Abstract
Self vs non-self discrimination by innate immune sensors is critical for mounting effective immune responses against pathogens, while avoiding harmful auto-inflammatory reactions against the host. Foreign DNA and RNA sensors must discriminate between self vs non-self nucleic acids, despite their shared building blocks and similar physicochemical properties. Recent structural and biochemical studies suggest that multiple steps of filament-like assembly are required for the functions of several nucleic acid sensors. Here, we discuss ligand discrimination and oligomerization of RIG-I-like receptors, AIM2-like receptors, and cGAS. We discuss how filament-like assembly allows for robust and accurate discrimination of self vs. non-self nucleic acids, and how these assemblies enable sensing of multiple distinct features in foreign nucleic acids, including structure, length and modifications. We also discuss how individual receptors differ in their assembly and disassembly mechanisms, and how these differences contribute to the diversity in nucleic acid specificity and pathogen detection strategies.
Recent structural and biochemical studies have demonstrated that filamentous assembly of intracellular nucleic acid sensors is required for their activity. Cadena and Hur describe the filament-like assembly of RIG-I-like receptors, AIM2-like receptors, and cGAS. Furthermore, they discuss how filament-like assembly allows for discrimination of self vs non-self nucleic acids.
Introduction
Self vs non-self-discrimination is fundamental to all immune functions. The last decade of studies showed that foreign nucleic acid sensing is a central mechanism by which the innate immune system detects pathogen infection. This is carried out by several nucleic acid sensors that are classified as pattern recognition receptors (PRRs). These nucleic acid sensors recognize features that are generally conserved among pathogens, so called pathogen-associated molecular patterns (PAMPs), and activate antiviral and pro-inflammatory signaling pathways. However, studies have also shown that, under various pathologic conditions, nucleic acid sensors can mis-construe self-derived nucleic acids as foreign, and that this misrecognition underlies a growing number of auto-inflammatory diseases. At the molecular level, this is not too surprising considering that all nucleic acids, regardless of their origin, are made up of the identical building blocks. This is unlike other immune receptors, such as TLR4 that recognizes lipid moieties that are normally absent in the host. Thus, this raises the question of exactly how nucleic acid sensors in the innate immune system accurately discriminate between self vs. non-self under normal physiological conditions.
In this review, we discuss three families of well-characterized nucleic acid sensors, namely RIG-I-like receptors (RLRs), AIM2-like receptors (ALRs) and cGAS (Fig. 1). We discuss their commonalities and differences with a focus on their filament-like multimerization properties. We discuss recent findings on how filament-like assemblies allow for robust and accurate discrimination between self vs. non-self nucleic acids, and how they enable sensing of multiple distinct features in foreign nucleic acids, including structure, length and modifications. We also discuss how individual receptors differ in their assembly and disassembly mechanisms, and how these differences contribute to the diversity in nucleic acid specificity and pathogen detection strategies.
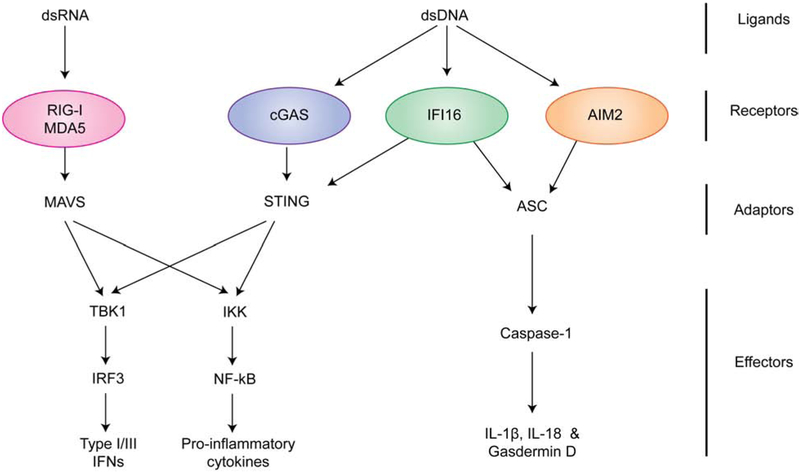
Figure 1.
Signaling pathways of RIG-I-like receptors (RLRs), cGAS, and AIM2-like receptors (ALRs). RLRs recognize dsRNA and activate MAVS, while cGAS recognizes dsDNA and activates STING. Activated MAVS and STING commonly stimulate TBK1 and IKK for production of type I/III interferons (IFNs) and other pro-inflammatory cytokines. As with cGAS, ALRs also recognize dsDNA, but the ligand engagement leads to assembly of inflammasomes, which include receptors, adaptor ASC and effector caspase-1. Within the assembled inflammasome, caspase-1 cleaves IL-1β, IL-18, and gasdermin D, which results maturation of the inflammatory cytokines and induction of pyroptosis. IFI16 was also reported to activate the interferon pathway through STING, albeit through a poorly understood mechanism.
RIG-I
Retinoic-acid inducible gene I (RIG-I) is a cytoplasmic sensor of viral dsRNA. RIG-I is the founding member of the RIG-I-like receptor (RLR) family, which also includes MDA5 (melanoma-differentiation-associated protein 5). RIG-I detects a broad range of viral RNAs during infection and activates an intracellular signaling pathway that leads to the production of type I interferons (Yoneyama et al., 2004). RIG-I consists of two N-terminal caspase activation and recruitment domains (CARDs), a central DExD/H-box helicase domain, and a C-terminal domain (CTD) (Fig. 2A). The CARDs are involved in downstream signal activation, while the helicase-CTD domains interact with dsRNA. In the resting state, the CARD signaling domain exists in an auto-repressed state, but it is released upon dsRNA and/or ATP binding (Kowalinski et al., 2011; Rawling et al., 2015). The released RIG-I CARDs form a homo-tetramer and interact with the CARD domain of the downstream adaptor molecule MAVS (mitochondrial antiviral-signaling protein) through a homotypic CARD-CARD interaction (Peisley et al., 2014a; Wu et al., 2014). This CARD-CARD interaction then triggers MAVS filament formation, which functions as a signaling platform to recruit further downstream signaling molecules including TRAFs, TBK1 and IKK. These molecules subsequently activate IRF3/7 and NF-kB to upregulate type I interferons and other antiviral molecules (Hou et al., 2011; Jiang et al., 2012; Wu and Hur, 2015).
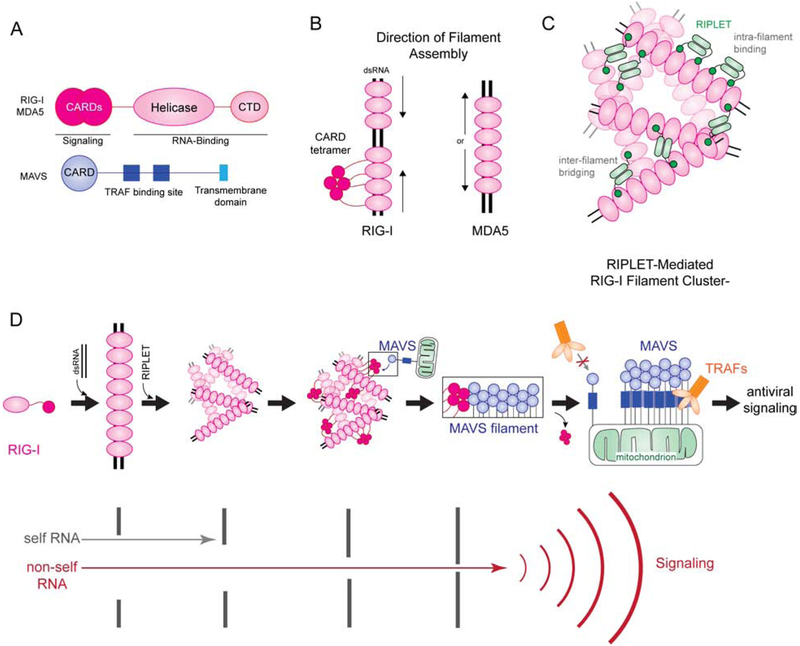
Figure 2.
Filament formation and higher order oligomerization of RIG-I-like receptors. A) Both RIG-I and MDA5 share the same domain architecture, consisting of CARDs, a helicase domain and a C-terminal domain (CTD). CARDs are responsible for signal activation, while helicase and CTD are for RNA binding. The downstream adaptor MAVS contains the N-terminal CARD, followed by ~400 residue-long linker containing TRAF binding sites and a C-terminal transmembrane domain (TM). MAVS CARD interacts with RLR CARDs, while TM anchors MAVS to mitochondria. B) RIG-I and MDA5 both form filaments on dsRNA, but their assembly mechanisms differ. RIG-I first binds the 5’ppp at the end of the dsRNA, then uses ATP hydrolysis to translocate to the interior. MDA5 binds the interior of the dsRNA then cooperatively forms a filament in a ATP-independent manner towards the end of the dsRNA. C) RIPLET uses bivalency to selectively bind RIG-I filaments (intra-filament binding). However, on long RIG-I filaments, RIPLET can induce higher order oligomerization by cross-bridging RIG-I filaments (inter-filament binding). This results in clustering of RIG-I filaments and further amplification of antiviral signaling. D) A model of how filament formation and higher order oligomerization of RIG-I and MAVS enable self vs. non-self discrimination and antiviral signaling. RIG-I can bind short dsRNA as a monomer, but RNA-binding is insufficient to activate downstream signaling. Filament formation along long dsRNA allows recruitment of RIPLET, which in turn bridges RIG-I filaments and conjugates K63-linked Ub chains. The high local receptor concentration and K63-polyUb together promote RIG-I CARD tetramer formation, which then acts as a nucleus to induce MAVS filament formation. MAVS filament then functions as a signaling platform to recruit and activate further downstream signaling molecules, such as TRAFs. The multiple steps of receptor oligomerization serve as independent check-points to filter out self RNAs and allow only non-self RNAs to activate downstream signaling. Once MAVS filament is nucleated, signal amplifies through polymerization of MAVS. Thus, the combination of multi-step oligomerization, both by the receptor and the signaling adaptor, ensures the accuracy and robustness of antiviral signaling in response to infection.
RIG-I senses viral dsRNA but discriminates between cellular and viral dsRNAs based on the combination of the RNA duplex structure and the 5’-triphosphate (5’ppp) moiety present in certain viral RNAs (Hornung et al., 2006; Pichlmair et al., 2006; Schlee et al., 2009). 5’ppp is present in all nascent transcripts, whether from the host or virus, but cellular RNAs typically undergo 5’-processing in the nucleus. This often results in replacement of 5’ppp by 5’p that cannot stimulate RIG-I (Ren et al., 2019). Some viral RNAs, however, are generated in the cytoplasm and can bypass 5’-processing, thereby being subjected to RIG-I detection. The structures of monomeric RIG-I in complex with dsRNA showed that the helicase forms a C-like structure around dsRNA, while the CTD caps the dsRNA end, directly recognizing the 5’ppp moiety (Lu et al., 2010; Wang et al., 2010).
In addition to 5’ppp and the duplex structure, RIG-I also recognizes dsRNA length for more accurate discrimination between self vs. non-self RNAs. Earlier studies showed that > 1kb dsRNA activates MDA5 but not RIG-I (Kato et al., 2006), which led to a model that MDA5 recognizes long dsRNA while RIG-I recognizes short dsRNA. However, a number of studies showed that RIG-I recognizes longer dsRNAs when compared among <~1 kb and provided at the same molar concentration, i.e. with the same concentration of 5’ppp (Binder et al., 2011; Patel et al., 2013). Even when the same mass concentration was used, where the available 5’ppp concentration decreases proportionally with dsRNA length, ~100–200 bp shows a significantly higher activity than 10–20 bp dsRNA (Cadena et al., 2019). These data suggest that both 5’ppp and duplex length play important roles, resulting in a bell-shaped dependence when comparing RNAs at the equivalent mass concentration. Direct association studies have not yet shown which physiologically relevant ligands are sensed by RIG-I during infection. This is also the case for the other receptors described below. Therefore, the duplex length discrimination hypothesis is based on studies using model dsRNA ligands (or dsDNA ligands for ALRs and cGAS). However, this duplex length-dependence can explain how different strains of Sendai virus (SeV) that generate defective viral genomes (DVGs) of different lengths activate RIG-I in a length-dependent manner (Strahle et al., 2006). Of note, a recent study reported that a short hairpin with ~10–14 bp stem was found sufficient to activate RIG-I, whereas ~10–14 bp dsRNA formed by two separate strands does not (Linehan et al., 2018). It is unclear exactly how the loop in the hairpin contributes to RIG-I activation, or if hairpin multimerization (Heinicke et al., 2009) plays a role.
How does RIG-I senses dsRNA length? The answer likely lies in the fact that dsRNA binding is not sufficient to activate RIG-I and that RIG-I CARD oligomerization is additionally required for MAVS activation. The helicase domain of RIG-I contains an active site for ATP binding and hydrolysis, which is important for RIG-I translocation and oligomerization on dsRNA (Myong et al., 2009; Patel et al., 2013; Peisley et al., 2013). While RIG-I binds 10–15 bp dsRNA end as a monomer (Jiang et al., 2011; Kowalinski et al., 2011; Luo et al., 2011), RIG-I forms a filamentous oligomer on longer dsRNA (Binder et al., 2011; Patel et al., 2013; Peisley et al., 2013). More specifically, RIG-I preferentially binds the dsRNA end as a monomer, but this triggers RIG-I ATP hydrolysis, which powers RIG-I translocation from the dsRNA end to the interior (Myong et al., 2009). RIG-I translocation then re-exposes the 5’ppp, allowing recruitment of another RIG-I molecule to the dsRNA end. Subsequently, iterations of end-recruitment and translocation of RIG-I leads to formation of filamentous oligomerization near the dsRNA end (Fig. 2B) (Peisley et al., 2013). Filament assembly of RIG-I then brings together CARDs from adjacent RIG-I molecules in close proximity, and consequently promotes CARD tetramerization (Fig. 2B) (Peisley et al., 2013). RIG-I filament formation is limited on >1 kb dsRNAs, because the assembly initiates from the dsRNA end to the interior, and the dsRNA ends become a limiting factor with very long dsRNA (Peisley et al., 2013). This explains why RIG-I signaling is inefficient with >1 kb dsRNA. RIG-I filament assembly may also provide an explanation as to how RIG-I senses RNA modifications present in the RNA interior distant from the dsRNA end (Hornung et al., 2006; Peisley et al., 2013; Uzri and Gehrke, 2009).
More recently, two additional mechanisms were reported to account for how RIG-I recognizes dsRNA in a length dependent manner. It has been known that RIG-I binds K63-linked polyUb chains, and that this non-covalent interaction promotes RIG-I CARD tetramerization for efficient MAVS activation (Peisley et al., 2014b; Zeng et al., 2010). While previous studies suggested that TRIM25 as a major E3 ligase responsible for generating K63-polyUb for RIG-I (Gack et al., 2007), a recent report (Cadena et al., 2019) showed that RIPLET, but not TRIM25, is required for RIG-I signaling and that the action of TRIM25 is limited to isolated RIG-I CARD under overexpression conditions. RIPLET is sufficient to bind RIG-I and conjugate K63-polyUb, and the conjugated Ub then serves as a source of K63-polyUb for RIG-I CARD tetramerization. Both RIPLET activities, RIG-I binding and ubiquitination, require dsRNA long enough to accommodate at least two RIG-I molecules (Cadena et al., 2019). This is because RIPLET, which exists as a dimer with two RIG-I-binding sites, requires bivalent binding for efficient recognition of RIG-I (Fig 2C). RIG-I filament formation satisfies this bivalency requirement, while monomeric RIG-I on short dsRNA does not. Additionally, RIPLET was also found to bridge RIG-I filaments and form RIG-I clusters (Cadena et al., 2019) (Fig. 2C). This clustering of RIG-I filaments places CARD in high local concentration, beyond what can be achieved within individual filaments, further promoting CARD tetramerization and MAVS stimulation. Thus, RIPLET utilizes a combination of intra-filament binding and inter-filament bridging to control RIG-I ubiquitination, RIG-I clustering and thus its signaling activity in a dsRNA length-dependent manner (Fig. 2D).
Altogether, viral RNA recognition and antiviral signaling by RIG-I is mediated by multiple steps of multimerization: first by dsRNA-mediated filament formation, second by RIPLET-dependent filament bridging, third by ubiquitin- and proximity-induced CARD tetramerization and finally recruitment of MAVS and induction of MAVS filament formation (Fig. 2D). In this model, each step of multimerization amplifies antiviral signaling, while rejecting RNAs that are less likely from pathogens. It also shows how multiple PAMPs, 5’ppp, dsRNA structure and dsRNA length, synergize to allow accurate self vs. non-self discrimination while ensuring robust signal amplification upon correct viral recognition. Whether there are yet-unidentified RNA features, for example more complex RNA structure or sequence, that can stimulate RIG-I oligomerization in a manner different from the current filament model remains to be tested. Multiple studies have shown that various RNAs besides the mid-long dsRNA with 5’ppp can activate RIG-I (Chen et al., in press; Chen et al., 2017; Saito et al., 2008; Xu et al., 2015). More detailed biochemical analysis of how these RNAs can induce RIG-I oligomerization would be an interesting area of future investigation. Finally, a few gain-of-function (GOF) mutations in RIG-I have been identified from patients with an auto-inflammatory disease, Singleton-Merten Syndrome (Ferreira et al., 2019; Jang et al., 2015). These GOF mutations were shown to constitutively activate RIG-I signaling, likely through mis-recognition of self RNAs. A Kinetic proof-reading mechanism was proposed to explain how wild-type RIG-I avoids self RNA recognition while GOF mutants do not (Chiang et al., 2018; Devarkar et al., 2018). However, which cellular RNAs are recognized by the GOF mutants and how these RNAs promote formation of signaling-competent RIG-I oligomers remains to be further investigated.
MDA5
MDA5 is a paralog of RIG-I, sharing the same domain architecture (Fig. 2A) and downstream signaling pathway (Fig. 1). As with RIG-I, MDA5 binds dsRNA in a sequence-independent manner, forms filaments along the length of the dsRNA, and utilizes proximity-induced tetramerization of the CARD signaling domain to activate MAVS (Wu et al., 2013). K63-linked poly-ubiquitination of MDA5 was also shown to assist CARD tetramerization (Jiang et al., 2012), although the responsible E3 ligase is TRIM65 not RIPLET (Lang et al., 2017). Despite the multiple shared features, MDA5 differs from RIG-I in RNA specificity and the mechanism of filament assembly and disassembly. These mechanistic differences make MDA5 and RIG-I recognize largely distinct groups of viruses and function as non-redundant receptors (Kato et al., 2006; Loo et al., 2008).
Unlike RIG-I, MDA5 filament formation is obligatory for high affinity interaction with dsRNA (RIG-I can bind dsRNA as a monomer, though this alone does not stimulate strong signaling) (Peisley et al., 2012; Peisley et al., 2011). Monomeric affinity for dsRNA is very low (Kd of ~1 μM) and high affinity interaction with dsRNA requires protein:protein interactions among the MDA5 protomers. Also unlike RIG-I, which forms filaments from the dsRNA end to the interior using ATP hydrolysis-driven translocation, the MDA5 filament nucleates directly from the dsRNA interior and propagates in a unidirectional manner (Peisley et al., 2012) (Fig. 2B). In fact, MDA5 does not recognize the dsRNA end (either the end structure or 5’ppp) and this difference is reflected in the differential positioning of CTD (Wu et al., 2013). Furthermore, MDA5 filament formation propagates solely through extensive protein-protein interactions between protomers and does not require ATP hydrolysis. ATP hydrolysis, instead, destabilizes the MDA5 filament, promoting its disassembly from filament termini (Peisley et al., 2012; Peisley et al., 2011).
How can these differences result in distinct dsRNA length specificity for MDA5 (> 1kb) and RIG-I (~100–200 bp) (Cadena et al., 2019; Kato et al., 2008)? First of all, the fact that MDA5 does not require the dsRNA end for filament formation means that the limiting amount of RNA end in long dsRNA does not limit MDA5 filament formation, as it does for RIG-I. Secondly, ATP-dependent end-disassembly of the MDA5 filament confers selective stabilization of long filaments, and induces rapid disassembly of filaments on short dsRNA (Peisley et al., 2012). This further shifts the length preference of MDA5 towards longer dsRNA, unlike RIG-I. Furthermore, de novo filament nucleation is extremely inefficient for MDA5, while filament elongation from an existing nucleus occurs rapidly. Thus, filaments on short dsRNA are difficult to re-form after complete disassembly, while shortened filaments on long dsRNA cycle between partial end disassembly and elongation, bypassing the slow nucleation step. Thus, MDA5 uses a combination of end disassembly and slow nucleation kinetics to “discard” short dsRNA rapidly (kinetic proof reading mechanism) (Peisley et al., 2012). Finally, MDA5 further uses this repetitive cycle of assembly and disassembly to repair filament discontinuities, which can be present because of multiple, internal nucleation events. As a result, ATP hydrolysis allows formation of longer, continuous filaments that more accurately reflect the length of the underlying dsRNA scaffold. Thus, MDA5 appears to utilize multiple mechanisms to ensure selective recognition of very long dsRNA (del Toro Duany et al., 2015). It is possible that the lack of the 5’ppp specificity requires MDA5 to employ more stringent length specificity for accurate discrimination between self vs. non-self dsRNAs.
In addition to dsRNA length, the integrity of the dsRNA duplex plays an additional role in regulating aberrant MDA5 activity against cellular dsRNAs. This was well demonstrated from the studies of common cellular dsRNAs formed by Alu (Ahmad et al., 2018; Chung et al., 2018). Alu is an abundant retroelement and often occurs in the inverted repeat (IR) configuration within 3’UTR of many mRNAs. IR-Alus can fold back and form ~300 bp Alu:Alu hybrids. Although this length is below the typical length required for optimal MDA5 activation, the dsRNA length threshold for MDA5 activation is dependent on the concentration of the dsRNA (i.e. subject to mass action law). Given that IR-Alus dsRNAs are abundantly present in the cytoplasm (constituting ~0.5 % of total non-ribosomal RNAs) (Ahmad et al., 2018), IR-Alus have the potential to stimulate MDA5. However, IR-Alus dsRNA does not normally activate wild-type MDA5 for at least two reasons (Ahmad et al., 2018). First, IR-Alus are imperfect duplexes with 20–30% sequence mismatches. Because MDA5 is sensitive to structural irregularities within dsRNA, it fails to form continuous filaments on IR-Alus. Second, IR-Alus (as well as other dsRNAs) are subject to post-transcriptional modification (adenosine to inosine, A-to-I) by the cellular enzyme ADAR1. A-to-I modification weakens Watson-Crick base-pairs and partially melts dsRNAs, the effects similar to those of mismatches. This further suppresses MDA5 filament formation and signaling on IR-Alus. Intriguingly gain-of-function (GOF) mutations in MDA5 relax these specificities of MDA5 (i.e. insensitive to mismatches and A-to-I modifications in dsRNA), leading to mis-recognition of IR-Alus and the pathogenesis of a spectrum of inflammatory disorders (Ahmad et al., 2018; Rice et al., 2014).
Thus, MDA5 filament formation provides a mechanism by which MDA5 can measure dsRNA length and structural integrity for robust self vs. non-self discrimination, but also explains how this discrimination fails under pathologic conditions. Whether there are additional levels of oligomerization of MDA5 filament, such as filament bridging shown for RIG-I by RIPLET, remains unclear. Polyubiquitination of MDA5 does not depend on RIPLET, but instead on another E3 ligase, TRIM65 (Lang et al., 2017). Immunofluorescence experiments of MDA5 and TRIM65 showed that they assemble into large aggregate-like structures upon dsRNA stimulation (Lang et al., 2017). It would be interesting to examine if the filament cross-bridging mechanism also applies to MDA5 and whether yet-unknown PAMPs can stimulate MDA5 filament formation or even a new type of MDA5 oligomerization for signal activation.
cGAS
Cyclic GMP-AMP synthase (cGAS) is a cytosolic sensor that detects dsDNA. cGAS contains the template-independent nucleotidyltransferase (NTase)-like domain and the N-terminal basic region (Fig. 3A). Upon dsDNA binding, cGAS generates cyclic GMP-AMP (cGAMP), which binds STING (Stimulator of interferon genes) to activate the type I IFN pathway (Ablasser et al., 2013; Gao et al., 2013; Sun et al., 2013) (Fig. 1). STING is a dimeric membrane-bound protein and resides on the surface of the ER in the resting state (Ishikawa and Barber, 2008). cGAMP binding triggers a conformational change in the STING dimer, and promotes its higher-order oligomerization for downstream signal activation (Ergun et al., 2019; Shang et al., 2019; Zhang et al., 2019). cGAMP binding also induces translocation of STING from ER to ERGIC/golgi compartment, which is also required for signal activation (Dobbs et al., 2015). In addition to activating the type I IFN pathway, cGAS can also activate an IFN-independent cell death pathway through STING-mediated lysosomal destabilization and consequent activation of inflammasomes (Gaidt et al., 2017). Finally, cGAS has been shown to play a role in cancer. DNA damage induces the translocation of cGAS into the nucleus where it impairs the recruitment of DNA repair-associated protein complexes and promotes tumorigenesis (Liu et al., 2018).
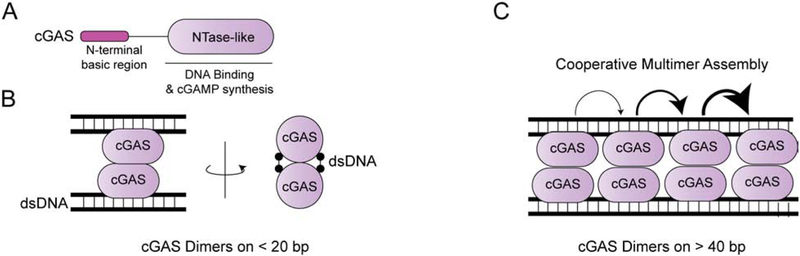
Figure 3.
cGAS forms ladder-like structures on long dsDNA. A) cGAS has an N-terminal basic region and an NTase-like domain. The latter is responsible for DNA binding and DNA-dependent cGAMP synthesis. B) on short DNA, the cGAS:DNA complex a 2:2 stoichiometry, with each cGAS monomer bridging two DNA strands. C) On longer DNA, a ‘founding’ cGAS dimer can promote binding of other cGAS dimers, allowing cooperative multimer assembly. These cGAS dimers arrange into ‘ladder-like’ structure. A direct dimer-to-dimer contact is unnecessary for the cooperative assembly.
cGAS recognizes DNA originated from multiple types of pathogens, including DNA virus (e.g. HSV), retrovirus (e.g. HIV) and intracellular bacteria (e.g. L. monocytogenes, M. tuberculosis) (Ablasser and Chen, 2019). However, studies showed that cGAS-STING pathways can be activated by RNA viruses with no DNA intermediate (Schoggins et al., 2014) or by other cellular stresses in the absence of infection (Gao et al., 2015; Gluck and Ablasser, 2019; Mackenzie et al., 2016; Thomas et al., 2017). For example, Dengue virus was found to activate cGAS, but the DNA ligand is not virus-derived, rather a host mitochondrial DNA leaked from damaged mitochondria (Aguirre et al., 2017). Exactly how Dengue virus infection causes mitochondrial DNA leakage is unclear. But the role of mitochondrial DNA as an endogenous source for cGAS activation has been shown in multiple other cases beside Dengue infection, for example during mitochondrial stress (West et al., 2015) and inflammasome activation (Aarreberg et al., 2019). Furthermore, recent studies showed that cGAS can also recognize mis-segregated nuclear genomes in the form of micronuclei (Harding et al., 2017; Mackenzie et al., 2017), or reverse transcription products of LINE1 retroelements under TREX1 or RNase H2 deficiency (Gao et al., 2015; Mackenzie et al., 2016; Thomas et al., 2017). As such, aberrant activation of cGAS has been shown to cause cellular senescence (Gluck and Ablasser, 2019) or inflammatory disease such as Aicardi-Goutieres syndrome (AGS) and systemic lupus erythematosus (SLE) (Gao et al., 2015; Mackenzie et al., 2016; Thomas et al., 2017).
These observations raise the question whether cGAS simply detects dsDNAs present in the cytoplasm and whether there is no additional mechanism to discriminate between self vs. non-self DNA. Multiple studies suggest that this is not the case. Instead, the activity of cGAS, in particular the human one, is strongly affected by dsDNA length. Multiple studies showed that cGAS requires at least ~100–200 bp dsDNA for cellular signaling activity and its activity increases continuously with DNA length even beyond this length (Andreeva et al., 2017a; Luecke et al., 2017; Zhou et al., 2018). This is presumably a mechanism to restrict unwanted cGAS activity against shorter DNA fragments that may become available during reverse transcription by endogenous retroviruses, nuclear damage, or mitochondrial damage (Ahn and Barber 2014; Kassiotis and Stoye 2016).
How does cGAS detect dsDNA structure and length? Crystal structures of cGAS in complex with DNA showed that cGAS interacts with dsDNA mostly through the DNA backbone, consistent with the sequence-independent recognition of DNA (Civril et al., 2013; Gao et al., 2013; Li et al., 2013). Note that dsDNA and dsRNA form distinct duplex structures (i.e. B vs. A form), and are thus recognized by distinct sets of sensors. DNA binding changes the conformation of cGAS in the active site, promoting the synthesis of cGAMP. Intriguingly, several crystal structures showed that cGAS adopts a dimeric configuration upon DNA binding, forming 2:2 complex (Li et al., 2013; Zhang et al., 2014). In this complex, two DNA molecules are sandwiched between two cGAS molecules, where each cGAS subunit utilizes two distinct surfaces to bind two DNA molecules (Fig. 3B). The dimeric interface and the two DNA binding sites are all required for high affinity binding (Li et al., 2013). More detailed studies showed that cGAS dimer formation, intrinsic catalytic activity, and DNA binding are inter-dependent (Hooy and Sohn, 2018). That is, DNA binding stimulates cGAS dimerization, and this in turn further stimulates cGAMP synthesis. Moreover, placement of two separate DNA molecules in close proximity within the 2:2 complex promotes additional cGAS dimer recruitment (Fig. 3C). This allows cooperative cGAS multimer assembly along the axis of DNA, albeit with little protein-protein contact along the DNA axis (Andreeva et al., 2017a; Hooy and Sohn, 2018). This filament-like assembly can explain how cGAS prefers long dsDNA for both binding and signaling. Intriguingly, monoubiquitination of cGAS by TRIM56 was shown to stabilize dimerization, thus promoting cGAMP production and signaling activity (Seo et al., 2018). Furthermore, a recent study showed that cGAS forms a phase separated condensate during DNA recognition (Du and Chen, 2018). These observations suggest that cGAS multimerization is more complex than a simple filament-like assembly on dsDNA, and are likely regulated by multiple factors in cells.
Several important questions remain to be addressed. While the ladder-like oligomerization of cGAS can explain how it measures naked dsDNA length, the purpose of cGAS’s ability to measure length, or if this relates to self vs non-self discrimination, remains unknown. Viral dsDNAs are typically sequestered in nucleocapsid or replication compartments, raising the question of what are the DNAs recognized by cGAS during infection and how cGAS gains access to them. There is also significant controversy as to the cellular location of cGAS. While it was initially thought to be exclusively located in the cytoplasm, nuclear (Liu et al., 2018) and membrane-bound species (Barnett et al., 2019) were also reported, and these were proposed to have various functions in suppressing homologous recombination and regulation of self DNA recognition. In addition to ubiquitination (Seo et al 2018), acetylation was also shown to inhibit cGAS activation (Dai et al., 2019). How these post-translational modifications are orchestrated to regulate cGAS activity remains unknown. Finally, multiple functions were proposed for the N-terminal basic domain, for example in DNA binding (Tao et al., 2017), phase separation (Du and Chen, 2018), dimer stabilization (Hooy and Sohn, 2018) and plasma membrane binding (Barnett et al., 2019). Future studies to integrate these seemingly conflicting and disparate observations are necessary for a comprehensive understanding of cGAS mechanism.
AIM2
Absent-in-melanoma 2 (AIM2) is another cytoplasmic receptor for dsDNA, and is the founding member of the AIM2-like receptor (ALR) family. Unlike cGAS, however, AIM2 is dispensable for the induction of IFN response (Gray et al., 2016), but it is important for activation of another inflammatory signaling pathway mediated by inflammasomes (Lugrin and Martinon, 2018). The inflammasome is a multi-component macromolecular signaling complex that assembles upon a variety of inflammatory stimuli including foreign DNA. Assembly of the inflammasome activates caspase-1, which processes the pro-inflammatory cytokines (i.e. IL-1β or IL-18) and the pore forming protein gasdermin D (Hauenstein et al., 2015; Lugrin and Martinon, 2018; Morrone et al., 2015). The gasdermin D pore then secrets the mature IL-1β or IL-18 and causes an inflammatory cell-death known as pyroptosis. AIM2 was also reported to have an inflammasome-independent function in restricting tumorigenesis (Man et al., 2015; Wilson et al., 2015). In colorectal cancers, AIM2 was found to suppress the activity of DNA-PK and Akt, thereby restricting overt cell proliferation and cancer growth (Wilson et al., 2015).
AIM2 has an N-terminal PYD domain (pyrin) and C-terminal HIN domain (hematopoietic expression, interferon-inducible, and nuclear localization) (Fig. 4A), which are responsible for signaling and DNA binding, respectively. The HIN domain binds double-stranded DNA with little sequence dependence. The structure of AIM2 HIN in complex with DNA showed that AIM2 utilizes a large basic patch of the protein to contact the dsDNA backbone (Jin et al., 2012). The PYD domain is similar to CARD in that it forms homo-oligomers and also interacts with the downstream signaling adaptor ASC (apoptosis-associated speck-like protein containing a CARD) through a homotypic PYD-PYD interactions (Fig. 4B–C). Analogous to how RIG-I/MDA5 activates MAVS by nucleating the MAVS CARD filament, AIM2 also activates ASC by nucleating ASC PYD filaments, a necessary step in the inflammasome assembly (Lu et al., 2014; Morrone et al., 2015).
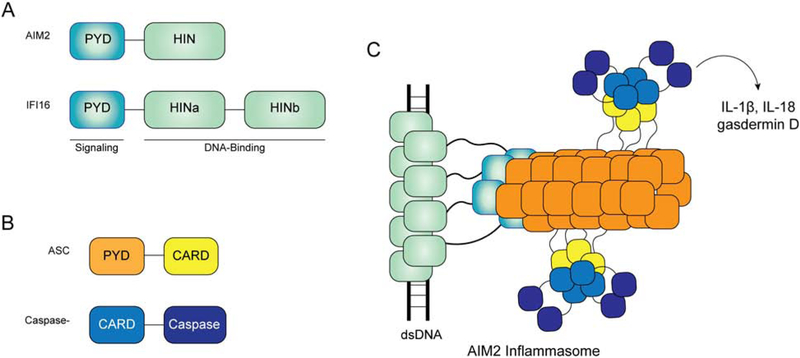
Figure 4.
Model of AIM-2-like receptor inflammasome assembly. A) AIM2 contains single PYD and HIN domains, while IFI16 contains one PYD but two HIN domains. B) domain architecture of the signaling adaptor ASC and effector caspase-1. Together with receptors in (A), ASC and caspase-1 constitute inflammasomes. C) The model of AIM2 inflammasome assembly. Upon dsDNA binding, the HIN domain of AIM2 coats the dsDNA, while the PYD forms an oligomer that seeds the filament formation of ASC PYD. ASC PYD filaments recruit the effector caspase-1 through CARD-CARD interactions and activates the caspase activity. The assembled inflammasome cleaves precursors of IL-1β, IL-18, and gasdermin D, leading to the maturation of the inflammatory cytokines and formation of the gasdermin D pore. IFI16 can also activate the inflammasome, likely though a similar mechanism as AIM2.
More detailed biochemical studies showed that both the DNA affinity of AIM2 and its inflammasome activity are dependent on dsDNA length, with the critical length of around 80 bp (Jin et al., 2012; Morrone et al., 2015). Similarly to RLRs and cGAS, AIM2 senses DNA length by assembling into filament-like structures along the length of dsDNA (Morrone et al., 2015) (Fig. 4C). Minimal oligomer assembly was found to require 6 protomers, while optimal oligomer assembly requires ~24 protomers (Morrone et al., 2015). As with cGAS, it is thought, while not proven, that this length-requirement contributes to the accuracy of self vs. non-self discrimination during normal physiological processes.
There are conflicting reports as to the protein domain requirement for AIM2 filament assembly. Lu et al (Lu et al., 2015) reported that the HIN domain alone can assemble into filaments. However, Morrone et al showed that HIN alone does not form ordered filaments, rather distributive ‘beads on a string’-like complexes on dsDNA (Morrone et al., 2015). Morrone et al suggested that PYD-PYD interactions drive filament formation and dsDNA binding. Intriguingly, at high protein concentration, AIM2 can form filaments in the absence of dsDNA. The dsDNA-free AIM2 filaments resemble brussel sprout-like structures, wherein the core-stems are formed by HIN domain filaments and multiple PYD clusters represent the ‘sprouts’ (Morrone et al., 2015). The assembly architecture of DNA-free AIM2 filament suggests that HIN domains do have an intrinsic affinity for each other. While the same filament architecture occurs in DNA-bound AIM2 filament remains unclear, these studies collectively support the model in which filament assembly is mediated by cooperative actions of HIN:dsDNA binding, HIN:HIN interaction and PYD:PYD interaction. The PYD oligomer then recruits the adaptor protein ASC through AIM2 PYD:ASC PYD interactions, nucleating the ASC filament formation (Fig. 4C). Finally, ASC recruits caspase-1 by CARD:CARD interactions to complete the inflammasome assembly (Cai et al., 2014; Lu et al., 2014; Morrone et al., 2015).
The mechanisms by which AIM2 utilizes multi-step filament assemblies for robust DNA detection and signal transduction parallels those of RLRs. Receptor filament formation allows detection of both the dsDNA structure and length, and couples the dsDNA-mediated filament formation to downstream signal activation. However, it still remains unclear how AIM2 is regulated in the absence of inflammatory stimuli. Some studies suggested an auto-inhibitory mechanism wherein the PYD domain is sequestered by the HIN domain (similar to the autoinhibition of CARDs for RIG-I) (Jin et al., 2012; Jin et al., 2013). On the other hand, others proposed that low basal concentration is sufficient to prevent spontaneous AIM2 oligomerization in the absence of DNA (Morrone et al., 2015). It is also unclear how AIM2 functions as a tumor suppressor in the inflammasome-independent manner (Man et al., 2015; Wilson et al., 2015) and if AIM2 oligomerization is also involved in this process. More detailed molecular mechanisms remain to be investigated.
IFI16
IFI16 is another DNA sensor in the AIM2-Like Receptor family. As with AIM2, IFI16 contains an N-terminal PYD domain and C-terminal HIN domains (Fig. 4A). Unlike AIM2, however, IFI16 contains nuclear localization signals and is found in both nucleus and cytosol, depending on the cell type and environmental stimuli, such as viral infection (Li et al., 2012; Veeranki and Choubey, 2012). IFI16 was shown to detect DNA from many DNA viruses, including HSV-1, HCMV, and KSHV (Ansari et al., 2015) (Kerur et al., 2011; Unterholzner et al., 2010). IFI16 also recognizes retroviruses (e.g. HIV-1) (Jakobsen et al., 2013; Monroe et al., 2014) and intracellular bacterial DNA (e.g. L. monocytogenes (Hansen et al., 2014; Unterholzner et al., 2010). Additionally, IFI16 was implicated in autoimmune diseases, such as amyotrophic lateral sclerosis (ALS), presumably through host DNA mis-recognition. More recently, nuclear DNA damage was shown to activate IFI16 and DNA damage response factors ATM and PARP-1, which together trigger NF-kB-mediated inflammatory response (Dunphy et al., 2018).
Multiple studies suggested that IFI16 detects viral DNA within the nucleus and promotes its transcriptional silencing (Johnson et al., 2014; Orzalli et al., 2013). IFI16 binding triggers recruitment of epigenetic silencing factors and other viral restriction factors, such as PML, Sp100 and ATRX, and suppresses RNA polymerase II recruitment (Merkl and Knipe, 2019; Merkl et al., 2018). However, IFI16 can also induce cytosolic innate immune signaling pathways. For example, IFI16 activates inflammasomes, presumably through a mechanism similar to that of AIM2 (Fig. 4C) (Kerur et al., 2011). IFI16 was reported to additionally stimulate a type I interferon signaling pathway through STING (Almine et al., 2017; Kerur et al., 2011; Orzalli et al., 2012; Unterholzner et al., 2010), although this activity appears independent of HIN domains and exclusively dependent on the PYD domain (Jonsson et al., 2017). Of note, the ability of IFI16 to activate STING or inflammasomes remains controversial, and detailed mechanisms of how nuclear IFI16 activates both remain unclear.
As with AIM2, IFI16 utilizes the HIN domains for sequence-independent dsDNA binding (Jin et al., 2012). Unlike AIM2, IFI16 contains two tandem HIN domains (HINa and HINb). Both HIN domains are required for IFI16 to bind DNA because individual HIN domains have low affinity for DNA (Jin et al., 2012; Unterholzner et al., 2010). While the footprint of individual HIN domain is ~ 16 bp, IFI16 prefers longer DNA (>100bp) and forms a filament along the length of DNA (Morrone et al., 2014). Single molecule analysis showed that a IFI16 molecule can track along exposed stretches of DNA to increase the chance of encountering other IFI16 molecules bound on the same DNA molecule (Stratmann et al., 2015). This leads to efficient filament nucleation through the PYD:PYD interaction (requiring ~10 IFI16 protomers), and provides a mechanism by which IFI16 measures dsDNA length (Stratmann et al., 2015).
It was proposed that this DNA length selectivity is the key to specific recognition of viral DNA against the background of chromatinized host genome in the nucleus (Stratmann et al., 2015). Consistent with this idea, IFI16 restricts gene expression from the naked transfected SV40 DNA, but not from infection of SV40 virus, which contains pre-chromatinized DNA in the virion (Orzalli et al., 2013). However, it remains unclear if the length of the accessible DNA region is sufficient to explain effective discrimination against the host DNA, given that sites of active transcription are highly accessible, and how these sites do not activate IFI16. Whether one-dimensional DNA scanning by IFI16 as a mechanism for filament nucleation allows detection of additional features in DNA beyond length, such as DNA modification or other DNA-bound factors, would be an interesting area of future research. Such analysis is required to gain more detailed insights into how IFI16 discriminates between self vs. non-self.
Concluding remarks and speculation
Here we described the mechanisms of receptor oligomerization by RLRs, ALRs, and cGAS and the role of oligomerization in ligand discrimination and signal activation. In all cases examined, receptor-ligand engagement is necessary, but insufficient for activating downstream immune functions. Receptor oligomerization, in particular filament or filament-like multimerization along the length of nucleic acids, whether through direct recruitment, translocation or one-dimensional diffusion, is additionally required. Filament-like assembly uniquely enables the innate immune receptors to detect nucleic acid structure, length and in some cases, nucleic acid integrity and modifications. In other words, filament-like assembly provides a mechanism to integrate signals from multiple disparate PAMPs that are dispersed across the body of long nucleic acids. However, differences among these receptors in their assembly and disassembly mechanisms endow them with different ligand specificities and diversify the strategies of viral detection. This diversification is likely the key to their collective function as a robust immune system that detects a broad range of foreign nucleic acids, either from pathogens or aberrant cellular processes.
Filament-like multimerization by nucleic acid sensors can be further modulated and augmented by other molecules in the host. These include RIPLET that clusters RIG-I filaments (Cadena et al., 2019), and HMGB and TFAM that bend DNA structure to promote cGAS nucleation (Andreeva et al., 2017b). The consequence of filament-like assemblies also varies, from acceleration of enzymatic activity (e.g. cGAS), proximity-induced oligomerization of signaling molecules (e.g. RLRs and ALRs) or avidity-dependent recruitment of further downstream effector molecules (i.e. RIPLET and MAVS). These various mechanisms for converting receptor oligomerization to downstream function highlights the versatility of filament-architecture as a signaling platform.
What are the outstanding questions? One common and important question remaining for most nucleic acid sensors is the identity of physiologically relevant ligands. So far, most studies have utilized model nucleic acids to examine the specificities of these receptors. As such, the studies to date have focused on the impact of duplex length. It is still unknown whether other features, for example complex secondary structures in single-stranded RNA/DNA, can activate these receptors, and whether recognition of these other structural elements is also mediated by filament-like assembly. In addition, further understanding of physiological ligands would help illuminate the mechanistic details of self vs non-self discrimination. While many studies utilize receptor pull-down to identify the RNA ligand, this strategy is often unsuitable for filament-like assemblies where multiple receptor proteins accumulate on a few molecules of nucleic acids (Hur, 2019). Oligomerization-specific methods, for example the one used in a recent study (Ahmad et al., 2018), are required for addressing this important question of ligand identity. With such knowledge, we would gain more comprehensive understanding of receptor multimerization in nucleic acid sensing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarreberg LD, Esser-Nobis K, Driscoll C, Shuvarikov A, Roby JA, and Gale M Jr. (2019). Interleukin-1beta Induces mtDNA Release to Activate Innate Immune Signaling via cGAS-STING. Mol Cell 74, 801–815 e806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, and Chen ZJ (2019). cGAS in action: Expanding roles in immunity and inflammation. Science 363. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, and Hornung V (2013). cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, et al. (2017). Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2, 17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, and Hur S (2018). Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, and Barber GN (2014). Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol 31, 121–126. [DOI] [PubMed] [Google Scholar]
- Almine JF, O’Hare CA, Dunphy G, Haga IR, Naik RJ, Atrih A, Connolly DJ, Taylor J, Kelsall IR, Bowie AG, et al. (2017). IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat Commun 8, 14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva L, Hiller B, Kostrewa D, Lassig C, de Oliveira Mann CC, Jan Drexler D, Maiser A, Gaidt M, Leonhardt H, Hornung V, et al. (2017a). cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398. [DOI] [PubMed] [Google Scholar]
- Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Jan Drexler D, Maiser A, Gaidt M, Leonhardt H, Hornung V, et al. (2017b). cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398. [DOI] [PubMed] [Google Scholar]
- Ansari MA, Dutta S, Veettil MV, Dutta D, Iqbal J, Kumar B, Roy A, Chikoti L, Singh VV, and Chandran B (2015). Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-beta Responses. PLoS Pathog 11, e1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, and Kagan JC (2019). Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction between Self- and Viral DNA. Cell 176, 1432–1446 e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, and Bartenschlager R (2011). Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I). J Biol Chem 286, 27278–27287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park JW, Oh SW, Fujita T, Hou F, Binder M, et al. (2019). Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 177, 1187–1200.e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, and Chen ZJ (2014). Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi S, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al. (in press). N6-methyladenosine modification controls circular RNA immunity. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, and Chang HY (2017). Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JJ, Sparrer KMJ, van Gent M, Lassig C, Huang T, Osterrieder N, Hopfner KP, and Gack MU (2018). Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol 19, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, et al. (2018). Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 172, 811–824 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, and Hopfner KP (2013). Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Huang YJ, He X, Zhao M, Wang X, Liu ZS, Xue W, Cai H, Zhan XY, Huang SY, et al. (2019). Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell 176, 1447–1460 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro Duany Y, Wu B, and Hur S (2015). MDA5-filament, dynamics and disease. Curr Opin Virol 12, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarkar SC, Schweibenz B, Wang C, Marcotrigiano J, and Patel SS (2018). RIG-I Uses an ATPase-Powered Translocation-Throttling Mechanism for Kinetic Proofreading of RNAs and Oligomerization. Mol Cell 72, 355–368 e354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, and Yan N (2015). STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe 18, 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, and Chen ZJ (2018). DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, Jakobsen MR, Nevels MM, Bowie AG, and Unterholzner L (2018). Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-kappaB Signaling after Nuclear DNA Damage. Mol Cell 71, 745–760 e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun SL, Fernandez D, Weiss TM, and Li L (2019). STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell 178, 290–301. e10. [DOI] [PubMed] [Google Scholar]
- Ferreira CR, Crow YJ, Gahl WA, Gardner PJ, Goldbach-Mansky R, Hur S, de Jesus AA, Nehrebecky M, Park JW, and Briggs TA (2019). DDX58 and Classic Singleton-Merten Syndrome. J Clin Immunol 39, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. (2007). TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920. [DOI] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, O’Duill F, Schmid-Burgk JL, Hoss F, Buhmann R, et al. (2017). The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell 171, 1110–1124.e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, and Chen ZJ (2015). Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A 112, E5699–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. (2013). Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck S, and Ablasser A (2019). Innate immunosensing of DNA in cellular senescence. Curr Opin Immunol 56, 31–36. [DOI] [PubMed] [Google Scholar]
- Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, and Stetson DB (2016). The AIM2-like Receptors Are Dispensable for the Interferon Response to Intracellular DNA. Immunity 45, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, et al. (2014). Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J 33, 1654–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, and Greenberg RA (2017). Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenstein AV, Zhang L, and Wu H (2015). The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr Opin Struct Biol 31, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, and Bevilacqua PC (2009). RNA dimerization promotes PKR dimerization and activation. J Mol Biol 390, 319–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooy RM, and Sohn J (2018). The allosteric activation of cGAS underpins its dynamic signaling landscape. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. (2006). 5’-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang QX, and Chen ZJ (2011). MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S (2019). Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu Rev Immunol 37, 349–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, and Barber GN (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, Laustsen A, Hansen K, Ostergaard L, Fitzgerald KA, et al. (2013). IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A 110, E4571–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M-A, Kim EK, Now H, Nguyen NTH, Kim W-J, Yoo J-Y, Lee JJ, Jeong Y-M, Kim C-H, Kim O-H, et al. (2015). Mutations in DDX58, which Encodes RIG-I, Cause Atypical Singleton-Merten Syndrome. Am J Hum Genet 96, 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, and Marcotrigiano J (2011). Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, and Chen ZJ (2012). Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. (2012). Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Smith P, Jiang J, and Xiao TS (2013). Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem 288, 13225–13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, and Chandran B (2014). IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog 10, e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson KL, Laustsen A, Krapp C, Skipper KA, Thavachelvam K, Hotter D, Egedal JH, Kjolby M, Mohammadi P, Prabakaran T, et al. (2017). IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat Commun 8, 14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G, and Stoye JP (2016). Immune responses to endogenous retroelements: Taking the bad with the good. Nat Rev Immunol 16, 207–219. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, and Akira S (2008). Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205, 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, and Chandran B (2011). IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, and Cusack S (2011). Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147, 423–435. [DOI] [PubMed] [Google Scholar]
- Lang X, Tang T, Jin T, Ding C, Zhou R, and Jiang W (2017). TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med 214, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Diner BA, Chen J, and Cristea IM (2012). Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A 109, 10558–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, and Li P (2013). Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan MM, Dickey TH, Molinari ES, Fitzgerald ME, Potapova O, Iwasaki A, and Pyle AM (2018). A minimal RNA ligand for potent RIG-I activation in living mice. Sci Adv 4, e1701854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, Jiang Y, Fei Y, Zhu C, Tan R, et al. (2018). Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136. [DOI] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. (2008). Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman E, and Wu H (2015). Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, and Egelman EH (2014). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, and Li P (2010). The structural basis of 5’ Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain. Structure 18, 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke S, Holleufer A, Christensen MH, Jønsson KL, Boni GA, Sørensen LK, Johannsen M, Jakobsen MR, Hartmann R, and Paludan SR (2017). cGAS is activated by DNA in a length-dependent manner. EMBO Rep 18, 1707–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin J, and Martinon F (2018). The AIM2 inflammasome: Sensor of pathogens and cellular perturbations. Immunol Rev 281, 99–114. [DOI] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, and Pyle AM (2011). Structural insights into RNA recognition by RIG-I. Cell 147, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Lettice L, Tarnauskaite Z, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, et al. (2016). Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35, 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, et al. (2015). Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 162, 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, and Knipe DM (2019). Role for a Filamentous Nuclear Assembly of IFI16, DNA, and Host Factors in Restriction of Herpesviral Infection. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, Orzalli MH, and Knipe DM (2018). Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, and Greene WC (2014). IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, and Sohn J (2015). Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun 6, 7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, and Sohn J (2014). Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A 111, E62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner K-P, and Ha T (2009). Cytosolic Viral Sensor RIG-I Is a 5′-Triphosphate–Dependent Translocase on Double-Stranded RNA. Science 323, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, and Knipe DM (2013). Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A 110, E4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, and Knipe DM (2012). Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109, E3008–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JR, Jain A, Chou YY, Baum A, Ha T, and Garcia-Sastre A (2013). ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep 14, 780–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Jo M, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, and Hur S (2012). Kinetic Mechanism for Viral dsRNA Length Discrimination by MDA5 Filament. Proc Natl Acad Sci U S A 109, E3340–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Bin W, Orme-Johnson M, Liu M, Walz T, and Hur S (2011). Cooperative Assembly and Dynamic Disassembly of MDA5 Filaments for Viral dsRNA Recognition. Proc Natl Acad Sci U S A 108, 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, and Hur S (2014a). Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Xu H, Chen ZJ, and Hur S (2014b). Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Wu B, Yao H, Walz T, and Hur S (2013). RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell 51, 573–583. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schultz DE, Tan CP, Naslund TI, Liljestrom P, Weber F, and Reis e Sousa C (2006). RIG-I–Mediated Antiviral Responses to Single-Stranded RNA Bearing 5’-Phosphates. Science 314, 997–1001. [DOI] [PubMed] [Google Scholar]
- Rawling DC, Fitzgerald ME, and Pyle AM (2015). Establishing the role of ATP for the function of the RIG-I innate immune sensor. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Linehan MM, Iwasaki A, and Pyle AM (2019). RIG-I Selectively Discriminates against 5’-Monophosphate RNA. Cell Rep 26, 2019–2027 e2014. [DOI] [PubMed] [Google Scholar]
- Rice GI, del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, et al. (2014). Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, and Gale M Jr. (2008). Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. (2009). Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. (2014). Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, and Jung JU (2018). TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun 9, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhang C, Chen ZJ, Bai XC, and Zhang X (2019). Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle L, Garcin D, and Kolakofsky D (2006). Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351, 101–111. [DOI] [PubMed] [Google Scholar]
- Stratmann SA, Morrone SR, van Oijen AM, and Sohn J (2015). The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex. Elife 4, e11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Zhang XW, Jin J, Du XX, Lian T, Yang J, Zhou X, Jiang Z, and Su XD (2017). Nonspecific DNA Binding of cGAS N Terminus Promotes cGAS Activation. J Immunol 198, 3627–3636. [DOI] [PubMed] [Google Scholar]
- Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, Macia A, Crow YJ, and Muotri AR (2017). Modeling of TREX1-Dependent Autoimmune Disease using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell Stem Cell 21, 319–331 e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. (2010). IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11, 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzri D, and Gehrke L (2009). Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol 83, 4174–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranki S, and Choubey D (2012). Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol Immunol 49, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. (2010). Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. (2015). Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, et al. (2015). Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med 21, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, and Hur S (2015). How RIG-I like receptors activate MAVS. Curr Opin Virol 12, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, and Hur S (2013). Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289. [DOI] [PubMed] [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, and Hur S (2014). Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell 55, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Mercado-Lopez X, Grier JT, Kim WK, Chun LF, Irvine EB, Del Toro Duany Y, Kell A, Hur S, Gale M Jr., et al. (2015). Identification of a Natural Viral RNA Motif That Optimizes Sensing of Viral RNA by RIG-I. MBio 6, e01265–01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, and Fujita T (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5, 730–737. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Hou F, Adhikari A, Xu M, and Chen ZJ (2010). Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shang G, Gui X, Zhang X, Bai XC, and Chen ZJ (2019). Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam CA, and Chen ZJ (2014). The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep 6, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, Gray NS, Mekalanos JJ, and Kranzusch PJ (2018). Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 174, 300–311.e311. [DOI] [PMC free article] [PubMed] [Google Scholar]