The protein kinase BIN2 interacts with and phosphorylates INDUCER OF CBF EXPRESSION1 under cold stress, accelerating its proteasome-mediated degradation to negatively regulate freezing tolerance.
Abstract
Cold acclimation is a crucial strategy for plant survival at freezing temperatures. C-REPEAT BINDING FACTOR (CBF) genes are rapidly and transiently induced by low temperature and play important roles in cold acclimation. However, the mechanism underlying the attenuation of CBF expression during the later stages of the cold stress response is obscure. Here, we show that the protein kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2) interacts with and phosphorylates INDUCER OF CBF EXPRESSION1 (ICE1) in Arabidopsis (Arabidopsis thaliana) under prolonged cold stress, facilitating the interaction between ICE1 and the E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 and thereby promoting ICE1 degradation. The kinase activity of BIN2 is inhibited during the early stages of the cold stress response and is subsequently restored, suggesting that BIN2 mainly downregulates ICE1 abundance when CBF expression is attenuated. A loss-of-function mutation of ICE1 partially suppresses the cold-induced expression of CBFs and compromises the enhanced freezing tolerance of bin2-3 bil1 bil2. These findings reveal an important role for BIN2 in fine-tuning CBF expression, and thus in balancing plant growth and the cold stress response.
INTRODUCTION
Cold stress, including chilling stress (0°C to 15°C) and freezing stress (<0°C), causes significant damage to plants (Agarwal et al., 2006; Shi et al., 2018). Deciphering the mechanisms underlying the plant response to cold stress could facilitate the breeding of cold-tolerant plant varieties. A series of mechanisms help plants adapt to cold stress, including cold acclimation, a process in which freezing tolerance increases in plants that have been pre-exposed to low but nonlethal temperatures (Thomashow, 1999; Chinnusamy et al., 2007).
The C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 (CBF/DREB1) transcription factors play key roles in cold acclimation in plants. CBF transcription factors directly activate a series of Cold-Regulated (COR) genes that increase freezing tolerance (Stockinger et al., 1997; Jia et al., 2016; Zhao et al., 2016). In Arabidopsis (Arabidopsis thaliana), CBF gene expression is strongly induced within 15–30 min after cold treatment and peaks at ∼1–3 h after cold treatment, followed by a rapid decline (Novillo et al., 2004). Therefore, the temporal expression patterns of CBF genes can be divided into the off, on, and attenuated stages before and after exposure to cold stress (Zhao et al., 2017).
Several transcription factors positively or negatively regulate CBF expression under cold stress (Shi et al., 2018). For instance, INDUCER OF CBF EXPRESSION (ICE1) and CALMODULIN BINDING TRANSCRIPTION ACTIVATORs are positive regulators that function upstream of CBFs (Chinnusamy et al., 2003; Doherty et al., 2009; Kim et al., 2013). By contrast, MYB15, ETHYLENE INSENSITIVE3, and PHYTOCHROME-INTERACTING FACTOR3 (PIF3), PIF4, and PIF7 are negative regulators of CBF expression (Agarwal et al., 2006; Lee and Thomashow, 2012; Shi et al., 2012; Jiang et al., 2017). Thus, CBF expression is tightly regulated.
Phosphorylation mediated by protein kinases has been extensively studied, as it is one of the most important and ubiquitous posttranslational modifications. Protein kinase-mediated phosphorylation modulates many cellular processes, including the activities of enzymes, signal transduction, transcription, and metabolism. Over 800 and 1,400 kinases are encoded in the genomes of Arabidopsis and rice (Oryza sativa), respectively. Emerging evidence indicates that these enzymes are involved in plant growth and development and responses to abiotic and biotic stress (Dardick et al., 2007; Lehti-Shiu et al., 2009; Xu and Zhang, 2015).
Recent studies have indicated that several protein kinases play positive or negative roles in plant responses to cold stress (Guo et al., 2018; Shi et al., 2018). OPEN STOMATA1 (OST1), a member of the SNF1-related protein kinase family, positively regulates freezing tolerance in plants (Ding et al., 2015, 2018). Cold-activated OST1 phosphorylates ICE1 and attenuates its interaction with the E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1), enhancing the stability of ICE1 and hence freezing tolerance (Ding et al., 2015). In addition, OST1 phosphorylates BASIC TRANSCRIPTION FACTOR3 proteins, the β-subunits of a nascent polypeptide-associated complex, and facilitates their interaction with CBF proteins in response to cold stress, thereby enhancing CBF protein stability (Ding et al., 2018).
We recently demonstrated that the protein phosphatase CLADE E GROWTH-REGULATING2 is a crucial regulator of the cold-induced activation of OST1 (Ding et al., 2019). Two independent groups showed that MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3) and MPK6 phosphorylate and destabilize ICE1 in response to cold stress in Arabidopsis (Li et al., 2017b; Zhao et al., 2017). By contrast, in rice, OsMPK3 phosphorylates ICE1 and disrupts the OsICE1-OsHOS1 interaction, thereby stabilizing ICE1 (Zhang et al., 2017). In addition to MPK3 and MPK6, we recently identified the plasma membrane-localized receptor-like cytoplasmic kinase COLD-RESPONSIVE PROTEIN KINASE1 as a negative regulator of the cold signaling pathway. Cold-activated COLD-RESPONSIVE PROTEIN KINASE1 phosphorylates 14-3-3 proteins and promotes their import from the cytosol into the nucleus, where they interact with and destabilize CBF proteins, thereby reducing freezing tolerance in plants (Liu et al., 2017, 2018). Finally, the GSK3-like kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2) negatively regulates freezing tolerance in plants (Eremina et al., 2016; Li et al., 2017a) by a largely unknown molecular mechanism.
In this study, we show that in Arabidopsis, BIN2 interacts with and phosphorylates ICE1, which promotes the interaction between ICE1 and HOS1, thereby accelerating HOS1-mediated ICE1 degradation. In addition, the kinase activity of BIN2 is inhibited at the early stage of cold treatment (within 1 h) and subsequently recovers. These findings suggest that BIN2 promotes the degradation of ICE1, mainly during the stage of the cold stress response in which CBF gene expression is attenuated, thus providing a strategy for balancing plant growth and cold stress responses.
RESULTS
BIN2 Interacts with ICE1 In Vitro and In Vivo
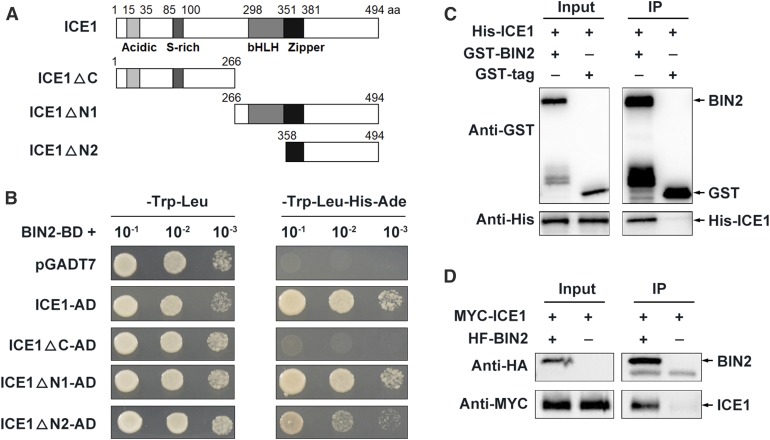
To investigate the role of BIN2 in plant responses to cold stress, we performed a yeast two-hybrid (Y2H) screen using BIN2 as the bait and identified ICE1 as a candidate partner of BIN2. Because ICE1 is a basic helix-loop-helix transcription factor that positively regulates plant freezing tolerance (Chinnusamy et al., 2003), we hypothesized that BIN2 would participate in the cold stress response through interacting with ICE1. Using the Y2H system, we verified their interaction and, through truncation analysis, confirmed that the C terminus of ICE1 is responsible for this interaction (Figure 1A).
Figure 1.
BIN2 Interacts with ICE1 In Vitro and In Vivo.
(A) and (B) BIN2 interacts with ICE1 in a Y2H assay, and the C terminus of ICE1 is required for this interaction. The empty vector pGADT7 was used as a negative control.
(C) BIN2 interacts with ICE1 in a pull-down assay. His-tagged ICE1 was incubated with GST-agarose bound with GST-BIN2 or GST proteins, and the immunoprecipitated proteins were detected with anti-His antibody.
(D) BIN2 interacts with ICE1 in a Co-IP assay. A construct harboring MYC-ICE1 was cotransformed with HF-BIN2 or HA-FLAG into Arabidopsis protoplasts and expressed for 13 h at 22°C. Total proteins were extracted and incubated with HA-agarose. The immunoprecipitated proteins were detected with anti-MYC antibody.
Two close homologs of BIN2, BIN2-LIKE1 (BIL1) and BIL2 (Jonak and Hirt, 2002), also interacted with ICE1 in yeast (Saccharomyces cerevisiae) cells (Supplemental Figure 1). In an in vitro pull-down assay, recombinant His-ICE1 bound to glutathione S-transferase (GST)-BIN2, but not to GST (Figure 1C). Furthermore, a coimmunoprecipitation (Co-IP) assay of Arabidopsis protoplasts co-expressing MYC-ICE1/HA-FLAG-BIN2 (abbreviated as “HF-BIN2” hereafter) or MYC-ICE1/HA-FLAG showed that BIN2 associated with ICE1 in plant cells (Figure 1D). A bimolecular fluorescence complementation assay indicated that BIN2 interacted with ICE1 in the nucleus and was not affected by cold stress (Supplemental Figure 1B). These results demonstrate that BIN2 interacts with ICE1 in vitro and in vivo.
BIN2 Phosphorylates ICE1
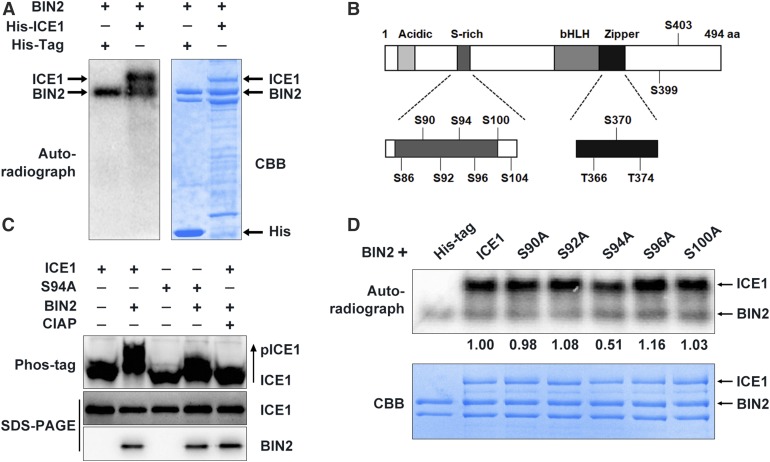
Because BIN2 is a protein kinase, we investigated whether it phosphorylates ICE1. In vitro phosphorylation assays revealed that ICE1 was indeed phosphorylated by BIN2 and its two homologs, BIL1 and BIL2 (Figure 2A; Supplemental Figure 2A). We then analyzed the conserved BIN2 phosphorylation motifs (Ser/ThrXXXSer/Thr; Wang et al., 2002; Ryu et al., 2007, 2010) in ICE1 and identified an N-terminal Ser-enriched region of ICE1 (amino acids 85–100; Figure 2B) that could be phosphorylated by BIN2. To determine whether these conserved amino acids in ICE1 are phosphorylated by BIN2, we mutated Ser90, Ser92, Ser94, Ser96, and Ser100 to Ala to mimic nonphosphorylated forms of ICE1 and performed in vitro phosphorylation assays. ICE1 and all mutated forms were phosphorylated by BIN2; however, the amount of phosphorylated ICE1S94A was only half that of phosphorylated ICE1, whereas the four other mutations did not markedly affect ICE1 phosphorylation (Figure 2C). Mass spectrometry assays further confirmed that Ser94 of ICE1 was phosphorylated by BIN2 in vitro (Supplemental Figures 2B and 2C).
Figure 2.
BIN2 Phosphorylates ICE1 In Vitro and In Vivo.
(A) BIN2 phosphorylates ICE1 in vitro. BIN2 plus His-tag proteins were used as a control. Autoradiograph (right) and a Coomassie brilliant blue (CBB)-stained gel (left).
(B) The 12 conserved putative BIN2 phosphorylation sites in ICE1.
(C) In vitro phosphorylation assays of the phosphorylation of ICE1 and its mutated forms by BIN2. An autoradiograph (top) and Coomassie brilliant blue (CBB)-stained gel (bottom) are shown. The IOD of the band was quantified using the software Image-Pro Plus (Media Cybernetics) to calculate the relative radiation intensity (IODICE1/IODWT).
(D) Phos-tag assays showing the phosphorylation status of ICE1 and ICE1S94A (S94A) in Arabidopsis protoplasts. Plasmids of the indicated combinations were transformed into Arabidopsis protoplasts and expressed at 22°C for 13 h before total protein extraction. The phosphorylation status of ICE1 was examined in a Phos-tag gel assay (top, anti-MYC). The levels of ICE1 (middle, anti-MYC) and BIN2 (bottom, anti-HA) proteins are shown in the SDS-PAGE gels.
To determine whether BIN2 phosphorylates ICE1 in planta, we expressed MYC-ICE1/HF-BIN2 or MYC-ICE1 alone in Arabidopsis protoplasts and performed phos-tag mobility shift assays. The amount of a slow-migrating form of ICE1 increased in protoplasts when co-expressed with BIN2; however, incubating the protein samples with calf intestinal alkaline phosphatase (CIAP) eliminated the slow-migrating form of ICE1 (Figure 2D), indicating that ICE1 is phosphorylated by BIN2 in planta. We also examined the effect of BIN2 on the phosphorylation of the mutated form MYC-ICE1S94A and found that the phosphorylation of MYC-ICE1S94A by BIN2 was considerably reduced, although not fully abolished, as compared with wild-type ICE1 protein (Figure 2D). These results demonstrate that ICE1S94 is a major phosphorylation site of BIN2.
BIN2 Destabilizes ICE1 and Attenuates Its Transcriptional Activity
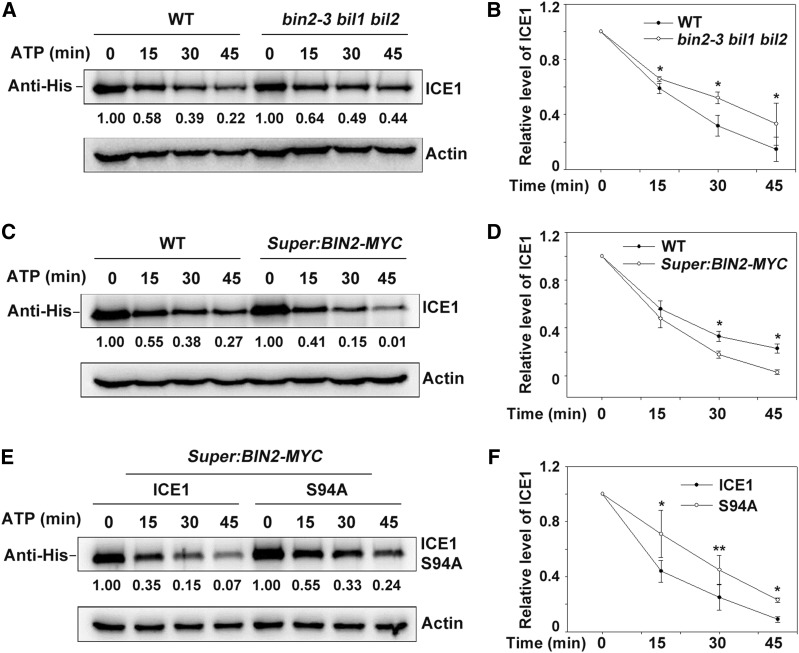
ICE1 is degraded by the 26S proteasome pathway (Dong et al., 2006; Ding et al., 2015). To investigate whether BIN2 affects ICE1 stability, we performed a cell-free degradation assay. Purified recombinant His-ICE1 proteins were incubated with total proteins extracted from wild type, bin2-3 bil1 bil2 triple mutant, and BIN2-overexpressing transgenic plants (Super:BIN2-MYC) supplemented with 10 mM of ATP at 22°C for various periods of time. The degradation rate of ICE1 was much higher in the wild-type than in bin2-3 bil1 bil2 plants but was lower than in BIN2-overexpressing plants (Figures 3A to 3D). These results indicate that BIN2 destabilizes ICE1 in vitro. In addition, ICE1S94A was more stable than ICE1 when incubated with total proteins extracted from transgenic Super:BIN2-MYC plants (Figures 3E and 3F), suggesting that BIN2-mediated phosphorylation of ICE1 is required for its degradation.
Figure 3.
BIN2 Promotes ICE1 Degradation.
(A) and (B) Mutation of BIN2 and its homologs attenuates ICE1 degradation in a cell-free degradation assay. Equal amounts of total proteins extracted from wild type (WT) and bin2-3 bil1 bil2 triple mutant plants (grown on half strength MS for 14 d) were combined with recombinant His-ICE1 and 10 mM ATP. ICE1 was detected using anti-His antibody, and Actin was used as the internal control. The IOD values of ICE1 and Actin were quantified using the software Image-Pro Plus (Media Cybernetics); the relative protein abundance of ICE1 (IODICE1/IODActin) was calculated and set in relation to IODICE1/IODActin at 0 min. Representative blot (A) and statistical analysis (B).
(C) and (D) Overexpression of BIN2 promotes ICE1 degradation in a cell-free degradation assay. The experiment was performed using wild-type (WT) and Super:BIN2-MYC plants with recombinant His-ICE1. The relative protein abundance of ICE1 was calculated as described for (A). Representative blot (C) and statistical analysis (D).
(E) and (F) The mutant protein ICE1S94A (S94A) is more stable than wild-type ICE1 in a cell-free degradation assay. The experiment was performed using Super:BIN2-MYC plants with recombinant His-ICE1 or His-ICE1S94A proteins. The relative protein abundance of ICE1 was calculated as described for (A). Representative blot (E) and statistical analysis (F).
In (B), (D), and (F), data are means ± sd of three independent experiments. *P < 0.05, **P < 0.01 (Student’s t test).
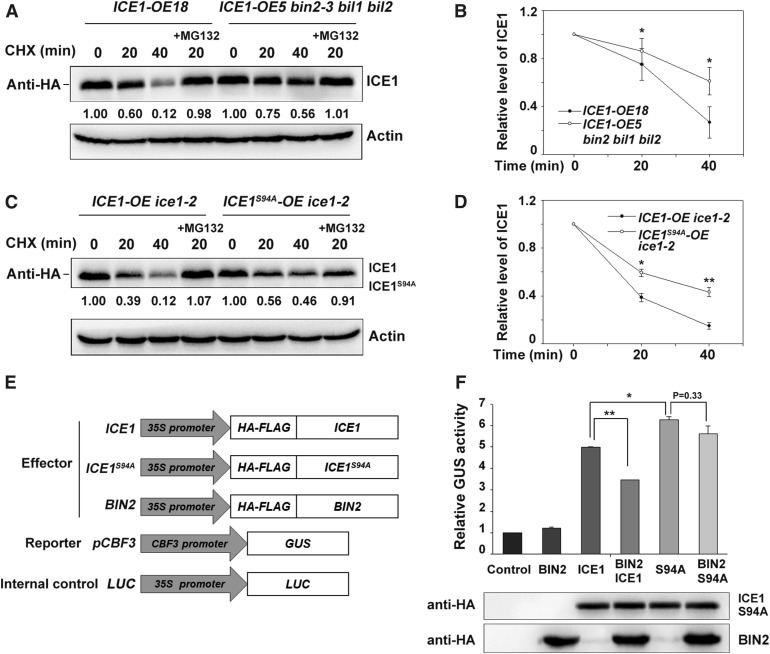
We then investigated the stability of ICE1 in planta in the presence of the protein synthesis inhibitor cycloheximide. Consistent with the results of the in vitro cell-free degradation assays, ICE1 was more stable in the bin2-3 bil1 bil2 triple mutant than in the wild-type background (Figures 4A and 4B; Supplemental Figures 3A and 3B). Moreover, ICE1S94A was degraded much more slowly than wild-type ICE1 in transgenic plants expressing ICE1 or ICE1S94A (Figures 4C and 4D). The addition of MG132, a 26S proteasome inhibitor, dramatically suppressed ICE1 degradation (Figures 4C and 4D). These results indicate that BIN2 phosphorylates ICE1 and promotes its proteasome-mediated degradation.
Figure 4.
BIN2 Destabilizes ICE1 and Attenuates Its Transcriptional Activity through Phosphorylation.
(A) and (B) ICE1 is more stable in the bin2-3 bil1 bil2 triple mutant than in the wild type in vivo. Two-week-old ICE1-OE18 and ICE1-OE5/bin2-3 bil1 bil2 plants were treated with 100 μM of cycloheximide (CHX) in the absence or presence of 50 μM of MG132 for the indicated period. Total proteins were extracted and subjected to immunoblot analysis. ICE1 was detected with anti-HA antibody, and Actin was used as the internal control. The IOD values of ICE1 and Actin were quantified using the software Image-Pro Plus (Media Cybernetics); the relative protein abundance of ICE1 (IODICE1/IODActin) was calculated and set in relation to IODICE1/IODActin at 0 min. Representative blot (A) and quantitative analysis (B). Each bar represents the mean ± sd of three independent experiments. *P < 0.05 (Student’s t test).
(C) and (D) The mutant protein ICE194A is more stable than wild-type ICE1 in vivo. The experiment was performed using ICE1-OE/ice1-2 and HF-ICE194A-OE/ice1-2 plants. The relative protein abundance of ICE1 was calculated as described for (A). Representative blot (C) and quantitative analysis (D). Data are the means ± sd of three independent experiments. *P < 0.05, **P < 0.01 (Student’s t test). CHX, cycloheximide.
(E) and (F) BIN2 attenuates ICE1-mediated activation of CBF3 expression. The reporter construct pCBF3:GUS and the internal control 35S:Luc with the indicated effector constructs shown in (E) were co-transformed into Arabidopsis protoplasts; protoplasts transformed with CBF3:GUS and 35S:Luc were used as a control. Relative GUS activity (GUS/Luc) indicates the CBF3 expression level; the control value was set to 1.00 (F, upper). Protein levels of ICE1, ICE1S94A, and BIN2 (F, lower). Data are the mean ± sd values of three independent experiments. *P < 0.05, **P < 0.01 (Student’s t test).
Because ICE1 is an important transcription factor regulating CBF gene expression in plants under cold stress (Chinnusamy et al., 2003), we performed transient transactivation assays in Arabidopsis protoplasts to investigate whether BIN2 affects the transcriptional activity of ICE1. In this assay, the CBF3 promoter was fused to the β-glucuronidase (GUS reporter gene (Ding et al., 2015). Plasmids harboring ICE1 and BIN2 driven by the 35S promoter served as effectors, and the empty vectors were used as controls (Figure 4E). We cotransformed Arabidopsis protoplasts with various combinations of effectors and the reporter construct. GUS activity was higher in protoplasts transformed with ICE1 as compared with the empty vector control (Figure 4F), suggesting that ICE1 indeed activates CBF3 expression, as reported previously by Chinnusamy et al. (2003) and Ding et al., 2015. Protoplasts expressing BIN2 alone were indistinguishable from the control; however, co-expression of BIN2 with ICE1 significantly reduced ICE1-activated CBF3 expression (Figure 4F). Furthermore, ICE1S94A induced GUS activity more strongly than wild-type ICE1 (Figure 4F). When co-expressed with BIN2, the effect of ICE1S94A on GUS activity appeared to be slightly inhibited, but the difference was not significant (Figure 4F).
An electrophoretic mobility shift assay (EMSA) indicated that ICE1 bound to the CBF3 promoter, as reported previously by Chinnusamy et al. (2003). Furthermore, the binding affinity of ICE1 was dramatically reduced after being phosphorylated by BIN2 in vitro (Supplemental Figure 3C). These results demonstrate that BIN2 negatively regulates the transcriptional activity of ICE1 in planta, at least partially via phosphorylation.
Freezing Tolerance of bin2-3 bil1 bil2 Partially Depends on ICE1
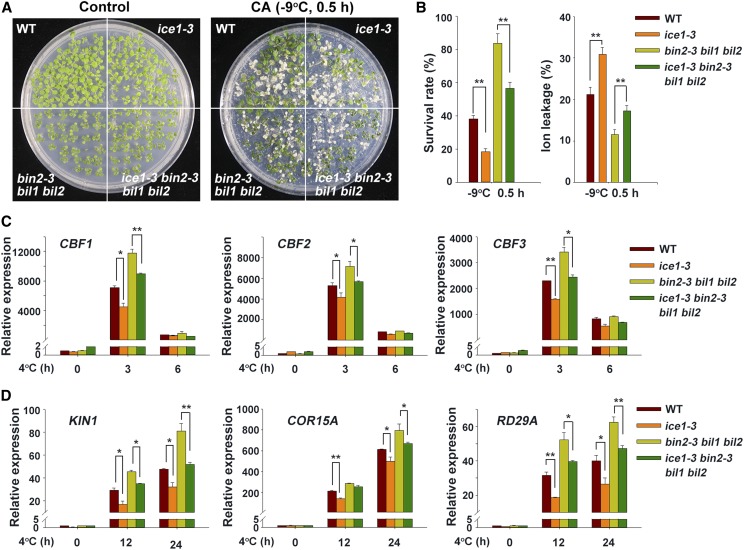
We then explored the genetic interaction between BIN2 and ICE1. As the bin2-3 bil1 bil2 mutant is in the Ws-2 background, and the available ice1-2 knockout mutant is in the Col background (Ding et al., 2015), we used clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) technology to generate a knock-out ice1 mutant in the Ws-2 background, named ice1-3 (Supplemental Figure 4A). A 37-bp deletion in this mutant, between 40 bp and 78 bp in exon 1, resulted in a frameshift mutation and thus premature translation termination of ICE1 (Supplemental Figure 4A).
Next, we crossed ice1-3 with the ICE1-overexpression lines to generate ice1-3/ICE1-OE plants (Supplemental Figures 4B and 4C) and performed freezing tolerance assays. The ice1-3 mutant showed substantially reduced freezing tolerance as compared with the wild type, which is consistent with the findings for ice1-2 (Ding et al., 2015). Overexpression of ICE1 rescued the freezing-sensitive phenotype of ice1-3 (Supplemental Figures 4D and 4E). Ion leakage, a marker of plasma membrane damage caused by stress, was negatively associated with the survival rates of these plants under freezing stress (Supplemental Figure 4E). Furthermore, the expression of CBFs and downstream CORs, including KIN1, COR15A, and RD29A, was strongly reduced in ice1-3 as compared with the wild type, and the expression of these genes was restored to wild-type levels by overexpressing ICE1 (Supplemental Figure 5).
We then crossed ice1-3 with bin2-3 bil1 bil2 to generate the bin2-3 bil1 bil2 ice1-3 quadruple mutant and subjected it to freezing tolerance assays. The bin2-3 bil1 bil2 ice1-3 mutant displayed reduced freezing tolerance as compared with bin2-3 bil1 bil2, although, as expected, it was not as sensitive to freezing stress as ice1-3 (Figures 5A and 5B), because BIN2 also phosphorylates BZR1 to modulate plant freezing tolerance (Li et al., 2017a).
Figure 5.
Mutation of ICE1 Partially Rescues the Phenotype of bin2-3 bil1 bil2.
(A) Phenotypes of acclimated wild-type, ice1-3, bin2-3 bil1 bil2, and ice1-3 bin2-3 bil1 bil2 plants after freezing treatment. The plants were grown on half strength MS plates at 22°C for 2 weeks followed by a 4°C treatment for 3 d and then a −9°C treatment for 0.5 h. At least three plates were used per treatment. Photographs show representative plants after recovery. WT, wild type.
(B) Survival rates and ion leakage of plants after the freezing assays described in (A). The data are the mean ± sd values of three independent experiments. **P < 0.01 (Student’s t test) indicates a significant difference between ice1-3 and wild type or bin2 bil1 bil2 and ice1-3 bin2 bil1 bil2. WT, wild type.
(C) and (D) Expression of CBF genes (C) and CBF target genes (D) in wild type, ice1-3, bin2 bil1 bil2, and ice1-3 bin2 bil1 bil2 plants under cold stress. Two-week–old plants grown on half strength MS plates at 22°C were treated at 4°C for the indicated period. Total RNA was extracted from the plants and subjected to reverse transcription-quantitative PCR analysis. The expression levels of the genes in the wild type at 22°C were set to 1.00. Data are the means ± sd. *P < 0.05, **P < 0.01 (Student’s t test). At least three independent experiments were performed with similar results. WT, wild type.
We then conducted multivariate analysis using ICE1 and BIN2/BIL1/BIL2 as the fixed factors and the survival rate and ion leakage as the dependent variable. The interactive effect between ICE1 and BIN2, BIL1, and BIL2 was statistically significant (Supplemental Table 1). In addition, CBF and COR expression levels were significantly lower in bin2-3 bil1 bil2 ice1-3 than in bin2-3 bil1 bil2 under cold stress (Figures 5C and 5D). These results suggest that BIN2 acts upstream of ICE1 in the regulation of CBF expression, thereby modulating freezing tolerance.
BIN2 Promotes the Interaction between ICE1 and HOS1
Given that ICE1 is degraded by the E3 ubiquitin ligase HOS1 (Dong et al., 2006) and that BIN2 enhances the ubiquitin-mediated degradation of ICE1 (Figures 3 and 4), we next examined whether BIN2 affects the interaction between HOS1 and ICE1. We extracted total proteins from Arabidopsis protoplasts expressing BIN2-GFP/HF-HOS1/MYC-ICE1 or HF-HOS1/MYC-ICE1, immunoprecipitated them with anti-HA agarose beads, and performed immunoblot analysis with anti-GFP, anti-HA, and anti-MYC antibodies. HOS1 associated with ICE1, which is consistent with previous findings (Dong et al., 2006; Ding et al., 2015), and their interaction was enhanced when co-expressed with BIN2 (Figures 6A and 6B). The interaction between HOS1 and ICE1 was attenuated by OST1 (Supplemental Figure 6), which served as the positive control (Ding et al., 2015). By contrast, the strength of the interaction between HOS1 and ICE1S94A was the same regardless of the presence of BIN2 (Figures 6C and 6D). These results suggest that BIN2 phosphorylates ICE1, enhancing its interaction with HOS1 and thereby destabilizing ICE1.
Figure 6.
BIN2 Promotes the Interaction between ICE1 and HOS1.
(A) and (B) Co-IP assays were performed as described for Figure 1C, and plasmids harboring BIN2-GFP, MYC-ICE1, and HA-FLAG-HOS1 were used. The IOD value of ICE1 was quantified using the software Image-Pro Plus (Media Cybernetics) to calculate the relative protein abundance of co-immunoprecipitated ICE1 (IODIP/IODINPUT). The IODIP/IODINPUT was set in relation to the group without the addition of BIN2-GFP and represents the interaction strength between ICE1 and HOS1. Data are the means ± sd of three independent experiments. *P < 0.05 (Student’s t test).
(C) and (D) BIN2 does not promote the interaction between ICE1S94A and HOS1 in vivo. Co-IP assays were performed as in (A), but MYC-ICE1S94A was used instead of MYC-ICE1-. The relative protein abundance of co-immunoprecipitated ICE1 was calculated as described in (A).
(E) Schematic diagram of the pull-down assay shown in (D) and (E).
(F) BIN2 promotes the interaction between ICE1 and HOS1 in vitro. The relative protein abundance of co-immunoprecipitated ICE1 was calculated as described in (A).
(G) BIN2 does not promote the interaction between ICE1 S94A and HOS1 in vitro. The relative protein abundance of co-immunoprecipitated ICE1 was calculated as described in (A).
This conclusion was further supported by the results of in vitro pull-down assays. GST-BIN2 recombinant protein was incubated with His-ICE1 in phosphorylation reaction buffer supplemented with or without 0.1 mM of ATP at 30°C for 30 min. Myelin basic protein (MBP)–HOS1 (immunoprecipitated by MBP beads) was then incubated with phosphorylated or nonphosphorylated His-ICE1 in pull-down buffer at 4°C for 2 h, followed by immunoblot analysis (Figure 6E). The amount of ICE1 that was immunoprecipitated by HOS1 was much higher in the BIN2-phosphorylated system than in the nonphosphorylated system (Figure 6F). By contrast, the degree of interaction between ICE1S94A and HOS1 was similar in the presence and absence of ATP (Figure 6G). These data, combined with the results of the Co-IP assays, demonstrate that the BIN2-mediated phosphorylation of ICE1 facilitates the association between ICE1 and HOS1.
BIN2 Kinase Activity Decreases during the Early Stages of the Cold Stress Response
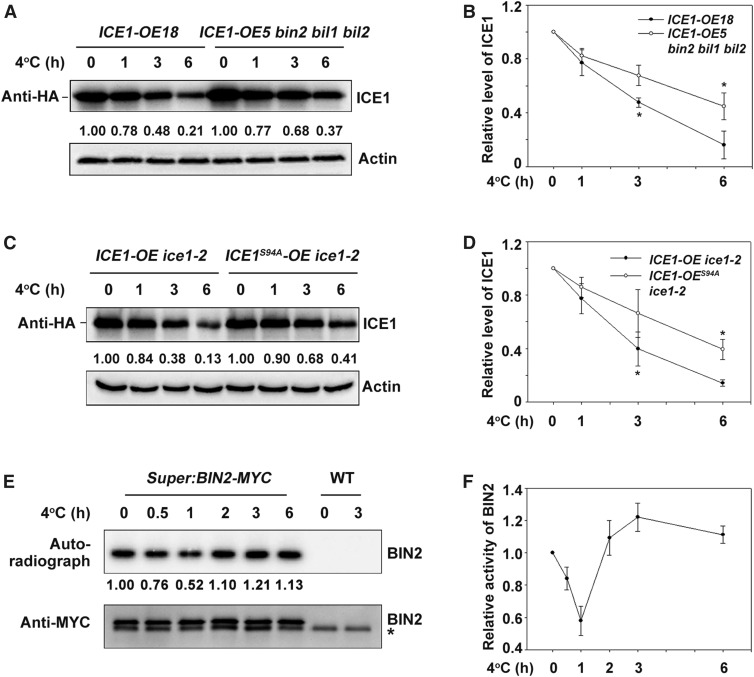
To determine when BIN2 executes its kinase function on ICE1 during cold stress, we compared the status of ICE1 protein in ICE1-OE and ICE1-OE/bin2-3 bil1 bil2 transgenic plants before and after cold treatment. ICE1 was indeed degraded after cold treatment, as previously reported (Figures 7A and 7B; Dong et al., 2006), and ICE1 was more stable in the bin2-3 bil1 bil2 mutant than in the wild type after 3 h and 6 h of cold treatment (Figures 7A and 7B; Supplemental Figures 7A and 7B). Overexpression of ICE1 enhanced the freezing tolerance of Arabidopsis, as reported previously byChinnusamy et al. (2003) and Miura et al. (2007, 2011), and ICE1-OE/bin2-3 bil1 bil2 plants were more freezing-tolerant than ICE1-OE (Supplemental Figures 7C to 7E). These results are consistent with the finding that ICE1 was more stable in bin2-3 bil1 bil2 than in the wild type under cold treatment (Figures 7A and 7B; Supplemental Figures 7A and 7B).
Figure 7.
The Kinase Activity of BIN2 Is Regulated by Cold Stress.
(A) and (B) Mutation of BIN2 and its homologs attenuates ICE1 degradation during cold stress. Total proteins from ICE1-OE18 transgenic plants or bin2-3 bil1 bil2/ICE1-OE5 (grown on half strength MS for 2 weeks) treated at 4°C for the indicated period were extracted and subjected to immunoblot analysis. ICE1 was detected with anti-HA antibody. Actin was used as the internal control. The IOD values of ICE1 and Actin were quantified using the software Image-Pro Plus (Media Cybernetics) to calculate the relative protein abundance of ICE1 (IODICE1/IODActin) in relation to IODICE1/IODActin at 0 h. Representative picture is shown in (A), and quantitative analysis is shown in (B). Data are the means ± sd of three independent experiments. *P < 0.05 (Student’s t test).
(C) and (D) ICE1S94A is more stable than wild-type ICE1 during cold stress. The experiment was performed using ICE1-OE/ice1-2 and ICE194A-OE/ice1-2 plants. The relative protein abundance of ICE1 was calculated as described in (A). Representative blot (C) and quantitative analysis (D). Data are the means ± sd of three independent experiments. *P < 0.05 (Student’s t test).
(E) and (F) In-gel kinase assays of BIN2. Total proteins were extracted from Super:BIN2-MYC transgenic plants treated at 4°C for the indicated period and incubated with anti-MYC agarose. A portion of the immunoprecipitated protein sample was used to perform an in-gel kinase assay using the common substrate MBP as a substrate (top), and a portion was used for the immunoblot assay (bottom). BIN2 kinase activity was detected by autoradiography, and BIN2 protein levels were detected with anti-MYC antibody. The IOD values of BIN2 kinase in the autoradiograph and protein abundance were quantified using the software Image-Pro Plus (Media Cybernetics), and the relative kinase activity was calculated as IODAuto/IODMYC. The relative kinase activity was set in relation to IODAuto/IODMYC at 0 h. Representative blot (E) and quantitative analysis (F). Data are the means ± sd of three independent experiments. *, nonspecific bands. WT, wild type.
We also crossed Super:BIN2-MYC with ICE1-OE ice1-2 to generate BIN2- and ICE1-overexpressing lines and compared the phosphorylation status of ICE1 between the BIN2- and ICE1-overexpressing lines and ICE1-OE ice1-2. After cold treatment, the phosphorylation level of ICE1 was higher in the BIN2- and ICE1-overexpressing lines than in ICE1-OE ice1-2, suggesting that BIN2 increases ICE1 phosphorylation in vivo during cold stress (Supplemental Figure 7E). In addition, consistent with previous results (Zhao et al., 2017), ICE1S94A was more stable than wild-type ICE1 during cold stress (Figures 7C and 7D).
We further examined the kinase activity of BIN2 by performing in-gel kinase assays using Super:BIN2-MYC transgenic plants. The kinase activity of BIN2 on the common substrate MBP notably decreased by 1 h of cold treatment, followed by an increase in response to further cold treatment (Figures 7E and 7F). These results suggest that the kinase activity of BIN2 is inhibited soon after exposure to cold treatment, but subsequently recovers.
DISCUSSION
CBF genes are the best-studied genes that orchestrate cold tolerance responses in plants. CBFs are rapidly and dramatically induced by cold stress (Gilmour et al., 1998; Liu et al., 1998). A series of positive regulators that promote CBF gene expression have been identified (Chinnusamy et al., 2003; Doherty et al., 2009; Shi et al., 2018), and a number of negative regulators of this process have also been described by Agarwal et al. (2006), Lee and Thomashow (2012), Shi et al. (2012), and Jiang et al. (2017). CBF-overexpressing transgenic plants show severe dwarfism (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000; Achard et al., 2008). Therefore, the strict regulation of CBF gene expression is crucial for maintaining the balance between cold tolerance and plant growth.
It is thought that plants use minimal energy under cold stress once sufficient CBF transcription has been induced. Indeed, the induction of CBF genes by cold is attenuated after ∼3 h of cold treatment (Supplemental Figure 7F; Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Novillo et al., 2004). Although this phenomenon was first observed when CBFs were discovered (Jaglo-Ottosen et al., 1998; Liu et al., 1998), the molecular mechanism by which plants achieve this CBF expression pattern under cold stress remains unclear. This study demonstrates that BIN2 plays an important role in this process (Figure 8).
Figure 8.
Proposed Model of the Role of BIN2 in Modulating Cold-Stress Responses through Regulating ICE1 Stability in Arabidopsis.
At the early stage of the cold stress response, BIN2 kinase activity decreases and OST1 kinase is activated, which coordinately stabilize ICE1, thereby promoting maximum CBF expression. At the attenuated stage, BIN2 activity increases, which promotes the phosphorylation of ICE1 and in turn facilitates the HOS1-mediated degradation of ICE1. As a result, ICE1-regulated CBF expression is attenuated.
We found that the transcription factor ICE1 is a substrate of BIN2 (Figures 1 and 2) and that BIN2 facilitates the degradation and inhibits the transcriptional activity of ICE1 by phosphorylation to downregulate CBF gene expression (Figures 3 to 7). Most importantly, we showed that the kinase activity of BIN2 decreases substantially in plants upon exposure to cold conditions (∼1 h at 4°C) and subsequently recovers (after 2 h at 4°C). In accordance with this finding, in vivo protein analysis of ICE1 indicated that BIN2 mainly downregulates ICE1 abundance after 3 h at 4°C (Figures 7A to 7D). Notably, our experiments also indicated that BIN2 regulates CBF gene expression partially through ICE1, which is consistent with the previous finding that BIN2 negatively regulates CBF expression via the transcription factors BZR1 and CESTA (Eremina et al., 2016; Li et al., 2017a). Taken together, we conclude that cold stress releases the inhibitory effects on transcription factors ICE1, BZR1, and CESTAs mediated by BIN2. This process allows CBF genes to be induced to the maximum extent during the early stages of the cold stress response, thereby enhancing freezing tolerance. Cold stress also inhibits the activities of these transcription factors, ensuring that there is no runaway expression of CBFs during the attenuated stages (Figure 7F). It would be interesting to explore the subtle relationship between BIN2 and these basic helix-loop-helix transcription factors by generating multiple-order mutants of bin2 bil1 bil2 ice1 with bzr1, cesta, and their homologs. This would require substantial effort, as both BZR1 and CESTA have several homologs that function redundantly (Yin et al., 2005; Eremina et al., 2016).
Another interesting phenomenon is that our in vitro phosphorylation assays showed that Ser94 of ICE1, an established phosphorylation site for MPK3 and MPK6 (Li et al., 2017b; Zhao et al., 2017), is also phosphorylated by BIN2 (Figure 2). MPK3 and MPK6 do not appear to influence the interaction between ICE1 and the E3 ligase HOS1 (Li et al., 2017b), whereas BIN2 enhances their interaction and Ser94 of ICE1 mediates this enhancement. Therefore, these protein kinases act on ICE1 via different mechanisms.
In animals, sequential phosphorylation by GSK3 and MAP kinases negatively regulates Smad1, Smad5, and Smad8 activity during neural development. MAPKs perform primed phosphorylation of Smad1, Smad5, and Smad8, enabling GSK3s to further phosphorylate these targets; only after phosphorylation by GSK3s is Smad1, Smad5, and Smad8 degraded by the 26S proteasome (Fuentealba et al., 2007; Eivers et al., 2008, 2009). We propose that a similar scenario involving BIN2 and MPK3 and MPK6 occurs in planta in which Ser94 acts as the primed phosphorylation site. MPK3 and MPK6 are activated during the early stages of the cold stress response and phosphorylate ICE1 first (Li et al., 2017b; Zhao et al., 2017). This allows ICE1 to be efficiently phosphorylated by BIN2 (Figure 2), whose activity is restored when CBF expression is attenuated (Figure 7). As a consequence, further phosphorylation of ICE1 by BIN2 leads to HOS1-mediated degradation of ICE1 (Figure 6). This scenario could also explain the low phosphorylation status of ICE1 in plants under warm conditions (Li et al., 2017b; Zhao et al., 2017), even when BIN2 shows high kinase activity (Figures 7E and 7F). In addition to ICE1, several other common targets are also phosphorylated by BIN2 and MPK3 and MPK6, such as the brassinosteroid signaling protein BES1 (Yin et al., 2002; Vert and Chory, 2006; Kang et al., 2015) and SPEECHLESS (Lampard et al., 2008; Casson and Hetherington, 2012; Gudesblat et al., 2012). Future research should explore whether this coordination between BIN2 and MPK3 and MPK6 contributes to other aspects of plant growth, development, and environmental stress responses.
METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown under a 16-h-light/8-h-dark photoperiod with fluorescent white light at 60 μE m−2 s−1 (F17T8/TL841 bulb; Philips) at 21 ± 2°C on 0.8% (w/v) agar plates containing half strength Murashige and Skoog (MS) medium (normal conditions). The Super:BIN2-MYC and ICE1-OE ice1-2 transgenic plants used in this study were described previously by Li et al. (2017a, 2017b). ICE1-OE, ICE1-OE bin2-3 bil1 bil2, and ICE1S94A-OE ice1-2 transgenic plants were generated as follows. The coding sequence (CDS) of the ICE1 or ICE1S94A mutant form was cloned into the SalI and KpnI sites of the pCM1307 vector harboring HA-FLAG (Ding et al., 2015), and then transformed into Arabidopsis plants through Agrobacterium tumefaciens (Agrobacterium)-mediated transformation by the floral dip method (Clough and Bent, 1998). The specific primers used are listed in Supplemental Table 2.
CRISPR/Cas9-Generated Plant Materials
To generate the ice1-3 mutant, two ICE1-specific motifs were selected as the targets for Cas9 to mutate ICE1. Vector construction and mutant identification were performed as described in Xing et al. (2014), with primers listed in Supplemental Table 2. The target motif was cloned into the pHSE401 vector. The constructs were then introduced into Ws-2 wild-type plants through Agrobacterium-mediated transformation by the floral dip method (Clough and Bent, 1998). The Cas9-free mutant was screened as described previously by Jia et al. (2016). The T3 transgenic seeds were screened on half strength MS medium containing 25 mg/L of hygromycin, and only the non-hygromycin–resistant plants were harvested and used for subsequent experiments.
Freezing Tolerance Assays
Freezing tolerance assays were performed as described previously by Shi et al. (2012) with minor modifications. Arabidopsis seedlings were grown under normal conditions for 2 weeks and subjected (or not) to 4°C treatment for 3 d before being subjected to a freezing assay in a freezing chamber (RuMED4001). The freezing program began at 1°C and dropped by 1°C per hour until reaching the temperature specified in the figure legends. After freezing treatment, the seedlings were shifted to 4°C and stored in the dark for 12 h before being transferred to normal conditions for ∼4 d. After recovery, seedlings that could still grow new leaves were scored as surviving seedlings.
Ion Leakage Assays
Ion leakage assays were performed as described in Shi et al. (2012). After the freezing experiments, seedlings were collected into tubes containing 8 mL of deionized water and shaken for 15 min at 22°C. Then, their electrical conductivities (EC) were measured, giving S1. The samples were boiled for at least 15 min and shaken at 22°C for another 20 min before their EC were measured again, giving S2. The EC of deionized water was set as S0. Ion leakage was calculated as: .
RNA Extraction and RT-qPCR
Total RNAs were extracted from Arabidopsis plants using TRIzol reagent (Thermo Fisher Scientific), followed by reverse transcription using HiScript II Q RT SuperMix (Vazyme). RT-qPCR was performed using a SYBR Green PCR Master Mix Kit (cat. no. RR820A; Takara). The reaction components were mixed and the reactions performed using the standard protocol (model no. 7500 Fast Real-Time PCR System; Applied Biosystems). The relative expression levels were calculated as described previously by Huang et al. (2010), and the specific primers used are listed in Supplemental Table 2.
Y2H Assay
The Y2H assays were performed as described previously by Wang et al. (2011). BIN2 was cloned into the EcoRI and SalI sites of the pGBKT7 bait vector and transformed into yeast strain AH109 (Clontech), followed by transformation with a wild-type Arabidopsis cDNA library generated by Shanghai OE Biotech. The yeast transformants were screened on SD/Leu/Trp/His selective medium. The pGADT7 clones harboring ICE1 and its truncated forms were described previously by Ding et al. (2015). The yeast transformants were screened on SD/Leu/Trp/His/Ade selective medium.
Purification of Recombinant Proteins
The construction and purification of fusion protein were performed as reported by Wang et al. (2011). His-ICE1, GST-BIN2, GST-BIL1, GST-BIL2, and MBP-HOS1 were purified as described previously by Ding et al. (2015) and Li et al. (2017b). The construction and purification of GST-BIN2, BIL1, and BIL2 were conducted as follows: BIN2, BIL1, and BIL2 cDNA was amplified and cloned into the BamHI and SalI sites of the pGEX4T-1 vector and transformed into Escherichia coli (BL21). The proteins were expressed at 16°C for 12 h using 0.5 mM of IPTG and purified using Glutathione-agarose (71502076-EG; GE Healthcare).
In Vitro Pull-Down Assay
The pull-down assays were performed as described previously by Wang et al. (2011). Glutathione-agarose containing 5 μg of GST or GST-BIN2 was incubated with 5 μg of His-ICE1 in PBS buffer (0.2% [v/v] TWEEN-20) at 4°C for 2 h. The Glutathione-agarose was washed at least five times using PBS buffer (0.2% [v/v] TWEEN-20). The proteins eluted from the agarose were detected with anti-GST antibody (AbM59001-2H5-PU; Beijing Protein Innovation) and anti-His antibody (AbM59012-18-PU; Beijing Protein Innovation).
Co-IP Assay
Co-IP assays were performed as described in Ding et al. (2015). The CDS of BIN2 was cloned into the SalI and KpnI sites of the pCM1307 vector with HA-FLAG to generate HF-BIN2 and MYC-ICE1 as previously in Li et al. (2017b). The plasmids were transformed into Arabidopsis protoplasts using combinations described in the figure legends. After 13 h, total proteins were extracted with IP buffer containing 50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 1% (v/v) Triton X-100, 2% (v/v) Nonidet P-40, 5 mM of dithiothreitol (DTT), and protease inhibitor cocktail (Roche), followed by incubated with anti-HA agarose for 2 h at 4°C. The agarose was washed at least five times with IP buffer before SDS-PAGE. The immunoprecipitated proteins were analyzed with anti-MYC antibodies (cat. no. M4439; Sigma-Aldrich).
Bimolecular Fluorescence Complementation Assay
The CDS of BIN2 was cloned into the SalI and KpnI sites of pFGC-nYFP, the CDS of truncated ICE1 (1 to 266 amino acids) was cloned into the XbaI and BamHI sites of pFGC-nYFP to generate ICE1ΔC-cYFP (Hu et al., 2013), and ICE1-cYFP was constructed in Li et al. (2017b). The plasmids were transformed into Arabidopsis protoplasts with the combinations described in the Supplemental Figure 1 legend. After 13 h, protoplasts with or without further cold treatment for 1 or 2 h were examined under a model no. LSM-710 Confocal Microscope (excitation and emission wavelengths were 488 and 550 nm, respectively; Zeiss).
In Vitro Phosphorylation Assay
The in vitro phosphorylation assay was performed as described in Ding et al. (2015). Recombinant GST-BIN2, BIL1, and BIL2 and recombinant His-ICE1 proteins were added to the reaction buffer (20 mM of Tris-HCl at pH 7.5, 10 mM of MgCl2, 100 mM of NaCl, 25 mM of ATP, 1 μCi [γ-32P] ATP, and 1 mM of DTT) at an enzyme/substrate ratio of 1:10 and incubated at 30°C for 30 min. The reactions were stopped by adding SDS sample loading buffer. The phosphorylated ICE1 was visualized by autoradiography after SDS-PAGE.
Phos-tag Mobility Shift Assay
The Phos-tag mobility shift assay (NARD Institute) was performed as described previously by Mao et al. (2011). Briefly, total proteins extracted from plants or protoplasts were separated in an 8% (w/v) SDS-PAGE gel containing 50 mM of Phos-tag and 200 mM of MnCl2. After electrophoresis, the gel was washed three times with transfer buffer (50 mM of Tris and 40 mM of Gly) containing 10 mM of EDTA for 10 min each time. The gel was washed with transfer buffer for an additional 10 min before being transferred to a polyvinylidene fluoride membrane. The CIAP (Takara) or λ-phosphatase (New England Biolabs) treatments were conducted by adding CIAP or λ-phosphatase with their reaction buffer to total proteins as described by the manufacturer before SDS-PAGE.
Cell-Free Protein Degradation Assay
The cell-free protein degradation assay of ICE1 was performed as described previously by Liu et al. (2010) and Wang et al. (2013). Briefly, total proteins were extracted from 14-d–old seedlings in degradation buffer (50 mM of Tris-MES at pH 8.0, 500 mM of Suc, 1 mM of MgCl2, 10 mM of EDTA at pH 8.0, and 5 mM of DTT. Recombinant His-ICE1 protein and 10 mM of ATP were added to the extracts. After incubating for the indicated time, ICE1 protein was detected by immunoblotting with anti-His antibodies.
Immunoblot Assay
Immunoblot assays were performed as described previously by Shi et al. (2012). Briefly, total proteins were extracted from seedlings that were treated as described in the figure legends of Figures 4A, 4C, 7A, and 7C, and Supplemental Figures 3A and 7A using protein extraction buffer (50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 1% [v/v] Triton X-100, 2% [v/v] Nonidet P-40, 5 mM of DTT, and protease inhibitor cocktail [Roche]). HF-ICE1 was detected with an anti-HA antibody (cat. no. H3663; Sigma-Aldrich). Actin detected with an anti-Actin antibody (cat. no. BE0037; EASYBio) was used as a loading control.
Transient Transactivation Assay
The pCBF3:GUS and HF-ICE1 vectors were generated previously by Ding et al. (2015). BIN2 CDS was cloned into the pCM1307 vector with HA-FLAG to generate HF-BIN2. pCBF3:GUS and 35S:Luc were cotransformed with the vectors described in the figure legends of Figure 4F into Arabidopsis protoplasts and incubated for 16 h in the dark. 35S:Luc was used as an internal control. GUS and luciferase (Luc) activity were measured as described in Shi et al. (2012), and relative GUS activity (GUS/Luc) was calculated to reflect transcriptional activity.
EMSA
EMSA was performed using a LightShift Chemiluminescent EMSA Kit (cat. no. 20,148; Thermo Scientific) as described in Chinnusamy et al. (2003) and Ding et al. (2015). Briefly, purified His-ICE1 fusion protein was incubated with GST-BIN2 in kinase reaction buffer (20 mM of Tris-HCl at pH 7.5, 10 mM of MgCl2, 100 mM of NaCl, and 1 mM of DTT) with or without 25 mM ATP at 30°C for 30 min before being subjected to EMSA. The binding reaction was performed at 25°C for 15 min in a thermal cycler (Bio-Rad). The biotin-labeled probes used in this assay were described by Chinnusamy et al. (2003) and Ding et al. (2015).
In-Gel Kinase Assays
In-gel kinase assays were performed as described in Fujii et al. (2007), with some modifications. Briefly, total proteins were extracted from Super:BIN2-MYC transgenic or wild-type plants after cold treatment (as described in the figure legend of Figure 7E) with protein extraction buffer (5 mM of EDTA, 5 mM of EGTA, 5 mM of DTT, 25 mM of NaF, 1 mM of Na3VO4, 20% [v/v] glycerol, 1 mM of phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail [Roche], and 50 mM of HEPES-KOH at pH 7.5). The proteins were immunoprecipitated with Anti-c-MYC magnetic beads (cat. no. 20,168; Thermo Fisher Scientific) and separated on an SDS-PAGE gel containing 0.3 mg/mL of purified MBP as the substrate. The gel was washed three times with SDS washing buffer (1 mM of DTT, 5 mM of NaF, 0.1 mM of Na3VO4, 0.5 mg/mL of BSA, 0.1% [v/v] Triton X-100, and 25 mM of Tris-HCl at pH 7.5) for 20 min each time. The gel was washed twice with renaturation buffer (2 mM of DTT, 5 mM of NaF, 0.1 mM of Na3VO4, and 25 mM of Tris-HCl at pH 7.5) for 20 min each time. The gel was then incubated in renaturation buffer for 12 h, followed by incubation in renewed renaturation buffer for 1 h at 4°C and in kinase reaction buffer (2 mM of EGTA, 12 mM of MgCl2, 1 mM of DTT, 0.1 mM of Na3VO4, and 25 mM of HEPES-KOH at pH 7.5) for 30 min. The gel was incubated in 30 mL of renewed kinase reaction buffer containing 60 μCi [γ-32P] of ATP and 9 μL of cold ATP (1 mM) at room temperature for 2 h and washed five times with washing buffer (5% [w/v] trichloroacetic acid and 1% [w/v] sodium pyrophosphate) for 30 min each time. Radioactivity was detected using a model no. Typhoon 9410 Variable Mode Imager (Molecular Dynamics).
Statistical Analysis
The integrated optical density (IOD) values of indicated signals were quantified using the software Image-Pro Plus (Media Cybernetics; the raw data had seven significant figures). The relative radiation of ICE1 in Figure 2C was calculated as IODICE1/IODWT. The relative protein levels of ICE1 (IODICE1/IODActin) in Figures 3, 4A to 4D, 6A to 6D, and to D7A to 7D and Supplemental Figures 3A, 3B, 6, 7A, and 7B were calculated as IODICE1/IODActin and set in relation to IODICE1/IODActin at 0 min. The relative kinase activity in Figures 7E and 7F was calculated as IODAuto/IODMYC and set in relation to IODAuto/IODMYC at 0 h. The relative fluorescence intensity in Supplemental Figure 1B was set in relation to the fluorescence intensity without cold treatment. All representative pictures are shown with two digits after the decimal point for proper display.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: BIN2 (AT4G18710), ICE1 (AT3G26744), HOS1 (AT2G39810), CBF1 (AT4G25490), CBF2 (AT4G25470), CBF3 (AT4G25480), KIN1 (AT5G15960), COR15A (AT2G42540), RD29A (AT5G52310), BIL1 (AT2G30980), BIL2 (AT1G06390), OST1 (AT4G33950).
Supplemental Data
Supplemental Figure 1. ICE1 interacts with BIN2 and its homologs.
Supplemental Figure 2. BIN2, BIL1, and BIL2 phosphorylate ICE1 in vitro.
Supplemental Figure 3. BIN2 decreases ICE1 stability and transcriptional activity.
Supplemental Figure 4. The CRISPR/Cas9-generated ice1-3 mutant exhibits increased sensitivity to freezing stress.
Supplemental Figure 5. Expression of CBF genes and CBF target genes in the ice1-3 mutant.
Supplemental Figure 6. OST1 attenuates the interaction between ICE1 and HOS1.
Supplemental Figure 7. Mutation of BIN2 and its homologs attenuates ICE1 degradation during cold stress.
Supplemental Table 1. Multivariate analysis indicates an interactive effect between ICE1 and BIN2/BIL1/BIL2.
Supplemental Table 2. List of primer sequences used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Tonglin Mao for providing the bin2-3 bil1 bil2 mutant seeds, Zhen Li for helping with the liquid chromatography-tandem mass spectrometry analysis, and Michael F. Thomashow for critically reading the article and discussing it with the authors. This work was supported by the National Key Scientific Research Project (2015CB910203), the Ministry of Agriculture of China for Transgenic Research (2016ZX08009002), and the National Natural Science Foundation of China (31730011 and 31330006).
AUTHOR CONTRIBUTIONS
S.Y. directed the project; K.Y., H.L., and S.Y. designed the experiments; K.Y. and H.L. performed the experiments; K.Y., H,L., Y.D., Y.S., C.S., Z.G., and S.Y. discussed and interpreted the data; K.Y., H.L., Y.D., and S.Y. wrote the article.
References
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M., Hao Y., Kapoor A., Dong C.H., Fujii H., Zheng X., Zhu J.K. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281: 37636–37645. [DOI] [PubMed] [Google Scholar]
- Casson S.A., Hetherington A.M. (2012). GSK3-like kinases integrate brassinosteroid signaling and stomatal development. Sci. Signal. 5: pe30. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J.K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12: 444–451. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dardick C., Chen J., Richter T., Ouyang S., Ronald P. (2007). The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 143: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Jia Y., Shi Y., Zhang X., Song C., Gong Z., Yang S. (2018). OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 37: e98228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32: 278–289. [DOI] [PubMed] [Google Scholar]
- Ding Y., Lv J., Shi Y., Gao J., Hua J., Song C., Gong Z., Yang S. (2019). EGR2 phosphatase regulates OST1 kinase activity and freezing tolerance in Arabidopsis. EMBO J. 38: e99819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in Cold-Regulated gene expression and freezing tolerance. Plant Cell 21: 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivers E., Demagny H., De Robertis E.M. (2009). Integration of BMP and Wnt signaling via vertebrate Smad1/5/8 and Drosophila Mad. Cytokine Growth Factor Rev. 20: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eivers E., Fuentealba L.C., De Robertis E.M. (2008). Integrating positional information at the level of Smad1/5/8. Curr. Opin. Genet. Dev. 18: 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina M., Unterholzner S.J., Rathnayake A.I., Castellanos M., Khan M., Kugler K.G., May S.T., Mayer K.F.X., Rozhon W., Poppenberger B. (2016). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 113: E5982–E5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba L.C., Eivers E., Ikeda A., Hurtado C., Kuroda H., Pera E.M., De Robertis E.M. (2007). Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131: 980–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124: 1854–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16: 433–442. [DOI] [PubMed] [Google Scholar]
- Gudesblat G.E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., Russinova E. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14: 548–554. [DOI] [PubMed] [Google Scholar]
- Guo X., Liu D., Chong K. (2018). Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 60: 745–756. [DOI] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., Yu D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li J., Bao F., Zhang X., Yang S. (2010). A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 154: 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- Jia Y., Ding Y., Shi Y., Zhang X., Gong Z., Yang S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212: 345–353. [DOI] [PubMed] [Google Scholar]
- Jiang B., Shi Y., Zhang X., Xin X., Qi L., Guo H., Li J., Yang S. (2017). PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 114: E6695–E6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C., Hirt H. (2002). Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 7: 457–461. [DOI] [PubMed] [Google Scholar]
- Kang S., Yang F., Li L., Chen H., Chen S., Zhang J. (2015). The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant Physiol. 167: 1076–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291. [DOI] [PubMed] [Google Scholar]
- Kim Y., Park S., Gilmour S.J., Thomashow M.F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75: 364–376. [DOI] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Thomashow M.F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu M.D., Zou C., Hanada K., Shiu S.H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ding Y., Shi Y., Zhang X., Zhang S., Gong Z., Yang S. (2017b). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 43: 630–642.e4. [DOI] [PubMed] [Google Scholar]
- Li H., Ye K., Shi Y., Cheng J., Zhang X., Yang S. (2017a). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 10: 545–559. [DOI] [PubMed] [Google Scholar]
- Liu J., Shi Y., Yang S. (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60: 780–795. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Jia Y., Ding Y., Shi Y., Li Z., Guo Y., Gong Z., Yang S. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14–3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66: 117–128. [DOI] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ohta M., Nakazawa M., Ono M., Hasegawa P.M. (2011). ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J. 67: 269–279. [DOI] [PubMed] [Google Scholar]
- Novillo F., Alonso J.M., Ecker J.R., Salinas J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Cho H., Kim K., Hwang I. (2010). Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 29: 283–290. [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23: 623–637. [DOI] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., Yang S. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. (2013). Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 27: 681–688. [DOI] [PubMed] [Google Scholar]
- Wang Z., Meng P., Zhang X., Ren D., Yang S. (2011). BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 67: 1081–1093. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang S. (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20: 56–64. [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li J., Li F., Liu H., Yang W., Chong K., Xu Y. (2017). OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev. Cell 43: 731–743. [DOI] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J.K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171: 2744–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Wang P., Si T., Hsu C.C., Wang L., Zayed O., Yu Z., Zhu Y., Dong J., Tao W.A., Zhu J.K. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43: 618–629.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]