Abstract
Taxus chinensis is a well-known gymnosperm with great ornamental and medicinal value. Its purple red brown heartwood (HW) has many attributes such as straight texture, high density, mechanical strength, rich elasticity and corrosion resistance that is highly prized commercially. T. chinensis sapwood (SW), in comparison, lacks these important traits. At present, little is known about the differences of metabolites between the SW and HW in T. chinensis. Widely targeted metabolic profiling was performed to analyze the metabolic profiles of HW and SW in T. chinensis using Liquid Chromatography-Electrospray Ionization-Mass Spectrometry (LC-EI-MS). A total of 607 metabolites were detected in HW and SW. Among them, 146 metabolites were significantly higher, and 167 metabolites significantly lower, in HW as compared to SW. These differential metabolites were mainly involved in metabolic pathways and biosynthesis of secondary metabolites, such as flavonoids, flavone and flavonol, phenylpropanoids and antibiotics. Moreover, 71 flavonoids and isoflavones were found to be significantly different between HW and SW. Our results show the difference of components between the HW and SW, which has potential significance to further elucidate the mechanism of HW color formation. The results will provide insight into the metabolites associated with wood color formation and useful information for understanding the metabolites associated with wood quality.
Subject terms: Bioinorganic chemistry, Metabolomics
Introduction
Wood, the secondary xylem of trees, is one of the most important source of materials and energy in the world1–3. It is important to human life, for example, wood can be as fuel for cooking as well as raw material for buildings2,3. In addition, it is also important to industrial, energy and environmental fields, such as renewable feedstock for pulp, biofuels, biomass energy and carbon sink2–4. Many tree species can be divided into sapwood (SW) and heartwood (HW) (Celedon and Bohlmann, 2018). HW is usually the dead inner wood, and reserve materials are converted into HW substances, while SW is the living, outermost portion wood that contains reserve materials5. Chemical composition plays an important role in wood properties, affecting physical and mechanical properties, natural durability, color and utilization of wood6,7. Compared to the SW, HW has an important economic value due to the natural decay resistance, wood color, wood fragrance and pharmaceutical substances8.
Wood color is an important trait for wood quality in the forest industry. It plays an important role in the processing of wood products, such as furniture, wood carving and building industries etc. It has been shown that the formation of wood color is due to the existence of secondary metabolites in the HW, including stilbenes, flavonoids and other phenolic compounds5, but the components of these secondary metabolites have not been investigated, and how these secondary metabolites affect wood color formation remain largely unknown. Predominately, research in wood color has mainly been focused on functional improvement through chemical methods9–11. The molecular genetic mechanisms of wood color formation are poorly understood. If the mechanism of wood color formation is understood, it may be possible to control wood color in trees via molecular genetics.
Taxus chinensis, belongs to the Taxus family, also known as yew, and is a well-known gymnosperm with great ornamental and medicinal value12. The bark of T. chinensis can produce paclitaxel, which has been widely used in the treatment of lung, ovarian and breast cancer13–15. In addition, the wood of Taxus has high commercial value because of its good aesthetic appearance, straight texture, high density, mechanical strength, rich elasticity, corrosion resistance, white yellowish SW and purple red brown colored HW. However, little is known about the metabolites variation between the SW and HW in T. chinensis.
Metabolomics is a valuable approach for the high-throughput and comprehensive study of complex metabolic compositions, and has been widely applied in plants16–20. Mass spectrometry methods have been used for detection and quantification of metabolites16–18. With the aim to investigate the metabolites variation between the SW and HW in T. chinensis, widely targeted metabolomics approach using liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed. The metabolites in SW and HW were identified, the differences in metabolite profile in SW and HW compared. Our results are potentially useful for the further elucidation of the mechanism of HW color formation. The results will provide insight into the molecular mechanisms of wood formation and useful information for improved wood quality.
Results and Discussion
Metabolic profiling of heartwood and sapwood based on LC-MS/MS
SW and HW were collected from the branch from an approximately 30-year-old of T. chinensis (Pilger) Rehd. The outer wood tissue with a pale yellow color is defined as SW and the central tissue with a red or dark-brown color is characterized as HW (Fig. 1). In order to investigate the components of HW and SW in T. chinensis, widely targeted metabolic profiling was performed to analyze the metabolic profiles of HW and SW in T. chinensis by using the Liquid Chromatography-Electrospray Ionization-Mass Spectrometry. Metabolomics data of HW and SW were processed using System Software Analyst (Version 1.6.3). Metabolites were quantitatively analyzed following collection of secondary data using the MRM model, as a result, a total of 607 metabolites were identified in HW and SW, including 52 lipids, 98 organic acids and their derivatives, 43 nucleotides and their derivatives, 132 flavonoids, 55 amino acids and their derivatives, 36 alkaloids, 93 phenylpropanoids, 12 vitamins, 17 terpenes, 21 carbohydrates and 48 others (supplementary file 1). These metabolites are involved in the most of primary and secondary metabolisms. Among these metabolites, the most abundant metabolites are the flavonoids, suggesting the flavonoids play a role in the process of the wood color formation.
Figure 1.
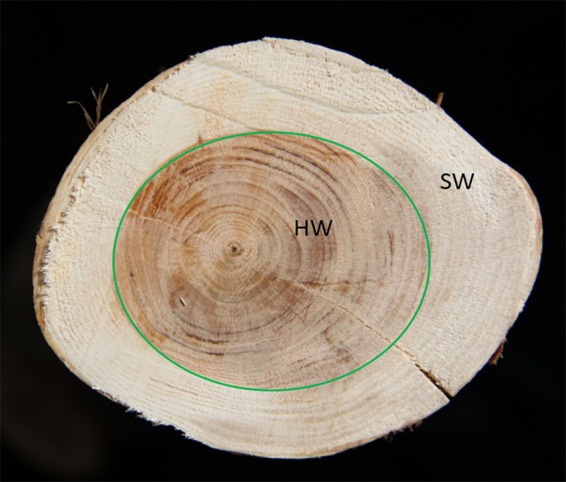
The heartwood and sapwood in T. chinensis (Pilger) Rehd. (HW: heartwood, SW: sapwood).
Principal component analysis (PCA) and cluster analysis of HW and SW
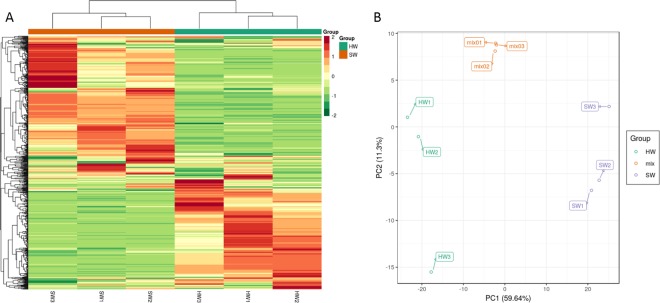
In order to investigate the HW and SW metabolic differences and multivariate, mixed samples of the HW and SW were prepared and performed to metabolic profiling using the Liquid Chromatography-Electrospray Ionization-Mass Spectrometry. Principal component analysis (PCA) is used to reveal the differences in metabolic profiles between the HW and SW. In the PCA plot (Fig. 2A), the mixed samples grouped together, indicating that the mixed samples had similar metabolic profiles. PCA clearly grouped these samples into distinct clusters, indicating significant differences in metabolites between the HW and SW. Hierarchical cluster analysis (HCA) of the metabolites of the HW and SW was performed. The results of all detected metabolites are shown in a heatmap (Fig. 2B), which indicates the significant differences in the relative abundance of metabolites between HW and SW. The metabolite profile of the HW and SW are clearly divided into two main clusters based on the differences in accumulation patterns on the heatmap, suggesting a clear variation in terms of the metabolites abundance in the HW and SW.
Figure 2.
(A) Heatmap of the metabolites in heartwood and sapwood; (B) PCA score plot of the metabolites in the heartwood and sapwood.
Differential metabolite analysis based on partial least squares-discriminant analysis (PLS-DA)
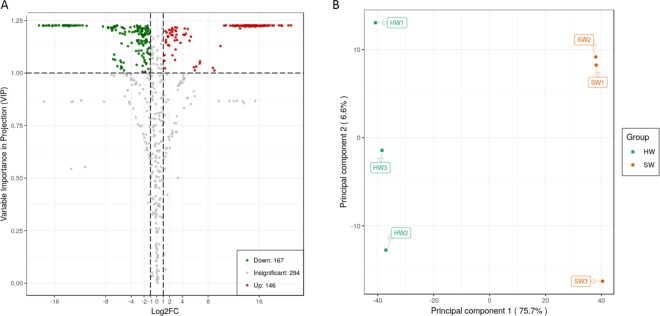
To obtain the metabolic differences between the HW and SW, the PLS-DA models was used to screen differential compounds between two groups of samples (Fig. 3). Moreover, a fold-change score ≥2 or ≤0.5 among the metabolites with a VIP value >1 was used to identify differential metabolites. The screening results have been illustrated using Volcano plots (Fig. 3). A total of 313 significant differences of metabolites were identified, of these differential metabolites there were 146 metabolites significantly higher and 167 metabolites significantly lower between the HW and SW (supplementary file 2). The top 30 metabolites that were higher in HW are shown in Table 1, and among them, 11 flavonoids were found to be significantly different between the HW and SW. These metabolites may be considered to be the representative differential metabolites for the HW and SW, influencing their wood properties especially the wood color.
Figure 3.
(A) The volcano plot of the differential metabolites in the heartwood and sapwood. Green dots represent down-regulated metabolites, red spots represent up-regulated metabolites and gray represent insignificant difference metabolites. (B) PLS-DA model plot of the differential metabolites in the heartwood and sapwood.
Table 1.
The top 30 significant differences of metabolites in heartwood and sapwood.
| Number | Compounds | Class | VIP | Fold_Change | Log2FC |
|---|---|---|---|---|---|
| 1 | C-pentosyl-apigenin O-p-coumaroylhexoside | Flavone | 1.227 | 2051851.852 | 20.968 |
| 2 | β-Caryophyllene | Terpene | 1.227 | 1548148.148 | 20.562 |
| 3 | Dehydrocorydaline | Alkaloids | 1.224 | 786666.667 | 19.585 |
| 4 | Ayanin | Flavonol | 1.227 | 708148.148 | 19.434 |
| 5 | Xanthotoxol | Phenylpropanoids | 1.227 | 555925.926 | 19.085 |
| 6 | Vitamin A | Vitamins and derivatives | 1.226 | 314444.444 | 18.262 |
| 7 | Ethyl 3,4-Dihydroxybenzoate | Organic acids and derivatives | 1.227 | 266296.296 | 18.023 |
| 8 | Schizandrin B | Phenylpropanoids | 1.227 | 200740.741 | 17.615 |
| 9 | N-Methyltryptamine | Alkaloids | 1.227 | 165925.926 | 17.340 |
| 10 | Ellagic acid | Polyphenol | 1.225 | 162481.481 | 17.310 |
| 11 | O-methylChrysoeriol 8-C-hexoside | Flavone | 1.226 | 131111.111 | 17.000 |
| 12 | Malvidin 3-O-glucoside (Oenin) | Anthocyanins | 1.226 | 130888.889 | 16.998 |
| 13 | Crocetin | Terpene | 1.226 | 119666.667 | 16.869 |
| 14 | Chrysin O-hexoside | Flavone | 1.226 | 119259.259 | 16.864 |
| 15 | 16-Hydroxy hexadecanoic acid | Lipids | 1.227 | 115148.148 | 16.813 |
| 16 | Malvidin 3-O-galactoside | Anthocyanins | 1.225 | 101592.593 | 16.632 |
| 17 | 4-Hydroxy-3,5-diisopropylbenzaldehyde | Organic acids and derivatives | 1.226 | 100814.815 | 16.621 |
| 18 | Prehelminthosporolactone | Others | 1.224 | 100555.556 | 16.618 |
| 19 | Ethyl cinnamate | Phenylpropanoids | 1.226 | 81074.074 | 16.307 |
| 20 | Luteolin O-hexosyl-O-pentoside | Flavone | 1.223 | 80074.074 | 16.289 |
| 21 | O-methylChrysoeriol 7-O-hexoside | Flavone | 1.226 | 75259.259 | 16.200 |
| 22 | O-methylChrysoeriol 5-O-hexoside | Flavone | 1.224 | 69925.926 | 16.094 |
| 23 | LysoPC 16:2 (2n isomer) | Lipids | 1.225 | 68037.037 | 16.054 |
| 24 | 1,10-decanediol | Alcohols | 1.224 | 59000.000 | 15.848 |
| 25 | 8-Methoxypsoralen | Phenylpropanoids | 1.226 | 54333.333 | 15.730 |
| 26 | Psoralen | Phenylpropanoids | 1.224 | 51148.148 | 15.642 |
| 27 | Myricetin | Flavonol | 1.226 | 50074.074 | 15.612 |
| 28 | Glabridin | Flavonoid | 1.227 | 48370.370 | 15.562 |
| 29 | Camptothecin | Alkaloids | 1.227 | 47222.222 | 15.527 |
| 30 | Phytocassane C | Terpene | 1.226 | 46925.926 | 15.518 |
Metabolic pathway analysis of differential metabolites
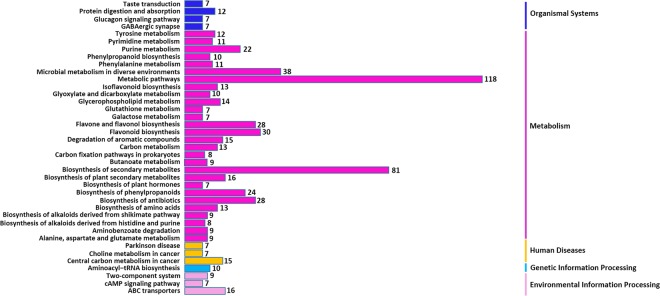
To obtain the pathway information of differential metabolites, the differential metabolites between the HW and SW were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). The results are shown in Fig. 4, these differential metabolites are mainly involved in metabolic pathways and biosynthesis of secondary metabolites, such as flavonoids, flavone and flavonol, phenylpropanoids and antibiotics etc. Flavonoids, flavone and flavonol were included in phenylpropanoids, which are a large class of plant secondary metabolites derived from phenylalanine in plants. In addition to flavonoids, it also includes monolignols, phenolic acids, stilbenes, and coumarins21–23. As we know, cellulose, hemicellulose and lignin are the major components of wood, but they do not exhibit color. It has been shown that the wood color is due to the existence of colored extractives contained in the wood. These colored extractives turn into dark color from a pale color by oxidation, polymerization and polymerization with wood main components over time24. Previous studies have shown that the extractives of some important colored woods were the flavonoids25,26, suggesting that the flavonoid metabolites identified in this study may explain the difference in wood color between the HW and SW in T. chinensis.
Figure 4.
The distribution of metabolic pathways of the differential metabolites.
Flavonoids identified in HW
Wood color is one of the important properties related to the wood utilization24–26. Flavonoids as the major pigment molecules are widely studied in plants and it has been shown that the flavonoids play a crucial role in the formation of wood color24–26. So the flavonoids in HW were selected for further analysis. The results are shown in Table 2, among 313 differential metabolites, we identified a total of 71 flavonoids and isoflavones, including 28 flavones, 3 anthocyanins, 16 flavonols, 11 flavonoids, 8 flavanones and 5 isoflavones. 42 flavonoids were higher and 29 flavonoids lower in HW compared to SW. Among them, the top 5 metabolites that were higher in HW were C-pentosyl-apigenin O-p-coumaroylhexoside, Ayanin, O-methylChrysoeriol 8-C-hexoside, malvidin 3-O-glucoside and chrysin O-hexoside, but the top 5 metabolites significantrly lower in HW compared to SW were 2′-hydroxydaidzein, 8-C-hexosyl chrysoeriol O-hexoside, cyanidin 3-O-malonylhexoside, isorhamnetin 5-O-hexoside and isorhamnetin 3-O-glucoside. These differential flavonoids metabolites show the difference of components between the HW and SW in T. chinensis, but the mechanism for the difference of components between the HW and SW is currently unknown and needs to be further investigated. Moreover, how these differential metabolites affect the wood color of HW also need to be further investigated.
Table 2.
A list of flavonoid metabolites identified in heartwood and sapwood.
| Compounds | Class | VIP | Fold_Change | Log2FC | Regulate |
|---|---|---|---|---|---|
| Cyanidin 3-O-malonylhexoside | Anthocyanins | 1.227 | 0.00 | −14.63 | down |
| O-methylChrysoeriol 5-O-hexoside | Flavone | 1.224 | 69925.93 | 16.09 | up |
| Chrysoeriol 5-O-hexoside | Flavone | 1.226 | 0.00 | −14.29 | down |
| Chrysin O-hexoside | Flavone | 1.226 | 119259.26 | 16.86 | up |
| O-methylnaringenin C-pentoside | Flavone | 1.207 | 5.66 | 2.50 | up |
| O-methylChrysoeriol 8-C-hexoside | Flavone | 1.226 | 131111.11 | 17.00 | up |
| Ayanin | Flavonol | 1.227 | 708148.15 | 19.43 | up |
| O-methylChrysoeriol 7-O-hexoside | Flavone | 1.226 | 75259.26 | 16.20 | up |
| Isorhamnetin O-hexoside | Flavonol | 1.226 | 0.00 | −14.31 | down |
| Selgin O-hexosyl-O-hexoside | Flavone | 1.226 | 25481.48 | 14.64 | up |
| Luteolin O-hexosyl-O-pentoside | Flavone | 1.223 | 80074.07 | 16.29 | up |
| Apigenin O-hexosyl-O-pentoside | Flavone | 1.223 | 4907.41 | 12.26 | up |
| Chrysin 5-O-glucoside (Toringin) | Flavone | 1.031 | 71.33 | 6.16 | up |
| Luteolin 3′,7-di-O-glucoside | Flavone | 1.226 | 0.00 | −12.42 | down |
| Isorhamnetin 5-O-hexoside | Flavonol | 1.227 | 0.00 | −14.51 | down |
| Chrysoeriol 7-O-hexoside | Flavone | 1.227 | 0.00 | −14.34 | down |
| C-hexosyl-isorhamnetin O-hexoside | Flavone | 1.227 | 32814.81 | 15.00 | up |
| 6-C-hexosyl chrysoeriol O-hexoside | Flavone | 1.227 | 38962.96 | 15.25 | up |
| C-pentosyl apigenin O-salicyloyl hexoside | Flavone | 1.223 | 38629.63 | 15.24 | up |
| C-pentosyl-apigenin O-p-coumaroylhexoside | Flavone | 1.227 | 2051851.85 | 20.97 | up |
| 8-C-hexosyl chrysoeriol O-hexoside | Flavone | 1.226 | 0.00 | −15.69 | down |
| Tricin 5-O-β-guaiacylglycerol | Flavone | 1.173 | 39.99 | 5.32 | up |
| Tricin O-rhamnoside | Flavone | 1.225 | 12848.15 | 13.65 | up |
| Apigenin C-hexosyl-O-rutinoside | Flavone | 1.057 | 2.27 | 1.19 | up |
| Tricin 7-O-hexoside | Flavone | 1.225 | 36296.30 | 15.15 | up |
| Tricin 4′-O-β-guaiacylglycerol | Flavone | 1.212 | 8200.00 | 13.00 | up |
| Tricin O-eudesmic acid | Flavone | 1.225 | 21555.56 | 14.40 | up |
| Di-O-methylquercetin | Flavonol | 1.157 | 0.32 | −1.63 | down |
| Kaempferol 7-O-rhamnoside | Flavonol | 1.210 | 0.21 | −2.25 | down |
| Chrysoeriol | Flavone | 1.095 | 0.36 | −1.47 | down |
| Kaempferol 3-O-rutinoside (Nicotiflorin) | Flavonol | 1.154 | 0.19 | −2.40 | down |
| Naringenin | Flavanone | 1.182 | 17.86 | 4.16 | up |
| Apigenin | Flavone | 1.202 | 0.30 | −1.72 | down |
| Malvidin 3-O-galactoside | Anthocyanins | 1.225 | 101592.59 | 16.63 | up |
| Malvidin 3-O-glucoside (Oenin) | Anthocyanins | 1.226 | 130888.89 | 17.00 | up |
| Xanthohumol | Flavanone | 1.225 | 7825.93 | 12.93 | up |
| Myricetin | Flavonol | 1.226 | 50074.07 | 15.61 | up |
| Isorhamnetin 3-O-neohesperidoside | Flavonol | 1.156 | 0.14 | −2.80 | down |
| Isorhamnetin | Flavonol | 1.144 | 13.00 | 3.70 | up |
| 7-O-Methyleriodictyol | Flavanone | 1.073 | 0.23 | −2.09 | down |
| Kaempferol 3-O-robinobioside (Biorobin) | Flavonol | 1.182 | 0.19 | −2.38 | down |
| Isohemiphloin | Flavone | 1.226 | 3637.04 | 11.83 | up |
| Hesperetin 7-rutinoside (Hesperidin) | Flavanone | 1.093 | 0.24 | −2.07 | down |
| Naringenin chalcone | Flavanone | 1.181 | 20.49 | 4.36 | up |
| Aromadedrin (Dihydrokaempferol) | Flavonol | 1.225 | 35185.19 | 15.10 | up |
| 3-Hydroxyflavone | Flavonol | 1.225 | 36000.00 | 15.14 | up |
| Liquiritigenin | Flavanone | 1.225 | 0.00 | −10.98 | down |
| Biochanin A | Isoflavone | 1.062 | 0.26 | −1.92 | down |
| 2′-Hydroxydaidzein | Isoflavone | 1.227 | 0.00 | −15.93 | down |
| 2′-Hydroxygenistein | Isoflavone | 1.226 | 45962.96 | 15.49 | up |
| Afzelechin (3,5,7,4′-Tetrahydroxyflavan) | Flavanone | 1.013 | 0.03 | −5.15 | down |
| 3,7-Di-O-methylquercetin | Flavonol | 1.062 | 7.16 | 2.84 | up |
| Prunetin | Isoflavone | 1.043 | 0.32 | −1.66 | down |
| Tricetin | Flavone | 1.150 | 0.20 | −2.29 | down |
| Rhamnetin (7-O-methxyl quercetin) | Flavonol | 1.227 | 16259.26 | 13.99 | up |
| Fustin | Flavonol | 1.200 | 0.19 | −2.43 | down |
| Rotenone | Isoflavone | 1.226 | 35777.78 | 15.13 | up |
| Homoeriodictyol | Flavanone | 1.150 | 0.34 | −1.55 | down |
| 4,2′,4′,6′-Tetrahydroxychalcone | Flavone | 1.187 | 23.28 | 4.54 | up |
| Narcissoside | Flavonoid | 1.106 | 0.14 | −2.83 | down |
| Orientin | Flavonoid | 1.226 | 23074.07 | 14.49 | up |
| 5,7-Dihydroxychromone | Flavonoid | 1.088 | 0.49 | −1.03 | down |
| Herbacetin | Flavonoid | 1.172 | 0.23 | −2.13 | down |
| Isorhamnetin 3-O-glucoside | Flavonoid | 1.227 | 0.00 | −14.36 | down |
| Farrerol | Flavonoid | 1.092 | 0.14 | −2.84 | down |
| Puararin | Flavonoid | 1.225 | 4048.15 | 11.98 | up |
| Diosmin | Flavonoid | 1.225 | 2848.15 | 11.48 | up |
| Tectochrysin | Flavonoid | 1.226 | 44629.63 | 15.45 | up |
| Tectorigenin | Flavonoid | 1.226 | 26111.11 | 14.67 | up |
| Glabridin | Flavonoid | 1.227 | 48370.37 | 15.56 | up |
| Chalcone | Flavonoid | 1.224 | 10122.22 | 13.31 | up |
Materials and Methods
Plant materials
The wood samples were collected from the branch of an approximately 30-year-old T. chinensis (Pilger) Rehd. in May 2015 from Liangdang County (106°25′E, 33°41′N) in Shanxi province of China. The HW and SW was separated on the basis of color, the inner wood was HW and the outer SW. Three independent biological replicates were tested for each sample. Samples were dried naturally and ground to a fine powder for usage.
Sample preparation for metabolite profiling
The HW and SW was separated by Tungsten steel knife. The dried HW and SW samples were ground using a mixer mill (MM 400, Retsch). The powder about 100 mg was extracted with 1.0 mL 70% methanol overnight at 4 °C, then the extraction was centrifuged at 10,000 g for 10 min. The extracts were absorbed and filtrated before LC-MS analysis. The mixed samples of the HW and SW as a quality control were prepared according to the above methods.
Liquid chromatographic mass spectrometry
LC-ESI-MS/MS system (HPLC, Shim-pack UFLC SHIMADZU CBM30A system, MS, Applied Biosystems 6500 Q TRAP) were used to analyze the sample extracts. HPLC analyses used Waters ACQUITY UPLC HSS T3 C18 (1.8μm, 2.1 mm*100 mm) column.
solvent system of mobile phase was 0.04% acetic acid in water and 0.04% acetic acid in acetonitrile; the gradient program was as follows: 95:5 V/V at 0 min, 5:95 V/V at 11.0 min, 5:95 V/V at 12.0 min, 95:5 V/V at 12.1 min, 95:5 V/V at 15.0 min; the temperature of the column was 40 °C; the injection volume was 2 μl and the flow rate was 0.4 ml per minute.
The MS parameter was set as described previously16,18. In brief, the temperature of ESI source was 500 °C; the voltage of ion spray was 5500 V; ion source gas I, gas II and curtain gas were set at 55, 60, and 25.0 psi, respectively; the collision gas was set to high. QQQ scans were acquired as MRM experiments with collision gas set to 5 psi. Declustering potential and collision energy for individual MRM transitions was performed with further optimization.
Qualitative and quantitative analysis of metabolites
Based on the MVDB V2.0 Database of Wuhan Maiteville Biotechnology Co., Ltd. (Wuhan, China) and the metabolite information public database27–31, the qualitative analysis was performed according to the secondary spectrum information. The isotopic signal is removed during analysis, including K+, Na+, NH4+ and other fragment ions of large molecular weight substances. The quantitative analysis of metabolites was based on the MRM mode as described previously16,18. In the MRM mode, the mass spectrum peak of each different color represented a metabolite. The characteristic ions for each metabolite were filtrated through the triple quadrupole mass spectrometer to obtain the signal strengths. Integration of chromatographic peaks was carried out using MultiQuant. In order to ensure the qualitative and quantitative accuracy, the mass spectrum peaks detected in different samples of each metabolite were corrected based on retention time and peak type according to the method described by Fraga et al.32.
Statistical analysis
Before analysis, the raw metabolic data were normalized by the method described previously33, and the normalized data were log2 transformed before they were used for further analysis. Hierarchical clustering analysis (HCA), principle component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) have been used to analyze the multivariate and differences of metabolites by soft R (www.r-project.org/) according to the previous study described34,35. Based the results of PLS-DA, a fold-change ≥2 or ≤0.5 among the metabolites with a VIP (variable importance in project) value >1 was used to identify differential metabolites. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to annotate the differential metabolites and analyze metabolic pathways36.
Conclusions
Wood color is one of the important factors related to wood property. It is critical to understand the chemical compositions that determine the wood color formation. The HW of T. chinensis has high commercial value for its purple red brown color and texture density. To the best of our knowledge, the difference of components between the HW and SW in T. chinensis have not been previously investigated. In this study, the components of the HW and SW in T. chinensis have been analyzed using widely targeted metabolic profiling. A total of 607 metabolites were detected in HW and SW. Among them, 146 metabolites were found significantly higher and 167 metabolites significantly lower in HW as compared to SW. These differential metabolites were mainly involved in metabolic pathways and biosynthesis of secondary metabolites, such as flavonoids, flavone and flavonol, phenylpropanoids and antibiotics. Moreover, the flavonoids in HW associated with wood color were identified. The results provide insight into the metabolites associated with wood color formation and may be useful for understanding the metabolites associated with wood quality.
Supplementary information
Acknowledgements
The work was supported by the National Key Research & Development Program of China (Grant Number 2017YFD0600205) and the Natural Science Foundation of China (Grant Numbers 31700584 and 31670676).
Author contributions
F.S. analyzed the data and performed the experiments; F.S. wrote the manuscript; D.Q. and F.S. designed the experiment; X.L. and W.M. performed sample collection and sample preparation; L.Z., J.G. and I.W. assisted in analyzing the data. All authors have read and approved the version of manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53839-2.
References
- 1.Chiang V. From rags to riches. Nat Biotechnol. 2002;20:557–558. doi: 10.1038/nbt0602-557. [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 3.Hinchee M, et al. Short-rotation woody crops for bioenergy and biofuels applications. In Vitro Cell Dev Bio Plant. 2009;45:619–629. doi: 10.1007/s11627-009-9235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarkanen KV. Renewable resources for the production of fuels and chemicals. Science. 1976;191:773–776. doi: 10.1126/science.191.4228.773. [DOI] [PubMed] [Google Scholar]
- 5.Celedon JM, Bohlmann J. An extended model of heartwood secondary metabolism informed by functional genomics. Tree Physiol. 2018;38:311–319. doi: 10.1093/treephys/tpx070. [DOI] [PubMed] [Google Scholar]
- 6.Spicer R. Symplasmic networks in secondary vascular tissues: parenchyma distribution and activity supporting long-distance transport. J Exp Bot. 2014;65:1829–1848. doi: 10.1093/jxb/ert459. [DOI] [PubMed] [Google Scholar]
- 7.Miranda I, Sousa V, Ferreira J, Pereira H. Chemical characterization and extractives composition of heartwood and sapwood from Quercus faginea. PLoS One. 2017;12:e0179268. doi: 10.1371/journal.pone.0179268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampe, A. & Magel, E. New insights into heartwood and heartwood formation. (Springer, Berlin, German, 2013).
- 9.George B, Suttie E, Merlin A, Deglise X. Photodegradation and photostabilisation of wood—The state of the art. Polym Degrad Stab. 2005;88:268–274. doi: 10.1016/j.polymdegradstab.2004.10.018. [DOI] [Google Scholar]
- 10.Sun QF, et al. Preliminary observations of hydrothermal growth of nanomaterials on wood surfaces. Wood Sci Technol. 2014;48:51–58. doi: 10.1007/s00226-013-0570-7. [DOI] [Google Scholar]
- 11.Yao Q, et al. One-step solvothermal deposition of ZnO nanorod arrays on a wood surface for robust superamphiphobic performance and superior ultraviolet resistance. Sci Rep. 2016;6:35505. doi: 10.1038/srep35505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wani MC, et al. McPhailPlant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J Am Chem Soc. 1971;9:2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- 13.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 14.Craag GM, Scheparat SA, Suffness M, Grever MR. The taxol supply crisis. New NCI policies for handling the large-scale production of novel natural product anticancer and anti-HIV agents. J Nat Prod. 1993;56:1657–1668. doi: 10.1021/np50100a001. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz SB. How to make taxol from scratch. Nature. 1994;367:593–594. doi: 10.1038/367593a0. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, et al. Identification of Nutritional Components in Black Sesame Determined byWidely Targeted Metabolomics and Traditional Chinese Medicines. Molecules. 2018;23:E1180. doi: 10.3390/molecules23051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu F, et al. Characterization of eleutheroside B metabolites derived from an extract of Acanthopanax senticosus Harms by high-resolution liquid chromatography/quadrupole time-of-flight mass spectrometry and automated data analysis. Biomed. Chromatogr. 2012;26:1269–1275. doi: 10.1002/bmc.2688. [DOI] [PubMed] [Google Scholar]
- 18.Wu KX, et al. A Comparative Metabolomics Analysis Reveals the Tissue-Specific Phenolic Profiling in Two Acanthopanax Species. Molecules. 2018;23:E2078. doi: 10.3390/molecules23082078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons H, O’Gorman A, Brennan L. Metabolomics as a tool in nutritional research. Curr Opin Lipidol. 2015;26:30–34. doi: 10.1097/MOL.0000000000000140. [DOI] [PubMed] [Google Scholar]
- 20.Meijón M, et al. Exploring natural variation of Pinus pinaster Aiton using metabolomics: is it possible to identify the region of origin of a pine from its metabolites? Mol Ecol. 2016;25:959–976. doi: 10.1111/mec.13525. [DOI] [PubMed] [Google Scholar]
- 21.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 22.Noel JP, Austin MB, Bomati EK. Structure function relationships in plant phenylpropanoid biosynthesis. Curr Opin Plant Biol. 2005;8:249–253. doi: 10.1016/j.pbi.2005.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, et al. Phenylpropanoids, flavonoids, and terpenoids from Artemisia ordosica Krasch. Magn Reson Chem. 2019;57:326–330. doi: 10.1002/mrc.4846. [DOI] [PubMed] [Google Scholar]
- 24.Yazaki Y. Wood Colors and their Coloring Matters: A Review. Nat Prod Commun. 2015;10:505–12. [PubMed] [Google Scholar]
- 25.Surowiec I, Nowik W, Trojanowicz M. Identification of “insoluble” red dyewoods by high performance liquid chromatography-photodiode array detection (HPLC-PDA) fingerprinting. J Sep Sci. 2004;27:209–216. doi: 10.1002/jssc.200301612. [DOI] [PubMed] [Google Scholar]
- 26.Robertson, A. & Whalley, W. B. The chemistry of the “insoluble red” woods. VI. Santalin and santarubin. Journal of the Chemical Society. 2794–2801 (1954).
- 27.Smith CA, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 28.Horai H, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 29.Afendi FM, et al. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- 30.Wishart DS, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moco S, et al. A liquid chromatography-mass spectrometry-based metabolome database for tomato. Plant Physiol. 2006;141:1205–1218. doi: 10.1104/pp.106.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraga CG, Clowers BH, Moore RJ, Zink EM. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal Chem. 2010;82:4165–4173. doi: 10.1021/ac1003568. [DOI] [PubMed] [Google Scholar]
- 33.Zhu G, et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell. 2018;172:249–261. doi: 10.1016/j.cell.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Thévenot EA, et al. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J Proteome Res. 2015;14:3322–35. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, et al. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chemistry. 2016;199:8–17. doi: 10.1016/j.foodchem.2015.11.113. [DOI] [PubMed] [Google Scholar]
- 36.Kanehisa M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:353–361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.