Abstract
G protein-coupled receptors (GPCRs) transduce physiological and sensory stimuli into appropriate cellular responses and mediate the actions of one-third of drugs. GPCR structural studies have revealed the general bases of receptor activation, signalling, drug action and allosteric modulation, but so far cover only 13% of non-olfactory receptors. We broadly surveyed the receptor modifications/engineering and methods used to produce all available GPCR crystal and cryo-EM structures and present an interactive resource integrated in GPCRdb (www.gpcrdb.org) to assist users in designing constructs and browsing appropriate experimental conditions for structure studies.
Introduction
G protein-coupled receptors (GPCRs) form the largest family of cell surface receptors, with over 800 members. GPCRs respond to a wide variety of extracellular signals, influencing most branches of human physiology. 34% of all drugs approved by the FDA act on over 100 GPCRs, and clinical trials are currently exploring 300 new GPCR agents and 66 additional orphan GPCRs1. The study of GPCR structure-function relationships and their application to rational drug design was long limited by a paucity of high-resolution molecular structures, but breakthroughs in protein engineering and crystallisation techniques have led to a surge of structures in the past decade (Fig. 1). Today, 271 structures of 53 distinct receptors (plus four species orthologues) have been reported across all four major GPCR classes A, B1, C and F (http://gpcrdb.org/structure/statistics). Moreover, for classes A and B1, structures of active states are also available, including complexes with an effector G protein2–5 or β–arrestin6 (for class A only). This wealth of GPCR crystal and cryogenic electron microscopy (cryo-EM) structures have provided many new functional insights7. Direct structural data are increasingly combined with complementary information from nuclear magnetic resonance (NMR), double electron-electron resonance (DEER), and electron paramagnetic resonance (EPR); these techniques have shed valuable insights into the dynamic conformational landscape of receptors8–11 and into their binding interfaces with G proteins5, 12 and arrestin6.
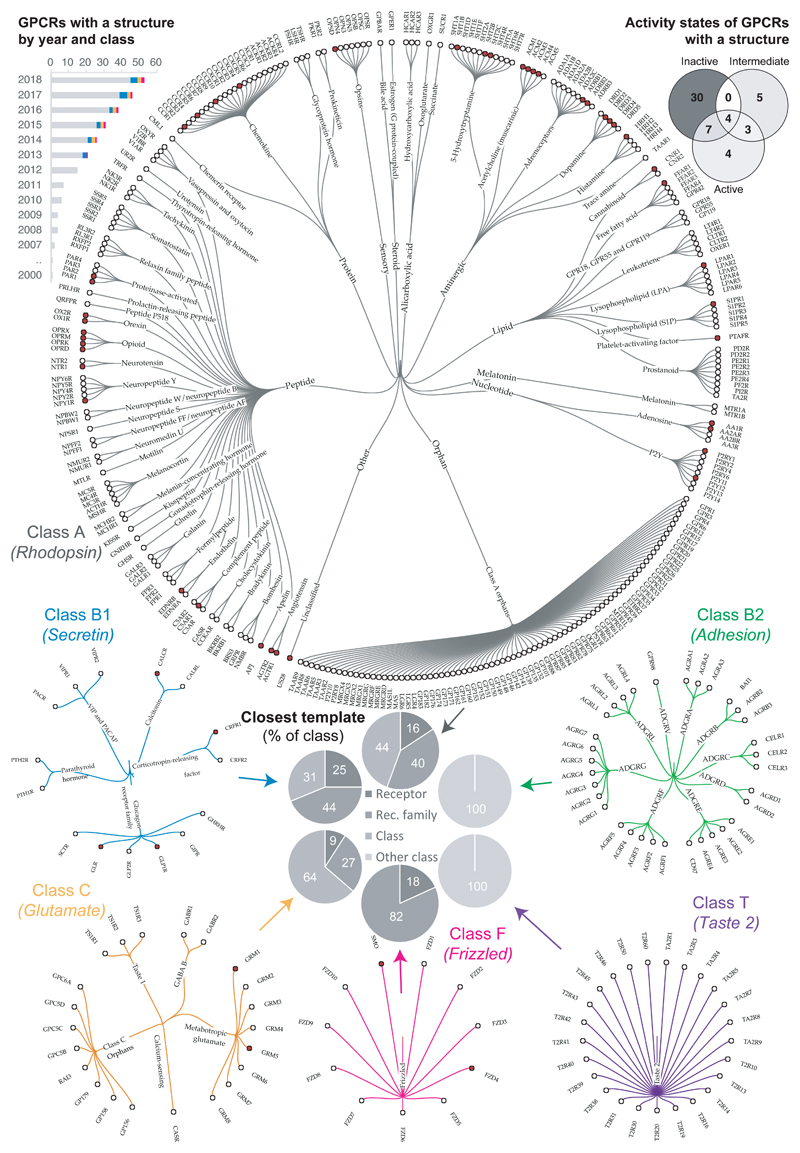
Figure 1.
Progress and space yet to cover in structural characterisation of GPCRs. The progress as number of solved structures by GPCR class and year is shown in the upper left histogram (data from http://gpcrdb.org/structure/statistics). The receptors within each GPCR class that have a solved structure are mapped with a red circle in trees that group receptors by families sharing endogenous ligands; class A has a second level of grouping of ligand types. To evaluate how well these available structures represent all GPCRs, the pie charts display the percentage of receptors with an experimental structure (“Receptor”), and those that currently possess a structural template belonging to the same receptor family, the same class or another class. The upper right Venn diagram depicts the distribution of available conformational states for receptors with solved structure, illustrating the poor representation of active and intermediate receptor states (data from http://gpcrdb.org/structure).
However, there are still no structures for the vast majority –87%– of GPCRs, and classes B2 and T lack structures altogether (while having homology to class B1 and A, respectively). Furthermore, for the majority (52%) of GPCRs there are no close structural templates –from same receptor family with a shared physiological ligand– that would enable reliable homology modelling (Fig. 1). Another limitation is the poor representation of active and intermediate receptor states, which are only available for 18 and 12 receptors, respectively (Fig. 1). Part of the challenge of obtaining GPCR structures is the requirement for protein engineering and the need for tailored protein biochemistry techniques, which have been mastered by only a handful of laboratories worldwide.
In order to expand our knowledge of GPCR function and promote the design of new drugs, it is fundamental to ease the generation of novel GPCR structures, especially of valuable complexes, for example signalling pathway-biased agonists13 and allosteric modulators14, and with signalling effector proteins (e.g. different classes of G proteins, arrestins, and kinases). To expedite this goal, here we provide a comprehensive analysis of all GPCR structure constructs obtained to date. We explore what can be learned from the successful protein engineering approaches, and investigate which experimental methods and reagents display the largest utility or have gained popularity recently in GPCR structural biology efforts.
A resource for GPCR structure constructs and experiments
To facilitate structure determination of GPCRs on a wider scale, we present our results in an interactive online resource, implemented in the GPCR community hub GPCRdb, currently serving ~2,500 distinct monthly users with reference data, interactive analysis and visualisation and experiment design tools. This resource will assist researchers in engineering GPCR constructs and selecting appropriate experimental conditions for crystallography and cryo-EM structure studies. Furthermore, researchers that use GPCR structures for their studies can compare them in a way that is not possible using ‘unprocessed’ data from the PDB or scientific articles. For example, researchers can now select the optimal structural template for their receptor of interest by comparing the underlying construct engineering/integrity or experimental conditions in which the structure was obtained. The analysis can then be extended through the integrated tools for druggable binding sites15–17 and G protein interfaces18.
Our interactive online resource is integrated as a new ‘Structure Constructs’ section in GPCRdb (Fig. 2)19, 20. This unique ‘structure construct databank’ integrates all the GPCR structures in the RCSB Protein Data Bank21 and residue annotations from SIFTS22; plus an extensive manual literature annotation of mutation effects and terminal inserts that aid protein production but are cleaved off before structure determination (Supplementary Note). Furthermore, this resource also holds a comprehensive collection of experimental methods and reagents for GPCR structure determination, amenable to systematic browsing and comparison. It will continue to increase in size and utility as more structures are determined.
Figure 2.
Snapshots from the online resource for GPCR crystallography and cryo-EM. Construct design tool: The Construct design tool allows generation of complete structure constructs for any human GPCR based on data on all available structures and stabilising mutations. The first section (left) presents three application modes: Truncation/Fusion scan, Mutation scan and Custom constructs; the snake plot (right) maps construct modifications onto a 2D sequence view; and the table (bottom) lists all designed constructs and their modifications. Structure view: The single-structure entry view (accessible from the Construct alignments) details all protein engineering, methods and reagents used to determine a given structure. Construct alignments: The Construct alignments has four –Browser, Wild type, Construct, and Sequence– alternative views that can be combined to compare the modifications in any set of constructs, as well as the structure determination method, resolution and release date. Experiment browser: The Experiment Browser tabulates methods and reagents for protein expression, purification, and structure determination from GPCR structure publications. Truncation, fusion and loop deletion sites: These pages visualise the frequencies for the specific truncation, fusion and loop deletion sites used in all structures, organised by receptor and class. Stabilising mutation analyser: The Stabilising Mutation Analyser shows stabilising mutations along with e.g. sequence conservation, helix propensity, hydrophobicity or functional receptor sites. Mutation browser: The Mutation browser shows all mutations and their reported effects in constructs for structural determination. Mutation substitution matrix: The Mutation substitution matrix shows the frequency of amino acid pairwise substitutions across all unique constructs for structural determination.
GPCR Construct Engineering
Common GPCR construct engineering strategies
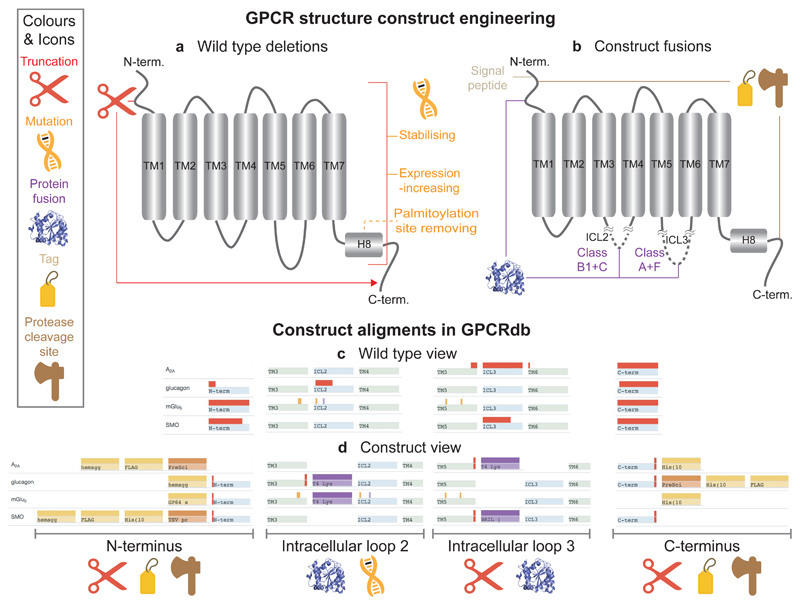
GPCR crystal structures obtained hitherto share common protein engineering strategies. The vast majority of structures have been obtained using N-/C-terminal truncations and mutations predominantly to increase protein stability (and, thus, crystal quality), but also to increase expression or to reduce heterogeneity due to glycosylation and palmitoylation (Fig. 3a). Furthermore, nearly all GPCR structure constructs have been fused to other proteins and peptides to facilitate crystallisation and protein production (Fig. 3b). These modifications can be viewed and compared across receptors in the Construct alignments (http://gpcrdb.org/construct/) page of the database. The user can switch between wild type or construct views (Figs. 3c-d) of the protein and focus on specific selections (e.g. receptors with an N-terminal fusion and no deletions in the third intracellular loop (ICL3)). A third, ‘browser view’ allows further focusing by, for example, structure determination method, resolution or release date. Finally, a fourth view provides a sequence alignment detailing truncations, mutations and inserts in the protein construct. Our analysis below of these ‘construct alignments’ reveals that many modifications have common features and recur at the same positions, suggesting that sufficient templates are now available to allow for data-driven design of new targets of interest.
Figure 3.
Common GPCR structure construct modifications and alignment of constructs for structural determination. (a-b) Types and location of common construct modifications and (c-d) their representation in the Construct Alignments, exemplified by the A2A, glucagon, mGlu5, and Smoothened receptors (representative of the GPCR classes A, B1, C and F; respectively).
Active-state crystal structures require an intracellular binding partner
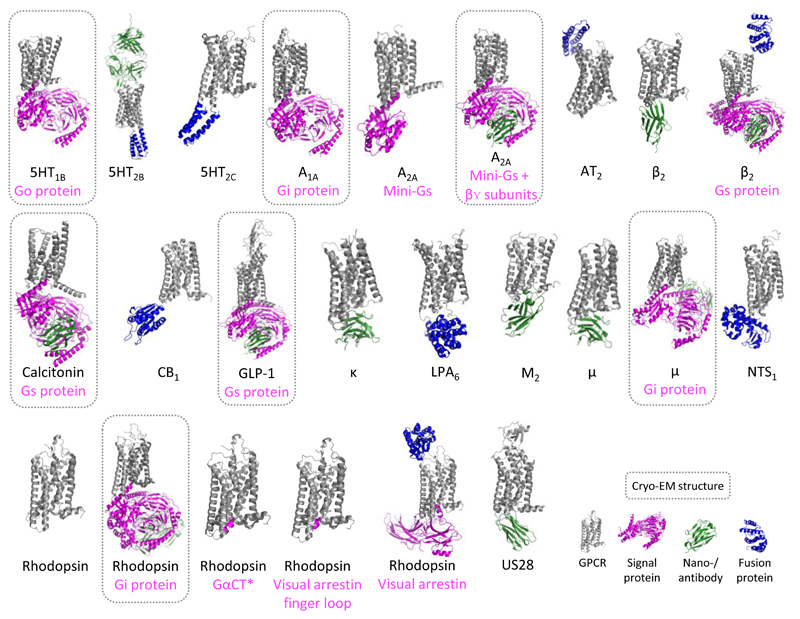
Structures of activated GPCRs are still relatively rare (Fig. 1), with only 16 class A and two class B receptors (Fig. 4 and Supplementary Table 1). Our analyses of truncations, fusions and mutations (below) reveal that active structures generated by crystallography have been obtained using very similar construct engineering as the inactive structures. For example, only three stabilising mutations are distinct for the active or intermediate structures (Supplementary Table 2). This suggests that solving active states requires more than simply transferring working construct design principles. Indeed, all active state structures –with the exception of (rhod)opsin– have required an intracellular bound (9 out of 13) and/or fused (4 out of 13) protein to lock them in an active open conformation. These auxiliary proteins include signalling partners such as a G protein (or a fragment thereof) in the A2A, β2 adrenergic, opsin, calcitonin and GLP-1 receptors, or arrestin in (rhod)opsin; as well as nano-/antibodies in the β2 adrenergic, μ- and κ-opioid, and M2 muscarinic receptors (Fig. 4). Fused proteins have all been placed in the third intracellular loop, joining –and presumably locking– transmembrane helices 5 and 6 (TM5-6), which undergo large movements upon activation (Fig. 4). Several recent developments to extend the ability to obtain stable protein complexes4, 5, in nanobody/antibody production23, and in structure determination by cryo-EM (see below) are expected to make GPCR active-state and signalling complexes more accessible, e.g. to obtain complexes with new effectors. The progress in the structural coverage of GPCRs can be followed in the GPCRdb Structure statistics (http://gpcrdb.org/structure/statistics), and the utilised ligands and auxiliary (including nanobodies) and fusion proteins are included in the Structure Browser (http://gpcrdb.org/structure).
Figure 4.
Active-state GPCR structures, including complexed proteins and cryo-EM structures. The figure shows the structures of active-state GPCRs, including 16 class A and two class B receptors (Calcitonin and GLP-1). With the only exception of one (rhod)opsin structure, all active state structures required a intracellular signalling protein, nano-/antibody or fusion protein. Notes: For rhodopsin bound to a Gt protein C-terminal fragment (GαCT), human GLP-1, opioid μ, and US28 receptors only one of the multiple similar structures available are shown (Supplementary Table 1).
Cryo-EM requires little protein modification
The field of cryo-EM has advanced rapidly24. The use of phase-plates in cryo-EM imaging has increased signal-to-noise ratio25 and allowed for structures at near-atomic detail of membrane proteins with the best resolution of 2.7 Å26 and of 3.3 Å for the GPCR complexes27. Cryo-EM is only applicable to large proteins/complexes, but recently the lower size limit was pushed to 64 kDa upon structural determination of haemoglobin28. While the current technology is unlikely to be suitable for structure determination of seven-transmembrane GPCR monomers, it has delivered structures of eight distinct GPCR-G protein complexes since 2017: calcitonin/Gs3, GLP-1/Gs27, 29, A1A/Gi30, A2A/miniGs25, rhodopsin/Gi5, rhodopsin/miniGo31, μ opioid-Gi4 and 5-HT1B/miniGo32 (Fig. 4).
The engineering of GPCR constructs for cryo-EM is minimal compared to crystallography due to lower requirements of conformational homogeneity and stability. GPCR cryo-EM constructs tend to have small truncations in the N-terminus, where the native signal peptide was replaced with that of haemagglutinin (or GP67 signal peptide in A2A/miniGs25) to enhance receptor expression. The most recent Gi complexes (rhodopsin/Gi5, μ opioid-Gi4 and A1A/Gi30) include purification tags (His and FLAG tags) at both N- and C-termini. Furthermore, the rabbit GLP-1 also features a truncated C-terminus and three point mutations (http://gpcrdb.org/construct), while rhodopsin/Gi5 and μ opioid-Gi4 also feature truncations at the C-terminus. Cryo-EM constructs usually do not require insertion of fusion proteins – the current exceptions are A2A/miniGs (with TrxA), rhodopsin/Gi (with apocytochrome b562RIL (BRIL)), and A1A/Gi (with 22 residues from the M4 muscarinic receptor), all in the N-terminus. All these structures were solved in complex with a G protein, and nanobodies/antibodies contributing to the size and stability of the overall complex.
Fusion sites are often transferrable across GPCRs
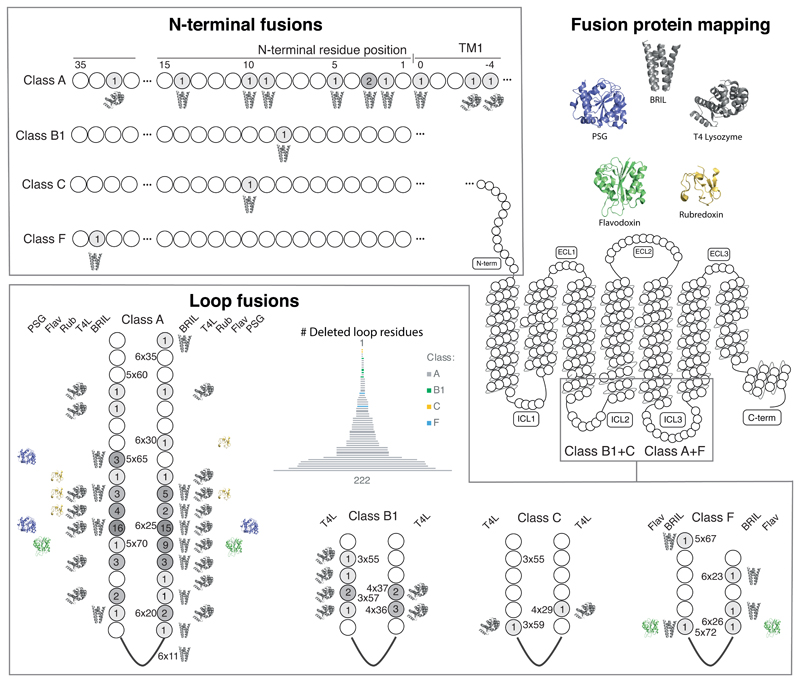
The majority of GPCR crystal structures have been obtained fused to readily crystallisable proteins, which replace flexible/disordered domains and increase the available surface for crystal packing33. Our analysis of all fusions (http://gpcrdb.org/construct/analysis#fusions) shows that the splice sites are predominantly located in the second (for class B/C) and third (for class A/F) intracellular loops (Fig. 5). Notably, the number of replaced residues in these loops spans from none to over 200. Recently, fusions have also been placed in the N-termini of ten receptors spanning all classes. To compare the fusion sites across receptors, we have assigned generic residue positions using the GPCRdb numbering, which extends the earlier Ballesteros-Weinstein scheme by correcting for helix bulges and constrictions (34 and Supplementary Note). This reveals that many fusion sites have been reused for several receptors and are found in coherent clusters within class A (5x65-71 and 6x24-27) and B1 (3x55-58 and 4x36-37) GPCRs. The most frequent fusion sites are found in the ICL3 of class A GPCRs –positions 5x69 and 6x25– which have been used in 16 and 15 unique receptors, respectively, which represents over one third of all (43) class A receptors that have been structurally characterised.
Figure 5.
Mapping of fusion proteins and fusion sites in all GPCR structure constructs. Mapping of fusion proteins shows that BRIL and T4L have been incorporated across a wide range of positions, in contrast to rubredoxin, PSG and flavodoxin. The number of receptors that have utilised a given fusion site (include linkers) are shown within each position in greyscale. Residue positions in the loops are indexed with the GPCRdb generic residue numbering scheme34, whereas N-terminal fusion sites represent their lengths (no. residues) after truncation (Supplementary Note).
Five different fusion proteins have been used so far in GPCR crystallography (Fig. 5). T4-Lysozyme (T4L) and BRIL exhibit the widest applicability, spanning 29 and 22 fusion sites, respectively, suggesting that they often constitute the first choices. In contrast, rubredoxin (CCR5, apelin, and P2Y1 receptors), glycogen synthase (CB1 and OX1-2 receptors), and flavodoxin (CB1 receptor) have only been used in a limited number of cases and at very specific fusion sites in ICL3. Receptors with short loops have, in some cases, been fused to an auxiliary protein without any deletion. In these cases, the use of linkers is more frequent, the most common being one or more iterations of the GS sequence motif –which can serve either to cap an α-helix or provide a flexible and polar spacer. A more exhaustive experimental evaluation is still needed to explore whether less-exploited fusion proteins can provide unique uses at specific targets and positions, and to optimise linker length in receptors with short intracellular loops.
Stabilising mutations have residue bias
Stabilising GPCR mutations have been generated by conceptually very different approaches, including alanine scanning, structure-based design and divergent evolution. We analyzed mutations across these methods to, where possible, deduce common patterns and rationale for their stabilising effect. We first analysed how frequently a specific amino acid has been substituted across all GPCRs (Fig. 6a and http://gpcrdb.org/construct/analysis#mutations). Notably, the 150 unique mutations (with respect to receptor and position) reveal a wide spectrum of amino acid substitutions. Among the wild type residues, G/L/S/T/V have been replaced ten or more times, whereas P/Q/W only one or two times. The most frequently introduced amino acid is, as expected, alanine; whereas several amino acids –H/T and D/P/R/S – have been introduced none and one time, respectively. This shows that although stabilised GPCRs share the canonical residue alphabet they have a different residue usage, i.e. amino acid composition, than wild type receptors. This is consistent with specialised roles of the mutated residues in conforming to evolutionary constraints, molecular structure and protein function - and analogous to two different languages that use the same alphabet but combine and use the letters in unique words with a specific meaning. Deciphering the ‘thermostability’ language may help to focus alanine scans and formulate rational mutagenesis strategies exploiting a wider set of amino acids.
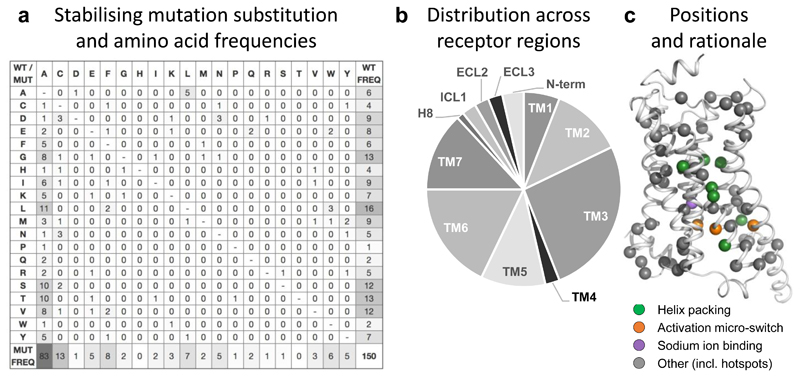
Figure 6.
Substitution frequencies, structural mapping and rationale of stabilising mutations. (a) Amino acid substitution matrix for all 150 stabilising GPCR mutations. The frequencies represent the number of distinct GPCRs and residue positions (merging multiple structures or orthologues) to optimally reflect the transferability across the GPCR superfamily. The different usage of the amino acid alphabet among wild type (rightmost) and mutant (bottom) residues, respectively, is shown with grey-scale. Similarly, grey-scaling is also used to illustrate the success of alanine mutants (leftmost) when substituting mainly similar (small and/or aliphatic hydrophobic) amino acids. (b) Distribution of stabilising mutations (http://gpcrdb.org/construct/stabilisation) across receptor segments, spanning all seven transmembrane helices, the N-terminus, three loops and helix 8. (c) Mapping of all stabilising mutations and their structural rationale onto the β2-adrenoceptor (PDB: 3SN6) using the GPCRdb generic residue indexing34.
The largest group of stabilising mutations comes from alanine scanning (59% when including five A=>L), a ‘brute force’ technique that typically does not have a pre-defined rationale. We analysed all 83 alanine mutations across the complete construct sequences to determine whether the success has been different among amino acid types. We found that the most frequently substituted residues are L, S and T (10-11 mutations), closely followed by G and V (eight mutations); which all represent small amino acids. In contrast, the least substituted residues are large and/or polar: D, H, N, P, W (one mutation) and E, Q, R (two mutations), with one exception, C. This pattern correlates well with traditional evolutionary substitution matrices, but not with an increased helical propensity35 as could be expected as GPCRs large consists of helices including the transmembrane domain, helix 8 (H8) and often ICL1-2. Alanine has the highest propensity to form α-helices whereas G and P have the second lowest and lowest propensity, respectively. This could explain the many substitutions of glycine residues, which also induces the most backbone flexibility, but not the single replacement of P and most of the amino acids with intermediate helix propensity. Prolines almost exclusively appear in transmembrane helical segments of GPCRs when they are important for receptor structure and/or function; therefore, it is expected that mutation of a proline would not be well-tolerated. Indeed, the only proline mutant in crystallised constructs (P6x42a / P6x47b in GLP-1) showed an impaired activity compared to wild type (Ext. Data Fig. S2 in36). In summary, while alanine scanning is a successful stabilisation method, more thorough analyses that factor in the wild type residue (e.g. the most frequently mutated small amino acids L, S, T, G and V), positions shown to stabilise multiple receptors/families (next section) and local environment (e.g. consensus contact networks in the transmembrane bundle37) of each mutation could assist in focusing this technique.
Stabilisation across receptors/families
We next investigated the receptor topology and specific receptor positions for all 150 stabilising mutations to elucidate crucial receptor regions and positions shown to stabilise multiple receptors/families. We found that the stabilising mutations are distributed across all seven transmembrane helices, as well as the N-terminus, loops and H8 (Fig. 6b-c). In the transmembrane helices, TM3 features the largest number of mutations, whereas TM1 and TM4 have the lowest. This trend reflects their relative positions in the transmembrane bundle, wherein TM3 is central and packs to four other helices, whereas TM1 and TM4 are more peripheral and interact primarily with only two other TM helices. To facilitate an efficient and clustered residue position-specific investigation, we developed a Stabilising Mutation Analyser, which can group mutations in different receptors by a common generic residue position and wild type and/or mutant amino acids (http://gpcrdb.org/construct/stabilisation). The tool also maps mutations to known functional receptor sites and calculates how the amino acid substitution changes sequence conservation, helix propensity and hydrophobicity. Leveraging this tool, we uncovered 25 positions that have stabilised more than one receptor and 13 also stabilised different receptor families (Supplementary Table 2 and Fig. 6c)
We proceeded to investigate the underlying rationale of the re-used stabilisation sites. The first of such mutations, 3x41W, has been used in six GPCRs. 3x41W packs to TM3-538, including the highly conserved P5x50 in TM5. A second mutation, 3x34A, seems to have an indirect effect, by allowing W4x50 in TM4 to pack between the transmembrane bundle instead of facing the membrane interface. A third position, 6x48, contains a highly conserved aromatic residue that packs to TM3, TM6 and TM7 (I3x40, F6x44 and T7x41). While classes A, B2 and C have primarily a W at this position (e.g. 68/16/3% WFY in class A), class B1 contains mainly F, Y or alternatively an E, which instead forms a charged/polar interaction to residues in TM2, TM5 and TM7 (2x53, 5x44, and 7x45, respectively). Stabilising construct mutations in this region have been placed in 6x48 (apelin and glucagon receptors), 3x40 (β1-adrenoceptor and apelin receptor), 6x47 (GLP-1) and 7x41 (adenosine A2A and neurotensin 1 receptors). Furthermore, a sodium ion binding site is present in the transmembrane bundle of several class A receptor structures, immediately below the transmembrane ligand binding pocket39. The sodium ion acts as a negative allosteric modulator by stabilising the inactive conformation, whereas its binding site collapses upon activation. Hence, the mutation in the sodium ion binding site D7x49N in P2YR1 and P2YR12 weakens the inactive state and stabilises intermediate state structures.
Certain GPCR residues –activation switches– rearrange their side chains to form unique stabilising contacts in inactive or active states40–42. For instance, R3x50, part of the conserved DRY motif of TM3, interacts with E3x49A in the inactive state and upon activation swings towards the transmembrane bundle core to form the ‘ceiling’ of the G protein binding site2. This switch has been modulated by the mutations R3x50L and E3x49A to stabilise agonist-bound and active state-like NTS1 structures; and also by the nearby mutation L6x37A in the A2A and NTS1 inactive/intermediate state structures. In the active state, another switch, Y5x58 swings towards the transmembrane bundle to form a polar contact with R3x50 in the DRY motif. Y5x58F/A mutations have been used to stabilize β1 and FFA1 inactive state structures. An additional activation switch, Y7x53ax57b in the NPxxY motif of TM7, forms, in the inactive state, hydrophobic and aromatic contacts with TM2 (2x43ax50b) and, in class B1, also a hydrogen bond to T6x37ax42b. In active state class A structures, this residue swings into the G protein site (above R3x50) to form hydrophobic contacts to TM3 (3x43 and 3x46) and a direct or water-mediated hydrogen bond to the backbone of TM6. Thermostabilising mutations of these motifs have been introduced in 7x53 (β2-adrenoceptor and CRFR1) and 3x46 (CB1). Finally, Y/K6x30ax35b in TM6 has been mutated to alanine in most inactive state structures of class B1 receptors to allow TM6 to pack tighter with TM3 and TM5 on the intracellular side. In the inactive state of the glucagon receptor, K6x30ax35b forms a salt-bridge to D6x28ax33b (all class B GPCRs have a proximal negatively charged residue), and in the intermediate state GLP-1 structure36, K6x30 swings towards the backbone of the last residue in TM7 (7x56ax60b).
Understanding the rationale behind these mutations allows our construct design tool (Fig. 2 and below) to suggest stabilising mutations – also for the many receptors lacking a close template (Fig. 1).
N-/C-terminal truncations
Truncation of flexible regions in the N- and C-termini of GPCRs aids the formation of well-ordered diffracting crystals. To study the extent and variation of such deletions, we developed a Truncation sites analysis tool (http://gpcrdb.org/construct/analysis#truncations) that maps the truncated (shaded) and preserved (solid) N- and C-terminal segments. The lengths of the N- and C-termini are defined as the number of residues before TM1 and after H8, respectively. Strikingly, the length of preserved N-termini in class A GPCRs spans from none to 50 residues. About half of the receptors have been obtained without any N-terminal truncation or deletion of only the initial methionine start codon to avoid transcription. Class B1, C and F GPCRs have structured N-termini that comprise the binding domains of their physiological ligands, and such elements are typically either completely truncated or left unaltered. Full-length structures have been obtained for the classes B1 and F, but not C. Furthermore, constructs for the same receptor family or even receptor often also exhibit a large variation of N-terminal lengths, suggesting that the truncation sites may vary with other factors such as the choice of tags and signal peptides. Taken together, this shows that N-terminal truncations may be difficult to infer to a new target, but that on the other hand, there are often multiple viable sites.
Furthermore, we investigated how the putative flexibility or rigidity of N-termini is actually reflected in the truncations. To this end, the Truncation sites analysis tool displays a colour-coded map of flexible, rigid and context-dependent regions predicted using Dynamine43 (http://gpcrdb.org/construct/analysis#truncations). As presumed, the vast majority of the truncated segments in class A GPCRs are predominantly flexible. However, the 5-HT2C and CB1 receptors were truncated closely after segments predicted to be rigid indicating that it may be possible to preserve a larger part of their native termini. Also as expected, the preserved parts of the N-termini in class A GPCRs are predominantly rigid although there are several exceptions. Finally, inspection of the structures with the longest N-termini –with over 30 preserved residues– shows that five out of seven receptors contain a secondary structure element that could reduce flexibility (Supplementary Table 3). This suggests that preserving such elements may be advantageous for structure determination with both crystallography and cryo-EM. Taken together, the prediction of protein flexibility could complement the site/length-based rationale for N-terminal truncations, especially for targets without a close structural template.
Unlike the N-termini, the vast majority of preserved C-terminal segments are short (up to 12 residues), and exhibit clusters of recurring truncation sites demonstrating that these are transferrable across GPCRs. Furthermore, fewer class A GPCRs were obtained with a wild type C-terminus –a third– compared to N-terminus –a half. In the other end of the spectrum, four receptors –P2Y1, CCR5, calcitonin, and squid rhodopsin– have long C-termini (>30 residues), but only the longest (squid rhodopsin, 118 AAs) contains rigid segments. Class B1 GPCRs have a long H8 and a comparison of the constructs shows that the GLP-1, glucagon and CRFR1 receptors have sometimes been truncated before the C-terminus at 9, 12 and 19 positions into H8, respectively.
Online GPCR Construct Design Tool
We developed an online Construct Design Tool (Fig. 2) that allows generation of complete construct sequences for any GPCR and type of modifications, with the goal of supporting structural determination on a wider scale by reducing the total number of constructs that need to be experimentally screened. Up-to-date instructions and an explanatory video are integrated in the top of the tool page.
Application modes
A first application mode ‘Truncation/fusion scan’ allows users to enter several N-/C-terminal truncation and protein fusion start/end sites either automatically by defining a number of top-ranked sites or by manual inspection of the suggestions. Suggested N-/C-terminal truncation and fusion protein sites are ranked first grouped by target-template homology; i.e. those in the same receptor, receptor family or class; and then by their frequency (number of distinct GPCRs with a structure exploiting the given site). For long N-termini it is possible to only replace the signal peptide, which has been predicted using SignalP44. Suggested N- and C-termini truncation sites/lengths are defined as the number of preserved residues before the start of TM1 and after the end of H8, respectively (the data excludes termini with a fusion protein). Loop fusions are placed in the second and third intracellular loops for the classes B and C, and A and F, respectively (Fig. 6). If the user does not select a loop fusion, long ICL3 loops (>8 residues) are instead assigned suggested deletions from non-fused and N-terminally fused constructs. Suggested fusion sites can be filtered by the type of fusion protein used. Finally, in a Truncation/fusion scan, the rightmost part of the construct table has a section to “Add known stabilising mutations to all constructs”. This allows the user to manually enter mutations found to be stabilising in previous experiments to all constructs already listed in the table. If the target receptor already has a published structure containing such mutations they are also listed as suggestions. The tool generates constructs for all unique combinations of selected truncation/fusion sites.
A second application mode ‘Mutation scan’ first designs just one reference construct –which can include mutations with known stabilising effect– and then selects a number of stabilising mutations which are individually added to generate as many constructs. Predicted stabilising mutations can be either hand-picked or added in batch by specifying a fixed number of top-listed suggestions in a construct table column “Scan predicted mutations in separate constructs”. Predicted stabilising mutations are assigned priorities by their rationale (see below); some are distinct for inactive or active conformational states (and can be filtered before selection).
A third application mode ‘Custom constructs’ can be used to manually modify constructs generated from the Truncation/fusion scan or Mutation scan applications, or to design de novo constructs. For any application, N-/C-terminal inserts and mutations of glycosylation and palmitoylation sites must be selected individually. Finally, the sequence and site of any modification can be custom-defined by the user, for example to incorporate an in-house validated mutation, alternative tag or modified fusion protein at a specific residue position. Constructs can be inspected in a snake plot and saved in a spreadsheet for ordering of cDNAs or to resume the design later.
Suggested stabilising mutations
The suggestions of stabilising mutations span a number of specific design rules (http://files.gpcrdb.org/mutation_rules.html) which are both data- and rationale-driven and cover five overall concepts: (1) Homology: This concept infers a mutant position and amino acid if the target is the same receptor or a member of the same receptor family. For the classes B-F, which are smaller and have less data than class A, mutations will also be informed from any member of the same class. (2) Common mutations: These are mutations that have been utilised within several distinct receptor families, but not yet that of the selected receptor target. (3) Conservation: This concept introduces residues that are missing in the target but at least 70% conserved in the receptor family or class. For the positively charged residues H, K and R a lower conservation threshold (40%) is used in order to incorporate multiple position at the ends of the transmembrane helices wherefrom these residues can interact with the polar head groups of the cell membrane. Furthermore, the low-propensity residues G and P are treated separately (below) and C, which can form disulphide bridges, is excluded. (4) Helix propensity: This concept aims to increases helix propensity by replacing G and P residues that are present in the target receptor but poorly conserved in the receptor family or class to alanine. G residues within four positions from a helix end are preserved, as they can be crucial for the transition to a loop structure. (5) State switches: These residues form interactions that are unique for either inactive or active receptor states. The state selected by the user will be targeted by adding and removing residues with such interactions.
Application of the construct design tool to 5-HT2C
The construct design tool has already been used for some time by the GPCR Consortium (http://gpcrconsortium.org), academic labs, and companies whom we have assisted in setting up local versions to incorporate also proprietary data (http://docs.gpcrdb.org/local_installation.html). To demonstrate the tool, we tested the thermostability of 17 single-point mutants of the wild type serotonin receptor 5-HT2C (Supplementary Note). The melting temperature of the designed constructs was increased by at least two degrees in 71% (12/17) of the mutants (Supplementary Table 4). However, vastly different success rates were observed depending on the ligand tested (no ligand (apo): 53% (9/17) > antagonist (ritanserin): 18% (3/17) > in-house agonist (N-(4-bromo-2,5-dimethoxyphenethyl)-1-(2-methoxyphenyl)ethan-1-amine): 12% (2/17) > partial agonist (ergotamine): 6% (1/17)). The higher success rate for the apo form indicates that the ligands –which all have, in this case, a thermostabilising effect (45 and data not shown)–, may be masking the inherent effect of the mutation. However, the selection and, particularly, the combination of a more stringent selection of thermostabilising mutations would likely match or surpass the thermostabilising effect of the ligand. Furthermore, only one mutation in our set –V185I4x56– increased the stability in two ligand experiments, in line with the ligand-specificity of stabilising mutations often observed in previous studies46. This limited assessment of our prediction of thermostabilising mutations can be deemed encouraging, considering that the construct design tool typically suggests 30-50 stabilising mutations per construct. However, ongoing validation will lead to a refined prioritisation of the suggested mutations. This will also allow us to reduce the number of mutations that need to be tested experimentally, and hence also further improve the success rate.
Survey of Methods Used in GPCR Structure Determination
Our extensive annotation of experimental data from GPCR structure publications is made available in the GPCRdb Experiment Browser page (http://gpcrdb.org/construct/experiments). This tool allows for swift navigation and filtering of methods and reagents for protein expression, purification and preparation of samples for structure determination by crystallography or cryo-EM. Researchers can quickly and easily infer the most relevant conditions for their experiments from related receptors and keep track of the development of new methods and materials.
Methods and reagent utility and trends
Expression
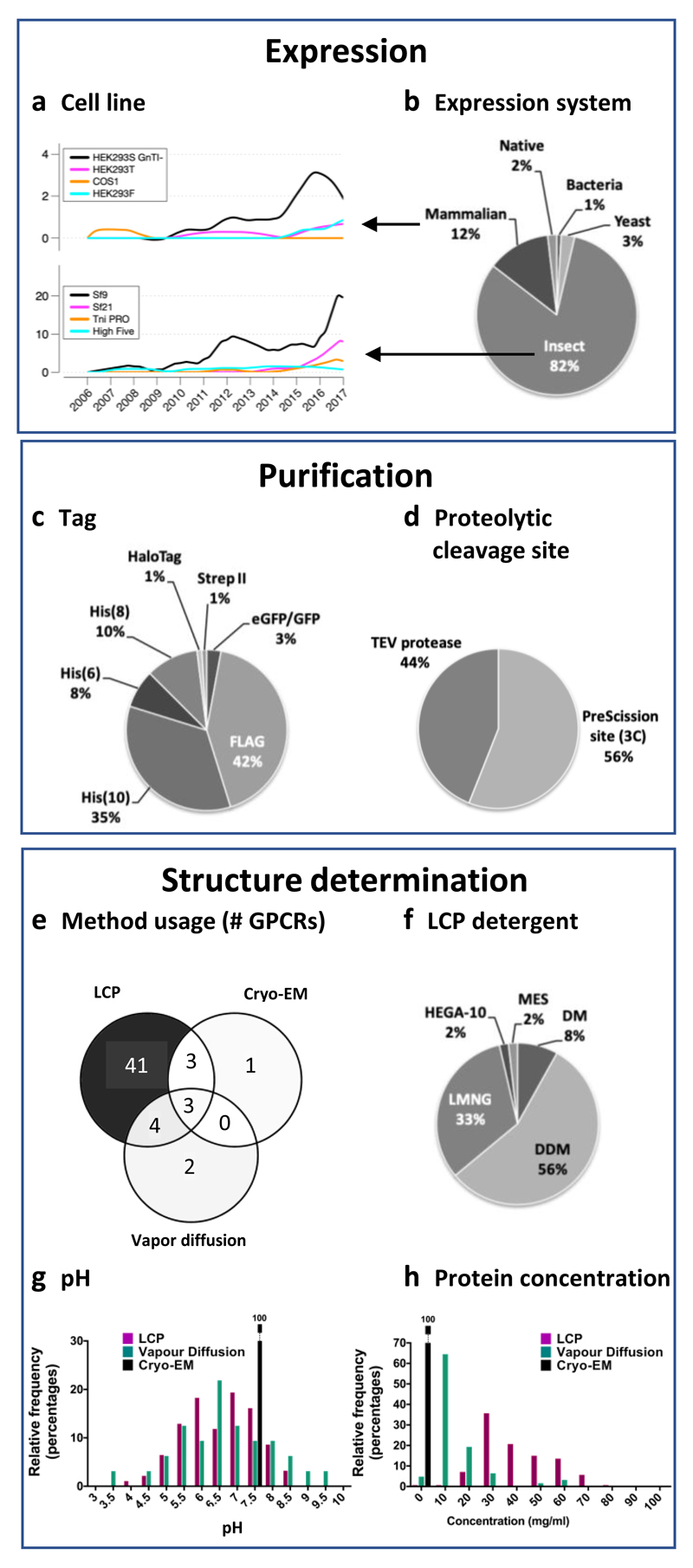
Currently, only bovine and squid rhodopsin have been obtained as native protein from natural sources. Most GPCRs have been obtained by recombinant expression in insect cells (Fig. 7a-b), predominantly in the Sf9 cell line, which was the expression system of choice in the first crystallised non-rhodopsin GPCRs (Fig. 7a, bottom). However, in the last two years, there has been an increase of GPCRs expressed in mammalian systems; for example the rhodopsin, CB1, smoothened, and the viral chemokine US28 receptors have also been expressed in different types of HEK293 cells, including the glycosylation-deficient GnTI-HEK 293 strain. GPCR-G protein complexes have been prepared by separate expression of the individual components (5-HT1B/miniGo32, A1A/Gi30, A2A/miniGs25, GLP-1/Gs129, rhodopsin/Gi5, rhodopsin/Go31 μ opioid-Gi4) or joint co-transfection (GLP-1/Gs227, calcitonin-Gs3). The hemagglutinin signal peptide is most frequently used to increase GPCR expression. Ligands are also often added at this stage to improve expression and protein stability47.
Figure 7.
Key expression, purification, and structure determination methods and reagents. (a-b) Expression systems: Trends in the use of mammalian and insect cell lines (a) and overall (all-time) usage of cell lines for recombinant expression of GPCRs (b). Purification: Usage of different affinity purification tags (c) and proteolytic cleavage sites (d). Crystallisation methods and conditions: Distribution of methods (e), LCP detergents (e), pH (g) and protein concentrations (h) used in GPCR structure determination.
Purification
His and FLAG tags are the most common purification tags used for GPCR structural biology experiments (Fig. 7c). Purification of GPCRs for structural studies is carried out mostly from protein solubilised in dodecyl-maltoside (see below) by immobilised-metal ion affinity chromatography (IMAC) –using a C-terminal poly-histidine tag– or by antibody affinity chromatography –using an N-terminal FLAG tag. Both tags can be used simultaneously to obtain a higher-quality samples48 and are usually later removed using proteases targeting engineered proteolytic cleavage sites (HRV 3C/PreScission or TEV, used with similar frequencies) (Fig. 7d). Some but not all laboratories use size-exclusion chromatography as an additional last purification step.
Structure determination
Over 90% of all GPCRs have been crystallised in lipidic mesophases (mostly cubic, but also in sponge phases) (Fig. 7e), using almost exclusively monoolein (9.9 MAG); only about 10 structures have been crystallised from other cubic phase lipids –9.7, 11.7, and 11.9 MAG. The vast majority of these structures have also been obtained using 10% cholesterol as an additive to the lipidic mesophases. Crystallisation from lipidic mesophases is achieved from protein solubilized in dodecyl-maltoside (DDM; ~60%), lauryl maltose-neopentyl glycol (LMNG; ~30%) and decyl-maltoside (DM; ~12%) (Fig. 7f), with cholesterol hemisuccinate (CHS) as an additive and PEG 400 as the most common precipitant (~70%) (Supplementary Table 5). Another strategy used recently is solubilisation in DDM and subsequent exchange of detergent to LMNG for crystallisation. HEGA-10 (decanoyl-N-Hydroxyethylglucamide) and MES (methyl ester sulfonate) have also been used, but only in a small number of cases.
Only about 13% of GPCRs (rhodopsin, and the β1 adrenergic, adenosine A2A, and neurotensin 1 receptors) have been crystallised by vapour diffusion (Fig. 7e). The conditions for crystallisation by vapour diffusion are more diverse (Supplementary Table 5) and include the use of harsher detergents (as OG and NG), alternative lipids (such as brain lipid extract), higher molecular weight PEGs, and alternative precipitants. Vapour diffusion can succeed at a broader pH range (Fig. 7g) and lower temperatures than LCP and also allows for lower protein concentration (Fig. 7h). Most of the receptors crystallised by vapour diffusion have also been crystallised in lipidic mesophases (Fig. 7e), usually yielding better-diffracting crystals. Hence, crystallisation from lipidic mesophases appears to be a more general method, while vapour diffusion has been the initial method of choice for a few selected targets that, in the end, seemed amenable to crystallisation in lipidic mesophases after optimisation.
Finally, cryo-EM requires a very low protein concentration (~1 mg/ml) compared to crystallisation (~20-80 mg/ml) (Fig. 7g) and allows for the use of mild detergents for solubilisation. Especially for GPCR complexes, which often require a complicated purification procedure and are hard to produce in large amounts, cryo-EM certainly is advantageous. The choice of detergent is quite limited and only mild detergents can keep the complex stable24. DM32, DDM supplemented with CHS4, 5, and LMNG without25 or with CHS27, 30 are used for solubilising cell membranes, and the complexes are purified in the last step using DM32, digitonin5, or LMNG without25 or with lipidic supplements3, 4, 27, 29, 30.
Directed evolution has improved GPCR expression
Receptor expression varies largely49, and insufficient protein yields hamper crystallisation screens, and X-ray diffraction and cryo-EM experiments. 42 mutations in GPCR crystal structure constructs have been reported to increase the surface expression, whereof only seven have also been reported to be thermostabilising (however this has not always been tested and the total number is likely to be higher). 32 of these mutations were identified using directed evolution method for increasing GPCR expression. In contrast, only four are mutations to alanine, suggesting that alanine scanning is not an efficient technique for increasing the surface expression. 17 mutations have introduced aliphatic residues (A: 4, L: 7, M: 1, V: 6), which have high helix propensity and especially L and V are known to stabilise helices through contact with the next helical turn50. Furthermore, nine mutations have introduced positively ionisable residues (R: 6, K: 2 and H: 1). Our structural investigation shows that these are located at the height of the membrane surface and hence are likely to anchor the receptor through salt bridges to the polar head groups. In all, this uncovers a distinct rationale for the most common type of receptor expression-increasing mutations, whereas others ought to be revisited when more such mutations have become available.
Removal of glycosylation and palmitoylation sites sometimes aid crystallisation
Removal of certain post-translational modifications can improve protein homogeneity and, therefore, the chances of crystal formation. Our Mutation Browser (http://gpcrdb.org/construct/mutations) provides a comprehensive annotated list of all construct mutations and their effect. Our analysis shows that 15 glycosylation sites have been removed from the extracellular interface of nine receptors. 13 mutations removed a glycosylated N residue, while two were of T, which is present in both O- and N-linked glycosylation consensus motifs (S/TS/TX1-10N and NXS/T, respectively). Glycosylations are often removed enzymatically during the protein purification steps. Furthermore, seven palmitoylation sites, which function as anchors to the cell membrane, have been deleted in or after H8 in four receptors. However, the fact that most structures have been obtained without removing post-translational modifications suggests that it is desirable, when possible, to preserve these sites, which can increase receptor expression and stability.
Supplementary Material
Summary.
We expect the lessons learnt from our extensive comparison of all available GPCR constructs and the accompanying online platform will assist the community in accelerating the determination and assessing the quality of GPCR structures. Given the role of these membrane proteins as major signalling mediators and therapeutic targets, we expect that our study will impact both our basic understanding of receptor-structure function, as well as our prospects to attain new templates for structure-based drug development.
Acknowledgements
Anna Tsolakou, Dalibor Milic and Kasper S. Harpsøe are acknowledged for help with data annotation, Isla Carson for development of the preliminary version of the Stabilising Mutation Analyser and Ching-Ju Tsai for input on the description of cryo-EM construct engineering and experiments. This work was supported in part by the ERC Starting Grant 639125 and Lundbeck Foundation grants R163-2013-16327 and R218-2016-1266 to D.E.G, the Swiss National Science Foundation grant CRSII2_160805 to X.D., the European Commisions Seventh Framework Program (FP7/2007-2013) grant 290605 (COFUND: PSI-FELLOW) to E.M, and the COST Action CM1207 (‘GLISTEN’).
Footnotes
Author Contributions
C.M, D.E.G., J.B. and T.F. made the construct analyses and figures; C.M. and V.I. developed the online resources; E.M. and C.M. annotated and analysed published experimental data. L.F.N. conducted the mutagenesis experiments; M.A.H. and R.C.S provided critical inputs on the project, manuscript writing, and data analysis; X.D. and D.E.G. drafted the paper and all authors commented on the draft. D.E.G. and X.D designed and D.E.G. managed the project.
Competing Interests Statement
The authors declare that they have no competing financial interests.
References
- 1.Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nature reviews Drug discovery. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [Landmark reference for GPCR drugs, target and indications describing recent drug discovery successes and new strategies in clinical trials.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang YL, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koehl A, et al. Structure of the micro-opioid receptor-Gi protein complex. Nature. 2018;558:547–552. doi: 10.1038/s41586-018-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y, et al. Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature. 2018;558:553–558. doi: 10.1038/s41586-018-0215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tautermann CS, Gloriam DE. Editorial overview: New technologies: GPCR drug design and function-exploiting the current (of) structures. Curr Opin Pharmacol. 2016;30:vii–x. doi: 10.1016/j.coph.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Eps N, et al. Conformational equilibria of light-activated rhodopsin in nanodiscs. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E3268–E3275. doi: 10.1073/pnas.1620405114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature. 2016;533:265–268. doi: 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- 12.Van Eps N, et al. Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:2383–2388. doi: 10.1073/pnas.1721896115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Violin JD, Crombie AL, Soergel DG, Lark MW. Biased ligands at G-protein-coupled receptors: promise and progress. Trends Pharmacol Sci. 2014;35:308–316. doi: 10.1016/j.tips.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Congreve M, Oswald C, Marshall FH. Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs. Trends Pharmacol Sci. 2017;38:837–847. doi: 10.1016/j.tips.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Munk C, Harpsoe K, Hauser AS, Isberg V, Gloriam DE. Integrating structural and mutagenesis data to elucidate GPCR ligand binding. Curr Opin Pharmacol. 2016;30:51–58. doi: 10.1016/j.coph.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isberg V, et al. GPCRdb: an information system for G protein-coupled receptors. Nucleic acids research. 2016;44:D356–364. doi: 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isberg V, et al. GPCRDB: an information system for G protein-coupled receptors. Nucleic acids research. 2014;42:D422–425. doi: 10.1093/nar/gkt1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flock T, et al. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandy-Szekeres G, et al. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic acids research. 2018;46:D440–D446. doi: 10.1093/nar/gkx1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munk C, et al. GPCRdb: the G protein-coupled receptor database - an introduction. Br J Pharmacol. 2016;173:2195–2207. doi: 10.1111/bph.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose PW, et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic acids research. 2017;45:D271–D281. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velankar S, et al. SIFTS: Structure Integration with Function, Taxonomy and Sequences resource. Nucleic acids research. 2013;41:D483–D489. doi: 10.1093/nar/gks1258. [Resource for residue-level mapping of UniProt (protein) and PDB (structure) entries also integrating annotations from many more major databases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchings CJ, Koglin M, Olson WC, Marshall FH. Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. Nature reviews Drug discovery. 2017;16:787–810. doi: 10.1038/nrd.2017.91. [DOI] [PubMed] [Google Scholar]
- 24.Renaud JP, et al. Cryo-EM in drug discovery: achievements, limitations and prospects. Nature reviews Drug discovery. 2018;17:471–492. doi: 10.1038/nrd.2018.77. [Reviews the recent advances in cryo-EM and provides an outlook of what to expect in the near future.] [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Nafria J, Lee Y, Bai X, Carpenter B, Tate CG. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. Elife. 2018;7:e35946. doi: 10.7554/eLife.35946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su X, et al. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science (New York, N.Y.) 2017;357:815–820. doi: 10.1126/science.aan0327. [DOI] [PubMed] [Google Scholar]
- 27.Liang YL, et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature. 2018;555:121–125. doi: 10.1038/nature25773. [DOI] [PubMed] [Google Scholar]
- 28.Khoshouei M, Radjainia M, Baumeister W, Danev R. Cryo-EM structure of haemoglobin at 3.2 A determined with the Volta phase plate. Nat Commun. 2017;8 doi: 10.1038/ncomms16099. 16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Draper-Joyce CJ, et al. Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature. 2018;558:559–563. doi: 10.1038/s41586-018-0236-6. [DOI] [PubMed] [Google Scholar]
- 31.Tsai CJ, et al. Crystal structure of rhodopsin in complex with a mini-Go sheds light on the principles of G protein selectivity. Sci Adv. 2018;4:eaat7052. doi: 10.1126/sciadv.aat7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Nafria J, Nehme R, Edwards PC, Tate CG. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. Nature. 2018;558:620–623. doi: 10.1038/s41586-018-0241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isberg V, et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [Generic numbering of receptor residues crucial for all GPCR structure-function studies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jazayeri A, et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature. 2017;546:254–258. doi: 10.1038/nature22800. [DOI] [PubMed] [Google Scholar]
- 37.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [Pioneering GPCR structure analysis uncovering common contact networks stabilising the receptor fold, and characteristic features of ligand binding and receptor activation.] [DOI] [PubMed] [Google Scholar]
- 38.Roth CB, Hanson MA, Stevens RC. Stabilization of the human beta2-adrenergic receptor TM4-TM3-TM5 helix interface by mutagenesis of Glu122(3.41), a critical residue in GPCR structure. J Mol Biol. 2008;376:1305–1319. doi: 10.1016/j.jmb.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White KL, et al. Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling. Structure. 2018;26:259–269 e255. doi: 10.1016/j.str.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends in Pharmacological Sciences. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter B, Tate CG. Active state structures of G protein-coupled receptors highlight the similarities and differences in the G protein and arrestin coupling interfaces. Current opinion in structural biology. 2017;45:124–132. doi: 10.1016/j.sbi.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Cilia E, Pancsa R, Tompa P, Lenaerts T, Vranken WF. The DynaMine webserver: predicting protein dynamics from sequence. Nucleic acids research. 2014;42:W264–270. doi: 10.1093/nar/gku270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 45.Peng Y, et al. 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell. 2018;172:719–730 e714. doi: 10.1016/j.cell.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proceedings of the National Academy of Sciences. 2008;105:10744–10749. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Stevens RC, Xu F. The importance of ligands for G protein-coupled receptor stability. Trends Biochem Sci. 2015;40:79–87. doi: 10.1016/j.tibs.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Milic D, Veprintsev DB. Large-scale production and protein engineering of G protein-coupled receptors for structural studies. Front Pharmacol. 2015;6:66. doi: 10.3389/fphar.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lv X, et al. In vitro expression and analysis of the 826 human G protein-coupled receptors. Protein Cell. 2016;7:325–337. doi: 10.1007/s13238-016-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo P, Baldwin RL. Origin of the different strengths of the (i,i+4) and (i,i+3) leucine pair interactions in helices. Biophys Chem. 2002;96:103–108. doi: 10.1016/s0301-4622(02)00010-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.