Abstract
Stingless bees are generalist pollinators distributed through the pantropical region. There is growing evidence that their wild populations are experiencing substantial decline in response to habitat degradation and pesticides. Policies for conservation of endangered species will benefit from studies focusing on genetic and molecular aspects of their development and behavior. The most common method for looking at gene expression is real-time quantitative polymerase chain reaction preceded by reverse transcription (RT-qPCR) of the mRNA of interest. This method requires the identification of reliable reference genes to correctly estimate fluctuations in transcript levels. To contribute to molecular studies on stingless bees, we used Frieseomelitta varia, Melipona quadrifasciata, and Scaptotrigona bipunctata species to test the expression stability of eight reference genes (act, ef1-α, gapdh, rpl32, rps5, rps18, tbp, and tbp-af) in RT-qPCR procedures in five physiological and experimental conditions (development, sex, tissues, bacteria injection, and pesticide exposure). In general, the rpl32, rps5 and rps18 ribosomal protein genes and tpb-af gene showed the highest stability, thus being identified as suitable reference genes for the three stingless bee species and defined conditions. Our results also emphasized the need to evaluate the stability of candidate genes for any designed experimental condition and stingless bee species.
Subject terms: Gene expression analysis, Transcription
Introduction
A global effort to sequence genomes of all living species on Earth is in progress1 with the aims of mitigating the impact of climate changes on biodiversity and preserving endangered species and ecosystems. In particular, the i5K consortium leads the arthropod genomics initiatives and emphasizes on sequencing insect genomes2. The expanded availability of genomic data during the last decade created a favorable condition for researchers to investigate a myriad of biological processes and their molecular bases, and explore the application of this knowledge to the areas of health, agriculture, industry, and ecology. In this context, more than thirteen bee species have had their genomes sequenced, among which most are from the main tribes of corbiculate Apidae: Apini (Apis cerana, A. dorsata, A. florea, A. mellifera), Euglossini (Eufriesea mexicana, Euglossa dilemma), Bombini (Bombus terrestris, B. impatiens) and Meliponini (Melipona quadrifasciata)3–8. Meliponini is the more diverse group of bees and comprises over 500 species of generalist pollinators9 distributed in the pantropical region10. The bees of this group have a non-functional sting and thus are called stingless bees. In the last years, the wild populations of stingless bees are experiencing substantial decline that has been attributed to several factors, including habitat degradation and exposure to toxic substances such as pesticides11–14.
The use of gene expression analysis tools has the potential of unravel how specific genes respond to disadvantageous environment conditions and thus allow us to infer the consequences on the organism physiology, development and survival. Thanks to the availability of genomic data, it is now possible to pinpoint the molecular bases of the impact of environmental changes on the stingless bee species through a wide range of techniques. Real-time quantitative PCR (qPCR) is an efficient, simple, and low-cost technique frequently used by molecular biologists to quantify gene expression. The calculation of the relative expression of a target-gene by qPCR is based on the use of reference gene(s) as endogenous control(s). Reference genes should display a constant expression through the experimental conditions, otherwise they would lead to unreliable transcriptional quantification15. Historically, the genes used as references for qPCR were those related to basic cell functions that were thought to be expressed at constant rates, the so called “housekeeping genes”. Nonetheless, several studies have shown that biotic and abiotic factors can affect the functioning of the cells or their basic machinery, which would impact the expression of the housekeeping genes. Reliable reference genes can be identified by algorithms such as geNorm16, NormFinder17, and BestKeeper18 that evaluate the stability of genes based on the variance of quantification cycle (Cq) values in each physiological or experimental condition.
Studies on how changes in the environment affect stingless bee populations are required to provide a foundation for the implementation of proper protection measures. Suitable genes for normalization in qPCR were identified in bee species of Apini19–24, Bombini25,26 and Euglossini27 (Supplementary Table S1). Information on stable genes in bees of Meliponini tribe is still missing. Aiming to contribute to molecular studies on Frieseomelitta varia, Melipona quadrifasciata, and Scaptotrigona bipunctata stingless bees, we tested the stability of eight genes previously used as references in insect studies19–21,23,28–33 and involved in protein biosynthesis (elongation factor 1-α, ribosomal protein S5, ribosomal protein S18, ribosomal protein L32), glucose metabolism (glyceraldehyde 3-phosphate dehydrogenase), transcription initiation (tata-box bind binding protein and tata-box binding protein associated factor) and cytoskeletal structure (actin). Here, the expression of these genes was characterized across physiological or experimental conditions related to development, sex, tissues, bacteria injection and pesticide exposure. The choice of the stingless bee species was based on the diversity of their life stories and social organization, on the availability of genomic and transcriptomic sequences for M. quadrifasciata4,34, F. varia34,35 (genome paper submitted) and S. bipunctata (in prep), and on the potential to explore (1) the effects of pesticides on stingless bee populations, (2) the immune response to bacterial challenge, and (3) the diversity of reproductive biology and caste determination mechanisms36. We used the RefFinder tool that compiles the results of four methods into a comprehensive ranking based on the stability of the candidate reference genes37. This method has been widely used and offer an effective integration of tools for identifying reference genes38–41. Our work contributes to studies that aim to investigate the molecular bases of events related to development, sex, tissues, immunity, and pesticide exposure in the three different models of stingless bee species here addressed.
Results
Primer evaluation and expression profiles of candidate reference genes
The genomic sequences of the eight candidate reference genes of F. varia, M. quadrifasciata and S. bipunctata were aligned and primers were designed to target similar exonic regions among the three species (Table 1, Supplementary Fig. S1). The specificity of amplification of the eight candidate reference genes was initially tested for each species by conventional PCR and qPCR using a pool of cDNA samples (including all physiological and experimental conditions tested in this study) and genomic DNA. The set of primers were designed to identify possible genomic DNA contamination, as they anneal to different exons and flank intronic regions (Supplementary Fig. S2). The fragments amplified by conventional PCR (amplicons) were visualized by electrophoresis on 2% agarose gels. Amplicons for all genes showed a single band, except for act and tbp (Supplementary Fig. S3). Specific amplification of the genes ef1-α, gapdh, rpl32, rps5, rps18 and tbp-af were subsequently confirmed by a single peak in melting curve analysis (Supplementary Fig. S4-6). In the PCR efficiency calculation, the candidate reference genes displayed high linear regression coefficients that were greater than to 0.99, except actin and tbp genes for the three species. The PCR amplification efficiency values of act and tbp were over 110% for all three species, and more than one peak was observed in the melting curve analysis (Supplementary Fig. S7-9). We tried different primer concentrations and annealing temperatures to optimize qPCR, but more than one peak in melting curve analysis was still observed for act and tbp (data not shown). Besides, act seems to have transcript variants in these bee species, making it a gene not suitable for use as a reference gene. Thus, act and tbp were discarded for further analysis in this work. PCR efficiency values of the other qPCR candidate genes ranged from 90% to 101.3% (Table 2).
Table 1.
Primer sequences, amplicon sizes and GenBank (NCBI database) accession numbers of candidate reference genes used for qPCR experiments with samples of F. varia, M. quadrifasciata, and S. bipunctata.
| Gene | Accession number F. varia | Accession number M. quadrifasciata | Accession number S. bipunctata | Primer sequence (5′-3′) | Amplicon size (bp) |
|---|---|---|---|---|---|
|
actin1 (act)* |
MN193732 | MN687953 | MN193747 |
F: CAAAGCAGGATTTGCAGGAG R: TAAAACGCCCCTTTTGCTTT |
135 |
|
elongation factor 1 alpha (ef1-α) |
MN193733 | MN687951 | MN193744 |
F: GACTGTCGAACGCAAGGAAG R: TCAACACACCGGTTTCAACA |
176 |
|
glyceraldehyde 3-phosphate dehydrogenase (gapdh) |
MN193734 | MN687952 | MN193745 |
F: GTTCAGTGAGCGTGATCCAA R: CTTTGCACCACCTTCCAAAT |
124 |
|
ribosomal protein l32 (rpl32) |
MN193737 | MN687948 | MN193741 |
F: CGTAGGCGTTTTAAGGGACA R: ACTCCGTGAGCAATCTCAGC |
173 |
|
ribosomal protein s5 (rps5) |
MN193738 | MN687947 | MN193740 |
F: TGTTGACAGGGGACAATCCT R: TGGCCTGATTTACTCGTCGT |
147 |
|
ribosomal protein s18 (rps18) |
MN193739 | MN687946 | MN193746 |
F: CGTGCTGGAGAATGTTCTGA R: ATTCGTTCCAAATCCTCACG |
179 |
|
tata-box binding protein (tbp) |
MN193735 | MN687950 | MN193743 |
F: CCCTCTTTTGCAACTCCACA R: GGATCTGCAGAAGCTGGTGT |
152 |
|
tata-box binding protein associated factor (tbp-af) |
MN193736 | MN687949 | MN193742 |
F: TGCTGGACAACCACTTTCTG R: GTGCGGCTAATGAAACCAAT |
143 |
Table 2.
Amplification efficiencies (E) and correlation coefficients (R2) obtained for primers used to amplify reference candidate genes in samples of F. varia, M. quadrifasciata and S. bipunctata. The annealing temperature was 60 °C for all primers and reactions.
| Gene | F. varia | M. quadrifasciata | S. bipunctata | |||
|---|---|---|---|---|---|---|
| E (%) | R2 | E (%) | R2 | E (%) | R2 | |
| act | 115.0 | 0.700 | 112.7 | 0.700 | 160.4 | 0.924 |
| ef1-α | 98.2 | 0.996 | 101.0 | 0.998 | 91.5 | 0.997 |
| gapdh | 97.0 | 0.997 | 95.2 | 0.997 | 90.2 | 0.996 |
| rpl32 | 98.9 | 0.998 | 95.5 | 0.993 | 93.8 | 0.994 |
| rps5 | 96.1 | 1.000 | 100.3 | 0.996 | 109.8 | 0.992 |
| rps18 | 101.3 | 0.999 | 96.9 | 0.995 | 95.4 | 0.995 |
| tbp | 312.3 | 0.994 | 218.9 | 0.880 | 257.9 | 0.774 |
| tbp-af | 99.7 | 0.992 | 96.8 | 0.996 | 92.9 | 0.994 |
Expression levels of the six candidate reference genes were verified in all samples of F. varia, M. quadrifasciata and S. bipunctata to obtain an overview of transcript abundance. A variable Cq value of all the candidates across the physiological and experimental conditions highlighted different expression levels and expression patterns for the three species (Fig. 1, Supplementary Data S1). In F. varia, Cq values varied from 13.66 (ef1-α in different developmental stages) to 29.17 (tbp-af among different tissues, organs and body parts). In M. quadrifasciata, the lowest Cq value was 14.75 (rps5 in development) and the highest was 32.32 (tbp-af in tissues, organs and body parts). In S. bipunctata, Cq values ranged from 12.60 (rps5 in bacterial injection) to 32.67 (tbp-af in tissues, organs and body parts) (Supplementary Data S1). The tbp-af gene showed a higher Cq geo mean in all experimental conditions analyzed for the three species (the Cq geo mean ranged from 15.43 to 26.31), except for pesticide exposure condition of F. varia, where gapdh showed the highest Cq geo mean (18.77) (Supplementary Data S1). The genes with highest Cq variation (standard deviation - SD) in all three species were ef1-α and gapdh: ef1-α showed highest Cq variation during F. varia and S. bipunctata development (±Cq SD 1.49 and 1.30, respectively), between sexes of the three bee species (±Cq SD ranged from 1.04 to 1.62), and after bacterial injection into the three bee species (±Cq SD ranged from 0.65 to 2.01); gapdh showed highest Cq variation during M. quadrifasciata development (±Cq SD 1.32), in tissues, organs and body parts of the three bee species (±Cq SD ranged from 2.84 to 4.58) and after pesticide exposure of the three bee species (±Cq SD ranged from 0.20 to 0.69) (Supplementary Data S1). These results show that ef1-α and gapdh were the least stable genes, but further validation was done using gene expression software tools.
Figure 1.
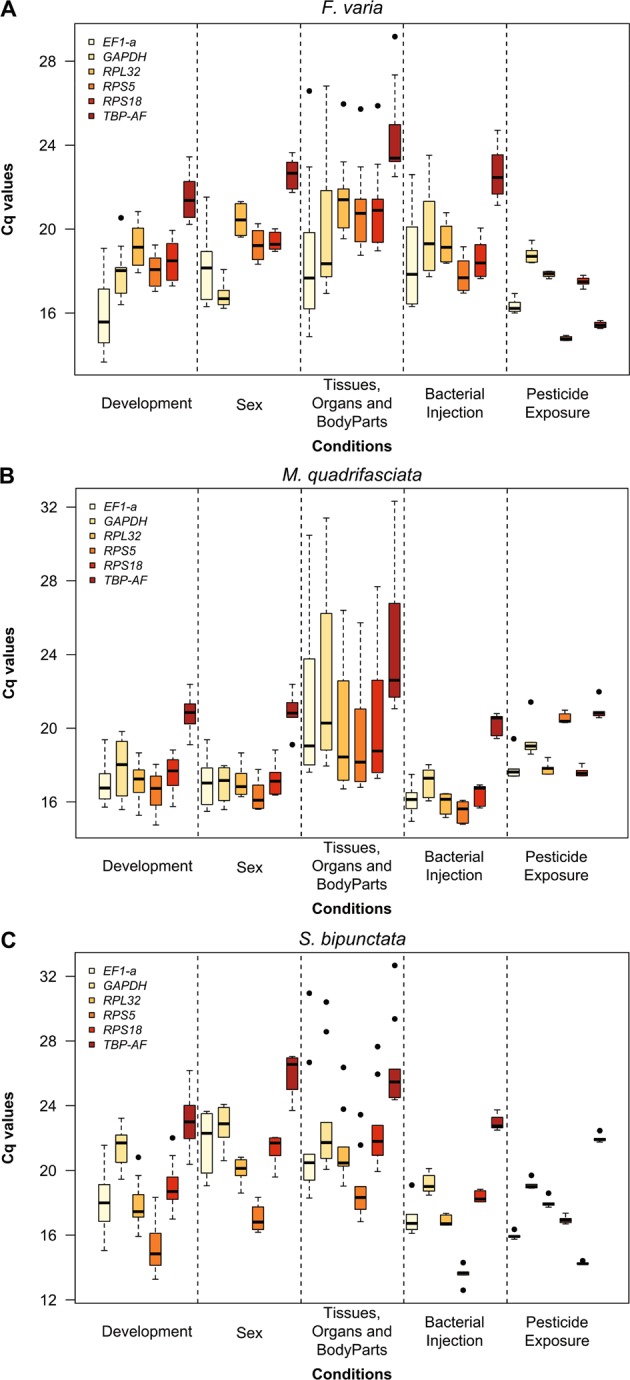
Expression levels (Cq value assessed by qPCR) of candidate reference genes across different experimental contexts using (A) Frieseomelitta varia, (B) Melipona quadrifasciata and (C) Scaptotrigona bipunctata. Box-plots show medians (horizontal lines), 25th to 75th percentiles (boxes), and interquartile ranges (whiskers). The dots indicate the outliers (replicated samples with Cq values above 50% of the interquartile ranges).
Stability of candidate reference genes
To determine the stability and rank the candidate reference genes, we used geNorm, NormFinder, Bestkeeper and delta-Ct. These programs are available at the web-tool RefFinder, which also calculated a comprehensive final overall ranking based on the results of these four different algorithms.
geNorm calculates expression stability value (M value) for a candidate reference gene based on the geometric mean of the SD of all studied genes in a pairwise comparison. The reference gene with the lowest M value should be the most stable gene and an M value under 1.5 is suggested by the geNorm software as a criterion for the selection of the reference gene(s)16. In our study, M values for all the candidate genes was lower than 1.5 for each physiological and experimental conditions and the three bee species, F. varia (Table 3), M. quadrifasciata (Table 4) and S. bipunctata (Table 5), thus indicating that all genes could act as potential reference genes. Like geNorm, NormFinder calculates stability values based on relative values, and the most stable reference genes are those exhibiting the lowest stability values17; however, a cut-off value is not suggested. Bestkeeper calculates the SD value and the coefficient of variation of each candidate gene. An SD greater than 1 indicates high variation of the expression of a gene and, consequently, its instability18. Our results demonstrate that some candidate genes are not stable. Gene expression varied most among the tissues, organs and body parts of all the bee species that were analyzed in this study (Tables 3, 4 and 5). The comparative delta-Ct method estimates the most stable reference gene using the SD means by pairwise comparison of two reference genes. An SD below 1 indicates stable gene expression. As observed in our results obtained with Bestkeeper, not all candidate reference genes were stable, and most of the gene expression variations were verified in the tissues, organs and body parts of the bee species (Tables 3, 4 and 5).
Table 3.
Ranking of six candidate reference genes for Frieseomelitta varia based on their expression stability according to analyses by geNorm, NormFinder, BestKeeper, and delta-Ct during development, between sexes, in tissues, and after bacterial injection and pesticide exposure.
| Condition | Reference Gene | geNorm | NormFinder | BestKeeper | delta-Ct | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Development | ef1-α | 0.506 | 5 | 0.851 | 5 | 1.489 | 6 | 0.98 | 5 |
| gapdh | 0.775 | 6 | 1.256 | 6 | 0.843 | 2 | 1.31 | 6 | |
| rpl32 | 0.132 | 1 | 0.066 | 1 | 0.892 | 4 | 0.54 | 1 | |
| rps5 | 0.290 | 4 | 0.387 | 4 | 0.673 | 1 | 0.67 | 4 | |
| rps18 | 0.132 | 1 | 0.146 | 3 | 0.891 | 3 | 0.56 | 2 | |
| tbp-af | 0.221 | 3 | 0.121 | 2 | 0.903 | 5 | 0.59 | 3 | |
| Sex | ef1-α | 0.745 | 6 | 1.398 | 6 | 1.471 | 6 | 1.42 | 6 |
| gapdh | 0.309 | 4 | 0.447 | 4 | 0.491 | 2 | 0.71 | 4 | |
| rpl32 | 0.120 | 3 | 0.065 | 3 | 0.548 | 4 | 0.53 | 3 | |
| rps5 | 0.085 | 1 | 0.043 | 1 | 0.563 | 5 | 0.50 | 1 | |
| rps18 | 0.409 | 5 | 0.715 | 5 | 0.375 | 1 | 0.81 | 5 | |
| tbp-af | 0.085 | 1 | 0.043 | 2 | 0.545 | 3 | 0.50 | 2 | |
| Tissues, organs and body parts | ef1-α | 1.153 | 6 | 1.389 | 6 | 2.892 | 5 | 1.56 | 6 |
| gapdh | 0.948 | 5 | 1.347 | 5 | 2.947 | 6 | 1.53 | 5 | |
| rpl32 | 0.200 | 3 | 0.852 | 4 | 1.390 | 1 | 1.02 | 4 | |
| rps5 | 0.078 | 1 | 0.556 | 3 | 1.552 | 3 | 0.89 | 2 | |
| rps18 | 0.078 | 1 | 0.539 | 2 | 1.543 | 2 | 0.89 | 1 | |
| tbp-af | 0.436 | 4 | 0.460 | 1 | 1.720 | 4 | 1.02 | 3 | |
| Bacterial injection | ef1-α | 0.880 | 6 | 1.222 | 6 | 2.013 | 6 | 1.26 | 6 |
| gapdh | 0.688 | 5 | 0.945 | 5 | 1.910 | 5 | 1.09 | 5 | |
| rpl32 | 0.084 | 1 | 0.542 | 2 | 0.843 | 3 | 0.72 | 2 | |
| rps5 | 0.117 | 3 | 0.649 | 4 | 0.789 | 1 | 0.77 | 4 | |
| rps18 | 0.084 | 1 | 0.555 | 3 | 0.887 | 2 | 0.72 | 1 | |
| tbp-af | 0.279 | 4 | 0.213 | 1 | 1.148 | 4 | 0.72 | 3 | |
| Pesticide exposure | ef1-α | 0.184 | 5 | 0.200 | 5 | 0.262 | 5 | 0.25 | 5 |
| gapdh | 0.217 | 6 | 0.260 | 6 | 0.334 | 6 | 0.28 | 6 | |
| rpl32 | 0.102 | 3 | 0.168 | 4 | 0.105 | 2 | 0.21 | 4 | |
| rps5 | 0.065 | 1 | 0.141 | 3 | 0.078 | 1 | 0.19 | 2 | |
| rps18 | 0.120 | 4 | 0.108 | 2 | 0.186 | 4 | 0.20 | 3 | |
| tbp-af | 0.065 | 1 | 0.043 | 1 | 0.115 | 3 | 0.17 | 1 | |
Table 4.
Ranking of six candidate reference genes for Melipona quadrifasciata based on their expression stability according to analyses by geNorm, NormFinder, BestKeeper, and delta-Ct during development, between sexes, in tissues, and after bacterial injection and pesticide exposure.
| Condition | Reference Gene | geNorm | NormFinder | BestKeeper | delta-Ct | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Stability | ||
| Development | ef1-α | 1.043 | 6 | 1.74 | 6 | 0.861 | 5 | 1.80 | 6 |
| gapdh | 0.665 | 5 | 1.21 | 5 | 1.321 | 6 | 1.34 | 5 | |
| rpl32 | 0.257 | 1 | 0.13 | 1 | 0.774 | 3 | 0.75 | 2 | |
| rps5 | 0.257 | 1 | 0.13 | 2 | 0.838 | 4 | 0.77 | 3 | |
| rps18 | 0.263 | 3 | 0.13 | 3 | 0.765 | 2 | 0.73 | 1 | |
| tbp-af | 0.382 | 4 | 0.35 | 4 | 0.677 | 1 | 0.87 | 4 | |
| Sex | ef1-α | 0.988 | 6 | 1.66 | 6 | 1.041 | 6 | 1.70 | 6 |
| gapdh | 0.629 | 5 | 0.97 | 5 | 0.759 | 5 | 1.14 | 5 | |
| rpl32 | 0.145 | 1 | 0.07 | 2 | 0.696 | 3 | 0.70 | 1 | |
| rps5 | 0.200 | 3 | 0.08 | 3 | 0.655 | 1 | 0.74 | 3 | |
| rps18 | 0.145 | 1 | 0.07 | 1 | 0.683 | 2 | 0.70 | 2 | |
| tbp-af | 0.444 | 4 | 0.70 | 4 | 0.701 | 4 | 0.94 | 4 | |
| Tissues, organs and body parts | ef1-α | 0.994 | 5 | 1.05 | 4 | 4.250 | 5 | 1.29 | 4 |
| gapdh | 1.164 | 6 | 1.39 | 6 | 4.580 | 6 | 1.51 | 6 | |
| rpl32 | 0.426 | 1 | 0.55 | 2 | 3.253 | 3 | 0.99 | 3 | |
| rps5 | 0.698 | 4 | 1.21 | 5 | 2.744 | 1 | 1.32 | 5 | |
| rps18 | 0.426 | 1 | 0.56 | 3 | 3.239 | 2 | 0.98 | 2 | |
| tbp-af | 0.583 | 3 | 0.33 | 1 | 3.632 | 4 | 0.89 | 1 | |
| Bacterial injection | ef1-α | 0.292 | 6 | 0.46 | 6 | 0.650 | 6 | 0.48 | 6 |
| gapdh | 0.197 | 5 | 0.19 | 5 | 0.638 | 5 | 0.31 | 5 | |
| rpl32 | 0.094 | 3 | 0.18 | 4 | 0.465 | 2 | 0.26 | 4 | |
| rps5 | 0.131 | 4 | 0.13 | 3 | 0.453 | 1 | 0.25 | 3 | |
| rps18 | 0.042 | 1 | 0.13 | 2 | 0.485 | 3 | 0.22 | 1 | |
| tbp-af | 0.042 | 1 | 0.12 | 1 | 0.497 | 4 | 0.23 | 2 | |
| Pesticide exposure | ef1-α | 0.306 | 5 | 0.28 | 3 | 0.516 | 5 | 0.43 | 5 |
| gapdh | 0.416 | 6 | 0.62 | 6 | 0.686 | 6 | 0.64 | 6 | |
| rpl32 | 0.117 | 1 | 0.22 | 2 | 0.214 | 2 | 0.34 | 2 | |
| rps5 | 0.140 | 3 | 0.32 | 5 | 0.237 | 3 | 0.39 | 4 | |
| rps18 | 0.117 | 1 | 0.30 | 4 | 0.186 | 1 | 0.37 | 3 | |
| tbp-af | 0.203 | 4 | 0.11 | 1 | 0.338 | 4 | 0.32 | 1 | |
Table 5.
Ranking of six candidate reference genes for Scaptotrigona bipunctata based on their expression stability according to analyses by geNorm, NormFinder, BestKeeper, and delta-Ct during development, between sexes, in tissues, and after bacterial injection and pesticide exposure.
| Condition | Reference Gene | geNorm | NormFinder | BestKeeper | delta-Ct | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Development | ef1-α | 0.485 | 5 | 0.718 | 5 | 1.295 | 6 | 0.84 | 5 |
| gapdh | 0.799 | 6 | 1.399 | 6 | 0.970 | 1 | 1.43 | 6 | |
| rpl32 | 0.182 | 1 | 0.091 | 1 | 1.036 | 2 | 0.57 | 2 | |
| rps5 | 0.266 | 3 | 0.221 | 3 | 1.152 | 4 | 0.67 | 3 | |
| rps18 | 0.182 | 1 | 0.091 | 2 | 1.067 | 3 | 0.57 | 1 | |
| tbp-af | 0.401 | 4 | 0.527 | 4 | 1.243 | 5 | 0.72 | 4 | |
| Sex | ef1-α | 0.991 | 6 | 1.251 | 6 | 1.624 | 6 | 1.33 | 6 |
| gapdh | 0.643 | 4 | 0.643 | 4 | 1.002 | 4 | 0.97 | 4 | |
| rpl32 | 0.228 | 1 | 0.343 | 3 | 0.575 | 1 | 0.80 | 2 | |
| rps5 | 0.819 | 5 | 1.212 | 5 | 0.715 | 3 | 1.28 | 5 | |
| rps18 | 0.228 | 1 | 0.114 | 1 | 0.713 | 2 | 0.73 | 1 | |
| tbp-af | 0.534 | 3 | 0.267 | 2 | 1.076 | 5 | 0.83 | 3 | |
| Tissues, organs and body parts | ef1-α | 1.223 | 6 | 1.561 | 6 | 3.106 | 6 | 1.69 | 6 |
| gapdh | 0.989 | 5 | 1.189 | 5 | 2.842 | 5 | 1.46 | 5 | |
| rpl32 | 0.341 | 1 | 0.752 | 3 | 1.630 | 2 | 1.04 | 3 | |
| rps5 | 0.341 | 1 | 0.981 | 4 | 1.586 | 1 | 1.15 | 4 | |
| rps18 | 0.414 | 3 | 0.537 | 2 | 1.889 | 3 | 1.00 | 2 | |
| tbp-af | 0.585 | 4 | 0.341 | 1 | 2.039 | 4 | 0.99 | 1 | |
| Bacterial injection | ef1-α | 0.608 | 6 | 0.897 | 6 | 0.763 | 6 | 0.92 | 6 |
| gapdh | 0.243 | 4 | 0.151 | 4 | 0.487 | 5 | 0.51 | 4 | |
| rpl32 | 0.055 | 1 | 0.027 | 2 | 0.279 | 1 | 0.43 | 2 | |
| rps5 | 0.450 | 5 | 0.884 | 5 | 0.343 | 3 | 0.91 | 5 | |
| rps18 | 0.055 | 1 | 0.027 | 1 | 0.286 | 2 | 0.42 | 1 | |
| tbp-af | 0.166 | 3 | 0.107 | 3 | 0.380 | 4 | 0.46 | 3 | |
| Pesticide exposure | ef1-α | 0.202 | 5 | 0.217 | 5 | 0.131 | 2 | 0.26 | 5 |
| gapdh | 0.229 | 6 | 0.251 | 6 | 0.202 | 6 | 0.28 | 6 | |
| rpl32 | 0.096 | 3 | 0.197 | 4 | 0.197 | 5 | 0.23 | 4 | |
| rps5 | 0.094 | 1 | 0.062 | 1 | 0.170 | 4 | 0.18 | 1 | |
| rps18 | 0.141 | 4 | 0.112 | 2 | 0.054 | 1 | 0.21 | 3 | |
| tbp-af | 0.094 | 1 | 0.141 | 3 | 0.163 | 3 | 0.20 | 2 | |
The comparison of the reference gene expression ranking of the four methods revealed variations in each physiological and experimental condition and bee species (Tables 3, 4 and 5). Even so, rpl32, rps5, rps18 and tbp-af were clearly among the top three stable reference genes in most analyses. It should be noted that Bestkeeper highlighted gapdh as a stable gene when we used samples of F. varia and S. bipunctata in the development condition, and also when we used F. varia males and females (sex condition) (Tables 3 and 5). The gene ef1-α was considered stable in M. quadrifasciata and S. bipunctata treated with pesticides by NormFinder (Table 4) and Bestkeeper (Table 5).
Comprehensive gene expression ranking and recommended reference genes
As the four programs (geNorm, NormFinder, BestKeeper, delta-Ct) showed different ranking orders of candidate reference genes, we used RefFinder to integrate the rankings and obtain a consensus result in order to recommend the best reference gene(s) for each physiological and experimental condition, and bee species. Based on the comprehensive ranking, rpls32 and rps18 were the most stable reference genes in the various developmental stages of F. varia, M. quadrifasciata and S. bipunctata (Table 6). In females and males (sex condition), the genes rpls32 and rps18 were highly stable in M. quadrifasciata and S. bipunctata, and the genes rps5 and tbp-af were the most stable in F. varia. In the different tissues, organs and body parts of the three bee species, rps18, rps5 and tbp-af were the most stable reference genes (rps18 and rps5 in F. varia, rps18 and tbp-af in M. quadrifasciata, rps5 and tbp-af in S. bipunctata). After bacterial injection, rps18 and rpl32 were highly stable in F. varia and S. bipunctata and rps18 and tbp-af were highly stable in M. quadrifasciata. After pesticide exposure, the genes tbp-af and rps5 were highly stable in F. varia and S. bipunctata and rpl32 and rps18 were highly stable in M. quadrifasciata.
Table 6.
Comprehensive ranking and best recommended pair of reference genes for accurate normalization of qPCR assays using samples of Meliponini bees under different experimental contexts. The comprehensive ranking is based on gene expression stability of each gene calculated by RefFinder using geNorm, NormFinder, BestKeeper and delta-Ct algorithms.
| Condition | Reference Gene | F. varia | M. quadrifasciata | S. bipunctata | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Comprehensive Ranking | Most stable | Comprehensive Ranking | Most stable | Comprehensive Ranking | Most stable | |||||
| Stability | Rank | Stability | Rank | Stability | Rank | |||||
| Development | ef1-α | 5.23 | 6 | rpl32, rps18 | 5.73 | 6 | rpl32, rps18 | 5.23 | 6 | rpl32, rps18 |
| gapdh | 4.56 | 5 | 5.23 | 5 | 3.83 | 4 | ||||
| rpl32 | 1.41 | 1 | 1.57 | 1 | 1.41 | 1 | ||||
| rps5 | 2.83 | 3 | 2.21 | 3 | 3.22 | 3 | ||||
| rps18 | 2.06 | 2 | 2.06 | 2 | 1.57 | 2 | ||||
| tbp-af | 3.08 | 4 | 2.83 | 4 | 4.23 | 5 | ||||
| Sex | ef1-α | 6.00 | 6 | rps5, tbp-af | 6.00 | 6 | rpl32, rps18 | 6.00 | 6 | rps18, rpl32 |
| gapdh | 3.36 | 5 | 5.00 | 5 | 4.00 | 4 | ||||
| rpl32 | 3.22 | 3 | 1.32 | 1 | 1.57 | 2 | ||||
| rps5 | 1.50 | 1 | 2.28 | 3 | 4.40 | 5 | ||||
| rps18 | 3.34 | 4 | 1.68 | 2 | 1.19 | 1 | ||||
| tbp-af | 1.86 | 2 | 4.00 | 4 | 3.08 | 3 | ||||
| Tissues, organs and body parts | ef1-α | 5.73 | 6 | rps18, rps5 | 4.47 | 5 | rps18, tbp-af | 6.00 | 6 | rps5, tbp-af |
| gapdh | 5.23 | 5 | 6.00 | 6 | 5.00 | 5 | ||||
| rpl32 | 2.63 | 3 | 2.06 | 3 | 2.06 | 3 | ||||
| rps5 | 2.06 | 2 | 3.16 | 4 | 2.00 | 1 | ||||
| rps18 | 1.41 | 1 | 1.86 | 1 | 2.45 | 4 | ||||
| tbp-af | 2.63 | 4 | 1.86 | 2 | 2.00 | 2 | ||||
| Bacterial injection | ef1-α | 6.00 | 6 | rps18, rpl32 | 6.00 | 6 | rps18, tbp-af | 6.00 | 6 | rps18, rpl32 |
| gapdh | 5.00 | 5 | 5.00 | 5 | 4.23 | 4 | ||||
| rpl32 | 1.86 | 2 | 3.13 | 4 | 1.41 | 2 | ||||
| rps5 | 2.63 | 3 | 2.45 | 3 | 4.40 | 5 | ||||
| rps18 | 1.57 | 1 | 1.57 | 1 | 1.19 | 1 | ||||
| tbp-af | 2.63 | 4 | 1.68 | 2 | 3.22 | 3 | ||||
| Pesticide exposure | ef1-α | 5.00 | 5 | tbp-af, rps5 | 4.40 | 5 | rpl32, rps18 | 3.98 | 5 | rps5, tbp-af |
| gapdh | 6.00 | 6 | 6.00 | 6 | 6.00 | 6 | ||||
| rpl32 | 3.13 | 3 | 1.68 | 1 | 3.94 | 4 | ||||
| rps5 | 1.57 | 2 | 3.66 | 4 | 1.41 | 1 | ||||
| rps18 | 3.13 | 4 | 1.86 | 2 | 2.21 | 3 | ||||
| tbp-af | 1.32 | 1 | 2.00 | 3 | 2.06 | 2 | ||||
It is important to highlight that RefFinder tool does not require PCR efficiency values to calculate gene stability and some concern was raised that the outputs of this tool would be thus biased when evaluating the best reference gene(s)42. We used geNorm, the most affected algorithm by different PCR efficiencies42, to compare the results from the geNormPLUS (qbasePLUS, version 3.043) and the geNorm from RefFinder. For all conditions and species, both geNormPLUS and geNorm considered the same three genes as the most stable with slight differences in the position of the genes in the ranking (Supplementary Table S2-4) (an exception was tbp-af in the sexes of S. bipunctata). Thus, for the set of primers and conditions we tested, PCR efficiency is unlikely to be relevant for the identification of the most stable genes.
Influence of reference gene choice on the relative expression of a target mRNA
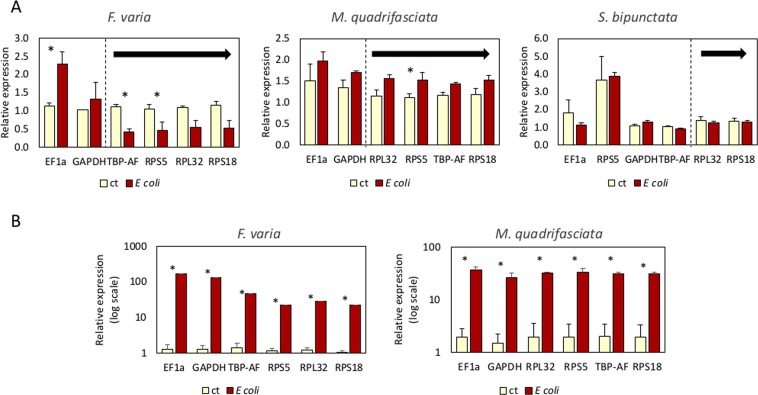
The four most stable and two least stable reference genes were used to normalize the expression of the immune deficiency (imd) and abaecin target genes in the bees submitted to bacterial infection. The extent of variation in imd and abaecin expression levels was then determined for each reference gene (Fig. 2). In F. varia, the relative expression of imd using the most stable genes was down-regulated after E. coli injection (tbp-af: most stable in NormFinder and rps5: most stable in BestKeeper, t-test, p < 0.05). However, when using an unstable gene, ef1-α, the result was the opposite: expression of imd was up-regulated after injection (t-test, p < 0.05). The use of the most stable reference genes in M. quadrifasciata resulted in up-regulation of imd expression (rps5: most stable in BestKeeper, t-test, p < 0.05). In S. bipunctata, imd relative expression did not differ between control and treatment when normalized with rpl32 and rps18, which were characterized as the most stable genes by each algorithm or in the comprehensive ranking. In general, expression of imd did not change substantially after bacterial injection, but the transcriptional levels of the antimicrobial gene abaecin was highly up-regulated compared to the non-injected controls. Normalization using all selected candidate reference genes showed the same profile (Fig. 2b). These results indicate the importance of choosing stable reference genes for normalization of target genes with slight expression changes after treatments.
Figure 2.
Validation of selected genes in Frieseomelitta varia, Melipona quadrifasciata and Scaptotrigona bipunctata. (A) Relative expression of imd was analyzed using the six selected reference genes for qPCR normalization in bees injected with Escherichia coli versus non-injected ones (control, ct). The first and the second most stable reference genes established for at least one of the algorithms (geNorm, NormFinder, BestKeeper, and delta-Ct) are indicated by arrows. Asterisks indicate p < 0.05 (t-test). Bars represent the means and standard errors of three biological replicates. (B) Relative expression (log scale) of abaecin was analyzed using the six selected reference genes for qPCR normalization in bees infected with Escherichia coli (E. coli) versus non-infected ones (control, ct). Asterisks indicate p < 0.05 (t-test). Bars represent the means and standard errors of three biological replicates.
Discussion
The identification of gold-standard reference genes is crucial to produce reliable qPCR results. The expression of reference genes is used to correct the fluctuations in the target gene expression levels caused by technical variations in the quantity of total RNA or in the cDNA synthesis. Since there is not a universal set of reference genes that are stably expressed in all organisms and under different physiological and experimental conditions, the stability of candidate genes must be tested. Here, we evaluated the stability of six reference genes (ef1-α, gapdh, rpl32, rps5, rps18 and tbp-af) in three stingless bee species during their development, in males and females, in tissues, organs and body parts, and after bacterial injection and pesticide exposure.
Our study showed the ribosomal protein genes rpl32, rps5 and rps18 as highly stable in each of the physiological and experimental conditions for the three stingless bee species and at least one of these genes was among the most stable genes for each condition tested. The genes rpl32 and rps18 were most frequent among the genes displaying high stability and were the most stable during development (F. varia, M. quadrifasciata and S. bipunctata), between sexes (M. quadrifasciata and S. bipunctata), and after bacterial injection (F. varia and S. bipunctata) and pesticide exposure (M. quadrifasciata) (Table 6). Ribosomal proteins are involved in translation and their genes have been found to be suitable reference genes for studies using bee species and other insects under several experimental conditions.
The rpl32 gene is highly stable during development19 and in nurses and foragers of A. mellifera (Hymenoptera)21, and in different tissues of Plutella xylostella (Lepidoptera)44. rps18 is among the most stable genes during the development of Tribolium castaneum (Coleoptera)45, Leptinotarsa decemlineata (Coleoptera)46 and Lipaphis erysimi (Hemiptera)47, and in nurses and foragers of A. mellifera19–24, and also after bacterial infection in A. mellifera20 and fungal infection in T. castaneum48. In addition to rpl32 and rps18, other ribosomal protein genes in insects are stably expressed during development, between tissues, and under different treatment conditions tested38,44,45,49,50.
We found the gene tbp-af as stably expressed in F. varia (between sexes and after pesticide exposure), M. quadrifasciata (among tissues and after bacterial injection), and S. bipunctata (among tissues and after pesticide exposure). The tbp-af gene codifies an evolutionarily conserved protein that composes the complex required for transcription initiation by RNA polymerase II51. tbp-af has also been shown to be stably expressed between tissues and after JH-treatment of A. mellifera19, and after exposure of Bemisia tabaci (Hemiptera) to the pesticides imidacloprid and buprofezin52.
Our results showed that the genes ef1-α and gapdh were the least stable genes in all tested conditions for F. varia and M. quadrifasciata. For S. bipunctata, the pair of least stable genes varied among different conditions: ef1-α and tbp-af were the least stable during development; ef1-α and gapdh were the least stable among tissues and after pesticide exposure; ef1-α and rps5 were the least stable between sexes and after bacterial injection. The protein codified by the gene ef1-α promotes chain elongation during polypeptide synthesis at the ribosome and gapdh codifies a key enzyme in the glycolytic pathway. ef1-α was also found to be the least stable gene in Bombus terrestris for target gene expression quantifications in tissues and after virus infection26, and also during development of the green peach aphid Myzus persicae (Hemiptera)41. gapdh was unstable after insecticide treatment of Plutella xylostella (Lepidoptera)44, during the development of Sesamia inferens (Lepidoptera)49, and in the green peach aphid under biotic and abiotic conditions41. In contrast, ef1-α was shown to be stably expressed across the tissues of A. mellifera19 and Bombus lucorum25,26, and during development and between tissues of P. xylostella (Lepidoptera)44 and during development of Diabrotica virgifera (Coleoptera)39. gapdh was also considered very stable in several studies with A. mellifera19–24. The different behavior of housekeeping genes among insects, and even among bee species (Table S1), underscores the need to evaluate them as reference genes for each species and physiological or experimental conditions.
The high variation of Cq values observed among tissues, organs and body parts for most candidate reference genes is due to the higher Cq values obtained for ovary samples in all species (Supplementary Data S2). The reason of these variations may be due to some heterogeneity of the samples. All three species belong to a group with variable reproductive capacities in workers, ranging from workers that always activate their ovaries to sterile workers53. Workers of Melipona and Scaptotrigona can lay reproductive haploid eggs, which will develop into males, and throphic eggs, which are larger eggs lacking the nucleus and that serve as food source to the queen53,54. Frieseomelitta workers are known to lack functional ovaries and be permanently sterile, however, a recent study on the morphology of adult worker ovaries revealed the presence of normal ovarioles, suggesting that these workers may retain some capacity to activate their ovaries53. The variety in worker reproduction capacity in these species, as well the variety of ovary activation/degradation grade among the workers of a given species, certainly reflects in the heterogeneity of tissues in the ovaries. Heterogeneous set of samples imposes extra challenges in the search for stable reference genes55. Yet, our results of geNormPLUS revealed average gene stability values (M) ≤ 0.5 (Supplementary Fig. 10), indicating that the reference genes are stable for this set of biological samples (fat body, head and ovaries). Other candidate reference genes should be evaluated only if average M-values are >1, that indicate low reference gene stability55.
All candidate reference genes were tested using RT-qPCR assay to evaluate their influence on the imd gene expression in F. varia, M. quadrifasciata and S. bipunctata infected with bacteria. imd is a member of the Imd pathway, one of the main pathways of immune response in insects56. The suitability of the reference genes for calculating the target gene (imd) expression levels was more evident in F. varia infected with bacteria. In this case, the use of highly stable reference genes (tbp-af and rps5) evidenced down-regulation of imd expression in infected bees whereas the use of one of the least stable genes (ef1-α) pointed to up-regulation of imd expression after infection (Fig. 2). Down-regulation of imd expression was also observed in A. mellifera infected with E. coli57. Our experiments also highlighted that in M. quadrifasciata and S. bipunctata, imd is mildly affected by bacterial injection, thus suggesting a distinct immune response in comparison to F. varia. Different responses to bacterial infection in the recognition and signaling genes from the immune pathways have also been observed between A. mellifera and Bombus terrestris, but the up-regulation of antimicrobial peptides such as abaecin, hymenoptaecin and defensin is always observed is these bees57–60. Yet, up-regulation of abaecin was detected in injected F. varia and M. quadrifasciata regardless of the candidate reference gene used to normalize abaecin expression levels. This result agrees with the observations that pronounced differences in expression levels of target gene are more easily detected and less prone to be affected by the choice of reference genes55.
This is the first study to validate reference genes for qPCR analysis in stingless bees and provides new resources for research on this diverse and important pollinator group. Recently, more than ten bee genomes have become available3,4,6 and allow the exploration of comparative genomic approaches to better understand the multifaceted aspects of bee biology and evolution. Yet, few studies evaluating candidate reference genes in bees were done (Table S1). Here, we used robust tools to evaluate the stability of the candidate reference genes in a comprehensive set of physiological and experimental conditions. Thus, our results have the potential to contribute to a wide range of molecular studies. In general, the ribosomal protein genes rpl32, rps18 and rps5, and tbp-af showed the highest stability in the stingless bees.
Material and Methods
Sample collection
Stingless bees
F. varia, M. quadrifasciata and S. bipunctata specimens were sampled directly from colonies located at the University of São Paulo, in Ribeirão Preto and in São Paulo, Brazil. We tested the candidate reference genes using different developmental stages, tissue (fat body), organ (ovaries) and body part (head), both sexes, and different experimental conditions, i.e., immune stimulation by bacterial injection and pesticide exposure. Details of sample collection are described below (see also Supplementary Data S2). For each species and condition, samples were stored in TRIzol ® Reagent (Invitrogen) and kept at −80 °C until RNA extraction. Whole individuals were processed for RNA extraction, except when tissues, organs and body parts were collected.
Developmental stages
F. varia, M. quadrifasciata and S. bipunctata workers were sampled throughout development including last instar larvae (defecating larvae, DL), pupae (white-eyed pupae, Pw), newly-emerged (NE) and foragers (FOR). DL have empty intestines61. Newly-emerged bees (<24 h-old) were obtained from pieces of brood combs taken from the nest and kept in an incubator (28 °C) for few days. Foragers were collected in the entrance of the nest carrying pollen in the hindlegs. For each species and developmental stage, we sampled three individuals for further analysis.
Tissues, organs and body parts
Fat body, head and ovaries from F. varia FOR, M. quadrifasciata NE and S. bipunctata NE were dissected in RNase-free 0.9% NaCl solution. Each sample of fat body or head was a pool obtained from three to six individuals of F. varia or S. bipunctata. Eighteen to 22 pairs of ovaries of F. varia and 11 to 13 pairs of S. bipunctata were pooled to compose each ovary sample. For M. quadrifasciata, only one individual was used to prepare each sample due to the bigger size of this stingless bee. Pooled (F. varia and S. bipunctata) or individual (M. quadrifasciata) samples were collected in triplicates.
Female vs Male
NE females of the three stingless bee species were compared with males. NE females and M. quadrifasciata males (<24 h-old) were collected as described above for NE females. Age-specific males were obtained by marking NE males on the thorax with a paint marker, returning them to experimental hives, and collecting them after seven days (F. varia) or after eight days (S. bipunctata). Three males were collected for each species.
Bacterial injection
Ten NE workers of F. varia, M. quadrifasciata and S. bipunctata in a total of 30 bees were separated in groups of five individuals. One group of each species was injected with a bacterial suspension of Escherichia coli DH5α whereas the other group did not receive any treatment (control group). F. varia workers were injected in the thorax with 1 µL of bacterial suspension in 0.9% NaCl (5 × 103 bacteria/µL) using a Nanofil 10 µL syringe (World Precision Instruments, Sarasota, FL, USA) and a needle with a 0.11 mm outer diameter. M. quadrifasciata workers were injected in the abdomen (between the 5th and 6th abdominal segments) with 1 µL of bacterial suspension (5 × 104 bacteria/µL) using a Hamilton syringe and a needle with a 0.47 mm outer diameter. S. bipunctata workers were injected in the thorax with 1 µL of bacterial suspension (5 × 103 bacteria/µL) using a Hamilton 701 N 10 µL syringe (Hamilton®, Reno, NV, USA) and a needle with a 0.47 mm outer diameter. The bacterial concentration used for each species was estimated based on the respective body weight. It is known that injection of 5 × 104 bacteria/µL is enough to activate the immune response in honey bees62; thus, we used the same concentration to inoculate M. quadrifasciata, which has a body weight equivalent to A. mellifera. F. varia and S. bipunctata are about 10-fold smaller than M. quadrifasciata and the bacterial concentration used for injections was 10-fold lower. The syringe and needle types used to administer the injections were chosen according to cuticle hardness (softer in F. varia). Only injection per se is enough to trigger the honey bee immune response60. Thus, immune stimulation was provoked by both injection and bacterial infection. Both injected and control groups of the three species were kept in the incubator at 28 °C with food (50% sucrose solution in water) and water ad libitum. The bees were individually collected (n = 3 for each injected and control group) six hours after the bacterial injection.
Treatment with imidacloprid pesticide
To produce a primary stock solution, 10 mg of imidacloprid (Sigma Aldrich) was added to 500 µL of acetone, with a subsequent dilution in distilled water to a concentration of 1000 ng active ingredient (a.i.)/µL. This aqueous stock solution was further diluted to prepare the solutions for topical applications. NE workers of F. varia and S. bipunctata received on the thorax a topical application of 1 µL of 25.2 ng a.i./µL, a dose tested in Scaptotrigona postica63, whereas NE workers of M. quadrifasciata received 1 µL of 2.01 ng a.i./µL, a dose also previously tested in these bees64. The control bees received topically 1 µL of water on the thorax. All the bees (n = 5 for each treatment) were maintained in Petri dishes under darkness in an incubator at 28 °C with food (50% sucrose solution in water) and water ad libitum. After 24 h, the bees were collected. At this time only alive and active bees were collected (n = 3 for each treated and control group).
RNA extraction and cDNA synthesis
Extraction of total RNA was performed according to the Trizol® manufacturer’s protocol. To increase total RNA precipitation of ovary samples, 1 µg of Molecular Biology Grade Glycogen (Sigma-Aldrich) was added to the isopropanol step and samples were kept at −20 °C for 24 to 48 hours. For tissue samples (fat body, head and ovary) 1 µg of total RNA was used for cDNA synthesis. For all the other samples (DL, Pw, NE, FOR, male, NE workers injected with E. coli and their non-injected controls, NE workers treated with pesticide and their non-treated controls), we used 3 µg of total RNA. cDNA synthesis was performed using SuperScript™ II (Invitrogen) and oligo(dT)12–18, except for the developmental stages (DL, Pw, RE, FOR) and the experiment of bacterial injection in F. varia for which SuperScript™ III (Invitrogen) and oligo(dT)20 were used. In both cases, cDNA synthesis was performed according to manufacturer’s protocol.
Primer design and validation by conventional PCR
The sequences of the genes act, ef1-α, gapdh, rpl32, rps5, rps18, tbp and tbp-af were retrieved from the recently sequenced genome of F. varia (The genome assembly was submitted to NCBI under BioProject PRJNA528016) and from the genome of M. quadrifasciata deposited in the Hymenoptera Genome Database4,65. S. bipunctata sequences were identified in a transcriptome dataset (not published). The structural organization of open reading frames and putative exon/intron splice sites of the candidate reference genes were inferred by the annotation tool Artemis version 16.0.066 (Supplementary Fig. S2). The sequences of the genes were deposited in the NCBI data bank (accession numbers in Table 1). Intron-spanning primers were designed based on F. varia gene sequences (Supplementary Fig. S2) using Primer3 software (bioinfo.ut.ee/primer3–0.4.0/). Melting temperatures between 60 °C and 61 °C, and amplicon length ranging from 100 to 190 bp were the restrictive parameters for primer selection. Other parameters were kept at the default setting. A maximum of two mismatches between the primer and the target sequence of M. quadrifasciata or S. bipunctata was allowed. We carefully checked the position of the mismatches in the primer sequence to avoid mismatches in the last five nucleotides of the 3′end. The only exception was the tbp-af forward primer for S. bipunctata, which contained a mismatch at the second last position67 (Supplementary Fig. S1). All genes, accession numbers, primer sequences and amplicon sizes used in this study are listed in Table 1.
Conventional PCRs were done to check amplification from complementary DNA (cDNA) and genomic DNA (gDNA) templates. Amplifications were carried out using 2x PCR Master Mix (Promega) and 0.6 pmol of each primer, under the following PCR regime: 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 45 s; and a final extension step of 72 °C for 7 min. The PCR products were checked by electrophoresis on 2% agarose gels stained with UniSafe Dye 20.000 × (Uniscience), visualized and documented in Kodak 1D Image Analysis program, version 3.6.2 (Eastman Kodak Co., Rochester, NY).
Real-time quantitative PCR
The real-time quantitative PCR (qPCR) assays were performed using the StepOnePlus™ Real-Time PCR System (Applied Biosystems). Amplifications were carried out in 15 μL reaction solutions containing 7.5 μL 2x qPCRBIO SyGreen Mix Separate-ROX (PCR Biosystems), 1.5 μL first-stranded cDNA (diluted 1:10), 6 ρmol/μL of each specific primer and 4.8 μL water. PCR conditions were 95 °C for 2 min followed by 40 cycles of 95 °C for 5 s and 60 °C for 25 s. The specificity of each pair of primers was checked by melting curve analysis (95 °C for 15 s, 60 °C for 1 min and a continuous raise in temperature to 95 °C at 0.3 °C/s ramp rate followed by 95 °C for 15 s). To check reproducibility, each assay was performed with technical triplicates for each of the three biological samples. PCR efficiency values (E) were calculated for each gene and bee species from the given slope after running standard curves and following the formula E = (10(−1/slope)−1)×100.
Evaluation of reference gene expression stability
To determine the stability of candidate reference genes we used RefFinder37. RefFinder is a web-based analysis tool that integrates four algorithms: geNorm16, NormFinder17, BestKeeper18, and delta-Ct68. Based on the rankings generated by each algorithm, a weight is assigned to each gene and geometric means of the gene weights are calculated for a comprehensive final ranking. Candidate genes having lower mean weights are considered transcriptionally stable and can be used as ideal reference genes.
Validation of reference gene selection
To evaluate each candidate reference gene for qPCR assays using F. varia, M. quadrifasciata and S. bipunctata samples, we measured the expression of the imd and abaecin gene in adult workers treated with E. coli and their respective untreated controls. imd gene encodes a protein that plays a role in the Imd pathway, which controls antibacterial defense57. Primer sequences for imd (forward 5′AAC AAC CGA TGC AAA ACC TG 3′ and reverse 5′ TCG TTG TTT TCG GTT CAT CA 3′) and for abaecin (F. varia forward 5′-GAA GGT AAC GAC GTT TAT TTT CG-3′, reverse 5′ TGG AAA CGG ATG TCG TTG TA 3′; M. quadrifasciata forward 5′ ATG CGC GAT ATT TGC GAT A 3′, reverse 5′TTT TCG GAT TGA ATG GTC CT 3′) were designed using Primer3 software, following the same parameters described in the previous section. The relative transcripts levels of imd and abaecin were calculated for each bee species using the 2−ΔΔCT method69 and the six candidate reference genes. Student’s t-test was performed to compare treated and control groups with significance reported for p < 0.05.
Supplementary information
Acknowledgements
This research was financially supported by FAPESP (Project Number 2016/06657-0), MCTIC/CNPq/Universal 14/2014 (Process number 454103/2014-0), FAPEMIG (CBB - PPM-00225-16) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) (Finance Code 001). The authors thank Prof. Dr. Isabel dos Santos and Msc. Sheina Koffler (University of São Paulo, São Paulo, Brazil) for providing the male samples of M. quadrifasciata, Ivan de Castro and Jairo de Souza (University of São Paulo, Ribeirão Preto, Brazil) for giving support to collect samples of F. varia and M. quadrifasciata, and Fernanda de Angeli Dalarmi for assistance with cDNA synthesis of F. varia.
Author contributions
A.P.L., T.S.D., L.T.A. and D.L.L. performed the experimental procedures. A.P.L., F.C.P.F. and T.S.D. performed the data analysis. F.M.F.N., M.M.G.B., Z.L.P.S. assisted in manuscript revising and provided helpful discussions. Z.L.P.S., M.M.G.B. and A.P.L. acquired funding. F.C.P.F. and A.P.L. wrote the manuscript. A.P.L. conceived and supervised the research. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53544-0.
References
- 1.Lewin HA, et al. Earth BioGenome Project: sequencing life for the future of life. Proc. Natl. Acad. Sci. USA. 2018;115:4325–4333. doi: 10.1073/pnas.1720115115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans JD, et al. The i5K initiative: Advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J. Hered. 2013;104:595–600. doi: 10.1093/jhered/est050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand P, et al. The nuclear and mitochondrial genomes of the facultatively eusocial orchid bee Euglossa dilemma. G3 (Bethesda). 2017;7:2891–2898. doi: 10.1534/g3.117.043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapheim KM, et al. Social evolution. Genomic signatures of evolutionary transitions from solitary to group living. Science. 2015;348:1139–1143. doi: 10.1126/science.aaa4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadd BM, et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsik CG, et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics. 2014;15:1–29. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park D, et al. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics. 2015;16:1. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michener, C. D. The Meliponini. in Pot-Honey: A legacy of stingless bees (eds Vit, P., Pedro, S. R. M. & Roubik, D.) 3–17, 10.1007/978-1-4614-4960-7_1 (Springer New York, 2013).
- 10.de Camargo JMF, de Menezes Pedro SR. Systematics, phylogeny and biogeography of the Meliponinae (Hymenoptera, Apidae): a mini-review. Apidologie. 1992;23:509–522. doi: 10.1051/apido:19920603. [DOI] [Google Scholar]
- 11.Del Sarto MCL, Oliveira EE, Guedes RNC, Campos LAO. Differential insecticide susceptibility of the Neotropical stingless bee Melipona quadrifasciata and the honey bee Apis mellifera. Apidologie. 2014;45:626–636. doi: 10.1007/s13592-014-0281-6. [DOI] [Google Scholar]
- 12.Jacob CRO, Soares HM, Carvalho SM, Nocelli RCF, Malaspina O. Acute toxicity of fipronil to the stingless bee Scaptotrigona postica Latreille. Bull. Environ. Contam. Toxicol. 2013;90:69–72. doi: 10.1007/s00128-012-0892-4. [DOI] [PubMed] [Google Scholar]
- 13.Lourenço CT, Carvalho SM, Malaspina O, Nocelli RCF. Oral toxicity of fipronil insecticide against the stingless bee Melipona scutellaris (Latreille, 1811) Bull. Environ. Contam. Toxicol. 2012;89:921–924. doi: 10.1007/s00128-012-0773-x. [DOI] [PubMed] [Google Scholar]
- 14.Valdovinos-Núñez GR, et al. Comparative toxicity of pesticides to stingless bees (Hymenoptera: Apidae: Meliponini) J. Econ. Entomol. 2009;102:1737–1742. doi: 10.1603/029.102.0502. [DOI] [PubMed] [Google Scholar]
- 15.Kozera B, Rapacz M. Reference genes in real-time PCR. J. Appl. Genet. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34–1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–15. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 19.Lourenço AP, Mackert A, Cristino ADS, Simões ZLP. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie. 2008;39:372–385. doi: 10.1051/apido:2008015. [DOI] [Google Scholar]
- 20.Scharlaken B, et al. Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 2008;8:1–10. doi: 10.1673/031.008.3301. [DOI] [Google Scholar]
- 21.Reim T, Thamm M, Rolke D, Blenau W, Scheiner R. Suitability of three common reference genes for quantitative real-time PCR in honey bees. Apidologie. 2013;44:342–350. doi: 10.1007/s13592-012-0184-3. [DOI] [Google Scholar]
- 22.Cameron RC, Duncan EJ, Dearden PK. Biased gene expression in early honeybee larval development. BMC Genomics. 2013;14:903. doi: 10.1186/1471-2164-14-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon K, Lee SH, Kim YH. Validation of quantitative real-time PCR reference genes for the determination of seasonal and labor-specific gene expression profiles in the head of Western honey bee, Apis mellifera. PLoS One. 2018;13:e0200369. doi: 10.1371/journal.pone.0200369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon K, Lee SH, Kim YH. Evaluation of reference genes for quantitative real-time PCR to investigate seasonal and labor-specific expression profiles of the honey bee abdomen. J. Asia. Pac. Entomol. 2018;21:1350–1358. doi: 10.1016/j.aspen.2018.10.014. [DOI] [Google Scholar]
- 25.Horňáková D, Matoušková P, Kindl J, Valterová I, Pichová I. Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 2010;397:118–120. doi: 10.1016/j.ab.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Niu J, Cappelle K, de Miranda JR, Smagghe G, Meeus I. Analysis of reference gene stability after Israeli acute paralysis virus infection in bumblebees Bombus terrestris. J. Invertebr. Pathol. 2014;115:76–79. doi: 10.1016/j.jip.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Boff S, Friedel A, Mara Mussury R, Roseti Lenis P, Raizer J. Changes in social behavior are induced by pesticide ingestion in a Neotropical stingless bee. Ecotoxicol. Environ. Saf. 2018;164:548–553. doi: 10.1016/j.ecoenv.2018.08.061. [DOI] [PubMed] [Google Scholar]
- 28.Schwarz RS, Evans JD. Single and mixed-species trypanosome and microsporidia infections elicit distinct, ephemeral cellular and humoral immune responses in honey bees. Dev. Comp. Immunol. 2013;40:300–310. doi: 10.1016/j.dci.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz RS, Moran NA, Evans JD. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA. 2016;113:9345–50. doi: 10.1073/pnas.1606631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teng X, Zhang Z, He G, Yang L, Li F. Validation of reference genes for quantitative expression analysis by real-time rt-PCR in four lepidopteran insects. J. Insect Sci. 2012;12:60. doi: 10.1673/031.012.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai Y, et al. Identification and validation of reference genes for quantitative real-time PCR in Drosophila suzukii (Diptera: Drosophilidae) PLoS One. 2014;9:e106800. doi: 10.1371/journal.pone.0106800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borges AA, Humann FC, Campos LAO, Tavares MG, Hartfelder K. Transcript levels of ten caste-related genes in adult diploid males of Melipona quadrifasciata (Hymenoptera, Apidae): a comparison with haploid males, queens and workers. Genet. Mol. Biol. 2011;34:698–707. doi: 10.1590/S1415-47572011005000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Judice CC, et al. Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol. Biol. 2006;15:33–44. doi: 10.1111/j.1365-2583.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 34.Woodard SH, et al. Genes involved in convergent evolution of eusociality in bees. Proc. Natl. Acad. Sci. 2011;108:7472–7477. doi: 10.1073/pnas.1103457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falcon T, et al. Exploring integument transcriptomes, cuticle ultrastructure, and cuticular hydrocarbons profiles in eusocial and solitary bee species displaying heterochronic adult cuticle maturation. PLoS One. 2019;14:e0213796. doi: 10.1371/journal.pone.0213796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartfelder K, et al. Physiological and genetic mechanisms underlying caste development, reproduction and division of labor in stingless bees. Apidologie. 2006;37:144–163. doi: 10.1051/apido:2006013. [DOI] [Google Scholar]
- 37.Xie F, Xiao P, Chen D. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues TB, Dhandapani RK, Duan JJ, Palli SR. RNA interference in the Asian longhorned beetle: identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues, T. B. et al. Validation of reference housekeeping genes for gene expression studies in Western corn rootworm (Diabrotica virgifera virgifera). PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 40.Gao XK, et al. Comprehensive evaluation of candidate reference genes for gene expression studies in Lysiphlebia japonica (Hymenoptera: Aphidiidae) using RT-qPCR. Gene. 2017;637:211–218. doi: 10.1016/j.gene.2017.09.057. [DOI] [PubMed] [Google Scholar]
- 41.Kang Z-W, et al. Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 2017;24:222–234. doi: 10.1111/1744-7917.12310. [DOI] [PubMed] [Google Scholar]
- 42.De Spiegelaere W, et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One. 2015;10:e0122515. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellemans J, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu W, et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae) Int. J. Biol. Sci. 2013;9:792–802. doi: 10.7150/ijbs.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toutges MJ, Hartzer K, Lord J, Oppert B. Evaluation of reference genes for quantitative polymerase chain reaction across life cycle stages and tissue types of Tribolium castaneum. J. Agric. Food Chem. 2010;58:8948–8951. doi: 10.1021/jf101603j. [DOI] [PubMed] [Google Scholar]
- 46.Shi X-Q, et al. Validation of reference genes for expression analysis by quantitative real-time PCR in Leptinotarsa decemlineata (Say) BMC Res. Notes. 2013;6:93. doi: 10.1186/1756-0500-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koramutla, M. K., Aminedi, R. & Bhattacharya, R. Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci. Rep. 6 (2016). [DOI] [PMC free article] [PubMed]
- 48.Lord JC, Hartzer K, Toutges M, Oppert B. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods. 2010;80:219–221. doi: 10.1016/j.mimet.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Lu Y, et al. Evaluation and validation of reference genes for SYBR Green qRT-PCR normalization in Sesamia inferens (Lepidoptera: Noctuidae) J. Asia. Pac. Entomol. 2015;18:669–675. doi: 10.1016/j.aspen.2015.08.002. [DOI] [Google Scholar]
- 50.Rajarapu SP, Mamidala P, Mittapalli O. Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis) Insect Sci. 2012;19:41–46. doi: 10.1111/j.1744-7917.2011.01447.x. [DOI] [Google Scholar]
- 51.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang P, Guo Y, Zhou X, Gao X. Expression profiling in Bemisia tabaci under insecticide treatment: indicating the necessity for custom reference gene selection. PLoS One. 2014;9:e87514. doi: 10.1371/journal.pone.0087514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luna-Lucena D, Rabico F, Simoes ZLP. Reproductive capacity and castes in eusocial stingless bees (Hymenoptera: Apidae) Curr. Opin. Insect Sci. 2019;31:20–28. doi: 10.1016/j.cois.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Vollet-Neto A, et al. Recent advances in reproductive biology of stingless bees. Insectes Soc. 2018;65:1–12. doi: 10.1007/s00040-018-0607-x. [DOI] [Google Scholar]
- 55.Hellemans, J. & Vandesompele, J. Selection of reliable reference genes for RT-qPCR analysis. In Methods in Molecular Biology (eds Biassoni, R. & Raso, A.) 1160, 19–26 (Springer New York, 2014). [DOI] [PubMed]
- 56.Keehnen NLP, Rolff J, Theopold U, Wheat CW. Insect antimicrobial defences: a brief history, recent findings, biases, and a way forward in evolutionary studies. Adv. In Insect Phys. 2017;52:1–33. doi: 10.1016/bs.aiip.2017.02.003. [DOI] [Google Scholar]
- 57.Evans JD, et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barribeau SM, et al. A depauperate immune repertoire precedes evolution of sociality in bees. Genome Biol. 2015;16:83. doi: 10.1186/s13059-015-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erler S, Popp M, Lattorff HMG. Dynamics of immune system gene expression upon bacterial challenge and wounding in a social insect (Bombus terrestris) PLoS One. 2011;6:e18126. doi: 10.1371/journal.pone.0018126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lourenço AP, Guidugli-Lazzarini KR, Freitas FCP, Bitondi MMG, Simões ZLP. Bacterial infection activates the immune system response and dysregulates microRNA expression in honey bees. Insect Biochem. Mol. Biol. 2013;43:474–482. doi: 10.1016/j.ibmb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Amaral IMR, et al. Circulating hemocytes from larvae of Melipona scutellaris (Hymenoptera, Apidae, Meliponini): Cell types and their role in phagocytosis. Micron. 2010;41:123–129. doi: 10.1016/j.micron.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Casteels-Josson K, Zhang W, Capaci T, Casteels P, Tempst P. Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire and required post-translational conversion of the precursor structures. J. Biol. Chem. 1994;269:28569–28575. [PubMed] [Google Scholar]
- 63.Soares HM, Jacob CRO, Carvalho SM, Nocelli RCF, Malaspina O. Toxicity of imidacloprid to the stingless bee Scaptotrigona postica Latreille, 1807 (Hymenoptera: Apidae) Bull. Environ. Contam. Toxicol. 2015;94:675–680. doi: 10.1007/s00128-015-1488-6. [DOI] [PubMed] [Google Scholar]
- 64.Costa, L. M. da, Grella, T. C., Barbosa, R. A., Malaspina, O. & Nocelli, R. C. F. Determination of acute lethal doses (LD50 and LC50) of imidacloprid for the native bee Melipona scutellaris Latreille, 1811 (Hymenoptera: Apidae). Sociobiology62 (2015).
- 65.Elsik CG, et al. Hymenoptera Genome Database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 2016;44:D793–800. doi: 10.1093/nar/gkv1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lefever S, Pattyn F, Hellemans J, Vandesompele J. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin. Chem. 2013;59:1470–1480. doi: 10.1373/clinchem.2013.203653. [DOI] [PubMed] [Google Scholar]
- 68.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:1–9. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.