Profiling of 11 genome-wide outputs of gene regulation in response to hypoxia reveals multiple aspects of nuclear and whole-cell epigenetic and posttranscriptional gene regulation in Arabidopsis.
Abstract
Gene regulation is a dynamic process involving changes ranging from the remodeling of chromatin to preferential translation. To understand integrated nuclear and cytoplasmic gene regulatory dynamics, we performed a survey spanning the epigenome to translatome of Arabidopsis (Arabidopsis thaliana) seedlings in response to hypoxia and reoxygenation. This included chromatin assays (examining histones, accessibility, RNA polymerase II [RNAPII], and transcription factor binding) and three RNA assays (nuclear, polyadenylated, and ribosome-associated). Dynamic patterns of nuclear regulation distinguished stress-induced and growth-associated mRNAs. The rapid upregulation of hypoxia-responsive gene transcripts and their preferential translation were generally accompanied by increased chromatin accessibility, RNAPII engagement, and reduced Histone 2A.Z association. Hypoxia promoted a progressive upregulation of heat stress transcripts, as evidenced by RNAPII binding and increased nuclear RNA, with polyadenylated RNA levels only elevated after prolonged stress or reoxygenation. Promoters of rapidly versus progressively upregulated genes were enriched for cis-elements of ethylene-responsive and heat shock factor transcription factors, respectively. Genes associated with growth, including many encoding cytosolic ribosomal proteins, underwent distinct histone modifications, yet retained RNAPII engagement and accumulated nuclear transcripts during the stress. Upon reaeration, progressively upregulated and growth-associated gene transcripts were rapidly mobilized to ribosomes. Thus, multilevel nuclear regulation of nucleosomes, transcript synthesis, accumulation, and translation tailor transient stress responses.
INTRODUCTION
The regulated expression of protein-coding genes in eukaryotes involves processes within the nucleus, including the remodeling of chromatin, recruitment of RNA polymerase II (RNAPII), cotranscriptional processing, and export of mature mRNA to the cytoplasm, where it may be translated, sequestered, or degraded. These processes are modulated by conditions that necessitate changes in metabolism and growth to maintain viability. In Arabidopsis (Arabidopsis thaliana), cellular hypoxia promotes the activation of anaerobic metabolism to enable ATP production for basic cellular processes. This involves the rapid activation of transcription of 49 hypoxia-responsive genes (HRGs) followed by the active translation of their mRNAs, while most other transcripts are sequestered from translation complexes until reaeration (Branco-Price et al., 2005, 2008; Juntawong et al., 2014; Sorenson and Bailey-Serres, 2014). The well-translated HRG mRNAs lack notable sequences or features that can explain their effective translation and avoidance of sequestration (Branco-Price et al., 2005; Juntawong et al., 2014; Sorenson and Bailey-Serres, 2014), leading to the hypothesis that their preferential translational reflects aspects of their nuclear regulation.
Transcriptional activation of many HRGs involves the evolutionarily conserved group VII ethylene response factor (ERFVII) transcription factors (TFs), which are required for survival under hypoxia (Xu et al., 2006; Hattori et al., 2009; Gibbs et al., 2011; Licausi et al., 2011; Yang et al., 2011; Abbas et al., 2015; Paul et al., 2016; Eysholdt-Derzsó and Sauter, 2017; Giuntoli and Perata, 2018). The five Arabidopsis ERFVIIs are stabilized under hypoxia due to attenuation of their oxygen-stimulated proteolysis via the Arg branch of the N-degron pathway (Gibbs et al., 2011; Licausi et al., 2011; Paul et al., 2016; White et al., 2017; Millar et al., 2019). These include three constitutively synthesized ERFVIIs (RELATED TO APETALA2.2 [RAP2.2], RAP2.3, and RAP2.12) shown to transactivate HRG promoters in protoplasts through a hypoxia-responsive promoter cis-element (HRPE) identified in ∼50% of the HRGs (Giuntoli et al., 2014; Gasch et al., 2016; Giuntoli and Perata, 2018). Two other ERFVIIs, HYPOXIA-RESPONSIVE ERF1 and 2 (HRE1/2), are upregulated by hypoxia along with ALCOHOL DEHYDROGENASE1 (ADH1) and PLANT CYSTEINE OXIDASE1/2 (PCO1/2), which facilitate anaerobic metabolism and catalyze turnover of the ERFVIIs, respectively (Giuntoli et al., 2014, 2017; Weits et al., 2014; White et al., 2017).
Transcription requires the binding of TFs to specific cis-elements typically located near the transcription start site (TSS) to facilitate the assembly of an RNAPII initiation complex. The depletion of nucleosomes near a TSS is controlled by ATP-dependent chromatin remodelers and can be influenced by interactions with the transcriptional apparatus. TF binding and RNAPII activity both influence and are influenced by specific modifications and variants of histones (Gates et al., 2017; Talbert and Henikoff, 2017), which thereby serve as a proxy of gene activity. For example, tri-methylated Histone 3-lysine 27 (H3K27me3) and H3K4me3 are prevalent in the bodies of genes transcribed at low and high levels, respectively (Asensi-Fabado et al., 2017; Gates et al., 2017). Acetylation of Histone H3 Lys residues (H3K9ac, H3K14ac) reduces interactions between DNA and histones and is associated with ongoing transcription (Gates et al., 2017), including genes actively transcribed during environmental stress in plants (Eberharter and Becker, 2002; Kim et al., 2008, 2012). A characteristic of heat-stress–activated genes of Arabidopsis is high levels of the Histone 2A variant H2A.Z at the first nucleosome within the gene body, a feature proposed to reduce the energy required to commence transcriptional elongation at elevated temperatures (Kumar and Wigge, 2010; Cortijo et al., 2017; Dai et al., 2017; Sura et al., 2017; Torres and Deal, 2019).
As transcription commences, the phosphorylation of specific residues within the heptad repeats of the carboxyl terminal domain (CTD) of RNAPII orchestrates interactions with factors that facilitate transcription-coupled histone modifications as well as the cotranscriptional 5′ capping, splicing, and polyadenylation of the nascent transcript (Hajheidari et al., 2013; Milligan et al., 2016). CTD phosphorylation at Ser 2 (Ser2P) demarks active elongation (Phatnani and Greenleaf, 2006). In animals, pausing of RNAPII downstream of the TSS is common among genes activated by heat stress (Jonkers and Lis, 2015). The mapping of the 5′ ends of nascent transcripts by Native Elongating Transcript sequencing indicated that elongation can be rate-limiting shortly after initiation in Arabidopsis seedlings (Zhu et al., 2018), although this was not discernable when nascent transcripts were surveyed by global nuclear run-on sequencing (Hetzel et al., 2016). Yet, both studies found that transcription becomes rate-limited as RNAPII pauses just beyond the site of cleavage and polyadenylation. After initiation, cotranscriptional intron splicing, polyadenylation, and export are regulated by environmental cues including hypoxia (Branco-Price et al., 2008; Mustroph et al., 2009; Juntawong et al., 2014; Niedojadło et al., 2016; van Veen et al., 2016; de Lorenzo et al., 2017).
To better understand the integration of nuclear and cytoplasmic regulation of gene activity in response to hypoxia and reoxygenation, we performed a multiomic study of histone dynamics, open regions of chromatin, HRE2 binding, and RNAPII–Ser2P distribution in Arabidopsis. Eight chromatin readouts were compared with the modulation of nuclear RNA (nRNA), polyadenylated RNA (polyA RNA), and polyadenylated ribosome-associated RNA. The integrated analysis of these data exposed patterns of histone modifications, dynamic regulation of chromatin accessibility, cis-element enrichment, and temporal regulation of nRNA that accompanied dynamics in mRNA abundance and translation. This analysis exposed multiple layers of gene regulation including rapid and complete upregulation of HRGs, progressive upregulation of heat stress-response genes, and incomplete downregulation of genes associated with growth, including ribosomal proteins (RPs). Both transcriptional elongation and nuclear export appear to be points of regulation in response to oxygen availability. The data set produced in this study provides a resource for evaluating epigenetic state and RNAPII activity through translation in a model organism.
RESULTS
A Multiscale Data Set for Analyzing Regulatory Dynamics from Chromatin to Translation
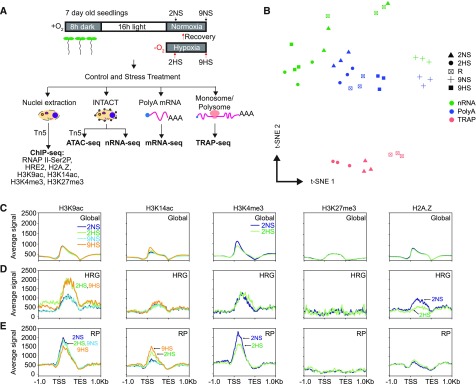
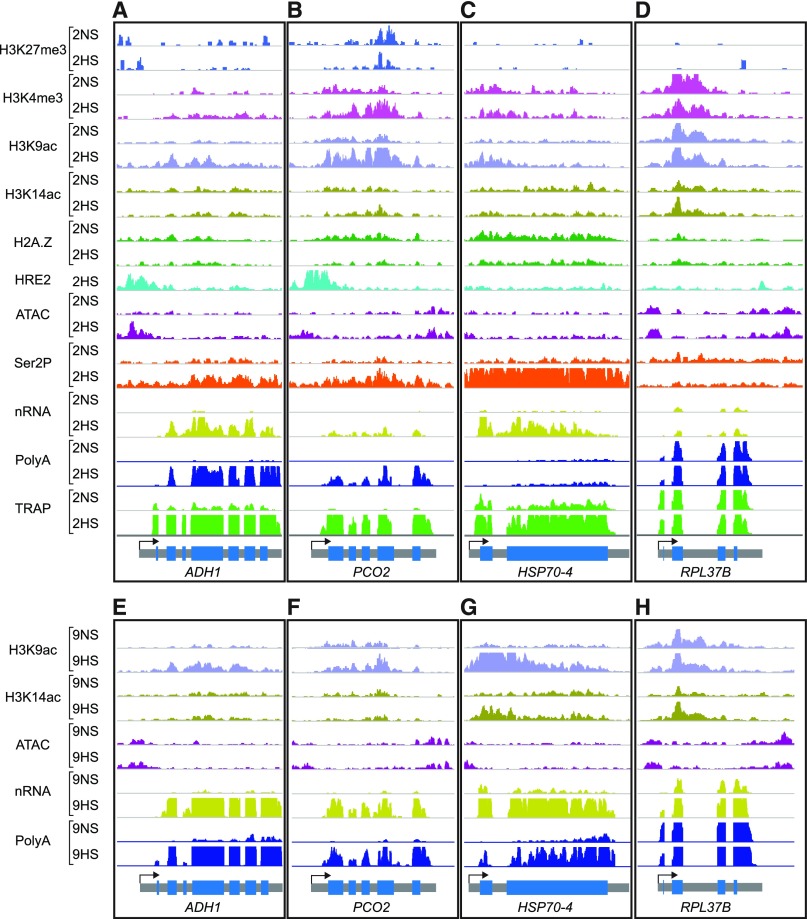
Here, we generated 11 distinct chromatin and RNA-based genome-scale data sets to evaluate gene regulatory dynamics in 7-day–old Arabidopsis seedlings grown under long-day light conditions that were treated with normoxia stress (NS; 2 h, 2NS; 9 h, 9NS), nonlethal hypoxic stress (HS; 2HS, 9HS), and reaeration (R; 2HS followed by 1 h to 2 h NS, R). All growth and treatment conditions were consistent with those of prior analyses (Figure 1A; Supplemental Figure 1A; Branco-Price et al., 2008; Mustroph et al., 2009; Juntawong et al., 2014). Chromatin immunopurification and sequencing (ChIP-seq) was used to survey the position and abundance of H3K9ac, H3K14ac, H3K4me3, H3K27me3, and H2A.Z along genes. The capture of nuclei by Isolation of Nuclei Tagged in Specific Cell Types (INTACT; Deal and Henikoff, 2010; Maher et al., 2018) was coupled with Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) to map regions of nucleosome-depleted chromatin (Buenrostro et al., 2013). We also used ChIP-seq to survey regions bound by RNAPII–Ser2P and the hypoxia-induced ERFVII HRE2. Finally, we assayed three transcript populations defined by the methods used for purification: ribosomal RNA (rRNA)-subtracted nuclear transcriptome obtained by INTACT (Reynoso et al., 2018b), polyA RNA selected by binding to oligo(dT) beads (transcriptome), and RNA obtained by Translating Ribosome Affinity Purification (TRAP and oligo[dT] selection, translatome; Zanetti et al., 2005; Juntawong et al., 2014). Each RNA population was reproducible and distinguishable when plotted by t-distributed stochastic neighbor embedding (t-SNE; Figures 1B and 1C; Supplemental Figure 1).
Figure 1.
Multiscale Chromatin and RNA Gene Regulatory Analyses of Control (Normoxic) and Hypoxic Arabidopsis Seedlings.
(A) Schematic of the experiments performed. Seven-day–old seedlings were subjected to control (normoxic; 2NS, 9NS), hypoxic stress (2HS, 9HS), or reoxygenation after 2HS (R) conditions. 2NS is ZT16, and 9NS is ZT1, with 1-h extended darkness. Vertical arrows indicate time of harvest. ChIP was performed to evaluate genomic regions bound by Ser2P, the Histone 2 variant H2A.Z, modified Histone 3 (H3K4me3, H3K27me3, H3K9ac, and H3K14ac), and the ERF TF, HRE2. INTACT purified nuclei were used for the ATAC and purification of nRNA. Ribosome-associated mRNA was obtained by TRAP.
(B) Individual replicate samples of nRNA, polyA RNA, and TRAP mRNA [TRAP] were compared by t-SNE.
(C) to (E) Distributions of histone modifications/variants across genic regions (C) for the core HRGs (n = 49; D) and cytosolic RPs (n = 246; E). Read distributions (RPKM * 1,000) are plotted from 1 kb upstream to 1 kb downstream of gene units defined by the TSS and TES.
Stress- and Growth-Associated Genes Differ in Chromatin Accessibility, Histone, Modifications, and RNA Modulation under Hypoxia
For the nucleosome-level evaluation, we plotted the histone data along each annotated protein-coding gene and compared global averages for each condition (Figure 1C; Supplemental Figure 2). H3 modifications associated with transcription (H3K9ac, H3K14ac, and H3K4me3) and stress-activated transcription (H2A.Z) were enriched just 3′ of the TSS and tapered off at the primary site of polyadenylation, the transcription end site (TES), whereas H3K27me3 was distributed more evenly across gene bodies. Hypoxia had little effect on these modifications at the global level, with the exception of a slight elevation of H3K14ac and reduction of H3K4me3 levels near the TSS.
We surveyed the dynamic changes in histone modification, abundance, and distribution for the induced and translated HRGs and stable but poorly translated cytosolic RP genes. H3K9ac significantly increased and H2A.Z decreased on HRGs at 2HS, with limited changes in these marks on RPs (Figures 1D and 1E; Supplemental Figures 1B and 3). H3K4me3 levels were notably reduced on the RPs, but not HRGs, in response to 2HS. When the stress was extended from 2 h to 9 h, H3K9ac significantly increased on 863 and 3,646 genes and H3K14ac on 2 and 1,216 genes after 2HS and 9HS, respectively (Supplemental Figure 1). A progressive increase of H3K14ac but not H3K9ac on RPs was accompanied by little change in steady-state transcript abundance (Supplemental Figures 1C, 3A, and 3C). These data indicate that these two gene cohorts undergo distinct epigenetic regulation in response to hypoxia.
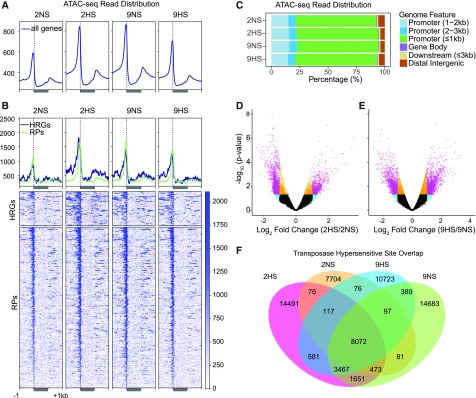
Next, we evaluated chromatin accessibility dynamics in genic regions by ATAC-seq (Figures 2A and 2B). Accessibility in 5′ promoter regions is associated with nucleosome depletion and the binding of transcriptional machinery (Lu et al., 2017; Maher, 2018; Sijacic et al., 2018), whereas accessibility in 3′ flanking regions is associated with the formation of chromatin loops and transcriptional termination (Ansari and Hampsey, 2005; Ozsolak et al., 2008). The ATAC-seq reads mapped almost exclusively within 1,000 bp 5′ of the TSS and ∼300 bp 3′ of the major TES of genes under all four conditions (Figure 2C). The average ATAC signal increased within the promoters of both HRGs and RPs (1.8- and 1.4-fold, respectively) after 2HS, yet ATAC reads showed expansion into more 5′ and 3′ flanking regions on the HRGs. Peak-calling of Tn5 hypersensitive insertion sites (THSs) across the genome identified constitutively present (8,072), non-stress–specific (22,387) and an even larger group of stress-specific THSs (25,795; Figures 2D to 2F; Supplemental Figure 2A), indicating that chromatin accessibility is readily influenced by the environment.
Figure 2.
Regions of Chromatin Accessibility Determined by ATAC-Seq Are Dynamically Regulated by HS.
(A) and (B) ATAC-seq read distribution of chromatin accessibility for protein-coding genes (A), HRGs and RPs (B).
(C) Distribution of THSs on genomic features for each condition.
(D) and (E) Volcano plots of the log2 FC in THSs identifiable under two conditions and the significance values of their differences. Genes indicated in pink meet the criteria of log2 FC value > |1|, 0.05 < P value.
(F) Overlap in the number of THSs identified for each of four conditions.
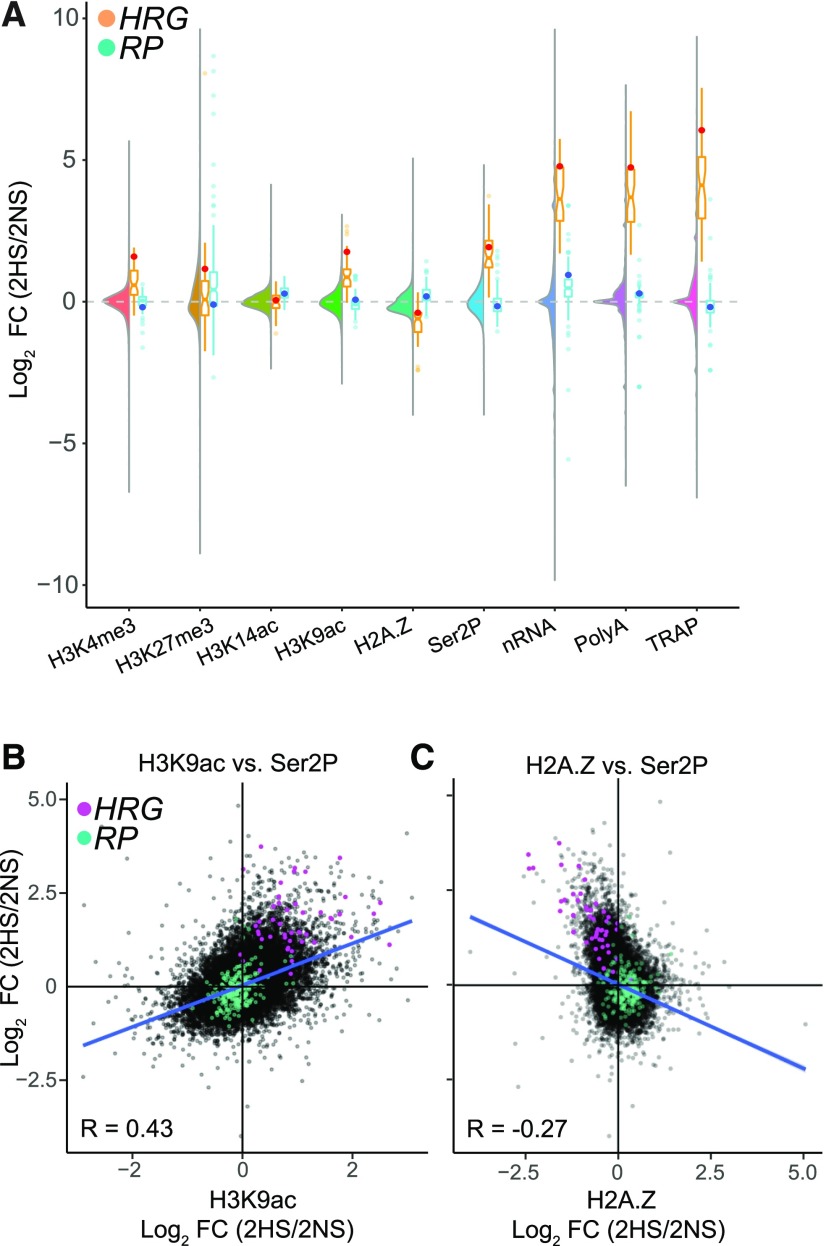
To complete our survey of gene activity, we compared the distribution of the log2 fold change (FC; 2HS/2NS) values of chromatin, RNAPII–Ser2P, and the four RNA populations at the genome-scale as well as for the HRGs and RPs (Figure 3A; Supplemental Figure 1A). We found that stress-induced RNAPII–Ser2P engagement was generally correlated with increased H3K9ac (global R = 0.43; HRGs R = 0.17) and H2A.Z eviction (global R = −0.27), particularly for the HRGs (R = −0.79; Figures 3B and 3C; Supplemental Figures 3B and 4A). RPs contrasted to the HRGs, displaying only minor changes in RNAPII–Ser2P association at 2HS but increased H3K14ac and nRNA levels at 9HS (Figure 3A; Supplemental Figures 3B and 3C). These results further demonstrate that the epigenetic regulation of stress-induced and growth-associated genes is distinct under hypoxia.
Figure 3.
Short-Term HS Promotes Pronounced Changes in Chromatin, Transcription, and Nascent Transcripts that Are Not Uniformly Reflected in Transcript Steady-State Abundance or Translation
Comparison of genome-wide data for chromatin and RNA for 2HS relative to 2NS.
(A) Combined violin and boxplot of log2 FC (2HS/2NS) of histone, RNAPII–Ser2P, and RNA outputs. Violin plot, all genes; boxplot for the HRGs plotted in orange with ADH1 depicted as a red dot and the cytosolic RPs plotted in blue, with RPL37B depicted as a dark blue dot.
(B) Comparison of log2 FC (2HS/2NS) between H2A.Z on the gene body and Ser2P on the gene body.
(C) Comparison of log2 FC (2HS/2NS) between H3K9ac and Ser2P on the gene body.
For (B) and (C), HRGs are plotted in pink and RPs in blue; Pearson’s coefficient of determination is shown for all genes.
Regulation at the Nuclear and Cytoplasmic Scales Can Be Concordant or Discordant
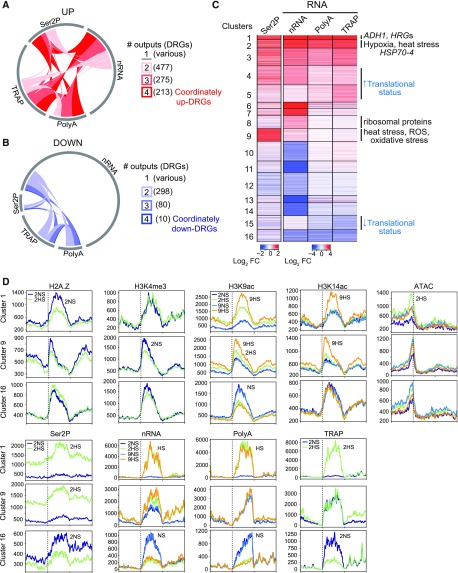
Next, we evaluated the generally presumed hypothesis that HRGs are coordinately transcribed and translated under hypoxia. To do so, we performed a global-scale systematic analysis of the RNAPII–Ser2P, nRNA, polyA RNA, and TRAP RNA data sets using coregulation and cluster analyses. When we investigated the dynamics of significantly up- and downregulated genes (up-DRGs and down-DRGs; |log2 FC| > 1; false discovery rate [FDR] < 0.05) in all four readouts of gene activity, we found that nRNA had the greatest (1,722) and polyA RNA the fewest (602) up-DRGs (Figure 4A). Only 213 genes were significantly upregulated in all four readouts. These coordinately up-DRGs included 38 of the 49 HRGs. A parallel analysis found that 72% of the polyA down-DRGs were significantly downregulated in at least one other data set, with only 10 coordinately down-DRGs (Figure 4B).
Figure 4.
Multiscale Analysis of Hypoxia-Regulated Genes Reveals Concordant and Discordant Patterns of Gene Regulation
Number of (A) up- and (B) down-RGs in response to 2 h of HS (|log2 FC| > 1; FDR < 0.05) identified in each of the four readouts related to RNA: Ser2P ChIP, nRNA, polyA RNA, and TRAP RNA. Arc of circle circumference indicated by the gray line represents the DRGs in each of the four readouts. Genes differentially regulated in two to four readouts were tabulated (number in parentheses) and depicted by links within the circle. Genes differentially regulated in only one readout are represented by unlinked gray lines.
(C) Heatmap showing similarly regulated genes based on four assays of gene activity. Partitioning around medoids clustering of 3,042 DRGs for the identification of cluster members and centers; 16 clusters. Selected enriched GO terms are shown on the right (P adjusted < 1.47e-09), |log2 FC| > 1, FDR < 0.05. Clusters displaying increased or decreased translational status (ribosome-associated [TRAP] relative to total abundance [polyA]) are shown.
(D) Average signals (RPKM * 1,000) of various chromatin and RNA outputs for genes of three clusters plotted from 1 kb upstream of the TSS to 1 kb downstream of the TES for the 2 h (2NS, 2HS) and 9 h (9NS, 9HS) time points. Signal scale of the graphs differ.
Gene transcripts can be nuclear-enriched (Yang et al., 2017), but there is limited knowledge of the coordination of differential regulation of nRNA and polyA mRNA populations under stress. Our pairwise comparison of nRNA and polyA RNA abundance at 2NS and 2HS identified transcripts that were enriched in either nRNA or polyA RNA (Supplemental Figure 5; Supplemental Figure 3). Under both conditions, transcripts enriched in nRNA included many associated with the cell cycle and gene silencing, whereas transcripts enriched in the polyA pool were associated with plastid and ribosome biogenesis and function. These data reveal a complex nuclear and cytoplasmic regulatory landscape, which we further explored by integrative analyses of the chromatin and RNA data sets.
Rapid and Coordinate Versus Progressive Upregulation Are Characterized by Distinct Patterns of Histone Alterations
To better identify genes with similar patterns of regulation, DRGs identified in at least one of the four data sets (n = 3,042) were clustered into 16 groups and evaluated by gene ontology (GO) term enrichment (Figure 4C; Supplemental Figure 4A). This analysis confirmed coordinately up-DRGs (clusters 1 to 3) and down-DRGs (cluster 16). Genes in the remaining clusters showed discordant regulation in one or more readout.
To better understand the regulatory variation at the chromatin, RNAPII, and RNA levels, we plotted the average signal value for each data type for each cluster along genic regions (Figure 4D; Supplemental Figure 6). This analysis revealed that the rapid and complete upregulation of cluster-1 to -3 genes at 2HS was accompanied by enhanced chromatin accessibility, H2A.Z eviction, and H3K9ac and RNAPII–Ser2P elevation across the gene body. The concomitant rise in TRAP RNA indicated that these transcripts were translated in proportion to their increased abundance, as shown for the HRGs by high-resolution ribosome footprint analysis (Juntawong et al., 2014). Not unexpectedly, the first two clusters were enriched for genes associated with decreased oxygen (cluster 1, P adjusted < 6.73e−22) and general stress (cluster 2, <1.06e−21; Supplemental Figure 4A). Genome browser views of ADH1 and PCO2 illustrate the coordinate upregulation of HRGs in all four assays of gene activity (Figures 5A and 5B). These examples also illustrate that individual genes undergo distinct changes in the chromatin and transcript readouts.
Figure 5.
Genome Browser View of Normalized Read Coverage of Histone, ATAC-seq, RNAPII–Ser2P, HRE2-ChIP, and RNA Outputs for Representative Genes
(A) to (D) Samples under 2NS and 2HS conditions.
(E) to (H) Samples under 9NS and 9HS conditions.
(A) and (E) ADH1 (Cluster 1), (B) and (F) PCO2 (Cluster 1), (C) and (G) HSP70-4 (Cluster 2), and (D) and (H) RPL37B (Cluster 9). The maximum read scale value used for chromatin-based and RNA-based outputs is equivalent for all genes depicted. At the bottom, the transcription unit is shown in gray with the TSS marked with an arrow. PCO2 shows a stronger decline in H2A.Z than ADH1 in response to hypoxia at 2HS (PCO2, −1.15 log2 FC; ADH1, −0.33 log2 FC) and provides an example of elevated H3K4me3 levels under HS.
Genes of clusters 4 to 8 were not upregulated to the same extent from RNAPII engagement through translation, indicating they are regulated by multiple mechanisms that are influenced by hypoxia. For cluster 4, the rise in nRNA levels coincided with increased H3K9ac and RNAPII–Ser2P, which is indicative of increased transcription, but was not accompanied by a concomitant rise in polyA or TRAP RNA (Figure 4C; Supplemental Figure 6). This incomplete upregulation could reflect nuclear retention or cytoplasmic destabilization of these transcripts. Clusters 6 to 8 genes were elevated only in nRNA at 2HS, suggesting these transcripts are nascent or remain in the nucleus. Of these, cluster 8 displayed little change in H3K9ac but a rise in H3K14ac as well as nRNA at 2HS and/or 9HS (Supplemental Figures 6 and 7), which is consistent with the regulation of the many RPs enriched in this cluster (Figure 3; Supplemental Figures 3B and 3C). RPL37B is shown as a representative RP (Figure 5D), as its regulation by hypoxia was followed previously (Sorenson and Bailey-Serres, 2014).
One of the most distinct gene regulatory patterns was observed for cluster 9, which showed a dramatic increase in RNAPII–Ser2P engagement that was not accompanied by a rise in nRNA or polyA RNA. This could reflect an increase in RNAPII engagement (initiation) without the completion of transcript synthesis or elevated rates of transcription accompanied by high cytoplasmic turnover. Compared with cluster-1 genes, cluster-9 genes had lower H2A.Z at 2NS, which was only slightly reduced at 2HS (Figure 4D; Supplemental Figures 6, 7A, and 7B). The association of cluster-9 genes with H3K4me3, a mark typical of actively transcribed genes, was significantly lower than for cluster-1 genes at 2HS (Supplemental Figures 6, 7C, and 7D). By 9HS, the promoter accessibility, H3K9ac, H3K14ac, nRNA, and polyA RNA abundance of cluster-9 genes increased (Figure 4D; Supplemental Figure 8), indicating that they are progressively activated by hypoxia. Cluster-9 genes were enriched for GO terms heat (<6.20e−13) and oxidative stress (<8.23e−11), which were also prevalent in cluster-2 (heat [7.74e−14]; oxidative stress [<1.13e−12]). Cluster 2 was distinguishable from cluster 9 in terms of induced Ser2P engagement, which produced nRNA and translated RNA. HEAT SHOCK PROTEIN 70-4 (HSP70-4), for example, displayed a 3.4-fold increase in RNAPII–Ser2P across the gene body region accompanied by increased nRNA and TRAP RNA levels but a delayed rise in polyA RNA abundance (Figure 5C). The pronounced early engagement of RNAPII–Ser2P followed by delayed elevation of polyA RNA levels indicates that genes of cluster 9 are progressively upregulated by hypoxia.
Highly Downregulated Genes Undergo Distinct Stimulus-Regulated Histone Modifications
A survey of the down-DRG genes (clusters 10 to 16) demonstrated stimulus-driven dynamics of chromatin, RNAPII, and RNAs with two dominant patterns: coordinate downregulation (cluster 16) and incomplete downregulation (clusters 10 to 15; Figure 4A). Cluster 10 to 14 genes were primarily distinguished by a significant reduction in nRNA at 2HS that was generally accompanied by increased H2A.Z and H3K14ac and decreased H3K9ac (Figure 4C; Supplemental Figure 6). Cluster-15 and -16 genes stood out as strongly and coordinately reduced in RNAPII–Ser2P engagement and at least two of the three RNA readouts at 2HS (Figure 4D; Supplemental Figure 6). These clusters had strongly reduced chromatin accessibility, elevation of H2A.Z, and a sharp decline in H3K9ac, all of which were antithetical to the changes observed for the continuously up-DRGs (Supplemental Figures 7A and 7C). Clusters 15 and 16 were enriched for GO categories including root development (cluster 15, <3.7e−8) and regulation of transcription and RNA biosynthesis (cluster 16, <7.78e−7). Thus, H2A.Z presence across a gene body, limited chromatin accessibility near the TSS, and reduced RNAPII engagement are signatures of restricted transcription under hypoxia.
Prolonged Hypoxia Promotes Variations in Nuclear Regulation of Growth-Associated Genes
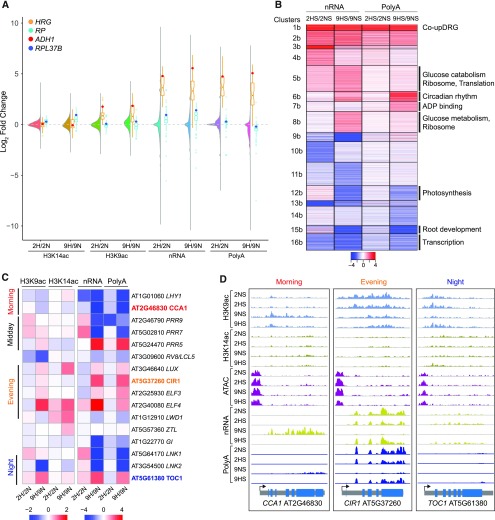
As the seedling’s response to sublethal HS changes over time (Branco-Price et al., 2008), we also evaluated changes in chromatin accessibility, H3K9ac, H3K14ac, nRNA, and polyA RNA at 9HS relative to aerated seedlings that underwent the same light treatment and other manipulations (Figures 2A to 2F, 6A, and 6B; Supplemental Figure 9; Supplemental Figure 1A). As observed for short-term hypoxia, there were differences in chromatin accessibility and THSs between 9HS and 9NS treatment. Additional study is required to determine if the distinction between 2NS and 9HS reflects diurnal regulation or an effect of a 1-h extended night corresponding to the 9NS time point. Of the 581 THSs observed under both 2HS and 9HS, 30 were associated with the promoter regions of the 49 HRGs. H3K9ac and H3K14ac levels were progressively yet distinctly altered for the HRG and RP cohorts (Figure 6A; Supplemental Figure 3). Over 350 genes were progressively upregulated, displaying a rise in H3K9ac, nRNA, and polyA RNA at 2HS and 9HS (clusters 1b to 2b; 38 HRGs). The remainder were incompletely up- (clusters 4b to 8b) or downregulated (clusters 9b to 11b), indicating that the regulatory patterns of genes within these clusters changed as hypoxia was prolonged. Many genes with elevated nRNA levels at 9HS were enriched for H3K14ac near their TSS (clusters 2b to 8b; Supplemental Figure 9), as exemplified by the RPs (enriched in clusters 5b and 8b). The increase in nRNA and H3K14ac without an accompanying rise in polyA RNA suggests that these genes are transcriptionally active but the nascent transcript remains in the nucleus, or the mature (polyA) transcript is rapidly degraded. Other genes demonstrated dynamic up- (cluster 6b) or downregulation between 2HS and 9HS (clusters 8b to 11b). The downregulation at 9HS was pronounced for genes related to photosynthesis (cluster 8b, <5.8e−7 and <1.5e−6), root development (cluster 10b, <1.7e−6), and transcription (cluster 11b, <7.6e−6; Supplemental Figure 4B). A key finding was that temporal regulation of nuclear and polyA transcript abundance during hypoxia can be distinct.
Figure 6.
Prolonged Hypoxia Leads to Further Variation in Epigenetic, Transcriptional, and Posttranscriptional Regulation.
(A) Combined violin and boxplot of H3K9ac and H3K14ac, nRNA, and polyA RNA data. Violin plots of log2 FC (9HS/9NS) data for all genes. Boxplots are data for the 49 HRGs plotted in orange with ADH1 depicted as a red dot and data for the cytosolic RPs plotted in blue, with RP37B depicted as a dark blue dot.
(B) Heatmap showing similarly regulated genes based on two dynamics in nRNA and polyA RNA. Partitioning around medoids clustering of 3,897 differentially regulated genes; 16 clusters. Selected enriched GO terms are shown at right (P adjusted < 1.37E-06), |log2 FC| > 1, FDR < 0.05.
(C) Heatmap of chromatin and RNA readouts of select circadian-regulated genes under short and prolonged HS. Heatmap scales between chromatin and RNA-based readouts differ.
(D) Brower views of a morning (CCA1), evening (CIR1, CIRCADIAN1), and night (TOC1) expressed gene. RNA scale of TOC1 is threefold lower than the scale used for CCA1 and CIR1.
A number of genes associated with phasing of the circadian clock were perturbed when hypoxia was prolonged (cluster 6b; circadian rhythm <7.9e−10). Upon closer examination, we found this included dampened upregulation of several morning genes and prolonged elevation of evening gene mRNAs, which normally peak toward the end of the light cycle when the stress was initiated (Figure 6C). Examples of this perturbation include the morning-expressed clock regulators CIRCADIAN CLOCK ASSOCIATED1 (CCA1; cluster 16b) and LATE ELONGATED HYPOCOTYL (cluster 15b), with dampened mRNA accumulation, and night-expressed TIMING OF CAB EXPRESSION (TOC1) with elevated polyA RNA levels after 9HS as compared with 9NS (Figures 6C and 6D). Thus, hypoxia imposed at the end of the light cycle delays or arrests circadian phasing.
Many Coordinately Hypoxia-Upregulated Genes Are Targets of ERFVII Regulation
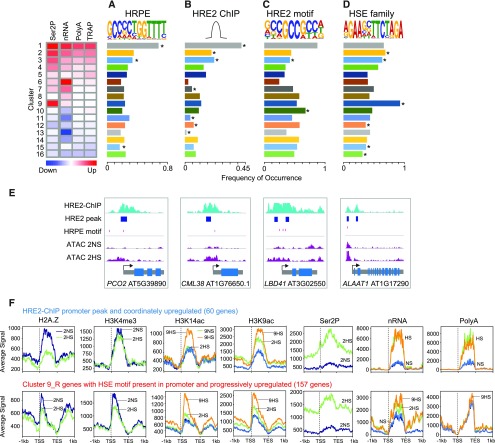
We hypothesized that the coordinately up-DRGs might be transcriptionally activated by the low-oxygen stabilized ERFVIIs. Motif scans for the HRPE within promoters (500 bp 5′ of the TSS) across the genome confirmed significant enrichment of this cis-motif in the cluster 1 genes resolved in the 2HS analysis (Figure 7A; Supplemental Figure 2), including genes with strong coordinate upregulation.
Figure 7.
Regulation of Hypoxic Responses by Two ERFVII and HSF TF Families
(A) to (D) Motif and peak enrichment in the promoter regions (1 kb upstream to 500 bp downstream of the TSS) of genes in clusters identified in Figure 4C for (A) HRPE, (B) HRE2-ChIP peaks, (C) HRE2-motif, and (D) heat shock elements (HSEs). Frequency of motif and peak occurrence was calculated as the number of motifs per number of genes in a cluster. Clusters with significant enrichment were identified by two-tailed Fisher’s exact test P < 0.05.
(E) Genome browser view of select HRGs (PCO2; CALMODULIN-LIKE38, CML38; LOB-DOMAIN CONTAINING PROTEIN41, LBD41; ALANINE AMINO TRANSFERASE1, ALAAT1). The locations of HRE2-ChIP peaks and HRPEs are depicted as a horizontal blue box and vertical red line, respectively.
(F) Average signal (RPKM * 1,000) of chromatin and RNA readouts for HRE2-bound genes with coordinate upregulation and genes with progressive upregulation (cluster 9) containing promoter HSEs. Average signal is plotted from 1 kb upstream of the TSS to 1 kb downstream of the TES.
The ERFVII HRE2 is encoded by an HRG. To gain insight into the role of HRE2, ChIP-seq was performed on 2HS seedlings overexpressing a stabilized version of this protein. Over 75% of the ∼1,800 promoter bound peaks identified mapped within 1 kb upstream of TSSs, with clear enrichment of binding within 500 bp upstream of the TSS (Supplemental Figure 10; Supplemental Figure 2A; Supplemental File 1), as evident in the promoters of ADH1 and PCO2 (Figures 5A and 5B; Supplemental Figures 10C and 10D). The binding of HRE2 to promoter regions was highest for cluster 1, but also significantly enriched for genes of clusters 2 and 3 of the 2HS analysis (Figure 7B). Overall, HRE2 peaks were present in 28% of the 213 coordinately up-DRGs (Supplemental Figure 1).
We anticipated that HRE2 might be associated with the HRPE known to be transactivated by RAP2.12, 2.2, and 2.3 (Bui et al., 2015; Gasch et al., 2016). However, de novo motif discovery using the HRE2 peak regions yielded an overrepresented multimeric 5′-GCC-3′ element (P value <1e−351; Figure 7C; Supplemental Figure 2) that resembles the GCC-rich EBP box bound by RAP2.12 in an in vitro DNA affinity purification and sequencing assay (O’Malley et al., 2016) and by RAP2.3 based on ChIP-quantitative (q)PCR and electrophoretic mobility shift assays (Büttner and Singh, 1997; Gibbs et al., 2014). The HRE2/GCC motif was enriched in cluster 1 but also was more prevalent than the HRPE in other clusters (Figure 7C). Our data strongly suggest that HRE2 binds to promoter regions containing a GCC-rich EBP-like box. Yet, in over 50% of the 213 coordinately up-DRGs, an HRPE is also present (Supplemental Figure 1). Collectively, these observations support the conclusion that many of the coordinately up-DRGs are transcriptionally activated by one or more ERFVII.
Hypoxic-Stress Progressively Activates Genes in Heat and Oxidative Stress Networks
The regulatory variation of oxidative stress and heat response genes displayed by clusters 2 to 3 and 9 genes at 2HS (Figure 4C) was notable. Anoxia upregulates heat-stress associated genes (Banti et al., 2010; Loreti et al., 2005), and both the onset of hypoxia and reoxygenation trigger a reactive oxygen species (ROS) burst associated with signal transduction (Chang et al., 2012). Elevation of RNAPII–Ser2P without a commensurate increase in polyA RNA was characteristic of HSPs, RESPIRATORY BURST OXIDASE D, and transcriptional co/activators associated with heat and oxidative stress (i.e. MULTIPROTEIN BRIDGING FACTOR 1C, DEHYDRATION-RESPONSIVE2A, and ZINC FINGER PROTEIN10 and 12; Supplemental Figure 11). We also noted that the transcript levels of several HEAT SHOCK FACTOR (HSF) transcriptional activators were elevated in TRAP RNA at 2HS (HSFA2, HSFA4A, and HSFA7A). We considered that the progressive upregulation of activity observed for cluster-9 genes may be mediated by HSFs and associated with certain epigenomic features. Indeed, motif-enrichment analysis confirmed that HSF cis-elements (HSEs) were significantly enriched in promoters of cluster-9 genes (85% of genes) and to a lesser extent in cluster-2 and -3 genes (Figure 7D). Moreover, progressively up-DRGs with a 5′ HSE motif were distinguishable from the continuously upregulated HRE2-bound DRGs in four ways (Figure 7F): (1) an overall 5′ bias in modified nucleosomes, (2) more 5′ localized H2A.Z with less dramatic eviction at 2HS, (3) reduced H3K4me3 and increased H3K14ac that preceded the rise in H3K9ac, and (4) a delay in the increase in polyA RNA until 9HS. Thus, the regulatory variations for progressively up-DRGs with promoter-localized HSEs are distinct from those of the coordinately up-DRGs bound by HRE2 under hypoxia.
Nuclear-Regulated Stress-Responsive Genes Are Highly Translated upon Reaeration
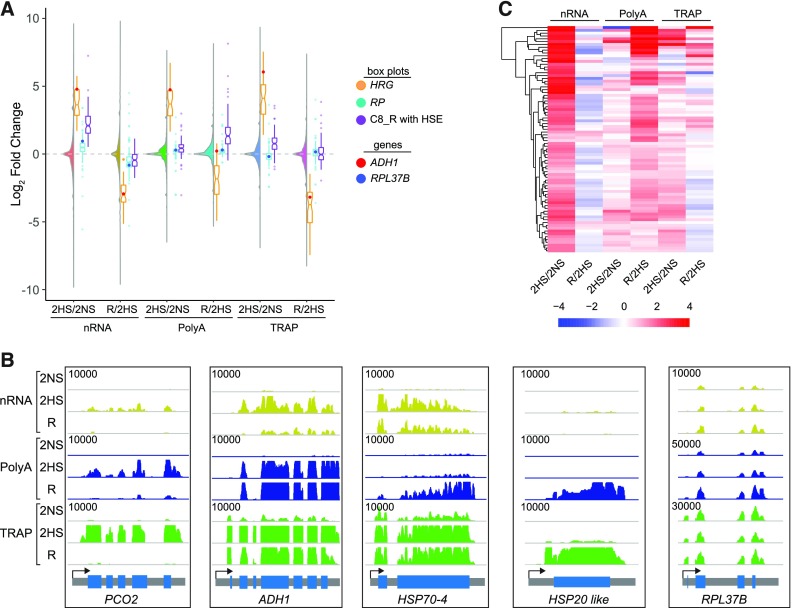
The discovery of genes with RNAPII–Ser2P engagement accompanied by increased nRNA but little to no polyA accumulation at 2HS (cluster 6; Figure 4C) suggests that some genes are poised for cytosolic upregulation upon release from oxygen deprivation. By evaluating dynamics in chromatin and RNA upon reaeration, the HRGs, RPs, and heat-responsive genes again showed distinct patterns of regulation (Figures 8A and 8B; Supplemental Figures 12 and 3). The coclustering of all genes differentially expressed under hypoxia (2HS/2NS) or reaeration (R/2HS) identified cluster 8_R as upregulated in nRNA at 2HS but not in polyA RNA until R (Figures 8A and 8B; Supplemental Figure 12A). These genes were enriched for GO categories associated with heat (4.55e−31), high light intensity (1.92e−33), protein folding (8.91e−33), and ROS (9.03e−25). Notably, over 50% of cluster 8_R genes had an HSE. The expression patterns of these HSE-containing genes contrasted to those of the HRGs and RPs, where rapid progression toward non-stress conditions was observed in all three RNA readouts, as shown by the genome browser views of five representative genes (Figures 8B and 8C). The expression pattern demonstrated by HSE-containing genes may be physiologically relevant, as reaeration is associated with a rapid restoration of protein synthesis and a ROS burst. Together, our data demonstrate functional nuclear segregation of a subset of newly synthesized RNAs from cytoplasmic-localized ribosomes in response to oxygen availability.
Figure 8.
Reaeration Promotes the Accumulation and Translation of Polyadenylated Heat-Responsive Transcripts that Are Nucleus-Enriched under Hypoxia
(A) Combined violin and boxplot of stress-recovery dynamics of hypoxia-induced nuclear-enriched transcripts. All genes shown as a violin; HRGs shown in orange, with ADH1 as a red dot; RPs shown in blue with RPL37B as a blue dot; heat shock element-containing cluster 8_R genes (C8_R, from Supplemental Figure 12) shown in purple.
(B) Genome browser view (RPKM * 1,000) of stress recovery dynamics of HRGs (PCO2, ADH1), heat-responsive genes (HSP70-4, HSP20 like), and a representative RP (RPL37B).
(C) Hierarchical clustering of heat shock element-containing cluster 8 R genes in response to hypoxia (2HS/2NS) and reoxygenation (R/2HS).
DISCUSSION
Gene Activity Dynamics in Response to HS Involves the Regulation of Chromatin, Transcription, and Posttranscriptional Processes
Our genome-wide measurements of chromatin state and intermediary steps of gene expression in seedlings exposed to sublethal HS and reaeration identified time-dependent chromatin and transcript dynamics not appreciated by routine transcriptomics. HS modulated the (1) position and degree of open chromatin near the TSS, (2) modification of H3 lysines, (3) eviction of H2A.Z, (4) engagement of RNAPII-Ser2P, and (5) abundance of nuclear, polyadenylated, and ribosome-associated gene transcripts. The results illustrate clear distinctions between steady-state nuclear and polyA transcriptomes, as shown with similar methods for rice (Oryza sativa; Reynoso et al., 2018a). The data also identified dynamics in chromatin accessibility and confirmed translational dynamics in response to hypoxia and reaeration (Branco-Price et al., 2005, 2008; Mustroph et al., 2009; Juntawong et al., 2014).
The monitoring of RNAPII–Ser2P engagement as a proxy for transcriptional elongation revealed that global levels of transcription were not dramatically dampened by 2HS (Figure 1C), although there was considerable regulation of individual genes (Figures 1C and 4C). Remarkably, less than 10% of all DRGs were coordinately up- or downregulated from transcriptional elongation through translation at 2HS. Three patterns of discontinuous gene regulation were apparent: (1) progressive upregulation, characterized by high RNAPII–Ser2P engagement and delayed elevation of polyA RNA, as observed for heat and oxidative stress genes; (2) incomplete downregulation, characterized by maintained RNAPII–Ser2P engagement and increased nRNA abundance with limited change in polyA RNA, as observed for RPs; and (3) enhanced or reduced translational status, measured by comparing TRAP RNA and polyA RNA abundance (Branco-Price et al., 2008; Mustroph et al., 2009; Juntawong et al., 2014). We also identified variations in nuclear regulation (chromatin accessibility, histone modification, RNAPII engagement, and nRNA abundance) that foreshadowed changes in polyA RNA abundance and translation. Finally, the evaluation of the three transcript populations after brief reaeration demonstrated a rapid rebound to homeostasis in the nRNA of the continuously up-DRGs, discontinuously up-DRGs, and discontinuously down-DRGs. In summary, these data illuminate remarkable complexity in the transcriptional and posttranscriptional regulation of genes associated with stress responses and growth.
Coordinate Upregulation of HRGs Is Characterized by Histone Modification, Histone Variant Eviction, and Increased Chromatin Accessibility
Hypoxia had marked effects on the position, composition, and modifications of specific histones of nucleosomes. The genes that were coordinately upregulated from transcription through ribosome association included the HRGs, which are critical for survival under low-oxygen stress (Mustroph et al., 2009, 2010; Gibbs et al., 2011; Giuntoli et al., 2014; Gasch et al., 2016; Giuntoli and Perata, 2018). The strong and coordinate upregulation of HRGs was characterized by eviction of H2A.Z, a rise in gene body H3K9ac, promoter accessibility, RNAP-Ser2P engagement, and nRNA, polyA, and ribosome-associated mRNA levels (Figure 9). The cotranscriptional neutralization of a positive charge on the N terminus of H3K9 by histone acetyltransferases is a reliable signature of active transcription (Eberharter and Becker, 2002) and is common for stress-activated genes (Kim et al., 2008; Zhou et al., 2010; Kim et al., 2012). In animals, H3K9ac is promoted by H3K4me3 and stimulates the release of poised RNAPII complexes (Jonkers and Lis, 2015). H3K4me3 generally rose on HRGs in response to 2HS, but was not as strongly correlated with RNAPII–Ser2P engagement as H3K9ac (Figure 3B; Supplemental Figure 4D). The stress-activated transcription coincided with eviction of H2A.Z from nucleosomes that were broadly distributed across the bodies of HRGs at 2NS. This observation is consistent with the finding that H2A.Z eviction from nucleosomes is a prerequisite for the activation of temperature-responsive genes (Kumar and Wigge, 2010; Cortijo et al., 2017; Dai et al., 2017; Sura et al., 2017; Torres and Deal, 2019).
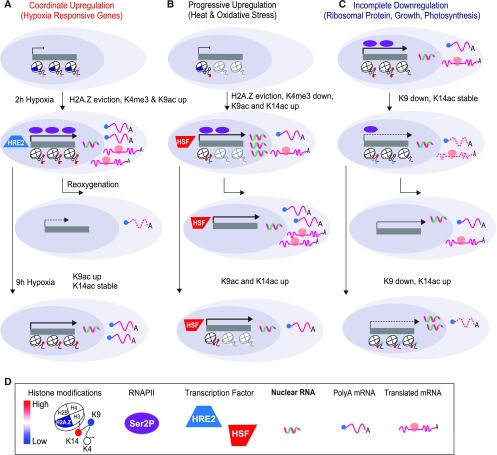
Figure 9.
Three Patterns of Dominant Nuclear Regulation in Response to Two Durations of HS and Reaeration
(A) Coordinate upregulation from chromatin to translation of HRGs characterized by hypoxia-triggered H2A.Z eviction and both H3K4me3 and H3K9ac elevation. Over half of the genes with this pattern of regulation showed HRE2 binding by ChIP. Transcript abundance and translation declines upon reoxygenation.
(B) Progressive upregulation of genes associated with heat and oxidative stress responses characterized by a strong 5′ bias in histone marks, hypoxia-triggered H2A.Z eviction, H3K9ac elevation, and H3K4me3 reduction. Many are known targets of HSFs. These genes are transcribed at low levels under normoxia (nonstress conditions), with evidence of transcriptional activation in response to brief hypoxia (2 h of hypoxia), as shown by elevated RNAPII–Ser2P binding, nRNA, and TRAP RNA. However, the elevation in polyA RNA was limited until the stress was prolonged (9 h hypoxia). Transcript abundance increases upon reoxygenation.
(C) Compartmentalized downregulation of many genes associated with growth and photosynthesis, including RP genes characterized by maintained RNAPII–Ser2P and nRNA levels under the stress, reduced H3K9ac, and progressive acquisition of H3K14ac. Transcript abundance and translation are restored upon reoxygenation. Cartoons illustrate histone modifications as defined by the key. HSF transcriptional activator; Ser2P, RNAPII with the Ser2P modification.
(D) Figure key for distinct outputs of gene regulation. Levels of assocation (high, red; low, blue) of Histone H3 modifications (H3K4me3, H3K9ac, H3K14ac) and the histone variant H2A.Z are depicted. Icons depicting binding by the transcription factors HRE2 and HSF, and RNAPII-Ser2P. Accumulation of nulcear, whole-cell, and ribosome associated RNA populations is also depicted.
H2A.Z deposition, eviction, and the impact on transcriptional regulation are complex and not fully understood. Deposition requires ACTIN-RELATED PROTEIN6, a component of the SWR1 remodeling complex, and other factors in Arabidopsis (Asensi-Fabado et al., 2017; Kumar, 2018). Yet disruption of H2A.Z deposition in an actin-related protein6 mutant had limited effect on regulation of HRGs under aerated growth conditions (Torres and Deal, 2019). Other chromatin remodeling proteins or specific TFs could be important. In response to small increases in temperature, displacement of H2A.Z is facilitated by HSF1A during the activation of the heat response network (Cortijo et al., 2017). H2A.Z eviction from HRGs could involve ERFVIIs. Indeed, the hypoxia-stabilized ERFVII RAP2.2 interacts with the SWI–SNF remodeler “BRAHMA” and contributes to H2A.Z eviction associated with increased chromatin accessibility (Efroni et al., 2013; Vicente et al., 2017). Many of the HRGs displayed increased chromatin accessibility in their proximal promoter regions and gene bodies. Moreover, 69 of the more than 200 coordinately up-DRGs had one or more HRPE within 1 kb of their TSS, and many were bound by HRE2 at 2HS (Figure 7E; Supplemental Figure 1). RAP2.12 is stabilized and localizes to the nucleus as external oxygen levels decline below 10 kPa (Kosmacz et al., 2015). The rise in nuclear levels of ERFVIIs under hypoxia may coordinate RNAPII initiation, facilitating H2A.Z eviction to promote productive elongation coupled with H3K9 acetylation. The interaction of ERFVII with the ATP-dependent chromatin remodeling machinery could also contribute to the expansion of regions of accessible chromatin to facilitate high levels of transcriptional activation. The uniform upregulation from transcription to translation of the up-DRGs with an HRPE- and/or HRE2-bound promoter is reminiscent of the production of translationally competent mRNAs during nutrient starvation in yeast (Saccharomyces cerevisiae) that is associated with specific TFs (Zid and O’Shea, 2014). We hypothesize that transcriptional activation by the ERFVIIs promotes the fidelity of cotranscriptional processing, export, and translation of their targets under hypoxia.
Genes Associated with Heat and Oxidative Stress Are Progressively Upregulated by Hypoxia
HS distinctly activated genes associated with heat and oxidative stress, despite treatment at ambient temperature. The progressive upregulation of cluster-9 genes after 2HS (Figure 4C; Supplemental Figure 8) became more evident when their activity was compared at 2HS and 9HS using two chromatin and two RNA readouts (Figure 7F). A significant proportion of these genes had HSEs within their promoters (Figures 4C and 7D; Supplemental Figures 6 and 8). Their pattern of regulatory variation included a 5′ bias in histone marks and moderate to very high RNAPII–Ser2P engagement at 2HS accompanied by elevated nRNA and TRAP RNA levels, but limited amounts of full-length polyA RNA (Figure 8).
Previously, heat-stress–responsive genes were recognized as highly induced in Arabidopsis seedlings directly transferred to anoxia (Loreti et al., 2005; Banti et al., 2010). A brief pretreatment with heat stress increased the resilience to anoxia of wild-type seedlings but not loss-of-function hsfa2 or hsf1a hsf1b mutant seedlings (Banti et al., 2008). This is enigmatic because high temperatures do not typically precede flooding in natural environments, but the availability of HSPs could reduce cellular damage under extreme stress conditions such as anoxia. Indeed, HSPs were plentiful in cluster 9 after 2HS. The limited synthesis of ATP-dependent chaperones after brief hypoxia due to nuclear sequestration of their transcripts could minimize demands on limited energy reserves. However, as the stress is prolonged or upon reaeration, the rapid synthesis of chaperones and proteins that provide protection from ROS could aid survival. For example, the upregulation of RESPIRATORY BURST OXIDASE D contributes to the survival of hypoxia, submergence, and postsubmergence recovery (Baxter-Burrell et al., 2002; Pucciariello et al., 2012; Gonzali et al., 2015; Yeung et al., 2018).
The nucleosome dynamics of cluster-2 and -9 genes after 2HS included similar modifications, with notable exceptions (Figures 4C and 4D; Supplemental Figures 6 and 11). Genes of both clusters underwent a loss of H2A.Z near the TSS of the gene body in conjunction with increased H3K9ac, but cluster-9 genes showed a loss of H3K4me3 proximal to their TSS. This was not evident for cluster-2 or other coordinately up-DRGs. The decrease in H3K4me3 may be an indication of nonproductive RNAPII–Ser2P engagement, as this modification is associated with RNAPII elongation (Ding et al., 2012; Kwak and Lis, 2013). Intriguingly, there was significant enrichment of HSEs in the promoters of both rapidly and progressively upregulated genes (Figure 7D).
We predict that the recruitment of specific TFs together with chromatin-modifying enzymes affects histone or RNAPII CTD modifications that influence the rate of RNAPII elongation, termination, or posttranscriptional processing, thereby tuning the temporal production of stress-induced transcripts. The progressive upregulation of heat and oxidative stress networks could be mediated by the upregulation of TF genes encoding HSFs (i.e. HSFB2B), ZINC FINGER PROTEIN10, and others during the early hours of the stress. Notably, the level of polyA mRNA accumulation and translation of these mRNAs were enhanced upon reaeration (Figure 8A). Our data sets provide an opportunity for further meta-analyses of heat and oxidative stress gene regulatory networks.
Genes Associated with Major Cellular Processes Continue To Be Transcribed During Hypoxia
Our study sheds light on the transcriptional and RNA regulation of genes associated with growth (Figure 8). Our prior comparisons of the transcriptome and translatome determined that hypoxia limits the investment of energy into the production of abundant cellular machinery such as ribosomes. Polyadenylated mRNAs encoding RPs and other abundant proteins are maintained but poorly translated under HS (Branco-Price et al., 2008, 2005; Juntawong et al., 2014; Sorenson and Bailey-Serres, 2014). The stress-limited translation of RPs mRNAs is associated with their sequestration in OLIGOURIDYLATE BINDING PROTEIN1C (UBP1C)-containing macromolecular complexes (Sorenson and Bailey-Serres, 2014). Upon reaeration, RP mRNAs rapidly reassociate with polysomes. UBP1C is nucleocytoplasmic (Branco-Price et al., 2008, 2005; Juntawong et al., 2014; Sorenson and Bailey-Serres, 2014). As the immunopurification of UBP1C and associated mRNAs does not discriminate between nuclear and cytoplasmic UBP1C-associated mRNAs, this study raises the interesting possibility that the association of UBP1C with transcripts in the nucleus contributes to their nuclear retention.
Here, we considered on a global scale if RP or other translationally limited mRNAs are synthesized during hypoxia. Our RNAP-Ser2P and nRNA data demonstrate that genes encoding RPs, metabolic enzymes, and some components of the photosynthetic apparatus are synthesized and retained as partially or completely processed transcripts in the nucleus during hypoxia (Figure 6), a situation readily reversed by reaeration (Figure 8A). The finding that housekeeping gene transcription can continue is consistent with results of traditional nuclear run-on transcription assays performed on fully submerged maize (Zea mays; Fennoy et al., 1998). The transcriptionally active genes showed progressive increases in H3K14ac but not H3K9ac between 2HS and 9HS. The limited increase in their polyA RNAs could be due to limited cleavage and polyadenylation or increased turnover. Similar uncoupling of the H3K9ac and H3K14ac marks associated with active transcription but limited polyA RNA accumulation was reported for promoters deemed inactive but primed for future activation in pluripotent embryonic stem cells of mice (Karmodiya et al., 2012). The incomplete downregulation of housekeeping genes may poise cells for the observed rapid recovery upon reaeration.
Our genome-scale analysis demonstrated that transcripts were retained within the nucleus during hypoxia, which is consistent with the results of visualization studies of specific mRNAs in Arabidopsis and lupine (Niedojadło et al., 2016). mRNAs associated with the cell cycle are reportedly enriched in the nucleus under control conditions in Arabidopsis (Yang et al., 2017). This was confirmed in our comparison of the nRNA and polyA RNA transcriptomes under both control and hypoxia stress (Supplemental Figure 5). These data support a scenario wherein the nuclear compartmentalization of abundant transcripts contributes to energy conservation by limiting translation, a mechanism similar to the cytoplasmic sequestration associated with the RNA binding protein UBP1C. Nuclear retention and cytoplasmic sequestration could provide two reservoirs of transcripts that can be deployed upon reaeration, enabling rapid restoration of cellular processes. Indeed, our data provide evidence that nuclear-retained transcripts increase in translation status upon reaeration (Figure 8A).
Thus, the comparative monitoring of nRNA, polyA, and TRAP RNA populations strongly support the conclusion that the nuclear export and subsequent accumulation and translation of mRNAs is dynamically regulated by stress in plants. Factors such as chromatin accessibility, RNAPII elongation and termination, and histone modification (i.e. H3K14ac) could contribute to the dynamics in nuclear transcript abundance. Circumstantial evidence of altered cotranscriptional processing includes hypoxia-promoted intron retention (Juntawong et al., 2014) and alternative polyadenylation site selection (de Lorenzo et al., 2017). Although there is no clear evidence of a general disruption of splicing or polyadenylation during hypoxia (Koroleva et al., 2009; Juntawong et al., 2014; de Lorenzo et al., 2017), these processes could be altered by changes in chromatin, RNAPII elongation, or association with certain proteins. The RNA helicase eIF4AIII (Koroleva et al., 2009) and several RNA binding proteins (UBP1A/B/C and CML38) form macromolecular complexes that are present in the nucleus but are primarily cytoplasmic under hypoxia (Sorenson and Bailey-Serres, 2014; Lokdarshi et al., 2016). Further experimentation is required to better understand the controlled dynamics in transcript synthesis, processing, and export during hypoxia stress.
An ERFVII Hierarchy Drives Transcriptional Activation in Response to Hypoxia
In this study, we performed a genome-wide analysis of the cis-element targets of an ERFVII. ChIP-seq of the hypoxia-induced ERFVII HRE2 identified a multimeric 5′-GCC-3′ cis-element that is distinct from the HRPE that is enriched in HRG promoters and activated by ERFVIIs (Gasch et al., 2016). This is consistent with the observation that neither HRE1 nor HRE2 interacted with a 33-bp promoter fragment containing the HRPE in a Yeast-1 hybrid assay or provided significant transactivation via the HRPE in Arabidopsis protoplasts (Gasch et al., 2016). The HRE2 binding motif strongly resembles the double GCC motif of the EBP box that was first defined for an ERF in tobacco (Nicotiana tabacum; Sato et al., 1996). Previously, RAP2.3 was shown to bind to the ABSCISIC ACID INSENSITIVE5 promoter in a region with two EBP boxes; transactivation of the ABSCISIC ACID INSENSITIVE5 promoter in protoplasts by all three constitutively expressed ERFVIIs (RAP2.2/2.3/2.12) was dependent on the EBP box (Gibbs et al., 2014).
Collectively, our observations support earlier conclusions that members of the two ERFVII subgroups are not functionally redundant (Gibbs, 2014; Gasch et al., 2016). Yet, the situation remains enigmatic. The HRPE contains a sequence resembling a double 5′-GCC-3′. Promoter regions with high scores for the HRPE and HRE2 motif coincided in 40 of the 213 coordinately up-DRGs, 13 of which were bound by HRE2 (Supplemental Figure 1; Supplemental File 1). Moreover, the Arabidopsis ADH1 promoter region contains both of these motifs and is sufficient for hypoxic upregulation (Gasch et al., 2016). The current model is that the low-oxygen–mediated stabilization of the constitutively expressed RAP-type ERFVIIs is responsible for rapid transactivation of HRGs including HRE2 (van Dongen and Licausi, 2015; van Dongen and Licausi, 2015,Voesenek and Bailey-Serres, 2013), which is synthesized and stabilized under hypoxia (Gibbs et al., 2011). We propose that HRE2 production and binding to promoters via the multimeric GCC element (HRE2 motif) is important for the continued upregulation of HRGs, as an hre1 he2 double mutant fails to maintain the upregulation of these genes after 4 h of anoxia (Licausi et al., 2010). This process may also enable cells to circumvent the inhibition of RAP2.12 activity by its interaction with the HYPOXIA RESPONSE ATTENUATOR (Giuntoli et al., 2014). Further analyses are required to comprehend the independent or overlapping roles of the RAP-type and HRE ERFVIIs in the temporal activation of transcription during hypoxia.
Mammals regulate transcriptional activity in response to hypoxia in a manner remarkably analogous to that of Arabidopsis (Watson et al., 2010). HRGs are activated by the two-subunit Hypoxia Inducible Factor (HIF) TF complex, which includes the oxygen-destabilized HIF1A subunit (LaGory and Giaccia, 2016). Similar to the three RAP-ERFVIIs of Arabidopsis, HIF1A is a constitutively synthesized protein that is destabilized in the presence of oxygen. A group of prolylhydroxlases catalyze the oxygen-dependent hydroxylation of specific residues of HIF1A that triggers its ubiquitylation and proteasome-mediated turnover. In animals, the chromatin landscape is also regulated through regional accessibility (Buck et al., 2014) and the posttranslational modification of histones (Chervona and Costa, 2012) in an oxygen-sensitive manner. HIF signaling itself directly facilitates histone modification of target genes through interactions with chromatin-modifying enzymes (Watson et al., 2010); several histone-modifying enzymes are directly targeted by HIF transcriptional activation, and HIF1A expression itself is regulated by histone acetylation (Kim et al., 2007). Moreover, specific histone-modifying enzymes function as direct oxygen sensors through the mediation of their activity based on cellular oxygen levels in animals and likely plants (Gibbs et al., 2018; Batie et al., 2019; Chakraborty et al., 2019), confirming the integration of oxygen sensing with epigenetic regulation. The low-oxygen–stabilized transcriptional regulators and the coordination of their function with chromatin modifications in Arabidopsis is evidence of convergence in gene regulation in response to hypoxia in plants and animals. This may extend beyond transcriptional regulation, as cytoplasmic sequestration and selective translation of mRNAs are also observed in plants and metazoa (Blais et al., 2004; Branco-Price et al., 2008; Young et al., 2008; Juntawong et al., 2014). It remains to be explored if hypoxia influences the synthesis and accumulation of nascent transcripts associated with growth in animals, as demonstrated in this multiomic analysis of Arabidopsis.
This multilevel investigation into gene regulatory processes revealed unexpectedly complex regulation of gene activity within the nucleus and cytoplasm that fine-tunes cellular and physiological acclimation to transient hypoxia, providing a model for a response to an acute stress. A key finding is that there are rapid changes in the epigenome in response to short-term HS that continue as the stress becomes more severe. Coordinate upregulation from chromatin accessibility to translation was observed for over 200 HRGs. Extreme stress-responsive and growth-associated genes showed more discontinuous upregulation of nascent transcript production, export to the cytoplasm, and recruitment to ribosomes. Each of these broadly defined gene cohorts showed a distinct pattern of epigenetic, nuclear, and cytoplasmic transcript regulation upon reaeration. Future investigations can consider roles of specific TF and their interactions with RNAPII, transcriptional coactivators, the chromatin landscape, and the posttranscriptional apparatus in directing the nuanced nuclear regulation uncovered in this study.
METHODS
Genetic Material
The following Arabidopsis (Arabidopsis thaliana) genotypes were used for genome-wide experiments: Col-0, Histone modification ChIP-seq, RNAPII ChIP-seq; pHTA11:HTA11-3xFLAG (Cortijo et al., 2017), H2A.Z ChIP-seq; pUBQ10:NTF/pACT2:BirA (Sullivan et al., 2014), ATAC-seq, nRNA-seq, polyA mRNA-seq; p35S:HF-RPL18 (Zanetti et al., 2005), TRAP sequencing, mRNA-seq; and HRE ChIP-seq p35S:C2A-HRE2-HA (Gibbs et al., 2011).
Plant Growth and Treatment
Arabidopsis seeds were sterilized by incubation in 70% (v/v) ethanol for 5 min, followed by incubation in 20% (v/v) bleach and 0.01% (v/v) TWEEN-20, and washed three times in double distilled water for 5 min, in triplicate. Sterilized seeds were placed onto 1× Murashige & Skoog medium (1.0× MS salts, 0.4% [w/v] Phytagel; Sigma-Aldrich), and 1% [w/v] Suc, pH 5.7) in 9-cm2 Petri dishes and stratified by incubation at 4°C for 3 d in complete darkness. After stratification, the plates were placed vertically into a growth chamber (Percival) with a 16-h light/8-h dark cycle at ∼120 μmol photons·s−1·m-2 (cat. no. 21770; Sylvania), at 23°C for 7 d. For HS, seedlings were removed from the growth chamber at Zeitgeber time (ZT) 16 and subjected to HS by bubbling argon gas into sealed chambers in complete darkness for 2 or 9 h at 24°C. The chamber setup was as described by Branco-Price et al. (2008). Oxygen partial pressure in the chamber was measured with a NeoFox Sport O2 sensor and probe (Ocean Optics). Hypoxia ([O2] < 2%) was attained within 15 min of initiation of the stress. Control samples were placed in an identical chamber that was open to ambient air under the same light and temperature conditions. For reaeration, seedlings were identically subjected to HS for 2 h and subsequently placed into the control chamber for 1 h (nRNA and polyA) or 2 h (TRAP) after the stress. Tissue was rapidly harvested into liquid N2 and stored at −80°C.
ChIP
Seedlings were grown on Petri dishes and treated as described in the section "Plant Growth and Treatment". For the HRE2 ChIP experiments, seedlings were pretreated by flooding with 10 mL of 100 μM of Calpain inhibitor IV (American Peptide) and 1% (v/v) DMSO for 2 h before the hypoxia treatment. For all genotypes, after control or hypoxia treatment, seedlings were immediately cross-linked in 1% (v/v) formaldehyde nuclei purification buffer (NPB: 20 mM of 3-(N-Morpholino)propanesulfonic acid at pH 7.0, 40 mM of NaCl, 90 mM of KCl, 2 mM of EDTA, and 0.5 mM of EGTA) in a vacuum chamber under house vacuum for 10 min. The reaction was quenched by adding 5M of Gly to reach a concentration of 125 mM, followed by vacuum infiltration for 5 min. This was followed by three washes in 30 mL of double distilled water. The seedlings were blotted dry and pulverized under liquid N2. To isolate nuclei, 0.5-g tissue was thawed to 4°C in 10 mL of NPB containing 0.5 mM of spermidine, 0.2 mM of spermine, and 1× Plant Protease Inhibitor Cocktail (cat. no. P9599; Sigma-Aldrich). Nuclei were pelleted by centrifugation in 4°C at 1200g for 10 min. The nuclei were washed in 10 mL of NPB and 0.1% (v/v) Triton X-100 (NPBt) and pelleted by centrifugation in 4°C at 1200g for 10 min in triplicate. After the final NPBt wash, the supernatant was aspirated and the nuclei pellet was resuspended in 120 μL of NPB and lysed with the addition of 120 μL of 2× nuclei lysis buffer (100 mM of Tris at pH 8.0, 20 mM of EDTA, 2% [w/v] SDS, and 2× Plant Protease Inhibitor Cocktail) by vortexing for 2 min at 24°C. The chromatin was sheared into 200- to 600-bp fragments by sonication (Diagenode) with 33 cycles of 30 s ON and 30 s OFF at 4°C. The sample was cleared by centrifugation at 16,000g at 4°C for 2 min and the supernatant was diluted fivefold with dilution buffer (16.7 mM of Tris at pH 8.0, 167 mM of NaCl, 1.1% [v/v] Triton X-100, and 1.2 mM of EDTA). The entire chromatin fraction was precleared by incubation with uncoupled Protein G Dynabeads (Thermo Fisher Scientific) for 30 min, followed by collection of the supernatant. Three-hundred microliters of the input chromatin (1 mL for HRE2) was incubated with 3 μL of undiluted anti-H3K4me3 (cat. no. ab8580; Abcam), anti-H3K27me3 (cat. no. 07-449; EMD Millipore), anti-H3K9ac (cat. no. ab4441; Abcam), anti-H3K14ac (cat. no. ab52946; Abcam), anti-p-CTD RNAPII (cat. no. ab5095; Abcam), anti-FLAG (cat. no. F1804; Sigma-Aldrich; 1:100 dilution), or anti-HA (cat. no. h3663; Sigma-Aldrich; 1:333 dilution) antibodies for 4 h (overnight for HRE2) while rocking at 4°C. Protein G Dynabeads (30 μL) were washed in dilution buffer, added to the chromatin fraction and allowed to incubate for 2 h while rocking at 4°C. Beads were magnetically captured and washed sequentially in 1 mL for 5 min with four buffers: low NaCl wash buffer (20 mM of Tris at pH 8.0, 150 mM of NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, and 2 mM of EDTA), high NaCl wash buffer (20 mM of Tris at pH 8.0, 500 mM of NaCl, 0.1% [w/v] SDS, 1% [v/v] Triton X-100, and 2 mM of EDTA), LiCl wash buffer (10 mM of Tris at pH 8.0, 250 mM of LiCl, 1% [w/v] sodium deoxycholate, 1% [v/v] Nonidet P-40, and 1 mM of EDTA), and standard TE buffer (10 mM of Tris at pH 8.0, and 1 mM of EDTA). The beads were then washed twice with 10 mM of Tris at pH 8.0, and resuspended in 25 μL of tagmentation reaction mix (10 mM of Tri at pH 8.0, 5 mM of MgCl2, and 10% [v/v] dimethylformamide) containing 1 μL of Tagment DNA Enzyme (Illumina) and incubated at 37°C for 1 min. Beads were washed twice with low NaCl wash buffer and then once in standard TE buffer. The chromatin was eluted from the beads by heating for 15 min at 65°C in elution buffer (100 mM of NaHCO3 and 1% [w/v] SDS) and reverse cross-linked by the addition of 20 μL of 5-M NaCl with heating at 65°C overnight. After reverse cross-linking, Proteinase K (0.8 units; New England Biolabs) was added and the sample was incubated at 55°C for 15 min. The final tagged ChIP-DNA sample was purified using MinElute columns (Qiagen) according to the manufacturer’s instructions and eluted with 14 μL of EB.
ChIP-seq Library Preparation
Library preparation for short-read sequencing (ChIP-seq) with tagmentation was performed according to Schmidl et al. (2015), with minor modifications. Final library enrichment was performed in a 50-μL reaction containing 12 μL of ChIP DNA, 0.75-μM primers, and 25 μL 2× NEBNext PCR Master Mix. To determine the appropriate amplification cycle number, a qPCR was performed on 1 μL of tagmented ChIP DNA in a 10-μL reaction volume containing 0.15-μM primers, 1× SYBR Green (Thermo Fisher Scientific), and 15 μL of 2× NEBNext PCR Master Mix (New England Biolabs) with the following program: 72°C for 5 min, 98°C for 30 s, 24 cycles of 98°C for 10 s, 63°C for 30 s, 72°C for 30 s, and a final elongation at 72°C for 1 min. Libraries were amplified for n cycles, where n is equal to the rounded-up Cq value determined in the qPCR. Amplified libraries were purified and size-selected using SPRI AMPure XP beads (Beckman). AMPure XP beads were added at a 0.7:1.0 bead/sample ratio, and the remaining DNA was recovered by the addition of AMPure XP beads at a 2.0:1.0 bead/sample and eluted in 13 μL of EB. Quantification of the final libraries was performed with Quant-iT PicoGreen (Thermo Fisher Scientific), and library fragment distribution was evaluated with the model no. 2100 Bioanalyzer (Agilent Technologies) using the high sensitivity DNA chip. Final libraries were multiplexed to >5-nM final concentration and sequenced on the HiSeq 3000/4000 at the UC Davis DNA Technologies Core.
Isolation of Nuclei Tagged in Specific Cell Types
INTACT was performed according to Deal and Henikoff (2010) with modifications. Frozen pulverized tissue (0.5 g) of pUBQ:NTF/pACT2:BirA seedlings was thawed in 10 mL of cold NPB buffer and filtered through 30-µm filters (Sysmex). Nuclei were pelleted by centrifugation at 1200g for 7 min at 4°C. The nuclear pellet was resuspended and washed twice in NPBt and resuspended in 1 mL of NPB. M280 Streptavidin Dynabeads (10 μL; Invitrogen) washed with 1 mL of NPB and resuspended in the original volume were added to the nuclei, and the sample was allowed to rock for 30 min at 4°C. The bead-nuclei mixture was diluted to 14 mL with NPBt and incubated with rocking for 30 s at 4°C. The beads were magnetically collected, the supernatant was removed, and the beads washed twice with NPBt and resuspended in 1 mL of NPBt. A 25-μL fraction was removed to quantify nuclei by adding 1 μL of 1.0 µg/μL Propidium Iodide, followed by incubation on ice for 5 min before visualization and quantification with a C-Chip Hemocytometer (Incyto) under a fluorescence microscope.
ATAC
Guided by Maher et al. (2018), 50,000 INTACT-purified nuclei from root tissue were magnetically captured. The supernatant was aspirated and any remaining NPB was removed, and the nuclei were resuspended in 50 μL of transposition mix (1× TD buffer, 2.5 μL of TDE1 transposase) and incubated for 30 min at 37°C with occasional mixing. The transposed DNA was purified with MinElute PCR purification columns (Qiagen), and the purified DNA was eluted in 11 μL of EB. For library construction, a reaction was prepared to amplify the sample via a two-step process. The reaction was assembled containing transposed DNA (10 μL), 5 μM of Ad1.1, and an indexing primer (Buenrostro et al., 2013), 1× NEBNext High Fidelity PCR mix (New England Biolabs), cycled in the following program: 72°C 5 min; 98°C 30 s; 3 cycles of 98°C 10 s, 63°C 30s, 72°C 1 min; 4°C and the samples were placed on ice. Then a qPCR was performed using an aliquot of the amplified library with the following program: 98°C for 30 s; 20 cycles of 98°C for 10 s, 63°C for 30 s, 72°C for 1 min. The original PCR was resumed for n additional cycles, where n is the cycle at which the reaction was at 1/3 of its maximum fluorescence intensity. Amplified DNA was then purified with MinElute PCR purification columns (Qiagen) and eluted in 20 μL of EB. Amplified libraries were purified and size-selected for fragments between 200 bp and 600 bp using SPRI AMPure XP beads (Beckman). AMPure XP beads were added at a 0.55:1.0 bead/sample ratio, and the remaining DNA was recovered by the addition of AMPure XP beads at a 1.55:1.0 bead/sample and eluted in 13 μL of EB. Library concentration was determined using a NEBNext (New England Biolabs) library quantification kit. Final libraries were multiplexed to >5 nM final concentration and sequenced on the HiSeq 3000/4000 at the UC Davis DNA Technologies Core.
INTACT Followed by nRNA Extraction
Guided by Reynoso et al., (2018a) and (2018b), 50,000 INTACT-purified nuclei were collected magnetically, and the supernatant was removed and resuspended in 20 μL of NPB. nRNA was extracted and purified using a RNeasy Micro kit (Qiagen) and eluted in 20 μL of water. The RNA was DnaseI-digested with 1 μL (2U/μL) of Turbo DNaseI (Thermo Fisher Scientific) and incubated for 30 min at 37°C. DNaseI was inactivated by adding EDTA to 15 mM, and heated at 75°C for 10 min, centrifuged at 2000g for 5 min, and transferred to a new tube. The RNA was purified using AMPure XP beads and eluted in 15 μL of water. The rRNA was depleted from the sample using a plant Ribo-Zero rRNA Removal Kit (Illumina) according to the manufacturer’s instructions.
TRAP Sequencing
TRAP of mRNA-ribosome complexes was performed following the procedure of Mustroph et al. (2009). Briefly, pulverized tissue (1 mL) from the 35S:HF-RPL18 genotype was added to 5 mL of polysome extraction buffer (200 mM of Tris at pH 9.0, 200 mM of KCl, 25 mM of EGTA, 35 mM of MgCl2, 1% [w/v] PTE, 1 mM of DTT, 1 mM of PMSF, 100 μg/mL of cycloheximide, and 50 μg/mL of chloramphenicol) containing 1% (v/v) detergent mix (20% [w/v] polyoxyethylene[23]lauryl ether, 20% [v/v] Triton X-100, 20% [v/v] Octylphenyl-polyethylene glycol, and 20% [v/v] Polyoxyethylene sorbitan monolaurate 20) and homogenized with a glass homogenizer on ice. The homogenized mixture was allowed to stand for 10 min on ice and centrifuged at 16,000g at 4°C for 15 min. After centrifugation, the supernatant was transferred to a new tube and filtered through one layer of Miracloth (Millipore) to produce the clarified extract.
Protein G Dynabeads (50 μL; Thermo Fisher Scientific) were prewashed twice with 1.5 mL of wash buffer-T (WB-T; 200 mM of Tris at pH 9.0, 200 mM of KCl, 25 mM of EGTA, 35 mM of MgCl2, 5 mM of PMSF, 50 μg/mL of cycloheximide, and 50 μg/mL of chloramphenicol) and resuspended in the original volume. The beads were added to the clarified tissue extract and incubated at 4°C for 2 h with gentle rocking. The beads were collected magnetically, the supernatant was removed, and the beads were gently resuspended in 6 mL of WB-T for 5 min at 4°C with rocking. This wash step was repeated two additional times, after which the beads were resuspended in 1 mL of WB-T and transferred to a new tube and the supernatant was removed.
RNA was purified from the sample and the reserved clarified extract by adding 105 μL of lysis/binding buffer (100 mM of Tris at pH 8.0, 500 mM of LiCl, 10 mM of EDTA, 1% [w/v] SDS, 5 mM of DTT, 1.5% [v/v] Antifoam A, and 5 μL/mL 2-mercaptoethanol), followed by vortexing for 5 min. The samples were incubated at room temperature for 10 min, centrifuged at 13,000g for 10 min, and transferred to a new tube. PolyA RNA selection was performed by adding 1 μL of 12.5-μM biotin-20nt-dT oligos (Integrated DNA Technologies) to 200 μL of the TRAP sample, followed by incubation at room temperature for 10 min. In a separate tube, 20 μL of magnetic streptavidin beads (New England Biolabs) were washed with 200 μL of lysis/binding buffer. The lysate was added to the washed beads and incubated at room temperature for 10 min with gentle agitation. The beads were magnetically separated and washed sequentially with wash buffer A (10 mM of Tris at pH 8.0, 150 mM of LiCl, 1 mM of EDTA, and 0.1% [w/v] SDS), wash buffer B (10 mM of Tris at pH 8.0, 150 mM of LiCl, and 1 mM of EDTA), and low salt buffer (20 mM of Tris at pH 8.0, 150 mM of NaCl, and 1 mM of EDTA). After the washes, the pellet was resuspended in 16 μL of 10 mM Tris at pH 8.0 containing 1 mM of 2-Mercaptoethanol and heated at 80°C for 2 min. The beads were magnetically collected, the supernatant was transferred to a new tube, the polyA+ RNA selection process was repeated, and the samples combined in a new tube before storage at −80°C.
Nuclear, PolyA, and TRAP-RNA Isolation and Library Construction
Total RNA was extracted from 50 mg of frozen pulverized tissue using Trizol (Life Sciences), treated with DNaseI, purified using AMPure XP beads, and eluted in 50 μL of water and polyA selected. PolyA RNA selection was performed using Oligo (dT)25 Dynabeads by two rounds of magnetic capture (Townsley et al., 2015). TRAP of mRNA-ribosome complexes was performed following the procedure of Mustroph et al. (2009). TRAP RNA extraction (Townsley et al., 2015) was performed with minor modifications, and polyA RNA was selected as described by Townsley et al (2015). RNA-seq library preparation for nRNA, polyA, and TRAP RNA was performed according to Kumar et al. (2012).
Bioinformatic Analyses
All libraries were sequenced in excess of 5 million reads per sample. For all high-throughput outputs, short reads were trimmed using FASTX-Toolkit (Hannon Lab) to remove barcodes, and short and low-quality reads were filtered before alignment to the TAIR10 genome with the programs BowTie2/TopHat2 (Langmead and Salzberg, 2012; Kim et al., 2013). Read quality reports were generated using the software “FastQC” (Babraham Bioinformatics). For all data sets excluding ATAC-seq and HRE2 ChIP-seq, counting of aligned reads was performed on entire transcripts (Lawrence et al., 2013) using the latest Araport11 annotation (201606; Cheng et al., 2017), and reads per kilobase of transcript per million mapped reads (RPKM) values were calculated. Differentially expressed genes were determined by “edgeR,” |FC| > 1, FDR < 0.01. TRAP data were not normalized to total cellular polysome levels as in one of our prior analyses (Branco-Price et al., 2008, 2005).
Data Visualization and File Generation
“Bigwig” software files for Integrated Genome Viewer (Thorvaldsdottir et al., 2012) visualization were generated from aligned “bam” files and normalized by RPKM values using the “deepTools” command “bamCoverage” with the “-normalizeUsingRPKM” specification, with the Arabidopsis Blacklisted Chromatin regions (Supplemental Figure 5; Supplemental File 2) masked. Within the Integrated Genome Viewer, all chromatin-based outputs were normalized to the same scale and all RNA outputs were normalized to a separate scale.
Peak Calling
For ChIP-seq and ATAC-seq data sets, peak calling was performed using independent replicates in combination as input for the program “HOMER” using the “findPeaks” command with the specification “-gsize 1.118e8” (Heinz et al., 2010). For downstream analyses, peaks identified from each time point comparison were combined and counting was performed on individual replicates. Peaks were annotated using the HOMER command “annotatePeaks.pl.” Differential regulation of peaks was performed using the software “edgeR.”
Motif Discovery
HRE2 peaks identified by HOMER command “findPeaks” were used as input for the HOMER command “findMotifsGenome.pl.” The matrix for the top HRE2 motif (P < 1e-69) was used as input the MEME command “ceqlogo” to generate the motif logo.
DNA Affinity Purification and Sequencing Motif Analysis
Sequences of promoter regions spanning 1 kb upstream to 500 bp downstream of the TSS were extracted for genes within each cluster. The promoter sequences were compared against each TF, per TF family, using motif matrices identified by O’Malley et al. (2016). The number of significant motifs identified within the promoters of each cluster were quantitated and normalized to the number of genes within each cluster.
t-SNE
For t-SNE dimensionality reduction, a principal component analysis was performed using RPKM values for each replicated data set and time point. The values for principal components 1 to 10 were then used as input for t-SNE dimensionality reduction, and two-dimensional output was plotted with the software “ggplot2.”
Wilcoxon Signed-Rank Test
A Wilcoxon signed rank sum test was performed by comparing the RPKM values of one genomic output for each gene within a cluster to the genes of another cluster. Each cluster was compared individually to all other clusters to determine significance, P < 0.05.
Fisher’s Exact Test
Fisher’s exact test was performed by comparing the number of motifs/peaks associated with genes within a cluster to the total number of motifs/peaks identified for all DRGs.
Bioinformatic Tools
Analyses were performed with “Bioconductor R” packages, particularly the Next Generation Sequencing analysis software of “systemPipeR” (Girke, 2014). Programs used within Bioconductor included: BiocParallel (Morgan et al., 2019a), BatchJobs (Bischl et al., 2015), GenomicFeatures (Lawrence et al., 2013), GenomicRanges (Lawrence et al., 2013), edgeR (McCarthy et al., 2012), gplots (Warnes et al., 2009), ggplot2 (Wickham, 2009), RColorBrewer (Neuwirth, 2011), Dplyr (Wickham et al., 2015), biomaRt (Durinck et al., 2009), ChIPseeker (Yu et al., 2015), rtracklayer (Lawrence et al., 2009), Rtsne (Krijthe, 2015), Pheatmap (Kolde, 2012), e1071 (Meyer et al., 2015), cluster (Maechler et al., 2019), ShortRead (Morgan et al., 2009), clusterProfiler (Yu et al., 2012), and Rsamtools (Morgan et al., 2019b). The publicly available UNIX/Python/Perl packages used included TopHat (Kim et al., 2013), FASTX-Toolkit (Gordon and Hannon, 2010), FastQC (Andrews et al., 2010), MEME (Bailey et al., 2009), HOMER (Heinz et al., 2010), deepTools (Ramírez et al., 2016), CIRCOS (Krzywinski et al., 2009), BEDtools (Quinlan and Hall, 2010), and SAMtools (Li, 2011).
Statistical Analysis
Statistical analysis of cluster motif enrichment was performed by two-tailed Fisher’s exact test comparing motif frequency for individual genes of each cluster; P < 0.05. Statistical significance of cluster values was determined for each genomic output using a Wilcoxon signed rank test for values of all genes within each cluster; P < 0.05.
Accession Numbers
The Arabidopsis genes examined in this study include: ADH1, AT1G77120; PCO2, AT5G39890; HSP70-4, AT3G12580; RPL37B, AT1G52300; LHY1, AT1G01060; CCA1, AT2G46830; PRR9, AT2G46790; PRR7, AT5G02810; PRR5, AT5G24470; RV8, AT3G09600; LUX, AT3G46640; CIR1, AT5G37260; ELF3, AT2G25930; ELF4, AT2G40080; LWD1, AT1G12910; ZTL, AT5G57350; GI, AT1G22770; LNK1, AT5G64170; LNK2, AT3G54500; TOC1, AT5G61380; CML38, AT1G76650; LBD41, AT3G02550; ALAAT1, AT1G17290; HSP20 like, AT1G53540.
The data sets generated and analyzed in this study are available in the National Center for Biotechnology Information Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122804.
Supplemental Data
Supplemental Figure 1. Expanded overview of the multiscale chromatin and RNA gene regulatory analyses.
Supplemental Figure 2. Average signal plots and heat maps for the global distribution of reads of chromatin and RNA data.
Supplemental Figure 3. Multilevel evaluation of histone and gene regulation activity of the HRGs and RPs.
Supplemental Figure 4. Evidence of cooperative association between H2A.Z eviction and H3K9ac and the relationship between histone H3 modification and RNAPII engagement.
Supplemental Figure 5. Genome-Wide comparison of nRNA to polyA RNA demonstrates nuclear and cytoplasmic enrichment of transcripts under two conditions.
Supplemental Figure 6. Histones, RNAPII–Ser2P, and RNA levels of genes that are coregulated in response to HS.
Supplemental Figure 7. Quantitative analysis of dynamics in histone modifications of coregulated gene clusters after 2 h of HS.
Supplemental Figure 8. Progressive regulation of variations in gene activity in response to hypoxia.
Supplemental Figure 9. Histone and gene activity comparisons of genes coregulated after 9 h of HS.
Supplemental Figure 10. Genome-Wide binding dynamics of HRE2 determined by ChIP-seq.
Supplemental Figure 11. Genome browser view of normalized read coverage of histone, ATAC-seq, RNAPII–Ser2P, HRE2-ChIP, and RNA outputs for representative genes of cluster 9.
Supplemental Figure 12. Reoxygenation after hypoxia promotes global changes in chromatin accessibility and RNA populations.
Supplemental Figure 13. Regulation of variations in gene activity in response to hypoxia and reoxygenation.
Supplemental Data Set 1. Survey of histones, RNAPII, HRE2, and three RNA subpopulations in response to two durations of HS.
Supplemental Data Set 2. Tabulation of ATAC and HRE2-ChIP peaks on chromatin.
Supplemental Data Set 3. nRNA enrichment and GO analysis.
Supplemental Data Set 4. GO analysis for clusters identified from two durations of HS and reoxygenation.
Supplemental Data Set 5. List of arabidopsis blacklisted chromatin (ABC).
Supplemental File 1. BED File of HRE2 ChIP-seq peaks.
Supplemental File 2. BED File of arabidopsis blacklisted chromatin (ABC).
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Mauricio Reynoso, Roger Deal, Marko Bajic, Seung-Cho Lee, Maureen Hummel, Lauren Dedow, Sonja Winte, Jérémie Bazin, Thomas Girke, Dawn Nagel, and Thomas Eulgem for helpful discussions. This work was supported by the U.S. National Science Foundation (grants IOS-1121626, IOS-1238243, and MCB-1716913) and the U.S. Department of Agriculture, National Institute of Food and Agriculture - Agriculture and Food Research Initiative (grant 2011-04015 to J.B.-S.).
AUTHOR CONTRIBUTIONS
T.A.L. and J.B.-S. designed research; T.A.L. performed research; T.A.L. and J.B.-S. analyzed data; T.A.L. and J.B.-S. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abbas M., Berckhan S., Rooney D.J., Gibbs D.J., Vicente Conde J., Sousa Correia C., Bassel G.W., Marín-de la Rosa N., León J., Alabadí D., Blázquez M.A., Holdsworth M.J. (2015). Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25: 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S., et al. (2010). FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Ansari A., Hampsey M. (2005). A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19: 2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensi-Fabado M.-A., Amtmann A., Perrella G. (2017). Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta. Gene Regul. Mech. 1860: 106–122. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banti V., Loreti E., Novi G., Santaniello A., Alpi A., Perata P. (2008). Heat acclimation and cross-tolerance against anoxia in Arabidopsis. Plant Cell Environ. 31: 1029–1037. [DOI] [PubMed] [Google Scholar]
- Banti V., Mafessoni F., Loreti E., Alpi A., Perata P. (2010). The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 152: 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batie M., Frost J., Frost M., Wilson J.W., Schofield P., Rocha S. (2019). Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363: 1222–1226. [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A., Yang Z., Springer P.S., Bailey-Serres J. (2002). RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028. [DOI] [PubMed] [Google Scholar]
- Bischl B., Lang M., Mersmann O., Rahnenführer J., Weihs C. (2015). BatchJobs and BatchExperiments: Abstraction mechanisms for using R in Batch environments. J. Stat. Softw. 64: 1–25. [Google Scholar]
- Blais J.D., Filipenko V., Bi M., Harding H.P., Ron D., Koumenis C., Wouters B.G., Bell J.C. (2004). Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 24: 7469–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K.A., Jang C.J.H., Larive C.K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56: 743–755. [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kawaguchi R., Ferreira R.B., Bailey-Serres J. (2005). Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 96: 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.J., Raaijmakers L.M., Ramakrishnan S., Wang D., Valiyaparambil S., Liu S., Nowak N.J., Pili R. (2014). Alterations in chromatin accessibility and DNA methylation in clear cell renal cell carcinoma. Oncogene 33: 4961–4965. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui L.T., Giuntoli B., Kosmacz M., Parlanti S., Licausi F. (2015). Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236: 37–43. [DOI] [PubMed] [Google Scholar]
- Büttner M., Singh K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94: 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A.A., et al. (2019). Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363: 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R., Jang C.J.H., Branco-Price C., Nghiem P., Bailey-Serres J. (2012). Transient MPK6 activation in response to oxygen deprivation and reoxygenation is mediated by mitochondria and aids seedling survival in Arabidopsis. Plant Mol. Biol. 78: 109–122. [DOI] [PubMed] [Google Scholar]
- Cheng C.-Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D. (2017). Araport11: a complete reannotation of the Arabidopsis thalianareference genome. Plant J 89 (4): 789–804. [DOI] [PubMed] [Google Scholar]
- Chervona Y., Costa M. (2012). The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic. Biol. Med. 53: 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo S., Charoensawan V., Brestovitsky A., Buning R., Ravarani C., Rhodes D., van Noort J., Jaeger K.E., Wigge P.A. (2017). Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in Arabidopsis. Mol. Plant 10: 1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Bai Y., Zhao L., Dou X., Liu Y., Wang L., Li Y., Li W., Hui Y., Huang X., Wang Z., Qin Y. (2017). H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in Arabidopsis. Mol. Plant 10: 1274–1292. [DOI] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18: 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo L., Sorenson R., Bailey-Serres J., Hunt A.G. (2017). Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell 29: 1262–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Ndamukong I., Xu Z., Lapko H., Fromm M., Avramova Z. (2012). ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 8: e1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4: 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A., Becker P.B. (2002). Histone acetylation: A switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Han S.-K., Kim H.J., Wu M.-F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eysholdt-Derzsó E., Sauter M. (2017). Root bending is antagonistically affected by hypoxia and ERF-mediated transcription via auxin signaling. Plant Physiol. 175: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennoy S.L., Nong T., Bailey-Serres J. (1998). Transcriptional and post-transcriptional processes regulate gene expression in oxygen-deprived roots of maize. Plant J. 15: 727–735. [DOI] [PubMed] [Google Scholar]
- Gasch P., Fundinger M., Müller J.T., Lee T., Bailey-Serres J., Mustroph A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates L.A., Foulds C.E., O’Malley B.W. (2017). Histone marks in the ‘driver’s seat’: Functional roles in steering the transcription cycle. Trends Biochem. Sci. 42: 977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., et al. (2018). Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 9: 5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Lee S.C., Isa N.M., Gramuglia S., Fukao T., Bassel G.W., Correia C.S., Corbineau F., Theodoulou F.L., Bailey-Serres J., Holdsworth M.J. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T. (2014). systemPipeR: NGS workflow and report generation environment. University of California, Riverside https://github.com/tgirke/systemPipeR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B., Lee S.C., Licausi F., Kosmacz M., Oosumi T., van Dongen J.T., Bailey-Serres J., Perata P. (2014). A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 12: e1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B., Licausi F., van Veen H., Perata P. (2017). Functional balancing of the hypoxia regulators RAP2.12 and HRA1 takes place in vivo in Arabidopsis thaliana plants. Front. Plant Sci. 8: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B., Perata P. (2018). Group VII ethylene response factors in Arabidopsis: Regulation and physiological roles. Plant Physiol 176: 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S., Loreti E., Cardarelli F., Novi G., Parlanti S., Pucciariello C., Bassolino L., Banti V., Licausi F., Perata P. (2015). Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 1: 15151. [DOI] [PubMed] [Google Scholar]
- Gordon A., Hannon G.J. (2010). FASTX-Toolkit. FASTQ/A short-reads preprocessing tools. http://hannonlab.cshl.edu/fastx_toolkit/.
- Hajheidari M., Koncz C., Eick D. (2013). Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 18: 633–643. [DOI] [PubMed] [Google Scholar]
- Hattori Y., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel J., Duttke S.H., Benner C., Chory J. (2016). Nascent RNA sequencing reveals distinct features in plant transcription. Proc. Natl. Acad. Sci. USA 113: 12316–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I., Lis J.T. (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L. (2012). H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics 13: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-M., To T.K., Ishida J., Matsui A., Kimura H., Seki M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53: 847–856. [DOI] [PubMed] [Google Scholar]
- Kim J.-M., To T.K., Ishida J., Morosawa T., Kawashima M., Matsui A., Toyoda T., Kimura H., Shinozaki K., Seki M. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49: 1580–1588. [DOI] [PubMed] [Google Scholar]
- Kim S.-H., Jeong J.-W., Park J.A., Lee J.-W., Seo J.H., Jung B.-K., Bae M.-K., Kim K.-W. (2007). Regulation of the HIF-1α stability by histone deacetylases. Oncol. Rep. 17: 647–651. [PubMed] [Google Scholar]
- Kolde R. (2012). Pheatmap: Pretty heatmaps. R package version 61. https://www.rdocumentation.org/packages/pheatmap/versions/0.6.1.
- Koroleva O.A., Calder G., Pendle A.F., Kim S.H., Lewandowska D., Simpson C.G., Jones I.M., Brown J.W.S., Shaw P.J. (2009). Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: Fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell 21: 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmacz M., Parlanti S., Schwarzländer M., Kragler F., Licausi F., Van Dongen J.T. (2015). The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 38: 1094–1103. [DOI] [PubMed] [Google Scholar]
- Krijthe J.H. (2015). Rtsne: T-distributed stochastic neighbor embedding using Barnes-Hut implementation. R package version 0. 13. https://github.com/jkrijthe/Rtsne.
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). CIRCOS: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Ichihashi Y., Kimura S., Chitwood D.H., Headland L.R., Peng J., Maloof J.N., Sinha N.R. (2012). A high-throughput method for Illumina RNA-Seq Library preparation. Front. Plant Sci. 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V. (2018). H2A.Z at the core of transcriptional regulation in plants. Mol. Plant 11: 1112–1114. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Kwak H., Lis J.T. (2013). Control of transcriptional elongation. Annu. Rev. Genet. 47: 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGory E.L., Giaccia A.J. (2016). The ever-expanding role of HIF in tumour and stromal biology. Nat. Cell Biol. 18: 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9 (4): 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Gentleman R., Carey V. (2009). rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. (2013). Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F., Kosmacz M., Weits D.A., Giuntoli B., Giorgi F.M., Voesenek L.A.C.J., Perata P., van Dongen J.T. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422. [DOI] [PubMed] [Google Scholar]
- Licausi F., van Dongen J.T., Giuntoli B., Novi G., Santaniello A., Geigenberger P., Perata P. (2010). HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62: 302–315. [DOI] [PubMed] [Google Scholar]
- Lokdarshi A., Conner W.C., McClintock C., Li T., Roberts D.M. (2016). Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 170: 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E., Poggi A., Novi G., Alpi A., Perata P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137: 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Hofmeister B.T., Vollmers C., DuBois R.M., Schmitz R.J. (2017). Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 45: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. (2019). cluster: Cluster Analysis Basics and Extensions. R package version 2.1.0. https://cran.r-project.org/package=cluster.
- Maher K.A., et al. (2018). Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30: 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D., Dimitriadou E., Hornik K., Weingessel A., Leisch F. (2015). e1071: Misc Functions of the Department of Statistics (e1071), TU Wien, 2014. R package version 1.6. https://CRAN.R-project.org/package=e1071.
- Millar A.H., Heazlewood J.L., Giglione C., Holdsworth M.J., Bachmair A., Schulze W.X. (2019). The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 70: 119–151. [DOI] [PubMed] [Google Scholar]
- Milligan L., Huynh-Thu V.A., Delan-Forino C., Tuck A., Petfalski E., Lombraña R., Sanguinetti G., Kudla G., Tollervey D. (2016). Strand-specific, high-resolution mapping of modified RNA polymerase II. Mol. Syst. Biol. 12: 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R. (2009). ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25: 2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M., Obenchain V., Lang M., Thompson R., Turaga N. (2019. a). BiocParallel: Bioconductor facilities for parallel evaluation. R package version 1.18.1, https://github.com/Bioconductor/BiocParallel.
- Morgan M., Pagès H., Obenchain V., Hayden N. (2019. b). Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package version 2.0.1, http://bioconductor.org/packages/Rsamtools.
- Mustroph A., Lee S.C., Oosumi T., Zanetti M.E., Yang H., Ma K., Yaghoubi-Masihi A., Fukao T., Bailey-Serres J. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152: 1484–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J.H., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwirth E. (2011). RColorBrewer: Colorbrewer palettes. R package version 1. https://CRAN.R-project.org/package=RColorBrewer.
- Niedojadło J., Dełeńko K., Niedojadło K. (2016). Regulation of poly(A) RNA retention in the nucleus as a survival strategy of plants during hypoxia. RNA Biol. 13: 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G., Zhang X., Song J.S., Fisher D.E. (2008). Chromatin structure analyses identify miRNA promoters. Genes Dev. 22: 3172–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.V., Iyer S., Amerhauser C., Lehmann M., van Dongen J.T., Geigenberger P. (2016). Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 172: 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H.P., Greenleaf A.L. (2006). Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20: 2922–2936. [DOI] [PubMed] [Google Scholar]
- Pucciariello C., Parlanti S., Banti V., Novi G., Perata P. (2012). Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 159: 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. (2016). deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso M.A., Pauluzzi G.C., Kajala K., Cabanlit S., Velasco J., Bazin J., Deal R., Sinha N.R., Brady S.M., Bailey-Serres J. (2018a). Nuclear transcriptomes at high resolution using retooled INTACT. Plant Physiol. 176: 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso M., Pauluzzi G., Cabanlit S., Velasco J., Bazin J., Deal R., Brady S., Sinha N., Bailey-Serres J., Kajala K. (2018b). Isolation of nuclei in tagged cell types (INTACT), RNA extraction and ribosomal RNA degradation to prepare material for RNA-Seq. Bio Protoc. 8: e2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F., Kitajima S., Koyama T., Yamada Y. (1996). Ethylene-induced gene expression of osmotin-like protein, a neutral isoform of tobacco PR-5, is mediated by the AGCCGCC cis-sequence. Plant Cell Physiol. 37: 249–255. [DOI] [PubMed] [Google Scholar]
- Schmidl C., Rendeiro A.F., Sheffield N.C., Bock C. (2015). ChIPmentation: Fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12: 963–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P., Bajic M., McKinney E.C., Meagher R.B., Deal R.B. (2018). Changes in chromatin accessibility between Arabidopsis stem cells and mesophyll cells illuminate cell type-specifictranscription factor networks. Plant J. 94: 215–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson R., Bailey-Serres J. (2014). Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A.M., et al. (2014). Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030. [DOI] [PubMed] [Google Scholar]
- Sura W., Kabza M., Karlowski W.M., Bieluszewski T., Kus-Slowinska M., Pawełoszek Ł., Sadowski J., Ziolkowski P.A. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 29: 791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Henikoff S. (2017). Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18: 115–126. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J.T., Mesirov J.P. (2012). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E.S., Deal R.B. (2019). The histone variant H2A.Z and chromatin remodeler BRAHMA act coordinately and antagonistically to regulate transcription and nucleosome dynamics in Arabidopsis. Plant J. 99: 144–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley B.T., Covington M.F., Ichihashi Y., Zumstein K., Sinha N.R. (2015). BrAD-seq: Breath Adapter Directional sequencing: A streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front. Plant Sci. 6: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J.T., Licausi F. (2015). Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66: 345–367. [DOI] [PubMed] [Google Scholar]
- van Veen H., Vashisht D., Akman M., Girke T., Mustroph A., Reinen E., Hartman S., Kooiker M., van Tienderen P., Schranz M.E., Bailey-Serres J., Voesenek L.A.C.J., et al. (2016). Transcriptomes of Eight Arabidopsis thaliana Accessions Reveal Core Conserved, Genotype- and Organ-Specific Responses to Flooding Stress. Plant Physiol. 172: 668–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J., Mendiondo G.M., Movahedi M., Peirats-Llobet M., Juan Y.-T., Shen Y.-Y., Dambire C., Smart K., Rodriguez P.L., Charng Y.-Y., Gray J.E., Holdsworth M.J. (2017). The Cys-Arg/N-End rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27: 3183–3190.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek L.A.C.J., Bailey-Serres J. (2013). Flooding tolerance: O2 sensing and survival strategies. Curr. Opin. Plant Biol. 16: 647–653. [DOI] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M., Venables B. (2009). gplots: Various R programming tools for plotting data. R package version 2.1. https://CRAN.R-project.org/package=gplots.
- Watson J.A., Watson C.J., McCann A., Baugh J. (2010). Epigenetics, the epicenter of the hypoxic response. Epigenetics 5: 293–296. [DOI] [PubMed] [Google Scholar]
- Weits D.A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H.-M., Riegler H., Hoefgen R., Perata P., van Dongen J.T., Licausi F. (2014). Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.D., et al. (2017). Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 8: 14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysis.. (New York: Springer Science & Business Media; ). [Google Scholar]
- Wickham H., Francois R., Henry L., Müller K. (2015). dplyr: A grammar of data manipulation. R package version 0.4.3. http://CRAN.R-project.org/package=dplyr
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708. [DOI] [PubMed] [Google Scholar]
- Yang C.-Y., Hsu F.-C., Li J.-P., Wang N.-N., Shih M.-C. (2011). The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 156: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Wightman R., Meyerowitz E.M. (2017). Cell cycle control by nuclear sequestration of CDC20 and CDH1 mRNA in plant stem cells. Mol. Cell 68: 1108–1119.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung E., et al. (2018). A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 115: E6085–E6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.M., Wang S.-J., Gordan J.D., Ji X., Liebhaber S.A., Simon M.C. (2008). Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J. Biol. Chem. 283: 16309–16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., He Q.-Y. (2015). ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zanetti M.E., Chang I.-F., Gong F., Galbraith D.W., Bailey-Serres J. (2005). Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 138: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X., He K., Charron J.-B.F., Elling A.A., Deng X.W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72: 585–595. [DOI] [PubMed] [Google Scholar]
- Zhu J., Liu M., Liu X., Dong Z. (2018). RNA polymerase II activity revealed by GRO-seq and pNET-seq in Arabidopsis. Nat. Plants 4: 1112–1123. [DOI] [PubMed] [Google Scholar]
- Zid B.M., O’Shea E.K. (2014). Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]