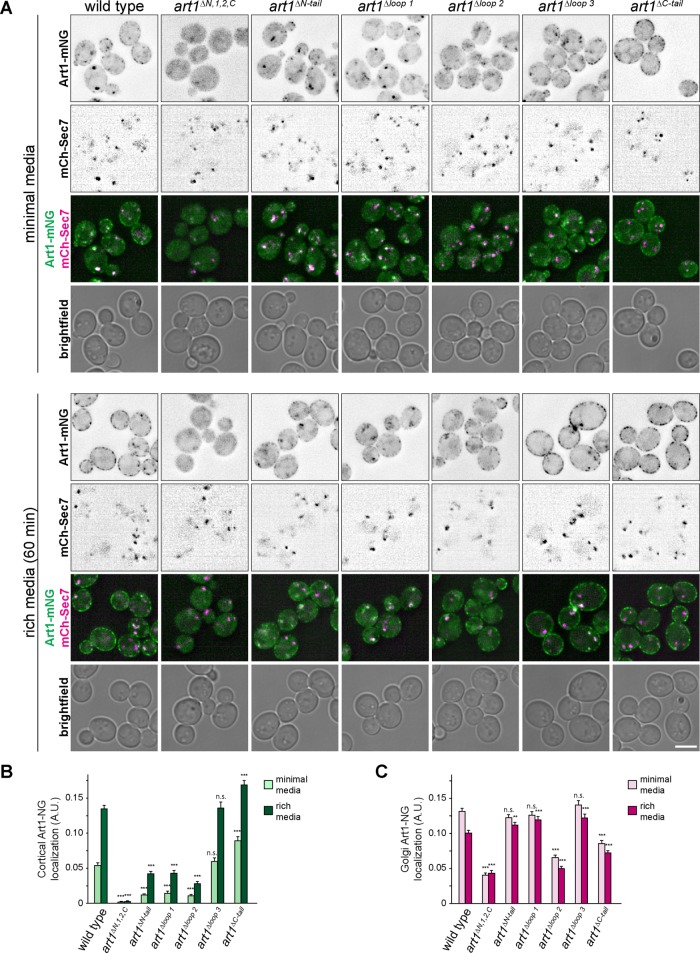
Art1 loops regulate its localization
To investigate the role the remaining loops and tails may be playing in Art1 regulation, we analyzed Art1 localization in these mutants (Figure 3). Art1 is predominantly localized to the Golgi but upon nutrient stimulation translocates to the PM (Lin et al., 2008; MacGurn et al., 2011; Martínez-Márquez and Duncan, 2018; Lee et al., 2019), presumably allowing Art1 to bind to its cargo, resulting in ubiquitylation and endocytosis. One such stimulus for Art1 PM association is shifting cells from minimal media (SCD) to rich media (YPD). Upon shifting to rich media, the intensity of Art1-mNG at the PM increased (Figure 3B and Supplemental Figure S4), with a concurrent decrease in intensity at the Golgi (Figure 3C and Supplemental Figure S4). By measuring the Art1-mNG intensity at both the PM and Golgi, we determined the localization of each Art1 loop mutant in minimal media and after 60 min in rich media (Figure 3).
FIGURE 3:
Localization of Art1 loop and tail mutants. (A) Localization of the indicated Art1-mNG loop and tail mutants in minimal media and after shifting to rich media for 60 min. Scale bar = 2 µm. (B) Quantification of PM localization of the indicated Art1 loop and tail mutants. (C) Quantification of Golgi localization of the indicated Art1 loop and tail mutants. Bars indicate 95% CI, n > 400 cells for each condition. ***, p < 0.001; **, p < 0.01 vs. WT at matched treatment.
The Golgi localization of Art1Δloop3-mNG in minimal media, and its ability to associate with the PM after treatment with rich media, was similar to WT Art1-mNG, demonstrating that its cargo-specific regulation occurs through a mechanism distinct from altered subcellular localization.
Art1ΔC-tail-mNG, even in minimal media, showed more PM localization than WT, and was accompanied by a decrease in Golgi localization (Figure 3). Thus, the Art1 C-tail promotes its Golgi localization, as its absence leads to increased Art1 on the PM.
Art1ΔN,1,2,C-mNG had a weaker association with the Golgi (Figure 3, A and C), resulting in only a diffuse cytosolic signal, despite being expressed to levels similar to WT Art1 (Figure 1F). It also showed completely undetectable PM localization (Figure 3, A and B). This decreased ability to localize to the PM, and thus bind PM cargo, is the likely mechanism explaining the defect in cargo endocytosis (Figure 2).
The loop model for other ARTs
Sequence analysis of the other ART paralogue groups (Art2/Art8, Art3/Art6, Art4/Art5/Art7) suggested that these ART family members also contain long, likely disordered insertions in their core arrestin domains. Art2 and Art8 were most striking and complex (Supplemental Figure S2A). Neither protein contains a clear signature for an arrestin-N domain, and the arrestin-C domain is conspicuously elongated to more than twice the length of the arrestin-C pfam model (Becuwe et al., 2012a). Our loop model, informed by sequence alignment and disorder predictions, resolves these peculiarities.
The Art4/5/7 and Art3/6 paralogous groups share a relatively simpler loop architecture, with evidence of one disordered loop in both N and C arrestin domains. Matches to the pfam arrestin-N domain in particular are often only partial across this subfamily, and ARTs 3, 4, 5, and 6 all contain a N-terminal, nonsignificant fragmentary match to the arrestin-N domain in addition to the main match in the center of the protein (Supplemental Figure S2). Between these core arrestin domain regions, many homologues are missing some or all of the predicted disordered regions (Supplemental Figure S2), consistent with the loop model.
Using Hxt6-GFP endocytosis to probe Art4 function (Nikko and Pelham, 2009; Ho et al., 2017), we tested the effects of deleting loop 1 and loop 2 in Art4. Art4Δloop 1 promotes Hxt6 endocytosis as well as WT Art4, while Art4Δloop 2 promotes modestly impaired, but still detectable Hxt6 endocytosis (Supplemental Figure S2D). As with the loops in Art1, extending either Art4 loop deletion N- or C- terminally by approximately 15 amino acids completely abrogated Hxt6 endocytosis, despite still being expressed (Supplemental Figure S2E). Whether the moderate defect in Art4Δloop 2 indicates that this loop is involved in substrate selection, localization, or some other process requires further analysis.
Art1 regulation by Pho85
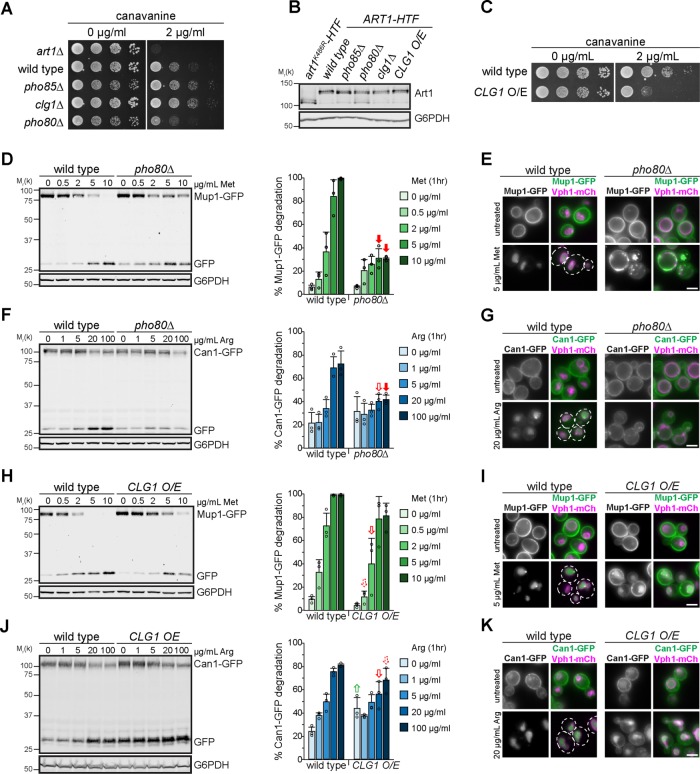
The effects of the large deletions of Art1 loops and tails on Art1 localization are consistent with the loops and tails participating in Art1 regulation. Previous work has demonstrated that Art1 is regulated by Npr1-mediated phosphorylation (MacGurn et al., 2011), on residues in loop 1 (Figure 1E). However, not all of the identified residues were dependent on Npr1, suggesting other kinases may regulate Art1 function. Thus, we tested various kinase knockout mutants for growth on canavanine and found that pho85Δ exhibited canavanine resistance (Figure 4A). Pho85, the yeast homologue of Cdk5 (Carroll and O’Shea, 2002), is a cyclin-dependent kinase, and 10 cyclins have been identified to direct Pho85 to its substrates (Measday et al., 1997; Moffat et al., 2000). Of the individual cyclin mutants, clg1Δ was canavanine resistant, and pho80Δ was canavanine hypersensitive (Figure 4A), whereas the remaining cyclin mutants grew like WT on canavanine (Supplemental Figure S5A), suggesting that Clg1 inhibits Art1 and Pho80 activates it. When the cyclin mutants were tested for growth on thialysine, clg1Δ and pho80Δ recapitulated their canavanine phenotypes, with clg1Δ being resistant and pho80Δ being hypersensitive (Supplemental Figure S5B). Importantly, Art1 expression and ubiquitylation was unaffected when PHO85, PHO80, or CLG1 were deleted (Figure 4B). Additionally, CLG1 overexpression caused cells to become canavanine hypersensitive (Figure 4C), consistent with Clg1 inactivating Art1.
FIGURE 4:
Pho85, and the cyclins Pho80 and Clg1, regulate Art1 activity. (A) Serial dilutions of the indicated null mutants spotted on synthetic media containing canavanine. (B) Immunoblot analysis of Art1-HTF expression in the indicated strains, detected with an α-FLAG antibody. (C) Serial dilutions of WT or cells overexpressing CLG1 spotted on synthetic media containing canavanine. (D) Immunoblot analysis of Mup1-GFP endocytosis induced with the indicated concentration of Met for 60 min. Band intensities were quantified and expressed as the mean % Mup1-GFP degradation. Individual experiments are represented as open circles. Error bars indicate 95% CI. n = 3. Filled arrow, p < 0.001; outlined arrow, p < 0.01; dashed arrow, p < 0.05 vs. WT at matched treatment. (E) Fluorescence microscopy of WT or pho80Δ cells expressing Mup1-GFP, with and without inducing endocytosis by treating with 5 µg/ml Met for 60 min. (F) As in D, except analyzing Can1-GFP endocytosis induced with the indicated concentration of Arg for 60 min. n = 3. (G) As in E, except analyzing Can1-GFP localization after inducing endocytosis with 20 µg/ml Arg for 60 min. (H) As in D, except comparing WT yeast to cells overexpressing CLG1. n = 3. (I) As in E, except comparing WT yeast to cells overexpressing CLG1. (J) As in F, except comparing WT yeast to cells overexpressing CLG1. n = 3. (K) As in G, except comparing WT yeast to cells overexpressing CLG1. Scale bar = 2 µm.
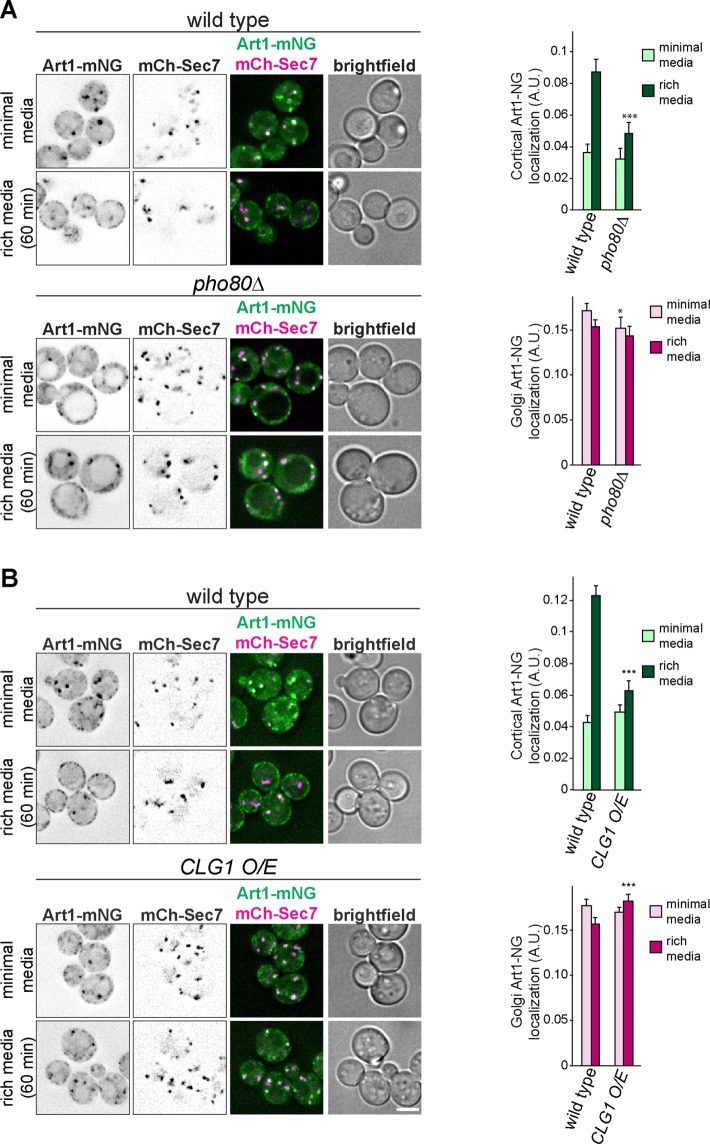
In the absence of PHO80, Art1-mediated endocytosis is impaired. While Mup1-GFP was almost completely degraded after 60 min of treatment with 5 or 10 µg/ml Met in WT cells, in pho80Δ only ∼40% of Mup1-GFP was degraded, even at higher Met concentrations (Figure 4D). Consistently, Mup1-GFP was completely sorted to the vacuole in WT cells after Met treatment, whereas in pho80Δ a portion of Mup1-GFP either was found on internal puncta, presumed to be endosomes, or was retained on the PM (Figure 4E). Importantly, this demonstrates that the defect in Mup1-GFP degradation was due to an endocytosis and vacuole sorting defect rather than a defect in vacuolar proteolysis. Analysis of Can1-GFP endocytosis revealed the same trend. Even at higher concentrations of Arg, Can1-GFP degradation was impaired in pho80Δ compared with WT cells (Figure 4F), and Can1-GFP was mostly retained on the PM in pho80Δ cells after treatment with Arg (Figure 4G), although some signal was observed on endosomes and (to a much lesser extent) in the vacuole lumen. Taken together with the hypersensitivity of pho80Δ cells to canavanine (Figure 4A), these data indicate that Pho80 positively regulates Art1, resulting in an Art1-mediated endocytosis defect in pho80Δ.
CLG1 overexpression resulted in a Mup1-GFP degradation defect (Figure 4H) and a delay in vacuolar sorting (Figure 4I). Similar effects were observed for Can1-GFP degradation and endocytosis (Figure 4, J and K). A striking increase in cargo endocytosis was not observed in clg1Δ (Supplemental Figure S5, C–F), despite clg1Δ cells exhibiting canavanine resistance. This may reflect the inherent differences in the sensitivity of these assays. Interestingly, there was no increase in constitutive, nonsubstrate induced endocytosis, as might be expected. However, that CLG1 overexpression caused an endocytosis defect is consistent with Clg1 activity inactivating Art1.
Pho80 and Clg1 affect Art1 localization
Because Pho80 and Clg1 activate and inactivate Art1, respectively, we next sought to understand the mechanism(s) by which they modulated Art1 function. Thus, we examined Art1 localization in the mutants. In pho80Δ cells grown in minimal media, Art1-mNG remained mostly localized at the Golgi, similar to WT. However, after an acute treatment with rich media, Art1-mNG was unable to efficiently translocate to the PM in the same way as in WT cells (Figure 5A), suggesting that the Art1-mediated endocytosis defect observed in pho80Δ cells is the result of Art1 failing to associate with the PM.
FIGURE 5:
Pho80 and Clg1 affect Art1 localization. (A) Localization of Art1-mNG in WT and pho80Δ yeast in minimal media and after shifting to rich media for 60 min. Right, quantification of Art1-mNG PM and Golgi localization; error bars are 95% CI, n > 250 cells for each condition. *, p < 0.05; ***, p < 0.001 vs. WT at matched treatment. (B) As in A, except comparing Art1-mNG in WT and cells overexpressing CLG1. Scale bar = 2 µm.
CLG1 overexpression caused a strong defect in Art1-mNG PM localization in rich media (Figure 5B), while in clg1Δ Art1-mNG mimicked its localization in WT cells (Supplemental Figure S5G), consistent with the cargo endocytosis phenotypes (Supplemental Figure S5, C–F). Therefore, Clg1 decreases Art1 activity by preventing or decreasing Art1 PM localization. Thus, both Clg1 and Pho80 regulate Art1 in a manner similar to some of the loops and tails mutants: by modulating its PM association.
Phosphorylation on loops and tails regulate Art1
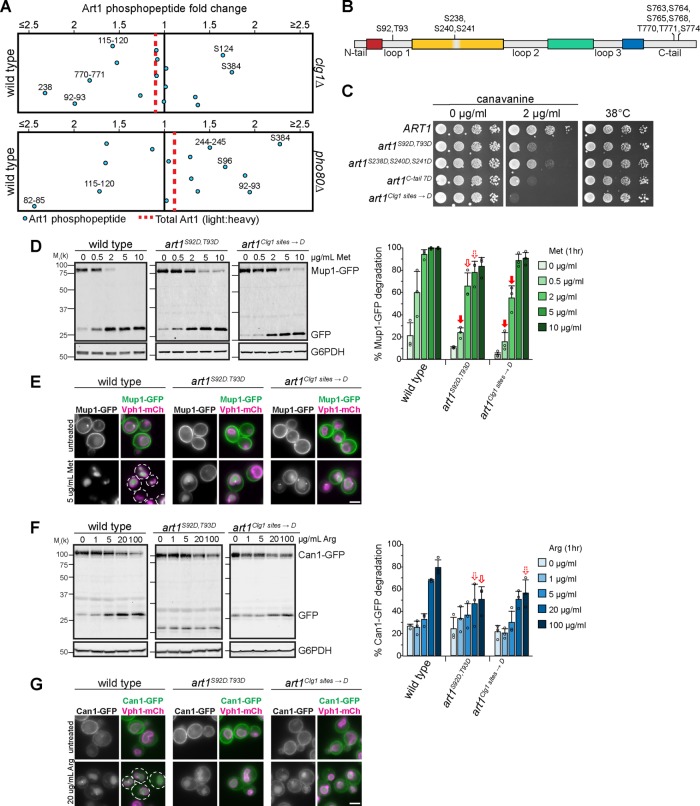
Considering that loop 1 in Art1 is a known target for Npr1-mediated phosphorylation (MacGurn et al., 2011), and that Clg1 and Pho80 can regulate Art1 localization reminiscent of the loop and tail mutants, we hypothesized that the loops and/or tails are phosphorylated in a Clg1- and Pho80-dependent manner. Thus, we identified the Clg1- and Pho80-mediated phosphorylated residues on Art1 using SILAC mass spectrometry (Figure 6A).
FIGURE 6:
Phosphorylation on loops and tails regulate Art1. (A) QMAPS depicting the fold change in phosphopeptide abundance of Art1 in (top) WT vs. clg1Δ or (bottom) WT vs. pho80Δ. (B) Schematic of Art1 showing the location of the phosphorylated residues that when mutated affect Art1 function. (C) Serial dilutions of cells expressing the indicated Art1 mutants spotted on synthetic media containing canavanine. (D) Immunoblot analysis of Mup1-GFP endocytosis induced with the indicated concentration of Met for 60 min. Band intensities were quantified and expressed as the mean % full-length Mup1-GFP remaining. Individual experiments are represented as open circles. Error bars indicate 95% CI. n = 3. Filled arrow, p < 0.001; outlined arrow, p < 0.01; dashed arrow, p < 0.05 vs. WT at matched treatment. (E) Fluorescence microscopy of Mup1-GFP, with and without inducing endocytosis by treating with 5 µg/ml Met for 60 min. (F) As in D, except analyzing Can1-GFP endocytosis induced with the indicated concentration of Arg for 60 min. n = 3. (G) As in E, except analyzing Can1-GFP localization after inducing endocytosis with 20 µg/ml Arg for 60 min. Scale bar = 2 µm.
In clg1Δ, Art1 phosphorylation was reduced at S92/T93, S238, and T770/T771 (Figure 6A), suggesting that Clg1 was responsible for these phosphorylation events. Residues S92/T93 are contained within loop 1. S238 falls within the “mini-loop,” and T770/T771 are in the C-terminal tail of Art1 (Figure 6B).
Mutating S92 and T93 to the phosphorylation mimetic Asp resulted in canavanine hypersensitivity (Figure 6C), consistent with the CLG1 overexpression phenotype. Similarly, mutating S238 (in combination with the nearby S240 and S241) also caused canavanine hypersensitivity, although less so than the S92D/T93D mutant. Mutating seven Ser residues on the C-tail (all within 15 amino acids of T770/T771) to Asp caused canavanine hypersensitivity (Figure 6C). Mutating T770/T771 alone did not affect growth on canavanine (Supplemental Figure S6A). Thus, similar to Npr1-mediated phosphorylation of loop 1 on Art1 (MacGurn et al., 2011) and the N-terminus of Art8 (Hovsepian et al., 2017), on the Art1 C-tail it is necessary to mutate phosphorylated residues identified by mass spectrometry as well as adjacent residues before a phenotype is revealed, presumably due to inexact phosphorylation of Art1 by its kinase. Notably, mutations to the phosphorylation-deficient Ala did not result in canavanine resistance (Supplemental Figure S6B).
Hence, three distinct and distant regions of Art1 are phosphorylated in a Clg1-dependent manner: loop 1, the “mini-loop,” and the C-tail (Figure 6B). While these data cannot determine whether all three of these regions are regulated simultaneously or whether they represent individually regulated subpopulations of Art1, combining the phosphorylation mimetic mutants of all three regions (termed “art1Clg1 sites→D”) resulted in an even greater canavanine sensitivity than the individual mutants (Figure 6C), demonstrating additive effects of phosphorylation at these regions of Art1. Notably, the steady-state expression level of each mutant was similar to WT, and all mutants were ubiquitylated (Supplemental Figure S6C). Importantly, while canavanine sensitivity is affected by these phosphorylation site mutants, the ability of Art1 to facilitate growth at an elevated temperature remains intact, suggesting that although Clg1-mediated phosphorylation can regulate substrate-mediated endocytosis of Can1, it does not block Art1-mediated PM quality control.
Analysis of Art1 phosphorylation in pho80Δ was consistent with Pho80 activating a downstream phosphatase or inactivating a downstream kinase. In pho80Δ, only phosphorylation at residues 82-STTS-85 was significantly decreased (Figure 6A). These sites have previously been shown to be phosphorylated by Npr1. Perplexingly, dephosphorylation at these sites results in canavanine resistance and Art1 activation (MacGurn et al., 2011), but pho80Δ cells are canavanine hypersensitive (Figure 4A). Intriguingly, multiple residues become hyperphosphorylated in pho80Δ, including S92/T93. The phosphorylation mimetic Asp mutants at S92/T93 resulted in canavanine hypersensitivity (Figure 6C), which is consistent with the pho80Δ phenotype (Figure 4A) and suggests that phosphorylation at S92/T93 can inactivate Art1 even when the Npr1 sites are dephosphorylated. Hence, Pho80 regulates Art1, but it does so indirectly, likely by activating another kinase or by inactivating a phosphatase that directly interacts with Art1. S384 was also hyperphosphorylated in pho80Δ (Figure 6A). However, neither mutating S384 to Ala or to Asp resulted in any change in canavanine sensitivity. Combining S384 mutants with S92/T93 mutants perfectly mimicked the S92/T93 mutant phenotypes (Supplemental Figure S6D), demonstrating that, while S384 is phosphorylated in a Pho80-dependent manner, it has no effect on Can1 endocytosis.
Consistent with the canavanine growth phenotypes, Mup1-GFP degradation was impaired in cells expressing art1S92D,T93D (to mimic pho80Δ) and art1Clg1 sites→D (to mimic CLG1 overexpression), and a portion of Mup1-GFP remained on the PM or, to a lesser extent, at endosomes (Figure 6, D and E). Also consistent with the growth on canavanine, Mup1-GFP degradation was like WT in cells expressing art1S92A,T93A and art1Clg1 sites→A, and no constitutive endocytosis was observed in the absence of Met treatment (Supplemental Figure S6, E and F). Further, cells expressing art1S238D,S240D,S241D or art1C-tail 7D showed more modest defects in Mup1-GFP degradation and vacuole sorting. Similar effects were observed with Can1-GFP endocytosis and degradation when art1 phosphorylation mutants were expressed (Figure 6, F and G, and Supplemental Figure S6, G and H). Thus, Clg1- and Pho80-dependent phosphorylation of Art1, although it is indirect, regulates cargo endocytosis.
Clg1 and Pho80 regulate Art1 via its subcellular localization (Figure 5), thus we tested whether Art1-mNG localization was altered when the Clg1- and Pho80-dependent phosphorylation sites were mutated. Cells expressing art1S92D,T93D and art1Clg1 sites→D demonstrated less PM localization in minimal media, and both had defects in the ability to associate with the PM after shifting to rich media (Figure 7). While art1Clg1 sites→D was able to increase its PM localization after shifting to rich media, the maximal level of PM localization was approximately the same as WT cells in minimal media. Thus, the PM association of Art1 phosphorylation mimetic mutants is defective. Consistent with all of the Ala mutants at Art1 phosphorylation sites phenocopying WT, no Ala mutant resulted in constitutive PM localization, and all were able to associate with the PM after acute shift to rich media (Supplemental Figure S6I). Taken together, these data suggest a mechanism by which Clg1- and Pho80-dependent phosphorylation regulates Art1 activity via its ability to associate with the PM.
FIGURE 7:
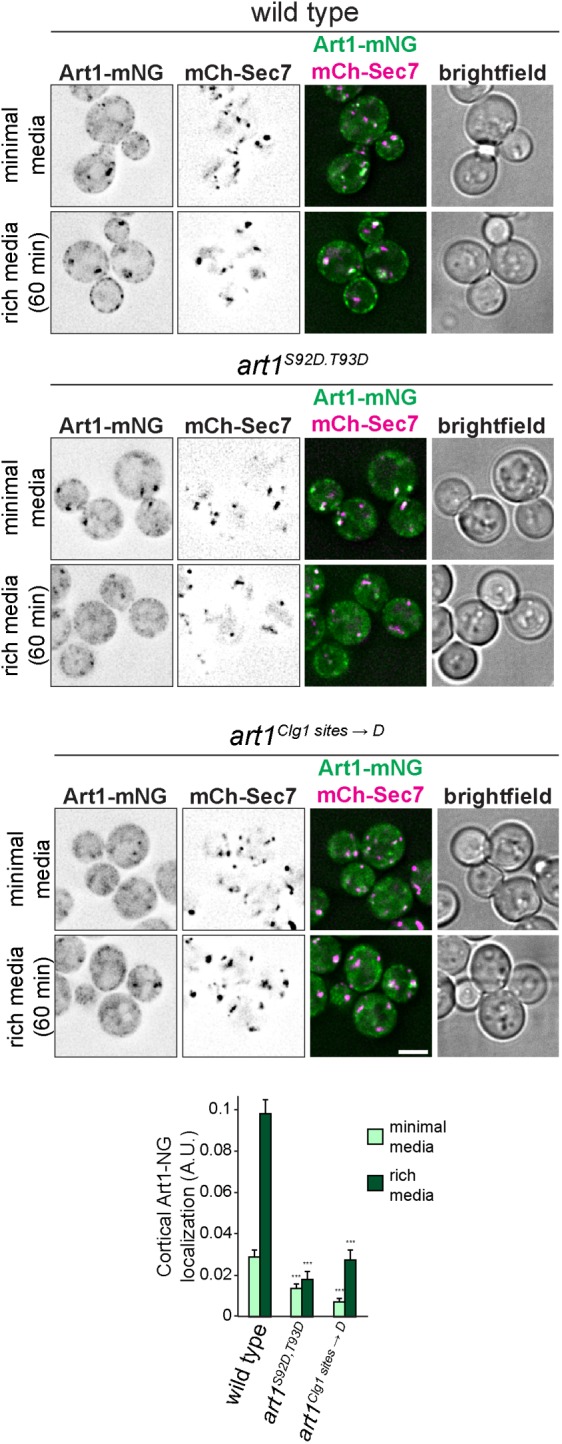
Phosphorylation affects Art1 localization. Localization of WT Art1-mNG, Art1S92D,T93D-mNG, or art1Clg1 sites → D-mNG in minimal media and after shifting to rich media for 60 min. Scale bar = 2 µm. Bottom, quantification of Art1 PM localization; error bars are 95% CI, n > 250 cells each condition; ***, p < 0.001 vs. WT at matched treatment.
DISCUSSION
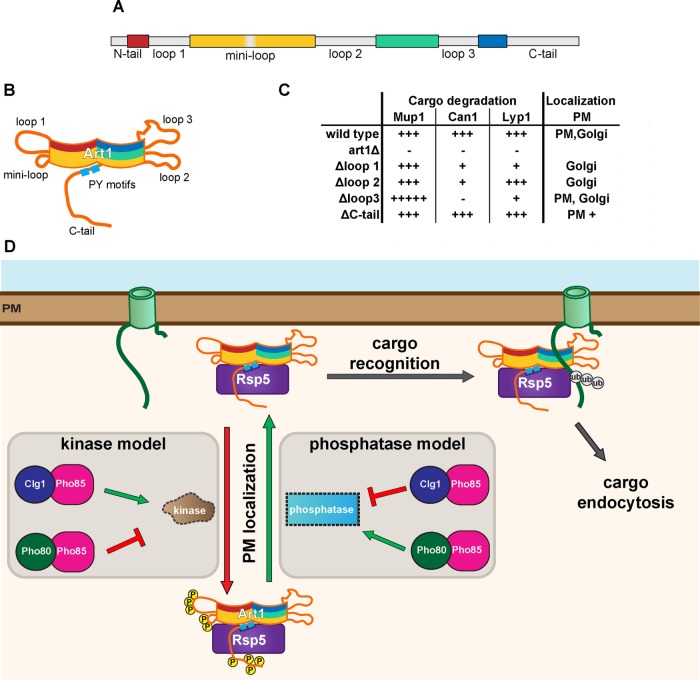
This work provides new insight into the mechanisms regulating Art1-mediated endocytosis. Specifically, we show that 1) Art1 forms an arrestin fold using the entire length of its sequence with interspersed and poorly conserved “loop” and “tail” insertions, rather than just its N-terminus; 2) the Art1 loops and tails regulate Art1 function via at least two separate mechanisms, by specifying substrate recognition and by controlling Art1 PM translocation; and 3) phosphorylation on the loop and tail regions regulates Art1 localization.
A new model for the Art1 arrestin fold
Our sequence analysis of Art1 fungal homologues suggests that Art1 contains multiple large loops, and N- and C-terminal tails, between conserved, arrestin fold–forming regions (Figure 1E). This is a striking deviation from previous structural predictions where only the N-terminal half of Art1 formed an arrestin domain (Lin et al., 2008; Becuwe et al., 2012a; Aubry and Klein, 2013). However, because the loops were not previously appreciated, these structural models for Art1 showed either an incomplete arrestin fold or required an analysis of only part of the Art1 sequence. However, our experimental data demonstrate that deleting the loops and tails retained the ability of Art1 to support growth at an elevated temperature (Figure 1G), and retained Rsp5 binding (Supplemental Figure S1D), providing evidence that the loop and tail regions are separate from the arrestin fold.
Structural modeling predicts that the loops are positioned in turns between β-strands or coils (Figure 1E and Supplemental Figure S1C), explaining how the loops can be removed without completely abolishing Art1 function. Importantly, most mutants where the loop deletions were extended into the conserved sequences of Art1 completely abolished Art1 function (Figure 1G), supporting the new structural prediction. This new model places Art1’s predicted substrate binding motif (Figure 1; Guiney et al., 2016) on the arrestin domain, rather than on a distal C-terminal region, consistent with other examples of arrestin domains binding to substrates (Aubry et al., 2009).
Analysis of other ART family members indicated that they, like Art1, also contain nonconserved loops and tails. Regions predicted to be disordered generally align well with sequences that are absent in the smaller fungal paralogues. Strikingly, these insertions occur at different positions relative to the conserved arrestin fold for each ART paralogue group (Art2/Art8, Art3/Art6, and Art4/Art5/Art7; Supplemental Figure S2). This suggests that large insertions can be tolerated at multiple places within the core arrestin domain without disrupting folding. We directly tested this hypothesis for Art4, and found that of its two predicted loops, the first loop could be deleted without compromising endocytosis of its substrate Hxt6, and that deleting the second loop partially impaired Hxt6 endocytosis (Supplemental Figure S2D). Given the similarity between Art4 and its paralogue Art7 (as well as with Art5, and more distantly, Art3 and Ar6), this helps corroborate the bioinformatic predictions for the broader ART family.
Considering the regulatory role of the tails and loops in Art1, this highlights potential regulatory regions in other ART family members as well. Interestingly, Art8 is regulated by PKA through phosphorylation on its N-terminus (Hovsepian et al., 2017), which is poorly conserved among fungi (Supplemental Figure S2A); and Art3 is regulated by calcineurin (O’Donnell et al., 2013) by dephosphorylating residues that map to other poorly conserved regions (Supplemental Figure S2B). Thus, it is possible that phosphorylation of poorly conserved loop regions interspersed between conserved arrestin fold regions is a recurring mode of regulation in the ART family. Notably, phosphorylation affects the ubiquitylation of Art4 and Art8 (Becuwe et al., 2012b; Hovsepian et al., 2017), but the phosphorylation events described here did not change the ubiquitylation status of Art1. Therefore, while phosphorylation of loop regions modulates ART function, the specific mechanisms through which this regulation is achieved varies.
Art1 loops and tails as regulatory elements
Art1 loops and tails contribute to its function through two distinct mechanisms: by dictating substrate specificity, and by regulating its localization. Loop 3 is required for Can1 degradation, but not for Mup1 (Figure 2). In fact, Mup1-GFP degradation occurs more completely at lower Met concentrations when loop 3 is deleted than with WT Art1. While previous models suggested that cargo specificity for ARTs occurs through changes in the permease (Guiney et al., 2016; Busto et al., 2018; Gournas et al., 2018), this is the first demonstration of adaptor-driven specificity. Considering the proximity of loop 3 to the basic patch on Art1 required for recognizing the N-terminal tail of its cargo (Figure 1E), it is possible that loop 3 is involved in cargo binding for select permeases. However, a more detailed biochemical and structural analysis, as well as a more complete inventory of Art1 cargoes, will be required to define the mechanism through which loop 3 contributes to substrate specificity.
The remaining loop and tail mutants had similar effects on both Mup1 and Can1, suggesting that they are involved in more general regulation of Art1. Although individually deleting the N-terminal tail, loop 1, and loop 2 had no detectable effect on cargo endocytosis (Figure 2), they did exhibit a reduced ability to associate with the PM (Figure 3), and hypersensitivity to canavanine (Figure 2A) and thialysine (Supplemental Figure S3D). Interestingly, combining the deletions in art1ΔN,1,2,C had a dramatic effect on both Can1 and Mup1 endocytosis (Figure 2), as well as Art1 PM association (Figure 3), suggesting that each loop and tail makes a minor contribution to Art1 regulation that is exacerbated in combination. Notably, art1ΔC alone increased Art1 activity. Therefore, the reduced activity of art1ΔN,1,2,C is likely through the combined effects of the N-terminal tail, loop 1, and loop 2 deletions.
Phosphoinhibition of Art1
Our data suggest that the previously identified Npr1-targeted residues on Art1 fall within loop 1 (MacGurn et al., 2011). Further, many of the Npr1-independent phosphorylation sites identified occurred on loops or the C-tail. Thus, we tested whether other kinases regulated Art1 through the phosphorylation of its loops and tails. Consequently, we identified Pho85 as a kinase that affects Art1 function, and further identified Pho80 and Clg1 as the two cyclins that mediate this regulation. However, regulation by Pho80-Pho85 is indirect, because in pho80Δ phosphorylation in loop 1 of Art1 was increased (Figure 6A). Additionally, Pho85 is a proline-directed kinase, but changes in phosphorylation at proline-directed sites were not detected in either clg1Δ or pho80Δ. While there is limited evidence that Pho85 can phosphorylate non–proline-directed sites (Zappacosta et al., 2006), it is likely that Clg1-Pho85 is acting in a signaling pathway upstream of Art1, rather than directly phosphorylating it. In this model, Clg1-Pho85 phosphorylates another (yet unidentified) downstream kinase or phosphatase that directly acts upon Art1 (Figure 8). Notably, in both clg1Δ and pho80Δ, phosphorylation was also modestly decreased at 82-STTS-85, residues previously identified as Npr1 targets (MacGurn et al., 2011). As previously hypothesized, there may be cross-talk between Npr1 and other kinases; in this case, Clg1. Nonetheless, the data here clearly show that Art1 activity is regulated by phosphorylation mediated by Pho80 and Clg1.
FIGURE 8:
Model of Art1 regulation. (A) Art1 schematic showing the conserved regions predicted to form the arrestin fold (colored), and the loop and tail regions (gray). (B) Cartoon depicting the predicted organization of the conserved arrestin fold and the localization of the loops. (C) Summary of the effects of Art1 loop region deletions. (D) New modes of Art1 regulation. In addition to the previously identified regulation through Npr1 or Ppz1/2, Art1 phosphorylation/dephosphorylation reactions can be mediated by Clg1-Pho85 and Pho80-Pho85 to regulate its activity. Phosphorylation of Art1 loops and the C-tail regulate its function by affecting its PM localization. When phosphorylated on loop 1, the “miniloop,” and the C-tail, Art1 remains in the cytosol or associated with the Golgi, rendering it inactive. Upon dephosphorylation, Art1 can associate with the PM, where it can recognize its substrate cargoes resulting in ubiquitylation and endocytosis.
The function of Clg1 is poorly defined. Despite belonging to the Pcl1/2 subfamily (Matsumoto and Wickner, 1993), which usually is involved in cell cycle control, mutating CLG1 alone results in no detectable defect in cell cycle progression, consistent with its constitutive expression throughout the cell cycle (Measday et al., 1997). More recently, Clg1 was shown to phosphorylate and degrade Sic1 and therefore promote autophagy (Yang et al., 2010). Notably, amino acid homeostasis is altered in clg1Δ (Mülleder et al., 2016). Thus, similar to the TORC1-Npr1 pathway that inactivates Art1 during starvation conditions (MacGurn et al., 2011), Clg1 may regulate Art1 in response to changes in nutrient levels. In this model, low nutrient/amino acid levels would cause Clg1 activation, which would in turn inactivate Art1, increasing the abundance of permeases on the PM. Identification of the input(s) that regulate Clg1 as well as a complete inventory of its downstream targets will be critical to understanding this signaling pathway.
In comparison, Pho80 function is much better defined. Pho80 is a critical component of the phosphate sensing pathway in yeast (Kaffman et al., 1994), is required for survival under various stress conditions (Huang et al., 2002), and is involved in Rim15-mediated nutrient sensing (Wanke et al., 2005). Although Art1 has not yet been implicated directly in either the phosphate response or Rim15 pathway, both are important metabolic pathways and modulation of PM permeases to control nutrient levels may be required for either cellular homeostasis or survival.
The increased phosphorylation of loop 1 observed in pho80Δ occurs at residues distinct from those identified as phosphorylated by Npr1 (Figure 6A; MacGurn et al., 2011). However, the phenotypes are similar. Phosphorylation mimetic mutations at S92/T93 result in Art1 inactivation and a failure to associate with the PM (Figures 6 and 7). Although the cellular inputs are different, the mechanism through which they attenuate Art1 function may be the same. In the structural model of Art1, loop 1 would extend distally from the N-terminal arrestin domain (Figure 1F and Supplemental Figure S1B). Thus, phosphorylation at any number of residues on this loop seems to prevent PM localization. This suggests that this region of Art1 interacts with either a PM resident protein or the lipids that constitute the PM, and that phosphorylation disrupts this interaction. Interestingly, the nonconserved “mini-loop,” which contains residues 238–241, is predicted to occur at a turn between two β-strands that is spatially adjacent to loop 1. Therefore, this region may also be involved in Art1 binding to the PM. While the structural model did not include the C-terminal tail, considering the effect on cargo endocytosis in the Asp mutants, and the effect on Art1 PM localization in the absence of the tail, the C-terminal tail likely regulates Art1 similarly, although it is currently unclear whether this occurs through a similar or distinct membrane binding region. Nonetheless, one aspect of Art1 regulation is its ability to properly localize to the membrane containing its substrates, which occurs via phosphorylation and dephosphorylation.
Recently, the Ppz phosphatases have been identified as regulators of Art1 (Lee et al., 2019). Intriguingly, one of the targets of Ppz1/2 is T93. Therefore, the possibility exists that either Pho80 or Clg1 regulate Art1 by acting on the Ppz proteins. Interestingly, T245 and T795 were also identified as Ppz targets that affect Art1 function. While these residues were not identified as Clg1- or Pho80-regulated sites in our study, T245 falls within the “mini-loop” of Art1, and T795 is on the C-tail. Thus, phosphorylation in these regions, regardless of the specific input, all regulates Art1.
This work paints a more complete picture of the molecular complexity underlying Art1 regulation during selective cargo ubiquitylation and endocytosis (Figure 8). For cargo to be sorted to the vacuole, it must first bind substrate, changing its conformation, and allowing Art1-Rsp5–mediated binding and ubiquitylation (Guiney et al., 2016; Gournas et al., 2017, 2018; Busto et al., 2018). Art1 binding to specific cargoes can be modulated by loop 3 in Art1. Further, ubiquitylation of cargo can be enhanced by Art1 PM association, which is controlled via phosphorylation. Multiple different signaling pathways, which include Clg1-Pho85, Pho80-Pho85, TORC1/Npr1, and Ppz1/2, affect Art1 phosphorylation in response to a diverse array of environmental cues. Therefore Art1, and likely the other ART proteins, are able to be finely tuned to remodel the composition of the PM and in turn, cellular homeostasis.
MATERIALS AND METHODS
Yeast strains and growth conditions
All yeast strains used in this study were generated from SEY6210 (Robinson et al., 1988) and are listed in Supplemental Table S1. Genetic knockouts were generated by replacing the entire open reading frame of the gene using PCR-mediated gene replacement (Wach et al., 1994; Longtine et al., 1998; Goldstein and McCusker, 1999), and multiple mutants were generated by crossing, sporulation, and tetrad dissection (Sherman, 2002). Chromosomally tagged strains were generated by PCR-mediated tagging using pFA6a-mCherry-kanMX (Malcova et al., 2016) to tag VPH1, pFA6a-GFP (Longtine et al., 1998) to tag MUP1, and pFA6a-LAP-eGFP-HIS3 (Adell et al., 2017) to tag CAN1. pRS305- and pRS306-based vectors (Sikorski and Hieter, 1989) were integrated after linearization with AflII or StuI, respectively, unless the inserted gene contained additional AflII or StuI sites, in which case the plasmid was linearized by PCR amplification.
Cells were grown at 26°C in synthetic media (0.17% [wt/vol] yeast nitrogen base, 0.5% [wt/vol] ammonium sulfate, 2% [wt/vol] glucose), and supplemented with adenine, uracil, Arg, Lys, Thr, Tyr, His, Leu, and Trp, unless otherwise specified. For acute YPD treatment, cells were grown in synthetic media, washed with dH2O, and then resuspended in YPD (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% [wt/vol] glucose) and incubated for the indicated time. For growth on canavanine, solid synthetic media (containing 2% agar) lacking Arg was supplemented with the indicated concentration of canavanine (Sigma) after autoclaving. Similarly, for growth on thialysine, solid synthetic media lacking Lys was supplemented with the indicated concentration of thialysine (Sigma) after autoclaving. For growth assays, 1:10 serial dilutions were spotted onto the indicated media using a frogger pinning apparatus and incubated at 26°C unless otherwise specified.
Plasmids
All plasmids used in this study are listed in Supplemental Table S2 and were generated using standard procedures and conditions. All restriction endonucleases were purchased from New England Biolabs. Plasmids containing ART1 included 536 base pairs of the 5′ UTR as the promoter and 502 base pairs of the 3′ UTR as the terminator. The HTF tag (MacGurn et al., 2011), consisting of a 6× His tag, a TEV cleavage site, and a 3× FLAG tag, contains a RIPGLINRGS linker between the C-terminus of Art1 and the 6× His tag. The mNeonGreen (mNG) tag (Shaner et al., 2013) adds a RIPGLINIF linker between Art1 and mNG. Both tags were added to ART1-containing plasmids using overlap extension (Ho et al., 1989). All ART1 point mutants were generated using overlap extension. Plasmids containing ART4 included 548 base pairs of the 5′ UTR as the promoter and the ADH1 terminator after the HTF tag. pEG166 included 596 base pairs of the MUP1 5′ UTR as the promoter and 279 base pairs of the 3′ UTR as the terminator, and introduced the HTF tag as an EagI-SacII fragment, containing a RPRIPGLINRGS linker between the C-terminus of Mup1 and the 6× His tag. pMB0268 was generated by inserting the CLG1 coding sequence immediately 3′ of the GPD (TDH3) promoter. pMB0408 was generated by ligating PGPDCLG1 into the MCS of pRS307, which was constructed by removing the CEN sequence from pRS317 via PCR. pMB0239 was generated by ligating the SEC7 coding sequence with 527 base pairs of the 5′ UTR and 303 base pairs of the 3′ UTR into pRS305. mCherry was inserted on the 5′ end of SEC7 by overlap extension and contains an SSGSGSS linker between mCherry and Sec7. All plasmids were verified by sequencing.
Endocytosis assays
For Mup1-GFP endocytosis, cells were grown at 30°C in synthetic media lacking Met, and were diluted multiple times to ensure that the culture never exceeded OD600 = 1 for more than 24 h. The indicated concentration of Met was added to initiate endocytosis, and cells were collected after 60 min for further analysis. Can1-GFP endocytosis was performed similarly except the synthetic media lacked Arg and endocytosis was initiated with the indicated concentration of Arg. The percent degradation was expressed as the signal for the full-length cargo divided by the total signal from the full-length cargo and free GFP bands, subtracted from 100%. For Hxt6-GFP endocytosis, the synthetic media contained 0.05% glucose and endocytosis was initiated with 5% glucose.
Protein extraction and immunoblotting
5 OD600 equivalents of yeast were incubated in 10% (vol/vol) trichloroacetic acid on ice overnight. Cells were pelleted, washed with cold acetone, and dried. Cells were then resuspended in 50 µl urea cracking buffer (50 mM Tris-HCl, pH 7.5, 8 M urea, 2% [wt/vol] SDS, 1 mM EDTA), lysed with glass beads by vortexing for 5 min at room temperature, then incubated at 42°C for 5 min. Sample buffer (50 µl 2×; 150 mM Tris-HCl, pH 6.8, 7 M urea, 10% [wt/vol] SDS, 24% [vol/vol] glycerol, 10% [vol/vol] β-mercaptoethanol, bromophenol blue) was added and the samples were vortexed again for 5 min at room temperature, then incubated at 42°C for 5 min. Proteins were separated by SDS–PAGE and transferred onto 0.45-µm nitrocellulose membranes (GE Healthcare) in transfer buffer (25 mM Tris, 192 mM glycine, 10% methanol). When analyzing Mup1-GFP or Can1-GFP, transfer buffer was supplemented with 0.06% SDS.
Membranes were blocked in blocking buffer (5% [wt/vol] fat-free milk in Tris-buffered saline with Tween [TBST; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20]) for 60 min. Primary antibodies were diluted in blocking buffer and incubated for 4 h at room temperature or overnight at 4°C. Membranes were washed three times for 10 min in TBST. Secondary antibodies were diluted in blocking buffer and incubated for 60 min at room temperature, and then membranes were washed three times for 10 min in TBST. Membranes were scanned using an Odyssey CLx imaging system (Li-Cor Biosciences) and analyzed using ImageStudio Lite version 5.2 (Li-Cor Biosciences) or ImageJ (National Institutes of Health).
The primary antibodies used were mouse anti-FLAG M2 (Sigma-Aldrich; F1804) 1:5000, rabbit anti-glucose-6-phosphate dehydrogenase (G6PDH; Sigma-Aldrich; A9521) 1:50,000, mouse anti-GFP (Roche; 11814460001) 1:5000 for detecting Can1-GFP, rabbit anti-GFP (TP401; Torry Pines Biolabs) 1:5000 for detecting Can1-GFP in Supplemental Figure S3C, mouse anti-GFP (Santa Cruz Biotechnology; sc-9996) 1:500 for detecting Mup1-GFP, mouse anti-Myc (9E10; Santa Cruz Biotechnology; sc-40) 1:3000, and rabbit anti-Rsp5 (Emr Lab) 1:10,000. The secondary antibodies used were goat anti-mouse 800 CW (Li-Cor Biosciences; 926-32210) 1:10,000 and goat anti-rabbit 680 LT (Li-Cor Biosciences; 926-68021).
Fluorescence microscopy
Microscopy was performed using a DeltaVision RT system (Applied Precision, Issaquah, WA), equipped with a DV Elite CMOS camera, a 100× objective, and a DV Light SSI 7 Color illumination system with Live Cell Speed Option (FITC for GFP and TRITC for mCherry/RFP). Image acquisition and deconvolution were performed using the provided DeltaVision software (softWoRx 6.5.2; Applied Precision, Issaquah, WA).
Quantification of Art1-mNG was conducted using a custom Python pipeline. Source code is available at github.com/elguiney/YeastMorphologyPypeline, commit 4fd7685. Briefly, edge detection using a Laplace of Gaussian kernel was used to define cell boundaries from brightfield images. For each cell, Golgi masks were defined based on a maximum intensity z-projection of the deconvolved mCherry-Sec7 signal, thresholded with Otsu’s method. Because Art1-mNG fluorescence at the Golgi was often directly adjacent to the mCherry-Sec7 marker, the Golgi mask regions were expanded (by ensuring that the local radius of the mask was always ≥7 pixels). A cell-cortex mask was defined as the region within 10 pixels of the cell border on the inside, and 5 pixels on the outside, that did not overlap with either a Golgi mask or an adjacent cell. Analysis was run in batches on all images from three independent biological replicates. After cell identification and mask generation, cells were cropped and randomized, and displayed for blinded visual inspection; cells where the cell boundary or the Golgi or cell-cortex masks were incorrectly identified were rejected from further analysis (typically fewer than 10% of cells were rejected). Finally, for each cell Art1-mNG enrichment in the Golgi and cell-cortex mask regions was calculated by subtracting the cytoplasmic background Art1-mNG fluorescence (as defined by regions in a cell not covered by a Golgi or cell-cortex mask), and then normalized to total Art1-mNG fluorescence in the cell. The number of cells analyzed is noted in each figure legend.
Immunoprecipitation
Anti-FLAG immunoprecipitation of Art1-HTF was performed by collecting 25 OD600 equivalents of yeast in mid–log (OD600 = 0.5–0.8) growth, followed by a wash in 20 ml of 10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 10 mM NEM, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM β-glycerophosphate, and 5 mM NaF. Cell pellets were collected and stored at −80°C for at least 12 h. Cell pellets were then thawed on ice and resuspended in 500 µl lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, 10 mM NEM, 1× cOmplete protease inhibitor cocktail (Roche), 10 mM β-glycerophosphate, 5 mM NaF). Cells were lysed by vortexing with 0.5 mm YZB zirconia beads (Yasui Kikai) for 30 s three times, with 30 s incubations on ice in between. Lysis buffer (500 µl) containing 0.1% Igepal CA-630 (USB) was added, and lysate incubated on ice for 10 min. Unbroken cells and the beads were removed by centrifugation at 500 × g for 10 min at 4°C, then lysates were cleared by centrifugation at 16,000 × g for 15 min at 4°C. The cleared lysates were incubated with preequilibrated EZview Red anti-FLAG M2 Affinity Gel (Sigma) at 4°C while rotating for 2 h. The resin was washed in lysis buffer three times, and the bound proteins eluted by incubating in elution buffer (100 mM Tris-HCl, pH 8.0, 1% SDS, 10 mM dithiothreitol [DTT]) at 95°C for 10 min.
Cargo ubiquitylation assay
The ubiquitylation assay was adapted from Guiney et al. (2016). Cells expressing pLZ555 were grown to early log phase and treated with 100 µM CuSO4 to induce myc-Ub expression. For Mup1-GFP–expressing strains, endocytosis was induced by adding 10 µg/ml Met for 10 min and 50 OD600 equivalents were collected. For Can1-GFP–expressing strains, endocytosis was induced by adding 100 µg/ml Arg for 30 min and 100 OD600 equivalents were collected. Cells were washed in 20 ml of 10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 10 mM NEM, 1 mM PMSF, and cell pellets frozen at –80°C. Cells were then thawed on ice and resuspended in 450 µl lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 20 mM NEM, 1× cOmplete protease inhibitor cocktail (Roche), 20 mM NaF, 1 mM DTT). Cells were lysed by vortexing with 0.5-mm zirconia beads (Biospec Products) for 6 min two times at 4°C, with a 6-min incubation on ice in between. Membranes were solubilized by adding 50 µl lysis buffer containing 10% Igepal CA-630, 5% deoxycholate, and 1% SDS and rotating at 4°C for 30 min. Unbroken cells and the beads were removed by centrifugation at 500 × g for 10 min at 4°C, then lysates were cleared by centrifugation at 16,000 × g for 15 min at 4°C. The lysates were precleared by incubating with preequilibrated IgG Sepharose 6 Fast Flow (GE Healthcare) for 60 min at 4°C while rotating. Precleared lysates were then incubated with GFP-nanobody conjugated to IgG Sepharose for 4 h at 4°C while rotating. The resin was washed in wash buffer (50 mM Tris-HCl, pH 7.4, 750 mM NaCl, 1 mM EDTA, 1 mM PMSF, 20 mM NEM, 1× cOmplete protease inhibitor cocktail [Roche], 20 mM NaF, 1 mM DTT, 0.5% SDS) two times, then once in lysis buffer. Bound proteins were eluted by adding a 1:1 mixture of urea cracking buffer and 2× sample buffer elution buffer and incubating at 42°C for 10 min.
Mass spectrometry
For analysis of Art1 phosphorylation, cells were grown to mid–log phase in synthetic media supplemented with adenine, uracil, Asp, Ile, Leu, Phe, Thr, Trp, Tyr, Val, His, and Pro, and either light Arg and Lys or heavy Arg (13C6 15N4; Sigma) and Lys (13C6 15N2; Sigma). Art1-HTF was immunoprecipitated as described above, except 250 OD600 equivalents were harvested and the lysis buffer additionally contained 1× PhosSTOP phosphatase inhibitor (Roche). Eluates were reduced with 10 mM DTT for 15 min at room temperature, then alkylated with 20 mM iodoacetamide. Heavy and light samples were mixed 1:1 and precipitated by adding three volumes of 50% acetone, 49.9% ethanol, and 0.1% acetic acid. Proteins were resuspended in 8 M urea, 50 mM Tris-HCl, pH 8.0, diluted with three volumes of dH2O, and digested overnight with 1 g trypsin at 37°C.
Phosphopeptides were enriched using Fe-NTA as described elsewhere (Bastos de Oliveira et al., 2018). Data were analyzed using both the Sorcerer-SEQUEST data analysis pipeline (Bastos de Oliveira et al., 2018) and MaxQuant for phosphopeptide identification, quantitation, and phosphorylation site localization. MS/MS spectra were also manually inspected for MS2 quality and to confirm phosphorylation site localization. The MaxQuant site localization score is included in the source data. Scatter plots were generated using a data analysis package courtesy of Haiyuan Yu (Cornell University).
Bioinformatic analysis
For analysis of Art1 conservation, homologues were retrieved from the NCBI refseq database with BLAST, and sequences annotated as “partial read” were removed. Art1 homologues were defined as proteins with a BLAST E-value < 0.1, and whose top BLAST hit in the S. cerevisiae proteome was also Art1. Likewise, Art2/8, Art3/6, and Art4/5/7 homologues were defined as proteins with a BLAST E-value < 0.1 when Art2, Art3, or Art4 were used as query, respectively; and whose top BLAST hit in the S. cerevisiae proteome was also Art2 or Art8, Art3 or Art6, or Art4, Art5, or Art7, respectively. Homologues were aligned with mafft, using the -linsi option (–maxiterate 1000 –localpair); phylogenetic trees were computed with FastTree (Price et al., 2010), using the Le-Gascuel substitution model and pseudocounts (options -lg and -pseudo); Art1, Art3/6, and Art4/5/7 multiple sequence alignments were rooted to TXNIP from Homo sapiens, and Art2/8 multiple sequence alignment was rooted to Uniprot ID Q6CG62 from Yarrowia lipolytica. Disorder predictions were calculated with Disopred3 (Jones and Cozzetto, 2015). Structural modeling of S. pombe Any1p and Art1ΔN,1,2,C was performed using iTASSER (Zhang, 2008; Roy et al., 2010; Yang et al., 2010). Statistical analysis. Statistical analysis of cargo endocytosis assays were conducted in R, using the aov, TukeyHSD, and emmeans packages; all treatments were analyzed with analysis of variance (ANOVA) followed by Tukey’s posttest, and all R-commands as well as exact p values can be found in Supplemental Table S4. Statistical analysis of Art1-mNG localization was conducted in Graphpad Prism 6 (Graphpad Software), using ANOVA followed by Kruskal-Wallis test with Dunn’s multiple comparisons test. Raw data used for statistical analysis can be found in Supplemental Table S3.
Supplementary Material
Acknowledgments
We thank Richa Sardana, Sho Suzuki, and Jeff Jorgensen for critical reading of the manuscript, Lu Zhu for plasmids, and Sudeep Banjade for the GFP-nanobody resin. Statistical consulting was provided by Stephen Parry of the Cornell Statistical Consulting Unit. M.G.B. was supported by a Sam and Nancy Fleming Research fellowship. This work was supported by a Cornell University Research Grant to S.D.E. and National Institutes of Health Grant no. R00GM-101077 to J.A.M.
Abbreviations used:
- ART
arrestin-related trafficking adaptor
- DTT
dithiothreitol
- mNG
mNeonGreen
- PM
plasma membrane
- SILAC
stable isotope labeling with amino acids in cell culture
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-08-0451) on October 16, 2019.
REFERENCES
- Adell MAY, Migliano SM, Upadhyayula S, Bykov YS, Sprenger S, Pakdel M, Vogel GF, Jih G, Skillern W, Behrouzi R, et al (2017). Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife , e31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Bussey H. (1986). Yeast arginine permease: nucleotide sequence of the CAN1 gene. Curr Genet , 587–592. [DOI] [PubMed] [Google Scholar]
- Alvarez CE. (2008). On the origins of arrestin and rhodopsin. BMC Evol Biol , 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Guetta D, Klein G. (2009). The arrestin fold: variations on a theme. Curr Genomics , 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Klein G. (2013). True arrestins and arrestin-fold proteins: a structure-based appraisal. In: Progress in Molecular Biology and Translational Science, ed. Luttrell LM, New York: Academic Press, 21–56. [DOI] [PubMed] [Google Scholar]
- Bastos de Oliveira FM, Kim D, Lanz M, Smolka MB. (2018). Quantitative analysis of DNA damage signaling responses to chemical and genetic perturbations. In: Genome Instability: Methods and Protocols, ed. Muzi-Falconi M, Brown GW, New York: Springer, 645–660. [DOI] [PubMed] [Google Scholar]
- Becuwe M, Herrador A, Haguenauer-Tsapis R, Vincent O, Léon S. (2012a). Ubiquitin-mediated regulation of endocytosis by proteins of the arrestin family. Biochem Res Int 2012, 242764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Léon S. (2012b). A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol , 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busto JV, Elting A, Haase D, Spira F, Kuhlman J, Schäfer-Herte M, Wedlich-Söldner R. (2018). Lateral plasma membrane compartmentalization links protein function and turnover. EMBO J , e99473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AS, O’Shea EK. (2002). Pho85 and signaling environmental conditions. Trends Biochem Sci , 87–93. [DOI] [PubMed] [Google Scholar]
- Dunn R, Hicke L. (2001). Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol Biol Cell , 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré S, Urban-Grimal D, Haguenauer-Tsapis R. (2004). Ubiquitin and endocytic internalization in yeast and animal cells. Biochim Biophys Acta Mol Cell Res , 89–111. [DOI] [PubMed] [Google Scholar]
- Fujita S, Sato D, Kasai H, Ohashi M, Tsukue S, Takekoshi Y, Gomi K, Shintani T. (2018). The C-terminal region of the yeast monocarboxylate transporter Jen1 acts as a glucose signal–responding degron recognized by the α-arrestin Rod1. J Biol Chem , 10926–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewska B, Kamin´ska J, Jesionowska A, Martin NC, Hopper AK, Z˙oła̧dek T. (2001). WW domains of Rsp5p define different functions: determination of roles in fluid phase and uracil permease endocytosis in Saccharomyces cerevisiae. Genetics , 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaddar K, Merhi A, Saliba E, Krammer E-M, Prévost M, André B. (2014). Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol Cell Biol , 4447–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast , 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gournas C, Gkionis S, Carquin M, Twyffels L, Tyteca D, André B. (2018). Conformation-dependent partitioning of yeast nutrient transporters into starvation-protective membrane domains. Proc Natl Acad Sci USA , E3145–E3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournas C, Prévost M, Krammer E-M, André B. (2016). Function and regulation of fungal amino acid transporters: insights from predicted structure. In: Yeast Membrane Transport, ed. Ramos J, Sychrová H, Kschischo M, Cham, Switzerland: Springer International Publishing, 69–106. [DOI] [PubMed] [Google Scholar]
- Gournas C, Saliba E, Krammer E-M, Barthelemy C, Prévost M, André B, Lemmon S. (2017). Transition of yeast Can1 transporter to the inward-facing state unveils an α-arrestin target sequence promoting its ubiquitylation and endocytosis. Mol Biol Cell , 2819–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson M, Mousset M, Wiame JM, Bechet J. (1966). I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta , 325–338. [DOI] [PubMed] [Google Scholar]
- Guiney EL, Klecker T, Emr SD. (2016). Identification of the endocytic sorting signal recognized by the Art1-Rsp5 ubiquitin ligase complex. Mol Biol Cell , 4043–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. (2007). Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol , 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haguenauer-Tsapis R, André B. (2004). Membrane trafficking of yeast transporters: mechanisms and physiological control of downregulation. In: Molecular Mechanisms Controlling Transmembrane Transport, Berlin, Heidelberg: Springer, 273–323. [Google Scholar]
- Hatakeyama R, Kamiya M, Takahara T, Maeda T. (2010). Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol Cell Biol , 5598–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Valdez-Taubas J, Pelham HRB. (2004). Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J , 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H-C, MacGurn JA, Emr SD, Glick BS. (2017). Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs. Mol Biol Cell , 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene , 51–59. [DOI] [PubMed] [Google Scholar]
- Hovsepian J, Defenouillère Q, Albanèse V, Váchová L, Garcia C, Palková Z, Léon S. (2017). Multilevel regulation of an α-arrestin by glucose depletion controls hexose transporter endocytosis. J Cell Biol , 1811–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Moffat J, Andrews B. (2002). Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol Cell Biol , 5076–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Cozzetto D. (2015). DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics , 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O’Shea EK. (1994). Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science , 1153–1156. [DOI] [PubMed] [Google Scholar]
- Keener JM, Babst M. (2013). Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic , 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. (2010). The ubiquitin code of yeast permease trafficking. Trends Cell Biol , 196–204. [DOI] [PubMed] [Google Scholar]
- Lee S, Ho H-C, Tumolo JM, Hsu P-C, MacGurn JA. (2019). Methionine triggers Ppz-mediated dephosphorylation of Art1 to promote cargo-specific endocytosis. J Cell Biol , 977–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S, Erpapazoglou Z, Haguenauer-Tsapis R, Lemmon S. (2008). Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol Biol Cell , 2379–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon S, Haguenauer-Tsapis R. (2009). Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp Cell Res , 1574–1583. [DOI] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. (2008). Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell , 714–725. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Iii AM, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- MacDonald C, Stringer DK, Piper RC. (2011). Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic , 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu P-C, Emr SD. (2012). Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem , 231–259. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Hsu P-C, Smolka MB, Emr SD. (2011). TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell , 1104–1117. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Hyvönen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. (1996). Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature , 646–649. [DOI] [PubMed] [Google Scholar]
- Malcova I, Farkasovsky M, Senohrabkova L, Vasicova P, Hasek J. (2016). New integrative modules for multicolor-protein labeling and live-cell imaging in Saccharomyces cerevisiae. FEMS Yeast Res , fow207. [DOI] [PubMed] [Google Scholar]
- Martínez-Márquez JY, Duncan MC. (2018). Investigation of Ldb19/Art1 localization and function at the late Golgi. PLoS One , e0206944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Wickner RB. (1993). CLG1, a new cyclin-like gene of Saccharomyces cerevisiae. Yeast , 929–931. [DOI] [PubMed] [Google Scholar]
- Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman AM, Andrews B. (1997). A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol , 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Huang D, Andrews B. (2000). Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog Cell Cycle Res , 97–106. [DOI] [PubMed] [Google Scholar]
- Mülleder M, Calvani E, Alam MT, Wang RK, Eckerstorfer F, Zelezniak A, Ralser M. (2016). Functional metabolomics describes the yeast biosynthetic regulome. Cell , 553–565.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Pelham HRB. (2009). Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic Cph Den , 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, Huang L, Thorner J, Cyert MS. (2013). A calcineurin-dependent switch controls the trafficking function of α-arrestin Aly1/Art6. J Biol Chem , 24063–24080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, Schmidt MC. (2019). AMPK-mediated regulation of alpha-arrestins and protein trafficking. Int J Mol Sci , 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One , e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol , 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc , 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Zhu L, Emr SD. (2018). Rsp5 Ubiquitin ligase–mediated quality control system clears membrane proteins mistargeted to the vacuole membrane. J Cell Biol , 234–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum . Nat Methods , 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (2002). Getting started with yeast. Methods Enzymol , 3–41. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast , 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, Virgilio CD. (2005). Regulation of G0 entry by the Pho80–Pho85 cyclin–CDK complex. EMBO J , 4271–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Geng J, Yen W-L, Wang K, Klionsky DJ. (2010). Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell , 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappacosta F, Collingwood TS, Huddleston MJ, Annan RS. (2006). A quantitative results-driven approach to analyzing multisite protein phosphorylation: the phosphate-dependent phosphorylation profile of the transcription factor Pho4. Mol Cell Proteomics , 2019–2030. [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics , 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, MacGurn JA, Liu M, Emr S. (2013). The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. eLife , e00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.