Abstract
Purpose
Quantitative imaging methods could improve diagnosis of rotator cuff degeneration, but the capability of quantitative MR and US imaging parameters to detect alterations in collagen is unknown. The goal of this study was to assess quantitative MR and US imaging measures for detecting abnormalities in collagen using an in vitro model of tendinosis with biochemical and histological correlation.
Method
36 pieces of supraspinatus tendons from 6 cadaveric donors were equally distributed into 3 groups (2 subjected to different concentrations of collagenase and a control group). Ultrashort echo time MR and US imaging measures were performed to assess changes at baseline and after 24 hours of enzymatic digestion. Biochemical and histological measures, including brightfield, fluorescence, and polarized microscopy, were used to verify the validity of the model and were compared with quantitative imaging parameters. Correlations between the imaging parameters and biochemically measured digestion were analyzed.
Results
Among the imaging parameters, macromolecular fraction (MMF), adiabatic T1ρ, T2*, and backscatter coefficient (BSC) were useful in differentiating between the extent of degeneration among the 3 groups. MMF strongly correlated with collagen loss (r=−0.81; 95% confidence interval [CI]: −0.90,−0.66), while the adiabatic T1ρ (r=0.66; CI: 0.42,0.81), T2* (r=0.58; CI: 0.31,0.76), and BSC (r=0.51; CI: 0.22,0.72) moderately correlated with collagen loss.
Conclusions
MMF, adiabatic T1ρ, and T2* measured and US BSC can detect alterations in collagen. Of the quantitative MR and US imaging measures evaluated, MMF showed the highest correlation with collagen loss and can be used to assess rotator cuff degeneration.
Keywords: rotator cuff tendon, tendinopathy, quantitative MRI, UTE, quantitative ultrasound
Introduction
Rotator cuff tendinopathy is common, but the non-invasive diagnosis of tendinosis and assessment of tendon quality remains a challenge. Clinical exams demonstrate poor specificity for tendinosis in part due to their inability to selectively test the cuff tissues [1]. On MR imaging, tendinosis is represented by an increase in signal intensity without morphologic disruption. However, signal intensity can increase in normal RCTs as a result of the magic angle effect [2]. On US imaging, tendinosis is characterized by a heterogeneous, ill-defined, and hypoechoic area in the tendon without a defect. Due to subjectivity in analyses of these imaging findings, sensitivity of these modalities for the diagnosis of tendinosis has been reported to range from 13–79%, with poor interobserver reliability [3, 4]. Quantitative methods would improve diagnosis and consequently benefit treatment planning and longitudinal follow-up.
In recent years, improvements in clinical imaging system hardware and signal processing have facilitated the implementation of quantitative imaging techniques. Promising quantitative MR imaging techniques include ultrashort echo time (UTE) imaging, which is well suited for the evaluation of short T2 structures, including RCTs [5]. In particular, the UTE magnetization transfer technique with two-pool modeling has been shown to be resistant to the magic angle effect, while maintaining the ability to distinguish between histologically normal and abnormal tendons [6, 7]. Similarly, acquisition of raw radiofrequency (RF) US data permits reliable calculation of fundamental ultrasonic parameters, including backscatter coefficient (BSC), which is sensitive to extracellular matrix changes [8].
An important consideration of quantitative imaging is the sensitivity of the technique to fundamental tissue properties, such as structure and composition. In RCT degeneration, alterations in collagen proportion and property are characteristic [9], but to our knowledge, the capability of quantitative MR and US imaging parameters to assess these changes is unknown. The purpose of this study was to assess the capability of quantitative MR and US imaging measures to detect abnormalities in collagen using an in vitro model of tendinosis with biochemical and histological correlation.
Materials and Methods
Study Design
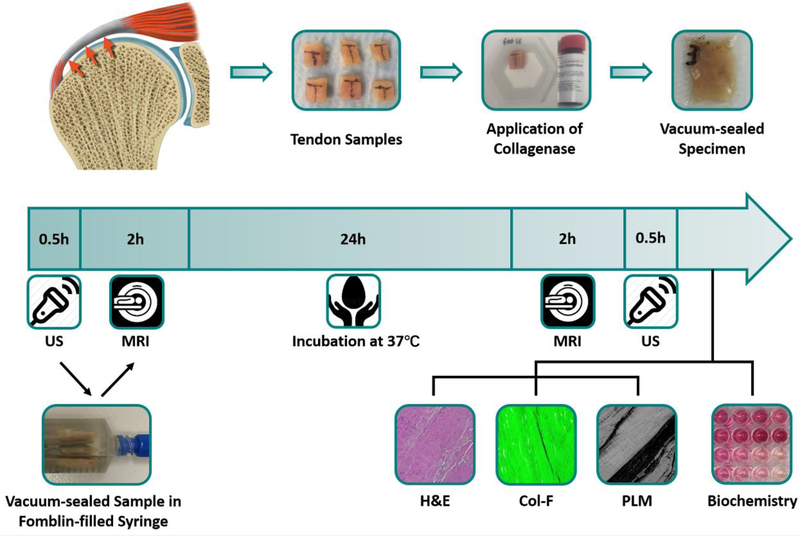
36 supraspinatus tendon samples were prepared from 6 specimens (mean age 52.8 years; range 36–64 years). Samples were equally distributed into three groups: (1) digestion with 600U collagenase, (2) digestion with 150U collagenase, and (3) undigested controls. 100 μl of solution of different concentrations (600U or 150U total) of collagenase type VII (C0773, Sigma-Aldrich Co., St. Louis, MO, USA) in 20 mM calcium acetate and 100 mM Tris, or vehicle control, were applied. The solution was applied on all sides of the sample, and samples were then vacuum sealed in small pouches for immediate MR and US scanning to establish baseline. After the MR and US scans, samples in the syringes were incubated at 37°C for 24 hours and imaging was repeated, followed by biochemical and histological testing. The experimental design is described in Figure 1.
Figure 1:
Study design. Supraspinatus tendon samples were harvested from cadaveric shoulder specimens, treated with different concentrations of collagenase, and vacuum-sealed. MR and US imaging were performed at baseline and again after 24 hours of incubation, followed by biochemical and histological analysis.
MR Imaging Protocol
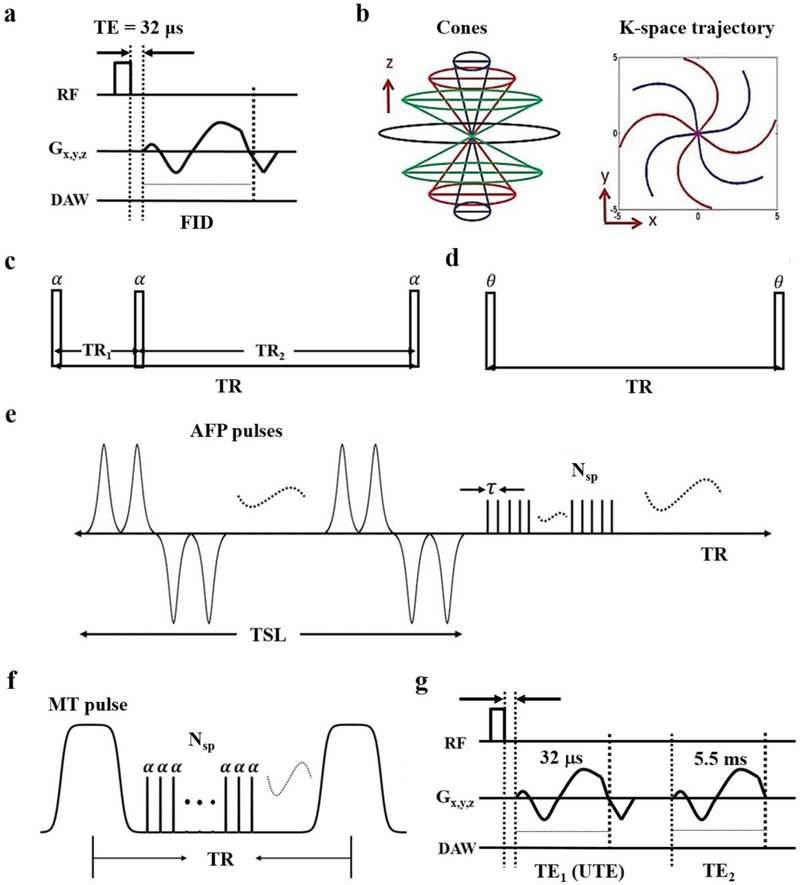
Using a clinical 3T scanner (MR750, GE Healthcare, Milwaukee, WI) and a homemade birdcage coil, a 3D UTE sequence was used (Figure 2a) with a cones readout (Figure 2b) [10]. The 3D UTE-Cones sequences included: 1) actual flip angle imaging and variable flip angle-based (AFI-VFA) T1 relaxation time (Figures 2c and 2d) [11], 2) adiabatic T1ρ relaxation time (Figure 2e) [12], 3) magnetization transfer (MT) imaging with two-pool modeling (Figure 2f) [13], and 4) T2* relaxation time (Figure 2g). Parameters are shown in Table 1 with total imaging time of 104 minutes. Using MATLAB (MathWorks, Natick, MA, USA), a region of interest (ROI) was drawn over the entire sample on the middle slice. Then, T1, adiabatic T1ρ, macromolecular fraction (MMF), and T2* were calculated and parametric maps were generated.
Figure 2.
UTE imaging. a) In the standard 3D UTE-Cones sequences, a short rectangular pulse is used for excitation followed by spiral sampling. b) Spiral trajectories are arranged with conical view ordering. c) The 3D UTE-Cones AFI sequence employs a pair of interleaved TRs (α=actual flip angle) for accurate B1 mapping, which followed by the VFA method with a single TR (θ is the nominal flip angle) provides accurate T1 measurements (d). e) The 3D adiabatic T1ρ UTE-Cones sequence used a train of AFP pulses, followed by 3D UTE-Cones acquisition. f) A Fermi pulse is used for MT preparation followed by 3D UTE-Cones acquisition. g) Dual-echo 3D UTE-Cones imaging consists of two 3D spiral samplings with two different TEs.
Table 1:
Quantitative MRI Sequences and Parameters
| Sequence | Sequence Parameters |
|---|---|
| AFI-VFA T1 | AFI: FOV = 4×4×4.5 cm3, ST = 1.6 mm, matrix = 192 × 192 × 28, TR1/TR2 = 20/100 ms, TE = 0.032 ms, FA = 45°, BW = 86.6 kHz, total time = 5 minutes VFA: FOV = 4×4×4.5 cm3; ST = 1.6 mm; matrix = 192 × 192 × 28; TR = 20 msl; TE = 0.032 ms; FA = 5°, 10°, 20°, and 30°; BW = 86.6 kHz, NEX=1, total time = 5 minutes |
| Adiabatic T1ρ | Adiabatic pulse: hyperbolic secant type 1 pulse with a duration of 6.048 ms, BW of 1.643 kHz and maximum B1 amplitude of 17 μT; FOV = 4×4×4.5 cm3; ST = 1.6 mm; matrix = 192 × 192 × 28; TR = 500 ms; TE = 0.032 ms; FA = 10°; TSL = 6, 12, 24, 36, 48, 72, and 96 ms; BW = 86.6 kHz, NEX =1, total time = 17.5 minutes |
| MT | FOV = 4×4×4.5 cm3; ST = 1.6 mm; matrix = 192 × 192 × 28; TR = 102 ms; TE = 0.032 ms; FA = 7°; MT pulse powers of saturation (FA = 400°, 600°, and 800°); MT offset frequency = 2, 5, 10, 20, and 50 kHz; BW = 86.6 kHz, NEX =1, total time = 22 minutes |
| T2* | FOV = 4×4×4.5 cm3; ST = 1.6 mm; matrix = 192 × 192 × 28; TR = 50 ms; TE = 0.032, 0.2, 0.4, 0.6, 0.8, 2.2, 4.4, 5.5, 6.6, 8.8, 11, 13, 15, and 22 ms; FA = 10°; BW = 86.6 kHz, NEX =1, total time = 44 minutes |
AFI-VFA T1 = actual flip angle and variable flip angle-based T1, MT = magnetic transfer, FOV = field of view, ST = slice thickness, TR = time-to-repetition, TE = time-to-echo, FA = flip angle, BW = bandwidth, NEX=number of excitations, TSL = time-to-spin lock
Ultrasound Imaging
Using a clinical scanner (VevoMD, FUJIFILM, Toronto, Canada) and an 18 MHz transducer (UHF22) B-mode images and RF data were acquired with ROIs drawn similar to the MR images. Attenuation coefficient (AC) and backscatter coefficient (BSC) were calculated as previously described using a reference phantom technique [8, 14]. The Nakagami parameter was estimated using the maximum likelihood estimator [15]. All quantitate US parameters were calculated for subregions of 15 wavelengths at 18 MHz. Parametric maps were also generated.
Biochemical Analyses
After all MR and US imaging was completed, one half of each sample was ground and washed in distilled water to fully dissolve the digested collagen fragments into solution. Suspensions were then centrifuged and hydroxyproline was quantified using an assay kit (K555–100, BioVision, Milpitas, CA) and spectrophotometer (SpectraMax 340PC, Molecular Devices, Menlo Park, CA). The biochemistry analysis was duplicated and averaged for each sample and digested collagen was defined as:
Histology and Analyses
The other half of each tissue sample was fixed, paraffin-embedded, sectioned, and stained with hematoxylin and eosin (H&E), picrosirius red, and Col-F (ImmunoChemistry Technologies, Bloomington, MN), which has an affinity for collagen and elastin. Image quantification was performed with ImageJ (Fiji, National Institutes of Health, Bethesda, MD). Fluorescence was defined as the mean signal intensity of the pixels with a gray scale threshold larger than 10 (range 0–255) in an ROI avoiding the tissue fissures. Optical retardation (Γ) was measured on samples stained with picrosirius red using a quantitative polarized light microscope system (OpenPolScope, Marine Biological Laboratory, Woods Hole, MA) [16]. Four randomly selected ROIs from the center and peripheral portions of the section were measured by a histotechnician, who was blinded to the MRI and US results.
Statistical Analyses
The Shapiro-Wilk test was used to assess normality. Quantitative MR and US measures at baseline and after treatment were compared using paired student’s t-tests. To account for baseline tendon differences, all endpoint imaging measures were normalized (Valueendpoint/Valuebaseline). Pearson’s correlations between the normalized parameters and biochemical results were performed. One-way ANOVA was used for comparison of measures with Tukey’s post-hoc test. P<0.05 was considered statistically significant. Statistical analyses were performed with SPSS (v19.0, SPSS Inc., Chicago, IL).
Results
Sample Preparation
All tendons were intact at the time of dissection without gross abnormality. Tendon morphology was comparable between baseline and post-incubation images, indicating that the use of high concentration collagenase in conjunction with a vacuum-sealing approach successfully minimized total water changes.
UTE-MR Imaging Measurements
Longitudinal relaxation time
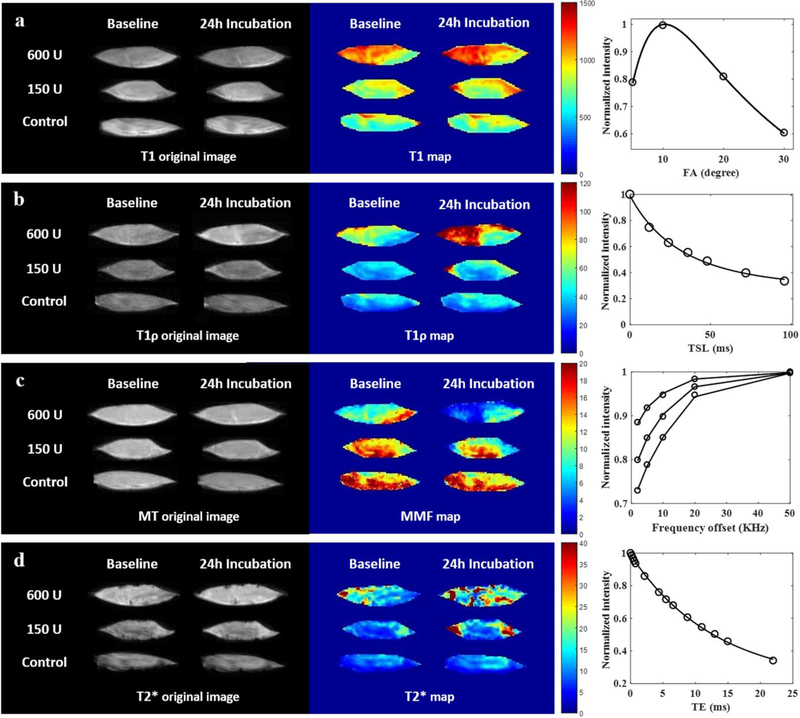
Excellent T1 fitting was achieved by using the AFI-VFA method. Quantitative biomarker measurements and normalized values are listed in Table 2 and Table 3. Representative T1 parametric maps are displayed in Figure 3a. Compared with baseline, the T1 values of the 600U (p=0.002) and 150U (p=0.033) samples were significantly increased after digestion, most notably at the peripheral regions, as noted on the parametric maps. No significant changes in T1 values were noted in the control samples after incubation. Similarly, no significant differences were found between normalized T1 values among the three groups.
Table 2:
UTE-MRI and US Measurements at Baseline and after 24 Hours of Incubation
| Parameter | 600 U (n = 12) | 150 U (n = 12) | Control (n = 12) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | 24h Incubation | P value | Baseline | 24h Incubation | P value | Baseline | 24h Incubation | P value | |
| UTE-MRI | |||||||||
| T1 (ms) | 779.3 ± 135.7 | 847.3 ± 150.2 | 0.0021* | 707.25 ± 117.7 | 746.4 ± 119.7 | 0.0329* | 744.3 ± 127.6 | 757.5 ± 123.9 | 0.1173 |
| T1ρ (ms) | 41.2 ± 9.1 | 53.1 ± 14.0 | 0.0004† | 36.7 ± 5.4 | 41.7 ± 6.9 | 0.0002† | 40 ± 6.4 | 40.2 ± 6.0 | 0.7104 |
| MMF (%) | 12.5 ± 2.2 | 9.0 ± 2.4 | 0.000008† | 13.7 ± 1.5 | 11.5 ± 1.2 | 0.0000001† | 13.1 ± 1.7 | 13.0 ± 1.4 | 0.5591 |
| T2* (ms) | 10.8 ± 4.1 | 16.5 ± 8.0 | 0.0074† | 8.9 ± 2.6 | 11.5 ± 2.3 | 0.00004† | 9.0 ± 2.7 | 9.5 ± 2.9 | 0.0227* |
| US | |||||||||
| AC (dB/cmMHz) | 2.56 ± 0.95 | 2.56 ± 0.96 | 0.9923 | 2.56 ± 0.86 | 2.78 ± 0.85 | 0.1149 | 2.55 ± 0.63 | 2.84 ± 0.88 | 0.1089 |
| BSC (dB) | −12.46 ± 4.48 | −17.51 ± 5.57 | 0.00001† | −14.53 ± 4.98 | −18.85 ± 4.94 | 0.0015† | −14.26 ± 4.97 | −14.66 ± 4.7 | 0.6183 |
| Nakagami | 0.74 ± 0.09 | 0.76 ± 0.1 | 0.2940 | 0.78 ± 0.1 | 0.76 ± 0.09 | 0.3152 | 0.7. 0.05 | 0.72 ± 0.08 | 0.3071 |
Note – Data presented in means ± standard deviation. P compares the values of the biomarkers before and after digestion.
indicates a significant difference compared with baseline (p<0.05).
indicates a significant difference compared with baseline (p< 0.005).
UTE-MRI = ultrashort echo time MRI. MMF = macromolecular fraction. AC = attenuation coefficient. BSC = backscatter coefficient.
Table 3:
Comparison of Normalized UTE-MRI and US Biomarkers among Different Treatment Groups
| Normalized Parameter | 600 U (n = 12) | 150 U (n = 12) | Control (n = 12) | ANOVA p Value | Post Hoc Test p Value | ||
|---|---|---|---|---|---|---|---|
| 600 U vs Control | 150 U vs Control | 600 U vs 150 U | |||||
| UTE-MRI | |||||||
| Normalized T1 | 1.09 ± 0.08 | 1.06 ± 0.08 | 1.02 ± 0.04 | 0.0715 | 0.0581 | 0.3818 | 0.5562 |
| Normalized T1ρ | 1.3 ± 0.18 | 1.14 ± 0.08 | 1.01 ± 0.05 | 0.000007† | 0.000004† | 0.0328* | 0.0073* |
| Normalized MMF | 0.71 ± 0.13 | 0.84 ± 0.04 | 0.99 ± 0.06 | 0.00000002† | 0.00000001† | 0.0002† | 0.0034† |
| Normalized T2* | 1.56 ± 0.48 | 1.33 ± 0.19 | 1.06 ± 0.06 | 0.0011† | −0.0007† | 0.0852 | 0.1565 |
| US | |||||||
| Normalized AC | 1.01 ± 0.2 | 1.1 ± 0.18 | 1.12 ± 0.22 | 0.4056 | 0.4170 | 0.9706 | 0.5538 |
| Normalized BSC | 1.51 ± 0.41 | 1.36 ± 0.39 | 1.06 ± 0.24 | 0.0129* | 0.0108* | 0.1105 | 0.5693 |
| Normalized Nakagami | 1.03 ± 0.08 | 0.98 ± 0.08 | 1.02 ± 0.08 | 0.2599 | 0.9998 | 0.3317 | 0.3227 |
Note – Data presented in means ± standard deviation.
indicates a significant difference compared with baseline (p<0.05).
indicates a significant difference compared with baseline (p< 0.005).
UTE-MRI = ultrashort echo time MRI. MMF = macromolecular fraction. AC = attenuation coefficient. BSC = backscatter coefficient.
Figure 3:
Supraspinatus tendon sample from a 48-year-old male donor. Representative morphologic UTE images and parametric maps at baseline and after 24 hours of incubation with different conditions are shown, as are the fitting curves for the parameters. a) After treatment, T1 values of the 3 groups only slightly changed, as shown by small differences in color mainly at the periphery of the samples. b) Adiabatic T1ρ values increased more for the 600U collagenase group than for the 150U collagenase group, while adiabatic T1ρ values of the control group were unchanged. c) Macromolecular fraction (MMF) maps showed drastically reduced values throughout the whole sample after digestion with 600U of collagenase, decreases only at the periphery of the sample after digestion with 150U of collagenase, and no appreciable change in the control group. d) T2* values increased throughout the entire sample in the 600U group and at the periphery of the sample in the 150U group, while the control group showed no apparent change.
Spin-lock relaxation time
Well-fitted signal decay curves and adiabatic T1ρ maps are shown in a representative sample in Figure 3b. The adiabatic T1ρ values of the sample after 600U of collagenase digestion were significantly increased (p<0.001). As evident on the adiabatic T1ρ map of the sample digested in 600U of collagenase, the dramatic color change at the periphery compared with the moderate color change in the center indicates a depth-dependent response. The adiabatic T1ρ value of the sample after 150U of collagenase digestion also increased significantly (p<0.001), but with a moderate degree of change, again with the most notable color change at the periphery of the sample. There was no significant change in the adiabatic T1ρ values of the control samples, with stable parametric maps at baseline and after 24 hours of incubation. The normalized adiabatic T1ρ values were significantly different among the three groups (p<0.001), and significant differences were observed between each of the groups on post-hoc pairwise comparisons.
Macromolecular fraction
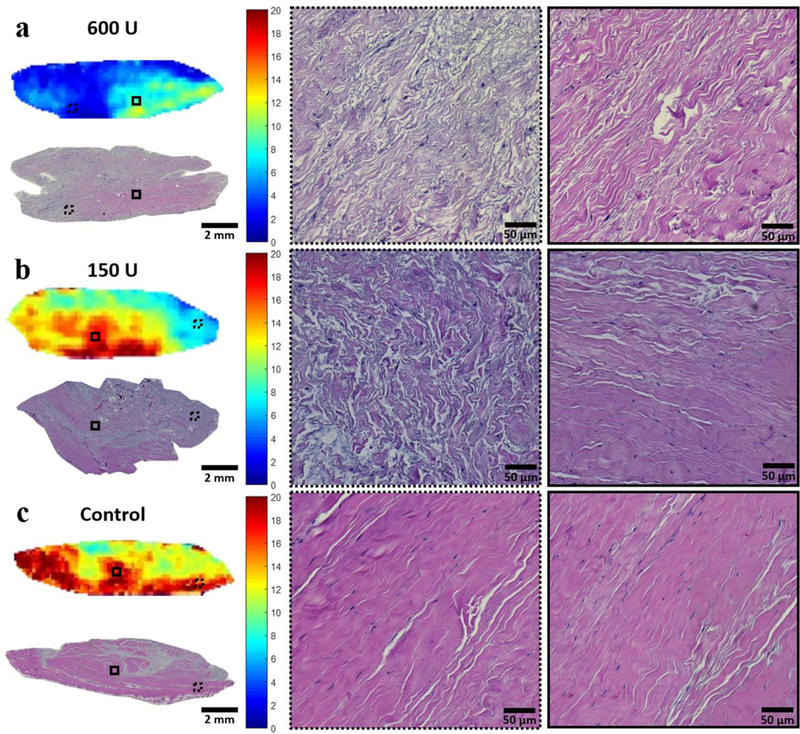
Excellent MT modeling curve fitting and MMF maps are shown in a representative sample in Figure 3c. The MMF of the 600U group significantly decreased from 12.5%±2.2 to 9.0%±2.4 (p<0.001) and the MMF of the 150U group significantly decreased from 13.7%±1.5 to 11.5%±1.2 (p<0.001), indicating loss of collagen protons as a result of collagenase digestion. Similar to the pattern on adiabatic T1ρ pixel maps, MMF pixel maps demonstrated a depth-dependent gradient of values after digestion. The samples subject to 600U of collagenase digestion experienced sufficient penetration to result in a decrease in MMF in the center, but there was still more digestion in the peripheral portions. Findings were also confirmed by the H&E histology results (Figure 4a). For the samples subject to 150U of collagenase digestion, the MMF reduction primarily occurred in the periphery, while the center was largely unaffected. Again, these findings were consistent with the histological observations of disorganized collagen fascicles at the periphery versus intact fascicles located at the center (Figure 4b). Again, the control samples did not show any notable changes on the parametric maps at baseline nor after incubation, and intact collagen fascicles were seen throughout the sample (Figure 4c). The normalized MMF values were significantly different among the three groups (p<0.001), and the significant differences were observed between each of the groups on post-hoc pairwise comparisons.
Figure 4:
Post-incubation MMF maps with photomicrograph correlation after hematoxylin and eosin staining from the same donor as shown in Figure 2. Magnifications of the periphery and center of the samples are shown in the dashed and solid ROIs, respectively. a) Low MMF in the 600U digested sample correlated with H&E stain, which showed separated and disorganized collagen fascicles both in the periphery and center of the sample. b) The peripheral portion of the 150U digested sample demonstrated low MMF, which correlated with disorganized collagen fascicles. Collagen fascicles were intact in the central portion. c) Control sample with high MMF correlated with intact collagen fascicles throughout the entire tendon.
Transverse relaxation time
The well-fitted signal decay curves and T2* maps are shown in a representative sample in Figure 3d. T2* values significantly increased after digestion (p=0.007, p<0.001), whereas no significant changes in the control samples were noted. T2* maps demonstrated similar, albeit less obvious, patterns to adiabatic T1ρ and MMF. A significant difference was found between normalized T2* values for the group subject to 600U of collagenase compared with controls (p<0.001).
Ultrasound Imaging
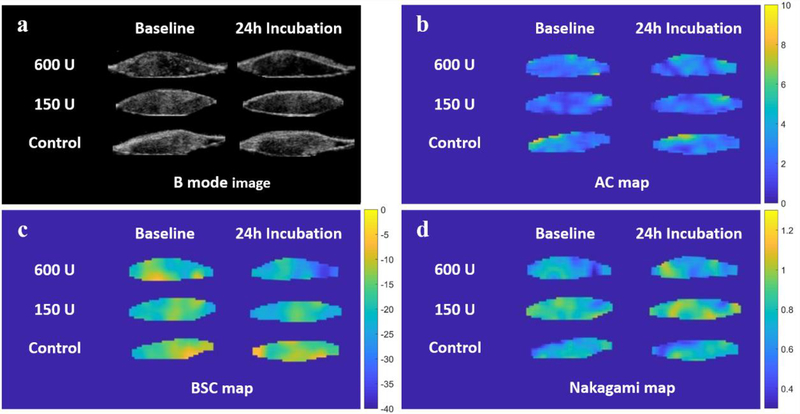
Representative US parametric maps are shown in Figure 5, and the original measurements and normalized values are listed in Table 2 and Table 3. The BSC values decreased significantly after digestion with 600U and 150U collagenase (p<0.001, p=0.001). The BSC maps also demonstrated a pattern of gradual color change from the periphery to the center of the sample after digestion, while the control pattern was consistent throughout the experiment. The 600U and control groups could be differentiated by normalized BSC (p=0.011). There were no significant changes in AC and Nakagami values after digestion, nor did the normalized values of these two parameters show any significant differences among the different treatment groups.
Figure 5:
Representative B-mode images and quantitative US parametric maps for the same donor as shown in Figure 2. a) Echogenicity of RCT samples was reduced after digestion with 600U and 150U of collagenase. b) Attenuation coefficient (AC) pixel maps showed no appreciable difference between baseline and after 24h of incubation. c) Backscatter coefficient (BSC) pixel maps showed that the most obvious change after incubation was in the sample incubated with 600U of collagenase enzyme. d) Nakagami pixel maps showed no clear difference or trend after digestion.
Biochemistry
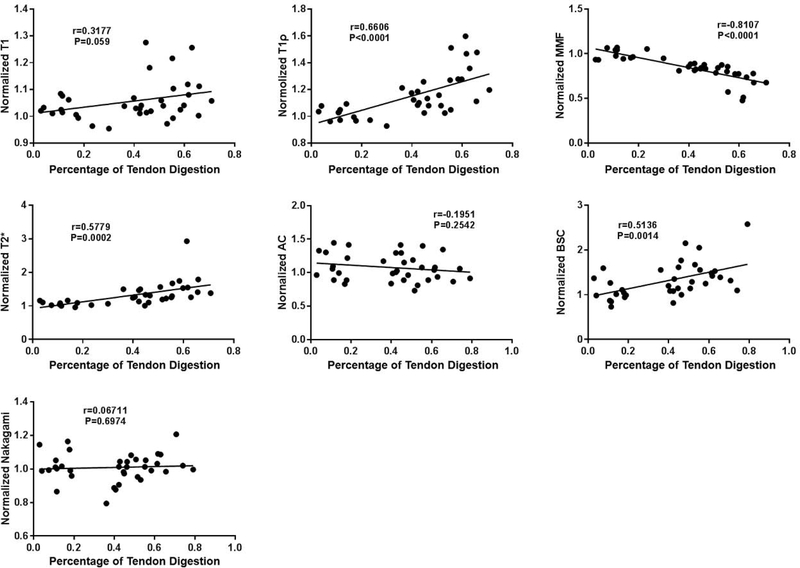
Biochemical analyses confirmed the efficiency of the RCT digestion model. 58.7% of collagen was digested using 600U of collagenase; 45.9% of collagen was digested using 150U of collagenase; and 13.5% of collagen was digested in the control group, likely due to endogenous collagenase. Significant differences in percentages of digestion were found among the three groups (Table 4). Correlations between the normalized parameters and the percentages of digestion are shown on Figure 6. The MMF strongly correlated with collagen loss (r=−0.81; 95% confidence interval [CI]: −0.90, −0.66), while the adiabatic T1ρ (r=0.66; CI: 0.42, 0.81), T2* (r=0.58; CI: 0.31, 0.76), and BSC (r=0.51; CI: 0.22, 0.72) moderately correlated with collagen loss. Correlations for the other parameters did not reach significance.
Table 4:
Comparison of Biochemistry and Histology Results Different Treatment Groups
| Biomarker | 600 U (n = 12) | 150 U (n = 12) | Control (n = 12) | ANOVA p Value | p Value of Post Hoc Test | ||
|---|---|---|---|---|---|---|---|
| 600U vs Control | 150U vs Control | 600U vs 150U | |||||
| Digested Percentage (%) | 58.7 ± 8.1 | 45.9 ± 6.3 | 13.5 ± 7.7 | 8E-16† | 5E-9† | 5E-9† | 0.0005† |
| Peripheral Fluorescence | 104.2 ± 39.3 | 127.7 ± 42.6 | 145.3 ± 31 | 0.0395* | 0.0311* | 0.5117 | 0.3077 |
| Peripheral Retardation (nm/μm2) | 11.6 ± 5.2 | 10.9 ± 4.3 | 15.8 ± 5.1 | 0.0059* | 0.0222* | 0.0097* | 0.9246 |
Note – Data in means ± standard deviation.
indicates a significant difference (p < 0.05).
indicates a significant difference (p< 0.005).
Whenever the ANOVA indicated a significant difference, the Tukey’s test was used to perform a post-hoc pairwise comparison of group means (significance level p=0.05).
Figure 6:
Significant positive correlations were observed between normalized adiabatic T1ρ, normalized T2*, normalized BSC, and tendon digestion percentage. Significant negative correlations were observed between normalized MMF and tendon digestion percentage. No significant correlations were seen between the other imaging biomarkers and tendon digestion percentage.
Histology
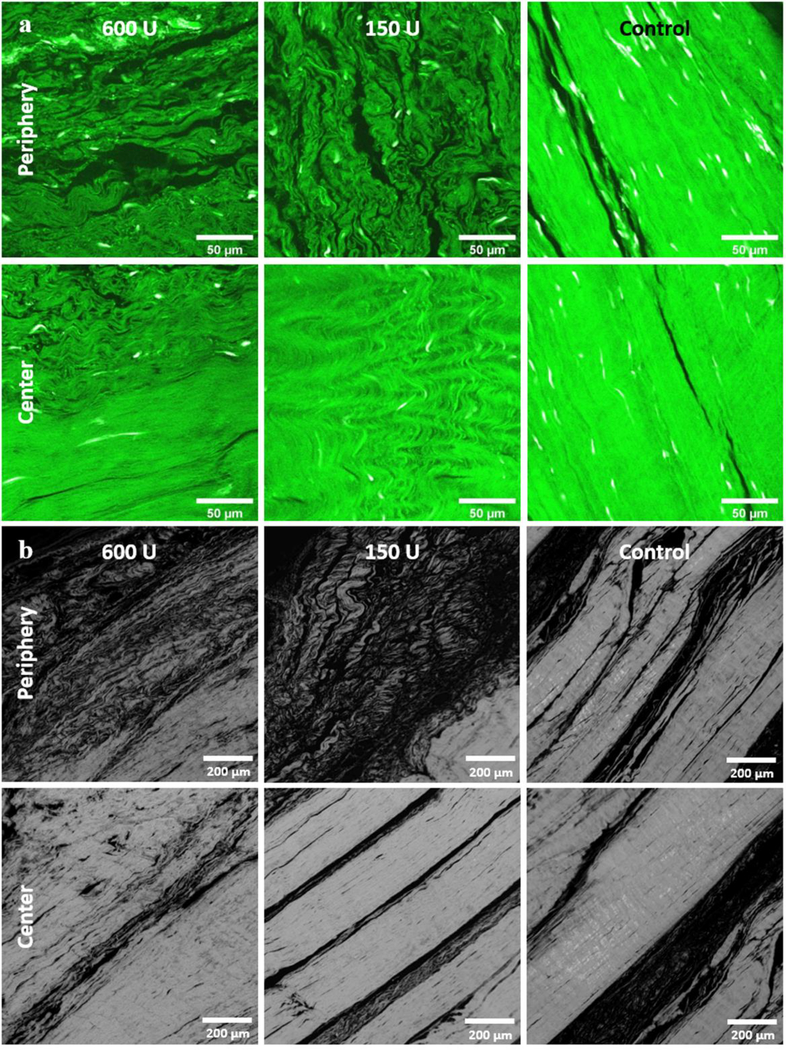
Representative Col-F and PLM images are shown in Figure 7, and their quantifications are listed in Table 4. In general, the fluorescence of Col-F was weak and retardation under PLM was low on the periphery of the digested samples. Relatively strong fluorescence and high retardation were demonstrated in the centers of the digested samples and throughout the control samples. In general, the morphology of the collagen bundles as demonstrated with Col-F and PLM was similar to that shown using the H&E stain at different spatial locations and with different treatments.
Figure 7:
Col-F staining and quantitative PLM results among the 3 digestion groups from the same donor as shown in Figure 2. a) Weak Col-F fluorescence is shown in the periphery of the 600U and 150U collagenase digested samples, while relatively strong fluorescence was present in the center of all the samples. Notably, the control sample had strong fluorescence at both the peripheral and central portions. b) Low retardation was observed in the periphery of the 600U and 150U samples, while relatively high retardation was observed in the centers of the digested samples and throughout the control samples.
Discussion
In this study, we used an in vitro model of tendon degeneration to assess the capability of quantitative MR and US imaging techniques for determining collagen degradation. In vitro models of degeneration using collagenase have been used before on tendons [17] and cartilage [18, 19], but unlike previous models which immersed their samples in solutions, we sought to minimize changes in water by utilizing a minimum amount of solution (100 μl) and vacuum-sealing our samples. The success of our model was confirmed with biochemical (hydroxyproline) and multiple histological assays, including brightfield (H&E), fluorescence (Col-F), and quantitative PLM. Similar to other groups, we noted a depth-dependent response with greater effect of tendon degeneration at the superficial portions [19]. This is due to the large size of the collagenase molecule (68,000–125,000 daltons), which limits penetration into the tendon substance. We applied collagenase to the surface, where the enzyme begins to cleave fibrillar collagen, degrading the extracellular matrix and loosening the collagen network. Over time, the looser network allows the collagenase molecule to increase its penetration and the reduced collagen strain uncovers additional cleavage sites that were previously sequestered [20]. The action of enzymatic degradation in collagen-rich tissues such as tendon and cartilage has been described as a “wavefront” [21].
We found that both quantitative MR and US imaging measures can be used to assess collagen degradation. In particular, the MMF generated with the 3D UTE-Cones-MT sequence showed the strongest correlations with collagen loss (r=−0.81). This complements the results from Zhu et al. that showed how MMF was useful for distinguishing between histologic grades of mild and severe tendinopathy [7]. One unique advantage of MT sequences is the insensitivity to the magic angle effect [6, 7], which makes it quite promising for evaluation of the RCT. Our results also demonstrated a moderate correlation of adiabatic T1ρ, measured with the 3D UTE-Cones-adiabatic T1ρ sequence, with collagen degradation (r=0.66). Although T1ρ is most commonly used to assess changes in proteoglycan content, our results support the findings from previous authors that T1ρ imaging lacks inherent tissue specificity [22]. However, T1ρ may remain sensitive to biologically meaningful changes. A moderate correlation of T2* with collagen degradation was also found (r=0.58). Previous authors have also measured the transverse relaxation times in RCT, including T2 [7, 23, 24] and T2* [23]. Of note, a major limitation of both T2 and T2* is their sensitivity to the magic angle effect [25], which is particularly problematic for highly anisotropic tissues such as tendon. It is known that RCTs can demonstrate a 6‐fold change in signal intensity [26] and a 300% change in T2 value [7] based purely on differences in orientation relative to the main magnetic field. As a result, the large range of transverse relaxation measurements diminishes clinical utility on an individual level [27].
Quantitative US measurements are considered efficient and reliable methods of tissue characterization [14]. Utilization of raw RF data is preferred since scanner-independent measurements can be generated, such as BSC. Recently, Byra et al. found that BSC demonstrated a correlation coefficient of −0.68 with histological quantification of collagen and myelin in human peripheral nerves [8]. In their study, AC and Nakagami parameter did not correlate well with histology findings. Similar to that study, we found a moderate correlation of BSC (r=0.51) and no significant correlation between AC or Nakagami parameter with collagen degradation in RCT.
From the clinical perspective, a noninvasive way to quantify tendon quality could potentially have a large impact on clinical practice. Tendon quality is currently assessed intraoperatively due to the shortcomings of routine clinical MR and US imaging. Multiple studies have shown that intraoperative determination of lower tendon quality increases surgical complexity and is a poor prognostic factor after rotator cuff repair [20, 21]. We envision that quantitative imaging techniques may be performed on select patients for whom surgery is being considered, for aiding in decision-making and surgical planning. For instance, knowledge of exceptionally poor tendon quality pre-operatively may allow the surgeon who is planning for rotator cuff repair to ensure a particular graft is available on-hand during the procedure [28]. However, for clinical translation, it is imperative that imaging times be as short as possible. We note that MR imaging parameters used in this study were not optimized for the in vivo condition since our study focused on cadaveric specimens. However, feasibility of translation of the MR sequences employed in this study has been previously demonstrated [7, 11, 12] and can be further optimized using advanced acceleration techniques such as parallel imaging or compressed sensing reconstruction. The tradeoff between speed and accuracy of quantification remains to be studied and will be a focus of our future studies.
Limitations include use of cadaveric tendon samples at room temperature. The absolute values of our results may therefore differ from the in vivo condition since many relaxation times, including T1, are temperature-dependent [29]. Changes in T1 also influence MT modeling and adiabatic T1ρ values. Additionally, tendinosis is represented by several other changes in addition to alterations in collagen, including cellular changes, glycosaminoglycan infiltration, and an increase in water and vascularity [30, 31]. We believe that controlled experiments are a necessary component of biomarker validation and understanding. Finally, analyses for the MR and US images included a global ROI over the whole tendon rather than subdividing into peripheral and central portions since we wished to maintain blinded analyses between modalities.
In conclusion, we found that MMF, adiabatic T1ρ, and T2* measured using 3D UTE-Cones MR sequences and US BSC could detect alterations in collagen using an in vitro model of tendinosis.
Highlights.
Controlled rotator cuff degeneration can be simulated in vitro using collagenase.
Both quantitative MRI and US can quantify the extent of rotator cuff degeneration.
MRI-derived macromolecular fraction shows the highest correlation with collagen loss.
Acknolwedgements
The authors acknowledge funding by the Veterans Affairs (Merit Awards I01CX001388 and I01RX002604), the National Institutes of Health (1R21AR073496, 1R01AR075825, 1R01AR062581, and T32EB005970), and the National Natural Science Foundation of China (81801673).
AUTHOR DECLARATION
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed.
We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs.
We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author and which has been configured to accept email from ericchangmd@gmail.com
Footnotes
Conflict of Interest
None
References
- [1].Lewis JS, Rotator cuff tendinopathy/subacromial impingement syndrome: is it time for a new method of assessment?, Br J Sports Med 43(4) (2009) 259–64. [DOI] [PubMed] [Google Scholar]
- [2].Buck FM, Grehn H, Hilbe M, Pfirrmann CW, Manzanell S, Hodler J, Magnetic resonance histologic correlation in rotator cuff tendons, J Magn Reson Imaging 32(1) (2010) 165–72. [DOI] [PubMed] [Google Scholar]
- [3].Roy JS, Braen C, Leblond J, Desmeules F, Dionne CE, MacDermid JC, Bureau NJ, Fremont P, Diagnostic accuracy of ultrasonography, MRI and MR arthrography in the characterisation of rotator cuff disorders: a systematic review and meta-analysis, Br J Sports Med 49(20) (2015) 1316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Robertson PL, Schweitzer ME, Mitchell DG, Schlesinger F, Epstein RE, Frieman BG, Fenlin JM, Rotator cuff disorders: interobserver and intraobserver variation in diagnosis with MR imaging, Radiology 194(3) (1995) 831–5. [DOI] [PubMed] [Google Scholar]
- [5].Chang EY, Du J, Chung CB, UTE imaging in the musculoskeletal system, J Magn Reson Imaging 41(4) (2015) 870–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ma YJ, Shao H, Du J, Chang EY, Ultrashort echo time magnetization transfer (UTE-MT) imaging and modeling: magic angle independent biomarkers of tissue properties, NMR Biomed 29(11) (2016) 1546–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu Y, Cheng X, Ma Y, Wong JH, Xie Y, Du J, Chang EY, Rotator cuff tendon assessment using magic-angle insensitive 3D ultrashort echo time cones magnetization transfer (UTE-Cones-MT) imaging and modeling with histological correlation, J Magn Reson Imaging 48(1) (2018) 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Byra M, Wan L, Wong JH, Du J, Shah SB, Andre MP, Chang EY, Quantitative Ultrasound and B-Mode Image Texture Features Correlate with Collagen and Myelin Content in Human Ulnar Nerve Fascicles, Ultrasound Med Biol (2019). [DOI] [PubMed] [Google Scholar]
- [9].Riley GP, Harrall RL, Constant CR, Chard MD, Cawston TE, Hazleman BL, Tendon degeneration and chronic shoulder pain: changes in the collagen composition of the human rotator cuff tendons in rotator cuff tendinitis, Ann Rheum Dis 53(6) (1994) 359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Du J, Diaz E, Carl M, Bae W, Chung CB, Bydder GM, Ultrashort echo time imaging with bicomponent analysis, Magn Reson Med 67(3) (2012) 645–9. [DOI] [PubMed] [Google Scholar]
- [11].Ma YJ, Zhao W, Wan L, Guo T, Searleman A, Jang H, Chang EY, Du J, Whole knee joint T1 values measured in vivo at 3T by combined 3D ultrashort echo time cones actual flip angle and variable flip angle methods, Magn Reson Med 81(3) (2019) 1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ma YJ, Carl M, Searleman A, Lu X, Chang EY, Du J, 3D adiabatic T1rho prepared ultrashort echo time cones sequence for whole knee imaging, Magn Reson Med 80(4) (2018) 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ma YJ, Chang EY, Carl M, Du J, Quantitative magnetization transfer ultrashort echo time imaging using a time-efficient 3D multispoke Cones sequence, Magn Reson Med 79(2) (2018) 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Han A, Andre MP, Erdman JW, Loomba R, Sirlin CB, O’Brien WD, Repeatability and Reproducibility of a Clinically Based QUS Phantom Study and Methodologies, IEEE Trans Ultrason Ferroelectr Freq Control 64(1) (2017) 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mohana Shankar P, A general statistical model for ultrasonic backscattering from tissues, IEEE Trans Ultrason Ferroelectr Freq Control 47(3) (2000) 727–36. [DOI] [PubMed] [Google Scholar]
- [16].Keikhosravi A, Liu Y, Drifka C, Woo KM, Verma A, Oldenbourg R, Eliceiri KW, Quantification of collagen organization in histopathology samples using liquid crystal based polarization microscopy, Biomed Opt Express 8(9) (2017) 4243–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bachmann E, Rosskopf AB, Gotschi T, Klarhofer M, Deligianni X, Hilbe M, Pfirrmann CWA, Snedeker JG, Fischer MA, T1- and T2*-Mapping for Assessment of Tendon Tissue Biophysical Properties: A Phantom MRI Study, Invest Radiol 54(4) (2019) 212–220. [DOI] [PubMed] [Google Scholar]
- [18].Lin PC, Reiter DA, Spencer RG, Sensitivity and specificity of univariate MRI analysis of experimentally degraded cartilage, Magn Reson Med 62(5) (2009) 1311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nissi MJ, Salo EN, Tiitu V, Liimatainen T, Michaeli S, Mangia S, Ellermann J, Nieminen MT, Multi-parametric MRI characterization of enzymatically degraded articular cartilage, J Orthop Res 34(7) (2016) 1111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Saini K, Cho S, Dooling LJ, Discher DE, Tension in fibrils suppresses their enzymatic degradation - A molecular mechanism for ‘use it or lose it’, Matrix Biol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moody HR, Brown CP, Bowden JC, Crawford RW, McElwain DL, Oloyede AO, In vitro degradation of articular cartilage: does trypsin treatment produce consistent results?, J Anat 209(2) (2006) 259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Menezes NM, Gray ML, Hartke JR, Burstein D, T2 and T1rho MRI in articular cartilage systems, Magn Reson Med 51(3) (2004) 503–9. [DOI] [PubMed] [Google Scholar]
- [23].Krepkin K, Bruno M, Raya JG, Adler RS, Gyftopoulos S, Quantitative assessment of the supraspinatus tendon on MRI using T2/T2* mapping and shear-wave ultrasound elastography: a pilot study, Skeletal Radiol 46(2) (2017) 191–199. [DOI] [PubMed] [Google Scholar]
- [24].Anz AW, Lucas EP, Fitzcharles EK, Surowiec RK, Millett PJ, Ho CP, MRI T2 mapping of the asymptomatic supraspinatus tendon by age and imaging plane using clinically relevant subregions, European journal of radiology 83(5) (2014) 801–5. [DOI] [PubMed] [Google Scholar]
- [25].Shao H, Pauli C, Li S, Ma Y, Tadros AS, Kavanaugh A, Chang EY, Tang G, Du J, Magic angle effect plays a major role in both T1rho and T2 relaxation in articular cartilage, Osteoarthritis Cartilage 25(12) (2017) 2022–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chang EY, Szeverenyi NM, Statum S, Chung CB, Rotator cuff tendon ultrastructure assessment with reduced-orientation dipolar anisotropy fiber imaging, AJR Am J Roentgenol 202(4) (2014) W376–8. [DOI] [PubMed] [Google Scholar]
- [27].Spencer RG, Pleshko N, How do statistical differences in matrix-sensitive magnetic resonance outcomes translate into clinical assignment rules?, J Am Acad Orthop Surg 21(7) (2013) 438–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gilot GJ, Attia AK, Alvarez AM, Arthroscopic repair of rotator cuff tears using extracellular matrix graft, Arthrosc Tech 3(4) (2014) e487–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Han M, Rieke V, Scott SJ, Ozhinsky E, Salgaonkar VA, Jones PD, Larson PE, Diederich CJ, Krug R, Quantifying temperature-dependent T1 changes in cortical bone using ultrashort echo-time MRI, Magn Reson Med 74(6) (2015) 1548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chard MD, Cawston TE, Riley GP, Gresham GA, Hazleman BL, Rotator cuff degeneration and lateral epicondylitis: a comparative histological study, Ann Rheum Dis 53(1) (1994) 30–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].de Mos M, van El B, DeGroot J, Jahr H, van Schie HT, van Arkel ER, Tol H, Heijboer R, van Osch GJ, Verhaar JA, Achilles tendinosis: changes in biochemical composition and collagen turnover rate, Am J Sports Med 35(9) (2007) 1549–56. [DOI] [PubMed] [Google Scholar]