Abstract
Staphylococcus aureus is a healthcare-associated pathogen that can harbour multiple antimicrobial resistance determinants and express multiple virulence factors e.g. Panton-Valentine Leukocidin (PVL). Unknown staphylococcal cassette chromosome mec (SCCmec) typing patterns were previously observed among 11% (n = 52) of methicillin-resistant S. aureus (MRSA) isolates; we further investigated these as well as the proportion of PVL, encoded by lukS/F-PV, in 761 S. aureus isolates from patients with a diagnosis of pneumonia/lower respiratory tract, skin/soft tissue, bone and joint infection. S. aureus isolates from blood culture were identified and antimicrobial susceptibility testing was performed using automated systems. Conventional PCR assays were used to identify the ccr and mec gene complexes in mecA-positive isolates with an unknown SCCmec type and screen for lukS/F-PV. Epidemiological data was used to classify isolates as healthcare- or community-associated infections. Antimicrobial susceptibility profiles according to SCCmec type and PVL were reported. Of the unknown SCCmec types, isolates were interpreted as type I-like (86%, 38/44), type II-like (9%, 4/44) and type III-like (5%, 2/44). Eight isolates did not produce definitive results. Of all MRSA isolates, majority were multidrug-resistant as indicated by their non-susceptibility to most antimicrobial agents; 92% were healthcare-associated. PVL was seen in 14% of the isolates (MRSA: 25%, MSSA: 75%); 56% were classified as healthcare-associated infection. The SCCmec typing method did not definitively classify all unknown isolates into clearly defined types. It showed that majority of these isolates were not the conventional types; untypeable elements appeared to be composite SCCmec elements, consisting of multiple ccr gene complexes. Majority of the MRSA isolates were non-susceptible to most antibiotics indicating that multiple resistance genes are present in our population. Furthermore, the proportion of PVL was low and more prevalent in MSSA.
Introduction
Staphylococcus aureus is a leading cause of community- and healthcare-associated infections. It is a pathogenic organism capable of causing a range of infections. This is due to its ability to express multiple virulence factors and harbour multiple antimicrobial resistance determinants [1, 2]. The Panton-Valentine Leukocidin (PVL) exotoxin is a pore-forming cytotoxic exoprotein encoded by the bicomponent lukS-PV and lukF-PV genes, which are carried on a bacteriophage [3]. PVL targets cells of the human immune system and causes cell death. Isolates of S. aureus harbouring PVL are often due to community-associated infections [4] and have been linked to more severe clinical manifestations such as necrotising pneumonia, severe bone and joint infections and skin and soft tissue infections often requiring surgical drainage [4, 5].
The PVL exotoxin is not produced by all S. aureus strains, but have been detected in both methicillin-susceptible S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) strains [3]. The clinical presentation of PVL-positive MSSA and MRSA is very similar. It has also been shown that sub-inhibitory concentrations of beta-lactams increases PVL toxin production in vitro and that other antimicrobials that inhibit protein synthesis (e.g. rifampicin, clindamycin and linezolid) should be considered during treatment [6].
Methicillin-resistance is harboured on a mobile genetic element called the staphylococcal chromosome cassette mec (SCCmec). This element is genetically diverse and many types, subtypes and variants have been reported [7]. The cassette consists of three main structural components, namely: i) the cassette chromosome recombinase (ccr) gene complex, ii) the mec gene complex and iii) the joining (J) regions [8, 9]. The ccr gene complex encodes for site-specific recombinases that are responsible for the excision and integration of the SCCmec element into the staphylococcal chromosome [2, 7, 9]. The ccr gene complex provides the SCCmec element with mobility and consequently facilitates its transfer to other staphylococcal species [9]. The mec complex is responsible for conferring methicillin resistance. The mec gene complex consists of: i) a mecA/C gene, ii) its regulatory genes, the mecR1 and the mecI genes encoding for a signal transducer protein and repressor protein respectively and iii) various insertion sequences [7, 10]. A combination of the ccr gene complex with the mec gene class is used to assign the specific type of SCCmec element. Currently, thirteen SCCmec types (I-XIII) based on complete sequence data have been defined in MRSA [2, 11–13]; International Working Group on the Staphylococcal Cassette Chromosome elements (IWG-SCC) (2015) Available online: http://www.sccmec.org).
Typing of the SCCmec element is of epidemilogical importance to understand the evolution of MRSA. There are several methods (i.e. restriction enzyme digestion, PCR assays) available to determine the SCCmec type. The International Working Group for SCCmec elements currently recommends the method by Kondo and colleagues (2007) [12]. The only drawback of this assay is that it consists of six multiplex-PCR assays. However, the first two assays are generally sufficient to classify the majority of SCCmec elements [9]. The assay by Kondo and colleagues (2007) can easily detect novel ccr and mec gene complex combinations. [2, 12].
Healthcare-associated MRSA infections have previously been associated with SCCmec types I, II or III whereas community-acquired MRSA infections are linked to smaller SCCmec types IV, V, VI or VII [14] but epidemiological data is required to make this conclusion. A more recent report however, states that the traditional classification of healthcare- and community-associated MRSA is no longer appropriate because there is a notable overlap of identical clones between these two groups [15]. Previous work characterised the isolates in this study to determine the circulating SCCmec types [16]. However, a number of isolates produced an unknown SCCmec type as indicated by unidentified banding patterns. This study aimed to resolve unidentified SCCmec types using an alternative method. In addition, selected S. aureus isolates have been screened for the presence of the lukS/F-PV gene. Epidemiological data was used to classify isolates as healthcare- and community-associated and specific antimicrobial susceptibility profiles according to SCCmec type and PVL were reported.
Methods
Case definition
A case of S. aureus bacteraemia was defined as the isolation of S. aureus from a blood culture. Isolates formed part of the GERMS-SA enhanced antimicrobial resistance surveillance study from January 2013 to January 2016. S. aureus isolates were from five sentinel centres in South Africa and sites represented six large academic hospitals from the Gauteng and the Western Cape provinces. A 21-day exclusion period was applied to avoid duplicate isolates of the organism from the same patient. In addition, the following clinical and epidemiological information were collected for each case: date of admission, previous hospitalisation or treatment and diagnosis. This information was used to define healthcare- and community-associated S. aureus infections. A case of community-associated bacteraemia was defined as a patient with S. aureus isolated from a blood culture specimen ≤ 48 hours of admission to a hospital and no contact within one year prior to the current episode of S. aureus infection with a healthcare facility (including prior surgery, dialysis and admission to a long term care facility). Healthcare-associated S. aureus infection was defined as a patient with S. aureus isolated >48 hours after admission or with any prior healthcare contact. The patient’s clinical diagnosis was used to select isolates for lukS/F-PV (i.e. PVL) screening.
Bacterial isolates and phenotypic assays
Isolates were submitted on Dorset transport media (Diagnostic Media Products, National Health Laboratory Service, South Africa) to the National Institute for Communicable Diseases (NICD), Johannesburg, South Africa. Each isolate was plated onto a 5% blood agar plate (Diagnostic Media Products, National Health Laboratory Service, South Africa) followed by organism identification and antimicrobial susceptibility testing using automated systems [(VITEK II system (bioMèrieux, Marcy-l'Etoile, France)/MALDI-TOF MS (Microflex, Bruker Daltonics, MA, USA) and MicroScan Walkaway system (Siemens, Sacramento, CA, USA), (Gram-positive panel PM33)], respectively. An isolate was phenotypically non-susceptible if it had an oxacillin MIC of >2 and a positive cefoxitin screening result. Interpretation of susceptibility was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [17].
Genotypic assays
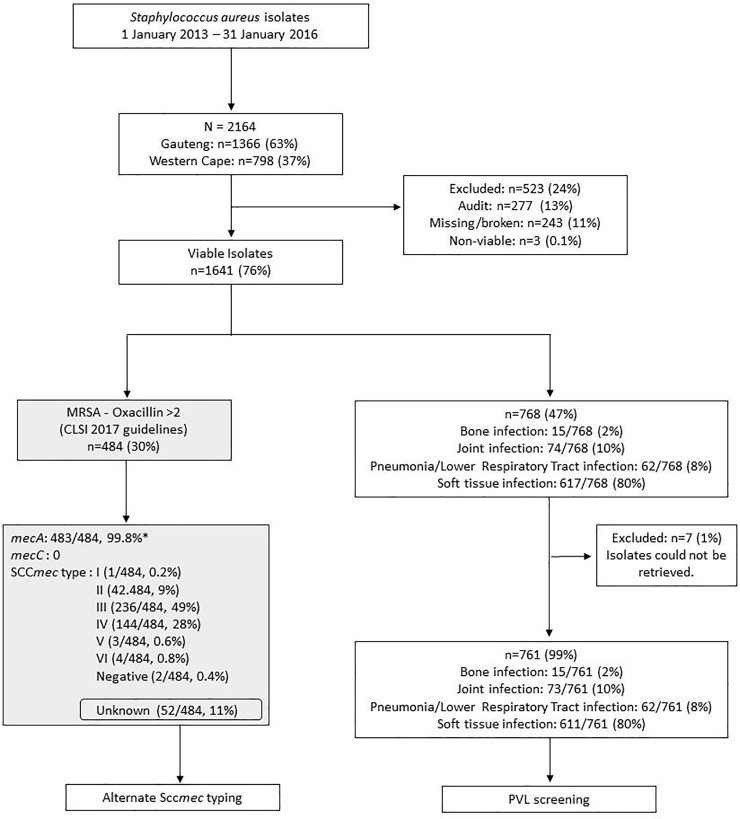
Total genomic DNA was extracted using a crude boiling method and used in the genotypic assays. Fig 1 shows how isolates were selected for the genotypic assays. Prior work involved the screening for methicillin-resistance determinants, mecA and mecC on 484 phenotypically confirmed MRSA isolates. These isolates were typed by multiplex PCR to determine the circulating SCCmec types I to VI as described previously [18].
Fig 1. Flowchart showing sample selection.
The grey boxes show results that have been published previously [18]. Audit cases were defined as those cases that were identified according to the public healthcare sector Corporate Data Warehouse (CDW) (which houses records of patient details and laboratory results) but not received for processing in the laboratory. * One isolate did not contain mecA or mecC and did not harbour a SCCmec element although it was phenotypically non-susceptible (oxacillin >2, cefoxitin screen positive). This is possible as explained previously [19] possibly due to excision of the cassette (SCCmec) and/or mecA/C gene drop-out.
SCCmec typing of unidentified banding patterns
An alternate SCCmec typing method was carried out by performing two conventional multiplex-PCR assays (i.e. ccr gene complex multiplex PCR assay detecting ccr gene complex 1 to 5 and the mec gene complex multiplex PCR assay detecting mec class A, B and C) using the G-Storm thermal cycler (Somerton Biotechnology Centre, Somerton, UK), the Qiagen Multiplex PCR kit (Qiagen, Nordrhein-Westfalen, Germany) and previously published primers [12]. The results of the ccr gene complex and the mec gene complex multiplex PCR assays were subsequently combined to assign a specific SCCmec type.
PCR screening for the lukS/F-PV gene
The presence of the lukS/F-PV gene was determined in MRSA and MSSA isolates from patients with a diagnosis of pneumonia or a lower respiratory tract infection, skin and soft tissue infection, bone infection and joint infection. Isolates were screened by a conventional PCR assay using the G-Storm thermal cycler, the Qiagen Multiplex PCR kit and previously published primers [20].
Statistical analysis
Descriptive analyses were performed using Stata version 14 (StataCorp LP, College Station, Texas, USA). Categorical data were summarised using absolute frequencies and percentages.
Ethical approval
Ethical clearance was obtained from the University of the Witwatersrand Human Research Committee (Protocol number M10464).
Results
SCCmec typing of unidentified banding patterns
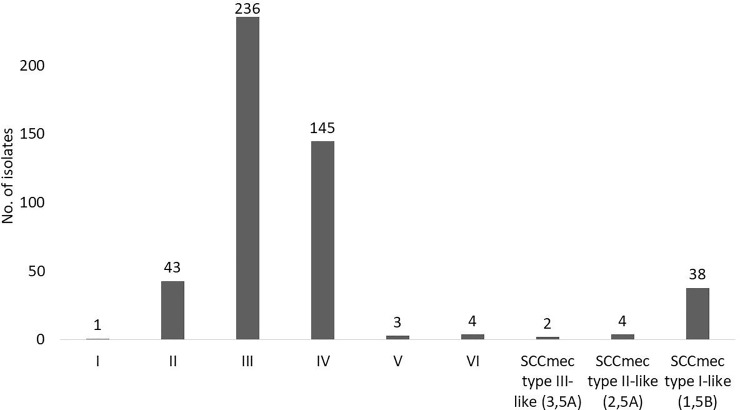
Only 51 isolates were tested with the ccr and the mec gene complex multiplex PCR assays, since a single isolate could not be retrieved from storage. A definite SCCmec type could not be determined for a three isolates, since all three of these isolates harboured two ccr gene complexes (ccr gene complex 1 and ccr gene complex 5) but was negative for all mec gene classes tested. A single isolate produced an indeterminate result and harboured either SCCmec type V or VII, since the assay by Kondo et al., 2007 [12] cannot distinguish between mec gene complex class C1 and C2. A further 44 isolates produced evidence of composite SCCmec elements as they harboured two ccr gene complexes and a single mec gene complex. These isolates were further interpreted as SCCmec type I-like (1B plus an additional ccr gene complex class 5) (86%, 38/44), SCCmec type II-like (2A plus an additional ccr gene complex class 5) (9%, 4/44) and SCCmec type III-like (3A plus an additional ccr gene complex class 5) (5%, 2/44). The SCCmec element type could clearly be assigned for two isolates only based on the ccr and mec gene complex combinations; these were SCCmec type II and IV. A new SCCmec combination (ccr gene complex class 5 and mec gene complex class A) was detected in a single isolate. Conclusive SCCmec types making use of both typing methods are seen in Fig 2.
Fig 2. Final SCCmec type distribution among MRSA isolates 2013–2016.
A total of 476 isolates are represented in this figure. One isolate could not be retrieved for testing, two isolates produced no SCCmec type, the mec class could not be identified for three isolates, one isolate could not be differentiated between types V or VII, and one isolate produced a new combination.
Epidemiological data were available for 463 of the 484 isolates that could be classified into healthcare- and community-associated S. aureus infection. Majority of these were healthcare-associated infections (92%, 428/463) followed by community-associated infections (8%, 35/463). Table 1 shows a breakdown of the SCCmec element types classified into healthcare- and community-associated infections.
Table 1. Final SCCmec type distribution among MRSA isolates from 2013 to 2016 and its association between origin of the infection (i.e. healthcare- or community associated).
| Healthcare-associated n/463 (%) | Community-associated n/463 (%) |
|
|---|---|---|
| SCCmec type I | 0 | 1 (0.2) |
| SCCmec type II | 43 (9) | 0 |
| SCCmec type III | 218 (47) | 9 (2) |
| SCCmec type IV | 116 (25) | 19 (4) |
| SCCmec type V | 2 (0.4) | 0 |
| SCCmec type VI | 3 (0.7) | 1 (0.2) |
| SCCmec type V or VII | 1 (0.2) | 0 |
| SCCmec type III-like | 2 (0.4) | 0 |
| SCCmec type II-like | 4 (0.9) | 0 |
| SCCmec type I-like | 33 (7) | 4 (0.9) |
| New combination | 1 (0.2) | 0 |
A single isolate could not be retrieved for testing, three isolates in which the mec gene class could not be detected were classified as healthcare-associated (n = 2) and community-associated (n = 1) and two isolates that produced no SCCmec banding pattern were classified as healthcare-associated.
Antimicrobial susceptibility testing and SCCmec types
Table 2 shows AST data classified by the predominant SCCmec element types. Majority of the isolates were multidrug-resistant as indicated by their non-susceptibility to most of the antimicrobial agents. The p-value was calculated between susceptible (S) and the non-susceptible (I and R) isolates collectively and was significant for all antimicrobial agents.
Table 2. Susceptibility of antimicrobial agents in MRSA isolates.
| Number of isolates for which MIC value was: | |||||
|---|---|---|---|---|---|
| Antimicrobial agents MIC range (μg/mL) | SCCmec type II (N = 43) | SCCmec type III (N = 236) | SCCmec type IV (N = 145) | SCCmec type I-like (1,5B) (N = 38) | p-value* |
| Ciprofloxacin | |||||
| ≤ 1 (S) | - | 3 | 13 | 32 | <0.001 |
| 2 (I) | - | - | 1 | 2 | |
| >2 (I) | 43 | 233 | 131 | 4 | |
| ≥4 (R) | - | - | - | - | |
| Clindamycin | |||||
| ≤0.5 (S) | 4 | 212 | 139 | 36 | <0.001 |
| 1–2 (I) | - | 3 | - | - | |
| >2 (I) | 39 | 21 | 6 | 2 | |
| ≥4 (R) | - | - | - | - | |
| Erythromycin | |||||
| ≤0.5 (S) | - | 1 | 61 | - | <0.001 |
| 1-4(I) | - | - | 5 | 1 | |
| >4 (I) | 43 | 233 | 79 | 37 | |
| ≥8 (R) | - | 1 | - | - | |
| Gentamicin | |||||
| ≤4 (S) | 35 | 3 | 31 | 3 | <0.001 |
| 8 (I) | 1 | - | 4 | 8 | |
| >8 (I) | 7 | 232 | 110 | - | |
| ≥16 (R) | - | - | - | - | |
| Rifampicin | |||||
| ≤ 1 (S) | 39 | 219 | 32 | 29 | <0.001 |
| 2 (I) | - | 3 | - | - | |
| >2 (I) | 3 | 14 | 112 | 1 | |
| ≥4 (R) | - | - | 1 | - | |
| Tetracycline | |||||
| ≤4 (S) | 39 | 6 | 31 | 34 | <0.001 |
| 8 (I) | 1 | 1 | - | 2 | |
| >8 (I) | 3 | 228 | 114 | 2 | |
| ≥16 (R) | - | - | - | - | |
| Trimethoprim/sulfamethoxazole | |||||
| ≤2/38 (S) | 38 | 9 | 28 | 37 | <0.001 |
| ≥4/76 | 4 | 225 | 106 | 1 | |
MIC: Minimum inhibitory concentration.
Interpretation of results was done according to CLSI guidelines [17]; S: susceptible, I: Intermediate resistance, R: Resistant.
*The p-value was calculated between susceptible (S) and the non-susceptible (I and R) isolates collectively.
Multidrug resistance was defined as non-susceptibility to at least one antimicrobial agents in three or more categories [21].
PCR screening for the lukS/F-PV gene
Of a total of 1641 isolates, 768 (47%) isolates were selected for screening for the lukS/F-PV gene based on the type of infection. Isolates from patients diagnosed with skin and soft tissue infection (80%, 617/768), pneumonia/lower respiratory tract infection (8%, 62/768), bone infection (2%, 15/768) or joint infection (10%, 74/768) were selected.
Seven isolates could not be retrieved and were therefore not tested for the presence of lukS/F-PV. The presence of the PVL toxin was seen in 14% (108/761) of the isolates tested; 25% (27/108) in MRSA and 75% (81/108) in MSSA. Majority of PVL-positive S. aureus cases were from the Gauteng (75%, 81/108) followed by the Western Cape province (25%, 27/108). Majority of these infections were from patients with skin and soft tissue infection (70%, 75/108), followed by pneumonia/lower respiratory tract infection (21%, 23/108), joint infection (8%, 9/108) and bone infection (1%, 1/108) (p<0.0001).
Eleven (41%) and 13 (48%) of 27 MRSA isolates that were PVL-positive were associated with SCCmec types III and IV respectively with just one isolate each for types I, V and VI (Table 3).
Table 3. Presence of SCCmec types and prevalence of the Panton-Valentine Leukocidin gene in MRSA isolates (n = 27).
| SCCmec type | Type of infection | PVL-positive strains (n, %) |
|---|---|---|
| II | Skin and soft tissue infection | 1 (4) |
| III | Skin and soft tissue infection | 8 (30) |
| Joint Infection | 2 (7) | |
| Lower respiratory tract infection | 1 (4) | |
| IV | Skin and soft tissue infection | 8 (30) |
| Joint Infection | 1 (4) | |
| Bone Infection | 1 (4) | |
| Lower respiratory tract infection | 3 (10) | |
| V | Joint Infection | 1 (4) |
| VI | Joint Infection | 1 (4) |
Table 4 shows the antimicrobial susceptibility profiles for PVL-positive S. aureus isolates according to patient’s diagnosis.
Table 4. Antimicrobial susceptibility profiles for PVL-positive S. aureus isolates according to patient’s diagnosis.
| Antimicrobial agents MIC range (μg/mL) | Patient’s diagnosis | |||
|---|---|---|---|---|
| Skin and soft-tissue infections n = 75 |
Pneumonia / Lower respiratory tract infection n = 23 |
Bone infection n = 1 |
Joint infection n = 9 |
|
| Oxacillin resistant /cefoxitin screen positive | 17 | 4 | 1 | 5 |
| Trimethoprim/sulfamethoxazole | ||||
| ≤2/38 (S) | 57 | |||
| >2.38 (R) | 1 | |||
| 4/76 (R) | 2 | |||
| >4/76 (R) | 15 | |||
| Clindamycin | ||||
| ≤0.25 (S) | 23 | 1 | 8 | |
| 0.5 (S) | 0 | 0 | 1 | |
| 1 (R) | 0 | 0 | 0 | |
| >2 (R) | 0 | 0 | 0 | |
| Daptomycin | ||||
| ≤1 (S) | 1 | 9 | ||
| Linezolid | ||||
| ≤1 (S) | 15 | 0 | 3 | |
| ≤2(S) | 0 | 0 | ||
| 2 (S) | 8 | 1 | 6 | |
| Rifampicin | ||||
| ≤0.5 (S) | 19 | |||
| ≤1 (S) | 1 | |||
| 2 (I) | 3 | |||
| >2 (R) | 0 | |||
S–Susceptible, I–Intermediate resistance, R–Resistant
Shaded grey boxes indicate that the antimicrobial agents is not the recommended treatment of choice.
Epidemiological data for five isolates were missing. Of 103 PVL-positive isolates, 56% (58/103) were classified as healthcare-associated S. aureus infection followed by 44% (45/103) that were community-associated infection.
Discussion
In the current study, MRSA accounted for 30% of the total staphylococcal bacteraemia infections. SCCmec element typing was performed on these MRSA isolates, but 11% could not be typed by the multiplex PCR assay described by Milheiriço and colleagues (2007) [22]. An alternate SCCmec typing method was employed to further classify untypeable SCCmec elements based on ccr and mec gene complex combinations. In addition, the association between antimicrobial susceptibility profiles, specific SCCmec types, the epidemiological point of acquisition (i.e. healthcare-associated vs. community-acquired infection) and the presence of the PVL toxin were reported.
The study showed that the majority of untypeable isolates harboured more than one ccr gene complex. This indicates the possibility of composite SCCmec elements circulating in MRSA strains in South Africa. Sequencing of the complete element would be required to determine if these elements represent a single SCCmec element with two ccr gene complexes or if the element consists of two separate integrated SCCmec elements [23]. It is also important to note that the method published by Kondo and colleagues (2007) [12] possesses some limitations. The ccr gene complex multiplex-PCR assay cannot detect ccr gene complex 6 to 9, hence SCCmec type X (ccr gene complex 7), SCCmec type XI (ccr gene complex 8), SCCmec XII (ccr gene complex 9) and SCCmec type XIII (ccr gene complex 9) would not have been detected. Furthermore, the mec gene complex multiplex-PCR assay cannot distinguish between mec class C1 and C2 and only detects the mec gene complex C resulting in not being able to distinguish between SCCmec type V (mec gene complex C2) and SCCmec type VII (mec gene complex C1). This assay also does not detect the mec gene complex E and therefore SCCmec type XI (mec gene complex E) would not be detected.
While this method did not definitively classify all the unknown isolates into clearly defined types, it showed that majority of these isolates were not the conventional SCCmec types. It should be noted that only two of the six multiplex-PCR assays were performed; however the first two assays are generally sufficient to classify the majority of SCCmec elements [9]. For this reason we performed only the two PCR assays. Types I, II and III are thought to be related to healthcare-associated infections and types IV, V and VI with community-associated infections [14]; this was the case for a large number of the isolates except for 116 SCCmec type IV isolates which were classified as healthcare-associated. A further few did not strictly behave as traditionally expected. A more recent report however, states that the traditional classification of healthcare-associated and community-associated MRSA is no longer appropriate because there is a notable overlap of identical clones between these two groups. This may confirm the limitation in defining healthcare-associated and community-associated infections based on SCCmec types and that traditionally considered community-associated isolates have the potential to establish themselves as healthcare-associated pathogens and vice versa. [15].
The results obtained demonstrate that majority of the S. aureus isolates displaying resistance to methicillin were non-susceptible to ciprofloxacin, erythromycin, gentamicin, rifampicin, tetracycline and trimethoprim/sulfamethoxazole while susceptible to clindamycin only, indicating that multiple resistance genes are present (Table 2). A previous 2010 Swiss study [24] showed some differences in their findings although the sample number was small (N = 78); 60% were resistant to ciprofloxacin compared to 90% in our findings, 33% were resistant to erythromycin compared to 87% in our study and 18% were resistant to gentamycin as compared to 83% in our study. Rifampicin and trimethoprim/sulfamethoxazole were resistant in none of the isolates from the Swiss study; however our findings showed that 30% and 75% were resistant respectively. Clindamycin displayed 15% resistance in our study and this was comparable to the Swiss study where 16% of the isolates were resistant. As in our study, erythromycin resistance was also quite high in 91.6% (n = 564) of the isolates in another study [25] but comparable to the Swiss study in another study (32.8%) [26]. This latter study looked at nasal colonisation with MRSA from the nasal secretion from children in day-care centres. Resistance to ampicillin was high (80%) but resistance to ciprofloxacin, clindamycin and tetracycline was low (7.1%, 7.1% and 4.3% respectively) [26]. The differences in the results between studies may be attributed to prescription practices which vary between hospitals and geographical locations.
A small American study (N = 60) showed that overall the healthcare-associated SCCmec type II isolates were resistant to more antimicrobials than the community-associated type IV isolates [27]. This was also seen in another small (N = 65) Polish study in 2013 [28]. The opposite was true in our study where type IV isolates were resistant to more antibiotics than the type II isolates. It should however be noted that in our study 116 SCCmec type IV isolates were classified as healthcare—associated infections and not community-associated infections.
Less than 50% of the isolates were screened for the presence of the PVL toxin as indicted by patient diagnosis; the presence of which was seen in 14% (n = 108/761) of the isolates tested; 25% (27/108) in MRSA and 75% (81/108) in MSSA. A 2012 Turkish study also showed a low proportion (2.2%, n = 10) of PVL-positive isolates, all of which were MSSA [4]. In the current study, 11 (41%) and 13 (48%) of 27 MRSA isolates that were PVL-positive were associated with SCCmec types III and IV respectively with just one isolate each for types I, V and VI. A previous study showed that 81.5% (22/27) PVL-positive isolates were associated with type IV elements but none of the type I, II or III elements detected were associated with the presence of the lukS/F-PV gene [29]. Another study that further characterised type IV isolates showed that these isolates were PVL-positive [30]. Isolates of S. aureus harbouring PVL are often because of community-associated infections [4]; however our findings showed that 56% were classified as healthcare-associated S. aureus infection followed by 44% that were community-associated infection. This was in contrast to a 2010 Swiss study where a high proportion (87%) of community-associated types IV and V among their PVL-positive strains was demonstrated [24]. A majority (48%) of the PVL-positive isolates were associated with SCCmec type IV (Table 3) followed by type III (41%) with only one each for types II, V and VI; this was consistent with the 2007 American study where 23 of 25 SCCmec type IV isolates were PVL-positive compared to the type II elements (n = 34) which did not harbour the gene [27], as well as with a 2013 Polish study where all the type IV isolates were PVL-positive (9/9) and none of the type II isolates (0/16) harboured the gene. In this latter study only three of 40 type III isolates were positive for PVL [28]. All of these differences highlight the evolving nature of S. aureus and variations in clones as a result of evolution is evident from one geographical region to another.
According to a recent publication [3] the recommended treatment of choice for PVL-positive S. aureus infections are as follows; skin and soft tissue infections: Trimethoprim / sulfamethoxazole, bone and joint infections: flucloxacillin combined with clindamycin for MSSA and daptomycin combined with linezolid for MRSA, and pneumonia: clindamycin, linezolid or rifampicin. When we further analysed the antimicrobial susceptibility profiles of the PVL-positive isolates, we found that the treatment of choice as recommended by Saeed et. al., [3] would be ideal for treating S. aureus skin and soft tissue infections and pneumonia as indicated by the high number of susceptible isolates to each of the antimicrobial agents. The ten PVL-positive isolates from patients with bone and joint infections were also susceptible to all antibiotics except for oxacillin. This is encouraging as it offers available treatment options.
This study aimed to resolve unidentified SCCmec types using an alternative method. While this method did not definitively classify all the unknown isolates into clearly defined types, it showed that majority of these isolates were not the conventional SCCmec types. Majority of the S. aureus isolates displaying resistance to methicillin were non-susceptible most antimicrobial agents indicating that multiple resistance genes are present in our population. The presence of PVL more prevalent in MSSA and was seen in small percentage of isolates indicating a low proportion of this exotoxin in our population.
Acknowledgments
We thank Cheryl Hamman, Jenna Allen, Rubeina Badat, Naseema Bulbulia, Rosah Mobokachaba, Gloria Molaba and Marshagne Smith for assistance with the laboratory work and Boniwe Makwakwa for assistance with the database.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was funded by the National Institute for Communicable Diseases and received no specific grant from any funding agency.
References
- 1.Price JR, Golubchik T, Cole K, Wilson DJ, Crook DW, Thwaites GE, Bowden R, Walker AS, Peto TEA, Paul J, Llewelyn MJ. Whole-genome-sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2014;58:609–18. 10.1093/cid/cit807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shore AC, Coleman DC. Staphylococcal cassette chromosome mec: recent advances and new insights. Int J Med Microbiol. 2013;303(6–7):350–9. 10.1016/j.ijmm.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 3.Saeed K, Gould I, Esposito S, Ahmad-Saeed N, Ahmed SS, Alp E, Bal AM, Bassetti M, Bonnet E, Chan M, Coombs G, Dancer SJ, David MZ, De Simone G, Dryden M, Guardabassi L, Hanitsch LG, Hijazi K, Kruger R, Lee A, Leistner R, Pagliano P, Righi E, Schneider-Barrus S, Skov RL, Tattevin P, Van Wamel W, Vos MC, Voss A. Panton–Valentine leukocidin-positive Staphylococcus aureus: a position statement from the International Society of Chemotherapy. International Journal of Antimicrobial Agents. 2018;51:16–25. 10.1016/j.ijantimicag.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 4.Gulmez D, Sancak B, Ercis S, Karakaya J, Hascelik G. Investigation of SCCmec types and Panton-Valentine leukocidin in community-acquired and nosocomial Staphylococcus aureus strains: comparing skin and soft tissue infections to the other infections. Mikrobiyol Bul. 2012;46(3):341–51. [PubMed] [Google Scholar]
- 5.Badiou C, Dumitrescu O, George N, Forbes ARN, Drougka E, Chan KS, Ramdani-Bouguessa N, Meugnier H, Bes M, Vandenesch M, Ettiene J, Hsu LY, Tazir M, Spiliopoulou I, Nimmo GR, Hulten KG, Lina G. Rapid detection of Staphylococcus aureus Panton-Valentine Leukocidin in Clinical Specimens by Enzyme-linked Immunosorbent Assay and Immunochromatographic Tests. J Clin Microbiol. 2010;48(4):1384–90. 10.1128/JCM.02274-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shields BE, Tschetter AJ, Wanat KA. Staphylococcus simulans: An emerging cutaneous pathogen. JAAD Case Rep. 2016;2(6):428–9. 10.1016/j.jdcr.2016.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of Staphylococcal Cassette Chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrobial agents and Chemotherapy 53: 4961–4967. 2009;53:4961–7. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker K, Ballhausen B, Köck R, Kriegeskorte A. Methicillin-resistance in Staphylococcus isolates: The "mec alphabet" with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages. International Journal of Medical Microbiology. 2014; 304:794–804. 10.1016/j.ijmm.2014.06.007 [DOI] [PubMed] [Google Scholar]
- 9.Turlej A, Hryniewicz W, Empel J. Staphylococcal cassette chromosome mec (SCCmec) classification and typing methods: an overview. Pol J Microbiol. 2011;60(2):95–103. [PubMed] [Google Scholar]
- 10.Ito T, Hiramatsu K, Tomasz A, De Lencastre H, Oerreten V, Holden MTG, Coleman DC, Goering R, Giffard PM, Skov RL, Zhang K, Westh H, O’Brien F, Tenover FC, Oliveira DC, Boyle-Vavra S, Laurent F, Kearns AM, Kreiswirth B, Ko KS, Grundmann H, Sollid JE, John JF Jr, Daum R, Soderquist B, Buist GGuidelines for reporting novel mecA gene homologues. Antimicrobial agents and Chemotherapy 2012;56:4997–9. 10.1128/AAC.01199-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2018;61:74–6. 10.1016/j.meegid.2018.03.013 [DOI] [PubMed] [Google Scholar]
- 12.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–74. 10.1128/AAC.00165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, F. L, Liu D, Xue H, Zhao X. Novel Type XII Staphylococcal Cassette Chromosome mec Harboring a New Cassette Chromosome Recombinase, CcrC2. Antimicrob Agents Chemother 2015;59(12):7597–601. 10.1128/AAC.01692-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganga R, Riederer K, Sharma M, Fakih MG, Johnson LB, Shemes S, Khatib R. Role of SCCmec type in outcome of Staphylococcus aureus bacteremia in a single medical center. J Clin Microbiol. 2009;47(3):590–5. 10.1128/JCM.00397-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: Blurring of the traditional definitions. Journal of Global Antimicrobial Resistance. 2016;6:95–101. 10.1016/j.jgar.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 16.Perovic O, Iyaloo S, Kularatne R, Lowman W, Bosman N, Wadula J, Seetharam S, Duse A, Mbelle N, Bamford C, Dawood H, Mahabeer Y, Bhola P, Abrahams S, Singh-Moodley A. Prevalence and Trends of Staphylococcus aureus Bacteraemia in Hospitalized Patients in South Africa, 2010 to 2012: Laboratory-Based Surveillance Mapping of Antimicrobial Resistance and Molecular Epidemiology. PLoS One. 2015;10(12). 10.1371/journal.pone.0145429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: twenty fourth informational supplement. 2017. [Google Scholar]
- 18.Perovic O, Singh-Moodley A, Govender NP, Kularatne R, Whitelaw A, Chibabhai V, Naicker P, Mbelle N, Lekalakala R, Quan V, Samuel C, Van Schalkwyk E, for GERMS-SA. A small proportion of community-associated methicillin-resistant Staphylococcus aureus bacteraemia, compared to healthcare-associated cases, in two South African provinces. Eur J Clin Microbiol Infect Dis. 2017;36(12):2519–32. 10.1007/s10096-017-3096-3 [DOI] [PubMed] [Google Scholar]
- 19.Singh-Moodley A, Marais E, Perovic O. Discrepancies in the identification of methicillin resistant Staphylococcus aureus and the absence of mecC in surveillance isolates in South Africa. South African Journal of Infectious Diseases. 2015;1(1):1–3. 10.1080/23120053.2015.1107256 [DOI] [Google Scholar]
- 20.Fosheim GE, Nicholson AC, Albrecht VS, Limbago BM. A Multiplex Real-time PCR Assay for Detection of Methicillin-Resistant Staphylococcus aureus and Associated Toxin Genes. J Clin Microbiol. 2011. 49(8):3071–3. 10.1128/JCM.00795-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2011. 18(3):268–81. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 22.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3374–7. 10.1128/AAC.00275-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of Staphylococcal Chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–7. 10.1128/AAC.00579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valsesia G, Rossi M, Bertschy S, Pfyffer GE. Emergence of SCCmec type IV and SCCmec type V methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leukocidin genes in a large academic teaching hospital in central Switzerland: external invaders or persisting circulators? J Clin Microbiol. 2010;48(3):720–7. 10.1128/JCM.01890-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David MZ, Glikman D, Crawford SE, Peng J, King KJ, Hostetler MA, Boyle-Vavara S, Daum RS. What is community-associated methicillin-resistant Staphylococcus aureus? J Infect Dis 2008;97 (9):1234–43. 10.1086/533502 [DOI] [PubMed] [Google Scholar]
- 26.de Carvalho SP, de Almeida JB, Andrade YMFS, da Silva LSC, de Oliveira AC, Nascimento FS, Campos GB, Oliveira MV, Timenetsky J, Marques LM. Community-acquired methicillin-resistant Staphylococcus aureus carrying SCCmec type IV and V isolated from healthy children attending public daycares in northeastern Brazil. Braz J Infect Dis 2017;21(4). 10.1016/j.bjid.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroney SM, Heller LC, Arbuckle J, Talavera M, Widen RH. Staphylococcal cassette chromosome mec and Panton-Valentine leukocidin characterization of methicillin-resistant Staphylococcus aureus clones. J Clin Microbiol. 2007;45(3):1019–21. 10.1128/JCM.01706-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szczuka E, Grabska K, Trawczynski K, Bosacka K, Kaznowski A. Characterization of SCCmec types, antibiotic resistance, and toxin gene profiles of Staphylococcus aureus strains. Acta Microbiologica et Immunologica Hungarica. 2013;60:261–70. 10.1556/AMicr.60.2013.3.3 [DOI] [PubMed] [Google Scholar]
- 29.Berglund C, Molling P, Sjoberg L, Soderquist B. Predominance of staphylococcal cassette chromosome mec (SCCmec) type IV among methicillin-resistant Staphylococcus aureus (MRSA) in a Swedish county and presence of unknown SCCmec types with Panton-Valentine leukocidin genes. Clin Microbiol Infect. 2005;11(6):447–56. 10.1111/j.1469-0691.2005.01150.x [DOI] [PubMed] [Google Scholar]
- 30.Ahmad N, Ruzan IN, Abd Ghani MK, Hussin A, Nawi S, Aziz MN, Maning N, Eow VL. Characteristics of community- and hospital-acquired meticillin-resistant Staphylococcus aureus strains carrying SCCmec type IV isolated in Malaysia. J Med Microbiol 2009;58(Pt 9):1213–8. 10.1099/jmm.0.011353-0 [DOI] [PubMed] [Google Scholar]