Abstract
Bactofilins are small β-helical proteins that form cytoskeletal filaments in a range of bacteria. Bactofilins have diverse functions from cell stalk formation in C. crescentus to chromosome segregation and motility in M. xanthus. However, the precise molecular architecture of bactofilin filaments has remained unclear. Here we revealed by sequence analysis and electron microscopy that in addition to wide distribution across bacteria and archaea, bactofilins are also present in a few eukaryotic lineages such as the Oomycetes. Cryo-EM analysis demonstrated that the sole bactofilin from Thermus thermophilus (TtBac) forms constitutive filaments that polymerise through end-to-end association of the β-helical domains. With the aid of a nanobody, we determined the near-atomic filament structure, showing that the filaments are non-polar. A polymerisation-impairing mutation enabled crystallisation and structure determination, while reaffirming the lack of polarity and the strength of the beta-stacking interface. To confirm the generality of the lack of polarity, we performed co-evolutionary analysis of a large set of sequences. Finally, we determined that the widely conserved N-terminal disordered tail of TtBac is responsible for direct binding to lipid membranes both on liposomes and in E. coli cells. Membrane binding is likely a common feature of these widespread but only recently discovered filaments of the prokaryotic cytoskeleton.
Keywords: prokaryotic cytoskeletons; evolution of the cytoskeleton; CcmA, β helical proteins; protein filaments
Introduction
Most bacteria and archaea contain functional protein filaments and have been collectively termed prokaryotic cytoskeletons1. Amongst those, cytomotive filaments, e.g., of the actin and tubulin types, are characterised by complex dynamics. Most prokaryotes, however, also encode protein filaments that do not belong to the actin and tubulin families and that instead act as molecular scaffolds2. Thanbichler and co-workers recognised in 2010 the existence of a conserved and widespread family of bacterial proteins that form constitutive and very stable filaments in vitro and which perform scaffolding roles in cells; they named these proteins bactofilins3.
Bactofilins contain a small domain of about 110 amino acids (PFAM domain PF04519), which has been recognised as being present in many bacterial genomes, often within multiple genes2. Most sequences consist of one bactofilin domain flanked by presumably disordered proline-rich tails at the N- and C-termini, although this is not universal: enterobacterial bactofilins, including Proteus mirabilis CcmA, have predicted transmembrane helices in the N-terminal tail3,4. Bactofilin filaments are very stable, being largely insensitive to pH, salt concentration and chelating agents and hence are always filamentous when purified from source or expressed heterologously3,5.
The structure of the conserved bactofilin domain was solved by solid state NMR6, with supporting evidence coming from sequence-based modelling and electron microscopy of the filaments7–9. The bactofilin domain has a right-handed β-helical fold, with 6 windings of ~17 amino acid residues producing triangular-shaped monomers that measure roughly 3 nm along the β-helical axis. However, so far, it has not proved possible to determine the filament structure of bactofilins.
Understanding of bactofilin function has been limited to a small number of examples. Caulobacter crescentus bactofilins BacA and BacB were identified in a localisation screen for proteins involved in stalk formation3. BacAB are expressed throughout the cell cycle and condense at the stalk-forming site within the cell at the onset of S phase. These bactofilins have been found to directly interact with the cell wall synthesis enzyme PbpC, and deletion of either the filaments or PbpC produces much shorter stalks. In the cells, BacA and BacB have been reported to form filaments or sheets close to the cell's inner membrane and over-expression deforms cells, making it possible that the filaments have intrinsic curvature and bind membranes. Some biochemical evidence has suggested that BacAB are peripheral membrane proteins3.
Other well-investigated bactofilins are BacM, N, O and P from Myxococcus xanthus. A bacM gene knock-out formed 'crooked' cells that have increased sensitivity to antibiotics5. BacNOP, in contrast, co-polymerise into filaments that recruit the ParABS chromosome segregation machinery to subpolar regions of the cells10. BacP has also been reported to be involved in type IV pilus localisation together with the GTPase SofG, both being important for the direction of motility of M. xanthus cells11.
Helicobacter pylori contains a single bactofilin called CcmA. A ccmA gene knock-out completely abolished the characteristic helical shape of the cells and a model has been put forward in which CcmA bactofilin filaments position lytic endopeptidases Csd1-3 in the periplasm, to remodel the shape-giving cell wall12,13. Bactofilins have also been described in Leptospira biflexa, with one of its bactofilin paralogues controlling the helical pitch of cells14.
Although not described as such then, an early sighting of a bactofilin was CcmA from Proteus mirabilis, where it is involved in cellular motility4, and a similar functional context was described for CcmA / bactofilin proteins in Vibrio parahaemolyticus15 and Bacillus subtilis16,17.
Clearly, more functional investigations will be needed and we hope to be able to facilitate this with the elucidation of the cryo-EM structure of the bactofilin filament from Thermus thermophilus. The filament is composed of domains stacked so that a continuous β-helical filament results. The subunits are arranged head-to-head, resulting in a filament that lacks polarity. This finding was confirmed by crystallography and co-evolutionary analysis. We show that the filaments bind directly to membranes in vitro and when heterologously expressed in E. coli cells, and that this interaction is enabled by a short conserved and hydrophobic motif in the N-terminal tail. Finally, we also show that polymerising bactofilins are not restricted to bacteria, with the visualisation of bactofilin filaments from Phytopthora infestans, an Oomycete eukaryote.
Results
Bactofilin domains are found across the tree of life
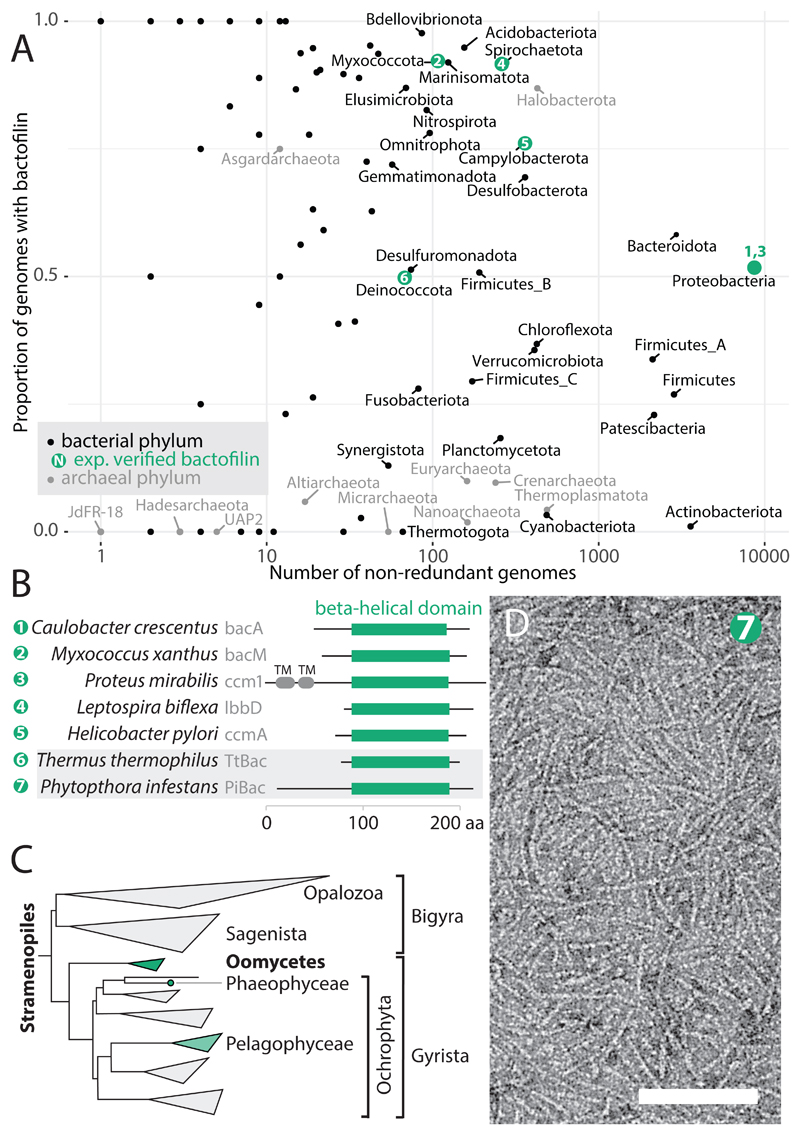
The presence of bactofilins has been reported and experimentally verified in several bacterial clades (Figures 1A and B). In order to assess further the distribution of the conserved bactofilin domain we searched for the presence of the conserved PF04519 / DUF 583 domain in a curated set of prokaryotic genomes18, annotated with a standardised phylogenomic taxonomy (GTDB v86) 19 (resulting tree: Supplementary Dataset D1). We found that bactofilin domains were present in 82 of the 114 phylum-level clades (Figure 1A). Thus, we conclude that bactofilins are very widely distributed within Bacteria. We also investigated the presence of bactofilin domains in the domain Archaea in the same way, finding that many archaeal genomes harbour bactofilin domains. Within the phylum Halobacterota more than 80% of genomes contained at least one bactofilin domain hit. It remains to be experimentally verified that these archaeal sequences encode polymerising bactofilin.
Figure 1. Bactofilins are highly conserved and widespread across bacteria and archaea, and also occur in some eukaryotic organisms.
A) Phylum level clades from the GTDB taxonomy v86 (Parks et al., 2018) of bacteria (black) and archaea (grey) are plotted as points with the number of non-redundant genomes in the clade against the proportion of genomes with at least one bactofilin hit. Bactofilin hits were HMMSEARCH results with PFAM PF04519, using an E-value cutoff of 1e-10 for bacteria, and 1e-6 for archaea, chosen so as to retain clusters of similar, convincing, sequences in each case. Pre-computed HMMSEARCH results were retrieved from the AnnoTree server (Mendler et al., 2018). Clades containing the validated bactofilins in (B) are shown in green, and numbered to correspond to (B). B) Domain schematic of selected experimentally investigated bactofilins. β helical domain (green) is shown as regions aligning to the β helical regions in C. crescentus BacA (Shi et al., 2015) and T. thermophilus TtBac structures. UniProt accessions for proteins shown, 1: Q9A753, 2: Q1CVJ5, 3: B4F0H9, 4: B0SPX3, 5: corresponds to GenBank CP001173.1 1607606:1607196 (Uniprot shows incorrect start codon see (Sycuro et al., 2010), 6: Q72HS6, 7: D0N980. C) A phylogenomic tree of the eukaryotic Stramenopile clade (Derelle et al., 2016). The subclades with genomes containing putative bactofilins are highlighted in green, the lighter shade of the Pelagophycaeae corresponds to a patchier distribution, see text. D) Negative stain electron micrograph showing filaments formed by recombinant bactofilin PiBac (PITG_07992) from the Oomycete Phytopthora infestans. The length of the scale bar is 100 nm.
While browsing the PFAM entry for the bactofilin domain PF04519 we noticed that several hits are listed within the domain Eukarya20. We found convincing bactofilin-like sequences in two taxonomic clusters, one within Stramenopiles (a deeply rooted eukaryotic clade, Figure 1C) and another within the Ascomycete fungi (Supplementary Dataset D2). We recombinantly expressed a putative bactofilin gene (Uniprot D0N980, PITG_07992, herein “PiBac”) from the Stramenopile plant pathogen Phytopthora infestans, a member of the Oomycete group, and found that it indeed forms bactofilin-like filaments (Figure 1D). Published data show that the mRNA encoding PiBac is expressed before and during spore formation (Supplementary Figure S1), and that the gene is conserved in context across the spore-forming oomycetes (Supplementary Figure S2) 21. Therefore, bactofilin domains retaining the ability to polymerise are conserved in eukaryotic genomes, are expressed, and likely play functional roles. The widespread distribution of polymerising bactofilin domains underscored the need for a fuller structural understanding of bactofilin polymers.
Bactofilin from Thermus thermophilus (TtBac) forms completely β-helical filaments
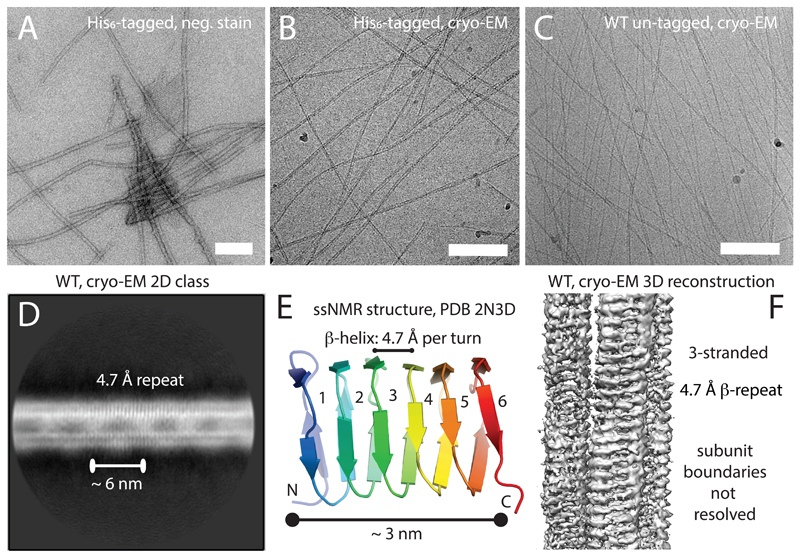
Bactofilin from Thermus thermophilus presented itself as a promising candidate for structural determination due to its short N- and C-terminal tails and the biological tractability of the source organism. Purification of recombinantly expressed His6-tagged TtBac (H6-TtBac, Supplementary Table T3) under denaturing conditions and subsequent refolding allowed visualisation of TtBac filaments by negative stain electron microscopy and confirmed the presence of a polymerising bactofilin in Thermus thermophilus (Figure 2A). Additionally, the H6-TtBac filaments recapitulated previously reported 2D sheets that seem to be an intrinsic consequence of fibrillar bactofilin assemblies.
Figure 2. Bactofilin from Thermus thermophilus forms filaments that show a continuous β-stacking repeat of 4.7 Å.
A) Hexahistidine-tagged and refolded bactofilin from Thermus thermophilus (H6-TtBac) forms filaments and 2D sheets as determined by negative stain electron microscopy (EM). B) The same protein shown using electron cryomicroscopy (cryo-EM). Note differing filament thicknesses caused by varying protofilament numbers. C) Untagged TtBac-WT protein expressed in E. coli and purified using a centrifugation protocol, imaged by cryo-EM, also showing protofilament numbers of 2-4. The length of the scale bar from A-C is 100 nm. D) 2D class average calculated using RELION (Scheres, 2012), showing a continuous repeat of 4.7 Å perpendicular to the filament axis. Note the upper strand shows stronger density, most likely because many filaments averaged have 3 protofilaments. Also note that there are fuzzy bridges visible between the protofilaments, roughly 6 nm apart. E) Ribbon representation of the Caulobacter cresentus BacA bactofilin structure determined by solid state NMR (ssNMR) (Shi et al., 2015). Taking D) into account, the structure suggests that bactofilin protofilaments are made by stacking the β-helical domains head-to-head and tail-to-tail. The 6 nm fuzzy bridges already suggest that the structure might not be polar as it is difficult to produce those repeats lengths from a 3 nm long β-helical domain otherwise. F) Preliminary RELION helical 3D reconstruction using the cryo-EM data (He and Scheres, 2017), revealing the structure of a 3-stranded TtBac filament. Because the individual domains show no features indicating their ends, the alignment does not converge and resolution remains low because the subunits cannot be registered correctly along the filament axis.
Visualisation by cryo-EM of both H6-TtBac and natively purified TtBac-WT filaments showed that the fibrils were prone to bundling and persisted unchanged over very wide ranges of pH and salt concentrations. Preparing the filaments at pH 11 allowed production of cryo-EM grids with largely un-bundled filaments that were amenable to structure determination but otherwise looked like filaments at more physiological pH values (Figures 2B & C).
Many 2D class averages showed two lines or protofilaments with twice as much signal in the top protofilament, suggesting that the majority of assemblies present in the grids were in fact made out of three protofilaments (Figure 2D). The class averages all showed strong vertical striations within the protofilaments, spaced 4.7 Å apart, the distance between β-strands in a sheet. This indicates that the bactofilin subunits arrange into continuous β-helical protofilaments, as suggested previously8,9. Surprisingly, the filaments also showed a weak ~ 6 nm repetitive feature visible between and in the protofilaments (Figure 2D), roughly twice the size of a BacA monomer6 (Figure 2E). This indicates that the repeating unit of the filament might be a head-to-head dimer making polarity within the filament impossible.
Subsequent 3D helical reconstruction of TtBac-WT filaments yielded a preliminary low-resolution structure that confirmed both the 3-strandedness and the β-helical nature of the subunits, but did not allow any atomistic description of the monomer (Figure 2F). The inability to reach higher resolution was mainly caused by the similarity between each β-helical winding, which impeded the correct positioning of the subunits along each protofilament.
A near atomic cryo-EM structure of the TtBac bactofilin filament bound to a nanobody shows it to be non-polar
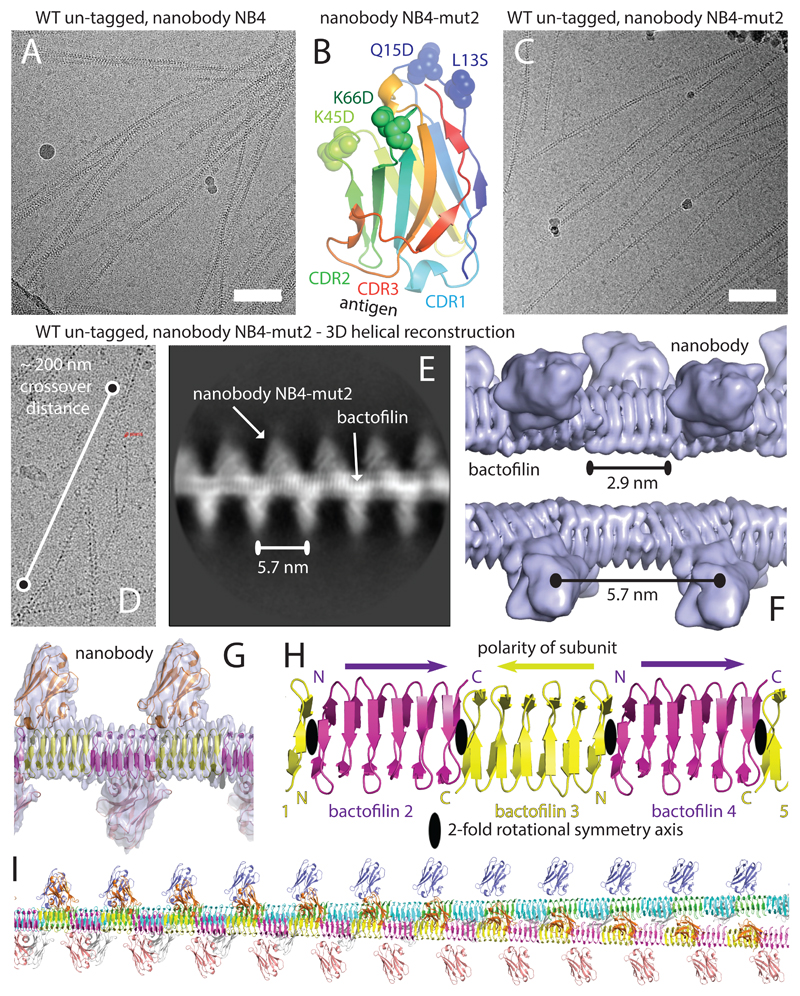
In an attempt to provide a solution to the computational problem of the subunit register along each protofilament, we raised Lama glama nanobodies against TtBac-WT. After selection of the suitable antibody clone NB4, cryo-EM of the wildtype filaments incubated with the anti-TtBac-WT nanobody showed fully decorated fibrils that formed superhelical structures unfit for structure determination (Figure 3A). As a result we designed 4 mutations at the opposite end from the CDR loops of the nanobody to counteract the bundling —L13S, Q15D, K45D, and K66D on NB4-mut2 — which almost completely abolished superhelicity (Figures 3B, C & D). Images were clear enough to directly derive approximate helical parameters for image reconstruction (Figure 3D). 2D class averages of the decorated filaments showed almost exclusively filaments with two protofilaments, a clear signal for each nanobody at a spacing of 5.7 nm and the two β-helical protofilaments (Figure 3E). The formation of higher order protofilament bundles is most likely prevented by the bulky presence of the nanobodies (cf. Figure 2F and 3F).
Figure 3. Cryo-EM structure of bactofilin from Thermus thermophilus (TtBac) shows a head-to-head non-polar filament.
A) In order to overcome the register problems highlighted in Figure 2D-E, a nanobody against TtBac was raised, purified and added to the filaments before cryo-EM imaging. The nanobody clearly binds but causes severe bundling. The length of the scale bar is 100 nm. B) Assuming that bundling is caused by the antigen-distal surface of the nanobody, four mutations were introduced, yielding NB4-mut2. C) Using the modified nanobody NB4-mut2 bundling is significantly reduced and cryo-EM images show many single filaments. The length of the scale bar is 100 nm. D) Estimation of the crossover distance (corresponding to a 180° twist of the filament) to over 2000 Å, indicating only very slowly twisting filaments of about 3 - 4 ° per 6 nm helical rise. E) Reference-free 2D class calculated using RELION, showing the altered appearance of the TtBac bactofilin filaments after addition of the NB4-mut2 nanobody (compare to Figure 2D). The nanobody densities appear at a repeat distance of 5.7 nm, clearly indicating a non-polar arrangement of the subunits in each protofilament. F) Preliminary low-resolution (5-6 Å) 3D reconstruction using RELION, showing clearly the continuous β-helical TtBac bactofilin filament pair and the nanobodies attached at 5.7 nm distances to each other. G) Final density with fitted (but not refined) atomic models at around 4 Å resolution. Correctness of the handedness of the structure was verified using the crystal structure fit of the nanobody atomic model. H) Ribbon representation of the arrangement of individual TtBac subunits in each protofilament. Subunits are arranged in a head-to-head-to-tail manner, with N- and C-termini coming together at alternating interfaces (N-C and C-C, leading to the 5.7 nm repeat of the nanobody binding). The two-fold axes at the N-N and C-C interfaces are nearly aligned, leading to a slowly twisting, overall helical filament. I) Atomic model of a longer stretch of the double-helical filament (see also Supplementary Movie M2). The entire double filament has additional 2-fold symmetry axes going between the two protofilaments.
Thus, by averaging ~346,000 helical segments and applying helical symmetry, the structure of TtBac-WT:NB4-mut2 filaments was solved to a nominal resolution of 3.4 Å (4.2 Å against atomic model, Figures 3F & G, Supplementary Table T1, Supplementary Figure S3, Supplementary Movie M1). Fitting the previously determined structure of BacA from C. crescentus6 revealed the close similarity between the two (Figure 3G). As in BacA, TtBac is made up of 6 right-handed windings of triangular parallel β-sheet structures with an exclusively hydrophobic core —a main contributor to the extreme stability of bactofilins— and two disordered terminal tails. Similar to the highly conserved glutamine and asparagine hydrogen bonds that give an increased sturdiness to amyloid fibres, bactofilins contain an outside bonding network comprised of glutamates and aspartates placed diametrically above or below lysines and arginines. The structural simplicity of the TtBac filaments means that amino acid composition alone reflects their three main characteristics: a hydrophobic core consisting of leucine, valine, or alanine (34 % of residues); a strong interaction network between surface charges residues, together comprising 28 % of amino acids; and a triangular arrangement of β-sheets with glycines (13 %) at each sharp corner.
Primarily, the cryo-EM structure provides insight into the filamentous arrangement of bactofilins, showing that the monomers are ordered in a non-polar fashion with alternating head-to-head (N-terminal to N-terminal, N-N) and tail-to-tail (C-terminal to C-terminal, C-C) interfaces (Figure 3H). This in turn produces 2-fold symmetry at each homoterminal interface. The interfaces themselves have a very small misalignment of their 2-fold axes, translating into a slight helical twist of 4.89° per dimer (Figure 3I, Supplementary Movie M2, Supplementary Figure S4).
The crystal structure of a TtBac polymerisation-impaired mutant confirms the non-polar filament architecture
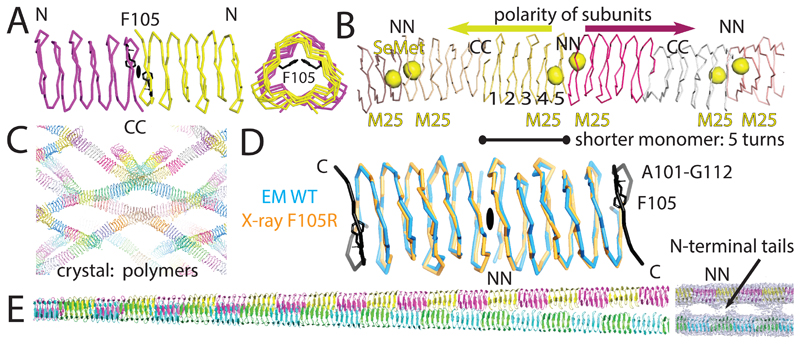
Although the filamentous structure converged to near-atomic resolution, the presence of anisotropy in the cryo-EM map, caused by the strong 4.7 Å repeat in a single direction made it difficult to confidently assert the exact conformation of amino acids and other characteristics of the structure. Therefore, in order to validate the cryo-EM reconstruction, we aimed to solve the structure of non-polymerising TtBac versions through X-ray crystallography. By introducing the mutation F105R9 into the C-terminal interface of TtBac we aimed to obstruct polymerisation at this interface while preserving the head-to-head N-N contacts and thus producing a dimer of TtBac(F105R) (Figure 4A). To further aid purification and crystallisation we added a C-terminal His6-tag and removed the first 10 N-terminal residues, which, according to the unresolved density in the 2D classes, seemed to be implicated in bundling (Figure 2D). Indeed, the easy handling of ΔN-TtBac(F105R) was in stark contrast to the difficult purification of TtBac-WT. Moreover, visualisation of the polymerisation-impaired and non-bundling mutant by negative stain EM showed a lack of filaments when tested at fairly high concentrations (~2 mg/ml; not shown).
Figure 4. A polymerisation-impaired TtBac mutant crystallises as non-polar filaments with reduced subunit length.
A) Based on previous studies (Zuckerman et al., 2015), F105 in TtBac was mutated to R in order to produce a version that does not polymerise under normal conditions. Normally, F105 interacts with itself across the C-C interface along the TtBac protofilament and changing this residue to charged arginine impairs normal filament formation. TtBac(F105R) can be purified easily, especially when the disordered N-terminal region is also removed. B) Crystal structure of ΔN-TtBac(F105R). The structure was solved by SeMet SAD phasing and the resulting signals of the single Met25 residue all lie very close to each other signalling the N-N interface and indicating non-polar head-to-head-to tail arrangement. C) The very large unit cell of the TtBac crystals contains several strands of filaments, formed from 32 molecules per asymmetric unit. D) The mutant crystallised as a distorted filament because the F105R mutation caused residues 101-112 to become disordered, allowing an entirely new C-C β-helical interface to form. This re-arrangement indicates the power of β-stacking interfaces as they are also occurring in amyloid structures, for example and that lead to the extraordinary stability of bactofilin filaments. E) Final atomic model of the double-helical TtBac bactofilin filament. Using subtraction (Bai et al., 2015), the nanobody density was removed and the atomic model refined against the crystal structure was used as guidance to produce a reliable atomic model that was real-space refined against the cryo-EM density. Note the position of the inter-protofilament density bridges, occurring at the locations of N-N interfaces, indicating that it is the N-terminal tails of TtBac that hold the two protofilaments together in double helical filaments.
The crystal structure of ΔN-TtBac(F105R) was solved by making use of the single N-terminal Met25 for selenomethionine (SeMet) SAD phasing (Figure 4B, Supplementary Table T2). In the solved structure, the un-mutated N-N interface was intact and head-to-head, as unequivocally shown by the pairs of anomalous selenium peaks. The map also showed that crystallisation occurred through formation of a C-C interface, leading to filaments in the crystals, with 32 bactofilin subunits per asymmetric unit of the crystals (Figure 4C). This unexpected polymer was formed by unwinding of the sixth β-helical turn, essentially flipping out the R105 residue that would otherwise have impaired polymerisation (Figure 4D). This not only suggests that F105R did indeed impair normal polymerisation but also serves to show the outstanding propensity of β-stacks to form, as for example in amyloid fibres.
Aside from confirming the lack of polarity within the protofilaments, the structure of ΔN-TtBac(F105R) also suggested that the formation of higher-order structures may well be caused by the N-terminal residues 1-10. The very large unit cell of the crystals contains well-dispersed filaments that do not form any type of doublets or bundles (Figure 4C).
The refined crystallographic model (Supplementary Table T2) was fitted into a new cryo-EM map generated by signal subtraction of the nanobody density22. We added the remaining residues and refined in reciprocal and real-space against the cryo-EM map to obtain a complete and reliable atomic model of the TtBac-WT bactofilin double filament (Figure 4E, Supplementary Table T1). The resulting map reaffirmed that the bridges between the protofilaments that make the double filaments are formed by N-terminal residues (Figure 4E, right).
Bactofilins form non-polar filaments as demonstrated by evolutionary coupling analysis
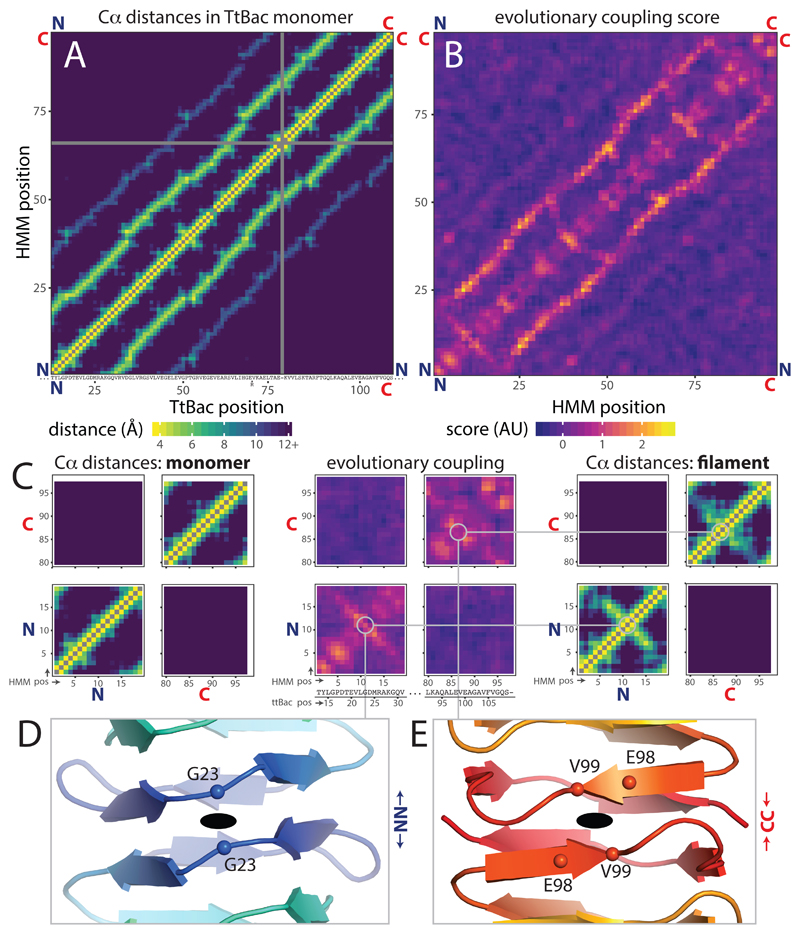
The high level of conservation between the monomer structures of TtBac and BacA (PDB ID 3N3D, RMSD 1.7 Å over 96 residues, sequence identity 35 %) suggested to us that the non-polar architecture seen for TtBac filaments may also be widely conserved. To investigate this, we aligned 12,646 putative bactofilin domain sequences and derived an evolutionary coupling score for each pair of residues in the alignment23. Visualisation of these scores as a heat map produced an overall view of co-evolution within the bactofilin sequence and was compared to calculated Cα distances in the TtBac monomer structure (Figures 5A & B). As shown by the second diagonal in Figure 5B, there is a high level of co-evolution, and high coupling scores, between amino acids ~17 residues apart, i.e. one turn of the β-helix, and a weaker line ~34 residues apart (two turns) is also visible. In other words, the β-helical structure of bactofilins, which enforces interactions between the residues exactly above and below, is conserved amongst a very large proportion of the aligned sequences.
Figure 5. Evolutionary sequence coupling analysis reveals conserved non-polar architecture of bactofilin filaments.
A) C alpha distance matrix for TtBac monomer structure within cryo-EM filament structure. The ~17 amino acid repeat becomes obvious. The TtBac sequence was aligned to PFAM HMM PF04519 using HMMALIGN, X-axis numbers are for TtBac sequence, insertions and gaps relative to HMM are indicated below axis, Y-axis is numbered in HMM coordinates. As are all other axes in the figure, except where noted. B) Evolutionary coupling scores calculated for an alignment of 12,646 bactofilin sequences. Scores were smoothed using a 3x3 Gaussian kernel. ~17 amino acid repeat is clear. C) Zoomed plots showing N-N, C-C and N-C interaction regions in distance matrix plot and evolutionary coupling matrix. (Left) zoom in to corners of (A). (Middle) zoom in to (B) with TtBac sequence added beneath. (Right) zoom as for A, but minimum distances between C alphas in filament model are plotted instead. Inter-subunit interactions appear in both the filament distance matrix and coupling matrix as diagonals going through the residues closest to the C2 axis. D) View along C2 axis at N-N inter-subunit interface of TtBac filament model. Chains are displayed in cartoon representation, coloured blue-red from N terminus to C terminus. C2 axis is shown as black oval. C alphas for residues closest to axes are shown as spheres and labelled. Grey lines connect panel to the relevant positions in C. E) As (D) for C2 axis at C-C inter-subunit interface.
As the strength of evolutionary coupling between amino acids is largely dictated by their physical distance, it is reassuring that the heat map of Cα distances for residues in the TtBac monomer in Figure 5A almost perfectly recapitulates the coupling scores observed in Figure 5B. However, the distances between TtBac residues in the monomer do not explain the co-evolution between homo-terminal amino acids observed in the lower left and upper right corners of the coupling heat map (Figures 5B and blown up in Figure 5C, middle). These scores can be explained, though, by taking into account the architecture of the TtBac filament as determined here (Figure 5C, right). In the TtBac filament it is precisely those residues with surprisingly high coupling scores that are brought closer together within the head-to-head and tail-to-tail polymer architecture. There is no sign of co-evolution corresponding to interaction between the C- and N-termini (top left/bottom right corners) as would be expected for a polar head-to-tail arrangement.
As the strong coupling scores exclusively indicate interaction between residues within each terminus (N-N and C-C, not N-C) it becomes clear that the non-polar arrangement of the TtBac filament must be highly conserved within bactofilins. The generality of the TtBac filament architecture extends to fine details such as minimal protofilament twist as the position of the orthogonal diagonal in the coupling heat map describing the homo-terminal interaction exactly matches the location of the two-fold symmetry seen in the cryo-EM structure (Figure 5D & E).
TtBac bactofilin binds to membranes in vitro and in situ, an activity facilitated by its N-terminal tail
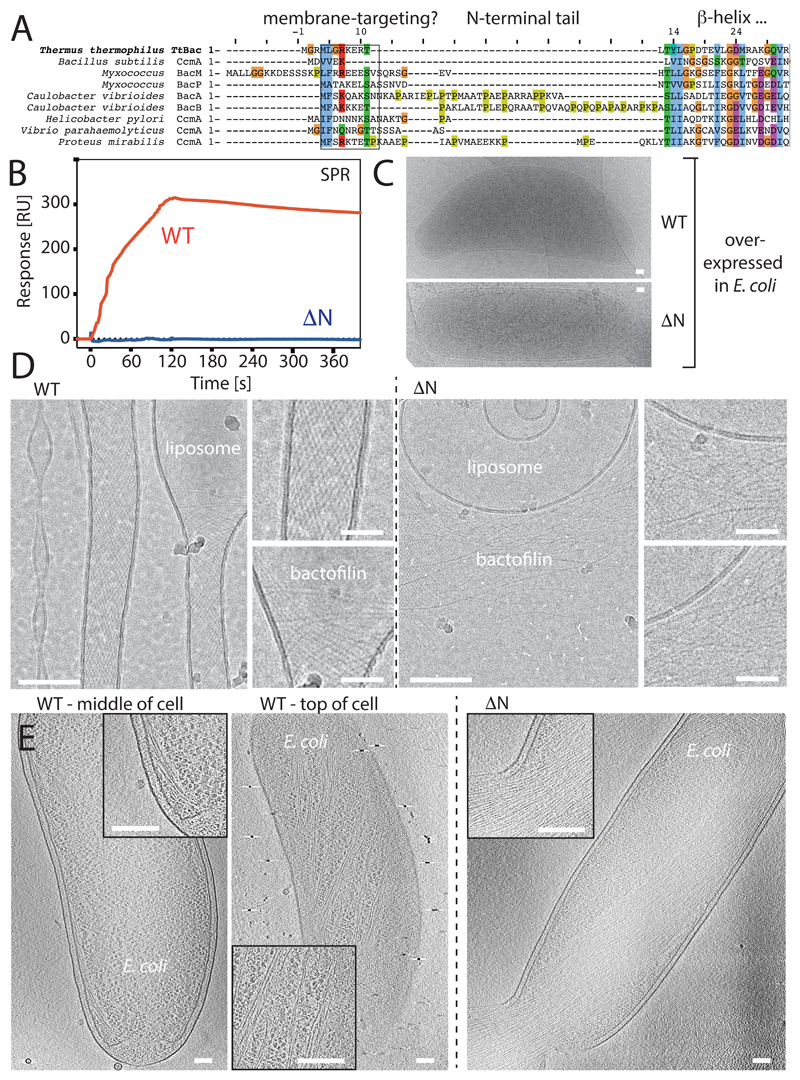
It has previously been shown that bactofilins are located close to membranes in cells in vivo3. Other filaments of prokaryotic cytoskeletons have been shown to directly interact with membranes, either through amphipathic helices or other small membrane targeting signals (MTS) 24–26. Because the N-terminal tail of TtBac was shown here to mediate interactions between the two protofilaments (Figure 4E), we hypothesised that it might contain hydrophobicity to interact directly with membranes. Indeed, when aligning a subset of well-characterised homologous bactofilin sequences, a conserved hydrophobic motif emerged at the very N-terminus of these sequences (Figure 6A).
Figure 6. TtBac bactofilin binds to lipid membranes through its N-terminal tail.
A) Multiple sequence alignment showing the N-terminal tails of bactofilins, including TtBac. A doublet of hydrophobic residues often followed by a positively charged residue is conserved across species. The fact that the N-terminal tails interact to form double helical filaments in our study (Figure 4E) and the occurance of unusual hydrophobicity in a disordered tail region prompted us to investigate membrane binding of TtBac in vitro and in situ. B) Surface Plasmon Resonance (SPR) traces of TtBac-WT and ΔN-TtBac proteins when added to liposome-coated L1 sensor chips (GE Healthcare Life Sciences). The unmodified protein clearly shows binding, whereas the mutant protein lacking the N-terminal tails shows no signal. C) When overexpressed in E. coli, TtBac leads to a morphological phenotype, it causes the cells to become crescent shaped and this effect disappears when over-expressing ΔN-TtBac under the same conditions. The length of the scale bar is 100 nm. D) When adding TtBac protein to pre-formed liposomes made from E. coli lipids, the protein was shown by subsequent cryo-EM imaging to deform and tubulate the liposomes. Note that single protofilaments appear to deform the liposomes from the outside (insets). When ΔN-TtBac was added to the same liposomes, no binding was observed and the liposomes were not deformed, consistent with the idea that the N-terminal tails of TtBac bactofilin, and presumably most bactofilins, are the membrane-targeting domains of these proteins. The length of the scale bars for the two large images is 100 nm and 25 nm for the four smaller ones. E) Investigating the overexpression system depicted in Figure 6C by electron cryotomography (cryo-ET). TtBac-WT (left two images, see also Supplementary Movie M3) clearly formed long bundles going around the cell close to the membrane, indicative of direct binding to the inner membrane of E. coli as no putative accessory protein were expected to be present. When doing the same experiment with ΔN-TtBac (right image, see also Supplementary Movie M4), a very large bundle of filaments was observed going through the centre of the cell, not close to the membrane, and impeding cell division. These results again indicated that it is the N-terminal tails of bactofilins that facilitate direct membrane binding. The length of the scale bars is 100 nm.
First we needed to show that TtBac does indeed bind to membranes directly. To investigate this hypothesis we used surface plasmon resonance (SPR) with liposomes (Figure 6B). TtBac-WT showed a strong signal, indicating liposome binding. Equally, when overexpressing TtBac in E. coli cells, we noted a strong phenotype of bent cells, in our experience a hallmark of filament formation directly on the inner membrane on one side of the cell's body (Figure 6C).
To demonstrate that the N-terminus of TtBac is necessary for membrane binding we produced an N-terminal deletion mutant ΔN-TtBac containing residues 11-123 (Supplementary Table T3). Both overexpression of the mutant in E. coli and incubation of the protein with liposomes and SPR showed that membrane binding and morphological changes were completely abolished (Figure 6B & C).
The binding of TtBac to membranes appears to include a demand for curvature, as seen by the morphological phenotype caused by overexpression of the full-length filamentous protein in E. coli (Figure 6C). Similarly, its interaction with liposomes in vitro is characterised by strong deformations or even tubulation (Figure 6D).
Finally, we also visualised TtBac-WT filaments in E. coli cells by cryo-ET, where they were visible as bundles circling the cell at an angle on or close to the inner membrane. The helicity confirms TtBac's preference for curvature. When doing the same experiment with ΔN-TtBac, all membrane proximity was abolished and the filaments formed one large bundle in the cytoplasm (that was so large that it also inhibited cell division) (Figure 6E, left and right, Supplementary Movies M3 & M4).
Discussion
We found bactofilins in eukaryotes, although, as far as we could tell, restricted to Stramenopiles and Ascomycete fungi. We believe that this may be the result of horizontal gene transfer from prokaryotes into these organisms27,28. It will be interesting to determine the function of bactofilins in eukaryotes as well as in archaea where they appear to be more widespread.
Our structural work showed bactofilin from Thermus thermophilus to be closely related to BacA from C. crescentus6; it polymerised into completely β-helical protofilaments that have head-to-head (N-N) and tail-to-tail (C-C) interfaces. This arrangement produces non-polar protofilaments and because the two-fold axes at the N-N and C-C interface are nearly aligned, the filament twists only very slightly, ~5° per dimer of ~6 nm length. The unanticipated lack of polarity8,9 makes the two ends of the filament equal and hence excludes all cytomotive mechanisms1. Bactofilins should therefore be classified as cytoskeletal, along with MreB, DivIVA, SepF and MinCD, all of which form non-polar filaments.
It is striking that the above list of cytoskeletal filaments contains exclusively cooperative filaments29 that bind directly to cell membranes from the inside. We showed here that bactofilins are no exception as they contain a conserved membrane targeting sequence (MTS) within their N-terminal tail.
From Figure 6D it is clear that the TtBac filaments form mostly single protofilaments when bound to membranes, indicating and confirming that the N-terminal tails function to bind membrane and not each other. We propose that the bona fide bactofilin filament is an almost straight, non-helical single protofilament, and that it is constitutively membrane bound (Supplementary Figure S6).
Both tomography and microscopy (Figure 6C, D & E) demonstrated that TtBac preferred curvature, another feature that it shares with MreB30, DivIVA31 and SepF32. Curvature preference seems to stem from the fact that they are not completely straight and this could also be a general mechanism of filament length restriction on flat, non-deformable membranes and/or curvature induction/sensing.
We firmly believe that bactofilins deserve greater attention as components of prokaryotic cytoskeletons. Armed with the analysis presented here demonstrating the widespread occurrence, biochemical stability, direct membrane binding, and non-polar filament architecture of these extraordinary proteins we hope to promote studies into the roles of bactofilins in the cell's biology. We anticipate further roles in morphogenesis and in processes where very stable membrane attachment is needed that cannot overlap with any of the other known systems. It also remains unclear how polymerisation, and with it membrane attachment, is regulated in cells and what role, if any, the C-terminal tails have.
Methods
Identification of bactofilins outside bacteria
We became interested in the possibility that bactofilins are present outside of the kingdom of bacteria after noting that PFAM (http://pfam.xfam.org) annotates several members of the PFAM 04519 (DUF 853, bactofilin) family in eukaryotes and archaea. This was investigated further by running a hmmsearch (HMMER3)33 with PFAM 04519 against the eukaryotic UniprotKB. The results included two taxonomic clusters of hits, one consisting of proteins found amongst the Stramenopiles, and the other of proteins within Ascomycete fungi. There were also hits in other eukaryotes, but these were either non-bactofilin repetitive sequences, or isolated hits within well-sequenced clades (in all these cases the best BLASTP matches were bacterial sequences). Clusters of hits within Stramenopiles and Ascomycetes, were aligned, HMMs were built and additional examples within UniprotKB were searched for. In both cases further examples were recovered from the relevant clades, but not outside them. To better understand the relationship between these putative eukaryotic bactofilins and the bacterial sequences, an alignment (using HMMALIGN) was made of putative bactofilin domains It comprised of bactofilins that are representative of sequence diversity within bacteria and archaea (found using the proGenomes set of representative prokaryotic genomes34 and HMMSEARCH with PFAM 04519), and all of the putative eukaryotic bactofilins, including the taxonomic singletons, and the additional sequences from the putative clusters. From the alignment a phylogeny was constructed using using the FastTree algorithm, after selecting informative columns using GBlocks35 (Supplementary Dataset D1). The phylogeny did not imply the existence of a common ancestor of eukaryotic bactofilins within eukaryotes. After inspecting alignments and trees three putative eukaryotic bactofilins were chosen to clone and express: one fungal protein (Q7RX79 from Neurospora crassa), and two Stramenopile proteins (D0N980 from Phytophthora infestans, and D7G0L1 from Ectocarpus siliculosis). Only D0N980 (PITG_07992) was successfully expressed and confirmed to be a bactofilin. It remains to be seen how many of the putative bactofilins outside the Oomycetes are polymerising bactofilins.
Protein cloning, expression and purification
The amino acid sequences of all proteins used in this study are listed in Supplementary Table T3.
PiBac Oomycetes bactofilin
To obtain a plasmid that codes for full length bactofilin protein from Phytophthora infestans (PiBac), the gene PITG_07992 (UniProtKB, D0N980) was synthesised (GenScript), amplified and cloned into the pHis17 plasmid using Gibson Assembly (New England Biolabs), with a stop codon before the C-terminal hexahistidine tag on the plasmid. C41(DE3) E. coli cells (Lucigen) were transformed with the resulting plasmid by electroporation. 40 ml 2xTY media supplemented with 100 μg/ml ampicillin were inoculated with a single colony from the plate, and were grown at 200 rpm, 37 °C overnight. The pre-culture was then used to inoculate 4 L 2xTY media with 100 μg/ml ampicillin. After reaching an OD600 of 0.6-1.0 at 200 rpm, 37 °C, the expression was induced with 1 mM isopropyl β-D-thiogalactoside for 4 h at the same temperature, and the cells were harvested by centrifugation. For purification, the entire pellet was resuspended in 150 ml Buffer D (50 mM Tris, 200 NaCl, 1 mM TCEP, pH 8) supplemented with DNase I, RNase A (Sigma) and EDTA-free protease inhibitor tablets (Roche). Cells were lysed by sonication, and the lysate was cleared by centrifugation at 10,000 rpm in a 45 Ti rotor (Beckman) for 30 minutes at 4 °C, followed by another centrifugation at 20,000 rpm in a 45 Ti rotor for 30 minutes at 4 °C. The supernatant was supplemented with 2% v/w PEG 8000 prior to centrifugation again at 40,000 rpm in the 45 Ti rotor for 30 minutes at 4 °C. Each pellet was resuspended in 5 ml Buffer D supplemented with 0.3 g/ml caesium chloride and 1 % v/v Triton X-100, and the pellet was solubilised overnight at 4 °C. The solubilised mixture was centrifuged in a TLA 100.3 rotor (Beckman) at 50,000 rpm for 15 minutes at 4 °C, the pellet was discarded and the supernatant was centrifuged in a TLA 100.3 rotor at 80,000 rpm for 5 hours at 4 °C. The white layer near the bottom of the centrifuge tube was extracted, resuspended in 3 ml Buffer D supplemented with 0.3 g/ml caesium chloride before centrifugation again in a TLA 100.3 rotor at 80,000 rpm overnight at 4 °C. The white layer containing filaments near the bottom of the centrifuge tube was extracted again and examined using negative stain electron microscopy (see below). Identity of the protein was confirmed by SDS-PAGE.
H6-TtBac 1-123
To obtain an E. coli expression plasmid for N-terminally His6-tagged full-length bactofilin protein from Thermus thermophilus (TtBac, MGSSHHHHHH-1-123), the gene coding for protein WP_011173792.1 (NCBI) was amplified from genomic DNA and was cloned into vector pET15b using Gibson Assembly (New England Biolabs). C41(DE3) E. coli cells (Lucigen) were transformed by electroporation. 60 ml 2xTY media supplemented with 100 μg/ml ampicillin were inoculated with a single colony from the plate, and were grown at 200 rpm, 37 °C overnight. The culture was then used to inoculate 6 L 2xTY media with 100 μg/ml ampicillin. After reaching an OD600 of 0.6-1.0 at 200 rpm, 37 °C, protein expression was induced with 1 mM isopropyl β-D-thiogalactoside for 4 h at the same temperature, and the cells were harvested by centrifugation. For purification, the entire pellet was resuspended in 200 ml Buffer A (50 mM Tris, 6 M guanidinium chloride, 1 mM TCEP, pH 7). Cells were disrupted at 25 kPSI in a cell disruptor (Constant Systems), and the lysate was cleared by centrifugation at 40,000 rpm in a 45 Ti rotor (Beckman) for 30 minutes at 25 °C. The cleared lysate was loaded onto a 5 ml HisTrap HP column (GE Healthcare), and was washed with stepwise increases of imidazole in Buffer A: 0, 20, 50, 200, 500 and 1000 mM. Eluted fractions was analysed by SDS-PAGE and those containing TtBac (mostly at 50 mM imidazole) were pooled and concentrated in Centriprep concentrators (10 kDa MWCO, Millipore) to 10 mg/ml. The H6-TtBac protein was refolded by a single step dialysis process into Buffer B (50 mM CAPS, 200 NaCl, 1 mM TCEP, pH 11) overnight at room temperature. The protein was stored at 4 °C.
TtBac-WT 1-123
To obtain an E. coli expression plasmid for untagged, full-length bactofilin protein from Thermus thermophilus (TtBac, 1-123), the gene coding for protein WP_011173792.1 (NCBI) was amplified from the above H6-TtBac pET15b plasmid and was cloned into plasmid pHis17 using Gibson Assembly (New England Biolabs) with a stop codon before the C-terminal tag on the plasmid. The plasmid encoding untagged TtBac was used to transform C41(DE3) E. coli cells (Lucigen) by electroporation. 40 ml 2xTY media supplemented with 100 μg/ml ampicillin were inoculated with a single colony from the plate, and were grown at 200 rpm, 37 °C overnight. The culture was then used to inoculate 4 L 2xTY media with 100 μg/ml ampicillin. After reaching an OD600 of 0.6-1.0 at 200 rpm, 37 °C, the expression was induced with 1 mM isopropyl β-D-thiogalactoside for 4 h at the same temperature, and the cells were harvested by centrifugation.
In order to isolate full length, unmodified, TtBac (TtBac-WT) we developed a protocol based on a series of gradient centrifugation steps adapted from previous work on Myxococcus xanthus BacM5. For purification, the entire pellet was resuspended in 150 Buffer B (50 mM CAPS, 200 NaCl, 1 mM TCEP, pH 11) supplemented with DNase I, RNase A (Sigma) and EDTA-free protease inhibitor tablets (Roche). Cells were lysed by sonication, and the lysate was cleared by centrifugation at 10,000 rpm in a 45 Ti rotor (Beckman) for 30 minutes at 4 °C, followed by another centrifugation at 20,000 rpm in the 45 Ti rotor for 30 minutes at 4 °C. The supernatant was supplemented with 2% v/w PEG 8000 prior to centrifugation at 40,000 rpm in the 45 Ti rotor for 30 minutes at 4 °C. Each pellet was resuspended in 5 ml Buffer B supplemented with 0.3 g/ml caesium chloride and 1 % v/v Triton X-100, and the pellet was solubilised overnight at 4 °C. The mixture was centrifuged in a TLA 100.3 rotor (Beckman) at 50,000 rpm for 15 minutes at 4 °C, the pellet was discarded and the supernatant was centrifuged in the TLA 100.3 rotor at 80,000 rpm for 5 hours at 4 °C. The white layer containing TtBac filaments near the bottom of the tube were extracted, resuspended in 3 ml Buffer B supplemented with 0.3 g/ml caesium chloride before centrifugation in the TLA 100.3 rotor at 80,000 rpm overnight at 4 °C. The layer containing TtBac filaments near the bottom of the centrifuge tube was extracted, and diluted with an equal volume of Buffer C (50 mM CAPS, 100 mM NaCl, 1 mM TCEP, 1 mM EDTA, pH 11) before being loaded onto a Superose 6 10/300 size exclusion column (GE Healthcare), equilibrated in Buffer C. The fractions containing TtBac filaments (in the void volume of the column) were pooled, and centrifuged in a TLA 100.3 rotor (Beckman) at 80,000 rpm for 30 minutes at 4 °C. The TtBac filaments formed a transparent pellet at the bottom of the tube, and were resuspended in Buffer C to 1-2 mg/ml and stored at 4 °C.
Nanobody NB4-mut2
Purified and refolded H6-TtBac filaments were sent to the Vlaams Instituut voor Biotechnologie’s Nanobody Core (VIB) for commercial camelid nanobody generation and selection. Frozen bacterial cultures containing individual nanobody genes in the pMECS vector were delivered by VIB. Genes encoding for each nanobody were amplified using colony PCR and cloned into the pHEN6c expression vector (VIB) using Gibson Assembly (New England Biolabs). The resulting constructs using pHEN6c encoded for the gene for PelB(leader)-nanobody-SSHHHHHH proteins.
For expression, the pHEN6c plasmids encoding for the nanobody proteins were used to transform WK6 E. coli cells (VIB, essential for this to work) by electroporation. 20 ml 2xTY media supplemented with 100 μg/ml ampicillin were inoculated with a single colony from the plate, and were grown at 200 rpm, 37 °C overnight. The culture was then used to inoculate 2 L 2xTY media with 100 μg/ml ampicillin. After reaching an OD600 of 0.6-1.0 at 200 rpm, 37 °C, the expression was induced with 1 mM isopropyl β-D-thiogalactoside for 4 h at the same temperature, and the cells were harvested by centrifugation.
The purification protocol followed recommendations from VIB. Since the plasmids encoded the gene for the PelB leader sequence, nanobody proteins were secreted into the periplasm. For purification, the entire pellet from 2 L culture was resuspended in 24 ml TES buffer (200 mM Tris, 500 mM sucrose, 0.5 mM EDTA, pH 8), with shaking at 4 °C for 1 hour. The mixture was supplemented with 36 ml TES/4 buffer (TES buffer diluted 4 times in water) and was mixed at 4 °C for 1 hour. The suspension was centrifuged in a 45 Ti rotor (Beckman) for 30 minutes at 40,000 rpm at 4 °C. The supernatant containing the periplasmic extract was sonicated before being loaded onto a 5 ml HisTrap HP column (GE Healthcare). The column was washed with stepwise increases of imidazole in buffer E (50 mM Tris, 200 mM NaCl, 1 mM TCEP, pH 8): 3, 200, 500 and 1000 mM. Elutions were analysed by SDS-PAGE and the fractions containing nanobodies (mostly at 200 mM imidazole) were pooled and concentrated in Centriprep concentrators (10 kDa MWCO, Millipore) to around 6 mg/ml. The concentrated proteins were buffer exchanged using a PD-10 desalting column (GE Healthcare) equilibrated with Buffer C (50 mM CAPS, 100 mM NaCl, 1 mM TCEP, 1 mM EDTA, pH 11) using the spin protocol.
To solve the bundling issue when nanobody proteins were added to bactofilin filaments, mutants were made to try to change selected surface amino acid residues into aspartic acids using nanobody 4 (NB4) as template, that had been selected by negative stain EM of TtBac and nanobody equimolar mixtures. The gene encoding NB4 mutant 2 (L13S, Q15D, K45D, K66D) was synthesised (GenScript), amplified and cloned into the pHEN6c expression vector. Nanobody mutant proteins were expressed and purified using the same protocol as the non-mutated nanobody proteins. The final concentration of NB4 mutant 2 was 6.8 mg/ml.
ΔN-TtBac(F105R)-H6
The required coding region was amplified from the H6-TtBac construct by PCR and was cloned into the plasmid pHis17 using Gibson Assembly (New England Biolabs), resulting in a C-terminal tag: GSHHHHHH. The point mutation was introduced by Q5 mutagenesis (NEB). The resulting plasmid was used to transform C41(DE3) E. coli cells (Lucigen) by electroporation. 12L cultures in 2xTY were inoculated and induced with 1 mM IPTG at an OD600 of 0.6 and further grown for 6 h at 37 °C. After harvesting of the cells, the pellet was dissolved in Buffer F (50 mM Tris/HCl, 200 mM NaCl, pH 7.5) and lysed by cell disruption at 35 kPSI (Constant Systems). The lysate was cleared by ultracentrifugation (2 h at 35,000 rpm in a 45 Ti rotor) and loaded onto a 5 ml HisTrap column. The bound fraction was eluted with 0.6 M imidazole, pH 7.0 and further purified by size exclusion on a Sephacryl S300 16/60 column (GE Healthcare) in Buffer G (20 mM CHES/NaOH, 250 mM NaCl, pH 9.5). Fractions were analysed by SDS-PAGE and the fractions containing ΔN-TtBac(F105R)-H6 were pooled and concentrated in Centriprep concentrators (10 kDa MWCO, Millipore) to around 12 mg/ml.
ΔN-TtBac 11-123
The required coding region was amplified from the H6-TtBac construct by PCR and was cloned into the plasmid pHis17 using Gibson Assembly (New England Biolabs), The plasmid was used to transform C41(DE3) E. coli cells (Lucigen) by electroporation. 12 L cultures in 2xTY were induced with 1 mM IPTG at an OD600 of 0.6 and further grown for 4 h at 37 °C. After harvesting of the cells, the cells were dissolved in the same buffer as for TtBac-WT 1-123 and spun for 30 min at 9k rpm in a Ti45 rotor (Beckman). The pellet was dissolved in the same buffer and from here onwards the same protocol as for TtBac-WT 1-123 was used.
Electron microscopy: negative stain
Continuous carbon grids were purchased from EMS (Electron Microscopy Sciences). After glow discharging, 3 µl of sample were applied, blotted and stained with fresh 2% uranyl acetate solution. Uranyl acetate was applied 1-3 times with wait times of up to 60 seconds. After air-drying, grids were imaged in an Thermo Fisher F20 electron microscope equipped with a Falcon 2 detector, operated at room temperature.
Electron microscopy: cryo-EM, helical reconstruction of TtBac-WT and with nanobody NB4-mut2
Initially, refolded and His6-tagged TtBac protein (H6-TtBac) was investigated using electron cryomicroscopy (cryo-EM) with the aim to obtain an atomic model of the filament. For this, 2 μl 60 μM H6-TtBac in Buffer C were added onto freshly glow-discharged Quantifoil Cu/Rh R2/2 holey carbon 200 mesh grids (Quantifoil). The grids were blotted for 2.5 s with a blotting force of -15, a drain time of 0.5 s, and were flash frozen in liquid nitrogen-cooled liquid ethane using an Thermo Fisher Vitrobot Mark IV. The Vitrobot chamber was set to 10 °C and 100 % humidity.
Grids were imaged at 300 kV using a Thermo Fisher Tecnai Polara G2 or Titan Krios microscope using a Falcon III detector (one dataset was collected at eBIC, Harwell, UK), using pixel sizes between 1.0 to 1.4 Å and average total doses of about 40 e-/Å2, distributed over 40-70 frames. Images were motion corrected using MOTIONCOR236 and CTF corrected using GCTF37. 2D classification analysis in RELION38 revealed that the filaments had varying widths, most likely caused by varying protofilament numbers that ranged from 2-4. We then switched to untagged and natively-purified TtBac-WT material, using the same vitrification and imaging conditions as above. The data showed very similar variations in protofilament number but it was possible to discern the 4.7 Å axial repeat caused by the β-stacking of the subunits in 2D classes obtained by processing in RELION, however, it proved impossible to go beyond 5 Å resolution when performing reconstructions in RELION, most likely because the filaments are smooth and the starts and ends of each subunit could not be determined during RELION's 3D refinement procedure. In order to overcome this issue, a nanobody was obtained, NB4, that showed very clear binding to the filaments as it changed their appearance and also led to a strong reduction in filaments with more than 2 protofilaments. Unfortunately, the filaments coated with NB4 tended to bundle heavily, impeding further analysis. Hence four mutations were introduced, L13S, Q15D, K45D and K66S, yielding NB4-mut2 nanobody that significantly reduced bundling and enabled image analysis and helical reconstruction of TtBac-WT bactofilin to near-atomic resolution. For the cryo-EM grid preparation with the nanobody, purified nanobody protein was added to TtBac filaments at a molar ratio of 1.2 : 1 (nanobody:TtBac). The final dataset used is summarised in Supplementary Table T1. It was collected on a Thermo Fisher Titan Krios microscope at 300 kV, using a Falcon III detector in electron counting mode with an estimated pixel size of 1.07 Å/pixel. 2130 good images were selected after MOTIONCOR2 and GTCF and from those around 456,000 helical segments were picked in RELION, 57 Å apart along the filament axis. 2D classification in RELION selected 346,000 good helical segments and these were used for helical reconstruction in RELION with helical parameters twist = 4.73° and rise = 57.48 Å39. Particle polishing and post-processing followed standard RELION procedures and led to a final map with a resolution determined from gold-standard two halves FSC calculations (FSC = 0.143) of 3.6 Å40. When inspecting the map this value hides that fact that the map has very anisotropic resolution, most likely caused by the dominant 4.7 Å β-stacking repeat along the filament axis and has poor resolution perpendicular to the filament axis. The previously determined B. subtilis BacA ssNMR structure (PDB 2N3D)6 was homology modelled into TtBac using the SWISSMODEL server and placed in the cryo-EM map, as was a SWISSMODEL of the NB4-mut2 nanobody. Placing these models produced excellent fit but the anisotropy of the map made atomic refinement difficult.
After the crystal structure of ΔN-TtBac(F105R)-H6 had been solved during the course of this study, a much better atomic model became available. And after performing signal substraction of the nanobody density from all 346,000 helical segment images in RELION22, an improved cryo-EM map of the two TtBac protofilaments was obtained using RELION with helical parameters twist = 4.89° and rise = 57.46 Å. Several cycles of manual model adjustment in MAIN and refinement with PHENIX.real_space_refine (with additional secondary structure restraints)41 produced the final model (Supplementary Table 1). Resolution was estimated to be 3.4 Å from half-map FSC analysis in RELION but FSC analysis against the atomic model yielded a lower value of 4.2 Å, caused by the strong resolution anisotropy of the map. The atomic coordinates have been deposited in the Protein Data Bank (PDB) with accession number 6RIB and the subtracted cryo-EM map has been deposited in EMDB with accession code EMD-4887.
Evolutionary sequence coupling analysis of filament formation
A large set of bactofilin sequences was obtained by searching the UniprotKB with the PFAM HMM PF04519 using HMMSEARCH with defaults. These 20,746 sequences were aligned to PF04519 using HMMALIGN. Inserts relative to the HMM were removed, and sequences with more than 5 % gaps relative to the HMM were discarded using the dcaTools package (gitlab.com/ducciomalinverni/dcaTools/), leaving 12,646 sequences. This alignment was used to infer a Direct Coupling Analysis (DCA) model of the bactofilin family using lbsDCA (gitlab.com/ducciomalinverni/lbsDCA/), an implementation of the asymmetric Pseudo-Likelihood Method for DCA23. lbsDCA reported an effective number of sequences after re-weighting with a 90 % identity cutoff of 5,770.4. The average-product corrected Frobenius norm DCA scores reported were scaled arbitrarily for plotting after smoothing of the heatmap with a 3x3 Gaussian kernel.
Crystal structure determination of ΔN-TtBac(F105R)-H6
ΔN-TtBac(F105R)-H6 protein was produced as described above and selenomethionine-substituted protein (SeMet) was obtained using published protocols42 with the same subsequent purification protocol as for the native protein, with the sole change that all buffers contained reducing agent TCEP at 1 mM. Initial crystallisation conditions were obtained using LMB's in-house high-throughput crystallisation facility43. All crystals were produced in MRC 2-drop crystallisation plates using 100 + 100 nl sitting drop setups and crystallisation experiments were performed at 19 °C. SeMet ΔN-TtBac(F105R)-H6 was crystallised using reservoir solution containing 6.7-7.3 v/v % 2-propanol, 0.17-0.19 M lithium sulfate, 0.1 M phosphate citrate, pH 3.7 and 0.4 M ammonium acetate as an additive. The crystals were cryo-cooled using 30% glycerol in reservoir solution. The crystals belonged to spacegroup I212121 and showed a very large unit cell (Supplementary Table T1). Three 360° datasets were collected at beamline I03 (Diamond Light Source, Harwell, UK) and merged together using CCP4 programs44 resulting in very high multiplicity and extending to about 4.0 Å resolution. 20-30 selenium sites were readily identified using SHELXD45 and subsequent phasing with PHASER46 and NCS averaging (operators deduced from SeMet sites within PHENIX) using DM resulted in interpretable electron density maps. It was determined that the number of recognisable subunits was 32 and both phasing and NCS averaging were adjusted to take this into account to obtain a final electron density map. The preliminary atomic model obtained from cryo-EM maps at 4-5 Å resolution was fitted manually, guided by the position of the single SeMet25 residue and its anomalous signal, and it was recognised that residues A101-G112 had become disordered, presumably caused by the F105R mutation used in the construct. Analysis then switched to the native protein. Native ΔN-TtBac(F105R)-H6 was crystallised using reservoir solutions as above and the crystals were cryo-cooled as above. The crystals again belonged to spacegroup I212121 with only slightly different cell constants. The crystals were isomorphous enough to enable simple rigid body refinement of all 32 chains since molecular replacement was not possible, presumably because of the smooth appearance of the protofilaments that makes it difficult to discerns the starts and ends of each subunit, very similar to the initial problems with cryo-EM image reconstruction. The structure was rebuilt manually in MAIN47 and refined with Phenix.refine48 (using NCS restraints and also real space refinement) for several cycles. Final statistics are summarised in Supplementary Table T1 and the resulting coordinates haven been deposited in the Protein Data Bank (PDB) with accession code 6RIA.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) data were collected using a BIAcore T200 instrument using a L1 Sensor Chip (GE Healthcare). Both reference and ligand channels were equilibrated in 50 mM CAPS, pH 8.0, 100 mM NaCl, 4 mM TCEP at 25 °C. Liposomes were prepared with E. coli Total Lipid Extract (Avanti Polar Lipids) using three freeze-thaw cycles Each cycle consists of two freeze-thaw events with liquid nitrogen followed by a single 10 minute water bath sonication. Liposomes at a lipid concentration of 3 mg/ml were captured onto the ligand surface at 10 µL/min to a level of ~ 1200 RU. To prevent non-specific binding the surfaces were passivated by injections of 0.2 mg/ml BSA with 1 mg/ml NSB (GE Healthcare Life Sciences) for 120 s prior to a 120 s injection at 30 µl/min of either TtBac-WT, ΔN-TtBac at 10 µM or buffer followed by dissociation for 300 s. After each measurement the surfaces were regenerated with a 30 s injection of 20 mM CHAPS. Data were doubly-referenced by subtraction of the reference channel data and from injections of buffer alone.
Bactofilin binding to liposomes
Liposomes were prepared with E. coli total lipid extract (Avanti Polar Lipids) using freeze-thaw cycles followed by sonication in buffer D (50 mM CAPS, 100 mM NaCl, 4 mM TCEP, pH 8.0). Images of liposomes together with native TtBac-WT and ΔN-TtBac were obtained by mixing 120 μM protein in buffer C with 2 mg/ml liposomes in buffer D. Of this mixture, 3 μl were applied onto freshly glow-discharged Quantifoil Cu/Rh R2/2 holey carbon 200 mesh grids (Quantifoil). The grids were blotted for 4 seconds with a blotting force of -15, a drain time of 0.5 s, and were flash frozen in liquid-nitrogen-cooled liquid ethane using an Vitrobot Mark IV (Thermo Fisher). The Vitrobot chamber was set to 10 °C and 100 % humidity. Grids were imaged in a Thermo Fisher F20 electron microscope equipped with a Falcon 2 detector, operated at cryogenic temperature.
Bactofilin overexpression in E. coli and electron tomography
Cells expressing TtBac-WT or ΔN-TtBac were mixed with 10 nm protein-A gold fiducials and plunge frozen on Quantifoil R2/2 holey carbon grids using a Vitrobot Mark IV (Thermo Fisher). Tomography data were collected on a Titan Krios electron microscope equipped with a Quantum imaging filter and K2 direct detector (both Gatan). Tilt series were acquired using SerialEM49 from 0° to ± 60° using a grouped dose symmetric tilt scheme50 with a 2° increment and a total dose of 160 e-/Å2. The pixel size was 5.44 Å, the target defocus 8 μm and the slit width of the energy filter 20 eV. Tomograms were reconstructed in IMOD51, using the SIRT algorithm.
Data availability
The data that support the findings of this study are available from the corresponding author, and we will provide all expression plasmids generated for this study, both upon request to Jan Löwe (jyl@mrc-lmb.cam.ac.uk). Atomic coordinates have been deposited in the Protein Data Bank with accession codes 6RIA and 6RIB. The cryo-EM volume has been deposited in the EMDB with accession code EMD-4887.
Statistics and reproducibility
The filaments shown in the micrograph displayed in Figure 1D were observed on all 23 images from 2 different grids. Similarly, the TtBac filaments shown in Figure 2A-C were observed ion almost all images in large data sets of more than 1000 images in total, from several grids. The nanobody-bound filaments were also seen in 5 sessions, with 2130 images collected for final 3D reconstruction. The SPR experiment shown in Figure 6B was run independently twice with similar results. The cells displayed in Figure 6C & E stem from a data set with 8 tomographic reconstructions each for the ΔN-TtBac and wild-type TtBac filaments and all show the same localisation of the filaments, either off or on the cell membrane, respectively. The binding and lack thereof of liposomes by TtBac and ΔN-TtBac filaments in Figure 7D was observed in more than 24 images and across 3 grids each. Regarding the data shown in Figure S1b, as per the original publication21, expression and differential expression calls were made with edgeR using TMM normalisation, a generalised linear model, and false discovery rate calculations based on the Benjamini-Hochberg method. The generalised linear model is two-sided.
Supplementary Material
Acknowledgements
We thank Howard S. Judelson (UC Riverside) for discussions regarding Oomycetes. We acknowledge Diamond Light Source for the cryo-EM facilities at the UK's national Electron Bio-imaging Centre (eBIC). We would like to thank Minmin Yu (MRC-LMB) for help with synchrotron data collection and staff at beamline I03 (Diamond Light Source, Harvell, UK). This work was funded by the Medical Research Council (U105184326 to JL) and the Wellcome Trust (202754/Z/16/Z to JL). James Wagstaff and Xian Deng were also supported by the Boehringer Ingelheim Fonds.
Footnotes
Author contributions
XD, AGL DKC, and JL performed protein purification experiments. XD, AGL, GC and JL performed cryo-EM data collection and image processing. AGL and JL performed crystallisation and crystallography. JW performed phylogeny and coupling analysis. VLH performed electron cryotomography experiments. SHLM and AG performed SPR experiments and analyses. AGL, JW and JL wrote the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Wagstaff J, Löwe J. Prokaryotic cytoskeletons: protein filaments organizing small cells. Nat Rev Microbiol. 2018;16:187–201. doi: 10.1038/nrmicro.2017.153. [DOI] [PubMed] [Google Scholar]
- 2.Lin L, Thanbichler M. Nucleotide-independent cytoskeletal scaffolds in bacteria. Cytoskeleton (Hoboken) 2013;70:409–423. doi: 10.1002/cm.21126. [DOI] [PubMed] [Google Scholar]
- 3.Kühn J, et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch MK, McHugh CA, Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi C, et al. Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR. Sci Adv. 2015;1:e1501087. doi: 10.1126/sciadv.1501087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kassem MM, Wang Y, Boomsma W, Lindorff-Larsen K. Structure of the Bacterial Cytoskeleton Protein Bactofilin by NMR Chemical Shifts and Sequence Variation. Biophys J. 2016;110:2342–2348. doi: 10.1016/j.bpj.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasa S, et al. β-Helical architecture of cytoskeletal bactofilin filaments revealed by solid-state NMR. Proc Natl Acad Sci U S A. 2015;112:E127–36. doi: 10.1073/pnas.1418450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuckerman DM, et al. The bactofilin cytoskeleton protein BacM of Myxococcus xanthus forms an extended β-sheet structure likely mediated by hydrophobic interactions. PLoS One. 2015;10:e0121074. doi: 10.1371/journal.pone.0121074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin L, Osorio Valeriano M, Harms A, Søgaard-Andersen L, Thanbichler M. Bactofilin-mediated organization of the ParABS chromosome segregation system in Myxococcus xanthus. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02015-z. 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulyha I, et al. Two small GTPases act in concert with the bactofilin cytoskeleton to regulate dynamic bacterial cell polarity. Dev Cell. 2013;25:119–131. doi: 10.1016/j.devcel.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Blair KM, et al. The Helicobacter pylori cell shape promoting protein Csd5 interacts with the cell wall, MurF, and the bacterial cytoskeleton. Mol Microbiol. 2018;110:114–127. doi: 10.1111/mmi.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sycuro LK, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson KM, Schwartz C, Wachter J, Rosa PA, Stewart PE. A widely conserved bacterial cytoskeletal component influences unique helical shape and motility of the spirochete Leptospira biflexa. Mol Microbiol. 2018;108:77–89. doi: 10.1111/mmi.13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79:240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Andari J, Altegoer F, Bange G, Graumann PL. Bacillus subtilis Bactofilins Are Essential for Flagellar Hook- and Filament Assembly and Dynamically Localize into Structures of Less than 100 nm Diameter underneath the Cell Membrane. PLoS One. 2015;10:e0141546. doi: 10.1371/journal.pone.0141546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopala SV, et al. The protein network of bacterial motility. Mol Syst Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendler K, Chen H, Parks DH, Hug LA, Doxey A. AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. BioRxiv. 2018;463455 doi: 10.1093/nar/gkz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parks DH, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 20.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ah-Fong AM, Kim KS, Judelson HS. RNA-seq of life stages of the oomycete Phytophthora infestans reveals dynamic changes in metabolic, signal transduction, and pathogenesis genes and a major role for calcium signaling in development. BMC Genomics. 2017;18:198. doi: 10.1186/s12864-017-3585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife. 2015;4 doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 24.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 25.Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci U S A. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarthy CG, Fitzpatrick DA. Systematic Search for Evidence of Interdomain Horizontal Gene Transfer from Prokaryotes to Oomycete Lineages. mSphere. 2016;1 doi: 10.1128/mSphere.00195-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16:1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 29.Ghosal D, Löwe J. Collaborative protein filaments. EMBO J. 2015;34:2312–2320. doi: 10.15252/embj.201591756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain S, et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. Elife. 2018;7 doi: 10.7554/eLife.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenarcic R, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gündoğdu ME, et al. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 2011;30:617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mende DR, et al. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 2017;45:D529–D534. doi: 10.1093/nar/gkw989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 36.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Ent F, Lockhart A, Kendrick-Jones J, Löwe J. Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Structure. 1999;7:1181–1187. doi: 10.1016/s0969-2126(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 43.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol. 2005;88:311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 46.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta Crystallogr D Biol Crystallogr. 2011;67:338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turk D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr D Biol Crystallogr. 2013;69:1342–1357. doi: 10.1107/S0907444913008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Hagen WJH, Wan W, Briggs JAG. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, and we will provide all expression plasmids generated for this study, both upon request to Jan Löwe (jyl@mrc-lmb.cam.ac.uk). Atomic coordinates have been deposited in the Protein Data Bank with accession codes 6RIA and 6RIB. The cryo-EM volume has been deposited in the EMDB with accession code EMD-4887.