Abstract
Extracellular vesicles, including microvesicles and exosomes, are lipidic membrane-derived vesicles that are secreted by most cell types. Exosomes, one class of these vesicles that are 30–100 nm in diameter, hold a great deal of promise in disease diagnostics, as they display the same protein biomarkers as their originating cell. For exosomes to become useful in disease diagnostics, and as burgeoning drug delivery platforms, they must be isolated efficiently and effectively without compromising their structure. Most current exosome isolation methods have practical problems including being too time-consuming and labor intensive, destructive to the exosomes, or too costly for use in clinical settings. To this end, this study examines the use of poly(ethylene terephthalate) (PET) capillary-channeled polymer (C-CP) fibers in a hydrophobic interaction chromatography (HIC) protocol to isolate exosomes from diverse matrices of practical concern. Initial results demonstrate the ability to isolate extracellular vesicles enriched in exosomes with comparable yields and size distributions on a much faster time scale when compared to traditional isolation methods. As a demonstration of the potential analytical utility of the approach, extracellular vesicle recoveries from cell culture milieu and a mock urine matrix are presented. The potential for scalable separations covering submilliliter spin-down columns to the preparative scale is anticipated.
Keywords: Exosomes, Extracellular vesicles, Fibers, HIC, Purification
1. Introduction
Exosomes are tiny lipid-bound vesicles, approximately 30–100 nm in diameter, that are secreted by most types of cells, including both normal and disease-state cells. They carry internal “cargo” molecules, such as nucleic acids and proteins, which are derived from their cell of origin. These biomarkers make exosomes a promising means of minimally invasive early disease diagnosis [1–4]. Once considered cellular debris, research has demonstrated that exosomes have multiple biological roles. They are found in many single- and multicellular eukaryotic organisms. In humans, exosomes may be involved in a myriad of normal physiological processes including cross-placental communication between the mother and fetus, fetal development, and bone calcification, as well as disease processes including metastasis, pathogenesis of thrombosis, diabetes, atherosclerosis, tumor growth, arthritis, and progression of neurodegenerative diseases [4, 5]. Exosomes can be found in most body fluids, including urine, saliva, amniotic fluid, semen, breast milk, plasma, and blood, making them a promising basis for the development of liquid biopsies [6, 7]. They have been shown to have unique microRNA (miRNA) signatures that could soon open the door for clinical and therapeutic applications [8]. Exosomes are being exploited for disease diagnostics [7, 9, 10], including cancer [11, 12], with potential biomarkers identified relative to a number of different types of cancers, including ovarian, lung, breast, prostate, and pancreatic cancer [11–14].
Despite these promising attributes, the analysis of exosomes is currently limited by the processes required to isolate them from body fluids [13, 15, 16]. In most cases, exosomes are isolated by differential centrifugation (DC), requiring the use of a high-speed centrifuge over several hours, including the sedimentation of other particulate debris and potentially impacting the integrity of the lipid bilayer membrane of the exosome. Other emerging exosome isolation techniques include density gradient centrifugation, size exclusion chromatography, ultrafiltration, polymer-based precipitation, immunological separation, and microfluidics techniques [13, 17–22]. Some of these methods are generic with respect to the specific types of exosomes that can be isolated, while others solely capture vesicles originating from specific cell types. Each of these exosome isolation techniques has benefits, but the aforementioned shortcomings are fairly universal [13, 16, 17]. Thus, opportunities exist for alternative separation methods ranging in use from clinical diagnostics to the isolation of larger lots from cell culture media, as would be required for drug delivery applications.
Exosome isolation methods are generally based on the size/hydrodynamic radii of the vesicles, i.e, centrifugation, filtration, and sieve-based approaches, or their affinity toward capture surfaces used in spin-down formats. However, methods relying on chemical separation and processing platforms, such as those employed in HPLC, have not been fully explored; yet, they may have many attractive features. Most of the common LC stationary phases used for chemical separations, such as porous silica beads, would not be practical, as exosomes would likely be excluded from the internal pore structures, and clogging would be a major operational problem. This study describes the use of capillary-channeled polymer (C-CP) fibers as stationary phases for the isolation and recovery of exosome-enriched populations of extracellular vesicles (referred to as “exosomes” from here forward) from culture media, buffer, and urine. C-CP fibers have been employed by Marcus and coworkers as stationary phases for protein separations via reversed phase, ion exchange, hydrophobic interaction, and affinity chromatographies [23–29]. These fibers are melt-extruded from commodity polymers (nylon 6, polypropylene, and poly(ethylene terephthalate) (polyester,PET)), having a unique cross-sectional profile consisting of eight “legs” on the periphery. When packed in column formats, the fibers inter-digitate to create massive numbers of 1–4 μm-wide channels that provide high permeability to fluid flow. The nonporous nature of the fiber surfaces (at least on the size scale of proteins) means that intraphase diffusion of solutes is prohibited. This combination of macro and micro characteristics results in the ability to affect protein separations at exceedingly high linear velocities (>50 mm/s) without the mass transfer limitations common to porous phases. These hydrodynamic advantages are complemented by a high degree of chemical separation diversity. In addition to the range modalities of separation that can be affected using different base polymers, an extensive tool box of simple surface modification approaches has also been developed. The fiber surfaces may be modified to affect high ligand densities for ion exchange (cationic and anionic) and affinity chromatography [28, 29]. Affinity separations include the use of protein A for IgG purification and quantification, biotin-streptavidin interactions, and chelates for immobilized metal affinity chromatography [30–32]. In total, these attributes make the utilization of the C-CP fibers for exosome isolation a promising alternative to traditional isolation methodologies.
This study describes the first successful use of the PET C-CP fibers to isolate and elute exosomes via a hydrophobic interaction chromatography (HIC) protocol. The procedure, first developed for protein separations [25], is readily implemented to isolate exosomes from host cell proteins and concomitant components present in cell culture media, phosphate buffer, and urine. The hydrophobic exosome surfaces adhere to the weakly ionized surfaces of the PET fibers, making HIC a selective method of exosome isolation. Use of an HIC approach, involving an inverse salt gradient for elution, is much preferred over a common RP method as organic solvents employed in RP might compromise the structural integrity of the vesicles or result in the loss of important species (e.g. proteins) adhered to the exosome surfaces. It is important to note that while hydrophobic substrates have been used in previous exosome assay methods such as the Qiagen exoEasy Kit, use of a truly chromatographic method holds the promise for higher throughput and sampling/analysis of other matrix components such as host cell proteins as retentates can be selectively eluted. Such advantages would be the same as argued in any case of SPE versus LC.
In order to investigate the ability of the C-CP fibers to isolate exosomes, Dictyostelium discoideum cells were used to generate generic extracellular vesicles (EVs, sometimes referred to as outer membrane vesicles). D. discoideum is not only simple and inexpensive to culture in the lab, but it is also a model organism used for studying cell signaling, the endocytic pathway, and generation of extracellular vesicles and exosomes [33, 34]. Under normal conditions, it is a common single-cell, soil-dwelling amoeba; however, under environmental stress, such as lack of water and nutrients, its cells can form multicellular aggregates, the formation of which require direct cell-to-cell communication [34]. The ease with which D. discoideum can be cultured, the prominent role of cellular communication in its life cycle, and its use as a model organism for EV research, make it a useful organism for generating EVs needed to investigate isolation techniques.
In this demonstration study, the efficacy and efficiency of the new HIC C-CP fiber exosome isolation methodology are compared to two commonly utilized isolation methods, standard differential centrifugation (DC), as it is the most widely used isolation method, and the exoEasy Maxi Kit (QIAGEN), as it is the most similar commercially available method to the proposed HIC C-CP fiber approach [35]. The kit method uses postcentrifugation, spin-down processing, and a membrane-based stationary phase to affect an “affinity” (presumably a hydrophobic interaction) binding step to isolate exosomes and other EVs from serum and plasma, or cell culture supernatants. There is no selectivity with regards to size or cellular origin of the EVs. It relies on generic characteristics of the vesicle surfaces to capture all forms of EVs in the sample. The implementation of a chromatographic (flow through) approach versus the spin-down, SPE approach, would seem to present a number of potential advantages. These attributes are highlighted herein. The utility of the C-CP fiber HPLC separation is further demonstrated by investigation of recovery of exosomes from simulated urine and standard cell culture media.
2. Materials and methods
2.1. Extracellular vesicle expression by Dictyostelium discoideum
Dictyostelium discoideum AX2 cells (provided by L. Temesvari, Clemson University) were grown and maintained axenically in HL5 medium (https://www.Dictybase.org) supplemented with 100 μg/mL ampicillin at room temperature in 25 mL culture flasks [36]. Cells were passaged at 70–90% confluency.
For EV expression, AX2 cells were used to inoculate 50 mL of HL5 media supplemented with 100 μg/mL ampicillin at a starting cell concentration of 5 – 10 × 105 cells/mL in a 250 mL Erlenmeyer flask. After inoculation, the flask was covered in aluminum foil to block out light and placed on a shaker (150 RPM, 22°C) for 48 hours [34].
2.2. Isolation of extracellular vesicles and exosomes via differential centrifugation and Qiagen exoEasy Kit
Two widely accepted methods of generic EV isolation, differential centrifugation, and the Qiagen exoEasy Maxi Kit, were chosen as benchmarks for comparison to the proposed PET C-CP HIC isolation method. These specific methods were chosen because differential centrifugation is one of the most widely used methods for EV isolation and the exoEasy Maxi Kit is most like the PET C-CP HIC isolation method in terms of being an EV/surface adhesion process. While differential centrifugation is the most utilized method of isolation, it can sediment host cell proteins along with EVs. The process is also time consuming, and requires expensive equipment [13, 17–19]. The Qiagen exoEasy Maxi kit uses a membrane-based affinity column to separate exosomes and other EVs from solutions, in much less time in comparison to differential centrifugation; however, the process still takes up to 30 minutes to complete and costs approximately $32 per sample [37]. Another shortcoming of the exoEasy method is the high carryover of host cell proteins along with the derived vesicles.
All resulting differential centrifugation and exoEasy kit isolations were resuspended in either PBS or Qiagen elution buffer and subsequently divided into two aliquots. One aliquot of each sample was used for nanoparticle tracking analysis (NTA) for the determination of vesicle concentration and size distribution, while the other was used as a concentrated sample during the initial testing of the HIC C-CP fiber isolation method.
2.3. Differential centrifugation
Differential centrifugation retrieval of D. discoideum derived EVs was conducted as previously described by Tatischeff et al., with slight modifications [38]. Due to the relatively recent introduction of D. discoideum in EV research, all centrifugation steps followed previous research completed by Tatischeff et al. [38], rather than standard mammalian cell exosome isolation ultracentrifugation protocols which require DC at ~ 100 000 × g. In short, centrifugation at low speeds (e.g., 12 000 × g) is used to isolate the bulk of the EVs, with the higher value employed to specifically isolate the exosome subset. In practice, the HIC separation serves the purpose of the ultracentrifugation step. All centrifugation steps performed below 12 000 × g were performed using an Eppendorf Centrifuge 5430R (Eppendorf, Hamburg, Germany). Centrifugations of 12 000 × g or more were performed using a Beckman Coulter Avanti J-26S XPI Centrifuge with a JA-25.50 rotor (Beckman Coulter, Brea, CA). The first centrifugation step was performed at 700 × g (5 min., 22°C) in a 50 mL conical centrifuge tube. After centrifugation, 45 mL of the supernatant was transferred to a new 50 mL conical centrifuge tube for further centrifugation, with the remaining 5 mL of supernatant saved for exosome isolation via the C-CP HIC method. The second centrifugation was performed at 2000 × g (10 min., 22°C.) The final centrifugation step was performed at 12 000 × g (30 min., 4°C.). The supernatant was carefully removed and the final pellet was resuspended in 400 μL of PBS and stored at 4°C.
2.4. Qiagen exoEasy Maxi Kit
The QIAGEN exoEasy Maxi kit (QIAGEN, Hilden, Germany) isolations were accomplished per the manufacturer’s instructions. All of the centrifugation steps required by the kit were performed using an Eppendorf Centrifuge 5430R. Briefly, 10 mL of the D. discoideum cell growth media prepared for exosome isolation were filtered using a 0.8 μm syringe filter. An additional 1 mL of the remaining media was filtered using a 0.8 μm syringe filter and set aside for exosome isolation via the C-CP HIC method. The resulting exosomes were eluted using 400 μL of the Qiagen XE elution buffer and stored at 4°C.
2.5. Hydrophobic interaction chromatography (HIC) method
The PET C-CP fibers were produced by the Materials Science and Engineering Department, Clemson University. All solvents were purchased from EMD (EMD Millipore, Billerica, MA). Ammonium sulfate ((NH4)2SO4) and all other chemicals and proteins were purchased from Sigma-Aldrich (St. Louis, MO). Deionized water (DI-H2O) was secured from a Milli-Q water system. The chromatographic exosome separations were performed on a Dionex Ultimate 3000 HPLC system, LPG-3400SD Quaternary pump, and MWD-3000 UV-vis absorbance detector (Thermo Fisher Scientific, Waltham, MA). A Rheodyne model 8125 low dispersion injector with 20 and 60 μL injection loops was used for sample injections.
The PET C-CP fiber microbore columns (column length: 200 mm, id: 0.762 mm PEEK tubing, 450 fibers) prepared as described previously [26], were used for the exosome separations. After flushing the column with Buffer C (10 mM potassium phosphate buffer; pH = 7.4), it was equilibrated with Buffer A (1.8 M (NH4)2SO4 solution dissolved in PBS; pH = 7.4). As previously described in research regarding antibody purification using HIC, appropriate amounts of organic additives (such as methanol and acetonitrile) in the elution buffer demonstrate improved protein recovery, while maintaining their respective bioactivities [39–41]. As suggested previously in this report regarding the case in a potential RP exosome separation, use of high organic solvent content may have delitereous effects on the integrity of the exosomes. As such, 30% acetonitrile (ACN) v/v dissolved in PBS was employed as buffer B. As applied, the exosomes elute at a composition of ~15% ACN. A mobile phase flow rate of 0.5 mL/min and a 20 min gradient from 100% buffer A to 100% buffer B was used for exosome separation. Briefly, samples were injected onto the column during the high-salt (buffer A) mobile phase. Under these conditions, latent HCPs and exosomes are adsorbed to the PET fiber media, with the gradient subsequently eluting species of increasingly greater hydrophobicity. UV absorbance at 216 nm was monitored as a means of detecting the eluting species (proteins and exosomes). Based on the detector response reflecting their elution, purified exosomes were collected postcolumn.
2.6. HIC elution and isolation of exosomes from simulated urine and standard cell culture media
In order to demonstrate the HIC C-CP column isolation of exosomes from body fluids, and to assess the potential quantitative aspects of the isolation method, previously isolated EVs were spiked into a simulated urine matrix (194 g urea, 6 g CaCl2, 11 g Mg2SO4, and 80 g NaCl in 1 L of DI-H2O), spiked with myoglobin (Myo), α-chymotrypsinogen A (Chymo), ribonuclease A (Ribo), BSA, and lysozyme (Lyso) (0.1 mg/mL each), as simple representatives of the variety of proteins present in urine. HIC isolation of the vesicles from the simulated urine was followed by quantification based on the integrated peak areas of the eluted exosomes. The gradient baseline absorbance, obtained by running the gradient with no vesicles injected, was subtracted from the vesicle-spiked separation chromatograms.
2.7. Extracellular vesicle characterization via NTA
NTA was performed using a Nanosight (Malvern, Worcestershire, UK) NS500 with a 532 nm laser and 565 nm long pass cut off fluorescent filter (Center for Nanotechnology in Drug Delivery, UNC Eshelman School of Pharmacy). Samples were diluted to a concentration between 1 × 108 and 5 × 108 particles/mL with 20 nm filtered PBS. For each sample, particles moving under Brownian motion were recorded on video five times for 40 s each. Hydrodynamic diameters were calculated using the Stokes-Einstein equation.
2.8. Transmission electron microscopy fixation, staining, and imaging
Transmission electron microscopy (TEM) imaging was used to verify the physical size of single vesicles. D. discoideum (isolated via differential centrifugation) extracellular vesicles and human urine standard exosomes (Galan Laboratory Supplies, North Haven, CT) were fixed in 4% paraformaldehyde for 1 hour. Immediately following fixation, 5 μL of each sample was applied to 200 mesh formvar carbon-coated copper grids (Electron Microscopy Sciences, Hatfield, PA) and allowed to adsorb for 5 minutes. Each sample grid was negatively stained with 5 μl of 2% Uranyl Acetate for 5 minutes. Grids were then washed 3 times in 10 μl of distilled water for 5 minutes each and allowed to dry for imaging. All TEM images were obtained using a Hitachi H-7600 TEM (Hitachi, Tokyo, Japan) in the Clemson University Electron Microscopy Facility.
2.9. Scanning electron microscope fixation and imaging
The capture of intact exosomes onto the C-CP fiber surfaces was confirmed by scanning electron microscopy (SEM) imaging. Proprietary PET C-CP fiber-packed tips were produced according to a previously described method [42]. The fiber surfaces were wetted by flushing with 1 mL of H2O, and then rinsed in 2M (NH4)2SO4 in PBS. A 100 μL of each sample (DC-derived EVs, milieu or 2M (NH4)2SO4) were mixed with 2M (NH4)2SO4, followed by flushing through the tip (300 × g for 5 min). The prepared fibers were fixed in 1% osmium tetroxide for 1 hour and washed three times for 3 minutes each in distilled water to remove any excess osmium tetroxide. Next, each sample was washed in a 6 step gradient of ethanol-distilled water solutions starting at 50% ethanol and ending at 100% ethanol for 3 minutes each. An additional 100% ethanol wash step was performed to ensure that all water had been removed from the sample. Finally, each sample was washed in a 50–50 hexamethyldisilazane (HMDS)-ethanol solution for 3 minutes and dried in 100% HMDS overnight. Samples were sputter coated with platinum at 70 mTorr argon for 2 minutes using a Hummer 6.2 Sputtering system (Anatech USA, Union City, CA). SEM imaging was performed on a Hitachi S-4800 at 5.0 kV (Hitachi, Tokyo, Japan).
3. RESULTS AND DISCUSSION
3.1. HIC isolation of exosomes
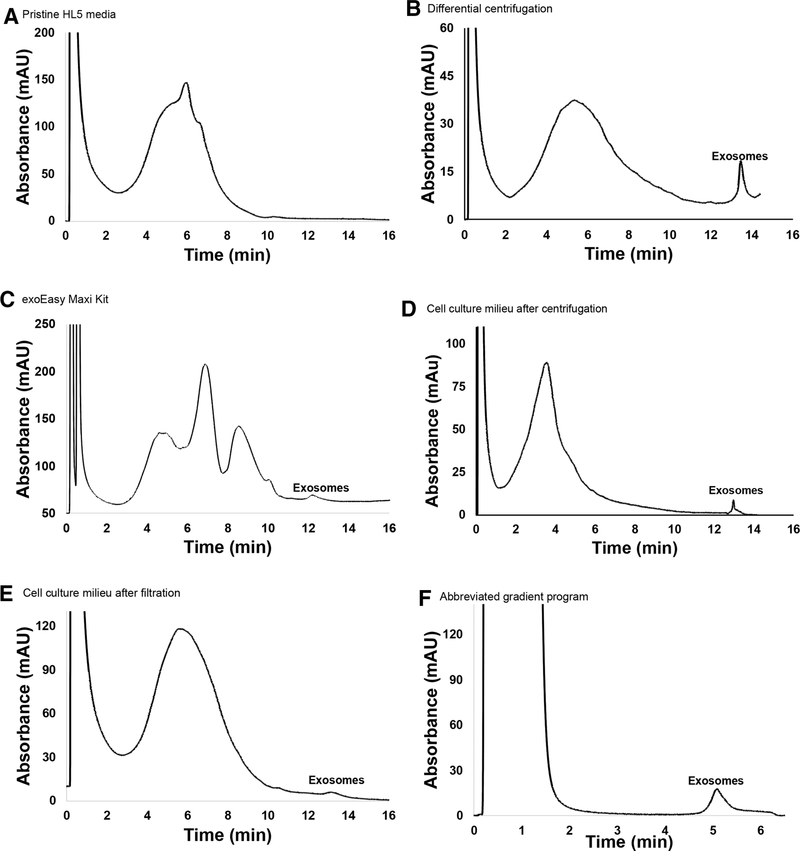
In order to determine whether or not exosomes could be isolated via HIC using the PET C-CP fibers, aliquots of the D. discoideum-derived EVs that had been previously isolated using the benchmark techniques were run on the PET C-CP columns with a mobile phase flow rate of 0.5 mL/min and a 20 minute gradient from 100% buffer A (1.8 M (NH4)2SO4 in PBS) to 100% buffer B (30% v/v acetonitrile in PBS). Figure 1 shows the resulting chromatograms derived from pristine HL5 media and four different isolation lots. In Fig. 1A, pristine HL5 was injected in order to establish a baseline chromatogram of the media components. The detector response shows a broadly eluting peak from ~2–10 minutes, composed of a myriad of proteins derived from yeast extract and peptone, various salts, and sugars. In Fig. 1B, EVs previously isolated via differential centrifugation were injected onto the fiber column. The resulting HIC chromatogram displays two major peaks, a very broad band between 2 and 11 minutes, and a fairly sharp feature reflecting more strongly retained (hydrophobic) species with an elution time of ~13–13.5 mins. In Fig. 1C, EVs previously isolated via the exoEasy kit were injected for HIC separation. The resulting chromatogram displays four prominent peaks. In this case, the first three broad peaks eluting within the same same time frame of 2–11 minutes bands in Fig. 1A and B. As in Fig. 1B, a discrete feature eluting in the ~13–13.5 min window is seen. Based on the structure of the respective chromatograms (Fig. 1A–C), it is not unreasonable to suggest that the peaks eluting between 2 and 11 minutes represent remaining HL5 media components, host cell (D. discoideum) proteinaceous and genetic material, salts, and other small molecules left behind during the previous isolation procedures, with the later eluting (13–13.5 min) smaller, sharper peak representing exosomes. One cannot rule out that there may be some EV-related material in the broad elution band, but one would expect that such species would be very hydrophobic in comparison to other media and cellular byproducts.
Figure 1.
Representative HIC chromatograms of exosome isolation using using PET C-CP fibers. Separations were performed with a mobile phase flow rate of 0.5 mL/min, 60 μL aliquot injections, and a 20 min gradient from 100% buffer A (1.8 M (NH4)2SO4 solution dissolved in PBS; pH = 7.4) to 100% buffer B (30% acetonitrile v/v dissolved in PBS). (A) Baseline chromatogram of pristine HL5 media. (B) D. discoideum-derived EVs previously isolated via differential centrifugation. (C) D. discoideum-derived EVs previously isolated via the Qiagen exoEasy Maxi Kit. (D) D. discoideum-derived EVs following centrifugation to remove cells and large debris. (E) D. discoideum-derived EVs following filtration through a 0.8 μm filter to remove cells and large debris. (F) Abbreviated gradient program (injection at A (0.8 M (NH4)2SO4 solution dissolved in PBS; pH = 7.4), gradient initiated at t = 2 min., gradient to 100% buffer B (30% acetonitrile v/v dissolved in PBS) in 10 min.
Indeed, simple Bradford assays revealed very high protein/amino acid content in these bands (data not shown). Further confirmation of the latter assignment is presented in particle tracking data and SEM images presented in subsequent sections. These results suggest that the PET C-CP HIC method is effective at separating the population of exosomes from other chemical species inherent in the spent cell media. Indeed, the presence of the broad concomitant elution bands in the HIC chromatograms reflects the nonspecificity and carryover toward proteins, etc. that exists in the DC and exoEasy isolates. It is believed that the far greater abundance of such species (based on the integrated absorbance of the bands) for the exoEasy case versus the DC is due to nonspecific binding to the hydrophobic matrix versus the differential centrifugation where proteins and EVs are more discretely segregated. HIC results prove that the exosome fractions can indeed be readily isolated from those otherwise undesirable components, as in the protein carryover in the exoEasy. Removal of such proteins would require further processing for both the DC and exoEasy isolation methods. The eluted exosome fractions from these isolations were collected and saved for NTA.
To more realistically compare the efficiencies of the C-CP fiber method with the benchmark methods, D. discoideum cell cultures were processed in similar fashions in terms of removing whole cells and cellular debris. In the case of DC, simple centrifugation is the first step, while macroscale debris is filter-removed prior to exoEasy Kit isolation. The resulting culture milieu solutions, having the exosomes in their native (relatively dilute) concentrations, were then subjected to HIC on the PET C-CP fiber columns using the previous gradient. The chromatogram of the centrifuged D. discoideum cell culture milieu (Fig. 1D), displays two prominent peaks at elution times of 2–11 and approximately 13 minutes, respectively. The differences in retention time (<5% relative) and shapes among the exosome eluents may be due to slight differences in the surface chemistries of the exosomes based on how they were processed prior to the HIC separation, especially solvent composition. This is certainly a point for further investigation. As before, the broader peak is attributed to various media components and cellular metabolites and debris, with the smaller second peak attributed to more hydrophobic species, likely exosomes. The relative responses for the concomitant species and the exosomes make sense as there is apt to be more debris and proteinaceous material, and lower exosome concentrations, in the milieu sample than the DC isolate suspended in PBS. The exosome peak from the milieu reflects their more dilute concentration in this stage of media processing. The corresponding HIC chromatogram taken of the filtered cell culture media is structurally the same as those seen in Fig. 1B–D, though with a lower relative exosome yield versus the other media components. This suggests that there may be some exosome loss in the initial filtration process in the exoEasy kit protocol, which is not unreasonable as the filter material itself is composed of both hydrophobic and hydrophilic layers.
As in any new chromatographic method, there are many aspects of the separation that can be optimized. The ability to improve the HIC method throughput is illustrated in Fig. 1F, where the initial DC milieu sample was injected at a reduced salt concentration of 0.8 M ammonium sulfate. In this way, the components making up the broad elution peak are not retained on the column. Following passage of the injection peak, the reverse salt gradient was then initiated, with clean elution of the exosomes. A simple comparison to the analogous full gradient (Fig. 1B) reveals that there is little or no difference in the exosome recovery for the abbreviated gradient method. Thus, there is an expectation that further improvements in throughput may be realized. Certainly, larger column formats would also improve processing speeds.
3.2. Comparison of vesicle recoveries for the different isolation methodologies
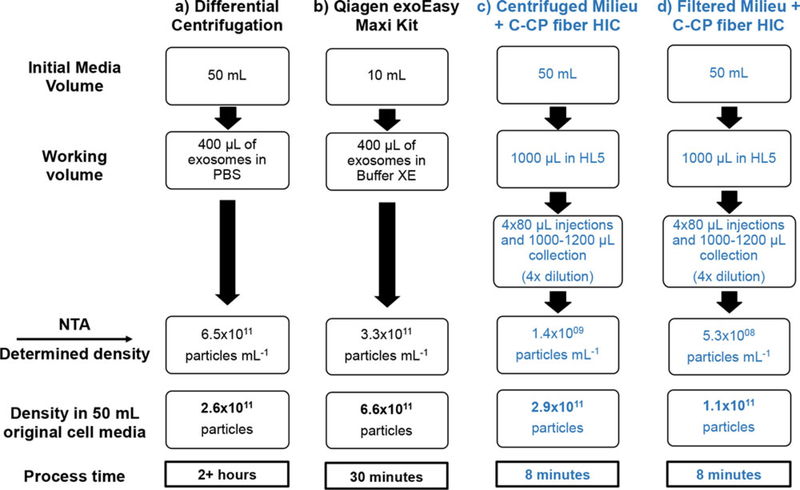
NTA is a widely accepted method for the evaluation of EV concentration and size distribution, and so was used to characterize the EV isolation from the DC and exoEasy kit procedures and those of the exosome fractions from the HIC isolations. Figure 2 provides a graphic overview of the procedural steps and NTA-determined particle concentrations for each EV isolation protocol. The initial entries into the flow charts (starting milieu volumes) were chosen based on previous experience in the use of DC and exoEasy methods, with the ultimate particle densities normalized to a common 50 mL volume sample.
Figure 2.
Comparison of NTA-determined particle population characteristics following the various isolation methodologies. All values were normalized to number of particles (exosomes/particles) derived from 50 mL of starting cell culture. Reported values are averages from triplicate isolations. (A) EVs isolated via differential centrifugation. (B) EVs isolated via Qiagen exoEasy Maxi Kit. (C) Cell culture media cleared of cells and large debris via centrifugation, followed by exosome isolation via PET C-CP HIC. (D) Cell culture media cleared of cells and large debris via filtration, followed by exosome isolation via PET C-CP HIC.
Since all particles counted during the NTA may not be exosomes (protein aggregates, other classes of extracellular vesicles, including microvesicles, and other cellular debris would also be counted), the resulting values (presented as particles-per-mL) in Fig. 2 should be compared to one another on an order-of-magnitude basis as opposed to absolute values. When compared in this manner, it can be seen that the differential centrifugation (Fig. 2A) and exoEasy kit (Fig. 2B) yield comparable exosome/particle recoveries. Figure 2 also shows that the PET C-CP HIC exosome isolation method, regardless of whether the cell culture was first centrifuged (Fig. 2C) or filtered (Fig. 2D) to remove cells and large debris, yields the same order of magnitude values as the benchmark methods following the initial debris removal steps. As a further point of reference, recent frontal analysis (breakthrough) studies indicate that the binding capacity for the present ~100 μL volid volume columns is ~2.9 × 1010 (data not shown).
The particle size distributions, as measured by NTA, were also very similar across all of the isolated samples, regardless of the isolation method (Table 1). In general, the D. discoideum EV size distributions were very similar to those previously reported by Tatischeff et al. [38]. Histograms of the NTA size distributions (representative data included as Supporting Information Fig. 1) each contained a prominant high concentration peak on the lower range of the size distribution scale ranging from ~90 to 160 nm, typically representing ~70% of the total population. Beyond this, a minor, larger-sized fraction is also seen in each distribution, most prominently in the DC-generated populations. These populations reflect protein and exosome agglomerates, as well as other classes of extracellular vesicles, including microvesicles, which would be most expected in the case of differential centrifugation. Such populations would also reflect the time-lag associated with sampling, packaging, and shipping of the materials for NTA characterization. Performing NTA/SEM/TEM measurements immediately following EV isolation may help remediate the level of potential vesicle or protein aggregation.
Table 1.
Comparison of EV size population characteristics for the different isolation procedures as determined by NTA
| Isolation method | Mean (nm) | Mode (nm) | Standard deviation (nm) | 10th Percentile (nm) | 90th Percentile (nm) |
|---|---|---|---|---|---|
| Differential centrifugation | 183 | 143 | 66.3 | 118.5 | 257 |
| Qiagen exoEasy Maxi kit | 154 | 97.1 | 80.2 | 87.6 | 263 |
| Centrifuged milieu + PET C-CP fibers | 155 | 121 | 60.1 | 106 | 228 |
| Filtered milieu + PET C-CP fibers | 142 | 108 | 60.4 | 87.6 | 206 |
Values represent averages for triplicate isolations.
As a final point of comparison, Fig. 2 also demonstrates the isolation time required for each of the processes. Differential centrifugation is the most time-consuming method, requiring more than 2 hours to perform (Fig. 2A). In comparison, the exoEasy kit requires approximately 30 minutes (Fig. 2B), while the PET C-CP HIC method can be affected in only 8 minutes, including four successive supernate injections and one chromatographic elution step. The C-CP fiber-based method is appreciably faster than either benchmark method, a major asset with regards to its potential usefulness for exosome isolation in a clinical setting. Indeed, the HIC method could be easilly affected in less than 3 minutes with the use of a step-gradient program as suggested in simple terms in Fig. 1F.
3.3. Imaging of exosomes from solution and adsorbed onto C-CP fibers
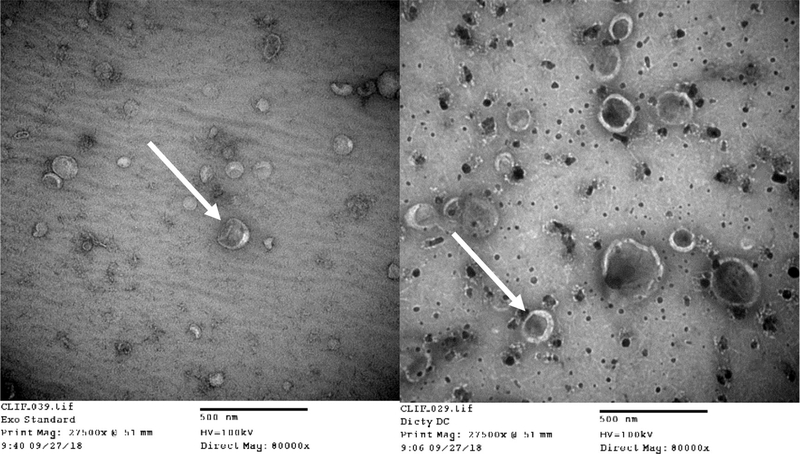
While the size distributions of the D. discoideum EVs were very similar to those previously reported by Tatischeff et al. [38], the standard method of verifying the presence of EVs or exosomes generally includes NTA size distribution results in conjunction with Western Blot verification of the presence of known exosomal protein markers, and a TEM micrograph to visualize and verify sizes of individual vesicles. However, this trio of verification methodologies is for mammalian cell-derived exosomes, where exosomal marker proteins have been verified and antibodies to these proteins are commercially available. To this end, no commercial antibodies exist for D. discoideum EV surface protiens. Therefore, we rely herein upon the resultant NTA data in combination with TEM micrographs of the commercially available exosome standards as compared to D. discoideum EVs resulting from the differential centrifugation method employed for isolation throughout this study (Fig. 3). The resulting micrographs show that the sizes and general morphology of the standard exosomes and those EVs derived from D. discoideum are very similar. Therefore, we contend that the initial differential centrifugation method employed resulted in the isolation of D. discoideum exosome-enriched EVs. These purified EVs were subsequently utilized to produce the TEM images.
Figure 3.
TEM images of EVs immobilized on copper grids. (Left) Standard exosomes derived from human urine (Right) D. discoideum-derived EVs previously isolated via differential centrifugation. Arrows are inserted to highlight the similar cup-like shape and size traits of the two EV species.
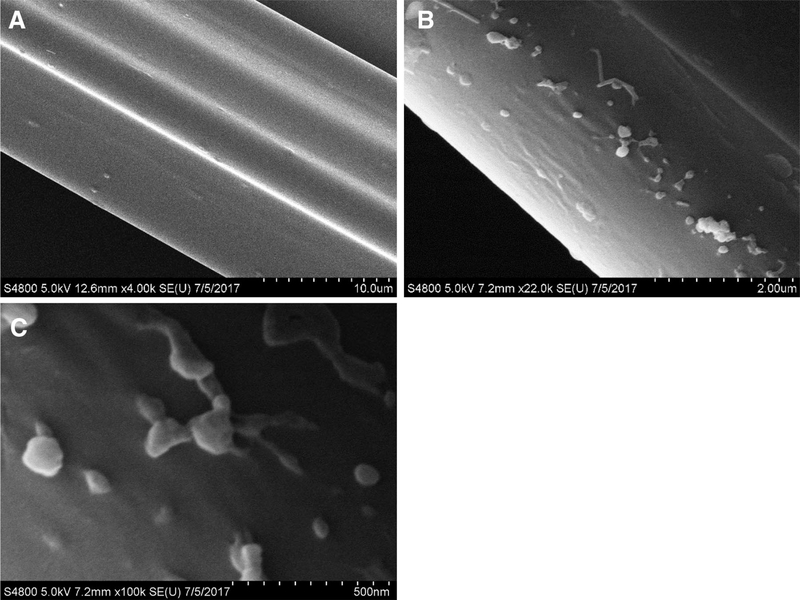
SEM images were collected to verify the presence and integrity of the exosomes and to investigate how they interact with the PET C-CP fiber channels. EVs that had been isolated from D. discoideum cell culture media via differential centrifugation and re-suspended in PBS were added to 2M ammonium sulfate chromatographic mobile phase and then each spun through PET C-CP fiber micropipette tips using the described SPE technique [30, 42]. In addition, 2M ammonium sulfate was spun through a separate tip as a non-vesicle control. In each case, an aqueous wash step was employed following exposure to remove any accumulated media particulates. As seen in Fig. 4A, the fibers exposed only to the salt media show a very smooth surface, with multiple channels of the same fiber seen at this 10 μm-scale micrograph. Passage of the DC-derived media through the fibers results in the adsorption of vesicle material as seen in Fig. 4B (2.0 μm-scale). A large number of vesicles are present as individual entities, but some appear as aggregates. It remains to be confirmed whether or not the agglomerates were formed on the fiber surface or originate in the milieu. Further magnification of the DC-derived aliquot (Fig. 4C, 500 nm scale) shows exquisite detail of individual exosomes, with diameters of 100 nm or less, on the PET fiber surfaces with some neighboring exosomes touching one another, perhaps stretching or elongating as they adhere to the fiber. This morphology could be early signs of aggregation or exosome fusion, though it is unclear at this point whether this is a native process or indeed caused by the fiber surface itself. While the SEM imaging demonstrates the phenomenon of the immobilization of the exosomes on the fiber surfaces, it also suggests that the fiber platform might be a viable means of studying exosome agglomeration or other phenomena.
Figure 4.
SEM images of exosomes immobilized on PET C-CP fibers. (A) 2M ammonium sulfate control (no exosomes exposed). (B) D. discoideum-derived EVs previously isolated via differential centrifugation. (C) Higher magnification image of exosomes depicted in micrograph B, showing detail of exosome interactions with each other and the fiber surface.
3.4. Isolation of exosomes via HIC from a simulated urine matrix
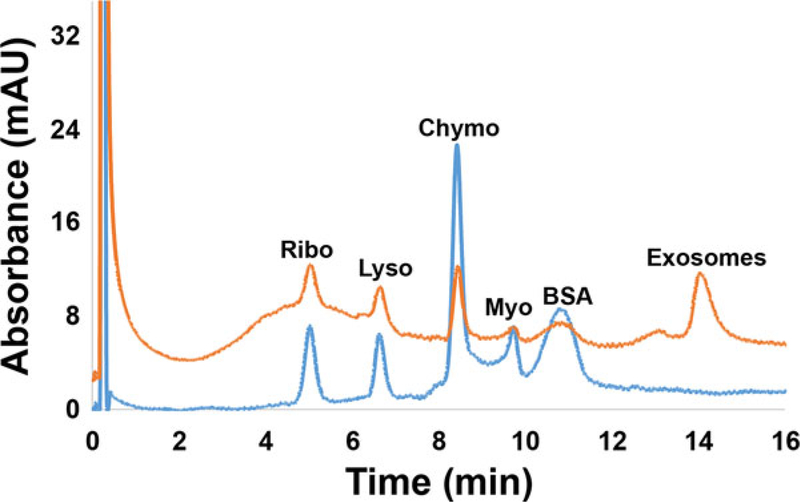
Following verification that exosomes could be isolated directly from D. discoideum culture milieu, the ability to isolate exosomes from synthetic urine (an ability needed for diagnostic applications) was investigated. For this experiment, EVs that had been previously isolated via differential centrifugation were spiked into a simulated urine matrix that was also spiked with the model proteins Myo, Chymo, Ribo, BSA, and Lyso, each at a concentration of 0.1 mg/mL. In this way, isolation from both urine matrix components and concomitant proteins is demonstrated using the same HIC gradient as employed in the Fig. 1 separations. Figure 5 (lower portion) shows the HIC chromatogram of the simulated urine matrix to establish baseline elution times for the matrix components and the spiked proteins. The five proteins are very well resolved in this case, with the other matrix species eluting as a band over the 8–10 min elution window. As seen in the upper chromatogram of Fig. 5, when the simulated urine was spiked 50:50 with DC-isolated EVs and subsequently run on the C-CP columns, HIC revealed the expected peaks for the spiked proteins along with an additional, later-eluting prominent peak attributed to the exosomes. Importantly, the added proteins appear as discrete peaks superimposed on a broader peak previously attributed to remnant proteins and debris associated with the spiked EVs following their DC-isolation (Fig. 1B). (The 0.1 mg/mL concentrations spiked here are clearly higher than those present from the spiked, equal-volume, milieu isolate.) The ability to cleanly separate exosomes from other species in a urine matrix suggests that C-CP fiber HIC (or simple SPE [42]) may be a promising method of isolation in a clinical setting. In fact, recent efforts into exosome isolation from patient urine samples have posed no new challenges in the capture or elution processes.
Figure 5.
PET C-CP fiber HIC chromatograms of simulated urine matrix spiked at 0.1 mg/mL concentration of model proteins (blue chromatogram (lower) and a 50:50 mixture of simulated urine and DC-isolated EVs (red chromatogram (upper)). Separations were performed with a mobile phase flow rate of 0.5 mL/min, 20 μL aliquot injections, and a 20 min gradient from 100% buffer A (1.8 M (NH4)2SO4 solution dissolved in PBS; pH = 7.4) to 100% buffer B (30% acetonitrile v/v dissolved in PBS).
3.5. Potential for exosome quantification
In order to make exosome retrieval easier to verify and potentially quantify, a simple measure of recovery is needed. It is most common to quantify exosomes based on lysing the vesicles and quantifying the total protein content via Western blot separations or a Bradford-type assay, or counting/sizing particles via NTA. Of course, this assumes no protein carryover in the isolation step, the protein content in each exosome is the same, and no extraneous cellular debris is present. Hook and coworkers have employed a surface plasmon resonance approach to quantification [43], wherein target proteins are captured onto a gold substrate through a surface immobilized antibody. Solution concentrations were extrapolated from first principles relationships, but not verified by any external standards.
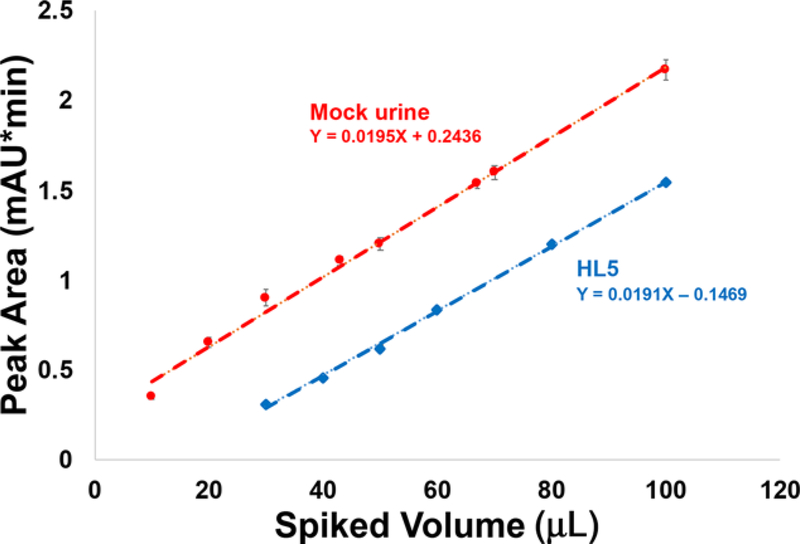
In the context of the HPLC isolation step, UV-vis absorbance at 216 nm is a viable approach. This is a common wavelength used in protein chromatography, and fortuitously, the exosomes “absorb” at this wavelength. To be clear, the phenomenon occurring here is likely scattering from the macrobodies and not discrete molecular band absorbance. As such, responses would also be related to differences in particle diameters as well as concentration. The fact that the exosomes are derived from the same source minimized such effects. To establish a quantitative relationship between exosome concentration and the measured integrated absorbance (reflective of scattering), increasingly larger aliquots of EVs (previously isolated via DC) were added to 100 μL aliquots of the simulated urine and the culture milieu (HL5) matrices, and the integrated absorbance values for the peaks eluting between 13 and 14 minutes were recorded. As shown in Fig. 6, a direct proportionality exists, suggesting quantitative recovery of exosomes from the C-CP fiber separation method. While the robustness of the method as employed here requires far greater evaluation, the results plotted in Fig. 6 represent a total of 24 and 15 injections of the respective samples (urine and media), each on a single column, without showing deleterious effects regarding the analytical precision. This suggests insignificant carryover or irreversible binding of the exosome solutes. The number of cycles that milieu-derived samples might be run is a more stringent and practical metric that is currently under review.
Figure 6.
Analytical response curves for separations of mixtures of 100 μL of the test matrices (simulated urine and HL5 media) and the designated volumes of DC-isolated EVs. Separations were performed with a mobile phase flow rate of 0.5 mL/min, 20 μL aliquot injections, and a 20 min gradient from 100% buffer A to 100% buffer B. Error bars reflect the standard deviation across n = 3 injections.
Importantly, the slopes of the two response curves are virtually identical, implying that the purity of the isolated fractions is very consistent between the two matrix forms. Note, we are in no position to place firm concentration values on the results of these experiments, as no appropriate certified reference materials exists. Sources of quantified exosomes will be investigated to provide more definitive figures of metrics. Based on the NTA values presented in Table 1, the working range presented in Fig. 6 is ~0.1 – 7 × 109 particles for the 20 μL injections. Accordingly, it is not difficult to imagine limits of detection for this method to be approximately 0.05 × 109 particles. Of course, UV-vis absorbance postcolumn quantification could also be applied for preparative purposes. One could also envision other LC-compatible detection methods, for example, multiangle light scatter (MALS), which would also provide size information for the eluting particles.
4. Concluding remarks
In order for basic research and potential application of exosomes to continue to grow, lower cost, more time-efficient methods of exosome isolation are needed. This report introduces a promising new isolation platform using PET C-CP fibers as stationary phases for HIC isolation of exosomes. The method isolates exosome populations of similar number density and size distribution as currently accepted isolation methods involving advanced centrifugation or SPE. The fiber platform is inexpensive ($5 USD per column that can be used >20 times on the analytical scale), while providing comparatively high throughput. Additionally, the versatility of the C-CP fibers will allow for the addition of antibodies, surface chemistries, and other isolation modalities to transform the generic (hydrophobic interaction) exosome isolation method demonstrated here into a type-specific method. In an alternative approach, the fibers may be used in spin-down column format (i.e., micropipette tips) to isolate exosomes from body fluids such as urine [44], providing high purity, highthroughput, and cost-effective exosome isolation for potential clinical use. Selective capture can be augmented with spectroscopic or visual imaging as means of verifying the presence of target exosome species on the fiber surfaces.
Much fundamental work remains toward realizing the practical utility of the method on the clinical, analytical, and preparative scales. Each of these areas will pose challenges and require evaluation regarding selectivity, robustness, loading capacity, throughput, and column structure/operation parameters. Comparison to the use of size exclusion chromatography would be relevant in this regard. Many basic challenges toward this end remain. Most importantly, verification of the biological efficacy of the HIC-isolated exosomes must be done. Multiple mammalian cell lines wherein Western blot procedures can be implemented will be key in this regard. Understanding of the potential causes of agglomeration is also very important. Ultimately, the potential impact of exosomes in modern medicine is a tremendous driving force for these continued efforts.
Supplementary Material
Acknowledgments
Financial support for the chromatography development efforts from the National Science Foundation, Division of Chemistry under grant CHE-1608663 is gratefully acknowledged. Financial support for the exosome isolation efforts from the Eppley Foundation for Scientific Research is gratefully acknowledged. The Gibson Foundation, the ITOR Biorepository, and the Greenville Hospital System are gratefully acknowledged. Matthew Haney and the Nanomedicines Characterization Core Facility at The University of North Carolina at Chapel Hill are gratefully ackowledged for completion of the Nanosight analysis. Special thanks to George Wetzel, Clemson University Electron Microscopy Facility, for assistance with EM.
Abbreviations
- ACN
acetonitrile
- C-CP
capillary-channeled polymer
- Chymo
α-chymotrypsinogen A
- DC
differential centrifugation
- EV
extracellular vesicle
- HCPs
host cell proteins
- HIC
hydrophobic interaction chromatography
- HMDS
hexamethyldisilazane
- Lyso
lysozyme
- miRNA
microRNA
- Myo
myoglobin
- NTA
nanoparticle tracking analysis
- PET
poly(ethylene terephthalate)
- Ribo
ribonuclease A
Footnotes
The authors have declared no conflict of interest.
Additional supporting information may be found online in the Supporting Information section at the end of the article.
5 References
- [1].Lässer C, Eldh M, Lötvall J, J. Vis. Exp 2012, 59, 3037–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, Polchi A, Tancini B, Emiliani C, Genes 2013, 4, 152–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Raposo G, Stoorvogel W, J. Cell Biol 2013, 200, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rashed MH, Bayraktar E, Helal GK, Abd-Ellah MF, Amero P, Chavez-Reyes A, Rodriguez-Aguayo C, Int. J. Mol. Sci 2017, 18, 538–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yanez-Mo M, Siljander PRM, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NHH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-t’ Hoen ENM, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MHM, De Wever O, J. Extracell. Vesicles 2015, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vlassov AV, Magdaleno S, Setterquist R, Conrad R, Biochim. Biophys. Acta Gen. Subj 2012, 1820, 940–948. [DOI] [PubMed] [Google Scholar]
- [7].Li M, Zeringer E, Barta T, Schageman J, Cheng AG, Vlassov AV, Philos. Trans. R. Soc. B Biol. Sci 2014, 369, 20130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee Y, El Andaloussi S, Wood MJA, Hum. Mol. Genet 2012, 21, R125–R134. [DOI] [PubMed] [Google Scholar]
- [9].Bellingham SA, Coleman BM, Hill AF, Nucleic Acids Res. 2012, 40, 10937–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L, BMC Genomics 2013, 14, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taylor DD, Gercel-Taylor C, Gynecol. Oncol 2008, 110, 13–21. [DOI] [PubMed] [Google Scholar]
- [12].Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH, Clin. Lung Cancer 2009, 10, 42–46. [DOI] [PubMed] [Google Scholar]
- [13].Li P, Kaslan M, Lee SH, Yao J, Gao ZQ, Theranostics 2017, 7, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].An MR, Lohse I, Tan ZJ, Zhu JH, Wu J, Kurapati H, Morgan MA, Lawrence TS, Cuneo KC, Lubman DM, J. Proteome Res 2017, 16, 1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, Soria CE, Oquin S, Bonebreak CM, Saracoglu E, Skog J, Kuo WP, Biol. Chem 2013, 394, 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Furi I, Momen-Heravi F, Szabo G, Ann. Transl. Med 2017, 5, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, Hendrix A, J. Extracell. Vesicles 2014, 3, 24858–24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jeppesen DK, Hvam ML, Primdahl-Bengtson B, Boysen AT, Whitehead B, Dyrskjøt L, Ørntoft TF, Howard KA, Ostenfeld MS, J. Extracell. Vesicles 2014, 3, 25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J, Chiesi A, Methods 2015, 87, 46–58. [DOI] [PubMed] [Google Scholar]
- [20].Yang F, Liao XZ, Tian Y, Li GY, Biotechnol. J 2017, 12, 8. [DOI] [PubMed] [Google Scholar]
- [21].Böing AN, van der Pol E, Grootemaat AE, Coumans FAW, Sturk A, Nieuwland R, J. Extracell. Vesicles 2014, 3, 23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ, Heusermann W, Wiklander OPB, Hallbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtio J, Smith CIE, Wood MJA, Andaloussi SEL, Nanomed. Nanotechnol. Biol. Med 2015, 11, 879–883. [DOI] [PubMed] [Google Scholar]
- [23].Marcus RK, Davis WC, Knippel BC, LaMotte L, Hill TA, Perahia D, Jenkins JD, J. Chromatogr. A 2003, 986, 17–31. [DOI] [PubMed] [Google Scholar]
- [24].Nelson DK, Marcus RK, J. Chromatogr. Sci 2003, 41, 475–479. [DOI] [PubMed] [Google Scholar]
- [25].Stanelle RD, Marcus RK, Anal. Bioanal. Chem 2009, 393, 273–281. [DOI] [PubMed] [Google Scholar]
- [26].Randunu JM, Marcus RK, Anal. Bioanal. Chem 2012, 404, 721–729. [DOI] [PubMed] [Google Scholar]
- [27].Schadock-Hewitt AJ, Pittman JJ, Stevens KA, Marcus RK, J. Appl. Polym. Sci 2013, 128, 1257–1265. [Google Scholar]
- [28].Jiang L, Marcus RK, Anal. Chim. Acta 2017, 954, 129–139. [DOI] [PubMed] [Google Scholar]
- [29].Jiang L, Marcus RK, Anal. Chim. Acta 2017, 977, 52–64. [DOI] [PubMed] [Google Scholar]
- [30].Schadock-Hewitt AJ, Marcus RK, J. Sep. Sci 2014, 37, 495–504. [DOI] [PubMed] [Google Scholar]
- [31].Trang HK, Marcus RK, J. Pharm. Biomed. Anal 2017, 142, 49–58. [DOI] [PubMed] [Google Scholar]
- [32].Jiang L, Marcus RK, Anal. Bioanal. Chem 2015, 407, 939–951. [DOI] [PubMed] [Google Scholar]
- [33].Lavialle F, Deshayes S, Gonnet F, Larquet E, Kruglik SG, Boisset N, Daniel R, Alfsen A, Tatischeff I, Int. J. Pharm 2009, 380, 206–215. [DOI] [PubMed] [Google Scholar]
- [34].Annesley SJ, Chen S, Francione LM, Sanislav O, Chavan AJ, Farah C, De Piazza SW, Storey CL, Ilievska J, Fernando SG, Smith PK, Lay ST, Fisher PR, BBA-GEN Subj. 2014, 1840, 1413–1432. [DOI] [PubMed] [Google Scholar]
- [35].QIAGEN, exoEasy Maxi Kit Handbook, Hilden, Germany, 2015. [Google Scholar]
- [36].Fey P, Dodson RJ, Basu S, Chisholm RL, Protocols 2013, 983, 59–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, Noerholm M, PLoS One 2015, 10, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tatischeff I, Larquet E, Falcón-Pérez JM, Turpin P-Y, Kruglik SG, J. Extracell. Vesicles 2012, 1, article 19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bobaly B, Beck A, Veuthey JL, Guillarme D, Fekete S, J. Pharm. Biomed. Anal 2016, 131, 124–132. [DOI] [PubMed] [Google Scholar]
- [40].Fekete S, Veuthey JL, Beck A, Guillarme D, J. Pharm. Biomed. Anal 2016, 130, 3–18. [DOI] [PubMed] [Google Scholar]
- [41].Bobaly B, D’Atri V, Beck A, Guillarme D, Fekete S, J. Pharm. Biomed. Anal 2017, 145, 24–32. [DOI] [PubMed] [Google Scholar]
- [42].Burdette CQ, Marcus RK, Analyst 2013, 138, 1098–1106. [DOI] [PubMed] [Google Scholar]
- [43].Rupert DLM, Lasser C, Eldh M, Block S, Zhdanov VP, Lotvall JO, Bally M, Hook F, Anal. Chem 2014, 86, 5929–5936. [DOI] [PubMed] [Google Scholar]
- [44].Manard BT, Jones SMH, Marcus RK, Proteomics- Clin. Apps 2015, 9, 522–530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.