Rhizobia form symbiotic associations with legumes that lead to the formation of nitrogen-fixing nodules. Sulfur-containing molecules play a crucial role in nitrogen fixation; thus, the rhizobia inside nodules require large amounts of sulfur. Rhizobia can assimilate both inorganic (sulfate) and organic (sulfonates) sources of sulfur. However, very little is known about rhizobial sulfur metabolism during symbiosis. In this report, we show that sulfonate utilization by Bradyrhizobium diazoefficiens is important for symbiotic nitrogen fixation in both soybean and cowpea. The symbiotic defect is probably due to increased sensitivity to oxidative stress from sulfur deficiency in the mutant strain defective for sulfonate utilization. The results of this study can be extended to other rhizobium-legume symbioses, as sulfonate utilization genes are widespread in these bacteria.
KEYWORDS: nodulation, nitrogen fixation, sulfonates, soybean, Bradyrhizobium, symbiosis
ABSTRACT
Sulfur (S)-containing molecules play an important role in symbiotic nitrogen fixation and are critical components of nitrogenase and other iron-S proteins. S deficiency inhibits symbiotic nitrogen fixation by rhizobia. However, despite its importance, little is known about the sources of S that rhizobia utilize during symbiosis. We previously showed that Bradyrhizobium diazoefficiens USDA110T can assimilate both inorganic and organic S and that genes involved in organic S utilization are expressed during symbiosis. Here, we show that a B. diazoefficiens USDA110T mutant with a sulfonate monooxygenase (ssuD) insertion is defective in nitrogen fixation. Microscopy analyses revealed that the ΔssuD mutant was defective in root hair infection and that ΔssuD mutant bacteroids showed degradation compared to the wild-type strain. Moreover, the ΔssuD mutant was significantly more sensitive to hydrogen peroxide-mediated oxidative stress than the wild-type strain. Taken together, these results show that the ability of rhizobia to utilize organic S plays an important role in symbiotic nitrogen fixation. Since nodules have been reported to be an important source of reduced S used during symbiosis and nitrogen fixation, further research will be needed to determine the mechanisms involved in the regulation of S assimilation by rhizobia.
IMPORTANCE Rhizobia form symbiotic associations with legumes that lead to the formation of nitrogen-fixing nodules. Sulfur-containing molecules play a crucial role in nitrogen fixation; thus, the rhizobia inside nodules require large amounts of sulfur. Rhizobia can assimilate both inorganic (sulfate) and organic (sulfonates) sources of sulfur. However, very little is known about rhizobial sulfur metabolism during symbiosis. In this report, we show that sulfonate utilization by Bradyrhizobium diazoefficiens is important for symbiotic nitrogen fixation in both soybean and cowpea. The symbiotic defect is probably due to increased sensitivity to oxidative stress from sulfur deficiency in the mutant strain defective for sulfonate utilization. The results of this study can be extended to other rhizobium-legume symbioses, as sulfonate utilization genes are widespread in these bacteria.
INTRODUCTION
Rhizobia, belonging to both alpha- and betaproteobacteria, form nitrogen-fixing symbioses with legume plants (1–3). Rhizobium-legume interactions are mediated by specific chemical signaling and lead to the formation of specialized structures, called nodules, in which the rhizobia fix atmospheric nitrogen in exchange for carbon from the plant (1–4). The formation of nodules on legumes is a multistage process, involving bacterial growth and multiplication in the rhizosphere, the recognition and infection of root hairs, bacterial growth inside infection threads, release of the bacteria into symbiosomes within the cytoplasm of the host root cells, and their subsequent differentiation and development into N2-fixing bacteroids (1, 5, 6).
Once inside the nodules, rhizobia are dependent on the plant for their mineral nutrition (4). Several studies have used rhizobial auxotrophs to gain insights into the type of nutrient sources that the legumes provide to the bacterial symbiont. These studies have defined the genetic mechanisms for the utilization of carbon (7), nitrogen (4, 8–10), and phosphorus (11, 12) sources by rhizobia. In contrast, and despite its essential importance, only a few studies have examined sulfur (S) exchange between the legume host and its nodule bacteria.
A better understanding of S metabolism during symbiotic nitrogen fixation is needed, as S-containing metabolites play an important role in the initiation and maintenance of nodulation as well as the efficiency of nitrogen fixation (13). S deficiency led to inhibition of nitrogen fixation in pea nodules, whereas growth and nitrogen fixation were increased by S supplementation (14, 15). Nitrogenase, the enzyme responsible for nitrogen fixation, contains elevated amounts of S, in the form of FeS clusters (16). In addition, low-potential electrons are transferred from Fe-S-containing ferredoxin or flavodoxin to nitrogenase (17). Apart from the S in nitrogenase, nodules contain elevated amounts of glutathione and/or homoglutathione that can alleviate oxidative stress (18) and are required for proper development of root nodules (19). Glutathione is also important for the microsymbiont, as rhizobial mutants defective in glutathione synthesis are severely impaired in nodulation and nitrogen fixation (20). In accordance with the importance of S in symbiotic nitrogen fixation, a nodule-specific sulfate transporter was shown to be required for nitrogen fixation in Lotus japonicus (21) indicating that the host-plant has evolved specialized mechanisms for providing S to the microsymbiont. It has been suggested that nodules are the primary source of assimilated S in nodulated legumes (22).
Bacteria can assimilate S from both inorganic and organic forms, and the metabolic pathways for S assimilation, and their regulation, have been studied extensively using Escherichia coli as a model (23–26). In contrast, few studies have examined S metabolism in rhizobia, and those that have, largely focused on assimilation of sulfate, one of the main inorganic forms of S (27–29). However, little is known about assimilation of S from organic compounds and the mechanisms that regulate it. A better understanding of the mechanisms of organic S metabolism is important, as the host plant converts the inorganic sulfate from soils into organic forms (30). Additionally, from an energetics perspective, it seems logical that bacteria would prefer to assimilate organic S-containing compounds (mainly sulfonates and S esters), as free sulfate requires at least 2 ATPs for its assimilation into the host cell via adenosine 5′-phosphosulfate (APS) and phosphoadenosine 5′-phosphosulfate (PAPS) (24).
We and others previously showed that in rhizobia, sulfonates play a role in symbiosis. For example, a mutant of Azorhizobium caulinodans ORS578 defective in a gene for sulfonate metabolism formed nodules that were ineffective for nitrogen fixation (31). Putative organic S utilization genes were shown to be expressed by Bradyrhizobium japonicum (now renamed Bradyrhizobium diazoefficiens) USDA 110T (32) (here, USDA110) in soybean nodules (33–35) and in Sesbania rostrata nodules by A. caulinodans ORS278 (36). We previously demonstrated that USDA110 assimilated sulfonates as sole S sources, and the sulfonate utilization genes were expressed at high levels in soybean nodules (37).
The aim of this study was to determine the role of sulfonate utilization in later stages of nodulation and symbiotic nitrogen fixation. By using a green fluorescent protein (GFP)-marked strain and microscopic observations, we now show that the ΔssuD mutant is defective in symbiosome formation and nitrogen fixation in soybean and cowpea. We also show that the ΔssuD mutant is more sensitive to oxidative stress and discuss the possible roles that sulfonate assimilation and S nutrition play in oxidative stress tolerance during rhizobium-plant interactions.
RESULTS
The sulfonate monooxygenase mutant is defective in symbiotic nitrogen fixation.
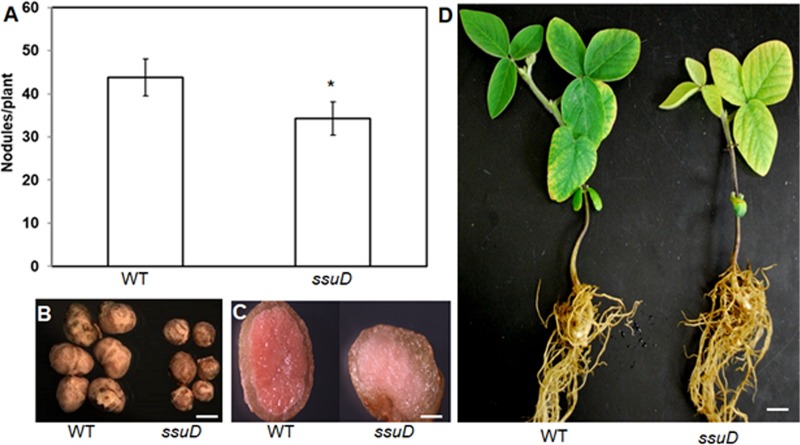
Results from our previous study demonstrated that ssuD was required for utilization of aliphatic sulfonates as a sole S source by USDA110 but was not required for initial nodule formation (37). To determine if sulfonate utilization might be important at later stages of symbiosis, we inoculated soybean with the wild-type USDA110 and ΔssuD strains and analyzed nodule formation and symbiotic function at 40 days postinoculation (dpi). Results of this analysis showed that the ΔssuD mutant formed fewer and smaller nodules than the wild-type (WT) strain (Fig. 1A and B). Visual inspection of dissected nodules showed that those formed by the WT strain were redder than those formed by the ΔssuD mutant (Fig. 1C). Furthermore, the leaves of plants inoculated with the ΔssuD mutant showed symptoms of N deficiency compared to those of the WT-inoculated plants (Fig. 1D). In accordance with these visual observations, the leaves of plants inoculated with the ΔssuD mutant contained significantly less chlorophyll and had lower total shoot dry weights than the WT-inoculated plants but had more than the uninoculated control plants (Fig. 2). These results indicate that nodules formed by the ΔssuD mutant are defective in symbiotic nitrogen fixation with soybean compared to those formed by the WT strain.
FIG 1.
The ΔssuD mutant strain of USDA110 shows ineffective symbiotic nitrogen fixation with soybean. (A) Soybean plants inoculated with the ΔssuD mutant formed fewer nodules than the WT strain. (B) The nodules of the ΔssuD mutant were much smaller than those formed by the WT. Bar, 1 mm. (C) Cross section of the nodules showing decreased leghemoglobin (red) in nodules of the ΔssuD mutant compared to that in the WT. Bar, 100 μm. (D). Soybean plants inoculated with the ΔssuD mutant show stunted growth and symptoms of nitrogen deficiency (yellow leaves), compared to full growth and green leaves in the WT-inoculated plants. Bar, 1.5 cm. *, P < 0.01 versus the WT.
FIG 2.
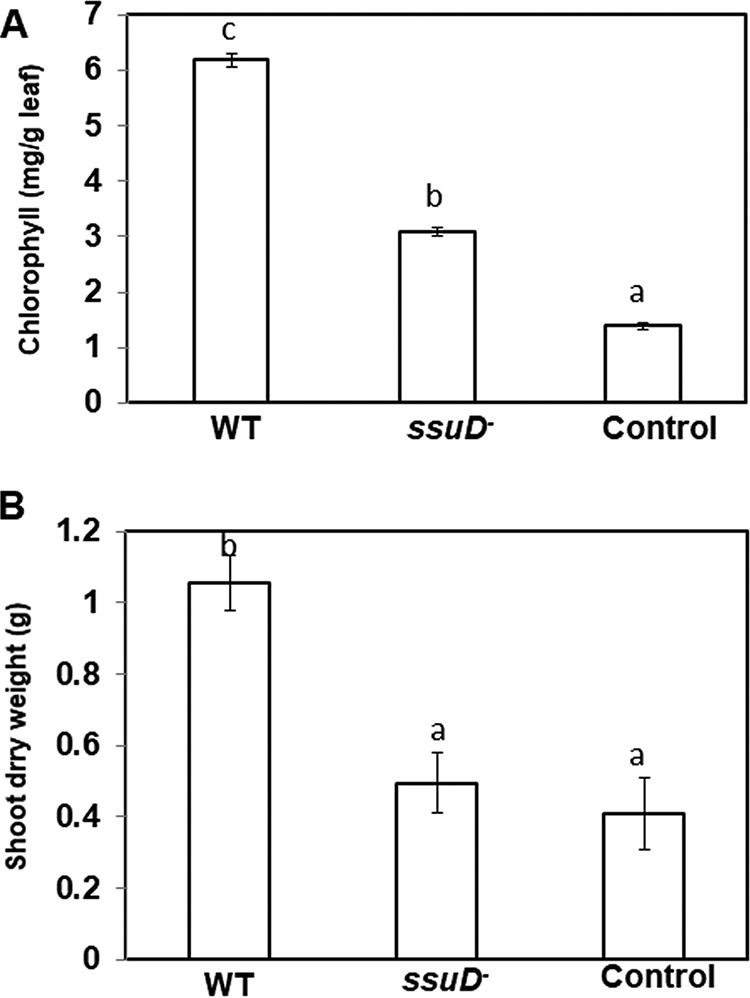
Chlorophyll content and growth promotion of soybean inoculated with either USDA110 WT or the ΔssuD mutant strains. (A) Chlorophyll levels in the leaves of uninoculated or inoculated soybean plants. The plants inoculated with the ΔssuD mutant showed significantly less chlorophyll than those inoculated with the WT strain but more than the uninoculated plants. (B) Shoot dry weights of plants inoculated with the ΔssuD mutant were similar to those of the uninoculated plants and significantly less than the WT inoculated plants. The results are the means ± standard deviations (SDs) from three independent observations, with six plants each. Bars with different lowercase letters are significantly different (P < 0.05) from each other.
ΔssuD mutant is impaired in root hair infection and survival in nodules.
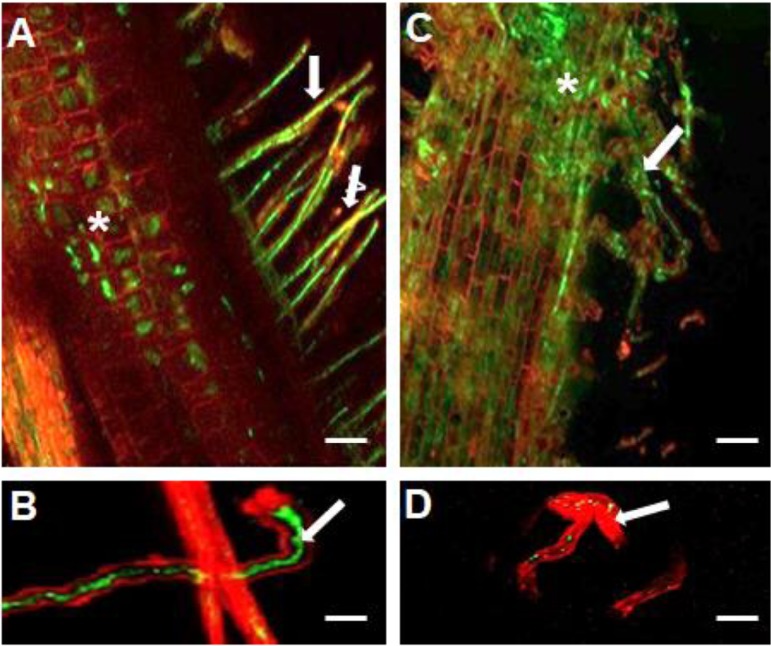
To determine the potential reasons for this observed symbiotic defect, the ΔssuD mutant and the WT strains were marked with pHC60 that constitutively expresses green fluorescent protein (GFP) (38). The GFP-marked strains were inoculated onto soybean seedlings, and colonization was studied using confocal microscopy. As expected, the WT strain colonized the root hairs and cortical cells and formed infection threads (Fig. 3A and B). In contrast, while the ΔssuD mutant was present on the root surface (Fig. 2C), it was only occasionally detected inside root hairs (Fig. 3D).
FIG 3.
Confocal laser scanning microscopy of colonization and root hair infection of soybean by GFP-marked USDA110 WT and ΔssuD mutant strains. (A) The WT strain colonizing the root hairs (arrows) and central root tissues (*). (B) Root hair infection thread formed by the WT strain. (C) The ΔssuD mutant showed diffuse colonization of root hairs (arrows) and the main root surface (*). (D) A few GFP-expressing ΔssuD mutant bacteria can be seen within the infected root hair (arrow). Bars, 100 μm for A and C, 30 μm for B and D.
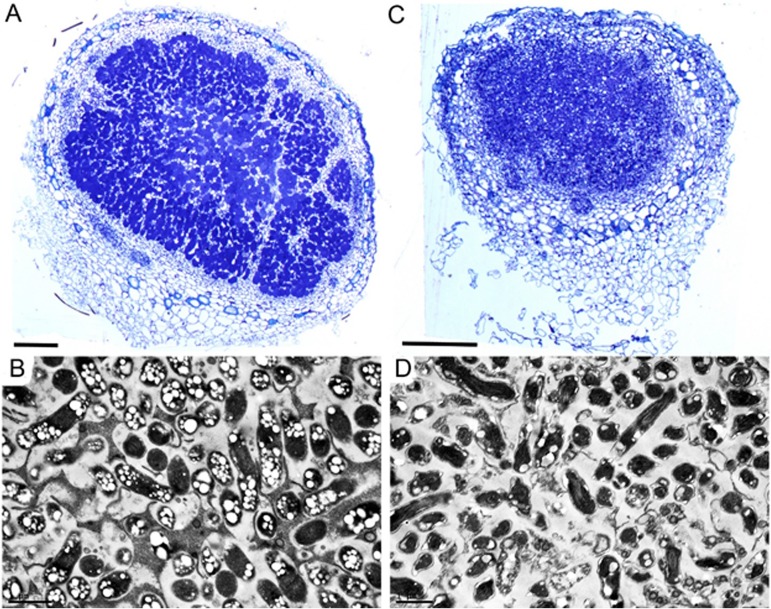
To further study the symbiotic interaction, nodules were fixed, sectioned, and examined using light and transmission electron microscopy (TEM). The nodules containing the WT strain showed central zone cells containing bacterioids that were densely labeled with toluidine blue (Fig. 4A). In contrast, the nodules formed by the ΔssuD mutant showed very diffuse toluidine blue staining, indicating reduced bacterial infection of the central zone cells and potentially reduced symbiosome formation (Fig. 4B). Further analyses using TEM on nodules induced by the WT strain showed the presence of symbiosomes containing densely packed bacteroids that contained polyhydroxybutyrate (PHB) granules (Fig. 4C), whereas the nodules produced by the ΔssuD mutant contained bacteroids that were likely not fully formed or were degraded, and these bacteroids also contained very little obvious PHB (Fig. 3D).
FIG 4.
Microscopic analysis of soybean nodules formed by USDA110 and the ΔssuD mutant strains. (A) Light micrograph of a transverse section of a nodule formed by the WT strain showing intense toluidine blue staining of bacteria. (B) Transmission electron micrograph (TEM) of WT nodule showing elongated bacteroids containing polyhydroxybutyrate granules in symbiosomes. (C) Light micrograph of nodule formed by the ΔssuD mutant showing diffuse toluidine blue staining. (D) TEM of a nodule formed by the ΔssuD mutant showing degraded bacteroids. Bars, 200 μm.
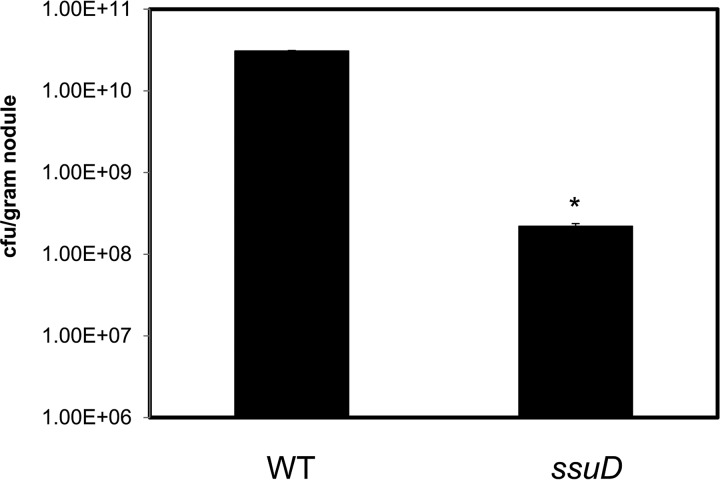
To determine if the ΔssuD mutant was affected in its survival/persistence within the nodule, we determined the viability of bacteroids by reisolation and cell counts. The numbers of ΔssuD mutants per gram of nodule were 100-fold less than that of the WT strain (Fig. 5). Taken together, these results suggest that ssuD could be involved in early as well as late symbiotic interactions between USDA110 and soybean.
FIG 5.
Nodules infected by the ΔssuD mutant contain less viable bacteria than those by the WT USDA110. Significantly fewer CFU of the ΔssuD mutant were reisolated from nodules than of the WT strain. Three plants were used, and 10 nodules from each plant were weighed, pooled, homogenized, and used for CFU determination. *, P < 0.01 versus WT.
The ΔssuD mutant is sensitive to oxidative stress.
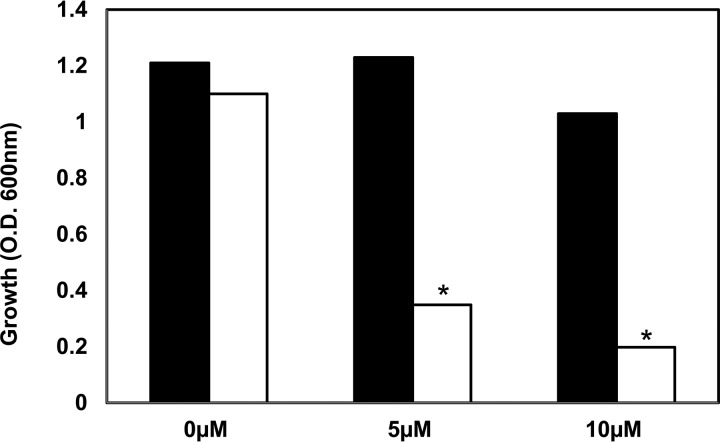
It is well known that rhizobia encounter oxidative stress during root hair infection as well as in the nodules (39, 40). S limitation can also lead to oxidative stress in E. coli (26), and H2O2 can oxidize the thiol in cysteine to sulfonate (41). To determine the role of oxidative stress in the symbiotic defect of the ΔssuD mutant, we compared the H2O2 sensitivity of the ΔssuD mutant and the WT strains. The growth of the ΔssuD mutant was significantly reduced in the presence of 5 and 10 μM H2O2 compared to the growth of the WT (Fig. 6). These results suggest that ssuD may also be involved in tolerance to oxidative stress and that the defects in root hair infection and nodule survival by the ΔssuD mutant could also be due to higher sensitivity to reactive oxygen species.
FIG 6.
The ΔssuD mutant is more sensitive to H2O2 than the WT USDA110. The growth of the ΔssuD mutant was inhibited in the presence of 5 μM and 10 μM H2O2, but the WT strain was unaffected by these concentrations. *, P < 0.01 versus WT.
The ΔssuD mutant is defective in symbiotic nitrogen fixation with cowpea.
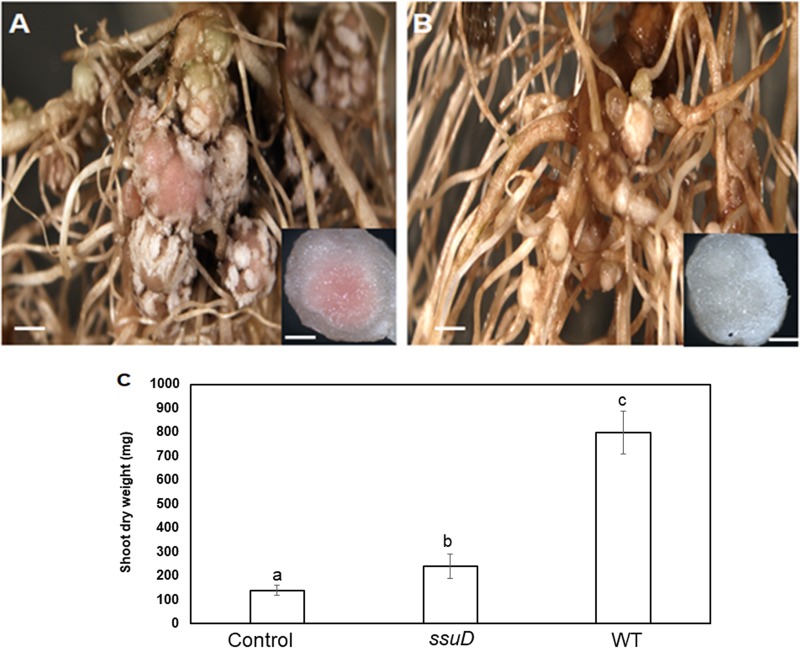
USDA110 forms symbioses with hosts other than soybean, such as cowpea (Vigna unguiculata). An earlier report suggested that bll7011, the periplasmic sulfonate-binding protein, was involved in host-specific adaptation by USDA110 (42). To determine if the role of ssuD in the USDA110 symbiosis extends to other host plants, we inoculated the WT and ΔssuD mutant onto cowpea seedlings and examined the symbiosis at 30 dpi. Similar to the results obtained with soybean, the WT strain formed large pink nodules (Fig. 7A) on the cowpea host, whereas the ΔssuD mutant formed many small and white nodules (Fig. 7B), typical of an ineffective symbiosis. Measurement of shoot dry weights showed that plants inoculated with the ΔssuD mutant had significantly less dry biomass (P < 0.01) than those inoculated with the WT strain and were similar to the uninoculated control plants. These results demonstrate that the ΔssuD mutant was defective in symbiotic nitrogen fixation with cowpea as well as soybean.
FIG 7.
The ΔssuD mutant is defective in symbiosis with cowpea. (A) The WT strain formed effective nodules on cowpea. Bar, 1 mm. Inset, nodule showing leghemoglobin (pink). Bar, 100 μm. (B) The ΔssuD mutant formed ineffective nodules. Bar, 1 mm. Inset, nodule showing lack of leghemoglobin (white). Bar, 100 μm. (C) Shoot dry weight of plants inoculated with the ΔssuD mutant was similar to that of the uninoculated control plant and significantly less than the plants inoculated with WT. The results are means ± SDs from three independent observations with four plants each. Bars with different lowercase letters are significantly different (P < 0.05) from each other.
DISCUSSION
We previously reported that USDA110 uses the sulfonate monooxygenase gene ssuD (bll7010) to metabolize alkane sulfonates as sole source of S. In this study, we show that the ssuD gene in USDA110 is required for an effective nitrogen-fixing symbiosis with both soybean and cowpea. While the ΔssuD mutant strain was able to form nodules on both of the tested hosts, the nodules were small, white, and less effective in N2 fixation than the nodules formed by the WT strain.
S is required in relatively large amounts during the nitrogen fixation process, as it is a required cofactor for the enzyme nitrogenase as well for the FeS clusters of many proteins involved in electron transport (13). S limitation leads to decreases in symbiotic nitrogen fixation, whereas supplementation of S can enhance the symbiosis (14, 15). A few studies have determined the mechanism of S exchange between the plant and rhizobial symbiont. Proteomic analysis identified a sulfate transporter that showed high expression in the symbiosome membrane of Lotus japonicus (43). Moreover, an L. japonicus mutant defective in a symbiotic sulfate transporter (SST1) showed a significant decrease in production of leghemoglobin, the Fe protein of nitrogenase, as well as in the rate of nitrogen fixation (21). In addition, Kalloniati et al. (22) reported that Mesorhizobium loti-induced N2-fixing nodules on L. japonicus are likely to be a major site for S assimilation in this legume.
Although the symbiotic defect of the SST1 mutant suggests that sulfate could be a major S form exchanged through the symbiosome membrane (21), it is possible that the infected plant cell could metabolize sulfate into organic forms that are then utilized by rhizobia. This is supported by observations that cysteine auxotrophs of Sinorhizobium meliloti formed effective nodules, indicating that the host plant provided cysteine or other organic S molecules (44). Our results show that the ability of USDA110 to utilize sulfonates as an S source is important for survival inside the host legume and that sulfonates could be a source of S within root nodules.
Analysis done using a GFP-marked strain revealed that the ΔssuD mutant showed impaired root hair infection compared to the WT strain (Fig. 3). However, the mutant was not completely defective, as the infection did lead to eventual nodule formation. It is likely that the growth of the ΔssuD mutant inside the root hairs was slower than the WT strain. Light microscopy and TEM observations showed that the ΔssuD mutant strain did not form intact symbiosomes and that bacterioids were likely to be degraded inside the nodules (Fig. 4). This is further supported by the viable cell counts from the nodules that showed that significantly fewer CFU were isolated from the ΔssuD mutant-induced nodules than from the WT-induced nodules (Fig. 5).
Limitation of S leads to oxidative stress in E. coli (26), and S-containing metabolites, such as glutathione, play an important role in tolerance to oxidative stress. It is likely that the inability of the ΔssuD mutant to utilize sulfonate could result in S starvation within the plant cells, leading to increased oxidative stress. It is also possible that ssuD may be involved in S homeostasis of oxidation of cysteine to sulfonic acid that can occur during rhizobium-host interactions. In fact, it was previously shown that sulfenylated proteins are present during various stages of the S. meliloti-Medicago truncatula symbiosis (49). It is clearly important for nitrogen fixation, as cysteine oxidation to sulfonic acid is postulated to be an important means of inactivating FixK2, a major regulator of microaerobic respiration in B. japonicum (41). In accordance with such a role, the ΔssuD mutant showed significantly higher sensitivity to H2O2 than did the WT (Fig. 6).
An earlier report by Koch et al. (42) suggested that the sulfonate utilization operon bll7011-7008 could be important for host-specific adaptation, as it showed higher induction in the nodules of siratro (Macroptilium atropurpureum) than in nodules of soybean and cowpea. In contrast, however, our results suggest that sulfonate utilization plays an important role in N2 fixation by USDA110 in symbiosis with both soybean and cowpea. Genes annotated as involved in sulfonate assimilation are widespread in other legume-nodulating rhizobia, such as Sinorhizobium (45) as well as rhizobia listed in GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The widespread presence of these genes suggests that the ability of rhizobia to utilize sulfonate is likely to be important for the efficiency of symbiotic nitrogen fixation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The USDA110 and the ssuD:kan strains and E. coli DH5α were maintained on yeast mannitol (YM) medium (46) and LB agar, respectively. The ssuD:kan insertion mutant was constructed by disrupting ssuD with a kanamycin resistance cassette in the same orientation and double-crossover recombination (37). Antibiotics were used as follows: kanamycin (100 μg/ml), tetracycline (70 μg/ml).
Marking of strains with green fluorescent protein.
A plasmid expressing GFP (pHC60) (38) was transferred from E. coli into the wild-type (WT) and ssuD mutant strains by triparental conjugation. Transconjugants were selected on YM medium, without yeast extract, containing 70 μg/ml tetracycline (to counterselect auxotrophic E. coli). The presence of plasmid in strains was confirmed by direct fluorescence microscopy.
Nodulation and plant growth promotion.
Soybean and cowpea plants were inoculated essentially as described previously (37). Seeds were surface sterilized with bleach, washed with sterile water six times, and germinated on petri plates containing sterile moist paper towels. Seedlings, free of any visual contamination, were transferred to pots containing sterile vermiculite and supplied with sterile nitrogen-free nutrient solution containing 2 mM MgSO4 as the sulfur source, which is sufficient for both plant and bacterial growth (37). Plants were inoculated with approximately 106 cells/ml of the WT or ΔssuD mutant strains and incubated in a plant growth chamber at 25°C with a 16-h light/8-h dark cycle. Uninoculated plants served as negative controls. The total numbers of nodules on plants were counted 40 days postinoculation (dpi), and their dry weight was determined. Nitrogen fixation effectiveness was determined by measuring chlorophyll content of leaves, using acetone extraction and absorption at 663 nm and 645 nm as described previously (47), and by quantifying shoot dry weight. At least six plants were used for each treatment, and the experiment was repeated thrice.
Enumeration of viable bacteria inside the nodules.
The viability of bacteroids in nodules formed by the WT and ΔssuD mutant strains was determined. Nodules were harvested, weighed, and surface sterilized using dilute (5%) bleach. Nodules were washed multiple times with sterile water and crushed using a wood applicator. The crushed nodule suspension was serially 10-fold diluted and plated on YM agar plates, and total CFU were counted.
Microscopic studies of plant colonization and nodulation.
The colonization of soybean by the GFP-marked WT and ΔssuD mutant strains was determined using confocal laser scanning microscopy (CLSM) as described previously (48). Replicate plants were germinated and inoculated with GFP-marked strains as described above. Plants were examined by using CLSM at weekly intervals following inoculation. Roots were stained with 100 ng ml−1 propidium iodide for 5 min. Free-hand sectioning was performed on propidium iodide-stained roots, and fluorescence was observed using a Leica TCS SP2 CLSM (Leica Microsystems, Bannockburn, IL), using a wavelength of 488 nm for excitation of both GFP and propidium iodide. The fluorescence of GFP and propidium iodide was observed in two specific emission windows of 500 to 550 nm (GFP) and 640 to 700 nm (propidium iodide) as described previously (48).
For light and transmission electron microscopy, nodules were fixed with glutaraldehyde, embedded in LR white, sectioned, and examined under a light and electron microscope as described previously (48). The light microscopy sections (1 μm) were viewed under a Zeiss Axiophot 2 optical microscope, and the ultrathin sections for TEM were viewed using a JEOL JEM 1400 transmission electron microscope.
Tolerance to hydrogen peroxide.
The WT and ΔssuD mutant strains were grown to mid-log phase in YM medium, and 10-μl aliquots of mid-log-phase cultures were subcultured in fresh YM medium containing 0, 5, 10, or 50 μM H2O2. After incubation overnight at 30°C, tolerance to H2O2 was determined by measuring the optical density at 600 nm (OD600).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by IOS-0820005 from The National Science Foundation to M.J.S. and by Research Growth Initiative, UW Milwaukee, to P.G.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01552-19.
REFERENCES
- 1.Oldroyd GE, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 2.Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, Estrada-de Los Santos P, Gross E, Dos Reis FB, Sprent JI, Young JP, James EK. 2011. Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol Plant Microbe Interact 24:1276–1288. doi: 10.1094/MPMI-06-11-0172. [DOI] [PubMed] [Google Scholar]
- 3.Poole PS, Ramachandran V, Terpolilli J. 2018. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16:291–303. doi: 10.1038/nrmicro.2017.171. [DOI] [PubMed] [Google Scholar]
- 4.Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 5.Verma D. 1992. Signals in root nodule organogenesis and endocytosis of rhizobium. Plant Cell 4:373–382. doi: 10.1105/tpc.4.4.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broughton WJ, Jabbouri S, Perret X. 2000. Keys to symbiotic harmony. J Bacteriol 182:5641–5652. doi: 10.1128/jb.182.20.5641-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yurgel SN, Kahn ML. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol Rev 28:489–501. doi: 10.1016/j.femsre.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Sadowsky MJ, Rostas K, Sista PR, Bussey H, Verma D. 1986. Symbiotically defective histidine auxotrophs of Bradyrhizobium japonieum. Arch Microbiol 144:334–339. doi: 10.1007/BF00409881. [DOI] [Google Scholar]
- 9.Poole PS, Allaway D. 2000. Carbon and nitrogen metabolism in Rhizobium. Adv Microb Physiol 43:117–163. doi: 10.1016/S0065-2911(00)43004-3. [DOI] [PubMed] [Google Scholar]
- 10.Hosie AH, Allaway D, Galloway CS, Dunsby H, Poole PS. 2002. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched chain transporters (Bra/LIV) of the ABC family. J Bacteriol 184:4071–4080. doi: 10.1128/jb.184.15.4071-4080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Niemi TS, Kahn ML, McDermott TR. 1997. P metabolism in the bean-Rhizobium tropici symbiosis. Plant Physiol 113:1233–1242. doi: 10.1104/pp.113.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botero LM, Al-Niemi TS, McDermott TR. 2000. Characterization of two inducible phosphate transport systems in Rhizobium tropici. Appl Environ Microbiol 66:15–22. doi: 10.1128/aem.66.1.15-22.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becana M, Wienkoop S, Matamoros MA. 2018. Sulfur transport and metabolism in legume root nodules. Front Plant Sci 9:1434. doi: 10.3389/fpls.2018.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Wood AP, McGrath SP. 1999. Effect of sulfur nutrition on growth and nitrogen fixation of pea (Pisum sativum L.). Plant Soil 212:207–217. doi: 10.1023/A:1004618303445. [DOI] [Google Scholar]
- 15.Varin S, Cliquet J-B, Personeni E, Avice J-C, Lemauviel-Lavenant S. 2010. How does sulphur availability modify N acquisition of white clover (Trifolium repens L.)? J Exp Bot 61:225–234. doi: 10.1093/jxb/erp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubio LM, Ludden PW. 2008. Biosynthesis of the iron‐molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 17.Carter KR, Rawlings J, Orme-Johnson WH, Becker RR, Evans HJ. 1980. Purification and characterization of a ferredoxin from Rhizobium japonicum bacteroids. J Biol Chem 255:4213–4223. [PubMed] [Google Scholar]
- 18.Matamoros MA, Clemente MR, Sato S, Asamizu E, Tabata S, Ramos J, Moran JF, Stiller J, Gresshoff PM, Becana M. 2003. Molecular analysis of the pathway for the synthesis of thiol tripeptides in the model legume Lotus japonicus. Mol Plant Microbe Interact 16:1039–1046. doi: 10.1094/MPMI.2003.16.11.1039. [DOI] [PubMed] [Google Scholar]
- 19.Frendo P, Harrison J, Norman C, Hernández Jiménez MJ, Van de Sype G, Gilabert A, Puppo A. 2005. Glutathione and homoglutathione play a critical role in the nodulation process of Medicago truncatula. Mol Plant Microbe Interact 18:254–259. doi: 10.1094/MPMI-18-0254. [DOI] [PubMed] [Google Scholar]
- 20.Harrison J, Jamet A, Muglia CI, Van de Sype G, Aguilar OM, Puppo A, Frendo P. 2005. Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J Bacteriol 187:168–174. doi: 10.1128/JB.187.1.168-174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, Stougaard J, Kawaguchi M, Miyamoto A, Suganuma N, Udvardi MK. 2005. The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17:1625–1636. doi: 10.1105/tpc.104.030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalloniati C, Krompas P, Karalias G, Udvardi MK, Rennenberg H, Herschbach C, Flemetakis E. 2015. Nitrogen-fixing nodules are an important source of reduced sulfur, which triggers global changes in sulfur metabolism in Lotus japonicus. Plant Cell 27:2384–2400. doi: 10.1105/tpc.15.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kredich NM. 1996. Biosynthesis of cysteine, p 514–527. In Neidhardt FC, Curtiss R III, Ingraham JL (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 24.Kertesz MA. 1999. Riding the sulfur cycle–metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol Rev 24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 25.van der Ploeg J, Eichhorn E, Leisinger T. 2001. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch Microbiol 176:1–8. doi: 10.1007/s002030100298. [DOI] [PubMed] [Google Scholar]
- 26.Gyaneshwar P, Paliy O, McAuliffe J, Popham D, Jordan M, Kustu S. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J Bacteriol 187:1074–1090. doi: 10.1128/JB.187.3.1074-1090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tate R, Riccio A, Iaccarino M, Patriarca AJ. 1997. A cysG mutant of Rhizobium etli pleiotropically defective in sulfate and nitrate assimilation. J Bacteriol 179:7343–7350. doi: 10.1128/jb.179.23.7343-7350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snoeck C, Verreth C, Hernández-Lucas I, Martínez-Romero E, Vanderleyden J. 2003. Identification of a third sulfate activation system in Sinorhizobium sp. strain BR816: the CysDN sulfate activation complex. Appl Environ Microbiol 69:2006–2014. doi: 10.1128/aem.69.4.2006-2014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu ZJ, Cao YQ, Long WJ, Long ZD, Chen G, Ma QS, Wu B. 2008. Isolation and characterization of an operon involved in sulfate and sulfite metabolism in Sinorhizobium fredii. FEMS Microbiol Lett 282:89–99. doi: 10.1111/j.1574-6968.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 30.Koprivova A, Kopriva S. 2014. Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Front Plant Sci 5:589. doi: 10.3389/fpls.2014.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki S, Aono T, Lee KB, Suzuki T, Liu CT, Miwa H, Wakao S, Iki T, Oyaizu H. 2007. Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata–Azorhizobium caulinodans ORS571 symbiosis. Appl Environ Microbiol 73:6650–6659. doi: 10.1128/AEM.01514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delamuta JR, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martínez-Romero E, Hungria M. 2013. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol 63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- 33.Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer HM, Hennecke H. 2007. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact 20:1353–1363. doi: 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
- 34.Mesa S, Hauser F, Friberg M, Malaguti E, Fischer HM, Hennecke H. 2008. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol 190:6568–6579. doi: 10.1128/JB.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delmotte N, Ahrens CH, Knief C, Qeli E, Koch M, Fischer HM, Vorholt JA, Hennecke H, Pessi G. 2010. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10:1391–1400. doi: 10.1002/pmic.200900710. [DOI] [PubMed] [Google Scholar]
- 36.Tsukada S, Aono T, Akiba N, Lee KB, Liu CT, Toyazaki H, Oyaizu H. 2009. Comparative genome-wide transcriptional profiling of Azorhizobium caulinodans ORS571 grown under free-living and symbiotic conditions. Appl Environ Microbiol 75:5037–5046. doi: 10.1128/AEM.00398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugawara M, Shah GR, Sadowsky MJ, Paliy O, Speck J, Vail AW, Gyaneshwar P. 2011. Expression and functional roles of Bradyrhizobium japonicum genes involved in the utilization of inorganic and organic sulfur compounds in free-living and symbiotic conditions. Mol Plant Microbe Interact 24:451–457. doi: 10.1094/MPMI-08-10-0184. [DOI] [PubMed] [Google Scholar]
- 38.Cheng HP, Walker GC. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180:5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos R, Hérouart D, Puppo A, Touati D. 2000. Critical protective role of bacterial superoxide dismutase in rhizobium-legume symbiosis. Mol Microbiol 38:750–759. doi: 10.1046/j.1365-2958.2000.02178.x. [DOI] [PubMed] [Google Scholar]
- 40.Shaw SL, Long SR. 2003. Nod factor inhibition of reactive oxygen efflux in a host legume. Plant Physiol 32:2196–2204. doi: 10.1104/pp.103.021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesa S, Reutimann L, Fischer HM, Hennecke H. 2009. Posttranslational control of transcription factor FixK2, a key regulator for the Bradyrhizobium japonicum-soybean symbiosis. Proc Natl Acad Sci U S A 106:21860–21865. doi: 10.1073/pnas.0908097106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch M, Delmotte N, Rehrauer H, Vorholt JA, Pessi G, Hennecke H. 2010. Rhizobial adaptation to hosts, a new facet in the legume root nodule symbiosis. Mol Plant Microbe Interact 23:784–790. doi: 10.1094/MPMI-23-6-0784. [DOI] [PubMed] [Google Scholar]
- 43.Wienkoop S, Saalbach G. 2003. Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131:1080–1090. doi: 10.1104/pp.102.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbas BA, Vineetha KE, Prasad CK, Vij N, Hassani R, Randhawa GS. 2002. Symbiotic characteristics of cysteine and methionine auxotrophs of Sinorhizobium meliloti. Indian J Exp Biol 40:1121–1130. [PubMed] [Google Scholar]
- 45.Sugawara M, Epstein B, Badgley BD, Unno T, Xu L, Reese J, Gyaneshwar P, Denny R, Mudge J, Bharti AK, Farmer AD, May GD, Woodward JE, Médigue C, Vallenet D, Lajus A, Rouy Z, Martinez-Vaz B, Tiffin P, Young ND, Sadowsky MJ. 2013. Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol 14:R17. doi: 10.1186/gb-2013-14-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent JM. 1970. A manual for practical study of root-nodule bacteria. Blackwell Science, Oxford, UK. [Google Scholar]
- 47.Huang Z, Jiang D, Yang Y, Sun J, Jin S. 2004. Effects of nitrogen deficiency on gas exchange, chlorophyll fluorescence, and antioxidant enzymes in leaves of rice plants. Photosynthetica 42:357–364. doi: 10.1023/B:PHOT.0000046153.08935.4c. [DOI] [Google Scholar]
- 48.Mitra S, Mukherjee A, Wiley-Kalil A, Das S, Owen H, Reddy PM, Ané JM, James EK, Gyaneshwar P. 2016. A rhamnose-deficient lipopolysaccharide mutant of Rhizobium sp. IRBG74 is defective in root colonization and beneficial interactions with its flooding-tolerant hosts Sesbania cannabina and wetland rice. J Exp Bot 67:5869–5884. doi: 10.1093/jxb/erw354. [DOI] [PubMed] [Google Scholar]
- 49.Oger E, Marino D, Guigonis JM, Pauly N, Puppo A. 2012. Sulfenylated proteins in the Medicago truncatula-Sinorhizobium meliloti symbiosis. J Proteomics 75:4102–4113. doi: 10.1016/j.jprot.2012.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.