Abstract
Adeno-associated virus (AAV) capsid libraries have generated improved transgene delivery vectors. We designed an AAV library construct, iTransduce, that combines a peptide library on the AAV9 capsid with a Cre cassette to enable sensitive detection of transgene expression. After only two selection rounds of the library delivered intravenously in transgenic mice carrying a Cre-inducible fluorescent protein, we flow sorted fluorescent cells from brain, and DNA sequencing revealed two dominant capsids. One of the capsids, termed AAV-F, mediated transgene expression in the brain cortex more than 65-fold (astrocytes) and 171-fold (neurons) higher than the parental AAV9. High transduction efficiency was sex-independent and sustained in two mouse strains (C57BL/6 and BALB/c), making it a highly useful capsid for CNS transduction of mice. Future work in large animal models will test the translation potential of AAV-F.
Keywords: adeno-associated virus vector, AAV vector, gene therapy, gene delivery, central nervous system, transduction efficiency, AAV capsid library
Graphical Abstract
Introduction
While adeno-associated virus 9 (AAV9) vectors have shown remarkable potential for delivery to the CNS after systemic delivery, resulting in clinical success in pediatric patients with spinal muscular atrophy,1 systemic injection of high doses of AAV vectors can lead to induction of a T cell response that can eliminate transduced cells.2 In monkeys, there is one report in which high systemic doses of an AAV9-like vector resulted in toxicity and death of the animal, which was attributed to systemic inflammation.3 A recent phase I clinical trial using high-dose AAV9 to treat muscular dystrophy was placed on hold by the US Food and Drug Administration (FDA) due to an immune reaction after vector infusion in one patient. The reason high doses are required is due to the relatively low efficiency of AAV on a per-vector genome copy basis to provide adequate transgene expression in a substantial number of target cells. Thus, developing new AAV capsids that allow more efficient transduction at lower doses should result in better therapeutic efficacy while lowering safety issues, such as immunotoxicity.
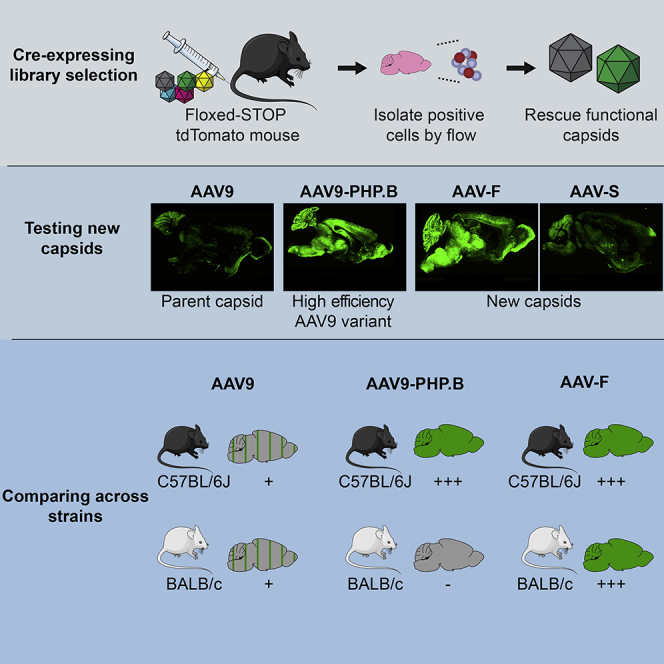
A promising approach to efficient delivery of transgenes to target cells is via a process of submitting a pool, or library, of AAV vector capsids variants to an in vivo selection process, a veritable “survival of the fittest” approach.4, 5, 6, 7, 8 AAV library approaches that use random oligomer nucleotides to insert short (6–9 aa) random peptides into an exposed region on the capsid surface have demonstrated success in identifying new AAV capsid variants with unique properties such as enhanced transduction of target tissues.9,10 One major limitation of AAV libraries is that the end readout of the selection process does not always differentiate capsids that mediate functional transgene expression from those that do not. AAV transduction is a process involving multiple steps, from cell receptor binding and entry to nuclear transport, second-strand synthesis, and finally gene and protein expression.11 A recent advance on the conventional AAV library approach, called CREATE, engineered a Cre-sensitive AAV genome that enabled selectively isolated capsids that have successfully trafficked to the nucleus in the context of a Cre-expressing transgenic animal.12 In this study, we describe a capsid selection system called iTransduce in which we also utilize the power of the Cre/loxP system. Instead of using Cre transgenic mice, we engineered the AAV to encode capsids with peptide inserts, along with a Cre-expression cassette. We then performed selection in mice with a Cre-sensitive fluorescent reporter to enable selection of capsids that mediate the entire process of transduction, including transgene expression. We show that in vivo selection of the library can result in the identification of an AAV capsid that mediates remarkable transduction efficiency of the CNS.
Results
Design of iTransduce—An Expression-Based AAV Library
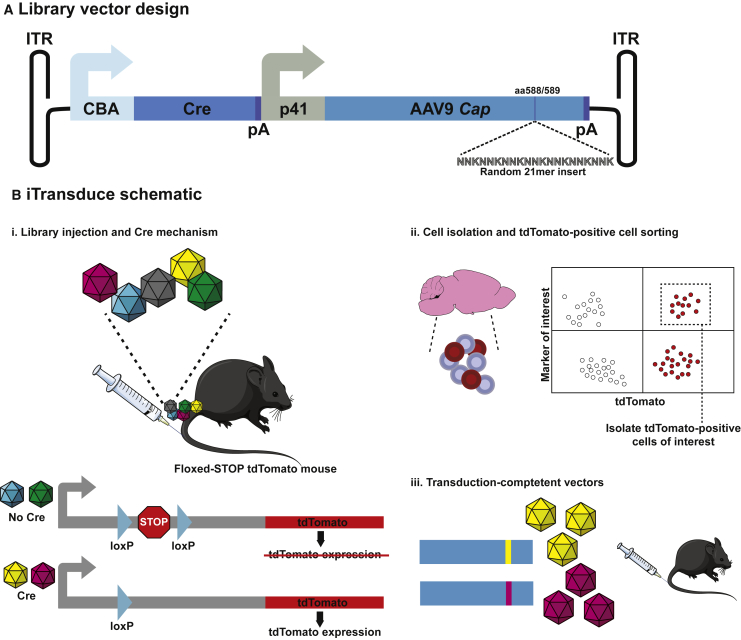
First, we constructed an AAV library plasmid that consisted of an AAV2 inverted terminal repeat (ITR)-flanked expression cassette comprised of a chicken β-actin (CBA)-driven Cre recombinase and a p41 promoter-driven AAV9 capsid (schematic in Figure 1A). Pseudorandom 21-base nucleotides were inserted between AAV9 VP1 nucleotides encoding amino acids 588/589 via PCR. Before viral packaging, we sequenced this plasmid library using low-depth next-generation sequencing (NGS) and confirmed the presence of 21-mer inserts in the vast majority of plasmids and the lack of variant bias (data not shown). We then packaged the capsid library and performed NGS to validate that the vector creation process maintained a sufficient diversity for selection. iTransduce relies on each unique capsid carrying both its own cap gene as well as a Cre-expressing construct (Figure 1B). Transgenic mice (Ai9) carrying a floxed-STOP tdTomato cassette are injected intravenously with the AAV library (Figures 1Bi). Those capsids that successfully transduce cells enable tdTomato expression in any target organ or cell type (without being dependent on the availability of specific Cre transgenic mouse lines); these tdTomato-positive cells can then be flow sorted from the tissue of interest (optionally, alongside cell-specific markers; Figure 1Bii). Viral DNA rescued from these cells should correspond to capsid variants that can effectively overcome all of the extracellular and intracellular biological barriers to transgene expression (Figure 1Biii).
Figure 1.
iTransduce Library for Selection of Novel AAV Capsids Capable of Efficient Transgene Expression in Target Tissue
(A) Two-component system of the library construct. (1) Cre recombinase is driven by a minimal chicken β-actin (CBA) promoter. (2) p41 promoter-driven AAV9 capsid with random heptamer peptide is inserted between amino acids 588 and 589, cloned downstream of the Cre cassette. (B) Selection strategy. (Bi) The iTransduce library comprised of different peptide inserts expressed on the capsid (represented by different colors) is injected intravenously (i.v.) into an Ai9 transgenic mouse with a loxP-flanked STOP cassette upsteam of the tdTomato reporter gene, inserted into the Gt(ROSA)26Sor locus. AAV capsids able to enter the cell of interest but that do not functionally transduce the cell (no Cre expression) do not turn on tdTomato expression. Capsids that can mediate functional transduction (express Cre) will turn on tdTomato expression. (Bii) Cells are isolated from the organ of interest (e.g., brain), and transduced cells are sorted for tdTomato expression and optionally cell markers. (Biii) Capsid DNA is PCR amplified from the sorted cells, cloned back to the library vector, and repackaged for another round of selection. DNA sequencing analysis is utilized after each round to monitor the selection process.
Selection of New AAV9 Capsid Variants Using Transgene Expression to Identify Functional Capsids
To test the iTransduce library strategy, we performed a proof-of-concept selection to attempt to isolate AAV capsid variants with the ability to transduce brain cells after systemic injection. 1.27 x 1011 vector genomes (vg; 5 × 1012 vg/kg) of the library were injected intravenously through the tail vein of one adult male and one female Ai9 mouse, and 3 weeks postinjection the mice were killed; sections of liver, brain, spleen, and kidney were cut and immunostaining of tdTomato was performed. We readily detected tdTomato-positive cells (likely hepatocytes) in the liver, with scattered tdTomato-positive cells (both neurons and astrocytes) in the brain as well as the other organs (Figure S1A). We also tested whether we could rescue the cap DNA containing the 21-base insert by PCR, and we detected specific bands in 10 organs/tissues in mice injected with the library, but not in control, non-injected mice (Figure S1B). We pooled the remaining brain tissue from the female and male mice, and DNA was extracted, initially from total brain tissue for the first round of selection. We amplified cap DNA containing the 21-mer insert and analyzed its identity and read counts using NGS. We observed substantial enrichment of specific peptides in the library population harvested from brain tissue following a single round of selection when compared to the unselected library and with the variants retrieved from liver (Figure S1C; data not shown). Next, we isolated the insert-containing region of viral DNA and re-cloned it back into the AAV plasmid backbone and repackaged capsids (“brain-enriched capsid library”) for a second round of selection.
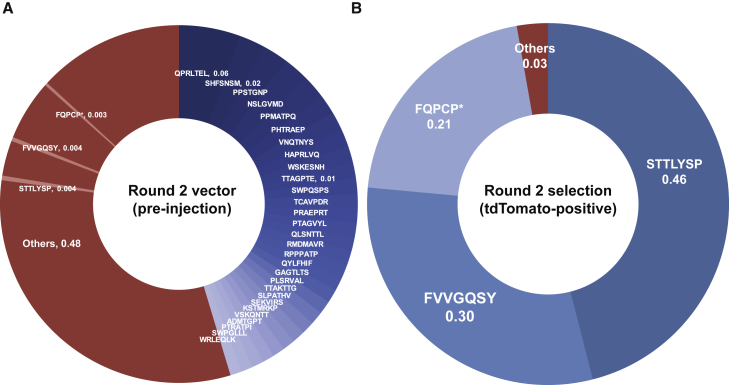
For the second round of selection, two Ai9 mice (one male, one female) were injected with the library rescued from round 1 (dose of 1.91 × 1010 vg, 7.64 × 1011 vg/kg) and sacrificed after 3 weeks. Prior to injection, sequence containing the variant region was amplified and sequenced by NGS to ensure that a preexisting bias had not been introduced into the vector pool (Figure 2). The brain tissue was dissociated to obtain a cell suspension for sorting tdTomato-positive cells by flow cytometry. We flow sorted 3,834 tdTomato-positive cells (0.043% of the initial cell suspension), which were indicative of successful transduction (Figure S2). Viral DNA from tdTomato-sorted cells was amplified and sequenced as previously done (Figure 2). Viral DNA isolated from tdTomato-positive cells showed 97% of reads represented by just three peptides, STTLYSP, FVVGQSY, and FQPCP∗ (where ∗ indicates a stop codon) (Figure 2). We selected two of these, STTLYSP (termed AAV-S) and FVVGQSY (termed AAV-F), for functional evaluation in vivo (we excluded evaluation of FQPCP∗, as it was likely a product of cross-packaging). As seen in Figure 2, both of these sequences were detectable in the round 2 library at low levels (∼0.4% of reads for each variant), but they were highly enriched in the brain after selection.
Figure 2.
Identification of AAV-S and AAV-F after Two Rounds of In Vivo Selection for Brain Transduction
Donut charts indicate the frequency of particular peptide inserts determined by NGS. (A) Chart of round 2 vector sequenced after production but before intravenous injection. (B) Chart of peptide frequency appearing in iTransduce isolation after round 2 injection. “Others” indicates sequence variants appearing as less than 1% of the total pool (in A, variants isolated after the round 2 screen are also highlighted, appearing at less than 1%). The asterisk (*) indicates a stop codon.
AAV-F Capsid Mediates Efficient Transgene Expression in the Murine CNS
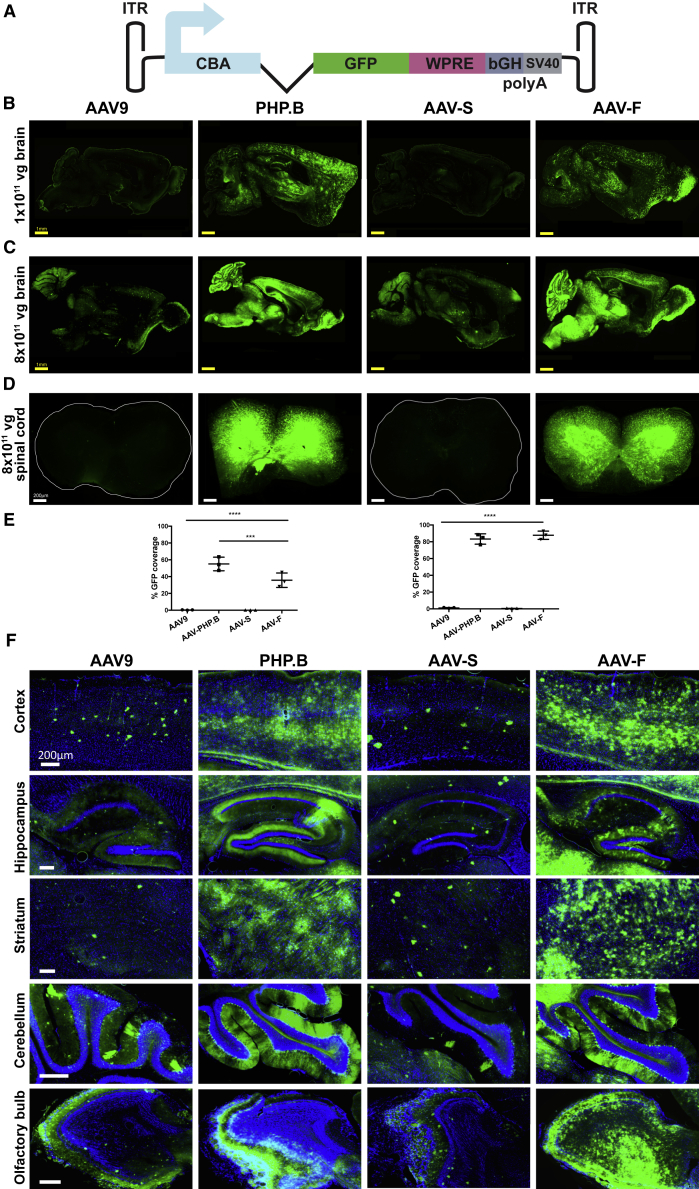
To test whether these peptides displayed on the capsid of AAV could mediate efficient transgene expression by an AAV vector, we produced these capsids (AAV-S and AAV-F), packaging a single-stranded CBA promoter-driven GFP expression cassette (Figure 3A). For comparison, we included the parental AAV9 vector and AAV9-PHP.B, the most widely studied AAV9 variant with a 7-mer peptide insertion (TLAVPFK) generated by directed evolution.12 All vectors produced well and gave slightly lower production efficiencies than did AAV9 (Table 1). Adult male C57BL/6J mice were injected via the lateral tail vein with a low dose or a high dose (1 × 1011 vg and 8 × 1011 vg of vector, respectively; approximately 4 × 1012 and 3.2 × 1013 vg/kg) of one of the following vectors: AAV9, AAV9-PHP.B, AAV-S, and AAV-F (n = 3 each). Three weeks postinjection, mice were killed and organs harvested for endogenous (unstained) GFP fluorescence analysis. We quantitated the percent coverage of the GFP signal in serial sagittal brain sections (three sections were analyzed per animal). Remarkably, AAV-F demonstrated a 119-fold (p < 0.0001) and 68-fold (p = 0.0004) increased GFP fluorescence coverage compared to the parental AAV9 vector at 1 × 1011 vg and 8 × 1011 vg, respectively (Figures 3B, 3C, 3E, and 3F; Figure S3). AAV9 and AAV-S displayed similar GFP coverage levels (Figures 3B, 3C, 3E, and 3F). AAV9-PHP.B gave slightly higher GFP coverage at the low dose compared to AAV-F, while similar levels of GFP coverage were observed at the 8 × 1011 vg dose (Figures 3B, 3C, 3E, and 3F). Similar to AAV9-PHP.B, AAV-F transduced the spinal cord with remarkable efficiency (Figure 3D). Most areas of the brain were effectively targeted by AAV-F, and a robust GFP signal was observed in the cortex, hippocampus, striatum, cerebellum, and olfactory bulb (Figure 3F).
Figure 3.
AAV-F Efficiently Transduces the Brain of Mice after Systemic Injection
(A) Single-stranded AAV-GFP expression cassette used to compare the transduction potential of capsids. ITR, inverted terminal repeat; CBA, hybrid CMV enhancer/chicken β-actin promoter; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; pA, poly(A) signals (both SV40 and bovine growth hormone derived). (B) Representative low-magnification images of whole-brain sagittal sections from C57BL/6 mice (males) transduced with 1 × 1011 vg (low dose) of AAV9, AAV9-PHP.B, AAV-S, or AAV-F. (C) Representative images of sagittal section of brains after injection of 8 × 1011 vg (high dose) of each vector in C57BL/6 males. (D) Example sections of spinal cords transduced by each of the four vectors administered intravenously at the higher dose (8 × 1011 vg/mouse). (E) Quantitation of native GFP expression from each vector by the percentage of sections covered by fluorescence at low (left panel) and high (right panel) doses. (F) Multiregional comparison of transduction in the brain at the higher dose. ***p < 0.001; ****p < 0.0001 after one-way ANOVA with Tukey’s multiple comparison test (n = 3 mice/group).
Table 1.
Production Efficiency of AAV Capsids
| AAV Capsid | Titer (vg/mL) | Production Efficiency (vg/Cell) |
|---|---|---|
| AAV9 | 3.20 × 1013 (±2.55 × 1013) | 3.06 × 104 (±1.39 × 104) |
| AAV-F | 5.51 × 1012 (±4.23 × 1012) | 9.57 × 103 (±8.00 × 103) |
| AAV-S | 1.88 × 1013 (1.38 × 1013) | 2.09 × 104 (±5.65 × 103) |
All capsids packaged a single-stranded AAV2 ITR-flanked AAV-CBA-GFP-WPRE transgene cassette.
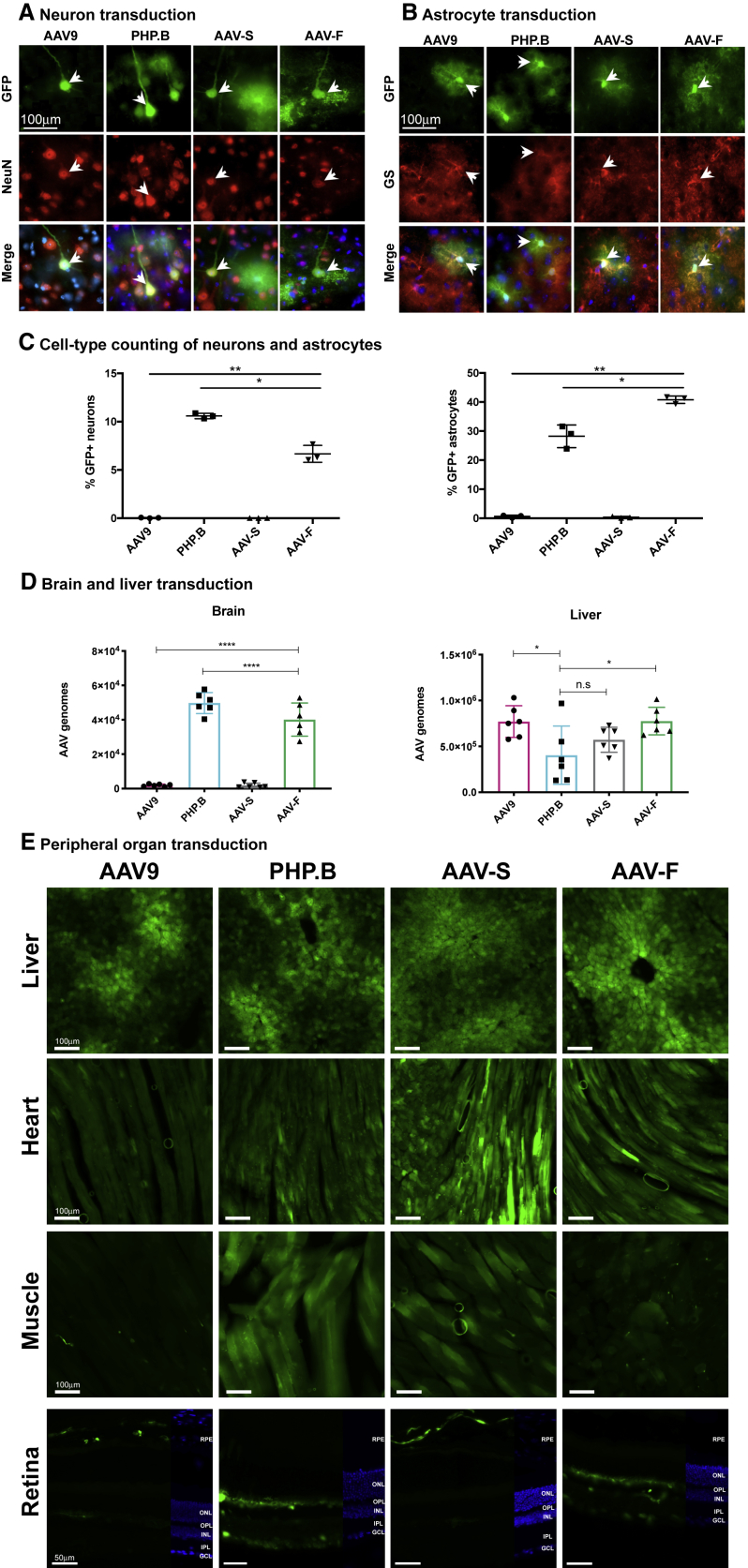
In order to get a detailed appreciation of the cell types being targeted by AAV-S and AAV-F, we next performed a series of co-immunostaining with GFP and markers of neurons (NeuN), astrocytes (glutamine synthetase [GS]), microglia (Iba-1), and oligodendrocytes (Olig2). AAV-F and AAV-S, similar to the other two reference vectors, mainly transduced neurons and astrocytes (none of the variants appeared to effectively transduce microglial or oligodendroglial cells; Figures 4A and 4B; data not shown). Stereological quantitation of neurons and astrocytes in the cortex at the 1 × 1011 vg dose confirmed the efficient transduction potential of AAV-F as compared with conventional AAV9 by a factor of 65 in astrocytes and 171 in neurons, while the difference between AAVS and AAV9 was not significant (the percent of GFP-positive astrocytes was 0.63% ± 0.24% for AAV9 and 0.36% ± 0.15% for AAV-S, and the percent of GFP-positive neurons was 0.039% ± 0.0.02% for AAV9 and 0.029% ± 0.002% for AAV-S; all ± numbers represent SEM). Of note, AAV-F targeted significantly more astrocytes (40.78% ± 0.73%) than AAV9-PHP.B (28.21% ± 0.25%), and the reverse was true for neurons (6.67% ± 0.5% for AAV-F and 10.59% ± 0.16% for AAV9-PHP.B; Figure 4C), suggestive of a slightly different tropism between those two vectors in mice. In addition, AAV-F transduced a variety of neuronal sub-types, including excitatory (CamKII-positive) and inhibitory (GAD67-positive) cortical neurons, dopaminergic neurons in the striatum (expressing tyrosine hydroxylase [TH]), Purkinje neurons in the cerebellum (calbindin positive), and motor neurons in the spinal cord (expressing the choline acetyltransferase [ChAT] marker; Figure S4A). Consistent with the stereological counts in the cortex (Figure 4C) and with the images of the high dose of AAV-F versus AAV9 (Figure 3F), we observed efficient transduction of neurons and astrocytes with AAV-F and not AAV9 at the dose of 1 × 1011 vg/mouse in the striatum, hippocampus, and cerebellum (Figure S4B).
Figure 4.
AAV Vector Comparison of Neuron and Astrocyte Transduction and Biodistribution
High-magnification images of AAV9-, AAV-PHP.B-, AAV-S-, and AAV-F-transduced cells (GFP positive) after co-immunostaining with markers for (A) neurons (NeuN) and (B) astrocytes (glutamine synthetase [GS]). Merged cells also include nuclear staining by DAPI. (C) Stereological evaluation of the percentage of transduced cortical astrocytes and neurons after intravenous (i.v.) delivery of 1 × 1011 vg of each vector. p < 0.0001 by one-way ANOVA. One (*) and two (**) asterisks represent the significant differences between each vector group after Tukey’s multiple comparisons test (n = 3 mice/group). (D) Biodistribution of vectors in the brain and liver as measured by qPCR of vector genomes, normalized by GAPDH genomic DNA levels (input DNA). (E) Transduction of AAV9, AAV9-PHP.B, AAV-S, and AAV-F in peripheral organs following intravenous administration at 8 × 1011 vg in C57BL/6 males. Retinal images: RPE, retinal pigment epithelium; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. ****p < 0.0001 (n = 3 mice/group, one-way ANOVA with Tukey’s multiple comparisons test).
To better understand whether the high levels of GFP transgene expression in the brain with AAV-F corresponded to higher levels of AAV genomes in the brain, we isolated vector and murine genomic DNA from liver and brain and performed qPCR on the 8 × 1011 vg-dosed mice. AAV-F displayed a 20-fold enhancement (p < 0.0001) in AAV genomes in the brain compared to AAV9 (Figure 4D). As reported, AAV9-PHP.B had a much higher (25-fold) amount of AAV genomes in the brain compared to AAV9, while AAV-S had a low level, similar to the GFP fluorescence data (Figure 3). PHP.B showed expression levels in the liver that were slightly lower than AAV9 and AAV-F (although not AAV-S; Figure 4D). AAV-F showed levels in the liver similar to AAV9, and AAV-S showed a lower, but nonsignificant trend downward. We also examined the biodistribution of AAV-F compared to AAV9 in several other organs, including skeletal muscle, heart, and spinal cord (Figure S5). We observed no significant difference between the two vectors in muscle and heart (although an upward trend was seen with respect to AAV-F in the heart). As would be expected from expression in the brain, AAV-F showed significantly higher expression levels in the spinal cord compared to AAV9 (p < 0.05, t test).
To investigate transgene expression in peripheral organs after systemic injection, we analyzed GFP fluorescence in the liver, heart, skeletal muscle, and retina (Figure 4E). Not surprisingly, all vectors transduced liver efficiently. In the heart and skeletal muscle, AAV-S yielded higher numbers of bright GFP signal/section than did AAV9 and AAV-F. All capsids mediated low transduction of the retina, as expected with intravenous delivery. Interestingly, while AAV-9 and AAV-S did not transduce the neuronal retina, consistent expression was observed in the retinal pigment epithelium (RPE). Both AAV9-PHP.B and AAV-F transduced multiple layers in the retina, most notably cells in the ganglion cell layer (GCL). GFP expression was also observed in the inner nuclear layer (INL), with substantial expression shown in the outer plexiform layer (OPL), which may reflect bipolar or inhibitory horizontal cell transduction.
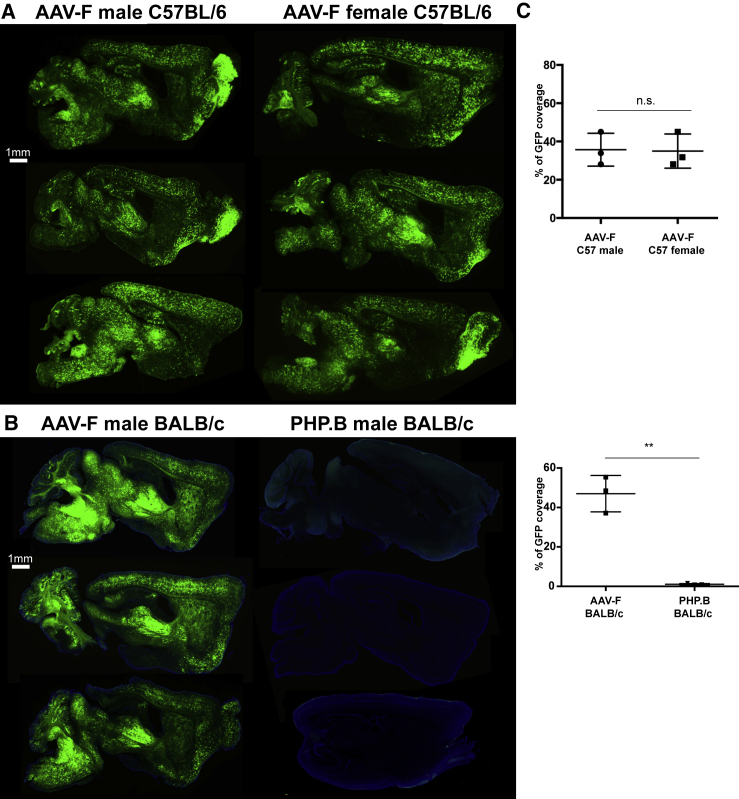
Since transduction by AAV vectors in mice can vary substantially between sex and mouse strain, we examined whether the most efficient capsid, AAV-F, could efficiently transduce brain after systemic injection of female C57BL/6 as well as male BALB/ mice. Mice were injected with 1 × 1011 vg (4 × 1012 vg/kg), and we observed robust and efficient transduction independent of strain or sex (Figure 5). These results contrasted with the lack of efficacy of AAV9-PHP.B at transducing the CNS after systemic delivery in the BALB/c strain, as previously reported (Figures 5B and 5C).13,14
Figure 5.
AAV-F Mediates High Transduction in Male and Female C57BL/6 Mice and Also in BALB/c Mice
(A) Representative images of GFP signal across sagittal brain sections in male and female mice (n = 3) transduced by AAV-F at 1 × 1011 vg/mouse. (B) Sagittal brain sections of male BALB/c mice injected with AAV-F (left) or AAV9-PHP.B (right) at 1 × 1011 vg/mouse. DAPI (blue) is provided as a counterstain alongside GFP (green) to visualize PHP.B-treated brain sections. (C) Quantitation of native GFP expression from each vector by the percentage of sections covered by fluorescence. **p < 0.01. Unpaired t test (n = 3 mice/group).
While iodixanol density gradient purification removes most empty capsids, to examine whether an excess of empty capsids in AAV-F could explain its increased biodistribution to the brain compared to AAV9, we performed transmission electron microscopy (TEM) on two separate preparations of AAV-F and compared them to two independent preparations of AAV9. As seen in Figure S6, we observed no significant difference in empty capsid levels between AAV-F and AAV9 (mean 4.63% ± 1.99% versus 5.2% ± 2.18% empty capsids for AAV9 and AAV-F respectively; mean ± SD; p = 0.54, unpaired t test).
Next, we tested whether AAV-F would have utility as a vector for CNS transduction via other routes of administration. We first tested AAV-S and AAV-F to mediate transgene expression in the brain after direct hippocampal injection of adult C57BL/6 mice. We found that both capsids achieve a widespread expression of GFP after direct injection, primarily in neurons (Figure S7). Intrathecal injection of AAV vectors to transduce the spinal cord has shown promise to treat this compartment. One drawback is limited spread of the vector to the brain after lumbar injection of vector. We compared AAV9 and AAV-F after bolus intrathecal injection of vector into the lumbar region of the spinal cord in adult C57BL/6 mice. Three weeks postinjection, mice were killed and spinal cords and brains analyzed. Remarkably, AAV-F resulted in much more intense GFP expression throughout the spinal cord compared to AAV9, transducing both white and gray matter. Meanwhile AAV9 transduction was mainly restricted to the white matter. Strikingly, we also detected transduction of astrocytes and neurons in the brain of mice injected with AAV-F, but not AAV9 (Figure S8).
AAV-F Mediates Enhanced Transduction of Human Neurons
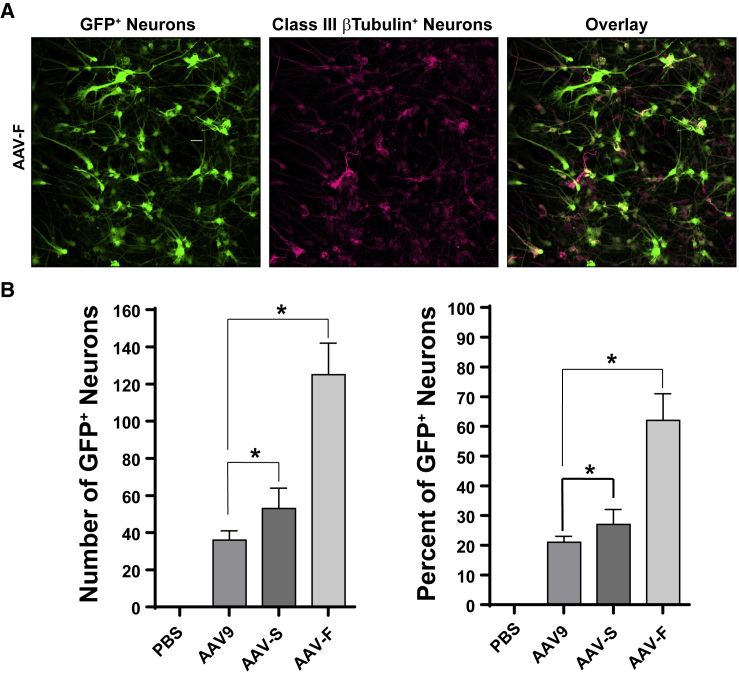
Because the selection was performed in mice, we analyzed whether the robust transduction characteristics of AAV-F also translated to human cells. Primary human stem-cell-derived neurons were transduced with equal doses of AAV9, AAV-S, and AAV-F, all encoding GFP. One week later they were fixed, stained with class III β-tubulin, and analyzed for the percentage of GFP-positive neurons. Remarkably, AAV-F transduced 62% of neurons, 3-fold higher than AAV9 (p < 0.05) (Figure 6). AAV-S yielded a slight, yet statistically significant (p < 0.05) increase in transduction efficiency over AAV9 (Figure 6A).
Figure 6.
AAV-F Mediates Higher Transduction Efficiency than AAV9 in Human Cortical Neurons
(A) GFP expression in fetal-derived primary human neurons, transduced by AAV-F. Neurons were co-labeled with an antibody to β-tubulin to quantify transduction. (B) Quantitation of transduction efficiency of human neurons by AAV9, AAV-S, and AAV-F. *p < 0.05.
Discussion
Using the iTransduce system, we have isolated a new AAV capsid, AAV-F, which mediates highly efficient transgene expression in the murine CNS (two strains tested). This capsid also mediated robust transduction of primary human neurons.
Interestingly, using expression-based selection, we identified three peptide clones (STTLYSP, FVVGQSY, and FQPCP∗) that represented 97% of NGS reads. Since FQPCP∗ had a stop codon, we reasoned that this genome was cross-packaged in another capsid during production, a phenomenon noted to occur with AAV libraries.15 This may also be the case for STTLYSP (AAV-S) (Figure 2). AAV-S was not a defective vector, however, as it mediated robust transduction of peripheral organs (Figure 4E). Because it had such a high production efficiency (Table 1), it may have had a propensity to be cross-packaged. Alternatively, AAV-F (FVVGQSY) was extremely efficient at transduction and was one of only two prospective candidates from the NGS (Figure 2). Both of these candidates were detectable at very low levels in the round 2 library pool (Figure 2); as such, we could confirm that their enrichment was not due to a preexisting bias. We do acknowledge that flow-cytometry-based selection of tdTomato-positive cells and subsequent PCR amplification from this cell population may have skewed the peptide population. The apparent “false positives” of AAV-S and FQPCP∗ may have resulted from our use of relatively high AAV plasmids/cell (∼100) during library production in 293T cells, and in the future lower amounts may decrease this issue. However, we consider identifying one highly efficient capsid (AAV-F) out of three candidates to be a successful selection.
Since we performed a demonstration of the iTransduce system using an agnostic approach to cell type (whole brain), it is not surprising that AAV-F was highly tropic to astrocytes and neurons, cells that are transduced by AAV9. In future studies we will combine cell-specific promoters to drive Cre expression from the AAV library vector to isolate capsids that can transduce cells that are refractory to conventional AAV vector transduction.
In addition to the potential of the iTransduce system to select clinical candidate AAV vectors, it may be used to identify vectors for use as research tools. The recently identified AAV-PHP.B capsid has served as an efficient vector to genetically modify the murine brain.12 However, it does not transduce BALB/c- or BALB/c-related mouse lineages.13,14,16 Interestingly, we observed robust transduction of BALB/c and C57BL/6 murine brain after intravenous injection of AAV-F. This indicates that the mechanism of enhanced transduction over AAV9 differs between AAV-PHP.B and AAV-F. It also enables AAV-F to be utilized as an efficient tool for CNS research in the popular BALB/c strain (https://www.labome.com/method/Laboratory-Mice-and-Rats.html). Additionally, we show that AAV-F can also mediate robust transgene expression in the CNS after both direct and intrathecal bolus injection.
While the results with AAV-F are very promising in mice, the true translational potential of this vector remains to be observed. Future studies in larger animals need to be carried out to test whether AAV-F has potential clinically. Dose escalation studies should be performed to test for dose-related toxicity of AAV-F, as was observed with PHP.B in NHPs.14 Other strategies may be employed in the near future to increase the chances of translational probability. First, doing iterative rounds of selection in different species (e.g., mice, then rats) may allow better cross-species translation of the transduction efficiency. This could be done in mice followed by floxed STOP tdTomato transgenic rats.17 Alternatively, in the future, direct selection of the iTransduce library could be performed in transgenic marmosets.18,19
Materials and Methods
AAV Library Construction
iTransduce Plasmid: pAAV-CBA-Cre-mut-p41-Cap9del
We constructed the iTransduce library backbone plasmid called pAAV-CBA-Cremut-p41-Cap9del, containing two expression cassettes in cis: (1) the CBA-Cremut, in which we introduced a mutant Cre cDNA (CCG→CCT encoding the Pro15 amino acid to eliminate the AgeI site initially present) under the ubiquitous promoter CBA; and (2) the p41-Cap9del composed of the AAV9 capsid gene under the AAV5 p41 promoter (residues 1680–1974 of GenBank: AF085716.1) and splicing sequences of the AAV2 rep gene (similar as described in Deverman et al.12).
Both the mutant Cre cDNA (Cremut) flanked by KpnI and SalI restriction sites and the p41-CAP9(del)-poly(A) fragment were synthesized by GenScript and cloned into a puC57 backbone. We constructed first the pAAV-CBA-Cremut-poly(A) plasmid by subcloning the mutant Cre cDNA (fragment KpnI-blunt, SalI) in place of EGFP-woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) in our pAAV-CBA-WPRE backbone (opened with NcoI-blunt/SalI, which eliminated the EGFP-WPRE fragment initially present). We then introduced the p41-Cap9del fragment harboring a K449R mutation in the Cap9 sequence, allowing the creation of a unique XbaI site, and in which the 447-bp Cap sequence between XbaI and AgeI restriction sites had been deleted.
Plasmid for Generating Random 7-mer Peptide Cap Fragments and Subcloning the CAP9 Fragments Retrieved by PCR: pUC57-Cap9-XbaI/KpnI/AgeI
We first had the 447-bp region of the AAV9 capsid sequence between the XbaI/AgeI sites (sequence that is missing in pAAV-CBA-Cremut-p41-Cap9del) synthesized by GenScript (Piscataway, NJ, USA) and subcloned into a pUC57-Kan plasmid. This capsid sequence contains the same removal of the EarI site in this 447-bp region wild-type (WT) AAV9 cap, which allows restriction digest removal of WT AAV9 as described by Deverman et al.12 This also creates a unique KpnI site. The cap fragment of pUC57-Cap9-XbaI/KpnI/AgeI was used to generate the initial library of random 21-mer nucleotide sequences (encoding for 7-mer peptides) inserted between nucleotides encoding amino acids 588 and 589 of AAV9 VP3. We used a strategy similar to that of Deverman et al.12 Briefly, pUC57-Cap9-XbaI/KpnI/AgeI served as template to amplify the cap DNA and insert random 21-mer sequences using a forward and reverse primer. The primer information was as follows: XF-extend (5′-GTACTATCTCTCTAGAACTATTAACGGTTC-3′) and reverse primer 588iRev (5′-GTATTCCTTGGTTTTGAACCCAACCGGTCTGCGCCTGTGCXMNNMNNMNNMNNMNNMNNMNNTTGGGCACTCTGGTGGTTTGTG-3′), in which the MNN repeat refers to the randomized 21-mer nucleotides (purchased from IDT). XF-extend and 588iRev were used in a PCR with Phusion polymerase (NEB) and pUC57-Cap9-XbaI/KpnI/AgeI as template. The 447-bp PCR product was digested with XbaI and AgeI overnight at 37°C and then gel purified (QIAGEN). Similarly, pAAV-CBA-Cre-mut-p41-Cap9del was digested with XbaI and AgeI and gel purified. Next, a ligation reaction (1 h at room temperature) with T4 DNA ligase (NEB) was performed using a 3:1 cap insert-to-vector molar ratio. The subsequent ligated plasmid was called pAAV-CBA-Cre-mut-p41-Cap9-7-mer and contained a pool of plasmids with random 7-mer peptides inserted in the cap gene between nucleotides encoding 588 and 589 of AAV9 VP3.
This plasmid (pUC57-Cap9-XbaI/KpnI/AgeI) was also used as our recipient plasmid for subcloning the CAP9 fragments amplified by PCR from brain tissue. We removed an upstream KpnI site in the pUC57 plasmid by digestion with SacI and NsiI and ligation. This allowed the KpnI site in the capsid fragment to be unique.
Rep Expression Plasmid
We constructed a rep expression plasmid called pAR9-Cap9-stop/AAP/Rep using a similar strategy to that as Deverman et al.12 The entire cDNA was synthesized by GenScript and cloned into a pUC57-Kan plasmid. Stop codons were inserted in place of start codons for VP1, VP2, and VP3 so that no parental AAV9 capsids were produced, while maintaining AAP and rep expression.
AAV Library Production and Purification
For each production, we plated 15-cm tissue culture dishes (usually 10-15 dishes total) with 1.5 × 107 293T cells/dish. The next day cells were transfected using the calcium phosphate method, with the adenovirus helper plasmid (pAdΔF6, 26 μg per plate), rep plasmid (pAR9-Cap9-stop/AAP/Rep, 12 μg per plate), and ITR-flanked AAV library (pAAV-CBA-Cre-mut/p41-Cap9-7-mer, 1 μg per plate) to induce production of AAV. The day after transfection, medium was changed to DMEM containing 2% fetal bovine serum (FBS). AAV was purified from the cell lysate using iodixanol density-gradient ultracentrifugation. Buffer exchange to PBS was done using Zeba spin columns (7 K molecular weight cutoff [MWCO]; Thermo Fisher Scientific), and further concentration was performed using Amicon Ultra 100 kDa MWCO ultrafiltration centrifugal devices (Millipore). Vectors were stored at −80°C until use. We quantified AAV genomic copies (vg) in AAV preparations using TaqMan qPCR with ITR-sequence-specific primers and probes.20,21
NGS of Library
NGS was performed on the plasmid AAV9 library pool, as well as following packaging of capsids. Sequencing was also performed following PCR rescue of the cap fragment (either from brain tissue or from isolated tdTomato-positive cells sorted by flow cytometry). For each round of selection, viral DNA corresponding to the insert-containing region was amplified by PCR using Q5 polymerase from New England Biolabs (forward primer, 5′-AATCCTGGACCTGCTATGGC-3′; reverse primer, 5′-TGCCAAACCATACCCGGAAG-3′). Unique barcode adapters were annealed to each sample, and samples were sequenced on an Illumina MiSeq (150 bp reads). Approximately 50–100,000 reads per sample were analyzed. Sequence output files were quality checked initially using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and analyzed on a program custom written in Python. Briefly, sequences were binned based on the presence or absence of insert; insert-containing sequences were then compared to a baseline reference sequence, and error-free reads were tabulated based on incidences of each detected unique insert. Inserts were translated and normalized.
Animals
All animal experiments were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care following guidelines set forth by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We used adult age (8–10 weeks old) Ai9 (strain no. 007909), C57BL/6 (strain no. 000664), and BALB/c (strain no. 000651) mice, all from The Jackson Laboratory (Bar Harbor, ME, USA). All animals were euthanized 3 weeks postinjection, perfused transcardially, and tissues were harvested and either fixed in 4% paraformaldehyde in PBS, snap frozen in liquid nitrogen, or dissociated for flow cytometry.
In Vivo Selection of Brain-Tropic Capsids
Ai9 mice were injected intravenously (tail vein) with the dose in vg indicated in the Results section, and 3 weeks postinjection, mice were euthanized and tissue was harvested.
Mice were deeply anesthetized by isoflurane and decapitated. For round 1, the brain was rapidly dissected and two coronal sections (2 mm thick) were harvested. One section was used for extracting whole-brain DNA (DNeasy blood and tissue kits, QIAGEN, Hilden, Germany). The other coronal section was fixed in 4% PFA and paraffin embedded for immunohistology (tdTomato-positive cells were detected after each round of selection by diaminobenzidine [DAB] staining using a rabbit anti-red fluorescent protein [RFP] antibody from Rockland Immunochemicals). For round 2, brain tissue was then cut with a razorblade into 1-mm3 pieces and neural cells were isolated by papain dissociation (papain dissociation system, Worthington Biochemical), according to the manufacturer’s instructions. Following dissociation, myelin was removed (Myelin Removal Beads II, human, mouse, rat from Miltenyi) and the tdTomato-positive cells were sorted by a S3e cell sorter (Bio-Rad). Cells were sorted by first setting gates to exclude cellular debris and select for singlets only. Cell suspensions from an AAV9-PHP.B-Cre-injected Ai9 (positive control) and a PBS-injected Ai9 mouse (negative control) were used to set gates to sort tdTomato-positive and tdTomato-negative cells. After sorting, the tdTomato-positive cells were immediately pelleted by centrifugation, and DNA was extracted using the Arcturus PicoPure DNA extraction kit (Thermo Fisher).
After DNA extraction, the Cap9 inserts (containing the 21-mer sequence encoding the 7-mer peptides) were amplified using the following primers: Cap9_Kpn/Age_For: 5′-AGCTACCGACAACAACGTGT-3′ and Cap9_ Kpn/Age_Rev: 5′- AGAAGGGTGAAAGTTGCCGT-3′ (Phusion high-fidelity PCR kit, New England Biolabs). The amplicons were then purified (Monarch PCR and DNA cleanup kit, New England Biolabs), digested by KpnI, AgeI, and BanII, and the Cap9 KpnI-AgeI fragments (144 bp) were agarose gel purified (Monarch DNA gel extraction kit, New England Biolabs) before ligation in the pUC57-Cap9-XbaI/AgeI/KpnI plasmid (opened with KpnI and AgeI and dephosphorylated with calf inositol phosphatase, New England Biolabs). The ligation products were transformed into electrocompetent DH5α bacteria (New England Biolabs), and the entire transformation was grown overnight in Lysogeny broth (LB)-ampicillin medium. pUC57-Cap9-XbaI/AgeI/KpnI plasmid was purified by Maxi Prep (QIAGEN). Plasmid was digested by XbaI/AgeI to release the 447-bp cap fragment, which was gel purified and ligated with similarly cut pAAV-CBA-Cre-mut/p41-Cap9del for the next round of AAV library production.
AAV rep/cap Plasmids Containing AAV-F and AAV-S Peptide Inserts for Vector Production
To create rep/cap plasmids encoding AAV9 capsids displaying the peptide insert of interest for production of vectors encoding a transgene of interest (e.g., GFP), we digested an AAV9 rep/cap plasmid with BsiWI and BaeI, which removes a fragment flanking the VP3 amino acid 588 site for peptide sequence insertion. Next, we ordered a 997-bp dsDNA fragment from Integrated DNA Technologies (IDT, Coralville, IA, USA), which contains overlapping Gibson homology arms with the BsiWI/BaeI cut AAV9 as well as the 21-mer nucleotide sequence encoding the peptide of interest in frame after amino acid 588 of VP3. Lastly, we performed Gibson assembly using the Gibson Assembly master mix (NEB, Ipswich, MA, USA) to ligate the peptide-containing insert into the AAV9 rep/cap plasmid. Sequence integrity of the plasmid was determined by whole-plasmid sequencing.
AAV Vectors for Transduction Analysis
For each production, we plated 15-cm tissue culture dishes with 1.5 × 107 293T cells/dish. The next day cells were transfected using the calcium phosphate method, with the adenovirus helper plasmid (pAdΔF6, 26 μg per plate), rep/cap plasmid (AAV9, AAV-F, AAV-S, 12 μg per plate), and ITR-flanked transgene cassette plasmid (single-stranded AAV-CBA-GFP-WPRE,22 10 μg per plate) to induce production of AAV. The day after transfection, medium was changed to DMEM containing 2% FBS. AAV was purified from the cell lysate using iodixanol density-gradient ultracentrifugation. Buffer exchange to PBS was done using Zeba spin columns (7 kDa MWCO; Thermo Fisher Scientific), and further concentration was performed using Amicon Ultra 100 kDa MWCO ultrafiltration centrifugal devices (Millipore). Vectors were stored at −80°C until use. We quantified AAV genomic copies in AAV preparations using TaqMan qPCR with bovine growth hormone (BGH) poly(A)-sequence specific primers and probe.23
Animal Euthanasia and Tissue Harvesting
Mice (strain indicated in each figure) were slowly injected via the lateral tail vein with 200 μL of the tested AAV vector diluted in sterile PBS (low dose, 4 × 1012 vg/kg; high dose, 3.2 × 1013 vg/kg) before gently finger clamping the injection site until bleeding stopped. Three weeks after injection, mice were euthanized and perfused transcardially with sterile cold PBS. Next, the brain was longitudinally bisected into two hemispheres. One hemisphere was postfixed in 15% glycerol/4% paraformaldehyde diluted in PBS for 48 h, followed by 30% glycerol for cryopreservation for another 48–72 h. The other half was fresh frozen for isolation of genomic and vector DNA. For the high-dose cohort, a small piece of heart, muscle (gastrocnemius), and the retina were also processed for immunohistology. We have made three independent preparations of AAV-S, AAV-F, and AAV9 (Table 1). The transduction results in mice were from one preparation of each vector; however, we have replicated these results in two more independent experiments.
Immunohistology and High-Magnification Imaging of AAV-CBA-GFP-Transduced Neural and Neuronal Cell Populations
Coronal floating sections (40 μm) were cut using a cryostat microtome. After rinsing off the glycerol in Tris-buffered saline (TBS) buffer, cryosections were permeabilized with 0.5% Triton X-100 (AmericanBio) in TBS for 30 min at room temperature and blocked with 5% normal goat serum (or normal donkey serum) and 0.05% Triton X-100 in TBS for 1 h at room temperature. Primary antibodies were incubated overnight at 4°C in 2.5% normal goat serum and 0.05% Triton X-100 in TBS, while Alexa Fluor 488- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Baltimore, MD, USA) were incubated for 1 h the next day. Primary antibodies used for this study were: chicken anti-GFP (Aves Labs, Tigard, OR, USA); mouse anti-NeuN (Millipore, Burlington, VT, USA); rabbit anti-GS (Abcam, Cambridge, MA, USA); rabbit anti-Olig2 (Millipore, Burlington, VT, USA); rabbit anti-Iba1 (Wako, Japan); rabbit anti-CamKII (Abcam, Cambridge, MA, USA); mouse anti-GAD67 (Millipore, Burlington, VT, USA); rabbit anti-ChAT (Millipore, Burlington, VT, USA); mouse anti-calbindin (Abcam, Cambridge, MA, USA); and rabbit anti-TH (Novus Biologicals, Littleton, CO, USA). Sections were mounted with Vectashield mounting medium with DAPI (Vector Laboratories, Burlingame, VT, USA).
To identify the neural cell types transduced by each vector and investigate the various neuronal subtypes targeted by AAV-F, a Zeiss Axio Imager Z epifluorescence microscope equipped with AxioVision software and a ×60 objective was used to take high-resolution images showing colocalization between GFP and each cell marker.
Imaging and Quantification of Global GFP Signal Coverage
To quantify the overall native GFP fluorescence signal in brain and liver sections, a robotic slide scanner virtual slide microscope VS120 (Olympus) was used to image the entire batch of slides on one go using an Olympus UPLSAPO ×10 objective. In order to reduce variability, the entire batch of slides was imaged in one session. The initial exposure time for GFP was set up so that the fluorescent signal was neither undersaturated nor oversaturated across all experimental groups and remained unchanged throughout the entire batch scan. The order of the slides was randomized and remained blinded until final statistical analysis. The Olympus cellSens Standard software was then used to analyze the percentage of GFP coverage in each brain section. A region of interest (ROI) was initially defined using the “ROI-polygon” tool, and we quantified the GFP-positive area within this initial ROI, after applying a similar detection threshold on the GFP channel for all of the slides analyzed (the threshold was set at a similar level for the analysis of all mouse brain sections, but a different threshold was applied for the analysis of all mouse liver sections and all rat brain sections). The percentage of GFP-positive area accordingly to the total surface of the ROI was then calculated. The autofluorescence signal was taken into account in our analysis, as we set the threshold for EGFP fluorescence intensity above the autofluorescence level (making sure that only the signal from AAV-GFP-transduced cells was taken into account). In addition, we drew each ROI for each brain section avoiding the very edges of the section, as those could also present with a high level of autofluorescence. The ventricular space was also excluded from our analysis. Finally, three technical replicates (brain sections) were measured per mouse and all measurements were done blinded until the final step of the analysis.
Stereology-Based Quantitative Analyses of the Percentages of Transduced Astrocytes and Neurons
Stereology-based studies were performed as previously described,24,25 after co-staining the brain sections for GFP and NeuN (neuronal marker) or GFP and GS (pan-astrocytic marker). We did not include microglia and oligodendrocytes in this analysis, as those cell types were not transduced to an appreciable amount. Stereological evaluation of the percentages of AAV-transduced neurons and astrocytes was done blindly after de-identification of the vector initially injected, using a motorized stage of an Olympus BX51 epifluorescence microscope equipped with a DP70 digital CCD camera, an X-Cite fluorescent lamp, and the associated CAST stereology software version 2.3.1.5 (Olympus, Tokyo, Japan). The cortex was initially outlined under the ×4 objective. Random sampling of the selected area was defined using the optical dissector probe of the CAST software. To evaluate the percentage of AAV9-, AAV9-PHP.B-, AAV-S-, and AAV-F-transduced astrocytes or neurons, the stereology-based counts were performed under the ×20 objective, with a meander sampling of 10% for the surface of cortex for the “high transduction” AAVs, and 20% for “low transduction” AAVs (considering the infrequency of GFP-positive cells in those cases). For each counting frame, the total numbers of astrocytes (GS-positive cells) or neurons (NeuN-positive cells) were evaluated, and, among each of those populations, the percentages of GFP-positive cells were determined. Only glial and neuronal cells with a DAPI-positive nucleus within the counting frame were considered.
Vector Genome Quantification in the Brain and Liver
One brain hemisphere and a small piece of liver were fresh frozen for AAV genome isolation for vector genome biodistribution. For the fresh-frozen brain and liver samples, we isolated genomic and AAV vector DNA from 10 mg of tissue using the DNeasy blood and tissue kit (QIAGEN) according to manufacturer’s instructions. DNA was quantitated using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). Next, using 50 ng of genomic DNA as template, we performed a TaqMan qPCR using probe and primers to the poly(A) region of the transgene expression cassette (same assay used to titer the purified AAV vectors). To ensure equal genomic DNA input for each sample, we performed a separate qPCR on each sample using a TaqMan probe and primer set that detects GAPDH genomic DNA (Thermo Fisher Scientific, assay ID Mm01180221_g1, gene symbol Gm12070). For each organ/tissue, we adjusted the AAV vector genome copies for each sample by taking into account any differences in GAPDH threshold cycle (Ct) values using the following formula: (AAV vector genome copies)/(2ΔCt). The ΔCt value was calculated by the following formula: GAPDH Ct value of sample of interest − average GAPDH Ct value of sample that had the lowest amount of GAPDH (highest Ct value). Data were expressed as AAV vector genomes per 50 ng of genomic DNA.
Human Neuron Transduction
Primary human fetal neural stem cells (NSCs) were obtained from the Birth Defects Research Laboratory (University of Washington, Seattle, WA, USA) in full compliance with the NIH ethical guidelines. The isolation procedure has been detailed previously26 with slight modifications. Briefly, brain tissue was incubated in 0.25% trypsin, DNase (90 U/mL), diluted in Hank’s balanced salt solution (HBSS) for 45 min. Tissue was titrated, transferred into 4°C heat-inactivated FBS, and centrifuged at 500 × g for 20 min. The pellet was resuspended in NSC complete media consisting of X-VIVO 15 (without phenol red and gentamicin; Lonza) supplemented with 10 μg of basic fibroblast growth factor (Life Technologies), 100 μg of epidermal growth factor (Life Technologies), 5 μg of leukemia inhibitory factor (Millipore), 60 ng/mL N-acetylcysteine (Sigma-Aldrich), 4 mL of neural survival factor-1 supplement (Lonza), 5 mL of 100× N-2 supplement (Life Technologies), 100 U of penicillin, 100 μg/mL streptomycin (Life Technologies), and 2.5 μg/mL Fungizone (Life Technologies). Supernatants were then filtered through a 40-μm cell strainer (Corning Life Sciences). Neurospheres larger than 40 μm in diameter were dissociated with Accutase (10 min). Neural differentiation medium consisted of 1× neurobasal medium, 2% B-27 serum-free supplement, and 2 mM GlutaMAX-I supplement (all from Invitrogen) and supplemented with human recombinant brain-derived neurotrophic factor (BDNF) (10 ng/mL; PeproTech).
Differentiating NSCs were grown in chamber slides in differentiation media for 2 weeks and then treated with the indicated AAV vector encoding GFP (7 × 109 vg/well added, 150 vg/cell). One week after transduction, cells were fixed with 4% paraformaldehyde and permeabilized with 0.05% Triton X-100 (Sigma-Aldrich) in 1× PBS (Invitrogen). Cells were stained with a primary monoclonal antibody (TU-20) to neuron-specific class III β-tubulin (1:50; Abcam). Secondary antibodies conjugated to Alexa Fluor 594 (diluted 1:200; Invitrogen) were added for 1 h, followed by DAPI for 30 min. The slides were then mounted with a ProLong antifade reagent (Invitrogen). Maximum projection images were generated from captured z stacks using the Nikon A1R confocal microscope.
The z stacks were loaded in Imaris, and the surface module was used to render the images into 3D volumes. GFP+ neurons were counted (under channel 1, green), as were class III β-tubulin-positive neurons (under channel 2, red). Using the Imaris colocalization module, the population of neurons double positive for the above was determined.
Statistics
Statistical analysis of data was performed using GraphPad Prism software (version 8.00). A one-way ANOVA test followed by a Tukey’s multiple comparisons test was performed, across the different groups AAV9, AAV9-PHP.B, AAV-S, and AAV-F. A value of p < 0.05 was considered to be statistically significant. Results are shown as the mean ± SEM. Similar analyses were performed to analyze the biodistribution of AAV genomes in brain and liver, as well as transduction of human neurons.
Author Contributions
E.H. and C.A.M. conceived of the study and designed the experiments. K.S.H., J.C.M., T.B., R.E.B., T.P.S., R.R., Y.G., C.N., D.M., S.D., S.H.R., M.J.C., J.N., E.H., and C.A.M. performed experiments and analyzed data. M.S.-E. provided technical advice on the design of the library plasmids. The manuscript was written by E.H., C.A.M., and K.S.H. with input from all of the authors.
Conflicts of Interest
K.S.H., E.H., and C.A.M. have filed a provisional patent application for iTransduce and the newly described vectors. C.A.M. has a financial interest in Chameleon Biosciences, Inc., a company developing an enveloped adeno-associated virus (AAV) vector platform technology for repeated dosing of systemic gene therapy. C.A.M.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Acknowledgments
We thank Jeyashree Natasan and Adrian Volak for assistance with vector production. We thank the MGH Quantitative Real-Time PCR Core Facility for their services. We thank Maria Ericsson of the Harvard Medical School Electron Microscopy Facility for generating the images of purified preparations of AAV-F and AAV9 capsids. This work was supported in part by NIH/NIDCD grant R01DC017117-01A1 (to C.A.M.) and NIH/NIA grant R00AG047336-05 (to E.H.). The Laboratory of Developmental Biology was supported by NIH award number 5R24HD00083 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.10.007.
Contributor Information
Eloise Hudry, Email: ehudry@mgh.harvard.edu.
Casey A. Maguire, Email: cmaguire@mgh.harvard.edu.
Supplemental Information
References
- 1.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 2.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 3.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulk N.K., Pekrun K., Charville G.W., Maguire-Nguyen K., Wosczyna M.N., Xu J., Zhang Y., Lisowski L., Yoo B., Vilches-Moure J.G. Bioengineered viral platform for intramuscular passive vaccine delivery to human skeletal muscle. Mol. Ther. Methods Clin. Dev. 2018;10:144–155. doi: 10.1016/j.omtm.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray S.J., Blake B.L., Criswell H.E., Nicolson S.C., Samulski R.J., McCown T.J., Li W. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol. Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tse L.V., Klinc K.A., Madigan V.J., Castellanos Rivera R.M., Wells L.F., Havlik L.P., Smith J.K., Agbandje-McKenna M., Asokan A. Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. USA. 2017;114:E4812–E4821. doi: 10.1073/pnas.1704766114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koerber J.T., Maheshri N., Kaspar B.K., Schaffer D.V. Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles. Nat. Protoc. 2006;1:701–706. doi: 10.1038/nprot.2006.93. [DOI] [PubMed] [Google Scholar]
- 9.Körbelin J., Sieber T., Michelfelder S., Lunding L., Spies E., Hunger A., Alawi M., Rapti K., Indenbirken D., Müller O.J. Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. mol. ther. 2016;24:1050–1061. doi: 10.1038/mt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sallach J., Di Pasquale G., Larcher F., Niehoff N., Rübsam M., Huber A., Chiorini J., Almarza D., Eming S.A., Ulus H. Tropism-modified AAV vectors overcome barriers to successful cutaneous therapy. Mol. Ther. 2014;22:929–939. doi: 10.1038/mt.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry G.E., Asokan A. Cellular transduction mechanisms of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:54–60. doi: 10.1016/j.coviro.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzaki Y., Tanaka M., Hakoda S., Masuda T., Miyata R., Konno A., Hirai H. Neurotropic properties of AAV-PHP.B are shared among diverse inbred strains of mice. Mol. Ther. 2019;27:700–704. doi: 10.1016/j.ymthe.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The eunrotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 2018;26:664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nonnenmacher M., van Bakel H., Hajjar R.J., Weber T. High capsid-genome correlation facilitates creation of AAV libraries for directed evolution. Mol. Ther. 2015;23:675–682. doi: 10.1038/mt.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Igarashi H., Koizumi K., Kaneko R., Ikeda K., Egawa R., Yanagawa Y., Muramatsu S., Onimaru H., Ishizuka T., Yawo H. A novel reporter rat strain that conditionally expresses the bright red fluorescent protein tdTomato. PLoS ONE. 2016;11:e0155687. doi: 10.1371/journal.pone.0155687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J.E., Zhang X.F., Choi S.H., Okahara J., Sasaki E., Silva A.C. Generation of transgenic marmosets expressing genetically encoded calcium indicators. Sci. Rep. 2016;6:34931. doi: 10.1038/srep34931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki E., Suemizu H., Shimada A., Hanazawa K., Oiwa R., Kamioka M., Tomioka I., Sotomaru Y., Hirakawa R., Eto T. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 20.D’Costa S., Blouin V., Broucque F., Penaud-Budloo M., François A., Perez I.C., Le Bec C., Moullier P., Snyder R.O., Ayuso E. Practical utilization of recombinant AAV vector reference standards: focus on vector genomes titration by free ITR qPCR. Mol. Ther. Methods Clin. Dev. 2016;5:16019. doi: 10.1038/mtm.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aurnhammer C., Haase M., Muether N., Hausl M., Rauschhuber C., Huber I., Nitschko H., Busch U., Sing A., Ehrhardt A., Baiker A. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- 22.György B., Meijer E.J., Ivanchenko M.V., Tenneson K., Emond F., Hanlon K.S., Indzhykulian A.A., Volak A., Karavitaki K.D., Tamvakologos P.I. Gene transfer with AAV9-PHP.B rescues hearing in a mouse model of Usher syndrome 3A and transduces hair cells in a non-human primate. Mol. Ther. Methods Clin. Dev. 2018;13:1–13. doi: 10.1016/j.omtm.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire C.A., Balaj L., Sivaraman S., Crommentuijn M.H., Ericsson M., Mincheva-Nilsson L., Baranov V., Gianni D., Tannous B.A., Sena-Esteves M. Microvesicle-associated AAV vector as a novel gene delivery system. Mol. Ther. 2012;20:960–971. doi: 10.1038/mt.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudry E., Martin C., Gandhi S., György B., Scheffer D.I., Mu D., Merkel S.F., Mingozzi F., Fitzpatrick Z., Dimant H. Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 2016;23:380–392. doi: 10.1038/gt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dashkoff J., Lerner E.P., Truong N., Klickstein J.A., Fan Z., Mu D., Maguire C.A., Hyman B.T., Hudry E. Tailored transgene expression to specific cell types in the central nervous system after peripheral injection with AAV9. Mol. Ther. Methods Clin. Dev. 2016;3:16081. doi: 10.1038/mtm.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watters A.K., Rom S., Hill J.D., Dematatis M.K., Zhou Y., Merkel S.F., Andrews A.M., Cena J., Potula R., Skuba A. Identification and dynamic regulation of tight junction protein expression in human neural stem cells. Stem Cells Dev. 2015;24:1377–1389. doi: 10.1089/scd.2014.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.