Camelina allopolyploid genomes originated through recurrent hybridization between two paleogenomes; descending dysploidies in Camelina were accompanied by complex chromosome shattering.
Abstract
Complexes of diploid and polyploid species have formed frequently during the evolution of land plants. In false flax (Camelina sativa), an important hexaploid oilseed crop closely related to Arabidopsis (Arabidopsis thaliana), the putative parental species as well as the origin of other Camelina species remained unknown. By using bacterial artificial chromosome–based chromosome painting, genomic in situ hybridization, and multi-gene phylogenetics, we aimed to elucidate the origin and evolution of the polyploid complex. Genomes of diploid camelinas (Camelina hispida, n = 7; Camelina laxa, n = 6; and Camelina neglecta, n = 6) originated from an ancestral n = 7 genome. The allotetraploid genome of Camelina rumelica (n = 13, N6H) arose from hybridization between diploids related to C. neglecta (n = 6, N6) and C. hispida (n = 7, H), and the N subgenome has undergone a substantial post-polyploid fractionation. The allohexaploid genomes of C. sativa and Camelina microcarpa (n = 20, N6N7H) originated through hybridization between an auto-allotetraploid C. neglecta–like genome (n = 13, N6N7) and C. hispida (n = 7, H), and the three subgenomes have remained stable overall since the genome merger. Remarkably, the ancestral and diploid Camelina genomes were shaped by complex chromosomal rearrangements, resembling those associated with human disorders and resulting in the origin of genome-specific shattered chromosomes.
INTRODUCTION
Polyploidization is an important speciation mechanism, particularly in ferns and angiosperms. New polyploid species form repeatedly, and go extinct or return to the genetically diploid state via a process described as (re)diploidization. Assuming that early angiosperm divergence was predated by a whole-genome duplication (Jiao et al., 2011), the polyploidization–diploidization cycles mark speciation and cladogenesis of angiosperms for the past ∼300 million years (Clark and Donoghue, 2017).
When encountering polyploidy in extant plant genomes, two key factors immediately emerge: the way in which polyploids are formed and their age. Both autopolyploidy (genome doubling within a species) and allopolyploidy (genome merger due to interspecies hybridization) gave rise to new clades and species, many of which thrived and still exist today (Parisod et al., 2010; Jiao et al., 2011; Garsmeur et al., 2014; Marcussen et al., 2014; Landis et al., 2018). The merging of reduced and unreduced gametes in diploids forms triploids or allotetraploids, and additional polyploid genomes can arise by backcrossing to the diploids or hybridization with other congeneric polyploids. This network of parental and hybrid-derived species of different ploidies represents a polyploid complex (Stebbins, 1971).
Polyploid complexes exhibit different levels of completeness and maturity (Stebbins, 1971) as genomes of diploid progenitors, as well as polyploid species, further evolve or eventually become extinct. The continuous extinction and genome evolution including rediploidization hamper the identification of parental (sub)genomes and inter-genome comparisons. A number of model and crop polyploid complexes, such as Brassica (Nagaharu and Nagaharu, 1935; Wang et al., 2011; Yang et al., 2016), Capsella (Douglas et al., 2015), Cardamine (Mandáková et al., 2013, 2019), Fragaria (Edger et al., 2019), Gossypium (Paterson et al., 2012; Hu et al., 2019), Nicotiana (Renny-Byfield et al., 2013; Sierro et al., 2014), Spartina (Salmon et al., 2005), Tragopogon (Symonds et al., 2010), and Triticum (Marcussen et al., 2014; El Baidouri et al., 2017), have been investigated over the years. Significant progress in high-throughput whole-genome sequencing of these and other diploid and polyploid genomes has greatly advanced our understanding of post-polyploid genome evolution. Despite recent technological advances, analysis of complex polyploid genomes remains challenging (Kyriakidou et al., 2018), especially when the parental genomes are extinct or cannot be easily identified.
Among crucifer species (Brassicaceae), Brassica is the most researched polyploid complex composed of three main diploid (2n = 16, 18, and 20; BB, CC, and AA genome) and three allotetraploid (2n = 34, 36, and 38; BBCC, AABB, and AACC) genomes (Nagaharu and Nagaharu, 1935). Because of its worldwide economic importance, the genus Brassica has become one of the most important models in plant polyploidy research (Mason and Snowdon, 2016; Gaebelein and Mason, 2018). Recently, false flax (Camelina sativa), an increasingly popular crucifer oilseed crop, was suggested to have an allohexaploid origin (Hutcheon et al., 2010; Kagale et al., 2014), similar to rediploidized Brassica genomes. Although both Brassica and Camelina polyploid complexes include allohexaploid genomes and genomic resources available for both genera allow for insightful comparisons, the extant as well as ancestral Camelina genomes remain virtually unknown.
The genus Camelina, represented by seven or eight species (Brock et al., 2019), belongs to one of the most karyologically variable crucifer genera, with chromosome numbers ranging from 2n = 12 to 40 (2n = 12, 14, 16, 26, 28, 32, 36, 38, and 40) and a threefold genome size variation (BrassiBase, https://brassibase.cos.uni-heidelberg.de/; Hutcheon et al., 2010; Brock et al., 2018). C. sativa (2n = 40), is an ancient oilseed crop with a newfound application as an aviation biofuel and omega-3–rich feedstock. Two species, Camelina microcarpa (2n = 40) and Camelina rumelica (2n = 26), are worldwide weeds. Three other species, Camelina hispida (2n = 14), Camelina laxa (2n = 12), and Camelina neglecta (2n = 12), occur in relatively restricted ranges in Europe and western Asia. Because of variable chromosome numbers in diploids (2n = 12 and 14) and uncorrelated numbers in polyploids (2n = 26 and 40), the evolution of the Camelina polyploid complex was a conundrum until recently. Hutcheon et al. (2010) revealed that three single-copy nuclear genes are present as three paralogous copies in the genome of C. sativa and suggested that the species most likely had an allohexaploid origin. Later genome sequencing of the C. sativa genome corroborated its allohexaploid origin by revealing three minimally diverged subgenomes of an unknown identity and origin (Kagale et al., 2014).
Recent developments on plant-based biofuels have spurred an increased interest in C. sativa as an oilseed crop. With high levels of long-chain hydrocarbons, C. sativa oil is well suited as an aviation biofuel, which has proven efficacious in jet aircraft with a 75% reduction in CO2 emissions over traditional petroleum jet fuels (Shonnard et al., 2010). Seed oil content in C. sativa ranges between 36 and 47% and has oil yields of 540 to 1410 kg/ha, which is higher than soybean (Glycine max) oil yield, but comparable to that of rapeseed (Brassica napus; Moser, 2012). Moreover, genetic resources for C. sativa have grown considerably in recent years and include efficient transformation protocols (Lu and Kang, 2008; Ruiz-Lopez et al., 2014), an available reference genome (Kagale et al., 2014), molecular genetic characterizations (Vollmann et al., 2005; Gehringer et al., 2006), and CRISPR/Cas-based technology. Through genome editing, C. sativa lines have been produced with increased oleic acid (Jiang et al., 2017; Morineau et al., 2017), decreased very long chain fatty acids (Ozseyhan et al., 2018), and increased oil content (YIELD10; Waltz, 2018). Furthermore, its close phylogenetic relationship to Arabidopsis (both Arabidopsis and Camelina are classified as members of the tribe Camelineae; Beilstein et al., 2006, 2008; Nikolov et al., 2019) gives C. sativa a unique ability to benefit from the ongoing developments in the world’s most well-studied plant model.
Given C. sativa’s rising importance as a plant model for biofuel development (Iskandarov et al., 2014), high-value molecule factory (Augustin et al., 2015; Iven et al., 2016; Augustin et al., 2017), and omega-3–rich feedstock, surprisingly little is known about its evolutionary history and subgenome origins. Little genetic diversity existing in currently available C. sativa cultivars (Vollmann et al., 2005; Brock et al., 2018; Luo et al., 2019) limits the effectiveness of traditional breeding programs. To this end, the identification of diploid progenitors, wild relatives, and the elucidation of hybridization histories are necessary requisites for further improvement of Camelina genomes for seed oil production.
Herein, we aimed to reconstruct the origin of the allohexaploid genome of C. sativa and elucidate the identity of its three subgenomes (Kagale et al., 2014) by comparative chromosome painting (CCP), genomic in situ hybridization (GISH), and a set of nuclear gene markers. By including all known diploid and polyploid Camelina species, we expected to (1) identify probable parental genomes of C. sativa and other Camelina polyploids, (2) elucidate evolutionary relationships among all Camelina genomes and species, (3) analyze subgenome (in)stability in the polyploid genomes, and (4) uncover mechanisms driving genome evolution in Camelina.
RESULTS
Camelina Diploids Exhibit Highly Reshuffled Genome Structures
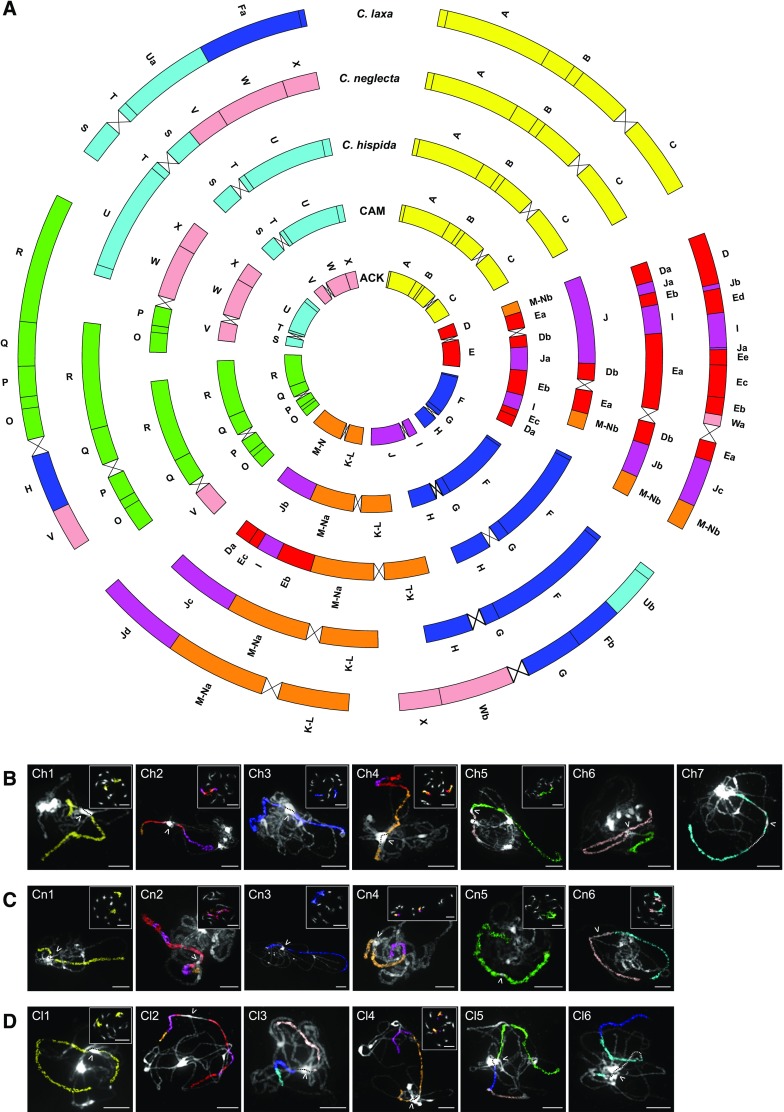
All analyzed plants of C. hispida had seven chromosome pairs (n = 7), whereas six pairs were counted in C. laxa and C. neglecta (n = 6; Figure 1). Since the ancestral crucifer karyotype (ACK, n = 8, AK1 to AK8; Figure 1; Supplemental Figure 1; Lysak et al., 2016) has been considered the ancestral genome of Camelina and the tribe Camelineae, genome structures of the investigated Camelina species are described here in relation to the eight chromosomes and 22 genomic blocks (GBs; Supplemental Data Set 1A) of ACK. In the analyzed diploid species, the used chromosome-specific Arabidopsis bacterial artificial chromosome (BAC) contigs, unambiguously hybridized either to one pachytene bivalent or one pair of mitotic metaphase chromosomes. In C. hispida, three of seven chromosome pairs (Ch1, Ch3, and Ch7) retain the ancestral structure as in ACK (AK1, AK3, and AK7, respectively). Chromosomes Ch2 and Ch4 show highly reshuffled combinations of GBs contributed by three different ancestral chromosomes (AK2, AK4, and AK5). Chromosomes Ch5 and Ch6 resemble ancestral chromosomes AK6 and AK8 modified by a whole-arm translocation (Figure 1; Supplemental Data Set 1B). Three chromosomes in C. neglecta (Cn1, Cn3, and Cn5) have the ancestral-like structure of AK1, AK3, and AK6, respectively. The remaining three chromosomes combine blocks of two (Cn4 and Cn6) or three ancestral chromosomes (Cn2), respectively (Figure 1; Supplemental Data Set 1C). In C. laxa, only chromosome Cl1 resembles the ancestral chromosome (AK1), whereas the remaining chromosomes consist of reshuffled blocks from two (Cl4 and Cl6), three (Cl3 and Cl5), or four (Cl2) ancestral chromosomes (Figure 1; Supplemental Data Set 1D).
Figure 1.
Comparative Genome Structures of Camelina Diploids.
(A) Circos graphic displaying chromosomal collinearity between ACK (n = 8; innermost circle), CAM (n = 7), C. hispida (n = 7), C. neglecta (n = 6), and C. laxa (n = 6). Color coding and capital letters (A to X) correspond to eight chromosomes and 22 GBs of the ACK, respectively. See Figure 2 and Supplemental Data Sets 1A to 1E for boundaries of GBs.
(B) to (D) Chromosomes of C. hispida (see [B]; Ch1 to Ch7), C. neglecta (see [C]; Cn1 to Cn6), and C. laxa (see [D]; Cl1 to Cl6) revealed by comparative painting using Arabidopsis BAC contigs as painting probes on pachytene and mitotic chromosome spreads. Chromosomes were counterstained with DAPI. The fluorescence of painting probes was captured as grey scale photographs and pseudocolored to match the eight AK chromosomes. Arrowheads point to centromeres. Bars = 10 µm.
The Origin of the Ancestral Camelina Genome
The ancestral Camelina genome (CAM; n = 7, chromosomes CAM1 to CAM7; Figures 1 and 2; Supplemental Data Set 1E) has been inferred based on the following premises: (1) it has descended from the older ACK; (2) its chromosome number equalled n = 7, as most present-day Camelina genomes are based on or were derived from seven chromosomes; and (3) two CAM chromosomes (CAM2 and CAM4), combining GBs of ancestral chromosomes AK2, AK4, and AK5, were identified in all Camelina diploids.
Figure 2.
Collinear Genome/Chromosomal Relationships between ACK (n = 8), CAM (n = 7), and Modern Diploid Genomes of C. hispida (n = 7), C. neglecta (n = 6), and C. laxa (n = 6).
Boundaries of individual GBs are specified as Arabidopsis genes (The Arabidopsis Information Resource, www.arabidopsis.org; see also Supplemental Tables 1 to 5).
Chromosomes CAM1, CAM3, CAM5, CAM6, and CAM7 structurally mirror chromosomes AK1, AK3, AK6, AK8, and AK7 of ACK, respectively. Chromosome CAM4 likely originated by a reciprocal translocation involving a major part of the bottom arm of AK4 [∼5.1 Mb in the A. thaliana genome; breakpoint within block J, in BAC clone T14G11 (AC002341)] and a minor part of the bottom arm of chromosome AK5 [∼1.53 Mb in GB M-N; breakpoint between BACs F17J16 (AL163527)/F25L23 (AL356014)]. The resulting translocation chromosome consists of AK5-derived GBs K-L and M-Na, and block Jb contributed by AK4. The second translocation product, bearing blocks I, Ja, and M-Nb, fused with chromosome AK2. The resulting CAM2 chromosome underwent a complex chromosome shattering (i.e., several subsequent translocations, peri- and paracentric inversions) and bears highly reshuffled blocks D (Da, Db) and E (Ea, Eb, Ec) from AK2, I and Ja from AK4, and M-Nb from AK5.
Evolution of CAM during the Divergence of Diploid Camelina Species
The parsimoniously inferred structure of the ancestral Camelina genome allowed us to reconstruct how the modern diploid genomes originated (Figures 1 and 2; Supplemental Figures 2 to 5; Supplemental Data Sets 1B to 1D). In C. hispida, chromosomes Ch5 and Ch6 originated by a whole-arm reciprocal translocation involving the upper arms of CAM5 (∼3.94 Mb, blocks O and P) and CAM6 (∼2.55 Mb, block V), with both breakpoints in the (peri)centromeric regions. The resulting translocation chromosomes Ch5 and Ch6 consist of GBs V, Q, and R and O, P, W, and X, respectively. Chromosomes Ch2 and Ch4 consist of blocks J, Db, Ea, and Nb and K-L, M-Na, Eb, I, Ec, and Da, respectively (Figure 3). These chromosomes originated by a reciprocal translocation between CAM2 (∼7.76 Mb, blocks Eb, I, Ec, and Da) and CAM4 (∼5.1 Mb, block Jb), involving breakpoints Ja/Eb [T14G11 (AC002341)/T6C23 (AC013289)] and M-Na/Jb [F17J16 (AL163527)/T14G11 (AC002341)]. The breakpoints Ja/Eb and M-Na/Jb can be considered as evolutionarily re-used as they previously played a role in the origin of chromosomes CAM2 and CAM4 from AK2, AK4, and AK5 (Supplemental Data Set 1E).
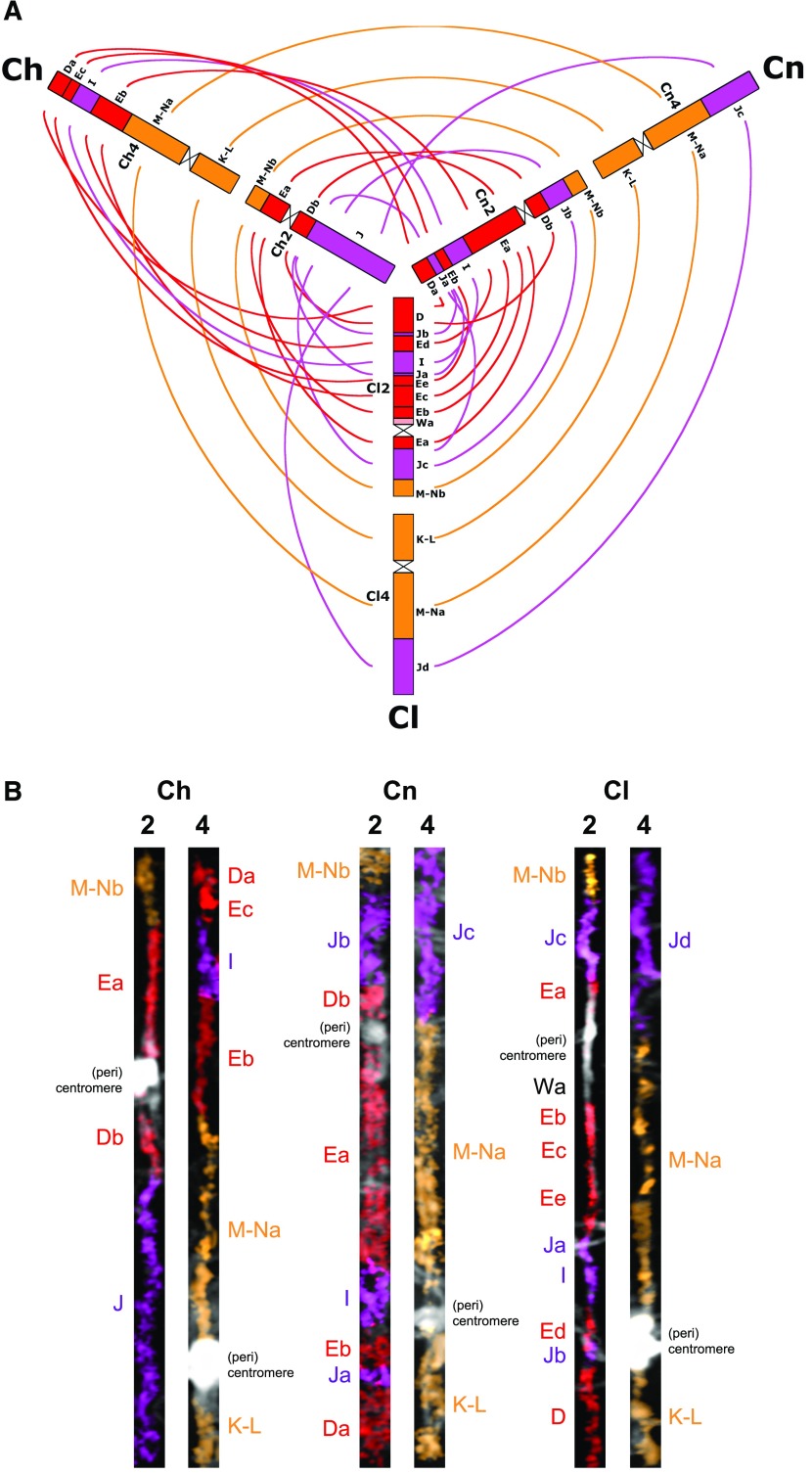
Figure 3.
Comparison of the Two Shattered Chromosomes in Camelina Diploids.
(A) Three-way comparison of GBs on chromosomes 2 and 4 in C. hispida (Ch, chromosomes Ch2 and Ch4), C. neglecta (Cn, Cn2, and Cn4), and C. laxa (Cl, Cl2 and Cl4). Color coding and capital letters correspond to three AK chromosomes (AK2, red; AK4, violet; AK5, orange) and six GBs (D, E, I, J, K-L, and M-N), respectively.
(B) Straightened pachytene chromosomes of C. hispida, C. neglecta, and C. laxa painted using differentially labeled Arabidopsis BAC contigs corresponding to six complete GBs. The fluorescence of painting probes was pseudocolored to match the corresponding AK chromosomes.
In C. neglecta, chromosome number reduction from n = 7 to n = 6 was mediated by an end-to-end translocation between chromosomes CAM6 and CAM7 (Figures 1 and 2; Supplemental Figure 3; Supplemental Data Set 1C). Breakpoints occurred in the (sub)telomeric regions of the upper arms of both chromosomes, and the end-to-end translocation event led to the origin of the fusion chromosome Cn6 (∼21.37 Mb; blocks U, T, S, V, W, and X). The Cn6 centromere came from CAM5, whereas the CAM6 centromere was eliminated during/after the fusion. Chromosome Cn2 is homeologous to CAM2; however, its structure was altered by massive chromosome shattering (Figure 3).
Unlike C. neglecta, descending dysploidy from n = 7 to n = 6 in C. laxa was not mediated by a single end-to-end translocation. The genome of C. laxa (Figures 1 and 2) underwent a complex five-step rearrangement involving four ancestral chromosomes (CAM3, CAM5, CAM6, and CAM7; Supplemental Figures 4 and 5; Supplemental Data Set 1D): (1) An end-to-end translocation between CAM3 and CAM5, with breakpoints in the (sub)telomeres of the upper and bottom arm, respectively, was accompanied by the elimination of the CAM5 centromere. (2) A whole-arm translocation between the upper arms of the fusion chromosome CAM3/5 (∼10.39 Mb, blocks F and G) and chromosome CAM6 (∼2.5 Mb, block V). (3) A reciprocal translocation between CAM3/6 [∼6.45 Mb, block Fa, breakpoint between BACs K24M9 (AP001303) and MVE11 (AB026654)] and CAM7 [∼3.14 Mb, block Ub, breakpoint between BACs F11C18 (AL049607) and F10N7 (AL021636)]. (4) Chromosome CAM3/5/6 was altered by an ∼3.42-Mb pericentric inversion involving block H and became C. laxa chromosome Cl5. (5) Chromosome CAM3/6/7 lost an ∼610-kb segment (Wa) through an unequal translocation with Cl2 and became chromosome Cl3. Chromosome Cl2 is homeologous to CAM2, however its structure was altered by massive chromosome shattering (Figure 3).
Genome Structure and Origin of the Allotetraploid C. rumelica
All the analyzed plants of C. rumelica had 26 chromosomes (n = 13; Supplemental Figure 6). The unambiguous identification of all GBs in four copies on mitotic chromosomes and in two copies on pachytene spreads, confirmed the assumed tetraploid origin of the species. To elucidate the parentage of C. rumelica, we performed GISH with differently combined genomic DNAs (gDNAs) of the diploid species. Only gDNA of C. hispida and C. neglecta hybridized to 14 and 12 chromosomes, respectively (Supplemental Figure 6). Thus, GISH strongly suggested an allotetraploid origin of C. rumelica (2n = 26, N6N6HH) from hybridization between genomes related to C. hispida (2n = 14, HH) and C. neglecta (2n = 12, N6N6).
Combining GISH and CCP, chromosomes of the two subgenomes were unequivocally identified: chromosomes Cr1 to Cr6 were assigned to the N6 subgenome (C. neglecta, n = 6), whereas chromosomes Cr7 to Cr13 belong to the H subgenome (C. hispida, n = 7). The absence of inter-subgenomic chromosomal translocations points to subgenome stability since the allopolyploidization event.
All but one (Cr1) chromosome in the N6 subgenome are highly reshuffled. The origin of chromosome Cr2 can be reconstructed by inferring a paracentric inversion (∼10.98 Mb, blocks GBs Ja, Eb, I, Ec, and Da) followed by a pericentric inversion (∼6.67 Mb, blocks Ea and D) on the ancestral CAM2 chromosomes (Supplemental Figure 7). This suggests that the origin of the allotetraploid genome of C. rumelica occurred prior to chromosome shattering forming the modern chromosome Cn2 in C. neglecta (Supplemental Figure 3). Although the N6 subgenome chromosomes Cr3, Cr4, Cr5, and Cr6 are reshuffled by complex C. rumelica–specific translocations and inversions, associations of GBs specific for the C. neglecta fusion chromosome Cn6 (CAM6 + CAM7) are still detectable within chromosomes Cr4 (blocks Ua+Xc) and Cr5 (blocks S+V+Wa+Ub+Xb; Supplemental Figures 6 and 7; Supplemental Data Set 1F).
Except for chromosomes Cr8 and Cr10, the remaining five chromosomes of the H subgenome are structurally identical with chromosomes of C. hispida, including the C. hispida–specific translocation chromosomes Ch5 (Cr11) and Ch6 (Cr12; Supplemental Figures 6 and 7; Supplemental Data Set 1F). Chromosomes Cr8 and Cr10 do not exhibit C. hispida–specific association of GBs (Supplemental Figure 2). Instead, the chromosomes have the ancestral structure of CAM2 and CAM4, modified by a single paracentric inversion on Cr8 (∼10.98 Mb, blocks Ja, Eb, I, Ec, and Da), which was identical in both subgenomes (compare with Cr2 in the N6 subgenome). These results suggest that chromosomes Ch2 and Ch4 in the parental C. hispida-like genome were shuffled only after the origin of C. rumelica.
C. sativa and C. microcarpa: Allohexaploid Origin and Prevailing Subgenome Stasis
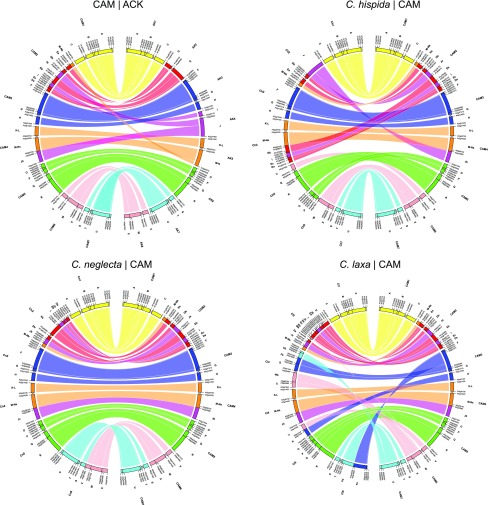
All the analyzed plants of C. sativa and C. microcarpa had 40 chromosomes (n = 20; Figure 4). The identification of all GBs in 6 and 12 copies on meiotic (pachytene) and mitotic chromosomes, respectively, confirmed the hexaploid status of both genomes. The gDNA probes of C. hispida (HH) and C. neglecta (NN) hybridized to 14 (HH) and 26 (NNNN) chromosomes, respectively, in both C. sativa and C. microcarpa (Figure 4). No inter-subgenomic translocations were identified by GISH, suggesting that the parental subgenomes remained stable since the allopolyploidization event. GISH results provide convincing evidence that the allohexaploid C. sativa and C. microcarpa (N6N6N7N7HH) genomes originated from hybridization between a not yet identified tetraploid C. neglecta–like genome (N6N6N7N7; subgenomes G1 and G2) and the diploid C. hispida–like genome (HH; subgenome G3). The alternative assignment of genomes N6 and N7 as subgenomes G1 and G2, and the H genome as subgenome G3, follows the designations introduced by Kagale et al. (2014).
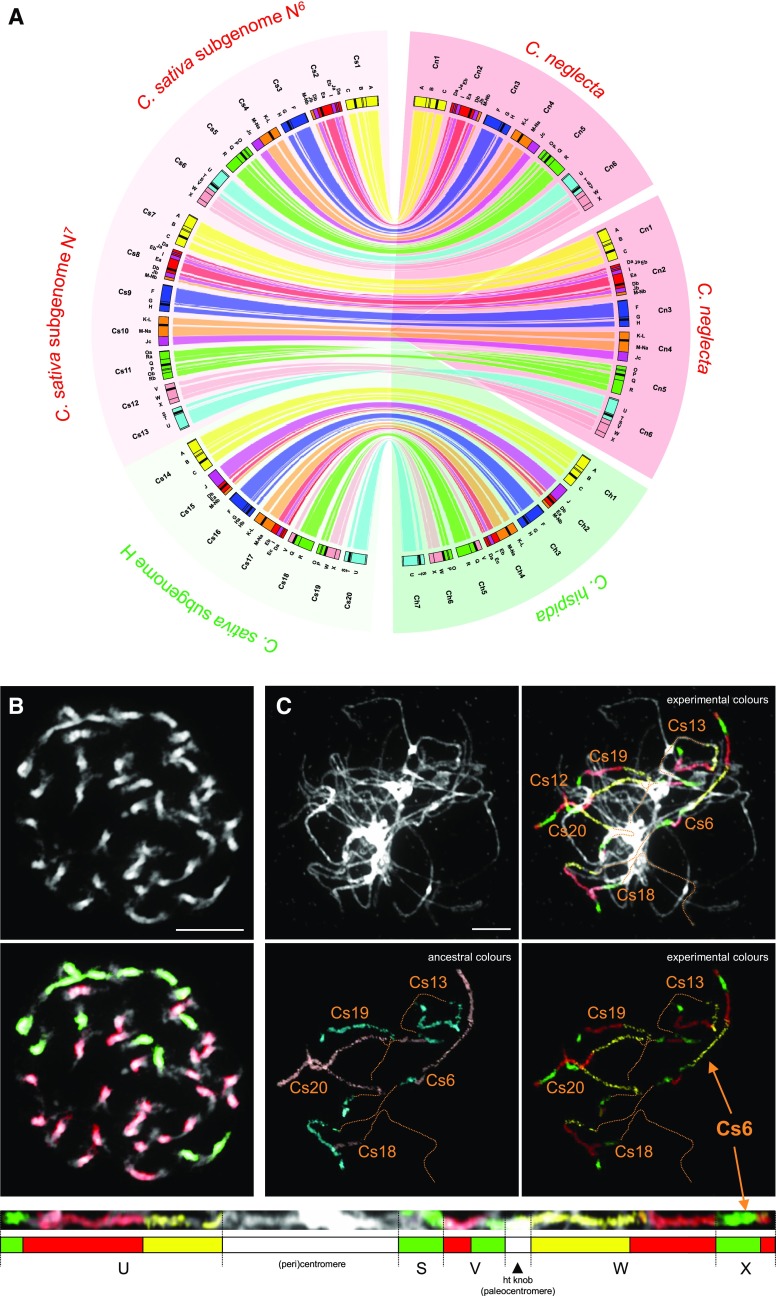
Figure 4.
Genome Structure of C. sativa.
(A) Circos graphic displaying chromosomal collinearity between 20 chromosomes (Cs1 to Cs20) of the three C. sativa subgenomes (N6N7H, i.e., G1-G2-G3, left half of the circle) and 2 × 6 chromosomes of C. neglecta (Cn1 to Cn6) and seven chromosomes of C. hispida (Ch1 to Ch7, right half of the circle). Color coding and capital letters correspond to eight chromosomes and 22 GBs of ACK, respectively. See Supplemental Tables 2, 3, and 7 for boundaries of all GBs.
(B) GISH of gDNA of C. neglecta (red fluorescence) and C. hispida (green fluorescence) to 40 mitotic chromosomes in C. sativa. GISH revealed 26 chromosomes (N6N6 and N7N7) labeled by the C. neglecta probe, whereas 14 chromosomes were contributed by C. hispida (HH).
(C) CCP on pachytene chromosomes of C. sativa. Painting probes for AK7 and AK8 revealed the structure of the fusion chromosome Cs6. Note that due to the whole-genome triplication the used BAC contigs also labeled homeologous regions on other Cs chromosomes. Differentially labeled painting probes are shown in experimental colors (red, green, and yellow fluorescence) and pseudocolored in turquoise and pink, respectively, following the color coding for AK chromosomes. Chromosomes were counterstained with DAPI. Bars = 10 μm.
The structure of the C. sativa genome was described by Kagale el al. (2014) and is further specified herein. Subgenomes G1 (n = 6, N6) and G2 (n = 7, N7) mirror the genome structure of C. neglecta including the highly reshuffled chromosome Cn2 (Cs2 and Cs8, blocks M-Nb, Jb, Db, Ea, I, Eb, Ja, and Da; Figures 3 and 4). As the chromosome Cn2 originated through complex chromosome shattering, its repeated origins in the two C. sativa subgenomes are highly improbable. Thus, the presence of two Cn2 chromosomes in subgenomes G1 and G2 represents the strongest support for C. neglecta–like genome being the progenitor of both subgenomes (Figure 4; Supplemental Figure 8; Supplemental Data Set 1G). C. neglecta–specific fusion chromosome Cn6 (CAM6 + CAM7; blocks U, T, S, V, W, and X) was identified in subgenome G1 (chromosome Cs6), whereas ancestral chromosomes CAM6 and CAM7 are homeologous to Cs12 and Cs13 in subgenome G2 (Figure 4; Supplemental Figure 8). This fact, together with the number of chromosomes in the two subgenomes (n = 6 and n = 7 in subgenome G1 and G2, respectively), argues for an auto-allotetraploid C. neglecta–related parental genome formed by hybridization between two (sibling) taxa of C. neglecta distinguished by chromosome numbers (n = 6, N6 and n = 7, N7). Except for an ∼10.67-Mb paracentric inversion on chromosome Cs11 [breakpoints within blocks O, between BACs F5I10 (AF195115) and F6N23 (AF058919), and R, T30N20 (AL365234)], subgenomes G1 and G2 did not experience subgenome-specific rearrangements and remained stable. As subgenome G3 (n = 7, H) resembles genome of C. hispida, including C. hispida–specific chromosomes Ch2 (Cs15), Ch4 (Cs17), Ch5 (Cs18), and Ch6 (Cs19), its structure provides conclusive evidence for the parentage of C. hispida. A single centromere repositioning on Cs16 [the Ch3 centromere, originally located between blocks G and H, was shifted ∼1.54 Mb into block H, i.e., between BACs F16F14 (AC007047) and T24I21 (AC005825)] differentiates the H subgenome from the parental C. hispida genome (Figure 4; Supplemental Figure 8; Supplemental Data Set 1G).
Altogether, our data suggest that the hexaploid genome of C. sativa (n = 20, N6N7H) originated through hybridization between the auto-allotetraploid C. neglecta (N6N7, n = 13) and diploid C. hispida (H, n = 7); the odd chromosome number in the inferred tetraploid C. neglecta was formed by hybridization between the ancestral (n = 7, N7) and derived C. neglecta–like genome (n = 6, N6), respectively.
To detect differences between hexaploid genomes of the cultivated C. sativa and its wild relative C. microcarpa, painting probes following the structure of 20 C. sativa chromosomes were identified in C. microcarpa. All 20 chromosome pairs were perfectly collinear in both hexaploid species (Supplemental Figure 8; Supplemental Data Set 1G). The identical chromosome number, subgenome composition, and chromosome structure indicate a common origin and recent genetic divergence of C. microcarpa and C. sativa.
Phylogenetic Analysis Further Supports the Allopolyploid Origin and Parentage of the Camelina Polyploids
To elucidate the phylogenetic relationships among the Camelina species, we provide additional support of parentage for polyploids and test possible hybridization events with 48 single-copy nuclear genes leveraged for the reconstruction of species trees and generation of phylogenetic networks. Four sets of data, with selective inclusion/exclusion of polyploid species, were analyzed (Supplemental Table 1): set 1 included all tested Camelina species, whereas the remaining data sets contained the diploids and (1) C. rumelica (set 2), (2) C. rumelica and C. sativa (set 3), and (3) C. rumelica and C. microcarpa (set 4).
Species trees were inferred for each set of data using the multi-species coalescent model in ASTRAL (Supplemental Figure 9) and maximum pseudo-likelihood estimate implemented in Maximum Pseudo-likelihood for Estimating Species Trees (MP-EST; Supplemental Figure 10). Species trees based on sets 1, 2, and 3 had the same topology in both analyses. Two main clades were identified: the first clade included the diploid C. neglecta being sister to the hexaploid C. microcarpa and C. sativa, while the second clade comprised the tetraploid C. rumelica together with the diploids C. hispida and C. laxa. Species trees based on set 4 differed between the two models. Whereas the ASTRAL tree was congruent with trees based on sets 1 and 3, the MP-EST tree retrieved C. hispida to be sister to C. rumelica, and C. laxa as sister to the remaining Camelina species. In both analyses, some clades had low statistical support (bootstrap value < 70%) due to reticulate evolution resulting from hybridization and polyploidization.
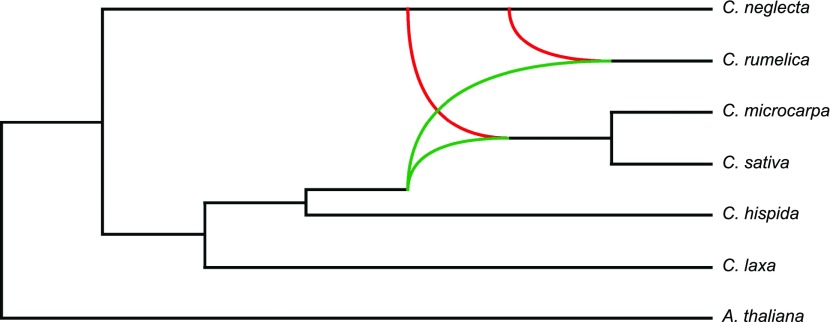
As the bifurcation branching pattern allowed in species trees cannot fully uncover complex evolutionary scenarios, we reconstructed phylogenetic networks in Phylonet testing for zero to three reticulation events. The most probable phylogenetic network, based on the all-species data (set 1) and assuming two hybridization events, is shown in Figure 5. These data suggest that C. laxa is the only species that did not participate in any hybridization event. The tetraploid C. rumelica originated through hybridization between diploid genomes closely related to C. hispida and C. neglecta. The second and independent hybridization between C. hispida (or closely related species) and C. neglecta led to the origin of a common hexaploid ancestor of C. sativa and C. microcarpa. The two independent hybridization events are also supported by the most probable phylogenetic networks for sets 3 and 4 (Supplemental Figure 11).
Figure 5.
Phylogenetic Relationships in the Genus Camelina Including the Origins of the Polyploid Genomes.
The tetraploid C. rumelica arose from hybridization between diploid genomes closely related to C. neglecta and C. hispida. A common allohexaploid ancestor of C. microcarpa and C. sativa originated from hybridization between a tetraploid C. neglecta–like genome and a diploid genome closely related to C. hispida. The species network based on set 1 (see Supplemental Table 1) was generated in Phylonet using the maximum pseudo-likelihood approach.
DISCUSSION
With detailed chromosomal analyses, we have characterized genomes of diploid Camelina species, identified genome-specific chromosomal signatures, and subsequently reconstructed the origin of Camelina allopolyploids. Our data corroborate the allohexaploid origin of C. sativa and C. microcarpa (Hutcheon et al., 2010) through a merger of three diploid (sub)genomes with seven or six chromosome pairs (Kagale et al., 2014).
By comparing genomes of the three extant diploid Camelina species (C. hispida, C. laxa, and C. neglecta), we have parsimoniously inferred the structure of an ancestral diploid Camelina genome (CAM, n = 7) preceding the divergence of diploid species. Although our CAM genome and the ancestral derivative of ACK genome, previously inferred from the C. sativa draft genome sequence (Kagale et al., 2014), are overall congruent, the CAM genome offers a higher precision of the ancestral genomic structures, including detailed characterization of chromosomal breakpoints. The CAM genome benefits from a more precise definition of ancestral GBs (Lysak et al., 2016) and incorporation of newly gained information on diploid genome structures. Furthermore, in derivative of ACK, descending dysploidy from n = 8 to n = 7 was interpreted as a fusion of chromosomes AK2 and AK4 (AK2/4) accompanied by several inversions. However, due to our analyses of diploid Camelina genomes, we were able to conclude that the descending dysploidy and formation of chromosomes CAM2 and CAM4 occurred through a chromothripsis-like rearrangement involving chromosomes AK2, AK4, and AK5.
We showed that all three extant Camelina polyploids originated through repeated hybridization between paleogenomes identical or closely related to C. hispida (H genome) and C. neglecta (N genome; Figures 5 and 6). While C. hispida was a long-recognized species, C. neglecta was recognized only recently (Brock et al., 2019) and more effort is needed to establish the full distribution range of the species as well as whether 2n = 12 is its exclusive chromosome number. The tetraploid C. rumelica (n = 13, N6H) originated through an interspecies hybridization between diploid genomes closely related to C. neglecta (n = 6, N6) and C. hispida (n = 7, H). Comparison of both C. rumelica subgenomes with the respective parental genomes revealed much more extensive shuffling of the recessive N subgenome and overall conservancy of the dominant H subgenome: post-polyploid genome reshuffling was restricted only to the N-subgenome chromosomes, whereas a single paracentric inversion was detected in the H subgenome.
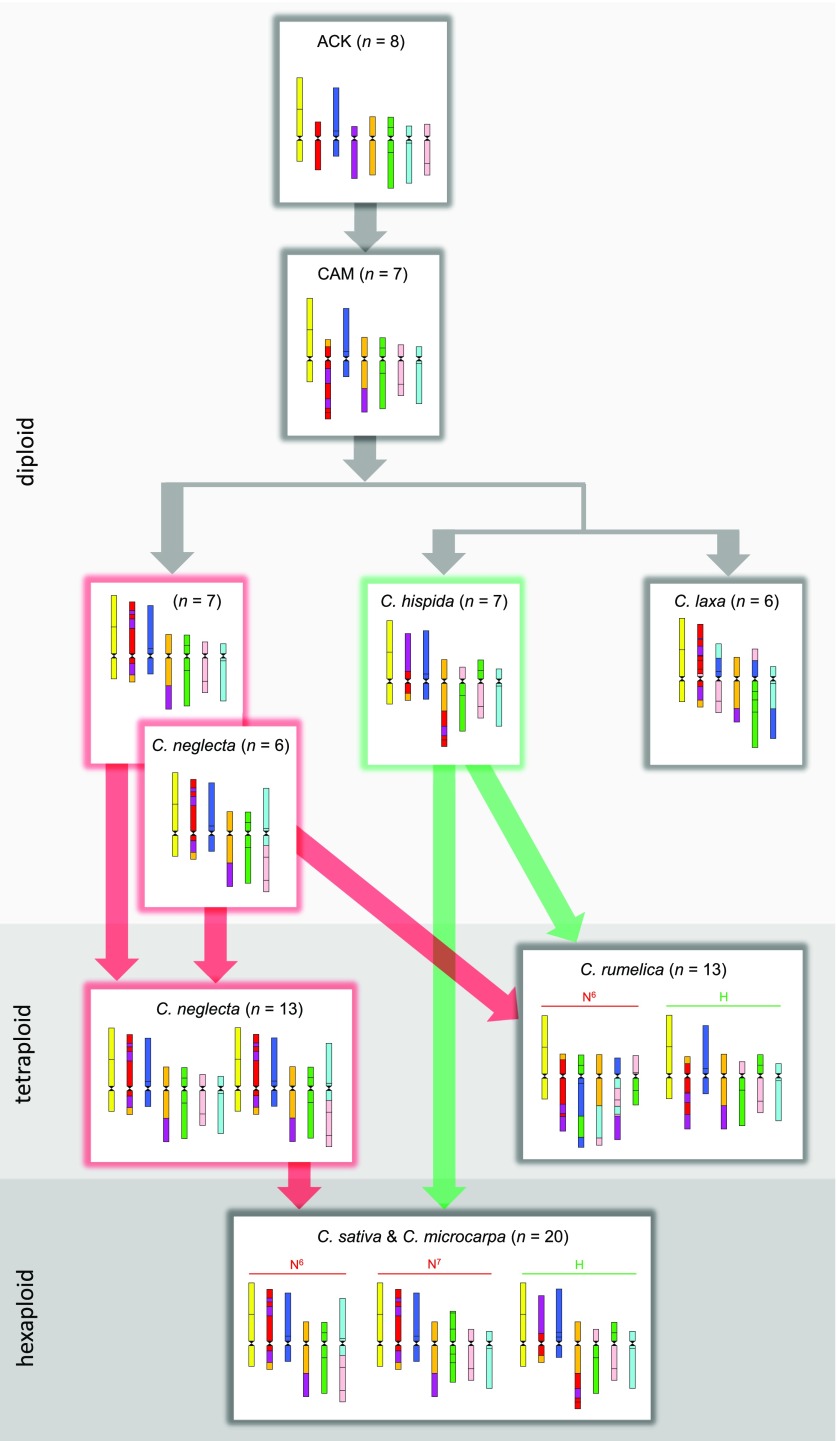
Figure 6.
Inferred Origins and Evolution of Camelina Genomes.
The CAM (n = 7) has evolved from an older genome with eight chromosomes (n = 8). The divergence of Camelina diploids was associated with species-specific genome reshuffling resulting in descending dysploidy in C. neglecta and C. laxa (both n = 6). Hybridization between two C. neglecta–like genomes with seven (n = 7, N7) and six (n = 6, N6) chromosomes led to the origin of an auto-allotetraploid C. neglecta–like genome (n = 13, N6N7). The allotetraploid genome of C. rumelica (n = 13, N6H) was formed upon hybridization between genomes related to C. neglecta (n = 6, N6) and C. hispida (n = 7, H). The allohexaploid progenitor genome of C. sativa and C. microcarpa (n = 20, N6N7H) originated through hybridization between the auto-allotetraploid C. neglecta–like genome (n = 13, N6N7) and C. hispida (n = 7, H).
The hexaploid genome of C. sativa (as C. microcarpa is genomically identical with C. sativa, both taxa are referred to as C. sativa hereafter) originated in two hybridization steps (Figure 6). The initial genome merger was either an intraspecific hybridization between two C. neglecta plants with deviating chromosome numbers (n = 7 and n = 6) or hybridization between closely related, recently diverged, populations or species differentiated by a n-1 descending dysploidy (N7, n = 7 and N6, n = 6). The tetraploid species has hybridized with the diploid C. hispida to form the allohexaploid progenitor genome of C. sativa and C. microcarpa (N6N7H, n = 20). In contrast to the C. rumelica genome, the two N and one H subgenomes in C. sativa remained remarkably stable since their merger: in the absence of any inter-subgenome rearrangements, only two intra-subgenome rearrangements differentiate subgenomes N7 and H from the parental ones (a paracentric inversion on chromosome Cs11 and centromere repositioning on Cs16). As the N subgenome in C. rumelica was restructured, whereas two N subgenomes in C. sativa remained conserved, we conclude that the allotetraploid C. rumelica originated earlier than the N6N7 tetraploid and the allohexaploid genome of C. sativa.
The absence of inter-subgenome chromosomal rearrangements mediated by homeologous recombination in C. sativa, C. microcarpa, and even in the older C. rumelica genome (Figure 6; Kagale et al., 2014; this study) corroborates the lacking subgenome dominance and fractionation bias in C. sativa (Kagale et al., 2014). Thus, Camelina allopolyploids can be put on the list of allopolyploid plant genomes with a general subgenome stability, such as Arabidopsis suecica (Novikova et al., 2017), Capsella bursa-pastoris (Douglas et al., 2015), Cardamine flexuosa (Mandáková et al., 2014), Cucurbita species (Sun et al., 2017), or teff (Eragrostis tef; VanBuren et al., 2019). It was proposed that the subgenome differences in the composition, densities, and regulation of transposable elements (TEs; Freeling et al., 2012; Bird et al., 2018; Edger et al., 2019) might be the primary cause of biased subgenome fractionation. If parental genomes show a comparable diversity of TEs and/or have generally low proportion of TEs, it can be hypothesized that post-polyploid genome fractionation will be unbiased. Although this is attractive, more factors, such as homeologous exchanges causing aberrant meiotic pairing and reduced fertility (Gaeta and Chris Pires, 2010), may play role in promoting long-term subgenome stability.
Evolution of Camelina Genomes Was Accompanied by Chromosome Shattering
Chromosome shattering, or chromothripsis, was recently discovered by cancer genome sequencing (Stephens et al., 2011). It is defined as an extensive intra-chromosomal cut-and-reassembly process accompanied by a high density of breakpoints and is known to be involved in both cancer and congenital diseases (Forment et al., 2012; Maher and Wilson, 2012; Korbel and Campbell, 2013). Multiple mechanisms of chromothripsis have been discussed, including evidence that abortive apoptosis, telomere erosion, mitotic errors, micronucleus formation, p53 inactivation, repetitive sequence, fragile site, or particular DNA conformations may stimulate chromothripsis occurrence (Ivkov and Bunz, 2015; Fukami et al., 2017; Pellestor and Gatinois, 2019). The evidence that plants can undergo the same extreme chromosome shattering observed in some human cancers and developmental syndromes is rather scarce and not yet conclusive. Tan et al. (2015) showed that Arabidopsis embryos resulting from a cross between a mutant with weakened centromeres and a wild-type plant underwent chromothripsis causing a shattered chromosome. In an alloplasmic wheat line (Triticum aestivum), multiple recombination events between two nonhomologous chromosomes generated a shattered chromosome built of alternating segments of the two chromosomes (Zhang et al., 2008). In the present study, using fine-scale CCP, we uncovered a complex, chromothripsis-like rearrangement as a key mechanism underlaying the formation of the shattered chromosome CAM2 in the ancestral CAM genome. The origin of CAM2 was associated with descending dysploidy from n = 8 in ACK to n = 7 in CAM preceding the species diversification in Camelina. Later, CAM2 underwent further independent species-specific chromosome shattering in C. laxa (Cl2) and C. neglecta (chromosome Cn2). Our data provide new evidence for chromothripsis-like events in naturally occurring plant genomes. As chromothripsis represents a mechanism of rapid and profound genome shattering, it might have the potential for rapid appearance of evolutionary novelties beneficial to their carriers. Conversely, the complex and abrupt nature of such catastrophic events may limit their offspring transmission and fixation. It can be hypothesized that some instances of chromosome shattering in plants occur in individuals (hybrids) heterozygous for gametocidal genes, that is, genes whose absence in gametes leads to chromosomal breakages and reduced gametic fitness. Although these chromosomal aberrations are frequently too severe, causing full gametic sterility, some semilethal rearrangements can become fixed after fertilization (Endo, 1990; Nasuda et al., 1998).
Descending Dysploidies in Camelina Were Mediated by Unusually Complex Chromosomal Rearrangements
The diploid Camelina genomes provide us with valuable insight into shuffling of an ancestral genome during species divergence. In Camelina diploids, descending dysploidy from n = 8 (ACK) to n = 7 (CAM), as well as dysploidy from n = 7 (CAM) to n = 6 (C. laxa), was mediated by complex chromosomal rearrangements, including chromothripsis-like events involving numerous breakages, inversions, shuffling of GBs, and a centromere loss. As CAM and all other ancestral genomes in lineage I/clade A have descended from ACK (Lysak et al., 2016), patterns of descending dysploidy can be compared across the clade. In contrast to Camelina, the reduction from n = 8 to n = 7 in Neslia paniculata (Camelineae; Lysak et al., 2006) was mediated by a single Robertsonian-like translocation, and a single end-to-end chromosome fusion in Boechera (Boechereae; Mandáková et al., 2015) and Descurainia (Descurainieae; Mandáková et al., 2017). Similarly, descending dysploidy from n = 8 to n = 6 in Hornungia alpina (Descurainieae; Lysak et al., 2006) was mediated by a single end-to-end chromosome fusion and nested chromosome insertion, respectively, and similar dysploidy in Smelowskia altaica (Smelowskieae; Mandáková et al., 2017) and Turritis glabra (without tribal assignment; Lysak et al., 2006) were mediated by a single end-to-end chromosome fusion and Robertsonian-like translocation, respectively. Genomes of the two n = 6 Camelina species were formed through descending dysploidy with contrasting patterns. In C. neglecta, dysploidy from CAM was mediated by a single end-to-end chromosome fusion, whereas three fusion chromosomes in C. laxa originated by chromosome shattering. The number of breakpoints generated during chromothripsis-related dysploidies in CAM and C. laxa exceeds even the breakpoints during the most extreme descending dysploidy from n = 8 in ACK to n = 5 in Arabidopsis, mediated by a single end-to-end fusion and two Robertsonian-like translocations (Lysak et al., 2006, 2016).
The Domesticated C. sativa Descended from the Weedy C. microcarpa
Here, we show identical subgenome composition and chromosome structure in C. microcarpa and C. sativa. These findings further validate previous findings showing C. sativa as being domesticated from the wild hexaploid C. microcarpa (Brock et al., 2018). The identification of C. microcarpa as the pre-domesticate allows for further studies in the process of redomestication and introgression of traits via breeding. However, little is known about the process of domestication in this taxon, although future population genetic studies in C. microcarpa may elucidate the demographic history of C. sativa. Hybrid formation between C. microcarpa and C. sativa has been observed (Séguin-Swartz et al., 2013; Martin et al., 2018) and may prove to be a valuable tool in generating novel cultivars of C. sativa with drought tolerance, disease resistance, and earlier flowering.
Camelina as a Model for Polyploidy and Evolutionary Studies
While Camelina and Arabidopsis are closely related and belong to the same crucifer clade (lineage I or clade A), the mesohexaploid Brassiceae belongs to lineage II or clade B (Franzke et al., 2011). Thus, the unrivalled Arabidopsis genomic resources can be more easily exploited for studies of (post-)polyploid genome evolution in Camelina than in Brassica. Moreover, due to the extinction of Brassiceae progenitor genomes and progressed rediploidization of Brassica genomes, these (sub)genomes cannot be compared with parental diploid genomes as it is feasible in the Camelina polyploid complex. Identifying diploid parental genomes of the tetraploid and hexaploid Camelina species opens the door not only for studies of (post-)polyploid genome evolution but also for creating lines of C. sativa with improved traits and potential resynthesis of polyploid Camelina genomes. With the ever-rising interest in false flax for aviation biofuels, production of high-value molecules, and as an omega-3–rich feedstock, the time is ripe to develop Camelina as a model system for the field of polyploidy research.
CONCLUSIONS
All three extant Camelina polyploids originated trough recurrent hybridization between paleogenomes identical or closely related to C. hispida (H genome) and C. neglecta (N genome). Whereas the allotetraploid C. rumelica genome shows signs of post-polyploid subgenome-specific fractionation, the three subgenomes in the allohexaploid C. sativa genome remained conserved since the genome merger. Remarkably, descending dysploidy preceding the origin of the ancestral CAM genome was accompanied by complex chromosome shattering involving numerous breakages, shuffling of GBs, and a centromere loss. Equally, the origins of fusion chromosomes in the C. laxa were mediated by chromosome shattering. The genome-shattering alterations, resembling those associated with human disorders, have been reported rarely in naturally occurring plant species. The identical genome structure of C. sativa and C. microcarpa points to their common origin and corroborates C. microcarpa as the wild hexaploid pre-domesticate of C. sativa. Identifying diploid parental genomes of the tetraploid and hexaploid Camelina species opens the door for creating lines of C. sativa with improved traits and potential resynthesis of polyploid Camelina genomes and allows for adopting Camelina as a model system for polyploidy research.
METHODS
Plant Material
The list of investigated accessions and their origin is provided in Supplemental Table 2. The experimental plants were grown from seeds in a growth chamber under the following conditions: temperature 21/18°C, daylength 16/8 h, light intensity 150 µmol/m2/s. Young inflorescences were collected and fixed in freshly prepared fixative (ethanol:acetic acid, 3:1) overnight, transferred into 70% (v/v) ethanol, and stored at −20°C until use. Young leaves were used directly or dried in silica gel. gDNA was extracted from harvested leaves of 22 Camelina accessions (Supplemental Table 2) using a NucleoSpin Plant II kit (Macherey-Nagel).
Chromosome Preparations
Chromosome spreads from young fixed flower buds, containing immature anthers, were prepared according to a published protocol (Mandáková and Lysak, 2016a). Briefly, selected flower buds were rinsed in distilled water (twice for 5 min) and citrate buffer (10 mM sodium citrate, pH 4.8; twice for 5 min), and digested in 0.3% (w/v) cellulase, cytohelicase, and pectolyase (all from Sigma-Aldrich) in citrate buffer at 37°C for 3 h. After digestion, individual anthers were dissected on a microscopic slide in 20 μL of 60% (v/v) acetic acid, spread on the slide, and placed on a metal hot plate (50°C) for ∼30 s, and fixed in freshly prepared fixative (ethanol:acetic acid, 3:1). Suitable slides containing well-spread pachytene and/or mitotic chromosomes were post-fixed in freshly prepared 4% (v/v) formaldehyde in distilled water for 10 min and air dried. Preparations were treated with 100 mg/mL RNase (AppliChem) in 2× SSC at 37°C for 60 min, and 0.1 mg/mL pepsin (Sigma-Aldrich) in 0.01 M HCl at 37°C for 5 min, and then post-fixed in 4% (v/v) formaldehyde in 2× SSC for 10 min, washed in 2× SSC twice for 5 min, and dehydrated in an ethanol series (70, 90, and 100% [v/v], 2 min each).
DNA Probes
In total, 674 chromosome-specific BAC clones of Arabidopsis (Arabidopsis thaliana) grouped into contigs according to eight chromosomes and 22 GBs (A to X) of the ACK (Supplemental Data Set 1B; Lysak et al., 2016) were used. To determine fine-scale chromosome structures, uncover species-specific chromosome rearrangements, and precisely characterize breakpoints, after initial CCP experiments, some BAC contigs were split into shorter subcontigs. In the case of highly reshuffled chromosome regions, individual differentially labeled BAC clones were used. gDNAs of Camelina hispida, Camelina laxa, and Camelina neglecta were used as probes in GISH experiments in Camelina polyploids (Camelina microcarpa, Camelina rumelica, and Camelina sativa). Individual BAC clones and gDNAs were labeled with biotin-dUTP, digoxigenin-dUTP, or Cy3-dUTP by nick translation as described previously (Mandáková and Lysak, 2016b). In CCP experiments, the concentration of each labeled BAC clone DNA varied from 100 ng per BAC clone/slide in diploids (C. hispida, C. laxa, and C. neglecta), to 300 ng in tetraploid C. rumelica, and up to 500 ng in hexaploids (C. microcarpa and C. sativa). For GISH experiments, 150 ng of each gDNA per slide were used. Labeled BAC clones or gDNAs were pooled following a given experimental design, ethanol precipitated, dried using a desiccator, and dissolved in 20 μL of 50% (v/v) formamide and 10% (w/v) dextran sulfate in 2× SSC per slide.
Fluorescence in Situ Hybridization, Microscopy, and Image Processing
For CCP and fluorescence in situ hybridization, 20 μL of the hybridization mix was pipetted on a suitable slide and immediately denatured on a hot plate at 80°C for 2 min. For GISH, 20 μL of the probe was denatured in a microfuge tube at 90°C for 10 min, placed on ice for 10 min, pipetted on a slide, and denatured on a hot plate at 80°C for 2 min. Hybridization was performed in a moist chamber at 37°C for 32 to 48 h. Post-hybridization washing was performed in 20% formamide in 2× SSC at 42°C. The immunodetection of hapten-labeled probes was performed as described by Mandáková and Lysak (2016b) as follows: biotin-dUTP was detected by avidin–Texas Red (Vector Laboratories) and amplified by goat anti-avidin–biotin (Vector Laboratories) and avidin–Texas Red; digoxigenin-dUTP was detected by mouse anti-digoxigenin (Jackson ImmunoResearch) and goat anti-mouse–Alexa Fluor 488 (Invitrogen). Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 2 μg/mL) in Vectashield. The preparations were photographed using an Axioimager Z2 epifluorescence microscope (Zeiss) with a CoolCube camera (MetaSystems). Images were acquired separately for all four fluorochromes using appropriate excitation and emission filters (AHF Analysentechnik). The four monochromatic images were pseudocolored and merged using Photoshop CS (Adobe Systems) and cropped using ImageJ (National Institutes of Health) software. Pachytene chromosomes were straightened using the straighten-curved-objects plugin in ImageJ (Kocsis et al., 1991). Circular visualizations of chromosome-scale pseudomolecules were prepared using Circos (Krzywinski et al., 2009).
To identify chromosomes from individual subgenomes in Camelina polyploids, CCP was combined with GISH (CCP/GISH; Mandáková et al., 2014). After initial CCP experiment, hybridization patterns were analyzed and photographed, and painting probes were removed by washing the slides in 2× SSC and 20% (v/v) formamide in 2× SSC at 42°C (5 min each). Subsequently, the slides were post-fixed in 4% (v/v) formaldehyde in 2× SSC for 10 min, washed in 2× SSC (twice for 5 min), and dehydrated in an ethanol series (70, 90, and 100% [v/v], 2 min each). The subsequent denaturation, hybridization and immunodetection steps were performed as described above.
Phylogenetic Analysis
In total, 48 nuclear markers (Supplemental Data Set 1C) targeting coding regions were selected based on Stockenhuber et al. (2015). Candidate markers were analyzed using BLAST searches against the genome sequence of C. sativa (PRJNA264159; Kagale et al., 2014) to identify conserved primer binding sites. The Access Array System (Fluidigm) was used to generate PCR amplicons for all the 48 markers, following the manufacturer’s protocol. PCR products were subsequently sequenced on the Illumina-MiSeq platform (2 × 300-bp paired-end reads). The quality check of raw reads was performed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Poor-quality reads and Illumina adaptors were removed using Trimmomatic 0.36 (Bolger et al., 2014). The HybPiper 1.3.1 pipeline (Johnson et al., 2016) was used to extract haplotypes of homeologous sequences for each gene in all accessions analyzed. The set of python scripts performed three main steps: (1) reads were searched and sorted according to a target gene, (2) assembled contigs were aligned to the reference genome, and (3) potential paralogous sequences were identified. Four sets of data were generated (Supplemental Table 1): set 1 included haplotypes of all tested species and accessions, while sets 2 to 4 included consensus sequences for all diploid species and haplotype sequences for partial taxon sampling of Camelina polyploids; only C. rumelica in data set 2, C. rumelica and C. sativa in data set 3, and C. rumelica and C. microcarpa in data set 4. Arabidopsis, which is generally accepted to be paraphyletic to core Camelina species (Nikolov et al., 2019), was selected as the outgroup in the phylogenetic study.
Multiple alignments were generated for each marker using MAFFT v.7.407 (Katoh and Standley, 2013), using default settings, automatically trimmed using Phyutility 2.2 (Smith and Dunn, 2008) and manually checked in Geneious 11.1.5. (https://www.geneious.com). Maximum likelihood (ML) trees were inferred using RAxML 8.2.12 (Stamatakis, 2014) with GTR+G substitution model and rapid bootstrap with 100 replicates. The best ML gene trees were compared against 100 random trees using the Shimodaira–Hasegawa test implemented in RAxML to test sufficient phylogenetic signal in alignments, whereby only trees that passed the test were used in the subsequent step. RAxML gene trees are shown in Supplemental File. Low support branches (bootstrap < 10%) of gene trees were collapsed using the Newick utilities 1.6 (Junier and Zdobnov, 2010) to get higher accuracy of species tree analyses. The ASTRAL 5.6.3 (Rabiee et al., 2019) and MP-EST method implemented in the STRAW: Species TRee Analysis Web server (Shaw et al., 2013) were used to estimate species trees from a set of gene trees and bootstrap replicates. The branch support values of species trees were generated using 100 bootstrap replicates.
As a classical species tree cannot explain reticulate types of evolution, we analyzed potential hybridization scenarios in Phylonet 3.6.9 (Wen et al., 2018). The maximum pseudo-likelihood approach was used, enabling testing of hybridization events under the influence of incomplete lineage sorting and is not as computationally exhaustive as ML method. The following settings were used: maximum number of reticulations (hybridizations), 0 to 3; number of optimal networks, 10; and gene trees bootstrap threshold, 50. The obtained phylogenetic networks were visualized and manipulated in Dendroscope 3.5.10 (Huson and Scornavacca, 2012).
Accession Numbers
Sequence data from this work can be found under the accession numbers listed in Supplemental Table 3.
Supplemental Data
Supplemental Figure 1. Structure of the ancestral Camelina genome (CAM, n = 7) and its origin from Ancestral Crucifer Karyotype (ACK, n = 8).
Supplemental Figure 2. Genome structure and the origin of C. hispida (n = 7) from CAM.
Supplemental Figure 3. Genome structure and the origin of C. neglecta (n = 6) from CAM.
Supplemental Figure 4. Genome structure and the origin of C. laxa (n = 6).
Supplemental Figure 5. Genome structure and the origin of C. laxa (n = 6).
Supplemental Figure 6. Genome structure of C. rumelica.
Supplemental Figure 7. Genome structure and the origin of C. rumelica.
Supplemental Figure 8. Genome structure and the origin of C. sativa and C. microcarpa.
Supplemental Figure 9. Species trees of all data sets generated in ASTRAL.
Supplemental Figure 10. Species trees of all data sets generated in MP-EST.
Supplemental Figure 11. The most probable networks of all data sets generated in Phylonet.
Supplemental Table 1. Scheme of data sets used in the phylogenetic analyses.
Supplemental Table 2. Collection data of Camelina species used in the present study.
Supplemental Table 3. List of used markers with gene IDs and lengths of theoretical amplicons.
Supplemental Data Set 1A. Structure of the eight chromosomes and 22 genomic blocks (GBs, A-X) of Ancestral Crucifer Karyotype (ACK).
Supplemental Data Set 1B. Genome structure of C. hispida.
Supplemental Data Set 1C. Genome structure of C. neglecta.
Supplemental Data Set 1D. Genome structure of C. laxa.
Supplemental Data Set 1E. Genome structure of the inferred ancestral Camelina genome (CAM).
Supplemental Data Set 1F. Genome structure of C. rumelica.
Supplemental Data Set 1G. Genome structure of C. sativa and C. microcarpa.
Supplemental File. Maximum likelihood gene trees for all four sets.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Petra Hloušková and Martin Krzywinski for their assistance with the Circos software package and construction of the hive plot. We are grateful to Herbert Hurka, Isobel Parkin, and Murat Ünal for providing seeds. CEITEC’s core facilities Plant Sciences and Genomics are acknowledged for the cultivation of experimental plants and DNA sequencing, respectively. Access to computing and storage facilities owned by parties and projects contributing to the National Grid Infrastructure MetaCentrum provided under the programme Projects of Large Research, Development, and Innovations Infrastructures (CESNET LM2015042) was greatly appreciated. This work was supported by a research grant from the Czech Science Foundation (grant 17-13029S) and by the CEITEC 2020 project (grant LQ1601).
AUTHOR CONTRIBUTIONS
T.M., J.R.B., and M.A.L. conceived and designed the study. T.M. and M.P. performed the experiments. I.A.A. contributed to the interpretation of results. T.M., M.P., J.R.B., and M.A.L. wrote the article. All authors read and approved the final article.
References
- Augustin J.M., Higashi Y., Feng X., Kutchan T.M. (2015). Production of mono- and sesquiterpenes in Camelina sativa oilseed. Planta 242: 693–708. [DOI] [PubMed] [Google Scholar]
- Augustin M.M., Shukla A.K., Starks C.M., O’Neil-Johnson M., Han L., Holland C.K., Kutchan T.M. (2017). Biosynthesis of Veratrum californicum specialty chemicals in Camelina sativa seeds. Plant Biotechnol. Rep. 11: 29–41. [Google Scholar]
- Beilstein M.A., Al-Shehbaz I.A., Kellogg E.A. (2006). Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93: 607–619. [DOI] [PubMed] [Google Scholar]
- Beilstein M.A., Al-Shehbaz I.A., Mathews S., Kellogg E.A. (2008). Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: Tribes and trichomes revisited. Am. J. Bot. 95: 1307–1327. [DOI] [PubMed] [Google Scholar]
- Bird K.A., VanBuren R., Puzey J.R., Edger P.P. (2018). The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 220: 87–93. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock J.R., Dönmez A.A., Beilstein M.A., Olsen K.M. (2018). Phylogenetics of Camelina Crantz. (Brassicaceae) and insights on the origin of gold-of-pleasure (Camelina sativa). Mol. Phylogenet. Evol. 127: 834–842. [DOI] [PubMed] [Google Scholar]
- Brock J.R., Mandáková T., Lysak M.A., Al-Shehbaz I.A. (2019). Camelinaneglecta (Brassicaceae, Camelineae), a new diploid species from Europe. PhytoKeys 115: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.W., Donoghue P.C.J. (2017). Constraining the timing of whole genome duplication in plant evolutionary history. Proc. Biol. Sci. 284: 20170912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G.M., et al. (2015). Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc. Natl. Acad. Sci. USA 112: 2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P.P., et al. (2019). Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51: 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M., Murat F., Veyssiere M., Molinier M., Flores R., Burlot L., Alaux M., Quesneville H., Pont C., Salse J. (2017). Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol. 213: 1477–1486. [DOI] [PubMed] [Google Scholar]
- Endo T.R. (1990). Gametocidal chromosomes and their induction of chromosome mutations in wheat. Jpn. J. Genet. 65: 135–152. [Google Scholar]
- Forment J.V., Kaidi A., Jackson S.P. (2012). Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 12: 663–670. [DOI] [PubMed] [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116. [DOI] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Fukami M., Shima H., Suzuki E., Ogata T., Matsubara K., Kamimaki T. (2017). Catastrophic cellular events leading to complex chromosomal rearrangements in the germline. Clin. Genet. 91: 653–660. [DOI] [PubMed] [Google Scholar]
- Gaebelein R., Mason A.S. (2018). Allohexaploids in the genus Brassica. Crit. Rev. Plant Sci. 37: 422–437. [Google Scholar]
- Gaeta R.T., Chris Pires J. (2010). Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol. 186: 18–28. [DOI] [PubMed] [Google Scholar]
- Garsmeur O., Schnable J.C., Almeida A., Jourda C., D’Hont A., Freeling M. (2014). Two evolutionarily distinct classes of paleopolyploidy. Mol. Biol. Evol. 31: 448–454. [DOI] [PubMed] [Google Scholar]
- Gehringer A., Friedt W., Lühs W., Snowdon R.J. (2006). Genetic mapping of agronomic traits in false flax (Camelina sativa subsp. sativa). Genome 49: 1555–1563. [DOI] [PubMed] [Google Scholar]
- Hu Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51: 739–748. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. (2012). Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Hutcheon C., Ditt R.F., Beilstein M., Comai L., Schroeder J., Goldstein E., Shewmaker C.K., Nguyen T., De Rocher J., Kiser J. (2010). Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol. 10: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskandarov U., Kim H.J., Cahoon E.B. (2014). Camelina: an emerging oilseed platform for advanced biofuels and bio-based materials In Plants and Bioenergy, McCann M.C., Buckeridge M.S., and Carpita N.C., eds (Berlin: Springer; ), pp. 131–140. [Google Scholar]
- Iven T., Hornung E., Heilmann M., Feussner I. (2016). Synthesis of oleyl oleate wax esters in Arabidopsis thaliana and Camelina sativa seed oil. Plant Biotechnol. J. 14: 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkov R., Bunz F. (2015). Pathways to chromothripsis. Cell Cycle 14: 2886–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.Z., Henry I.M., Lynagh P.G., Comai L., Cahoon E.B., Weeks D.P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ. 2016. HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant. Sci. 4: apps.1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T., Zdobnov E.M. (2010). The Newick utilities: High-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26: 1669–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., et al. (2014). The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 5: 3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsis E., Trus B.L., Steer C.J., Bisher M.E., Steven A.C. (1991). Image averaging of flexible fibrous macromolecules: The clathrin triskelion has an elastic proximal segment. J. Struct. Biol. 107: 6–14. [DOI] [PubMed] [Google Scholar]
- Korbel J.O., Campbell P.J. (2013). Criteria for inference of chromothripsis in cancer genomes. Cell 152: 1226–1236. [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidou M., Tai H.H., Anglin N.L., Ellis D., Strömvik M.V. (2018). Current strategies of polyploid plant genome sequence assembly. Front. Plant Sci. 9: 1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J.B., Soltis D.E., Li Z., Marx H.E., Barker M.S., Tank D.C., Soltis P.S. (2018). Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 105: 348–363. [DOI] [PubMed] [Google Scholar]
- Lu C., Kang J. (2008). Generation of transgenic plants of a potential oilseed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Rep. 27: 273–278. [DOI] [PubMed] [Google Scholar]
- Luo Z., Brock J., Dyer J.M., Kutchan T., Schachtman D., Augustin M., Ge Y., Fahlgren N., Abdel-Haleem H. (2019). Genetic diversity and population structure of Camelina sativa spring panel. Front. Plant Sci. 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Berr A., Pecinka A., Schmidt R., McBreen K., Schubert I. (2006). Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Mandáková T., Schranz M.E. (2016). Comparative paleogenomics of crucifers: Ancestral genomic blocks revisited. Curr. Opin. Plant Biol. 30: 108–115. [DOI] [PubMed] [Google Scholar]
- Maher C.A., Wilson R.K. (2012). Chromothripsis and human disease: Piecing together the shattering process. Cell 148: 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Kovarík A., Zozomová-Lihová J., Shimizu-Inatsugi R., Shimizu K.K., Mummenhoff K., Marhold K., Lysak M.A. (2013). The more the merrier: Recent hybridization and polyploidy in cardamine. Plant Cell 25: 3280–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A. (2016a). Chromosome preparation for cytogenetic analyses in Arabidopsis. Curr. Protoc. Plant Biol. 1: 43–51. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A. (2016b). Painting of Arabidopsis chromosomes with chromosome-specific BAC clones. Curr. Protoc. Plant Biol. 1: 359–371. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Marhold K., Lysak M.A. (2014). The widespread crucifer species Cardamine flexuosa is an allotetraploid with a conserved subgenomic structure. New Phytol. 201: 982–992. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Pouch M., Harmanová K., Zhan S.H., Mayrose I., Lysak M.A. (2017). Multispeed genome diploidization and diversification after an ancient allopolyploidization. Mol. Ecol. 26: 6445–6462. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Schranz M.E., Sharbel T.F., de Jong H., Lysak M.A. (2015). Karyotype evolution in apomictic Boechera and the origin of the aberrant chromosomes. Plant J. 82: 785–793. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Zozomová-Lihová J., Kudoh H., Zhao Y., Lysak M.A., Marhold K. (2019). The story of promiscuous crucifers: Origin and genome evolution of an invasive species, Cardamine occulta (Brassicaceae), and its relatives. Ann. Bot. 124: 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T., Sandve S.R., Heier L., Spannagl M., Pfeifer M., Jakobsen K.S., Wulff B.B., Steuernagel B., Mayer K.F., Olsen O.A.; International Wheat Genome Sequencing Consortium (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- Mason A.S., Snowdon R.J. (2016). Oilseed rape: Learning about ancient and recent polyploid evolution from a recent crop species. Plant Biol (Stuttg) 18: 883–892. [DOI] [PubMed] [Google Scholar]
- Morineau C., Bellec Y., Tellier F., Gissot L., Kelemen Z., Nogué F., Faure J.D. (2017). Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15: 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.L., Lujan-Toro B.E., Sauder C.A., James T., Ohadi S., Hall L.M. (2018). Hybridization rate and hybrid fitness for Camelina microcarpa Andrz. ex DC (♀) and Camelina sativa (L.) Crantz (Brassicaceae) (♂). Evol. Appl. 12: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser B.R. (2012). Biodiesel from alternative oilseed feedstocks: Camelina and field pennycress. Biofuels 3: 193–209. [Google Scholar]
- Nagaharu U., Nagaharu N. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Nasuda S., Friebe B., Gill B.S. (1998). Gametocidal genes induce chromosome breakage in the interphase prior to the first mitotic cell division of the male gametophyte in wheat. Genetics 149: 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov L.A., Shushkov P., Nevado B., Gan X., Al-Shehbaz I.A., Filatov D., Bailey C.D., Tsiantis M. (2019). Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 222: 1638–1651. [DOI] [PubMed] [Google Scholar]
- Novikova P.Y., et al. (2017). Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol. Biol. Evol. 34: 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozseyhan M.E., Kang J., Mu X., Lu C. (2018). Mutagenesis of the FAE1 genes significantly changes fatty acid composition in seeds of Camelina sativa. Plant Physiol. Biochem. 123: 1–7. [DOI] [PubMed] [Google Scholar]
- Parisod C., Holderegger R., Brochmann C. (2010). Evolutionary consequences of autopolyploidy. New Phytol. 186: 5–17. [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. [DOI] [PubMed] [Google Scholar]
- Pellestor F., Gatinois V. (2019). Chromothripsis, a credible chromosomal mechanism in evolutionary process. Chromosoma 128: 1–6. [DOI] [PubMed] [Google Scholar]
- Rabiee M., Sayyari E., Mirarab S. (2019). Multi-allele species reconstruction using ASTRAL. Mol. Phylogenet. Evol. 130: 286–296. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S., Kovarik A., Kelly L.J., Macas J., Novak P., Chase M.W., Nichols R.A., Pancholi M.R., Grandbastien M.A., Leitch A.R. (2013). Diploidization and genome size change in allopolyploids is associated with differential dynamics of low- and high-copy sequences. Plant J. 74: 829–839. [DOI] [PubMed] [Google Scholar]
- Ruiz-Lopez N., Haslam R.P., Napier J.A., Sayanova O. (2014). Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J. 77: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A., Ainouche M.L., Wendel J.F. (2005). Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14: 1163–1175. [DOI] [PubMed] [Google Scholar]
- Séguin‐Swartz G., Nettleton J.A., Sauder C., Warwick S.I., Gugel R.K. (2013). Hybridization between Camelina sativa (L.) Crantz (false flax) and North American Camelina species. Plant Breed. 132: 390–396. [Google Scholar]
- Shaw T.I., Ruan Z., Glenn T.C., Liu L. (2013). STRAW: Species TRee Analysis Web server. Nucleic Acids Res. 41: W238-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonnard D.R., Williams L., Kalnes T.M. (2010). Camelina-derived jet fuel and diesel: Sustainable advanced biofuels. Environ. Prog. Sustain. Energy 29: 382–392. [Google Scholar]
- Sierro N., Battey J.N., Ouadi S., Bakaher N., Bovet L., Willig A., Goepfert S., Peitsch M.C., Ivanov N.V. (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 5: 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.A., Dunn C.W. (2008). Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24: 715–716. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G.L. (1971). Chromosomal Evolution in Higher Plants.. (London: Edward Arnold; ). [Google Scholar]
- Stephens P.J., et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockenhuber R., Zoller S., Shimizu-Inatsugi R., Gugerli F., Shimizu K.K., Widmer A., Fischer M.C. (2015). Efficient detection of novel nuclear markers for Brassicaceae by transcriptome sequencing. PLoS One 10: e0128181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., et al. (2017). Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita Genomes. Mol. Plant 10: 1293–1306. [DOI] [PubMed] [Google Scholar]
- Symonds V.V., Soltis P.S., Soltis D.E. (2010). Dynamics of polyploid formation in Tragopogon (Asteraceae): Recurrent formation, gene flow, and population structure. Evolution 64: 1984–2003. [DOI] [PubMed] [Google Scholar]
- Tan E.H., Henry I.M., Ravi M., Bradnam K.R., Mandakova T., Marimuthu M.P., Korf I., Lysak M.A., Comai L., Chan S.W. (2015). Catastrophic chromosomal restructuring during genome elimination in plants. eLife 4: e06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R., Wai C.M., Pardo J., Yocca A.E., Wang X., Wang H., Chaluvadi S.R., Bryant D., Edger P.E., Bennetzen J.L., Mockler T.C., Michael T.P. (2019). Exceptional subgenome stability and functional divergence in allotetraploid teff, the primary cereal crop in Ethiopia. bioRxiv (preprint) 10.1101/580720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmann J., Grausgruber H., Stift G., Dryzhyruk C., Lelley T. (2005). Genetic diversity in Camelina germplasm as revealed by seed quality characteristics and RAPD polymorphism. Plant Breed. 124: 446–453. [Google Scholar]
- Waltz E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36: 6–7. [DOI] [PubMed] [Google Scholar]
- Wang X., et al. ; Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wen D., Yu Y., Zhu J., Nakhleh L. (2018). Inferring phylogenetic networks using PhyloNet. Syst. Biol. 67: 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., et al. (2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48: 1225–1232. [DOI] [PubMed] [Google Scholar]
- Zhang P., Li W., Friebe B., Gill B.S. (2008). The origin of a “zebra” chromosome in wheat suggests nonhomologous recombination as a novel mechanism for new chromosome evolution and step changes in chromosome number. Genetics 179: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]