Abstract
Background and Aims
Auxin response factors (ARFs) as transcription activators or repressors have important roles in plant growth and development, but knowledge about the functions of wheat ARF members is limited. A novel ARF member in wheat (Triticum aestivum), TaARF4, was identified, and its protein function, haplotype geographic distribution and allelic frequencies were investigated.
Methods
Tissue expression of TaARF4 was analysed by real-time PCR. Sub-cellular localization was performed using green fluorescent protein (GFP)-tagged TaARF4. Ectopic expression of TaARF4-A in arabidopsis was used to study its functions. Electrophoretic mobility shift assays (EMSAs), chromatin immunoprecipitation (ChIP) analyses and gene expression were performed to detect TaARF4 target genes. A dCAPS (derived cleaved amplified polymorphic sequence) marker developed from TaARF4-B was used to identify haplotypes and association analysis between haplotypes and agronomic traits.
Key Results
TaARF4-A was constitutively expressed and its protein was localized in the nucleus. Ectopic expression of TaARF4-A in arabidopsis caused abscisic acid (ABA) insensitivity, shorter primary root length and reduced plant height (PH). Through expression studies and ChIP assays, TaARF4-A was shown to regulate HB33 expression which negatively responded to ABA, and reduced root length and plant height by repressing expression of Gretchen Hagen 3 (GH3) genes that in turn upregulated indole-3-acetic acid content in arabidopsis. Association analysis showed that TaARF4-B was strongly associated with PH and root depth at the tillering, jointing and grain fill stages. Geographic distribution and allelic frequencies suggested that TaARF4-B haplotypes were selected in Chinese wheat breeding programmes. An amino acid change (threonine to alanine) at position 158 might be the cause of phenotype variation in accessions possessing different haplotypes.
Conclusions
Ectopic expression and association analysis indicate that TaARF4 may be involved in root length and plant height determination in wheat. This work is helpful for selection of wheat genotypes with optimal root and plant architecture.
Keywords: ABA, association analysis, auxin response factor, haplotypes, IAA, plant growth, root, Triticum aestivum
INTRODUCTION
Auxin, a plant hormone, plays important roles in plant growth and development (Guilfoyle, 2007; Guan et al., 2017; Luo et al., 2018). According to a model of the auxin-mediated signalling pathway in arabidopsis (Dharmasiri et al., 2005; Chapman and Estelle, 2009) at low auxin level, auxin response factor (ARF) binds to an auxin response element (AuxRE) together with Aux/IAA. Aux/IAA represses ARF activity and auxin-responsive gene expression. With high auxin concentrations, auxin receptor E3 ubiquitin ligases TIR1/AFB and Aux/IAA form a complex with auxin (Gray et al., 2001; Tan et al., 2007; Calderón Villalobos et al., 2012). The complex initiates degradation of the Aux/IAA proteins by the 26S proteasome, then de-represses ARF activity and enables auxin-responsive gene activation (Zenser et al., 2001; Kepinski and Leyser, 2005).
Previous studies demonstrated that ARFs as transcription factors could bind to TGTCTC-containing AuxREs in the promoters of auxin-responsive genes to activate or repress their expression (Ulmasov et al., 1997, 1999a; M. Zhang et al., 2017). ARF proteins are conserved, and most of them consist of three domains, i.e. an N-terminal DNA-binding domain (DBD), variable middle regions as activation (AD) or repression (RD) domains, and a C-terminal domain (CTD) (Tiwari et al., 2003; Korasick et al., 2014). The ARF DBD is a plant-specific B3 type that binds to AuxREs. Its nuclear magnetic resonance (NMR) solution structure has also been determined (Yamasaki et al., 2004). The variable middle regions determine whether it is a transcription activator or a repressor. The AD is enriched in glutamine (Q), serine (S) and leucine (L), whereas the RD is enriched in serine (S), proline (P), leucine (L) and/or glycine (G) (Chandler, 2016; Nanao et al., 2014). The CTD is a dimerization domain for combining with ARFs or Aux/IAA proteins (Kim et al., 1997; Ulmasov et al., 1999b).
The ARF family in arabidopsis contains 22 members and one pseudogene (ARF23) (Guilfoyle and Hagen, 2007; Remington et al., 2004), and rice contains 25 ARF members (Wang et al., 2007; Zhang et al., 2018). The functions of ARFs in arabidopsis were revealed mainly by study of arf mutants. AtARF2 has roles in seed size, fertility, senescence and hormone cross-talk (Okushima et al., 2005; Promchuea et al., 2017). AtARF3 functions in pattern development of floral meristems and reproductive organs (Sessions et al., 1997), while AtARF5 works in embryo axis formation and vascular tissue development (Hardtke and Berleth, 1998). AtARF7 is involved in aerial tissue growth (Harper et al., 2000), AtARF8 functions in fruit initiation (Goetz et al., 2006) and AtARF19 is active in hormone cross-talk (Li et al., 2006). Some AtARFs have redundant roles in plant development (Hardtke et al., 2004). However, research into ARF family members in wheat is lacking.
Wheat is a staple food crop worldwide, and its root and plant architectures strongly affect grain yield (Li et al., 2016). As a hexaploid species, wheat has a large and complex genome (AABBDD) that poses a challenge for gene discovery and determining gene function (International Wheat Genome Sequencing Consortium, 2014; Luo et al., 2017; Zhao et al., 2017; Appels et al., 2018; Ling et al., 2018). Association analysis is an efficient approach to reveal relationships between genes and traits (Bradbury et al., 2007; Wang et al., 2016). Pyramiding elite alleles through marker-assisted selection (MAS) greatly enhances the efficiency of wheat breeding (Collard and Mackill, 2008). Thus, finding elite alleles and developing functional markers are fundamental to genetic improvement of the crop.
In this study, three copies of TaARF4 were isolated from wheat and characterized. Ectopic expression of TaARF4 in arabidopsis resulted in reduced HB33 expression and response to abscisic acid (ABA). TaARF4-overexpressiong plants also had shorter primary roots and reduced plant height; it is proposed that this is due to repressing Gretchen Hagen 3 (GH3) gene expression that mediates indole-3-acetic acid (IAA) homeostasis. Association analysis showed that TaARF4-B was strongly associated with plant height (PH) and root depth of wheat. The geographic distribution and allelic frequencies demonstrated that TaARF4-B haplotypes were selected in the history of Chinese wheat breeding.
MATERIALS AND METHODS
Identification of TaARF4 and construction of a phylogenetic tree
Using the URGI website tBLASTn program and arabidopsis ARF2 N-terminal DBD as a query sequence, we isolated an ARF member (named TaARF4) with cDNA (AK335756.1) and protein (CDM85878.1) sequence accession numbers in GenBank; nothing was known about its function. The full-length cDNAs of the homoeologous genes TaARF4-A, TaARF4-B and TaARF4-D were cloned by reverse transcription–PCR (RT–PCR) from wheat cultivar Hanxuan 10. ARF family members were identified from different plant species by a protein BLAST search in the NCBI database. The full-length sequence was used to construct a phylogenetic tree by the maximum likelihood method through the Molecular Evolutionary Genetics Analysis (MEGA) software version 5.2.
Expression pattern analysis of TaARF4 in wheat
Hanxuan 10, a drought- and heat-tolerant common wheat cultivar, was used for genomic sequence isolation of TaARF4 and gene expression pattern analysis. Root and leaf tissues from 2-week-old seedlings and different tissues at the flowering stage (stem, leaf and spike, and root tissues from different depths) were sampled for spatio-temporal expression pattern analysis. Two-week-old wheat seedlings were sprayed with 50 μm ABA solution, then whole plants were sampled for ABA-induced expression analysis at 0, 1, 3, 6, 12, 24 and 48 h after treatment. Total RNAs were extracted using Trizol reagent (Invitrogen, 15596-018). cDNA was synthesized with a reverse transcription kit (TIANGEN, KR104). SYBR Premix Ex Taq (TaKaRa, DRR820A) was used for real-time PCR on an ABI QuantStudio® 7 Flex analyser according to the manufacturer’s instructions. Because the nucleotide sequences of TaARF4-A, TaARF4-B and TaARF4-D were highly conserved, it was difficult to distinguish them; therefore, a common primer pair for all three copies was designed by Primer Premier 5 software (Supplementary Data Table S1). Glyceraldehyde phosphate dehydrogenase (GAPDH) was used as the internal control. Three biological replicates and three technical replicates were assayed for each sample.
Sub-cellular localization of TaARF4 protein and production of TaARF4–GFP transgenic lines
The full-length cDNA of TaARF4-A was cloned into the modified vector pCAMBIA1300 at the XbaI and SpeI sites under control of the 35S promoter and with a green fluorescent protein (GFP) tag. Primers are listed in Supplementary Data Table S2. The constructs and empty vectors were separately transfected into Agrobacterium tumefaciens strain GV3101 by electroporation. For transient expression, Agrobacterium was cultured in 5 mL of YEB medium supplemented with 100 mg mL–1 rifampicin, 50 mg mL–1 kanamycin then infiltrated into the abaxial side of 5-week-old Nicotiana benthamiana leaves. The Agrobacterium-infected tobacco plants were grown in a greenhouse for 3 d before GFP fluorescence in leaves was observed with a confocal laser scanning microscope (Zeiss LSM700). At the same time, stable transgenic arabidopsis lines were produced by infection with Agrobacterium by the floral dip method (Clough and Bent, 1998). Transformants were selected on agar plates containing 50 μg mL–1 hygromycin B. GFP fluorescence in homozygous T3 generation plants was detected in 1-week-old transgenic plant roots by a Zeiss LSM700 microscope, and DAPI (4’,6-diamidino-2-phenylindole) was used to stain nuclei.
Purification of TaARF4-Δ protein and electrophoretic mobility shift assays (EMSAs)
The TaARF4-A N-terminus containing the DBD (TaARF4-Δ, amino acids 1–350) was fused with a glutathione S-transferase (GST) tag and expressed in Escherichia coli BL21 cells. The recombinant protein was induced by 0.2 mm isopropyl-β-d-thiogalactoside (IPTG), and the E. coli were incubated at 18 °C for 12 h. TaARF4-Δ protein was purified by glutathione–Sepharose 4B (GE Healthcare, 52-2303-00). P3 containing an inverted repeat of AuxRE was used as a probe. Cold (unlabelled) P3, cold mutated P3, biotin-labelled P3, biotin-labelled mutated P3 and their reverse complementary sequences were synthesized and annealed. EMSA was performed using a LightShift® Chemiluminescent EMSA Kit (Thermo Scientific, 20148) according to the manufacturer’s instructions. Briefly, probes and protein in binding buffer were incubated at room temperature for 20 min. The binding reaction mixes were separated in 5 % native polyacrylamide gels. DNA fragments in the gels were transferred to nitrocellulose membranes. After cross-linking, the membranes were incubated in blocking buffer, and then transferred to conjugate/blocking buffer. After washing, membranes were incubated in Substrate Equilibration Buffer, followed by Substrate Working Solution (Revzin, 1989). Finally, the membranes were photographed with a CCD camera.
Phenotypic analyses of transgenic arabidopsis
ABA sensitivity in seed germination.
Seeds of the wild type (Columbia, Col) and transgenic lines were sterilized and placed on Murashige and Skoog (MS) medium (Sigma-Aldrich, M5519) and MS medium supplemented with 0.5 μm ABA. After incubation for 2 d at 4 °C, the plates were transferred to a growth chamber with a 23 h light (21 °C)/1 h darkness (19 °C) cycle. Numbers of seedlings with cotyledon greening were scored after cultivation for 1 week.
Root length and plant height.
Seeds for root growth assays were sown on MS plates and grown in a chamber with 23 h light (21 °C)/1 h darkness (19 °C) for 5 d. Seedlings were then transferred to MS medium vertical culture with root tips placed at the same level. Photographs were taken after 7 d, and new growth parts of the primary roots were measured and analysed by Image J software. The growth rate for each genotype was calculated. At least 15 plants from three Petri dishes were measured for each experiment, and three independent biological experiments were performed. Two-month-old seedlings growing in a forest soil:vermiculite (1:1) mixture with 16 h light (21 °C)/8 h darkness (19 °C) in a greenhouse were used to assess plant height. At least 15 seedlings were measured from three pots for each experiment, and three independent biological experiments were performed.
Detection of gene expression
Total RNAs were extracted from 2-week-old Columbia and transgenic arabidopsis; cDNA synthesis and real-time PCR were performed as mentioned above. HB33, GH3.2 and GH3.5 gene expression patterns were determined using Tubulin as the control. Primers are listed in Supplementary Data Table S1.
Chromatin immunoprecipitation (ChIP) analyses
Chromatin immunoprecipitation was carried out on 2 g of 2-week-old transgenic arabidopsis plants (GFP and TaARF4–GFP) according to the method of Saleh et al. (2008). Briefly, after DNA and protein were cross-linked with 1 % formaldehyde, chromatin was isolated, and DNA was sheared on ice by sonicating five times for 15 s at 1 min intervals using an LTRASONIC PROCESSOR-500. Salmon sperm DNA/protein A agarose (Millipore, 16-157) and anti-GFP antibody (Abcam, AB290) were used to precipitate the DNA and protein complex. Then DNA and protein were reverse cross-linked, and DNA was precipitated. The precipitates were separately dissolved in 500 μL of TE to carry out real-time PCR using 5 μL of ChIP product for each one. Three independent biological experiments, each with three technical replicates, were performed. Primers are shown in Supplementary Data Table S3.
Free IAA extraction and analysis
Three independent replicates were performed. For each replicate, about 200 mg of 2-week-old fresh seedling tissue were ground in liquid nitrogen, weighed and IAA was extracted at –20 °C for 24 h with 2 mL of cold methanol. Antioxidant and internal standard (2H2-IAA, CDN Isotopes) were added and samples were purified using an Oasis MAX solid-phase extract cartridge (150 mg 6 mL–1; Waters), and analysed by a UPLC-MS/MS system (ACQUITY UPLC; Waters and Quattro Premier XE; Waters) as previously described (Wang et al., 2015). This part of the work was conducted on a plant hormone platform at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences.
Gene sequence polymorphism and functional marker development
Thirty-two wheat accessions with wide variation identified by simple sequence repeat (SSR) markers were chosen to sequence TaARF4 fragments for analysis of polymorphisms. Genome-specific primers were designed for amplification of TaARF4-A, TaARF4-B and TaARF4-D fragments from the A, B and D genomes (Supplementary Data Table S4). Purified PCR products were ligated into pEASY-Blunt vectors and transformed into E. coli top 10 competent cells. Positive clones were selected and sequenced with an ABI 3730 DNA Analyzer. Sequence polymorphism was analysed by SeqMan software. Derived cleaved amplified polymorphic sequence (dCAPS) primers for target genes were developed based on dCAPS Finder 2.0 software (http://helix.wustl.edu/dcaps/dcaps.html). The genotypes of the wheat accessions were identified as follows: first, TaARF4 genes in A, B and D genomes were amplified using genome-specific primers; second, 1 μL of PCR product as template was subjected to a second round of PCR; third, the second-round PCR products were digested by restriction endonucleases and electrophoresed in 4 % agarose gels.
Wheat populations used in the study
Five wheat germplasm populations were used for different research purposes. Accession names in the populations are listed in Supplementary Data Table S5. Population 1 consisted of 150 doubled haploid (DH) lines derived from the cross Hanxuan 10 × Lumai 14. This population was employed for gene mapping and analysis of the effects of different haplotypes. Population 2 consisted of 262 accessions mainly from the Northern Winter Wheat Zone and Yellow and Huai River Valleys Facultative Wheat Zone, and was used for association analysis between agronomic traits and genotypes. Population 3 consisted of 323 accessions from the same region and was used for association analysis between root traits and haplotypes. Population 4 consisting of 157 landraces and Population 5 consisting of 348 modern cultivars from all of ten major wheat zones of China were used to determine the temporal and spatial distributions of haplotypes.
Agronomic and root traits
Population 1 and Population 2 were planted at Shunyi (40°23′N, 116°56′E) and Changping (40°13′N, 116°13′E), Beijing, over 3 years (2010–2012) for measurement of plant height under two water regimes, drought-stressed (DS) and well-watered (WW). The amounts of rainfall during the three growing seasons were 131, 180 and 158 mm, respectively. DS plots were rain-fed, while WW plots were irrigated with 750 m3 ha–1 (75 mm) at the pre-overwintering, booting, flowering and grain fill stages when the amounts of rainfall were insufficient during each corresponding period. A greenhouse covered with polythene at the flowering stage to increase temperature and simulate heat stress (HS) was used at Shunyi. Seven agronomic traits, i.e. PH, length of penultimate internode (LPI), spike length (SL), 1000 grain weight (TGW), number of spikes per plant (NSP), number of spikelets per spike (NSS) and numbers of grain per spike (NGS), were measured under both water regimes, with and without heat stress. Population 3 was planted in PVC tubes, and root phenotypes were recorded at the seedling, tillering, jointing and grain fill stages. Population 5 for investigation of agronomic traits was planted at Luoyang (34°61′N, 112°45′E) in Henan province in 2002 and 2005, and at Shunyi, Beijing in 2010.
Association analysis
Population structure was examined by software Structure v2.3.2 (B. Zhang et al., 2017). Association analysis was performed by the mixed linear model (MLM) in TASSEL 5, in which population structure parameter Q was used. Associations at P < 0.05 were considered significant. Statistical analyses were conducted using SPSS 16.0 software.
Phosphorylated site prediction
TaARF4 protein sequences were submitted to the NetPhos 3.1 Server website (http://www.cbs.dtu.dk/services/NetPhos/). Serine, threonine or tyrosine phosphorylation sites were predicted, and both generic and kinase-specific predictions were performed.
RESULTS
Cloning and structural analysis of TaARF4 genes
Three full-length cDNA sequences of TaARF4 were cloned from wheat cultivar Hanxuan 10, and named TaARF4-A, TaARF4-B and TaARF4-D according to their genomic origins. The three TaARF4 genes were highly similar in sequence, encoding 797, 796 and 796 amino acids, respectively, and very similar in protein structure identity at 99.08 %. Like other ARF family members, TaARF4s consisted of a DBD at the N-terminus, an RD that is usually enriched in serine (S), proline (P), leucine (L) and/or glycine (G) (SPL-RD) in the middle region, and a CTD (Supplementary Data Fig. S1A). The phylogenetic tree (Supplementary Data Fig. S1B) showed that the TaARF4s were more similar to ARF4s in monocotyledons than those in arabidopsis.
Expression pattern of TaARF4 in wheat tissues
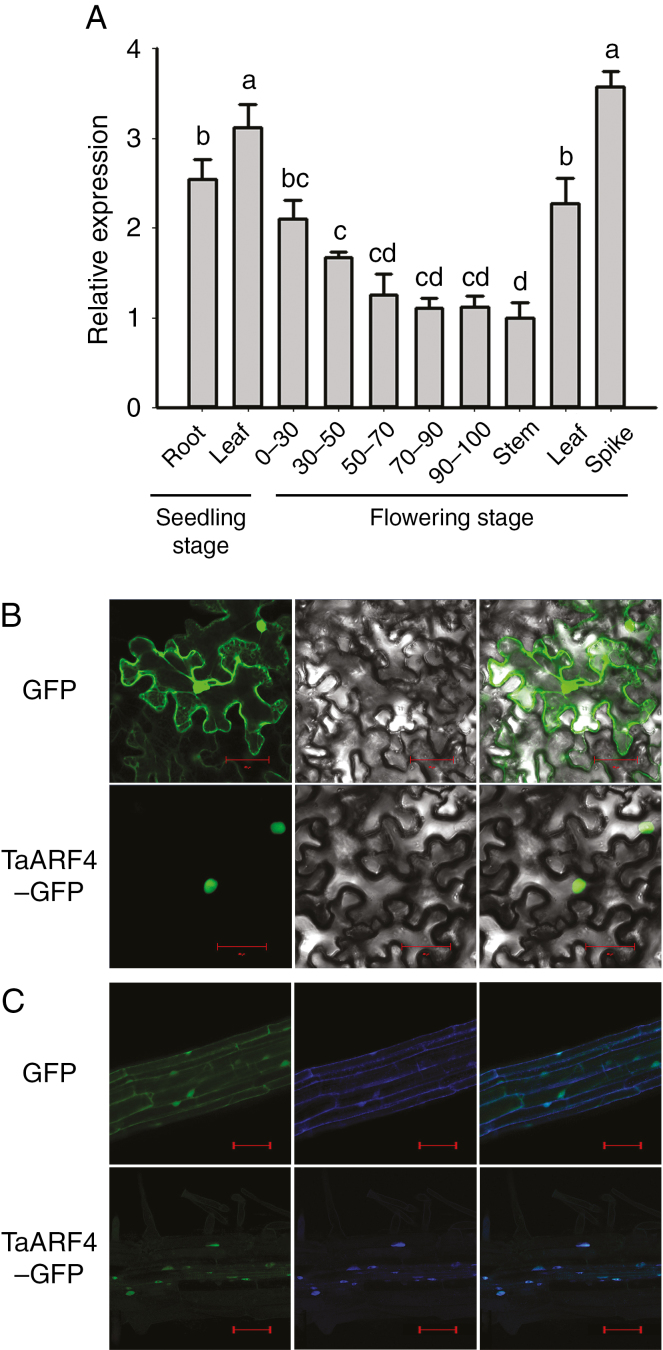
Real-time PCR was performed to identify expression patterns of the TaARF4 genes. TaARF4 was constitutively expressed in wheat tissues (roots and leaves of seedlings; roots, stems, leaves and spikes at flowering). The highest expression level was detected in the spikes (Fig. 1A), and in roots expression levels were lower at greater depths. Constitutive expression of TaARF4 genes suggested that they might have roles in growth of all tissues.
Fig. 1.
Expression patterns of TaARF4 and sub-cellular localization of TaARF4 protein. (A) Tissue expression patterns of TaARF4 were detected by real-time PCR. GAPDH was used as the control. Three independent biological experiments, each with three technical replicates, were carried out. Error bars are 2 × s.e. ‘0–30’ indicates the root section from the ground to 30 cm depth at flowering stage; ‘30–50’ indicates the root section from 30 to 50 cm; ‘50–70’ indicates the root section from 50 to 70 cm; ‘70–90’ indicates the root section from 70 to 90 cm; and ‘90–100’ indicates the root section from 90 to 100 cm. Significant differences were calculated based on ANOVA. (B) GFP and TaARF4–GFP were transiently expressed in transfected tobacco leaf cells. Green fluorescence images are on the left, bright field images are in the middle and merged images are on the right. Scale bars = 20 μm. (C) GFP and TaARF4–GFP were stably expressed in transgenic arabidopsis roots. Green fluorescence images are on the left, DAPI-stained images are in the middle and merged images are on the right. Scale bars = 100 μm.
Subcellular localization of TaARF4 protein
Fluorescence was detected in all cell parts of tobacco epidermal leaf cells and root cells of T3 transgenic arabidopsis lines with the empty vector carrying the GFP gene, whereas TaARF4–GFP was specifically distributed in the nucleus (Fig. 1B, C).
TaARF4 protein binds to the AuxRE cis-element in vitro
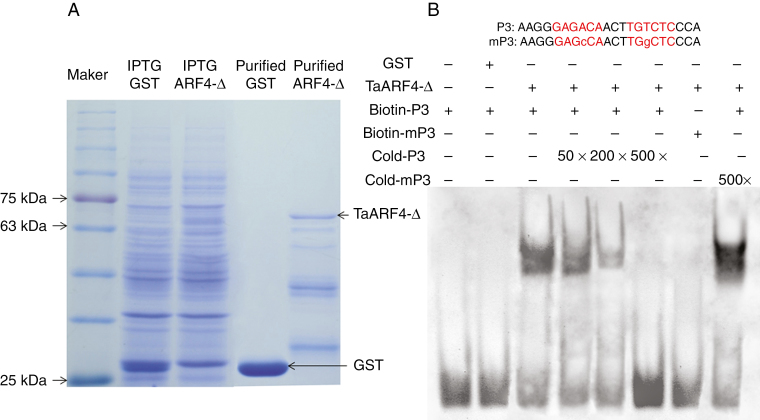
The promoters of auxin response genes share a consensus sequence (TGTCTC) known as the AuxRE. EMSAs were performed to test whether TaARF4 binds to AuxRE cis-acting elements in vitro. Since we could not purify the entire TaARF4 protein, the TaARF4 N-terminus with the DBD (TaARF4-Δ, amino acids 1–350) of the corresponding cDNA was cloned into a pGEX-4T1 vector (GST tag). Expression and purification of protein were carried out using E. coli BL21 cultures (Fig. 2A). A DNA-binding band detected with addition of TaARF4-Δ and biotin-labelled P3 probes had slower migration compared with the free probe (Fig. 2B, lane 3), whereas TaARF4-Δ could not bind to biotin-labelled mutated P3 probes (Fig. 2B, lane 7), and no target band was visible in the GST control (Fig. 2B, lane 2). Along with the gradual increase in cold P3 probe concentrations, the biotin-labelled DNA-binding band was diminished (Fig. 2B, lanes 4–6). The cold mutated P3 (mP3) probe did not compete with labelled P3 probes (Fig. 2B, lane 8). These results showed that the TaARF4 N-terminal DBD binds to AuxRE in vitro.
Fig. 2.
Expression and purification of TaARF4-Δ protein and electrophoretic mobility shift assays (EMSAs). (A) Expression and purification of TaARF4-Δ protein in E. coli strain BL21. Purified GST and TaARF4-Δ protein are marked by arrows. Lane 1 is a pre-stained protein marker. Lane 2 contains the entire E. coli protein suspension that was transfected by an empty pGEX-4T1 vector after IPTG induction. Lane 3 contains an entire E. coli protein suspension, which was transfected by pGEX-4T1 vector fused with TaARF4-Δ after IPTG induction. Lane 4 contains purified GST protein, and lane 5 contains purified TaARF4-Δ. (B) EMSA of purified TaARF4-Δ protein and P3 probe. Probe P3 and mutant P3 (mP3) sequences are shown above the lanes. The cis-element regions are shown in red script. Lower case letters indicate mutated bases. Lane 1 contains only biotin-labelled P3 probe. Lane 2 contains biotin-labelled P3 probe and GST protein. Lane 3 contains biotin-labelled P3 probe and TaARF4-Δ protein. Lane 4 contains biotin-labelled P3 probe and TaARF4-Δ protein with 50× cold (unlabelled) P3 probe. Lane 5 contains biotin-labelled P3 probe and TaARF4-Δ protein with 200× cold P3 probe. Lane 6 contains biotin-labelled P3 probe and TaARF4-Δ protein with 500× cold P3 probe. Lane 7 contains biotin-labelled mP3 probe and TaARF4-Δ protein. Lane 8 contains biotin-labelled P3 probe and TaARF4-Δ protein with 500× cold mP3 probe. Upper bands show TaARF4-Δ protein bound to biotin-labelled P3 probe; lower bands show free probe.
Overexpression of TaARF4 enhances ABA resistance in arabidopsis
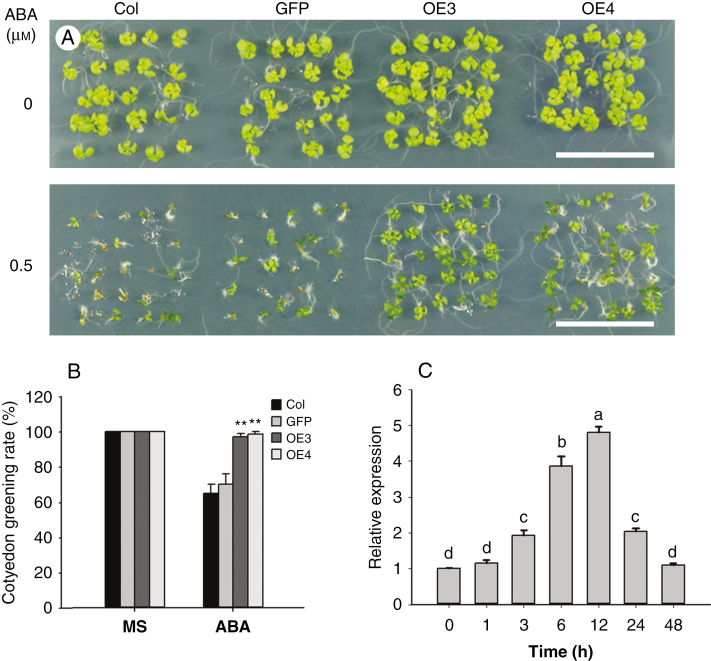
TaARF4-A, TaARF4-B and TaARF4-D have highly similar sequences, therefore TaARF4-A was chosen to represent all three TaARF4 genes for characterization of functions. TaARF4-A in a vector in which its expression was driven by the 35S promoter was transformed into arabidopsis. The Col and empty vector line (GFP) were used as controls. All six transgenic lines showed higher TaARF4 expression levels than the controls (Supplementary Data Fig. S2). We selected transgenic lines OE3 and OE4 to investigate stay green in cotyledons in response to ABA. Under normal conditions (MS medium), there was no significant difference between the controls and TaARF4 overexpression lines. Following culture on MS medium with 0.5 μm ABA for 7 d, the cotyledon greening ratio was about 67 % for the controls, compared with about 97 % for the two overexpression lines (Fig. 3A, B). We also detected the expression level of TaARF4 under ABA treatment. As shown in Fig. 3C, the transcription level of TaARF4 was upregulated by ABA and reached its highest level on exposure to ABA for 12 h, a level which was about 5-fold that of the non-treated control. At 48 h, the expression level declined to a similar level to that of the control. These results indicated that overexpression of TaARF4 led to resistance to ABA, and that TaARF4 negatively regulates ABA response in arabidopsis.
Fig. 3.
Overexpression of TaARF4 enhances arabidopsis resistance to ABA. (A) Cotyledon greening of Columbia (Col), GFP and two TaARF4-OE lines on MS or MS supplemented with 0.5 μm ABA. After sterilization, seeds were imbibed on MS medium plates with or without ABA and cultured in a growth chamber for 7 d. Scale bar = 2 cm. (B) Statistical analyses of cotyledon greening rates. There were three independent experiments, each with three replicates (in different plates), and in each replicate at least 25 seeds were counted. Error bars are 2× s.e. *P < 0.05, **P < 0.01 (Student’s t-test). (C) Expression patterns of TaARF4 following ABA treatment were detected by real-time PCR. Two-week-old wheat seedlings were sprayed with 50 μm ABA solution, and whole plants were harvested at 0, 1, 3, 6, 12, 24 and 48 h after treatment. GAPDH was used as the control. Three independent biological experiments, each with three technical replicates, were carried out. Error bars are 2 × s.e. Significant differences were calculated based on ANOVA.
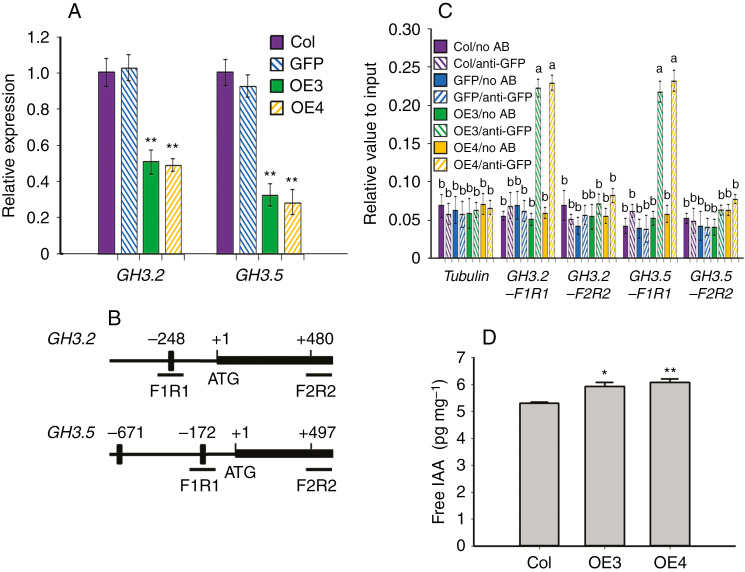
TaARF4 takes part in ABA response by binding to the HB33 promoter and regulating its expression
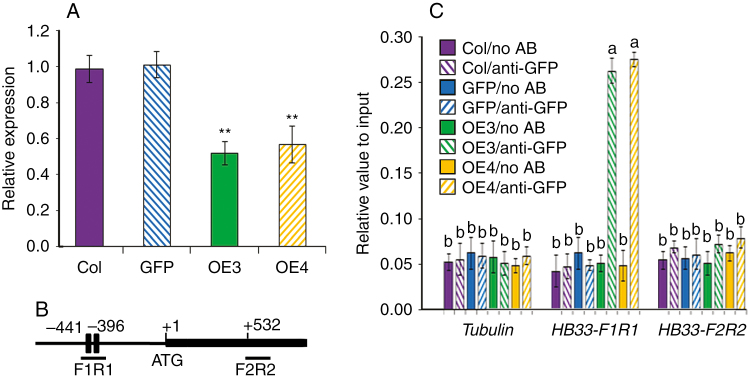
It was reported that lots of homeobox gene family members took part in abiotic stress response and one member HB33 had an important role in ABA response (Söderman et al., 1996; Tan and Irish, 2006; Wang et al., 2011; Bhattacharjee et al., 2016). Therefore, we analysed HB33 expression in controls and TaARF4-OE transgenic plants. Expression was lower in TaARF4-OE plants than in the controls (Fig. 4A). There are two AuxREs in the HB33 promoter region, one at –441 bp in the forward direction and the other in the reverse direction at –396 bp (Fig. 4B). ChIP assays were carried out to determine whether TaARF4 could directly bind to the HB33 promoter regions in vivo. GFP antibody was used for precipitations of protein and DNA. DNA abundance relative to the input was detected by real-time PCR. As shown in Fig. 4C, TaARF4–GFP bound to the HB33 promoter region, where there are two AuxRE cis-elements (HB33-F1R1), but could not bind to the Tubulin gene promoter region and HB33 coding region (HB33-F2R2) that lack AuxRE. These results indicated that TaARF4 can take part in the ABA pathway by binding to the HB33 promoter region to regulate HB33 expression.
Fig. 4.
HB33 expression in TaARF4-OE plants and ChIP assay on the promoter of HB33. (A) Expression of HB33 was negatively regulated by TaARF4. Expression of HB33 was detected in 2-week-old seedlings of Col, GFP and two TaARF4-OE plants. Three biologically independent experiments, each with three technical replicates, were performed. Error bars are 2 × s.e. *P < 0.05, **P < 0.01 (Student’s t-test). (B) Bars indicate putative TaARF4-binding sites of HB33. The ATG translation start site is indicated at position +1. Primers used for ChIP PCR are listed in Supplementary Data Table 3. (C) ChIP assay of TaARF4 binding to the promoter of HB33. Col and empty GFP seedlings were used as negative controls. Two transgenic lines (TaARF4-GFP-OE3 and TaARF4-GFP-OE4) and GFP antibody were used for the ChIP assay. Real-time PCR was carried out to show DNA abundance relative to input. Three biological replicates were performed, and each biological replicate had three technical replicates. Error bars are 2 × s.e. Significant differences were calculated based on ANOVA.
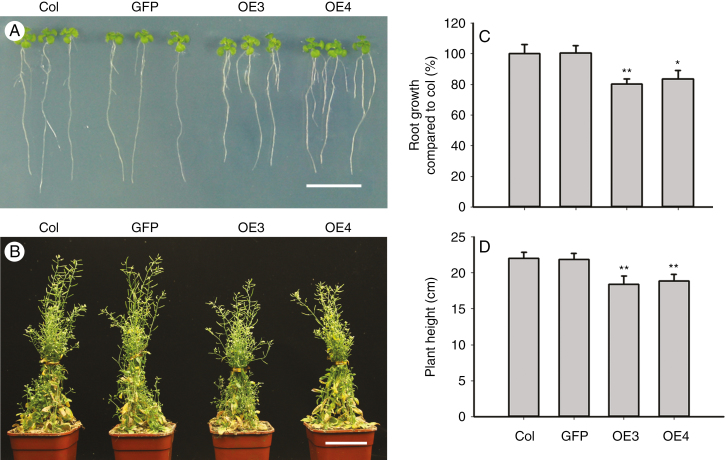
Overexpression of TaARF4 in arabidopsis led to shorter primary roots and plant height
Since many arf mutants were reported to affect plant development, we investigated the morphological characteristics of TaARF4-overexpressing arabidopsis plants. When 5-day-old seedlings overexpressing TaARF4 were transferred to MS medium in vertical orientation for 7 d, they developed shorter primary roots and longer lateral roots than Col and transgenic GFP controls (Fig. 5A). Primary root growth of the transgenic lines was reduced to about 82 % of that of the controls (Fig. 5C). The PHs of 2-month-old transgenic lines planted in soil were about 85 % of that of the controls (Fig. 5B, D). Collectively, these data indicate that TaARF4 has roles in root growth and PH.
Fig. 5.
Overexpression of TaARF4 in arabidopsis changes root architecture and plant height. (A) Overexpression of TaARF4 produces shorter primary roots and longer lateral roots than the control. Five-day-old seedlings were transferred to vertical culture and photographed after 7 d. Scale bar = 1 cm. (B) Overexpression of TaARF4 in arabidopsis caused shorter plant height than the control. Two-month-old plants growing in soil were used to compare plant heights. (C) Relative primary root growth of seedlings of GFP, OE3 and OE4 compared with Col. Three independent experiments were performed and at least 20 seedlings were measured in each experiment. Scale bar = 5 cm. Error bars are 2 × s.e. (D) Statistical analyses of plant height. Three independent experiments; in each experiment at least 20 plants were counted. Error bars are 2 × s.e. *P < 0.05, **P < 0.01 (Student’s t-test).
TaARF4 binds to the GH3 promoters, and regulates GH3 gene expression and IAA homeostasis in arabidopsis
Three kinds of genes respond to auxin, namely Aux/IAA, SAUR and GH3 (Nemhauser et al., 2006). The GH3 gene was first discovered as an auxin response gene in Glycine max (Hagen et al., 1984). It was shown to conjugate various amino acids to jasmonic acid (JA) and auxin, facilitating hormone activation, storage or transport, and helping to maintain hormone homeostasis (Staswick et al., 2005). However, the relationship of TaARF4 and GH3 is not clear. We therefore checked GH3 gene expression and IAA homeostasis of TaARF4-overexpressing arabidopsis plants. Expression levels of GH3.2 and GH3.5 were lower in TaARF4-OE plants compared with the controls (Fig. 6A). In the GH3.2 promoter region there is one AuxRE at –248 bp; and in the GH3.5 promoter region, there are two AuxREs at –172 bp and –671 bp (Fig. 6B). ChIP assays using GFP antibody were carried out to determine whether TaARF4 could directly bind to the GH3 promoter regions in vivo. Real-time PCR results indicated that TaARF4–GFP bound to the AuxRE regions, not the coding regions. In the absence of the GFP antibody, we could not detect TaARF4–GFP bound to DNA fragments (Fig. 6C). Free IAA contents of OE3 and OE4 were 12.1 and 14.9 % higher than in the Col wild type (Fig. 6D). Thus, it is proposed that TaARF4 can bind to the promoter region of GH3 genes to repress GH3 gene expression, and further repress free IAA changing into other forms, leading to higher free IAA content in OE plants. This suggests that the shorter primary root length and reduced height of TaARF4-overexpressing arabidopsis plants might be due to higher free IAA content, which inhibits apical dominance.
Fig. 6.
GH3 gene expression in TaARF4-OE plants and ChIP assay on the promoters of GH3 genes. (A) Expression of GH3 genes was negatively regulated by TaARF4. Expression of GH3.2 and GH3.5 was detected in 2-week-old seedlings of Col, GFP and two TaARF4-OE plants. Three biologically independent experiments, each with three technical replicates, were performed. Error bars are 2 × s.e. *P < 0.05, **P < 0.01 (Student’s t-test). (B) Bars indicate putative TaARF4-binding sites of GH3 genes. The ATG translation start site is indicated at position +1. Primers used for ChIP PCR are listed in Supplementary Data Table 3. (C) ChIP assay of TaARF4 binding to the promoter of GH3 genes. Col and empty GFP seedlings were used as the negative control. Two transgenic lines (TaARF4-GFP-OE3 and TaARF4-GFP-OE4) and GFP antibody were used for the ChIP assay. Real-time PCR was carried out to show DNA abundance relative to input. Three biological replicates were performed, and each biological replicate had three technical replicates. Error bars are 2 × s.e. Significant differences were calculated based on ANOVA. (D) Free IAA levels in 2-week-old seedlings. Three independent replicates were performed. Error bars are 2 × s.e. *P < 0.05, **P < 0.01 (Student’s t-test).
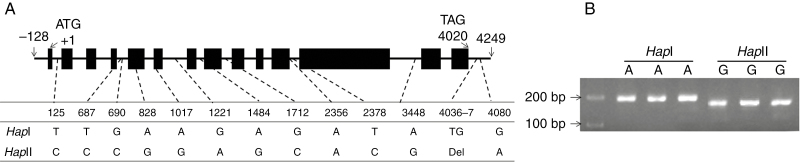
Sequence polymorphism and genetic mapping
Genome-specific primers were designed based on the polymorphism detected in the flanking regions of TaARF4 genes in three genomes. Using these primers, TaARF4-A, TaARF4-B and TaARF4-D genomic fragments were separately cloned from A, B and D genomes; their lengths were 4584, 4375 and 4123 bp, respectively. We sequenced genomic fragments from 32 diverse accessions, and no nucleotide polymorphism was detected in TaARF4-A or TaARF4-D. However 13 variants of TaARF4-B formed two haplotypes (Fig. 7A). Among the variants, only one single nucleotide polymorphism [SNP (A/G)] at 1017 bp resulted in an amino acid change (threonine to alanine). A dCAPS marker was developed based on an SNP (A/G) at 828 bp (Fig. 7B). The dCAPS marker was then used to genotype Population 1 (150 DH lines). TaARF4-B was mapped on chromosome 3B and was flanked by markers P3622.4 and P2076 (Supplementary Data Fig. S3A). One quantitative trait locus (QTL) affecting PH was earlier mapped in this marker interval (Wu et al., 2010). The genetic distances between two markers and TaARF4-B were 6.24 and 4.79 cM, respectively. The location of TaARF4-B on chromosome 3B was validated using diploid wild relative species and nulli-tetrasomic lines of Chinese Spring (Supplementary Data Fig. S3B).
Fig. 7.
Nucleotide polymorphisms and functional marker development for TaARF4-B. (A) Twelve single nucleotide polymorphisms and one deletion were detected in the TaARF4-B genomic region. Black rectangles indicate exons. (B) A dCAPS marker was developed based on SNP-828 (A/G). Using SacI restriction endonuclease, a 192 bp PCR product amplified from accessions with SNP-828G (HapII) can be digested to 168 and 24 bp, whereas the 192 bp PCR product amplified from accessions with SNP-828A (HapI) could not be digested. A 100 bp DNA ladder is shown on the left.
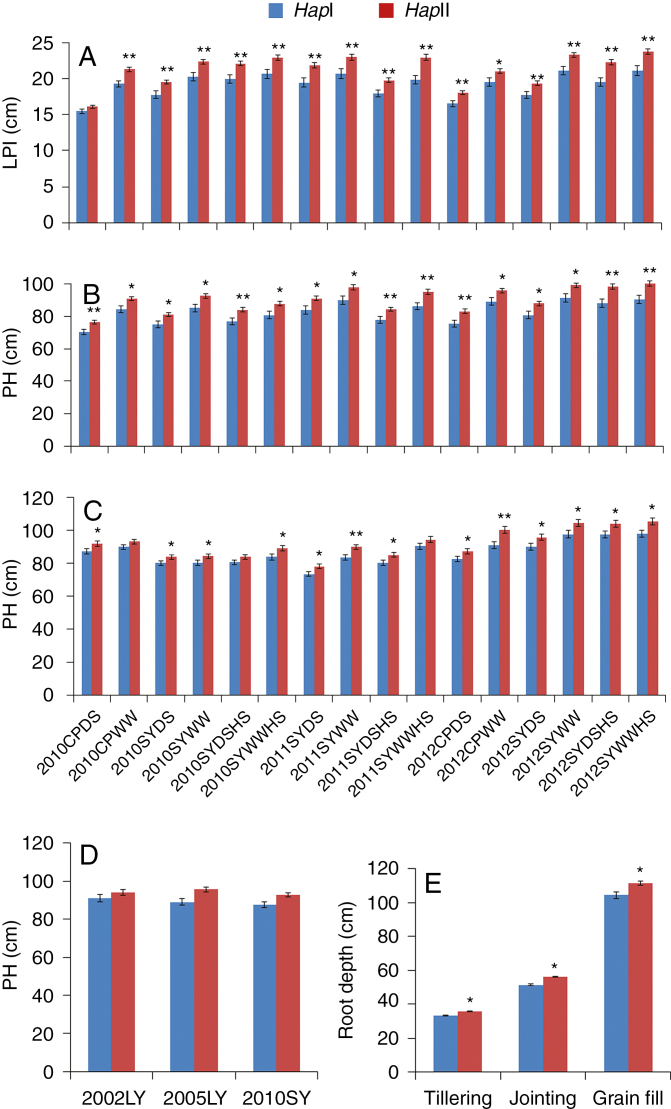
Association analysis of TaARF4-B haplotypes and agronomic traits
To investigate the phenotypic effects of the two TaARF4-B haplotypes, Population 2 consisting of 262 accessions was used for association analysis of a range of agronomic traits. Among 16 environments (years × sites × water regimes × heat treatment), significant associations between TaARF4-B and LPI were identified in 14 environments (except 2010CPDS and 2012CPWW). TaARF4-B was strongly associated with PH in 14 environments (except 2011SYWW and 2012SYWW) (Table 1). The phenotypic variation in LPI explained by TaARF4-B ranged from 2.44 to 10.54 %, and the phenotypic variation in PH explained by TaARF4-B ranged from 4.26 to 8.82 %. Accessions with HapI had significantly shorter LPI than those with HapII in all the environments except 2010CPDS. HapI reduced LPI by 1.47–2.75 cm compared with HapII (Fig. 8A). Accessions possessing HapI had significantly shorter PH than those with HapII in all 16 environments. HapI reduced PH by 5.97–10.35 cm compared with HapII (Fig. 8B).
Table 1.
TaARF4-B haplotypes associated with agronomic traits in 16 environments
| Year | Site | Environment | LPI | PH | ||
|---|---|---|---|---|---|---|
| P-value | PVE (%) | P-value | PVE (%) | |||
| 2010 | CP | DS | n.s. | 2.44 | 0.0247* | 7.16 |
| CP | WW | 0.0022*** | 6.00 | 0.0374* | 5.26 | |
| SY | DS | 0.0185* | 7.42 | 0.0472* | 6.27 | |
| SY | WW | 0.0105* | 5.42 | 0.0288* | 4.64 | |
| SY | DS–HS | 0.0184* | 7.55 | 0.0401* | 5.53 | |
| SY | WW–HS | 0.0115* | 6.76 | 0.0361* | 6.52 | |
| 2011 | SY | DS | 0.0327* | 8.32 | 0.0498* | 6.21 |
| SY | WW | 0.0161* | 7.28 | n.s. | 5.34 | |
| SY | DS–HS | 0.0148* | 8.31 | 0.0305* | 5.89 | |
| SY | WW–HS | 5.79E-04*** | 7.75 | 0.0130* | 4.88 | |
| 2012 | CP | DS | 0.0129* | 6.21 | 0.0141* | 7.20 |
| CP | WW | n.s. | 3.85 | 0.0406* | 4.88 | |
| SY | DS | 0.0204* | 3.59 | 0.0422* | 4.26 | |
| SY | WW | 0.0126* | 5.82 | n.s. | 4.75 | |
| SY | DS–HS | 0.0075** | 10.19 | 0.0107* | 8.12 | |
| SY | WW–HS | 0.0046*** | 10.54 | 0.0143* | 8.82 |
LPI, length of penultimate internode; PH, plant height; n.s., not significant; *P < 0.05, **P < 0.01 and ***P < 0.001; PVE, phenotypic variation explained. The environments were at Changping (CP) and Shunyi (SY) under well-watered (WW), drought-stressed (DS) and/or heat-stress (HS) conditions in 2010–2012.
Fig. 8.
Phenotypic comparisons of two TaARF4-B haplotypes. (A) LPI (length of penultimate internode) comparisons of two TaARF4-B haplotypes in Population 2 in 16 environments. (B) PH (plant height) comparisons of two TaARF4-B haplotypes in Population 2 in 16 environments. (C) PH comparisons of two TaARF4-B haplotypes in Population 1 in 16 environments. (D) PH comparisons of two TaARF4-B haplotypes in Population 5 in three environments. (E) Root depth comparisons of two TaARF4-B haplotypes in Population 3 at tillering, jointing and grain fill stages. Error bars are 2 × s.e. *P < 0.05, **P < 0.01 (Student’s t-test).
The effects of TaARF4-B haplotypes were confirmed in two other populations. Accessions possessing TaARF4-B HapI in Population 1 also had significantly shorter PH than those with HapII in 13 of the 16 environments. HapI reduced PH by 3.67–9.10 cm compared with HapII (Fig. 8C). Similar results were obtained for Population 5, where HapI reduced PH from 5.23 to 6.67 cm relative to HapII (Fig. 8D). The results suggested that the TaARF4 haplotype affected plant growth, and TaARF4 HapI was considered a superior allele for reducing PH.
Association analysis between TaARF4-B haplotypes and root phenotype
We also performed an association analysis between TaARF4-B haplotypes and root depth phenotype at the seedling, tillering, jointing and grain fill stages. Significant associations were identified between haplotypes and root depth at tillering (P = 0.035, PVE = 4.50 %), jointing (P = 0.020, PVE = 5.48 %) and grain fill (P = 0.015, PVE = 5.94 %) stages. HapI accessions had shallow roots, which were 2.46 cm shorter compared with those of HapII at tillering, 4.71 cm at jointing and 7.00 cm at grain fill stages (Fig. 8E).
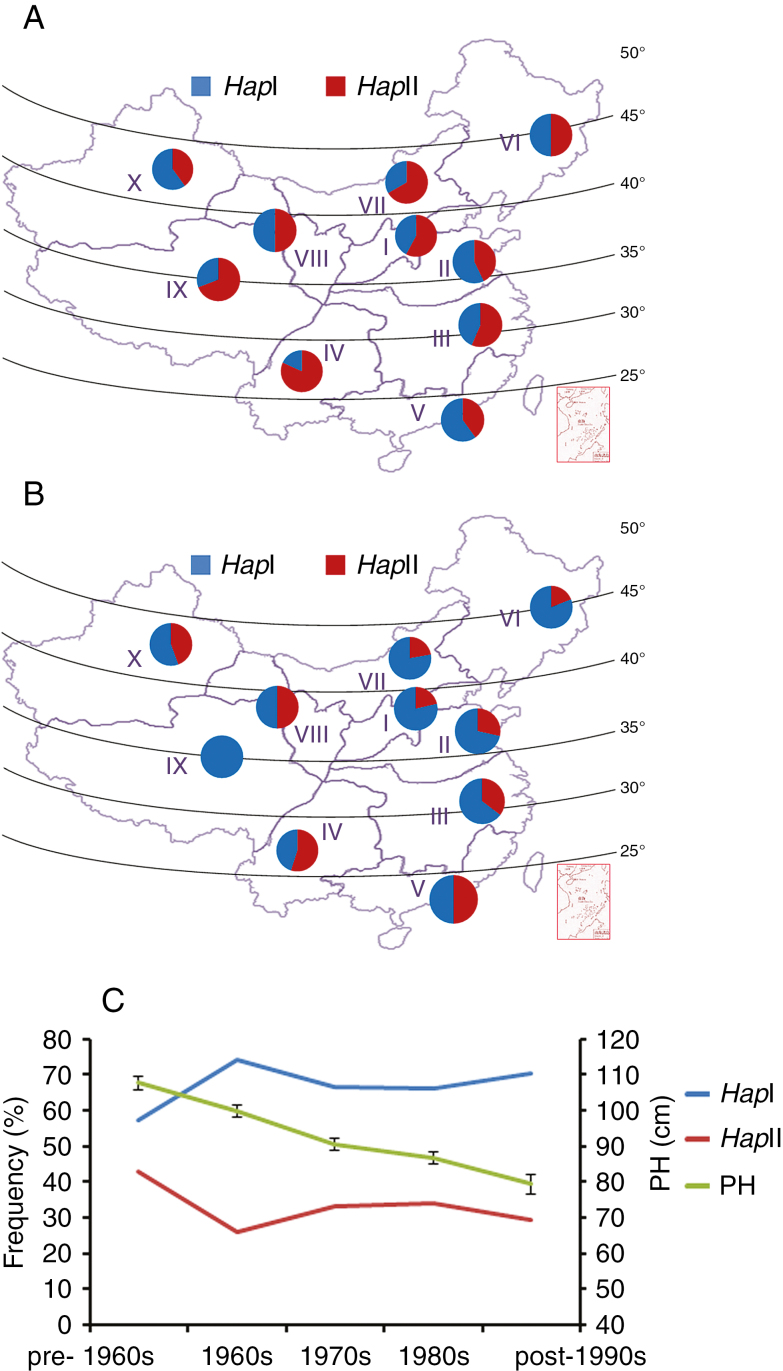
Geographic distribution of TaARF4-B haplotypes in ten Chinese wheat zones
Chinese wheat production regions are divided into ten ecological zones based on growing season and ecological conditions. Landraces (Population 4) and modern cultivars (Population 5) were used to investigate the distribution of TaARF4-B haplotypes (Fig. 9A, B). For the landraces, HapII was the dominant haplotype in seven zones [except Zone II (43 %), Zone V (40 %) and Zone X (40 %)]. The average frequency of HapII was 56 %. For modern cultivars, HapI was the dominant haplotype in almost all zones, with the only exception being Zone IV (45 %). The average frequency of HapI was 67 %. From Chinese landraces to modern cultivars, the frequencies of HapI increased across eight zones [with the exception of Zone V (from 60 to 50 %) and Zone X (from 44 to 40 %)]. Comparisons of landraces and modern cultivars indicated that TaARF4-B HapI was positively selected in wheat breeding.
Fig. 9.
Haplotype distribution and frequency change of TaARF4-B. (A) Distribution of two TaARF4-B haplotypes in 157 landraces of ten Chinese major wheat zones. (B) Distribution of two TaARF4-B haplotypes in 348 modern cultivars of ten Chinese major wheat zones. I, Northern Winter Wheat Region; II, Yellow and Huai River Valley Winter Wheat Region; III, Low and Middle Yangtze River Valley Winter Wheat Region; IV, South-western Winter Wheat Region; V, Southern Winter Wheat Region; VI, North-eastern Spring Wheat Region; VII, Northern Spring Wheat Region; VIII, North-western Spring Wheat Region; IX, Qinghai–Tibet Spring–Winter Wheat Region; X, Xinjiang Winter–Spring Wheat Region. (C) Haplotype frequency change of TaARF4-B in Chinese wheat breeding programmes. The left y-axis shows frequencies of TaARF4-B in Population 5 over decades. The right y-axis shows PH in Population 5 over decades. Accessions released in the pre-1960s, 1960s, 1970s, 1980s and post-1990s are numbered 37, 55, 102, 106 and 34, respectively. Thirteen accessions with unknown release dates were excluded. Error bars are 2 × s.e.
TaARF4-B haplotypes were selected during wheat breeding
Wheat breeding is a process of accumulating favourable haplotypes, and leaves footprints in genomes. In order to verify further that selection had occurred for a particular haplotype, we determined haplotype frequency changes according to cultivar release dates. PH continually declined from pre-1960s to post-1990s, whereas the frequency of HapI increased from 57.1 % pre-1960s to 74.1 % in the 1960s (Fig. 9C). After the 1960s, the frequency of HapI remained stable while PH continued to decline, implying selection of reduced height based on other genes.
DISCUSSION
Optimizing root and plant architecture is regarded as an important objective for wheat breeding. Therefore, it is essential to understand the molecular mechanisms of root growth and plant development. Here, we identified a novel ARF gene family member, TaARF4, and demonstrated that TaARF4 overexpression in arabidopsis caused shorter primary root length and PH. We propose that this effect occurs by repressing GH3 gene expression to mediate IAA homeostasis. Meanwhile, association analysis results showed that variation in TaARF4-B was significantly associated with root depth and PH. Therefore, TaARF4 may be an important gene resource for regulating wheat growth, and dCAPS markers of TaARF4 might be useful for the selection of wheat genotypes with optimal plant architecture.
ARFs and their target genes
ARFs as transcription activators or repressors participate in the auxin pathway, and affect auxin-responsive gene expression (Hagen and Guilfoyle, 2002; Weijers et al., 2018). Identification of the target genes for ARFs is a major step in studying their function. Previous research by ChIP, EMSA and gene expression analyses proved that Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation (Krogan et al., 2014). ATHB8 (Arabidopsis thaliana Homeobox Gene 8) is a target of ARF5/MP which regulates pre-procambial cell state acquisition by auxin signalling in arabidopsis leaves (Donner et al., 2009). DRN (DORNROSCHEN) is another target of ARF5/MP in the arabidopsis embryo (Cole et al., 2009). PLT (PLETHORA) transcription requires ARF5/MP and NPH4/ARF7 to mediate embryonic root patterning (Aida et al., 2004). In rice, CRL1 (crown rootless 1) is a target of ARF in crown root formation (Inukai et al., 2005). Thus, ARFs target different genes and have different functions. Our research indicates that overexpression of TaARF4 leads to ABA insensitivity by targeting HB33, and reduces root length and PH by targeting GH3 genes.
TaARF4, GH3s and IAA homeostasis
Indole-3-acetic acid, as the major auxin form in plants, plays an important role in plant growth (Benková et al., 2003; Mockaitis and Estelle, 2008). Biosynthesis, catabolism and conjugation of IAA significantly affect plant development (Ljun et al., 2002; González-Lamothe et al., 2012). IAA has many conjugate forms, such as ester conjugates with sugars, and amide conjugates with amino acids (Pencik et al., 2009). IAA amide forms IAA-Ala, IAA-Leu and IAA-Phe are forms that store IAA, whereas IAA-Asp and IAA-Glu are degradation precursors (Ludwig-Müller, 2011). These forms maintain IAA homeostasis. GH3s have roles in multiple processes (Ludwig-Müller et al., 2009). There are 19 GH3 family members in arabidopsis, and at least seven members (GH3.2–GH3.6, GH3.9 and GH3.17) are encoded by group II genes and can catalyse IAA amide synthesis (Staswick et al., 2005). In this study, we showed that TaARF4 overexpression in arabidopsis led to low GH3 gene expression and higher free IAA content. The high free IAA that inhibits apical dominance also reduced primary root length and PH.
TaARF4-B haplotypes and phosphorylation status
Thirteen variants were identified among two haplotypes of TaARF4-B, but only one amino acid change was discovered, threonine to alanine at amino acid position 158. This amino acid is located in a conserved DBD. Threonine can be phosphorylated whereas alanine is a dephosphorylated site. Using the NetPhos 3.1 Server, we predicted phosphorylated sites in TaARF4 and found that this site can be phosphorylated by DNA-dependent protein kinase (Supplementary Data Fig. S4). This predicts that TaARF4-B HapI might be phosphorylated at amino acid position 158, whereas HapII cannot be phosphorylated at this site. In plants, phosphorylation of transcription factors is an important regulatory mechanism, especially in hormone signalling pathways (Hill, 2015). For example in arabidopsis, the phosphorylation status of BZR1/2 affects target gene expression in the brassinosteroid (BR) signalling pathway (Wang et al., 2012). In the ABA signalling pathway, the phosphorylation status of RAV1 affects expression of ABI3, ABI4 and ABI5 (Feng et al., 2014). Similarly, the phosphorylation status of ARF2 is important to ARF2 function. In the BR signalling pathway, ARF2 can be phosphorylated by BIN2, resulting in loss of DNA binding and repression activities (Vert et al., 2008). Low potassium triggers a Ser689 phosphorylation in ARF2 and relieves its repression on HAK5 (Zhao et al., 2016). So different TaARF4-B haplotypes in wheat accessions may lead to a difference in TaARF4-B phosphorylation status, which in turn may lead to different root depth and PH.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: structural analysis and phylogenic tree of TaARF4 protein and homologous ARF proteins. Figure S2: semi-quantitative PCR detecting TaARF4-A expression levels in arabidopsis. Figure S3: mapping of TaARF4-B on wheat chromosomes. Figure S4. potential phosphorylation sites in TaARF4-B predicted by the NetPhos 3.1 Server. Table S1: primers used for real-time PCR. Table S2: primers used for vector construction. Table S3: primers used for ChIP assays. Table S4: primers used for genomic fragment isolation, sequencing and marker development. Table S5: accession names in Populations 2, 3, 4 and 5.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Robert A. McIntosh (Plant Breeding Institute, University of Sydney, Australia) for revising the manuscript. This study was supported by the National Key R&D Program of China (2016YFD0100605); the National Natural Science Foundation of China (31461143024) and the Agricultural Science and Technology Innovation Program, Chinese Academy of Agricultural Sciences (CAAS-XTCX2016019).
LITERATURE CITED
- Aida M, Beis D, Heidstra R, et al. . 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Appels R, Eversole K, Feuillet C, et al. . 2018. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: 661–673. [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, et al. . 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Khurana JP, Jain M. 2016. Characterization of rice homeobox genes, OsHOX22 and OsHOX24, and over-expression of OsHOX24 in transgenic Arabidopsis suggest their role in abiotic stress response. Frontiers in Plant Science 7: 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, et al. . 2012. A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nature Chemical Biology 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW. 2016. Auxin response factors. Plant, Cell & Environment 39: 1014–1028. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. 2009. Mechanism of auxin-regulated gene expression in plants. Annual Review of Genetics 43: 265–285. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. 2009. DORNRÖSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136: 1643–1651. [DOI] [PubMed] [Google Scholar]
- Collard BCY, Mackill DJ. 2008. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Transactions of the Royal Society: Biological Sciences 363: 557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Donner TJ, Sherr I, Scarpella E. 2009. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves. Development 136: 3235–3246. [DOI] [PubMed] [Google Scholar]
- Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF. 2014. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. The Plant Journal 80: 654–668. [DOI] [PubMed] [Google Scholar]
- Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM. 2006. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. The Plant Cell 18: 1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R, El Oirdi M, Brisson N, Bouarab K. 2012. The conjugated auxin indole-3-acetic acid–aspartic acid promotes plant disease development. The Plant Cell 24: 762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- Guan C, Wu B, Yu T, et al. . 2017. Spatial auxin signaling controls leaf flattening in Arabidopsis. Current Biology 27: 2940–2950 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T. 2007. Plant biology: sticking with auxin. Nature 446: 621–622. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Current Opinion in Plant Biology 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. 2002. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Molecular Biology 49: 373–385. [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt A, Guilfoyle T. 1984. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162: 147–153. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO Journal 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, et al. . 2004. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131: 1089–1100. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, et al. . 2000. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. The Plant Cell 12: 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. 2015. Post-translational modifications of hormone-responsive transcription factors: the next level of regulation. Journal of Experimental Botany 66: 4933–4945. [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, et al. . 2005. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell 17: 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. 1997. Protein–protein interactions among the Aux/IAA proteins. Proceedings of the National Academy of Sciences, USA 94: 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, et al. . 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proceedings of the National Academy of Sciences, USA 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Yin X, Ckurshumova W, Berleth T. 2014. Distinct subclades of Aux/IAA genes are direct targets of ARF5/MP transcriptional regulation. New Phytologist 204: 474–483. [DOI] [PubMed] [Google Scholar]
- Li B, Liu D, Li Q, et al. . 2016. Overexpression of wheat gene TaMOR improves root system architecture and grain yield in Oryza sativa. Journal of Experimental Botany 67: 4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JS, Dai XH, Zhao YD. 2006. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiology 140: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Ma B, Shi X, et al. . 2018. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557: 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljun K, Hul AK, Kowalczyk M, et al. . 2002. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Molecular Biology 50: 309–332. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. 2011. Auxin conjugates: their role for plant development and in the evolution of land plants. Journal of Experimental Botany 62: 1757–1773. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Julke S, Bierfreund NM, Decker EL, Reski R. 2009. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytologist 181: 323–338. [DOI] [PubMed] [Google Scholar]
- Luo L, Zeng J, Wu H, Tian Z, Zhao Z. 2018. A molecular framework for auxin-controlled homeostasis of shoot stem cells in Arabidopsis. Molecular Plant 11: 899–913. [DOI] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, et al. . 2017. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. 2008. Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology 24: 55–80. [DOI] [PubMed] [Google Scholar]
- Nanao MH, Vinos-Poyo T, Brunoud G, et al. . 2014. Structural basis for oligomerization of auxin transcriptional regulators. Nature Communications 5: 3617–3624. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. 2006. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A. 2005. AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. The Plant Journal 43: 29–46. [DOI] [PubMed] [Google Scholar]
- Pencik A, Rolcik J, Novák O, et al. . 2009. Isolation of novel indole-3-acetic acid conjugates by immunoaffinity extraction. Talanta 80: 651–655. [DOI] [PubMed] [Google Scholar]
- Promchuea S, Zhu Y, Chen Z, Zhang J, Gong Z. 2017. ARF2 coordinates with PLETHORAs and PINs to orchestrate ABA-mediated root meristem activity in Arabidopsis. Journal of Integrative Plant Biology 59: 30–43. [DOI] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. 2004. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiology 135: 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revzin A. 1989. Gel electrophoresis assays for DNA–protein interactions. Biotechniques 7: 346–355. [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. 1997. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- Söderman E, Mattsson J, Engstrom P. 1996. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. The Plant Journal 10: 375–381. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, et al. . 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell 17: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan QKG, Irish VF. 2006. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiology 140: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, et al. . 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. 2003. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1997. ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999a. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences, USA 96: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999b. Dimerization and DNA binding of auxin response factors. The Plant Journal 19: 309–319. [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proceedings of the National Academy of Sciences, USA 105: 9829–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, et al. . 2015. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 112: 4821–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DK, Pei KM, Fu YP, et al. . 2007. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24. [DOI] [PubMed] [Google Scholar]
- Wang L, Hua D, He J, et al. . 2011. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genetics 7: e1002172. doi: 10.1371/journal.pgen.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Liu S, et al. . 2016. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nature Genetics 48: 1233–1241. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. 2012. Brassinosteroid signaling network and regulation of photomorphogenesis. Annual Review of Genetics 46: 701–724. [DOI] [PubMed] [Google Scholar]
- Weijers D, Nemhauser J, Yang Z. 2018. Auxin: small molecule, big impact. Journal of Experimental Botany 69: 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang Z, Chang X, Jing R. 2010. Genetic dissection of the developmental behaviours of plant height in wheat under diverse water regimes. Journal of Experimental Botany 61: 2923–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, et al. . 2004. Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. The Plant Cell 16: 3448–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proceedings of the National Academy of Sciences, USA 98: 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Xu W, Liu X, et al. . 2017. Functional conservation and divergence among homoeologs of TaSPL20 and TaSPL21, two SBP-box genes governing yield-related traits in hexaploid wheat. Plant Physiology 174: 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Hu X, Zhu M, Xu M, Wang L. 2017. Transcription factors NF-YA2 and NF-YA10 regulate leaf growth via auxin signaling in Arabidopsis. Scientific Reports 7: 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yu H, Yu H, et al. . 2018. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. The Plant Cell 30: 1461–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Zou C, Li K, et al. . 2017. The Aegilops tauschii genome reveals multiple impacts of transposons. Nature Plants 3: 946–955. [DOI] [PubMed] [Google Scholar]
- Zhao S, Zhang ML, Ma TL, Wang Y. 2016. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. The Plant Cell 28: 3005–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.