Invasive species have been a worldwide problem for many years. However, the potential for microorganisms to become invasive is relatively underexplored. As the tools to study bacterial communities become more affordable, we are able to perform large-scale studies and examine bacterial communities in higher resolution than was previously practical. This study looked at the potential for bacteria to colonize both boat surfaces and bilge water. We describe the bacterial communities on boats in 20 shipping ports in five regions around the world, describing how these microorganisms were similar to microorganisms found in port water. This suggests that the water influences the bacterial community of a boat and that microorganisms living on a boat could be moved from place to place when the boat travels.
KEYWORDS: microbial community composition, bilge water, vessels, port water microbes, microbial communities, ships
ABSTRACT
In the past, ballast water has been a key vector in the ship-mediated dispersal of invasive species. Here, we evaluate the potential for port microorganisms to enter and colonize the hull and bilge water of ships. Due to the small size and ubiquitous nature of bacteria, they also have the potential to be spread through hull fouling and bilge water discharge. The goal of this study was to identify the extent to which the boat microbial community is shaped by the microbial community in the port water where the boat spends most of its time. Here, we compared the microbial communities of the hull and bilge compartments of 20 boats to those of the port water in 20 different ports in five regions around the world. We found that there was a significant difference in microbial diversity between boat and port microbial communities. Despite these differences, we found that Cyanobacteria were present at high abundances in the bilge water of most vessels. Due to the limited light in the bilge, the presence of Cyanobacteria suggests that port microorganisms can enter the bilge. Using source-tracking software, we found that, on average, 40% of the bilge and 52% of the hull microbial communities were derived from water. These findings suggest that the bilge of a vessel contains a diverse microbial community that is influenced by the port microbial community and has the potential to serve as an underappreciated vector for dispersal of life.
IMPORTANCE Invasive species have been a worldwide problem for many years. However, the potential for microorganisms to become invasive is relatively underexplored. As the tools to study bacterial communities become more affordable, we are able to perform large-scale studies and examine bacterial communities in higher resolution than was previously practical. This study looked at the potential for bacteria to colonize both boat surfaces and bilge water. We describe the bacterial communities on boats in 20 shipping ports in five regions around the world, describing how these microorganisms were similar to microorganisms found in port water. This suggests that the water influences the bacterial community of a boat and that microorganisms living on a boat could be moved from place to place when the boat travels.
INTRODUCTION
The ability of organisms to enter specific compartments of boats and colonize boat surfaces has contributed to the spread of invasive species. Ballast water is one of the most widely recognized routes for the transport of invasive species (1–5). Invasive species often outcompete native species for habitat and/or resources, costing an estimated $120 billion annually (6). In contrast to many studies on invasive macroscopic species, few studies have evaluated the ability of microorganisms to colonize ships and the environmental impact of moving microorganisms around the world. Microorganisms are present in most environments (7) and are an important consideration in ballast water management. Waterborne pathogens are a growing concern worldwide (8–10). In one study, Vibrio cholerae was found in ballast samples from 93% of ships sampled (9). Over 30 unique pathogens (8) and viruses (11) have been detected in the ballast water of ships.
The International Maritime Organization (IMO) was put in place by the United Nations to develop international maritime laws and shipping regulations (12). According to the IMO, ships are required to undergo a ballast water exchange at least 200 nautical miles from shore at a depth of more than 200 m (13). The rationale is that coastal organisms will not survive in the higher-salinity open ocean (8). When ballast water exchange is impossible, ships are required to discharge fewer than 10 viable organisms per m3 of ballast water and to ensure that indicator microbes that are discharged do not exceed specified limits. These limits require that there be <1 CFU of V. cholerae per 100 ml of water, <205 CFU of Escherichia coli per 100 ml of water, and <100 CFU of intestinal enterococci per 100 ml of water (13). This can be carried out by chemically treating ballast water with an onboard ballast water treatment system.
While bacteria can be transported in ballast water (8, 14), they may be transported on ships in additional ways, including on the boat’s hull or in bilge water (15, 16). Very few studies have investigated the microbial diversity of bilge water. Biofilms can form both on boat surfaces and in the bilge and offer bacteria protection and a constant environment (7). Biofilms can cause biofouling, which is estimated to cost the U.S. Navy $56 million annually (17). Bilge water collects in the bottom of boats and comes from rainwater runoff, from splash over the side of the boat, or through small leaks in the propeller seal. The bilge is typically located near the engine and also collects waste fuel and oil that leaks from the engine. Boats with inboard engines and drive shafts are most likely to leak pollutants into their bilge compartments (18). When the water reaches a certain level, the bilge water is pumped into the environment. According to the IMO, ships are required to clean environmental contaminants from the bilge before disposal but are not required to remove biological organisms (19). It is possible that pathogens, invasive bacteria, or single-celled eukaryotes are being released into new environments when bilge water is discharged.
In this study, we compared the microbial community compositions of bilge water, boat surfaces, and port water from 20 different ports in five regions around the world (Asia, Europe, the U.S. East Coast, the U.S. West Coast, and the U.S. Great Lakes). The microbial community composition was measured on two places on the boats: (i) boat surfaces (“swab” samples) and (ii) bilge water. Both port water and boat samples were collected in each of the 20 locations. Samples were collected from boats that spend the majority of their time in the same port. The goal of this research is to characterize the microbial communities of shipping vessels and ports, expanding on the limited literature on microorganisms in bilge water and investigating the extent to which the bacterial community of a boat is influenced by the port water in which it spends the majority of its time. We hypothesized that the microbial communities on boats throughout the world are influenced by the microbial communities in the water and that bilge water and swabs will reflect the water microbial community.
RESULTS
Overall bacterial community diversity.
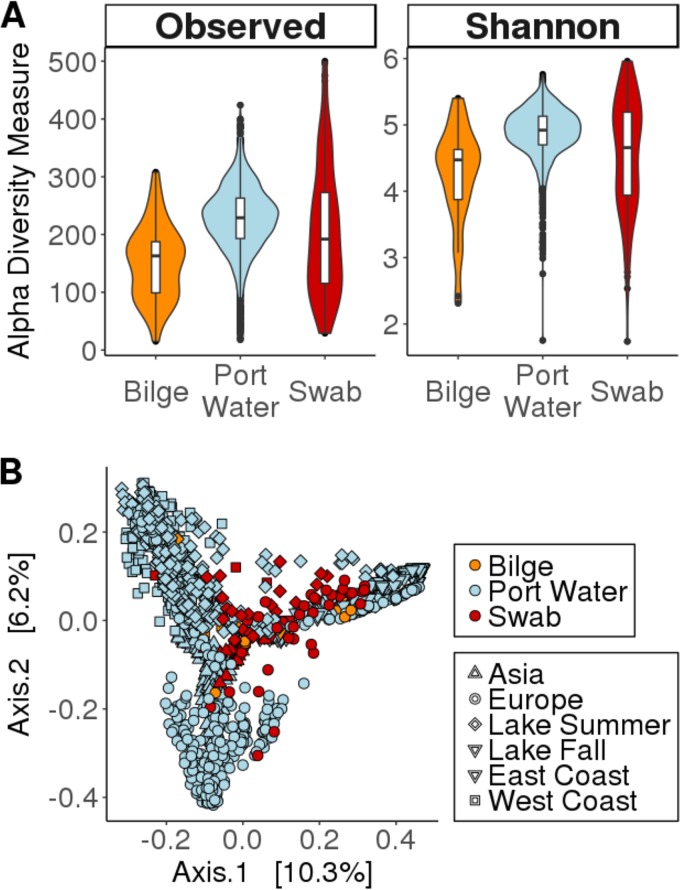
To quantify the similarities between boat and port water bacterial communities, we investigated similarities in alpha diversity between our three types of samples: swab, bilge, and port water. We observed that for both the Shannon index and observed species, the mean level of diversity for the port water samples is higher than that for the bilge and swab samples (Fig. 1A). The mean and median Shannon diversity indices for port water are 4.87 and 4.92, respectively. Swab samples have the largest range of any sample type, at a Shannon diversity index of 4.23. To determine if there was a significant difference in alpha diversity, one-way analysis of variance (ANOVA) was performed, which indicated a significant difference between sample types for both diversity metrics (P values of <2e−10 for both) (see Table S2 in the supplemental material). To determine between which sample types there was a significant difference, we performed a Tukey honestly significant difference (HSD) post hoc test. This test revealed a significant difference for all pairwise comparisons of sample types for both diversity metrics (all P values of <0.001 for all) (Table S3). The results of the Tukey HSD tests confirmed qualitative observations from the alpha diversity plots by showing significant differences in the alpha diversity between each pair of sample types for both metrics. We constructed a principal-component analysis (PCoA) plot to visualize the similarities and differences in microbial community compositions between port water and boat samples (Fig. 1B). We did this to test our hypothesis that the microbial community on the boat reflects the microbial community in the water and to help us to better understand similarities and differences between the bacterial communities in port waters and on the boats that we sampled. Our PCoA plot shows that the boat samples (bilge and swab) clustered near port water samples, which indicates that the boat samples are similar to the port water samples on some level. However, the samples cluster most tightly with samples of the same type, which suggests that the microbial community on boats has some characteristics that are distinct from those of the microbial community in port water and unique to the boat samples. Permutational multivariate analysis of variance (PERMANOVA) was performed to test for significant differences in the microbial community compositions between sample types. Results from the PERMANOVA showed a significant difference between each pairwise comparison of sample types (Table S4), which suggests that there are key differences in the bacterial community compositions of our different sample types.
FIG 1.
(A) Alpha diversity of sample types shows a diverse bacterial community in each of the sample types. Port water has the most diverse microbial community on average, followed by swab and bilge bacterial communities, respectively. This suggests that there is a diverse microbial community living on the boat surface (swab samples) as well as in the bilge compartment. (B) PCoA plot of the microbial community showing the boat samples clustering more closely with similar sample types than with their respective regions. This suggests that the bacterial community of the boat has unique characteristics that are distinct from those of the bacterial community of the port water.
Differential abundances of ASVs between boat and port water microbial communities.
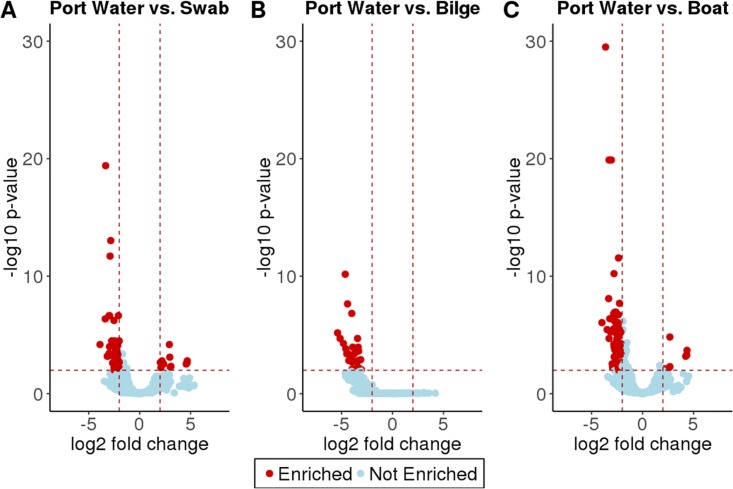
To explain the observed differences in the boat sites, we used DESeq2 to find specific amplicon sequence variants (ASVs) that were enriched in certain sample types. For the full enrichment tables, including adjusted P values, see the data in Tables S5 to S7. When we compared port water samples to swab samples, DESeq2 identified 51 ASVs enriched in port water and 8 ASVs enriched in swab samples relative to port water (Fig. 2A and Table S5). Proteobacteria, Bacteroidetes, Cyanobacteria, and Actinobacteria were most abundant in the port water samples. These taxa showed a substantial log2-fold change, which was between 2.0- and 3.8-fold higher in the port water than on the boat. The ASVs enriched in the swab samples were classified as members of the Proteobacteria and Actinobacteria. These taxa all showed log2-fold changes of between 2.3 and 4.6.
FIG 2.
Differential abundance comparisons of the microbial community in port water to the microbial communities in different areas of the boat. (A) Comparison of port water versus swab (boat surface) samples showing 51 ASVs enriched in the port water and 8 ASVs enriched in the swab samples. (B) Comparison of port water versus bilge water showing 38 ASVs enriched in the port water and no ASVs enriched in the bilge, suggesting that very few taxa are positively selected for in the bilge. (C) Comparison of port water versus boat samples (bilge and swab) showing 80 ASVs enriched in the port water and 9 ASVs enriched in the boat samples.
We compared the enrichment of microbial taxa in port water to that in the bilge samples. DESeq2 analysis revealed that no ASVs were enriched in bilge and that 38 ASVs were enriched in port water relative to bilge (Fig. 2B and Table S6). The taxa enriched in port water were classified as belonging to the phyla Cyanobacteria, Bacteroidetes, and Proteobacteria. We expect Cyanobacteria to be enriched in port water relative to the bilge due to the lack of sunlight in the bilge, which would limit the growth of Cyanobacteria in the bilge compartment. These taxa showed significant log2-fold changes of between 3.1 and 5.5 in port water relative to the bilge. This finding suggests that very few taxa are consistently found in bilge water in different ports in different regions of the world. This could be due to the variability between microbial communities of ports or differences in boat management (cleaning and maintenance, etc.). These results suggest that there are no ASVs that are globally found in the bilge water of all boats.
We hypothesized that microbes from the port water can colonize the surfaces of boats and enter the bilge water. It is possible that taxa from the port water may enter the bilge compartment through leakage in the propeller seal and/or splashes from overlying port water. However, these taxa would not necessarily be enriched in the bilge relative to the port water.
For these DESeq comparisons, ASVs identified as being differentially abundant must be present in the sample type of interest across most of the locations sampled. However, since the samples in this study were from diverse environments around the world, there was a high level of variation in the microbial communities. There were observable differences in the microbial communities between port water and bilge water when comparing smaller sample sets and grouping ASVs by taxonomic classification. For example, on average, the following phyla were present at higher abundances in bilge samples than in port water samples for the specified locations (Table S7): Actinobacteria (Duluth, MN, in the fall; Green Bay, WI, in the fall; Keweenaw, MI, in the summer; Los Angeles, CA; Martigues, France; Naples, Italy; Oakland, CA; Rotterdam, Netherlands; Seattle, WA; and Singapore), Bacteroidetes (Duluth in the fall, Duluth in the summer, Green Bay in the fall, and Green Bay in the summer), Firmicutes (New York, NY), and Planctomyces (Green Bay in the fall; Green Bay in the summer; Keweenaw in the summer; Rotterdam; and Venice, Italy), among others. This suggests that there is not a universal set of ASVs that are selected for in the bilge; however, in certain locations, it appears that certain ASVs are selected for in the bilge relative to port water.
We also did a third comparison looking for enrichment between all boat samples and port water. DESeq2 analysis revealed that there were 9 ASVs that were enriched in our boat samples and 80 ASVs that were enriched in port water relative to boats (Fig. 2C and Table S8). The ASVs enriched in boats were classified as belonging to the phyla Proteobacteria, Bacteroidetes, and Cyanobacteria. These taxa all showed log2-fold changes of 2.0 to 4.3. See the supplemental material for the full enrichment table.
Pathogens in bilge water and on boat surfaces.
Due to previous work showing the presence of pathogens in ballast water, we also evaluated our samples for abundances of common pathogens (10). We looked for the genera that have been previously found in ballast water (8, 10, 20, 21). We found that the following genera were present in our samples: Aeromonas (22, 23), Klebsiella (24), Legionella (25), Mycobacterium (26), Pseudomonas (27), Ralstonia (28), Staphylococcus (29), Stenotrophomonas (30), Vibrio (23), and Yersinia (31). We did not find any sequences of the following genera at a relative abundance of greater than 0.5%: Bartonella, Borrelia, Campylobacter, Cryptosporidium, Escherichia, Helicobacter, Listeria, and Salmonella.
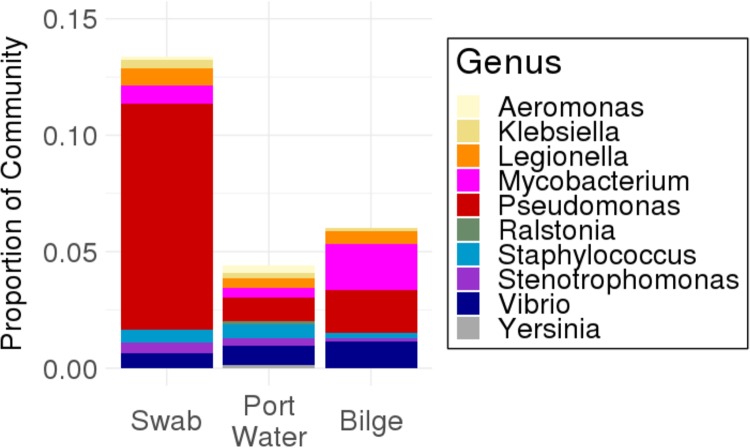
Our port water samples had 10 unique genera or putative pathogens, while our bilge and swab samples presented 7 and 8 genera, respectively (Fig. 3). The overall abundance of these genera was highest in the swab samples (13.4% of all reads were from genera associated with pathogens); port water (4.4% abundance) and bilge (6.0% abundance) have comparatively similar abundances of pathogens. The abundances of these genera are highly variable across each of our sampling locations (Table S9 and Fig. S1 and S2). The presence of these genera in both bilge and swab samples shows us that relatives of waterborne pathogens were present both in the port water and on the boat. This suggests that there may be a mechanism by which bacterial life in the port water is able to colonize the boat.
FIG 3.
Average relative abundances of genera frequently associated with pathogens in each sample type. Relative abundances were variable between locations and sample types (see Fig. S1 and S2 in the supplemental material for more information). This shows that genera closely related to waterborne pathogens are found in our boat samples at higher abundances than in the water.
Differential abundance analysis with DESeq2 did not show any enrichment of sequences related to pathogens in one sample type over the others or in the boat samples relative to the port water samples. Similar to our other comparisons, since we are comparing samples from diverse locations, for an ASV to be significantly enriched on the boat versus in the water, it would have to be enriched across all of our regions for an enrichment to be identified by DESeq2.
Relationship between bacterial communities in port water and bilge water.
Despite the significant difference in the microbial community compositions between port water and boat samples, we wanted to further explore our hypothesis that the bacterial community of port water influences the bacterial community living on the boat. In our differential abundance data, we saw that Cyanobacteria are often one of the dominant phyla in port water. However, since the bilge compartment is dark and previous studies have shown substantial decreases in Cyanobacteria in ballast tanks due to their photosynthetic nature (32), we would not expect to find Cyanobacteria growing in bilge water. Therefore, we hypothesized that if cyanobacterial 16S rRNA reads were present in the bilge at measurable levels, they would indicate that there must be a source of entry of port water containing Cyanobacteria into the bilge. The entry of Cyanobacteria into the bilge could be through leakage around the propeller seal or splashes of port water onto the boat.
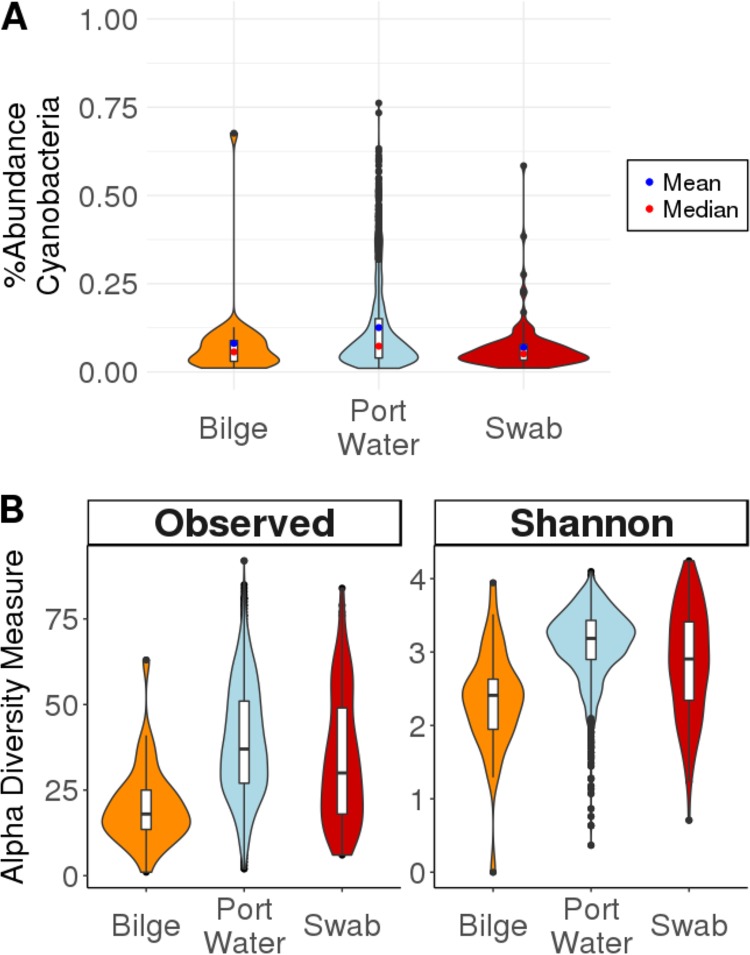
Our results demonstrate that Cyanobacteria were present on boat surfaces and in bilge water at relatively high abundances (Fig. 4A). The average relative abundance of Cyanobacteria in port water was the highest (12.6% ± 76.2%). However, relatively high abundances of Cyanobacteria were also found in the bilge water (8.1% ± 67.8%) and in the swab samples (7.0% ± 58.5%) (Table S9). This shows that while the proportion of Cyanobacteria present in our samples was variable, on average, a significant portion of the microbial community in each sample type was made up of Cyanobacteria. This supports the idea that there is a source of entry of port water into the bilge compartment.
FIG 4.
(A) Relative abundances of Cyanobacteria in each of our sample types. While there is a large amount of variation in the abundances of Cyanobacteria between our different sample types and in each of the different locations, on average, each of our sample types is made up of a relatively high proportion of Cyanobacteria. (B) Alpha diversity of Cyanobacteria in each sample type. This shows that there is a diverse population of Cyanobacteria in each of the sample types. Since Cyanobacteria would be unable to grow in the dark bilge compartment, this suggests that there is a source of entry of Cyanobacteria into the bilge compartment from water.
To further investigate the diversity of the cyanobacterial populations present in our samples, we subset our data to include only Cyanobacteria and measured alpha diversity using two metrics, observed ASVs and Shannon diversity (Fig. 4B). The port water samples had the largest range, at a Shannon diversity index of 4.28; the port water samples also had the highest mean and median Shannon diversity indices (3.10 and 3.19, respectively) out of the three sample types. While the port water samples contained the most diverse community of Cyanobacteria, there was also a relatively high mean diversity of the population of Cyanobacteria in bilge and swab samples (Shannon diversity indices of 3.01 and 2.93, respectively). These data indicate that there is a diverse and abundant community of Cyanobacteria in the bilge water and on the boat surfaces that were sampled for this study. To test if there was a difference in the diversity of the cyanobacterial communities between these sites, ANOVA was performed, which showed significant differences (P values of <0.001 for both observed ASVs and Shannon diversity) (Table S10). A Tukey HSD post hoc test was done to determine between which pairs of sample types there was a significant difference. The results of our post hoc test showed a significant difference between all sample types for Shannon diversity; for observed species, there was a significant difference between all comparisons except for the swab-port water comparison (Table S11). For this study, we collected samples at only one time point, so we were unable to track changes in the abundances of Cyanobacteria to determine if the detected Cyanobacteria were growing. However, the presence of port water-associated microbes in the bilge and swab samples suggests that the boat microbial community is, in part, derived from water.
Quantification of the proportion of water as a source in boat microbiomes.
Our results so far provide support for the hypothesis that the bacterial community of port water influences the bacterial community on the boat. To more finely investigate the ability of microbes from port water to colonize boat surfaces and bilge water, we looked for similarities between the boat samples and port water using SourceTracker2. SourceTracker uses latent Dirichlet allocation and Gibbs sampling to determine how much of the microbial community in a sink sample is derived from various sources (33, 34). We trained our SourceTracker model using port water from each location as a source and boat samples (bilge and boat surface) as sinks. The model then classified which proportion of each sink sample appeared to be derived from each of our 20 sources. The output from SourceTracker identifies the proportion of the microbial community derived from each of the known sources as well as the proportion of the community that is derived other, unknown sources.
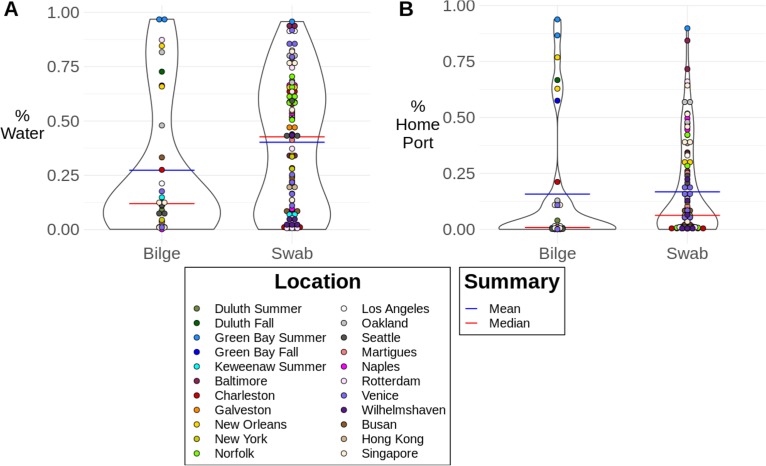
The SourceTracker model indicated that the port water bacterial community contributed a substantial portion of the bacterial community found on the two boat sites sampled in this study. In our data set, the mean percentage of the swab community recognized as being derived from port water was higher than the percentage of the bilge water community recognized as being derived from port water (Fig. 5A). On average, 52% of the swab microbial community and 40% of the bilge water microbial community were recognized as being derived from port water by the SourceTracker model (Fig. S5 and Table S12).
FIG 5.
(A) Proportions of bilge and swab samples classified by our SourceTracker model as being derived from the water microbial community (sum of the proportions of all individual locations identified in a sample). We hypothesize that the proportion of the sample described as being derived from water is variable due to other factors that also influence the bacterial community of the boat, not described by this model. (B) Proportions of bilge and swab samples classified as being derived from the microbial community of water from the correct home port where the samples were taken. The proportion identified as being derived from water from the home port was variable; we expect that this variation is linked to additional factors (other than port water) that contribute to the bacterial diversity on a boat, not accounted for in this study.
To more finely test the accuracy of this model, we looked at the proportion of the microbial community in each sample recognized as being derived from the port where boat was located during sample collection (home port). Our models indicated that between 0% and 93.9% of the microbial community in the boat samples matched the microbial community in the home port, whereas between 0.12% and 96.8% of the bilge water microbial community was described as being derived from water from the home port. Using the output of proportions from SourceTracker (Fig. S4), we looked at how often it listed the home port as being the source of the largest proportion of the microbial community for the boat samples. Excluding the unknown portion of the microbial community, the home port microbial community was correctly listed as the highest proportion in the SourceTracker output in 17/37 (45.9%) bilge samples and 62/121 (51.2%) swab samples (Table 1).
TABLE 1.
Numbers of boat samples with the highest percentages of the bacterial community classified as coming from the correct home port by our SourceTracker modela
| Location (season) | No. of samples classified as coming from the correct home port |
|
|---|---|---|
| Bilge | Swab | |
| Baltimore | 4 | |
| Busan | 4 | |
| Charleston | 1 | 2 |
| Duluth (fall) | 1 | |
| Duluth (summer) | 2 | 1 |
| Galveston | 1 | |
| Green Bay (fall) | 1 | |
| Green Bay (summer) | 2 | 2 |
| Hong Kong | 5 | |
| Keweenaw (summer) | 2 | 5 |
| Los Angeles | 3 | |
| Martigues | 1 | 2 |
| Naples | 4 | |
| New Orleans | 2 | 2 |
| New York | 1 | |
| Norfolk | 2 | |
| Oakland | 4 | |
| Rotterdam | 3 | |
| Seattle | 2 | |
| Singapore | 7 | |
| Venice | 1 | 3 |
| Wilhelmshaven | 5 | |
| Totalb | 17/37 | 62/121 |
Our SourceTracker model suggests that the boat bacterial community has some level of similarity with the port water in which it spends most of its time.
Total shows the total number of correctly classified samples/total number of samples. The total percentages of correctly classified samples were 45.9% for bilge samples and 51.2% for swab samples.
These results demonstrate that the amount of the port water community that is on these boats is highly variable. The microbial communities of some of our boats showed very high similarity to those of port water and/or the home port. Samples from Green Bay; Los Angeles; Baltimore, MD; New Orleans, LA; Singapore; Venice; Norfolk, VA; and Oakland showed >75% similarity of the boat microbial community to that of port water (Fig. 5). Additionally, some samples also showed >75% similarity to the community of the home port; these samples were from Green Bay, New Orleans, and Baltimore (Fig. 5). To confirm the accuracy of the SourceTracker model, we used the model to classify the port water samples on which we had trained the model. The SourceTracker model accurately classified, on average, 92% of the microbial community in the port water samples as being derived from water, suggesting that this model was accurate for classifying water communities. Interestingly, in most samples, there was a substantial amount of the boat microbial community that could not be classified as being derived from port water, and thus, between 3.19% and 99.9% of the boat microbial communities were classified as being derived from an unknown source. These results demonstrate that while the boat bacterial community may be influenced by the port water bacterial community, there are other factors that also contribute to shaping the boat bacterial community, which may not have been measured or controlled for in this study.
DISCUSSION
In this study, we compared similarities and differences in the bacterial communities of boats across 20 distinct locations worldwide. This study is among the largest global studies of boat-associated microbial diversity to date. Our goal was to determine the extent to which microbial communities from port water influence the microbial communities on boat surfaces and in bilge water.
A few studies have looked at the microbial communities of ballast water and in shipping ports. Several studies support the presence of fecal indicator bacteria and pathogens in ballast water and have warned about the transfer of harmful organisms by ships; these studies have been done in Singapore, the Mediterranean Sea, the North Sea, and the Port of Houston (8, 21, 35–37). Other studies have used next-generation sequencing to look at abundances of pathogens in ballast water; these studies have suggested that exchanging ballast water at sea is not effective for removing bacteria from ballast tanks (8, 14).
Since it has been well documented that harmful bacteria have been found in ballast water on ships, we wanted to determine if other places on the boat can also be a reservoir for pathogens and other aquatic organisms. While microbial communities in ballast water have been the focus of several studies, comparatively few studies have focused on microbial communities of bilge water. The limited studies done on bilge water have focused on preventing further pollution by removing hydrocarbons and other contaminants before discharging the water back into the environment (18, 38). To our knowledge, the microbial community of bilge water had not been characterized in detail and compared to the microbial community of port water prior to this study.
Our results indicate that boat bacterial communities were significantly different from port water bacterial communities. This is in line with a recent study of biofilms on submerged surfaces (including ships’ hulls), which showed that the microbial communities of the biofilms were more similar to each other than to the microbial community of the water column. E. coli and V. cholerae were quantified at relatively high abundances in the marine biofilm samples, suggesting that biofilms, including those on ships, can be an overlooked reservoir for pathogens (39). Additionally, similar to previous studies on ballast water (8, 14, 20, 21, 36, 40, 41), we have found that both bilge water and boat surface bacterial communities contain a diverse set of organisms related to pathogenic microbes. This finding suggests that bacterial life on boats is not limited to ballast tanks; bacterial communities from the water can exist on any ship, even if it lacks a ballast tank.
To better understand the potential of port water to enter the bilge of a vessel, we examined the abundance and diversity of Cyanobacteria in boat samples. For the most part, Cyanobacteria are obligate phototrophs (42). Since the bilge compartment is typically dark, we expected that the presence of Cyanobacteria in the bilge water would be indicative of a source outside the vessel, presumably port water. We have found that bilge water contains an abundant and diverse community of Cyanobacteria. This suggests that the cyanobacterial community in bilge water most likely had its origin in port water and that bacterial communities from port water can colonize boats, leading to similarities in the boat and port water microbial communities.
To further clarify the extent to which the boat bacterial community was influenced by port water, we used the program SourceTracker to determine the proportion of the boat microbial community that reflected the port water microbial community. Our analysis showed that some of the boat samples had a very high proportion of the community that reflected the port water, while others exhibited very little similarity to the port water community. Despite this variability, both bilge samples and boat surface samples demonstrated port water as a substantial source of the boat microbial community. The ports with the highest percentages of port water microbes in boat communities were Green Bay, Duluth, Singapore, and New Orleans. In order for SourceTracker to accurately differentiate between sources, there must be sufficient differences in the microbial communities between locations. Work by Ghannam et al. (using the same port samples as the ones used in this study) showed the ability of supervised machine learning to accurately differentiate between ports using the port water microbial community (R. B. Ghannam, L. G. Schaerer, T. M. Butler, and S. M. Techtmann, unpublished data).
Many key differences were found in the port water microbial community that may give insights into the biosignature taxa that could be identified in boat samples. For example, each sample type for the Green Bay samples had significant abundances of Cyanobacteria, including some samples with high abundances of Microcystaceae, a family of Cyanobacteria which contains some of the organisms responsible for harmful algal blooms (43). In the 2013 State of the Bay report, Lower Green Bay and Fox River had been classified as areas of concern, due to increased eutrophication (44). It seems that water problems due to cyanobacterial blooms have been on the rise in the past several years (45), so it is not surprising that we found relatively high abundances of Microcystaceae (family) and other types of Cyanobacteria in our Green Bay samples. Our results indicate the microbial communities of boats are substantially influenced by the port water microbial communities to which they are exposed. This expands on the work done on the microbial communities in ballast water, which have shown that ships can unintentionally move invasive species (1, 3–5, 46–48) and pathogens (8, 10, 20) in ballast water if care is not taken to prevent invasions. Unlike in ballast water, where the water is intentionally taken onto the ship and released in a controlled manner to maintain stability, the sites examined in this study pick up water and organisms in a passive manner. Furthermore, the release of these organisms and water from these sites is typically done in an uncontrolled manner. Our results indicate that the microbial communities of boats are strongly impacted by the water through which they pass, and even in sites with passive exposure to port microbial communities, there are substantial signatures of the port microbial communities.
Moving forward, there are still several questions left which need to be answered. While our SourceTracker analysis quantified some of the sources of the boat microbial community, some questions remain. One of the advantages of SourceTracker is that it allows for an “unknown” category when quantifying the sources of samples. Our results indicate that, on average, 62.7% of the microbial community is derived from an unknown source (all sample types). This unknown portion of the microbial community must be quantified to determine additional sources from which microbes are able to influence the microbial communities on boats. It is possible for some other potential sources to be major contributors to the boat microbial community. While boats that spend the majority of their time in the home port were used for this study, water and microbes from other locations outside the ports could complicate this analysis and contribute to the unknown portion of the microbial community. Additionally, our models were built based on samples taken from a single time point, and growth of microbes on the boat could contribute to the unknown portion, further complicating our results. However, despite these limitations, our results demonstrate that port water is a source of the microbial communities on boats. It is also possible that microbes found on dust in the air as well as soil microbes carried by humans may shape the microbial community on a vessel. More work is needed to further quantify sources of the unknown portion of the microbial community on boats. This will help us to further clarify what determines the microbial community of boat samples. Additionally, classifying the unknown portion of the microbial community will help us investigate the potential for vessels to disperse microbes from other biomes as well as carry water microbes between ports.
This study also shows that bilge water reflects a variable proportion of the microbial community in port water. This means that the bilge microbial community of one boat may be representative of the water microbial community, while the bilge microbial community of a boat in another port may be minimally representative of the water microbial community. We already know that releases of large amounts of untreated ballast water can lead to the introduction of invasive species and the spread of pathogenic bacteria. While our work demonstrates the potential for port microbes to enter and colonize a vessel, more work is required to determine the persistence of these microbes from place to place. This would confirm the potential for bilge water to serve as an underappreciated mechanism for the dispersal of organisms. Further studies are also needed to quantify how much bilge water is currently released from vessels and to determine what concentration of organisms can be safely discharged without adverse effects on the environment. Future work will determine if current water management regulations are adequate or if they need to be changed to prevent further introduction of invasive species and spread of microbes through shipping.
MATERIALS AND METHODS
Sample collection.
Samples were collected from 20 different shipping ports on three continents during summer 2017 (in the United States, Seattle, WA; Los Angeles, CA; Oakland, CA; Duluth, MN; Green Bay, WI; Keweenaw, MI; Baltimore, MD; Charleston, SC; Galveston, TX; New Orleans, LA; New York, NY; and Norfolk, VA; in Europe, Martigues, France; Naples, Italy; Rotterdam, Netherlands; Venice, Italy; and Wilhelmshaven, Germany; and in Asia: Busan, South Korea; Hong Kong; and Singapore). Additionally, samples from ports in the Great Lakes region (Keweenaw Peninsula, MI; Duluth, MN; and Green Bay, WI) were also collected during the fall of 2017. At each port, approximately 30 surface water samples were taken at a range of sites throughout each port (see Table S1 in the supplemental material). For the port water samples, 1 liter of water was filtered through a glass fiber prefilter and a 0.2-μm-pore-size polyether sulfone (PES) postfilter using a peristaltic pump. The filters were stored in Zymo RNA/DNA shield (Zymo Research Corporation, Irvine, CA) for transport back to the laboratory, where they were stored at −80°C until processing. A more comprehensive analysis of the port water bacterial communities and the differences between locations has been described by Ghannam et al. (unpublished).
In addition to port water samples, samples of bilge water and boat surfaces were also collected from the vessels used for port water sample collection in each port location. To the best of our knowledge, all of the boats sampled for this study spent the majority of their time in the home port. The boats chosen for sampling as part of this study were primarily fishing vessels and local research vessels, which are used primarily in the port area and surrounding local waters. These boats were chosen to represent a range of smaller vessels with inboard and outboard engines (Table S1). Bilge water samples were collected from the bilge compartment on each boat; approximately 250 ml of bilge water was collected and filtered through a glass fiber prefilter and a 0.2-μm PES filter. On each boat, three sites from the external surfaces were sampled (hull, transom, and deck). Boat surface samples were collected using Puritan sterile polyester-tipped swabs. For the three sites on each boat, three swabs were collected and placed into a single tube. All swab samples were collected just above the waterline. All samples (filters and swabs) were stored in Zymo RNA/DNA shield (Zymo Research Corporation, Irvine, CA) for shipment to the laboratory. Upon return to the laboratory, samples were frozen at −80°C until further analysis was performed.
DNA extraction and sequencing library preparation.
DNA extractions were performed on one-half of each filter (both the glass fiber prefilter and the 0.2-μm PES postfilter), and the other half was stored at −80°C as an archive. All three of the swabs were used for extraction, due to the expected low biomass of these samples. DNA extractions were performed with the ZymoBIOMICS DNA Microprep kit (Zymo Research Corporation, Irvine, CA). Filter halves were cut into small pieces and put into bead tubes according to the ZymoBIOMICS protocol. The manufacturer’s protocol was followed, with the following exceptions. Samples were processed in a homogenizer for 200 s at 5 m/s and then centrifuged at 12,000 × g for 1 min. The remaining steps were performed according to the manufacturer’s specifications. 16S rRNA sequencing libraries were prepared according to a modified version of the Illumina 16S rRNA metagenomic sequencing library preparation protocol. Briefly, an initial PCR was performed using Thermo Scientific Phusion Flash PCR master mix to amplify the V4-V5 region of the 16S rRNA gene using the primers 515YF and 926R (49). The primers 515Y and 926R have been shown to amplify bacterial and archaeal 16S rRNA genes and some eukaryotic 18S rRNA genes. PCR products from the first round of amplification were purified using AxyPrep Mag PCR clean-up beads according to the Illumina 16S rRNA protocol. A second short-cycle (8-cycle) PCR was performed to add Illumina adaptors and indices for multiplexed sequencing. Distinct 12-bp Golay barcodes were added to the amplicons for each sample. Samples were pooled to result in roughly similar amounts of PCR product for each sample. The pool of 16S rRNA gene products was then diluted to 4 nM and sequenced using the Illumina MiSeq system. Sequencing was done using a v3 600-cycle reagent kit to produce a 2-by-300 paired-end run.
16S rRNA sequence analysis.
Raw 16S rRNA sequencing reads were demultiplexed by using the Illumina MiSeq system. Overlapping paired-end reads were merged, quality filtered, and cleansed of the internal standard (phiX) through the DADA2 (divisive amplicon denoising algorithm) package in R (57). Amplicon sequence variants (ASVs) were then inferred. To account for differences in error rates between each of the three separate sequencing runs, error rates were inferred for each run independently (from >100 million bases). Denoised reads were then merged, and ASVs were assigned using the SILVA v132 data set.
Statistical analysis of 16S rRNA reads.
The majority of analyses were performed in R (50). Diversity analysis was performed using the phyloseq package (51). To begin, our data were rarefied using the “rarefy_even_depth” function in phyloseq to a minimum sample size of 1,014 reads. Alpha diversity was measured using the “estimate_richness” function in phyloseq; the Shannon and observed ASV metrics were used.
To determine if there was sufficient replication in our data set to detect a statistical difference, power analysis was performed using the “pwr” package in R (58). Power analysis was done to determine if we had sufficient statistical power for ANOVA between the categories of port water, boat surfaces (swab), and bilge water. Power analysis showed that for three categories, a sample size of 36.7 was required for a significance level of 0.05 and a power of 0.8. Since we have 1,340 port water samples, 121 swab samples, and 37 bilge samples, this indicates that we have sufficient statistical power for ANOVA. To test the hypothesis that there was a statistically significant difference in richness and evenness between our samples, one-way ANOVA was performed on observed species and Shannon diversity. Base R (50) was used to perform a Tukey HSD post hoc test to determine if there was a significant difference between sample types.
We also used phyloseq to examine the changes in community composition. PCoA plots were used to visualize differences in the community composition. PCoA analysis was done with a Bray-Curtis dissimilarity matrix. PERMANOVA was performed to look for statistically significant differences between pairwise comparisons of sample types. PERMANOVAs were performed in R using the Adonis function in the vegan package (52). Sequences related to chloroplasts and mitochondria were removed from our data set before looking at abundances of pathogens and Cyanobacteria.
To understand the potential for the water bacterial community to influence the bacterial community on the boat, the ASV table was subset to include only reads classified as Cyanobacteria. Diversity analyses similar to those described above were performed on the subset ASV table containing only Cyanobacteria. Similar approaches were used to understand the diversity of pathogens: we subset the ASV table to include only members of the following genera: Aeromonas, Klebsiella, Legionella, Mycobacterium, Pseudomonas, Ralstonia, Staphylococcus, Stenotrophomonas, Vibrio, and Yersinia (above a 0.5% abundance). These taxa were chosen because they have been previously found in ballast water.
The PROPER package in R was used to determine the replication needed for sufficient statistical power to detect significant differences with DESeq analysis (53). The replication in our sample set provides a power of 0.82 at a significance level of 0.01. We used the package DESeq2 (54) to calculate the enrichment of specific taxa in each of the sample types. A nonrarefied ASV table was subset to include only two of the sample types (port water, bilge water, or boat surface), and DESeq2 analysis was performed. ASVs were considered enriched if they had a log2-fold change of >2 and an adjusted P value of <0.01. Three comparisons were performed using DESeq2: port water versus boat (bilge and swab), port water versus swab, and port water versus bilge.
We used SourceTracker2 (55) to determine the similarity of our boat samples to the water as well as to look for bacterial signatures unique to each location. SourceTracker uses latent Dirichlet allocation and Gibbs sampling to deconvolute the mixing proportions of a sink sample into the source components. This approach has been used previously to understand the sources of microbes on surfaces in the built environment (33, 34, 56). Our port water samples were used as sources, and our boat (boat surface and bilge) samples were used as sinks. For our ASV table, we used the same table that we had already rarified in phyloseq, so we did not rarefy our data in SourceTracker. The SourceTracker model was trained with each of the 20 ports as different sources. Therefore, the mixing proportions identified in our sink samples included the predicted mixing proportion for all 20 ports in each boat sample. To determine the proportion of the port water bacterial community in the boat samples, any proportion that was classified into one of the 20 ports was considered derived from water. For the classification of home port, we considered only the proportion that was identified as being derived from the port in which the boat was used for sampling.
Data availability.
Raw reads were deposited in the SRA (BioProject accession numbers PRJNA542897 for boat bacterial communities and PRJNA542890 and PRJNA542685 for port bacterial communities).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge funding from the DARPA YFA program (D16AP00146).
We also acknowledge all of the captains and crews of the vessels involved in this work. Especially, we thank Gian Marco Luna, Grazia Quero, Thorsten Brinkhoff, and Stanley Lau for provision of the vessels and help in sample collection. We also thank Jamey Anderson and Chris Pinnow from the MTU Great Lakes Research Center for help in sample collection in the Great Lakes ports.
L.G.S. processed environmental samples, analyzed the data, and wrote the manuscript; R.B.G. collected samples, analyzed data, and wrote the manuscript; T.M.B. collected samples and helped to write the manuscript; and S.M.T. collected samples, oversaw the research, and wrote the manuscript.
We have no competing interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01804-19.
REFERENCES
- 1.Carlton JT. 1985. Transoceanic and interoceanic dispersal of coastal marine organisms: the biology of ballast. Oceanogr Mar Biol Annu Rev 23:313–371. [Google Scholar]
- 2.Carlton JT, Thompson JK, Schemel LE, Nichols FH. 1990. Remarkable invasion of San Francisco Bay (California, USA) by the Asian clam Potamocorbula amurensis. I. Introduction and dispersal. Mar Ecol Prog Ser 66:81–94. doi: 10.3354/meps066081. [DOI] [Google Scholar]
- 3.Harbison G, Volovik S. 1994. The ctenophore, Mnemiopsis leidyi, in the Black Sea: a holoplanktonic organism transported in the ballast water of ships, p 25–36. In Proceedings of the National Oceanic and Atmospheric Administration Conference and Workshop on Nonindigenous Estuarine and Marine Organisms. US Government Printing Office, Washington, DC. [Google Scholar]
- 4.Mills EL, Leach JH, Carlton JT, Secor CL. 1993. Exotic species in the Great-Lakes—a history of biotic crises and anthropogenic introductions. J Great Lakes Res 19:1–54. doi: 10.1016/S0380-1330(93)71197-1. [DOI] [Google Scholar]
- 5.Vinogradov M, Shushkina E, Musaeva E, Sorokin PY. 1989. A newly acclimated species in the Black Sea: the ctenophore Mnemiopsis leidyi (Ctenophora: Lobata). Oceanology 29:220–224. [Google Scholar]
- 6.Pimentel D, Zuniga R, Morrison D. 2005. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ 52:273–288. doi: 10.1016/j.ecolecon.2004.10.002. [DOI] [Google Scholar]
- 7.Madigan MT, Clark DP, Stahl D, Martinko JM. 2010. Brock biology of microorganisms, 13th ed Benjamin Cummings, San Francisco, CA. [Google Scholar]
- 8.Brinkmeyer R. 2016. Diversity of bacteria in ships ballast water as revealed by next generation DNA sequencing. Mar Pollut Bull 107:277–285. doi: 10.1016/j.marpolbul.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz GM, Fofonoff PW, Carlton JT, Wonham MJ, Hines AH. 2000. Invasion of coastal marine communities in North America: apparent patterns, processes, and biases. Annu Rev Ecol Syst 31:481–531. doi: 10.1146/annurev.ecolsys.31.1.481. [DOI] [Google Scholar]
- 10.Sharma S, Sachdeva P, Virdi JS. 2003. Emerging water-borne pathogens. Appl Microbiol Biotechnol 61:424–428. doi: 10.1007/s00253-003-1302-y. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Aw TG, Teal TK, Rose JB. 2015. Metagenomic investigation of viral communities in ballast water. Environ Sci Technol 49:8396–8407. doi: 10.1021/acs.est.5b01633. [DOI] [PubMed] [Google Scholar]
- 12.United Nations. 5 August 2019, accession date Oceans and the law of the sea. United Nations, New York, NY: http://www.un.org/en/sections/issues-depth/oceans-and-law-sea/. [Google Scholar]
- 13.International Maritime Organization. 2017. International convention for the control and management of ships’ ballast water and sediments (BWM). International Maritime Organization, London, United Kingdom: http://www.imo.org/en/About/Conventions/ListOfConventions/Pages/International-Convention-for-the-Control-and-Management-of-Ships'-Ballast-Water-and-Sediments-(BWM).aspx. Accessed 17 October 2019. [Google Scholar]
- 14.Lymperopoulou DS, Dobbs FC. 2017. Bacterial diversity in ships’ ballast water, ballast-water exchange, and implications for ship-mediated dispersal of microorganisms. Environ Sci Technol 51:1962–1972. doi: 10.1021/acs.est.6b03108. [DOI] [PubMed] [Google Scholar]
- 15.Dunne WM. 2002. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15:155–166. doi: 10.1128/cmr.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olivera NL, Commendatore MG, Delgado O, Esteves JL. 2003. Microbial characterization and hydrocarbon biodegradation potential of natural bilge waste microflora. J Ind Microbiol Biotechnol 30:542–548. doi: 10.1007/s10295-003-0078-5. [DOI] [PubMed] [Google Scholar]
- 17.Schultz MP, Bendick JA, Holm ER, Hertel WM. 2011. Economic impact of biofouling on a naval surface ship. Biofouling 27:87–98. doi: 10.1080/08927014.2010.542809. [DOI] [PubMed] [Google Scholar]
- 18.Cappello S, Santisi S, Calogero R, Hassanshahian M, Yakimov MM. 2012. Characterisation of oil-degrading bacteria isolated from bilge water. Water Air Soil Pollut 223:3219–3226. doi: 10.1007/s11270-012-1103-y. [DOI] [Google Scholar]
- 19.International Maritime Organization. 1983. International convention for the prevention of pollution from ships. International Maritime Organization, London, United Kingdom: http://www.imo.org/en/About/conventions/listofconventions/pages/international-convention-for-the-prevention-of-pollution-from-ships-(marpol).aspx. Accessed 17 October 2019. [Google Scholar]
- 20.Drake LA, Meyer AE, Forsberg RL, Baier RE, Doblin MA, Heinemann S, Johnson WP, Koch M, Rublee PA, Dobbs FC. 2005. Potential invasion of microorganisms and pathogens via ‘interior hull fouling’: biofilms inside ballast water tanks. Biol Invasions 7:969–982. doi: 10.1007/s10530-004-3001-8. [DOI] [Google Scholar]
- 21.Emami K, Askari V, Ullrich M, Mohinudeen K, Anil AC, Khandeparker L, Burgess JG, Mesbahi E. 2012. Characterization of bacteria in ballast water using MALDI-TOF mass spectrometry. PLoS One 7:e38515. doi: 10.1371/journal.pone.0038515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altwegg M, Geiss HK. 1989. Aeromonas as a human pathogen. Crit Rev Microbiol 16:253–286. doi: 10.3109/10408418909105478. [DOI] [PubMed] [Google Scholar]
- 23.Dumontet S, Krovacek K, Svenson SB, Pasquale V, Baloda SB, Figliuolo G. 2000. Prevalence and diversity of Aeromonas and Vibrio spp. in coastal waters of southern Italy. Comp Immunol Microbiol Infect Dis 23:53–72. doi: 10.1016/S0147-9571(99)00059-4. [DOI] [PubMed] [Google Scholar]
- 24.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bopp CA, Sumner JW, Morris GK, Wells JG. 1981. Isolation of Legionella spp. from environmental water samples by low-pH treatment and use of a selective medium. J Clin Microbiol 13:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaerewijck MJM, Huys G, Palomino JC, Swings J, Portaels F. 2005. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol Rev 29:911–934. doi: 10.1016/j.femsre.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Vaz-Moreira I, Nunes OC, Manaia CM. 2012. Diversity and antibiotic resistance in Pseudomonas spp. from drinking water. Sci Total Environ 426:366–374. doi: 10.1016/j.scitotenv.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Ryan MP, Adley CC. 2014. Ralstonia spp.: emerging global opportunistic pathogens. Eur J Clin Microbiol Infect Dis 33:291–304. doi: 10.1007/s10096-013-1975-9. [DOI] [PubMed] [Google Scholar]
- 29.Efstratiou MA, Mavridou A, Richardson SC, Papadakis JA. 1998. Correlation of bacterial indicator organisms with Salmonella spp., Staphylococcus aureus and Candida albicans in sea water. Lett Appl Microbiol 26:342–346. doi: 10.1046/j.1472-765x.1998.00345.x. [DOI] [PubMed] [Google Scholar]
- 30.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7:514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 31.Arvanitidou M, Stathopoulos G, Constantinidis T, Katsouyannopoulos V. 1995. The occurrence of Salmonella, Campylobacter and Yersinia spp. in river and lake waters. Microbiol Res 150:153–158. doi: 10.1016/S0944-5013(11)80050-9. [DOI] [PubMed] [Google Scholar]
- 32.Ng C, Le TH, Goh SG, Liang L, Kim Y, Rose JB, Yew-Hoong KG. 2015. A comparison of microbial water quality and diversity for ballast and tropical harbor waters. PLoS One 10:e0143123. doi: 10.1371/journal.pone.0143123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin HD, Pei ZH, Martinez KA II, Rivera-Vinas JI, Mendez K, Cavallin H, Dominguez-Bello MG. 2015. The first microbial environment of infants born by C-section: the operating room microbes. Microbiome 3:59. doi: 10.1186/s40168-015-0126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prussin AJ, Marr LC. 2015. Sources of airborne microorganisms in the built environment. Microbiome 3:78. doi: 10.1186/s40168-015-0144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David M, Gollasch S, Cabrini M, Perkovič M, Bošnjak D, Virgilio D. 2007. Results from the first ballast water sampling study in the Mediterranean Sea—the Port of Koper study. Mar Pollut Bull 54:53–65. doi: 10.1016/j.marpolbul.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 36.Dobbs FC, Goodrich AL, Thomson FK, Hynes W. 2013. Pandemic serotypes of Vibrio cholerae isolated from ships’ ballast tanks and coastal waters: assessment of antibiotic resistance and virulence genes (tcpA and ctxA). Microb Ecol 65:969–974. doi: 10.1007/s00248-013-0182-7. [DOI] [PubMed] [Google Scholar]
- 37.Joachimsthal E, Ivanov V, Tay S-L, Tay J-H. 2004. Bacteriological examination of ballast water in Singapore Harbour by flow cytometry with FISH. Mar Pollut Bull 49:334–343. doi: 10.1016/j.marpolbul.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 38.Sivaraman C, Ganguly A, Nikolausz M, Mutnuri S. 2011. Isolation of hydrocarbonoclastic bacteria from bilge oil contaminated water. Int J Environ Sci Technol 8:461–470. doi: 10.1007/BF03326232. [DOI] [Google Scholar]
- 39.Shikuma NJ, Hadfield MG. 2010. Marine biofilms on submerged surfaces are a reservoir for Escherichia coli and Vibrio cholerae. Biofouling 26:39–46. doi: 10.1080/08927010903282814. [DOI] [PubMed] [Google Scholar]
- 40.Dobbs FC, Rogerson A. 2005. Ridding ships’ ballast water of microorganisms. Environ Sci Technol 39:259a–264a. doi: 10.1021/es053300v. [DOI] [PubMed] [Google Scholar]
- 41.Seiden JM, Way C, Rivkin RB. 2010. Microbial hitchhikers: dynamics of bacterial populations in ballast water during a trans-Pacific voyage of a bulk carrier. Aquat Invasions 5:13–22. doi: 10.3391/ai.2010.5.1.3. [DOI] [Google Scholar]
- 42.Beck C, Knoop H, Axmann IM, Steuer R. 2012. The diversity of cyanobacterial metabolism: genome analysis of multiple phototrophic microorganisms. BMC Genomics 13:56. doi: 10.1186/1471-2164-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchis D, Carrasco D, Quesada A. 2004. The genus Microcystis (Microcystaceae/Cyanobacteria) from a Spanish reservoir: a contribution to the definition of morphological variations. Nova Hedwig 79:479–495. doi: 10.1127/0029-5035/2004/0079-0479. [DOI] [Google Scholar]
- 44.Qualls T, Harris HJB, Harris V. 2013. State of the bay: the condition of the Bay of Green Bay/Lake Michigan. University of Wisconsin Sea Grant Institute, Madison, WI. [Google Scholar]
- 45.Whaley K. 2014. Can what happened in Toledo occur in Wisconsin? Experts have warned for years of water problems facing Great Lakes. wpr.org, Madison, WI. [Google Scholar]
- 46.Cariton JT, Geller JB. 1993. Ecological roulette: the global transport of nonindigenous marine organisms. Science 261:78–82. doi: 10.1126/science.261.5117.78. [DOI] [PubMed] [Google Scholar]
- 47.Carlton JT. 1996. Marine bioinvasions: the alteration of marine ecosystems by nonindigenous species. Oceanography 9:36–43. doi: 10.5670/oceanog.1996.25. [DOI] [Google Scholar]
- 48.International Maritime Organization. 5 August 2019, accession date International Maritime Organization ballast water management—the control of harmful invasive species. International Maritime Organization, London, United Kingdom: http://www.imo.org/en/MediaCentre/HotTopics/BWM/Pages/default.aspx. [Google Scholar]
- 49.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 50.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 51.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oksanen J, Blanchet FG, Kindt R, Legendre P, O’hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2010. Vegan: community ecology package. R package version 1.17-4 http://cran.r-project.org.
- 53.Wu H, Wang C, Wu ZJ. 2015. PROPER: comprehensive power evaluation for differential expression using RNA-seq. Bioinformatics 31:233–241. doi: 10.1093/bioinformatics/btu640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vazquez-Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen J. 1988. Statistical power analysis for the behavioral sciences, 2nd ed Lawrence Erlbaum Associates, New York, NY: http://www.utstat.toronto.edu/~brunner/oldclass/378f16/readings/CohenPower.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads were deposited in the SRA (BioProject accession numbers PRJNA542897 for boat bacterial communities and PRJNA542890 and PRJNA542685 for port bacterial communities).