Abstract
Different tissue types are characterized by varying stiffness and biochemical ligands. Increasing substrate stiffness has been shown to trigger Yes-associated protein (YAP) translocation from cytoplasm to the nucleus, yet the role of ligand density in modulating mechanotransduction and stem cell fate remains largely unexplored. Using polyacrylamide hydrogels coated with fibronectin as a model platform, we showed stiffness-induced YAP translocation occurs only at intermediate ligand densities. At low or high ligand densities, YAP localization is dominated by ligand density independent of substrate stiffness. We further showed ligand density-induced YAP translocation requires cytoskeleton tension and αVβ3-integrin binding. Finally, we demonstrate that increasing ligand density alone can enhance osteogenic differentiation regardless of matrix stiffness. Together, the findings from the present study establish ligand density as an important parameter for modulating stem cell mechanotransduction and differentiation, which is mediated by integrin clustering, focal adhesion and cytoskeletal tension.
Keywords: ligand density, YAP, mechanotransduction, stem cells, hydrogel
Graphical Abstract
Introduction
Stem cell fate is modulated by physical stimuli from the surrounding extracellular matrix (ECM) through a process of mechanotransduction [1], through which stimuli are transduced into biochemical signals. Transcriptional regulator Yorkie-homologue YAP (Yes-associated protein) has been identified as a mechanical rheostat of the cell, where YAP localization and activity correlates with ECM stiffness [2]. High stiffness triggers YAP translocation to the nucleus and activation and further downstream signaling [2]. Stiffness-induced YAP nuclear translocation is accompanied by changes in cytoskeletal F-actin and focal adhesions [3, 4]. Inhibiting cytoskeletal tension abolishes stiffness-induced YAP translocation [2]. Furthermore, YAP regulates how cells sense matrix stiffness through controlling transcription of focal adhesion molecules [5]. These adhesions are comprised of a number of proteins that are recruited to integrin transmembrane proteins: the site of contact with the ECM and the primary mechanosensors of the cell. Integrin receptors of the cells adhere to the ECM through biochemical ligands within the ECM proteins.
As cell adhesion serves as the interface between cells and their niche, it is essential to incorporate biochemical ligands to allow cells to sense matrix stiffness. Previous mechanotransduction studies have employed hydrogel substrates of tunable stiffness to mimic ECM, and substrates were functionalized with various ECM proteins such as type I collagen [6–9], fibronectin [2, 5, 10–15], and laminin [11, 16, 17]. Varying ligand density has been shown to alter cell motility [15, 18] and spreading [9, 14, 15, 18–20] as a function of hydrogel stiffness. However, the trends reported in previous studies were inconsistent. While some report increasing ligand density resulted in increased cell area [14, 15, 19, 20], others have observed a biphasic relationship [9, 18]. One factor that contributes to the inconsistent findings from these studies is that they employ different conjugation protocols without directly characterizing the actual amount of incorporated proteins. As such, the protein density vary across different studies, which makes it difficult to directly compare the trend. Furthermore, previous ligand studies often focused on fully differentiated cell types such as various epithelial cell lines [2, 8, 11–13]. How ligand density modulate stem cell mechanotransduction and fate remains largely unclear.
Using a polyacrylamide hydrogel platform with enhanced conjugation efficiency of biochemical ligands, here we seek to elucidate how varying ligand density modulates mechanotransduction and differentiation of mesenchymal stem cells. We chose fibronectin as the model biochemical ligand given its wide use as biochemical ligand for previous mechanotransduction studies [2, 5, 10–15] and robust efficacy in supporting cell adhesion [21]. To assess whether ligand density may impact stem cell mechanotransduction, human mesenchymal stem cells (hMSCs) were cultured on soft (3 kPa) or stiff (38 kPa) polyacrylamide hydrogels conjugated with tunable fibronectin density. To probe the molecular machineries involved in ligand density-induced mechanotransduction in stem cells, MSCs were characterized by imaging the localization of YAP, F-actin, and focal adhesion proteins. Here we report that YAP translocation and stem cell differentiation not only depend on substrate stiffness, but can also be directly modulated by ligand density. We further demonstrate that ligand density-induced YAP translocation is mediated through integrin binding, focal adhesion formation, and cytoskeletal tension.
Results
Biochemical ligand density alters YAP translocation and F-actin formation
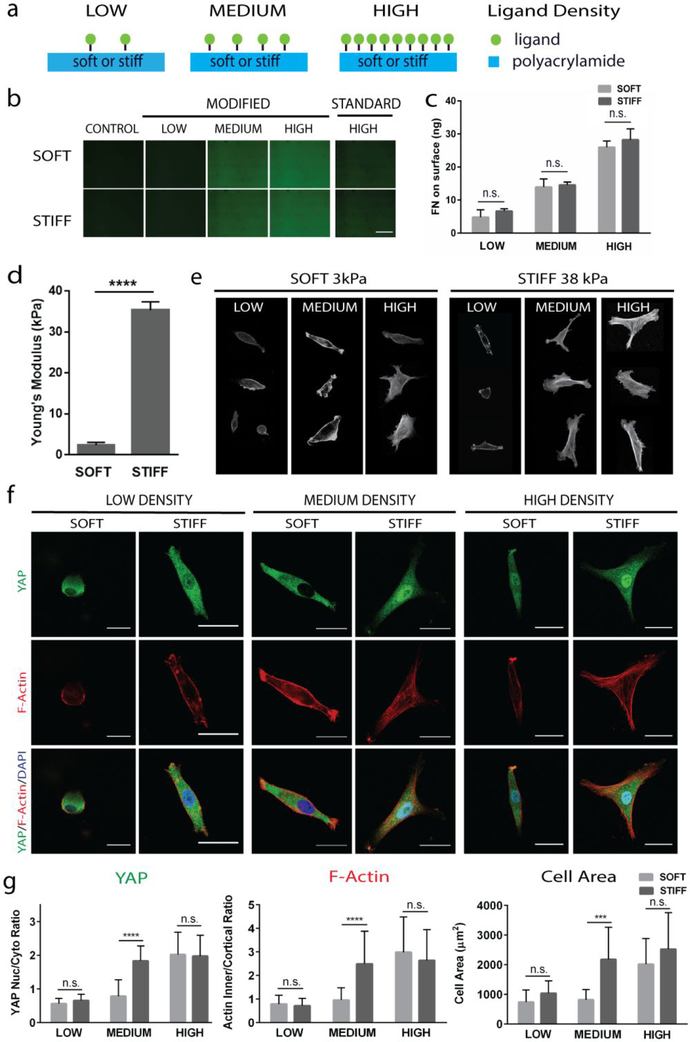
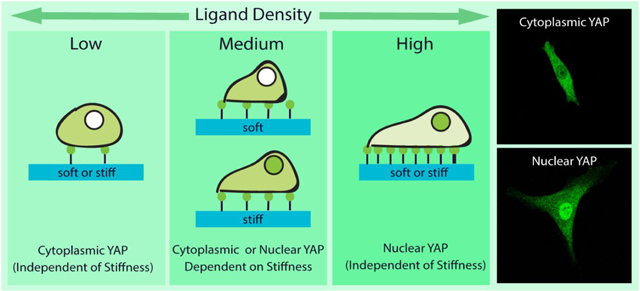
Previous studies have reported that biochemical ligand density alters cell morphology, but how it alters YAP mechanotransduction and the underlying mechanisms remain unclear. To assess the effect of varying ligand density on mechanotransduction of hMSCs, we chose to measure YAP translocation and F-actin formation. To enhance the conjugation efficiency of biochemical ligands to the polyacrylamide hydrogel substrate, we modified the conventional method of fabricating polyacrylamide hydrogels. Specifically, we introduced primary amine groups onto polyacrylamide hydrogel surface, which has higher binding efficiency with the nitrophenyl azide group in sulfo-SANPAH than the amide group in unmodified polyacrylamide. We verified the incorporation of ligand by imaging gels conjugated with fluorescently-labelled fibronectin (Fig. 1b). The amount of actually incorporated fibronectin was quantified using ELISA (Fig. 1c), which confirmed the increase in ligand density. At any given ligand density, no significant differences in ligand incorporation was observed between soft and stiff hydrogels (Fig. 1c). Hydrogels were fabricated with young’s modulus of 3 kPa (designated as soft) or 38 kPa (designated as stiff) (Fig. 1d) to represent a range of stiffnesses relevant for lineage-specific differentiation. Previous mechanotransduction studies of hMSCs have shown that soft substrates (1–5 kPa) promote the adipogenic differentiation [22] whereas stiff substrates (25–40 kPa) promote osteogenesis [23]. We cultured bone marrow-derived human mesenchymal stem cells (hMSCs) on these soft and stiff polyacrylamide hydrogel surfaces coated with low, intermediate, and high fibronectin densities (Fig. 1a, e). Regardless of substrate stiffness, increasing ligand density led to increase in cell spreading and F-actin cable formation (Fig. 1e, f). Interestingly, stiffness-induced YAP nuclear translocation was only observed at medium ligand density. At low or high ligand density, YAP localization is dominated by ligand density regardless of substrate stiffness, also shown using the fibronectin binding motif, RGD peptide (Fig. S1). Specifically, low ligand density invariably led to cytoplasmic YAP, whereas high ligand density always resulted in YAP translocation to nucleus (Fig. 1f, g; Fig. S2). A similar trend was observed in F-actin cable formation and cell spread area in response to varying ligand density and substrate stiffness (Fig. 1f, g). We devised a method to assess the amount of F-actin cables by calculating the ratio between the outer, cortical region of the cells, which we define by a 2μm outer perimeter, and the region inside of this cortical region where F-actin cables form (Fig. S3).
Figure 1: Stiffness-induced YAP translocation occurs only at intermediate ligand densities, not at low or high densities.
(a) Schematic illustrating experimental set up, (b) images of fluorescently-labeled fibronectin surfaces across ligand densities for our modified protocol and the standard protocol with high ligand density for comparison (Scale bar, 500 μm), (c) quantification of fibronectin incorporated on hydrogel surfaces with ELISA assay, (d) quantification of stiffness on polyacrylamide gel substrates, (e) cell morphology across a range of ligand densities at each stiffness, as shown by F-actin staining, (f) YAP localization (green) and F-Actin arrangement (red) on soft and stiff gels coated with low and high densities of fibronectin (Scale bars, 30 μm), and (g) quantification of YAP and F-Actin localization and cell areas. ****p<0.0001.
Ligand density-induced YAP nuclear translocation requires cytoskeletal tension
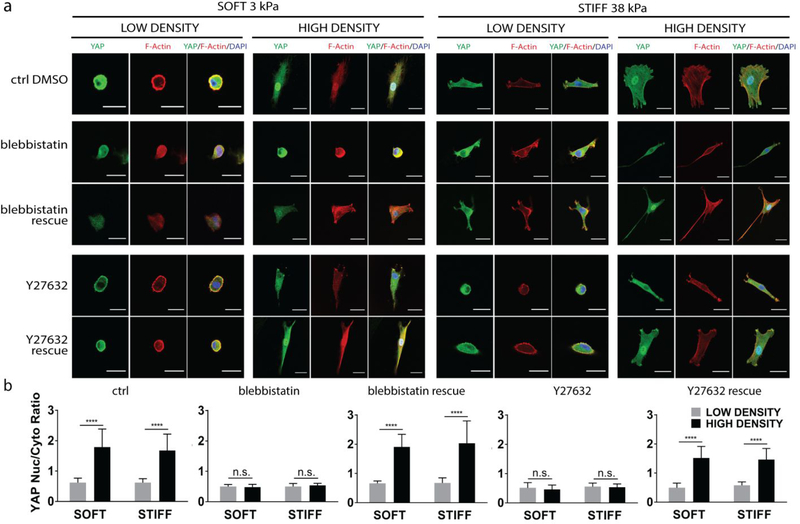
Given the F-actin formation correlated well YAP translocation as a function of ligand density (Fig. 1e–g), we next tested the effect of blocking cytoskeletal tension in ligand density-induced YAP translocation. For stiffness-induced YAP nuclear translocation, YAP activity was shown to require actin cytoskeletal tension, and inhibiting cytoskeleton tension abrogates stiffness-induced YAP activity [2]. Whether ligand density-induced YAP translocation relies on the same mechanism remains unknown. To block cytoskeleton tension, hMSCs were treated with blebbistatin, a myosin II ATPase inhibitor. Blebbistatin treatment abolished the ligand density-induced differences in YAP nuclear translocation and F-actin cable formation (Fig. 2a, b). Regardless of the ligand density or substrate stiffness, YAP was sequestered in the cytoplasm when hMSCs were treated with blebbistatin (Fig. 2a, b). We further demonstrate this process is reversible. Removal of blebbistatin restored ligand density-induced changes in F-actin and YAP translocation. To corroborate this observation, we also treated hMSCs with Y-27632, an inhibitor for Rho-associated kinase (ROCK). The same trend was observed (Fig. 2a, b). Together, these results provide clear evidence that cytoskeletal tension is required for ligand-induced YAP translocation.
Figure 2: Ligand density-induced YAP nuclear translocation requires cytoskeletal tension.
(a) YAP (green) and F-Actin (red) localization after the disruption and subsequent rescue of cytoskeletal tension through addition and removal of blebbistatin and with ROCK-inhibitor Y27632. hMSCs were cultred on soft and stiff substrates with low and high ligand densities. Scale bar: 30 μm. (b) Quantification of YAP nuclear localization, ****p<0.0001.
Increasing ligand density leads to increasing focal adhesion formation regardless of substrate stiffness
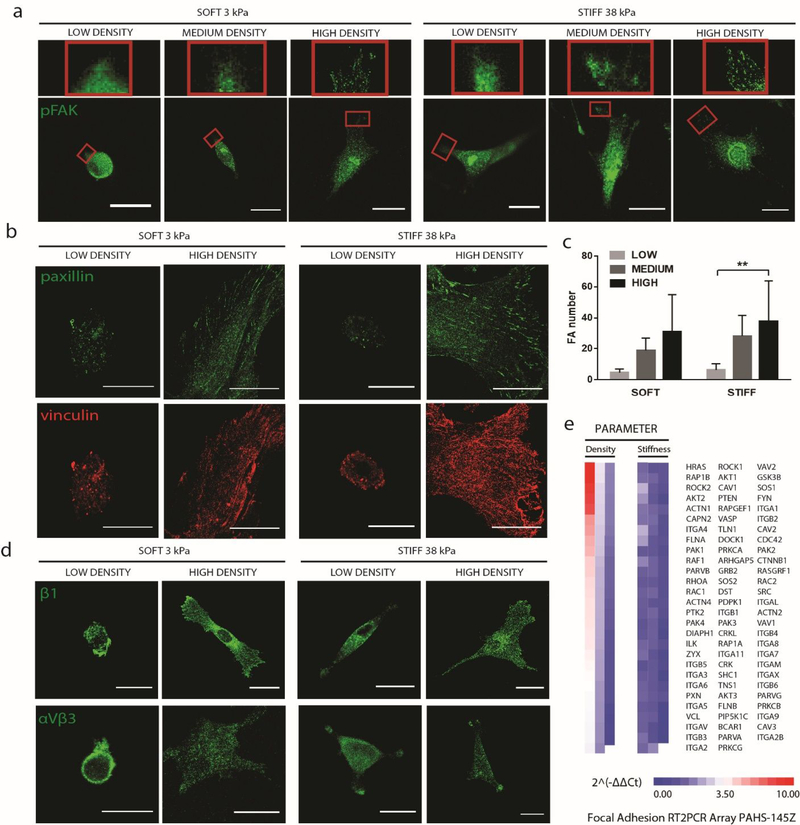
Cell adhesions are required for stiffness sensing [12, 22] and integrin-growth factor crosstalk has been shown to modulate stem cell fate [24]. Increasing focal adhesions generates more intracellular tension, and has been shown to promote osteogenesis [24–27]. To understand how altered ligand density translates into changes in cytoskeletal tension and YAP localization, we next examined the localization and expression of key adhesion proteins in response to changes in ligand density, using confocal microscopy to image adhesions at the bottom surface of the cells (Fig. S4a). Regardless of substrate stiffness, low ligand density led to minimal focal adhesion formation, as shown by the diffuse localization of phospho-focal adhesion kinase (pFAK), paxillin and vinculin. In contrast, high ligand density led to more punctae formation, indicating enhanced focal adhesion formation (Fig. 3a, b). This trend was verified through quantification of the number of punctae throughout the cells using an automated image analysis (Fig. 3c; Fig. S4b). We next examined the localization of β1 integrin and αVβ3 integrins, which are the transmembrane proteins in direct contact with the ligands. A similar trend was observed, with increased punctae formation in response to increasing ligand density regardless of substrate stiffness (Fig. 3d). Recent evidence also suggests an interplay between YAP translocation and focal adhesion gene expressions [5]. We assayed for changes in focal adhesion genes using a PCR array of 84 genes. Varying ligand density resulted in much more dramatic changes in gene expression patterns of focal adhesion genes than varying substrate stiffness, highlighting ligand density has a more dominant role in focal adhesion formation (Fig. 3e). A few examples of genes that upregulates substantially in response to increasing ligand density include GTPase HRAS, an important enzyme in the MAPK/ERK pathway; RAP1B, a GTP-binding protein that regulates integrin-mediated signaling; and ROCK2, a regulator of actin stress fiber formation and focal adhesion formation (Fig. 3e).
Figure 3: Increasing biochemical ligand density leads to increased focal adhesion punctae formation regardless of matrix stiffnesses.
hMSCs were cultured on soft and stiff substrates with low or high ligand densities. (a) Immunostaining of pFAK, a focal adhesion marker;(b) immunostaining of paxillin (green) and vinculin (red); (c) quantification of focal adhesion punctae; and (d) immunostainings of β1- and αVβ3- integrins; (e) PCR array analyses of genes related to focal adhesion expressed by hMSCs cultured on stiff substrates at low and high fibronectin densities. Scale bars, 30 μm.
Ligand density-induced YAP translocation requires αVβ3-integrin adhesion
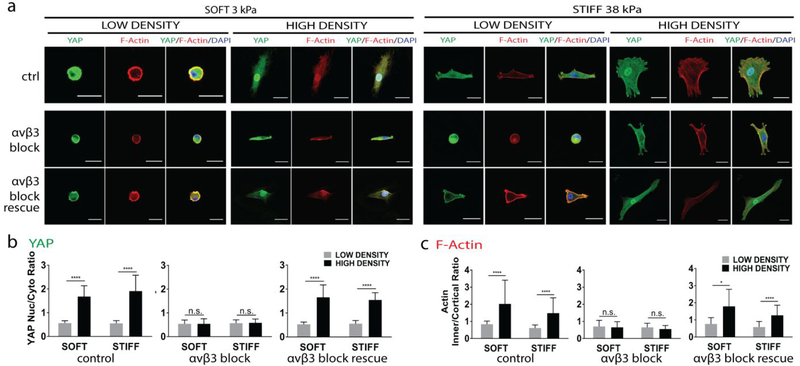
As ligand density-induced YAP translocation is accompanied by integrin clustering (Fig. 3d), we next sought to determine which integrin subunits are required. A recent study suggested that α5β1-integrin governs cell adhesion, while αVβ3-integrin enables mechanosensing [28]. To test whether αVβ3-integrin adhesion is required for ligand density-induced YAP translocation, antibodies were added to block αVβ3, and were subsequently removed to test whether the process can be rescued. Blocking αVβ3-integrin abolished YAP nuclear translocation across all groups regardless of ligand densities or substrate stiffness. Removing the blocking antibody restored the ligand density-induced YAP translocation, suggesting this process is reversible (Fig. 4a, b). Morphology of F-actin exhibits similar trend– blocking αVβ3-integrin led to cortical actin, and removal antibody restored F-actin cable formation. (Fig. 4a, c). These results indicate that ligand density-induced YAP translocation requires αVβ3-integrin adhesion.
Figure 4. Ligand density-induced YAP nuclear translocation and F-actin formation requires αVβ3-integrin binding.
(a) Immunostainings of YAP (green) and F-Actin (red) localization on hydrogels with varying ligand density (low vs. high) and varying stiffnesses (3 kPa and 38 kPa). αVβ3-integrin binding was blocked and rescued. Scale bars: 30 μm. Quantification of ligand induced YAP (b) and F-Actin localization (c) in response to αVβ3-integrin blocking and rescue, ****p<0.0001.
Increasing ligand-density enhances osteogenic differentiation of hMSCs regardless substrate stiffness
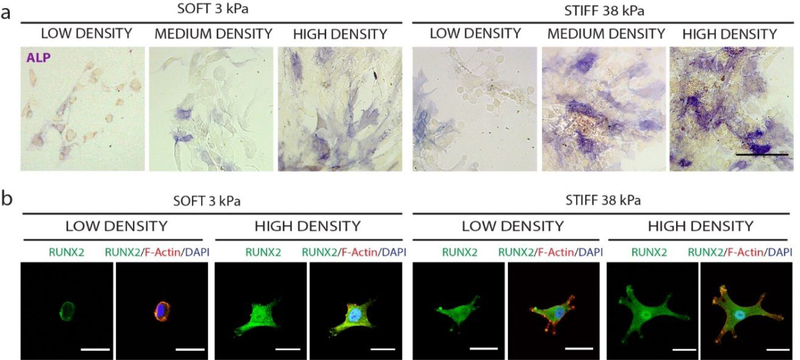
YAP nuclear localization has been shown to enhance osteogenic differentiation [2]. We then examined the effects of varying ligand density on osteogenic differentiation of hMSCs when cultured on soft or stiff substrates. Osteogenic differentiation was assessed by staining of early osteogenic markers alkaline phosphatase (ALP) and runt-related transcription factor 2 (RUNX2). Regardless of substrate stiffness, increasing ligand density more cells staining positive for ALP (Fig. 5a) and enhanced RUNX2 expression (Fig. 5b). In addition to staining, we also performed real time qRT-PCR to quantify the expressions of two early bone markers, ALP and RUNX2 (Fig. S5). Gene expression data showed a similar trend, with increasing ligand density leading to an increase in the expression level of osteogenesis markers. The difference, however, was not statistically significant. This may be due to the relative early time point chosen (day 3).
Figure 5: Increasing biochemical ligand density enhances osteogenic differentiation of hMSCs regardless of matrix stiffness.
Increasing ligand density enhances expression of early osteogenic markers (a) ALP (Scale bar, 100 μm) and (b) and RUNX2 (green), indicated by nuclear localization (Scale bar, 30 μm).
Discussion
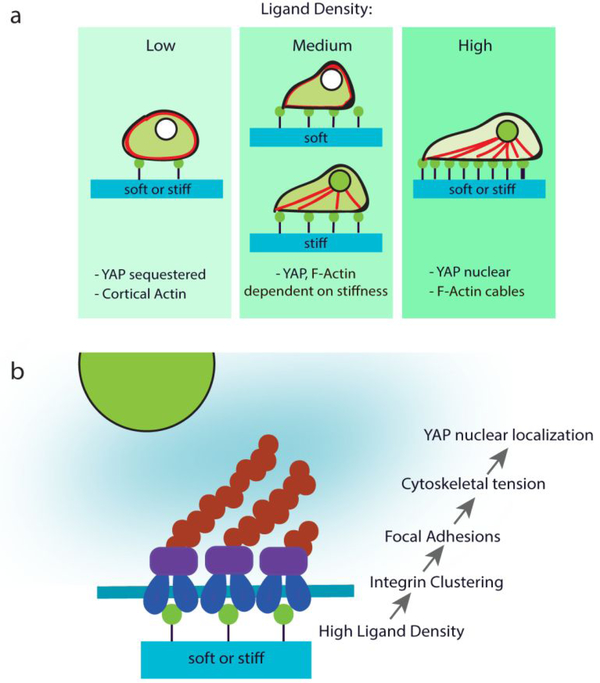
Previous mechanotransduction studies have largely focused on the effects of varying substrate stiffness on cell fates [2, 22, 23], but the role of ligand density on mechanotransduction remain largely unexplored. Extracellular matrix contains various biochemical ligands such as fibronectin, type I collagen, and laminin [29]. Different tissue types contain different types and density of ligands, and differ substantially in tissue stiffness. Using fibronectin as a model biochemical ligand, here we demonstrate that varying ligand density alone can directly impact how hMSCs feel the mechanical cues from the matrix, and modulate differentiation of hMSCs without altering substrate stiffness. Our work highlights the importance of taking ligand density into consideration for mechanotransduction studies. Importantly, our results showed stiffness-induced YAP translocation is only valid at intermediate ligand density for fibronectin coated polyacrylamide hydrogels (Fig. 1). In contrast, at low or high fibronectin density, YAP translocation is dominated by ligand density regardless of substrate stiffness (Fig. 1). We further demonstrate that ligand density-induced YAP translocation requires cytoskeletal tension (Fig. 2) and αVβ3-integrin adhesion (Fig. 4), which is a shared mechanism through which substrate stiffness induce YAP translocation (Fig. 6).
Figure 6: Schematic summary.
(a) Ligand density alters actin and YAP localization and (b) ligand-induced YAP nuclear localization is mediated through integrin clustering, focal adhesions, and cytoskeletal tension.
Previous studies have reported seemingly contradictory findings about the relationship between ligand density and cell spreading and stiffness [9, 14, 15, 18–20]. While some report increasing ligand density increased cell spreading [14, 19], others report a biphasic relationship with cell area increasing with increasing ligand density until an optimum and then decreasing [9, 15, 18]. These studies use different ligand conjugation protocols and characterize the ligand coatings with different methods. Most of these studies only reported the concentration of ligand solution used for coating the hydrogel surface, but did not direct quantify the actual amount of protein successfully conjugated. As such, it is very difficult to directly compare the results from the different studies. Given the varying conjugation efficiency from different studies, the inconsistent trend reported from different studies are likely due to variation of ligand density conjugated on the hydrogel substrate, as suggested by the results from our study. To facilitate comparing research findings from various research groups, it is important for future mechanotransduction studies to directly quantify the amount of the incorporated ligand density while interpreting their results.
A recent study has shown changes in protein tethering may also impact mechanosensing of stem cells [6]. In our study, we do not expect protein tethering to be a contributing factor for the observed ligand density-induced YAP nuclear translocation. To validate this, we assessed the effect of increasing ligand density using a short peptide, RGD, which would be less affected by potential changes in protein tethering. Our results indeed showed increasing RGD dosage from 0.1 mM to 2.5 mM led to translocation of YAP from cytoplasm to nucleus without altering hydrogel stiffness (Fig. S1). These results remove protein tethering as a confounding factor that may contribute to the ligand density-induced YAP nuclear translocation. Recent studies have also explored potential mechanisms underlying stiffness-induced YAP nuclear translocation. Perinuclear stress fibers were found to be required to compress the nucleus, opening nuclear pores and driving YAP nuclear translocation [30, 31]. It is likely that ligand density-induced YAP nuclear translocation require perinuclear stress as well, and would be of interest for future mechanistic investigations.
Increasing substrate stiffness [23] has been shown to enhance hMSC spreading and actomyosin contractility, which in turn promoted osteogenesis. One important finding from the present study is that varying ligand density on 2D substrate can directly modulate stem cell differentiation without changing substrate stiffness (Fig. 5). This finding could potentially be harnessed for enhancing osteogenesis through priming cells on high fibronectin ligand density substrates, and may be used to guide rational scaffold design to enhance stem cell-based bone formation. We chose to focus on early bone markers only in response to varying ligand density. This is because long term culture would be necessary to assess mature bone markers. Since these cells can produce new extracellular matrix within a few days, the initial ligand density would be quickly masked by the newly deposited ECM cues. As such, we intentionally chose to focus on short term studies over a few days to avoid confounding factors from newly deposited ECM cues. In the present study, we have chosen fibronectin as the biochemical ligand given its abundance in ECM, important role in tissue pathology and wound healing, [21] and wide use as cell adhesive ligand in previous mechanotransduction studies. In addition to fibronectin, the ECM also contain other important proteins including collagen I, collagen IV, laminin, elastin [29]. The ratio and compositions of these ECM proteins vary across different tissue types and during morphogenesis. In addition, different ECM proteins engages different integrin subunit types [32]. While the present study focuses on the fibronectin as a model ligand, the principles that arise from this study may be broadly applicable to other types of ECM proteins. Future studies can further examine the effect of varying ECM ligand types and ratios on mechanotransduction and stem cell differentiation.
In summary, the present study validate ligand density as a niche cue that directly modulates mechanotransduction and differentiation of adult stem cells. Ligand density-induced YAP translocation requires cytoskeleton tension and αVβ3-integrin adhesion, which are also shared by stiffness-induced mechanotransduction. Increasing ligand density alone can enhance osteogenesis of hMSCs even when cultured on soft substrates. Our results highlight that mechanotransduction not only depends on substrate stiffness, but also on ligand density. Findings from the present study not only fill in a gap of knowledge in mechanobiology, but also shed lights into harnessing ligand density as a parameter to enhance desirable stem cell fates and tissue regeneration.
Materials and Methods
Hydrogel Fabrication
Polyacrylamide hydrogels were fabricated by adapting a previously reported protocol [33, 34] to incorporate primary amine end groups, for the purpose of enhancing protein conjugation efficiency. In brief, 2-aminoethyl methacrylate (Aldrich 516155, 15 mM in deionized water) was added to hydrogel precursor solution containing acrylamide (Sigma A4058,40% (v/v)) and N,N′-methylenebisacrylamide (Sigma M1533, 2% (v/v)). Soft or stiff hydrogels were fabricated by maintaining acrylamide concentration constant (8% (v/v)) while varying the concentration of bis-acrylamide (0.08 or 0.48% (v/v)). To initiate photocrosslinking, photoinitiator 2-Hydroxy-1-[4-(2-hydroxyethoxy) phenyl]-2-methyl-1-propanone (Irgacure 2959, Ciba, 0.05% (w/v)) was used. Hydrogel precursor solution (65 μl) were loaded between two round glass coverslips (15 mm in diameter) and exposed to ultraviolet light (365 nm, 4 mW/cm2, 5 min) to form a hydrogel substrate with thickness of ~ 370 μm. The hydrogel surface was then modified with sulfo-SANPAH (Life Technologies 22589, 0.83 mg/ml in PBS) and exposed to light (365 nm, 4 mW/cm2, 5 min). To incorporate biochemical ligand, hydrogel substrates were washed with PBS and incubated overnight at 37 °C in fibronectin (BD Biosciences 354008) diluted in PBS with varying concentrations of 1 μg/mL (low), 7 μg/mL(intermediate), and 10 μg/mL (high). RGD-incorporated hydrogels were fabricated according to previously reported methods [7]. In brief, acrylated-PEG3400-Maleimide (Laysan Bio, Inc.) was reacted with CRGDS peptide (Bio Basic) and incorporated at 0.1 or 2.5 mM into the hydrogel precursor solution described above.
Mechanical testing
The stiffness of polyacrylamide hydrogels was measured using an Instron 5944 materials testing system (Instron Corporation, Norwood, MA). Hydrogels were immersed in PBS at room temperature and placed on custom-made aluminum compression plates lined with PTFE to minimize friction. Before each test, a preload of approximately 2 mN was applied. The upper plate was then lowered at a rate of 1% strain/sec to a maximum strain of 30%. Load and displacement data were recorded at 100 Hz. The Young’s modulus was calculated using linear curve fit of the stress vs strain curve with a strain range of 10−20%.
Characterization of protein conjugation
To visualize the incorporation of ligand on the hydrogel, fluorescently-labeled protein (Hylite 488, Cytoskeleton FNR02-A) was conjugated to the hydrogel at the various ligand densities. Hydrogel surfaces were visualized under low magnification fluorescent imaging.
To quantify protein incorporation, hydrogels with various ligand density and stiffness were punched out using a 6-mm biopsy punch and placed in wells of a 96-well plate. Fibronectin was assayed using a human fibronectin ELISA kit (Thermo Fisher Scientific BMS2028). Hydrogel substrates were incubated with 1:100 biotin-conjugated anti-human Fibronectin antibody, washed 6 times for 10 min in 12-well plate to ensure thorough washing, incubated with 1:200 streptavidin-HRP, washed another 6 times, incubated with a substrate solution where a colored product was formed proportionally to the amount of human fibronectin present on the hydrogel. The reaction was terminated through addition of acid. Absorbance of hydrogels was measured at 450 nm in the 96-well plate. A standard curve was determined according to the kit protocol to calculate the actual amount of fibronectin contained on hydrogel surfaces.
Cell culture
hMSCs (Lonza) were cultured in growth medium comprised of Dulbecco’s Modified Eagle Medium (Gibco), fetal bovine serum (10% v/v, Gibco), penicillin-streptomycin (1% v/v, ThermoFisher Scientific), and recombinant human fibroblast growth factor-basic (10 ng/mL, Peprotech). For all mechanotransduction characetetization studies, passage 6 hMSCs were plated at 2,500 cells/cm2 onto the hydrogels, and cultured 6 hours before analyzed by immunofluorescence staining. For osteogenic studies cells were seeded at 30,000 cells/ cm2 and cultured for in osteogenic media comprised of DMEM, FBS (10%), penicillin-/streptomycin (1% v/v, ThermoFisher Scientific), dexamethasone (100 nM), ascorbic-2-phosphate (50 μg/mL, Sigma), and beta glycerol phosphate (10 mM, Sigma). Runx2 expression was assessed after 3 days and ALP expression was assessed after 5 days.
Inhibitor experiments
For inhibitor studies, cells were seeded on substrates and allowed to adhere for 3 hours. Blebbistatin (50 μM, Abcam ab120425) or Y27632 (10 μM, Abcam ab120129) was diluted in fresh culture medium and applied to the culture for 2 hours prior to cell fixation. For rescue experiments, culture media with inhibitors were replaced with growth medium and allowed to recover for 2 hours prior to cell fixation. Integrin blocking experiments were conducted by incubating the cells in growth medium in the presence of αVβ3-integrin (25 μg/mL, Abcam ab78614) for 1 hour at 37C and then seeded onto the substrates. For integrin blocking rescue experiments, culture medium was replaced and cells were allowed to recover for 4 hours. The overall length of culture for all inhibitor studies was 5 hours.
Immunofluorescence imaging
Cells were fixed using 4% paraformaldehyde/PBS for 15 min at room temperature, washed three times with washing buffer (0.1% Tween-20/PBS, 5 min), and permeabilized with 1% Triton X-100/PBS for 30 min. Samples were incubated in blocking buffer (3% BSA, 2% goat serum in PBS) for 30 min, then incubated with various primary antibodies including mouse anti-YAP (Santa Cruz Biotechnology, sc-101199), mouse anti-αVβ3-integrin (Abcam ab78614), mouse anti-β1-integrin (Abcam ab24693), rabbit anti-paxillin (Abcam ab32084), rabbit anti-pFAK (Abcam ab81298), mouse anti-RUNX2 (Abcam ab76956) overnight at 4 °C on a shaker. All antibodies were diluted in buffer at 1:100 except YAP antibody was diluted at 1:300. After washing, samples were incubated with corresponding secondary antibodies including Alexa 488 Goat-anti-mouse (Invitrogen A11001), rhodamine goat-anti-rabbit (Millipore AP132), rhodamine-phalloidin (Sigma P1951) for 1 h at room temperature on a shaker. All secondary antibodies were diluted at 1:300. Cell nucleus counter stain was performed using Hoeschst nuclear stain (Cell Signaling Technology 4082S, 2 ug/mL). Samples were washed with washing buffer (three times, 5 min per wash) before being imaged using a confocal microscope (40x oil immersion, Leica SP8 confocal system). All images were processed using open-source Fiji software [35, 36].
Gene Expression
Cells were cultured for 48 hours in growth medium for focal adhesion gene expression analysis and for 3 days in osteogenic medium for osteogenic marker analysis. RNA was extracted using an RNeasy mini kit (Qiagen 74106). cDNA was synthesized using SuperScript III First-Strand Synthesis (Invitrogen 18080–051). For focal adhesions, RT-PCR was performed on cDNA using RT2 Profiler™ PCR Array of human focal adhesions (Qiagen PAHS-145ZA-2) in thermocyclers (Stratagene Mx3000p). A total of three experimental groups were examined including: (1) low ligand density/stiff hydrogels, (2) high ligand density/stiff hydrogels, and (3) high ligand density/soft hydrogels. Both groups 2 and3 display nuclear localization of YAP (Fig. 1f). The following cycling parameters were used: 1 cycle at 95 °C for 10 min; 45 cycles at 95 °C for 15 s and 60 °C for 1 min. The internal panel of housekeeping genes provided by the manufacture was used for normalization. To compare the relative impact of increasing the stiffness versus increasing ligand density, ddCt values were calculated for these two comparisons. Genes showing minimal expressions in all the experiments (with Ct values ranging between 35 and 40) were not included in the analyses. Relative fold of change in gene expressions were reported as 2^(-ddCt) values, and plotted in a heatmap using a statistical software (R-project) as previously reported [37]. For osteogenic markers, qRT-PCR was performed following the manufacturer’s protocol using Power SYBR Green PCR Master Mix (Life Technologies). All samples underwent 40 cycles on an Applied Biosystems 7900 Real-Time PCR System. The relative expression level of target genes was determined and plotted as 2^(-ddCt) values. Target gene expression was first normalized to an endogenous housekeeping gene (glyceraldehyde 3-phosphate dehydrogenase, GAPDH), followed by a second normalization to the expression level measured in day 0 control cells. The primers used are provided here in the 5’ to 3’ direction: GAPDH – ATGGGGAAGGTGAAGGTCG, GGGGTCATTGATGGCAACAATA; ALP – ACCACCACGAGAGTGAACCA, CGTTGTCTGAGTACCAGTCCC; RUNX2 – TGGTTACTGTCATGGCGGGTA, TCTCAGATCGTTGAACCTTGCTA.
Image analysis
To characterize YAP localization in a quantitative manner, we employed a method [13] which reports the ratio of nuclear YAP intensity vs. cytoplasm YAP intensity. In brief, a region of interest (ROI) in the nucleus and a region of interest of equal area in the cytoplasm immediately adjacent to the nucleus were selected. The nuclear region was defined using Hoechst staining. The fluorescence intensity of YAP staining within the nucleus ROI and the cytoplasm ROI were then quantified. Results are reported as the ratio of fluorescence intensity within nucleus vs. fluorescence intensity in cytoplasm (Fig. S2). To characterize the degree of F-actin polymerization in individual cells, images of phalloidin-stained cells were imported into Fiji software (Fig. S3a). The outer perimeter of the cell was selected as a region of interest (ROI) by thresholding the image and using the wand selection tool (Fig. S3b). The selection was then shrunk radially by 2 μm to define an inner perimeter (Fig. S3c). This allows us to calculate the integrated intensity within the inner region. The integrated intensity of the cortical region was calculated by subtracting the integrated intensity of the inner region from the integrated intensity from the total cell area. The final result was reported as the ratio between the integrated intensity within the inner region to the integrated intensity from the outer cortical region. The number of focal adhesions was quantified following a previously reported method [5]. The adhesion proteins were stained and imaged on a confocal microscope. The adhesions were imaged at a z-position in which the staining was in focus (Fig. S4a), as shown from our results (Fig. S4a). Single cross-section from this z-position was used for data analysis. In brief, confocal images acquired at the same magnification and exposure were imported into ImageJ software. Background was first subtracted using a sliding paraboloid and rolling ball. The contrast in images was then automatically adjusted (0.35 saturation) using the Clahe plug in. Images were binarized using the threshold command and the resultant particles were analyzed given dimensional constraints (size: 0.3–15; circularity: 0.00–0.99).
Statistical Analysis
Data are presented as mean ± standard deviations. For comparisons, data were analyzed with GraphPad Prism using one-way ANOVA by Tukey’s multiple comparisons test or two-way ANOVA by Bonferroni’s multiple comparisons test. Confidence intervals were kept at 95%, and P-values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
The authors acknowledge NIH R01DE024772 (F.Y.), NSF CAREER award CBET-1351289 (F.Y.), California Institute for Regenerative Medicine Tools and Technologies Award RT3-07804 (F.Y.), the Stanford Bio-X Interdisciplinary Initiative Program (F.Y.), the Stanford Child Health Research Institute Faculty Scholar Award (F.Y.), the Bio-X fellowship from the Stanford Bio-X program (A.S. and S.L.) and NIH T32GM008412 (A.S.) for support.
Abbreviations
- YAP
Yes-Associated Protein
- ECM
extracellular matrix
Footnotes
Supporting Information. Density effect of RGD-peptide on YAP translocation, methods of YAP and F-actin quantification, further confocal analysis of conditions, and RT-PCR analysis of osteogenic study.
References
- 1.Humphrey JD; Dufresne ER; Schwartz MA, Mechanotransduction and Extracellular Matrix Homeostasis. Nat. Rev. Mol. Cell Biol 2014, 15, p. 802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dupont S; Morsut L; Aragona M; Enzo E; Giulitti S; Cordenonsi M; Zanconato F; Le Digabel J; Forcato M; Bicciato S; Elvassore N; Piccolo S, Role of YAP/TAZ in Mechanotransduction. Nature. 2011, 474, 179–83. [DOI] [PubMed] [Google Scholar]
- 3.Aragona M; Panciera T; Manfrin A; Giulitti S; Michielin F; Elvassore N; Dupont S; Piccolo S, A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013, 154, 1047–1059. [DOI] [PubMed] [Google Scholar]
- 4.Das M; Ithychanda S; Qin J; Plow EF, Mechanisms of Talin-Dependent Integrin Signaling and Crosstalk. Biochim. Biophys. Acta 2013, 1838, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nardone G; Oliver-De La Cruz J; Vrbsky J; Martini C; Pribyl J; Skladal P; Pesl M; Caluori G; Pagliari S; Martino F; Maceckova Z; Hajduch M; Sanz-Garcia A; Pugno NM; Stokin GB; Forte G, YAP Regulates Cell Mechanics by Controlling Focal Adhesion Assembly. Nat. Commun 2017, 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trappmann B; Gautrot JE; Connelly JT; Strange DGT; Li Y; Oyen ML; Cohen Stuart MA; Boehm H; Li B; Vogel V; Spatz JP; Watt FM; Huck WTS, Extracellular-Matrix Tethering Regulates Stem-Cell Fate. Nat. Mater 2012, 11, 642–649. [DOI] [PubMed] [Google Scholar]
- 7.Wen JH; Vincent LG; Fuhrmann A; Choi YS; Hribar KC; Taylor-Weiner H; Chen S; Engler AJ, Interplay of Matrix Stiffness and Protein Tethering in Stem Cell Differentiation. Nat. Mater 2014, 13, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valon L; Marin-Llaurado A; Wyatt T; Charras G; Trepat X, Optogenetic Control of Cellular Forces and Mechanotransduction. Nat. Commun 2017, 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engler A; Bacakova L; Newman C; Hategan A; Griffin M; Discher D, Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophys. J 2004, 86, 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadden WJ; Young JL; Holle AW; McFetridge ML; Kim DY; Wijesinghe P; Taylor-Weiner H; Wen JH; Lee AR; Bieback K; Vo B; Sampson DD; Kennedy BF; Spatz JP; Engler AJ; Choi YS, Stem Cell Migration and Mechanotransduction on Linear Stiffness Gradient Hydrogels. PNAS. 2017, 114, 5647–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman CD; Isenberg BC; Chua SG; Wong JY, Extracellular Matrix Type Modulates Cell Migration on Mechanical Gradients. Exp. Cell Res 2017, 359, 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elosegui-Artola A; Bazellieres E; Allen MD; Andreu I; Oria R; Sunyer R; Gomm JJ; Marshall JF; Jones JL; Trepat X; Roca-Cusachs P, Rigidity Sensing and Adaptation through Regulation of Integrin Types. Nat. Mater 2014, 13, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elosegui-Artola A; Oria R; Chen Y; Kosmalska A; Perez-Gonzalez C; Castro N; Zhu C; Trepat X; Roca-Cusachs P, Mechanical Regulation of a Molecular Clutch Defines Force Transmission and Transduction in Response to Matrix Rigidity. Nat. Cell Biol 2016, 18, 540–548. [DOI] [PubMed] [Google Scholar]
- 14.Rape AD; Zibinsky M; Murthy N; Kumar S, A Synthetic Hydrogel for the High-Throughput Study of Cell-ECM Interactions. Nat. Commun 2015, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peyton SR; Putnam AJ, Extracellular Matrix Rigidity Governs Smooth Muscle Cell Motility in a Biphasic Fashion. J. Cell Physiol 2005, 204, 198–209. [DOI] [PubMed] [Google Scholar]
- 16.Lee J; Abdeen AA; Tang X; Saif TA; Kilian KA, Matrix Directed Adipogenesis and Neurogenesis of Mesenchymal Stem Cells Derived from Adipose Tissue and Bone Marrow. Acta Biomater. 2016, 42, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert PM; Havenstrite KL; Magnusson KEG; Sacco A; Leonardi NA;Kraft P; Nguyen NK; Thrun S; Lutolf MP; Blau HM, Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science. 2010, 329, 1078–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudet C; Marganski WA; Kim S; Brown CT; Gunderia V; Dembo M; Wong JY, Influence of Type I Collagen Surface Density on Fibroblast Spreading, Motility, and Contractility. Biophys. J 2003, 85, 3329–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel S; Tsang J; Harbers GM; Healy KE; Li. S, Regulation of Endothelial Cell Function by GRGDSP Peptide Grafted on Interpenetrating Polymers. J. Biomed. Mater. Res. A 2007, 83A, 423–433. [DOI] [PubMed] [Google Scholar]
- 20.Lee JP; Kassianidou E; MacDonald JI; Francis MB; Kumar S N-Terminal Specific Conjugation of Extracellular Matrix Proteins to 2-Pyridinecarboxaldehyde Functionalized Polyacrylamide Hydrogels. Biomaterials. 2016, 102, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zollinger AJ; Smith ML, Fibronectin, the Extracellular Glue. Matrix Biol. 2017, 60–61, 27–37. [DOI] [PubMed] [Google Scholar]
- 22.Huebsch N; Arany PR; Mao AS; Shvartsman D; Ali OA; Bencherif AA; Rivera-Feliciano J; Mooney DJ, Harnessing Traction-Mediated Manipulation of the Cell/Matrix Interface to Control Stem-Cell Fate. Nat. Mater 2010, 9, 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engler AJ; Sen S; Sweeney HL; Discher DE, Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- 24.Dalby MJ; Garcia AJ; Salmeron-Sanchez M, Receptor Control in Mesenchymal Stem Cell Engineering. Nat. Rev. Mater 2018, 3, 1–14. [Google Scholar]
- 25.McBeath R; Pirone DM; Nelson CM; Bhadriraju K; Chen CS, Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 2004, 6, 483–495. [DOI] [PubMed] [Google Scholar]
- 26.Kilian KA; Bugarija B; Lahn BT; Mrksich M, Geometric Cues for Directing the Differentiation of Mesenchymal Stem Cells. PNAS. 2010, 107, 4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalby MJ; Gadegaard N; Tare R; Andar A; Riehle MO; Herzyk P; Wilkinson CDW; Oreffo ROC, The Control of Human Mesenchymal Cell Differentiation using Nanoscale Symmetry and Disorder. Nat. Mater 2007, 6, 997–1003. [DOI] [PubMed] [Google Scholar]
- 28.Roca-Cusachs P; Gauthier NC; Del RA; Sheetz MP, Clustering of alpha(5)beta(1) Integrins Determines Adhesion Strength Whereas alpha(v)beta(3) and Talin Enable Mechanotransduction. PNAS. 2009, 106, 16245–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frantz C; Stewart KM; Weaver VM, The Extracellular Matrix at a Glance. J. Cell Science, 2010. 123: p. 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiu J; Aries L; Lin Z; Vogel V, Nanopillar Force Measurements Reveal Actin-Cap-Mediated YAP Mechanotransduction. Nat. Cell Biol 2018, 20, 262–271. [DOI] [PubMed] [Google Scholar]
- 31.Elosegui-Artola A; Andreu I; Beedle AEM; Navajas D; Garcia-Manyes S; Roca-Cusachs P, Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell. 2017, 171, 1–14. [DOI] [PubMed] [Google Scholar]
- 32.Humphries JD; Byron A; Humphries MJ, Integrin Ligands at a Glance. J. Cell Sci 2006, 119, 3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelham RJ; Wang Y, Cell Locomotion and Focal Adhesions are Regulated by Substrate Flexibility. PNAS. 1997, 94, 13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tse JR; Engler AJ, Preparation of Hydrogel Substrates with Tunable Mechanical Properties. Curr. Protoc. Cell Biol 2010, 10, 1–16. [DOI] [PubMed] [Google Scholar]
- 35.Schindelin J; Arganda-Carreras I; Frise E; Kaynig V, Longair M; Pietzsch T; Preibisch S, Rueden C; Saalfeld S; Schmid B; Tinevez JY; White DJ; Hartenstein V; Eliceiri K; Tomancak P; Cardona A, Fiji: an Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindelin J; Rueden CT; Hiner MC; Eliceiri KW, The ImageJ Ecosystem: An Open Platform for Biomedical Image Analysis. Mol. Reprod. Dev 2015, 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team, R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.