The lack of a shade-induced hypocotyl elongation response in Cardamine hirsuta results from the enhanced repressor activity of the phytochrome A photoreceptor.
Abstract
Plants have evolved two major ways to deal with nearby vegetation or shade: avoidance and tolerance. Moreover, some plants respond to shade in different ways; for example, Arabidopsis (Arabidopsis thaliana) undergoes an avoidance response to shade produced by vegetation, but its close relative Cardamine hirsuta tolerates shade. How plants adopt opposite strategies to respond to the same environmental challenge is unknown. Here, using a genetic strategy, we identified the C. hirsuta slender in shade1 mutants, which produce strongly elongated hypocotyls in response to shade. These mutants lack the phytochrome A (phyA) photoreceptor. Our findings suggest that C. hirsuta has evolved a highly efficient phyA-dependent pathway that suppresses hypocotyl elongation when challenged by shade from nearby vegetation. This suppression relies, at least in part, on stronger phyA activity in C. hirsuta; this is achieved by increased ChPHYA expression and protein accumulation combined with a stronger specific intrinsic repressor activity. We suggest that modulation of photoreceptor activity is a powerful mechanism in nature to achieve physiological variation (shade tolerance versus avoidance) for species to colonize different habitats.
INTRODUCTION
Understanding how plants colonize different habitats requires identifying the genetic differences underlying physiological variation between species. In this work, we focus on angiosperm responses to changes in light produced by nearby vegetation, perception of which alerts the plant to potential resource competition by other plants. Nearby vegetation is perceived as changes in light parameters: whereas sunlight has a high red light (R) to far-red light (FR) ratio (R:FR > 1.1), proximity to vegetation lowers this ratio (Smith, 1982). Because vegetation specifically reflects FR, proximity to other plants initially results in a mild reduction in R:FR (<0.7) due to the FR enrichment. Eventually, when the vegetation canopy closes, sunlight is filtered by photosynthetic tissues, strongly reducing the intensity of the PAR (400–700 nm, which includes blue and R) while marginally affecting FR. As a result, R:FR resulting from natural canopy shade typically drops to lower values (<0.05; Smith, 1982; Casal, 2012; Martínez-García et al., 2014; de Wit et al., 2016). In the laboratory, both vegetation proximity and canopy shade can be simulated by providing plants grown under white light (W; high R:FR) varying amounts of supplemental FR (W+FR; low or very low R:FR) while maintaining total PAR, a treatment known as simulated shade (Casal, 2012; Roig-Villanova and Martínez-García, 2016).
Plants have two main strategies to acclimate to vegetation proximity and shade: avoidance or tolerance. In the early stages of development, shade-avoider species invest energy into promoting elongation to overgrow their neighbors as part of the so-called shade avoidance syndrome (SAS). By contrast, shade-tolerant plants adopt other physiological and metabolic responses to adapt to a highly conservative utilization of resources, commonly accompanied by very low growth rates (i.e., do not involve promotion of elongation growth; Smith, 1982; Valladares and Niinemets, 2008).
Analyses of the shade-avoider Arabidopsis (Arabidopsis thaliana) laid the basis for our knowledge of the genetic components and mechanisms involved in the regulation of the SAS (Martínez-García et al., 2010; Casal, 2012; Roig-Villanova and Martínez-García, 2016). The shade signal is perceived by the phytochrome photoreceptors: phytochrome B (phyB) and phyA have major and antagonistic roles (respectively) in hypocotyl elongation, the most conspicuous Arabidopsis response to low R:FR (Mathews, 2010; Casal, 2012). Lowering the R:FR to resemble either vegetation proximity or canopy shading deactivates phyB in wild-type seedlings, resulting in the hypocotyl elongation promotion. By contrast, phyA accumulates and is strongly activated under very low R:FR to prevent excessive seedling elongation (Martínez-García et al., 2014; Yang et al., 2018). Consistent with this, Arabidopsis phyB-deficient mutants display constitutive shade responses under high R:FR, whereas phyA mutant seedlings show enhanced hypocotyl elongation only under very low R:FR conditions, which indicates that phyA antagonizes phyB activity under these specific canopy shade conditions (Yanovsky et al., 1995; Casal et al., 2014; Martínez-García et al., 2014; Yang et al., 2018).
SAS responses are mainly initiated because of the interaction of active phytochromes with PHYTOCHROME INTERACTING FACTORs (PIFs), eventually triggering rapid changes in the expression of dozens of genes that implement the SAS responses. Genetic analyses in Arabidopsis indicate that PIFs, which are basic helix-loop-helix transcription factors, have a role in positively regulating the shade-triggered hypocotyl elongation. The active form of phyB interacts with PIFs and inhibits their transcriptional activity (Martinez-Garcia et al., 2010; Casal, 2012). After exposure to shade, the proportion of active phyB decreases and PIF activity increases. Enhanced PIF binding to G-boxes of auxin biosynthetic genes (e.g., YUCCA genes) then promotes their expression, which results in a rapid (1–4 h) increase in free indole-3-acetic acid (IAA) that is required for the promotion of shade-induced hypocotyl elongation (Tao et al., 2008; Hornitschek et al., 2012; Li et al., 2012; Bou-Torrent et al., 2014). In addition, nuclear-pore complex components and chloroplast-derived signals also prevent an excessive response to shade, providing additional regulatory levels of this response (Gallemí et al., 2016; Ortiz-Alcaide et al., 2019).
There are, however, still major gaps in understanding the genetic and molecular regulation of SAS and, by extension, shade-tolerance traits. Comparative analyses using shade-avoiding and shade-tolerant species is expected to identify regulators of traits associated with shade tolerance habits (Gommers et al., 2013). Indeed, a comparative transcriptomic approach using two Geranium species with divergent petiole responses to shade unveiled components that might suppress growth in the shade-tolerant species (Gommers et al., 2017, 2018). The use of related species that are amenable for genetic analyses is expected to push this effort further to find regulatory components used in nature to modulate these divergent responses. This is what we are addressing in this work.
Comparing Arabidopsis and its close relative Cardamine hirsuta to understand the genetic basis for trait diversification between species is a powerful strategy to understand the evolution of morphological traits. Key to this approach is the wide morphological and physiological diversity between these species, such as differences in leaf morphology and seed dispersal mechanism among others (Barkoulas et al., 2008; Hay et al., 2014; Vlad et al., 2014; Hofhuis et al., 2016; Vuolo et al., 2016). Like Arabidopsis, C. hirsuta has a short generation time, small size, inbreeding habit, abundant progeny, and ease of large-scale cultivation (Hay et al., 2014; Hay and Tsiantis, 2016). It is a diploid species with a small genome and eight chromosomes that has been completely sequenced (Gan et al., 2016). Genetic transformation by floral dipping, a dense genetic map, and chemically mutagenized populations provide the tools to identify the genetic components and molecular mechanisms underlying diversification or morphology and response to environment (Hay and Tsiantis, 2016). C. hirsuta is an invasive herbaceous plant that can grow in open sun but is often found in shaded or semishaded areas. Indeed, C. hirsuta does not need much light to grow, and their stems become purplish (likely to prevent oxidative damage) in strong sun (http://edis.ifas.ufl.edu/pdffiles/EP/EP51100.pdf; http://practicalplants.org/wiki/Cardamine_hirsuta; http://www.asturnatura.com/especie/cardamine-hirsuta.html; http://dnr.wi.gov/topic/Invasives/documents/classification/LR_Cardamine_hirsuta.pdf; https://www.wildfooduk.com/edible-wild-plants/hairy-bittercress/). These observations are consistent with C. hirsuta being shade-tolerant (Bealey and Robertson, 1992). In agreement, whereas seedlings of Arabidopsis elongate in response to shade, those of C. hirsuta are unresponsive to the same stimulus (Hay et al., 2014).
The divergent hypocotyl response to shade of Arabidopsis and C. hirsuta species led us to take a comparative approach to understand the genetic basis of the evolution of this physiological trait. We found that C. hirsuta has acquired a highly efficient phyA-dependent pathway that represses hypocotyl elongation and other SAS-associated responses when exposed to simulated shade. After complementing Arabidopsis phyA mutant plants with endogenous or C. hirsuta phyA molecules, we concluded that these two photoreceptors are not exchangeable. Differences in phyA intrinsic activity hence contribute to a different response of C. hirsuta and Arabidopsis to shade exposure.
RESULTS
C. hirsuta Seedlings Perceive Low R:FR but Do Not Elongate
A recent study revealed that different species of the Tradescantia genus with divergent tolerance to shade showed clear differences in maximum quantum efficiency of photosystem II (Fv/Fm) upon variations of the growth light (Benkov et al., 2019). In particular, the sun-resistant T. sillamontana (a succulent growing in semidesert regions of Mexico and Peru, hence adapted to high light intensities) was more tolerant to changes in irradiation intensity (i.e., showed a more constant Fv/Fm) than the shade-tolerant T. fluminensis (habitant of tropical rainforests and other shaded areas in southeastern Brazil and hence adapted to grow under low light intensities).
Using a similar experimental system, we aimed to confirm whether C. hirsuta is a shade-tolerant plant compared with Arabidopsis (a broadly accepted shade-avoider). Indeed, when wild-type seedlings of these two species (ChWT and AtWT) were transferred from normal W to conditions in which PAR was first increased 10-fold (high light [HL]) and then reduced fivefold relative to W (low light [LL]) or vice versa, Fv/Fm changes were much more pronounced in ChWT (Supplemental Figure 1A). The lower capacity of ChWT to adapt to intense irradiation was confirmed by the bleaching symptoms (e.g., lower chlorophyll contents) observed in ChWT (but not in AtWT) upon transferring to HL (Supplemental Figure 1B). ChWT seedlings only showed a better performance than AtWT when transferred from W to LL. Rapid light curve analysis confirmed that ChWT was better able to maintain its level of photosynthetic activity under LL conditions than AtWT (Supplemental Figure 1C), as expected for a shade-tolerant plant (Han et al., 2015).
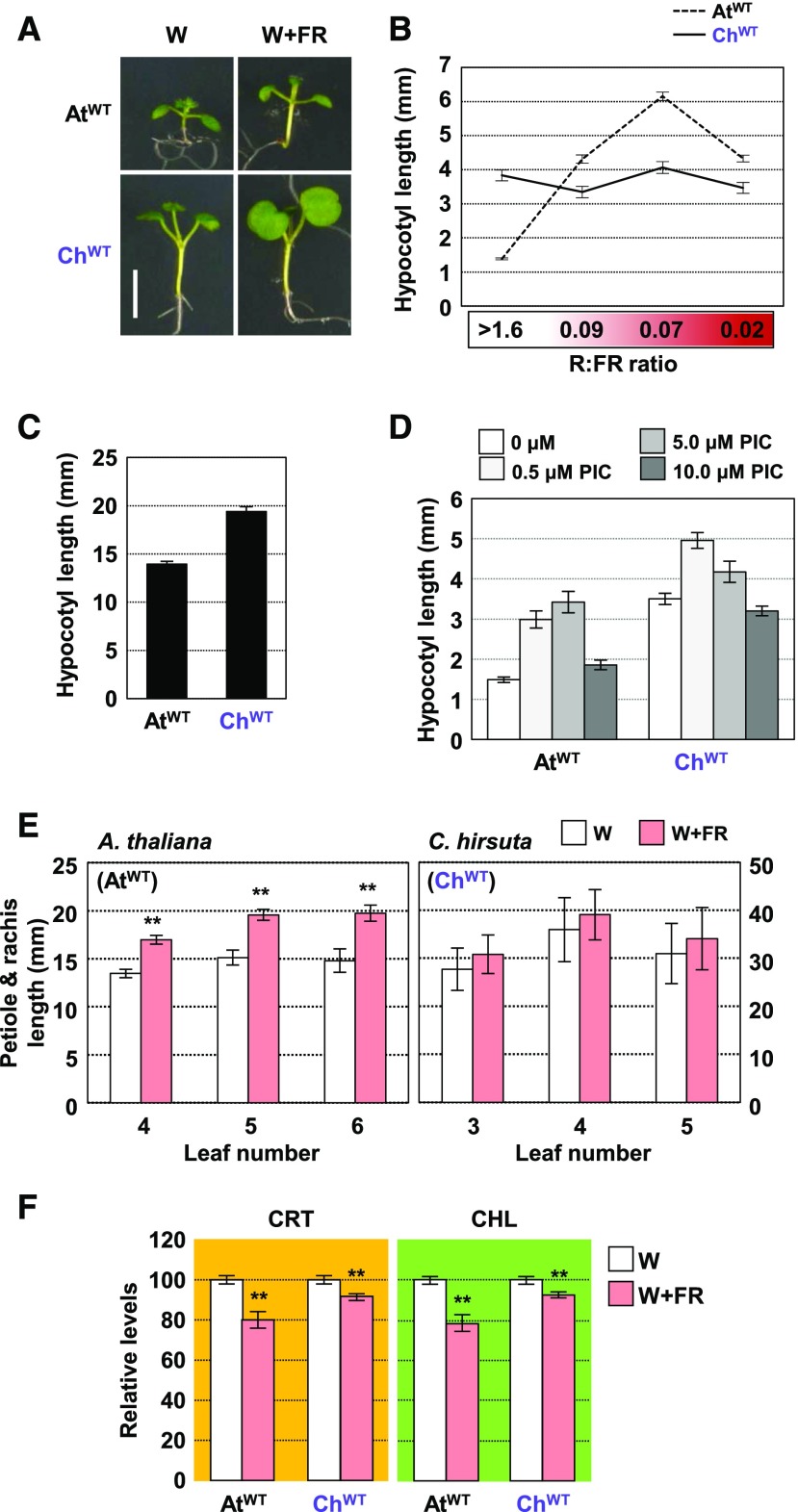
Besides differentially responding to decreased light quantity, plant species from open habitats show a stronger elongation response to reduced R:FR (i.e., light quality) compared with those from woodland shade habitats (Smith, 1982; Gommers et al., 2017). Further supporting the conclusion that C. hirsuta tolerates shade, ChWT failed to elongate their hypocotyls when exposed to a range of low R:FR treatments (i.e., W+FR) that mimic vegetation proximity (intermediate or low R:FR; 0.09–0.07) and canopy shade (very low R:FR; 0.02; Figure 1; Supplemental Figure 2). W-grown ChWT hypocotyls, as well as cotyledons, are substantially longer than those of AtWT growing under the same conditions. ChWT hypocotyls were also longer than those of AtWT when growing in the dark (Figure 1C), indicating that C. hirsuta is overall bigger than Arabidopsis. More importantly, when treated with growth stimulants, such as gibberellic acid (Hay et al., 2014) or picloram (a synthetic auxin), hypocotyls of both species elongate (Figure 1D). We therefore concluded that the elongation of C. hirsuta hypocotyls is not generally compromised, arguing against the possibility that this species displays a constitutive SAS phenotype.
Figure 1.
Arabidopsis and C. hirsuta Differ in the Hypocotyl Elongation Response to Neighboring Vegetation.
(A) Phenotype of representative seedlings of AtWT and ChWT after 3 d grown in W and retained in W (left panels) or transferred to W+FR (R:FR of 0.02; right panels) until day 7. Bar = 5 mm.
(B) Hypocotyl length of day-7 AtWT and ChWT seedlings grown for the last 4 d under the indicated R:FR.
(C) Hypocotyl length of day-4 AtWT and ChWT seedlings grown in darkness.
(D) Hypocotyl length of day-7 AtWT and ChWT seedlings grown under W in medium supplemented with increasing concentrations of picloram (PIC).
(E) Petiole and rachis length of 3-week-old leaves of AtWT and ChWT plants grown for the last 7 d under the indicated R:FR.
(F) Carotenoid (CRT) and chlorophyll (CHL) levels of AtWT and ChWT seedlings grown in W and W+FR (as detailed in [A]).
Values are means and se of three to five independent samples. Asterisks indicate significant differences (**, P < 0.01) relative to W-grown plants.
In Arabidopsis, exposure to simulated shade also triggers the elongation of leaf petioles. We quantified the elongation response of the petiole and rachis in 2-week-old ChWT and AtWT plants subjected to 7 d of high (W) or low (W+FR) R:FR. In agreement with previous studies (Kozuka et al., 2010; Sasidharan et al., 2010; de Wit et al., 2015), shade-treated AtWT leaves showed substantially longer petioles than those of plants grown under W. Petiole and rachis length in ChWT, however, was similar in leaves from plants grown under W or W+FR (Figure 1E; Supplemental Figure 3). These results together suggest that elongation responses to low R:FR are dramatically arrested in C. hirsuta plants.
C. hirsuta Shows Other Attenuated Responses to Shade
Beyond elongation responses, low R:FR triggers a reduction in the levels of photosynthetic pigments (i.e., carotenoids and chlorophylls; Roig-Villanova et al., 2007; Cagnola et al., 2012; Bou-Torrent et al., 2015). While these pigments were also significantly reduced in shade-treated ChWT seedlings (Figure 1F), the decrease was less prominent than in AtWT. These results indicated that not all SAS responses are equally compromised in C. hirsuta.
We next used RNA sequencing (RNA-seq) to compare the genome-wide expression patterns of 7-d-old AtWT and ChWT whole seedlings in W versus 1 h of simulated shade (W+FR; Figure 2). Incorporating knowledge about gene orthology, 432 differentially expressed genes (DEGs) were categorized as rapidly regulated by shade in one species or in both. Plotting the W+FR versus W fold change in C. hirsuta against the same ratio in Arabidopsis resulted in a linear regression equation with a slope of 0.54 (Figure 2B), which supported that shade-modulated changes in gene expression are also attenuated in C. hirsuta compared with Arabidopsis. In Arabidopsis, shade treatment induced 246 genes (fold change > 1.5, P < 0.05) and repressed 58 genes (fold change ≤ 1.5, P < 0.05). In C. hirsuta, this same treatment induced 181 genes and repressed 54 genes (Supplemental Figure 4A; Supplemental Data Sets 1 to 4). From the set of induced DEGs, 102 responded in both species. They included several of the well-known shade-marker genes in Arabidopsis and other species, such as ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2 (ATHB2), BRASSINOSTEROID-ENHANCED EXPRESSION1, BES1-INTERACTING MYC-LIKE1, LONG HYPOCOTYL IN FR1, and XYLOGLUCAN ENDOTRANSGLYCOSYLASE7 (Ueoka-Nakanishi et al., 2011; Karve et al., 2012; Cifuentes-Esquivel et al., 2013; Procko et al., 2014).
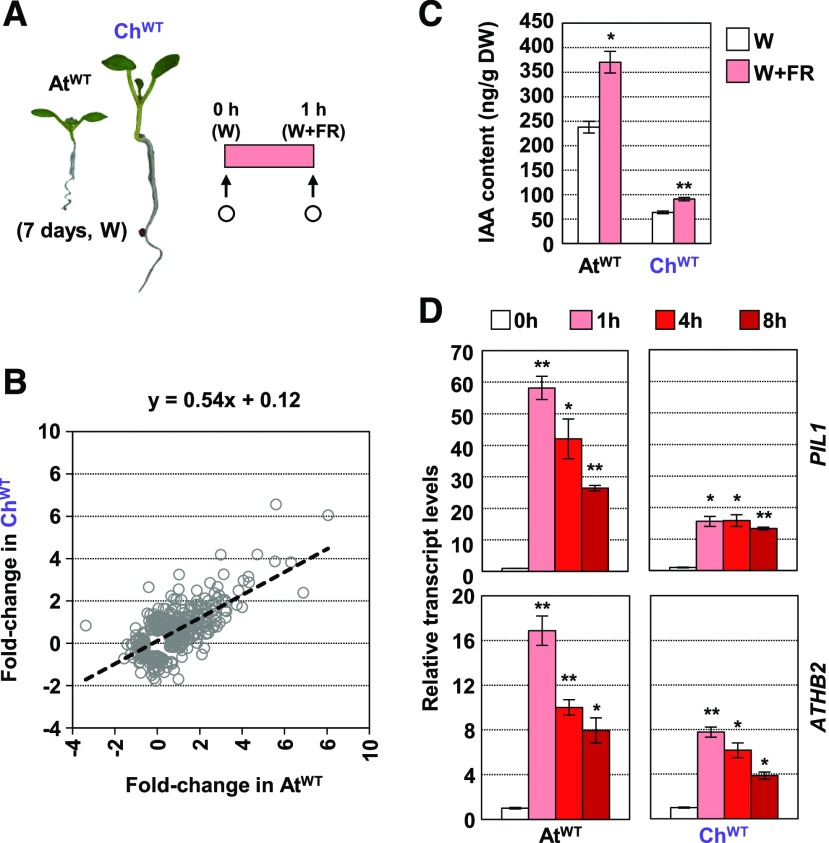
Figure 2.
Arabidopsis and C. hirsuta Seedlings Respond to Neighboring Vegetation by Altering Gene Expression.
(A) RNA-seq was performed with RNA extracted from AtWT and ChWT seedlings that were grown in W for 7 d and then treated for 1 h with W+FR (R:FR = 0.02). White circles indicate the moment of harvesting for RNA extraction. Three independent biological replicates were used for each genotype and treatment.
(B) Correlation between log-transformed fold change of 432 DEGs in AtWT and ChWT. The estimated regression equation is shown at the top of the graph.
(C) IAA content in AtWT and ChWT seedlings grown and harvested as indicated in (A). Whole seedlings were collected and lyophilized to measure IAA levels. Data are presented as means and se of three (AtWT) or four (ChWT) biological replicates. DW, dry weight.
(D) Effects of W+FR treatment on PIL1 and ATHB2 expression in AtWT and ChWT seedlings (R:FR = 0.02). W-grown day-7 seedlings of Col-0 and Oxford were treated for 0, 1, 4, and 8 h with W+FR. Transcript abundance, normalized to EF1α, is shown. Values are means and se of three independent RT-qPCR biological replicates relative to values at 0 h for each species.
In (C) and (D), asterisks indicate significant differences (**, P < 0.01 and *, P < 0.05) relative to 0-h samples.
Gene Ontology (GO) and MapMan-Bin (MMB) functional prediction of these upregulated gene groups indicated that terms related to auxin were significantly overrepresented (Supplemental Data Sets 5 and 6), suggesting an early role for auxins in both Arabidopsis and C. hirsuta. Indeed, W+FR treatment for 1 h increased auxin (IAA) levels not only in AtWT, as published (Tao et al., 2008; Hornitschek et al., 2012; Bou-Torrent et al., 2014; Hersch et al., 2014), but also in whole ChWT seedlings (Figure 2C).
Using public transcriptomic data, we identified a group of 13 genes whose expression was induced in Arabidopsis wild-type seedlings but not in mutants that do not accumulate auxins (shade avoidance3-2 and pif7-1) after 1 h of shade treatment (Tao et al., 2008; Li et al., 2012; Bou-Torrent et al., 2014). Based on our RNA-seq data, the expression of these genes was significantly upregulated in AtWT and, to a lower extent, ChWT seedlings (Supplemental Figure 5), consistent with the observed increase in IAA content in both species. Since only Arabidopsis elongates in response to shade exposure, either the observed early changes in gene expression and auxin levels are not reflecting the differences in hypocotyl growth between these species or the elongation is a consequence of differential later events.
In our RNA-seq analyses, 55 and 49 DEGs were specifically repressed in either AtWT or ChWT seedlings, respectively, and just 3 genes were repressed in both species. Regarding upregulated genes, 142 and 79 DEGs were specifically induced either in AtWT or ChWT, respectively (Supplemental Figure 4A). GO and MMB functional prediction of the 142 DEGs specific for AtWT showed genes related to several aspects of plant development, whereas the 79 DEGs specifically induced in ChWT showed enrichment for genes related to the photosynthetic machinery. Particularly, C. hirsuta rapidly responds by inducing the expression of genes encoding components of both PSI and PSII, the NADH dehydrogenase-like complex (involved in chlororespiration), and both small and large subunits of plastidial ribosomes (Supplemental Figure 5B; Supplemental Data Sets 5 and 6). Whether these rapid changes are maintained after prolonged exposure to shade or have any functional relevance is unknown. Nonetheless, these transcriptome differences support that the two mustard species employ alternative strategies to adapt to plant proximity and shade that go further from the modulation of elongation growth.
Comparative approaches have been used before to investigate the differential responses to shade of related species. Transcriptomic analyses using two Geranium species that display divergent shade-induced petiole elongation (G. pyrenaicum as a shade avoider or responsive and G. robertianum as a shade tolerant) identified a series of 31 upregulated genes that included a number of candidate regulators of differential shade avoidance (Gommers et al., 2017). In these two species, putatively orthologous transcript groups (OMCL) were defined, and the best BLAST hit with the Arabidopsis transcriptome was used to name Geranium OMCL groups (Gommers et al., 2017). When we compared our lists of shade-regulated genes with the Geranium OMCLs differentially regulated after 2 h of low R:FR in the petioles, we found that the number of genes upregulated in both shade-tolerant and shade-avoider species was higher for the AtWT/ChWT pair than between the Geranium species (Supplemental Figure 4C; Supplemental Data Sets 7and 8). GO analyses did not identify any function from the lists of genes specifically induced in either G. pyrenaicum or G. robertianum. Overlap was very limited between the sets of repressed genes. Together, the contrasting rapid shade-induced gene expression changes might either support differences in the early molecular mechanisms between the Geranium and mustard groups or just reflect the differences in tissues (whole seedlings versus leaf petioles) and/or shade and growth conditions (continuous light versus photoperiod) between experiments.
We also analyzed the changes in gene expression of PIF3-LIKE1 (PIL1) and ATHB2, two typical shade-marker genes, in response to longer (up to 8 h) exposure to low R:FR. Expression of PIL1 and ATHB2 was rapidly induced in both mustard seedlings after simulated shade exposure. However, the relative induction of the expression of these genes was attenuated in ChWT compared with AtWT (Figure 2D). Together, our results indicate that C. hirsuta seedlings sense plant proximity and respond molecularly and metabolically to it; however, this signal does not promote hypocotyl elongation in C. hirsuta as it does in the shade-avoider Arabidopsis.
Shade-Induced Elongation in C. hirsuta Is Repressed
To explain the hypocotyl elongation differences between Arabidopsis and C. hirsuta, we hypothesized two mutually exclusive mechanisms: (1) uncoupling: shade perception is specifically unplugged from the endogenous mechanisms of control of hypocotyl elongation; and (2) suppression: there are mechanisms that strongly suppress the shade-induced elongation of hypocotyls. To distinguish between these possibilities, a genetic screening looking for C. hirsuta seedlings with long hypocotyls under simulated shade (>6 mm long) was performed, using an ethyl methane sulfonate-mutagenized population (Vlad et al., 2014). If suppression mechanisms exist, then loss-of-function mutants that unleash shade-induced hypocotyl elongation might be recovered. Indeed, from the various long-hypocotyl seedlings identified, we focused on two slender in shade (sis) mutants, shown to be recessive and allelic. After backcrossing these mutants twice with the ChWT plants, homozygous mutants had slightly longer hypocotyls in W than the wild type and had very long hypocotyls under W+FR. We named the mutants as sis1-1 and sis1-2 (Figure 3). These results indicated that (1) loss-of-function (recessive) mutations support the “suppression” mechanisms in C. hirsuta to establish shade tolerance and (2) a single gene, SIS1, is able to repress the elongation response to shade in C. hirsuta.
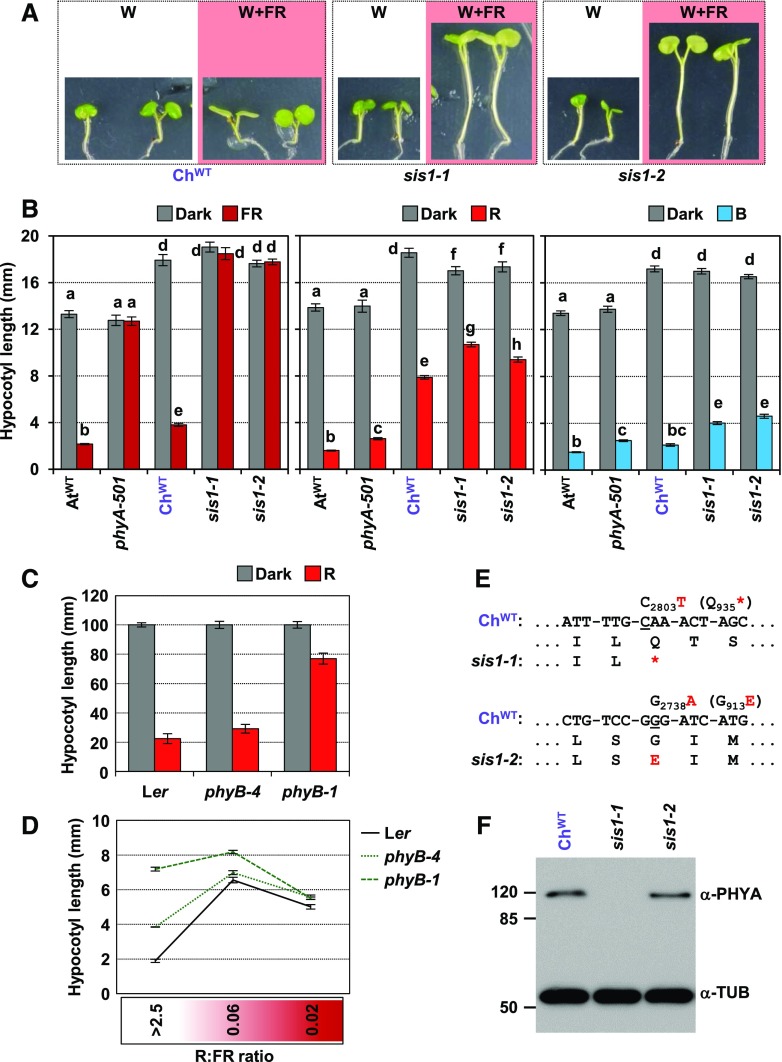
Figure 3.
Mutant sis1 Seedlings of C. hirsuta Are Deficient in phyA Activity.
(A) Phenotypes of representative seedlings of ChWT, sis1-1, and sis1-2 after 3 d grown in W and retained in W (white panels) or transferred to W+FR (R:FR of 0.02; pink panels) until day 7. All panels are to the same scale.
(B) Hypocotyl length of AtWT, phyA-501 (Arabidopsis), ChWT, sis1-1, and sis1-2 (C. hirsuta) lines grown for 4 d in darkness (Dark) or under monochromatic FR (2.6 µmol m−2 s−1), R (38.9 µmol m−2 s−1), and blue light (B; 1.9 µmol m−2 s−1).
(C) Hypocotyl length of Arabidopsis Landsberg erecta (Ler), phyB-4, and phyB-1 seedlings grown for 4 d in darkness or under monochromatic R (40.6 µmol m−2 s−1).
(D) Hypocotyl length of Arabidopsis Ler, phyB-4, and phyB-1 seedlings under the indicated R:FR. Seedlings were grown for 2 d in W (R:FR > 2.5) and then kept in W (R:FR > 2.5) or transferred to W+FR (R:FR of 0.06 or 0.02) until day 7.
(E) Schematic diagram of the lesions found in the ChPHYA gene in the sis1-1 and sis1-2 alleles compared with the wild-type sequence (ChWT) and the predicted changes in the amino acid sequence.
(F) Immunoblot detection of phyA and tubulin with mouse monoclonal anti-phyA (073D) and anti-TUB antibodies in extracts of etiolated seedlings of ChWT, sis1-1, and sis1-2 lines.
As a first step to explore SIS1 identity, we determined whether light perception was altered in sis1 mutants by analyzing hypocotyl length after deetiolation under monochromatic lights. We noticed that ChWT seedlings were quite hyposensitive to R compared with AtWT (Figure 3B), suggesting that an attenuated phyB signaling might result in a constitutive SAS hypocotyl response, causing the observed suppression of the shade-induced hypocotyl elongation. Considering the relationship between the attenuated responsiveness to R and the strength of the shade-induced hypocotyl elongation of the weak phyB-4 and strong phyB-1 Arabidopsis mutant seedlings (Figures 3C and 3D), the hyposensitivity to R observed in ChWT might contribute but is not enough to fully suppress the shade-induced hypocotyl elongation in this species. Therefore, additional components are required to establish the shade-tolerant hypocotyl habit in C. hirsuta. Indeed, mutant sis1 seedlings, although slightly hyposensitive to R and blue light, were fully blind to FR compared with ChWT seedlings (Figure 3B).
A very similar pattern of response was also shown by Arabidopsis phyA-deficient phyA-501 seedlings (Figure 3B; Li et al., 2011), which suggested that sis1 seedlings might be deficient in phyA activity or signaling. Sequencing of the C. hirsuta PHYA (ChPHYA) gene from sis1-1 and sis1-2 plants showed point mutations (transitions) that introduced either a nonsense mutation in Gln-935 (in sis1-1) or a missense mutation in the conserved Gly-913 (in sis1-2; Figure 3E; Supplemental Figure 6A). Immunoblot analyses using a specific monoclonal antibody against phyA (073D) indicated that only sis1-1 was lacking phyA (Figure 3D). Consistent with this, C. hirsuta lines with reduced activity of phyA by overexpressing an RNA interference construct directed toward the ChPHYA gene (line 35S:RNAi-ChPHYA) also resulted in a sis phenotype (Supplemental Figures 6B to 6D). Together, these results indicated that sis1 are C. hirsuta phyA-deficient mutants (for clarity, we will keep the sis1 mutant name in this article to distinguish it from the phyA mutants from Arabidopsis). They also suggested that shade tolerance in C. hirsuta might be caused by the existence of a phyA-dependent suppression mechanism that represses the hypocotyl elongation response to shade.
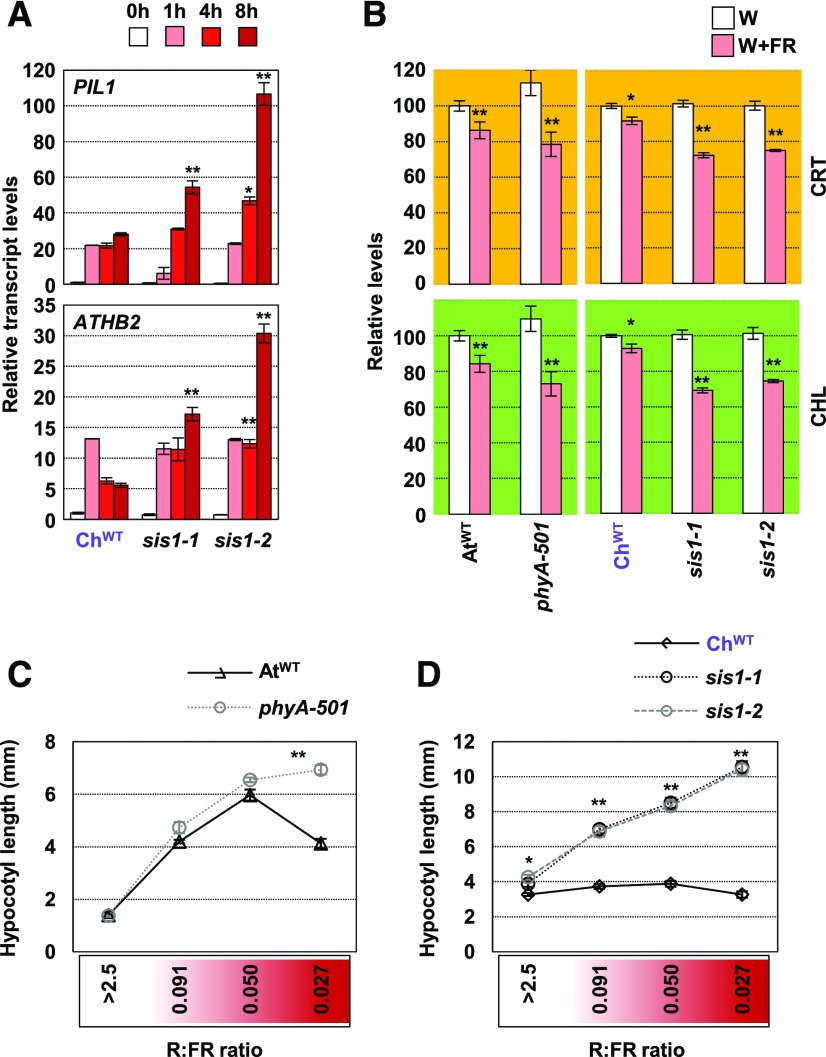
Molecular analyses showed that the relative induction of PIL1 and ATHB2 expression was enhanced in both sis1 mutants compared with ChWT seedlings after more than 4 h of simulated shade exposure (Figure 4). This relatively late effect of ChPHYA absence (sis1) on gene expression is consistent with what was observed in Arabidopsis phyA mutants (Ciolfi et al., 2013). We also measured the levels of photosynthetic pigments (carotenoids and chlorophylls) after long-term exposure to low R:FR in wild-type and phyA-deficient Arabidopsis and C. hirsuta seedlings. Simulated shade triggered a stronger decrease in the accumulation of these pigments in phyA-501, sis1-1, and sis1-2 seedlings compared with wild-type controls (Figure 4B), hence indicating that phyA represses this trait in both species, likely to avoid exaggerated losses of photosynthetic pigments in response to vegetation proximity and shade.
Figure 4.
C. hirsuta sis1 Seedlings Are Impaired in Their Tolerance to Plant Proximity.
(A) Effects of W+FR treatment on PIL1 and ATHB2 expression in ChWT sis1-1 and sis1-2 seedlings. Seedlings were grown as in Figure 2D. Transcript abundance, normalized to EF1α, is shown. Values are means and se of three independent RT-qPCR biological replicates relative to values at 0 h for each genotype. Asterisks indicate significant differences (**, P < 0.01) relative to 0-h samples.
(B) Carotenoid (CRT) and chlorophyll (CHL) levels of AtWT and phyA-501 Arabidopsis and ChWT, sis1-1, and sis1-2 C. hirsuta seedlings grown in W and W+FR (as detailed in Figure 1A). Values are means and se of five independent samples. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) relative to W-grown plants.
(C) and (D) Hypocotyl length of day-7 AtWT, phyA-501 (Arabidopsis; [C]) and ChWT, sis1-1, and sis1-2 (C. hirsuta; [D]) seedlings grown for the last 4 d under the indicated R:FR. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) relative to the corresponding wild-type plant grown under the same R:FR. In (D), asterisks apply for both sis1 mutants.
PhyA represses the shade-induced hypocotyl elongation in Arabidopsis caused by the deactivation of phyB only under conditions that mimic closed canopies (i.e., under very low R:FR; Yanovsky et al., 1995; Casal et al., 2014; Martínez-García et al., 2014). Indeed, Arabidopsis phyA-deficient mutants behaved almost like AtWT seedlings under various shade-mimicking conditions except for the lowest R:FR tested (Figure 4C). By contrast, C. hirsuta sis1 mutants behaved differently than ChWT under all the low R:FR applied (Figure 4D), indicating that phyA has a broader role in suppressing the shade-induced hypocotyl elongation in C. hirsuta than in Arabidopsis.
C. hirsuta Has Higher phyA Activity Than Arabidopsis
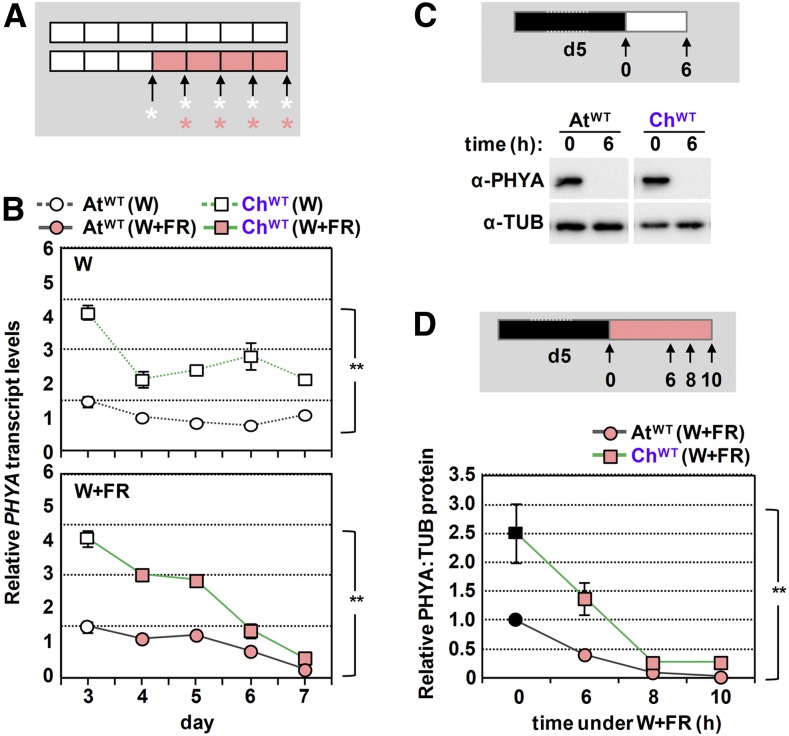
Our results suggested the possibility that phyA activity is higher in the shade-tolerant C. hirsuta than in the shade-avoider Arabidopsis. Higher phyA activity can be achieved by at least two alternative and nonexclusive ways: higher phyA levels and/or higher specific (intrinsic) activity of the photoreceptor. To analyze these possibilities, we first aimed to compare PHYA expression levels in AtWT and ChWT seedlings. Data extracted from our RNA-seq experiment indicated that the expression of several commonly used reference genes, such as EF1α or YLS8 (Hornitschek et al., 2009; Kohnen et al., 2016; Gallemí et al., 2017), was within the same range (Supplemental Table 1). Then, we quantified PHYA expression levels in AtWT and ChWT seedlings growing under W or W+FR (Figure 5) using primers that recognize the sequences of the target gene (PHYA) and three normalizer genes (EF1α, SPC25, and YLS8) in both species (Supplemental Figure 7). Expression of PHYA was significantly higher in C. hirsuta than in Arabidopsis seedlings (two-way ANOVA tests, P < 0.05) in seedlings of different ages grown under W or W+FR conditions (Figure 5B).
Figure 5.
C. hirsuta Seedlings Have Higher phyA Levels Than Those of Arabidopsis.
(A) Cartoon showing the design of the experiment. AtWT and ChWT, grown as in Figure 1A, were harvested at the indicated times of W or W+FR treatments (asterisks) for RNA extraction.
(B) Evolution of PHYA transcript levels in AtWT and ChWT seedlings grown as detailed in (A). Primers used (Supplemental Figure 8A) allow quantifying and comparing expression levels by RT-qPCR between both species. PHYA transcript abundance was normalized to three reference genes (EF1α, SPC25, and YLS8). Values are means and se of three independent RT-qPCR biological replicates relative to PHYA transcript levels of day-3 Arabidopsis seedlings. Two-way ANOVA showed that PHYA levels are significantly different (**, P < 0.01) between species under either W or W+FR.
(C) Immunoblot detection of phyA and tubulin with the antibodies indicated in Figure 3C in extracts of AtWT and ChWT seedlings grown as detailed at the top of the panel: 5-d-old etiolated seedlings were exposed to W light, and material was harvested before and after 6 h of W exposure (arrows).
(D) Evolution of relative phyA protein levels (PHYA:TUB) in AtWT and ChWT seedlings exposed to simulated shade, as detailed at the top of the panel: 5-d-old etiolated seedlings were exposed to W+FR light, and material was harvested before and after 6, 8, and 10 h of simulated shade exposure (arrows). Values are means and se of four independent biological replicates relative to PHYA:TUB levels of etiolated AtWT seedlings. Two-way ANOVA showed that relative PHYA levels under W+FR are significantly increased (**, P < 0.01) in C. hirsuta over Arabidopsis.
Higher expression of PHYA in C. hirsuta might result in higher phyA protein levels, contributing to an increased phyA activity in this species. Our immunoblot analyses showed that PHYA protein levels were significantly higher in C. hirsuta than in Arabidopsis etiolated seedlings (Figure 5D). More importantly, whereas PHYA levels almost disappear after 6 h of W exposure in both species, C. hirsuta seedlings maintained higher PHYA levels than Arabidopsis when exposed to W+FR for 6 to 10 h (Figures 5C and 5D). Together, these results support that PHYA levels in C. hirsuta are generally higher than in Arabidopsis seedlings, even under shade conditions. This observation is consistent with the strongest difference in hypocotyl length under W of wild-type and phyA-deficient seedlings from C. hirsuta compared with Arabidopsis (Figures 4C and 4D; Supplemental Figure 6D; Martínez-García et al., 2014). Furthermore, transgenic overexpression of PHYA has been shown to attenuate shade-triggered hypocotyl elongation in Arabidopsis seedlings and stem elongation in other species (Heyer et al., 1995; Robson et al., 1996; Roig-Villanova et al., 2006).
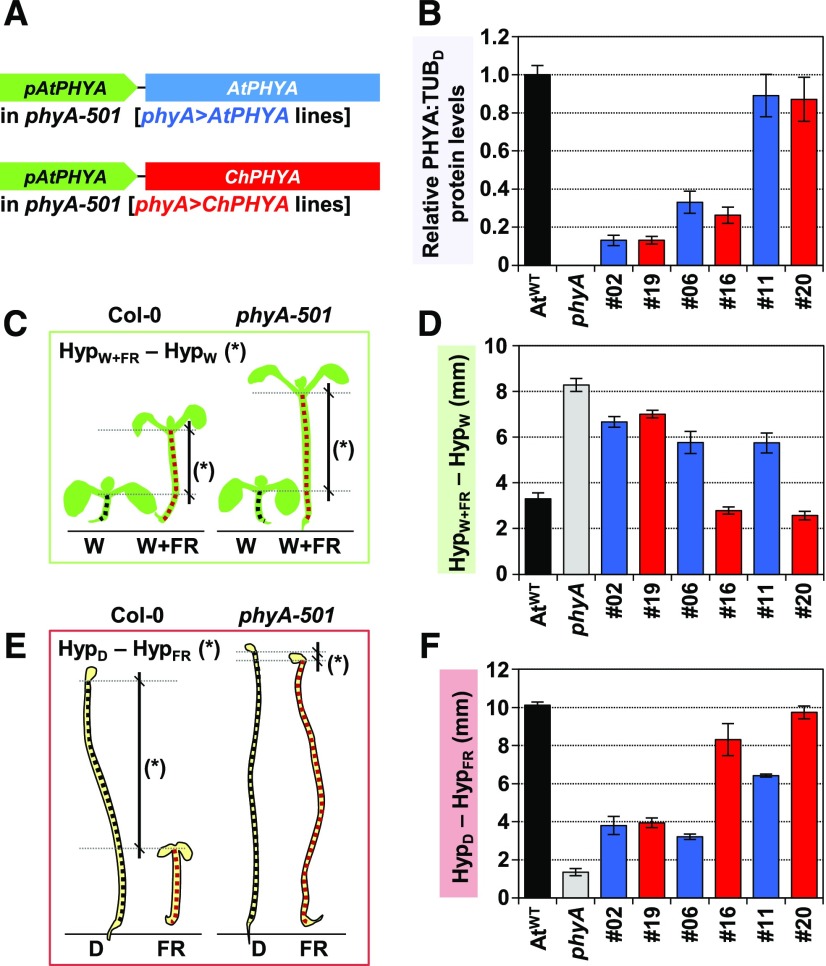
To compare AtphyA and ChphyA specific (intrinsic) activities, complementation analyses of the Arabidopsis phyA-501 mutant were performed with the AtPHYA or ChPHYA gene under the control of the endogenous promoter of AtPHYA (pAtPHYA:AtPHYA or pAtPHYA:ChPHYA, respectively). The resulting lines were named as phyA>AtPHYA and phyA>ChPHYA (Figure 6). We obtained a total of five independent phyA>AtPHYA lines and seven independent phyA>ChPHYA lines with different transcript and protein levels (Supplemental Figure 8). To estimate PHYA protein levels, we used etiolated seedlings, as phyA is photolabile. Because PHYA expression is repressed by light via phyA and phyB (Cantón and Quail, 1999), RNA was extracted from seedlings either grown in the dark or under W+FR (Supplemental Figure 8A). PHYA expression in seedlings grown in these two conditions correlated positively in both phyA>AtPHYA (R2 = 0.79) and phyA>ChPHYA (R2 = 0.79) lines (Supplemental Figure 8B). The slope of these equations, however, was significantly higher (P < 0.05) for phyA>AtPHYA (7.49) than for phyA>ChPHYA (2.81). Specifically, phyA>AtPHYA and phyA>ChPHYA lines with comparable PHYA expression levels in the dark showed lower PHYA expression under simulated shade when complemented by ChPHYA (phyA>ChPHYA) compared with AtPHYA (phyA>AtPHYA). These results pointed to a stronger activity for the ChphyA protein in repressing its own (PHYA) expression.
Figure 6.
ChphyA Has a Stronger Activity Than AtphyA in Repressing Shade-Induced Hypocotyl Elongation.
(A) Cartoon detailing the constructs used to complement Arabidopsis phyA-501 mutant plants.
(B) Relative PHYA:TUB in etiolated seedlings of AtWT, phyA-501, and selected phyA>AtPHYA (blue bars) and phyA>ChPHYA (red bars) complementation lines. Seedlings were grown as indicated in Supplemental Figure 8. Values are means and se of four independent biological replicates relative to PHYA:TUB levels of etiolated AtWT seedlings.
(C) Cartoon illustrating how phyA activity in simulated shade was established as differences in hypocotyl length between simulated shade- and W-grown seedlings (HypW+FR-HypW). Seedlings were grown for 2 d under W and then for another 5 d under W or W+FR (R:FR = 0.02), when hypocotyls were measured.
(D) HypW+FR-HypW in seedlings of AtWT, phyA-501, and selected phyA>AtPHYA (blue bars) and phyA>ChPHYA (red bars) complementation lines. Values are the difference of means of hypocotyl length between seedlings grown under W+FR (HypW+FR) and under W (HypW). se were propagated accordingly.
(E) Cartoon illustrating how phyA activity in deetiolation was established as differences in hypocotyl length between dark- and FR-grown seedlings (HypD-HypFR). Seedlings were grown as indicated in Figure 3B.
(F) HypD-HypFR in seedlings of AtWT, phyA-501, and selected phyA>AtPHYA (blue bars) and phyA>ChPHYA (red bars) complementation lines. Values are the difference of means of hypocotyl length between seedlings grown in the dark (HypD) and under FR (HypFR). se were propagated accordingly.
In (C) and (E), mutant phyA-501 seedlings have no phyA activity.
For the comparison of AtphyA and ChphyA activities, we initially studied their effects on the promotion of the shade-induced hypocotyl elongation in transgenic lines. AtWT and phyA-501 seedlings were incorporated as controls. In these experiments, the difference in hypocotyl length between seedlings grown under W+FR versus W (HypW+FR-HypW) provided values indicative of the complementation level (or phyA biological activity) for the response analyzed. Consequently, in these analyses, the lower the HypW+FR-HypW value, the higher the phyA activity. Opposite to that observed with transcript levels (Supplemental Figure 8C), HypW+FR-HypW correlated well with ChPHYA but not with AtPHYA protein levels (Supplemental Figure 8D). These results together indicate that the two photoreceptors are not fully exchangeable and suggest different intrinsic qualities (i.e., biological activity) between the phyA receptors of Arabidopsis and C. hirsuta.
When lines with comparable PHYA protein levels were selected (Figure 6B), the response to shade (HypW+FR-HypW) was more strongly attenuated by ChPHYA (Figures 6C and 6D). As an additional way to test for phyA activity, we estimated hypocotyl elongation in seedlings etiolated (HypD) and deetiolated under monochromatic FR (HypFR). In this case, the higher the difference between these two values (HypD-HypFR), the stronger the activity of phyA. Similar to the shade-response analyses, ChphyA showed a stronger activity than AtphyA in deetiolating seedlings under FR (Figures 6E and 6F). A good correlation between these two phyA-mediated responses was also found when all the lines were considered together (Supplemental Figure 8E), reinforcing our interpretation that ChphyA is intrinsically more active than AtphyA.
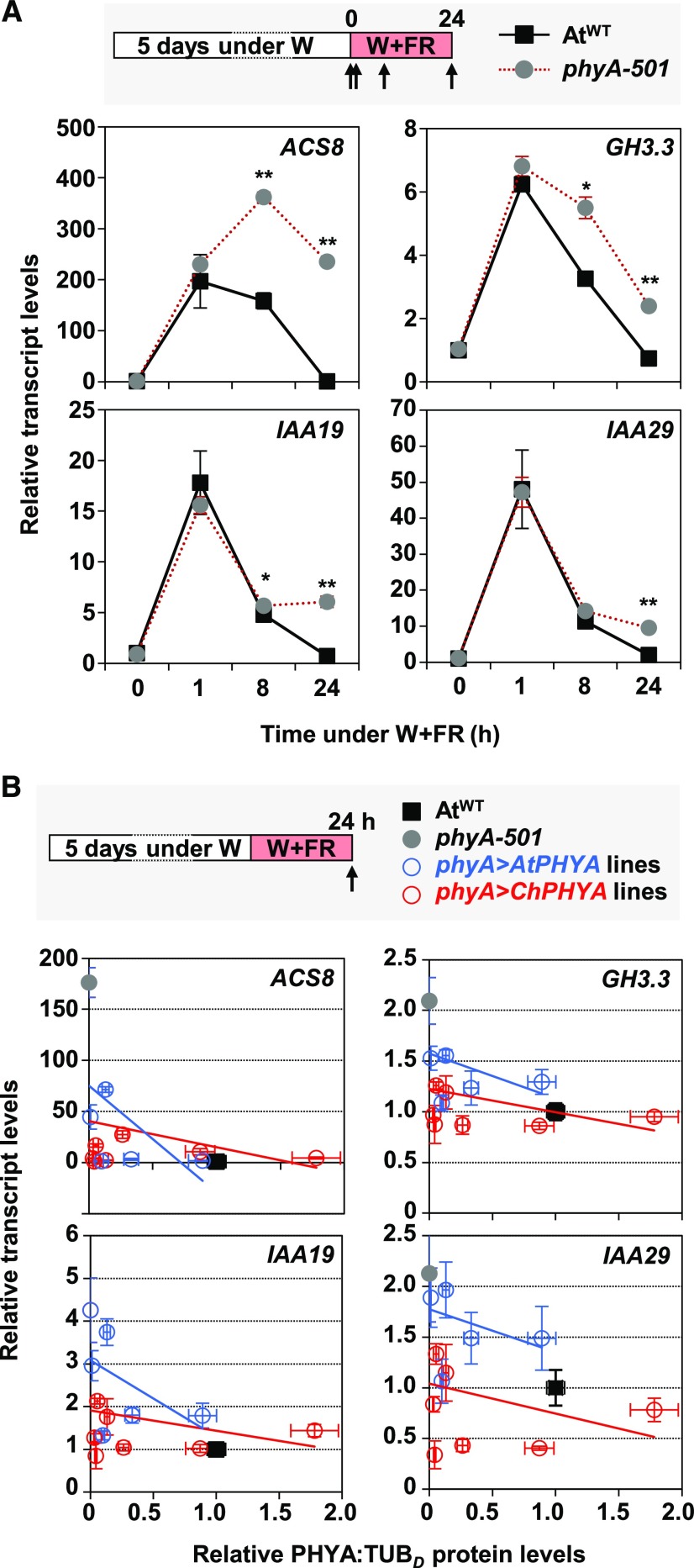
The expression of dozens of auxin-responsive genes is repressed by phyA after just 1 h of very low R:FR treatment (Yang et al., 2018). As an additional and complementary test of phyA biological activity different from hypocotyl elongation, we evaluated the repressive effect of AtphyA and ChphyA on the expression of these genes. First, we selected 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE8 (ACS8), GRETCHEN HAGEN3.3 (GH3.3), INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19), and IAA29, four auxin-responsive genes described as repressed phyA targets (Yang et al., 2018). As expected, the shade-induced expression of these genes was attenuated in AtWT compared with phyA-501 seedlings, but under our shade conditions, the differences were most obvious after long exposure to W+FR (Figure 7).
Figure 7.
ChphyA Has a Stronger Activity Than AtphyA in Repressing Shade-Induced Expression of ACS8, GH3.3, IAA19, and IAA29 Genes.
(A) Effects of phyA in the shade-induced expression of ACS8, GH3.3, IAA19, and IAA29. W-grown day-5 seedlings of AtWT and phyA-501 were treated for 0, 1, 8, and 24 h with W+FR (R:FR = 0.02), when material was harvested for RNA extraction, as indicated at the top of the panel. Transcript abundance, normalized to EF1α, is shown. Values are means and se of three independent RT-qPCR biological replicates relative to values at 0 h for AtWT. Asterisks indicate significant differences (**, P < 0.01 and *, P < 0.05) between phyA-501 and AtWT seedlings exposed for the same time to W+FR.
(B) Correlation between ACS8, GH3.3, IAA19, and IAA29 expression and relative levels of PHYA protein in the seedlings of AtWT, phyA-501, and phyA>AtPHYA (blue lines and dots) and phyA>ChPHYA (red lines and dots) complementation lines. Gene expression was quantified in W-grown day-5 seedlings exposed to W+FR (R:FR = 0.02) during 24 h, as indicated at the top of the panel. Transcript abundance was normalized to EF1α. Relative phyA protein levels (PHYA:TUB; data already shown in Supplemental Figure 8) were estimated in etiolated seedlings. Values are means and se of three independent RT-qPCR biological replicates relative to values of AtWT. The estimated regression lines for the phyA>AtPHYA (blue line) and phyA>ChPHYA (red line) complementation lines are shown for each correlation.
The expression of the same genes was next quantified in seedlings from the various phyA>AtPHYA and phyA>ChPHYA lines grown for 24 h under W+FR. When plotting transcript levels of phyA target genes as a function of PHYA expression in these lines, the clouds of data corresponding to phyA>ChPHYA lines (red) were separated from those of phyA>AtPHYA lines (blue; Figure 7B). Importantly, the expression of all phyA target genes tested was overall lower in phyA>ChPHYA than in phyA>AtPHYA, indicating that ChphyA repressed more efficiently gene expression than AtphyA (Figure 7B). Consistent with this conclusion, the expression of these and other phyA target genes (Yang et al., 2018) was attenuated in shade-induced seedlings of ChWT compared with AtWT (Supplemental Figure 9). Together, these data further support that ChphyA is intrinsically more active than AtphyA.
DISCUSSION
Currently, the genetic basis of shade tolerance is poorly understood. To address this open question, we have focused on comparative analyses of the hypocotyl response to shade in young seedlings of two related mustards, Arabidopsis and C. hirsuta. Shade avoidance and tolerance are ecological concepts originated from the natural habitats of plant species (Callahan et al., 1997). Hence, defining the shade habit of a species is difficult because shade tolerance is not an absolute value but a relative concept; indeed, plants may exhibit different strategies during the juvenile and adult phases of their lives (Valladares and Niinemets, 2008). Despite the uncertainty, Arabidopsis is generally considered as a shade avoider and it is a model broadly used to study the SAS hypocotyl response, but there is little information referring to its physiological shade-responsiveness habit. C. hirsuta, by contrast, has been previously described as a shade-tolerant species whose hypocotyls are unresponsive to shade (Bealey and Robertson, 1992; Hay et al., 2014), but little is known about other shade-response mechanisms. Here, we confirm that, as expected for a shade-tolerant species, C. hirsuta showed a much better capacity to acclimate to LL than to HL compared with Arabidopsis (Supplemental Figure 1). Most strikingly, C. hirsuta seedlings failed to elongate in response to simulated proximity or canopy shade (Figure 1). Such a dramatic hypocotyl elongation response compared with Arabidopsis makes these two related species good candidates for comparative analyses of divergent responses to shade.
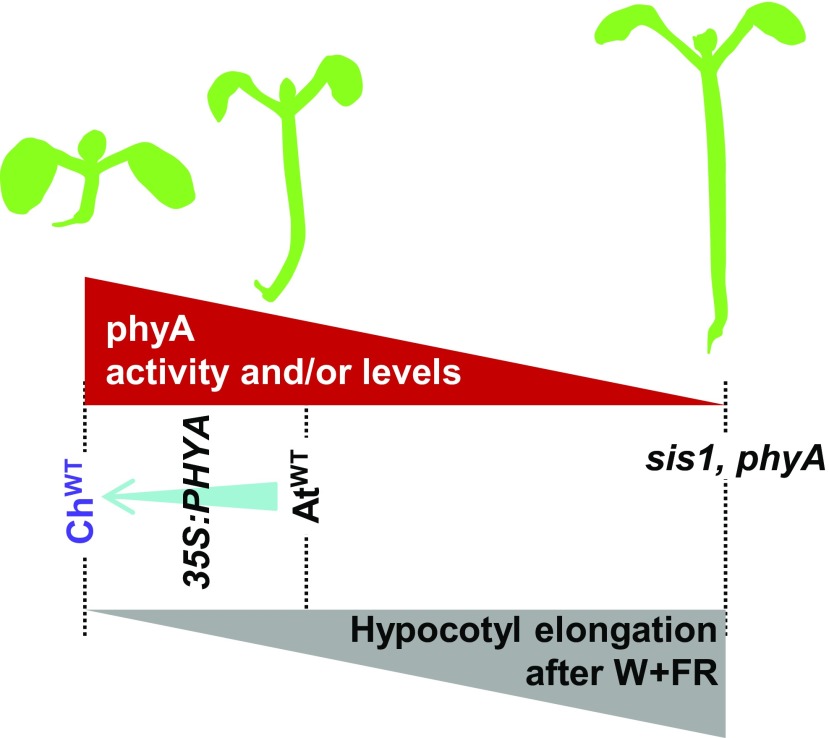
Our comparative and genetic analyses suggest that the absence of a shade-induced hypocotyl elongation in C. hirsuta is not caused by defects in the rapid biosynthesis of auxin in seedlings (Figure 2). Although we cannot exclude local defects in auxin biosynthesis (e.g., in hypocotyls) that might be masked by collecting whole seedlings, our conclusion is consistent with the lack of effect of phyA on the rapid shade-induced biosynthesis of auxin (Yang et al., 2018). On the contrary, we favor that the differences in hypocotyl elongation between these species is the result of a suppression mechanism sustained by the stronger activity of the ChphyA photoreceptor, likely enhanced by the attenuated ChphyB activity (Figure 3B). A stronger intrinsic (specific) repressor activity of ChphyA would result in a strong suppression of the elongation of C. hirsuta seedlings when exposed to shade (Figure 8). The underlying mechanism likely relies, at least partly, upon suppression of auxin signaling via phyA directly binding and stabilizing AUX/IAA proteins, as has been shown in Arabidopsis (Yang et al., 2018). In this scenario, ChphyA seems to suppress not auxin biosynthesis but signaling more strongly than AtphyA, as deduced from the results with transgenic lines (Figure 7B) but also from the stronger repression in shade of auxin-responsive genes with a putative role in auxin signaling (e.g., several IAA and SAUR genes) detected in ChWT compared with AtWT (Supplemental Figure 9).
Figure 8.
Model of How Increased phyA Activity in C. hirsuta Might Implement the Shade Tolerance of Hypocotyl Elongation.
Increases in phyA activity caused by the constitutive overexpression of PHYA also attenuate the shade-induced hypocotyl elongation in transgenic plants, and it results in partially tolerant Arabidopsis seedlings.
AtphyA and ChphyA might achieve different activities by changes in particular residues that could alter susceptibility to posttranslational modifications. For instance, phyA stability, Pfr-to-Pr reversion rate upon shade treatment, and/or interaction with protein partners (e.g., PIF1/PIF3, FHY1/FHL, and AUX/IAA) affect phyA activity in Arabidopsis (Kim et al., 2004; Seo et al., 2004; Dieterle et al., 2005; Genoud et al., 2008; Oka et al., 2012; Sheerin et al., 2015). These intrinsic differences might be also enhanced by changes in protein abundance of phyA and/or other components in its signaling pathway specifically acting in light-grown seedlings (see below). Comparison of the amino acid sequences of AtphyA and ChphyA, however, did not point to any obvious specific residue or region that could be responsible for the observed intrinsic differences in activity (Supplemental Figure 10). This is an issue that would need future research.
The genetic mechanisms underlying physiological evolution remain largely unknown, but changes in the timing, location, and levels of gene expression (i.e., cis-regulatory evolution of key genes) have caused much of morphological evolution changes (Carroll, 2008). Our data on PHYA expression and PHYA protein levels (Figure 5) agree with this view, but they go a step beyond by showing that differences in protein (ChphyA and AtphyA) intrinsic activities also contribute to differential responses to shade (Figures 6 and 7). As both components (levels versus intrinsic activity) are intimately connected (e.g., phyA represses its own expression in a light-dependent manner), at this stage it is difficult to quantify the specific contribution of each one. Moreover, additional components might contribute: while we show that phyA is a central component of a range of regulators that can be modulated in nature to implement shade tolerance, the observation that none of the phyA>ChPHYA lines display a shade-tolerant habit (Supplemental Figure 8D) strongly suggests that additional downstream components of the shade-regulatory network are also participating in suppressing this response in C. hirsuta (e.g., differences in phyB activity). Indeed, it cannot be excluded that the mutant screen, despite identifying an important regulator, did not establish the causal difference between the two species in terms of shade-induced hypocotyl elongation. Nonetheless, our results unveil the importance of modulating photoreceptor activity as a powerful evolutionary mechanism in nature to achieve physiological variation between species, hence enabling the colonization of new, different habitats. In addition, searching for variability in phyA function could provide a suitable tool to modify the impact of neighbors’ cues in crops to minimize yield losses.
METHODS
Plant Material and Plant Growth Conditions
Plants of Arabidopsis (Arabidopsis thaliana) Col-0 (AtWT) phyA-501 (in the Col-0 background), phyB-1, phyB-4 (both phyB-deficient lines are in the Landsberg erecta background), and Cardamine hirsuta, of the reference Oxford accession (ChWT), have been described (Reed et al., 1993; Hay et al., 2014; Martínez-García et al., 2014). Plant growth conditions have been described elsewhere (Martínez-García et al., 2014; Gallemí et al., 2016). Normal light conditions refer to W produced by cool-white vertical fluorescent tubes (PAR of 20–24 µmol m−2 s−1). LL and HL conditions corresponded to PAR values of 4 and 200 µmol m−2 s−1, respectively. Shade treatments in seedlings were provided by enriching W (R:FR of 2.5) with different intensities of FR LEDs (730-nm peak; Philips Greenpower Research modules) to produce the indicated R:FR (0.091–0.021) without altering PAR. Light spectra are presented in Supplemental Figure 2. For estimating petiole and rachis length, rosette plants were grown under a long-day (16 h of light, 8 h of dark) photoperiod, in which W was generated by cool-white horizontal fluorescent tubes (PAR of ∼100 µmol m−2 s−1, R:FR of 3.0); for shade treatments, W was supplemented with FR (W+FR, PAR of ∼100 µmol m−2 s−1, R:FR of 0.05). Fluence rates were measured with a Spectrosense2 meter associated with a four-channel sensor (Skye Instruments), which measures PAR (400–700 nm) and 10-nm windows in the blue (464–473 nm), R (664–673 nm), and FR (725–734 nm) regions (Gallemí et al., 2017). Light spectra were generated using a Flame Model Spectrometer with Sony Detector (FLAME-S; Ocean Optics).
Hypocotyl, Petiole, and Rachis Measurements
For hypocotyl measurement, ∼30 seeds of each genotype were germinated on the plates for observing the seedling phenotype and at least 20 seedlings were measured for quantification of hypocotyl length. All experiments were repeated at least three times with consistent results. Hypocotyl measurements from all the different experiments were averaged ((Supplemental Data Set 9). For petiole measurement, ∼30 seeds of each genotype were germinated under continuous W. One week later, 20 seedlings in a similar stage of development were transferred to individual pots and moved to a long-day growth chamber (R:FR of 3.0). After 1 week, half of the rosette plants stayed under W and the other half were moved to a W+FR shelf (R:FR of 0.05). After 1 week of differential R:FR treatment, leaves were harvested and petiole was measured; in the case of complex leaves from C. hirsuta, rachises were measured, covering the distance from the base of the leaf to the base of the main leaflet (Supplemental Figure 3). At least eight leaves were measured for quantification of petiole and rachis length for each leaf number. Experiments were repeated four times with consistent results. Petiole and rachis measurements from all four experiments were averaged ((Supplemental Data Set 9).
Photosynthetic Pigment Quantification and Chlorophyll Fluorescence
Whole 7-d-old seedlings of the indicated genotypes grown under W or W+FR (Figures 1 and 4) or transferred to HL conditions (Supplemental Figure 1B) were harvested, ground in liquid nitrogen, and the resulting powder was used for quantification of chlorophylls and carotenoids spectrophotometrically or by HPLC, as described (Bou-Torrent et al., 2015).
Fluorescence measurements were performed on seedlings grown under different light regimes using a MAXI-PAM fluorometer (Heinz Walz). For every measurement, the whole cotyledons of seven seedlings were considered. Fv/Fm was calculated as (Fm − Fo)/Fm, where Fm and Fo are the maximum and minimum fluorescence of dark-adapted samples, respectively. For dark acclimation, plates were incubated for at least 30 min in darkness to allow the full relaxation of photosystems. Rapid light curves were constructed with 10 incremental steps of actinic irradiance (E; 0, 1, 21, 56, 111, 186, 281, 396, 531, and 701 μmol photons m−2 s−1). For each step, the effective quantum yield of PSII (ΔF/Fm′) was monitored every 1 min, and relative electron transport rate was calculated as E × ΔF/Fm′. The light response was characterized by fitting iteratively, using MS Excel Solver, the model of Platt et al., (1980) to relative electron transport rate versus E curves. The fit was very good in all the cases (r > 0.98).
Expression Analyses by RT-qPCR and RNA-Seq
RNA was extracted from whole seedlings of Arabidopsis and C. hirsuta (grown as detailed in each experiment, three biological replicates per time point, each biological replicate composed of 30–40 seedlings) using commercial kits (RNAeasy Plant Mini kit, Qiagen; or the semiautomatic Maxwell SimplyRNA kit, Promega). For real-time qPCR analysis, 2 μg of RNA was reverse-transcribed using the M-MLV Reverse Transcriptase (Invitrogen) or Transcriptor First Strand cDNA synthesis (Roche). Reference genes used were UBQ10, EF1α, SPC25, and/or YLS8 ((Supplemental Table 2).
For RNA-seq analyses, quantification of gene expression was performed as indicated elsewhere (Gan et al., 2016) and detailed in the Supplemental Data. From the lists of genes, we selected as differentially expressed those whose fold change was significantly (adjusted P < 0.05) higher than 1.5 (Supplemental Data Sets 1 and 3) or lower than 0.67 (Supplemental Data Sets 2 and 4) in seedlings treated for 1 h with W+FR compared with those grown under W in either C. hirsuta (Supplemental Data Sets 1 and 2) or Arabidopsis (Supplemental Data Sets 3 and 4).
GO and MapMan Analysis
A strict synteny-based approach was used to identify conserved orthologs between the two species. The Arabidopsis orthologs of the C. hirsuta genes were used for getting the GO term annotations and MMBs. The GO term annotations for Arabidopsis genes, used as a reference, were obtained from the Gene Ontology Consortium (http://www.geneontology.org/; Ashburner et al., 2000). The results are presented as Supplemental Data Set 5. For the MMB analyses, each list of genes was submitted to the Mercator gene function prediction pipeline (Lohse et al., 2014), which annotates the query genes with the hierarchical ontology MMBs (Thimm et al., 2004; Klie and Nikoloski, 2012). Based on these MMB annotations, exact Fisher’s tests for function enrichment within the six groups of DEGs were performed and interpreted (Supplemental Data Set 6).
Protein Extraction and Immunoblot Analysis
Methods for extracting and detecting phyA protein levels in Arabidopsis or C. hirsuta seedlings (Martínez-García et al., 1999; Gallemí et al., 2017) are as follows. Protein extracts from C. hirsuta seedlings analyzed in Figure 3 and Supplemental Figure 6 were prepared following the direct extract protocol (Martínez-García et al., 1999) with the modifications described below. Extracts were prepared from ChWT, sis1, and RNAi-ChPHYA seedlings germinated and grown in the dark for 4 d. Ten seedlings per genotype were harvested in the dark and extracted in 1.5-mL microfuge tubes containing 300 μL of Laemmli buffer supplemented with protease inhibitors (10 µg/mL aprotinin, 1 µg/mL E-64, 10 µg/mL leupeptin, 1 µg/mL pepstatin A, and 100 µM PMSF). These extracts were prepared in duplicate and similar results were observed. Plant material was ground using disposable grinders in the Eppendorf tube at room temperature until the mixture was homogeneous (usually less than 15 s). Once all the samples were prepared, tubes were placed in boiling water for 3 min. Tubes were centrifuged in a microfuge at maximum speed (13,000g, 10 min) immediately before loading. Fifteen microliters of each extract, equivalent to ∼0.5 seedlings, was loaded per lane in an SDS-8% PAGE device.
Protein extracts analyzed in Figure 5 were prepared from AtWT and ChWT seedlings grown as indicated in the figure legend. Extracts were obtained from four biological replicates. Protein extracts analyzed in Figure 6 were prepared from AtWT, phyA-501, phyA>AtPHYA, and phyA>ChPHYA seedlings germinated and grown in the dark for 4 d, as described (Gallemí et al., 2017). Extracts were obtained from three biological replicates. Each biological replicate was obtained from ∼100 seedlings. Protein concentration in these extracts was determined using the Pierce BCA Protein Assay kit (catalog number 23225). Five or 7.5 µg of each extract was loaded per lane in an SDS-8% PAGE device.
Immunoblot analyses of PHYA and TUB were performed at the same time with the antibodies (073D, commercial anti-TUB) and dilutions indicated elsewhere (Martínez-García et al., 2014). Anti-mouse horseradish peroxidase-conjugated antibody (Promega) was used as a secondary antibody. ECL or ECL-plus chemiluminescence kits (GE Healthcare) were used for detection. Signal was visualized and quantified using the ChemiDoc Touch Imaging System (Bio-Rad).
Hormone Analyses
Hormone extraction and analysis were performed as described (Durgbanshi et al., 2005) with a few modifications. Briefly, 0.02 g of dry tissue (∼150 AtWT seedlings and 100 ChWT seedlings) was extracted in 1 mL of ultrapure water after spiking with 50 ng of [2H2]IAA in a ball mill (MillMix20, Domel). After centrifugation at 4000g at 4°C for 10 min, supernatants were recovered and pH adjusted to 3 with 30% (v/v) acetic acid. The water extract was partitioned twice against 2 mL of diethyl ether, and the organic layer was recovered and evaporated under vacuum in a centrifuge concentrator (Speed Vac, Jouan). Once dried, the residue was resuspended in a 10:90 methanol:water solution by gentle sonication. The resulting solution was filtered through 0.22-µm polytetrafluoroethylene membrane syringe filters (Albet) and directly injected into an ultraperformance LC system (Acquity SDS, Waters). Chromatographic separations were performed on a reverse-phase C18 column (gravity, 50 × 2.1 mm, 1.8-µm particle size, Macherey-Nagel) using a methanol:water (both supplemented with 0.1% acetic acid) gradient at a flow rate of 300 μL/min. IAA was quantified with a triple quadrupole mass spectrometer (Micromass) connected online to the output of the column though an orthogonal Z-spray electrospray ion source.
Data Availability
The Illumina RNA-seq reads are available from the website http://chi.mpipz.mpg.de/assembly. Source code of BAMLINK is available at http://chi.mpipz.mpg.de/software. The data that support the findings of this study are also available from the corresponding author on request.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or the C. hirsuta (http://chi.mpipz.mpg.de/assembly) databases under the following accession numbers: AtATHB2 (At4g16780), ChATHB2 (CARHR223400), AtPIL1 (At2g46970), ChPIL1 (CARHR142340), AtUBQ10 (At4g05320), AtPHYA (At1g09570), ChPHYA (CARHR009540), ACS8 (At4g37770), GH3.3 (At2g23170), IAA19 (At3g15540), IAA29 (At3g15540), AtEF1α (At5g60390), ChEF1α (CARHR274060 and CARHR274080), SPC25 (At2g39960), ChSPC25 (CARHR134880 and CARHR134890), YLS8 (At5g08290), and ChYLS8 (CARHR204840).
Supplemental Data
Supplemental Figure 1. Photosynthetic-related responses of A. thaliana and C. hirsuta seedlings to changing light conditions.
Supplemental Figure 2. Light spectra of the treatments used in this study.
Supplemental Figure 3. Longitudinal length of A. thaliana and C. hirsuta leaves respond differently to simulated shade.
Supplemental Figure 4. A. thaliana and C. hirsuta seedlings change gene expression differently in response to simulated shade.
Supplemental Figure 5. The expression of a set of shade-induced but auxin-dependent genes, identified in A. thaliana, is also shade-induced in C. hirsuta.
Supplemental Figure 6. Reduction of phyA activity in C. hirsuta seedlings results in a sis phenotype.
Supplemental Figure 7. Partial alignment of ChPHYA/AtPHYA, ChEF1α/AtEF1α, ChSPC25/AtSPC25 and ChYLS8/AtYLS8 sequences.
Supplemental Figure 8. Strategies to compare biological activity between AtphyA and ChphyA in transgenic lines.
Supplemental Figure 9. The expression of a set of shade-induced phyA-repressed genes, identified in A. thaliana, is attenuated in C. hirsuta.
Supplemental Figure 10. Alignment of C. hirsuta and A. thaliana phyA amino acid sequences.
Supplemental Table 1. RPKM of eight genes commonly used for normalizing in RT-qPCR analyses.
Supplemental Table 2. Primers used in this work.
Supplemental Data Set 1. Bioset of up-regulated genes in C. hirsuta seedlings in response to simulated shade.
Supplemental Data Set 2. Bioset of down-regulated genes in C. hirsuta seedlings in response to simulated shade.
Supplemental Data Set 3. Bioset of up-regulated genes in A. thaliana seedlings in response to simulated shade.
Supplemental Data Set 4. Bioset of down-regulated genes in A. thaliana seedlings in response to simulated shade.
Supplemental Data Set 5. Results of Venn diagrams of the GO categorization.
Supplemental Data Set 6. Functional enrichment groups based on the MapMan-Bin analyses.
Supplemental Data Set 7. Bioset of shade-regulated OMCL groups in Geranium pyrenaicum petioles in response to simulated shade.
Supplemental Data Set 8. Bioset of shade-regulated OMCL groups in Geranium robertianum petioles in response to simulated shade.
Supplemental Data Set 9. Summary of statistical tests.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ester Botterweg and M. Rosa Rodríguez (Centre for Research in Agricultural Genomics [CRAG]) for their technical support; Victor González and Martí Bernardo (Bioinformatics Core unit, CRAG) for help in statistical analyses; Peter Quail (Plant Gene Expression Center) for providing the anti-phyA antibody; Fernando Valladares (National Museum of Natural History, Madrid, Spain) for discussions about the shade habit of Arabidopsis and C. hirsuta; and Charlotte Gommers (CRAG) for comments on the article. Support for this work was provided by the Spanish Ministerio de Economía y Competitividad (MINECO; FPI program predoctoral fellowship to M.J.M.-C.), the Agència d’Ajuts Universitaris i de Recerca (AGAUR-Generalitat de Catalunya; FI program predoctoral fellowship to S.P.), and La Caixa Foundation (INPhINIT fellowship LCF/BQ/IN18/11660004 to L.C.). Support was also provided by an International CRAG “Severo Ochoa” postdoctoral program fellowship and a postdoctoral contract funded by the European Commission (H2020-MSCA-IF-2017, Proposal 797473 to J.M.-R.), and by a Marie Curie postdoctoral contract funded by the European Commission and a CRAG short-term fellowship (FP7-PEOPLE-IEF-2008, Proposal 237492 to C.T.). Our research is supported by the Biotechnology and Biological Science Research Council (grant BB/H006974/1), by the Max Planck Society (core grant to M.T.), by MINECO-FEDER (grants BIO2017-85316-R and BIO2017-84041-P), and AGAUR (grants 2017-SGR1211, 2017-SGR710, and Xarba to J.F.M.-G. and M.R.C.). We also acknowledge the support of MINECO for the “Centro de Excelencia Severo Ochoa 2016-2019” (award SEV-2015-0533) and by the CERCA Programme/Generalitat de Catalunya.
AUTHOR CONTRIBUTIONS
J.F.M.-G. conceived the original research plan and directed and coordinated the study; A.H., H.J., X.G., and M.T. performed RNA-seq and analyzed the data; A.G.-C. analyzed auxin levels; L.M. and M.R.-C. measured and analyzed photosynthetic parameters and pigment levels; M.J.M.-C., S.P., C.T., J.M.-R., P.P.-A., and I.R.-V. performed all the other experiments; all authors analyzed their data and discussed the results; J.F.M.-G. wrote the article with revisions of M.R.-C. and contributions and/or comments of all other authors.
References
- Ashburner M., et al. (2000). Gene Ontology: Tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141. [DOI] [PubMed] [Google Scholar]
- Bealey C.E., Robertson P.A. (1992). Coppice management for pheasants In Ecology and Management of Coppice Woodlands, Buckley G.P., ed (Dordrecht: Springer; ), pp. 193–210. [Google Scholar]
- Benkov M.A., Yatsenko A.M., Tikhonov A.N. (2019). Light acclimation of shade-tolerant and sun-resistant Tradescantia species: Photochemical activity of PSII and its sensitivity to heat treatment. Photosynth. Res. 139: 203–214. [DOI] [PubMed] [Google Scholar]
- Bou-Torrent J., Galstyan A., Gallemí M., Cifuentes-Esquivel N., Molina-Contreras M.J., Salla-Martret M., Jikumaru Y., Yamaguchi S., Kamiya Y., Martínez-García J.F. (2014). Plant proximity perception dynamically modulates hormone levels and sensitivity in Arabidopsis. J. Exp. Bot. 65: 2937–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J., Toledo-Ortiz G., Ortiz-Alcaide M., Cifuentes-Esquivel N., Halliday K.J., Martinez-García J.F., Rodriguez-Concepcion M. (2015). Regulation of carotenoid biosynthesis by shade relies on specific subsets of antagonistic transcription factors and cofactors. Plant Physiol. 169: 1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnola J.I., Ploschuk E., Benech-Arnold T., Finlayson S.A., Casal J.J. (2012). Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol. 160: 1110–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan H.S., Pigliucci M., Schlichting C.D. (1997). Developmental phenotypic plasticity: Where ecology and evolution meet molecular biology. BioEssays 19: 519–525. [DOI] [PubMed] [Google Scholar]
- Cantón F.R., Quail P.H. (1999). Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol. 121: 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. (2008). Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Casal J.J. (2012). Shade avoidance. The Arabidopsis Book 10: e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Candia A.N., Sellaro R. (2014). Light perception and signalling by phytochrome A. J. Exp. Bot. 65: 2835–2845. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Esquivel N., Bou-Torrent J., Galstyan A., Gallemí M., Sessa G., Salla Martret M., Roig-Villanova I., Ruberti I., Martínez-García J.F. (2013). The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 75: 989–1002. [DOI] [PubMed] [Google Scholar]
- Ciolfi A., Sessa G., Sassi M., Possenti M., Salvucci S., Carabelli M., Morelli G., Ruberti I. (2013). Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol. 163: 331–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M., Keuskamp D.H., Bongers F.J., Hornitschek P., Gommers C.M.M., Reinen E., Martínez-Cerón C., Fankhauser C., Pierik R. (2016). Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 26: 3320–3326. [DOI] [PubMed] [Google Scholar]
- de Wit M., Ljung K., Fankhauser C. (2015). Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208: 198–209. [DOI] [PubMed] [Google Scholar]
- Dieterle M., Bauer D., Büche C., Krenz M., Schäfer E., Kretsch T. (2005). A new type of mutation in phytochrome A causes enhanced light sensitivity and alters the degradation and subcellular partitioning of the photoreceptor. Plant J. 41: 146–161. [DOI] [PubMed] [Google Scholar]
- Durgbanshi A., Arbona V., Pozo O., Miersch O., Sancho J.V., Gómez-Cadenas A. (2005). Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. J. Agric. Food Chem. 53: 8437–8442. [DOI] [PubMed] [Google Scholar]
- Gallemí M., Galstyan A., Paulišić S., Then C., Ferrández-Ayela A., Lorenzo-Orts L., Roig-Villanova I., Wang X., Micol J.L., Ponce M.R., Devlin P.F., Martínez-García J.F. (2016). DRACULA2 is a dynamic nucleoporin with a role in regulating the shade avoidance syndrome in Arabidopsis. Development 143: 1623–1631. [DOI] [PubMed] [Google Scholar]
- Gallemí M., Molina-Contreras M.J., Paulišić S., Salla-Martret M., Sorin C., Godoy M., Franco-Zorrilla J.M., Solano R., Martínez-García J.F. (2017). A non-DNA-binding activity for the ATHB4 transcription factor in the control of vegetation proximity. New Phytol. 216: 798–813. [DOI] [PubMed] [Google Scholar]
- Gan X., et al. (2016). The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2: 16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers C.M., Keuskamp D.H., Buti S., van Veen H., Koevoets I.T., Reinen E., Voesenek L.A., Pierik R. (2017). Molecular profiles of contrasting shade response strategies in wild plants: Differential control of immunity and shoot elongation. Plant Cell 29: 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers C.M., Visser E.J., St. Onge K.R., Voesenek L.A., Pierik R. (2013). Shade tolerance: When growing tall is not an option. Trends Plant Sci. 18: 65–71. [DOI] [PubMed] [Google Scholar]
- Gommers C.M.M., Buti S., Tarkowská D., Pěnčík A., Banda J.P., Arricastres V., Pierik R. (2018). Organ-specific phytohormone synthesis in two Geranium species with antithetical responses to far-red light enrichment. Plant Direct 2: e00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Jiang J., Li H., Song A., Chen S., Chen F. (2015). The differential response of two Chrysanthemum cultivars to shading: Photosynthesis, chloroplast, and sieve element-companion cell ultrastructure. HortScience 50: 1192–1195. [Google Scholar]
- Hay A., Tsiantis M. (2016). Cardamine hirsuta: A comparative view. Curr. Opin. Genet. Dev. 39: 1–7. [DOI] [PubMed] [Google Scholar]
- Hay A.S., et al. (2014). Cardamine hirsuta: A versatile genetic system for comparative studies. Plant J. 78: 1–15. [DOI] [PubMed] [Google Scholar]
- Hersch M., Lorrain S., de Wit M., Trevisan M., Ljung K., Bergmann S., Fankhauser C. (2014). Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 6515–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer A.G., Mozley D., Landschütze V., Thomas B., Gatz C. (1995). Function of phytochrome A in potato plants as revealed through the study of transgenic plants. Plant Physiol. 109: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofhuis H., et al. (2016). Morphomechanical innovation drives explosive seed dispersal. Cell 166: 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve A.A., Jawdy S.S., Gunter L.E., Allen S.M., Yang X., Tuskan G.A., Wullschleger S.D., Weston D.J. (2012). Initial characterization of shade avoidance response suggests functional diversity between Populus phytochrome B genes. New Phytol. 196: 726–737. [DOI] [PubMed] [Google Scholar]
- Kim J.I., Shen Y., Han Y.J., Park J.E., Kirchenbauer D., Soh M.S., Nagy F., Schäfer E., Song P.S. (2004). Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16: 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klie S., Nikoloski Z. (2012). The choice between MapMan and Gene Ontology for automated gene function prediction in plant science. Front. Genet. 3: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnen M.V., Schmid-Siegert E., Trevisan M., Petrolati L.A., Sénéchal F., Müller-Moulé P., Maloof J., Xenarios I., Fankhauser C. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28: 2889–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T., Kobayashi J., Horiguchi G., Demura T., Sakakibara H., Tsukaya H., Nagatani A. (2010). Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 153: 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Nagel A., Herter T., May P., Schroda M., Zrenner R., Tohge T., Fernie A.R., Stitt M., Usadel B. (2014). Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37: 1250–1258. [DOI] [PubMed] [Google Scholar]
- Martínez-García J.F., Gallemí M., Molina-Contreras M.J., Llorente B., Bevilaqua M.R., Quail P.H. (2014). The shade avoidance syndrome in Arabidopsis: The antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLoS One 9: e109275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia J.F., Galstyan A., Salla-Martret M., Cifuentes-Esquivel N., Gallemí M., Bou-Torrent J. (2010). Regulatory components of shade avoidance syndrome. Adv. Bot. Res. 53: 65–116. [Google Scholar]
- Martínez-García J.F., Monte E., Quail P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20: 251–257. [DOI] [PubMed] [Google Scholar]
- Mathews S. (2010). Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22: 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Ono Y., Toledo-Ortiz G., Kokaji K., Matsui M., Mochizuki N., Nagatani A. (2012). Arabidopsis phytochrome A is modularly structured to integrate the multiple features that are required for a highly sensitized phytochrome. Plant Cell 24: 2949–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Alcaide M., Llamas E., Gomez-Cadenas A., Nagatani A., Martínez-García J.F., Rodríguez-Concepción M. (2019). Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving HY5 and abscisic acid. Plant Cell 31: 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Gallegos C.L., Harrison W.G. (1980). Photoinhibition of photosynthesisin natural assemblages of marine phytoplankton. Journal of Marine Research 38: 687–701. [Google Scholar]
- Procko C., Crenshaw C.M., Ljung K., Noel J.P., Chory J. (2014). Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 165: 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson P.R., McCormac A.C., Irvine A.S., Smith H. (1996). Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat. Biotechnol. 14: 995–998. [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P.F., Martínez-García J.F. (2006). Identification of primary target genes of phytochrome signaling: Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., Martínez-García J.F. (2007). Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 26: 4756–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Martínez-García J.F. (2016). Plant responses to vegetation proximity: A whole life avoiding shade. Front. Plant Sci. 7: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Staal M., Elzenga J.T., Yokoyama R., Nishitani K., Voesenek L.A., Pierik R. (2010). Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 154: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. (1982). Light quality, photoperception, and plant strategy. Annu. Rev. Plant Physiol. 33: 481–518. [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Ueoka-Nakanishi H., Hori N., Ishida K., Ono N., Yamashino T., Nakamichi N., Mizuno T. (2011). Characterization of shade avoidance responses in Lotus japonicus. Biosci. Biotechnol. Biochem. 75: 2148–2154. [DOI] [PubMed] [Google Scholar]
- Valladares F., Niinemets U. (2008). Shade tolerance, a key plant feature of complex nature and consequences. Annu. Rev. Ecol. Evol. Syst. 39: 237–257. [Google Scholar]
- Vlad D., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Vuolo F., Mentink R.A., Hajheidari M., Bailey C.D., Filatov D.A., Tsiantis M. (2016). Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 30: 2370–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Xie F., Jiang Y., Li Z., Huang X., Li L. (2018). Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell 44: 29–41.e24. [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Casal J.J., Whitelam G.C. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: Weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18: 788–794. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Illumina RNA-seq reads are available from the website http://chi.mpipz.mpg.de/assembly. Source code of BAMLINK is available at http://chi.mpipz.mpg.de/software. The data that support the findings of this study are also available from the corresponding author on request.