Abstract
Gastric chief cells differentiate from mucous neck cells and develop their mature state at the base of oxyntic glands with expression of secretory zymogen granules. After parietal cell loss, chief cells transdifferentiate into mucous cell metaplasia, designated spasmolytic polypeptide-expressing metaplasia (SPEM), which is considered a candidate precursor of gastric cancer. We examined the range of microRNA (miRNA) expression in chief cells and identified miRNAs involved in chief cell transdifferentiation into SPEM. Among them, miR-148a was strongly and specifically expressed in chief cells and significantly decreased during the process of chief cell transdifferentiation. Interestingly, suppression of miR-148a in a conditionally immortalized chief cell line induced up-regulation of CD44 variant 9 (CD44v9), one of the transcripts expressed at an early stage of SPEM development, and DNA methyltransferase 1 (Dnmt1), an established target of miR-148a. Immunostaining analyses showed that Dnmt1 was up-regulated in SPEM cells as well as in chief cells before the emergence of SPEM in mouse models of acute oxyntic atrophy using either DMP-777 or L635. In the cascade of events that leads to transdifferentiation, miR-148a was down-regulated after acute oxyntic atrophy either in xCT knockout mice or after sulfasalazine inhibition of xCT. These findings suggest that the alteration of miR-148a expression is an early event in the process of chief cell transdifferentiation into SPEM.
Keywords: SPEM, miRNA, CD44 Variant 9, miR-148a, Transdifferentiation, Metaplasia, Gastric, DNMT1, Plasticity
Abbreviations used in this paper: DAPI, 4′,6-diamidino-2-phenylindol; DMEM, Dulbecco minimum essential medium; DNMT, DNA methyltransferase; GFP, green fluorescent protein; GIF, gastric intrinsic factor; GSII, Griffonia simplicifolia lectin II; IM, intestinal metaplasia; miRNA, microRNA; PCR, polymerase chain reaction; SPEM, spasmolytic polypeptide-expressing metaplasia
Graphical abstract
See editorial on page 189.
Summary.
Following parietal cell loss, chief cells transdifferentiate into mucous cell metaplasia, designated spasmolytic polypeptide-expressing metaplasia (SPEM). Induction of SPEM was associated with loss of miR-148a. Loss of miR-148a is an early step in chief cell transdifferentiation.
In the stomach mucosa, gastric chief cells are located at the base of oxyntic glands and express secretory zymogens. Chief cells differentiate from mucous neck cells in the lower half of corpus glands without cell division and remain in a fully differentiated state under normal conditions with a lifetime of more than 60 days.1 Previous studies demonstrated that some transcription factors, including XBP1 and MIST1, are required for the differentiation from mucous neck cells into chief cells and the maintenance of chief cells.2, 3, 4 On the other hand, parietal cell loss and inflammation induce chief cells to transdifferentiate into mucous cell metaplasia, designated spasmolytic polypeptide-expressing metaplasia (SPEM), with the loss of zymogen granules and the formation of Muc6-containing mucous granules.2, 5 SPEM is considered a likely precursor lineage for intestinal metaplasia (IM) development,3, 6 and these metaplasias are possible precursor lesions of gastric cancer. However, the regulatory mechanisms for the chief cell transdifferentiation process have not been fully elucidated.
MicroRNAs (miRNAs) are critical post-transcriptional regulators of gene expression.7, 8 MiRNAs are involved in the developmental process of various organs as well as cancer progression.9, 10 Dysregulation of miRNAs has been reported in human gastric cancer11 and Helicobacter pylori–induced gastritis,12, 13 contributing to gastric epithelial cell proliferation. We previously reported an analysis of miRNAs in laser capture microdissected human chief cells, SPEM cells, and IM cells, suggesting that miR-30a down-regulation and miR-194 up-regulation were related to metaplasia progression through regulation of the transcription factors HNF4γ and NR2F2.14 However, it remains unclear whether miRNAs are involved in the initiation of SPEM development.
Chief cell transdifferentiation and the transition to SPEM cells occur through series of ordered events. Our group and others have identified a number of events that chief cells undergo to transdifferentiate from zymogen secreting cells into mucous secreting metaplastic cells. Acute oxyntic atrophy induces an early loss of the chief cell maturation-specifying transcription factor, Mist1,15, 16, 17 and up-regulation of the specific splice variant of CD44, CD44 variant 9 (CD44v9).17, 18, 19 CD44v9 is an activator and a stabilizer of xCT, a cystine transporter, which promotes adaptation to reactive oxygen species and cellular stress.18, 19, 20 The increased cellular stress is associated with an increase in autophagy, which is necessary for breaking down the zymogen granules.21 Importantly, the process of transdifferentiation can be arrested at different stages,17, 19, 21 suggesting that chief cell transdifferentiation occurs through a set of stepwise events that are coordinated and maintained in a defined order.
Here we investigated the influence of miRNAs on the initiation of chief cell transdifferentiation into SPEM cells. We performed miRNA profiling specifically on mouse chief cells and compared miRNA expression with that in conditionally immortalized mouse chief cell and SPEM cell lines. Interestingly, several miRNAs were highly expressed in normal chief cells but down-regulated in SPEM cells. Among them, miR-148a was the most highly expressed miRNA by more than 10-fold in chief cells. Loss of miR-148a was associated with the early initiation of chief cell transdifferentiation. In addition, the loss of miR-148a led to up-regulation of an early SPEM marker, CD44 variant 9, and one of its target genes, DNA methyltransferase 1 (Dnmt1). The loss of miR-148a was found early during the chief cell transdifferentiation process, preceding up-regulation of CD44v9. These findings suggest that miR-148a is an early regulator in reprogramming chief cells during transdifferentiation into SPEM.
Results
MicroRNA Profile of In Vivo Mouse Chief Cells and Immortalized Chief Cell and Spasmolytic Polypeptide-Expressing Metaplasia Cell Lines
In a previous study, our group reported the miRNA profile of human SPEM and IM in comparison with chief cells from normal stomach.14 In those studies, we identified miRNAs related with human IM progression; however, no miRNAs related to SPEM development were confirmed. We therefore sought to investigate miRNAs during the initiation of SPEM development by using mouse models. First, to profile miRNAs from mouse chief cells, we crossed Mist1CreERT2/+mice with R26RmTmG reporter mice. After tamoxifen injection, immunostaining analyses of these mouse stomachs showed that most of green fluorescent protein (GFP)-positive cells were chief cells at the base of glands, with only occasional labeled cells in the isthmus regions (Figure 1A).1 We sorted GFP-positive cells from 2 mice and performed miRNA sequencing (Figure 1B). Forty-three miRNAs were highly expressed in mouse chief cells (read values >500 in both mice) (Table 1). Among them, miR-148a-3p was the most highly expressed miRNA (read values >150,000) in chief cells, more than 10-fold higher than other highly expressed miRNAs such as miR-375-3p, let-7 family (let-7b-5p, let-7c-5p, let-7f-5p and let-7a-5p), and miR-200b-3p (read values >8000). Interestingly, these miRNAs have already been reported as down-regulated in human gastric cancer tissues and related to gastric cancer progression.22, 23, 24, 25
Figure 1.
miRNAs related to SPEM development. (A) Immunofluorescence staining in Mist1CreERT2/+;R26RmTmG/+ mice. Membrane-associated GFP was expressed in GIF-positive chief cells and occasionally in the isthmus regions at 10 days after tamoxifen injection. (B) Schemes of miRNA sequencing experiments. (left) miRNAs from sorted chief cells of Mist1-mTmG mice. (right) miRNAs from ImChief cells and ImSPEM cells. (C) Fifteen miRNAs that were both highly expressed in sorted chief cells (red) and down-regulated in ImSPEM cells (yellow) compared with ImChief cells (blue). The read values for these miRNAs were more than 500, and fold changes of expression between ImSPEM cells and ImChief cells were more than 5. miRNA sequencing analyses detected 2 different let-7f-5p as up-regulated miRNAs in chief cells. Top one was at chromosome X, and another was at chromosome 13 (Table 1).
Table 1.
MicroRNAs Highly Expressed in Mouse Chief Cells
| Accession | Chromosome | Strand | Start | End | Gene ID | Sample 1 | Sample 2 | Entrez ID |
|---|---|---|---|---|---|---|---|---|
| mmu-miR-148a-3p | chr6 | – | 51219828 | 51219849 | MI0000550_1 | 157846.9654 | 154942.4573 | 387166 |
| mmu-miR-375-3p | chr1 | – | 74947235 | 74947256 | MI0000792_1 | 35248.20084 | 21476.74894 | 723900 |
| mmu-let-7b-5p | chr15 | + | 85537755 | 85537776 | MI0000558_1 | 25949.91154 | 27815.42445 | 387245 |
| mmu-let-7c-5p | chr16 | + | 77599917 | 77599938 | MI0000559_1 | 15987.66912 | 22193.04491 | 387246 |
| mmu-let-7c-5p | chr15 | + | 85537046 | 85537067 | MI0000560_1 | 14821.59409 | 20835.45521 | 723966 |
| mmu-let-7f-5p | chrX | + | 1.48E+08 | 1.48E+08 | MI0000563_1 | 13039.97967 | 15870.56644 | 387253 |
| mmu-miR-7a-5p | chr13 | – | 58494202 | 58494224 | MI0000728_2 | 11223.58539 | 6231.293692 | 723902 |
| mmu-miR-200b-3p | chr4 | – | 1.55E+08 | 1.55E+08 | MI0000243_1 | 9422.302927 | 10591.6593 | 387243 |
| mmu-let-7a-5p | chr13 | – | 48633608 | 48633629 | MI0000556_2 | 8746.256843 | 12009.56271 | 387244 |
| mmu-miR-7a-5p | chr7 | + | 86033181 | 86033203 | MI0000729_1 | 8740.205588 | 4823.681713 | 723884 |
| mmu-let-7a-5p | chr9 | + | 41344815 | 41344836 | MI0000557_1 | 8363.615366 | 11563.89665 | 723965 |
| mmu-let-7f-5p | chr13 | – | 48633258 | 48633279 | MI0000562_2 | 8230.526464 | 11138.08047 | 387252 |
| mmu-miR-192-5p | chr19 | + | 6264857 | 6264877 | MI0000551_1 | 7084.120318 | 7057.18444 | 387187 |
| mmu-miR-21-5p | chr11 | – | 86397622 | 86397643 | MI0000569_2 | 5868.145795 | 7696.271615 | 387140 |
| mmu-miR-26a-5p | chr10 | + | 1.26E+08 | 1.26E+08 | MI0000706_1 | 5001.537708 | 3946.313897 | 723962 |
| mmu-miR-26a-5p | chr9 | + | 1.19E+08 | 1.19E+08 | MI0000573_1 | 5001.537708 | 3946.313897 | 387218 |
| mmu-let-7i-5p | chr10 | – | 1.22E+08 | 1.22E+08 | MI0000138_2 | 4517.566115 | 5195.810144 | 387251 |
| mmu-miR-99a-5p | chr16 | + | 77599185 | 77599206 | MI0000146_1 | 3504.250029 | 3589.631416 | 387229 |
| mmu-miR-30d-5p | chr15 | – | 68172819 | 68172840 | MI0000549_2 | 2757.120297 | 2825.520615 | 387228 |
| mmu-miR-7b-5p | chr17 | + | 56382440 | 56382462 | MI0000730_1 | 2657.301111 | 1373.046719 | 723883 |
| mmu-miR-200a-3p | chr4 | – | 1.55E+08 | 1.55E+08 | MI0000554_1 | 2465.225664 | 2397.499834 | 387242 |
| mmu-miR-200c-3p | chr6 | – | 1.25E+08 | 1.25E+08 | MI0000694_1 | 2239.877318 | 2418.092972 | 723944 |
| mmu-miR-182-5p | chr6 | – | 30115962 | 30115986 | MI0000224_2 | 2006.965359 | 1888.583689 | 387177 |
| mmu-miR-1a-3p | chr18 | – | 10785483 | 10785504 | MI0000652_1 | 1755.906123 | 63.24694981 | 723959 |
| mmu-miR-125a-5p | chr17 | + | 17967781 | 17967804 | MI0000151_1 | 1663.648576 | 1174.480463 | 387235 |
| mmu-let-7g-5p | chr9 | + | 1.06E+08 | 1.06E+08 | MI0000137_1 | 1585.004257 | 1562.052254 | 387249 |
| mmu-miR-183-5p | chr6 | – | 30119711 | 30119732 | MI0000225_2 | 1533.581369 | 2018.018941 | 387178 |
| mmu-miR-151-3p | chr15 | – | 73085250 | 73085270 | MI0000173_1 | 1308.233185 | 1660.600129 | 387169 |
| mmu-miR-215-5p | chr1 | + | 1.87E+08 | 1.87E+08 | MI0000974_1 | 1215.975967 | 3101.305644 | 387211 |
| mmu-miR-127-3p | chr12 | + | 1.11E+08 | 1.11E+08 | MI0000154_2 | 1188.753604 | 600.1103493 | 387146 |
| mmu-miR-143-3p | chr18 | – | 61808853 | 61808873 | MI0000257_1 | 1025.412731 | 771.4655906 | 387161 |
| mmu-miR-378-3p | chr18 | – | 61557492 | 61557512 | MI0000795_1 | 1020.875345 | 2281.30322 | 723889 |
| mmu-miR-423-5p | chr11 | – | 76891624 | 76891646 | MI0004637_2 | 1001.21424 | 1177.423 | 751519 |
| mmu-miR-27b-3p | chr13 | + | 63402068 | 63402088 | MI0000142_2 | 860.560016 | 781.7616481 | 387221 |
| mmu-miR-30c-5p | chr1 | + | 23298553 | 23298575 | MI0000548_1 | 852.9982668 | 620.7028672 | 723964 |
| mmu-miR-30c-5p | chr4 | – | 1.2E+08 | 1.2E+08 | MI0000547_2 | 848.4606843 | 617.0253704 | 387227 |
| mmu-miR-92a-3p | chr14 | + | 1.15E+08 | 1.15E+08 | MI0000719_2 | 739.5680038 | 1023.717545 | 751549 |
| mmu-let-7e-5p | chr17 | + | 17967330 | 17967351 | MI0000561_1 | 680.5837996 | 718.5145194 | 387248 |
| mmu-let-7d-5p | chr13 | – | 48631447 | 48631468 | MI0000405_2 | 603.4510756 | 628.792413 | 387247 |
| mmu-miR-30a-5p | chr1 | + | 23279113 | 23279134 | MI0000144_1 | 586.8142113 | 556.7203464 | 387225 |
| mmu-miR-320-3p | chr14 | + | 70843364 | 70843385 | MI0000704_2 | 580.7647177 | 527.3028558 | 723838 |
| mmu-miR-25-3p | chr5 | – | 1.39E+08 | 1.39E+08 | MI0000689_1 | 552.0292875 | 714.1023224 | 723926 |
| mmu-miR-200a-5p | chr4 | – | 1.55E+08 | 1.55E+08 | MI0000554_2 | 547.491972 | 686.8912993 | 387242 |
To investigate miRNAs related to metaplasia development, we examined miRNA expression profiles for conditionally immortalized mouse chief cell (ImChief) and SPEM cell (ImSPEM) lines, previously established from Immortomice.26 ImChief cells express chief cell markers such as pepsinogen C (Pgc) and Mist1 and produce characteristic zymogen granules, although they do not express gastric intrinsic factor (GIF). In contrast, ImSPEM cells express SPEM-specific markers such as Tff2 and He4 and some intestinalized markers such as Cftr and PigR. We extracted total RNAs from ImChief cells and ImSPEM cells and performed miRNA sequencing. We detected 87 miRNAs down-regulated (P < .01, with read values for ImChief cells >500 and fold-change >5) and 7 miRNAs up-regulated (P < .01, with read values for ImSPEM cells >500 and fold-change >5) in ImSPEM cells compared with ImChief cells (Tables 2 and 3). From these 2 different sequencing studies, we identified 15 miRNAs that were both highly expressed in sorted chief cells and down-regulated in ImSPEM cells compared with ImChief cells (Figure 1C) as candidate miRNAs related to SPEM development. This group of miRNAs included miR-148a-3p, miR-200 family members (miR-200a-3p, miR-200a-5p, miR-200b-3p, and miR-200c-3p), miR-30 family members (miR-30a-5p and miR-30c-5p), and let-7 family members (let-7f-5p and let-7i-5p).
Table 2.
MicroRNAs Down-regulated in ImSPEM Cells Compared With ImChief Cells
| Feature | ImSPEM cells_1 | ImSPEM cells_2 | ImSPEM cells_3 | ImChief cells_1 | ImChief cells_2 | ImChief cells_3 | ImSPEM cells/ImChief cells |
|---|---|---|---|---|---|---|---|
| mmu-miR-141-5p | 0 | 0 | 0 | 1005 | 1576 | 1593 | 0 |
| mmu-miR-141-3p | 60.5 | 75.5 | 84 | 147862 | 151753 | 220628.5 | 0.000423 |
| mmu-miR-200c-3p | 7 | 6 | 3 | 8326.5 | 13315 | 12489.5 | 0.000469 |
| mmu-miR-205-5p | 145 | 148 | 197 | 233099 | 355470 | 318724 | 0.00054 |
| mmu-miR-205-3p | 5 | 5 | 3 | 4571 | 5175 | 5850 | 0.000834 |
| mmu-miR-203-3p | 8 | 11 | 11 | 4932 | 6346 | 6065 | 0.00173 |
| mmu-miR-672-5p | 1 | 2 | 4 | 1015 | 1430 | 1390 | 0.001825 |
| mmu-miR-676-3p | 2 | 2 | 2 | 682 | 735 | 856 | 0.00264 |
| mmu-miR-429-3p | 60 | 69 | 73 | 18745.5 | 22299.5 | 23313.5 | 0.003139 |
| mmu-miR-146a-5p | 136.5 | 163.5 | 172.5 | 28169.5 | 67458.5 | 42172 | 0.003429 |
| mmu-miR-200a-3p | 42.5 | 40.5 | 46 | 9297 | 12749 | 13597.5 | 0.003619 |
| mmu-miR-200b-3p | 239 | 269 | 316 | 37915 | 48814.5 | 56526 | 0.005752 |
| mmu-miR-135b-5p | 6 | 16 | 17 | 1674.67 | 2275.83 | 2806.67 | 0.005772 |
| mmu-miR-200a-5p | 12 | 5 | 3 | 624 | 1067 | 745 | 0.00821 |
| mmu-miR-183-5p | 1153 | 1362 | 1308 | 109600 | 114757 | 173820 | 0.009601 |
| mmu-miR-183-3p | 11 | 14 | 13 | 1045 | 1147 | 1645 | 0.009904 |
| mmu-miR-182-5p | 15006 | 12308 | 14206 | 1002426 | 1136518 | 1453717 | 0.011557 |
| mmu-miR-96-5p | 98 | 112 | 92 | 6537 | 7080 | 10821 | 0.012358 |
| mmu-miR-193b-3p | 67 | 108 | 94 | 3006 | 2259 | 5077.5 | 0.026009 |
| mmu-miR-31-3p | 24 | 82 | 48 | 1569 | 1446 | 2632 | 0.027271 |
| mmu-miR-31-5p | 6738 | 5724 | 7552 | 182407 | 217916 | 244071 | 0.031059 |
| mmu-miR-582-3p | 32 | 21 | 48 | 844 | 905 | 1241 | 0.033779 |
| mmu-miR-421-3p | 167 | 192 | 226 | 4175 | 4981 | 5907 | 0.038837 |
| mmu-miR-147-3p | 26 | 18 | 28 | 427 | 604 | 599 | 0.044172 |
| mmu-miR-222-3p | 2279 | 1489 | 2261 | 32959 | 50742 | 47026 | 0.046119 |
| mmu-miR-210-5p | 39 | 46 | 64 | 684 | 1217 | 1212 | 0.047864 |
| mmu-miR-27b-3p | 58480 | 65172.5 | 74847.5 | 1248916 | 1245684 | 1584584 | 0.048662 |
| mmu-miR-181b-5p | 563 | 509.5 | 742.67 | 10536.83 | 12984.17 | 13485.67 | 0.04905 |
| mmu-miR-181a-5p | 7652 | 7487 | 8969 | 142888 | 140394 | 203123 | 0.049564 |
| mmu-miR-148a-3p | 8590.5 | 9828.5 | 9697.5 | 140196 | 181813 | 223713.5 | 0.051522 |
| mmu-miR-148a-5p | 67 | 66 | 78 | 1008 | 1140 | 1544 | 0.057151 |
| mmu-miR-298-5p | 770 | 630 | 794 | 10712 | 10690 | 15097 | 0.060111 |
| mmu-miR-21a-3p | 480 | 1124 | 901 | 7385 | 10894 | 16739 | 0.071535 |
| mmu-miR-101b-3p | 142.5 | 211 | 182 | 1863.5 | 2280 | 2870.5 | 0.076347 |
| mmu-miR-27b-5p | 82 | 85 | 100 | 965 | 1248 | 1267 | 0.076724 |
| mmu-miR-126a-5p | 212 | 406 | 383 | 3065 | 4248 | 5532 | 0.077929 |
| mmu-miR-1843a-5p | 85 | 103 | 117 | 1180 | 1189 | 1462 | 0.079614 |
| mmu-miR-365-3p | 64 | 129 | 103 | 1071 | 917 | 1658 | 0.081185 |
| mmu-miR-221-3p | 2572 | 2973 | 3118 | 27309 | 32974 | 39637 | 0.086699 |
| mmu-miR-29b-3p | 151.67 | 274 | 211 | 1543.33 | 2317 | 2887.33 | 0.094354 |
| mmu-miR-30c-5p | 2801 | 3116 | 3936 | 29133.67 | 30398.33 | 40878 | 0.098128 |
| mmu-miR-92a-3p | 5585 | 4445.33 | 5511.67 | 42970.67 | 47225.33 | 66885.67 | 0.098942 |
| mmu-miR-194-5p | 165 | 136 | 242 | 1465 | 2072 | 1948 | 0.098997 |
| mmu-miR-425-5p | 346 | 436 | 499 | 3521 | 4204 | 4789 | 0.102365 |
| mmu-miR-19b-3p | 301.5 | 369.5 | 309.5 | 2310.17 | 3555.5 | 3663.5 | 0.102895 |
| mmu-miR-221-5p | 607 | 297 | 339 | 3409 | 3600 | 5063 | 0.102966 |
| mmu-miR-374b-5p | 55 | 100 | 130 | 744 | 914 | 1025 | 0.106224 |
| mmu-miR-126a-3p | 53 | 49 | 76 | 387 | 561 | 665 | 0.110353 |
| mmu-miR-17-5p | 101 | 130 | 130 | 830 | 1150.83 | 1259.83 | 0.111397 |
| mmu-miR-328-3p | 160 | 152.5 | 210.5 | 1513 | 1110.5 | 1946.5 | 0.114442 |
| mmu-miR-130b-3p | 119 | 112.5 | 176 | 629.5 | 1266.5 | 1656 | 0.114724 |
| mmu-miR-19a-3p | 57.5 | 63.5 | 55.5 | 363.83 | 552.5 | 615.5 | 0.115222 |
| mmu-miR-484 | 1241 | 1134 | 1489 | 9205 | 9226 | 14465 | 0.117461 |
| mmu-miR-20a-5p | 84 | 113 | 84 | 570 | 774 | 887.33 | 0.125934 |
| mmu-miR-23b-3p | 1793.5 | 3092 | 2818.5 | 17506 | 18406.5 | 25082 | 0.126306 |
| mmu-miR-30a-5p | 22727.5 | 24771 | 26421.5 | 164380 | 193723.5 | 225365.3 | 0.126691 |
| mmu-miR-30e-5p | 10724.5 | 10780.5 | 11758.5 | 70581 | 89679 | 98109.33 | 0.128744 |
| mmu-miR-192-5p | 2938 | 2785 | 3313.5 | 20230 | 21444 | 27916.5 | 0.129852 |
| mmu-let-7f-5p | 22306.05 | 28289.77 | 35971.93 | 174340.2 | 270159 | 221880.1 | 0.129908 |
| mmu-miR-210-3p | 846 | 949 | 1217 | 6351 | 7870 | 8930 | 0.130102 |
| mmu-miR-22-3p | 107186 | 109978 | 146919 | 737758 | 925662 | 1022961 | 0.135529 |
| mmu-miR-107-3p | 182.5 | 186.33 | 225.67 | 1261.83 | 1418 | 1703.83 | 0.135617 |
| mmu-let-7i-5p | 7772 | 9401 | 12263.5 | 59123 | 79414.5 | 77297.5 | 0.136384 |
| mmu-miR-191-5p | 13618 | 16310 | 18084 | 97568 | 100312 | 151097 | 0.137579 |
| mmu-miR-98-5p | 514.27 | 1025.47 | 1211.27 | 4932.67 | 7444.4 | 7119.93 | 0.141099 |
| mmu-miR-34b-5p | 444.5 | 548 | 642 | 3035.5 | 4005.5 | 4381 | 0.143101 |
| mmu-miR-93-5p | 4297 | 4794 | 5926 | 31000 | 32748 | 40799 | 0.143639 |
| mmu-let-7b-3p | 63 | 87 | 108 | 539 | 443 | 811 | 0.143893 |
| mmu-let-7j | 980.5 | 966.5 | 1326 | 6196 | 8255.5 | 8213.5 | 0.144408 |
| mmu-miR-99b-3p | 104 | 119 | 123 | 625 | 822.5 | 854 | 0.150337 |
| mmu-miR-30a-3p | 357 | 445 | 506.5 | 2347.5 | 2619.5 | 3139.5 | 0.161414 |
| mmu-miR-24-3p | 3038 | 3982 | 3287 | 16617 | 22764 | 24253 | 0.161973 |
| mmu-miR-27a-3p | 4684 | 6406.5 | 7145.5 | 31267 | 36340 | 43419.5 | 0.164249 |
| mmu-miR-34b-3p | 347 | 405 | 567 | 2219 | 2505 | 3299 | 0.164402 |
| mmu-miR-342-3p | 179 | 159 | 162 | 899 | 732 | 1409 | 0.164474 |
| mmu-miR-1839-5p | 194 | 235 | 293 | 1243 | 1427 | 1642 | 0.16744 |
| mmu-miR-130a-3p | 2613 | 2714.5 | 3310 | 13909.5 | 17149.5 | 20256 | 0.168323 |
| mmu-miR-140-3p | 616 | 798 | 807 | 3466 | 4682 | 5039 | 0.168423 |
| mmu-miR-29a-3p | 5652.5 | 5820 | 6999 | 28115.5 | 38135.5 | 42291.83 | 0.170177 |
| mmu-miR-450b-5p | 310 | 290 | 330 | 1263 | 1804 | 2195 | 0.176739 |
| mmu-miR-34c-5p | 19682.5 | 20120 | 26912 | 100916.5 | 127993 | 144827 | 0.178507 |
| mmu-miR-351-5p | 2316 | 1802 | 1999 | 9210 | 9920 | 15121 | 0.178593 |
| mmu-miR-34c-3p | 126 | 327 | 317 | 1091 | 1488 | 1633 | 0.182811 |
| mmu-miR-28a-3p | 157 | 207 | 307 | 942 | 1264 | 1400 | 0.186079 |
| mmu-miR-28a-5p | 611 | 711 | 929 | 3248 | 4056 | 4324 | 0.193584 |
| mmu-miR-186-5p | 5336 | 5697 | 6401 | 23436 | 30215 | 36028 | 0.194404 |
| mmu-let-7d-3p | 387 | 589 | 647 | 2297 | 2030 | 3951 | 0.196062 |
Table 3.
MicroRNAs Up-regulated in ImSPEM Cells Compared With ImChief Cells
| Feature | ImSPEM cells_1 | ImSPEM cells_2 | ImSPEM cells_3 | ImChief cells_1 | ImChief cells_2 | ImChief cells_3 | ImSPEM cells/ImChief cells |
|---|---|---|---|---|---|---|---|
| mmu-miR-10b-5p | 255958.5 | 248754.5 | 263925 | 69.5 | 107.5 | 118.5 | 2601.144 |
| mmu-miR-199a-5p | 1767 | 1715 | 2536 | 6 | 8 | 18 | 188.0625 |
| mmu-miR-199a-3p mmu-miR-199b-3p |
2801 | 6977 | 7968 | 47 | 48 | 58 | 115.9869 |
| mmu-miR-344d-3p | 587 | 491 | 675 | 3 | 6 | 11 | 87.65 |
| mmu-miR-10a-5p | 60241.5 | 55707.5 | 59273 | 3125.5 | 3297.5 | 5008.5 | 15.328 |
| mmu-miR-155-5p | 887 | 1304 | 1593 | 164 | 199 | 235 | 6.327759 |
| mmu-miR-181c-5p | 561 | 590 | 657 | 213 | 267 | 328 | 2.237624 |
MiR-148a Expression in Mouse Stomach Assessed by In Situ Hybridization
Because it was by far the most highly expressed miRNA species in chief cells, we focused our subsequent studies on miR-148a. To examine the distribution of miR-148a expression, we performed in situ hybridization analyses for miR-148a (Figure 2). In normal stomach, miR-148a was strongly expressed in the bases of corpus glands, deep to Griffonia simplicifolia lectin II (GSII)-positive mucous neck cells, and surface cells and mucous neck cells showed no or very low expression of miR-148a (Figure 2A). MiR-148a expression was localized in the cytoplasm, especially in the basal side of cells. In contrast to the corpus, the antrum and the duodenum showed little or no expression of miR-148a. Importantly, dual immunostaining with in situ hybridization demonstrated that miR-148a–positive cells expressed the chief cell markers Mist1 and GIF (Figure 2B). Examination of Mist1CreERT2/+;R26RtdTomato/+ mice also showed that Mist1-positive chief cells have strong miR-148a expression (Figure 2C). These data suggest that miR-148a is strongly and specifically expressed in chief cells in the gastric corpus.
Figure 2.
miR-148a expression in normal mouse stomach. (A) Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (green) and DAPI (blue). miR-148a was strongly expressed in the base of corpus glands deep to GSII-positive mucous neck cells. miR-148a expression was localized in the cytoplasm. No or little expression of miR-148a was seen in antrum and duodenum. (B) Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (blue) and Mist1 or GIF (green). miR-148a–positive cells expressed Mist1 and GIF. (C) Fluorescence in situ hybridization for miR-148a using Mist1CreERT2/+;R26RtdTomato/+ mice. miR-148a (green), tdTomato (red), and GSII (blue). miR-148a was expressed in tdTomato-positive chief cells.
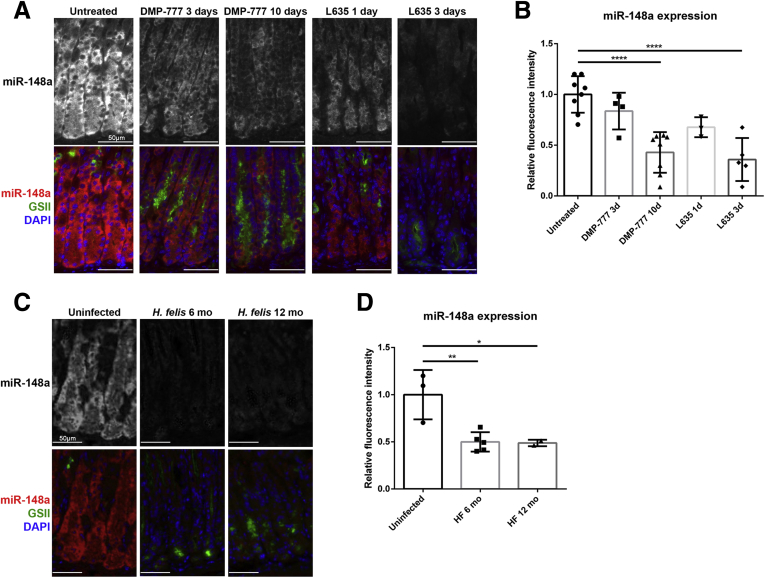
Next, to evaluate the alteration of expression of miR-148a during the development of SPEM, we examined in situ hybridization analyses in 2 mouse models of acute oxyntic atrophy (administration of either DMP-777 or L635). Compared with normal chief cells, miR-148a was down-regulated in GSII-positive SPEM cells in mice after 10 days of DMP-777 treatment or 3 days of L635 treatment (Figure 3A). Importantly, miR-148a was down-regulated in chief cells without GSII expression after 3 days of DMP-777 treatment or 1 day of L635 treatment. Fluorescence intensity was measured for quantitation, and miR-148a expression was decreased significantly in mice with 10 days of DMP-777 treatment or 3 days of L635 treatment (Figure 3B). We also examined miR-148a expression by in situ hybridization in mice infected with Helicobacter felis, and mice infected with H felis for 6 months and 12 months showed decreased levels of miR-148a expression (Figure 3C and D). These findings suggest that the down-regulation of miR-148a could be involved in the chief cell transdifferentiation into SPEM cells.
Figure 3.
miR-148a expression during chief cell transdifferentiation into SPEM cells. (A) Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (green) and DAPI (blue). miR-148a was significantly down-regulated in GSII-positive SPEM cells developed in mice after 10 days of DMP-777 treatment or 3 days of L635 treatment as well as in chief cells without GSII expression after 3 days of DMP-777 treatment or 1 day of L635 treatment, compared with normal chief cells. (B) Quantitation of relative miR-148a staining intensity in chief cells and transdifferentiating chief cells at the base of the gland. One-way analysis of variance, P < .0001. Bonferroni multiple comparisons, ****P < .0001. (C) Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (green) and DAPI (blue). miR-148a was significantly decreased in GSII-positive SPEM cells developed in mice 6 months or 12 months after H felis infection. (D) Quantitation of relative miR-148a staining intensity in chief cells and H felis infection induced SPEM cells at the base of the gland. One-way analysis of variance, P = .0081. Bonferroni multiple comparisons, *P < .05, **P < .01.
MiR-148a Expression in Human Stomach Assessed by In Situ Hybridization
To assess whether the alteration in miR-148a expression was related to SPEM development in humans, we performed in situ hybridization analyses for miR-148a using human stomach tissue (Figure 4). The human stomach tissue section shown in Figure 4 conveniently contained normal corpus glands and SPEM glands side-by-side, allowing for clear comparison of expression between them. The miR-148a was exclusively expressed in chief cells located at the base of the normal corpus glands below the GSII-positive mucous neck cells. The SPEM glands, marked with GSII-positive staining to the base of the glands, had no or very low expression of miR-148a. This finding confirmed the down-regulation of miR-148a expression in SPEM in human stomach.
Figure 4.
miR-148a expression in human stomach. Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (green) and DAPI (blue). miR-148a was strongly expressed at the base of the normal glands below the GSII-positive mucous neck cells. The SPEM glands, marked with GSII-positive staining to the base of the glands, had no or little expression of miR-148a.
Involvement of MiR-148a in Initiation of Chief Cell Transdifferentiation
ImChief cells and ImSPEM cells grow continuously at the permissive temperature of 33°C because of their expression of temperature-dependent T antigen. However, at the non-permissive temperature (39°C), the temperature-sensitive T antigen misfolds and thus no longer immortalizes the cells. Thus, ImChief cells and ImSPEM cells become more primary-like at the non-permissive temperature.26 To investigate the role of miR-148a in chief cell plasticity, we examined these cell lines incubated at the non-permissive temperature for 72 hours. First, to confirm the expression levels of miR-148a in ImChief cells and ImSPEM cells, the expression of miR-148a was detected by microRNA assay, and miR-148a expression in ImSPEM cells was significantly lower than in ImChief cells (Figure 5A).
Figure 5.
Involvement of miR-148a in initiation of chief cell transdifferentiation. (A) Expression of miR-148a in ImChief cells and ImSPEM cells by microRNA assay. Mann-Whitney U test, *P < .05. (B) Expression of miR-148a by microRNA assay in ImChief cells treated with control inhibitor or miR-148a inhibitor. Mann-Whitney U test, *P < .05. (C) Expression of genes related with SPEM cells (He4, Sox9, Cd44v9, and Clu) or chief cells (Pgc and Mist1) in ImChief cells treated with control inhibitor or miR-148a inhibitor by quantitative PCR. Mann-Whitney U test, *P < .05. (D) Expression of miR-148a target genes in ImChief cells treated with control inhibitor or miR-148a inhibitor by quantitative PCR. Mann-Whitney U test, *P < .05.
To investigate whether down-regulation of miR-148a is related to SPEM development, we examined the influence of inhibitors for miR-148a in ImChief cells. ImChief cells were transfected with miR-148a inhibitors and then incubated at 33°C overnight, followed by incubation at 39°C for 72 hours. The expression of miR-148a was significantly down-regulated in ImChief cells treated with the inhibitor (Figure 5B). Known chief cell and SPEM cell marker expression was examined by quantitative reverse transcription polymerase chain reaction (PCR), and ImChief cells treated with miR-148a inhibitor showed significant up-regulation of the variant 9 splice isoform of Cd44 (Cd44v9), an early marker of SPEM,18, 19 although Tff2 expression was not detected (Figure 5C). The expression of Sox9 and Clusterin (Clu), other SPEM-associated markers, was increased, but not significantly (Figure 5C). The expression of chief cell specific genes, such as Mist1 and Pgc, was not significantly changed. These data suggest that down-regulation of miR-148a could be involved in initiation of chief cell transdifferentiation.
DNA Methyltransferase 1 Up-regulation in Chief Cells During Chief Cell Transdifferentiation via Down-regulation of MiR-148a
We have previously examined genes related to the emergence of SPEM by investigating microarray assay for mRNA expression for RNA extracted from SPEM regions in gastrin-deficient mice treated with DMP-777 for 1 day or 3 days.27 To investigate the direct linkage of changes in miRNA expression with the emergence of SPEM, we compared the candidate miR-148a target gene list with previous mRNA microarray data (Table 4). Interestingly, Ccnf, Dgcr8, Dnmt1, Kat7, and Rcc2 were identified as possible targets of miR-148a during the transdifferentiation process of chief cells into SPEM cells; all were predicted by 3 different miRNA databases: microRNA.org, TargetScan, and miRDB.
Table 4.
Candidate MicroRNAs and their Targeted Genes Related to SPEM Development
| Candidate miRNAs (miRNA seq data) | Possible targeted genes up-regulated in early stage of SPEM (Nozaki et al27) |
|---|---|
| miR-148a | Ccnf Dgcr8 Dnmt1 Kat7 Rcc2 |
| let-7f/7i | Limd2 Rdh10 Thoc1 Myo1f |
| miR-200b/200c | Rdh10 Dnajc5 Pou6f1 BC037034 |
| miR-192 | Ereg Thoc1 |
| miR-200a | Csnk2a1 E2f3 Nme1 Sdf2 Sft2d2 Src |
| miR-182 | Nup155 Rasa1 |
| miR-183 | (-) |
| miR-27b | Plekhh1 Nek6 Litaf Gpd1 Nfx1 Timm8a1 |
| miR-30a/30c | Sdad1 Vat1 Xpo1 Arl6ip5 Mboat1 Poldip3 |
To confirm the miR-148a regulation of these possible target genes, mRNA expression was examined in ImChief cells transfected with control inhibitor or miR-148a inhibitor. As expected from miRNA database prediction, most of the candidate target genes showed up-regulation of mRNA expression with miR-148a inhibition, including Ccnf, Dgcr8, Dnmt1, and Rcc2 (Figure 5D). Among the putative target genes, we focused on Dnmt1, one of the DNA methyltransferases that is essential for the maintenance of DNA methylation. Several studies have validated that miR-148a directly targets Dnmt1 3′-untranslated region in various cells, including gastric cancer cells, contributing to tumor progression.28, 29, 30 To confirm the expression of Dnmt1 in mouse stomach, we performed immunostaining analyses. In normal corpus, Dnmt1 was strongly expressed in isthmus cells and mucous neck cells (Figure 6A). Dnmt1-positive isthmus cells were positive for Ki67, but Dnmt1-positive mucous neck cells were negative for Ki67. In contrast, chief cells and parietal cells showed little or no detectable expression of Dnmt1. In mice with DMP-777 or L635 treatment, Dnmt1 was clearly up-regulated in SPEM cells as well as in chief cells before SPEM development (Figure 6B and C), and 40%–60% Dnmt1-positive cells were also proliferative (Figure 6B and D). These results suggest that the up-regulation of Dnmt1 in chief cells via miR-148a down-regulation could be involved in the initiation of chief cell transdifferentiation.
Figure 6.
Up-regulation of Dnmt1 in chief cells during chief cell transdifferentiation via down-regulation of miR-148a. (A) Immunofluorescence staining in the corpus of untreated mice. Left panel: GSII (green), Dnmt1 (red), and GIF (blue). Middle panel: GSII (green), Dnmt1 (red), and H/K-ATPase (blue). Right panel: GSII (green), Dnmt1 (red), and Ki67 (blue). Dnmt1 was strongly expressed in isthmus cells and neck cells. Dnmt1-positive isthmus cells were positive for Ki67, but Dnmt1-positive mucous neck cells were negative for Ki67. (B) Immunofluorescence staining in the corpus of mice treated by DMP-777 for 3 days or 10 days and L635 for 1 day or 3 days. Upper panel: GSII (green), Dnmt1 (red), and GIF (blue). Dnmt1 was up-regulated in SPEM cells of mice treated with DMP-777 for 10 days or L635 for 3 days as well as in chief cells of mice treated with DMP-777 for 3 days or L635 for 1 day. Lower panel: Ki67 (green), Dnmt1 (red), and GIF (blue). Ki67 was expressed in Dnmt1-positive SPEM cells of mice treated with DMP-777 for 10 days or L635 for 3 days as well as in Dnmt1-positive chief cells of mice treated with DMP-777 for 3 days or L635 for 1 day. (C) Quantitation of Dnmt1-positive chief cells. Percent of GIF-positive chief cells per corpus glands that are Dnmt1-positive. One-way analysis of variance, P = .0003. Bonferroni multiple comparisons, **P < .01, ***P < .001. (D) Quantitation of Ki67 in Dnmt1-positive chief cells. Percent of Dnmt1-positive chief cells per corpus glands that are Ki67-positive.
MiR-148a Down-regulation During the Chief Cell Transdifferentiation Process
Chief cell transdifferentiation occurs through coordinated stepwise events, and the process of transdifferentiation can be arrested at different steps. Our group recently showed that inhibition of the cystine transporter xCT by sulfasalazine treatment or xCT knockout arrests chief cell transdifferentiation process and prevents development of SPEM in mouse models of acute oxyntic atrophy.19 It was also shown that xCT deficiency blocks chief cell transdifferentiation process at a distinct step where the initiating step of Mist1 loss in chief cell reprogramming is not affected, but up-regulation of CD44v9 and autophagy that occur during chief cell transdifferentiation are blocked. To investigate where in this process miR-148a down-regulation occurs, we used these mouse models of xCT deficiency.19 In mice treated with sulfasalazine only, miR-148a was expressed normally in chief cells, whereas co-treatment with sulfasalazine and 3 days of L635 resulted in loss of miR-148a (Figure 7A and B). In xCT knockout mice, untreated stomach exhibited similar expression of miR-148a as in wild-type mice, with expression in chief cells at the gland base, whereas 3 days of L635 treatment resulted in a significant down-regulation of miR-148a expression (Figure 7C and D). These results suggest that, like Mist1, the loss of miR-148a cannot be rescued with xCT deficiency, and miR-148a down-regulation most likely occurs early in the initiating step of chief cell transdifferentiation.
Figure 7.
miR-148a down-regulation during chief cell transdifferentiation process. (A) Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (green) and DAPI (blue). miR-148a was expressed in chief cells treated with xCT inhibitor, sulfasalazine, only. With 3 days of L635 co-treatment with sulfasalazine, miR-148a expression was lost. (B) Quantitation of relative miR-148a staining intensity. Mann-Whitney U test, *P < .05. (C) Fluorescence in situ hybridization for miR-148a (red) and immunofluorescence staining for GSII (green) and DAPI (blue). In xCT knockout (KO) mice, miR-148a was expressed in chief cells. With 3 days of L635, miR-148a expression was decreased in xCT KO mice. (D) Quantitation of relative miR-148a staining intensity. Mann-Whitney U test, *P < .05.
Discussion
The gastric corpus mucosa contains 6 types of differentiated cells: parietal cells, chief cells, surface mucous cells, mucous neck cells, tuft cells, and enteroendocrine cells. All cells originate from stem cells in the isthmus region, but mucous neck cells can function as an intermediate precursor for chief cells.16 The differentiation process from mucous neck cells into chief cells involves dynamic alterations, including granule changes from mucous to serous, the alteration of apical-basal cell shape organization, and the expansion of rough endoplasmic reticulum.16, 31 MIST1 and its upstream gene XBP1 are transcription factors that regulate the cellular architecture maturation in chief cells.16 In addition, parietal cell loss induces the transdifferentiation of chief cells into mucous cell metaplasia, designated SPEM.32 In this study, we have demonstrated the involvement of miR-148a in the process of transdifferentiation of chief cells into SPEM. In situ hybridization analyses showed that miR-148a was specifically expressed in normal chief cells compared with mucous neck cells. In addition, miR-148a was significantly down-regulated in chief cells during SPEM development. Therefore, miR-148a could be involved in chief cell maturation and maintenance as well as plasticity.
The extremely high expression of several miRNAs in chief cells suggested that miRNAs could also be involved in chief cell maturation and maintenance in addition to chief cell plasticity. The report that XBP1 can directly induce the transcription of miR-148a supports this hypothesis.33 However, we did not find that miR-148a can increase chief cell factors in ImChief cells (data not shown). These results suggest that the down-regulation of miR-148a might lead chief cells toward transdifferentiation into SPEM cells. To understand the role of miRNAs on chief cell maturation, other experiments such as the use of mouse models with the deletion of miR-148a specifically in chief cells will be needed.
To investigate chief cell plasticity, we performed experiments using the ImChief cell line that we previously established.26 The ImChief cell line allows studies to be conducted in an in vitro model that resembles in vivo chief cells. The ImChief cell line also allows for the study of chief cell factors, which are difficult to study with gastric cancer cell lines and mouse models, although ImChief cells are not functionally fully mature.26 In this study, we also showed that ImChief cells can be transfected effectively with miRNA inhibitors. The list of miRNAs down-regulated during chief cell transdifferentiation included miR-148a and members of the miR-30, miR-200, and let-7 families. Among them, we examined the role of miR-148a in chief cell plasticity. A number of studies have shown that miR-148a is down-regulated in human cancer tissues, including gastric cancer.28, 34, 35, 36, 37 These previous investigations have demonstrated that miR-148a down-regulation could contribute to human gastric cancer progression by directly targeting MMP7, DNMT1, CCK-BR, and ROCK1.38, 39, 40, 41, 42 This study suggests a role of miR-148a on the normal physiology of gastric chief cells.
DNA methylation is a major modulator of gene expression. In mammalian cells, methylation is catalyzed by DNMTs that cooperatively establish tissue-specific methylation patterns. DNMT1 is responsible for the maintenance of DNA methylation during replication.43 Previous reports demonstrated that aberrant DNA methylation is induced in human and Mongolian gerbil gastric mucosa by H pylori infection,44, 45, 46 suggesting that regulation of gene expression by DNA methylation could be related with the initiation of metaplasia and neoplasia development. However, the up-regulation of DNMTs has not been observed in human or gerbil gastric mucosa with H pylori infection.45, 46 In this report, we demonstrated that Dnmt1 is up-regulated in chief cells during chief cell transdifferentiation via the down-regulation of miR-148a.
Previous studies have demonstrated that miR-148a directly targets the 3′-untranslated region of DNMT1 in various cancer cell lines.28, 29, 30 In contrast, overexpression of DNMT1 leads to hypermethylation of the promoter region of miR-148a in some cancer cell lines, including gastric cancer lines.28, 47 Thus, the regulatory network between miRNAs and epigenetic pathways seems to be important to organize gene expression in cancer biology. In this study, Dnmt1 was up-regulated in chief cells by the suppression of miR-148a especially at early stages of SPEM development. We have previously noted that transdifferentiating chief cells up-regulate a number of proteins involved in unwinding DNA, which is consistent with a need to reprogram the cell transcriptome.27 The fact that transdifferentiating chief cells show Dnmt1 up-regulation indicates that the induction of DNA methylation could be necessary for the transdifferentiation of chief cells. To identify methylated genes related with SPEM development, further detailed methylation analyses of chief cells and SPEM cells such as a global methylation analysis are needed at different stages of transdifferentiation.
Interestingly, the down-regulation of miR-148a correlated not only with its target gene, DNMT1, but also with the up-regulation of CD44v9, an early SPEM cell marker, in chief cells. CD44v9 expression is observed in human gastric cancer tissues and has been implicated as a cancer stem cell marker.48, 49 Importantly, SPEM is also associated with the up-regulation of CD44s as well as CD44v9,18, 50 whereas CD44v9 is not expressed in any cells in the normal gastric corpus mucosa. CD44v9 functions by blocking the stress signaling induced by reactive oxygen species via the interaction with the cystine transporter xCT.19, 20 Our recent report identified xCT as required for the SPEM development and chief cell reprogramming after injury and showed that xCT deficiency blocks the transdifferentiation process.19 Targeting xCT arrested chief cell reprogramming at a specific step where the transdifferentiation was initiated, marked by loss of Mist1, but up-regulation of autophagy did not occur. Investigation of miR-148a expression at this early step in chief cell transdifferentiation confirmed that miR-148a down-regulation occurs early in the initiation of chief cell reprogramming.
Metaplasia is considered to be a precursor lesion for gastric cancer; however, the mechanisms of how metaplasia develops in stomach are not fully elucidated. The mouse models of induced acute parietal cell loss such as DMP-777, L635, and high-dose tamoxifen have shown that chief cells can give rise to SPEM cells via a transdifferentiation process.21, 32, 51 Recent analyses suggest that parietal cell loss alone is not sufficient to induce metaplasia,52 and another signaling process, such as the interleukin 33/interleukin 13 cytokine signaling network, is required for SPEM development.17 The results from this study indicate that post-transcriptional regulation by miR-148a could be involved in the chief cell transdifferentiation by regulating DNMT1 expression and epigenetic change. Moreover, the miR-148a down-regulation triggered in response to mucosal injury occurred very early in the chief cell transdifferentiation process. The miR-148a inhibition in vitro using ImChief cell lines also supported this notion, because miR-148a inhibition alone was sufficient to induce up-regulation of CD44v9 transcripts in chief cells. The rapid down-regulation of miR-148a, especially from such a high level of expression in normal chief cells, would be expected to result in a cascade of targeted events, and the early change in miR-148a expression suggests that miR-148a down-regulation could play a crucial role in the initiation of chief cell reprogramming.
Materials and Methods
Mice
Eight-week-old C57BL/6 mice were used for all mouse experiments. For drug treatment experiments, 3 mice were used per group. DMP-777 and L635 treatment and dosage were conducted as previously described.15, 32 Briefly, DMP-777 was dissolved in 1% methylcellulose and administered by oral gavage (350 mg/kg) once a day for 3 or 10 consecutive days. L635 was dissolved in deionized DNA and RNA-free water and administered by oral gavage (350 mg/kg) once a day for 1 or 3 consecutive days. Mist1CreERT2/+ mice were crossed with R26RmTmG or R26RtdTomato mice, and 5 mg tamoxifen was administered to these mice subcutaneously every other day for 3 total doses.
Archival sections of stomach from H felis infected, sulfasalazine treated, and xCT knockout mice were obtained from previous study.15, 19 Regular mouse chow and water ad libitum were provided during experiments in a temperature-controlled room with 12-hour light-dark cycles. All treatment maintenance and care of animals in these studies followed protocols approved by the Institutional Animal Care and Use Committees of Vanderbilt University.
Cell Lines
Mouse chief cell line (ImChief cell) and SPEM cell line (ImSPEM cell) were established in a previous study.26 These cell lines were maintained in a 1:1 mixture of Ham’s F-12 and Dulbecco minimum essential medium (DMEM) containing 10% fetal bovine serum, 8 μg/mL insulin/transferrin/selenium solution, 1 μg/mL hydrocortisone, 100 U/mL penicillin and streptomycin, 100 μg/mL MycoZap Plus-PR, 1 ng/mL epidermal growth factor, 1 ng/mL basic fibroblast growth factor, and 5 U/mL interferon-γ. For ongoing maintenance, these cells were incubated and passaged at the permissive temperature (33°C). For analyses of experiments, cells were incubated at the non-permissive temperature (39°C) for 72 hours.
Total RNA Reparation From In Vivo Chief Cells and Chief Cell or Spasmolytic Polypeptide-Expressing Metaplasia Cell Lines for MicroRNA Sequencing
Mist1CreERT2/+ mice were crossed with R26RmTmG reporter mice, and 2 Mist1CreERT2/+;R26RmTmG/+ mice were used for chief cell isolation. Mice were killed 10 days after tamoxifen injection. The stomachs were removed and opened along the greater curvature. Stomachs were rinsed with ice-cold phosphate-buffered saline without Ca and Mg, and the antrums were removed with a razor blade and discarded. The corpus mucosa was separated from the serosa by dragging a cell scraper along the muscle layer. The corpus mucosa was minced with scissors and digested in the buffer containing advanced DMEM/F12, 5% fetal bovine serum, 1 mg/mL collagenase type Ia, and DNase I at 37°C. Advanced DMEM/F12 supplemented with 10 μmol/L Y-27632 and 1 mmol/L dithiothreitol was added to stop the reaction. These cells were resuspended in cold TrypLE Express supplemented with Y-27632 and incubated at 37°C. After stopping reaction, cells were resuspended in advanced DMEM/F12 with 1% fetal bovine serum and DNase I supplemented with Y-27632. Before cell sorting, cells were incubated with 1.0 μg/mL 4′,6-diamidino-2-phenylindol (DAPI). Cells were sorted by using a BD fluorescence-activated cell sorting Aria III (BD Biosciences, San Jose, CA) and initially segregated from debris by using forward scatter and side scatter properties of the 488-nm laser. Single cells were selected using the voltage pulse geometries of the forward scatter diode and side scatter photomultiplier tube detectors. Dead cells were excluded on the basis of their DAPI staining. GFP-positive chief cells were sorted directly into TRIzol (Invitrogen, Carlsbad, CA) using a 100-μm nozzle, followed by total RNA extraction according to the manufacturer’s instructions.
Total RNA extraction from chief cell lines and SPEM cell lines for miRNA sequencing and ImChief cells and ImSPEM cells grown at the non-permissive temperature were collected from 80% confluent T-75 dish. Cells were trypsinized, washed with phosphate-buffered saline twice, and pelleted. Total RNA was extracted by using the miRVana miRNA Isolation Kit according to manufacturer’s instructions.14
MicroRNA Library Preparation, Sequencing, and Data Analysis
Total RNA from each sample was processed through an RNA library preparation protocol using NEBNext Small RNA Library Prep Set for Illumina (New England BioLabs Inc, Ipswich, MA) according to manufacturer’s protocol. Briefly, 3′ adapters were ligated to total input RNA, followed by hybridization of multiplex SR RT primers and ligation of multiplex 5′ SR adapters. Reverse transcription was done using ProtoScript II RT for 1 hour at 50°C. Immediately after reverse transcription reaction, PCR amplification was performed for 15 cycles using LongAmp Taq 2X master mix. Illumina indexed primers were added to uniquely barcode each sample. Post-PCR material was purified by using QIAquick PCR purification kit (Qiagen Inc, Valencia, CA). Size selection of small RNA was done using 3% agarose free gel cassettes on Pippin prep instrument (Sage Science Inc, Beverly, MA). Post-size selection concentration and quality of the libraries were assessed by using Qubit 2.0 Fluorometer (Invitrogen) and DNA 1000 chip on Agilent 2100 Bioanalyzer (Applied Biosystems, Carlsbad, CA), respectively. Accurate quantification for sequencing applications was performed by using the quantitative PCR-based KAPA Biosystems Library Quantification kit (Kapa Biosystems, Inc, Woburn, MA). Each library was diluted to a final concentration of 1.25 nmol/L and pooled in equimolar ratios before clustering. Single End sequencing (50 base pairs) was performed to generate approximately 15 million reads per sample on an Illumina HiSeq2500 sequencer (Illumina, Inc, San Diego, CA).
Post-processing of the sequencing reads from miRNA-seq experiments from each sample was performed as per the Genomic Services Laboratory unique in-house pipeline. Briefly, quality control checks on raw sequence data from each sample were performed by using FastQC (Babraham Bioinformatics, London, UK). Raw reads were imported on a commercial data analysis platform AvadisNGS (Strand Scientifics, CA). Adapter trimming was done to remove ligated adapter from 3′ end of the sequenced reads, with only one mismatch allowed; poorly aligned 3′ ends were also trimmed. Sequences shorter than 15 nucleotides length were excluded from further analysis. Trimmed reads with low qualities (base quality score less than 30, alignment score less than 95, mapping quality less than 40) were removed. Filtered reads were then used to extract and count the small RNA, which was annotated with miRNAs from the miRBase release 20 database. The quantification operation carries out measurement at the gene level and at the active region level. Active region quantification considers only reads whose 5′ end matches the 5′ end of the mature miRNA annotation. For comparison of ImChief and ImSPEM cells, samples were grouped as identifiers, and the differential expression of miRNA was calculated on the basis of their fold change observed between different groups; P value of differentially expressed miRNAs was estimated by implementing z-score calculations using Benjamini Hochberg FDR corrections of 0.05.53
Immunofluorescence Staining
Mouse stomachs were fixed in 4% paraformaldehyde overnight and transferred into 70% ethanol for paraffin embedding. Five-micrometer paraffin-embedded sections were used for all immunofluorescence staining. Sections were deparaffinized, rehydrated, and submitted to antigen retrieval using Target Retrieval solution (Dako North America, Inc, Carpinteria, CA) in a pressure cooker. Blocking was performed by using Protein Block Serum-Free (Dako North America, Inc) for 1 hour 30 minutes at room temperature. The primary antibody incubation was performed in Antibody Diluent with Background Reducing Components (Dako North America, Inc) overnight at 4°C. Primary antibodies used were as follows: rat anti-Cd44v9 (1:25,000), goat anti-GIF (1:2000), rabbit anti-Mist1 (1:300), mouse anti-HK-ATPase (1:10,000), rat anti-Ki67 (1:50), and rabbit anti-Dnmt1 (1:200). Fluorescent second antibodies (1:500) and Alexa-488 and 647-conjugated GSII (1:2000) were incubated for 1 hour at room temperature. After incubation with DAPI for 5 minutes, slides were mounted with ProLong Gold Antifade Reagent (Invitrogen). Fluorescence imaging was analyzed by using an Axio Imager 2 microscope (Carl Zeiss AG, Oberkochen, Germany) or a Versa S200 automated slide scanner (Leica Biosystems, Buffalo Grove, IL) in the Vanderbilt Digital Histology Shared Resource.
Fluorescence In Situ Hybridization for MiR-148a
Stomachs of wild-type mice were fixed in 4% paraformaldehyde overnight and transferred into 70% ethanol for paraffin embedding. Five-micrometer paraffin-embedded sections were deparaffinized and rehydrated. Stomachs of Mist1CreERT2/+;R26RtdTomato/+ mice were fixed in 4% paraformaldehyde overnight and embedded in OCT compound. Ten-micrometer sections were cut from OCT-embedded blocks, mounted on SuperFrost Plus slides, allowed to air-dry for 15 minutes, and then rinsed for 3 × 3 minutes. In situ hybridization process and TSA Plus fluorescence system were performed as the manufacturer’s protocol from Exiqon (South Korea). Briefly, incubation with proteinase-K (2 μg/mL) was performed for 10 minutes at 37°C. Then blocking of endogenous peroxidase activity was performed by using peroxidase block (Dako) for 10 minutes at room temperature. In situ hybridization was performed by using locked nucleic acid probes (25 nmol/L) for 1 hour at 60°C. After washing slides in side scatter buffers, blocking was conducted by using 1% sheep serum for 15 minutes at room temperature. Anti-digoxigenin-POD antibody (1:400) was applied to slides and incubated for 1 hour at room temperature. To detect digoxigenin, TSA Plus Cy5 substrate (1:200) was applied to slides and incubated for 10 minutes at room temperature. After incubation with Alexa-488 conjugated GSII (1:2000) for 30 minutes and DAPI for 5 minutes, slides were mounted with ProLong Gold Antifade Reagent (Invitrogen).
To examine the localization of miRNAs and proteins, a combination analysis with in situ hybridization for miRNAs and immunofluorescence staining for proteins was performed. Five-micrometer paraffin-embedded sections were deparaffinized, rehydrated, and submitted to antigen retrieval using Target Retrieval solution (Dako North America, Inc) in a pressure cooker. Blocking of endogenous peroxidase activity, in situ hybridization, protein blocking with sheep serum, incubation with anti-digoxigenin-POD, and incubation with TSA plus Cy5 were performed as noted above. Then slides were blocked by using Protein Block Serum-Free (Dako North America, Inc) and incubated with primary and secondary antibodies as in the regular immunofluorescence staining method.
MicroRNA Transfection Experiments
ImChief cells were transfected with 100 nmol/L miRNA inhibitors (miR-148a or control inhibitor) by using Lipofectamine 2000 reagent (Invitrogen) at 33°C, incubated at 33°C for 24 hours, and then cultured at 39°C for 72 hours. Total RNA was extracted from ImChief cells or ImSPEM cells with TRIzol (Invitrogen) according to the manufacturer’s instructions. For quantitative reverse transcription PCR of mRNA, 1 μg total RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI) and then reverse-transcribed with Superscript III reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed with SYBR Green by using specific primers (Table 5) in triplicate using an ABI StepOnePlus Real-Time PCR System (Applied Biosystems). A set of PCR primers for TATA-box-binding protein (Tbp) gene was used as an endogenous control and reference for verification of sufficient cDNA in the reaction. Each sample was collected from at least 3 biological replicate experiments. The miRNA expression in cell lysates was analyzed by using TaqMan MicroRNA assays according to the manufacturer’s instructions. U6 was used as an endogenous control. Statistical significance (P < .05) of the differences in the expression levels was determined with Mann-Whitney U test.
Table 5.
Primer Sequences for Quantitative PCR
| Primer name | Sequence |
|---|---|
| Tbp F | CAAACCCAGAATTGTTCTCCTT |
| Tbp R | ATGTGGTCTTCCTGAATCCCT |
| He4 F | TGCCTGCCTGTCGCCTCTG |
| He4 R | TGTCCGCACAGTCCTTGTCCA |
| Sox9 F | CACGGAGCAGACGCACATCT |
| Sox9 R | TCTCGCTTCAGGTCAGCCTT |
| Cd44v9 F | GGAGATCAGGATGACTCCTTCT |
| Cd44v9 R | AGTCCTTGGATGAGTCTCGATC |
| Clu F | CCAGCCTTTCTTTGAGATGA |
| Clu R | CTCCTGGCACTTTTCACACT |
| Pgc F | TCTAACGGCGGGCAGATT |
| Pgc R | AGGTACTGGGCAGGCATGAC |
| Mist1 F | GCTGACCGCCACCATACTTAC |
| Mist1 R | TGTGTAGAGTAGCGTTGCAGG |
| Dnmt1 F | AGATGCCATCACCCAAAAAG |
| Dnmt1 R | TCATCGATGCTCACCTTCTG |
| Tff2 F | TGCTTTGATCTTGGATGCTG |
| Tff2 R | GGAAAAGCAGCAGTTTCGAC |
| Ccnf F | CACACCAGCCTGTCCATCTATG |
| Ccnf R | ACGAGGTCACTGTAGGAGAAGC |
| Dgcr8 F | GATGGTGTGACTTACGGATCTGG |
| Dgcr8 R | TAGGCTTCTCCTCAGAGGTCTG |
| Kat7 F | AGGAAAAGGTGGCTGAACTCAGG |
| Kat7 R | GTCAGGTTTTCCAAGAGAGGCTC |
| Rcc2 F | GTGGTTCGAGATGTAGCCTGTG |
| Rcc2 R | AGGCACCATCTCATCCTTCTGC |
Image Quantitation
For miR-148a expression, experimental groups contained 3–8 mice. Images were analyzed with CellProfiler by using total intensity divided by the area detected, and all graphs and statistics were completed in GraphPad Prism (San Diego, CA). The intensity and area were calculated by masking out all areas except the base of the gland. More than 100 glands of proximal stomach corpus were taken from each mouse at ×20 objective for quantification. For acute and chronic mouse model of SPEM, one-way analysis of variance and Bonferroni multiple comparisons test were used to calculate statistical significance (P < .05). For xCT deficiency mouse model, the Mann-Whitney U test was used to determine statistical significance (P < .05).
To perform the quantitation of the percentage of Dnmt1-positive cells or Ki67 and Dnmt1 double-positive cells in chief cells or SPEM cells, 30 corpus glands were analyzed for each sample. Three samples were analyzed per group (untreated, DMP-777 treatment for 3 days, DMP-777 treatment for 10 days, L635 treatment for 1 day, and L635 treatment for 3 days). Chief cells were detected as GIF-positive cells. Counting was performed in slides digitally imaged with a Leica Versa 200 scanner (Vanderbilt Digital Histology Shared Resource) or in overlaid fluorescence images using Adobe Photoshop (San Jose, CA). One-way analysis of variance and Bonferroni multiple comparisons test were used to calculate statistical significance (P < .05).
Footnotes
Author contributions T.S. and Y.S. performed experiments, analyzed data, wrote the manuscript, and prepared figures. E.C., N.P., and C.P.P. performed experiments, analyzed data, and edited the manuscript. J.R.G. developed experimental design, analyzed data, and edited the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding These studies were supported by grants from Department of Veterans Affairs Merit Review Award IBX000930 and NIHRO1 DK071590 and RO1 DK101332 (to J.R.G) and from DODW81XWH-17-1-0257, AACR17-20-41-CHOI, and NIHP30 DK058404 (to E.C). T.S. was the recipient of JSPS Postdoctoral Fellowships for Research Abroad. C.P.P. was supported by NIHNRSA Predoctoral Fellowship (F31 DK104600). This work was supported by core resources of the Vanderbilt Digestive Disease Center (NIHP30 DK058404), Translational Pathology Shared Resource (NCI/NIH Cancer Center Support Grant 2P30 CA068485-14), and imaging supported by the Vanderbilt Digital Histology Shared Resource supported by a VA Shared Instrumentation grant (1IS1BX003097).
References
- 1.Weis V.G., Petersen C.P., Weis J.A., Meyer A.R., Choi E., Mills J.C., Goldenring J.R. Maturity and age influence chief cell ability to transdifferentiate into metaplasia. Am J Physiol Gastrointest Liver Physiol. 2016;312:G67–G76. doi: 10.1152/ajpgi.00326.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldenring J.R., Nam K.T., Mills J.C. The origin of pre-neoplastic metaplasia in the stomach: chief cells emerge from the Mist. Exp Cell Res. 2011;317:2759–2764. doi: 10.1016/j.yexcr.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lennerz J.K.M., Kim S., Oates E.L., Huh W.J., Dherty J.M., Tian X., Bredemeyer A.J., Goldenring J.R., Lauwers G.Y., Shin G.Y., Mills J.C. The transcription factor MIST1 is a novel human gastric chief cell marker whose expression is lost in metaplasia, dysplasia and carcinoma. Am J Pathol. 2010;177:1514–1533. doi: 10.2353/ajpath.2010.100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredemeyer A.J., Geahlen J.H., Weis V.G., Huh W.J., Zinselmeyer B.H., Srivatsan S., Miller M.J., Shaw A.S., Mills J.C. The gastric epithelial progenitor cell niche and differentiation of the zymogenic (chief) cell lineage. Dev Biol. 2009;325:211–224. doi: 10.1016/j.ydbio.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills J.C., Sansom O.J. Reserve stem cells: differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci Signal. 2015;8:re8. doi: 10.1126/scisignal.aaa7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldenring J.R., Nam K.T., Wang T.C., Mills J.C., Wright N.A. Spasmolytic polypeptide-expressing metaplasia and intestinal metaplasia: time for reevaluation of metaplasias and the origins of gastric cancer. Gastroenterology. 2010;138:2207–2210. doi: 10.1053/j.gastro.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A., Slack F.J. Oncomirs: microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Ueda T., Volinia S., Okumura H., Shimizu M., Taccioli C., Rossi S., Alder H., Liu C.G., Oue N., Yasui W., Yoshida K., Sasaki H., Nomura S., Seto Y., Kaminishi M., Calin G.A., Croce C.M. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima K., Isomoto H., Inoue N., Nakayama T., Hayashi T., Nakayama M., Nakao K., Hirayama T., Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128:361–370. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 13.Zabaleta J. MicroRNA: a bridge from H pylori infection to gastritis and gastric cancer development. Frontiers in Genetics. 2012;3:294. doi: 10.3389/fgene.2012.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sousa J.F., Nam K.T., Petersen C.P., Lee H.J., Yang H.K., Kim W.H., Goldenring J.R. miR-30-HNF4gamma and miR-194-NR2F2 regulatory networks contribute to the upregulation of metaplasia markers in the stomach. Gut. 2016;65:914–924. doi: 10.1136/gutjnl-2014-308759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weis V.G., Sousa J.F., LaFleur B.J., Nam K.T., Weis J.A., Finke P.E., Ameen N.A., Fox J.G., Goldenring J.R. Heterogeneity in mouse SPEM lineages identifies markers of metaplastic progression. Gut. 2013;62:1270–1279. doi: 10.1136/gutjnl-2012-302401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramsey V.G., Doherty J.M., Chen C.C., Stappenbeck T.S., Konieczny S.F., Mills J.C. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 17.Petersen C.P., Meyer A.R., De Salvo C., Choi E., Schlegel C., Petersen A., Engevik A.C., Prasad N., Levy S.E., Peebles R.S., Pizarro T.T., Goldenring J.R. A signalling cascade of IL-33 to IL-13 regulates metaplasia in the mouse stomach. Gut. 2018;67:805–817. doi: 10.1136/gutjnl-2016-312779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada T., Ishimoto T., Seishima R., Tsuchihashi K., Yoshikawa M., Oshima H., Oshima M., Masuko T., Wright N.A., Furuhashi S., Hirashima K., Baba H., Kitagawa Y., Saya H., Nagano O. Functional role of CD44v-xCT system in the development of spasmolytic polypeptide-expressing metaplasia. Cancer Science. 2013;104:1323–1329. doi: 10.1111/cas.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer A.R., Engevik A.C., Willet S.G., Williams J.A., Zou Y., Massion P.P., Mills J.C., Choi E., Goldenring J.R. Cystine/glutamate antiporter (xCT) is required for chief cell plasticity after gastric injury. Cell Mol Gastroenterol Hepatol. 2019;8:379–405. doi: 10.1016/j.jcmgh.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimoto T., Nagano O., Yae T., Tamada M., Motohara T., Oshima H., Oshima M., Ikeda T., Asaba R., Yagi H., Masuko T., Shimizu T., Ishikawa T., Kai K., Takahashi E., Imamura Y., Baba Y., Ohmura M., Suematsu M., Baba H., Saya H. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Willet S.G., Lewis M.A., Miao Z.F., Liu D., Radyk M.D., Cunningham R.L., Burclaff J., Sibbel G., Lo H.G., Blanc V., Davidson N.O., Wang Z.N., Mills J.C. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J. 2018;37(7) doi: 10.15252/embj.201798311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu X., Zhu H., Liu S., Tao G., Jin J., Chu H., Wang M., Tong N., Gong W., Zhao Q., Qiang F., Zhang Z. Expression and prognostic value of microRNA-26a and microRNA-148a in gastric cancer. J Gastroenterol Hepatol. 2017;32:819–827. doi: 10.1111/jgh.13533. [DOI] [PubMed] [Google Scholar]
- 23.Tsukamoto Y., Nakada C., Noguchi T., Tanigawa M., Nguyen L.T., Uchida T., Hijiya N., Matsuura K., Fujioka T., Seto M., Moriyama M. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 24.Tang H., Deng M., Tang Y., Xie X., Guo J., Kong Y., Ye F., Su Q., Xie X. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]
- 25.Motoyama K., Inoue H., Nakamura Y., Uetake H., Sugihara K., Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–2340. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 26.Weis V.G., Petersen C.P., Mills J.C., Tuma P.L., Whitehead R.H., Goldenring J.R. Establishment of novel in vitro mouse chief cell and SPEM cultures identifies MAL2 as a marker of metaplasia in the stomach. Am J Physiol Gastrointest Liver Physiol. 2014;307:G777–G792. doi: 10.1152/ajpgi.00169.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nozaki K., Ogawa M., Williams J.A., LaFleur B.J., Ng V., Drapkin R.I., Mills J.C., Konieczny S.F., Nomura S., Goldenring J.R. A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology. 2008;134:511–521. doi: 10.1053/j.gastro.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu A., Xia J., Zuo J., Jin S., Zhou H., Yao L., Huang H., Han Z. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med Oncol. 2012;29:2701–2709. doi: 10.1007/s12032-011-0134-3. [DOI] [PubMed] [Google Scholar]
- 29.Gailhouste L., Gomez-Santos L., Hagiwara K., Hatada I., Kitagawa N., Kawaharada K., Thirion M., Kosaka N., Takahashi R.U., Shibata T., Miyajima A., Ochiya T. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58:1153–1165. doi: 10.1002/hep.26422. [DOI] [PubMed] [Google Scholar]
- 30.Wu T., Qu L., He G., Tian L., Li L., Zhou H., Jin Q., Ren J., Wang Y., Wang J., Kan X., Liu M., Shen J., Guo M., Sun Y. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553–11566. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karam S.M., Leblond C.P. Dynamics of epithelial cells in the corpus of the mouse stomach: III—inward migration of neck cells followed by progressive transformation into zymogenic cells. Anat Rec. 1993;236:297–313. doi: 10.1002/ar.1092360204. [DOI] [PubMed] [Google Scholar]
- 32.Nam K.T., Lee H.-J., Sousa J.F., Weis V.G., O'Neal R.L., Finke P.E., Romero-Gallo J., Shi G., Mills J.C., Peek R.M., Konieczny S.F., Goldenring J.R. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139:2028–2037. doi: 10.1053/j.gastro.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho Y.M., Kim T.M., Hun Kim D., Hee Kim D., Jeong S.W., Kwon O.J. miR-148a is a downstream effector of X-box-binding protein 1 that silences Wnt10b during adipogenesis of 3T3-L1 cells. Exp Mol Med. 2016;48:e226. doi: 10.1038/emm.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Q., He M., Ma M.T., Wu H.Z., Yu Z.J., Guan S., Jiang L.Y., Wang Y., Zheng D.D., Jin F., Wei M.J. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol Rep. 2016;35:1425–1432. doi: 10.3892/or.2015.4502. [DOI] [PubMed] [Google Scholar]
- 35.Cao H., Liu Z., Wang R., Zhang X., Yi W., Nie G., Yu Y., Wang G., Zhu M. miR-148a suppresses human renal cell carcinoma malignancy by targeting AKT2. Oncol Rep. 2017;37:147–154. doi: 10.3892/or.2016.5257. [DOI] [PubMed] [Google Scholar]
- 36.Liu L., Ye J.X., Qin Y.Z., Chen Q.H., Ge L.Y. Evaluation of miR-29c, miR-124, miR-135a and miR-148a in predicting lymph node metastasis and tumor stage of gastric cancer. International Journal of Clinical and Experimental Medicine. 2015;8:22227–22236. [PMC free article] [PubMed] [Google Scholar]
- 37.Peng L., Liu Z., Xiao J., Tu Y., Wan Z., Xiong H., Li Y., Xiao W. MicroRNA-148a suppresses epithelial-mesenchymal transition and invasion of pancreatic cancer cells by targeting Wnt10b and inhibiting the Wnt/beta-catenin signaling pathway. Oncol Rep. 2017;38:301–308. doi: 10.3892/or.2017.5705. [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto N., Naito Y., Oue N., Sentani K., Uraoka N., Zarni Oo H., Yanagihara K., Aoyagi K., Sasaki H., Yasui W. MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Science. 2014;105:236–243. doi: 10.1111/cas.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J., Guo X., Xia J., Shan T., Gu C., Liang Z., Zhao W., Jin S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31:879. doi: 10.1007/s12032-014-0879-6. [DOI] [PubMed] [Google Scholar]
- 40.Yu B., Lv X., Su L., Li J., Yu Y., Gu Q., Yan M., Zhu Z., Liu B. MiR-148a functions as a tumor suppressor by targeting CCK-BR via inactivating STAT3 and Akt in human gastric cancer. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng B., Liang L., Wang C., Huang S., Cao X., Zha R., Liu L., Jia D., Tian Q., Wu J., Ye Y., Wang Q., Long Z., Zhou Y., Du C., He X., Shi Y. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 42.Li B., Wang W., Li Z., Chen Z., Zhi X., Xu J., Li Q., Wang L., Huang X., Wang L., Wei S., Sun G., Zhang X., He Z., Zhang L., Zhang D., Xu H., El-Rifai W., Xu Z. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017;410:212–227. doi: 10.1016/j.canlet.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perdigoto C.N., Valdes V.J., Bardot E.S., Ezhkova E. Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med. 2014;4(2) doi: 10.1101/cshperspect.a015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maekita T., Nakazawa K., Mihara M., Nakajima T., Yanaoka K., Iguchi M., Arii K., Kaneda A., Tsukamoto T., Tatematsu M., Tamura G., Saito D., Sugimura T., Ichinose M., Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12(Pt 1):989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 45.Niwa T., Tsukamoto T., Toyoda T., Mori A., Tanaka H., Maekita T., Ichinose M., Tatematsu M., Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–1440. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 46.Nakajima T., Yamashita S., Maekita T., Niwa T., Nakazawa K., Ushijima T. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int J Cancer. 2009;124:905–910. doi: 10.1002/ijc.24018. [DOI] [PubMed] [Google Scholar]
- 47.Long X.R., He Y., Huang C., Li J. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis. Int J Oncol. 2014;44:1915–1922. doi: 10.3892/ijo.2014.2373. [DOI] [PubMed] [Google Scholar]
- 48.Go S.I., Ko G.H., Lee W.S., Kim R.B., Lee J.H., Jeong S.H., Lee Y.J., Hong S.C., Ha W.S. CD44 variant 9 serves as a poor prognostic marker in early gastric cancer, but not in advanced gastric cancer. Cancer Res Treat. 2016;48:142–152. doi: 10.4143/crt.2014.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau W.M., Teng E., Chong H.S., Lopez K.A., Tay A.Y., Salto-Tellez M., Shabbir A., So J.B., Chan S.L. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630–2641. doi: 10.1158/0008-5472.CAN-13-2309. [DOI] [PubMed] [Google Scholar]
- 50.Bertaux-Skeirik N., Wunderlich M., Teal E., Chakrabarti J., Biesiada J., Mahe M., Sundaram N., Gabre J., Hawkins J., Jian G., Engevik A.C., Yang L., Wang J., Goldenring J.R., Qualls J.E., Medvedovic M., Helmrath M.A., Diwan T., Mulloy J.C., Zavros Y. CD44 variant isoform 9 emerges in response to injury and contributes to the regeneration of the gastric epithelium. J Pathol. 2017;242:463–475. doi: 10.1002/path.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leushacke M., Tan S.H., Wong A., Swathi Y., Hajamohideen A., Tan L.T., Goh J., Wong E., Denil S., Murakami K., Barker N. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat Cell Biol. 2017;19:774–786. doi: 10.1038/ncb3541. [DOI] [PubMed] [Google Scholar]
- 52.Burclaff J., Osaki L.H., Liu D., Goldenring J.R., Mills J.C. Targeted apoptosis of parietal cells is insufficient to induce metaplasia in stomach. Gastroenterology. 2017;152:762–766. doi: 10.1053/j.gastro.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]