Arabidopsis is anchored by a carotenoid-derived metabolite.
Abstract
Anchor roots (ANRs) arise at the root-shoot junction and are the least investigated type of Arabidopsis root. Here, we show that ANRs originate from pericycle cells in an auxin-dependent manner and a carotenogenic signal to emerge. By screening known and assumed carotenoid derivatives, we identified anchorene, a presumed carotenoid-derived dialdehyde (diapocarotenoid), as the specific signal needed for ANR formation. We demonstrate that anchorene is an Arabidopsis metabolite and that its exogenous application rescues the ANR phenotype in carotenoid-deficient plants and promotes the growth of normal seedlings. Nitrogen deficiency resulted in enhanced anchorene content and an increased number of ANRs, suggesting a role of this nutrient in determining anchorene content and ANR formation. Transcriptome analysis and treatment of auxin reporter lines indicate that anchorene triggers ANR formation by modulating auxin homeostasis. Together, our work reveals a growth regulator with potential application to agriculture and a new carotenoid-derived signaling molecule.
INTRODUCTION
Carotenoids are common isoprenoid pigments synthesized by all photosynthetic organisms and many heterotrophic bacteria and fungi (1–4). They are essential constituents of the photosynthetic apparatus (5), as well as a source for biologically important compounds such as retinoids (6), the phytohormones abscisic acid (ABA) (7) and strigolactone (SL) (8), and the recently discovered plant regulatory metabolite zaxinone (9). All of these derivatives arise by virtue of the extended, conjugated double bond system that makes carotenoids prone to oxidative cleavage (10). This reaction yields carbonyl products called apocarotenoids (2, 3) and can be catalyzed by enzymes from the carotenoid cleavage dioxygenase (CCD) family, which break defined C-C double bonds by inserting molecular oxygen (10). Arabidopsis CCDs are divided into nine-cis-epoxycarotenoid cleavage dioxygenases (NCED2, NCED3, NCED5, NCED6, and NCED9) that form the ABA precursor xanthoxin from the cleavage of 9-cis-epoxycarotenoids, and CCDs with different substrate and regiospecificities (10, 11). The latter group includes CCD1, which forms a plentitude of C13, C10, and C8 volatiles from different apocarotenoids and C40-carotenoids (12); CCD4, which cleaves all-trans-β-carotene into β-ionone (C13) and β-apo-10′-carotenal (C27) (13); the SL biosynthesis enzyme CCD7 (MAX3), which breaks 9-cis-β-carotene into β-ionone (C13); and CCD8 (MAX4), which converts 9-cis-β-apo-10′-carotenal into the SL biosynthesis intermediate carlactone (8). CCD8 can also cleave all-trans-β-apo-10′-carotenal into the ketone β-apo-13-carotenone (d’orenone) but with low activity (14). Nonenzymatic cleavage, which occurs at each double bond in the carotenoid backbone, is another important route for apocarotenoid formation (15). This process is triggered by reactive oxygen species (ROS) and can also yield signaling molecules, such as the plant stress signal β-cyclocitral, which is formed by singlet oxygen (1O2), mediating gene responses to ROS (16) and plant root growth (17). In addition to monocarbonyls, carotenoid cleavage can yield dialdehyde products (diapocarotenoids), as shown for several plant and cyanobacterial CCDs that cleave multiple double bonds within carotenoids or target apocarotenoids (18, 19). The question of whether diapocarotenoids are also regulatory metabolites has not yet been answered.
Plant root systems provide anchorage and are the primary site for water and nutrient uptake (20, 21). Arabidopsis is an ideal model plant to study root development because of its genetic tractability and its fast-growing and relatively simple root system. Arabidopsis has three highly characterized types of roots: (i) a primary root initiated in embryogenesis; (ii) lateral roots (LRs) that form from the primary root and other LRs; and (iii) adventitious roots, which emerge from non-root tissues, such as stem, leaves, and hypocotyl (20, 21). Anchor roots (ANRs) (22) constitute a fourth type of roots. They emerge from the collet (23), a region located just below the root-hypocotyl junction, and have remained largely uncharacterized.
The development of LRs occurs in a series of well-documented stages. They are positioned through a process that involves gene expression oscillation (24) mediated by an unidentified carotenoid-derived signal (25). In Arabidopsis, LRs initiate from xylem pole pericycle cells, which form the cell layer between the vascular bundle and the endodermis. These cells divide to produce LR primordia, which continue to grow until the new roots emerge by pushing through the outer layers of the originating root (26). The plant hormone auxin plays a central role in root development (27, 28). For instance, the auxin signaling component MP/ARF5 regulates embryonic root development (29), and the auxin-related transcription factors ARF7 and ARF19 are pivotal for LR initiation (30).
In addition to auxin, the carotenoid-derived phytohormones, SL and ABA, have been shown to regulate different aspects of root development (31, 32). To explore the biological functions of diapocarotenoids and to identify new carotenoid derivatives involved in Arabidopsis root development, we tested the activity of diapocarotenoids, which had either been previously identified as CCD products or were structurally predicted to result from carotenoids. We discovered that ANR formation requires carotenoid biosynthesis and is triggered by a previously unidentified diapocarotenoid that we called “anchorene.” To characterize the role of anchorene, we present the first comprehensive analysis of ANR development.
RESULTS
Anchorene is a specific regulator of ANR development
To identify new carotenoid-derived signals involved in root development, we checked the activity of six previously identified or predicted diapocarotenoids with carbon numbers ranging from C9 to C15 (Diapo1 to Diapo6; fig. S1A). Diapo1 (C9) is the expected product formed upon CCD8 cleavage of all-trans-β-apo-10′-carotenal (14). Diapo2 (C10) results from cleaving the C7, C8 and C15, C15′ double bonds in different carotenoids and is produced by cyanobacterial retinal-forming enzymes from C30 apocarotenoids (19). Diapo3 (C10) is a structural isomer of Diapo2 and a predicted cleavage product that can be formed by cutting the C11, C12 and C11′, C12′ bonds in almost all plant carotenoids (Fig. 1A and fig. S1B). Diapo4 (C12) results from cleaving the C7, C8 and C13′, C14′ bonds in many carotenoids and is formed by CCD1 from apo-10′-lycopenal (22). Diapo5 (C15) results from cleaving the C7, C8 and C11′, C12′ bonds in many carotenoids and is formed by CCD1 from apo-10′-lycopenal (22). Diapo6 (C14) is a common CCD1 product formed by cleaving the C19, C10 and C9′, C10′ bonds in many carotenoids (18). We treated seedlings with either 5 or 25 μM each compound and determined the length of primary roots 7 days post-stratification (dps). At lower concentrations, we did not observe notable effects with any of the compounds. At the higher concentration, application of Diapo2 and Diapo5 led to a severe decrease (approximately 80%) in primary root growth (fig. S2A). Diapo1, Diapo3, and Diapo4 showed weak inhibition of primary root length; however, the most notable effect of Diapo3 was the promotion of ANR formation (fig. S2A). On the basis of this activity, we named Diapo3 anchorene.
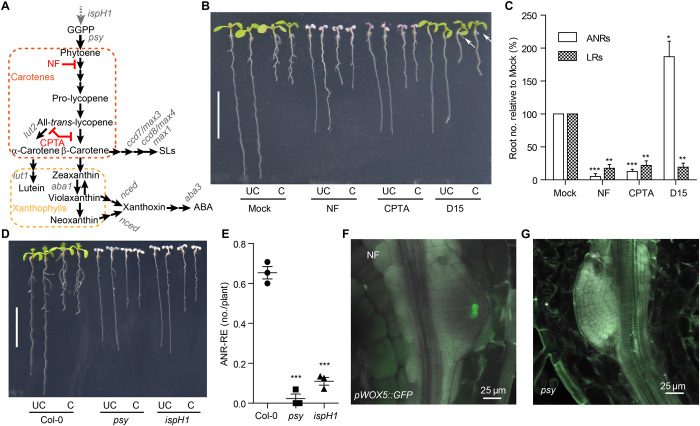
Fig. 1. Anchorene promotes ANR formation.
(A) Proposed production of anchorene (AR) (12,12′-diapocarotene-12,12′-dial) from β-carotene by oxidative cleavage; all-trans-β-carotene is taken as an example for the C-atom numbering. (B) AR promotes ANR formation in a concentration-dependent manner. Bottom: Representative seedlings treated with the indicated AR concentration. Scale bar, 1 cm. Top: Quantification of ANR no. as percentage (means ± SD) of seedlings with zero, one, or two ANRs (three independent replicates) corresponding to the treatments in the bottom panel. UC, uncut root apical meristems (RAMs); C, cut RAMs. (C) Two opposing ANRs are formed at the collet. White and red arrows in the right panel indicate primary ANRs and secondary ANRs (or LRs), respectively. (D) ANRs are initiated from the pericycle cells of the root. pDR5::nls-YFP was used for confocal microscopy imaging. The yellow fluorescent protein (YFP) signal is indicated by green, and SCRI Renaissance 2200 staining is indicated by purple. Ep, epidermis; C1, cortex layer 1; C2, cortex layer 2; En, endodermis; P, pericycle; ARI, ANR initiation site; ARP, ANR primordia; RH, root hair. Photo credit: K.-P.J. and S.A.-B., KAUST (B and C) and T.T.X. and I.B., KAUST (D).
Excision of the root apical meristem (RAM) triggers ANR formation (fig. S2B) (22). To test the effect of anchorene under these conditions, we applied different concentrations of this compound to Arabidopsis seedlings with or without RAM excision. About 9% of control Col-0 seedlings developed ANRs under normal conditions compared to about 50% upon RAM excision (Fig. 1B). The effect of 5 μM anchorene was comparable to that of RAM excision, triggering the formation of ANRs in 55% of the seedlings. Higher anchorene concentrations (10 and 20 μM) enhanced this ratio to 97 and 100%, respectively (Fig. 1B). There was also an increase in the number of seedlings that developed two ANRs from 0% in the control to approximately 80% upon application of 20 μM anchorene (Fig. 1B). Using a wide range of concentrations, we established a dose-response curve in the presence and absence of the RAM. The effect of anchorene was dose dependent in both cases (fig. S2C). Next, we investigated the effect of anchorene on LR formation and primary root length using different concentrations. As shown in fig. S2 (D and E), anchorene inhibited the growth of primary roots in a concentration-dependent manner but did not affect the number of LRs. To determine whether the anchorene effect on ANRs is caused by inhibiting primary root growth, we applied the compound locally either on shoots and collets or on root tips (fig. S2F). As shown in fig. S2 (G to I), application of anchorene on shoots and collets, but not on root tips, promoted ANR formation. By contrast, the inhibitory effect on primary root growth required local application and was not observed upon treating collets with anchorene. Together, these results indicate that anchorene effects on ANR formation and primary root growth are independent.
To test the specificity of anchorene, we evaluated the effect of structurally similar compounds on ANR formation. The application of Diapo2, a structural isomer of anchorene (fig. S1A) with a different position of one of the two methyl groups, gave only a modest increase in ANR formation, even at the relatively high concentration of 25 μM (fig. S2J). However, this concentration caused a pronounced inhibition of primary root growth, which may be the reason for the stimulatory effect on ANR formation. Modification of anchorene’s structure by reducing the aldehyde groups to alcohols or converting the aldehydes into acids or acid-ethyl esters resulted in a loss of activity (figs. S1C and S2J). These data suggest that anchorene promotion of ANR formation requires specific structural features found only in anchorene, among the compounds tested.
To understand how anchorene exerts its activity, we characterized its effect on ANR development using stereo and confocal microscopy. ANRs arise in the collet region characterized by the presence of dense root hairs (Fig. 1C) (23). Arabidopsis seedlings form one or two ANRs opposite each other, mirroring the positions of the cotyledons (Fig. 1C). In contrast, LRs emerge in much higher numbers at alternating positions (33). ANRs themselves can branch, forming secondary ANRs or LRs (Fig. 1C). The cellular pattern of ANR primordia in the collet indicates that they originate, similar to LRs, from the xylem pole pericycle. Analysis of the yellow fluorescent protein (YFP) signal in a pDR5::nlsYFP auxin reporter line (34), which has been used to localize LR initiation sites, indicated the pericycle origin of ANRs (Fig. 1D).
To track ANR development, we used a pDR5::LUC line, which marks LR prebranch sites (24, 25). We observed a clear LUC signal in the collet as early as 3 dps (fig. S3A), suggesting that this line can also be used to visualize ANR primordia. Consistent with anchorene effect on ANR formation, the application of this compound intensified the LUC signal in the collet (fig. S3B). To acquire more knowledge about ANR development and the role of anchorene in this process, we evaluated the green fluorescent protein (GFP) signal in a pDR5rev::GFP marker line in the presence and absence of anchorene from 2 to 6 dps. Both mock- and anchorene-treated seedlings developed clear GFP signals in the collet at 3 dps (fig. S3C). In the following 2 days, this signal faded in mock pDR5rev::GFP seedlings and disappeared completely at 6 dps, which indicates that these seedlings establish ANR primordia but the ANRs do not emerge. In contrast, anchorene-treated pDR5rev::GFP seedlings maintained a strong GFP signal in the collet, and ANRs emerged at 6 dps (fig. S3C).
A carotenoid-derived metabolite is required for normal ANR formation
To understand the role of carotenoids in ANR development, we monitored ANR formation under carotenoid-deficient conditions. Arabidopsis seedlings grown on media frequently do not form emerged ANRs, which impeded the characterization of factors affecting ANR formation. Therefore, we monitored ANR numbers after RAM excision (ANR-RE) to stimulate ANR development (fig. S2B) (22). We used these measurements as an indicator of ANR formation capacity. We first measured ANR formation upon treatment with norflurazon (NF) and 2-(4-chlorophenylthio)-triethylamine hydrochloride (CPTA) (4), which block phytoene desaturation and lycopene cyclization, respectively (Fig. 2A). We also investigated the effect of D15, a CCD inhibitor (25). These three compounds have been shown to reduce LR initiation (Fig. 2, B and C) (25), suggesting that a CCD product is required for LR development. Both NF and CPTA strongly reduced ANR-RE, while D15 promoted this process (Fig. 2, B and C). These results suggest that carotenoids are necessary for proper ANR and LR development but that each root type is regulated by a specific apocarotenoid. To test this hypothesis, we asked whether anchorene can restore LR capacity in D15-treated seedlings. Anchorene had no significant effect on LR formation after D15 treatment. Furthermore, it inhibited LR capacity in untreated seedlings (fig. S4A), providing evidence that anchorene is not the carotenoid-derived signal required for LR capacity. The positive effect of D15 on ANR-RE may be caused by increased carotenoid levels or by its inhibitory effect on primary root growth (25). Confirming the carotenoid dependency of ANR formation, the carotenoid-deficient mutants, ispH1 (35) and psy (4), disrupted in plastid isoprenoid and phytoene biosynthesis, respectively (Fig. 2A), exhibited a great reduction in ANR-RE compared to wild type (Fig. 2, D and E).
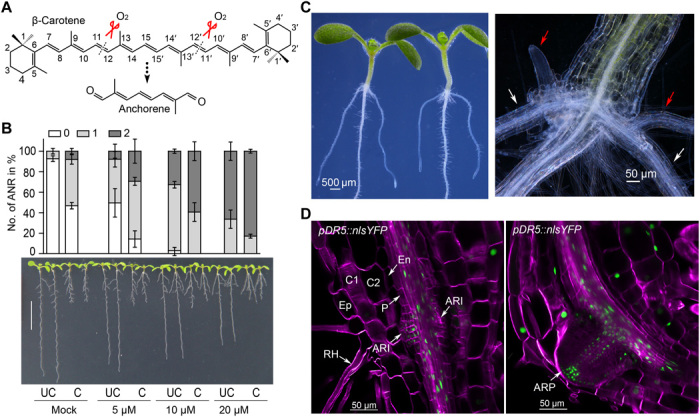
Fig. 2. Carotenoid deficiency leads to reduced ANR formation.
(A) Schematic of plant carotenoid biosynthesis. The gray dotted arrow indicates the upstream steps; mutants are shown in gray italics; reactions inhibited by NF and CPTA are depicted in red. Representative seedlings (B) and quantification data (C) show the effect of NF, CPTA, and D15 treatment on ANRs and LRs formation in Col-0 seedlings. ANRs and LRs no. were quantified after RAM excision; data are presented as means ± SD (three independent replicates). *P < 0.05, **P < 0.01, and ***P < 0.001, by two-tailed paired Student’s t tests. (D) Comparison of ANR formation between Col-0 and psy and ispH1 mutants. (E) ANR-RE quantification in Col-0, psy, and ispH1 seedlings. Data are presented as means ± SD (three independent replicates); ***P < 0.001, by two-tailed paired Student’s t test. Representative 5 dps NF-treated pWOX5::GFP (F) or psy mutant (G) seedlings have initiated ANR primordium. Photo credit: K.-P.J. and S.A.-B., KAUST (B and D) and A.J.D. and P.N.B., Duke University (F and G).
CPTA inhibits the synthesis of both α- and β-carotene, the precursors for the two separate branches of plant carotenoid biosynthesis (Fig. 2A) (36). The mutants lut1 (37) and lut2 (38) that are affected in α-carotene and lutein formation, respectively, were indistinguishable from wild type with regard to ANR-RE (fig. S4B). This indicates that the α-branch is not the primary source for the ANR signal. Next, we asked whether ABA or SL is the apocarotenoid signal required for ANR formation. We observed an increase in ANR-RE in the SL-deficient ccd8/max4 (39) and max1 (40) mutants and an inhibitory effect of the SL analog GR24 on ANR-RE (fig. S4, C and D), indicating a negative role of SL in ANR formation. However, we did not observe this increase in SL-deficient ccd7/max3 seedlings (fig. S4C). We did not detect a difference in ANR-RE in the ABA-deficient mutants, aba1 (41) and aba3 (42), or upon ABA application (fig. S4, E and F), excluding a role of ABA in ANR development. We also examined ANR-RE in nced and other ccd mutants—i.e., nced2, nced3, nced5, nced6, nced9, ccd1, and ccd4—but did not observe a significant difference in ANR-RE (fig. S4G), indicating that the corresponding enzymes are unnecessary or work redundantly in this regard.
To determine at which developmental stage the apocarotenoid signal regulates ANR development, we examined the effect of NF on the pDR5::LUC marker line. In NF-treated plants, we detected a clear collet LUC signal (fig. S3B), suggesting that carotenoids are not required for ANR initiation. This assumption is corroborated by the detection of ANR primordia in pWOX5::GFP (43) seedlings upon treatment with NF and in the carotenoid-deficient psy mutant (Fig. 2, F and G).
Anchorene is an endogenous signal that rescues ANR formation under carotenoid deficiency
To determine whether anchorene is the carotenoid-derived metabolite required for ANR formation, we asked whether it can rescue the ANR-RE reduction caused by inhibiting carotenoid biosynthesis. Seedlings cotreated with anchorene and either NF or CPTA were similar to untreated seedlings and did not show a reduction in ANR-RE, as observed upon treatment with NF or CPTA alone (Fig. 3, A and B). Similarly, anchorene application completely restored wild-type ANR-RE in the psy mutant (Fig. 3, C and D). These results indicate that anchorene is sufficient to promote ANR formation under carotenoid-deficient conditions and exclude the possibility that reduction in ANR-RE observed in carotenoid-deficient seedlings is an indirect consequence of albinism. We also examined the effect of NF alone and in combination with anchorene on pDR5rev::GFP seedlings. We did not see a significant reduction in GFP signal after NF application; however, the combined NF/anchorene treatment clearly enhanced the GFP signal (fig. S5), suggesting that anchorene stimulated growth of ANR primordia after initiation.
Fig. 3. AR is an endogenous metabolite.
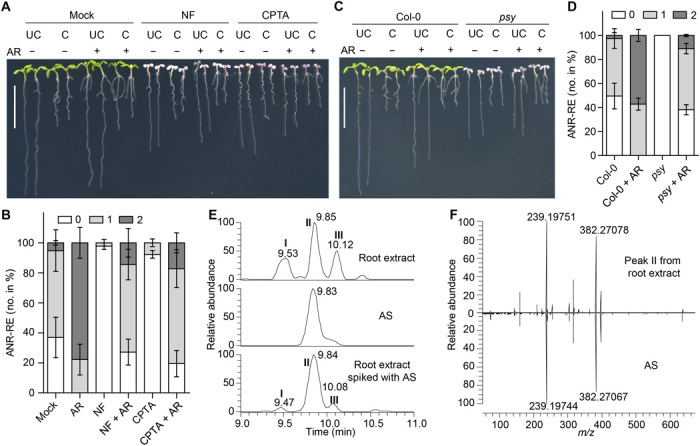
Representative seedlings (A) or ANR-RE quantification (B) of NF- and CPTA-treated seedlings exposed to AR. Representative seedlings (C) or ANR-RE quantification (D) of psy mutant seedlings treated with AR. “−” and “+” in (A) and (C) indicate the absence or presence of AR; in (B) and (D), data are presented as the percentage (means ± SD) of seedlings with zero, one, or two ANRs (three independent replicates). NF (1 μM), CPTA (100 μM), and AR (20 μM) were used. Scale bars, 1 cm. (E) Liquid chromatography mass spectrometry (LCMS) identification of endogenous AR in Arabidopsis root extract. Extracted ion chromatograms of AR from Arabidopsis root extract (top), AR standard (AS; middle), and Arabidopsis root extract spiked with AS (bottom). Peak II indicates endogenous AR (top) or endogenous AR spiked with AS (bottom). Peaks I and III represent AR isomers. (F) Endogenous AR (peak II) from Arabidopsis root extract and AS displayed identical pattern of product ion spectra. m/z, mass/charge ratio. Photo credit: K.-P.J. and S.A.-B., KAUST (A and C).
To determine whether anchorene is a natural plant metabolite, we developed an extraction, derivatization and liquid chromatography–mass spectrometry (LCMS) protocol for carotenoid-derived diapocarotenoids. We identified endogenous anchorene from Arabidopsis root and shoot tissues based on its precise match with the mass, chromatographic retention time, and product ion spectrum of the authentic anchorene standard (Fig. 3, E and F). We also detected two potential anchorene isomers identified by their exact mass and product ion spectra (Fig. 3E and fig. S6, A and B). The relative amounts of the three isomers differ between roots and shoots, with the anchorene peak being the most pronounced peak in roots (fig. S6A). This pattern is consistent with anchorene’s role in root development. Quantitative analysis showed about fourfold higher anchorene content in shoots compared to roots (0.08 ± 0.003 pmol/mg versus 0.02 ± 0.001 pmol/mg dry weight) (fig. S6C). Confirming its carotenoid origin, anchorene content decreased continuously after NF application (fig. S6, D and E). The formation of anchorene from carotenoids requires the cleavage of the C11═C12 and C11′═C12′ double bonds (Fig. 1A). This process could be initiated by NCEDs that cleave the C11═C12 double bond in 9-cis-epoxycarotenoids and 9-cis-zeaxanthin (7), leading to 9-cis-apo-11-carotenoids (C15), e.g., the ABA precursor xanthoxin, and all-trans-apo-12′-apocarotenoids, such as all-trans-3-OH-β-apo-12′-carotenal (OH-Apo12′; C25; fig. S7A). To test whether all-trans-apo-12′-apocarotenoids are further cleaved at the C11═C12 bond to form anchorene in planta, we measured anchorene content and quantified ANRs in Arabidopsis seedlings after feeding with OH-Apo12′ or the longer apocarotenoid all-trans-3-OH-β-apo-10′-carotenal (OH-Apo10′; C27), which we used as a control (fig. S7B). Arabidopsis seedlings fed with OH-Apo12′ contained much higher anchorene levels and developed more ANRs compared to untreated or OH-Apo10′ fed seedlings (fig. S7, C and D). These results suggest that all-trans-apo-12′-apocarotenoids, which can be formed by NCEDs, are likely precursors of anchorene in planta (fig. S7A).
Anchorene promotes ANR formation by modulating auxin distribution
Anchorene’s promotion of the GFP signal in the DR5 marker line suggested the involvement of auxin in ANR formation. Therefore, we investigated the role of ARF7 and ARF19, two auxin-responsive transcription factors required for LR initiation (28, 30). The arf7arf19 double mutant did not form any ANRs after anchorene application, even after RAM excision (Fig. 4A and fig. S8A). Moreover, confocal microscopy examination revealed that arf7arf19 double mutant lacks ANR primordia (Fig. 4B), suggesting that ARF7 and ARF19 are crucial for ANR initiation. Next, we investigated the effect of the auxin analog 1-naphthaleneacetic acid (NAA) and the auxin efflux transport inhibitor 1-N-naphthylphthalamic acid (NPA) on ANR formation. As shown in Fig. 4C and fig. S8B, treatment with NAA greatly increased ANR formation under normal conditions and upon RAM excision, while NPA treatment completely blocked ANR-RE. These results demonstrate that auxin signaling is required for ANR initiation and that auxin transport is essential for ANR formation.
Fig. 4. AR promotes ANR formation by regulating auxin distribution.
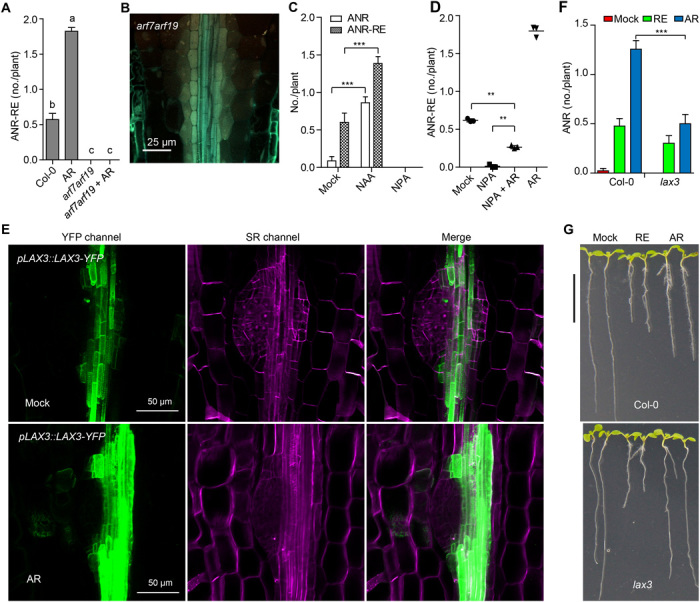
(A) Effect of AR on ANR-RE in Col-0 and arf7arf19 seedlings. (B) Confocal microscopy imaging of the collet region in a representative arf7arf19 seedling. (C) Effect of NAA and NPA on ANR-RE. (D) AR partially rescued ANR-RE in NPA-treated seedlings. (E) A representative confocal microscopy image of the collet region to show the AR effect on pLAX3::LAX3-YFP expression during ANR primordia formation. The YFP channel is shown in green, and the SCRI Renaissance 2200 staining is shown in purple. ANR quantification (F) and a representative picture (G) to show the seedlings of Col-0 and lax3 under normal (mock), RAM excision (RE), and AR application conditions. In (A), (C), and (F), data are presented as means ± SEM from one representative experiment. In (A) (n = 49, 51, 44, and 50), different letters denote significant differences (one-way ANOVA with Tukey’s multiple comparisons test, P < 0.05). In (C) (n = 25, 27, 31, 28, 25, and 27) and (F) (n = 43, 44, 39, 45, 43, and 48), ***P < 0.001 by two-tailed Student’s t test. In (D), data are presented as means ± SD (three independent replicates); **P < 0.01, by two-tailed paired Student’s t test. Photo credit: A.J.D. and P.N.B., Duke University (B); T.T.X. and I.B., KAUST (E); and K.-P.J. and S.A.-B., KAUST (G).
To determine whether anchorene acts by modulating auxin transport, we first evaluated its effect in the presence of NPA. Anchorene partially rescued the negative impact of NPA on ANR-RE (Fig. 4D and fig. S8C). Furthermore, anchorene largely rescued the loss of gravitropism caused by disruption of auxin transport in NPA-treated seedlings (fig. S8, D and E). Using the pPIN3::PIN3-GFP marker line, we also investigated the effect of anchorene on the auxin efflux carrier PIN3, which plays an important role in LR initiation and emergence (26). Application of anchorene led to a significant increase in PIN3 levels (fig. S8, F and G). However, the pin3 mutant did not show altered ANR-RE (fig. S8H), which is likely due to redundancy of the PINs in regulating auxin transport. Next, we used the auxin transporter marker line pLAX3::LAX3-YFP, which has been described to have a role during LR emergence (44, 45), to monitor the effect of anchorene on auxin influx. As shown in Fig. 4E, anchorene increased the expression levels of pLAX3::LAX3-YFP in root vascular tissues neighboring the ANR initiation site. Furthermore, the lax3 mutant (45) was less responsive to anchorene treatment as compared to wild type (Fig. 4, F and G). These results suggest that anchorene promotes ANR formation by modulating both auxin influx and efflux.
Next, we performed RNA sequencing (RNA-seq) on collet tissues isolated from seedlings after treatment with anchorene, NPA, or RAM excision. NPA and anchorene treatment affected the expression level of 3355 overlapping genes and exerted the opposite effect on 2791 (83%) of them (fig. S9, A and C, and dataset S1). This is consistent with the opposite effects of NPA and anchorene on ANR development. In contrast, RAM excision and anchorene treatment led to a similar response in the expression level of 1459 genes and caused opposite effects in only 36 genes (fig. S9, B and D, and dataset S1), which is in line with their common role in triggering ANR development. Biological process (BP) Gene Ontology term analysis showed that many genes up-regulated by both anchorene treatment and RAM excision are related to auxin metabolism (fig. S9E and dataset S2), providing further evidence for a role of auxin in ANR development, and that anchorene regulates ANR development mainly by modulating auxin homeostasis. For example, TAA1 and YUC7, two genes up-regulated by anchorene, are key enzymes involved in tryptophan-dependent auxin biosynthesis (fig. S9F) (46). Many genes that were up-regulated by anchorene and down-regulated by NPA are related to RNA methylation and RNA modification (fig. S9E and dataset S2), which indicates that anchorene may also regulate gene activity at a posttranscriptional level.
ANR formation and anchorene contents are regulated by nutrient availability
Plants modulate architecture and growth of their roots according to soil texture and nutrient availability. To explore the effect of soil on ANR formation, we compared the number of ANRs in seedlings grown in organic or sandy soil. A total of 63 ± 17.5% of seedlings grown in sand formed ANRs at 8 dps compared to only 4 ± 3.5% of those grown in organic soil (Fig. 5, A and B). Next, we analyzed the elemental composition of the sandy and organic soil and found that they have quite different levels of many nutrients, including phosphorus and nitrogen (table S1). Therefore, we determined the effect of phosphorus and nitrogen deficiency on ANR formation. Lack of phosphorus did not affect ANR formation. In contrast, exposing Arabidopsis seedlings to nitrogen deficiency led to an obvious increase in the average of ANR number per plant (Fig. 5, C and D). Nitrogen deficiency also led to much higher amounts of anchorene (Fig. 5E), indicating that nitrogen availability regulates the level of anchorene, which points to a role of this metabolite in Arabidopsis response to nitrogen deficiency.
Fig. 5. AR and ANR formation are triggered by nitrogen deficiency.
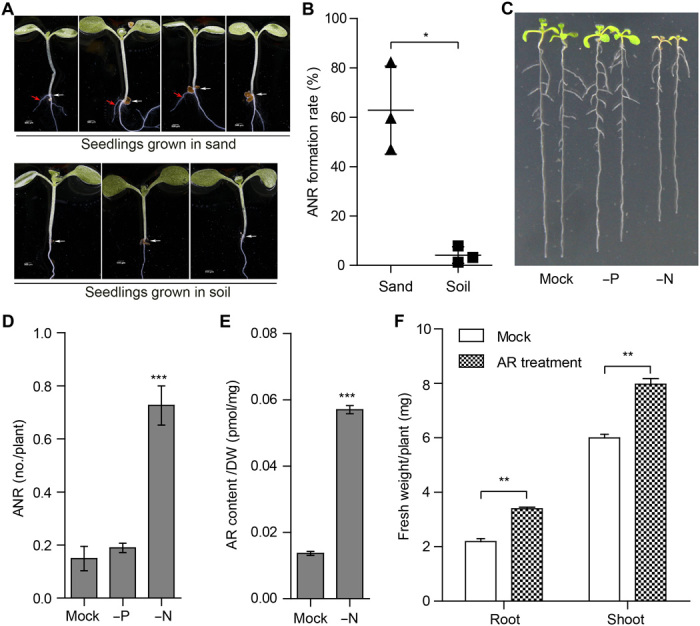
(A) Representative 8-day-old sand- or soil-grown seedlings. White and red arrows indicate collets and ANRs, respectively. (B) ANR formation rate of seedlings grown in sand and soil. Representative seedlings (C) and ANR no. counting (D) of 10-dps Col-0 seedlings grown in agar plates with Hoagland (mock), phosphorus deficiency (−P), or nitrogen deficiency (−N) media. (E) AR contents of the root tissues of seedlings grown in the plates with Hoagland or −N media. (F) AR treatment increases root and shoot biomass of Arabidopsis seedlings. Seventeen-day-old seedlings with or without AR treatment were used for biomass analysis. In (B), (D), and (F), data are presented as means ± SD from three, four, and three independent experiments, respectively, and paired two-tailed Student’s t test was used; in (E), data are presented as means ± SEM from one representative experiment, and unpaired two-tailed Student’s t test was used (n = 6 and 3). *P < 0.05, **P < 0.01, and ***P < 0.001. Photo credit: K.-P.J. and S.A.-B., KAUST (A and C).
Last, we investigated the effect of anchorene treatment on plant growth by measuring root and shoot biomass in 17-day-old anchorene-treated seedlings. Treatment with anchorene for 1 week led to increased ANR number, a wider root system, and enhanced root (about 50%) and shoot (about 30%) fresh biomass (Fig. 5F and fig. S10A). We also tested the effect of anchorene on rice roots and observed a notable induction in root length (fig. S10, B to D), indicating the potential of this metabolite in promoting plant growth.
DISCUSSION
In this study, we investigated the development of ANRs, the least characterized Arabidopsis root type, and demonstrate that their formation is triggered by a carotenoid-derived signal. Moreover, we show that anchorene is the signal that regulates ANR development. To our knowledge, anchorene is the first reported diapocarotenoid with a specific regulatory function. Because of their instability and reactivity, diapocarotenoids have attracted little attention and have mainly been studied as precursors of pigments, such as crocin (47). Hence, the identification of anchorene is expected to facilitate the discovery of further diapocarotenoid-based plant regulatory compounds and unravel new functions of carotenoid-derived metabolites.
Anchorene is a specific inducer of ANR formation, as shown by the inactivity of its isomer and derivatives, i.e., the corresponding dialcohol, diacid, and diethyl ester (fig. S1, A and C). Unlike LRs, ANRs originate from the collet, which emerges from embryonic tissue. Therefore, ANR development is fundamentally different from that of LRs, which initiate in the differentiation zone nearer the root tip (23). Although both ANRs and LRs originate from the pericycle, the collet pericycle forms only one or two ANRs located opposite each other (Fig. 1, C and D), while the primary root pericycle continuously develops alternating LRs. Anchorene also significantly inhibits primary root growth. However, local application of anchorene demonstrated that the effects of anchorene on ANR formation and primary root growth are independent processes.
Anchorene is a synthetic compound that could arise in planta from carotenoid cleavage. Our work demonstrates that ANR development requires a carotenoid-derived signal (Fig. 2) and that anchorene exerts the function of this signal in carotenoid-deficient seedlings (Fig. 3, A to D). Further, we show that anchorene is a natural Arabidopsis metabolite and that the content of this compound decreased upon application of the carotenoid biosynthesis inhibitor NF (Fig. 3, E and F, and fig. S6, D and E). However, the question of how anchorene is produced from carotenoids remains elusive. Theoretically, anchorene can arise by cleaving C11═C12 and C11′═C12′ double bonds in all carotenoids starting from ζ-carotene in the carotenoid biosynthesis pathway (fig. S1B). In contrast to ABA and SL, which derive from specific 9-cis-carotenoids (7, 8), anchorene can be formed from all carotenoids with a continuously conjugated, trans-configured central moiety (C11 to C11′; Fig. 1A).
Several CCDs from plants (18), fungi (48), and cyanobacteria (19) produce diapocarotenoids, either by repeated cleavage of carotenoid substrates or by specifically targeting apocarotenoids. However, enzymatic studies on Arabidopsis CCDs do not support the formation of anchorene by a single CCD. None of the Arabidopsis CCDs are capable of performing cleavage at both of the C11═C12 and C11′═C12′ double bonds in vitro (3). Nevertheless, NCEDs could catalyze a first step in anchorene formation since they cleave the C11═C12 double bond in 9-cis–configured epoxycarotenoids and zeaxanthin (7). This cleavage activity leads to all-trans-apo-12′-carotenoids that can be immediate precursors of anchorene. Our feeding experiment indicates that OH-Apo12′ may be a specific precursor of anchorene in Arabidopsis (fig. S7). However, we did not detect a reduction in ANR-RE in any single nced or ccd mutant (fig. S4, C and G), which might be due to functional redundancy of the five NCEDs and four CCDs present in Arabidopsis. Carotenoids are also cleaved nonenzymatically and catalyzed by ROS (16). If this is the case, then the specificity of anchorene activity may be regulated by a receptor rather than by anchorene biosynthesis.
Our study reveals a central role of auxin in ANR development. The auxin-responsive transcription factors ARF7 and ARF19, which are key regulators of LR initiation (28, 30), are also indispensable for ANR initiation (Fig. 4, A and B). Moreover, the auxin analog NAA strongly increases ANR-RE, while the auxin transport inhibitor NPA impedes the formation of ANRs (Fig. 4C), suggesting that auxin content and distribution are both important for ANR-RE.Using auxin-responsive marker lines, we showed that the application of anchorene increased the levels of auxin reporters both in ANR primordia and the surrounding tissue (figs. S3 and S5), suggesting that anchorene triggers ANR formation by modulating auxin transport and levels. Anchorene treatment resulted in a significant increase at the protein level of PIN3 and LAX3, auxin efflux and influx transporters, respectively (Fig. 4E and fig. S8, F and G). Consistent with these results, transcriptome analysis showed that anchorene application increased the transcript levels of many auxin biosynthesis genes, including the two key genes in tryptophan-dependent auxin biosynthesis, TAA1 and YUC7 (fig. S9).
Root systems are important not only for anchoring plants in soil but also for absorbing water and nutrients. We observed an increase in ANR number upon using nutrient-poor sandy soil (Fig. 5, A and B). Moreover, we found higher anchorene levels and increased ANR formation under nitrogen-deficient conditions (Fig. 5, C to E), suggesting that anchorene production can be regulated by nutrient availability. In addition, we observed a growth-promoting effect of this metabolite in both Arabidopsis and rice (Fig. 5F and fig. S10). These results indicate that ANRs may improve nutrient uptake and indicate a potential of anchorene for applications in agriculture or horticulture. It is worth mentioning that anchorene is a commercially available compound used as a building block for the manufacturing of different carotenoids on an industrial scale (49), which makes its application quite feasible.
MATERIALS AND METHODS
Chemicals
Diapo1, Diapo2, Diapo4, Diapo5, and Diapo6 were synthesized by Buchem (The Netherlands). Diapo3 (anchorene) was first custom-synthesized by Buchem (The Netherlands) and was later synthesized together with D6-anchorene and anchorene derivatives according to the protocol (see below). All diapocarotenoids were dissolved in acetone to make 10 mM stock solutions. For the screening experiments, each chemical was diluted in half-strength Murashige and Skoog (MS) media [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] to reach indicated concentrations of 25 and 5 μM. Stock solutions for D15 (100 mM), NF (10 mM) (Chem Service), and CPTA (50 mM) [obtained from the laboratory of L. E. Sieburth at the University of Utah (Salt Lake City, UT)] were prepared in dimethyl sulfoxide. D15 (125 μM), NF (1 μM), and CPTA (100 μM) working solutions were obtained by diluting the corresponding stock solutions in the aforementioned MS medium. GR24 (Chiralix, The Netherlands) was prepared in acetone. ABA, NAA, and NPA were purchased from Sigma-Aldrich, and their stock solutions were all prepared in water at 1 mM concentration.
Plant materials and growth conditions
Col-0 was used as wild type, unless otherwise noted. Mutants psy, isph1, lut1, lut2, ccd1, ccd4, ccd7, ccd8, nced2, nced3, nced5, nced6, nced9, aba1, and aba3 were described previously (25). The mutants pin3 to pin4 were acquired from Arabidopsis Biological Resource Center stock center. The arf7arf19 (CS24625) mutants were acquired from the European Arabidopsis Stock Centre and as descripted previously (30). pDR5::nlsYFP (34), pWOX5::GFP (43), pDR5::LUC (24), pDR5rev::GFP (50), pPIN3::PIN3-GFP (51), and pLAX3::LAX3-YFP (45) transgenic marker lines were all as described previously. All mutants and transgenic line information can be found in table S2.
Sterilized Col-0 and mutant seeds were kept at 4°C in darkness for 3 days to stimulate seed germination and then sown on half-strength MS [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] plates supplemented with the indicated compounds. Plates were vertically grown in Percival growth chambers under long day (16-hour light/8-hour dark; 22°C; 60% relative humidity; light density, 4000 lux) light-emitting diode (LED) white light [Hyperikon 16W LED Light Bulb A21, 16 W (100 W equivalent), CRI92, 1620 lumens, and 4000 K (daylight glow)] conditions. Light fluorescence rates were measured using a digital lux meter (PeakTech 5025).
ANR, LR, and primary root phenotyping assays
ANRs were counted in seedlings vertically grown on half-strength MS [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] media 8 dps using a dissection microscope. To investigate ANR-RE, RAM of seedlings was excised using sterile scalpels at 5 dps and then grown for another 3 days before counting protocol for quantifying LR capacity has been described previously (25). Specifically, RAM of seedlings were excised at 8 dps and then grown for another 3 days before counting the number of emerged LRs. For determining the effect of anchorene on LRs, the number of emerged LRs was counted using a dissection microscope at 8 dps. Primary root length was measured using the publicly available ImageJ software (http://rsbweb.nih.gov/ij/) after taking digital photographs.
For the quantification of ANR formation under OH-Apo10′ and OH-Apo12′ treatment conditions, the sterilized Arabidopsis seeds were first exposed to light for 24 hours and then plated to half-strength MS [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] media plates with indicated chemicals; the plates were kept under darkness (22°C) for another 2 days and then exposed to long day light conditions (16-hour light/8-hour dark; 22°C; 60% relative humidity; light density, 4000 lux) for another 7 days to count the ANR emergence.
Confocal microscopy
To characterize ANR initiation and primordium, the ClearSee protocol was applied as described previously (52). Briefly, seedlings exposed to various treatments were fixed using 4% paraformaldehyde dissolved in phosphate-buffered saline (PBS) for 30 min. Fixed seedlings were washed twice with PBS and then immersed in ClearSee solution [10% (w/v) xylitol, 15% (w/v) sodium deoxycholate, and 25% (w/v) urea in water]. After incubating the seedlings in ClearSee solution in the dark at room temperature for 2 to 3 days, laser scanning confocal microscopy (Zeiss LSM 510 microscope) was used to visualize the roots. To determine pDR5Rev::GFP fluorescence at the collet region, live seedlings were directly used for laser scanning confocal microscopy (Zeiss LSM 510 microscope) examination.
For fluorescence intensity quantification of pDR5Rev::GFP and pPIN3::PIN3-GFP marker lines, ImageJ software (http://rsbweb.nih.gov/ij/) was used after taking confocal microscopy photos. All calculations are background subtracted.
To monitor ANR emergence, 3 to 4 dps pDR5::nlsYFP and pLAX3::LAX3-YFP Arabidopsis seedlings were embedded in 8% agarose and sectioned with a vibratome (Leica VT1000S vibratome). Sections were immediately stained with SCRI Renaissance 2200 (53) for 5 min to visualize cell walls. Images were acquired using a Zeiss LSM 880 with Airyscan using an Objective C-Apochromat 40×/1.2 DIC M27 water immersion. YFP fluorescence was excited with a 514 nm laser and detected with 519- to 620-nm wavelength, and the emission was recorded at 570 nm. For SCRI Renaissance, fluorescence was excited with a 405 nm laser and detected at 410- to 507-nm, and the emission was recorded at 459 nm. Images were processed with Zeiss ZEN software and Adobe Photoshop.
Luciferase assay
Luciferase activity was assayed as previously described (24). Briefly, 1 ml of 5 mM potassium luciferin (Gold Biotechnology) dissolved in water was directly applied to pDR5::LUC seedlings grown vertically on half-strength MS plates. The luciferin solution was allowed to dry for 5 to 10 min in the dark at room temperature. Seedlings were then imaged using a Lumizone CA automated Chemiluminescence system. Seven-minute exposure times were used.
Qualitative and quantitative identification of anchorene using LCMS
Freeze-dried and ground Arabidopsis seedlings tissues (30 to 40 mg) were extracted using 1 ml of acetonitrile with antioxidant (0.1% butylated hydroxytoluene) for 15 min in an ultrasonic bath (Branson 5510EDTH; 25°C), followed by centrifugation for 8 min at 13,000 rpm at 4°C. The collected supernatant was dried using a concentrator (Labconco RapidVap System). The extract was derivatized (fig. S11B) according to the protocol described previously with minor modification (54). Derivatization solution (50 μl) containing derivatization reagent (10 mg/ml) (N2,N2,N4,N4-tetraethyl-6-hydrazineyl-1,3,5-triazine-2,4-diamine) (Chemspace) in 1% formic acid methanol was added in the extract for the incubation at 37°C for 15 min. Then, the sample solution was diluted to 150 μl with 1% formic acid in methanol and filtered by a 0.22-μm filter before LCMS analysis.
The qualitative analysis of derivative anchorene were performed on Ultra High Performance Liquid Chromatography (UHPLC) Q Exactive Plus MS. Chromatographic separation was achieved on an Acquity UPLC BEH C18 column (100 mm by 2.1 mm; 1.7 μm; Waters) using a mobile phase consisting of water:acetonitrile (90:10, v:v; A) and acetonitrile:isopropanol (90:10, v:v; B), both containing 0.2% formic acid. A gradient was applied, starting with 20% B and increasing to 100% B over 15 min. A concentration of 100% B was maintained for 3 min. To equilibrate the column before the next run, the mobile phase was adjusted back to 20% B in 1 min, and this concentration was maintained for 3 min before the next sample injection. A flow rate of 0.2 ml/min and a column temperature of 35°C were maintained throughout the run. The eluent of the column was introduced to the mass spectrometer using heated-electrospray ionization in positive mode. The injection volume was 15 μl. The conditions of the mass spectrometer were set as follows: resolution, 280,000; automatic gain control, 3 × 106; maximum injection time, 150 ms; sheath gas flow, 40 arbitrary units; auxiliary gas flow, 10 arbitrary units; spray voltage, 4.0 kV; capillary temperature, 350°C; and auxiliary gas heater temperature, 400°C. The quantitative analysis of derivative anchorene was carried out on HPLC QTRAP tandem MS. Chromatographic separation was achieved on an Acquity UPLC CSH C18 column (50 mm by 2.1 mm; 1.7 μm; Waters) using a mobile phase consisting of water:acetonitrile (95:5, v:v; A) and pure acetonitrile (B), both containing 0.2% formic acid. A gradient was applied, starting with 10% B and increasing to 40% B over 5 min. Then, 15% B was increased within 10 min, followed by an increase of 45% B over 2 min. Next, 100% B was maintained for 10 min. To equilibrate the column before the next run, the mobile phase was adjusted back to 10% B in 1 min, and this concentration was maintained for 8 min before the next sample injection. A flow rate of 0.15 ml/min and a column temperature of 40°C were maintained throughout the run. The eluent of the column was introduced to the mass spectrometer using turbo spray ion source in positive mode. The injection volume was 10 μl. The conditions of the mass spectrometer were set as follows: curtain gas, 30; ionspray voltage, 5 kV; temperature, 400°C; ion source gas 1, 30; ion source gas 2, 40; declustering potential, 55; entrance potential, 10; collision energy, 25; and collision cell exit potential, 10. For derivative anchorene, Q1 mass (Da), 635.5 and Q3 mass (Da), 239.2 were used; for derivative D6-anchorene, Q1 mass (Da), 641.5 and Q3 mass (Da), 239.2 were used.
RNA-seq material preparation and data analysis
For anchorene and NPA treatment, Col-0 seedlings were vertically grown on half-strength MS [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] plates supplemented with 20 μM anchorene or 1 μM NPA for 5 days. RAM excision treatment was applied by excising the RAM from 4.5 dps Col-0 seedlings, and samples were collected 12 hours after excision. Col-0 seedlings treated with acetone were used as mock. About 2-mm-long sections at the collet region were sampled, and sections from approximately 50 seedlings were used for each RNA sample. Total RNA was extracted using Direct-zol RNA MiniPrep Plus (200 preps) with Zymo-Spin (Zymo Research). One hundred nanograms of total RNA was used for RNA HiSeq 4000. For each treatment, we collected three RNA samples isolated from three independent experiments.
Before the analysis of RNA-seq data, the adaptor sequences and low-quality ends of sequenced reads were trimmed using Trimmomatic v0.32 (55) and quality-checked using FastQC v0.11.3 (56). To quantify the expression level of genes, the remaining reads were pseudo-aligned to the publicly available TAIR10 Arabidopsis thaliana transcriptome (release 34) using Kallisto v0.43.0 (57). The estimated read counts and calculated transcripts per million were subsequently passed to sleuth v0.28.1 (58) for differential expression analysis. Significantly differentially expressed genes were identified on the basis of a cutoff of fold change > 1.5 and q value < 0.05. Gene ontology and enrichment analysis was conducted using clusterProfiler (59). RNA-seq data can be accessed at National Center for Biotechnology Information (NCBI) via BioProject ID PRJNA489360.
ANR phenotyping in sand and soil
Silver sand (VWR) and soil (Asdcofert.com) were used for ANR phenotyping experiments. Col-0 seeds were kept in 4°C for 3 days to ensure uniform germination and then sown in pots with sand or soil. The pots were kept under long-day photoperiod conditions (16-hour light/8-hour darkness, 22°C, and 60% humidity) for 8 days. Then, forceps were used to gently take the seedlings from the sand or soil. Seedlings were then washed using water before counting the ANRs under a microscope.
Nutrient element analysis in sand and soil
For micronutrient elements analysis, inductively coupled plasma optical emission spectrometry (ICP-OES) was used to measure elemental levels of Fe, K, Mg, Mn, P, and Zn. Specifically, homogenized dry soil (50 to 100 mg) and sand (100 to 200 mg) samples were dissolved in the vessels with 8 ml of nitric acid together with negative control, which only include nitric acid and samples spiked with all the standard (6 ml of nitric acid + 2 ml of standards mixture). Samples were digested using microwave digestion. After the digestion, ddH2O was added for a final volume of 25 ml. Samples were centrifuged, and the supernatant was collected for ICP-OES analysis. For the C, H, N, and S elemental analysis, 5 to 20 mg of homogenized dry soil and sand samples were analyzed with a CHNS Elemental Analyzer (LECO), and sulfanilamide was used as a standard.
Phosphorus- and nitrogen-deficient growing conditions for Arabidopsis
Col-0 seedlings were grown vertically in Hoagland medium (60) [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] plates for 10 or 12 dps in Percival growth chambers under long day (16-hour light/8-hour dark; 22°C; 60% relative humidity; light density, 4000 lux) LED white light [Hyperikon 16W LED Light Bulb A21, 16 W (100-W equivalent), CRI92, 1620 lumens, 4000 K (daylight glow)] conditions. Ten-dps seedlings were used for ANR phenotyping, and 12-dps seedling root tissues were used for anchorene measurement. For phosphorus-deficient medium, we removed the KH2PO4 and added equal content of potassium by adding KCl in Hoagland solution; for nitrogen-deficient medium, we removed the NH4NO3 in Hoagland solution.
Anchorene bioassay on Arabidopsis and rice growth
Col-0 seedlings were grown in vertical half-strength MS [with 0.5% sucrose + 1% agar and MES (0.5 g/liter) (pH 5.7)] plates with or without anchorene application (20 μM) for 1 week. We selected well-growing mock (without anchorene treatment) and anchorene-treated seedlings and transferred them to new half-strength MS agar media plates. Twelve seedlings per plate were grown vertically for another 10 days. After 10 days, roots and shoots were separately collected as one technical replicate from each plate. Four technical replicates for each treatment were quantified in each experiment, and three independent experiments were conducted.
Sterilized Nipponbare rice seeds were kept at 30°C in darkness for 1 day to stimulate germination and transferred to Petri dishes with half-strength MS media [with MES (0.5 g/liter) (pH 5.7)] and autoclaved filter paper. Petri dishes were incubated in darkness for another 2 days at 26°C and then exposed to light for 1 day in Percival growth chamber (12-hour light/12-hour dark, 26°C, 55% humidity, and 500 μmol m−2 s−1). Uniform rice seedlings were transferred to hydroponic culture (Hoagland) supplied with acetone (mock) or 20 μM anchorene. Seedlings were grown under the same growth conditions for another 6 days, and solutions (mock; 20 μmol of anchorene) were replaced every 2 days.
Statistical analyses
For ANR no. and ANR-RE quantification, data are presented as means ± SD when they are from three biological replicates, and data are presented as means ± SEM when they are from one representative experiment. The analyses were conducted using GraphPad Prism 5 and Microsoft Excel 2013. Statistical significance was determined by paired or unpaired two-tailed Student’s t tests when only comparing two groups or by one-way analysis of variance (ANOVA) with Tukey’s posttest where multi groups are need to be compared. Differences between groups were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank B. Schaefer for valuable discussions and M. Khalid for technical support. Funding: This work was supported by Baseline funding and the Research Grants Program-Round 4 (CRG4) from King Abdullah University of Science and Technology (KAUST) to S.A.-B., by the Arnold and Mabel Beckman Postdoctoral Fellowship to A.J.D., and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through grant GBMF3405) to P.N.B. Author contributions: K.-P.J., A.J.D., P.N.B., and S.A.-B. conceived the study. J.M., N.M.K., and X.G. performed anchorene LCMS analysis. G.C. and M.A. performed the RNA-seq analysis. T.T.X. and I.B. performed the longitudinal section and confocal microscopy examination for pDR5::nlsYFP and pLAX3::LAX3-YFP. E.S. and M.R. synthesized the anchorene derivatives. K.-P.J. and A.J.D. performed the other experiments and analyzed the data. K.-P.J., A.J.D., P.N.B., and S.A.-B. wrote the paper. Competing interests: S.A.-B., K.-P.J., and A.J.D. are authors on a patent on anchorene and its applications, published by KAUST, Thuwal, Saudi Arabia and Duke University, USA (WO 2019/180638 A1; 26 September 2019.) All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. All dialdehydes and their derivatives used in this study can be provided by KAUST pending scientific review and a completed material transfer agreement. Requests for the dialdehydes and their derivatives used in this study should be submitted to S.A.-B. RNA-seq data can be accessed at NCBI via BioProject ID PRJNA489360.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/11/eaaw6787/DC1
Supplementary Text
Fig. S1. Structures of diapocarotenoids and supposed precursors for anchorene.
Fig. S2. Effects of diapocarotenoid and its derivatives on Arabidopsis root development.
Fig. S3. Characterization of ANR development by different DR5 marker lines.
Fig. S4. Root formation under different treatments and in various mutants.
Fig. S5. Anchorene effects on DR5 expression in ANR primordia.
Fig. S6. Anchorene isomer identification and anchorene quantification.
Fig. S7. Conversion of OH-Apo12′ into anchorene in plants.
Fig. S8. Involvement of auxin signaling and distribution on ANR development.
Fig. S9. Transcriptomic change analysis of collet tissues upon different treatments by RNA-seq.
Fig. S10. Effect of anchorene on plant growth.
Fig. S11. The synthesis route and derivatization for anchorene.
Table S1. The nutrient element composition in Argo soil and Silver sand.
Table S2. Mutants and marker lines used in this study.
Dataset S1. Gene list (1.5-fold change) for different treatments in RNA-seq.
Dataset S2. BP enrichment for different treatments in RNA-seq.
REFERENCES AND NOTES
- 1.DellaPenna D., Pogson B. J., Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 57, 711–738 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Walter M. H., Strack D., Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 28, 663–692 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Nisar N., Li L., Lu S., Khin N. C., Pogson B. J., Carotenoid metabolism in plants. Mol. Plant 8, 68–82 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Moise A. R., Al-Babili S., Wurtzel E. T., Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 114, 164–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimot H., Uraqami C., Cogdell R. J., Carotenoids and photosynthesis. Subcell. Biochem. 79, 111–139 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Moise A. R., von Lintig J., Palczewski K., Related enzymes solve evolutionarily recurrent problems in the metabolism of carotenoids. Trends Plant Sci. 10, 178–186 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Schwartz S. H., Tan B. C., Gage D. A., Zeevaart J. A., McCarty D. R., Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S., The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Wang J. Y., Haider I., Jamil M., Fiorilli V., Saito Y., Mi J., Baz L., Kountche B. A., Jia K.-P., Guo X., Balakrishna A., Ntui V. O., Reinke B., Volpe V., Gojobori T., Blilou I., Lanfranco L., Bonfante P., Al-Babili S., The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 10, 810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giuliano G., Al-Babili S., von Lintig J., Carotenoid oxygenases: Cleave it or leave it. Trends Plant Sci. 8, 145–149 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Nambara E., Marion-Poll A., Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Ilg A., Yu Q., Schaub P., Beyer P., Al-Babili S., Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in Golden Rice endosperm suggests apocarotenoids as substrates in planta. Planta 232, 691–699 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Bruno M., Koschmieder J., Wuest F., Schaub P., Fehling-Kaschek M., Timmer J., Beyer P., Al-Babili S., Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J. Exp. Bot. 67, 5993–6005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alder A., Holdermann I., Beyer P., Al-Babili S., Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochem. J. 416, 289–296 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Schaub P., Wüst F., Koschmieder J., Yu Q., Virk P., Tohme J., Beyer P., Nonenzymatic β-carotene degradation in provitamin A-biofortified crop plants. J. Agric. Food Chem. 65, 6588–6598 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M., Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickinson A. J., Lehner K., Mi J., Jia K.-P., Mijar M., Dinneny J., Al-Babili S., Benfey P. N., β-cyclocitral is a conserved root growth regulator. Proc. Natl. Acad. Sci. U.S.A. 116, 10563–10567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilg A., Bruno M., Beyer P., Al-Babili S., Tomato carotenoid cleavage dioxygenases 1A and 1B: Relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoids and isoprenoid volatiles. FEBS Open Bio. 4, 584–593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherzinger D., Ruch S., Kloer D. P., Wilde A., Al-Babili S., Retinal is formed from apo-carotenoids in Nostoc sp. PCC7120: In vitro characterization of an apo-carotenoid oxygenase. Biochem. J. 398, 361–369 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellini C., Pacurar D. I., Perrone I., Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 65, 639–666 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Petricka J. J., Winter C. M., Benfey P. N., Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucas M., Swarup R., Paponov I. A., Swarup K., Casimiro I., Lake D., Peret B., Zappala S., Mairhofer S., Whitworth M., Wang J., Ljung K., Marchant A., Sandberg G., Holdsworth M. J., Palme K., Pridmore T., Mooney S., Bennett M. J., Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155, 384–398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheres B., Wolkenfelt H., Willemsen V., Terlouw M., Lawson E., Dean C., Weisbeek P., Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120, 2475–2487 (1994). [Google Scholar]
- 24.Moreno-Risueno M. A., Van Norman J. M., Moreno A., Zhang J., Ahnert S. E., Benfey P. N., Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Norman J. M., Zhang J., Cazzonelli C. I., Pogson B. J., Harrison P. J., Bugg T. D., Chan K. X., Thompson A. J., Benfey P. N., Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc. Natl. Acad. Sci. U.S.A. 111, E1300–E1309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilches-Barro A., Maizel A., Talking through walls: Mechanisms of lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 23, 31–38 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Kazan K., Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., De Smet I., Fukaki H., Beeckman T., Bennett M., Laplaze L., Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 18, 450–458 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Hardtke C. S., Berleth T., The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okushima Y., Overvoorde P. J., Arima K., Alonso J. M., Chan A., Chang C., Ecker J. R., Hughes B., Lui A., Nguyen D., Onodera C., Quach H., Smith A., Yu G., Theologis A., Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris J. M., Abscisic acid: Hidden architect of root system structure. Plants 4, 548–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandya-Kumar N., Shema R., Kumar M., Mayzlish-Gati E., Levy D., Zemach H., Belausov E., Wininger S., Abu-Abied M., Kapulnik Y., Koltai H., Strigolactone analog GR24 triggers changes in PIN2 polarity, vesicle trafficking and actin filament architecture. New Phytol. 202, 1184–1196 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Malamy J. E., Benfey P. N., Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., Long J. A., Meyerowitz E. M., Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Guevara-García A., San Román C., Arroyo A., Cortés M. E., de la Luz Gutiérrez-Nava M., León P., Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 17, 628–643 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyer P., Kröncke U., Nievelstein V., On the mechanism of the lycopene isomerase/cyclase reaction in Narcissus pseudonarcissus L. chromoplasts. J. Biol. Chem. 266, 17072–17078 (1991). [PubMed] [Google Scholar]
- 37.Tian L., DellaPenna D., Characterization of a second carotenoid β-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol. Biol. 47, 379–388 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Pogson B. J., Niyogi K. K., Björkman O., DellaPenna D., Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. U.S.A. 95, 13324–13329 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorefan K., Booker J., Kaurogné H., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O., MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17, 1469–1474 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Booker J., Sieberer T., Wright W., Williamson L., Willett B., Stirnberg P., Turnbull C., Srinivasan M., Goddard P., Leyser O., MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8, 443–449 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Xiong L., Lee H., Ishitani M., Zhu J.-K., Regulation of osmotic stress-responsive gene expression by theLOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277, 8588–8596 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Xiong L., Ishitani M., Lee H., Zhu J.-K., The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13, 2063–2083 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T., Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Péret B., De Rybel B., Casimiro I., Benková E., Swarup R., Laplaze L., Beeckman T., Bennett M. J., Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 14, 399–408 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Swarup K., Benková E., Swarup R., Casimiro I., Péret B., Yang Y., Parry G., Nielsen E., De Smet I., Vanneste S., Levesque M. P., Carrier D., James N., Calvo V., Ljung K., Kramer E., Roberts R., Graham N., Marillonnet S., Patel K., Jones J. D., Taylor C. G., Schachtman D. P., May S., Sandberg G., Benfey P., Friml J., Kerr I., Beeckman T., Laplaze L., Bennett M. J., The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y., Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frusciante S., Diretto G., Bruno M., Ferrante P., Pietrella M., Prado-Cabrero A., Rubio-Moraga A., Beyer P., Gomez-Gomez L., Al-Babili S., Giuliano G., Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, 12246–12251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medina H. R., Cerdá-Olmedo E., Al-Babili S., Cleavage oxygenases for the biosynthesis of trisporoids and other apocarotenoids in Phycomyces. Mol. Microbiol. 82, 199–208 (2011). [DOI] [PubMed] [Google Scholar]
- 49.B. Schaefer, Natural Products in the Chemical Industry (Springer, 2015). [Google Scholar]
- 50.Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J., Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Kleine-Vehn J., Ding Z., Jones A. R., Tasaka M., Morita M. T., Friml J., Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. U.S.A. 107, 22344–22349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurihara D., Mizuta Y., Sato Y., Higashiyama T., ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musielak T. J., Slane D., Liebig C., Bayer M., A versatile optical clearing protocol for deep tissue imaging of fluorescent proteins in Arabidopsis thaliana. PLOS ONE 11, e0161107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tie C., Hu T., Jia Z.-X., Zhang J.-L., Derivatization strategy for the comprehensive characterization of endogenous fatty aldehydes using HPLC-multiple reaction monitoring. Anal. Chem. 88, 7762–7768 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Andrews, A Fastqc, a Quality Control Tool for High Throughput Sequence Data, (2010); www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 57.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 58.H. J. Pimentel, N. Bray, S. Puente, P. Melsted, L. Pachter, Differential Analysis Of Rna-Seq Incorporating Quantification Uncertainty, (2016); bioRxiv 058164 [Preprint]. [DOI] [PubMed]
- 59.Yu G., Wang L.-G., Han Y., He Q.-Y., ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoagland D. R., Arnon D. I., The water-culture method for growing plants without soil. Circular Calif. Agr. Exp. Stat. 347, 1–32 (1950). [Google Scholar]
- 61.Jansen F., Kwestro M., Schmitt D., Lugtenburg J., Synthesis and characterization of all-E (12, 12′-13C2)-,(13, 13′-13C2)-,(14, 14′-13C2)-,(15, 15′-13C2)-and (20, 20′-13C2) astaxanthin. Recl. Trav. Chim. Pays Bas 113, 552–562 (1994). [Google Scholar]
- 62.van Wijk A. A., Lugtenburg J., Synthetic scheme for the preparation of 13C-labeled 2, 7-dimethylocta2, 4, 6-triene-1, 8-dial, the central part of carotenoids. European J. Org. Chem. 2002, 4217–4221 (2002). [Google Scholar]
- 63.Frederico D., Donate P. M., Constantino M. G., Bronze E. S., Sairre M. I., A short and efficient synthesis of crocetin-dimethylester and crocetindial. J. Org. Chem. 68, 9126–9128 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Azim E.-m., Auzeloux P., Maurizis J.-C., Braesco V., Grolier P., Veyre A., Madelmont J.-C., Synthesis of all-trans-beta-carotene retinoids and derivatives labelled with 14C. J. Label. Compd. Radiopharm. 38, 441–451 (1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/11/eaaw6787/DC1
Supplementary Text
Fig. S1. Structures of diapocarotenoids and supposed precursors for anchorene.
Fig. S2. Effects of diapocarotenoid and its derivatives on Arabidopsis root development.
Fig. S3. Characterization of ANR development by different DR5 marker lines.
Fig. S4. Root formation under different treatments and in various mutants.
Fig. S5. Anchorene effects on DR5 expression in ANR primordia.
Fig. S6. Anchorene isomer identification and anchorene quantification.
Fig. S7. Conversion of OH-Apo12′ into anchorene in plants.
Fig. S8. Involvement of auxin signaling and distribution on ANR development.
Fig. S9. Transcriptomic change analysis of collet tissues upon different treatments by RNA-seq.
Fig. S10. Effect of anchorene on plant growth.
Fig. S11. The synthesis route and derivatization for anchorene.
Table S1. The nutrient element composition in Argo soil and Silver sand.
Table S2. Mutants and marker lines used in this study.
Dataset S1. Gene list (1.5-fold change) for different treatments in RNA-seq.
Dataset S2. BP enrichment for different treatments in RNA-seq.