Tripartite symbioses between anaerobic ciliated protists and their intracellular archaeal and bacterial symbionts are not uncommon, but most reports have been based mainly on microscopic observations. Deeper insights into the function, ecology, and evolution of these fascinating symbioses involving partners from all three domains of life have been hampered by the difficulties of culturing anaerobic ciliates in the laboratory and the frequent loss of their prokaryotic partners during long-term cultivation. In the present study, we report the isolation of an anaerobic scuticociliate, strain GW7, which has been stably maintained in our laboratory for more than 3 years without losing either of its endosymbionts. Unexpectedly, molecular characterization of the endosymbionts revealed that the bacterial partner of GW7 is phylogenetically related to intranuclear endosymbionts of aerobic ciliates. This strain will enable future genomic, transcriptomic, and proteomic analyses of the interactions in this tripartite symbiosis and a comparison with endosymbioses in aerobic ciliates.
KEYWORDS: symbiosis; anaerobic ciliate; methanogen; hydrogenosome; “Candidatus Hydrogenosomobacter endosymbioticus,” sewage treatment reactor
ABSTRACT
A number of anaerobic ciliates, unicellular eukaryotes, intracellularly possess methanogenic archaea and bacteria as symbiotic partners. Although this tripartite relationship is of interest in terms of the fact that each participant is from a different domain, the difficulty in culture and maintenance of those host species with symbiotic partners has disturbed both ecological and functional studies so far. In this study, we obtained a stable culture of a small anaerobic scuticociliate, strain GW7. By transmission electron microscopic observation and fluorescent in situ hybridization with domain-specific probes, we demonstrate that GW7 possesses both archaeal and bacterial endosymbionts in its cytoplasm. These endosymbionts are in dependently associated with hydrogenosomes, which are organelle producing hydrogen and ATP under anaerobic conditions. Clone library analyses targeting prokaryotic 16S rRNA genes, fluorescent in situ hybridization with endosymbiont-specific probes, and molecular phylogenetic analyses revealed the phylogenetic affiliations and intracellular localizations of these endosymbionts. The endosymbiotic archaeon is a methanogen belonging to the genus Methanoregula (order Methanomicrobiales); a member of this genus has previously been described as the endosymbiont of an anaerobic ciliate from the genus Metopus (class Armophorea), which is only distantly related to strain GW7 (class Oligohymenophorea). The endosymbiotic bacterium belongs to the family Holosporaceae of the class Alphaproteobacteria, which also comprises several endosymbionts of various aerobic ciliates. For this endosymbiotic bacterium, we propose a novel candidate genus and species, “Candidatus Hydrogenosomobacter endosymbioticus.”
IMPORTANCE Tripartite symbioses between anaerobic ciliated protists and their intracellular archaeal and bacterial symbionts are not uncommon, but most reports have been based mainly on microscopic observations. Deeper insights into the function, ecology, and evolution of these fascinating symbioses involving partners from all three domains of life have been hampered by the difficulties of culturing anaerobic ciliates in the laboratory and the frequent loss of their prokaryotic partners during long-term cultivation. In the present study, we report the isolation of an anaerobic scuticociliate, strain GW7, which has been stably maintained in our laboratory for more than 3 years without losing either of its endosymbionts. Unexpectedly, molecular characterization of the endosymbionts revealed that the bacterial partner of GW7 is phylogenetically related to intranuclear endosymbionts of aerobic ciliates. This strain will enable future genomic, transcriptomic, and proteomic analyses of the interactions in this tripartite symbiosis and a comparison with endosymbioses in aerobic ciliates.
INTRODUCTION
Ciliated protists inhabit a wide variety of habitats, from aquatic to terrestrial and from aerobic to anaerobic environments. It has been estimated that 27,000 to 40,000 free-living ciliate species exist in the world (1). Based on the systematics of ciliated protists (2), Fenchel and Finlay (3) reviewed that free-living anaerobic ciliates have been found in at least eight orders: three of them consist of only anaerobic members, and the other five orders include both anaerobic and aerobic members. The former include the orders Armophorida (e.g., Metopus) and Plagiopylida (e.g., Trimyema), and the latter include the order Pleuronematida (e.g., Cyclidium) (see reference 3 and references therein).
Anaerobic ciliates, as well as trichomonads, chytrid fungi, and heterolobosean flagellates, possess hydrogenosomes within their cells, which are organelles homologous to mitochondria and produce hydrogen and ATP under anaerobic conditions (4). Actually, hydrogenosomes have been known to share an ancestry with mitochondria (5). Another feature of anaerobic ciliates is that not all but many species, belonging to distinct lineages, establish symbiotic relationships with two types of microorganisms: methanogenic archaea and bacteria (6–9). This tripartite relationship is of interest in terms of the fact that each participant is from a different domain of life. Despite a number of previous descriptions of endosymbionts in various anaerobic ciliates (10), the interactions between the hosts and partners from the three different domains have remained unclear.
A free-living anaerobic ciliate, Trimyema compressum (class Plagiopylea, order Plagiopylida), inhabits various anoxic aquatic environments and establishes a symbiotic association with a methanogenic archaeal and a bacterial endosymbiont (8, 11). In our previous studies, we obtained a laboratory culture of T. compressum strain S10 isolated from small-scale sewage treatment reactors and have maintained the strain for over 20 years in our laboratory (8, 12, 13). Because anaerobic ciliates have often lost their endosymbiotic microorganisms during laboratory maintenance (12, 14, 15), such stable culture strains of anaerobic ciliates are rare, with the exception of some host species (e.g., see references 16 and 17), although the usefulness of cultured strains has been well recognized. By using a cultured strain, physiological and metabolic profiles have been investigated, showing that both types of endosymbionts could positively affect the host’s fitness (8, 12). Recently, the complete genome sequence of the bacterial endosymbiont of T. compressum strain S10, strain TC1, was successfully determined (18). Genomic information on endosymbionts could give insights into their functions and the evolution of microbe-microbe symbiotic associations.
Based on the culture system for T. compressum (8, 12), we obtained a small anaerobic ciliate, strain GW7 (11). Since then, at the time of the writing of this paper, strain GW7 with endosymbiotic microorganisms has been maintained for more than 3 years in our laboratory. In this study, we characterized this anaerobic ciliate strain, GW7, and its endosymbionts by microscopic observations, molecular phylogenetic analyses, and fluorescent in situ hybridization (FISH).
RESULTS
Cultivation of an anaerobic ciliate, strain GW7.
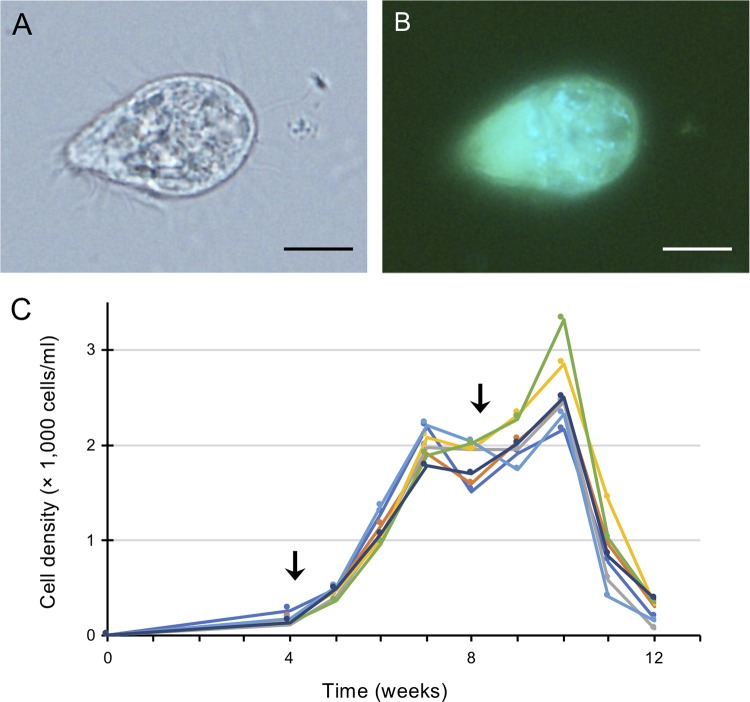
A medium based on one for methanogenic archaea, supplemented with a bacterial suspension as food, was used for culturing anaerobic ciliates (see Materials and Methods). After several passages of cultivation, we obtained a stable culture of a small anaerobic ciliate, strain GW7 (Fig. 1A). Inside the ciliate cells, blue-green autofluorescent cells, indicative of methanogenic endosymbionts harboring coenzyme F420 (10), were observed by epifluorescence microscopy (Fig. 1B). Cells of strain GW7 were 28.6 ± 2.7 μm long and 17.4 ± 2.3 μm wide (means ± standard deviations [SD]; n = 20). The growth property of this strain was then examined over 3 months (Fig. 1C). The maximum cell density in a 50-ml volume was 2,582 ± 386 cells/ml (mean ± SD; n = 7) 10 weeks after inoculation if 0.05 g food bacterium per vial was supplemented once a month. Under this culture condition, the doubling time of GW7 was 4.7 days during the exponential growth phase. Compared to the culture of T. compressum strain S10, the other strain cultured in our laboratory (8), strain GW7 was smaller and had a lower growth rate.
FIG 1.
An anaerobic scuticociliate strain, GW7. (A and B) Phase-contrast micrograph (A) and fluorescence micrograph (B) of a cell of strain GW7. Autofluorescence of coenzyme F420 (light blue) was detected. (C) Growth curve of strain GW7. The timing of supplementation of the food bacterium (0.05 g) is indicated by an arrow. Different-colored lines show each replicate (n = 7). Bars, 10 μm (A and B).
Molecular phylogenetic analysis of strain GW7.
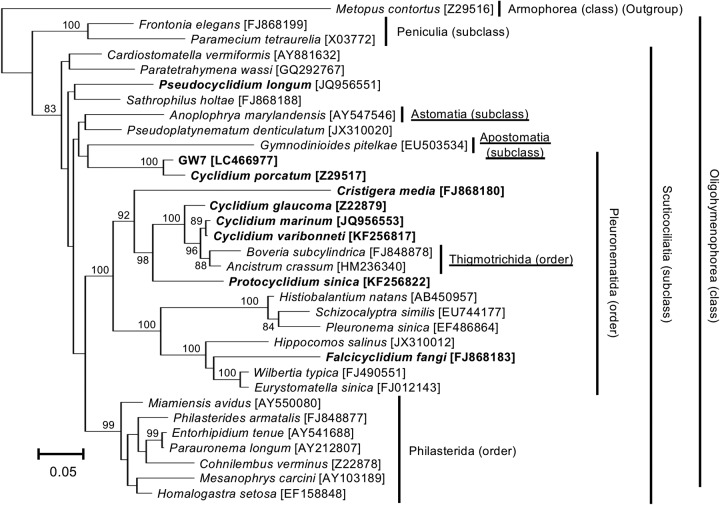
To obtain phylogenetic information on strain GW7, we performed clone library analysis targeting the eukaryotic 18S rRNA gene with DNA extracted from single ciliate cells. From two independent DNA samples, we obtained three and five clone sequences of the 18S rRNA gene of 1,685 bp (DDBJ/EMBL/GenBank accession no. LC466977 to LC466984). The pairwise differences between these clones were very small, at most 0.5%. All eight clones were subjected to BLASTN searches against the NCBI nt database. The top BLASTN hit for all clone sequences was the 18S rRNA gene sequence of an anaerobic ciliate, Cyclidium porcatum (DDBJ/EMBL/GenBank accession no. Z29517), with at most 96.9% sequence identity. Cy. porcatum is a freshwater anaerobic scuticociliate (class Oligohymenophorea) and harbors taxonomically unidentified methanogenic archaeal and bacterial endosymbionts (7). The class Oligohymenophorea includes the model organisms Paramecium and Tetrahymena, both of which are aerobic (2, 19). Phylogenetic analysis based on the 18S rRNA gene confirmed that the closest relative of strain GW7 was Cy. porcatum and that GW7 belonged to the class Oligohymenophorea (Fig. 2).
FIG 2.
Molecular phylogeny of an anaerobic scuticociliate strain, GW7. The tree displays a maximum likelihood (ML) phylogeny of ciliates belonging to the class Oligohymenophorea. An alignment of 1,524 nucleotide sites of the eukaryotic 18S rRNA gene was used. Accession numbers in the DDBJ/EMBL/GenBank DNA database are shown in square brackets. Bootstrap support values higher than 70% are depicted on or below the internal branches. Boldface type indicates the members of the family Cyclidiidae, which is a systematically problematic taxon. Other systematically problematic taxa are underlined.
Localization of endosymbionts within the host ciliate.
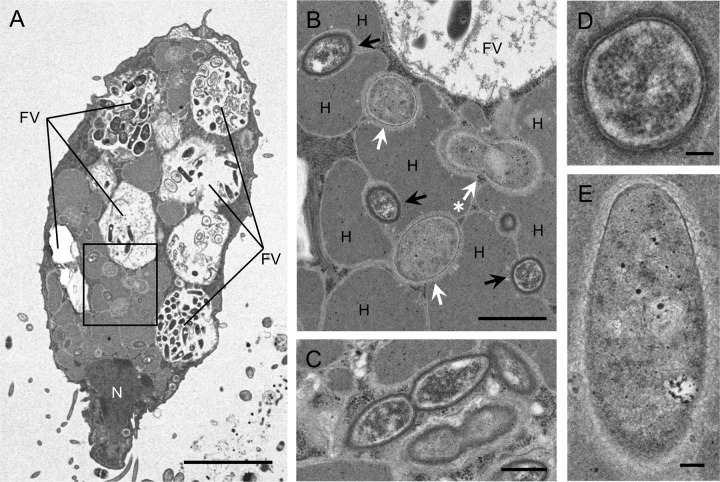
We examined the presence of endosymbiotic microorganisms and their localization inside the ciliated host cell by transmission electron microscopy (TEM) (Fig. 3). While food vacuoles were filled with microorganisms at various digestion steps, two different endosymbionts with a short rod shape were found to be localized in the cytoplasm of the host cell (Fig. 3A). Both endosymbionts were independently surrounded by hydrogenosomes, but direct contact between the endosymbionts was not observed (Fig. 3B). The observed subcellular localization of endosymbionts resembled the complex tripartite structure consisting of the host’s hydrogenosomes and methanogenic archaeal and bacterial endosymbionts reported in Cy. porcatum (7, 20). Dividing cells of both endosymbionts were found (Fig. 3B and C), indicating their adequate adaptation to the intracellular environment. Endosymbiotic cells of the electron-denser ones, probably methanogenic archaeal endosymbionts (see Discussion), were 0.9 to 1.4 μm long and 0.4 to 0.6 μm wide, with a discriminating cell wall structure (Fig. 3D). The other ones, probably bacterial endosymbionts, were 1.2 to 1.5 μm long and 0.4 to 0.8 μm wide, with an unclear cell membrane structure (Fig. 3E). These sizes were slightly different from those of the unidentified endosymbionts of Cy. porcatum (7).
FIG 3.
Transmission electron micrographs of an anaerobic scuticociliate strain, GW7. (A) Whole cell of strain GW7. (B) Enlarged view of the boxed region in panel A. Methanogenic archaeal and bacterial endosymbionts are indicated by black and white arrows, respectively. A dividing cell of the bacterial endosymbiont is indicated by a white arrow with an asterisk. (C) Dividing cells of the methanogenic archaeal endosymbiont. (D) Cross section of a methanogenic endosymbiont cell. The cell shows two discernible membranes. (E) Longitudinal section of a bacterial endosymbiont cell. Abbreviations: N, nucleus; H, hydrogenosome; FV, food vacuole. Bars, 5 μm (A), 1 μm (B), 500 nm (C), and 100 nm (D and E).
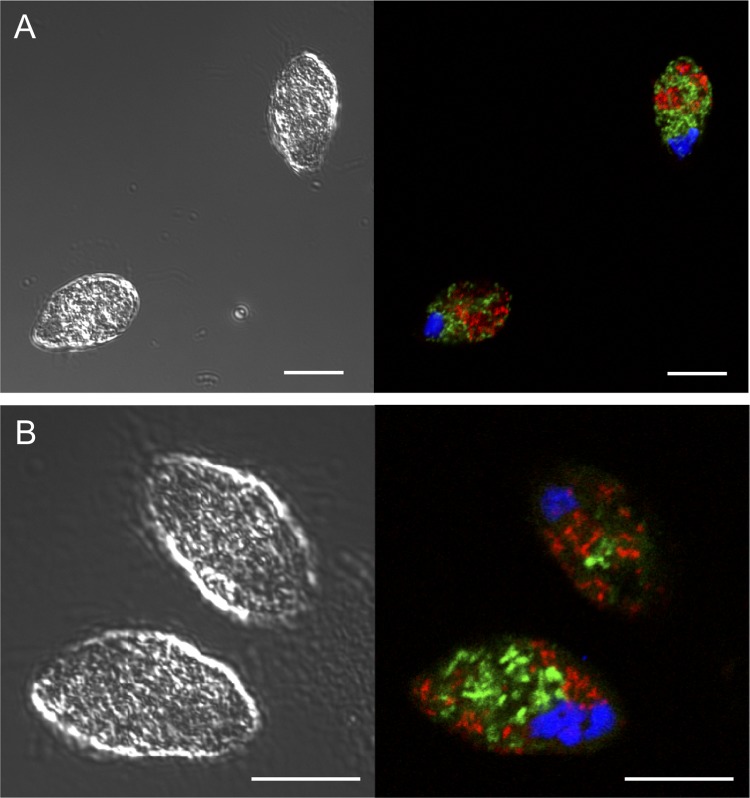
We then performed FISH analysis with universal archaeal and bacterial probes, confirming that the two endosymbionts inside the GW7 cell were indeed archaea and bacteria (Fig. 4A). As shown in TEM images (Fig. 3), these endosymbiotic microorganisms were localized not inside the nuclei, which were stained with 4′,6-diamidino-2-phenylindole (DAPI), but instead within the cytoplasm of the host cell (Fig. 4).
FIG 4.
Detection of endosymbionts in an anaerobic scuticociliate strain, GW7. Endosymbionts of strain GW7 were detected by FISH with universal and specific probes. Shown are differential interference contrast (left) and confocal laser scanning (right) micrographs. FISH was performed with EUB338 (Alexa 488) (green) and ARC915 (Alexa 568) (red) probes (A) and with GW7Met1 (Alexa 488) (green) and GW7Bac2 (Alexa 568) (red) probes (B). The host’s nucleus was stained with DAPI (blue). Formamide concentrations were 35% (A) and 20% (B). Bars, 10 μm.
Molecular identification of the endosymbionts.
To identify the two endosymbionts, clone library analyses targeting prokaryotic 16S rRNA genes were performed by using the universal archaeal and bacterial primer pairs (Table 1). From the two independent DNA samples used in the analysis of the host’s 18S rRNA, in total, 52 archaeal and 49 bacterial clones were obtained and partially sequenced. Their partial sequences were subjected to a BLASTN search against the NCBI 16S rRNA sequence database and clustered based on the top hit in the search. For the archaeal clones, the diversity among clones was very limited. They were classified into only two groups, both of which belong to the class Methanomicrobia: the first one (named Met1) consisted of 43 clones and their top BLASTN hit was Methanoregula formicica (DDBJ/EMBL/GenBank accession no. NR_102441), and the second one (Met2) consisted of nine clones and their top hit was Methanosaeta concilii (Methanothrix concilii) (DDBJ/EMBL/GenBank accession no. NR_102903).
TABLE 1.
PCR primers and FISH probes used in this study
| Purpose and target | Primer name | Fluorescent label | Formamide concn (%) | Sequence (5′–3′) | Reference(s) |
|---|---|---|---|---|---|
| PCR | |||||
| 18S rRNA of eukaryotes | 18S-42F | CTCAARGAYTAAGCCATGCA | 44 | ||
| 18S-1520R | CYGCAGGTTCACCTAC | 44 | |||
| 16S rRNA of bacteria | Eub11fa | TGRGTTTGATCMTGGCTYAG | 8 | ||
| Eub1511r | TGGDTACCTTGTTACGACTT | 8 | |||
| 16S rRNA of archaea | Ar109f | AMDGCTCAGTAACACGT | 46 | ||
| Ar1490Ra | GGHTACCTTGTTACGACTT | 45 | |||
| Colony PCR and sequencing | |||||
| Cloning vector pCR2.1 | pCR2.1f | GTAATACGACTCACTATAGGG | This study | ||
| pCR2.1r | ATTACGCCAAGCTTGGTACC | This study | |||
| Sequencing | |||||
| 18S rRNA of eukaryotes | EK-555F | AGTCTGGTGCCAGCAGCCGC | 44 | ||
| 16S rRNA of bacteria | Eub920r | CCGYCAATTCCTTTGAGTTT | 8 | ||
| 16S rRNA of archaea | Arc1000r | TCTCGCTCGTTGCCTGACT | 8, 47 | ||
| FISH | |||||
| 16S rRNA of endosymbiotic Methanoregula (Met1) of GW7 | GW7Met1 | Alexa 488 | 20 | CAGCCCGACTATCATTCAGCTG | This study |
| 16S rRNA of Methanosaeta spp. (used for detecting Met2) | MX825 | Alexa 488 | 0 | TCGCACCGTGGCCGACACCTAGC | 54 |
| 16S rRNA of the bacterial candidate (Bac1) of GW7 | GW7Bac1 | Alexa 568 | 0 | ACAATAACACTTGCTCACAA | This study |
| 16S rRNA of endosymbiotic “Candidatus Hydrogenosomobacter endosymbioticus” (Bac2) of GW7 | GW7Bac2 | Alexa 568 | 20 | CTCTGTTTCCAGAGCCCTCGAT | This study |
| 16S rRNA of the bacterial candidate (Bac3) of GW7 | GW7Bac3 | Alexa 568 | 0 | CCGACTGGTTACTTTCATAACC | This study |
| 16S rRNA of archaea | ARC915 | Alexa 488/Alexa 568 | 20/35 | GTGCTCCCCCGCCAATTCCT | 55 |
| 16S rRNA of most bacteria | EUB338 | Alexa 488/Alexa 568 | 20/35 | GCTGCCTCCCGTAGGAGT | 56 |
| 16S rRNA of Verrucomicrobia | EUB338III | Alexa 488 | 0 | GCTGCCACCCGTAGGTGT | 54 |
In the respective references, the primers are named Eub8f and 1490R.
Although no clone sequence of Lactococcus lactis, which was supplemented in the ciliate culture as food, was detected, the diversity among bacterial clones was greater than that of the archaeal clones (data not shown). In spite of such a high level of diversity, three groups based on top BLASTN hits were shared between the two independent DNA samples. The first bacterial group (named Bac1) consisted of 15 clones and their top hit was Limisphaera ngatamarikiensis (DDBJ/EMBL/GenBank accession no. NR_134756) of the phylum Verrucomicrobia, the second one (Bac2) consisted of seven clones and their top hit was Caedibacter caryophilus (DDBJ/EMBL/GenBank accession no. NG_041959) of the class Alphaproteobacteria, and the last one (Bac3) consisted of three clones and their top hit was Lyticum flagellatum (DDBJ/EMBL/GenBank accession no. NR_125566) of the class Alphaproteobacteria. The above-described two archaeal and three bacterial groups were considered candidate endosymbionts of GW7.
To examine which of the above-described candidates were true endosymbionts of strain GW7, FISH experiments were performed with specific probes for detecting each candidate. These probes were newly designed based on the partial clone sequences determined as described above (Table 1). A series of FISH experiments with confocal laser scanning microscopy (CLSM) demonstrated that the endosymbiotic methanogenic archaeon and bacterium were affiliated with Met1 and Bac2, respectively. Figure 4B shows a CLSM image of GW7 cells simultaneously detecting both endosymbionts with Met1- and Bac2-specific probes. For other candidate-specific probes, no positive signals were detected within the host cell (data not shown).
Phylogenetic analyses of the endosymbionts.
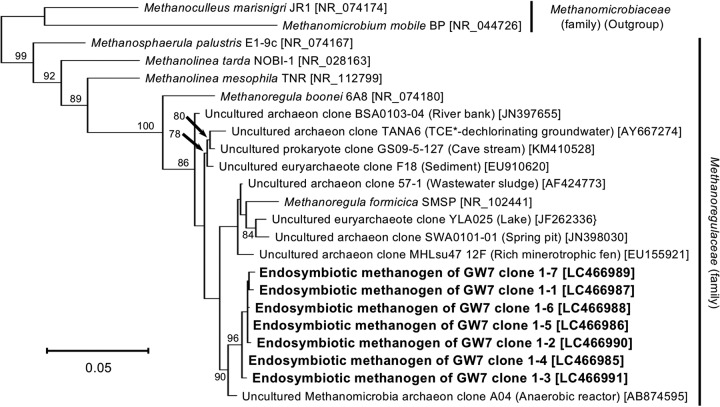
For obtaining phylogenetic information on the endosymbionts belonging to Met1 and Bac2, nearly full-length sequences of their 16S rRNA genes were determined and subjected to phylogenetic analyses. Figure 5 shows the phylogenetic relationships of seven endosymbiotic methanogenic archaeal clones (DDBJ/EMBL/GenBank accession no. LC466985 to LC466991) and their related species and uncultured clones. This phylogeny showed that the seven clones formed a monophyletic group, with a 96% bootstrap support value, and that the endosymbiotic methanogenic archaeon of strain GW7 belongs to the genus Methanoregula, with a 100% bootstrap support value. The sequence identities with its closely related described species were at most 97.3% with Methanoregula formicica (DDBJ/EMBL/GenBank accession no. NR_102441) and 95.6% with Methanoregula boonei (DDBJ/EMBL/GenBank accession no. NR_074180).
FIG 5.
Molecular phylogeny of the methanogenic archaeal endosymbiont of an anaerobic scuticociliate strain, GW7. The tree displays an ML phylogeny of seven clones of the endosymbiotic methanogenic archaeon derived from an anaerobic ciliate strain, GW7, together with related species/clones of the family Methanoregulaceae. An alignment of 1,298 nucleotide sites of the archaeal 16S rRNA gene was used. The origins or sources of isolation of species/clones are presented in parentheses. Accession numbers in the DDBJ/EMBL/GenBank DNA database are shown in square brackets. Bootstrap support values higher than 70% are depicted on or below the internal branches. *TCE, trichloroethene.
Figure 6 shows the phylogenetic position of the endosymbiotic bacterium. All the clones (DDBJ/EMBL/GenBank accession no. LC466992 to LC466998) showed a monophyletic relationship, with a 100% bootstrap support value, and were included in the family Holosporaceae of the class Alphaproteobacteria, with an 81% bootstrap support value. Interestingly, the genus Holospora, which comprises nuclear bacterial endosymbionts of aerobic ciliate Paramecium spp. (21), also belongs to this family. Furthermore, the order Holosporales (22, 23) includes a number of nuclear endosymbionts of various ciliates and amoebae, e.g., “Candidatus Gortzia,” Caedibacter, and so on (23–28). On the other hand, the 16S rRNA gene of the endosymbiont showed a lower sequence identity with those of the closely related described species: at most 87.3% with Holospora elegans (DDBJ/EMBL/GenBank accession no. BAUP01000039), 86.6% with Holospora obtusa (DDBJ/EMBL/GenBank accession no. X58198), and 85.2% with Caedibacter caryophilus (DDBJ/EMBL/GenBank accession no. X71837). Statistically supported monophyletic relationships and 16S rRNA sequence identity with the type genus Holospora (>81%) (23) indicated that the bacterial endosymbiont of strain GW7 was a novel member of the family Holosporaceae. Based on the localization within the host ciliated cell, for the bacterial endosymbiont of strain GW7, we propose a novel candidate genus and species, “Candidatus Hydrogenosomobacter endosymbioticus.”
FIG 6.
Molecular phylogeny of the bacterial endosymbiont of an anaerobic scuticociliate strain, GW7. The tree displays an ML phylogeny of seven clones of the endosymbiotic bacterium “Candidatus Hydrogenosomobacter endosymbioticus” derived from an anaerobic ciliate strain, GW7, together with related species/clones of the order Holosporales. An alignment of 1,333 nucleotide sites of the bacterial 16S rRNA gene was used. The origins or sources of isolation of species/clones are presented in parentheses. Accession numbers in the DDBJ/EMBL/GenBank DNA database are shown in square brackets. Bootstrap support values higher than 70% are depicted on or below the internal branches.
DISCUSSION
In this study, we report that (i) a stable laboratory culture of a small anaerobic scuticociliate, strain GW7, was obtained; (ii) its hydrogenosome-associated endosymbiotic methanogenic archaeon was a member of the genus Methanoregula; and (iii) its hydrogenosome-associated endosymbiotic bacterium was a Holospora-related alphaproteobacterium, “Ca. Hydrogenosomobacter endosymbioticus.” Taxonomic information on the endosymbionts provided us with a hint for morphologically distinguishing them in the TEM images. It has been reported for H. obtusa that this species was Gram stain negative, but the inner membrane was hardly visible in conventional thin sections (29). Probably, the bacterial endosymbiont of strain GW7, “Ca. Hydrogenosomobacter endosymbioticus,” was in the same situation, indicating that cells with an unclear cell membrane structure in the TEM image were the bacterial endosymbiont (Fig. 3E). On the other hand, more-electron-dense cells, whose cell wall structure was actually similar to that of Methanoregula boonei, with an S layer and a cytoplasmic membrane (30), were the methanogenic archaeal endosymbiont (Fig. 3D).
Hirakata et al. (9) reported that an endosymbiotic methanogenic archaeon of an anaerobic ciliate, Metopus sp., showed a 16S rRNA gene sequence identity of 99% with Methanoregula boonei. It was surprising for us that the endosymbiotic methanogenic archaeon of strain GW7 was a member of the genus Methanoregula because these host anaerobic ciliates are only distantly related to each other, as shown in Fig. 2. Although the endosymbiotic methanogen of Metopus sp. was not included in Fig. 5 due to its short read length produced by Illumina MiSeq, a sequence comparison between the methanogenic archaeal endosymbiont of GW7 and that of Metopus sp. (DDBJ/EMBL/GenBank accession no. LC062151) showed 7 to 9 differences out of 252 aligned sites. This difference was at a level similar to that against Methanoregula boonei (8 different sites) and higher than that against Methanoregula formicica (1 to 2 different sites) in the same alignment region. In addition, the phylogenetic analysis based on the short-length alignment showed that the closest described species of the methanogenic endosymbionts of strain GW7 and Metopus sp. were Methanoregula formicica and Methanoregula boonei, respectively (data not shown). Thus, the endosymbiotic methanogenic archaea of GW7 and Metopus sp. would be different at the species level.
Phylogenetic affiliation of strain GW7.
Although the homology search and phylogenetic analysis based on 18S rRNA gene sequences revealed that the closest relative of strain GW7 was Cy. porcatum, monophyletic relationships of neither the genus Cyclidium nor the family Cyclidiidae were supported in the molecular phylogeny. In addition, our phylogeny was inconsistent with the current, morphology-based taxonomy: the subclasses Apostomatia and Astomatia were nested within the subclass Scuticociliatia (Fig. 2). Phylogenetic uncertainty in this lineage has also been reported in previous phylogenetic research based on concatenated data for three rRNA genes, which generally provide higher taxonomic resolution than single-rRNA data (31, 32). It has been discussed that two main reasons would cause this uncertainty in this lineage (33, 34). First, scuticociliates are generally small and share similar morphological traits, resulting in many taxonomic misidentifications. Second, detailed information on some key taxa, including both morphological and molecular data, is still missing. These features would also make the revision of the existing classification and systematics difficult. For the above-mentioned reasons, we are not sure that GW7 is a member of the genus Cyclidium and/or the family Cyclidiidae; however, it is never doubtful that GW7 is a member of the subclass Scuticociliatia. By continuing to maintain strain GW7 in the laboratory, detailed taxonomic assignment of this strain might be accomplished in the future.
Non-ciliate-associated prokaryotes in culture.
At the timing of culture passage (1- to 2-month interval), we routinely confirmed the uniformity of the ciliate culture by microscopic examination. However, it also contained numerous unidentified free-living prokaryotes derived from the original sludge sample. This caused the prokaryotic diversity detected in our clone library analyses, as described in Results. Actually, the most abundant bacterial clone, Bac1, belonging to the phylum Verrucomicrobia, was demonstrated to be present in the culture independent of the ciliate cells by a clone-specific in situ hybridization experiment (data not shown). In addition, we attempted to cultivate bacterial constituents from both T. compressum S10 and GW7 cultures and succeeded in obtaining the isolate assigned to verrucomicrobial clone Bac1 (K. Takeshita and N. Shinzato, unpublished data). Considering the estimated large population of the clone in the ciliate culture, it might play an important role for the growth of the ciliate, for example, by providing some nutrients.
Possible role(s) of the endosymbionts.
It has generally been thought that anaerobic ciliates can acquire energy more efficiently by possessing endosymbiotic methanogenic archaea. This is because host ciliates can form more oxidized fermentation products since their endosymbiotic methanogenic archaea keep hydrogen partial pressure in the host cells very low by utilizing hydrogens for producing energy and methane (12, 35). Compared to methanogenic and archaeal endosymbionts, our understanding about functions of bacterial endosymbionts in anaerobic ciliates is still not enough. Physically direct contact of the bacterial endosymbiont with the host’s hydrogenosomes was observed in Cy. porcatum and strain GW7 (7) (Fig. 3B), indicating a direct interaction between the host ciliate and the bacterial endosymbiont via hydrogenosomes. In the case of methanogenic archaea, their localization to neighboring hydrogenosomes is very reasonable because it would maximize the acquisition of hydrogen, which is a small molecule produced by the organelle and spreads immediately. The reason why “Ca. Hydrogenosomobacter endosymbioticus” lives near hydrogenosomes might also be because it receives small molecules produced from the organelles effectively. It remains unknown what kinds of molecules “Ca. Hydrogenosomobacter endosymbioticus” receives from hydrogenosomes. One possible molecule is hydrogen. If so, this bacterial endosymbiont would act as a backup or would even be a competitor of the methanogenic archaeal endosymbiont. We found that the number of endosymbionts and their proportion vary among host cells (Fig. 4A and B); this might be caused by competition between the endosymbionts in each host cell. In addition, the development of a mutualistic interaction between the two endosymbionts seems unlikely in view of the frequent replacement of the methanogenic partner in the evolutionary history of the host ciliates. For instance, Trimyema ciliates have been reported to possess phylogenetically distinct, Methanobrevibacter- or Methanocorpusculum-related methanogens as endosymbionts (8, 36). On the other hand, Metopus spp. are known to have Methanocorpusculum- or Methanobacterium-related methanogens in the cytoplasm (37, 38). It is unclear whether such a symbiont switch had occurred in the lineage of the GW7 ciliate, and the extent of colonization of other anaerobic scuticociliates with their respective partners also remains to be investigated. Furthermore, genomic and transcriptomic analyses and quantitative comparison of these endosymbionts, as well as information on the host ciliate, would clarify the functions and their relationship in the future.
Ecological and evolutionary interests in the bacterial endosymbiont.
From phylogenetic analyses of host ciliates, it is evident that the appearance of hydrogenosomes and their subsequent adaptation to anaerobic environments have parallelly and repeatedly occurred, at least three times, in their evolution (39–41). The lineage including GW7 and Cy. porcatum was one of such adaptations that occurred; therefore, the stably cultured strain GW7 might be suitable for investigating anaerobic adaptation in ciliates. In addition, it is intriguing that “Ca. Hydrogenosomobacter endosymbioticus” is phylogenetically related to the bacterial endosymbionts of aerobic ciliate Paramecium spp. (Fig. 6). These facts indicate that adaptation to anaerobic environments in the host ciliate might force this bacterial endosymbiont to adapt there at the same time. For understanding this ecological and evolutionary issue, in addition to detailed studies with strain GW7, more extensive and intensive investigations of endosymbiotic interactions in various anaerobic ciliates are needed.
From another point of view, the difference in the subcellular localizations of bacterial endosymbionts between the anaerobic and aerobic ciliates is also of interest. “Ca. Hydrogenosomobacter endosymbioticus” has never been found within the host nuclei (Fig. 3 and 4), which different from bacterial endosymbionts of aerobic ciliate Paramecium spp. (21). One possibility is that “Ca. Hydrogenosomobacter endosymbioticus” has already lost genetic resources for invading the host nuclei during a long-term partnership with the host ciliate. Genome sequencing of “Ca. Hydrogenosomobacter endosymbioticus” and comparative analysis with related genomes (42, 43) might highlight candidate genes involved in endonuclear localization and anaerobic adaptation in this bacterial lineage.
Description of candidate taxa.
“Candidatus Hydrogenosomobacter” (Hy.dro.ge.no.so.mo.bac′ter. N.L. n. hydrogenosome, hydrogenosome; N.L. masc. n. bacter, a rod; N.L. masc. n. Hydrogenosomobacter, a rod living close to the hydrogenosome). Alphaproteobacteria, Holosporales. Only the type species “Ca. Hydrogenosomobacter endosymbioticus” is known, and its 16S rRNA gene sequence (DDBJ/EMBL/GenBank accession no. LC466993; 1,418 bp) shows percent identities of 87.2% with Holospora elegans (DDBJ/EMBL/GenBank accession no. BAUP01000039), 86.5% with H. obtusa (DDBJ/EMBL/GenBank accession no. X58198), and 85.1% with Cae. caryophilus (DDBJ/EMBL/GenBank accession no. X71837).
“Candidatus Hydrogenosomobacter endosymbioticus” (en.do.sym.bi.o.ti′cus. Gr. pref. endo-, within; N.L. adj. symbioticus, from the Greek biōtikos, living together; N.L. adj. endosymbioticus, living symbiotically within [another organism]). It is a short rod, 1.2 to 1.5 μm long and 0.4 to 0.8 μm wide. It lives in the cytoplasm, neighboring hydrogenosomes, of an anaerobic scuticociliate strain, GW7. Cultivation without the host ciliate is not possible so far. The host ciliate is maintained in the laboratory of Naoya Shinzato, University of the Ryukyus, Japan. Probe GW7Bac2 (5′-CTCTGTTTCCAGAGCCCTCGAT-3′) is available for specific detection of this species by FISH. It is the type species of the genus.
MATERIALS AND METHODS
Culture medium for anaerobic ciliates.
The medium for culturing anaerobic ciliates was based on one for methanogenic archaea but with some modifications. The ciliate culture medium contained the following: KH2PO4 at 0.3 g/liter, NaCl at 0.6 g/liter, MgCl2·2H2O at 0.1 g/liter, CaCl2·2H2O at 0.08 g/liter, NH4Cl at 1.0 g/liter, NaHCO3 at 1.0 g/liter, a 1.0% vitamin solution for DSM medium 141 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH), a 0.1% resazurin solution (1.0 g/liter), and a 0.1% trace element solution. The trace element solution contained the following: nitrilotriacetic acid at 12.8 g/liter, FeCl3·6H2O at 1.35 g/liter, MnCl2·4H2O at 0.1 g/liter, CoCl2·6H2O at 0.024 g/liter, CaCl2·2H2O at 0.1 g/liter, ZnCl2 at 0.1 g/liter, CuCl2·2H2O at 0.025 g/liter, H3BO3 at 0.01 g/liter, Na2MoO4·2H2O at 0.024 g/liter, NaCl at 1.0 g/liter, and NiCl2·6H2O at 0.12 g/liter. After dispensing the medium into culture vials and sparging them with a gas mixture of 80% N2 and 20% CO2, 1.0% volumes of each reductant solution (Na2S·9H2O at 8.0 g/liter and cysteine-HCl at 8.0 g/liter) and a food bacterial suspension (0.1 g/ml) were added (final concentration of the food bacterium of 0.001 g/ml or 2.8 × 108 cells/ml). The culture medium for the food bacterium Lactococcus lactis strain A1 was prepared according to methods reported in a previous study (8). Harvested food bacterial cells were stored at −20°C and resuspended in distilled water (DW) at the above-mentioned concentrations before use.
Isolation and maintenance of the ciliate.
The sludge samples containing ciliate cells were anaerobically collected from a large-scale sewage treatment reactor at Ginowan Sewage Treatment Center in Okinawa Prefecture, Japan, in July 2015. The procedure for isolating anaerobic ciliates was performed according to that for T. compressum described in a previous study (8). Culture vials were kept at 23°C without light. For maintaining the isolated ciliate strain, 0.1 to 1 ml of the culture, depending on the cell density, was transferred to a new culture vial (100 ml) containing 50 ml of fresh medium with food bacterial cells every 1 to 2 months.
The autofluorescence of coenzyme F420 inside the ciliate cells was detected by fluorescence microscopy with a U-MWBV2 fluorescence filter cube (excitation filter at 400 to 440 nm, emission filter at 475 nm, and dichromatic mirror at 455 nm; Olympus) after fixation with 4% formaldehyde.
For drawing growth curves, ciliate cells were fixed with 4% formaldehyde and directly counted with an optical plastic plankton counter (Matsunami Glass Ind.) every week after 1 month. Five hundred microliters of a food bacterial suspension (0.1 g/ml) was added every month.
Transmission electron microscopy.
The culture of strain GW7 was prefixed by adding an equal volume of 2× fixation solution (4% paraformaldehyde [PFA] and 4% glutaraldehyde [GA] in 0.1 M phosphate-buffered saline [PBS] [pH 7.4]) and incubating the culture at 4°C for 1 h. After three washes with 0.1 M phosphate buffer (PB) for 20 min each, postfixation was performed with 2% OsO4 in 0.1 M PB at 4°C for 2 h. The fixed cells were dehydrated in 50%, 70%, 90%, and 100% ethanol. The schedule was as follows: 50% and 70% for 20 min each at 4°C, 90% for 20 min at room temperature, and three times with 100% for 20 min each at room temperature. The samples were infiltrated with propylene oxide (PO) two times for 10 min and put into a 70:30 mixture of PO and resin (Quetol-812; Nisshin EM Co.) for 1 h. After PO was volatilized, the samples were transferred to fresh 100% resin and polymerized at 60°C for 48 h. Ultrathin sections (70 nm) were cut with a diamond knife by using an ultramicrotome (Ultracut UCT; Leica) and stained with 2% uranyl acetate at room temperature for 15 min on copper grids. After washing with DW, the sections were secondarily stained with a lead stain solution (Sigma-Aldrich Co.) at room temperature for 3 min. The stained sections were examined by using a transmission electron microscope (JEM-1400Plus; JEOL Ltd.) at an acceleration voltage of 100 kV.
DNA extraction, cloning, and Sanger sequencing.
Before DNA extraction, the ciliate culture passed through an 11-μm nylon net filter was washed three times with fresh culture medium. Single ciliate cells were transferred to new tubes with 9 μl of fresh medium under microscopy (CKX41; Olympus) with a micromanipulator (Transferman NK2; Eppendorf). After three cycles of freezing and thawing, 1 μl of lysozyme (10 mg/ml) was added to the suspension, and the mixture was incubated at 37°C for 30 min. Next, 2.5 μl of proteinase K (1 mg/ml) and 12.5 μl of extraction buffer (40 mM Tris-HCl [pH 8.0], 0.2 mM EDTA [pH 8.0], 1% Tween 20, and 0.2% Nonidet P-40) were added, and the mixture was incubated at 37°C for 30 min, followed by inactivation treatment of the enzymes by incubation at 95°C for 10 min.
The 18S rRNA gene of the ciliate and 16S rRNA genes of bacteria and archaea were amplified with primer sets 18S-42F and 18S-1520R (44), Eub11f and Eub1511r (8), and Ar109f and Ar1490R (45, 46), respectively (Table 1). PCR was conducted with TaKaRa Ex Taq (TaKaRa Bio) and its supplemented buffer system. The temperature profile for the eukaryotic 18S rRNA gene and archaeal 16S rRNA was as follows: (i) 95°C for 3 min; (ii) 35 cycles, with 1 cycle consisting of 95°C for 30 s, 53°C for 30 s, and 72°C for 2 min; and (iii) a final extension step at 72°C for 10 min. The temperature profile for bacterial 16S rRNA was as follows: (i) 95°C for 3 min; (ii) 10 cycles, with 1 cycle consisting of 95°C for 30 s, 56°C for 30 s, and 72°C for 1.5 min; (iii) 30 cycles, with 1 cycle consisting of 95°C for 30 s, 53°C for 30 s, and 72°C for 1.5 min; and (iv) a final extension step at 72°C for 10 min.
The PCR products of rRNA genes were cloned with the TOPO TA cloning kit (Invitrogen). Single colonies were picked up with sterile toothpicks and subjected to colony PCR with a cloning vector-specific primer set, pCR2.1f and pCR2.1r (Table 1). Colony PCR with TaKaRa Ex Taq was performed under the following temperature profile: (i) 95°C for 3 min; (ii) 30 cycles, with 1 cycle consisting of 95°C for 30 s, 60°C for 30 s, and 72°C for 2 min; and (iii) a final extension step at 72°C for 10 min. The PCR products of insert-positive colonies were cleaned up with exonuclease I and alkaline phosphatase (shrimp) (both from TaKaRa Bio). Sanger sequencing was performed by Macrogen Japan Corp. with sequencing primers EK-555F (44), Eub920r (8), and Ar1000r (8, 47) (Table 1). For all eight clones derived from the host, seven selected Met1 clones, and all seven Bac2 clones, cloning vector-specific primers pCR2.1f and pCR2.1r were also used for sequencing of full-length clones. The 7 Met1 clones were selected from 43 Met1 clones because partial sequences of these clones were not identical to each other. The sequence reads derived from the same colony were assembled with phredPhrap software (48, 49), followed by manual inspections. Calculation of sequence identity of prokaryotic 16S rRNA genes was performed with EzBioCloud (50).
Molecular phylogenetic analyses.
The clone sequences of the 18S rRNA gene of GW7 and 16S rRNA genes of bacterial and archaeal endosymbionts, as well as similar sequences retrieved from the NCBI nt database, were independently subjected to phylogenetic analysis. Multiple alignments were constructed by using SINA (SILVA Incremental Aligner) v1.2.11 (51), and gap-including and ambiguous sites in the alignments were then removed. Phylogenetic relationships were reconstructed with RAxML v8.2.3 (52) using the general time reversible + gamma (GTR+Γ) model of nucleotide substitution and the maximum likelihood (ML) method. The bootstrap values of 1,000 replicates for all internal branches were calculated with a rapid bootstrapping algorithm (53).
Fluorescence in situ hybridization.
Based on the results of clone library analyses, FISH probes for specifically detecting candidates of endosymbionts were newly designed. The specificity of the probes was checked with TestProbe 3.0 (https://www.arb-silva.de/search/testprobe/). A probe for specifically detecting Methanosaeta and universal archaeal and bacterial probes designed in the previous studies were also used. All probes used in this study are listed in Table 1.
Strain GW7 used in FISH analyses was cultured with medium without resazurin in order to minimize the level of autofluorescence. The ciliate culture was incubated for 5 min on ice and concentrated by centrifugation (110 × g for 5 min). Next, cells of GW7 were fixed with 4% paraformaldehyde in 10 mM PBS for 1 h on ice, followed by washing with 10 mM PBS. The fixed cells were picked up and loaded onto hybridization wells of a Teflon printed glass slide (Adcell; Funakoshi) under microscopy with a micromanipulator. After drying, the cells on the slide were dehydrated in 50%, 80%, and 100% ethanol for 5 min each and dried again. Prehybridization was performed by placing hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate) containing an appropriate concentration of formamide (Table 1) and 1% blocking reagent (Roche Diagnostics) onto each well and incubating the wells for 1 h at 46°C. Next, fluorescent probes (final concentration, 5 ng/μl each) were added to the buffer, and the slide was further incubated for 3 h at 46°C. Both hybridization steps were done in a moist chamber. After hybridization, the slide was incubated in fresh hybridization buffer for 20 min at 48°C, rinsed in DW cooled on ice for 5 s, and dried. The wells were mounted with Vectashield mounting medium (Vector Laboratories) containing DAPI (1 μg/ml). The hybridized ciliate cells were examined by CLSM using the Nikon C2 system (Nikon Instech). The obtained CLSM data were analyzed with NIS-Elements (Nikon Instech).
Accession number(s).
The nucleotide sequences of the 18S rRNA and 16S rRNA genes determined in this study have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence database under accession no. LC466977 to LC466998.
ACKNOWLEDGMENTS
We thank Atsushi Taira for maintaining the ciliate strain and Yumi Green for technical assistance.
This study was supported by JSPS KAKENHI grant no. 17H06979 (to Kazutaka Takeshita) and 17H01901 (to Naoya Shinzato).
Naoya Shinzato and Yoichi Kamagata designed the study. Takanori Yamada, Takashi Narihiro, and Naoya Shinzato sampled and isolated strain GW7. Kazutaka Takeshita, Yuto Kawahara, Michihiro Ito, and Naoya Shinzato performed experiments and data analyses. Kazutaka Takeshita and Naoya Shinzato wrote the manuscript.
REFERENCES
- 1.Foissner W, Chao A, Katz LA. 2008. Diversity and geographic distribution of ciliates (Protista: Ciliophora). Biodivers Conserv 17:345–363. doi: 10.1007/s10531-007-9254-7. [DOI] [Google Scholar]
- 2.Lynn DH. 2008. The ciliated protozoa: characterization, classification, and guide to the literature, 3rd ed Springer, Dordrecht, Netherlands. [Google Scholar]
- 3.Fenchel T, Finlay BJ. 2010. Free-living protozoa with endosymbiotic methanogens, p 1–11. In Hackstein JHP. (ed), Microbiology monographs 19, (endo)symbiotic methanogenic archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 4.van der Giezen M, Tovar J, Clark CG. 2005. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol 244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- 5.Muller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AGM, Martin WF. 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgcomb VP, Leadbetter ER, Bourland W, Beaudoin D, Bernhard JM. 2011. Structured multiple endosymbiosis of bacteria and archaea in a ciliate from marine sulfidic sediments: a survival mechanism in low oxygen, sulfidic sediments? Front Microbiol 2:55. doi: 10.3389/fmicb.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteban G, Guhl BE, Clarke KJ, Embley TM, Finlay BJ. 1993. Cyclidium porcatum n. sp.: a free-living anaerobic scuticociliate containing a stable complex of hydrogenosomes, eubacteria and archaeobacteria. Eur J Protistol 29:262–270. doi: 10.1016/S0932-4739(11)80281-6. [DOI] [PubMed] [Google Scholar]
- 8.Shinzato N, Watanabe I, Meng XY, Sekiguchi Y, Tamaki H, Matsui T, Kamagata Y. 2007. Phylogenetic analysis and fluorescence in situ hybridization detection of archaeal and bacterial endosymbionts in the anaerobic ciliate Trimyema compressum. Microb Ecol 54:627–636. doi: 10.1007/s00248-007-9218-1. [DOI] [PubMed] [Google Scholar]
- 9.Hirakata Y, Oshiki M, Kuroda K, Hatamoto M, Kubota K, Yamaguchi T, Harada H, Araki N. 2015. Identification and detection of prokaryotic symbionts in the ciliate Metopus from anaerobic granular sludge. Microbes Environ 30:335–338. doi: 10.1264/jsme2.ME15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackstein JHP. (ed). 2010. Microbiology monographs 19, (endo)symbiotic methanogenic archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 11.Shinzato N, Takeshita K, Kamagata Y. 2018. Methanogenic and bacterial endosymbionts of free-living anaerobic ciliates, p 37–53. In Hackstein JHP. (ed), Microbiology monographs 19, (endo)symbiotic methanogenic archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 12.Yamada K, Kamagata Y, Nakamura K. 1997. The effect of endosymbiotic methanogens on the growth and metabolic profile of the anaerobic free-living ciliate Trimyema compressum. FEMS Microbiol Lett 149:129–132. doi: 10.1016/S0378-1097(97)00068-2. [DOI] [Google Scholar]
- 13.Yamada K, Kamagata Y, Nakamura K, Inamori Y, Nakamura I. 1994. Selectivity of food bacteria for the growth of anaerobic ciliate Trimyema compressum. Arch Microbiol 161:229–233. doi: 10.1007/BF00248697. [DOI] [Google Scholar]
- 14.Wagener S, Pfennig N. 1987. Monoxenic culture of the anaerobic ciliate Trimyema compressum Lackey. Arch Microbiol 149:4–11. doi: 10.1007/BF00423128. [DOI] [Google Scholar]
- 15.Goosen NK, Wagener S, Stumm CK. 1990. A comparison of two strains of the anaerobic ciliate Trimyema compressum. Arch Microbiol 153:187–192. doi: 10.1007/BF00247819. [DOI] [Google Scholar]
- 16.Lewis WH, Sendra KM, Embley TM, Esteban GF. 2018. Morphology and phylogeny of a new species of anaerobic ciliate, Trimyema finlayi n. sp., with endosymbiotic methanogens. Front Microbiol 9:140. doi: 10.3389/fmicb.2018.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beinart RA, Beaudoin DJ, Bernhard JM, Edgcomb VP. 2018. Insights into the metabolic functioning of a multipartner ciliate symbiosis from oxygen-depleted sediments. Mol Ecol 27:1794–1807. doi: 10.1111/mec.14465. [DOI] [PubMed] [Google Scholar]
- 18.Shinzato N, Aoyama H, Saitoh S, Nikoh N, Nakano K, Shimoji M, Shinzato M, Satou K, Teruya K, Hirano T, Yamada T, Nobu MK, Tamaki H, Shirai Y, Park S, Narihiro T, Liu WT, Kamagata Y. 2016. Complete genome sequence of the intracellular bacterial symbiont TC1 in the anaerobic ciliate Trimyema compressum. Genome Announc 4:e01032-16. doi: 10.1128/genomeA.01032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagnes D, Roberts E, Lukeš J, Lowe C. 2012. The rise of model protozoa. Trends Microbiol 20:184–191. doi: 10.1016/j.tim.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Embley TM, Finlay BJ. 1993. Systematic and morphological diversity of endosymbiotic methanogens in anaerobic ciliates. Antonie Van Leeuwenhoek 64:261–271. doi: 10.1007/bf00873086. [DOI] [PubMed] [Google Scholar]
- 21.Fujishima M, Kodama Y. 2012. Endosymbionts in Paramecium. Eur J Protistol 48:124–137. doi: 10.1016/j.ejop.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Ferla MP, Thrash JC, Giovannoni SJ, Patrick WM. 2013. New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS One 8:e83383. doi: 10.1371/journal.pone.0083383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szokoli F, Castelli M, Sabaneyeva E, Schrallhammer M, Krenek S, Doak TG, Berendonk TU, Petroni G. 2016. Disentangling the taxonomy of Rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) sharing the cytoplasm of the ciliate protist Paramecium biaurelia. Appl Environ Microbiol 82:7236–7247. doi: 10.1128/AEM.02284-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beier CL, Horn M, Michel R, Schweikert M, Görtz HD, Wagner M. 2002. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl Environ Microbiol 68:6043–6050. doi: 10.1128/aem.68.12.6043-6050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boscaro V, Fokin SI, Schrallhammer M, Schweikert M, Petroni G. 2013. Revised systematics of Holospora-like bacteria and characterization of “Candidatus Gortzia infectiva,” a novel macronuclear symbiont of Paramecium jenningsi. Microb Ecol 65:255–267. doi: 10.1007/s00248-012-0110-2. [DOI] [PubMed] [Google Scholar]
- 26.Hess S, Suthaus A, Melkonian M. 2016. “Candidatus Finniella” (Rickettsiales, Alphaproteobacteria), novel endosymbionts of viridiraptorid amoeboflagellates (Cercozoa, Rhizaria). Appl Environ Microbiol 82:659–670. doi: 10.1128/AEM.02680-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potekhin A, Schweikert M, Nekrasova I, Vitali V, Schwarzer S, Anikina A, Kaltz O, Petroni G, Schrallhammer M. 2018. Complex life cycle, broad host range and adaptation strategy of the intranuclear Paramecium symbiont Preeria caryophila comb. nov. FEMS Microbiol Ecol 94:fiy076. doi: 10.1093/femsec/fiy076. [DOI] [PubMed] [Google Scholar]
- 28.Fokin SI, Serra V, Ferrantini F, Modeo L, Petroni G. 2019. “Candidatus Hafkinia simulans” gen. nov., sp. nov., a novel Holospora-like bacterium from the macronucleus of the rare brackish water ciliate Frontonia salmastra (Oligohymenophorea, Ciliophora): multidisciplinary characterization of the new endosymbiont and its host. Microb Ecol 77:1092–1106. doi: 10.1007/s00248-018-1311-0. [DOI] [PubMed] [Google Scholar]
- 29.Görtz HD, Ahlers N, Robenek H. 1989. Ultrastructure of the infectious and reproductive forms of Holospora obtusa, a bacterium infecting the macronucleus of Paramecium caudatum. J Gen Microbiol 135:3079–3085. doi: 10.1099/00221287-135-11-3079. [DOI] [Google Scholar]
- 30.Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB, Zinder SH. 2011. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol 61:45–52. doi: 10.1099/ijs.0.021782-0. [DOI] [PubMed] [Google Scholar]
- 31.Gao F, Katz LA, Song W. 2013. Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol Phylogenet Evol 68:55–63. doi: 10.1016/j.ympev.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Gao F, Gao S, Wang P, Katz LA, Song W. 2014. Phylogenetic analyses of cyclidiids (Protista, Ciliophora, Scuticociliatia) based on multiple genes suggest their close relationship with thigmotrichids. Mol Phylogenet Evol 75:219–226. doi: 10.1016/j.ympev.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 33.Gao F, Warren A, Zhang Q, Gong J, Miao M, Sun P, Xu D, Huang J, Yi Z, Song W. 2016. The all-data-based evolutionary hypothesis of ciliated protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci Rep 6:24874. doi: 10.1038/srep24874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao F, Huang J, Zhao Y, Li L, Liu W, Miao M, Zhang Q, Li J, Yi Z, El-Serehy HA, Warren A, Song W. 2017. Systematic studies on ciliates (Alveolata, Ciliophora) in China: progress and achievements based on molecular information. Eur J Protistol 61:409–423. doi: 10.1016/j.ejop.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Fenchel T, Finlay BJ. 1991. Endosymbiotic methanogenic bacteria in anaerobic ciliates: significance for the growth efficiency of the host. J Protozool 38:18–22. doi: 10.1111/j.1550-7408.1991.tb04788.x. [DOI] [Google Scholar]
- 36.Finlay BJ, Embley TM, Fenchel T. 1993. A new polymorphic methanogen, closely related to Methanocorpusculum parvum, living in stable symbiosis within the anaerobic ciliate Trimyema sp. J Gen Microbiol 139:371–378. doi: 10.1099/00221287-139-2-371. [DOI] [PubMed] [Google Scholar]
- 37.Embley TM, Finlay BJ, Brown S. 1992. RNA sequence analysis shows that the symbionts in the ciliate Metopus contortus are polymorphs of a single methanogen species. FEMS Microbiol Lett 97:57–62. doi: 10.1016/0378-1097(92)90363-S. [DOI] [PubMed] [Google Scholar]
- 38.Embley TM, Finlay BJ, Thomas RH, Dyal PL. 1992. The use of rRNA sequences and fluorescent probes to investigate the phylogenetic positions of the anaerobic ciliate Metopus palaeformis and its archaeobacterial endosymbiont. J Gen Microbiol 138:1479–1487. doi: 10.1099/00221287-138-7-1479. [DOI] [PubMed] [Google Scholar]
- 39.Embley TM, Finlay BJ. 1994. The use of small subunit rRNA sequences to unravel the relationship between anaerobic ciliates and their methanogenic endosymbionts. Microbiology 140:225–235. doi: 10.1099/13500872-140-2-225. [DOI] [PubMed] [Google Scholar]
- 40.van Hoek AH, van Alen TA, Sprakel VS, Leunissen JA, Brigge T, Vogels GD, Hackstein JHP. 2000. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol Biol Evol 17:251–258. doi: 10.1093/oxfordjournals.molbev.a026304. [DOI] [PubMed] [Google Scholar]
- 41.Hackstein JHP, Tielens AGM. 2010. Hydrogenosomes, p 175–206. In Hackstein JHP. (ed), Microbiology monographs 19, (endo)symbiotic methanogenic archaea. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 42.Dohra H, Suzuki H, Suzuki T, Tanaka K, Fujishima M. 2013. Draft genome sequence of Holospora undulata strain HU1, a micronucleus-specific symbiont of the ciliate Paramecium caudatum. Genome Announc 1:e00664-13. doi: 10.1128/genomeA.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dohra H, Tanaka K, Suzuki T, Fujishima M, Suzuki H. 2014. Draft genome sequences of three Holospora species (Holospora obtusa, Holospora undulata, and Holospora elegans), endonuclear symbiotic bacteria of the ciliate Paramecium caudatum. FEMS Microbiol Lett 359:16–18. doi: 10.1111/1574-6968.12577. [DOI] [PubMed] [Google Scholar]
- 44.López-García P, Philippe H, Gail F, Moreira D. 2003. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc Natl Acad Sci U S A 100:697–702. doi: 10.1073/pnas.0235779100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatamoto M, Imachi H, Ohashi A, Harada H. 2007. Identification and cultivation of anaerobic, syntrophic long-chain fatty acid-degrading microbes from mesophilic and thermophilic methanogenic sludges. Appl Environ Microbiol 73:1332–1340. doi: 10.1128/AEM.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imachi H, Sekiguchi Y, Kamagata Y, Loy A, Qiu YLL, Hugenholtz P, Kimura N, Wagner M, Ohashi A, Harada H. 2006. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol 72:2080–2091. doi: 10.1128/AEM.72.3.2080-2091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori K, Yamamoto H, Kamagata Y, Hatsu M, Takamizawa K. 2000. Methanocalculus pumilus sp. nov., a heavy-metal-tolerant methanogen isolated from a waste-disposal site. Int J Syst Evol Microbiol 50:1723–1729. doi: 10.1099/00207713-50-5-1723. [DOI] [PubMed] [Google Scholar]
- 48.Ewing B, Hillier L, Wendl M, Green P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 49.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194. doi: 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 50.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 54.Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 55.Stahl DA, Amann R. 1991. Development and application of nucleic acid probes, p 205–248. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 56.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]