The Pseudomonas syringae effector protein AvrRpm1 ADP ribosylates RIN4 proteins from Arabidopsis and soybean, which promotes association of RIN4 with EXO70E2 and suppression of callose deposition.
Abstract
The Pseudomonas syringae effector protein AvrRpm1 activates the Arabidopsis (Arabidopsis thaliana) intracellular innate immune receptor protein RESISTANCE TO PSEUDOMONAS MACULICOLA1 (RPM1) via modification of a second Arabidopsis protein, RPM1-INTERACTING PROTEIN4 (AtRIN4). Prior work has shown that AvrRpm1 induces phosphorylation of AtRIN4, but homology modeling indicated that AvrRpm1 may be an ADP-ribosyl transferase. Here, we show that AvrRpm1 induces ADP-ribosylation of RIN4 proteins from both Arabidopsis and soybean (Glycine max) within two highly conserved nitrate-induced (NOI) domains. It also ADP ribosylates at least 10 additional Arabidopsis NOI domain-containing proteins. The ADP-ribosylation activity of AvrRpm1 is required for subsequent phosphorylation on Thr-166 of AtRIN4, an event that is necessary and sufficient for RPM1 activation. We also show that the C-terminal NOI domain of AtRIN4 interacts with the exocyst subunits EXO70B1, EXO70E1, EXO70E2, and EXO70F1. Mutation of either EXO70B1 or EXO70E2 inhibited secretion of callose induced by the bacterial flagellin-derived peptide flg22. Substitution of RIN4 Thr-166 with Asp enhanced the association of AtRIN4 with EXO70E2, which we posit inhibits its callose deposition function. Collectively, these data indicate that AvrRpm1 ADP-ribosyl transferase activity contributes to virulence by promoting phosphorylation of RIN4 Thr-166, which inhibits the secretion of defense compounds by promoting the inhibitory association of RIN4 with EXO70 proteins.
INTRODUCTION
Plants are able to defend themselves from a wide range of disease-causing bacteria and do so in part by detecting the presence of type III secretion system effector proteins delivered by these pathogens (Martin et al., 2003). Two of these effectors, AvrB and AvrRpm1, are detected in Arabidopsis (Arabidopsis thaliana) by RPM1, a member of the nucleotide binding leucine-rich repeat (NLR) family of intracellular innate immune receptors (Bisgrove et al., 1994; Grant et al., 1995; Jones et al., 2016). Although it has been known for more than 20 years that RPM1 confers dual effector specificity, most research on this receptor has focused on the molecular basis of AvrB recognition, with less molecular information available on how AvrRpm1 is detected.
Recognition of AvrB by RPM1 requires a second host protein, AtRIN4 (Mackey et al., 2002). Upon delivery into the host, AvrB becomes acylated and associates with the plasma membrane (Nimchuk et al., 2000; Gao et al., 2011). There, it interacts with AtRIN4 and the protein kinase RIPK (Liu et al., 2011). This interaction leads to the autophosphorylation of RIPK and the subsequent phosphorylation of AtRIN4 at residues Thr-21, Ser-160, and Thr-166 (Liu et al., 2011). Phosphorylation of AtRIN4 Thr-166 is necessary and sufficient for activation of RPM1 (Chung et al., 2011).
Like the detection of AvrB, detection of AvrRpm1 also requires the presence of AtRIN4 (Mackey et al., 2002). Furthermore, expression of AvrRpm1 in rpm1 mutant Arabidopsis plants induces a modification of AtRIN4 (Mackey et al., 2002). This modification was observed as reduced band mobility when resolved by SDS-PAGE. When treated with calf intestine alkaline phosphatase, this shift in AtRIN4 size was eliminated, indicating that AvrRpm1 induces phosphorylation on AtRIN4 (Mackey et al., 2002). However, the specific AtRIN4 residues modified by AvrRpm1 have not been identified, and whether AvrRpm1 possesses kinase activity has not been determined. The possibility that AvrRpm1 induces a distinct or additional modification of AtRIN4 arose from computational structural modeling of AvrRpm1, which indicated that AvrRpm1 may contain a fold homologous to ADP-ribosyl transferases such as diphtheria toxin. Elimination of AvrRpm1 function by mutagenesis of putative catalytic residues supported this model (Cherkis et al., 2012).
Soybean (Glycine max) is also able to detect and respond to the presence of AvrB and AvrRpm1 (Ashfield et al., 1995). Unlike Arabidopsis, however, recognition of AvrB and AvrRpm1 in soybean is divided among two NLR receptors, RPG1b and RPG1r, respectively (Ashfield et al., 1995). The soybean genome contains four homologs of RIN4, designated GmRIN4a (Glyma03g19920), GmRIN4b (Glyma16g12160), GmRIN4c (Glyma18g36000), and GmRIN4d (Glyma08g46400; Supplemental Figure 1). Thus, the role of GmRIN4 in disease resistance might be subfunctionalized. In support of this idea, virus-induced gene silencing of GmRIN4a or GmRIN4b, but not GmRIN4c or GmRIN4d, suppresses AvrB recognition by RPG1b (Selote and Kachroo, 2010). In addition, recognition of AvrRpm1 by RPG1r can be suppressed by AvrRpt2, a Cys protease that cleaves all four GmRIN4 orthologs (Ashfield et al., 2014), consistent with findings from Arabidopsis (Ritter and Dangl, 1996; Mackey et al., 2003; Kim et al., 2005a). This result indicates that one or more of the GmRIN4s likely are required for RPG1r-mediated recognition of AvrRpm1.
In plants genetically unable to recognize them, type III effectors such as AvrRpm1 contribute to the ability of bacteria to proliferate and cause disease (Ritter and Dangl, 1995). This virulence activity is mediated in many cases via suppression of host defenses, including those triggered by microbe-associated molecular patterns (MAMPs). RIN4 negatively regulates the ability of Arabidopsis plants to produce cell wall fortifications, such as deposition of callose, in response to a prototypical MAMP consisting of a 22-amino acid peptide from the bacterial flagellin protein (flg22; Kim et al., 2005b). AvrRpm1 appears to promote the activity of RIN4, as transgenic expression of AvrRpm1 suppresses flg22-induced callose deposition (Kim et al., 2005b). Thus, AvrRpm1-induced modifications of RIN4 that contribute to activation of NLR receptors in Arabidopsis, soybean, and other plants may also underlie virulence activity in susceptible host plants that lack a responsive NLR.
With the working hypothesis that at least one GmRIN4 is required for RPG1r-mediated recognition of AvrRpm1 in soybean, we sought to elucidate the underlying molecular mechanisms. Specifically, we wanted to determine whether AvrRpm1 could induce modification of one or more of the GmRIN4 proteins, and if so, what these modifications might be. Here, we show that AvrRpm1 induces mono-ADP-ribosylation on AtRIN4, GmRIN4a, and GmRIN4b within their conserved nitrate-induced (NOI) domains. A D153A substitution in the C-terminal NOI domain of AtRIN4, which blocks ADP-ribosylation at this position, inhibits phosphorylation on Thr-166 and thereby inhibits activation of RPM1 by AvrRpm1. We also show that RIN4 proteins from both Arabidopsis and soybean associate with Arabidopsis exocyst subunits EXO70B1, EXO70E1, EXO70E2, and EXO70F1 and that these interactions are influenced by modification of Thr-166. Lastly, we show that mutation of EXO70B1 and EXO70E2 impairs flg22-induced callose deposition, indicating that AvrRpm1 likely inhibits callose deposition, at least in part, by enhancing the ability of RIN4 to bind to EXO70 proteins and interfere with EXO70-mediated secretion.
RESULTS
AvrRpm1 Modifies GmRIN4b in Planta
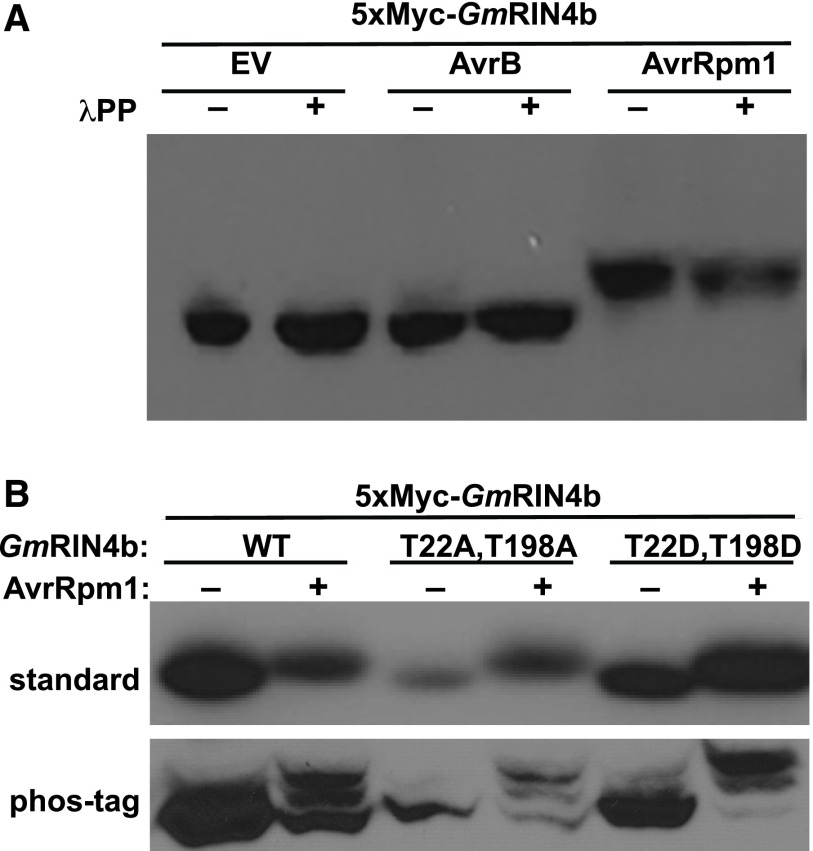
Prior work has shown that the GmRIN4b gene can complement the ability of an Arabidopsis rin4 mutant to recognize either AvrB or AvrRpm1 (Selote and Kachroo, 2010). This observation indicates that GmRIN4b is likely modified by these effectors. To test this hypothesis, we generated an N-terminal Myc-tagged GmRIN4b construct under control of a dexamethasone-inducible promoter in pTA7001 (Aoyama and Chua, 1997) and transiently coexpressed it in Nicotiana benthamiana along with transiently expressed AvrB, AvrRpm1, or an empty vector control (EV). To detect possible phosphorylation of GmRIN4b, we isolated total protein from N. benthamiana leaves and separated the proteins on a polyacrylamide gel supplemented with Phos-tag reagent (Kinoshita et al., 2006), followed by immunoblot analysis. The mobility of GmRIN4b was markedly reduced when coexpressed with AvrRpm1, but not when coexpressed with AvrB or EV (Figure 1).
Figure 1.
AvrRpm1 Induces a Size Shift in GmRIN4b That Is Insensitive to Protein Phosphatase Treatment.
(A) 5×Myc-GmRIN4b was transiently expressed in N. benthamiana in the presence of AvrB, AvrRpm1, or EV. Total protein lysate was incubated with (+) or without (−) λPP and separated on an 8% polyacrylamide gel supplemented with 50 μM Phos-tag reagent. Proteins were detected by immunoblot using anti-Myc antibodies. Similar results were obtained from three independent experiments.
(B) AvrRpm1-dependent modification of GmRIN4b occurs at a novel location. The wild-type, phosphodeficient (T22A T198A), or phosphomimic (T22D, T198D) alleles of GmRIN4b were transiently expressed in N. benthamiana in the presence of AvrRpm1. Total protein lysate was separated for 16 h on a 10-cm 8% polyacrylamide gel at low voltage (top) or separated on an 8% polyacrylamide gel supplemented with 50 μM Phos-tag reagent (bottom). Proteins were detected by immunoblot using anti-Myc antibodies. Similar results were obtained from three independent experiments. WT, wild type.
See also Supplemental Figure 1.
To determine whether the AvrRpm1-induced modification of GmRIN4b was phosphorylation, protein extracts were treated with lambda protein phosphatase (λPP) prior to being separated by SDS-PAGE. The posttranslational modification to GmRIN4b was not sensitive to λPP treatment (Figure 1A), indicating that the modification was most likely not phosphorylation.
Previous work with Arabidopsis RIN4 has shown that AvrB induces phosphorylation of AtRIN4 at Thr-21, Ser-160, and Thr-166 (Liu et al., 2011). Furthermore, substitution of Thr-166 with a phosphomimicking amino acid (T166E or T166D) induces an RPM1-mediated hypersensitive response in the absence of AvrB (Chung et al., 2011; Liu et al., 2011). Equivalent substitutions in GmRIN4b (T22D, T198D) induce an RPG1b-mediated hypersensitive response (Selote et al., 2013). To determine whether these residues are modified in the presence of AvrRpm1, GmRIN4b (T22A, T198A) or GmRIN4b (T22D, T198D) was transiently expressed in N. benthamiana in the presence of AvrRpm1 or EV. Total protein was harvested from leaves and resolved by SDS-PAGE on a gel supplemented with Phos-tag reagent. Regardless of the substitutions at these positions, GmRIN4b mobility was reduced in the presence of AvrRpm1 (Figure 1B). Additionally, multiple distinct bands were visible when GmRIN4b was expressed with AvrRpm1 (Figure 1B). Taken together, these results indicate that GmRIN4b is modified at multiple locations in the presence of AvrRpm1 and that these modifications are at residues that differ from those involved in AvrB-induced activation of RPG1b.
GmRIN4b Is ADP Ribosylated by AvrRpm1 in Vivo
To identify the modifications induced by AvrRpm1, and their locations on GmRIN4b, we performed immunoprecipitation of GmRIN4b followed by mass spectrometry. N-Terminal super yellow fluorescent protein (sYFP)-tagged GmRIN4b was coexpressed with EV or AvrRpm1 in N. benthamiana using a dexamethasone-inducible vector (Aoyama and Chua, 1997). Protein was allowed to accumulate for 3.5 h after dexamethasone induction, and the clarified total protein extract was incubated for 1 h at 4°C with GFP–Trap beads in order to immunoprecipitate sYFP-GmRIN4b. The washed beads were then subjected to on-bead trypsin digestion, and the recovered peptides were analyzed by liquid chromatography coupled to mass spectrometry (see “Methods”).
The mass spectral analyses obtained from two independent experiments ranged in coverage from 62 to 84%, depending on the experiment and sample (Figure 2; Supplemental Data Set 1). We screened these mass data for known posttranslational modifications, including phosphorylation on Ser, Thr, or Tyr and ADP-ribosylation on Cys, Asp, Glu, Lys, Asn, Arg, Ser, or Thr. Based on known phosphorylation sites induced by AvrB on AtRIN4, we first assessed whether GmRIN4b residues Thr-22 and Thr-198 were phosphorylated in the presence of AvrRpm1. These analyses revealed that neither Thr-22 nor Thr-198 was specifically phosphorylated in the presence of AvrRpm1. This is consistent with our observation that neither the T22A/D nor T198A/D substitution prevented modification of GmRIN4b in the presence of AvrRpm1 (Figure 1B) and the fact that Rpg1b, which responds to GmRIN4b with phosphomimicking residues at Thr-22 and Thr-198, does not respond to the presence of AvrRpm1.
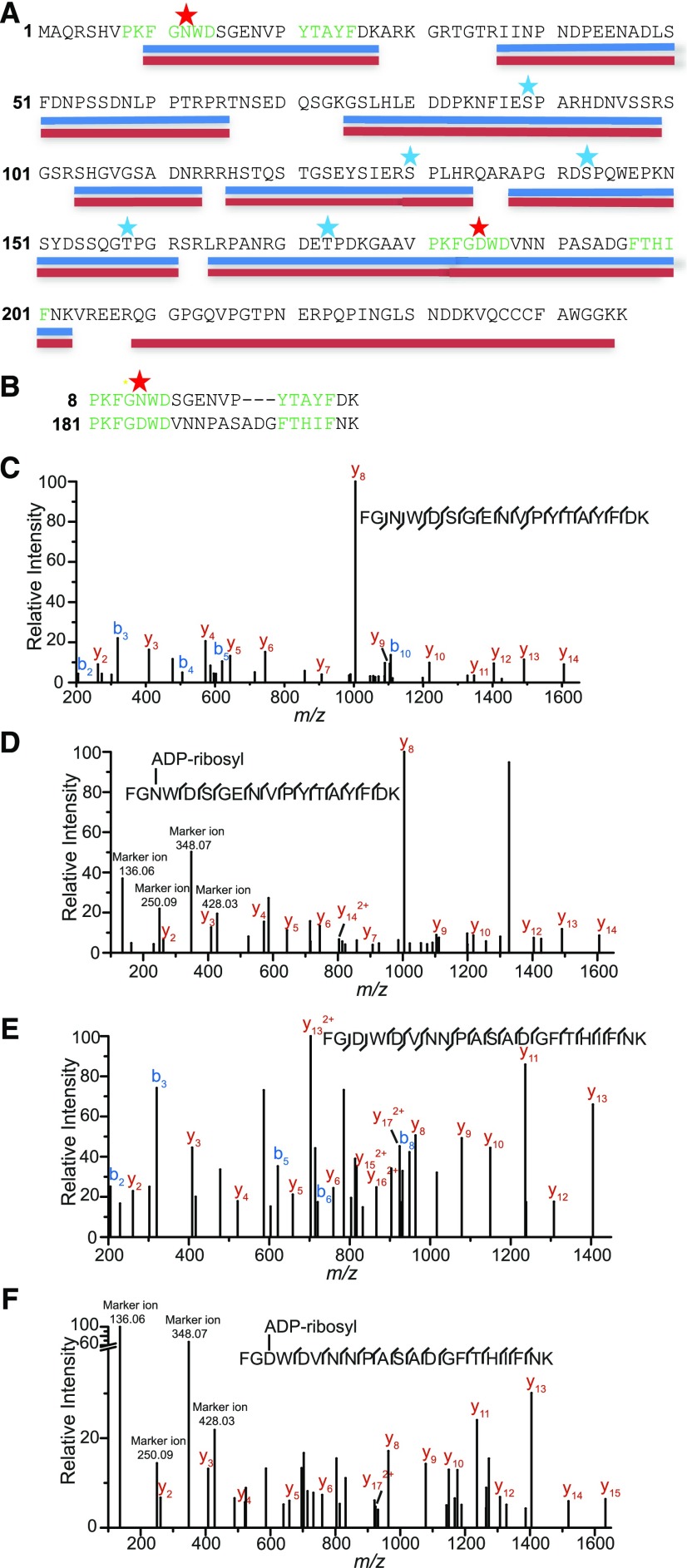
Figure 2.
GmRIN4b Is ADP Ribosylated in Vivo in the Presence of AvrRpm1.
sYFP-GmRIN4b was transiently expressed with AvrRpm1 or an EV control in N. benthamiana, and purified YFP-GmRIN4b protein was subjected to trypsin digestion prior to analysis by liquid chromatography–tandem mass spectrometry.
(A) GmRIN4b coverage map. Peptides detected in the negative control EV samples are underlined in blue. Peptides detected in the AvrRpm1-treated samples are underlined in red, with red stars indicating potentially ADP-ribosylated residues (only found in the AvrRpm1 samples; Supplemental Data Set 1). Blue stars indicate phosphorylated residues found in both EV and AvrRpm1-treated samples. The highly conserved PxFGxWD and F/YTxxFxK motifs of the NOI domains are highlighted in green.
(B) Alignment of the N- (top) and C-terminal (bottom) NOI domain peptides found to be modified by AvrRpm1. Star indicates position of ADP-ribosylation.
(C) Mass spectra resulting from fragmentation of the GmRIN4b N-terminal NOI domain peptide FGNWDSGENVPYTAYFDK, which was expressed in the absence of AvrRpm1.
(D) Mass spectra resulting from fragmentation of the GmRIN4b N-terminal NOI domain peptide FGNWDSGENVPYTAYFDK, which was expressed in the presence of AvrRpm1.
(E) Mass spectra resulting from fragmentation of the GmRIN4b C-terminal NOI domain peptide FGDWDVNNPASADGFTHIFNK, which was expressed in the absence of AvrRpm1.
(F) Mass spectra resulting from fragmentation of the GmRIN4b C-terminal NOI domain peptide FGDWDVNNPASADGFTHIFNK, which was expressed in the presence of AvrRpm1.
See also Supplemental Data Set 1.
Although we did not identify any phosphorylation sites that were specifically associated with AvrRpm1 treatment, we observed several residues on GmRIN4b that were phosphorylated in both EV and AvrRpm1-treated samples, including Ser-89, Ser-130, Ser-143, Thr-158, and Thr-173, indicating that GmRIN4b is phosphorylated by endogenous kinases in N. benthamiana (Figure 2; Supplemental Data Set 1).
The most notable of the modifications identified, however, were found on two peptides located within the N-terminal and C-terminal NOI domains of GmRIN4b: FGNWDSGENVPYTAYFDK (N-NOI) and FGDWDVNNPASADGFTHIFNK (C-NOI), respectively (Figure 2; Supplemental Data Set 1). Both of these peptides were found to be mono-ADP ribosylated only when expressed in the presence of AvrRpm1. Based on the fragmentation data of the N-NOI domain peptide (FGNWDSGENVPYTAYFDK), it was ADP ribosylated on the first Asn residue (Asn-12; Figure 2). Fragmentation data for the C-NOI domain peptide (FGDWDVNNPASADGFTHIFNK) revealed that it was ADP ribosylated at the first Asp residue (Asp-185); thus, both NOI domains are modified at the same relative position but on different amino acids (Asn versus Asp; Figure 2B). For the ADP-ribosylated versions of the peptides, we also observed characteristic marker ions resulting from fragmentation of the ADP-ribose moiety at m/z values of 136.06, 250.09, 348.07, and 428.03 (Rosenthal et al., 2015).
To confirm that AvrRpm1 induces ADP-ribosylation of GmRIN4b, we used an anti–pan-ADP-ribose binding reagent (anti-panADPR), which is a recombinant protein that selectively binds to mono- and poly-ADP–ribosylated proteins (Gibson et al., 2016). sYFP-tagged GmRIN4b was coexpressed with EV or AvrRpm1 in N. benthamiana, and total protein, isolated 4 h after induction, was immunoblotted with either anti-panADPR or anti-GFP. The anti-panADPR immunoblot showed a strong band corresponding in size to sYFP-GmRIN4b only in the samples coexpressed with AvrRpm1 (Figure 3A). In addition, we observed a second lower molecular weight band in the AvrRpm1 samples, even in the absence of GmRIN4b, indicating that AvrRpm1 can modify an endogenous N. benthamiana protein. These results confirm that AvrRpm1 possesses ADP-ribosylation activity.
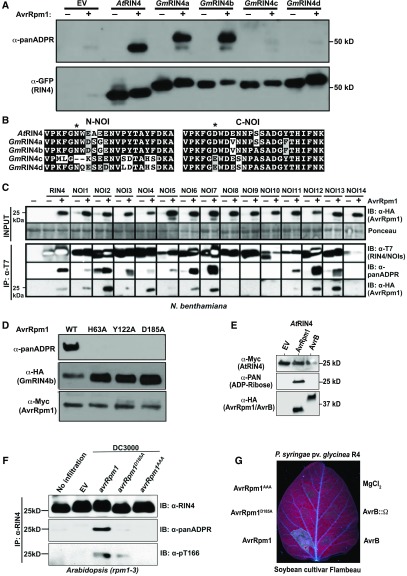
Figure 3.
AvrRpm1 ADP Ribosylates GmRIN4a, AtRIN4 and at Least 10 Other Arabidopsis NOI Domain-Containing Proteins.
(A) AvrRpm1 ADP ribosylates GmRIN4a, GmRIN4b, and AtRIN4. The indicated RIN4 proteins were fused to sYFP on their N termini and transiently expressed in N. benthamiana in the presence (+) and absence (−) of AvrRpm1. Total protein lysates were then analyzed by immunoblot using anti-GFP antibody or an anti-panADPR reagent. Similar results were obtained from three independent experiments.
(B) Sequences of the N-NOI domains of GmRIN4c and GmRIN4d are poorly conserved. The asterisk indicates the position of ADP-ribosylation in GmRIN4b. Note, in particular, the lack of a Trp adjacent to the predicted ribosylation site. The C-NOI domains, by contrast, are well conserved across all five RIN4 proteins.
(C) AvrRpm1 ADP ribosylates the majority of Arabidopsis NOI domain-containing proteins. AtRIN4 and 14 Arabidopsis NOI proteins, epitope tagged with the T7 peptide, were transiently expressed in N. benthamiana in the presence (+) or absence (−) of AvrRpm1. Immunoprecipitation with anti-T7 was performed followed by immunoblotting with anti-T7, anti-panADPR, or anti-HA. The expression of AvrRpm1 was confirmed in the input samples by immunoblot with anti-HA. All images in a single row are extracted from a single blot. The molecular weight of NOI domain-containing proteins ranges from 10 to 30 kD. Similar results were obtained from two independent experiments. IB, immunoblot; IP, immunoprecipitation.
(D) Mutation of putative catalytic site residues in AvrRpm1 inhibit ADP-ribosylation activity. HA-tagged GmRIN4b was transiently coexpressed with the indicated Myc-tagged AvrRpm1 mutant proteins in N. benthamiana. Total protein lysates were then analyzed by immunoblot to detect AvrRpm1, GmRIN4b, and ADP-ribose. Similar results were obtained from three independent experiments. WT, wild type.
(E) AvrB does not possess ADP-ribosylase activity. AtRIN4 was transiently expressed in N. benthamiana with the indicated proteins. Total protein lysates were then analyzed by immunoblot using anti-Myc antibody or an anti-panADPR reagent. Similar results were obtained from two independent experiments.
(F) Delivery of AvrRpm1 from P. syringae strain DC3000 induces ADP-ribosylation of AtRIN4 and phosphorylation of AtRIN4 T166. Arabidopsis rpm1-3 mutant plants were inoculated with 5 × 107 cfu/mL of P. syringae strains DC3000 (EV), DC3000 (avrRpm1), DC3000 (avrRpm1D185A), or DC3000 (avrRpm1AAA). Immunoprecipitations with anti-RIN4 were performed with tissue samples collected 12 h postinfection followed by immunoblots with anti-RIN4, anti-panADPR, and anti-pT166. Similar results were obtained from three independent experiments. IB, immunoblot; IP, immunoprecipitation.
(G) ADP-ribosylation activity is required for recognition of AvrRpm1 by soybean. The indicated proteins were delivered by P. syrinage pv glycinea Race 4 into unifoliate leaves of soybean cv Flambeau. The leaf was imaged under UV light to highlight areas of cell death. Both unifoliate leaves on three plants were inoculated, and all leaves showed the same pattern. R4, Race 4.
AvrRpm1 ADP Ribosylates Multiple NOI-Containing Proteins
Having established that the anti-panADPR reagent was an effective tool for detecting ADP-ribosylation of GmRIN4b, we used it to assess whether AvrRpm1 could induce ADP-ribosylation of sYFP-tagged versions of the other three GmRIN4 proteins and the Arabidopsis AtRIN4 protein. These analyses revealed strong modification of both sYFP-AtRIN4 and sYFP-GmRIN4a, but very little modification of sYFP-GmRIN4c and sYFP-GmRIN4d (Figure 3A). The relatively weaker modification of sYFP-GmRIN4c and sYFP-GmRIN4d may be a consequence of sequence differences in the N-NOI domains of GmRIN4c and GmRIN4d relative to GmRIN4a and GmRIN4b (Figure 3B). We observed, however, that the mobility of sYFP-GmRIN4c and sYFP-GmRIN4d was retarded in the presence of AvrRpm1 (Figure 3A), indicating that these two proteins may still be modified, but for unknown reasons the modification is not detected by the anti-panADPR reagent.
The specificity of AvrRpm1 for the NOI domains of RIN4 indicated that AvrRpm1 may be able to modify other NOI domain-containing proteins. We thus coexpressed in N. benthamiana AvrRpm1 and each of 14 additional Arabidopsis proteins (AtNOIs) that, aside from containing predicted NOI domains and C-terminal acylation motifs, lack any other similarity to RIN4. AvrRpm1-dependent ADP-ribosylation was observed for 10 of the 14 AtNOI proteins—NOI1, NOI2, NOI3, NOI4, NOI5, NOI6, NOI7, NOI11, NOI12, and NOI13—and AvrRpm1 coimmunoprecipitated with the same 10 AtNOI proteins (Figure 3C). Alignment of the NOI domains from these 14 proteins revealed that NOI9 and NOI14 have nonpreferred residues at the predicted ADP-ribosylation site (S and G, respectively), but it is not obvious why NOI8 and NOI10 were not modified (Supplemental Figure 2). In summary, AvrRpm1 associates with and ADP ribosylates most AtNOI proteins (Figure 3C).
To further support our conclusion that AvrRpm1 functions as an ADP-ribosyl transferase, we assessed putative ADP-ribosylation catalytic domain mutants of AvrRpm1 (Cherkis et al., 2012) for their ability to ADP ribosylate GmRIN4b. These AvrRpm1 mutants were dramatically reduced in their ADP-ribosylation activity (Figure 3D), consistent with their reduced virulence function and ability to activate RPM1 (Cherkis et al., 2012). We also assessed whether AvrB could induce ADP-ribosylation on AtRIN4, but observed no evidence for this (Figure 3E), indicating that AvrB likely possesses a different enzymatic activity.
To confirm that AvrRpm1 can induce ADP-ribosylation of AtRIN4 when delivered by P. syringae, we inoculated Arabidopsis rpm1-3 mutant plants with P. syringae strain DC3000 carrying EV or a plasmid expressing the wild-type avrRpm1 or catalytic site mutants. Native AtRIN4 protein was then immunoprecipitated and immunoblotted with α-panADPR reagent. These analyses revealed ADP-ribosylation of AtRIN4 by the wild type, but not mutant forms of AvrRpm1 (Figure 3F), indicating that AvrRpm1 delivered by P. syringae is able to induce ADP-ribosylation of native AtRIN4 protein.
Lastly, we assessed whether the ADP-ribosylation activity of AvrRpm1 is required to induced a resistance response on soybean (Figure 3G). Inoculation of soybean cv Flambeau, which expresses Rpg1-r, with P. syringae pv glycinea Race 4 expressing the wild-type AvrRpm1 induced a strong cell death response by 25 h postinoculation, while neither the D185A nor the catalytic site triple mutant induced a visible response. Thus, ADP-ribosylation activity is required for recognition of AvrRpm1 by both RPM1 and Rpg1-r.
ADP-Ribosylation of AtRIN4 by AvrRpm1 Is Required for Full Phosphorylation of Thr-166 and Modulates Activation of RPM1
Because Thr-166 of AtRIN4 contributes to activation of RPM1 by AvrRpm1 and because a phosphomimic mutation of AtRIN4 (T166D) is sufficient to activate RPM1 (Chung et al., 2011), we hypothesized that ADP-ribosylation of the AtRIN4 NOI domains is mechanistically related to phosphorylation of Thr-166. To test this hypothesis, we extended the previously defined RPM1-activation system in N. benthamiana (Chung et al., 2011). We confirmed that activation of RPM1 in N. benthamiana required both AtRIN4 and AvrRpm1 and that only the combination of all three proteins induced cell death (Supplemental Figure 3A). We then tested the ability of AvrRpm1 ADP-ribosyl transferase catalytic site mutants to elicit AtRIN4-dependent activation of RPM1. We observed that avrRpm1D185A retained partial activation of RPM1, while the more severe allele avrRpm1AAA was unable to activate RPM1 (Figure 4A). As a control in these experiments, we included the nonphosphorylatable AtRIN4T166A allele, which also retains partial activation of RPM1 in response to the wild-type AvrRpm1 (Figure 4A; Chung et al., 2011).
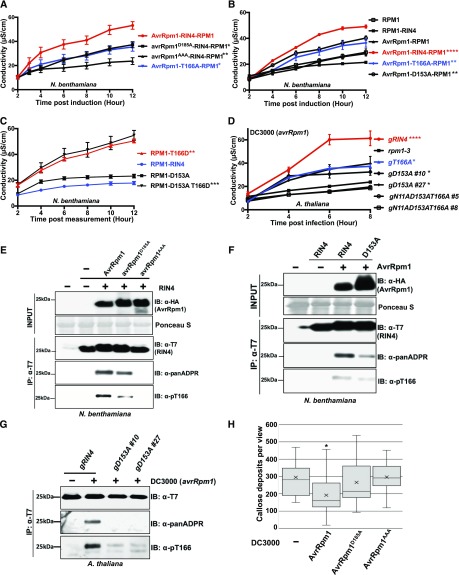
Figure 4.
ADP-Ribosylation of AtRIN4 by AvrRpm1 Is Required for AtRIN4 T166 Phosphorylation.
(A) Proposed AvrRpm1 ADP-ribosyl transferase catalytic residues are required for activation of RPM1 in N. benthamiana. The indicated AvrRpm1 mutants were coexpressed with genomic RPM1-myc and AtRIN4. The AtRIN4 T166A mutant was used as a control as it is known to partially suppress RPM1 activation. Cell death induced by RPM1 activation was monitored by conductivity measurements following induction of AvrRpm1 expression with 20 μM estradiol. Statistical analyses were performed by using one-way analysis of variance at 95% confidence comparing the values for the indicated AvrRpm1 and RIN4 mutant derivatives to the wild-type AvrRpm1-RIN4-RPM1 combination at 6 h postinduction. Asterisks represent statistical differences with the indicated P-values (****P < 0.0001, ***P < 0.005, **P < 0.001, *P < 0.05).
(B) D153 ribosylation site of AtRIN4 contributes to full AvrRpm1-mediated RPM1 activation in N. benthamiana. The wild-type AtRIN4, AtRIN4T166A (T166A), or AtRIN4D153A (D153A) were co-infiltrated with RPM1-myc and the wild-type AvrRpm1. Cell death was quantified by conductivity measurements as described in (A). Statistical analyses were performed as in (A), but using the wild-type RPM1-RIN4 in the absence of AvrRpm1 as the reference.
(C) Cell death mediated by the AtRIN4T166D (T166D) phosphomimic is epistatic to the loss of ribosylation at AtRIN4D153A (D153A). The quantification of cell death was monitored as in (A) and (B). Statistical analyses were performed as in (B).
(D) Ribosylation at AtRIN4D153 and phosphorylation of AtRIN4T166 contribute additively to AvrRpm1-mediated activation of RPM1 in transgenic Arabidopsis plants. P. syringae strain DC3000 (avrRpm1) was inoculated (5 × 107 cfu/mL) into leaves of transgenic Arabidopsis lines expressing genomic versions of AtRIN4 wild type (gRIN4), AtRIN4T166A (gT166A), AtRIN4D153A (gD153A), and AtRIN4N11AD153AT166A (gN11AD153AT166A) mutant plants. The rpm1-3 null mutant was used as a negative control. The quantification of cell death was monitored as in (A) and (B). Statistical analyses were performed as in (A), but using rpm1-3 as the reference.
(E) ADP-ribosylation activity of AvrRpm1 is required for full phosphorylation of AtRIN4T166. The ADP-ribosylation and phosphorylation of AtRIN4 in N. benthamiana were monitored by immunoprecipitation of AtRIN4 with anti-T7 followed by immunoblots with anti-T7, anti-panADPR, or anti-pT166. Co-infiltration of AvrRpm1 alleles and AtRIN4 was performed as in (A). Samples were collected 4 h posttreatment with 20 μM estradiol. This result represents one of four independent experiments. −, absence; +, presence; IB, immunoblot; IP, immunoprecipitation.
(F) ADP-ribosylation at AtRIN4D153 is required for full phosphorylation of Thr-166. N. benthamiana leaves were co-infiltrated with AvrRpm1 and AtRIN4 or AtRIN4D153A, as in (A). Samples were taken 12 h postinduction of AvrRpm1 with 20 μM estradiol. The ADP-ribosylation and phosphorylation of AtRIN4 in N. benthamiana were monitored by immunoprecipitation of AtRIN4 with anti-T7 followed by immunoblots with anti-T7, anti-panADPR, or anti-pT166. This result represents one of five independent experiments. −, absence; +, presence; IB, immunoblot; IP, immunoprecipitation.
(G) ADP-ribosylation at AtRIN4D153 is necessary for full phosphorylation of T166 in Arabidopsis. Transgenic plants expressing the wild-type AtRIN4 (gRIN4) and two independent T2 plants expressing AtRIN4D153A (gD153A #10 and #27) were inoculated with DC3000(avrRpm1) as in (D). Samples were collected 4 h postinfection before hypersensitive response developed. Immunoprecipitation of RIN4 proteins with anti-T7 was followed by immunoblots with anti-T7, anti-panADPR, and anti-pT166. −, absence; +, presence; IB, immunoblot; IP, immunoprecipitation.
(H) AvrRpm1 ADP-ribosylation catalytic residues are required for a virulence function of AvrRpm1 in Arabidopsis. Arabidopsis rpm1-3 mutant plants were inoculated with 5 × 107 cfu/mL of P. syringae strains DC3000 (EV), DC3000 (avrRpm1), DC3000 (avrRpm1D185A), or DC3000 (avrRpm1AAA). Callose accumulation from Arabidopsis leaves 18 h postinfection was analyzed by aniline blue staining. The box plot indicates mean (X), median (inner line), inner quartiles (box), outer quartiles (whiskers), and outliers (dots) for composite data from 16 samples (n = 16). Statistical significances compared with DC3000 (EV)-infected samples were determined by Student’s t test; *P < 0.05. This result was confirmed in three independent experiments. See also Supplemental Figure 2.
AtRIN4 is predicted to be ADP ribosylated by AvrRpm1 on Asp-153, which is located in the C-terminal portion of AtRIN4 (amino acids 142 to 211) previously demonstrated to be sufficient for RPM1 activation (Chung et al., 2011). To test the requirement of AtRIN4 Asp-153 in AvrRpm1-mediated RPM1 activation, we replaced this residue with Ala (D153A), which should block ADP-ribosylation at that site. This substitution reduced RPM1 activation to a level equivalent to AtRIN4T166A (Figure 4B). These results support a model in which ADP-ribosylation of Asp-153 promotes full phosphorylation on Thr-166, a modification previously demonstrated to be sufficient for RPM1 activation (Chung et al., 2011). To further examine the relationship between Asp-153 ribosylation and Thr-166 phosphorylation, we constructed cis double mutants of AtRIN4D153A with either nonphosphorylatable Thr-166, AtRIN4T166A (D153A T166A), or phosphomimic Thr-166, AtRIN4T166D (D153A T166D). Assessing effector-independent RPM1 activation indicated that the AtRIN4 D153A T166D double mutant retained full activation of RPM1 (Figure 4C). We generated Arabidopsis transgenic plants expressing native levels of AtRIN4D153A (D153A) or AtRIN4N11AD153AT166A (N11A D153A T166A) mutants in an RPM1-myc rpm1 rps2 rin4 background (Asn-11 in AtRIN4 is equivalent to Asn-12 in GmRIN4b). We activated RPM1 in leaves of these transgenics via delivery of AvrRpm1 from P. syringae. Conductivity measurements using two independent T2 transgenic lines expressing each AtRIN4 mutant demonstrated that AtRIN4D153A retained partial function in RPM1 activation, again at levels similar to that observed in plants expressing AtRIN4T166A (Figure 4D). AtRIN4N11AD153T166A, which cannot be ribosylated and cannot be phosphorylated on Thr-166, was a complete loss-of-function allele. These results indicate that ribosylation of Asp-153 and phosphorylation of Thr-166 act additively for complete RPM1 activation.
Arabidopsis transgenic lines expressing native levels of AtRIN4D153AT166D (D153A T166D) and AtRIN4N11AD153AT166D (N11A D153A T166D) displayed the dwarf phenotypes and diminution of RPM1 accumulation typical of RPM1 autoimmune responses previously attributed to AtRIN4T166D (gT166D; Supplemental Figure 3B; Chung et al., 2011). The combined results of Figures 4A to 4D and Supplemental Figures 3B and 3C indicate that AvrRpm1-mediated ribosylation of Asp-153 contributes to activation of RPM1. Furthermore, the epistasis of the gain-of-function T166D mutation over D153A, which blocks ADP-ribosylation, is consistent with the hypothesis that ADP-ribosylation of Asp-153 promotes phosphorylation of Thr-166 during activation of RPM1 by AvrRpm1.
As expected, the AtRIN4 N11 residue had no effect on AvrRpm1-mediated RPM1 activation, nor did N11A alter the RIN4 T166D RPM1 activation phenotype (Supplemental Figure 3CC). We confirmed that the partial loss of AvrRpm1-mediated RPM1 activation in RIN4 D153A was unlikely due to a structural defect, since this allele retains interaction with both AvrRpm1 and RPM1 in planta, as measured by coimmunoprecipitation (Supplemental Figure 3D). We also verified the expression of AvrRpm1, RPM1, and RIN4 alleles used in effector-dependent or -independent RPM1 activation assays in N. benthamiana, again noting that expression of AtRIN4 T166D alleles results in less RPM1 accumulation due to RPM1 activation (Supplemental Figures 3B and 3E; Chung et al., 2011).
We tested ADP-ribosylation and Thr-166 phosphorylation in N. benthamiana following co-infiltration of AtRIN4 with AvrRpm1, AvrRpm1D185A, or AvrRpm1AAA mutants. We observed that AvrRpm1 catalytic activity is required for both ADP-ribosylation of AtRIN4 and for full phosphorylation of AtRIN4 Thr-166 (Figure 4E).
We further tested whether the ability of AvrRpm1 to induce ADP-ribosylation is required for full phosphorylation of AtRIN4 Thr-166 in N. benthamiana (in the absence of RPM1) following transient expression of either the AtRIN4 wild type or D153A with the wild-type AvrRpm1. Following immunoprecipitation of AtRIN4 or D153A, we detected either RIN4 ADP-ribosylation or phosphorylated Thr-166 accumulation (using a phosphospecific antisera; Chung et al., 2011). Results displayed in Figure 4F demonstrate that AvrRpm1 induces less ADP-ribosylation and Thr-166 phosphorylation on the D153A derivative of AtRIN4 compared with the wild-type AtRIN4. Consistent with this observation, in the Arabidopsis transgenic RIN4 lines, we observed no ADP-ribosylation and drastically reduced Thr-166 phosphorylation in lines complemented with AtRIN4D153A compared with lines complemented with the wild-type AtRIN4 following delivery of AvrRpm1 from DC3000 (Figure 4G).
Lastly, we assessed whether AvrRpm1 ADP-ribosylation activity was required for a typical AvrRpm1-mediated virulence activity, namely, the suppression of callose deposition at sites of infection. We inoculated leaves of rpm1-3 mutant Arabidopsis with DC3000 (EV), (avrRpm1), (avrRpm1D185A), or (avrRpm1AAA) and counted callose depositions 18 h later. As displayed in Figure 4H, the wild-type AvrRpm1 suppressed callose deposition triggered by DC3000, and this suppression required AvrRpm1 catalytic activity. These results confirm that AvrRpm1-dependent ADP-ribosylation of RIN4, and likely other host NOI domain-containing targets, is relevant to its virulence activity (Belkhadir et al., 2004).
Taken together, results in Figures 4E to 4H indicate that AvrRpm1-dependent ADP-ribosylation of AtRIN4 D153 is required for full phosphorylation of AtRIN4 Thr-166 to drive RPM1 activation and for successful repression of plant defense responses in the absence of RPM1.
AtRIN4 Interacts with Multiple EXO70 Family Members via the C-Terminal NOI Domain
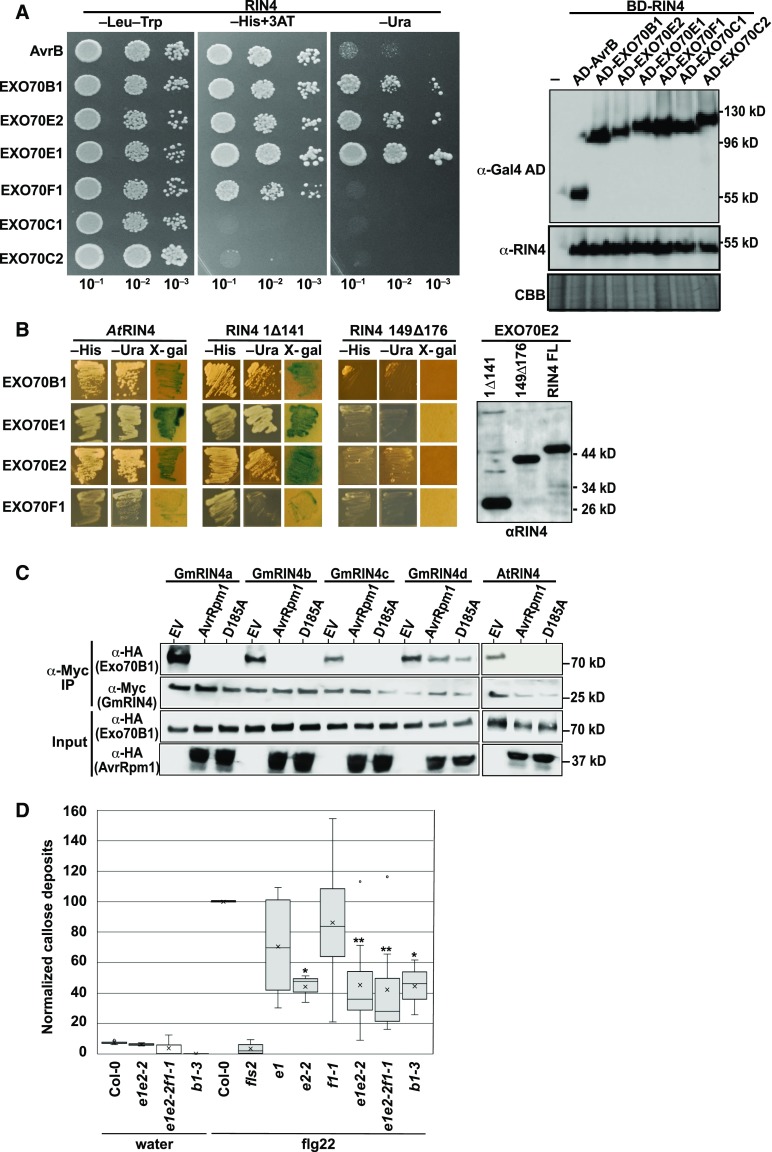
The Arabidopsis RIN4 protein can associate with the exocyst subunit EXO70B1 in an NOI domain-dependent manner (Sabol et al., 2017). EXO70B1 is a member of a large family of EXO70 proteins in Arabidopsis (23 family members in total; Supplemental Figure 4; Supplemental File; Cvrčková et al., 2012). To assess whether AtRIN4 can interact with other Arabidopsis EXO70 family members, we used yeast two-hybrid analysis to test interactions with 11 members distributed across the Arabidopsis EXO70 phylogenetic tree (Supplemental Figure 4). Of these 11 pairwise tests, four showed a robust interaction (EXO70B1, EXO70E1, EXO70E2, and EXO70F1; Figure 5A). To assess whether this interaction was mediated by either NOI domain in RIN4, we constructed two deletion derivatives of AtRIN4: one eliminating the first 141 amino acids, including N-NOI, and one in which only C-NOI (amino acids 149 to 176) was removed. The latter deletion eliminated the interaction with all four EXO70 proteins, while the former deletion did not, indicating that all of the RIN4-EXO70 interactions are mediated by C-NOI (Figure 5B).
Figure 5.
RIN4 Proteins Interact with EXO70 Proteins via the C-NOI Domain.
(A) AtRIN4 interacts with multiple Arabidopsis EXO70 family members in yeast. Shown are the results of a yeast two-hybrid assay, with yeast expressing the indicated protein fusions. The AvrB protein was used as a positive control for interaction with AtRIN4. Growth on −His and −Ura plates indicate positive interactions. The immunoblot on the right confirms protein expression of each bait and prey construct. CBB, Coomassie Brilliant Blue.
(B) C-NOI domain of AtRIN4 is required for interaction with Arabidopsis EXO70 proteins. 1Δ141 indicates deletion of AtRIN4 amino acids 1 to 141, which contains the N-NOI domain. 149Δ176 indicates deletion of amino acids 149 to 176, which contains the C-NOI domain. The anti-RIN4 immunoblot shows that all three RIN4 protein fusions were expressed in yeast.
(C) AvrRpm1 inhibits association of Arabidopsis EXO70B1 with Arabidopsis and soybean RIN4 proteins. The indicated proteins were coexpressed in N. benthamiana, and then RIN4 proteins were immunoprecipitated with anti-Myc antibody. EXO70B1 coimmunoprecipitated in the absence of AvrRpm1, but not in its presence. D185A indicates AvrRpm1 with the D185A substitution. This result represents one of three independent experiments. IP, immunoprecipitation.
(D) Leaves from Arabidopsis plants were infiltrated with water or 100 μM flg22, and callose deposits were enumerated after 16 h. Shown is a box plot indicating mean (X), median (inner line), inner quartiles (box), outer quartiles (whiskers), and outliers (dots) for composite data from 13 independent experiments (for flg22 treatments, n = 13 [ecotype Columbia-0 (Col-0)], 6 [fls2, exo70e1, exo70 f1-1], 8 [exo70e1exo70e2-2 and exo70e1exo70e2-2exo70f1-1], and 3 [exo70e2-2 and exo70b1-3]), with the number of callose deposits detected in flg22-infiltrated Col-0 normalized to 100 within each experiment. Significance for flg22-treated mutant plants compared with flg22-treated Col-0 plants were determined by Student’s t test (*P < 0.05, **P < 0.005).
See also Supplemental Figures 3 and 4.
Because the RIN4-EXO70 interaction is mediated by C-NOI and this domain is targeted by AvrRpm1, we tested whether AvrRpm1 could affect this interaction by transiently expressing Arabidopsis and soybean RIN4 proteins with Arabidopsis EXO70 proteins in N. benthamiana and performing coimmunoprecipitation analyses. We observed that all four soybean RIN4 proteins and AtRIN4 coimmunoprecipitated with Arabidopsis EXO70B1 in the absence of AvrRpm1 (Figure 5C). AtRIN4 also coimmunoprecipitated with EXO70E1 (Supplemental Figure 5). Coexpression with AvrRpm1 strongly reduced these interactions for AtRIN4, GmRIN4a, GmRIN4b, and GmRIN4c, but it had no effect on the interactions of EXO70 proteins with GmRIN4d. However, we also observed that the AvrRpm1D185A and AvrRpm1AAA mutants blocked RIN4-EXO70 interactions (Figure 5C; Supplemental Figure 5). This latter result indicates that AvrRpm1 and EXO70 compete for binding to the C-NOI domain and that when AvrRpm1 is overexpressed, it can block the interaction with EXO70. This may not be a biologically relevant result, considering that AvrRpm1 is likely delivered at much lower levels during a natural infection. Nevertheless, it points to a common interaction surface on RIN4. Perhaps related to this observation, the NLR protein RPS2, which is inhibited by RIN4 and is typically only weakly activated by AvrRpm1, is strongly activated by inactive mutants of AvrRpm1, perhaps through competition for binding to RIN4 (Kim et al., 2009; Cherkis et al., 2012).
EXO70 Proteins Contribute to flg22-Induced Callose Deposition in Cell Walls
Prior work has shown that transgenic overexpression of either AvrRpm1 or AtRIN4 in Arabidopsis suppresses flg22-induced callose secretion (Kim et al., 2005b). The observation that RIN4 proteins interact with EXO70 proteins raised the possibility that callose secretion may depend on EXO70 function and that RIN4 may regulate callose secretion via its interactions with EXO70. We therefore quantified the level of flg22-induced callose deposition in various single, double, and triple exo70 mutants (see Supplemental Figure 6 for details on the mutations). Figure 5D shows that knockout of either EXO70B1 or EXO70E2 significantly reduced callose deposition, while knockout of EXO70E1 or knockdown of EXO70F1 did not, indicating that a subset of EXO70 proteins contribute to callose deposition.
Mutation of AtRIN4 T166 Stabilizes RIN4-EXO70 Interactions
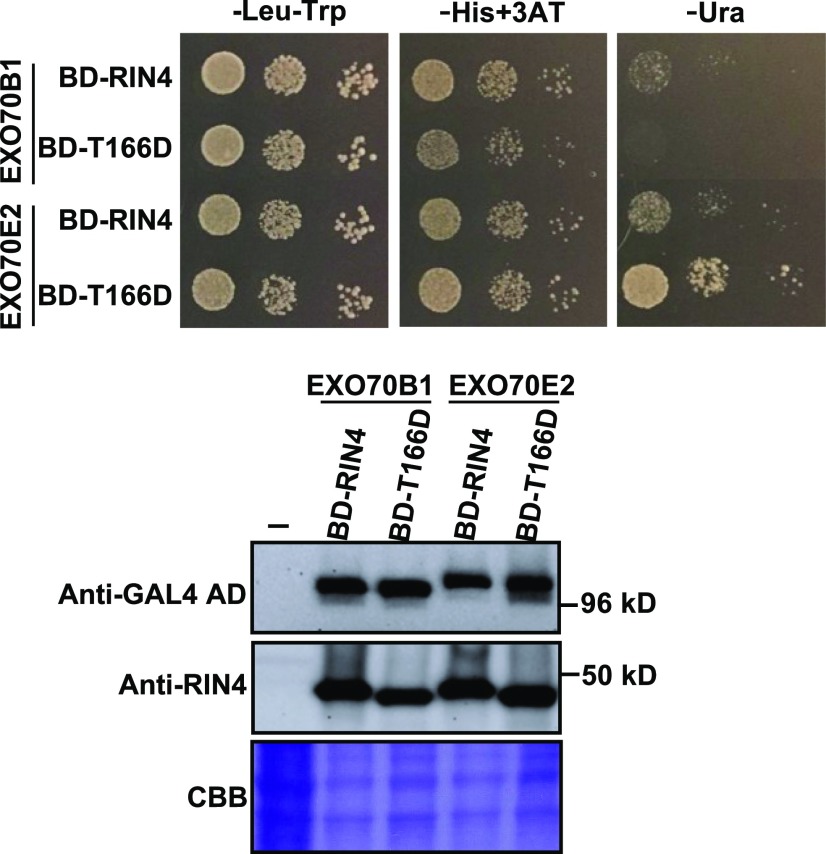
Because overexpression AvrRpm1 in Arabidopsis has the same effect on callose deposition as overexpression of AtRIN4, we hypothesized that AvrRpm1 may be suppressing callose deposition by enhancing the interaction of AtRIN4 with EXO70. However, transient overexpression of AvrRpm1 in N. benthamiana had the opposite effect, possibly due to the artificially high levels of AvrRpm1 resulting in competition for binding to the C-NOI domain of RIN4. To circumvent this issue, we used a yeast two-hybrid assay to test whether a T166D phosphomimic substitution in AtRIN4 would promote the AtRIN4-EXO70 interaction, using the rationale that since AvrRpm1 promotes Thr-166 phosphorylation, this might function to stabilize the AtRIN4-EXO70 interaction. These analyses showed that AtRIN4T166D interacted with EXO70E2 more strongly than did the wild-type AtRIN4 (Figure 6), supporting the model that AvrRpm1 suppresses callose deposition by enhancing the affinity of RIN4 for specific EXO70 proteins (Figure 7).
Figure 6.
T166D Phosphomimic Mutation in AtRIN4 Enhances RIN4 Interaction with EXO70E2.
A yeast two-hybrid assay was used to assess the impact of the T166D substitution on the interaction between AtRIN4 and EXO70B1 and EXO70E2. (Top) Enhanced growth on minus uracil (−Ura) plates indicates that the T166D substitution stabilizes the interaction. (Bottom) Immunoblots of protein extracts from the yeast strains growing on −Leu−Trp media, establishing equivalent expression levels in all four strains. Data shown were replicated with three independent yeast transformations for each combination of plasmids. 3AT, 3-amino-1,2,4-triazole; AD, activation domain; BD, binding domain; CBB, Coomassie Brilliant Blue.
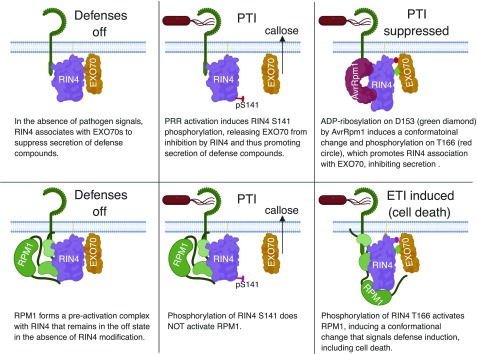
Figure 7.
Model for How ADP-Ribosylation of RIN4 Affects Resistance and Susceptibility.
Perception of pathogens by a cell surface receptor promotes phosphorylation of AtRIN4 S141 (Chung et al., 2014), which decreases the affinity of AtRIN4 for EXO70. Release of EXO70 promotes secretion of defense compounds such as callose. To combat this, P. syringae injects AvrRpm1, which phosphorylates AtRIN4 on Asp-153, promoting a conformational change in AtRIN4. This then enables phosphorylation on Thr-166, which enhances the affinity of AtRIN4 for EXO70, thus suppressing secretion of defense compounds. To guard against such suppression, plants have evolved NLR proteins that sense RIN4 modification. In this example, phosphorylation on Thr-166 activates RPM1. ETI, effector-triggered immunity; PTI, Pattern recognition receptor (PRR)-triggered immunity.
DISCUSSION
Our conclusion that AvrRpm1 functions as an ADP-ribosyl transferase is supported by the work of Cherkis et al. (2012), who generated computational models for AvrRpm1 using an alignment with the catalytic domains of several poly(ADP-ribose) polymerase (PARP)–containing proteins, including several members of the diphtheria toxin-like family of proteins as well as the human PARP-1 protein. These analyses showed that AvrRpm1 may contain a fold homologous to other ADP-ribosyl transferases. Significantly, mutation of the predicted catalytic residues in AvrRpm1 eliminated the ADP-ribosyl transferase activity (Figure 4A), which is consistent with the observation of Cherkis et al. (2012) that these mutations strongly attenuate AvrRpm1’s ability to activate RPM1. These data indicate that AvrRpm1 is indeed acting as an ADP-ribosyl transferase. However, we were unable to confirm this with purified proteins due to instability of recombinant AvrRpm1 protein (Cherkis et al., 2012).
The Pseudomonas effectors HopF1 and HopF2 both share a PARP domain fold that is similar to diphtheria toxin (Singer et al., 2004; Wang et al., 2010). Although the enzymatic activity of HopF1 has not been described, HopF2 has been shown to have ADP-ribosyl transferase activity and to directly ADP ribosylate Arabidopsis MAP Kinase Kinase5 (MKK5) at Arg-313 (Wang et al., 2010). This results in suppressing MKK5 kinase activity and inhibits pathogen-associated molecular pattern–induced MAP kinase activation (Wang et al., 2010). Interestingly, it has also been shown that HopF2 is capable of directly ADP ribosylating AtRIN4 in vitro; however, the residues modified were not identified (Wang et al., 2010; Wilton et al., 2010). They are presumably different from the residues modified by AvrRpm1, because HopF2 does not activate RPM1 and, unlike AvrRpm1, does interfere with AvrRpt2-induced cleavage of RIN4 and the resultant activation of RPS2 (Ritter and Dangl, 1996; Wilton et al. 2010). Notably, however, transgenic expression of HopF2 in Arabidopsis suppresses callose deposition induced by a type III secretion system–deficient mutant of P. syringae strain DC3000, and this ability is dependent on ADP-ribosyl transferase activity (Hurley et al., 2014). It is tempting to speculate that HopF2-mediated modification of RIN4 also serves to suppress EXO70 function.
Our mass spectrometry data indicate that GmRIN4b is ADP ribosylated twice, once within each NOI domain present in GmRIN4b: FGNWDSGENVPYTAYFDK and FGDWDVNNPASADGFTHIFNK (Figure 2; Afzal et al., 2013). The C-NOI domain of AtRIN4 has previously been shown to mediate the association of AtRIN4 with the exocyst subunits EXO70B1 and EXO70E1 at the plasma membrane (Sabol et al., 2017). The role of RIN4-interacting EXO70 proteins in the secretion of defense compounds is supported by our finding that EXO70B1 and EXO70E2 are required for full induction of callose by the flagellin-derived peptide flg22 (Figure 5). Because RIN4 is known to suppress flg22-induced callose deposition (Kim et al., 2005b), the finding that RIN4 physically associates with EXO70 indicates that this association acts to inhibit EXO70’s role in callose deposition. Thus, modification of RIN4 by AvrRpm1 should function to enhance this inhibitory activity (Figure 7). Consistent with this model, transgenic expression of AvrRpm1 also suppresses flg22-induced callose deposition (Kim et al., 2005b). We speculate that ADP-ribosylation of RIN4 functions to promote the association of RIN4 with EXO70 proteins, either by a direct effect of the ADP-ribose moiety or by promoting phosphorylation on Thr-166. In support of the latter, a T166D phosphomimic substitution in AtRIN4 enhanced its interaction with EXO70E2 in a yeast two-hybrid assay (Figure 6). However, the T166D substitution appeared to have a slightly deleterious effect on the interaction of AtRIN4 with EXO70B1, indicating that ADP-ribosylation on Asp-153 may be required to stabilize this interaction. Although further work is needed to solidify our model that ADP-ribosylation and/or Thr-166 phosphorylation function to enhance AtRIN4-EXO70 interactions, it is notable that knockout of EXO70B2 enhances the susceptibility of Arabidopsis to infection by P. syringae strain DC3000, indicating that EXO70 proteins play a role in basal defenses (Stegmann et al., 2012). It thus makes sense that EXO70 proteins may be direct or indirect targets of bacterial effectors.
Consistent with EXO70 proteins being targeted by effectors, EXO70B1 is known to associate with an NLR-family member named TN2 (Zhao et al., 2015), suggesting that EXO70B1 is being guarded. Indeed, knockout of EXO70B1 activates TN2-mediated cell death, at least in older plants grown under short days (Zhao et al., 2015). Under our growth conditions, we observed less exaggerated macroscopic cell death in 6-week-old exo70b1 plants and no symptoms yet in the 5-week-old plants used for measuring callose deposition.
EXO70-NOI domain interactions in the context of plant immunity have also recently been reported in rice (Oryza sativa). The rice NLR protein Pii-2 contains an NOI domain that appears to function as an integrated decoy to sense a fungal effector protein (Fujisaki et al., 2017). In that system, the Pii-2 NOI domain interacts with a rice EXO70 protein, OsEXO70-F3, that is targeted by the AVR-Pii effector protein from the fungus Magnaporthe oryzae. Mutations in the NOI core motif of Pii-2 (PxFGxWD) eliminated its interaction with OsEXO70-F3 and detection of AVR-Pii. Notably, AvrRpm1 modifies both RIN4 NOI domains within this core motif, on the Asn or Asp residue between the Gly and Trp (Figure 2). The finding that, similar to AvrRpm1 from P. syringae, an effector protein from M. oryzae perturbs an NOI-EXO70 interaction indicates that this type of perturbation is likely a virulence strategy of diverse pathogens.
Although our data strongly support our conclusion that ADP-ribosylation of AtRIN4 on Asp-153 promotes phosphorylation on Thr-166, we did not observe AvrRpm1-induced phosphorylation of GmRIN4b in our mass spectrometry analyses when these two proteins were overexpressed in N. benthamiana (Supplemental Data Set 1). We believe this may be because we did not also overexpress an appropriate kinase protein capable of this phosphorylation, such as RIPK. In support of this, we failed to detect a RIN4 size shift when we coexpressed soybean or Arabidopsis RIN4s with AvrB in N. benthamiana. Either RIPK function is not conserved in N. benthamiana or its native expression levels are not sufficient to modify the majority of the overexpressed RIN4 protein.
RIN4 cannot be the only target of AvrRpm1, however, because AvrRpm1 enhances the virulence of P. syringae even in a rin4 mutant background (Ritter and Dangl, 1995; Belkhadir et al., 2004). In addition to RIN4, there are 14 NOI domain-containing proteins found in Arabidopsis, which were initially identified from a screen searching for NOI genes. Aside from possessing an NOI domain, which includes two highly conserved PxFGxWD and F/YTxxFxK motifs, and a likely prenylation or palmitoylation sequence near their C termini, these proteins do not share homology with one another and their functions are unknown (Afzal et al., 2013). Our data show that AvrRpm1 can ADP ribosylate many of these proteins, which might then contribute to the enhanced virulence observed for Pseudomonas strains expressing avrRpm1 on susceptible Arabidopsis plants lacking AtRIN4 (Ritter and Dangl, 1995; Belkhadir et al., 2004).
Prior work in independent laboratories has shown that AvrRpm1 induces hyperphosphorylation of AtRIN4 (Mackey et al., 2002; Chung et al., 2011, 2014; Liu et al., 2011). Our observations that AvrRpm1 induces ADP-ribosylation of RIN4 and that this event is required for full phosphorylation, as well as the additive loss of function phenotype in transgenic plants expressing native levels of an AtRIN4 mutant that cannot be ribosylated at Asp-153 or phosphorylated at Thr-166, are consistent with a model in which ribosylation promotes subsequent phosphorylation. We speculate that ADP-ribosylation of AtRIN4 on Asp-153 induces a change in conformation and/or subcellular localization that enables or enhances phosphorylation of Thr-166 by endogenous Arabidopsis kinases such as RIPK and related RLCKs (Chung et al., 2011; Liu et al., 2011; Xu et al., 2017; Toruño et al., 2019). Under this model, maximal phosphorylation of T166 in response to AvrRpm1 would be dependent on ADP-ribosylation of Asp-153. Consistent with this prediction, a D153A substitution reduced AvrRpm1-induced phosphorylation of Thr-166 (Figure 4).
Alterations in conformation and/or localization resulting from Thr-166 phosphorylation may explain how RPM1 can detect two sequence unrelated effectors, AvrB and AvrRpm1 (Grant et al., 1995). AvrB also induces hyperphosphorylation on Thr-166 (Chung et al., 2011; Liu et al., 2011), and T166D substitutions are sufficient for activating RPM1 (Chung et al., 2011; Liu et al., 2011). The assumption that phosphorylation of Thr-166 alters conformation and/or localization of RIN4 is supported by the finding that phosphorylated Thr-166 inhibits phosphorylation on Ser-141, an event that is required to relieve inhibition of MAMP-triggered responses (Chung et al., 2014). Collectively, these data indicate that RPM1 is activated by an alteration of RIN4 that results from Thr-166 phosphorylation that is itself promoted by ADP-ribosylation within the C-NOI domain. In this context, it should be noted that AvrB also binds to C-NOI (Desveaux et al., 2007), and we speculate that AvrB modifies C-NOI in a way that promotes Thr-166 phosphorylation. AvrB contains a filamentation-induced by c-AMP domain, which is a domain known to posttranslationally modify proteins through AMPylation, UMPylation, phosphorylation, or phosphocholination (Garcia-Pino et al., 2014). AMPylation of C-NOI would be structurally similar to the addition of ADP-ribose.
In soybean, recognition of AvrB and AvrRpm1 is divided among two NB-LRR proteins, RPG1b and RPG1r, respectively (Ashfield et al., 1995). RPG1b and RPG1r are closely related paralogs, but they are distantly related to RPM1 (Ashfield et al., 2014). Phylogenetic analyses indicate that the ability to recognize AvrB and AvrRpm1 evolved independently in soybean and Arabidopsis (Ashfield et al., 2004, 2014). Because of this, it is plausible that RPG1b and RPG1r have evolved molecular mechanisms to detect AvrB and AvrRpm1 that differ from those used by RPM1. However, RPM1 and RPG1r both recognize the ADP-ribosylation activity of AvrRpm1 (Figure 3), and both require RIN4; thus, the molecular mechanism underlying recognition appears similar. Notably, AvrB does not possess ADP-ribosylation activity (Figure 3); thus, it is plausible that RPG1r directly senses ADP-ribosylation of RIN4, while RPG1b senses the specific modification catalyzed by AvrB (possibly AMPylation or UMPylation). This explanation makes sense in evolutionary terms, since the two RPG1 loci are the result of a recent duplication event in soybean (Ashfield et al., 2014). In this scenario, it is possible that the initial RPG1 locus encoded an NLR protein that responded to ADP-ribosylation of RIN4, as this appears to be the more ancestral specificity (Ashfield et al., 2014). Following gene duplication, the second RPG1 locus was maintained because it evolved a new specificity, enabling recognition of AvrB-modified GmRIN4, giving rise to RPG1b.
METHODS
Plant Materials and Growth Conditions
Nicotiana benthamiana seeds were sown in a 4:1:1 (v/v) ratio of Pro-Mix-B Biofungicide (Premier Horticulture) potting mix:vermiculite:perlite supplemented with Osmocote 14-14-14 slow-release fertilizer (w/w). Plants were grown in growth rooms under a 12-h light/12-h dark cycle (24°C d/22°C night). Illumination was provided by GE HI-LUMEN XL Starcoat 32-W fluorescent bulbs (a 50:50 mixture of 3500K and 5000K spectrum bulbs; 9-h d, ∼150 µmol/m2/s at the canopy surface). Plants were used for transient expression assays at 3 to 4 weeks of age.
For Arabidopsis (Arabidopsis thaliana) transgenic lines, all AtRIN4 mutants were expressed from the native promoter and transformed into an RPM1-myc rpm1 rps2 rin4 background. T2 lines segregating 3:1 upon Basta selection were confirmed and matched for expression of RPM1 and AtRIN4 alleles by immunoblot with anti-myc or anti-T7, respectively. Three plants with similar expression of RPM1 and AtRIN4 mutant alleles were used for conductivity measurements to monitor RPM1 activation upon P. syringae strain DC3000 (avrRpm1) infection. Previously generated transgenic lines in the same mutant background expressing the wild-type AtRIN4, AtRIN4T166A, or AtRIN4T166D were used to compare effector-dependent and -independent RPM1 activation with Arabidopsis transgenic plants generated in this study (Chung et al., 2011). Seeds from these lines were sown in a custom potting mix prepared from 4.5 parts sterilized Sungrow Professional Growing Mix (Sun Gro Horticulture), 2 parts white sterilized sand (Mellotts), 1 part Horticultural Perlite (Carolina Perlite), 250 PPM Peter’s special 20-20-20 (w/w) fertilizer (Everris NA), and 75 mg/ft3 Marathon Pesticide (1% granular, OHP). Plants were grown in growth chambers under a 9-h light/15-h dark cycle (21°C d/18°C night). Illumination was provided by Westinghouse cool-white fluorescent bulbs (F32T8/741/ECOMAX) at ∼100 µmol/m2/s at the canopy surface.
For Arabidopsis exo70 mutants, the following T-DNA insertion lines were used: exo70b1-3 (At5g58430; Zhao et al., 2015); exo70e1 (At3g29400), SALK_084145; exo70e2-2 (At5g61010), SK26782; and exo70f1-1 (At5g50380), SALK_139110. See Supplemental Figure 6 for locations of T-DNA insertions and verification of mRNA knockdown. All lines were obtained from the Arabidopsis Biological Resource Center at Ohio State University, except for exo70b1-3, which was provided by Dingzhong Tang (Zhao et al., 2015). Double and triple exo70 mutants were generated by crossing the relevant lines and selecting homozygous segregants in the F2 generation using PCR.
Arabidopsis exo70 mutant seed were sown in a 50:50 mix of ProMix BX (Premier Horticulture) and MetroMix 360 (Sungro Horticulture) with ∼35 g of Osmocote 14-14-14 (w/w) slow-release fertilizer per flat. Plants were grown in growth chambers under an 8-h light/16-h dark cycle (24°C d/17 to 18°C night). Illumination was provided by white T5 fluorescent bulbs with an intensity of ∼300 µmol/m2/s1 at canopy height.
Construction of Plasmid Clones
To generate cDNA clones of soybean RIN4 genes, total RNA was isolated from partially expanded trifoliate soybean (Glycine max) cv Williams-82 leaves using the Total RNA Spectrum kit (Sigma-Aldrich), and cDNA was generated using the High-Capacity cDNA RT kit (Applied Biosystems) following the manufacturers’ instructions. Specific GmRIN4 sequences were then amplified from this cDNA using the primers listed in Supplemental Table 1 using Phusion High-Fidelity DNA Polymerase (Thermo Fisher). PCR products were cloned into the Gateway entry vector pBSDONR P4rP2 (Qi et al., 2012) using the BP Clonase II kit (Thermo Fisher). Entry clones were then recombined with the dexamethasone-inducible vector pTA7001-Dest (Vinatzer et al., 2006) and N-terminal epitope tags 3× hemagglutinin (HA), 5×Myc, or sYFP in pBSDONR P1P4 (Qi et al., 2012) using the LR Clonase II kit (Thermo Fisher Scientific).
AtRIN4 and AtNOI genes were also cloned using the Gateway system (Chung et al., 2011). All AtRIN4 mutants were generated via site-directed mutagenesis by using genomic AtRIN4 as a template and cloned into the entry vector pDONR207 (Thermo Fisher Scientific; Chung et al., 2011). To introduce multiple mutation sites, single mutant entry clones of AtRIN4 (T166A or T166D) were used as a template. For AtNOI genes, all intron and exons were amplified and cloned into the entry vector pENTR-D-TOPO, followed by LR recombination into a destination vector containing a T7 epitope-tag at the N terminus under control of a 35S promoter.
To generate the AvrRpm1 triple mutant, site-directed mutagenesis was performed by using an AvrRpm1 H63A single mutant clone to amplify an N-terminal fragment with the Y122A mutation, and an AvrRpm1 D185A single mutant clone to amplify a C-terminal fragment with the Y122A mutation, followed by cloning the full-length triple mutant fragment into pENTR-D-TOPO vector (Invitrogen; Cherkis et al., 2012). All entry clones were then cloned into the pBAR-GW destination vector for AtRIN4 mutants and the pMDC7 destination vector for avrRpm1 mutants. All primers used for site-directed mutagenesis are listed in Supplemental Table 1.
To generate EXO70 clones for yeast two-hybrid analyses, entry clones for EXO70A1(G20002), EXO70B1(G21983), EXO70B2(G21656), EXO70D2(G24035), EXO70E2(G13181), EXO70G1(G22207), and EXO70H1(G19572) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Entry clones for EXO70C1, EXO70C2, EXO70E1, and EXO70F1 were generated by PCR with Pfu DNA polymerase (Stratagene), using Arabidopsis seedling cDNA as template and cloned into pCR-CCD-F. The primer pairs used for plasmid constructs are listed in Supplemental Table 1. These entry clones were then recombined into the destination vector pDEST22 using LR Clonase (Thermo Fisher Scientific).
To generate EXO70 clones for transient expression in N. benthamiana, the entry clones were recombined into the destination vector pCsVMV;HA-C-1300, which contains a Cassava vein mosaic virus promoter and 3×HA C-terminal epitope tag (Kim et al., 2013).
Agrobacterium-Mediated Transient Expression in N. benthamiana or Nicotiana glutinosa
The A. tumefaciens transformed with the pTA7001 constructs carrying the wild-type GmRIN4s, AvrB, AvrRpm1, or EV were in strain GV3101 (pMP90). Expression of the pSITE constructs carrying the phosphomimetic or phosphodeficient GmRIN4b alleles were in A. tumefaciens strain LBA440 (Selote et al., 2013). Liquid cultures of all Agrobacterium strains were initially grown overnight at 30°C with agitation in Luria-Bertani (LB) media supplemented with the appropriate antibiotics (50 μg/mL kanamycin and 50 μg/mL gentamicin for all pTA7001 constructs, or 100 μg/mL rifampicin, 100 μg/mL spectinomycin, and 50 μg/mL streptomycin for pSITE constructs).
For transient expression in N. benthamiana, the overnight cultures were subcultured the following morning at a dilution of 1:10 (v/v) in fresh Luria-Bertani media (without antibiotics) and grown for an additional 5 h at 30°C. The bacterial cells were pelleted by centrifugation at 4000 rpm for 5 min. The pellet was washed once in 10 mM MgCl2 before resuspending the Agrobacterium strains to an OD600 of 0.8 in 10 mM MgCl2. The strains harboring a GmRIN4 construct were mixed in a 1:1 (v/v) ratio with strains carrying an effector construct (or EV) directly before fully injecting the plant leaves. The infiltrated plants were then returned to the growth room. Transgene expression was induced within 36 h after infiltration by spraying plants with 50 μM dexamethasone. Tissue was collected within 3.5 h after induction and either directly prepared for analysis by immunoblot or mass spectrometry or immediately frozen in liquid nitrogen and stored at −80°C.
The N. benthamiana transient assay for RPM1 activation by AvrRpm1 and/or AtRIN4 alleles was performed by infiltrating the Agrobacterium at OD600 of 0.1, 0.3, and 0.3 for strains carrying the AvrRpm1 alleles, AtRIN4 alleles, or RPM1, and 0.1 for a strain carrying the viral p19 suppressor of genes silencing, respectively. To induce the expression of AvrRpm1 alleles, 20 μM β-estradiol was applied by spray onto the infiltrated leaves. For the immunoblots with anti-HA for AvrRpm1, anti-panADPR for AtRIN4 ribosylation, and anti-pT166 for AtRIN4 T166 phosphorylation, the same infiltration condition was applied for AvrRpm1 alleles and AtRIN4 alleles without RPM1.
For transient expression in N. glutinosa, the overnight cultures were pelleted by centrifugation and washed once in 10 mM MgCl2 before being resuspended in 10 mM MgCl2 supplemented with 100 μM acetosyringone. The bacterial suspension was incubated for 3 to 4 h at room temperature before it was diluted to an OD600 of 0.3. The strains with a GmRIN4 construct were mixed in a 1:1 (v/v) ratio with strains carrying an effector construct (or EV) before infiltration. Transgene expression was induced 40 h after infiltration by spraying plants with 50 μM dexamethasone with 0.02% (v/v) Silwet L-77 (OSi Specialties) surfactant added. Tissue was collected 4 h after induction and prepared for immunoblot analysis.
Immunoblot Analyses
Total protein was extracted from whole N. benthamiana or N. glutinosa leaves in 4 volumes (w/v) of lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 0.1% [v/v] Nonidet P-40, 1% [v/v] Plant Protease Inhibitor Cocktail [Sigma-Aldrich], and 1% [w/v] 2,2′-dithiodipyridine) and separated on 8% (w/v) polyacrylamide gels supplemented with 50 μM Phos-tag reagent (Wako Laboratory Chemical), or on 4 to 20% (w/v) Tris-Gly gradient gels (Bio-Rad). Proteins were transferred to a nitrocellulose membrane (GE Healthcare Life Sciences) and then detected with a 1:4000 (v/v) diluted peroxidase-conjugated anti-HA antibody (rat monoclonal; catalog no. 12013819001, Roche), a 1:4000 (v/v) diluted peroxidase-conjugated anti–c-Myc antibody (mouse monoclonal; catalog no. MA1-81357, Thermo Fisher Scientific), or a 1:2000 (v/v) diluted unconjugated anti-GFP antibody (mouse monoclonal; catalog no. NB600597, Novus Biologicals), which was detected using 1:5000 (v/v) diluted goat anti-mouse peroxidase-conjugated secondary antibody (catalog no. 31,460, Thermo Fisher Scientific). To detect ADP-ribosylated proteins, we used a 1:5000 (v/v) dilution of an anti-panADPR (catalog no. MABE1016, EMD Millipore). Immunoblots with anti–T7-horseradish peroxidase (HRP; catalog no. 69,048, Millipore Sigma) or anti–HA-HRP (catalog no. sc-7392 HRP, Santa Cruz) to detect AtRIN4 or AvrRpm1 alleles were performed as described in Chung et al. (2011). The immunoblots were visualized using the Clarity Western ECL Substrate (Bio-Rad) or the ECL prime (Amersham).
Immunoprecipitation of GmRIN4b, AtRIN4, and NOI Proteins
The N-terminally tagged sYFP-GmRIN4b was transiently expressed with AvrRpm1 in N. benthamiana. Total protein was extracted from whole N. benthamiana leaves in 4 volumes (v/w) of lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 0.1% (v/v) Nonidet P-40 [Sigma-Aldrich], 1% (v/v) Plant Protease Inhibitor Cocktail [Sigma-Aldrich], 1% (v/v) 2,2′-dithiodipyridine [Sigma-Aldrich], and 0.5 mM EDTA [Sigma-Aldrich]) using a ceramic mortar and pestle. The crude lysate was filtered through a double layer of Miracloth and then centrifuged at 10,000g and 4°C, for 5 min, twice to remove plant debris. The clarified extract was then incubated with 25 μL of GFP-Trap A (Chromotek) anti-GFP bead slurry for 1 h at 4°C with constant end-over-end tumbling. The beads were pelleted by centrifugation at 2500g and 4°C, for 2 min, and washed at least three times with 20× bed-volumes of wash buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 0.1% (v/v) Nonidet P-40, 1% (v/v) 2,2′-dithiodipyridine, and 0.5 mM EDTA).
To prepare GmRIN4b samples for mass spectrometry, an on-bead digestion method was used. Briefly, GmRIN4b-bound anti-GFP beads were washed with 50 mM ammonium bicarbonate (AMBIC, pH 8.0) at least three times and then incubated with 2× bed-volumes of 50 mM AMBIC with 50 mM DTT for 1 h at 60°C. The AMBIC plus DTT solution was removed, and then the beads were incubated in the dark with 2× bed-volumes of 50 mM AMBIC with 50 mM iodoacetamide for 1 h at room temperature. To digest the proteins, 250 ng of trypsin (Promega) was added to the beads, and then they were incubated overnight at 37°C. To collect the digested peptides, the samples were centrifuged at 3000g for 3 min, and the supernatant was transferred to a fresh Eppendorf tube. The beads were incubated with 2× bed-volumes of 60% acetonitrile with 0.1% trifluoroacetic acid for an additional 10 min at room temperature. The samples were centrifuged at 3000g for 3 min, and the supernatant was added to the new Eppendorf tube.
Immunoprecipitation or coimmunoprecipitation with anti-T7 antibody for AtRIN4 and AtNOI proteins was performed as described by Chung et al., (2011). Agrobacteria expressing AvrRpm1, RPM1, AtRIN4 alleles, and NOI proteins were infiltrated into N. benthamiana leaves at OD600 of 0.1, 0.3, 0.3, and 0.3, respectively. Leaf samples for immunoprecipitation or coimmunoprecipitation were collected 12 h postestradiol induction for the expression of AvrRpm1 alleles. Immunoprecipitation by anti-RIN4 with Arabidopsis tissue samples was done as described previously by Mackey et al., (2002).
Mass Spectrometry and Data Analysis
Tryptic peptides were injected into an Easy-nLC 100 HPLC system coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Specifically, peptide samples were loaded onto an Acclaim PepMapTM 100 C18 trap column (75 μm × 20 mm, 3 μm, 100 Å) in 0.1% (v/v) formic acid. The peptides were separated using an Acclaim PepMapTM RSLC C18 analytical column (75 μm × 150 mm, 2 μm, 100 Å) using an acetonitrile-based gradient (solvent A: 0% [v/v] acetonitrile and 0.1% [v/v] formic acid; solvent B: 80% [v/v] acetonitrile and 0.1% [v/v] formic acid) at a flow rate of 300 nL/min. A 30-min gradient was performed as follows: 0 to 0.5 min, 2 to 8% B; 0.5 to 24 min, 8 to 40% B; 24 to 26 min, 40 to 100% B; 26 to 30 min, 100% B, followed by re-equilibration to 2% B. Electrospray ionization was then performed with a nanoESI source at a 275°C capillary temperature and 1.9-kV spray voltage. The mass spectrometer was operated in data-dependent acquisition mode with mass range 400 to 2000 m/z. Precursor ions were selected for tandem mass spectrometry analysis in the Orbitrap with 3-s cycle time using higher energy collisional dissociation at 28% collision energy as described by Rosenthal et al. (2015). The intensity threshold was set at 5 × 104. The dynamic exclusion was set with a repeat count of 1 and exclusion duration of 30 s. The resulting data were searched in Protein Prospector (http://prospector.ucsf.edu/prospector/mshome.htm) against the G. max Rin4B sequence. Carbamidomethylation of Cys residues was set as a fixed modification. Protein N-terminal acetylation, oxidation of Met, protein N-terminal Met loss, pyroglutamine formation, phosphorylation on STY, and ADP-ribosylation on CDEKNRST were set as variable modifications. In total, three variable modifications were allowed. Trypsin digestion specificity with one missed cleavage was allowed. The mass tolerance for precursor and fragment ions was set to 10 ppm for both. Peptide and protein identification cutoff scores were set to 15 and 22, respectively. Marker ions (ADP-ribose fragments) at m/z 136.0632, 250.0940, 348.0709, and 428.0372 were used to confirm the existence of ADP-ribose on a given peptide (Rosenthal et al., 2015).
Yeast Two-Hybrid Analyses
The ProQuest yeast two-hybrid system (Invitrogen) was used. The full-length AtRIN4 coding sequence and the derivatives of AtRIN4 (RIN4 1Δ141 and RIN4 149Δ176) were cloned into the bait vector (pDEST32). EXO70 family genes from Arabidopsis (EXO70A1, EXO70B1, EXO70B2, EXO70C1, EXO70C2, EXO70E1, EXO70E2, EXO70F1, EXO70D2, EXO70G1, and EXO70H1) were cloned into the prey vector (pDEST22). The interaction between AtRIN4 and AvrB was used as positive control. The bait and prey vectors were cotransformed into yeast strain MaV203 in different combinations, and the transformants were selected on synthetic dextrose medium without Leu and Trp (SD−Leu−Trp). The positive clones were plated on SD−Leu−Trp−His medium including 25 mM 3-amino-1,2,4-triazole (Sigma-Aldrich), or SD−Leu−Trp−Ura medium. In addition, clones were subjected to X-Gal assays following the manufacturer’s instructions. For immunoblot detection of full-length and derivatives of AtRIN4 proteins, total protein was extracted from yeast as described previously (Horvath and Riezman, 1994). EXO70-GAL4 fusions were detected using anti-GAL4 monoclonal antibody (catalog no. 630402, Takara), while RIN4 fusions were detected using anti-RIN4 antibody (Mackey et al., 2002).
Coimmunoprecipitation Analyses
Protein coimmunoprecipitation analyses were performed as reported previously using GFP-Trap A (Chromotek) anti-GFP beads (Carter et al., 2019), except that Myc-Trap A (Chromotek) anti-Myc beads were used and incubation with beads was performed for only 1 h rather than overnight. Following incubation, beads were pelleted by centrifugation at 3000g and 4°C, for 2 min. Bead pellets were washed five times with 500 μL of immunoprecipitation wash buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 1 mM DTT, 1 mM EDTA, 1% (v/v) Nonidet P-40, and 0.1% (v/v) Triton X-100). After the last wash, the immune-complexes were resuspended in 40 μL of immunoprecipitation wash and 10 μL of 5× SDS loading buffer. Protein samples were denatured at 95°C for 10 min and were resolved on a 4 to 20% (v/v) gradient Precise Protein Gels (Thermo Fisher Scientific). Gels were run for 1 h at 150 V in 1× Tris/Gly/SDS running buffer. Total protein was transferred to a nitrocellulose membrane (GE Water and Process Technologies) for 45 min at constant amperage of 200 mA. Membranes were blocked with 5% (w/v) Difco skim milk (BD) for 1 h. Proteins were detected with 1:5000 (v/v) anti-panADPR (catalog no. MABE1016, EMD Millipore) or 1:5000 (v/v) HPR-conjugated anti-HA antibody (rat monoclonal; catalog no. 12013819001, Roche) or 1:5000 (v/v) diluted peroxidase-conjugated anti–c-Myc antibody (mouse monoclonal; catalog no. MA1-81357; Thermo Fisher Scientific) for 1 h. After 1 h, membranes were washed in 1× Tris-buffered saline, pH 7.5, solution containing 0.1% (v/v) Tween 20 three times for 10 min. Membranes were incubated with 1:5000 (v/v) HRP-conjugated goat anti-mouse or goat anti-rabbit antibody depending on primary antibodies for 1 h and then washed three times for 10 min in 1× Tris-buffered saline, pH 7.5, solution containing 0.1% (v/v) Tween 20. Protein bands were imaged using an Immuni-Star Reagents (Bio-Rad) or Supersignal West Femto Maximum Sensitivity Substrates (Thermo Fisher Scientific) and x-ray film.
Electrolyte Leakage Assays
Ion leakage measurements for hypersensitive response assays in N. benthamiana or Arabidopsis were performed as described previously (Chung et al., 2011). Leaf disks were collected 2 h postestradiol induction. Four leaf discs were sampled and submerged into 6 mL of double distilled water with three replicates per treatment (n = 12 leaf disks). Conductivity was measured using a conductivity meter (model 130, Orion) at the indicated time points. Error bars represent 2 × se. One-way ANOVA analyses were performed to calculate significant differences (Supplemental Data Set 2).
Callose Deposition Analyses
Leaves from ∼5-week-old plants were syringe infiltrated with 100 μM flg22 or distilled water and collected after 16 h. To visualize callose deposition, leaves were stained with aniline blue (Sigma-Aldrich) as described previously (Jin and Mackey, 2017). In short, leaves were cleared with lactophenol (mixture of 1 volume of 1:1:1:1 (v/v) glycerol, saturated phenol, lactic acid, and deionized water with 2 volumes of 95% ethanol) and washed with 50% (v/v) ethanol and then water. Callose was stained with 0.01% (w/v) aniline blue in 150 mM K2HPO4, pH 9.5. Stained leaves were mounted in 50% (v/v) glycerol and visualized with a Nikon eclipse 80i microscope. The number of callose deposits was counted using ImageJ software (http://rsbweb.nih.gov/ij/).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession number(s): AvrRpm1 (NP_114197), GmRIN4a (Glyma03g19920; NP_001235221), GmRIN4b (Glyma16g12160; NP_001239973), GmRIN4c (Glyma18g36000; NP_001235235), GmRIN4d (Glyma08g46400; NP_001235252), AtRIN4 (AT3G25070; NP_001325873), AtEXO70A1 (AT5G03540; NM_120434), EXO70B1 (AT5G58430; Q9FGH9); EXO70B2 (AT1G07000; NM_100573), EXO70C1 (AT5G13150; NM_121318), EXO70C2 (AT5G13990; NM_121402.2), EXO70D2 (AT1G54090; NM_104286.3), EXO70E1 (AT3G29400; AEE77581), EXO70E2 (AT5G61010; Q9FNR3), EXO70F1 (AT5G50380; AED95937), EXO70G1 (AT4G31540; NM_119303.3), EXO70H1 (AT3G55150; NM_115373.4), NOI1 (AT5G63270; AED97726), NOI2 (AT5G40645; AED94577), NOI3 (AT2G17660; AEC06662), NOI4 (AT5G55850; O22633), NOI5 (AT3G48450; AEE78418), NOI6 (AT5G64850; ABI49424), NOI7 (AT5G09960; ABD38888), NOI8 (AT5G18310; BAH19857), NOI9 (AT5G48500; AED95678), NOI10 (AT5G48657; AAO44036), NOI11 (AT3G07195; AAS76281), NOI12 (AT2G04410; AEC0583), NOI13 (AT4G35655; AEE86545), NOI14 (AT5G19473; AED92713).
Supplemental Data
Supplemental Figure 1. RIN4 is highly conserved between species.
Supplemental Figure 2. NOI domains from Arabidopsis NOI proteins.
Supplemental Figure 3. ADP-ribosylation of AtRIN4 on D153 promotes phosphorylation on T166.
Supplemental Figure 4. Phylogenetic tree of Arabidopsis EXO70 family members.
Supplemental Figure 5. Overexpression of AvrRpm1 blocks association of AtRIN4 with Arabidopsis EXO70 proteins independent of catalytic activity.
Supplemental Figure 6. Characterization of exo70 mutants described in this work.
Supplemental Data Set 1. GmRIN4b mass spectrometry summary data.
Supplemental Data Set 2. One-way ANOVA tables in support of Figure 4.
Supplemental Table. Primers used for this study
Supplemental File. Alignment used to generate phylogenetic tree in Supplemental Figure 4.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Laura Giese for technical support in the Mackey lab. This work was supported by the U.S. National Science Foundation (grants IOS-1551452 to R.W.I. and IOS-1758400 to J.L.D.) and by the Howard Hughes Medical Institute. J.L.D. is an Investigator of the Howard Hughes Medical Institute. Support was provided by the Rural Development Administration, Republic of Korea, Systems & Synthetic Agrobiotech Center (grant PJ01326904 to D.M.) and the National Institutes of Health (grant R01GM092772 to D.M.). In addition, support was provided by the Human Frontier Science Program (grant LT000607/2010-L to J.H.K.), and by a fellowship from the China Scholarship Council (201308430497 to Q.Z.).
AUTHOR CONTRIBUTIONS
R.W.I., T.J.R., D.M., E.H.C., and J.L.D. conceived the study. T.J.R., H.Z.K., J.T., E.-H.C., J.H.K., Q.Z., and M.S. created the methodology. T.J.R., H.Z.K., N.R., Y.Z., E.-H.C., J.H.K., Q.Z., and M.S. performed the research. T.J.R. and E.-H.C. wrote the article. R.W.I., D.M., and J.L.D. reviewed and edited the article. R.W.I., D.M., and J.L.D acquired the funding for this study.
Footnotes
Articles can be viewed without a subscription.
References
- Afzal A.J., Kim J.H., Mackey D. (2013). The role of NOI-domain containing proteins in plant immune signaling. BMC Genomics 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Ashfield T., Keen N.T., Buzzell R.I., Innes R.W. (1995). Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141: 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T., Ong L.E., Nobuta K., Schneider C.M., Innes R.W. (2004). Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T., Redditt T., Russell A., Kessens R., Rodibaugh N., Galloway L., Kang Q., Podicheti R., Innes R.W. (2014). Evolutionary relationship of disease resistance genes in soybean and Arabidopsis specific for the Pseudomonas syringae effectors AvrB and AvrRpm1. Plant Physiol. 166: 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Nimchuk Z., Hubert D.A., Mackey D., Dangl J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove S.R., Simonich M.T., Smith N.M., Sattler A., Innes R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.E., Helm M., Chapman A., Wan E., Restrepo Sierra A.M., Innes R., Bogdanove A.J., Wise R.P. (2019). Convergent evolution of effector protease recognition by Arabidopsis and barley. Mol. Plant Microbe Interact. 32: 550–565. [DOI] [PubMed] [Google Scholar]
- Cherkis K.A., Temple B.R., Chung E.H., Sondek J., Dangl J.L. (2012). AvrRpm1 missense mutations weakly activate RPS2-mediated immune response in Arabidopsis thaliana. PLoS One 7: e42633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.H., da Cunha L., Wu A.J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.H., El-Kasmi F., He Y., Loehr A., Dangl J.L. (2014). A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 16: 484–494. [DOI] [PubMed] [Google Scholar]
- Cvrčková F., Grunt M., Bezvoda R., Hála M., Kulich I., Rawat A., Zárský V. (2012). Evolution of the land plant exocyst complexes. Front. Plant Sci. 3: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D., Singer A.U., Wu A.J., McNulty B.C., Musselwhite L., Nimchuk Z., Sondek J., Dangl J.L. (2007). Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 3: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki K., Abe Y., Kanzaki E., Ito K., Utsushi H., Saitoh H., Bialis A., Banfield M.J., Kamoun S., Terauchi R. (2017). An unconventional NOI/RIN4 domain of a rice NLR protein binds host EXO70 protein to confer fungal immunity. bioRxiv. [Google Scholar]
- Gao Z., Chung E.H., Eitas T.K., Dangl J.L. (2011). Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 108: 7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pino A., Zenkin N., Loris R. (2014). The many faces of Fic: Structural and functional aspects of Fic enzymes. Trends Biochem. Sci. 39: 121–129. [DOI] [PubMed] [Google Scholar]
- Gibson B.A., Zhang Y., Jiang H., Hussey K.M., Shrimp J.H., Lin H., Schwede F., Yu Y., Kraus W.L. (2016). Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 353: 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Horvath A., Riezman H. (1994). Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10: 1305–1310. [DOI] [PubMed] [Google Scholar]
- Hurley B., Lee D., Mott A., Wilton M., Liu J., Liu Y.C., Angers S., Coaker G., Guttman D.S., Desveaux D. (2014). The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS One 9: e114921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Mackey D.M. (2017). Measuring callose deposition, an indicator of cell wall reinforcement, during bacterial infection in Arabidopsis. Methods Mol. Biol. 1578: 195–205. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Vance R.E., Dangl J.L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354: 1117. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Desveaux D., Singer A.U., Patel P., Sondek J., Dangl J.L. (2005a). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102: 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Geng R., Gallenstein R.A., Somers D.E. (2013). The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140: 4060–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.G., da Cunha L., McFall A.J., Belkhadir Y., DebRoy S., Dangl J.L., Mackey D. (2005b). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121: 749–759. [DOI] [PubMed] [Google Scholar]
- Kim M.G., Geng X., Lee S.Y., Mackey D. (2009). The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 57: 645–653. [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5: 749–757. [DOI] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.J., Coaker G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Holt B.F. III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Martin G.B., Bogdanove A.J., Sessa G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z., Marois E., Kjemtrup S., Leister R.T., Katagiri F., Dangl J.L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363. [DOI] [PubMed] [Google Scholar]
- Qi D., DeYoung B.J., Innes R.W. (2012). Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter C., Dangl J.L. (1995). The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol. Plant Microbe Interact. 8: 444–453. [DOI] [PubMed] [Google Scholar]
- Ritter C., Dangl J.L. (1996). Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal F., Nanni P., Barkow-Oesterreicher S., Hottiger M.O. (2015). Optimization of LTQ-Orbitrap mass spectrometer parameters for the identification of ADP-ribosylation sites. J. Proteome Res. 14: 4072–4079. [DOI] [PubMed] [Google Scholar]
- Sabol P., Kulich I., Žárský V. (2017). RIN4 recruits the exocyst subunit EXO70B1 to the plasma membrane. J. Exp. Bot. 68: 3253–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D., Kachroo A. (2010). RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 153: 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D., Robin G.P., Kachroo A. (2013). GmRIN4 protein family members function nonredundantly in soybean race-specific resistance against Pseudomonas syringae. New Phytol. 197: 1225–1235. [DOI] [PubMed] [Google Scholar]
- Singer A.U., Desveaux D., Betts L., Chang J.H., Nimchuk Z., Grant S.R., Dangl J.L., Sondek J. (2004). Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12: 1669–1681. [DOI] [PubMed] [Google Scholar]
- Stegmann M., Anderson R.G., Ichimura K., Pecenkova T., Reuter P., Žársky V., McDowell J.M., Shirasu K., Trujillo M. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24: 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño T.Y., Shen M., Coaker G., Mackey D. (2019). Regulated disorder: Posttranslational modifications control the RIN4 plant immune signaling hub. Mol. Plant Microbe Interact. 32: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer B.A., Teitzel G.M., Lee M.W., Jelenska J., Hotton S., Fairfax K., Jenrette J., Greenberg J.T. (2006). The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62: 26–44. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Hou S., Wang X., Li Y., Ren D., Chen S., Tang X., Zhou J.M. (2010). A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22: 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M., Subramaniam R., Elmore J., Felsensteiner C., Coaker G., Desveaux D. (2010). The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 107: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Luo X., Li W., Wang Z., Liu J. (2017). The bacterial effector AvrB-Induced RIN4 hyperphosphorylation is mediated by a receptor-like cytoplasmic kinase complex in Arabidopsis. Mol. Plant Microbe Interact. 30: 502–512. [DOI] [PubMed] [Google Scholar]
- Zhao T., Rui L., Li J., Nishimura M.T., Vogel J.P., Liu S., Zhao Y., Dangl J.L., Tang D. (2015). A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet 11 (1): e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]