The Arabidopsis (Arabidopsis thaliana) transcription factors SPL9 and DEWAX act antagonistically to regulate rhythmic CER1 expression and diurnal wax synthesis.
Abstract
Plant surface waxes form an outer barrier that protects the plant from many forms of environmental stress. The deposition of cuticular waxes on the plant surface is regulated by external environmental changes, including light and dark cycles. However, the underlying molecular mechanisms controlling light regulation of wax production are still poorly understood, especially at the posttranscriptional level. In this paper, we report the regulation of cuticular wax production by the miR156-SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9) module in Arabidopsis (Arabidopsis thaliana). When compared with wild-type plants, miR156 and SPL9 mutants showed significantly altered cuticular wax amounts in both stems and leaves. Furthermore, it was found that SPL9 positively regulates gene expression of the alkane-forming enzyme ECERIFERUM1 (CER1), as well as the primary (1-) alcohol-forming enzyme ECERIFERUM4 (CER4), to enhance alkane and 1-alcohol synthesis, respectively. Our results indicate that complex formation of SPL9 with a negative regulator of wax synthesis, DEWAX, will hamper SPL9 DNA binding ability, possibly by interfering with SPL9 homodimerization. Combined with their diurnal gene and protein expressions, this dynamic repression–activation transcriptional module defines a dynamic mechanism that may allow plants to optimize wax synthesis during daily cycles. These findings provide a regulatory framework for environmental signal integration in the regulation of wax synthesis.
INTRODUCTION
The surface of most aerial plant organs is covered with a coating of cuticular waxes that provides protection against multiple stress factors. Cuticular wax formation is tightly regulated by both developmental and environmental cues, which allows plants to adapt to changing environmental conditions during their life cycle (Shepherd and Wynne Griffiths, 2006; Samuels et al., 2008). For example, multiple plant species show higher cuticular wax synthesis in the light than in the dark (Baker, 1974; Giese, 1975; Shepherd et al., 1995; Go et al., 2014). However, it is unclear how wax synthesis is activated by light; nor is it clear whether light induction of wax functions as a beneficial environmental adaptation or as a developmental regulator (Shepherd and Wynne Griffiths, 2006).
Cuticular waxes comprise an admixture of very-long chain aliphatic compounds including primary alcohols, alkyl esters, alkanes, ketones, aldehydes, and secondary alcohols, which are mainly synthesized in the endoplasmic reticulum (ER; Bernard and Joubès 2013). Through characterizing the wax-deficient mutants and analyzing the transcriptome of stem epidermal peels, cuticular wax biosynthetic pathways in the model plant Arabidopsis (Arabidopsis thaliana) have been extensively studied (Jenks et al., 1995; Suh et al., 2005; Nawrath et al., 2013). In the epidermal cells, the wax precursor very long chain fatty acids (VLCFAs) are synthesized in serial pathways. First, C16 or C18 fatty acids are synthesized in the plastid from acetyl-CoA. Second, these LCFAs are converted to LCFA-CoAs by long chain acyl-CoA synthetases and then further elongated into VLCFA-CoAs (24 to 34 carbons in length) in the ER through fatty acid elongase (Bernard and Joubès 2013). Third, different classes of cuticular wax component are further produced from VLCFA-CoAs through enzymes either involved in the alkane-forming pathway or the alcohol-forming pathway (Aarts et al., 1995; Hannoufa et al., 1996; Rowland et al., 2006; Li et al., 2008). Finally, wax compounds are transported to the plasma membrane and secreted outside cells, and then deposited on the epidermis (Bernard and Joubès 2013).
Cuticular wax metabolic pathways are under a substantial amount of transcriptional regulation, especially by members of the MYB and AP2/EREBP families (Lee and Suh, 2015a). In Arabidopsis, WAX INDUCER1/SHINE1, which belongs to the AP2/EREBP-type transcription factor (TF) family, and its homologs SHN2 and SHN3, activates cuticular wax synthesis, improving plant drought tolerance (Aharoni et al., 2004; Broun et al., 2004). Recently, WRINKLED4 (WRI4, also a subfamily of AP2/EREBP TF) was reported to be involved in direct activation of genes in the wax synthesis pathway (Park et al., 2016). DEWAX, another AP2/ERF TF, negatively regulates wax production in Arabidopsis (Go et al., 2014). Besides AP2/EREBP families, cuticular wax biosynthesis is also regulated by MYB TFs, such as MYB16, MYB30, MYB96, MYB94, and MYB106 (Raffaele et al., 2008; Seo et al., 2011; Oshima et al., 2013; Lee and Suh, 2015b). The transcriptional regulation of wax metabolism by MYB96 and MYB30 was also found to participate in plant tolerance to abiotic and biotic stresses, respectively (Raffaele et al., 2008; Seo et al., 2011). Based on the above findings, it is clear that even though many TFs and their targets in wax synthesis have been identified, a more complete understanding of the transcriptional network regulating cuticular wax biosynthesis needs to be addressed (Lee and Suh, 2015a).
Apart from transcriptional regulation, cuticular wax biosynthesis is also regulated at the posttranscriptional level by small RNAs (sRNAs; Hooker et al. 2007; Lam et al. 2012, 2015). CER7 encodes an exoribonuclease, a core subunit of the RNA-processing/degrading exosome complex, which was first reported to regulate the waxes synthesis on the developing stems of Arabidopsis (Hooker et al. 2007). Through identifying the cer7 suppressor mutants, Lam et al. (2012) speculated that CER7-mediated exosomal degradation alters the levels of sRNA species, which in turn controls CER3 expression by gene silencing at the posttranscriptional level. Indeed, the authors further demonstrated that trans-acting small interfering RNAs (tasiRNAs), one type of plant sRNAs, directly control CER3 expression levels and regulate stem wax deposition (Lam et al. 2015). In addition to tasiRNAs, another type of sRNAs, micro RNAs (miRNAs), plays important roles in gene expression regulatory networks, and affects diverse aspects of plant growth and development at the posttranscriptional level (Borges and Martienssen, 2015). miR156 is one of the few miRNAs that is highly conserved within the plant kingdom (Chuck et al., 2007; Wang et al., 2009, 2011; Wu et al., 2009). miR156 targets members of the plant-specific SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) transcription factor gene family (Klein et al., 1996; Guo et al., 2008). In Arabidopsis, 10 SPL genes have miR156 binding sites either in the coding region or the 3′-untranslated region. They can be further classified into four taxonomic subgroups: SPL3/SPL4/SPL5, SPL9/SPL15, SPL2/SPL10/SPL11, and SPL6/SPL13 (Cardon et al., 1999; Wu and Poethig, 2006; Gandikota et al., 2007). The miR156-SPL9 module has been found to be involved in multiple biological processes, including phase transition, root and leaf development, and flowering as well as stress responses (Wu and Poethig, 2006; Wang et al., 2009; Gou et al., 2011; Cui et al., 2014; Rubio-Somoza et al., 2014; Yu et al., 2015a). Specifically, the miR156-SPL9 module is reported to regulate secondary metabolism. For example, SPL9 interacts with PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and decreases anthocyanin biosynthesis through directly regulating DIHYDROFLAVONOL 4-REDUCTASE (DFR) expression (Gou et al., 2011). Furthermore, Yu et al. (2015b) demonstrated that SPL9 directly binds to the Terpene Synthase21 (TPS21) gene promoter and activates its expression to regulate sesquiterpene production in Arabidopsis and Patchouli (Pogostemon cablin). To date, however, whether miR156 or SPLs are involved in wax synthesis is not known.
A previous study reported that DEWAX is a negative regulator of wax accumulation in the dark (Go et al., 2014). However, optimization of the daily light/dark responses would require an additional positive regulator in the light. In this paper, we have shown that SPL9 acts as a positive light-induced regulator of wax synthesis. Our research demonstrates that SPL9 controls CER1 expression, a rate limiting step in wax alkane synthesis. This is achieved by directly binding to GTAC motifs in the CER1 promoter. Our data also indicate that SPL9 and DEWAX act antagonistically to control CER1 expression via direct protein–protein interaction. The sophisticated combinatorial regulation exerted by the SPL9-DEWAX loop constitutes a key molecular mechanism mediating the light-dark on-off switch controlling wax synthesis.
RESULTS
The miR156-SPL9 Module Regulates Wax Synthesis
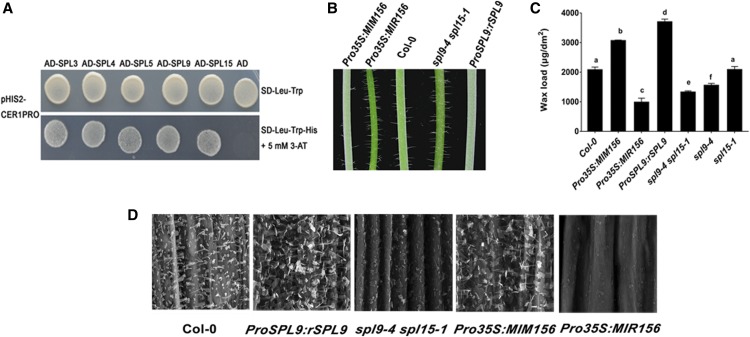
Alkanes are the major components of cuticular wax, and previous studies have shown that the alkane synthesizing gene CER1 is expressed in a diurnal cycle (Go et al., 2014). To identify factors responsible for light-regulated wax synthesis, we performed a yeast one-hybrid screen with CER1 promoter DNA with a prey library composed of ∼1500 transcription factor cDNAs of Arabidopsis (Mitsuda et al., 2010). Interestingly, this screen identified a positive clone encoding SPL9. Using a full-length cDNA of SPL9 inserted into a pGADT7 plasmid, we demonstrated that SPL9 interacted with the CER1 promoter in a yeast one-hybrid assay via the expression of the HIS reporter gene driven by the CER1 promoter (Figure 1A). SPL9 has been reported to be a miR156 target, and to participate in multiple plant developmental and secondary metabolic regulatory pathways. However, whether miR156 or SPL9 was involved in wax synthesis has not been demonstrated.
Figure 1.
miR156-SPL9 Module Regulates Wax Synthesis.
(A) Yeast one-hybrid assay to dissect the binding of SPLs to CER1 promoter DNA. For yeast one-hybrid experiment, CER1 promoter DNA region was fused to the HIS3 (auxotrophic marker) reporter gene in pHIS2 plasmid and tested for AD-SPLs binding. Empty pGADT7 vector was used as negative control.
(B) Glossy green and white waxy phenotypes of 6-week-old inflorescence stems of Pro35S:MIR156, Pro35S:MIM156, and SPL9 related lines compared with the wild type (Col-0). Pro35S:MIR156 and Pro35S:MIM156 are transgenic lines overexpressing miR156 and its artificial miRNA target mimic construct, respectively. ProSPL9:rSPL9 and spl9-4 spl15-1 represent SPL9 gain-of-function and loss-of-function plants, respectively.
(C) Cuticular wax amounts of inflorescence stems from 6-week-old wild-type, Pro35S:MIR156, Pro35S:MIM156, and SPL9 related Arabidopsis lines, which were grown under long-day conditions (16 h of light/8 h of dark). Cuticular waxes were extracted with hexane and analyzed by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate means from four replicate experiments, and their SD are shown. The significant differences between samples labeled with different lowercase letters were determined using ANOVA with a post hoc Tukey Honestly Significant Difference test.
(D) SEM images of epicuticular wax crystals on inflorescence stems of 6-week-old wild-type, Pro35S:MIR156, Pro35S:MIM156, and SPL9 related Arabidopsis lines. Bars = 5 μm.
We then obtained miR156- or SPL9-related genetic materials that were previously used in other studies (Gou et al., 2011), including Pro35S:MIR156, Pro35S:MIM156, ProSPL9:rSPL9, spl9-4 spl15-1. Pro35S:MIR156 is a transgenic line overproducing miR156, whereas Pro35S:MIM156 is a transgenic line in which miR156 activity is reduced via target mimicry (Franco-Zorrilla et al., 2007). Pro35S:MIM156 was used because miR156 is generated from eight separate loci in the Arabidopsis genome and it is difficult to obtain a complete knockout allele of miR156. ProSPL9:rSPL9 and spl9-4 spl15-1 were used as SPL9 gain-of-function and loss-of-function lines, respectively. ProSPL9:rSPL9 expresses a miR156-resistant SPL9 transcript; therefore, SPL9 was overexpressed. The spl9-4 spl15-1 double mutant was first chosen for phenotype analysis because SPL15 is the closest paralog of SPL9. As in previous studies, the SPL genes showed a high degree of functional redundancy in development, and single mutants were phenotypically normal as wild-type plants (Wang et al., 2008; Wu et al., 2009). We observed glossy green (Pro35S:MIR156 and spl9-4 spl15-1) or glaucous white (Pro35S:MIM156 and ProSPL9:rSPL9) phenotypes in the stems, respectively (Figure 1B).
Consistent with this observation, chemical analysis of wax revealed that total wax loads were increased by ∼50% and 70% on Pro35S:MIM156 and ProSPL9:rSPL9 stems, respectively, relative to the wild type. By contrast, a substantial decrease in total wax amount was observed on stems of Pro35S:MIR156 (∼47% of wild type, P < 0.01) and spl9-4 spl15-1 (∼63% of wild type, P < 0.01), relative to the wild type, with Pro35S:MIR156 displaying the least severe wax defect (Figure 1C).
Using scanning electron microscopy (SEM), we examined the deposition of epicuticular wax crystals on the stem surface of these lines. Relative to wild type, we observed significantly fewer epicuticular wax crystals on the Pro35S:MIR156 and spl9-4 spl15-1 stems, and a significantly higher density of epicuticular wax crystals on the Pro35S:MIM156 and ProSPL9:rSPL9 stems (Figure 1D). Together, these results indicated that the miR156-SPL9 module has a significant effect on the regulation of epidermal wax synthesis.
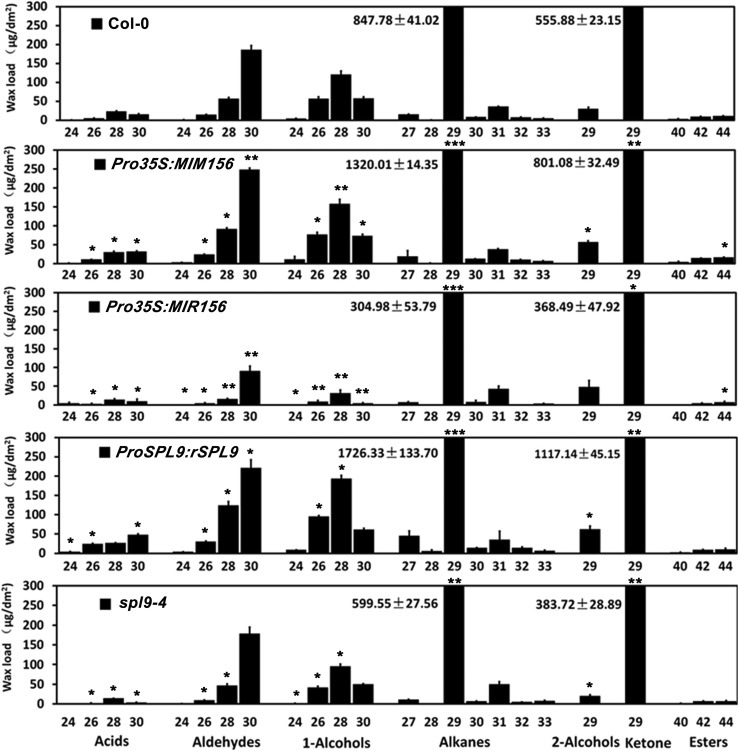
To elucidate the role of SPL9 or SPL15 in wax biosynthesis, we examined the stem wax contents of the spl9-4 or spl15-1 mutant. The total wax loads on spl9-4 stems were much lower than those in the wild type, whereas spl15-1 exhibit similar wax accumulation as the wild type (Figure 1C). Furthermore, the wax content of spl9-4 was significantly higher than that of the spl9-4 spl15-1 double mutant (Figure 1C), whereas the Pro35S:MIR156 overexpression transgenic line exhibited the lowest wax content. These data suggest that the production of wax is apparently coordinately regulated by multiple SPLs, with a major contribution of SPL9. Consistent with this idea, several other SPLs including SPL3, SPL4, SPL5, and SPL15 also could bind the CER1 promoter in yeast one-hybrid assays (Figure 1A). In the following research, we focused our analysis on SPL9. We found that essentially all individual wax constituents were increased in Pro35S:MIM156 and ProSPL9:rSPL9 stems relative to the wild type. Particularly, there was a prominent increase in the amounts of the C29 alkanes and C29 ketones in both Pro35S:MIM156 and ProSPL9:rSPL9. Conversely, almost all wax constituents were reduced in Pro35S:MIR156 and spl9-4 stems, especially the C29 alkanes and C29 ketones (Figure 2).
Figure 2.
Cuticular Wax Composition of Inflorescence Stems of 6-week-old Wild-Type, Pro35S:MIR156, Pro35S:MIM156, and SPL9 Related Arabidopsis Lines.
Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Each wax constituent is designated by carbon chain length and is labeled by chemical class along the x axis. Values shown are means ±sd (n = 4). The differential significance between transgenic or mutant lines and wild-type plants was determined with Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).
To test whether the miR156-SPL9 module regulates wax synthesis in the leaves, the total cuticular wax amounts and compositions on the surface of leaves of the above lines were determined. The total wax amounts on Pro35S:MIR156 and spl9-4 leaves were reduced to ∼30% and 60% of the wild-type level, respectively. While in Pro35S:MIM156 and ProSPL9:rSPL9 leaves, wax loads were increased by ∼60% and ∼150%, respectively, relative to the wild type (Supplemental Figure 1). The alterations observed in individual wax constituents were most prominent in the C26 and C28 primary alcohols, and the C29, C31, and C33 alkanes (Supplemental Figure 2). Consistent with the higher wax accumulation in ProSPL9:rSPL9 leaves, epicuticular wax crystals, which are not normally present on wild-type leaves when viewed using SEM, were observed on ProSPL9:rSPL9 leaf surfaces (Supplemental Figure 3). Overall, these results demonstrate that SPL9 positively regulates cuticular wax deposition on leaf and stem surfaces.
SPL9 Directly Regulates CER1 Expression, but Indirectly Regulates CER4 Expression
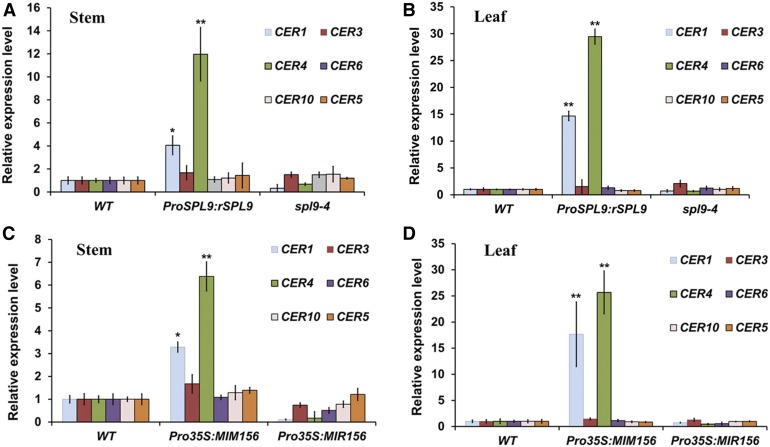
To investigate how SPL9 influences wax synthesis at the transcriptional level, RT-qPCR was used to analyze the expression of six representative genes previously associated with wax biosynthesis or transport. Among these genes, CER1 and CER3 (also known as WAX2 or YRE) as two components of a multiprotein enzyme complex catalyze the conversion of very long chain acyl-CoAs to very long chain alkanes, which are the most abundant (more than 50% in all organs) wax components in Arabidopsis (Bernard et al., 2012; Lee and Suh, 2015a). CER4 (also known as FAR3) encoding a fatty acyl-CoA reductase converts the VLCFAs into primary alcohols in the alcohol-forming pathway (Rowland et al., 2006). Interestingly, alkane and 1-alcohol amounts were much higher in the stems and leaves of ProSPL9:rSPL9 than in those of the wild type (Supplemental Tables 1 and 2), indicating both biosynthetic components are simultaneously affected in the two tissues. CER6 (also known as KCS6, encoding a 3-ketoacyl-CoA synthase) and CER10 (also known as ECR, encoding an enoyl-CoA reductase) are important for VLCFA elongation (Hooker et al., 2002; Zheng et al. 2005). CER5 (also known as ABCG12, encoding an adenosine triphosphate binding cassette transporter) is responsible for wax transportation to the epidermal surface (Pighin et al., 2004). Among these genes, CER1 and CER4 showed significantly enhanced expression (>twofold) in Pro35S:MIM156 and ProSPL9:rSPL9, but decreased expression in Pro35S:MIR156 and spl9-4, in both leaves and stems (Figure 3), which is consistent with the altered compositions of alkanes and primary alcohols we report for these lines. To demonstrate whether SPL9 could modulate CER1 and CER4 gene expression in vivo, we constructed the CER1 and CER4 promoter luciferase (LUC) reporter plasmids, and performed a transient expression assay in tobacco (Nicotiana benthamiana). The results suggested that coexpression of SPL9 protein strongly increased the LUC reporter gene activities driven by the endogenous promoters of CER1 and CER4 (Figure 4A). To test whether the differentially expressed CER1 and CER4 were direct targets of SPL9, we conducted experiments using transgenic lines expressing the rSPL9 chimeric protein fused to the glucocorticoid receptor sequence (rSPL9-GR), and exogenous dexamethasone (DEX) exposure to assess nuclear transport and biological activity (Gou et al., 2011). For time-course analysis of gene expression, we chose 0-, 10-, and 30-min and 1-, 3-, 6-, and 12-h treatments of transgenic plants with or without DEX. As CER1 has been reported to show diurnal rhythmic expression (Go et al., 2014), we calculated the gene expression ratio with and without DEX treatment to exclude other factors possibly affecting CER1 and CER4 expression. Interestingly, both CER1 and CER4 could be induced by DEX treatment, but the induction times showed large differences. CER1 could be induced within 10 min and lasted 12 h. However, CER4 was not induced until 6 h of DEX treatment (Supplemental Figure 4). This result indicates that CER1 expression can be rapidly induced by sudden changes of SPL9 activity. Moreover, CER1, but not CER4 gene expression, could be enhanced by SPL9 with exogenous exposure to cycloheximide (CHX) and DEX (Figure 4B), raising the possibility that induction of CER4 expression by SPL9 requires synthesis of additional downstream factors.
Figure 3.
Expression of Wax Synthesis Related Genes in the Stem and Leaf of Pro35S:MIR156, Pro35S:MIM156, and SPL9 Related Arabidopsis Lines.
For all the RT-qPCR experiments, the expression level in the wild type (WT) was set as 1. Expression of PP2A was used as internal control. Error bars indicate ±sd (n = 3). * and **Means differed significantly from WT at P < 0.05 and P < 0.01, respectively, according to Student’s t test.
(A) RT-qPCR analysis of wax biosynthetic genes from the stems of 5-week-old wild-type, ProSPL9:rSPL9, and spl9-4 plants.
(B) RT-qPCR analysis of wax biosynthetic genes from the leaves of 3-week-old wild-type, ProSPL9:rSPL9, and spl9-4 plants.
(C) RT-qPCR analysis of wax biosynthetic genes from the stems of 5-week-old wild-type, Pro35S:MIM156, and Pro35S:MIR156 plants.
(D) RT-qPCR analysis of wax biosynthetic genes from the leaves of 3-week-old wild-type, Pro35S:MIM156, and Pro35S:MIR156 plants.
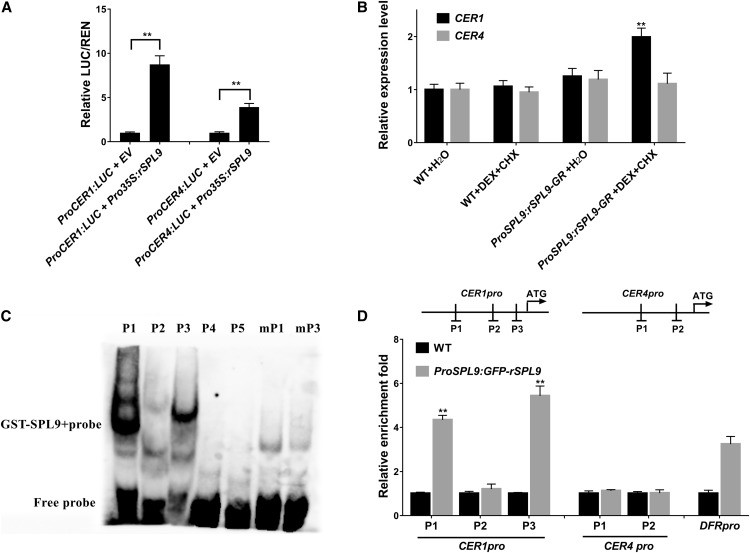
Figure 4.
SPL9 Directly Regulates CER1 Expression, but Indirectly Regulates CER4 Expression.
(A) SPL9 upregulates CER1 and CER4 expression in a transient dual-luciferase reporter system. The coinfiltrations of empty vector (EV) or Pro35S:rSPL9 with ProCER1:LUC or ProCER4:LUC reporters were simultaneously expressed in N. benthamiana leaves, and the plants were first incubated in dark for 12 h and then in light for 48 h. The relative luciferase luminescence intensities were quantitated using Renilla luciferase (REN) for normalization. Asterisks indicate significant differences between the indicated means with P < 0.01 by Student’s t test. Data are presented as the means of three independent assays. Error bars represent ±sd (n = 3).
(B) Induction of expression of CER1 and CER4 in ProSPL9:rSPL9-GR plants. Seven-day-old, long day–grown seedlings were treated with either water or DEX and CHX. Seedlings were harvested 6 h after treatment. Expression was normalized relative to that of PP2A. Each column represents the mean of three independent assays, each based on over 20 seedlings. Errors bars indicate ±sd (**P < 0.01, Student’s t test). WT, wild type.
(C) EMSAs to test binding of SPL9 to the putative GTAC motifs in the CER1 (P1-P3) and CER4 (P4 and P5) promoters, respectively. The GTAC motifs in P1 and P3 were mutated to AAAA in mP1 and mP3 to test the sequence specificity.
(D) ChIP-qPCR confirmed direct binding of SPL9 in the CER1 promoter. Diagram depicts the putative promoters of CER1 and CER4. PCR amplicons indicated as P1-P3 for the CER1 promoter and P1/P2 for the CER4 promoter were used for ChIP-qPCR. Chromatin from 3-week-old Arabidopsis seedlings expressing ProSPL9:GFP-rSPL9 (Gou et al., 2011) was extracted using anti-GFP antibodies. Wild-type seedlings were used as negative control. qPCR was used to quantify enrichment of SPL9 to promoter regions. SPL9 DNA binding ratio (as revealed by GFP enrichment in ChIP experiments) to the promoter regions was assayed. DFR promoter was used as a positive control. Each column represents the mean of three biological repeats. Error bars denote ±sd (**P < 0.01, Student’s t test).
SPL9 is known to preferentially recognize DNA motifs with the GTAC sequence (Yamasaki et al., 2009). We identified three and two typical SPL9 binding sites (GTAC) in the CER1 and CER4 promoters, respectively. To test whether SPL9 directly binds to these sequences, we conducted electrophoretic mobility shift assays (EMSAs). For these experiments, complementary oligonucleotides spanning the corresponding GTAC and flanking regions from the CER1 promoter (P1–P3) and the CER4 promoter (P4 and P5) were synthesized and labeled with biotin. As shown in Figure 4C, the recombinant GST-SPL9 protein caused a mobility shift of the two probes P1 and P3 in the CER1 promoter (Figure 4C). However, the P2 probe in the CER1 promoter and P4 and P5 probes in the CER4 promoter showed no mobility shift. Furthermore, the probes mP1 and mP3, in which the GTAC motif had been removed by mutations, did not show any shifted band when incubated with GST-SPL9 (Figure 4C), indicating that SPL9 bound specifically to GTAC motifs. To further determine whether SPL9 directly associates with CER1 promoter in vivo, a chromatin immunoprecipitation (ChIP) assay was performed using transgenic ProSPL9:GFP-rSPL9 lines (Gou et al., 2011). After the protein–DNA complexes were immunoprecipitated by anti–GFP antibody, the DNA fragments were quantified by qPCR. The promoter region of DFR (a gene known to be a direct SPL9 target, Gou et al., 2011) was amplified as a positive control, and the promoter of ACT2 was amplified as a negative control. Similar to the DFR positive control, the P1 and P3 regions of the CER1 promoter, which include the GTAC motif, were enriched in ProSPL9:GFP-rSPL9 when compared with the wild type (Figure 4D), consistent with the EMSA results. By contrast, there was no obvious enrichment for regions P2 in CER1 and P1 and P2 in CER4 promoters. These results confirmed that CER1 was a direct target of SPL9 (Figure 4D). Therefore, we suggest that SPL9-mediated direct regulation of CER1 and indirect regulation of CER4 expression represents a major regulatory pathway controlling epidermal wax biosynthesis.
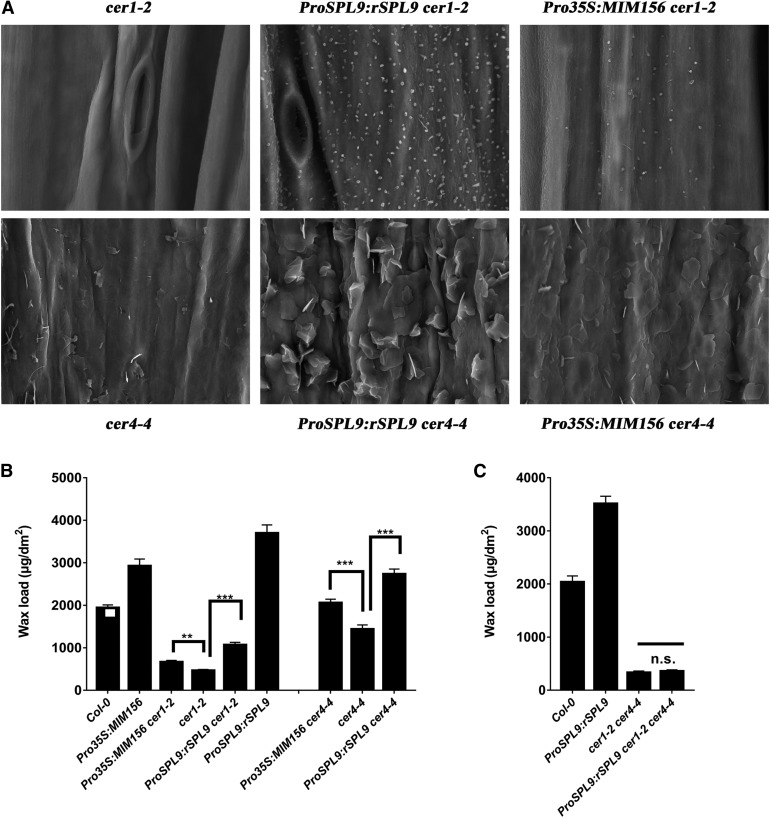
CER1 and CER4 are Independently Responsible for SPL9 Regulated Wax Synthesis
Although we have observed that both CER1 and CER4 are differentially expressed in miR156-SPL9 related mutants, whether changes in the expression of these genes is responsible for the altered wax phenotype is unknown. To shed light on this, we crossed the loss of function mutants cer1-2 and cer4-4 to ProSPL9:rSPL9 and Pro35S:MIM156, respectively, to create the genetic lines ProSPL9:rSPL9 cer1-2, ProSPL9:rSPL9 cer4-4, Pro35S:MIM156 cer1-2, and Pro35S:MIM156 cer4-4. As an initial observation, all of these lines exhibited a glossy phenotype comparable with cer1-2 and cer4-4 (Supplemental Figure 5). Next, we performed SEM analysis to observe the distributions of epicuticular wax crystals on the stem surface of these lines. Surprisingly, the stem surfaces of all these lines exhibited a much higher density of wax crystals than either cer1-2 or cer4-4, with especially higher crystal density on ProSPL9:rSPL9 cer1-2 and ProSPL9:rSPL9 cer4-4 (Figure 5A). To explore the basis for this, we examined the stem wax amounts and wax chemical compositions of these lines using gas chromatography with flame ionization detection (GC-FID). Interestingly, Pro35S:MIM156 cer1-2, Pro35S:MIM156 cer4-4, ProSPL9:rSPL9 cer1-2, and ProSPL9:rSPL9 cer4-4 showed significantly higher wax accumulation compared with the single cer1-2 or cer4-4 mutants, with the accumulation being higher on ProSPL9:rSPL9 cer1-2 and ProSPL9:rSPL9 cer4-4 than on Pro35S:MIM156 cer1-2 and Pro35S:MIM156 cer4-4 (Figure 5B). When the wax chemical compositions were considered, it was notable that in ProSPL9:rSPL9 cer1-2 or Pro35S:MIM156 cer1-2, the synthesis of primary alcohols was greatly increased compared with the cer1-2 single mutant (Supplemental Table 1). By comparison, in ProSPL9:rSPL9 cer4-4 and Pro35S:MIM156 cer4-4, the total alkane content was much higher than in cer4-4 (Supplemental Table 1). Interestingly, although the total wax loads in the stem of Pro35S:MIM156 cer4-4 and ProSPL9:rSPL9 cer4-4 lines were higher than in the wild-type plants, these two lines still display a glossy phenotype. Moreover, the data obtained for leaves was similar as in stems, in that ProSPL9:rSPL9 and Pro35S:MIM156 exhibited increased wax amounts in the cer1-2 and cer4-4 background, with specific upregulation of the primary alcohols or alkanes, respectively (Supplemental Figure 6; Supplemental Table 2). These results indicate that independent regulation of CER1 and CER4 by SPL9 are involved in the regulation of wax synthesis. Based on this, we constructed a cer1-2 cer4-4 double mutant, and ProSPL9:rSPL9 cer1-2 cer4-4 genetic line, and examined their wax phenotypes. Consistent with our hypothesis, the wax amount in ProSPL9:rSPL9 cer1-2 cer4-4 was the same as in cer1-2 cer4-4 (Figure 5C).
Figure 5.
CER1 and CER4 Are Independently Responsible for SPL9 Regulated Wax Synthesis.
(A) SEM images of epicuticular wax crystals on inflorescence stems of 6-week-old Arabidopsis crossing lines between 35S:rSPL9 or 35S:MIM156 and cer1-2 or cer4-4. Bars = 5 μm.
(B) Cuticular wax amounts of inflorescence stems from 6-week-old Arabidopsis crossing lines between 35S:rSPL9 or 35S:MIM156 and cer1-2 or cer4-4 grown in long-day conditions. Cuticular waxes were extracted with hexane and analyzed by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate ±sd (**P < 0.01, ***P < 0.001, Student’s t test) from four replicate experiments.
(C) Cuticular wax amounts of inflorescence stems from 6-week-old wild-type, cer1-2 cer4-4, and ProSPL9:rSPL9 cer1-2 cer4-4 mutants. Cuticular waxes were extracted with hexane and analyzed by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate ±sd from four replicate experiments; n.s., no significance by Student’s t test.
SPL9 Specifically Interacts with DEWAX In Vitro and In Vivo
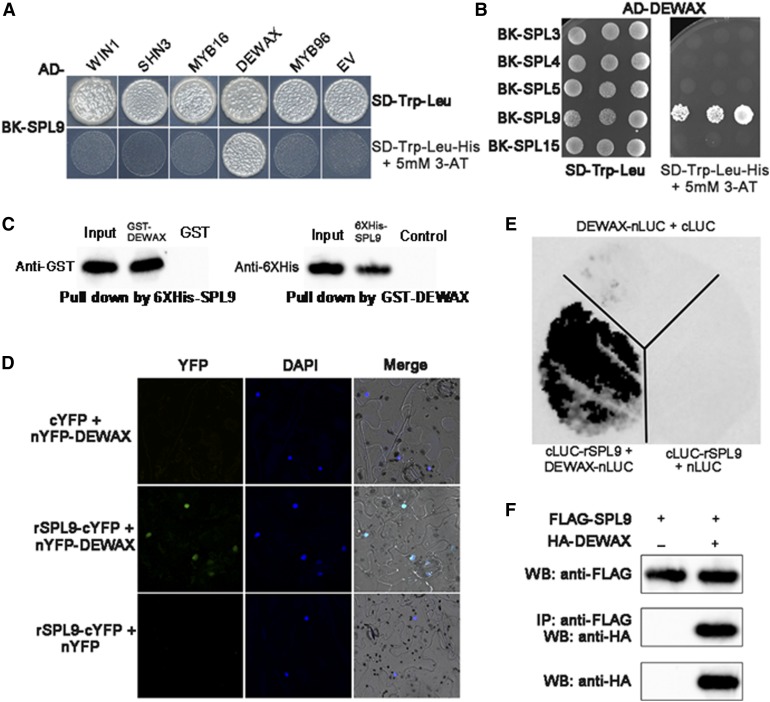
To investigate mechanisms by which the SPL9-dependent signaling cascade regulates wax synthesis, we performed a yeast two-hybrid experiment using SPL9 as the bait against a battery of genes, which have been reported to regulate wax synthesis in Arabidopsis. These genes included positive regulators WIN1, SHN3, MYB16, and MYB96 and a negative regulator DEWAX. SPL9 interacted specifically with DEWAX (Figure 6A). We next examined if DEWAX interact with other SPLs using yeast two-hybrid assays, and the results showed that DEWAX did not interact with SPL3, SPL4, SPL5, or SPL15 (Figure 6B), suggesting that DEWAX interacted specifically with SPL9. Furthermore, we mapped the domain of SPL9 required for the interaction with DEWAX. Y2H assays showed that the SPL9 N-terminal (1–160 amino acids), containing the DNA binding domain, bound to the DEWAX N-terminal (1–100 amino acids) but not to the DEWAX C-terminal (101–201 amino acids) containing the DNA binding domain (Supplemental Figure 7).
Figure 6.
SPL9 Specifically Interacts with DEWAX In Vitro and In Vivo.
(A) Yeast two-hybrid assay shows the specific interaction between SPL9 and DEWAX. SPL9 was in-frame fused to the GAL4 binding domain (BD) in pGBKT7, whereas WIN1, SHN3, MYB16, MYB96, and DEWAX were fused to the GAL4 activation domain (AD) in pGADT7. Transformed yeast cells were grown on SD-Trp-Leu (top). The direct protein interactions were assayed on a SD-Trp-Leu-His plate, supplemented with 5 mM 3-amino-1,2,4,-triazole (3-AT; bottom). The empty pGADT7 were used as negative control.
(B) Interaction analysis of DEWAX with other SPL family proteins (SPL3, SPL4, SPL5, and SPL15) using yeast two-hybrid assays.
(C) An in vitro pull-down assay shows the interaction between SPL9 and DEWAX. 6xHis-SPL9 protein was incubated with immobilized GST or GST-DEWAX protein, and immunoprecipitated fractions were detected by anti-6xHis antibody. Alternatively, GST-DEWAX protein was incubated with immobilized 6xHis-SPL9 or control protein, and immunoprecipitated fractions were detected by anti-GST antibody.
(D) BiFC analysis of the interaction between SPL9 and DEWAX. Blue and green fluorescence represent 4',6-diamidino-2-phenylindole (DAPI) and GFP signals, respectively. The empty nYFP or cYFP vector was used as negative control.
(E) BiLC demonstrates SPL9 interacts with DEWAX in vivo. The leaves of N. benthamiana were infiltrated with agrobacteria as indicated. Constructs were combined at a 1:1 ratio. The empty nLUC (N-terminal LUC) or cLUC (C-terminal LUC) vector was used as negative control.
(F) Co-IP assays showing that SPL9 physically associates with DEWAX in vivo. FLAG-SPL9 and HA-DEWAX proteins were transiently expressed in N. benthamiana. Protein was immunoprecipitated with anti-FLAG antibody, and the IP fraction was analyzed in a protein blot with anti-HA antibody. The input fraction was analyzed by immunoblotting using either anti-FLAG or anti-HA antibody.
Furthermore, the SPL9-DEWAX physical interaction was confirmed by in vitro pull-down assay using recombinant proteins purified from E. coli. GST-DEWAX was precipitated with 6xHiS-SPL9, but not with the control alone using Ni-NTA agarose. Similarly, 6xHis-SPL9 was precipitated with GST-DEWAX but not GST alone using anti-GST-beads (Figure 6C). To confirm the SPL9-DEWAX interaction in vivo, we performed bimolecular fluorescence complementation (BiFC) assays with N. benthamiana epidermal leaf cells transiently coexpressing nYFP-DEWAX and rSPL9-cYFP. Strong yellow fluorescent protein (YFP) fluorescence was observed in the nucleus of cells coexpressing nYFP-DEWAX and rSPL9-cYFP, but not in those coexpressing nYFP and rSPL9-cYFP or nYFP-DEWAX and cYFP (Figure 6D), suggesting a specific interaction of SPL9 with DEWAX. Furthermore, we performed a firefly LUC complementation (BiLC) assay. We fused rSPL9 to the C-terminal half of LUC (cLUC-rSPL9) and DEWAX to the N-terminal half of LUC (DEWAX-nLUC) and transiently introduced both fusion proteins into tobacco epidermal leaf cells. In the leaves infiltrated with combinations of Pro35S:nLUC and Pro35S:cLUC-rSPL9, or Pro35S:cLUC and Pro35S:DEWAX-nLUC, LUC activity was barely detectable. By contrast, leaves that coexpressed Pro35S:cLUC- rSPL9 and Pro35S:DEWX-nLUC produced a strong LUC signal (Figure 6E). In addition, BiLC experimental results provided evidence that the SPL9 N-terminal DNA binding domain and DEWAX N-terminal domain interacted in vivo (Supplemental Figure 8). Finally, we performed coimmunoprecipitation (Co-IP) experiments to confirm their interaction in vivo. To do this, we coexpressed FLAG-rSPL9 and HA-DEWAX fusion proteins in tobacco leaves. HA-DEWAX coimmunoprecipitated with FLAG-rSPL9 (Figure 6F). Taken together, our data suggest that SPL9 directly binds to DEWAX, both in vitro and in vivo.
DEWAX Inhibits SPL9 Activity in Regulating CER1 Expression
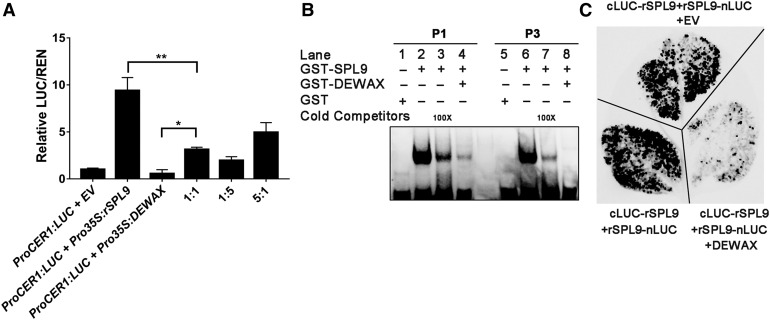
Although we demonstrated that SPL9 activated wax synthesis, and that SPL9 interacted with the negative regulator DEWAX, it was still uncertain how this interaction was involved in wax synthesis. In transient gene expression assays using the ProCER1:LUC construct as a reporter of Pro35:rSPL9 transcriptional activity, rSPL9 expression alone resulted in αν approximately eightfold activation of the reporter, as anticipated (Figure 7A). Consistent with a previous report (Go et al., 2014), coexpression of Pro35S:DEWAX alone inhibited CER1 expression. Interestingly, coexpression of the SPL9 and the DEWAX proteins led to significantly lower LUC reporter activity, than with SPL9 alone, in a dosage-dependent manner (Figure 7A), suggesting that DEWAX interacts with SPL9 and hampers its transcription activating activity. Additionally, although CER4 is an indirect target of SPL9, its regulation by the SPL9 and DEWAX interaction appears similar as with CER1 (Supplemental Figure 9).
Figure 7.
DEWAX Represses DNA Binding Ability of SPL9.
(A) DEWAX represses the SPL9 transcriptional activity on the CER1 promoter in the transient expression system. Pro35S:rSPL9 or Pro35S:DEWAX alone, or Pro35S:rSPL9/Pro35S:DEWAX in different ratios with the reporter ProCER1:LUC were coinfiltrated and simultaneously expressed in N. benthamiana leaves. The relative LUC activity in the coexpressed samples was calculated by normalizing the LUC values against REN. Averages from three biological replicates and their SD are shown. The values were statistically treated using Student’s t test (*P < 0.05, **P < 0.01).
(B) EMSAs show that SPL9-DEWAX protein interaction attenuates the DNA binding ability of SPL9 to the GTAC motifs in the promoter region of CER1. GST protein was used as negative control. Unlabeled cold competitor probes were used as positive control for attenuated DNA binding.
(C) DEWAX hampers SPL9 homodimer formation in a BiLC experiment. N. benthamiana leaves were infiltrated with agrobacteria as indicated.
Because DEWAX interacts with the SPL9 DNA binding domain (Supplemental Figures 7 and 8), it is possible that this interaction may interfere with SPL9 DNA binding activity. Indeed, in EMSAs, the signal generated by binding of SPL9 to the P1 and P3 probes of the CER1 promoter was dramatically decreased with increasing DEWAX level, even to a greater extent than observed with exogenous 100× cold competitor probes (Figure 7B), providing evidence that the interaction of the SPL9 DNA binding domain with the CER1 promoter is attenuated by DEWAX. We next examined how DEWAX regulates SPL9 activity. Intriguingly, BiLC assays showed that expression of DEWAX substantially repressed the formation of SPL9 homodimers (Figure 7C), demonstrating that DEWAX possibly inhibits SPL9 protein activity by reducing its DNA binding ability via the formation of nonfunctional heterodimers.
SPL9 Acts Downstream of DEWAX in Regulating Epidermal Wax Synthesis
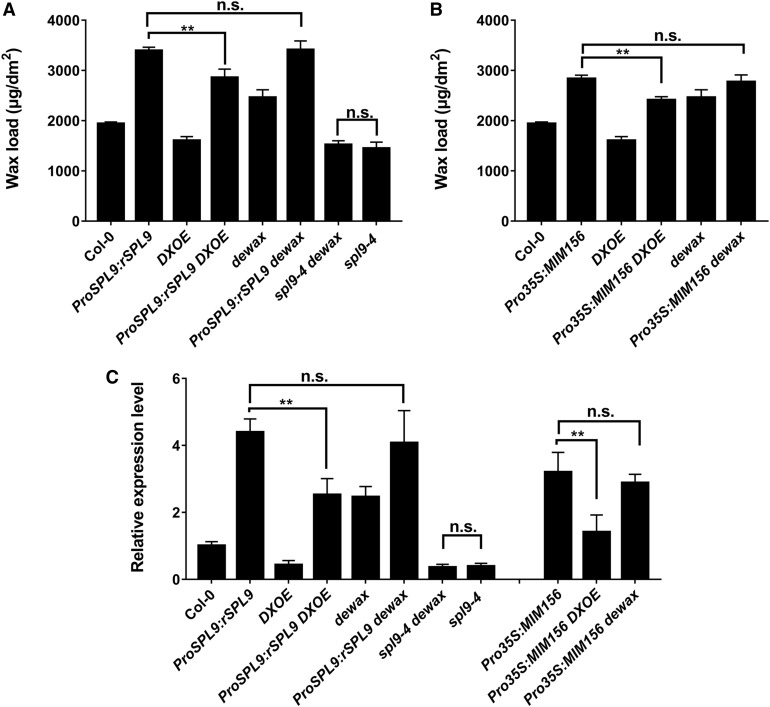
To further explore the antagonistic genetic relationship between SPL9 and DEWAX, we created a series of unique genetic lines by crossing spl9-4, ProSPL9:rSPL9 and Pro35S:MIM156 with the DEWAX loss of function mutant dewax or overexpression line DXOE and analyzed their wax compositions. As expected, when DEWAX was overexpressed in the ProSPL9:rSPL9 background, the stem wax content of the DXOE ProSPL9:rSPL9 double mutant was greatly decreased compared with ProSPL9:rSPL9 (Figure 8A). Similar results were also observed in the DXOE Pro35S:MIM156 line when compared with the Pro35S:MIM156 line (Figure 8B). In addition, although the dewax knockout mutant showed enhanced wax synthesis compared with the wild type, when dewax was placed in the ProSPL9:rSPL9 or Pro35S:MIM156 backgrounds, the wax contents of ProSPL9:rSPL9 dewax or Pro35S:MIM156 dewax lines exhibited no further increases in comparison with ProSPL9:rSPL9 or Pro35S:MIM156 (Figures 8A and 8B). More importantly, the double mutant spl9-4 dewax displayed a similarly reduced wax content phenotype as that of spl9-4, but different from the higher wax content phenotype of dewax (Figure 8A). These results lend further evidence that DEWAX suppresses SPL9 protein function. Consistent with the phenotype observed, CER1 expression ProSPL9:rSPL9Pro35S:MIM156 was reduced in the DXOE Pro35S:MIM156 and DXOE ProSPL9:rSPL9 lines compared with their parent lines Pro35S:MIM156 or ProSPL9:rSPL9, whereas CER1 expression was similarly reduced in spl9-4 dewax and spl9-4 compared with wild-type plants (Figure 8C). These results, combined with the above molecular data, support the idea that SPL9 acts downstream of DEWAX in regulating wax synthesis.
Figure 8.
SPL9 Acts Downstream of DEWAX in Wax Synthesis.
(A) Genetic relationship analysis of SPL9 and DEWAX in wax synthesis with double mutant phenotype analysis. Cuticular wax amounts of inflorescence stems from 6-week-old double mutants between Pro35S:rSPL9, spl9-4, dewax, and DXOE grown in long-day conditions were examined by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate ±sd (**P < 0.01, Student’s t test) from four replicate experiments; n.s. represents no significant difference.
(B) Genetic relationship analysis of miR156 and DEWAX in wax synthesis with double mutant phenotype analysis. Cuticular wax amounts of inflorescence stems from 6-week-old double mutants between Pro35S:MIM156 and dewax or DXOE grown in long-day conditions were examined by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate ±sd (**P < 0.01, Student’s t test) from four replicate experiments; n.s. represents no significant difference.
(C) RT-qPCR analysis of CER1 expression in the indicated mutants. Expression was normalized relative to that of PP2A. Expression level in the wild type (WT) was set as 1. Each column represents the mean of three independent assays, and the error bars indicate ±sd (**P < 0.01, Student’s t test); n.s. represents no significant difference.
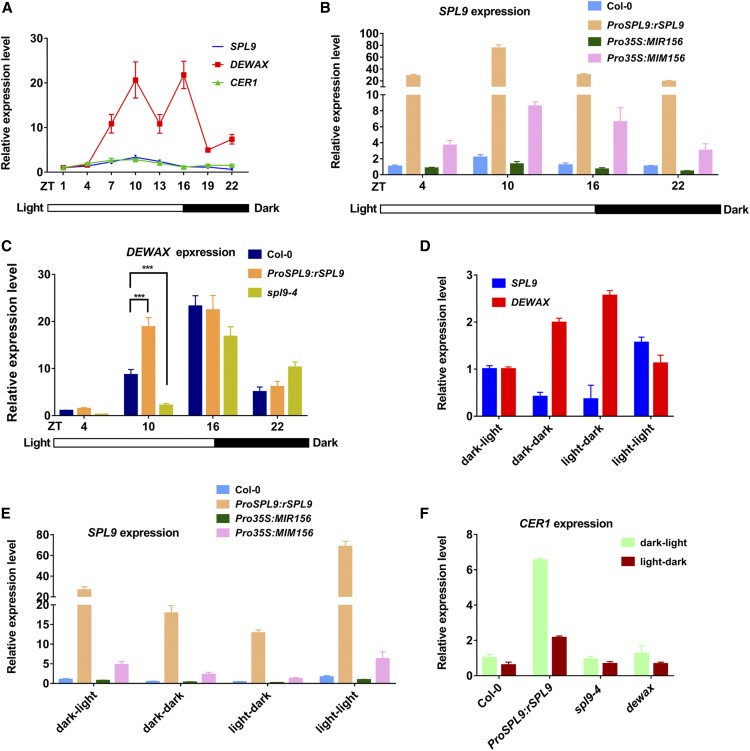
SPL9 and DEWAX Expression Are Regulated by Diurnal Cycle And Light/Dark Changes
A previous study has found that CER1 expression occurs in diurnal patterns and that DEWAX may inhibit its expression in the dark (Go et al., 2014). However, how CER1 expression is activated in light is unknown. With our findings that SPL9 interacts with DEWAX, and that SPL9 activates CER1 gene expression, we hypothesized that SPL9-DEWAX may form a regulatory circuit for light/dark-mediated wax synthesis. We analyzed 7 dold wild-type seedlings grown on Murashige and Skoog agar plates in long day (LD) conditions, collected the samples every 3 h in one diurnal cycle, and then examined the gene expression of CER1, SPL9, and DEWAX. In accordance with previous findings, CER1 expression was higher in the day and lower in the night. Interestingly, SPL9 also showed a diurnal expression pattern, with highest expression in the light at Zeitgeber time (ZT)10, similar to CER1, suggesting that SPL9 may be involved in the regulation of diurnal CER1 expression (Figure 9A). Furthermore, to explore whether SPL9 diurnal expression was regulated by miR156 at the posttranscriptional level, we examined the expression of two major MIR156 primary transcripts of MIR156A and MIR156C (pri-MIR156A and pri-MIR156C) and mature miR156 at four time points (ZT4, 10, 16, and 22) by RT-qPCR. The results suggested that the expression of pri-MIR156A and pri-MIR156C were decreased in the day and increased during the night, opposite to CER1 and SPL9 expression. Furthermore, the expression of mature miR156 was similar to that of its precursors, consistent with its speculated role in the negative regulation of SPL9 (Supplemental Figure 10). Surprisingly, SPL9 was still expressed diurnally in the miR156-resistant transgenic line ProSPL9:rSPL9 (Figure 9B). Additionally, in Pro35S:MIR156 and Pro35S:MIM156, the diurnal expression pattern of SPL9 was similar to the ztwild type, although the oscillation amplitude differed (Figure 9B). These results indicated that diurnal expression of SPL9 is partially independent of miR156 at the transcriptional level.
Figure 9.
Diurnal Cycles or Light/Dark Changes Affect SPL9 and DEWAX Gene Expression at Multiple Levels.
(A) Diurnal expression of CER1, SPL9, and DEWAX. Gene expression was analyzed in 7-d-old Arabidopsis wild-type seedlings, which were grown under long-day conditions (16 h of light/ 8 h of dark). The light is on at Zeitgeber time 0 (ZT0) and off at ZT16. Total RNA was extracted from at least 20 seedlings (every 3 h from ZT1) and subjected to RT-qPCR analysis. Errors bars indicate ±sd with at least three replicates.
(B) Diurnal expression of SPL9 is at least partially independent of miR156. Seven-day-old seedlings at four time points (ZT4, ZT10, ZT16, and ZT22) in four different genetic materials (Col-0, ProSPL9:rSPL9, Pro35S:MIR156, and Pro35S:MIM156) were collected, and SPL9 expression was analyzed by RT-qPCR. Error bars indicate ±sd with at least three replicates.
(C) SPL9 positively regulates DEWAX expression in the day. Seven-day-old seedlings at four time points (ZT4, ZT10, ZT16, and ZT22) in Col-0, ProSPL9:rSPL9, and spl9-4 were collected, and DEWAX expression was analyzed by RT-qPCR. Errors bars indicate ±sd with at least three replicates. ***P < 0.001, Student’s t test.
(D) Light/dark changes regulate SPL9 and DEWAX expression. For light-dark treatment, at ZT4, 7-d-old seedlings were placed at dark conditions (light-dark) and the controls were maintained at light (light-light). At ZT10, both plants were collected. For dark-light treatment, at ZT16, 7-d-old seedlings were placed at light conditions (dark-light) and the controls were maintained at dark (dark-dark). At ZT22, both plants were collected. Total RNAs were extracted from these seedlings collected from the above treatments, and SPL9 and DEWAX expression were analyzed by RT-qPCR. Errors bars indicate ±sd with at least three replicates.
(E) Light/dark changes altered SPL9 expression at least partially independently of miR156. The treatment processes were as indicated in (D). Seven-day-old seedlings from different genetic materials (Col-0, ProSPL9:rSPL9, Pro35S:MIR156, and Pro35S:MIM156) were collected, and SPL9 expression was analyzed by RT-qPCR. Error bars indicate ±sd with at least three replicates.
(F) SPL9 only regulates the expression amplitude of CER1 under light/dark change conditions. CER1 expression in Col-0, ProSPL9:rSPL9, spl9-4, and dewax is shown. Seven-day-old seedlings during light/dark change conditions were analyzed by RT-qPCR. Error bars indicate ±sd with at least three replicates.
In contrast with CER1 and SPL9, DEWAX expression oscillated twice a day, once in the day (ZT10) and again in the night (ZT16), consistent with a previous report (Go et al., 2014). Interestingly, SPL9 also showed highest expression at ZT10 (Figure 9A). Therefore, we assessed the expression of DEWAX in ProSPL9:rSPL9 and spl9-4. DEWAX expression was significantly elevated in ProSPL9:rSPL9 and decreased in spl9-4, especially at ZT10 (Figure 9C), suggesting that SPL9 was involved in regulating DEWAX expression in the day. By searching the DEWAX promoter region, we found two potential SPL9 binding sites (GTAC at −1264 to −1261 and −1366 to −1363 bp relative to the ATG start codon, respectively). ChIP-qPCR results showed that the P1 and P2 regions containing GTAC motifs in the DEWAX promoter were enriched in ProSPL9:GFP-rSPL9 as compared with the wild type (Supplemental Figure 11). Consistently, Y1H experiments demonstrated that SPL9 binds the 1.5-kb upstream promoter DNA with these two GTAC motifs, but does not bind the 1.2-kb upstream promoter DNA without these two motifs, supporting the idea that the two sites were required for SPL9 binding. To determine whether SPL9 modulates DEWAX expression, we performed transient expression assays with the DEWAX promoter LUC reporter in N. benthamiana. Coexpression of SPL9 strongly provoked the activity of the LUC reporter gene driven by the 1.5-kb promoter of DEWAX. Interestingly, the LUC reporter gene activity driven by the 1.2-kb promoter of DEWAX was expressed at a lower level, which is not activated by SPL9 (Supplemental Figure 11). In sum, these results indicate that SPL9 directly binds DEWAX promoter DNA and activates its gene expression in vivo. SPL9 and DEWAX may operate in a negative feedback loop important in moderating the light response in wax synthesis.
To test whether the diurnal expression patterns of SPL9 or DEWAX are regulated by the circadian clock, we performed artificially controlled light/dark change experiments. Wild-type plants were grown in long-day conditions for 7 d, and at ZT4 (light) the seedlings were kept in light or transferred to dark condition for 6 h. Both of the seedlings at ZT10 were collected and referred to as light–light and light–dark, respectively. Similarly, seedlings at ZT16 (dark) were held in dark or transferred to light conditions for 6 h. Both seedlings at ZT22 were collected and referred to as dark–dark and dark–light, respectively. As shown in Figure 9D, after the transfer, light treatment increased SPL9 expression and decreased DEWAX expression. By contrast, dark treatment increased DEWAX expression and decreased SPL9 expression. Importantly, the diurnal expression pattern of SPL9 (higher at ZT10 for light–light compared with dark–dark at ZT22) or DEWAX (higher at ZT22 for dark–dark compared with light–light at ZT10) was totally conversed, as SPL9 showed higher expression at ZT22 (dark–light) than at ZT10 (light–dark) and vice versa for DEWAX. The results indicated that the oscillating expression pattern of SPL9 or DEWAX was mainly controlled by light and dark exposure, but not the circadian clock. Furthermore, expression of pri-MIR156A and pri-MIR156C and mature miR156 during light/dark changes was examined by RT-qPCR. The results suggested that the expressions of pri-MIR156A, pri-MIR156C, and mature miR156 were inhibited by light and improved by dark treatment (Supplemental Figure 12). Similar to the above diurnal cycle results, when we examined the expression of SPL9 in ProSPL9:rSPL9, Pro35S:MIR156, and Pro35S:MIM156, the expression alterations still appeared (Figure 9E). These results implied that miR156-independent factor(s) may be involved in regulating SPL9 expression levels during changes from light to dark, and vice versa.
Finally, to test whether the light and dark changes of SPL9 or DEWAX expression influenced CER1 expression, we examined CER1 expression in ProSPL9:rSPL9, spl9-4, and dewax. CER1 expression still exhibited a strong response to light and dark conditions in these lines (Figure 9F), suggesting that SPL9 and DEWAX play a primary role in regulating the amplitude of light responsiveness of wax synthesis.
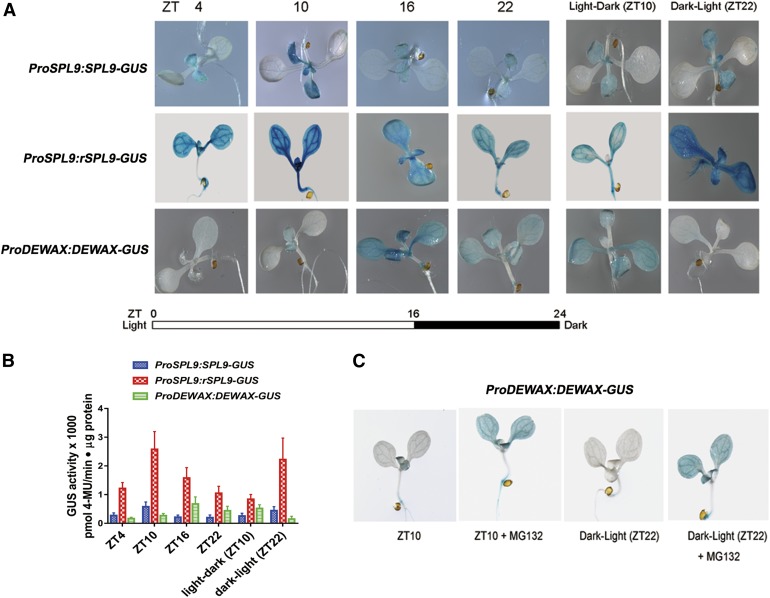
Both SPL9 and DEWAX Protein Expression Is Regulated by the Diurnal Cycle
The temporal expression patterns of SPL transcripts do not always reflect protein levels and function, as miR156 may regulate SPL genes through translational repression. To examine the diurnal expression of SPL9 protein, we took advantage of reporter lines ProSPL9:SPL9-β-glucuronidase (GUS) and ProSPL9:rSPL9-GUS containing miR156-sensitive or miR156-resistant genomic constructs of the gene fused to GUS, respectively. For comparison, we also constructed a transgenic line ProDEWAX:DEWAX-GUS to explore impacts of the in vivo protein level of DEWAX. Consistent with the gene expression patterns, GUS staining was higher in the light and lower in the dark in ProSPL9:SPL9-GUS. For ProDEWAX:DEWAX-GUS, the GUS staining was lower in the light and higher in the dark. ProSPL9:rSPL9-GUS still showed higher GUS staining in the light and lower in the dark (Figure 10A), consistent with miR156-independent gene expression alterations (Figure 9B). For the dark-to-light treatment of seedlings at ZT22, we observed higher expression of SPL9 and lower expression of DEWAX protein, in comparison with seedlings at Z22 in the normal growth conditions (Figure 10A). For the light-to-dark treatment seedlings at ZT10, we observed higher expression of the DEWAX and lower expression of the SPL9 protein, compared with the seedlings at Z10 in normal growth conditions. The results further demonstrated that light and dark conditions, but not circadian signals, regulate SPL9 and DEWAX protein levels. Moreover, to quantify GUS fusion protein levels, 4-methylumbelliferyl β-D-glucuronide (MUG) assays were performed. The quantification results were consistent with the histochemical staining experiments (Figure 10B). Finally, it should be noted that the expression levels of DEWAX in ZT10 and ZT16 were similar, but their protein levels showed significant differences (Figure 10A). This phenomenon prompted us to think that there may be a posttranslational mechanism that regulates DEWAX protein stability. Indeed, when the ProDEWAX:DEWAX-GUS seedlings were treated with the 26S proteasome inhibitor MG132, the GUS signals in ZT10 or dark-to-light samples were greatly increased (Figure 10C). The results suggested that light may regulate DEWAX protein level through protein degradation pathways.
Figure 10.
Diurnal Cycles or Light/Dark Changes Affect SPL9 and DEWAX Protein Accumulation.
(A) Diurnal expression of SPL9 and DEWAX protein levels revealed by GUS reporter lines. Histochemical detection of GUS activity driven by the native gene ProSPL9:SPL9-GUS or mutated miR156-resistant gene ProSPL9:rSPL9-GUS and the native protein ProDEWAX:DEWAX-GUS is shown. Seven-day-old seedlings were collected every 6 h from ZT4 in long-day conditions. For light-dark treatment, at ZT4, 7-d-old seedlings were placed at dark conditions and the controls were maintained at light. At ZT10, both plants were collected and GUS activity was stained. For dark-light treatment, at ZT16, 7-d-old seedlings were placed at light conditions and the controls were maintained at dark. At ZT22, both plants were collected and GUS activity was stained. The experiments were repeated three times, and similar results were obtained.
(B) MUG assays for the quantitative analysis of GUS activity in extracts of the samples illustrated in (A). Errors bars indicate ±sd with at least three replicates.
(C) DEWAX protein stability was regulated at the posttranslational level. For MG132 treatment, at ZT4, 7-d-old seedlings were placed on an MS plate with 50 μM MG132 and the controls were maintained on an MS plate. At ZT10, both plants were collected and GUS activity was measured. For dark-light treatment, at ZT16, 7-d-old seedlings were placed on an MS plate with 50 μM MG132 and the controls were maintained on an MS plate. Both plates were placed in light conditions. At ZT22, both seedlings were collected and GUS activity was measured. The experiments were repeated three times, and similar results were obtained.
SPL9-DEWAX Regulates Diurnal Wax Synthesis
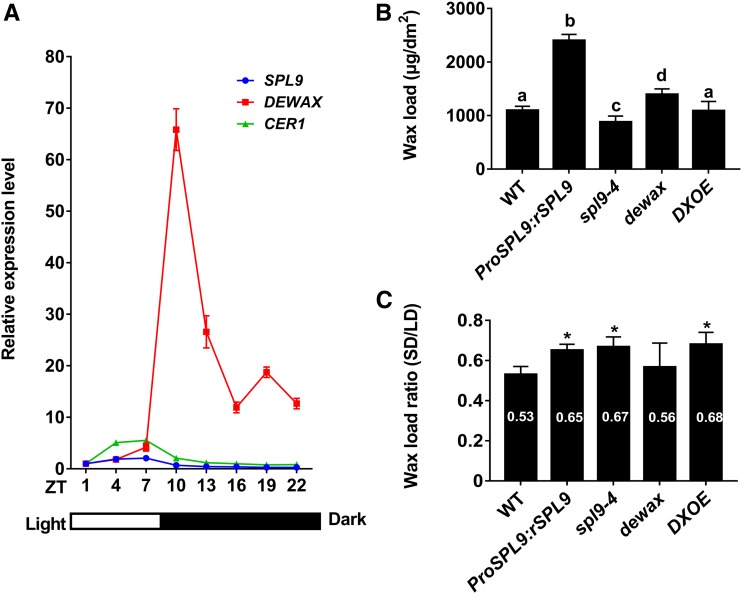
Although we have demonstrated that SPL9-DEWAX modulated diurnal CER1 expression, it is unknown whether this regulation affects diurnal wax synthesis. To address this question, it would theoretically be necessary to design an experiment to track surface wax accumulation in a diurnal manner (within 24 h). However, this might not be possible or practical. Some experiments by Jenks et al. (1994) with Sorghum indicate that measurable wax accumulation is delayed at least 24 h after induction by light. In Arabidopsis, it would likely be even more difficult since there is much less total wax than in Sorghum. Thus, we investigated the long-term effects of the length of day or night, or photoperiod, on wax accumulation. First, we examined the expression patterns of CER1, SPL9, and DEWAX under short day (SD; 8/16) conditions. Seven-day-old wild-type seedlings grown on MS-agar plates in SD conditions were collected every 3 h in one diurnal cycle, and the transcript levels were investigated by RT-qPCR. CER1 and SPL9 displayed a similar expression pattern as shown in LD conditions, i.e., higher in the day and lower in the night in SD conditions (Figure 11A). However, the expression levels of CER1 and SPL9 peaked between ZT4 and ZT7 under SD conditions, which advanced as compared with LD (the highest expression at ZT10). As for DEWAX, expression was significantly increased (over 64-fold) at the beginning of the night in SD and still maintained a higher level until light (from ZT10 to ZT22; Figure 11A). The altered expression pattern of CER1, SPL9, and DEWAX motivated us to investigate whether SD conditions change the wax accumulation. Indeed, the short daylength greatly reduced the total wax loads on the stems of wild-type plants (Figure 11B). In LD conditions, the total wax on the stems of wild type was ∼2067.97 ± 105.39 μg/dm2, whereas in SD conditions, it was ∼47% lower (∼1096.29 ± 28.08 μg/dm2). We also assessed the stem wax contents of mutant plants in SD conditions. Like in LD, ProSPL9:rSPL9 and dewax in SD conditions contain increased wax loads, whereas spl9-4 and DXOE plants have reduced wax loads compared with wild type (Figure 11B). The daylength also apparently affects the wax accumulation in these mutants since the wax loads in these mutants in SD conditions are far lower than in LD conditions. However, we noticed that the reduction rate in ProSPL9:rSPL9, spl9-4, and DXOE in SD conditions is markedly lower than in wild-type plants (Figure 11C). These results reinforce the idea that SPL9-DEWAX participate in the diurnal or daylength regulated wax accumulation.
Figure 11.
SPL9-DEWAX Is Involved in the Diurnal Regulation of Wax Synthesis.
(A) Diurnal expression of CER1, SPL9, and DEWAX genes in SD (8 h of light/16 h of dark) conditions. Gene expression was analyzed in 9-d-old Arabidopsis wild-type seedlings, which were grown under SD conditions. The light is on at Zeitgeber time 0 (ZT0) and off at ZT8. Total RNA was extracted from each sample (every 3 h from ZT1) and subjected to RT-qPCR analysis. Error bars indicate ±sd with at least three replicates, with each replicate containing at least 20 seedlings.
(B) Cuticular wax amounts of stems from ∼3-month-old wild-type, ProSPL9:rSPL9, spl9-4, dewax, and DXOE plants grown in SD conditions. Cuticular waxes were extracted with hexane and analyzed by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate ±sd from three replicate experiments with samples independently harvested on different days. Letters denote statistically significant differences between the indicated samples, as determined by ANOVA with post hoc Tukey Honestly Significant Difference.
(C) Total wax loads ratio between SD and LD conditions. Values from plants grown in SD conditions were divided by those from plants grown in LD conditions, and the data were analyzed using Student’s t test (*P < 0.05). Error bars indicate ±sd from biological triplicate experiments.
Collectively, our results showed that, on one hand, SPL9 and DEWAX protein levels exhibit strong diurnal shifts through transcriptional, posttranscriptional, and/or posttranslational regulation, and on the other hand, SPL9 protein activity was modulated by DEWAX interaction. The two mechanisms may operate together to moderate CER1 rhythmic gene expression and wax accumulation through the light/dark cycle.
DISCUSSION
Cuticular waxes form the plant’s outermost barrier between the plant and its environment, and previous studies show that the plant’s cuticle structure and chemical components can change substantially in response to different environmental stimuli (such as temperature, water, pathogen, and phytophagous insect), resulting in a modified cuticle better able to protect the plant (Shepherd and Wynne Griffiths, 2006; Bourdenx et al., 2011; Bernard and Joubès 2013; Lee and Suh, 2015a). And yet, the regulatory mechanisms that determine these precise cuticle responses remain mostly undefined. In this paper, we report a novel mechanism for wax synthesis regulation by the miR156-SPL9 module, expanding knowledge of the environmental regulation of the cuticle to the posttranscriptional level. These and downstream regulatory mechanisms were further described using molecular and genetic approaches, and these results generate key findings that shed light on the role of surface waxes and wax-related genes in the plant environmental response.
Roles of the miR156-SPLs Module in Wax Synthesis Regulation
sRNAs (21 to 24 nucleotide) are noncoding signaling molecules involved in multiple biological pathways (Samad et al., 2017). sRNAs can be classified into two categories including miRNAs and small interfering RNAs (siRNAs), based on their differences in the biogenesis and mode of action. miRNAs are processed from a single stranded noncoding RNA precursor that forms a hairpin structure. By contrast, tasiRNAs biogenesis requires an initial process of specific miRNA-mediated cleavage of their TAS RNA precursors through the role of RNA-DEPENDENT-RNA-POLYMERASE-6 (RDR6). The biogenesis of miRNA and tasiRNA also shares the same components, such as HUA ENHANCER1 and ARGONAUTE1 (Borges and Martienssen, 2015). Lam et al. (2015) demonstrated that tasiRNAs directly silence CER3 expression. In this study, we found that the miR156-SPL9 module directly regulates CER1 expression. Intriguingly, CER1, CER3/WAX2/YRE, and the cytochrome b5 isoforms constitute a multiprotein enzyme complex in the alkane-forming pathway (Chen et al., 2003; Rowland et al., 2007; Bourdenx et al., 2011; Bernard et al., 2012). Whether there is crosstalk between the two classes of small RNAs (miRNA and tasiRNA) in balancing CER1 and CER3 dosage for their complex formation in wax synthesis is now something to be considered further.
Previous studies have shown that CER1 is responsible for alkane biosynthesis and is closely associated with biotic and abiotic stress responses (Aarts et al., 1995; Bourdenx et al., 2011; Bernard et al., 2012). For example, CER1-overexpressing transgenic plants showed reduced cuticle permeability and drought tolerance. However, these plants also had increased susceptibility to microbial pathogens (Bernard et al., 2012). Our results demonstrate that the miR156-SPL9 module directly regulates CER1 expression and wax synthesis, primarily alkane synthesis. Microarray analysis or miRNA sequencing has provided evidence that miR156 expression is regulated by various abiotic stresses such as salt, drought, and cold (Sunkar and Zhu, 2004; Liu et al., 2008; Zhou et al., 2008; Lee et al., 2010) or biotic stresses (Joshi et al., 2016; Zhang et al., 2019). Yet, we do not know how miR156 perceives and responds to these environmental signals, and whether the miR156-SPL9-CER1 pathway defines a more commonly used or general stress response mechanism involved in plant tolerance to many other types of stress.
Transcriptional regulators of cuticle biosynthesis are classified mainly by several families of TFs including MYB and AP2/EREBP (Aharoni et al., 2004; Broun et al., 2004; Seo et al., 2011; Borisjuk et al., 2014). Our results here provide the first identification of a novel gene family, the SPLs in wax regulation. In plants, multiple SPLs are targeted by miR156s through cleavage and/or translational repression. In our results, the spl9-4 spl15-1 double mutant showed a lower wax load than the spl9 single mutant, and the Pro35S:MIR156 transgenic line exhibited the lowest wax amounts, which illustrated that members of the SPL family may act redundantly, or otherwise have overlapping functions, in regulating wax synthetic responses. Therefore, further experiments to analyze the regulatory effects of other genes in the SPL family are needed for a better understanding of their roles in wax biosynthesis.
Besides some redundancy as indicated here, genes in the SPL family also have distinct functions (Xu et al., 2016). SPL9 and SPL15 are closely related homologs, and yet they exhibit distinct mRNA expression patterns (Wang et al., 2009; Hyun et al., 2016). Indeed, Arabidopsis gene expression analysis (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) showed that the SPL9 transcript level is approximately three- to ninefold higher than SPL15 in epidermal stems, which is consistent with the more distinct wax defects exhibited by spl9-4 and spl15-1. These results suggest that SPL9 plays a larger role in epidermal wax synthesis than SPL15, perhaps determined by their different epidermal expression patterns. Furthermore, among the examined five SPL genes, only SPL9 could be shown to interact with DEWAX, which suggested that both gene expression and protein interaction difference led to SPL functional diversification from the other family members in wax synthesis regulation. Additionally, a recent paper demonstrated that there was a significant decrease in alkane levels in DEWAX2 (a DEWAX homolog) overexpression leaves when compared with the wild type (Kim et al., 2018). Whether DEWAX2 interacts with SPL9 remains to be determined.
Downstream Genes in Wax Synthesis Regulated by miR156-SPL9
In previous studies, transcription factors involved in wax regulation showed multiple targets (reviewed in Borisjuk et al., 2014; Lee and Suh, 2015a). However, there is no direct evidence or genetic analysis that reveals the causal relationship between these genes and those associated with wax synthesis. In this report, we examined a genetic line having one or more mutations, and transgenes, in various combinations to test the regulatory role of transcription factors on wax synthesis genes. The specific significance for a single wax component in cuticle function during environmental or developmental processes remains largely unknown. Because the cuticle wax synthesis pathways are tightly interconnected, it is challenging to examine the role of a single wax component using a loss of function approach (Goodwin et al., 2005). For example, in the cer1-2 mutant, besides the alkanes, the amounts of 1-alcohols (primarily regulated by CER4) were also decreased (Supplemental Table 1). Through genetic approaches, we were able to uncouple the CER1 and CER4 synthesis pathway through overexpression of SPL9. In the ProSPL9:rSPL9 cer1-2 line, the alkane contents were similar to cer1-2. However, the 1-alcohol contents were increased. Conversely, in the ProSPL9:rSPL9 cer4-4 line, the 1-alcohol contents were similar to cer4-4, whereas the alkane contents were increased. It is of great interest to note that the Pro35S:MIM156 cer4-4 and ProSPL9:rSPL9 cer4-4 lines show a glossy phenotype as in some wax deficient mutants, although their total wax loads are higher than in wild-type plants. These results suggest that the different crystalline organization but not the total content of the waxes determines the appearance of the wax phenotype. This type of observation has also been reported in several other studies (Rowland et al., 2006; Pascal et al., 2013, 2019; Haslam et al., 2015). In the ProSPL9:rSPL9 cer1-2 cer4-4 line, both the alkane and 1-alcohol contents were decreased, similar to cer1-2 cer4-4. These unique genetic materials shed light on gene control over wax metabolism, and ultimate composition.
At the molecular level, we demonstrated that SPL9 directly regulated CER1 expression, but also has a significant indirect effect on CER4 expression. Although the promoter region of CER4 contains a putative SPL9 binding cis-element, neither EMSA nor ChIP-qPCR experiments revealed SPL9 binding. Thus, the CER4 promoter is likely not the immediate target of SPL9. Furthermore, it should be noted that almost all wax components in the stems of ProSPL9:rSPL9 showed elevated levels in comparison with wild-type plants (Figure 2; Supplemental Table 1). Until now, at least tens of genes are reported to affect wax synthesis (Lee and Suh, 2015a). Although we did not examine all of the genes, it is possible that other wax synthesis-related genes are also differentially expressed in the mutant. In the future, systematic analysis of the transcriptome of the miR156-SPL9-related genetic materials may help identify the immediate transcription factor(s) governing the expression of CER4 and other wax biosynthetic genes as potential SPL9 targets.
Potential Upstream Factors Regulate miR156-SPL9
We observed that SPL9 protein levels oscillated in a diurnal manner, with higher levels in the light and lower levels in the dark, respectively, and could be tied directly to the rhythmic synthesis of wax. By contrast, DEWAX accumulated in the dark (and inhibited wax synthesis) and decreased after light illumination. The upstream regulatory factors of miR156-SPL9 or DEWAX are unknown. Light/dark cycles influence photosynthesis, and ultimately sugar metabolism in plants. Indeed, sugar levels could regulate miR156 abundance, as reported by several groups (Wahl et al., 2013; Yang et al., 2013; Yu et al., 2013). The expression of miR156 was repressed by exogenous sugar treatment, leading to an increase in SPL gene expression and an early juvenile-adult phase transition (Yang et al., 2013; Yu et al., 2013). In accordance with this result, mutants with reduced endogenous sugars abundance showed increased miR156 expression and delayed phase transition (Yang et al., 2013; Yu et al., 2013). Another set of experiments demonstrated the role of trehalose 6-phosphate (T6P) in repressing miR156 expression during flowering (Wahl et al., 2013). T6P is considered to be a signaling molecule because it is present at low concentrations in plant cells. Therefore, further research will be necessary to investigate whether sugar or T6P regulates wax synthesis upstream of miR156.
As sessile organisms that rely on sunlight as the main source of energy, plants have developed sophisticated systems to sense and respond to light cues. Interestingly, light has been reported to control miRNA accumulation and their biological function at multiple layers, such as miRNA gene transcription, miRNA biogenesis, and RNA-induced silencing complex activity (Cho et al., 2014; Tsai et al., 2014; Sanchez-Retuerta et al., 2018). Recently, Xie et al. (2017) demonstrated that several PHYTOCHROME INTERACTING FACTORs (PIFs) directly bound to the promoters of five MIR156 genes and repress their transcription. Their results established a direct link between the light–phytochrome–PIF pathways and miR156-SPL modules. Furthermore, it was shown that PIF4 integrates red light signaling and miRNA function through interacting with a miRNA biogenesis component DICER-LIKE1 and simultaneously regulates the expression of many miRNA genes during dark-to-red-light or red-light-to-dark transitions (Sun et al., 2018). Taken together, these data could provide a potential mechanistic explanation, at the molecular level, on how the diurnal light cycles might be involved in regulating the miR156-SPL module via genes involved in light signal transduction such as PIFs. The roles of such genes in wax synthesis await further investigation. It should also be noted that, in our results, we showed that even in ProSPL9:rSPL9 plants, the diurnal expression of SPL9 still exists (Figure 9), which may be independent of miR156. Thus, there may be an unknown direct regulator of SPL9 during the light response.
Protein Interaction Regulation of SPL9 Activity
SPL9 has been known to be integrated into diverse developmental and metabolic pathways through either transcript cleavage or translational repression by miR156 at the posttranscriptional level. However, whether its protein activity is regulated by other factors remains to be fully elucidated. As shown by our EMSA results, SPL9 specifically bound to the GTAC motifs in the CER1 promoter. Furthermore, the binding of SPL9 to the GTAC motif was repressed when the DEWAX protein was present, demonstrating that the DNA binding ability of SPL9 protein could be regulated by DEWAX. Interestingly, previous studies found that although DEWAX acts as a repressor of wax biosynthetic genes, it does not contain the ERF-associated repression motif. Our results provide a molecular explanation of DEWAX-mediated transcriptional repression. This hypothesis is also supported by the transient transcription assays in tobacco, which showed the DEWAX sequestering activity of the SPL9 protein. Homo- or heterodimerization of SPL9 and DEWAX might regulate the target genes in response to the development and ever-changing environment. Similarly, protein activity regulation through heterodimerization has also been found in other light signal transduction pathways (Hornitschek et al., 2009; Hao et al., 2012). For example, atypical basic helix-loop-helix (bHLH) transcription factors, LONG HYPOCOTYL IN FAR-RED1 and PHY RAPIDLY REGULATED1, could directly interact with PIF4/PIF5, and pair with them to form a non-DNA binding heterodimer. This regulatory mechanism in plants may help to prevent an exaggerated response to shade, affording plants the flexibly to adapt to changeable light conditions. Alternatively, it should be noted that DEWAX and DEWAX2 transcription factors are able to bind directly to the canonical GCC or GCC-like motifs of the CER1 promoter (Go et al., 2014; Kim et al., 2018). In our EMSA experiment, the DNA probes used contained no canonical GCC or GCC-like motifs, which preclude the possibility that DEWAX competes with SPL9 to bind the CER1 promoter in vitro. However, we cannot exclude the possibility that the DEWAX-DNA interaction could compete with SPL9 and prevent its binding to the CER1 promoter in vivo.
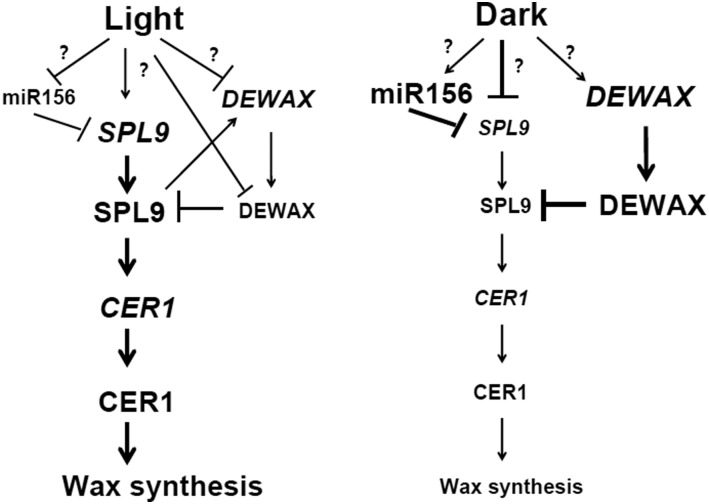
Working Model for SPL9-DEWAX Interaction
We report the regulation of SPL9 activity by the negative regulator DEWAX, and link diurnal cycles in plants to wax synthesis. DEWAX interferes with the formation of SPL9 homodimers by forming nonfunctional heterodimers with reduced DNA binding ability. If all these results together are considered, it is possible to propose a hypothetical model that explains the roles of SPL9 and DEWAX in mediating the diurnal regulation of wax synthesis (Figure 12). Under light conditions, SPL9 binds to GTAC motifs in the promoter of CER1 to regulate alkane synthesis, but also indirectly increases CER4 gene expression for 1-alcohol production through hitherto unknown mechanisms. Under dark conditions, DEWAX protein accumulates and competes with SPL9 to bind to the GTAC motifs present in the promoters of CER1, resulting in the downregulation of CER1. Our findings illustrate an elegant SPL9-DEWAX module that explains the antagonistic interactions between light and dark signaling pathways, ostensibly as a means to optimize wax synthesis in development and/or the environmental response in Arabidopsis.
Figure 12.
Proposed Working Model of the SPL9-DEWAX Signaling Module.
SPL9 and DEWAX gene and protein expression levels oscillated in a diurnal cycle, through multiple layers of transcriptional, posttranscriptional, and/or posttranslational regulations. During the day or in the light conditions, SPL9 expression was transcriptionally activated in a miR156-resistant manner, and miR156 and DEWAX expressions were inhibited. SPL9 also upregulated DEWAX expression in the day, forming a negative feedback loop that may be important in moderating the light response in wax synthesis. Furthermore, DEWAX protein was degraded through the 26S proteasome-dependent pathway in the light. During the night or in the dark conditions, miR56 and DEWAX expressions were induced and SPL9 expression was repressed. Furthermore, DEWAX directly interacts with SPL9 to form a heterodimer and inhibit its DNA binding ability. The diurnal gene or protein expression of SPL9 and DEWAX, and SPL9 protein activity regulation by DEWAX may operate together to modulate CER1 rhythmic gene expression and wax synthesis through the light/dark cycle.
In Arabidopsis, the total wax amount in the stems is over 10 times higher than that in the leaves (Suh et al. 2005), but the underlying mechanism of such organ-specific wax deposition remains unknown. Go et al. (2014) suggested that DEWAX may be responsible for the repression of wax biosynthesis in the leaves. In their study, the DEWAX expression levels were ∼10- to 75-fold higher in leaves than in stems. Conversely, SPL9 expression has been shown to be developmentally regulated, and to be higher in stems than in leaves (Wu et al., 2009). Thus, SPL9-DEWAX may function as a molecular switch in the integration of endogenous developmental cues and external signals, notably varying light levels, to coordinate plant development with epidermal wax synthesis.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) materials used in this study are of the Columbia (Col-0) genetic background. All plants were grown under long days (16-h light/8-h dark) at 23°C. The transgenic and mutant lines used in this study including spl9-4 spl15-1, Pro35S:MIR156, Pro35S:MIM156, ProSPL9:rSPL9, ProSPL9:rSPL9-GR, and ProSPL9:GFP-rSPL9 were previously described by Gou et al. (2011), and were kindly provided by Dr. Jia-Wei Wang. The reporter lines ProSPL9:SPL9-GUS and ProSPL9:rSPL9-GUS (Xu et al., 2016) were supplied by Scott Poethig. DXOE and dewax were kind gifts from Mi Chung Suh. The T-DNA insertion mutants spl9-4 (SAIL_150_B05), spl15-1 (SALK_074426), cer1-2 (SALK_014839), and cer4-4 (SALK_000575) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). For seedling experiments, seeds were sown on Murashige and Skoog agar medium containing 2% (w/v) Suc after surface sterilization and stratified at 4°C in the dark for 3 d. Then the seedlings were placed in a growth chamber and grown under white fluorescent light at light intensity of 100 µmol photons/m2/s at 23°C (16-h light/8-h dark) for 7 d before further treatment. For DEX induction, 1-week-old seedlings were sprayed with 10 μM DEX plus 10 μM CHX, 10 μM DEX, or water (mock), respectively.
Cuticular Wax Analysis
The cuticular wax composition of leaves and stems of 6-week-old plants was determined as described by Lü et al. (2009).
Constructs and Plant Transformation
DNA constructs used in the study were generated based on construction methods following the classic molecular biology protocols and Gateway technology (Invitrogen). For Gateway cloning, all the gene sequences were cloned into the pDONR207 vector (Gateway) and subsequently introduced into certain destination vectors. To generate Pro35S:HA-DEWAX or Pro35S:FLAG-rSPL9 fusion constructs, the coding DNA sequence of DEWAX or rSPL9 was cloned into pEarleyGate 201 or pEarleyGate 202, respectively. For ProDEWAX:DEWAX-GUS transgene, a genomic fragment spanning the 1.5-kb DEWAX promoter region upstream of the start codon and the DEWAX genomic region without stop codon was PCR amplified and cloned into the pMDC162 vector. For plant transformation, The pMDC162 harboring the ProDEWAX:DEWAX-GUS construct was introduced into Agrobacterium tumefaciens strain GV3101 by the freeze-thaw method and further transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998).
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted with the Plant RNeasy Mini kit (Qiagen). About 1 μg of total RNA was DNase I treated and further used for reverse transcription with Moloney murine leukemia virus reverse transcriptase (Promega). The cDNA was diluted to 100 μL with water, and 1 μL of the diluted cDNA was used for RT-qPCR with the SYBR Premix ExTaq kit (Takara, Japan) in a total volume of 25 μL on the Applied Biosystems 7500 real-time PCR system according to the manufacturer’s manual. The level of PP2A (AT1G13320) transcript was used as an internal control. The expression level of target genes are the ratio of expression in samples compared with the controls by the 2-ΔΔCt method. All the experiments were performed independently three times. All the primers used are listed in Supplemental Table 3. For mature miR156, a stem loop RT-qPCR was performed according to a previous protocol (Varkonyi-Gasic et al., 2007).
Yeast One-Hybrid and Yeast Two-Hybrid Assay
For yeast one-hybrid assay, the coding regions of SPL9 were PCR amplified and ligated to the pGADT7 vector (Clontech) to produce AD-SPL9, and a 1-kb promoter of CER1 was cloned into the pHIS2 vector (Clontech) to produce pHIS2-CER1. For yeast two-hybrid assay, the coding regions of DEWAX and SPL9 were PCR amplified from cDNAs and ligated to the pGADT7 and pGBKT7 vectors (Clontech) to generate pGADT7-DEWAX and pGBKT7-SPL9, respectively. Plasmids were cotransformed into yeast strain AH109 (Clontech) by the LiAc-PEG3350 method. Transformants were selected on SD-Leu-Trp plates. Interactions were tested on SD-Leu-Trp-His plates supplemented with 5 mM 3-amino-1,2,4,-triazole . Three independent clones for each transformation were tested.
ChIP-qPCR Assay
ChIP-qPCR assays was performed following the procedure described previously (Gou et al., 2011). About 2 g of harvested ProSPL9:rSPL9-GFP and the respective control (Col-0) samples were subjected to vacuum infiltration and cross-linked in fixation buffer [10 mM Tris–HCl (pH 8.0), 0.4 M Suc, 0.05% (v/v) Triton X-100, 1% (v/v) formaldehyde] for 10 min, followed by neutralization with 0.25 M Gly. Sonication was applied to break the genomic DNA into a size range from 200 to 1000 bp. Rabbit Anti-GFP antibody (use 5 µg antibody for 1 µg of chromatin, Abcam ChIP grade, ab290) and protein G plus agarose (Santa Cruz, sc-2002) were used for the immunoprecipitation. The cross-linking was reversed at 65°C overnight, and DNA was extracted. Finally, the GFP-specific enrichments of the fragments from CER1 promoters were analyzed by qPCR with specific primers, as shown in Supplemental Table 3. The values were standardized to the input DNA to obtain the enrichment fold. All samples for ChIP were prepared with three biological replicates (samples independently harvested on different days).
BiFC Analysis
For the BiFC assays, the full-length DEWAX was fused with N-terminal YFP (nYFP), and rSPL9 was fused with C-terminal YFP (cYFP; Walter et al., 2004). A. tumefaciens bacteria strain GV3101 was transformed with the above vectors or control empty vector and then coinjected into young Nicotiana benthamiana leaves. Fluorescence signals were observed with confocal microscopy (Leica TCS SP8).
Co-IP Analysis
Co-IP assay was performed as previously described (Gou et al., 2011). N. benthamiana leaves were infiltrated with A. tumefaciens harboring Pro35S:HA-DEWAX or Pro35S:FLAG-rSPL9 constructs.
Pull Down
To analyze in vitro protein interaction between SPL9 and DEWAX, Escherichia coli BL21 (DE3) cells were transformed with GST-DEWAX and 6xHis-SPL9, respectively. An E.coli strain expressing GST was used as a negative control. E. coli was cultivated overnight and then diluted 1:100 the next morning to grow another 3 h at 37°C. isopropyl β-D-1-thiogalactopyranoside (0.5 mM) was used to induce the expression of proteins at 16°C when the cells reached the logarithmic phase. Then the mixed 1 mL of E. coli (0.5 mL GST tagged plus 0.5 mL 6xHis tagged) cells were resuspended in phosphate buffered saline + Tween 20 and broken with a sonicator on ice. After centrifugation at 15,000 g for 30 min at 4°C, 20 μL nickel-nitrilotriacetic acid agarose (Bio-Rad) or glutathione-sepharose resin (GE Healthcare) was added to the supernatant mixture sample and rotated at 4°C for 3 h. The beads were washed with phosphate buffered saline + Tween 20 at least 4 times. Then SDS-PAGE gel-loading buffer was added to the beads and boiled for 5 min before electrophoresis. The protein was detected with anti-GST (Abmart M20007, 1/5000) or anti-6xHis (Abmart M30111, 1/5000) antibodies by immunoblot analysis.
Transient Transcription Dual-LUC Assays
For the Dual-LUC assays, the 1-kb promoter region of CER1 was cloned into the pGreenII 0800-LUC vector (Hellens et al., 2005). The Pro35S:rSPL9 or ProCER1:LUC constructs were introduced into Agrobacterium tumefaciens strain GV3101, and these GV3101 cells harboring Pro35S:rSPL9 or ProCER1:LUC were coinjected into N. benthamiana leaves and incubated at 25°C for 2 to 3 d. The Pro35S:REN gene (Renilla luciferase) in the vector was used as an internal control. The ratio of firefly luciferase to Renilla luciferase (LUC/REN) was measured using a Dual-Luciferase Reporter Assay System (Promega) to reflect the activity of the promoter under various conditions.
SEM Analysis
Cryogenic SEM was used to view epicuticular wax crystallization patterns. Stem (second internode above the rosette) samples were collected from 6-week-old plants. Samples were prepared and viewed by cryo-SEM as described by Lü et al. (2009).
BiLC Assays
The BiLC assay was performed as described (Chen et al., 2008). The full-length or truncated forms of SPL9 and DEWAX were cloned into pCAMBIA1300-nLUC or pCAMBIA1300-cLUC vector. Agrobacterium tumefaciens bacteria strain GV3101 carrying different constructs was coinfiltrated into N. benthamiana leaves. After infiltration, plants were incubated under dark conditions for 12 h and then transferred to light conditions for 48 h. A low-light-cooled charge-coupled device imaging apparatus (LAS 4000 mini) was used to capture the LUC image. In each analysis, three independent leaves were infiltrated and analyzed, and three biological replications were performed by different transformations at different days with similar results.
Recombinant Protein Purification
The full-length coding region of SPL9 or DEWAX was PCR amplified and cloned into the pGEX-2T vector (Amersham) to generate the pGEX-2T-SPL9 or pGEX-2T-DEWAX constructs, respectively. The plasmid was transformed into the E. coli BL21 (DE3) strain. Expression of the fusion protein was induced by 0.5 mM IPTG and incubated at 16°C overnight. The fusion protein was purified using glutathione-sepharose resin (GE Healthcare).
EMSA
For EMSAs, two complementary 50-bp length oligonucleotides containing the GTAC motif of the CER1 promoter were synthesized and labeled with biotin. Double strand DNA probes were obtained by annealing two complementary oligonucleotides. DNA gel mobility shift assay was performed using the EMSA kit (Beyotime) following the manufacturer’s protocol.
GUS Assays
For histochemical GUS staining, seedlings were pretreated in 90% (v/v) acetone and then soaked and kept in the GUS staining buffer [50 mM sodium phosphate (pH = 7.2), 10 mM EDTA, 2.5 mM potassium ferricyanide, 2.5 mM potassium ferrocyanide, and 0.1% (v/v) Triton X-100] containing 1 mg/ml 5-bromo-4-chloro-3-indolylglucuronide at 37°C in darkness for 3 h. The stained seedlings were decolorized in 70% (v/v) ethanol and photographed using a SMZ25 dissecting microscope (Nikon). Results shown are representative of 3-6 individual plants. For quantifications of GUS protein levels, MUG activity was determined with a fluorescence spectrophotometer as previously described (Wu and Poethig, 2006).
Statistical Analyses
For statistical analysis, one-way analysis of variance (ANOVA) or Student’s t test performed by GraphPad Prism 7.0 (GraphPad Software, www.graphpad.com) was used as specified in each figure and in Supplemental Data Set. Asterisks indicate statistical differences (*P < 0.05, **P < 0.01, ***P < 0.001). Different lowercase letters in the graphs indicate significant differences. Data represent mean values, and error bars are SD.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: SPL3 (AT2G33810), SPL4 (AT1G53160), SPL5 (AT3G15270), SPL9 (At2g42200), SPL15 (AT3G57920), DFR (At5g42800), DEWAX (At5g61590), CER1 (At1g02205), CER3 (At5g57800), CER4 (At4g33790), CER5 (AT1G51500), CER6 (AT1G68530), CER10 (AT3G55360), MIR156A (AT1G66783), MIR156C (AT4G31877), PP2A (At1g13320).
Supplemental Data
Supplemental Figure 1. Cuticular wax amounts of leaves from 5-week-old wild-type, Pro35S:MIR156, Pro35S:MIM156 and SPL9 related Arabidopsis mutants grown in long day conditions.
Supplemental Figure 2. Cuticular wax compositions of leaves of wild-type, Pro35S:MIR156, Pro35S:MIM156 and SPL9 related Arabidopsis mutants grown in long day conditions.
Supplemental Figure 3. SEM images of epicuticular wax crystals on leaves of 5-week-old wild-type and ProSPL9:rSPL9 plants grown in long-day conditions.
Supplemental Figure 4. RT-qPCR analysis of time course induction of CER1 and CER4 expression by dexamethasone (DEX) treatment in the ProSPL9:rSPL9-GR line.
Supplemental Figure 5. Glossy green and white waxy phenotypes of inflorescence stems of 6-week-old ProSPL9:rSPL9 cer1-2, ProSPL9:rSPL9 cer4-4, Pro35S:MIM156 cer1-2 and Pro35S:MIM156 cer4-4 plants and their parents.
Supplemental Figure 6. Cuticular wax amounts of leaves from 5-week-old ProSPL9:rSPL9 cer1-2, ProSPL9:rSPL9 cer4-4, Pro35S:MIM156 cer1-2, Pro35S:MIM156 cer4-4 plants and their parents grown in long-day conditions.
Supplemental Figure 7. The N-terminal part of SPL9 and the N-terminal part of DEWAX contribute to their interaction.
Supplemental Figure 8. SPL9 and DEWAX protein interaction domain analysis in BiLC experiment.
Supplemental Figure 9. DEWAX represses the SPL9 transcriptional activity on the CER4 promoter in the transient expression system.
Supplemental Figure 10. Diurnal expression of pri-MIR156A, pri-MIR156C and mature miR156.
Supplemental Figure 11. SPL9 directly regulates DEWAX expression.
Supplemental Figure 12. Light/dark changes altered expression of pri-MIR156A, pri-MIR156C and mature miR156.
Supplemental Table 1. Cuticular wax compositions in inflorescence stems of 6-week-old ProSPL9:rSPL9 cer1-2, ProSPL9:rSPL9 cer4-4, Pro35S:MIM156 cer1-2 and Pro35S:MIM156 cer4-4 plants and their parents grown in long day conditions.
Supplemental Table 2. Cuticular wax compositions in leaves of 5-week-old ProSPL9:rSPL9 cer1-2, ProSPL9:rSPL9 cer4-4, Pro35S:MIM156 cer1-2 and Pro35S:MIM156 cer4-4 plants and their parents grown in long day conditions.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set. Summary of statistical tests.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jia-Wei Wang (Shanghai Institutes for Biological Sciences), Mi Chung Suh (Chonnam National University), and Scott Poethig (University of Pennsylvania) for providing us with the seeds. This work was supported by the National Natural Science Foundation of China (NSFC) (31570186 to S.L. and 31400276 to R.-J.L.).
AUTHOR CONTRIBUTIONS
R.-J.L., L.-M.L., and S.L. designed the research; R.-J.L., L.-M.L., X.-L.L., and J.-C.K. performed the experiments and analyzed the data; R.-J.L., M.A.J., and S.L. wrote the article.
References
- Aarts M.G., Keijzer C.J., Stiekema W.J., Pereira A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7: 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E.A. (1974). The Influence of environment on leaf wax development in Brassica Oleracea Var. Gemmifera. New Phytol. 73: 955–966. [Google Scholar]
- Bernard A., Domergue F., Pascal S., Jetter R., Renne C., Faure J.D., Haslam R.P., Napier J.A., Lessire R., Joubes J. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Joubès J. (2013). Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog. Lipid Res. 52: 110–129. [DOI] [PubMed] [Google Scholar]
- Borges F., Martienssen R.A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk N., Hrmova M., Lopato S. (2014). Transcriptional regulation of cuticle biosynthesis. Biotechnol. Adv. 32: 526–540. [DOI] [PubMed] [Google Scholar]
- Bourdenx B., Bernard A., Domergue F., Pascal S., Leger A., Roby D., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Joubes J. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156: 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P., Poindexter P., Osborne E., Jiang C.Z., Riechmann J.L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon G., Hohmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104. [DOI] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Ben Chaabane S., Shah P., Poulsen C.P., Yang S.W. (2014). COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5: 5867. [DOI] [PubMed] [Google Scholar]
- Chuck G., Cigan A.M., Saeteurn K., Hake S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39: 544–549. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cui L.G., Shan J.X., Shi M., Gao J.P., Lin H.X. (2014). The miR156–SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80: 1108–1117. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Hohmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693. [DOI] [PubMed] [Google Scholar]
- Giese B.N. (1975). Effects of light and temperature on the composition of epicuticular wax of barley leaves. Phytochemistry 14: 921–929. [Google Scholar]
- Go Y.S., Kim H., Kim H.J., Suh M.C. (2014). Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor. Plant Cell 26: 1666–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S.M., Rashotte A.M., Rahman M., Feldmann K.A., Jenks M.A. (2005). Wax constituents on the inflorescence stems of double eceriferum mutants in Arabidopsis reveal complex gene interactions. Phytochemistry 66: 771–780. [DOI] [PubMed] [Google Scholar]
- Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A.Y., Zhu Q.H., Gu X., Ge S., Yang J., Luo J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8. [DOI] [PubMed] [Google Scholar]
- Hannoufa A., Negruk V., Eisner G., Lemieux B. (1996). The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. Plant J. 10: 459–467. [DOI] [PubMed] [Google Scholar]
- Hao Y., Oh E., Choi G., Liang Z., Wang Z.Y. (2012). Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 5: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam T.M., Haslam R., Thoraval D., Pascal S., Delude C., Domergue F., Fernández A.M., Beaudoin F., Napier J.A., Kunst L., Joubès J. (2015). ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 167: 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker T.S., Millar A.A., Kunst L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129: 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing M.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker T.S., Lam P., Zheng H., Kunst L. (2007). A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Richter R., Vincent C., Martinez-Gallegos R., Porri A., Coupland G. (2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 37: 254–266. [DOI] [PubMed] [Google Scholar]
- Jenks M.A., Rich P.J., Ashworth E.N. (1994). Involvement of cork cells in synthesis and secretion of filamentous epicuticular wax on Sorghum bicolor (L.) Moench. Int. J. Plant Sci. 155: 506–518. [Google Scholar]
- Jenks M.A., Tuttle H.A., Eigenbrode S.D., Feldmann K.A. (1995). Leaf Epicuticular Waxes of the Eceriferum Mutants in Arabidopsis. Plant physiology 108: 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R.K., Megha S., Basu U., Rahman M.H., Kav N.N. (2016). Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS One 11: e0158784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Go Y.S., Suh M.C. (2018). DEWAX2 transcription factor negatively regulates cuticular wax biosynthesis in Arabidopsis leaves. Plant Cell Physiol. 59: 966–977. [DOI] [PubMed] [Google Scholar]
- Klein J., Saedler H., Huijser P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250: 7–16. [DOI] [PubMed] [Google Scholar]
- Lam P., Zhao L., Eveleigh N., Yu Y., Chen X., Kunst L. (2015). The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 167: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P., Zhao L., McFarlane H.E., Aiga M., Lam V., Hooker T.S., Kunst L. (2012). RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 159: 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.B., Suh M.C. (2015a). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34: 557–572. [DOI] [PubMed] [Google Scholar]
- Lee S.B., Suh M.C. (2015b). Cuticular wax biosynthesis is up-regulated by the MYB94 transcription factor in Arabidopsis. Plant Cell Physiol. 56: 48–60. [DOI] [PubMed] [Google Scholar]
- Lee H., Yoo S.J., Lee J.H., Kim W., Yoo S.K., Fitzgerald H., Carrington J.C., Ahn J.H. (2010). Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 38: 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wu X., Lam P., Bird D., Zheng H., Samuels L., Jetter R., Kunst L. (2008). Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 148: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.H., Tian X., Li Y.J., Wu C.A., Zheng C.C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S., Song T., Kosma D.K., Parsons E.P., Rowland O., Jenks M.A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59: 553–564. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ikeda M., Takada S., Takiguchi Y., Kondou Y., Yoshizumi T., Fujita M., Shinozaki K., Matsui M., Ohme-Takagi M. (2010). Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 51: 2145–2151. [DOI] [PubMed] [Google Scholar]
- Nawrath C., Schreiber L., Franke R.B., Geldner N., Reina-Pinto J.J., Kunst L. (2013). Apoplastic diffusion barriers in Arabidopsis. The Arabidopsis Book 11: e0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y., Shikata M., Koyama T., Ohtsubo N., Mitsuda N., Ohme-Takagi M. (2013). MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.S., Go Y.S., Suh M.C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88: 257–270. [DOI] [PubMed] [Google Scholar]
- Pascal S., Bernard A., Sorel M., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Domergue F., Joubès J. (2013). The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 73: 733–746. [DOI] [PubMed] [Google Scholar]
- Pascal S., Bernard A., Deslous P., Gronnier J., Fournier-Goss A., Domergue F., Rowland O., Joubès J. (2019). Arabidopsis CER1-LIKE1 functions in a cuticular very-long-chain alkane-forming complex. Plant Physiol. 179: 415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin J.A., Zheng H., Balakshin L.J., Goodman I.P., Western T.L., Jetter R., Kunst L., Samuels A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704. [DOI] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Leger A., Joubes J., Miersch O., Huard C., Blee E., Mongrand S., Domergue F., Roby D. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544. [DOI] [PubMed] [Google Scholar]
- Rowland O., Zheng H., Hepworth S.R., Lam P., Jetter R., Kunst L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142: 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Zhou C.M., Confraria A., Martinho C., von Born P., Baena-Gonzalez E., Wang J.W., Weigel D. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24: 2714–2719. [DOI] [PubMed] [Google Scholar]
- Samad A.F.A., Sajad M., Nazaruddin N., Fauzi I.A., Murad A.M.A., Zainal Z., Ismail I. (2017). MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 8: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707. [DOI] [PubMed] [Google Scholar]
- Sanchez-Retuerta C., Suarez-Lopez P., Henriques R. (2018). Under a new light: Regulation of light-dependent pathways by non-coding RNAs. Front. Plant Sci. 9: 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Lee S.B., Suh M.C., Park M.J., Go Y.S., Park C.M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd T., Wynne Griffiths D. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171: 469–499. [DOI] [PubMed] [Google Scholar]
- Shepherd T., Robertson G.W., Griffiths D.W., Birch A.N.E., Duncan G. (1995). Effects of environment on the composition of epicuticular wax from kale and swede. Phytochemistry 40: 407–417. [Google Scholar]
- Suh M.C., Samuels A.L., Jetter R., Kunst L., Pollard M., Ohlrogge J., Beisson F. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139: 1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Li M., Zhou Y., Guo T., Liu Y., Zhang H., Fang Y. (2018). Coordinated regulation of Arabidopsis microRNA biogenesis and red light signaling through Dicer-like 1 and phytochrome-interacting factor 4. PLoS Genet. 14: e1007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.L., Li Y.H., Hsieh W.P., Lin M.C., Ahn J.H., Wu S.H. (2014). HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26: 2858–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E.F., Hellens R.P. (2007). Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V., Ponnu J., Schlereth A., Arrivault S., Langenecker T., Franke A., Feil R., Lunn J.E., Stitt M., Schmid M. (2013). Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749. [DOI] [PubMed] [Google Scholar]
- Wang J.W., Park M.Y., Wang L.J., Koo Y., Chen X.Y., Weigel D., Poethig R.S. (2011). miRNA control of vegetative phase change in trees. PLoS Genet. 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Liu Y., Wang H., Ma X., Wang B., Wu G., Wang H. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hu T., Zhao J., Park M.Y., Earley K.W., Wu G., Yang L., Poethig R.S. (2016). Developmental functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 21: 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xu M., Koo Y., He J., Poethig R.S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Niu Q.W., Ng K.H., Chua N.H. (2015a). The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83: 673–685. [DOI] [PubMed] [Google Scholar]
- Yu Z.X., Wang L.J., Zhao B., Shan C.M., Zhang Y.H., Chen D.F., Chen X.Y. (2015b). Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol. Plant 8: 98–110. [DOI] [PubMed] [Google Scholar]
- Yu S., Cao L., Zhou C.M., Zhang T.Q., Lian H., Sun Y., Wu J., Huang J., Wang G., Wang J.W. (2013). Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang Y., Wang S., Hao L., Wang S., Xu C., Jiang F., Li T. (2019). Characterization of genome-wide microRNAs and their roles in development and biotic stress in pear. Planta 249: 693–707. [DOI] [PubMed] [Google Scholar]
- Zheng H., Rowland O., Kunst L. (2005). Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Sutoh K., Zhu J.K., Zhang W. (2008). Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta 1779: 780–788. [DOI] [PubMed] [Google Scholar]