Recently, coevolutionary theory in macroorganisms has been advanced by the geographic mosaic theory of coevolution (GMC), which considers how geography and local adaptation shape coevolutionary dynamics. Here, we test GMC in an ancient symbiosis in which the ant Apterostigma dentigerum cultivates fungi in an agricultural system analogous to human farming. The cultivars are parasitized by the fungus Escovopsis. The ants maintain symbiotic actinobacteria with antibiotic properties that help combat Escovopsis infection. This antibiotic symbiosis has persisted for tens of millions of years, raising the question of how antibiotic potency is maintained over these time scales. Our study tests the GMC in a bacterial defensive symbiosis and in a multipartite symbiosis framework. Our results show that this multipartite symbiotic system conforms to the GMC and demonstrate that this theory is applicable in both microbes and indirect symbiont-symbiont interactions.
KEYWORDS: coevolution, geographic mosaic theory of coevolution, secondary metabolism
ABSTRACT
The geographic mosaic theory of coevolution (GMC) posits that coevolutionary dynamics go beyond local coevolution and are comprised of the following three components: geographic selection mosaics, coevolutionary hot spots, and trait remixing. It is unclear whether the GMC applies to bacteria, as horizontal gene transfer and cosmopolitan dispersal may violate theoretical assumptions. Here, we test key GMC predictions in an antibiotic-producing bacterial symbiont (genus Pseudonocardia) that protects the crops of neotropical fungus-farming ants (Apterostigma dentigerum) from a specialized pathogen (genus Escovopsis). We found that Pseudonocardia antibiotic inhibition of common Escovopsis pathogens was elevated in A. dentigerum colonies from Panama compared to those from Costa Rica. Furthermore, a Panama Canal Zone population of Pseudonocardia on Barro Colorado Island (BCI) was locally adapted, whereas two neighboring populations were not, consistent with a GMC-predicted selection mosaic and a hot spot of adaptation surrounded by areas of maladaptation. Maladaptation was shaped by incongruent Pseudonocardia-Escovopsis population genetic structure, whereas local adaptation was facilitated by geographic isolation on BCI after the flooding of the Panama Canal. Genomic assessments of antibiotic potential of 29 Pseudonocardia strains identified diverse and unique biosynthetic gene clusters in BCI strains despite low genetic diversity in the core genome. The strength of antibiotic inhibition was not correlated with the presence/absence of individual biosynthetic gene clusters or with parasite location. Rather, biosynthetic gene clusters have undergone selective sweeps, suggesting that the trait remixing dynamics conferring the long-term maintenance of antibiotic potency rely on evolutionary genetic changes within already-present biosynthetic gene clusters and not simply on the horizontal acquisition of novel genetic elements or pathways.
IMPORTANCE Recently, coevolutionary theory in macroorganisms has been advanced by the geographic mosaic theory of coevolution (GMC), which considers how geography and local adaptation shape coevolutionary dynamics. Here, we test GMC in an ancient symbiosis in which the ant Apterostigma dentigerum cultivates fungi in an agricultural system analogous to human farming. The cultivars are parasitized by the fungus Escovopsis. The ants maintain symbiotic actinobacteria with antibiotic properties that help combat Escovopsis infection. This antibiotic symbiosis has persisted for tens of millions of years, raising the question of how antibiotic potency is maintained over these time scales. Our study tests the GMC in a bacterial defensive symbiosis and in a multipartite symbiosis framework. Our results show that this multipartite symbiotic system conforms to the GMC and demonstrate that this theory is applicable in both microbes and indirect symbiont-symbiont interactions.
INTRODUCTION
The diversity of life is shaped by coevolution; the multitude of species on Earth engage in complex webs of antagonistic and mutualistic interactions that undergo reciprocal selection. Coevolution with symbiotic (i.e., “living together”) microorganisms can result in extraordinary evolutionary innovation, including the origin of eukaryotic cells (1), chloroplasts (2), fungal agriculture (3), and emergent properties (4, 5). Despite the ubiquity of microbial symbionts in nature, the coevolutionary and ecological theory to describe microbial interaction dynamics lags in comparison to the established theory surrounding macroorganisms. Indeed, the extent to which established ecological and evolutionary theory applies to microbes (and the extent to which microbes violate macroorganism-specific assumptions) remains debated (6). The evolution of local adaptation, for example, requires dispersal limitations because intraspecific gene flow from adjacent populations can disrupt local selection (7). The potential for microbial cosmopolitan dispersal and promiscuous horizontal gene transfer may require microbe-specific theories.
Coevolutionary theory has made recent advances through the geographic mosaic theory of coevolution (GMC) (8), a framework that goes beyond the traditionally localized view of coevolution. The GMC posits that coevolutionary dynamics are shaped by the following three geographic-scale components: selection mosaics, coevolutionary hot spots, and trait remixing (9, 10). GMC has also advanced coevolutionary theory by abandoning a common view that pairwise interactions are the primary drivers of reciprocal selection. Rather, it takes the perspective that seemingly “diffuse” multispecies interactions can be partitioned into subsets of fixed species-level interactions or variable population-level coevolved interactions. Whether the GMC is applicable to microbial interactions is unknown, as Thompson noted, “We do not know the extent to which microbes undergo geographic differentiation” (8).
The majority of empirical coevolutionary studies (including those surrounding the GMC) have focused on predator-prey relationships, host-symbiont interactions, or herbivory (11–14). However, studies examining the facilitating dynamics of symbiont-symbiont interactions are lacking despite their widespread ecological importance across biology (15). Symbiont-symbiont interactions within defensive symbioses are of particular interest, as the nature of these microbial communities offers a more structured framework to test local community dynamics (compared to more “open” systems, where the dispersal, inheritance, and specificity of interactions are often difficult to elucidate). Moreover, symbiont-symbiont interactions can indirectly impact hosts (e.g., by increasing disease virulence or shifting transmission specificity); understanding these ecological and evolutionary consequences is necessary for determining the forces shaping broader host-symbiont coevolution (15). The Apterostigma dentigerum fungus-growing ant–microbe symbiosis provides a unique opportunity to test components of the GMC among multiple microbial symbionts known to have geographic population structure (16–18). Ants of the subtribe Attina (including A. dentigerum; Fig. 1A) cultivate basidiomycetous fungi in an agricultural system that is analogous to human farming (19–21). Like nearly all human crops, these fungal cultivars have low genetic diversity (22) and are vulnerable to virulent pathogens (23). Specialized ascomycetous pathogens in the genus Escovopsis (Fig. 1C to E) form persistent and chronic infections throughout most of the life cycle of a given ant colony. If not controlled, the infection can rapidly overwhelm and consume entire fungal gardens (23, 24). Thus, Escovopsis parasites impose persistent and strong selection pressure. To combat Escovopsis infection, the ant host performs generalized defense behaviors, such as fungus garden weeding and grooming, to physically remove infected portions of the cultivar and/or Escovopsis spores (25). The ants also employ chemical defense through a symbiotic association with antibiotic-producing actinobacteria (Fig. 1B) (26–29). Although fungal cultivars are the primary target of Escovopsis (26), an indirect selection pressure is imposed on the ant host, which obligately depends on its fungal cultivar, and the defensive bacterial symbiont, which obligately depends on the ant (20, 30, 31). Interactions are summarized in Table S1.
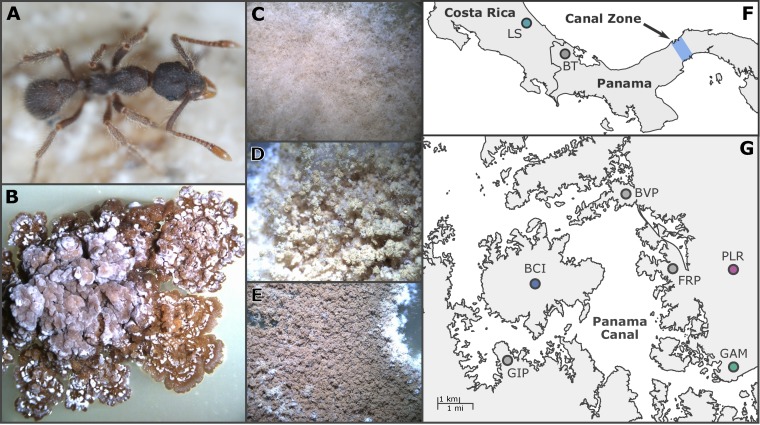
FIG 1.
The fungus-farming ant Apterostigma dentigerum, seen on top of its fungal garden (A), maintains symbiotic Pseudonocardia actinobacteria, shown here in axenic culture (B). Secondary metabolites from Pseudonocardia inhibit the growth of Escovopsis pathogens that directly consume the fungal garden. Three Escovopsis phylotypes, fuzzy brown (C), yellow (D), and brown (E), are known to infect A. dentigerum gardens. Pseudonocardia and Escovopsis were sampled from Central America (F), with focal populations residing in the Panama Canal Zone (G). Maps created in R using plotly.
The fungus gardens of A. dentigerum are host to at least three distinct lineages of the Escovopsis parasite, each with a distinct morphology (32, 33). Cultivar-Escovopsis in vitro inhibition assays indicate that antifungals produced by fungal cultivars are capable of inhibiting many Escovopsis lineages but are universally poor at the inhibition of the brown Escovopsis morphotype regardless of geographic location (16, 33, 34). Moreover, the brown Escovopsis morphotype appears to be one of the more common lineages found within A. dentigerum colonies. Given that A. dentigerum cultivars are seemingly ill-equipped to combat brown Escovopsis pathogens, we hypothesize that the symbiotic actinobacteria (Pseudonocardia) are instead coadapted to this specific parasite lineage. Our previous finding of congruent population genetic structure between Pseudonocardia symbionts and brown Escovopsis pathogens (17) may impact coevolution in this interaction by facilitating local adaptation (35, 36). In particular, the Pseudonocardia populations on Barro Colorado Island (BCI) and Gamboa Forest (GAM) in the Panama Canal Zone may be good candidates for local adaptation, as they contain high-identity sequence assemblages (17, 18) thought to be characteristic of recent periodic selection in bacteria (37).
The antibiotic potential of Pseudonocardia populations is determined by the acquisition and evolution of biosynthetic gene clusters that can reside on plasmids and therefore may be transiently acquired through horizontal transfer (38). Although the frequency and impact of these lateral exchanges are unclear in Pseudonocardia, recent studies of closely related Streptomyces actinobacteria suggest that these events are rare on evolutionary time scales (39, 40). Most antibiotics that are used clinically are sourced from actinobacteria that, in their natural contexts, use antibiotics to mediate antagonism (40, 41) and are often the connections of complex species-species interaction networks (40, 42). The biosynthetic gene clusters that assemble antibiotic secondary metabolites are highly diverse, reflecting the vast diversity of interactions they mediate (40, 43–45). Within insect-actinobacterium associations, including those involving the southern pine beetle (46), the solitary digger wasp (47, 48), and fungus-growing ants (18, 26, 43), actinobacteria readily associate with insect hosts to provide antibacterial and antifungal chemical defenses through their antibiotic secondary metabolites (43, 49, 50).
Here, we use antibiotic inhibition assays in combination with population genetic structure analyses and whole-genome sequencing to examine whether A. dentigerum-associated Pseudonocardia-Escovopsis interactions conform to a GMC. First, we establish whether Pseudonocardia-Escovopsis interactions are fixed across the Canal Zone of Panama (compared to cultivar-Escovopsis interactions) and identify the appropriate geographic scale of symbiont selection. Second, we describe the broad geographic differences in Pseudonocardia antibiotic potency across Central America and test for locally adapted populations. Finally, to shed light on the specific secondary metabolite pathways and genetic mechanisms conferring antibiosis, we genomically characterized 29 Pseudonocardia strains and examined the role biosynthetic gene clusters play in Pseudonocardia-Escovopsis coevolution.
RESULTS AND DISCUSSION
Establishing specificity.
A matrix bioassay utilizing 2,417 individual inhibition assays revealed a hierarchical pattern, namely, average inhibition against yellow and fuzzy brown Escovopsis isolates was significantly lower than that against brown Escovopsis isolates. Moreover, these inhibition averages correlated with the relative abundances of Escovopsis morphotypes in Central America (Fig. 2). Similarly, a test strain inhibition assay pairing a strain of each of three Escovopsis lineages (brown, yellow, and fuzzy brown) against 52 Pseudonocardia strains from across Panama and Costa Rica revealed significant hierarchical inhibition in paired t tests: namely, brown was inhibited more than yellow, and yellow was inhibited more than fuzzy brown (Fig. S1). Interestingly, while isolating Escovopsis pathogens from Peruvian Apterostigma species that maintain a different cultivar lineage from that of A. dentigerum, we failed to recover any brown Escovopsis isolates and obtained only seven fuzzy brown and two yellow isolates. This observation, in combination with previous cultivar-pathogen studies (32, 33), supports the hypothesis that cultivar antibiosis mediates yellow and fuzzy brown infection, while Pseudonocardia is specialized to suppress brown Escovopsis. These results suggest that yellow and fuzzy brown Escovopsis strains are experiencing a type of “rare morph advantage” (51, 52), whereby, due to lack of exposure, selection is toward Pseudonocardia inhibiting brown Escovopsis, the more common parasite, rather than toward evolving antibiotic activity against rarer strains.
FIG 2.
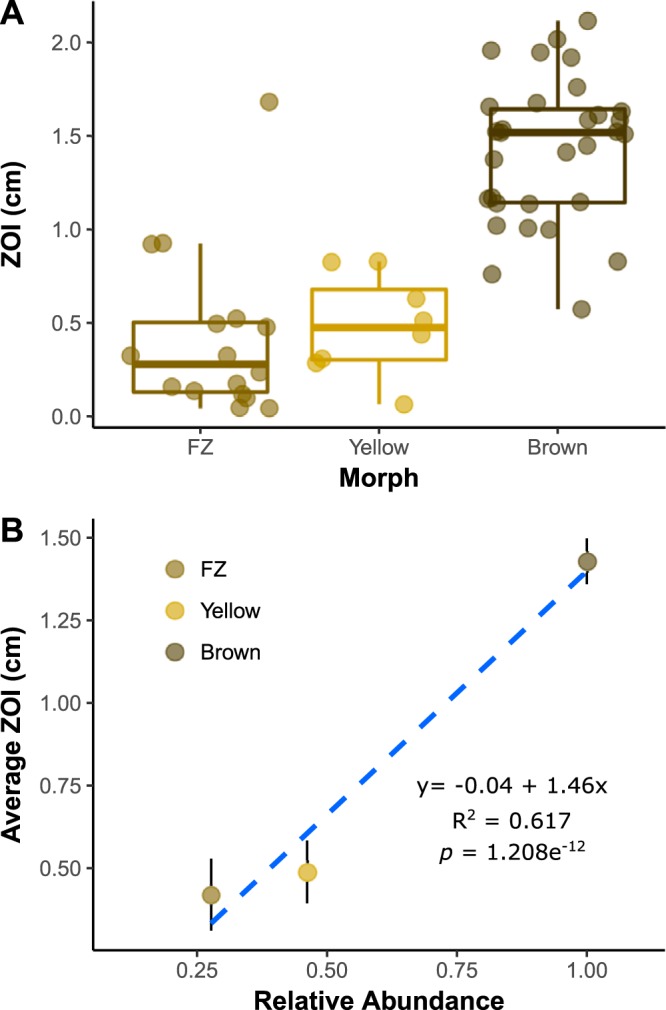
(A) Central American Pseudonocardia antibiotic inhibition of three Escovopsis pathogen lineages (fuzzy brown [FZ], yellow, and brown). Rare Escovopsis pathogen morphotypes experience less inhibition by Pseudonocardia (center, median; box, upper and lower quantiles; whiskers, 1.5× interquartile range). (B) Average zone of inhibition (ZOI) corresponds to the relative frequency at which each lineage is encountered in Central American Apterostigma dentigerum ant cultivars, with common brown Escovopsis parasites experiencing significantly greater inhibition than the rarer fuzzy brown and yellow parasites. The strength of inhibition corresponds to parasite morphotype abundance in nature, suggesting that parasites experience rare morph advantage. Blue indicates linear regression of individual data points from panel A error bars indicate standard error of the mean.
GMC assumptions.
A 6-by-6 inhibition assay pairing Panama Canal Zone strains of Pseudonocardia and brown Escovopsis isolated from six mature ant nests found that all bacterial strains exhibited the ability to inhibit strains of the garden pathogen and that there is variation in the degree of inhibition (Fig. S2). There was no significant difference (P = 0.8533) in zone of inhibition distance (or variance, P = 0.393) between within-colony pairings (mean, 0.882; standard error [SE], 0.209) and among-colony pairings (mean, 0.847; SE, 0.074). This pattern of bacterial inhibition is important for three reasons, as follows. (i) It further dispels an already disputed hypothesis in which free-living Pseudonocardia acquisition and subsequent symbiont selection occur within ant nests every generation (53, 54)—if ants frequently recruit free-living Pseudonocardia bacteria for antibiotics effective against pathogens, one would anticipate elevated parasite inhibition within colonies versus between colonies. In fact, within-colony versus between-colony average inhibitions and variances were nearly identical. (ii) Pseudonocardia inhibition contrasts with inhibition assays pairing A. dentigerum fungal cultivars and brown Escovopsis pathogens over the same geographic scale, in which no sign of cultivar-pathogen inhibition was observed in 35 of 36 combinations (16), despite known antibiosis to different sympatric Escovopsis lineages. This further supports an evolved specificity for Pseudonocardia (not cultivars) to combat the common brown Escovopsis lineage. (iii) Central to the GMC is the notion that coevolution occurs at the population scale and that coevolved interactions are variable and rarely fix within species (8). Our observed variation in Pseudonocardia-Escovopsis (brown) inhibition appears consistent with this postulate, whereas the cultivar-Escovopsis (brown) interactions more closely resemble fixed, species-level interactions. Moreover, these results help identify populations of ant nests (as opposed to individual nests) as the appropriate scale of selection.
GMC hypotheses.
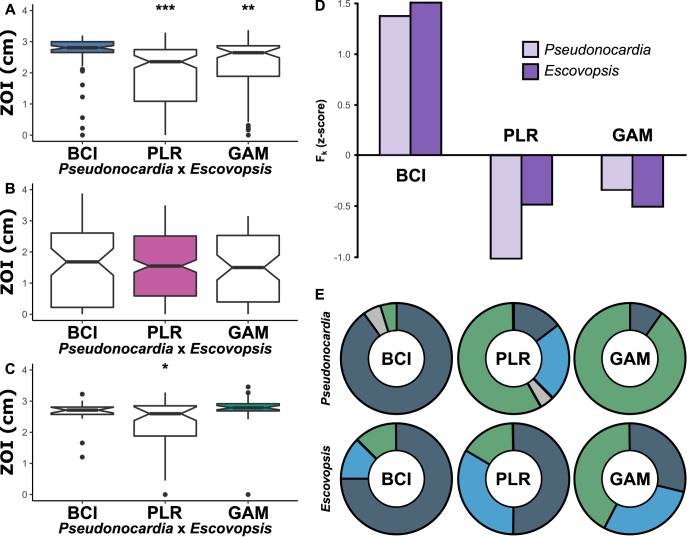
(i) Selection mosaics and hot/cold spots. Although identifying appropriate population-level variation meets an important assumption of the GMC, a thorough demonstration requires the observation of (i) geographic variation in the structure of selection on interactions (i.e., selection mosaics) and (ii) coevolutionary hot spots of local reciprocal selection embedded in a broader matrix of coevolutionary cold spots, where local selection is nonreciprocal (i.e., coevolutionary hot spots and cold spots) (8, 10, 55). Our matrix bioassay revealed these two components. Average inhibition by Panamanian Pseudonocardia was highest against Escovopsis from BCI (zone of inhibition [ZOI] = 1.50) and GAM (ZOI = 1.55), significantly lower against that from Pipeline Road (PLR) (ZOI = 0.980, P < 0.001), and lower still against that from La Selva Biological Station (LS), Costa Rica (ZOI = 0.317696176, P < 0.001), which suggests a geographic selection mosaic (Fig. 3).
FIG 3.
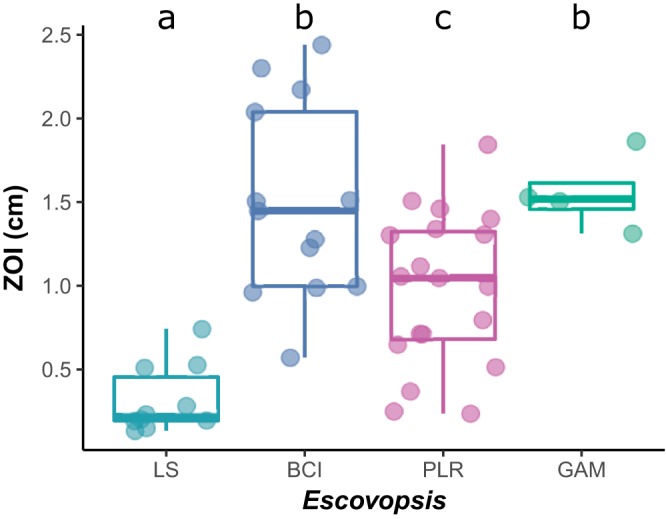
A mosaic of antibiotic inhibition by Panamanian Pseudonocardia to Central American populations of Escovopsis (x axis). The average zone of inhibition distance in petri plate bioassays is presented on the y axis (center, median; box, upper and lower quantiles; whiskers, 1.5× interquartile range). Letters indicate significantly different mean zone of inhibition (t test, P < 0.05). LS, La Selva Biological Station; BCI, Barro Colorado Island; PLR, Pipeline Road; GAM, Gamboa Forest.
Within-population versus across-population comparisons showed that the Pseudonocardia population on BCI is locally adapted (Fig. 4A)—the BCI × BCI ZOI (2.65 cm) was significantly greater than those in cross-population comparisons (BCI × PLR ZOI = 1.93, P < 0.001; BCI × GAM ZOI = 2.19, P = 0.007). Local adaptation occurs between species when the following two conditions are met: (i) reciprocal selection is imposed, and (ii) selection varies over space (9, 56, 57). Thus, local adaptation by one species to another is often regarded as a demonstration of the reciprocal selection necessary to demonstrate coevolution (58). While BCI Pseudonocardia isolates were locally adapted, PLR Pseudonocardia inhibition was similar regardless of parasite population (PLR × PLR ZOI = 1.539, PLR × BCI ZOI = 1.532, PLR × GAM ZOI = 1.435; Fig. 4B), and GAM Pseudonocardia isolates were locally adapted only in comparison to PLR (GAM × GAM ZOI = 2.750, GAM × BCI ZOI = 2.59, GAM × PLR ZOI = 2.23; P = 0.003; Fig. 4C). These comparisons are consistent with the GMC prediction that coevolutionary hot spots of reciprocal adaptation (BCI) are embedded within cold spots that lack reciprocal adaptation (GAM and PLR) (8).
FIG 4.
Reciprocal tests for local adaptation using Pseudonocardia from (A) Barro Colorado Island (BCI), (B) Pipeline Road (PLR), and (C) Gamboa Forest (GAM), with significant local adaptation on BCI indicated by a mean zone of inhibition that was significantly different from the other two pairings in the cross (x axes indicate Escovopsis location). (A to C) t test versus local pairing. *, P = 2.98E−3; **, P = 6.994E−4; ***, P = 8.668E−10; center, median; box, upper and lower quantiles; notches, 95% confidence; whiskers, 1.5× interquartile range; points, outliers. Population STRUCTURE analysis demonstrates that Escovopsis and Pseudonocardia populations on BCI are genetically distinct, having elevated Fk (D) and more than 75% of their isolates assigned to one genetic cluster (E).
(ii) Trait remixing. The third component of the GMC is the concept of “trait remixing,” which posits that genetic structuring across landscapes continuously shapes the coevolutionary process. Our population genetic analyses and genomic antibiotic potential characterizations of biosynthetic gene clusters revealed how genetic structure has shaped local adaptation. Local adaptation on BCI (Fig. 4A) was consistent with predictions that congruent population structure facilitates reciprocal adaptation (35, 36). Pseudonocardia and Escovopsis isolates represent distinct genetic populations on BCI, as they both had an elevated Fk (an Fst analog that estimates the divergence between each population and a theoretical ancestral population) value (Fig. 4D), and each symbiont had a large majority of isolates grouping to a single genetic population as defined using STRUCTURE software (Pseudonocardia, 90%; Escovopsis, 75%; Fig. 4E). Pseudonocardia isolates on GAM were similarly structured, with 90% of isolates grouping to a single STRUCTURE-defined population; however, GAM Escovopsis isolates were evenly distributed across 3 genetic populations (Fig. 4E). This lack of geographic structuring in GAM pathogens may explain why GAM Pseudonocardia isolates appear to be locally adapted to pathogens from PLR but not those from BCI (Fig. 4C). The PLR data lacked geographic structure for both Pseudonocardia and Escovopsis isolates (Fig. 4E), and no signal of local adaptation was observed in these comparisons (Fig. 4B). These results contrast with a seminal study of local adaptation across three lake populations of snails and their trematode parasites, in which local adaptation was observed in all three populations (35). In that study, lakes acted as natural barriers to gene flow and likely helped create the congruent host-parasite genetic structure that facilitated local adaptation. In our study, it appears that the asymmetries in local adaptation were shaped by the asymmetries in population genetic structure. Anthropogenic disturbances can also erect genetic barriers that facilitate local adaptation (59). The flooding of the Panama Canal, which created BCI approximately one hundred years ago, may have facilitated local adaptation by erecting a barrier to both ant dispersal and Escovopsis transmission, although periodic migration events undoubtedly occur (18). Similarly, GAM has an “island nature” that may facilitate partial local adaptation. GAM is nested within the town of Gamboa, surrounded by paved streets and homes in addition to being surrounded by water on three sides. These subtler anthropogenic barriers may be sufficient to restrict ant dispersal, resulting in the relatively distinct GAM Pseudonocardia clade, but no distinct clade for Escovopsis, which contains multiple genetic populations within GAM (Fig. 4E).
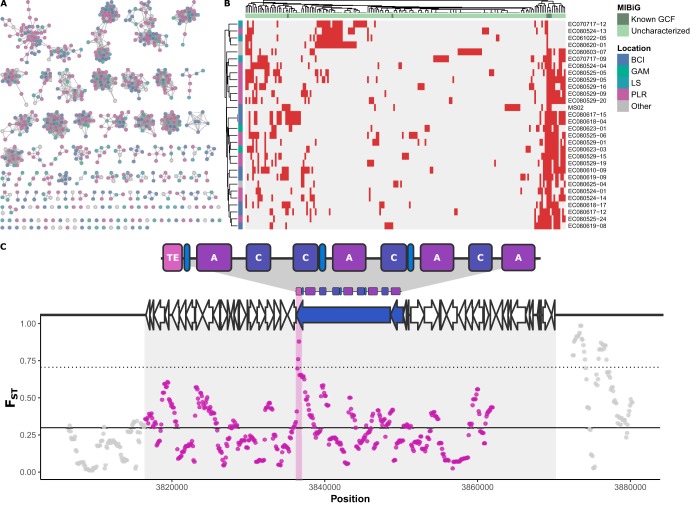
Genomic sequencing of 29 Pseudonocardia genomes identified 146 gene cluster families of secondary metabolite biosynthetic gene clusters. A total of 93 gene cluster families occurred within a single population, 80 of which were only found in a single genome (Fig. 5). Of the single-population gene cluster families found in more than one strain, BCI had ten unique gene cluster families, which included a siderophore, a nonribosomal peptide synthetase (NRPS), and a type 2 polyketide found in five or more strains (Fig. 5). PLR had three nonsingleton, population-specific gene cluster families, which included a nucleoside biosynthetic gene cluster found in four strains (Fig. 5). GAM- and PLR-specific biosynthetic gene clusters include a lasso peptide found only in isolates from GAM and the southern part of PLR. Finally, there was no significant Pearson correlation between biosynthetic gene cluster presence/absence and inhibition by location (Fig. S3). Tests for selective sweeps identified at least two biosynthetic gene clusters with significant Fst (fixation index, a measure of population differentiation due to genetic structure) values in BCI-mainland comparisons. A bacteriocin appears to have significant Fst values in the following three enzymatic regions: an acetyl-coenzyme A (CoA) carboxylase, a SufE (cysteine desufuration), and a sulfurtransferase (Fig. S4). Figure 5C shows an NRPS biosynthetic gene cluster that had an Fst peak in a thioesterase domain.
FIG 5.
Characterization of biosynthetic gene cluster families in symbiotic Pseudonocardia isolates spanning Panama and Costa Rica. (A) Each node represents a contiguous set of genes that are part of a secondary metabolite biosynthesis cluster. Edges represent BiG-SCAPE distances of 0.3 or below. Connected subgraphs correspond to gene cluster families. Nodes are colored by their source (see top right of panel B for legend). (B) Presence-absence map of gene cluster families. Known gene cluster families with examples in MIBiG are shown as dark green in the top annotation strip. Sample location is shown in the left annotation strip. Dendrograms for gene cluster families and samples are derived from Euclidean clustering. (C) Example of selective sweep within a biosynthetic gene cluster. Fst between BCI and other samples is shown as the y axis. Solid and dashed lines correspond to the Fst genome-wide average and two standard deviations above the genome-wide average, respectively (window, 1 kb; step size, 100 bp). A nonribosomal peptide synthetase (NRPS) biosynthetic gene cluster is shown with boundaries corresponding to the shaded region; windows within the gene cluster are shown in purple, and those flanking it are in gray. Gene and domain structure for the biosynthetic gene cluster are shown above. C, condensation domain; A, adenylation domain; TE, thioesterase domain; blue, NRPS genes; pink, TE domain.
The presence of several unique biosynthetic gene clusters on BCI appears to contribute to elevated inhibition in the island population, and a lack of certain biosynthetic gene clusters may contribute to lower inhibition in the Costa Rican LS population. Correlations with bioassay inhibition demonstrated that a particular biosynthetic gene cluster may be associated with either high or low inhibition (Fig. S3A); however, no pairing reaches significance by location, which suggests that the presence/absence of biosynthetic gene clusters is not driving local adaptation. At least two cross-population biosynthetic gene clusters appear to have undergone selective sweeps on BCI. A bacteriocin on BCI showed a significantly high Fst value compared to that of the mainland populations (Fig. S4). Bacteriocins are generally thought to be niche-defending compounds, so while they are unlikely to be the source of Escovopsis inhibition, they could be involved with host-Pseudonocardia specificity either directly or indirectly via the killing of potential invader actinobacterial strains. An NRPS biosynthetic gene cluster may also have undergone a selective sweep on BCI; an Fst peak occurs within the thioesterase domain that could potentially affect secondary metabolite production and concentrations by modulating a potentially rate-limiting step (60). These results suggest that adaptation at the local level is occurring within genes and not just through the acquisition of novel biosynthetic gene clusters.
Conclusions.
A powerful utility of the GMC is that it provides a framework whereby the historically artificial dichotomy that coevolution is either characterized by local pairwise reciprocal selection or diffuse multispecies interactions can be abandoned, and seeming diffuse associations can be partitioned into coevolving population-level interactions and fixed species-level interactions (8, 10). Indeed, studies of attine-symbiont coevolution often fall into this dichotomy, sometimes reaching the conclusion that diffuse multispecies interactions are likely to negate the opportunity for reciprocal adaptation (61) or that a single species interaction facilitates pairwise “red queen” dynamics (62). Here, we have shown that the seemingly diffuse association between Pseudonocardia isolates and three Escovopsis species conforms to a GMC framework in which the interactions of brown Escovopsis pathogens are variable and coevolving among populations, whereas those of yellow and fuzzy brown Escovopsis pathogens appear to be fixed at the species level. Finally, our results demonstrate that the GMC is applicable to microbes containing geographic population structure and for which selection pressure is imposed indirectly via symbiont-symbiont interactions.
MATERIALS AND METHODS
Symbiont collection and isolation.
From 2006 through 2010, we sampled Pseudonocardia and Escovopsis isolates associated with the fungus-growing ant Apterostigma dentigerum from La Selva Biological Station (LS) in Costa Rica and the following populations in the Canal Zone of Panama: Barro Colorado Island (BCI), Pipeline Road (PLR), Gamboa Forest (GAM), Frijoles Peninsula (FRP), and Buena Vista Peninsula (BVP). Ant colonies were collected aseptically in the field, using flame-sterilized forceps and spoons to prevent lateral transfer of symbionts between nests. John T. Longino’s key to Costa Rican Apterostigma was used to identify A. dentigerum species (http://ants.biology.utah.edu/genera/apterostigma/specieslist.html). Pseudonocardia isolations were conducted by first removing the ant’s head and forelegs to expose the mesosternal lobes, where the bacteria are concentrated. We then used fine, sterilized probes to scrape small tufts of bacterium onto chitin medium. For further details on Pseudonocardia isolation from A. dentigerum, see reference 17. To isolate garden parasites, we plated pieces of fungal cultivar onto potato-dextrose agar (PDA) medium and subcultured Escovopsis hyphae as they emerged (16). Escovopsis isolates were sampled from the same geographic range as Pseudonocardia in the Panama Canal Zone, but with additional sampling of Escovopsis from Apterostigma spp. from the Los Amigos Station in Peru (PU). A collection of 50 Pseudonocardia strains and 55 Escovopsis strains was generated. Each symbiont strain represents a unique ant colony.
Inhibition assays and analysis.
To quantify Escovopsis inhibition by Pseudonocardia, we performed inhibition assays pairing each of the two symbionts and measured the zone of inhibition (ZOI). The in vitro ZOI corresponds to fungus garden loss in higher attines (29) and thus provides a valuable proxy for the impact of Escovopsis in vivo. Petri plate bioassay experiments and statistical comparisons for local adaptation (63) were conducted as follows: Pseudonocardia cells were point inoculated in the center of a yeast malt extract agar (YMEA) plate and allowed to grow for 3 weeks, after which Escovopsis spores were inoculated at the edge of the plate. Plates were inspected twice a week, and when a clear ZOI had formed in a given pairing (typically within 2 to 3 weeks after fungus inoculation), the minimum ZOI was measured. Depending on time constraints, ZOI was either measured using calipers and recorded immediately or standardized digital photos of the inhibition assay were taken and the ZOI was subsequently measured using ImageJ (64). For ImageJ measurements, the digital scale was calibrated against a 10-cm line above the petri plate to ensure that the digital and analog measurements were consistent.
Three separate inhibition assays (referred to as “test strain,” “within colony,” and “large/matrix”) were conducted in 2008, 2009, and 2010, respectively. First, to obtain an initial assessment of the geographic variation in Pseudonocardia-Escovopsis interactions, we utilized a test strain (8) approach, in which 52 Pseudonocardia isolates spanning Costa Rica and Panama were paired with each of three Escovopsis pathogen lineages, namely, brown, yellow, and fuzzy brown. These Escovopsis lineages form distinct phylogenetic clades and are taken to represent species with brown, fuzzy brown, and yellow morphotypes corresponding to Escovopsis aff. multiformis, Escovopsis aff. cavatus, and Escovopsis sp. (yellow), respectively (32, 65). Second, to examine whether within-colony symbiont selection is occurring, we performed inhibition assays utilizing all pairings of Pseudonocardia and Escovopsis isolates from six mature ant colonies (36 total assays) in the Canal Zone of Panama, for which we isolated both symbionts from each nest and compared within-colony versus between-colony inhibition. Third, we conducted a large/matrix assay utilizing all pairing of 50 Pseudonocardia and 55 Escovopsis isolates. From this large matrix, we used average Pseudonocardia inhibition to test for differences among locations (i.e., selection mosaics) and differences among Escovopsis lineages, and we subsampled all interactions among BCI, PLR, and GAM (the most thoroughly sampled sites) to test for local adaptation. Paired t tests were used to assess differences in inhibition to Escovopsis test strains. We used two-tailed t tests for comparisons within colony versus cross-colony and within-population versus cross-population Escovopsis-Pseudonocardia pairings. P values were adjusted using Bonferroni’s correction to reduce type I error in multiple comparisons. F tests were performed to establish whether individual t tests should be performed with equal or unequal variances.
Population structure.
To describe the geographic population structure of Pseudonocardia, we utilized sequence data from 71 Pseudonocardia strains that were previously genotyped at six housekeeping loci using Sanger sequencing technology (17), and we subsequently whole-genome sequenced a subset of 29 strains using next-generation sequencing. Both data sets were subjected to Bayesian STRUCTURE analyses (66, 67), to assign genetic populations (K) to strains without prior information about sampling location (the whole-genome data set additionally used fineSTRUCTURE to speed up analysis). For the six-locus data set, we treated each polymorphic site (355 total) as an individual locus, and simulations were run with both no-admixture and admixture/linkage models. The admixture model assumes that each individual derives ancestry from only one population and is most appropriate for discrete populations. For the no-admixture model, we assumed a constant λ and correlated allele frequencies, which improves clustering for closely related populations. The linkage model allows for individuals to have mixed ancestry (i.e., admixture). Following Falush et al. (67), the linkage model assumed correlated allele frequencies, estimated λ, and treated polymorphic sites within genes as linked, with linkage being proportional to genetic distance in base pairs. K was selected by the highest likelihood score and by calculating the ΔK (68). All simulations were run for K = 1 to K = 20 with 20,000 iterations following a burn-in period of 10,000. Each run was repeated five times to assess consistency and facilitate ΔK calculations. For the three focal Panama Canal populations (GAM, PLR, and BCI) we also calculated Fk, an Fst analog that estimates the divergence between each population and a theoretical ancestral population. Escovopsis STRUCTURE output was taken from Gerardo and Caldera 2007 (16). Outputs from the 71-strain, 6-locus data set are presented in Fig. S3, and fineSTRUCTURE-determined populations from the subset of 29 whole genomes are color coded and presented in Fig. S4. The subset of fineSTRUCTURE genomic analysis similarly recovered six genetic populations, although fineSTRUCTURE, in contrast to the 6-locus STRUCTURE analysis, was able to separate out a northern region of BCI.
Biosynthetic potential.
To begin understanding the genetic mechanisms of antibiotic resistance to Escovopsis, we whole-genome sequenced a subset (n = 29) of the Pseudonocardia strains used in inhibition experiments and characterized the presence of conserved biosynthetic gene clusters. Sequencing of Pseudonocardia strains was performed by Duke University or Washington University in St. Louis. Pacific Biosciences (Pseudonocardia reference genome EC080625-04; Duke University) assemblies utilized Hierarchical Genome Assembly Process (HGAP) 1.4 (69), whereas Illumina (Washington University) genomes were assembled using Velvet (70). Prodigal v2.60 (71) was used for protein prediction, whereas Rfam (72) hidden Markov models and Infernal 1.1.1 (73) were used for rRNA prediction. Annotation of protein coding genes was through TIGRFam v15 (74), PFAM v29, the Kyoto Encyclopedia of Genes and Genomes (KEGG), actNOG (75) hidden Markov models, and HMMer 3.1 (76). In each genome, secondary metabolite clusters were identified by antiSMASH v4.0.2 (77). Biosynthetic gene clusters were grouped into families by BiG-SCAPE under hybrid, mix, and glocal modes (78). To assess the extent to which the presence/absence of specific secondary metabolite clusters contributes to local adaptation, biosynthetic gene clusters were grouped (80% nucleotide identity via nucmer [79] alignment and 50% coverage for each segment) and Pearson correlations were calculated between biosynthetic gene clusters with ZOI by location.
All Illumina genome assemblies were aligned to the PacBio EC080625-04 assembly using nucmer (79). Reference single-nucleotide polymorphism (SNP) positions covered by a contig for every genome were used for fineSTRUCTURE analysis to avoid missing data for any SNP positions. Multiple runs with various values of c (effective number of genomic segments with contiguous ancestry) and estimated population size had little effect on overall strain clustering, except for very high values of c merging neighboring clusters together. The Fst value was calculated in sliding windows across the reference Pseudonocardia genome EC080625-04. Briefly, reads for each genome were pooled to either BCI or non-BCI groups based on their sampling location. Group reads were mapped to EC080625-04 with bwa aln v0.7.17-r1188 (maximum edit distance of 0.01; first 100 subsequences as seed; maximum gap opens, 1; disallow long deletions within 12 bp at 3′ end; maximum gap extensions, 12) (80). SAMtools mpileup v1.7 (81) was used to call variants between the two groups (disabled per-base alignment quality; alignment minimum mapQ, 20; base minimum baseQ, 20). PoPoolation2 (82) was then used to calculate allele frequencies. Fst was calculated in sliding windows with windows:steps of 1:1, 1,000:100, and 10,000:1,000 bp (maximum coverage, 2%; minimum coverage, 50; minimum count, 6).
Supplementary Material
ACKNOWLEDGMENTS
This project was supported through National Institutes of Health (NIH) grants NIH U19 Al109673 and NIH U19 TW009872 and National Science Foundation grant MCB-0702025. Additional support was provided to M.G.C. through NIH National Research Service Award T32 GM008505.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01580-19.
REFERENCES
- 1.Margulis L. 1970. Origin of eukaryotic cells. Yale University Press, New Haven, CT. [Google Scholar]
- 2.Moreira D, Le Guyader H, Philippe H. 2000. The origin of red algae and the evolution of chloroplasts. Nature 405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 3.Mueller UG, Gerardo N. 2002. Fungus-farming insects: multiple origins and diverse evolutionary histories. Proc Natl Acad Sci U S A 99:15247–15249. doi: 10.1073/pnas.242594799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yannarell SM, Grandchamp GM, Chen S-Y, Daniels KE, Shank EA. 2019. A dual-species biofilm with emergent mechanical and protective properties. J Bacteriol 201:e00670-18. doi: 10.1128/JB.00670-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozano GL, Bravo JI, Garavito Diago MF, Park HB, Hurley A, Peterson SB, Stabb EV, Crawford JM, Broderick NA, Handelsman J. 2019. Introducing THOR, a model microbiome for genetic dissection of community behavior. mBio 10:e02846-18. doi: 10.1128/mBio.02846-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shade A, Dunn RR, Blowes SA, Keil P, Bohannan BJM, Herrmann M, Küsel K, Lennon JT, Sanders NJ, Storch D, Chase J. 2018. Macroecology to unite all life, large and small. Trends Ecol Evol 33:731–744. doi: 10.1016/j.tree.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Lenormand T. 2002. Gene flow and the limits to natural selection. Trends Ecol Evol 17:183–189. doi: 10.1016/S0169-5347(02)02497-7. [DOI] [Google Scholar]
- 8.Thompson JN. 2005. The geographic mosaic of coevolution. University of Chicago Press, Chicago, IL. [Google Scholar]
- 9.Thompson JN. 1994. The coevolutionary process. University of Chicago Press, Chicago, IL. [Google Scholar]
- 10.Thompson JN. 1999. Specific hypotheses on the geographic mosaic of coevolution. Am Nat 153:S1–S14. doi: 10.1086/303208. [DOI] [Google Scholar]
- 11.Brodie ED, Ridenhour BJ, Brodie ED. 2002. The evolutionary response of predators to dangerous prey: hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution 56:2067–2082. doi: 10.1111/j.0014-3820.2002.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 12.Parchman TL, Buerkle CA, Soria-Carrasco V, Benkman CW. 2016. Genome divergence and diversification within a geographic mosaic of coevolution. Mol Ecol 25:5705–5718. doi: 10.1111/mec.13825. [DOI] [PubMed] [Google Scholar]
- 13.Laine A-L. 2009. Role of coevolution in generating biological diversity: spatially divergent selection trajectories. J Exp Bot 60:2957–2970. doi: 10.1093/jxb/erp168. [DOI] [PubMed] [Google Scholar]
- 14.Bravo-Monzón AE, Ríos-Vásquez E, Delgado-Lamas G, Espinosa-García FJ. 2014. Chemical diversity among populations of Mikania micrantha: geographic mosaic structure and herbivory. Oecologia 174:195–203. doi: 10.1007/s00442-013-2748-y. [DOI] [PubMed] [Google Scholar]
- 15.Zélé F, Magalhães S, Kéfi S, Duncan AB. 2018. Ecology and evolution of facilitation among symbionts. Nat Commun 9:4869. doi: 10.1038/s41467-018-06779-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerardo NM, Caldera EJ. 2007. Labile associations between fungus-growing ant cultivars and their garden pathogens. ISME J 1:373–384. doi: 10.1038/ismej.2007.57. [DOI] [PubMed] [Google Scholar]
- 17.Caldera EJ, Currie CR. 2012. The population structure of antibiotic-producing bacterial symbionts of Apterostigma dentigerum ants: impacts of coevolution and multipartite symbiosis. Am Nat 180:604–617. doi: 10.1086/667886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mcdonald BR, Chevrette MG, Klassen JL, Horn HA, Caldera EJ, Wendt-Pienkowski E, Cafaro MJ, Ruzzini AC, Arnam EV, Weinstock GM, Gerardo NM, Poulsen M, Suen G, Clardy J, Currie CR. 2019. Biogeography and microscale diversity shape the biosynthetic potential of fungus-growing ant-associated Pseudonocardia. bioRxiv 10.1101/545640. [DOI]
- 19.Chapela IH, Rehner SA, Schultz TR, Mueller UG. 1994. Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science 266:1691–1694. doi: 10.1126/science.266.5191.1691. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Sosa-Calvo J, Horn HA, Pupo MT, Clardy J, Rabeling C, Schultz TR. 2018. Convergent evolution of complex structures for ant-bacterial defensive symbiosis in fungus-farming ants. Proc Natl Acad Sci U S A 115:10720–10725. doi: 10.1073/pnas.1809332115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yek SH, Boomsma JJ, Poulsen M. 2012. Towards a better understanding of the evolution of specialized parasites of fungus-growing ant crops. Psyche 2012:239392. doi: 10.1155/2012/239392. [DOI] [Google Scholar]
- 22.Mueller UG, Scott JJ, Ishak HD, Cooper M, Rodrigues A. 2010. Monoculture of leafcutter ant gardens. PLoS One 5:e12668. doi: 10.1371/journal.pone.0012668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Currie CR, Mueller UG, Malloch D. 1999. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci U S A 96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currie CR, Bot ANM, Boomsma JJ. 2003. Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos 101:91–102. doi: 10.1034/j.1600-0706.2003.12036.x. [DOI] [Google Scholar]
- 25.Currie CR, Stuart AE. 2001. Weeding and grooming of pathogens in agriculture by ants. Proc Biol Sci 268:1033–1039. doi: 10.1098/rspb.2001.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Currie CR, Scott JA, Summerbell RC, Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704. doi: 10.1038/19519. [DOI] [Google Scholar]
- 27.Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, Mueller UG, Sung G-H, Spatafora JW, Straus NA. 2003. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science 299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 28.Oh D-C, Poulsen M, Currie CR, Clardy J. 2009. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol 5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulsen M, Cafaro MJ, Erhardt DP, Little AEF, Gerardo NM, Tebbets B, Klein BS, Currie CR. 2010. Variation in Pseudonocardia antibiotic defence helps govern parasite-induced morbidity in Acromyrmex leaf-cutting ants. Environ Microbiol Rep 2:534–540. doi: 10.1111/j.1758-2229.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 31.Steffan SA, Chikaraishi Y, Currie CR, Horn H, Gaines-Day HR, Pauli JN, Zalapa JE, Ohkouchi N. 2015. Microbes are trophic analogs of animals. Proc Natl Acad Sci U S A 112:5119–5124. doi: 10.1073/pnas.1508782112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerardo NM, Mueller UG, Currie CR. 2006. Complex host-pathogen coevolution in the Apterostigma fungus-growing ant-microbe symbiosis. BMC Evol Biol 6:88. doi: 10.1186/1471-2148-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerardo NM, Jacobs SR, Currie CR, Mueller UG. 2006. Ancient host-pathogen associations maintained by specificity of chemotaxis and antibiosis. PLoS Biol 4:e235. doi: 10.1371/journal.pbio.0040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birnbaum SSL, Gerardo NM. 2016. Patterns of specificity of the pathogen Escovopsis across the fungus-growing ant symbiosis. Am Nat 188:52–65. doi: 10.1086/686911. [DOI] [PubMed] [Google Scholar]
- 35.Lively CM. 1999. Migration, virulence, and the geographic mosaic of adaptation by parasites. Am Nat 153:S34–S47. doi: 10.1086/303210. [DOI] [PubMed] [Google Scholar]
- 36.Lively CM, Dybdahl MF. 2000. Parasite adaptation to locally common host genotypes. Nature 405:679–681. doi: 10.1038/35015069. [DOI] [PubMed] [Google Scholar]
- 37.Whitaker RJ, Banfield JF. 2006. Population genomics in natural microbial communities. Trends Ecol Evol 21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. 2015. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci U S A 112:13150–13154. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald BR, Currie CR. 2017. Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8:e00644-17. doi: 10.1128/mBio.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chevrette MG, Currie CR. 2019. Emerging evolutionary paradigms in antibiotic discovery. J Ind Microbiol Biotechnol 46:257–271. doi: 10.1007/s10295-018-2085-6. [DOI] [PubMed] [Google Scholar]
- 41.Miller I, Chevrette M, Kwan J. 2017. Interpreting microbial biosynthesis in the genomic age: biological and practical considerations. Mar Drugs 15:165. doi: 10.3390/md15060165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adnani N, Chevrette MG, Adibhatla SN, Zhang F, Yu Q, Braun DR, Nelson J, Simpkins SW, McDonald BR, Myers CL, Piotrowski JS, Thompson CJ, Currie CR, Li L, Rajski SR, Bugni TS. 2017. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem Biol 12:3093–3102. doi: 10.1021/acschembio.7b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chevrette MG, Carlson CM, Ortega HE, Thomas C, Ananiev GE, Barns KJ, Book AJ, Cagnazzo J, Carlos C, Flanigan W, Grubbs KJ, Horn HA, Hoffmann FM, Klassen JL, Knack JJ, Lewin GR, McDonald BR, Muller L, Melo WGP, Pinto-Tomás AA, Schmitz A, Wendt-Pienkowski E, Wildman S, Zhao M, Zhang F, Bugni TS, Andes DR, Pupo MT, Currie CR. 2019. The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun 10:516. doi: 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevrette MG, Aicheler F, Kohlbacher O, Currie CR, Medema MH. 2017. SANDPUMA: ensemble predictions of nonribosomal peptide chemistry reveal biosynthetic diversity across Actinobacteria. Bioinformatics 33:3202–3210. doi: 10.1093/bioinformatics/btx400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubbendieck RM, Vargas-Bautista C, Straight PD. 2016. Bacterial communities: interactions to scale. Front Microbiol 7:1234. doi: 10.3389/fmicb.2016.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott JJ, Oh D-C, Yuceer MC, Klepzig KD, Clardy J, Currie CR. 2008. Bacterial protection of beetle-fungus mutualism. Science 322:63–63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaltenpoth M, Göttler W, Herzner G, Strohm E. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol 15:475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 48.Engl T, Kroiss J, Kai M, Nechitaylo TY, Svatoš A, Kaltenpoth M. 2018. Evolutionary stability of antibiotic protection in a defensive symbiosis. Proc Natl Acad Sci U S A 115:E2020–E2029. doi: 10.1073/pnas.1719797115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matarrita-Carranza B, Moreira-Soto RD, Murillo-Cruz C, Mora M, Currie CR, Pinto-Tomas AA. 2017. Evidence for widespread associations between neotropical hymenopteran insects and Actinobacteria. Front Microbiol 8:2016. doi: 10.3389/fmicb.2017.02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanArnam EB, Currie CR, Clardy J. 2017. Defense contracts: molecular protection in insect-microbe symbioses. Chem Soc Rev 47:1638–1651. doi: 10.1039/c7cs00340d. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi Y, Kawata M. 2013. Alternative trait combinations and secondary resource partitioning in sexually selected color polymorphism. Ecol Evol 3:2038–2046. doi: 10.1002/ece3.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi Y, Kawata M. 2013. A comprehensive test for negative frequency-dependent selection. Popul Ecol 55:499–509. doi: 10.1007/s10144-013-0372-7. [DOI] [Google Scholar]
- 53.Mueller UG, Dash D, Rabeling C, Rodrigues A. 2008. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution 62:2894–2912. doi: 10.1111/j.1558-5646.2008.00501.x. [DOI] [PubMed] [Google Scholar]
- 54.Mueller UG, Ishak H, Lee JC, Sen R, Gutell RR. 2010. Placement of attine ant-associated Pseudonocardia in a global Pseudonocardia phylogeny (Pseudonocardiaceae, Actinomycetales): a test of two symbiont-association models. Antonie Van Leeuwenhoek 98:195–212. doi: 10.1007/s10482-010-9427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomulkiewicz R, Drown DM, Dybdahl MF, Godsoe W, Nuismer SL, Pepin KM, Ridenhour BJ, Smith CI, Yoder JB. 2007. Dos and don’ts of testing the geographic mosaic theory of coevolution. Heredity (Edinb) 98:249–258. doi: 10.1038/sj.hdy.6800949. [DOI] [PubMed] [Google Scholar]
- 56.Parker MA. 1985. Local population differentiation for compatibility in an annual legume and its host-specific fungal pathogen. Evolution 39:713. doi: 10.2307/2408672. [DOI] [PubMed] [Google Scholar]
- 57.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. 2011. Host-parasite local adaptation after experimental coevolution of Caenorhabditis elegans and its microparasite Bacillus thuringiensis. Proc Biol Sci 278:2832–2839. doi: 10.1098/rspb.2011.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janzen DH. 1980. When is it coevolution? Evolution 34:611–612. doi: 10.1111/j.1558-5646.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- 59.Singer MC, Thomas CD. 1996. Evolutionary responses of a butterfly metapopulation to human- and climate-caused environmental variation. Am Nat 148:S9–S39. doi: 10.1086/285900. [DOI] [Google Scholar]
- 60.Hansen DA, Koch AA, Sherman DH. 2017. Identification of a thioesterase bottleneck in the pikromycin pathway through full-module processing of unnatural pentaketides. J Am Chem Soc 139:13450–13455. doi: 10.1021/jacs.7b06432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikheyev AS, Mueller UG, Boomsma JJ. 2007. Population genetic signatures of diffuse co-evolution between leaf-cutting ants and their cultivar fungi. Mol Ecol 16:209–216. doi: 10.1111/j.1365-294X.2006.03134.x. [DOI] [PubMed] [Google Scholar]
- 62.Poulsen M, Cafaro M, Boomsma JJ, Currie CR. 2005. Specificity of the mutualistic association between actinomycete bacteria and two sympatric species of Acromyrmex leaf-cutting ants. Mol Ecol 14:3597–3604. doi: 10.1111/j.1365-294X.2005.02695.x. [DOI] [PubMed] [Google Scholar]
- 63.Cafaro MJ, Poulsen M, Little AEF, Price SL, Gerardo NM, Wong B, Stuart AE, Larget B, Abbot P, Currie CR. 2011. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc Biol Sci 278:1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montoya QV, Sutta Martiarena MJ, Polezel DA, Kakazu S, Rodrigues A. 2019. More pieces to a huge puzzle: two new Escovopsis species from fungus gardens of attine ants. Mc 46:97–118. doi: 10.3897/mycokeys.46.30951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Mégraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 68.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 69.Chin C, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 70.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, Bateman A. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res 37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, Jensen LJ, von Mering C, Bork P. 2012. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res 40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. 2017. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Navarro-Muñoz J, Selem-Mojica N, Mullowney M, Kautsar S, Tryon J, Parkinson E, De Los Santos E, Yeong M, Cruz-Morales P, Abubucker S, Roeters A, Lokhorst W, Fernandez-Guerra A, Teresa Dias Cappelini L, Thomson R, Metcalf W, Kelleher N, Barona-Gomez F, Medema MH. 2018. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv 10.1101/445270. [DOI] [PMC free article] [PubMed]
- 79.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kofler R, Pandey RV, Schlötterer C. 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.