In 2016, there was a large-scale waterborne outbreak of campylobacteriosis in New Zealand, which was estimated to have affected over 5,000 people. This highlighted the need for a greater understanding of the sources of contamination of both surface and groundwater and risks associated with exposure to both drinking and recreational water. This study reports the prevalence and population structure of Campylobacter jejuni in six recreational waters of the Manawatu-Wanganui region of New Zealand and models the relationship between Campylobacter spp. and ruminant-associated Campylobacter and the parameters “sites,” “months,” and “river flow.” Here, we demonstrate that both low and high river flows, month of the year, and recreational sites could influence the Campylobacter isolation from recreational waters. The presence of genotypes associated with human infection allowed us to describe potential risks associated with recreational waters.
KEYWORDS: Campylobacter jejuni (C. jejuni), epidemiology, MLST, recreational water, ruminant, wild birds, public health
ABSTRACT
Campylobacter jejuni, a leading cause of gastroenteritis worldwide, has been frequently isolated from recreational rivers and streams in New Zealand, yet the public health significance of this is unknown. This study uses molecular tools to improve our understanding of the epidemiology and sources of Campylobacter in recreational waterways, with a view to preventing human infection. Epidemiological and microbiological data were collected between 2005 and 2009 from six high-use recreational waterways in the Manawatu-Wanganui region of the North Island. Campylobacter spp. and C. jejuni were isolated from 33.2% and 20.4% of 509 samples, respectively. Isolation of Campylobacter was observed in both low and high river flows. After adjusting for the confounding effects of river flow, there was a significantly higher likelihood of isolating Campylobacter in the winter month of June compared to January. A high diversity of C. jejuni multilocus sequence types was seen, with the most commonly isolated being the water rail-associated ST-2381 (19/91 isolates [20.9%]), ST-1225 (8/91 isolates [8.8%]), and ST-45 (6/91 isolates [6.6%]). The ST-2381 was found in all rivers, while the most commonly isolated ST from human cases in New Zealand, the poultry-associated strain ST-474, was isolated only in one river. Although the majority of Campylobacter sequence types identified in river water were strains associated with wild birds that are rarely associated with human disease, poultry and ruminant-associated Campylobacter strains that are found in human infection were also identified and could present a public health risk.
IMPORTANCE In 2016, there was a large-scale waterborne outbreak of campylobacteriosis in New Zealand, which was estimated to have affected over 5,000 people. This highlighted the need for a greater understanding of the sources of contamination of both surface and groundwater and risks associated with exposure to both drinking and recreational water. This study reports the prevalence and population structure of Campylobacter jejuni in six recreational waters of the Manawatu-Wanganui region of New Zealand and models the relationship between Campylobacter spp. and ruminant-associated Campylobacter and the parameters “sites,” “months,” and “river flow.” Here, we demonstrate that both low and high river flows, month of the year, and recreational sites could influence the Campylobacter isolation from recreational waters. The presence of genotypes associated with human infection allowed us to describe potential risks associated with recreational waters.
INTRODUCTION
Campylobacter is a Gram-negative, thermophilic and microaerophilic bacterium that causes approximately 5 to 14% of diarrheal illnesses worldwide (1, 2). Most of the identified Campylobacter species can cause human campylobacteriosis, but of these, Campylobacter jejuni and C. coli are the major cause of gastroenteritis, accounting for 95% of all reported human campylobacteriosis cases (1). This organism naturally inhabits the intestines of warm-blooded animals, including poultry, wild birds, and ruminants (3). Several epidemiological studies have shown these animals to be potential reservoirs for human infections (3–5). These studies also indicate that animal hosts are the main sources of food and water contamination, while the food chain route has been shown to be the predominant infection pathway for human campylobacteriosis, particularly via poultry meat (6–8). Sporadic cases of Campylobacter infection have been most commonly linked to food (9, 10), whereas outbreaks of campylobacteriosis have been associated with drinking contaminated water, including a large-scale outbreak in New Zealand in 2016 (11, 12), the accidental ingestion of water during recreational activities, and the consumption of poultry meat and raw milk (13–15). Hence, each potential exposure pathway needs to be studied in detail in order to increase our understanding of Campylobacter transmission dynamics and thus inform the design of effective, country-specific, prevention programs.
Campylobacteriosis is the most common zoonotic bacterial enteric disease in New Zealand, and over the last decade, this country has persistently reported one of the highest campylobacteriosis rates among developed countries (16–18). From 1980 (the first year of mandatory notification of cases) to 2006, the annual number of reported cases of campylobacteriosis increased steadily from a few hundred to peak at 15,873 (383 cases per 100,000 population) in 2006 (9, 19, 20). These cases accounted for 70% of all the notified enteric diseases in New Zealand, and resulted in an estimated cost in New Zealand dollars (NZD) of 75 million per year (21, 22). Campylobacteriosis cases may incur a further disease burden due to sequelae such as Guillain-Barré syndrome, with an average rate of 2.32 hospitalizations per 100,000 population per year (23, 24) in New Zealand. An investigation into the sources of human cases identified poultry as a major source of infection (17). Subsequently, in early 2007, the three major poultry suppliers implemented both voluntary and regulatory interventions to reduce poultry carcass contamination. In the 2 years subsequent to the interventions, there was a 59% reduction in reported human campylobacteriosis cases from 383 cases in 2006 to 157 cases per 100,000 population in 2008 (25). Nevertheless, the campylobacteriosis rate in New Zealand remains among the highest of the industrialized countries, indicating the need to understand the role of other sources of infection, including water and ruminants.
In New Zealand, thermophilic Campylobacter strains have been isolated from river water, streams, lakes, ponds, runoff water, and drinking water (26, 27). Savill and coworkers (28) found 60% of 30 recreational water sites were positive, with counts of Campylobacter reaching a most probable number (MPN) of 11 per 100 ml. Between December 1998 and February 2000, a large-scale survey was conducted in 25 freshwater recreational and drinking water supply sites distributed throughout New Zealand (27). This study showed that 60% of the 725 samples were positive for Campylobacter spp. with 48% of the positive samples identified as C. jejuni. Till et al. (19) also estimated that 5% of campylobacteriosis cases could be attributed to recreational water. Based on this freshwater survey and a quantitative risk assessment, new national recreational freshwater quality guidelines were developed in 2003. However, applying these guidelines to recreational rivers is complicated because many factors, including the rate of river flow, land use, animal access to the waterways, and surface runoff, could influence the bacterial risks to human health.
Campylobacter spp. only replicate in animals, and although they are frequently recovered from environmental water, they have not been demonstrated to multiply outside the host (1). Both C. jejuni and C. coli are thermophilic, growing preferentially at 42°C, and microaerophilic, requiring reduced O2 and increased CO2 (1). However, it has been suggested that Campylobacter spp. may persist in the environment by entering a viable but nonculturable (VBNC) state, and/or by forming a monospecies biofilm, colonizing preexisting biofilm (29, 30) or internalized within protozoa (31, 32). Even so, ambient temperatures, high oxygen concentrations, UV radiation, and desiccation may all decrease the survival of Campylobacter in the environment (1, 33–35). Therefore, the presence of Campylobacter in water is likely to indicate recent fecal contamination from either a point source (e.g., meat plant effluent) or nonpoint sources (e.g., agricultural runoff).
Several methodologies, including serotyping and pulsed-field gel electrophoresis (PFGE) have been developed for subtyping of C. jejuni (36, 37). However, multilocus sequence typing (MLST) has good discriminatory power and better reproducibility than other typing methods (38). The use of MLST for typing C. jejuni has also provided important insights into the population genetics of this organism (38, 39) and has helped to increase the understanding of transmission pathways for human campylobacteriosis (40). Diverse sequence types of C. jejuni have been isolated from rivers in New Zealand, and the majority of these sequence types identified as those associated with wild birds (41).
Previous investigations into Campylobacter in rivers showed evidence of seasonal differences in recovery of Campylobacter from water samples (42–44). Here, we report the findings of a longitudinal study of Campylobacter in six high-use recreational waterways in the Manawatu-Wanganui region during 2005 to 2009 (Fig. 1). The primary objectives of this study were (i) to assess the proportions of Campylobacter- and C. jejuni-positive water samples from each of the study sites and (ii) to determine the potential associations between the presence of both Campylobacter spp. and ruminant-associated C. jejuni and explanatory variables such as month, site of sample collection, and river flow rates. In addition, the population genetic structure of C. jejuni was assessed to explore possible animal sources of river-borne isolates and the potential of the strains present to cause human infection.
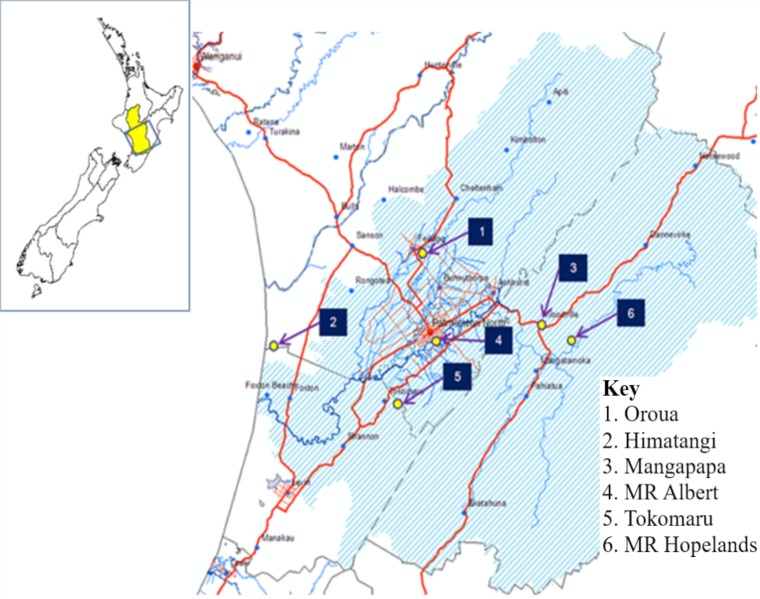
FIG 1.
The recreational swimming water study sampling sites within the Manawatu-Wanganui region of New Zealand. “MR Albert” refers to the Albert Street section of the Manawatu River, and “MR Hopelands” refers to the Hopelands Picnic Reserve section of the Manawatu River.
RESULTS
Thirty-three percent (169/509; 95% confidence interval [CI], 29.1%, 37.5%) of the water samples tested positive for presumptive Campylobacter spp. and 20.4% of samples (104/509; 95% CI, 17.0%, 24.2%) were confirmed by PCR as C. jejuni positive. The breakdown of positive samples across each of the six study sites is shown in Table 1. The proportion of presumptive Campylobacter-positive samples was significantly different between months (χ2 = 83.6, df = 11, P < 0.0001) and between sites (χ2 = 34.6, df = 5, P < 0.0001).
TABLE 1.
Total number of water samples collected from six recreational river sites in the Manawatu-Wanganui region of New Zealand and the numbers and percentages of presumptive Campylobacter spp. and C. jejuni in those samples
| Source site | No. of water samples | No. (%) of positive samples: |
|
|---|---|---|---|
| Presumptive Campylobactera | C. jejunib | ||
| Kaikokopu Stream, Himatangi | 85 | 22 (25.9) | 12 (14.1) |
| Mangapapa Stream | 87 | 45 (51.7) | 29 (33.3) |
| Manawatu River at Albert Street | 87 | 42 (48.3) | 27 (31.0) |
| Manawatu River at Hopelands | 85 | 22 (25.9) | 15 (17.6) |
| Oroua River | 87 | 22 (25.3) | 15 (17.2) |
| Tokomaru River | 78 | 16 (20.5) | 6 (7.7) |
| Total | 509 | 169 (33.2) | 104 (20.4) |
A sample was considered presumptive Campylobacter positive when samples cultured on mCCDA and BA produced colonies with typical Campylobacter morphology and at least one isolate was positive by the Campylobacter genus PCR.
A sample was considered C. jejuni positive when at least one isolate from that sample was confirmed as C. jejuni by PCR.
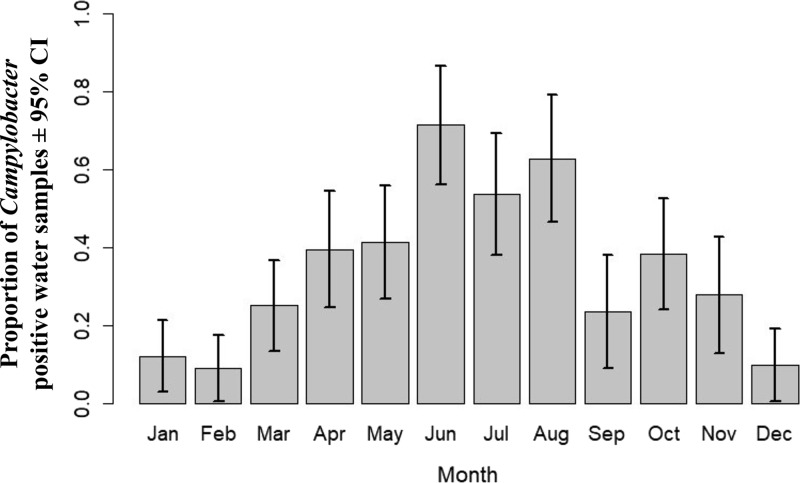
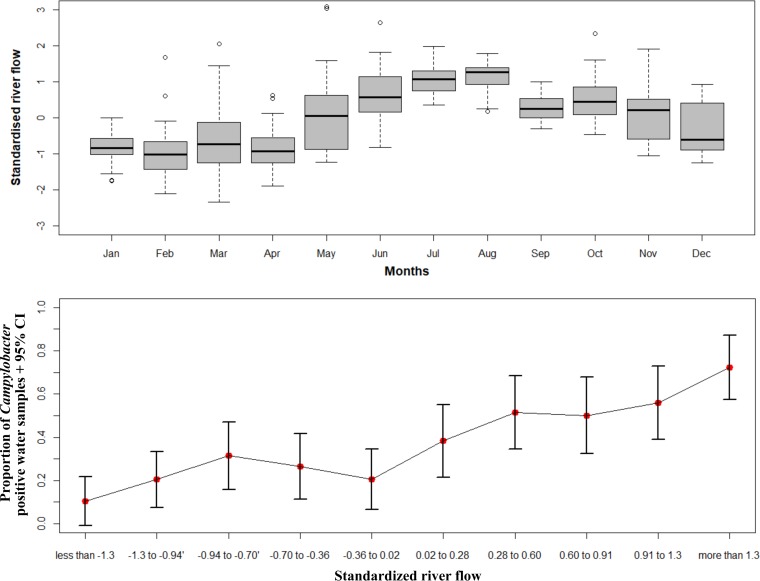
This study found a higher proportion of presumptive Campylobacter-positive samples in the winter months (June, July, and August) compared to the summer months (December, January, and February) (Fig. 2). Similarly, the upper graph in Fig. 3 illustrates that river flow rates were higher in winter months than summer months, with the greatest monthly range of flow rates being seen in March, (i.e., in early autumn). Our data showed higher proportions of presumptive Campylobacter-positive samples when standardized river flows were between the third and fourth deciles and above the fifth deciles compared to first decile (Fig. 3, bottom panel).
FIG 2.
A bar plot showing the monthly variations in presumptive Campylobacter-positive water samples from six recreational water sites in the Manawatu-Wanganui region combining the data collected between December 2005 and April 2009. The vertical lines represent the 95% confidence intervals.
FIG 3.
(Upper graph) Box-and-whiskers plot showing the temporal variation of standardized river flow rates (cubic meters per second) for four recreational water sites in the Manawatu-Wanganui region between December 2005 and April 2009. The top and bottom of the box represent the 25th and 75th percentiles, the horizontal line in the box shows the median value, and the two ends of the whiskers depict the minimum and maximum values of river flow rates. Round dots depict the outliers of the data. (Lower graph) The error bar plot illustrating the variation in proportion of presumptive Campylobacter-positive water samples (dot) across each decile of the standardized river flow rates for the four recreational water sites. The vertical lines represent the 95% confidence intervals.
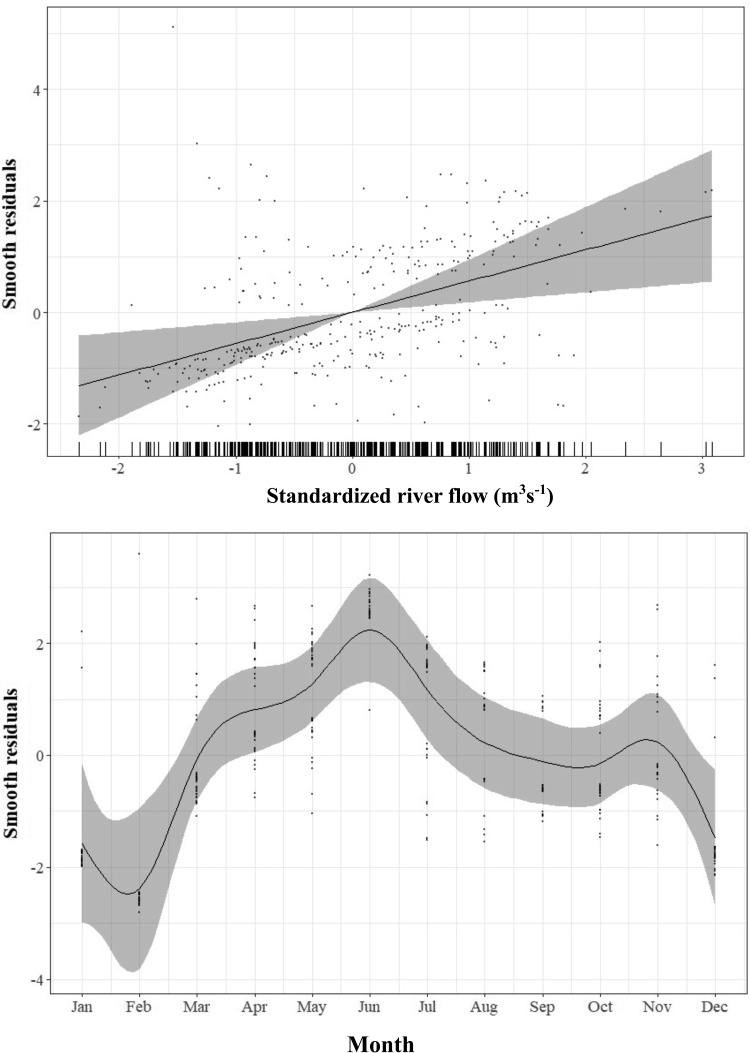
A linear association was observed between the response variable “Campylobacter spp. isolated” and “standardized river flow” (χ2 = 8.61; P = 0.003; estimated df [edf] = 1) using a logistic generalized additive model, when adjusted for the month and the sampling sites (Fig. 4, top panel). On examining the “river sites” variable, the model indicated that there are higher odds of detecting presumptive Campylobacter in water samples from the Mangapapa Stream (odds ratio [OR] = 5.05; 95% CI, 2.3, 11.1) and the Manawatu River at Albert Street (OR = 4.5; 95% CI, 2.05, 9.9) compared to the water samples from the Oroua River.
FIG 4.
A generalized additive model estimate of the probability of Campylobacter spp. in the water samples at different river flow rates (top panel) and in 12 months (bottom panel). The rugs on the x axis in the top panel indicate the individual data points. The line shows the fitted model: gray shading represents the 95% confidence intervals, and dots are the residuals.
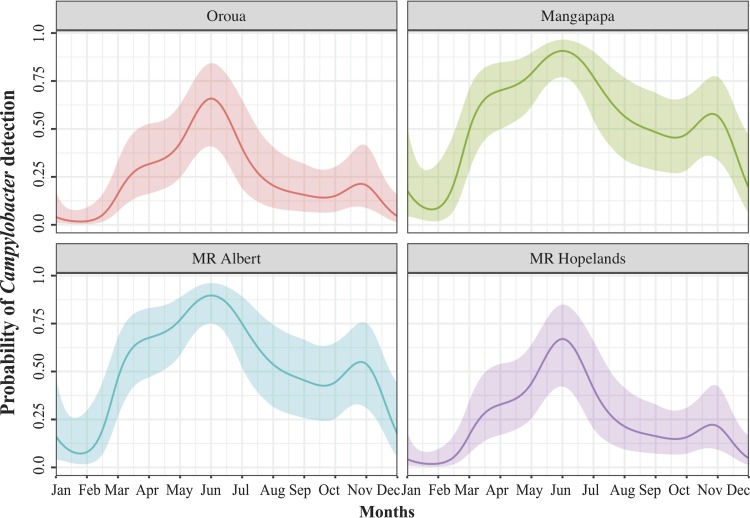
A generalized additive model also indicated a nonlinear relationship between probability of isolating Campylobacter spp. and “month” (χ2 = 44.4; P < 0.0001; edf = 8.37) (Fig. 4, bottom panel). The probability function curves in Fig. 5 show increased probabilities of isolation during the autumn and winter months, April to August. Figure 5 also highlights between-site differences, with lower probabilities of isolating Campylobacter from the Manawatu River at Hopelands and the Oroua River compared to the Mangapapa Stream and the Manawatu River at Albert Street.
FIG 5.
A model-fitted relationship of the probability of detecting presumptive Campylobacter-positive water samples in each month from four rivers, adjusted for site and river flow. The colored solid line with shading represents the mean probability and 95% confidence interval. “MR Albert” refers to the Albert Street section of the Manawatu River, and “MR Hopelands” refers to the Hopelands Picnic Reserve section of the Manawatu River.
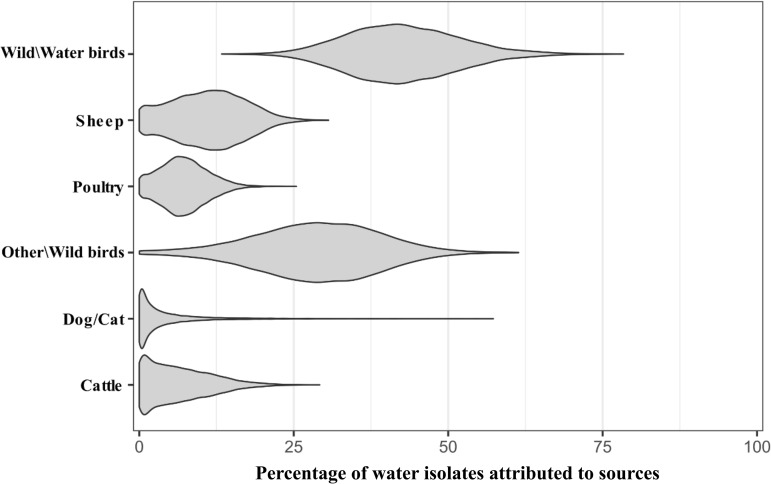
There were 86.2% (100/116) of C. jejuni isolates from 104 samples able to be typed using MLST, of which 91% (91/100) had a full MLST profile. In total, 12 different clonal complexes (CCs) and 49 different sequence types (STs) were identified. Approximately 49.5% (45/91) of the total C. jejuni isolates could not be assigned to a clonal complex (i.e., “undefined” [U/A]). Of the recognized clonal complexes and sequence types, the three most dominant genotypes are highlighted in Table 2. In this study, 90/91 (98.9%) isolates were attributed to various sources such as wild birds, ruminants, and poultry. Of these, 28/90 (31.1%) isolates were likely to be associated with the source “Wild water birds,” 27/90 (30.0%) with “Other wild birds,” 21/90 (23.3%) isolates with “Ruminant,” and 9/90 (10.0%) with “Poultry” when an arbitrary cutoff of 50% (40, 45, 46) was used to define the possible sources of STs (Table 2 and Fig. 6).
TABLE 2.
Clonal complexes, sequence types, housekeeping genes, total number and relative frequency of C. jejuni isolated from six recreational rivers, and most probable source of STs obtained from the island modela
| CC | ST | Housekeeping geneb |
Total no. | Relative frequency (%) | Most probable source of ST (%)c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | |||||
| 21 | 50 | 2 | 1 | 12 | 3 | 2 | 1 | 5 | 1 | 1.1 | Ruminant (73.4) |
| 422 | 2 | 1 | 5 | 3 | 2 | 5 | 5 | 3 | 3.3 | Ruminant (79.1) | |
| 3610 | 2 | 1 | 5 | 88 | 2 | 11 | 5 | 1 | 1.1 | Ruminant (84.9) | |
| 42 | 42 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | 1 | 1.1 | Ruminant (72.2) |
| 3676 | 1 | 307 | 3 | 4 | 5 | 9 | 3 | 1 | 1.1 | Ruminant (77.1) | |
| 45 | 25 | 4 | 7 | 10 | 1 | 1 | 7 | 1 | 1 | 1.1 | Poultry (69.7) |
| 45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | 6 | 7.0 | Ubiquitous | |
| 137 | 4 | 7 | 10 | 4 | 42 | 7 | 1 | 1 | 1.1 | Other wild birds (57.0) | |
| 3802 | 4 | 319 | 10 | 4 | 1 | 7 | 1 | 1 | 1.1 | Ubiquitous | |
| 48 | 38 | 2 | 4 | 2 | 2 | 6 | 1 | 5 | 1 | 1.1 | Ruminant (74.8) |
| 474 | 2 | 4 | 1 | 2 | 2 | 1 | 5 | 2 | 2.2 | Ubiquitous | |
| 61 | 61 | 1 | 4 | 2 | 2 | 6 | 3 | 17 | 1 | 1.1 | Ruminant (78.3) |
| 177 | 177 | 17 | 2 | 8 | 5 | 8 | 2 | 4 | 2 | 2.2 | Other wild birds (64.1) |
| 354 | 1517 | 8 | 10 | 149 | 2 | 11 | 12 | 6 | 1 | 1.1 | Ruminant (80.8) |
| 403 | 2026 | 10 | 1 | 16 | 19 | 10 | 5 | 7 | 1 | 1.1 | Ruminant (69.9) |
| 677 | 677 | 10 | 81 | 50 | 99 | 120 | 76 | 52 | 1 | 1.1 | Ubiquitous |
| 692 | 2584 | 2 | 1 | 57 | 26 | 127 | 29 | 35 | 1 | 1.1 | Ubiquitous |
| 3659 | 37 | 52 | 57 | 26 | 127 | 29 | 1 | 2 | 2.2 | Wild water birds (56.4) | |
| 3664 | 37 | 52 | 4 | 26 | 127 | 29 | 23 | 1 | 1.1 | Wild water birds (82.2) | |
| 1034 | 694 | 2 | 59 | 4 | 105 | 126 | 25 | 23 | 1 | 1.1 | Wild water birds (93.1) |
| 2378 | 2 | 15 | 4 | 48 | 356 | 25 | 23 | 1 | 1.1 | Wild water birds (67.6) | |
| 1275 | 1223 | 27 | 33 | 22 | 49 | 43 | 9 | 31 | 1 | 1.1 | Wild water birds (79.7) |
| 1225 | 27 | 33 | 22 | 49 | 43 | 7 | 31 | 8 | 9.0 | Poultry (72.1) | |
| 3657 | 27 | 33 | 22 | 104 | 134 | 7 | 31 | 1 | 1.1 | Wild water birds (80.0) | |
| 3661 | 27 | 33 | 22 | 49 | 134 | 7 | 31 | 1 | 1.1 | Ubiquitous | |
| 3662 | 27 | 33 | 22 | 49 | 43 | 110 | 31 | 3 | 3.3 | Wild water birds (74.3) | |
| 3674 | 27 | 33 | 22 | 49 | 43 | 350 | 31 | 1 | 1.1 | Wild water birds (79.7) | |
| U/Ad | 436 | 7 | 21 | 5 | 62 | 4 | 61 | 44 | 1 | 1.1 | Ruminant (85.9) |
| 526 | 2 | 15 | 4 | 27 | 13 | 80 | 23 | 1 | 1.1 | Wild water birds (59.6) | |
| 992 | 2 | 59 | 4 | 27 | 126 | 29 | 23 | 1 | 1.1 | Ubiquitous | |
| 996 | 2 | 29 | 84 | 48 | 131 | 25 | 57 | 1 | 1.1 | Wild water birds (95.9) | |
| 1030 | 37 | 4 | 4 | 48 | 13 | 25 | 57 | 1 | 1.1 | Wild water birds (90.9) | |
| 2347 | 2 | 4 | 4 | 105 | 10 | 25 | 57 | 1 | 1.1 | U/A | |
| 2354 | 37 | 4 | 4 | 48 | 13 | 25 | 23 | 1 | 1.1 | Wild water birds (67.5) | |
| 2381 | 175 | 251 | 216 | 282 | 359 | 293 | 102 | 19 | 20.9 | Other wild birds (50.7) | |
| 2619 | 191 | 251 | 216 | 282 | 359 | 293 | 214 | 2 | 2.2 | Wild water birds (60.0) | |
| 3538 | 47 | 2 | 4 | 2 | 6 | 5 | 17 | 1 | 1.1 | Ruminant (84.1) | |
| 3640 | 1 | 6 | 5 | 4 | 261 | 7 | 3 | 3 | 3.3 | Wild water birds (79.8) | |
| 3655 | 1 | 6 | 5 | 282 | 261 | 7 | 3 | 2 | 2.2 | Wild water birds (79.8) | |
| 3656 | 175 | 251 | 216 | 282 | 359 | 293 | 3 | 1 | 1.1 | Wild water birds (79.7) | |
| 3658 | 1 | 295 | 216 | 282 | 359 | 293 | 102 | 1 | 1.1 | Wild water birds (63.0) | |
| 3660 | 192 | 295 | 216 | 282 | 359 | 293 | 102 | 1 | 1.1 | Other wild birds (59.7) | |
| 3663 | 175 | 6 | 216 | 282 | 261 | 7 | 3 | 2 | 2.2 | Other wild birds (68.0) | |
| 3672 | 236 | 306 | 254 | 339 | 433 | 349 | 255 | 1 | 1.1 | Wild water birds (79.7) | |
| 3673 | 175 | 6 | 216 | 4 | 434 | 7 | 3 | 1 | 1.1 | Wild water birds (59.0) | |
| 3675 | 237 | 2 | 254 | 340 | 435 | 349 | 256 | 1 | 1.1 | Other wild birds (68.4) | |
| 3800 | 175 | 6 | 5 | 282 | 261 | 7 | 262 | 1 | 1.1 | Wild water birds (83.4) | |
| 3801 | 175 | 318 | 216 | 282 | 359 | 293 | 102 | 1 | 1.1 | Other wild birds (56.9) | |
| 3803 | 27 | 8 | 34 | 6 | 39 | 88 | 3 | 1 | 1.1 | Wild water birds (87.0) | |
| Total | 91 | 100.0 | |||||||||
The first (shaded), second (boldface), and third (italic) highest frequencies of sequence types obtained in this study are highlighted as described in parentheses.
Housekeeping genes: aspA, aspartase A; glnA, glutamine synthetase; gltA, citrate synthase; glyA, serine hydroxymethyltransferase; pgm, phosphoglucomutase; tkt, transketolase; and uncA, ATP synthase alpha subunit.
“Ubiquitous” indicates that sequence types that were not able to be assigned to a defined probable source as calculated probabilities using the island model were <50%.
U/A, undefined.
FIG 6.
Violin plot showing the overall source attribution (%) of the isolated sequence types of C. jejuni in water. The y axis represents various sources of Campylobacter, in which “Wild\Water birds” represents geese, wild ducks, and pukeko and “Other\Wild birds” represents starlings and other passerines.
The generalized additive model (GAM) showed no significant difference in the likelihood of finding ruminant-associated C. jejuni between months (F = 0.77, edf = 1.0, P = 0.78), when adjusted for all the river sites. However, closer examination of the relationship of ruminant-associated C. jejuni with the river sites showed a higher likelihood of finding ruminant-associated C. jejuni in Mangapapa (OR = 42.41; 95% CI, 2.18, 825.1), Manawatu River at Hopelands (OR = 34.0; 95% CI, 1.36, 849.59), Himatangi (OR = 24.45; 95% CI, 1.04, 574.46), and Manawatu River at Albert Street (OR = 23.41; 95% CI, 1.11, 495.46) than the river at Tokomaru.
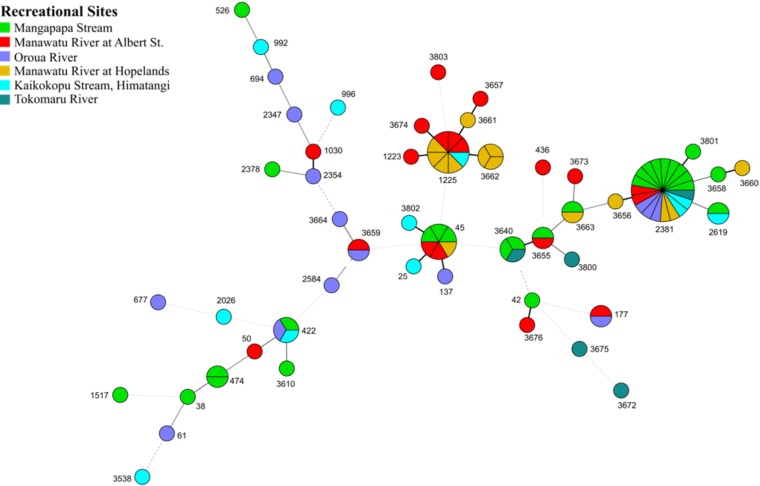
Figure 7 is a minimum-spanning tree showing the genetic relationships between STs of C. jejuni isolates from natural recreational water sites. Four major clusters were found to be related to each other. A distinct larger pie with adjoined smaller pies is a wild bird cluster, with the majority of them being ST-2381, while other smaller clusters of ST-45 and ST-1225 were related to humans and poultry. In addition, there is a widely dispersed cluster of many single isolates of different sequence types. Some of the STs have been found in both animals and humans: for example, ST-42 and ST-61. Not all the STs were isolated from all rivers. For example, ST-2381 was found in all rivers, while ST-61 was found only in Kaikokopu Stream, and ST-474 and ST-42 were present only in the Mangapapa Stream. The wild bird-associated STs were found more frequently in the Mangapapa Stream (10/26) and less frequently in the Manawatu River at Albert Street (2/26).
FIG 7.
Minimum-spanning tree of subtypes of C. jejuni found in the six recreational rivers in the Manawatu region of New Zealand. Each pie chart represents one subtype and the colors represent different recreational swimming sites. Larger pie charts indicate a greater number of isolates of that subtype present. The thicker connecting lines show greater similarities between subtypes. For example, a thick-black solid line between ST-2381 and ST-3656 illustrates that they are different in only one allele of seven housekeeping genes.
DISCUSSION
Data were collected from a 3-year longitudinal study in the Manawatu-Wanganui region to explore the temporal and spatial patterns of isolations of Campylobacter spp. and C. jejuni from river water, as well as the genetic population structure of C. jejuni in recreational water. The isolation of Campylobacter and C. jejuni in water samples from six recreational river sites (Table 1) shows the presence of fecal contamination in the rivers. Although genetic studies revealed that wild birds were the major sources of C. jejuni in river water, the presence of human and ruminant-associated strains indicate the possible dissemination of waterborne diseases via the recreational use of the river, and these risks to human health are highest in the winter months and at times of high river flow. Waterways may also be a vehicle for transmission of Campylobacter to and between animals.
The results of studies in other countries show wide variations in the isolation of Campylobacter from recreational river water. Some of these variations are due to differences in the isolation methods used, and some are related to differences in the river catchments. In New Zealand, Campylobacter isolation rates of 60% have been reported from recreational water (27, 28), while overseas studies report between 0 to 87.5% positive samples (47–49). In this analysis, Campylobacter spp. were found in 33.2% of the water samples and C. jejuni in 20.4% (n = 509). This isolation frequency is relatively low compared with the previous freshwater studies in New Zealand by Savill et al. and Till et al. (27, 28). These two studies were conducted in different geographic regions between 1998 and 2000, before the implementation of national freshwater recreational water quality guidelines (50). These guidelines have provided a driver for the avoidance of stock grazing nearby waterways plus the fencing of land near waterways to reduce direct fecal deposition into the water, which could be an influential factor in the differences in the isolation frequencies between their studies and ours (50). In addition, both Savill’s and Till’s studies utilized a most probable number method to detect and quantify the Campylobacter spp. in water samples, while in our study, Campylobacter was detected using selective media culture and PCR techniques (27, 28). Moore indicated that a direct PCR assay is a better method for detecting Campylobacter in water than traditional culture techniques due to its ability to detect viable but nonculturable (VBNC) Campylobacter in water (49). It is important to detect organisms in a VBNC state as they have been demonstrated to be able to revert to an infective state (29) and thus may present a potential risk for humans.
Our analysis has shown a marked monthly seasonal variation with a distinct peak in June (70%) (Fig. 2) and a second, marginally consistent peak in July and August. The regression analyses also showed a higher likelihood of obtaining Campylobacter spp. in June compared to January. This result is similar to that reported by studies that demonstrated a higher prevalence of Campylobacter spp. in winter months in rivers in the United Kingdom (43, 51). Therefore, it is likely that other sources are the cause of human campylobacteriosis during summer, as the peaks of Campylobacter in our study are discordant with the seasonal peak of human campylobacteriosis cases, which is in summer, in New Zealand (9, 51). These seasonal differences in Campylobacter isolation could be related to factors such as larger amounts of rainfall increasing the agricultural runoff during winter months (52) and the inability of Campylobacter spp. to survive in water in the summer months due to greater levels of UV radiation (35). A relationship between ruminant-associated C. jejuni and month was not established, though it should be noted that this analysis has relatively low study power (n = 70).
Heavy rainfall events initiate agriculture runoff, which leads to water contamination and increased river flow (52). In New Zealand, it is evident that the heaviest rainfall occurs during winter months (53), which would account for the higher river flows seen during those months (Fig. 3). Our analyses suggest that the likelihood of retrieving Campylobacter-positive water samples are mostly higher when rivers have flood flows rather than base flows, and runoff may be a cause of Campylobacter contamination in water. Contrary to our findings, Eyles et al. reported two main peaks of Campylobacter flux: one during high flow with moderate Campylobacter levels and another when flow is low but with high Campylobacter levels (54). They suggested that the abundance of Campylobacter in water during summer when river flow is low could be due to continuous fecal contamination of water (54). Nevertheless, it implies that high river flow is likely to be associated with increased Campylobacter contamination in river water.
Our study also identified that there were significant differences in the probability of obtaining Campylobacter-positive samples between different sampling sites. The lowest occurrences of Campylobacter were found in the Tokomaru River and the highest in the Mangapapa Stream. The generalized additive model also showed the variation in the Campylobacter distribution between sites. Other researchers have also observed the variations in the presence of Campylobacter spp. between locations and have suggested that these variations are attributed to differences between the catchments. For example, Kemp et al. examined the possible risk factors for the presence of Campylobacter spp. in water and found that soil types and farm types in the catchments are significant contributors to the differences (48). A large-scale freshwater study conducted in New Zealand over 15 months reported higher concentrations of Campylobacter spp. in water obtained from catchments containing ruminants than from other catchments (27). Among our study sites, more than 60% of the land in the Mangapapa catchment had been utilized for sheep, beef, and dairy farming, while the Tokomaru catchment contained more bush than farmland (Agribase, AsureQuality, 2012). As well as a higher incidence of Campylobacter spp., there was also a higher likelihood of finding ruminant-associated Campylobacter in the Mangapapa Stream (OR, 42.41). The two predominant genotypes isolated from human campylobacteriosis samples in the 2016 Havelock North outbreak were also able to be isolated from sheep in paddocks near the substandard drinking water bore (12) indicating that ruminant Campylobacter can be a potent source of human campylobacteriosis. This supports the hypothesis that land use within a river catchment needs to be considered when conducting risk assessments and fitting prediction models for the presence of Campylobacter in water.
In this study, 91 species-specific C. jejuni PCR-positive isolates from the six recreational waters were assigned to sequence types using multilocus sequence typing. Our results indicate the presence of diverse C. jejuni subtypes in recreational water. A large percentage of these isolates (49.5%) were assigned to unknown clonal complexes that also contain ST-2381, the most prevalent sequence type (21% of 91) recovered from all rivers. This result provides evidence that the ST-2381 is prevalent in the majority of New Zealand’s river water. This subtype has only been isolated from the native wild birds pukeko and takahē, but not from human cases of campylobacteriosis, implying that ST-2381 is possibly nonpathogenic to humans (55). In this study, 61.1% of the total sequence types recovered are STs that are known to be associated with wild birds (http://pubmlst.org/campylobacter/), which provides evidence that recreational water contamination is often due to the wildlife inhabiting the area, findings similar to those of Mughini-Gras et al. (56). However, the presence of C. jejuni belonging to CC-21, CC-42, CC-45, CC-48, CC-61, CC-177, CC-1517, CC-2026, and CC-677 represents the possibility of zoonotic transmission, because many sequence types of these clonal complexes have been isolated from human campylobacteriosis outbreaks such as the 2016 Havelock North outbreak, where members of CC-21 and CC-42 made up 72% of the isolates from human cases (12). Interrogation of the PubMLST website (http://pubmlst.org/campylobacter/) also shows evidence of these clonal complexes having been isolated from sporadic cases of human Campylobacter infections, ruminants, poultry, dogs, and various meat samples (lamb, chicken, and beef) in the United Kingdom, Canada, and the Netherlands (http://pubmlst.org/campylobacter/).
The geographic isolation and unique physical features of New Zealand might be responsible for the fact that ST-3656, ST-3659, ST-3664, and ST-2381 have only been isolated from New Zealand waters, as reported in the Campylobacter PubMLST database. Possibly, genetic mutation and/or recombination of C. jejuni might have led to a new sequence type arising in New Zealand (55) or, alternatively, these sequence types may either not have been present at the time of sampling, or were present only in very low numbers and thus unidentified in water samples from other countries. Further investigation is required before making any conclusive findings.
The population structure of C. jejuni from swimming sites demonstrated the dominance of wild bird strains of C. jejuni in these waters. Nonetheless, findings of livestock-associated strains of C. jejuni suggest that cattle and sheep may contribute to water contamination, and isolation of strains associated with human infections in the same region demonstrates that there is a risk of contracting campylobacteriosis from swimming in natural freshwaters. This study also highlights the spatial and temporal variations and effect of river flows in isolating Campylobacter spp. from recreational river water in the Manawatu-Wanganui region. The model will be useful to drinking water suppliers for generating an effective treatment plan by including measurements of river flow to aid in the determination of the risks of fecal contamination in the source water. In addition, public health officers can use this model to mitigate the Campylobacter exposure by assessing the risks, times, and sources of potential water contamination.
MATERIALS AND METHODS
Sources of Campylobacter data.
Data used for this study were obtained from the Manawatu Sentinel Surveillance Program New Zealand (MSSP). During the MSSP, water samples were collected from six waterways identified as high-use recreational swimming sites by the Horizons Regional Council. These waterways were the Mangapapa Stream, Woodville; Manawatu River, Hopelands Picnic Reserve, Hopelands; Oroua River, Timona Park, Feilding; Manawatu River, Albert Street, Palmerston North; Tokomaru River, Horseshoe Bend, Tokomaru; and Kaikokopu Stream, Himatangi Beach (Fig. 1). The rate of river flow for each sampling occasion was obtained from the Horizons Regional Council’s automatic flow recording system. There were no river flow data available for Tokomaru River and Kaikokopu Stream. In total, 509 water samples were collected over 40 months between December 2005 and April 2009, with one sample being taken per visit and one to two visits being made to each site per month.
Water samples were collected in 100-ml sterile bottles fortnightly from each site. The samples were transported in a cool box within 1 h to the Molecular Epidemiology and Public Health Laboratory (mEpiLab) at the Hopkirk Research Institute, Massey University. Campylobacter spp. were isolated from the water samples by using microaerobic culture techniques as previously described (24). The presence of grayish, flat, and moistened colonies on mCCDA (Fort Richard, Auckland, NZ) was considered confirmation of a presumptive Campylobacter-positive sample. From each of these samples, up to five colonies were selected for C. jejuni identification using a mapA gene PCR adapted from Stucki et al. and as described by Mullner et al. (24, 57). Isolates negative by the mapA PCR were tested by the Campylobacter genus PCR of Linton et al. (58). Those isolates confirmed as C. jejuni were typed by multilocus sequence typing based on the previously published method (38). The multilocus sequence typing alleles, sequence types, and clonal complexes (CCs) were assigned using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/) developed by Keith Jolley and hosted by the University of Oxford. Ruminant-associated C. jejuni data were obtained after running the island model as described by Wilson et al. (40).
Statistical analysis.
Two response variables were assessed during the analyses: the presence or absence of presumptive Campylobacter spp. within a water sample in general and the probability of being ruminant-associated C. jejuni in particular. The variables available within the MSSP database are shown in Table 3. Due to the different data available for the sampling sites, two different data sets were analyzed. Data set A comprised available information on all six sites and was used for descriptive analyses, whereas data set B contained only the four sites with river flow data and was used for estimating the magnitude of the relationship between variables using a generalized additive model (GAM). The descriptive analysis and GAM were conducted in R version 3.2.3 (R Development Core Team, 2011).
TABLE 3.
Information used for building logistic regression models to identify the relationship between the isolation of Campylobacter spp. from freshwater samples and various covariates
| Variable | Type of variable | Presence in: |
|
|---|---|---|---|
| Data set Aa | Data set Bb | ||
| Standardized river flow rate (m3 s−1) | Continuous | No | Yes |
| Source site | Categorical | ||
| Kaikokopu Stream, Himatangi | Yes | Noc | |
| Mangapapa Stream | Yes | Yes | |
| Manawatu River at Hopelands | Yes | Yes | |
| Manawatu River at Albert Street | Yes | Yes | |
| Oroua River | Yes | Yes | |
| Tokomaru River | Yes | Nod | |
| Months | Ordinal | ||
| January to December | Yes | Yes | |
| Presumptive Campylobacter | Dichotomous | ||
| Presence or absence | Yes | Yes | |
Data set A contained 509 water samples.
Data set B contained 344 water samples.
River flow data were not available for the Himatangi sampling sites.
River flow data were not available for the Tokomaru sampling sites.
Mean base flow rate of four rivers at the time of sampling was not normally distributed, and it varied considerably between sites and within a site over time. Therefore, the flow rates were log-transformed to normalize the data, and the log-transformed flow data were scaled using the Z-score to reduce the effect of between-site variations, thereby obtaining the standardized flow data (m3 s−1).
The percentage of water samples that tested positive for presumptive Campylobacter and C. jejuni were calculated for each site along with confidence intervals calculated using the formula described by Fleiss (59). Pearson’s chi-square tests were also performed to determine if there was any variation in distribution of presumptive Campylobacter-positive samples across the sampling sites and months. A box plot and an error bar plot were produced to show variation in the proportion of water samples positive for presumptive Campylobacter across months and standardized river flow rates, respectively. The box-and-whisker plot was produced to investigate the temporal variation of standardized river flow rates for four river sites.
We fitted a logistic generalized additive model (GAM) to evaluate the relationship of presence or absence of presumptive Campylobacter spp. in water samples with predictor variables, month, standardized river flow, and sample source sites. GAM employs a link function to relate each linear predictor to the mean response, as shown in equation 1
| (1) |
where θi is the mean response of the linear predictor and Yi is the response variable (1 for positive, 0 for negative) for each observation i, sourcei is the water sampling site, and fM and fF are smooth trends for month and flow effects, respectively. In this model, a logistic link and a binomial error term were used, and we assessed each variable’s contribution to explaining residual deviance through addition or dropping of each variable.
The predicted probability of detecting presumptive Campylobacter-positive water samples in each month from four source sites may then be computed as described by Hosmer and Lemeshow (60), using the probability of isolating Campylobacter (P), as shown in equation 2:
| (2) |
All food, animal, water, and environmental isolates in the MSSP C. jejuni surveillance database were used to estimate the ruminant-associated C. jejuni in our data. The food and environmental isolates in the database were classified into six sources: cattle, sheep, wild water birds (geese, wild ducks, and pukeko), other wild birds (starlings and other passerines), dogs or cats, and poultry (broiler chickens, spent layer hens, farmed ducks, and turkeys). Then, the asymmetric island model (40) was run using the iSource program (http://www.danielwilson.me.uk/iSource.html) to attribute water isolates to their most likely source, with the model providing estimates of the probability that each ST found in water came from one of the six sources. The probability that each ST was ruminant associated was determined by the sum of the probabilities that the ST came from either cattle or sheep. These proportions of C. jejuni attributable to ruminant origin were logit transformed and used as a response variable in the model. The relationship between ruminant-associated strains of C. jejuni and month of the year, adjusted for all the river sites, was investigated using a generalized additive model similar to that described above, with the outcome variable logit(rumi) related to θi, as shown in equation 3, via
| (3) |
where “rum” is “ruminant” and σ2 describes the variance of residuals.
C. jejuni population structure analysis.
Phylogenetic relationships between the C. jejuni isolates were described using a Kruskal’s algorithm-based minimum-spanning tree in BioNumerics version 6.1 software (Applied Maths NV, Sint-Martens-Latem, Belgium).
ACKNOWLEDGMENTS
The authors wish to acknowledge all the members of the Molecular Epidemiology and Public Health Laboratory (mEpiLab) Team and Horizons Regional Council for their valuable contributions in generating the data for the Manawatu Sentinel Surveillance Program. We also wish to thank the reviewers, whose suggestions have helped improve this paper.
This publication made use of the Campylobacter Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/), developed by Keith Jolley and sited at the University of Oxford (61). The development of this site has been funded by the Wellcome Trust.
REFERENCES
- 1.Nachamkin I, Szymanski CM, Blaser MJ (ed). 2008. Campylobacter. ASM Press, Washington, DC. [Google Scholar]
- 2.Cotruvo JA, Dufour A, Rees G, Bartram J, Carr R, Cliver DO, Craun GF, Fayer R, Gannon VPJ (ed). 2004. Waterborne zoonoses: identification, causes and control. IWA Publishing, London, United Kingdom. [Google Scholar]
- 3.Stanley K, Jones K. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J Appl Microbiol 94Suppl:104S–113S. doi: 10.1046/j.1365-2672.94.s1.12.x. [DOI] [PubMed] [Google Scholar]
- 4.French NP, Midwinter A, Holland B, Collins-Emerson J, Pattison R, Colles F, Carter P. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. J Appl Environ Microbiol 75:779–783. doi: 10.1128/AEM.01979-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong T, On S, Michie H. 2006. Campylobacter in New Zealand: reservoirs, sources and the labyrinth of transmission routes. N Z J Environ Health 29:1–6. [Google Scholar]
- 6.Corry JEL, Atabay HI. 2001. Poultry as a source of Campylobacter and related organisms. J Appl Microbiol 90:96S–114S. doi: 10.1046/j.1365-2672.2001.01358.x. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, Yamamoto S. 2009. Campylobacter contamination in retail poultry meats and by-products in the world: a literature survey. J Vet Med Sci 71:255–261. doi: 10.1292/jvms.71.255. [DOI] [PubMed] [Google Scholar]
- 8.Skarp C, Hänninen M-L, Rautelin H. 2016. Campylobacteriosis: the role of poultry meat. Clin Microbiol Infect 22:103–109. doi: 10.1016/j.cmi.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 9.ESR. 2011. Notifiable and other diseases in New Zealand: Annual Report 2010. Institute of Environmental Science and Research Ltd, Porirua, New Zealand. [Google Scholar]
- 10.Samuel MC, Vugia DJ, Shallow S, Marcus R, Segler S, McGivern T, Kassenborg H, Reilly K, Kennedy M, Angulo F. 2004. Epidemiology of sporadic Campylobacter infection in the United States and declining trend in incidence, FoodNet 1996–1999. Clin Infect Dis 38:S165. doi: 10.1086/381583. [DOI] [PubMed] [Google Scholar]
- 11.ESR. 2016. New Zealand Public Health Surveillance Report, December 2016. Institute of Environmental Science and Research Ltd, Porirua, New Zealand. [Google Scholar]
- 12.Gilpin B, ESR . 2016. Analysis of water, sediment, and animal faecal samples from Havelock North in August and September 2016. Institute of Environmental Science and Research Ltd, Porirua, New Zealand. [Google Scholar]
- 13.Evans M, Roberts R, Ribeiro C, Gardner D, Kembrey D. 1996. A milk-borne Campylobacter outbreak following an educational farm visit. Epidemiol Infect 117:457–462. doi: 10.1017/s0950268800059112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost JA, Gillespie IA, O'Brien SJ. 2002. Public health implications of Campylobacter outbreaks in England and Wales, 1995–9: epidemiological and microbiological investigations. Epidemiol Infect 128:111–118. doi: 10.1017/s0950268802006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schönberg-Norio D, Takkinen J, Hänninen ML, Katila ML, Kaukoranta SS, Mattila L, Rautelin H. 2004. Swimming and Campylobacter infections. J Emerg Infect Dis 10:1474–1477. doi: 10.3201/eid1008.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson N. 2005. A systematic review of the aetiology of human campylobacteriosis in New Zealand: report to the Food Safety Authority of New Zealand. New Zealand Food Safety Authority, Wellington, New Zealand. [Google Scholar]
- 17.Baker M, Wilson N, Edwards R. 2007. Campylobacter infection and chicken: an update on New Zealand’s largest ‘common source outbreak.’ N Z Med J 120:U2717. [PubMed] [Google Scholar]
- 18.Muellner P, Marshall JC, Spencer SEF, Noble AD, Shadbolt T, Collins-Emerson JM, Midwinter AC, Carter PE, Pirie R, Wilson DJ, Campbell DM, Stevenson MA, French NP. 2011. Utilizing a combination of molecular and spatial tools to assess the effect of a public health intervention. Prev Vet Med 102:242–253. doi: 10.1016/j.prevetmed.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Baker M, Sneyd E, Wilson N. 2007. Is the major increase in notified campylobacteriosis in New Zealand real? Epidemiol Infect 135:163–170. doi: 10.1017/S0950268806006583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ESR. 2007. Notifiable and other diseases in New Zealand Annual Report. Institute of Environmental Science and Research Ltd, Porirua, New Zealand. [Google Scholar]
- 21.NZFSA, Applied Economics Pty Ltd. 2010. The economic cost of foodborne disease in New Zealand. Applied Economics Pty Ltd, Sydney, Australia. [Google Scholar]
- 22.Scott WG, Scott HM, Lake RJ, Baker MG. 2000. Economic cost to New Zealand of foodborne infectious disease. N Z Med J 113:281–284. [PubMed] [Google Scholar]
- 23.Baker MG, Kvalsvig A, Zhang J, Lake R, Sears A, Wilson N. 2012. Declining Guillain-Barré syndrome after campylobacteriosis control, New Zealand, 1988–2010. Emerg Infect Dis 18:226. doi: 10.3201/eid1802.111126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullner P, Jones G, Noble A, Spencer SEF, Hathaway S, French NP. 2009. Source attribution of food borne zoonoses in New Zealand: a modified Hald model. Risk Anal 29:970–984. doi: 10.1111/j.1539-6924.2009.01224.x. [DOI] [PubMed] [Google Scholar]
- 25.Sears A, Baker MG, Wilson N, Marshall J, Muellner P, Campbell DM, Lake RJ, French NP. 2011. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. J. Emerg Infect Dis 17:1007–1015. doi: 10.3201/eid/1706.101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenraad P, Rombouts F, Notermans S. 1997. Epidemiological aspects of thermophilic Campylobacter in water-related environments: a review. Water Environ Res 69:52–63. doi: 10.2175/106143097X125182. [DOI] [Google Scholar]
- 27.Till D, McBride G, Ball A, Taylor K, Pyle E. 2008. Large-scale freshwater microbiological study: rationale, results and risks. J Water Health 6:443–460. doi: 10.2166/wh.2008.071. [DOI] [PubMed] [Google Scholar]
- 28.Savill MG, Hudson JA, Ball A, Klena JD, Scholes P, Whyte RJ, McCormick RE, Jankovic D. 2001. Enumeration of Campylobacter in New Zealand recreational and drinking waters. J Appl Microbiol 91:38–46. doi: 10.1046/j.1365-2672.2001.01337.x. [DOI] [PubMed] [Google Scholar]
- 29.Chaisowwong W, Kusumoto A, Hashimoto M, Harada T, Maklon K, Kawamoto K. 2012. Physiological characterization of Campylobacter jejuni under cold stress conditions: its potential for public threat. J Vet Med Sci 74:43–50. doi: 10.1292/jvms.11-0305. [DOI] [PubMed] [Google Scholar]
- 30.Reuter M, Mallett A, Pearson BM, van Vliet A. 2010. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol 76:2122–2128. doi: 10.1128/AEM.01878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Axelsson-Olsson D, Waldenström J, Broman T, Olsen B, Holmberg M. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl Environ Microbiol 71:987–992. doi: 10.1128/AEM.71.2.987-992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trigui H, Paquet VE, Charette SJ, Faucher SP. 2016. Packaging of Campylobacter jejuni into multilamellar bodies by the ciliate Tetrahymena pyriformis. Appl Environ Microbiol 82:2783–2790. doi: 10.1128/AEM.03921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inglis GD, McAllister TA, Larney FJ, Topp E. 2010. Prolonged survival of Campylobacter species in bovine manure compost. Appl Environ Microbiol 76:1110–1119. doi: 10.1128/AEM.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rollins D, Colwell R. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. J Appl Environ Microbiol 52:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obiri-Danso K, Paul N, Jones K. 2001. The effects of UVB and temperature on the survival of natural populations and pure cultures of Campylobacter jejuni, Camp. coli, Camp. lari and urease-positive thermophilic campylobacters (UPTC) in surface waters. J Appl Microbiol 90:256–267. doi: 10.1046/j.1365-2672.2001.01239.x. [DOI] [PubMed] [Google Scholar]
- 36.Lorenz E, Lastovica A, Owen RJ. 1998. Subtyping of Campylobacter jejuni Penner serotypes 9, 38 and 63 from human infections, animals and water by pulsed field gel electrophoresis and flagellin gene analysis. Lett Appl Microbiol 26:179–182. doi: 10.1046/j.1472-765x.1998.00311.x. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen EM, Engberg J, Fussing V, Petersen L, Brogren CH, On S. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J Clin Microbiol 38:3800–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJL, Urwin R, Maiden M. 2001. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingle KE, Colles FM, Falush D, Maiden M. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol 43:340–347. doi: 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet 4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter PE, McTavish SM, Brooks HJL, Campbell D, Collins-Emerson JM, Midwinter AC, French NP. 2009. Novel clonal complexes with an unknown animal reservoir dominate Campylobacter jejuni isolates from river water in New Zealand. Appl Environ Microbiol 75:6038–6046. doi: 10.1128/AEM.01039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones K. 2001. Campylobacters in water, sewage and the environment. J Appl Microbiol 90:68S–79S. doi: 10.1046/j.1365-2672.2001.01355.x. [DOI] [PubMed] [Google Scholar]
- 43.Obiri-Danso K, Jones K. 1999. Distribution and seasonality of microbial indicators and thermophilic campylobacters in two freshwater bathing sites on the River Lune in northwest England. J Appl Microbiol 87:822–832. doi: 10.1046/j.1365-2672.1999.00924.x. [DOI] [PubMed] [Google Scholar]
- 44.Vereen E Jr, Lowrance RR, Cole DJ, Lipp EK. 2007. Distribution and ecology of campylobacters in coastal plain streams (Georgia, United States of America). Appl Environ Microbiol 73:1395–1403. doi: 10.1128/AEM.01621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gras LM, Smid JH, Wagenaar JA, de Boer AG, Havelaar AH, Friesema IHM, French NP, Busani L, Pelt WV. 2012. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: a combined case-control and source attribution analysis. PLoS One 7:e42599. doi: 10.1371/journal.pone.0042599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müllner P, Spencer S, Wilson D, Jones G, Noble A, Midwinter A, Collins-Emerson JM, Carter P, Hathaway S, French NP. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Genet Evol 9:1311–1319. doi: 10.1016/j.meegid.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Van Dyke M, Morton V, McLellan N, Huck P. 2010. The occurrence of Campylobacter in river water and waterfowl within a watershed in southern Ontario, Canada. J Appl Microbiol 109:1053–1066. doi: 10.1111/j.1365-2672.2010.04730.x. [DOI] [PubMed] [Google Scholar]
- 48.Kemp R, Leatherbarrow A, Williams N, Hart C, Clough H, Turner J, Wright E, French N. 2005. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl Environ Microbiol 71:1876–1882. doi: 10.1128/AEM.71.4.1876-1882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore J, Caldwell P, Millar B. 2001. Molecular detection of Campylobacter spp. in drinking, recreational and environmental water supplies. Int J Hyg Environ Health 204:185–189. doi: 10.1078/1438-4639-00096. [DOI] [PubMed] [Google Scholar]
- 50.MfE/MoH. 2003. Microbiological water quality guidelines for marine and freshwater recreational areas. Ministry for the Environment and Ministry of Health, Wellington, New Zealand. [Google Scholar]
- 51.Abulreesh HH, Paget TA, Goulder R. 2006. Campylobacter in waterfowl and aquatic environments: incidence and methods of detection. Environ Sci Technol 40:7122–7131. doi: 10.1021/es060327l. [DOI] [PubMed] [Google Scholar]
- 52.Roig B, Delpla I, Baurès E, Jung AV, Thomas O. 2011. Analytical issues in monitoring drinking-water contamination related to short-term, heavy rainfall events. Trends Analyt Chem 30:1243–1251. doi: 10.1016/j.trac.2011.04.008. [DOI] [Google Scholar]
- 53.NIWA. 2010. New Zealand River Environment Classification Database. Ministry for the Environment, Wellington, New Zealand. [Google Scholar]
- 54.Eyles R, Niyogi D, Townsend C, Benwell G, Weinstein P. 2003. Spatial and temporal patterns of contamination underlying public health risk in the Taieri River, New Zealand. J Environ Qual 32:1820–1828. doi: 10.2134/jeq2003.1820. [DOI] [PubMed] [Google Scholar]
- 55.French N, Marshall J, Mohan V. 2011. New and emerging data on typing of Campylobacter spp. strains in animals, environmental matrices and humans. Technical report. Ministry of Agriculture and Forestry, Wellington, New Zealand. [Google Scholar]
- 56.Mughini-Gras L, Penny C, Ragimbeau C, Schets FM, Blaak H, Duim B, Wagenaar JA, de Boer A, Cauchie H-M, Mossong J, van Pelt W. 2016. Quantifying potential sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res 101:36–45. doi: 10.1016/j.watres.2016.05.069. [DOI] [PubMed] [Google Scholar]
- 57.Stucki U, Frey J, Nicolet J, Burnens AP. 1995. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J Clin Microbiol 33:855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linton D, Owen RJ, Stanley J. 1996. Rapid identification by PCR of the genus Campylobacter and five Campylobacter species enteropathogenic for man and animals. Res Microbiol 147:707–718. doi: 10.1016/s0923-2508(97)85118-2. [DOI] [PubMed] [Google Scholar]
- 59.Fleiss JL. 1981. Balanced incomplete block designs for inter-rater reliability studies. Appl Psychol Meas 5:105–112. doi: 10.1177/014662168100500115. [DOI] [Google Scholar]
- 60.Hosmer DW, Lemeshow S. 2000. Applied logistic regression, 2nd ed. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 61.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]