Abstract
NUT carcinoma (NC) shows very aggressive clinical behavior, occurs predominantly in the thorax and head and neck region of children and adults, and is defined by the presence of NUT (aka NUTM1) rearrangement, mostly BRD4-NUTM1 fusion resulting from t(15;19)(q13; p13.1). So-called “NUT variants” harbor alternate fusions between NUTM1 and BRD3, NSD3, ZNF532, or unknown partners. Rare cases of pediatric tumors with CIC-NUTM1 fusion were recently reported in somatic soft tissue, brain, and kidney. However, such cases have not been identified in adult patients and the presence of a fusion between CIC, characteristic of CIC-rearranged sarcoma, and NUTM1–a defining feature of NC–poses a diagnostic challenge. We herein report a case of malignant epithelioid neoplasm with myoepithelial features harboring CIC-NUTM1 fusion arising in soft tissue of the head in a 60-year-old man. Immunohistochemistry revealed strong expression of NUT, but only weak ETV4 staining and negativity for keratins, EMA, p40, CD99, and WT1. SMARCB1 expression was retained. Fluorescence in situ hybridization and targeted next-generation sequencing identified a CIC-NUTM1 fusion resulting from t(15;19)(q14;q13.2). In light of morphologic features that overlap with those of NC from typical anatomical sites we have seen previously, the tumor was best classified as falling within the NC spectrum rather than CIC-associated sarcoma. This case highlights the emerging diagnostic challenges generated by newly detected gene fusions of unknown clinical and biologic significance. Careful integration of cytogenetic, molecular, and immunohistochemical findings with morphologic appearances in the diagnostic workup of undifferentiated neoplasms is essential.
1 |. INTRODUCTION
NUT carcinoma (NC), a very aggressive type of poorly differentiated carcinoma, shows a wide anatomic distribution and is most frequently found in the chest and head and neck region along the midline. This tumor occurs in both children and adults with a wide age distribution (range, 0.1–82 years), a median age of 22 years, and a slight female predominance.1,2 NC is defined by NUTM1 rearrangements and roughly 75% of cases harbor BRD4-NUTM1 fusion resulting from t (15;19)(q14;p13.1).3 The remaining cases harbor either BRD3-NUTM1 fusion resulting from t(15;19)(q14;p34.2),4 NSD3-NUTM1 fusion resulting from t(8;15)(p12;q15),5 ZNF532-NUT,6 or NUTM1 fusion with an unknown partner gene.
CIC-rearranged round cell sarcoma, in contrast, is an aggressive tumor predominantly arising in somatic soft tissue of trunk and extremities in young adults with a mean age of 32 years (range, 6–81 years) and a slight male predominance.7 The identification of characteristic CIC rearrangements in a subset of round cell tumors lacking EWSR1 rearrangement in recent years led to the recognition of CIC-rearranged sarcomas as a distinct entity, separate from Ewing sarcoma.7–9 Most cases harbor CIC-DUX4 fusion resulting from either t(4;19) or t(10;19).7–9 CIC fusion genes (such as CIC-LEUTX) may also be seen in angiosarcoma, and CIC aberrations have been reported in oligodendroglioma, medulloblastoma, breast cancer, and others.10,11
Recently, cases of pediatric round cell tumors harboring CIC-NUTM1 fusions were identified in somatic soft tissue (N = 3, 2/3 in the head),12 brain (N = 2),13 or kidney (N = 1).31 In addition, rare cases of round cell tumors located in somatic soft tissue, visceral locations, and brain were shown to harbor NUTM1 rearrangement with fusion partners other than CIC, including BRD4 (N = 3), BRD3, BCORL1, and MXD1 (one case each) fusions14—several of which had a myoepithelial-like appearance, and it is possible that these cases further expand the spectrum of NC.
We report herein a case of malignant epithelioid neoplasm with CIC-NUTM1 fusion arising in somatic soft tissue of the head in an adult patient and highlight the morphologic, cytogenetic, molecular, and immunohistochemical features of what we favor to be an NC with a novel fusion gene.
2 |. MATERIALS AND METHODS
2.1 |. Immunohistochemistry
Immunohistochemical analysis was performed on 4-μm formalin-fixed paraffin-embedded (FFPE) tissue sections using the following antibodies and conditions: NUT (Cell Signaling, Danvers, MA; clone C52B1; 1:200), ETV4 (Santa Cruz, Santa Cruz, CA; clone 16; 1:50), p40 (Biocare, Pacheco, CA; clone BC28; 1:250), AE1/AE3 (Dako, Carpinteria, CA; clone AE1/AE3; 1:200), CD99 (Biolegend, San Diego, CA; clone O13; 1:250), WT1 (Dako; clone 6F-H2; 1:50), CAM5.2 (Becton Dickinson, Franklin Lakes, NJ; clone CAM5.2; 1:50), synaptophysin (Leica Biosystems, Buffalo Grove, IL; clone NCL-SYNAP-299; 1:100), NKX2.2 (Developmental Studies Hybridoma Bank, Iowa City, IA; clone 74.5A5; 1:25), PD-L1 (Cell Signaling; clone E1L3N; 1:100), and SMARCB1 (BD Biosciences; clone 25; 1:250). The Envision Plus detection system (Dako) was used for all antibodies. Appropriate positive and negative controls were used throughout.
2.2 |. Targeted next-generation sequencing
DNA extracted from FFPE tissue was subjected to targeted exon hybrid capture (Agilent, Santa Clara, CA) and next-generation sequencing using an Illumina HiSeq 2500 (Illumina, San Diego, CA). The exonic sequences of 447 cancer-associated genes were interrogated for mutations and copy number variations, and 191 introns across 60 genes were examined for gene rearrangements, using our institution’s targeted sequencing platform (OncoPanel). Bioinformatic detection of genomic aberrations is achieved using MuTect and GATK software for the detection of single nucleotide variants and small insertion-deletions, RobustCNV (an internally developed tool) for copy number analysis, and BreaKmer for large structural variations (details available upon request).15,16
2.3 |. Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) to confirm the targeted next-generation sequencing findings was performed on 5-μm-thick paraffin sections containing tumor nuclei using a homebrew BRD4-NUTMI Dual Color-Dual Fusion Translocation Probe at 15q14 and BRD4 at 19p13.1, a homebrew Break-Apart Probe specific for the 5′ and 3′ regions of NUTM1 at 15q14, and a homebrew Break-Apart Probe specific for the 5′ and 3′ regions of CIC at 19q13.2, according to standard protocols in our laboratory.
3 |. RESULTS
3.1 |. Clinical course
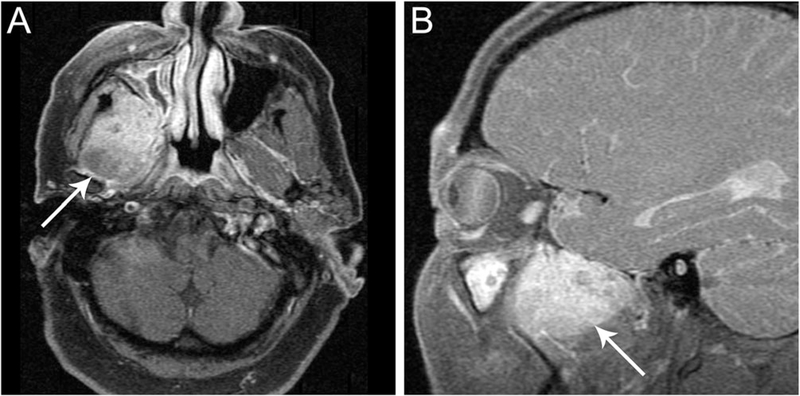
A 60-year-old man with no pertinent medical history presented with two months of right-sided jaw pain, referred otalgia, and numbness along the right side of his cheek and chin. Exam confirmed numbness along the maxillary and mandibular branches of the trigeminal nerve (V2/V3). The remainder of the oral and nasopharyngolaryngoscopic exam was normal. Magnetic resonance imaging demonstrated a 4.7 cm irregularly shaped mass in the right masticator space eroding the greater wing of the sphenoid bone and extending into the infratemporal fossa (Figure 1A,B) with no cervical adenopathy. The patient underwent resection of the tumor using a pre-auricular approach to the middle cranial fossa and subtemporal craniotomy and a 45, X,−Y, +6,t(15;19)(q14;p13) karyotype was reported. The patient was referred to our institution for treatment and the diagnosis of NC was made. He received one cycle of induction chemotherapy as a bridge to adjuvant concurrent chemoradiation with platinum-based therapy. Following definitive treatment, he started off-label adjuvant maintenance immunotherapy with pembrolizumab given the concern for minimal residual disease. He is currently free of disease and without symptoms 10 months after resection.
FIGURE 1.
Imaging findings of NUT variant with CIC-NUTM1 fusion. Magnetic resonance imaging revealed an infiltrative 4.7-cm mass in the right masticator space (A, B, arrows)
3.2 |. Morphology and Immunohistochemistry
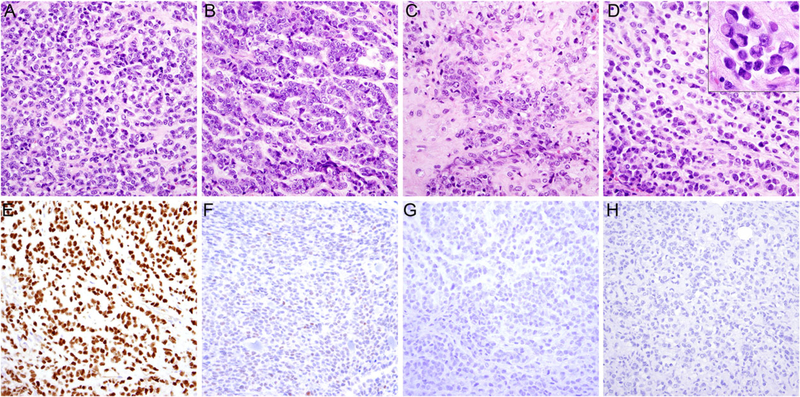
The resection specimen consisted of an ill-defined solid mass extending to resection margins. Histologically, the tumor consisted of a monomorphic proliferation of small, round-to-ovoid cells with variable amounts of eosinophilic cytoplasm. Tumor cells exhibited a range of growth patterns, including solid sheets (Figure 2A), strands (Figure 2B), embedded in a variably prominent chondroid (Figure 2C), or myxoid to hyalinized stromal background (Figure 2D). The tumor cells exhibited irregular and occasionally grooved, atypical nuclei with prominent single nuclei and scant cytoplasm, morphologically resembling myoepithelial carcinoma, and a subset of cells showed rhabdoid features with prominent eosinophilic cytoplasmic inclusions and eccentrically placed nuclei (Figure 2D, inset). Focal pleomorphism and necrosis were present; mitotic figures and apoptotic bodies were frequent. The tumor cells expressed NUT in a strong and diffuse fashion (Figure 2E) and multifocal weak ETV4 (Figure 2F). Staining for p40 (Figure 2G), AE1/AE3 (Figure 2H), and CD99, WT1, CAM5.2, synapto-physin, NKX2.2, and PD-L1 (<1% positivity in tumor cells) was negative. Expression of SMARCB1 was retained. Additional immunohistochemistry for pan-keratin, EMA, CK5/6, p63, and S100 previously performed at the referring institution was negative. However, staining for CK7 was focally positive.
FIGURE 2.
Histologic features of NUT variant with CIC-NUTM1 fusion. The tumor consisted of round to ovoid cells arranged in A, sheets or B, strands embedded in a variably prominent C, chondroid or D, myxoid to hyalinized stromal background. The tumor cells exhibited intermediate to large, irregular, and occasionally grooved, atypical nuclei with occasional rhabdoid inclusions (D, inset). Immunohistochemical staining was positive for E, NUT, F, multifocal weak for ETV4, and G, negative for p40 and H, AE1/AE3
3.3 |. Genetics
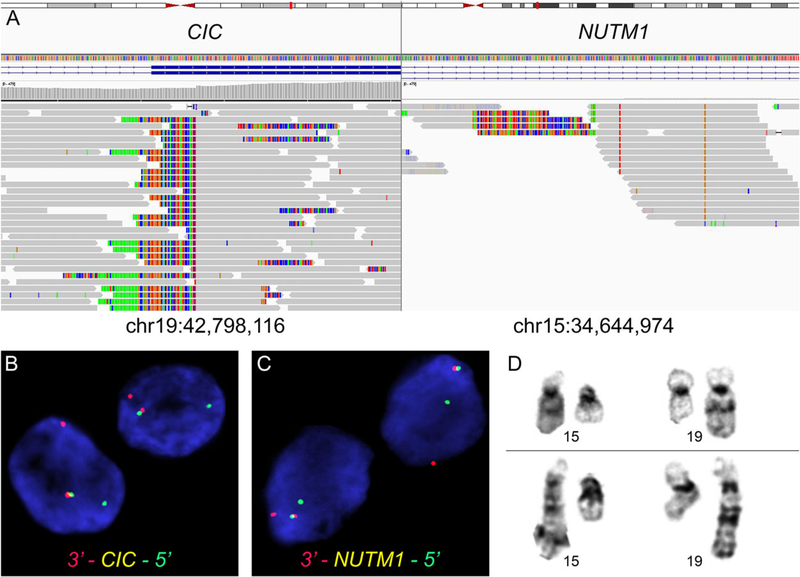
Targeted exome sequencing of 447 commonly mutated cancer-associated genes revealed no oncogenic mutations of known significance. Structural variant analysis (BreaKmer) confirmed the presence of a fusion of CIC at 19q13.2 and NUTM1 at 15q14 (Figure 3A). The breakpoints were mapped to CIC (chr19:42,798,116) and NUTM1 (chr15:34,644,974) with 23 split reads and 23 discordant reads, and were confirmed by manual review of the raw data. No other pathogenic single-nucleotide variants or copy number alterations were identified. Additional FISH analysis confirmed the rearrangements of both CIC and NUTM1 (Figure 3B,C). The breakpoint of derivative 19 of the t(15;19) was reinterpreted as 19q13.2 instead of 19p13.1 (Figure 3D).
FIGURE 3.
Molecular genetic and cytogenetic findings in NUT neoplasm with CIC-NUTM1 fusion. A, Targeted next-generation sequencing demonstrated the presence of a fusion between CIC exon 17 and NUTM1 intron 4. B, Fluorescence in-situ hybridization confirmed rearrangement of the CIC locus at chromosome 19q13.2 and C, the NUTM1 locus at chromosome 15q14 using break-apart probes (arrows). D, Partial GTG banding karyotype showing the presence of t(15;19)(q14;q13.2)
4 |. DISCUSSION
We report herein the first case of malignant epithelioid neoplasm with an alternate CIC-NUTM1 fusion likely representing a novel variant of NC in somatic soft tissue of an adult patient. NC follows an aggressive clinical course, with median overall survival of 6.7 months.1 Immunohistochemical staining for NUT has been shown to be highly specific for NC and is positive in the vast majority of cases,17 resulting from NUTM1 fusions with either BRD4, BRD3, NSD3, ZNF532, or unknown partners.6 As shown for the classic BRD4-NUTM1 fusion protein, the NUT portion of the oncogenic fusion protein binds to and activates a histone acetyl-transferase, p300, which is hypothesized to acetylate regional chromatin, recruiting more local BRD4-NUT and p300, leading to more local acetylation in a self-perpetuating manner.18,19 The creation by this process of large contiguous acetylated regions of chromatin enriched with BRD4-NUT and p300, called megadomains,20 is thought to sequester p300 away from prodifferentiation genes. As a result, genes required for differentiation are repressed. Targeted therapies aim at reversing this feed-forward mechanism and enabling the tumor cells to differentiate. Bromodomain inhibitors such as JQ1, which act as acetyl-histone mimics, and histone deacetylase inhibitors, which artificially increase acetylation, demonstrated efficacy both in vitro and in vivo21–23 and are being evaluated in clinical trials.
CIC-NUTM1 fusions are increasingly recognized in pediatric patients.12,31 The presence of a fusion between CIC—characteristic of (but by no means limited to) CIC-rearranged sarcoma—and NUTM1—a defining feature of NC—raises the question of whether to classify these neoplasms as sarcoma or carcinoma.
CIC-rearranged sarcomas behave in a clinically aggressive fashion, show poor response to Ewing sarcoma chemotherapy regimens, and 5-year overall survival rates of only 43% compared to 76% for patients with Ewing sarcoma, highlighting the importance of recognizing these tumors as a distinct entity.7 They are comprised of solid proliferations of round to ovoid cells in a variably prominent myxoid stroma. Most cases show a relatively monomorphic cytomorphology, but nuclear variability, mitoses, and necrosis are common. Immunohistochemistry highlights expression of ETV4 in 90% and WT1 in 95% of cases.24
The present case showed morphologic appearances of an undifferentiated carcinoma with a prominent myoepithelial appearance, that is, areas of chondromyxoid matrix and rhabdoid cytomorphology, and lacked ETV4 and WT1 expression which argues against a diagnosis of CIC-rearranged sarcoma. In line with our case, a previously reported neoplasm with CIC-NUTM1 fusion arising in the kidney showed only limited expression of WT1 (30% of cells) and negativity for ETV4,31 distinct from CIC-rearranged sarcoma.
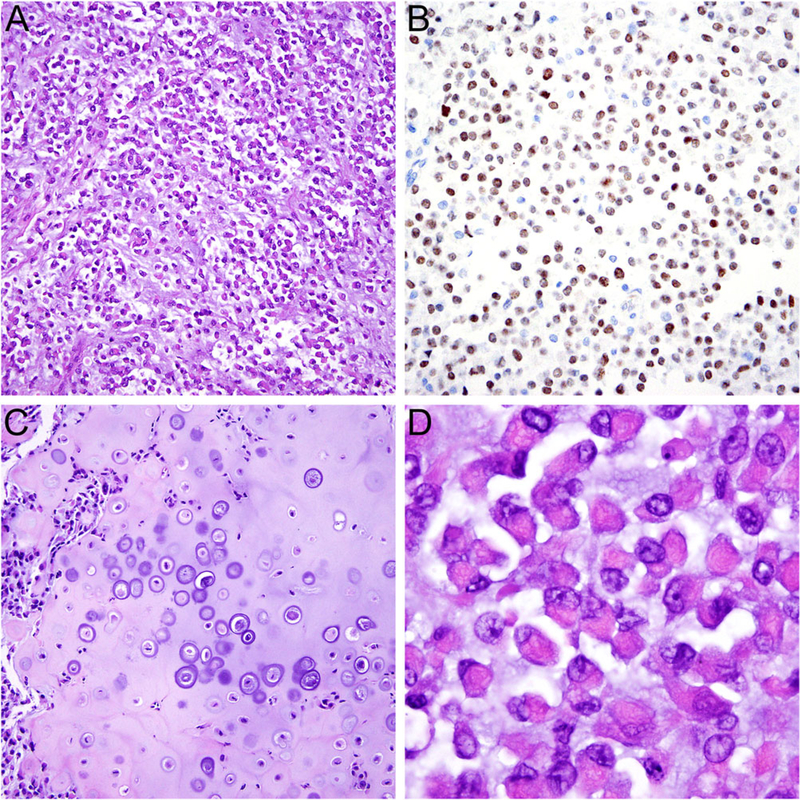
The absence of expression of keratins and p40 observed in our case does not necessarily favor sarcoma over carcinoma. In our experience, NC may occasionally lack expression of keratins25 or p40.26,27 Importantly, NC may occasionally demonstrate pronounced myoepithelial morphology, including the presence of chondroid differentiation, often when arising in the salivary glands,28 and has been mistaken for carcinoma ex pleomorphic adenoma. It is notable that the reported NSD3-NUTM1-positive tumor14 arising in the soft tissue of the leg of a 16-year-old boy, who was recently also seen at our institution, exhibited similar myoepithelial morphology (Figure 4), as did the BCORL1-NUTM1 fused tumor in the same case series. Thus, a myoepithelial appearance is within the spectrum of known NC morphology, and is supportive of this diagnosis.
FIGURE 4.
Histologic features of NUT variant with NSD3-NUTM1 fusion. A, The tumor consisted of round to ovoid cells mostly arranged in variably cohesive sheets embedded in a myxoid stroma. B, The tumor cells showed diffuse nuclear expression of NUT. C, Focal heterologous cartilage formation and D, areas with prominent rhabdoid cytomorphology were present, resembling features of myoepithelial carcinoma
On the other hand, there are some CIC-NUTM1-positive and likely other variant NUTM1-fusion tumors that are indisputably distinct from NC and better classified as sarcomas, including those of the central nervous system.13 Moreover, gene expression profiling performed in a large series of round cell sarcomas demonstrated that tumors with CIC-NUTM1 fusion (N = 3) clustered tightly with CIC-DUX4 and CIC-FOXO4-positive sarcomas and separate from NCs harboring BRD3/4-NUTM1 fusion, indicating a likely biologic relationship between these CIC-rearranged neoplasms.12
In summary, the present case of a CIC-NUTM1-rearranged malignant epithelioid neoplasm is best considered within the spectrum of NC, but exemplifies the emerging challenge in tumor classification in an era of next-generation sequencing. Tumors are not always reliably defined by driving fusion oncogenes; well-known examples of promiscuous fusion partner genes include EWSR1, HMGA2, FUS, and ALK,29,30 that are present in a large range of different cancers. Thus, molecular findings should be integrated with the clinicopathologic findings. Further studies in larger series are required to further define the biologic underpinnings and clinical behavior of this novel fusion variant, and its relation to other neoplasms with NUTM1 rearrangement.
ACKNOWLEDGMENTS
The authors would like to acknowledge the Profile project at Dana-Farber Cancer Institute (DFCI) and Brigham and Women’s Hospital, and the DFCI Oncology Data Retrieval System (OncDRS) for the aggregation, management, and delivery of the clinical and operational research data used in this project. The content is solely the responsibility of the authors. The authors thank Dr Aurelia Meloni-Ehrig, CSI Laboratories, Alpharetta, GA, for providing the tumor karyotype.
REFERENCES
- 1.Bauer DE,Mitchell CM,Strait KM, et al. 2012. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res. 18:5773–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chau NG,Hurwitz S,Mitchell CM, et al. 2016. Intensive treatment and survival outcomes in NUT midline carcinoma of the head and neck. Cancer. 122:3632–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.French CA,Miyoshi I,Kubonishi I,Grier HE,Perez-Atayde AR, Fletcher JA. 2003. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63:304–307. [PubMed] [Google Scholar]
- 4.French CA,Ramirez CL,Kolmakova J, et al. 2008. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 27:2237–2242. [DOI] [PubMed] [Google Scholar]
- 5.French CA,Rahman S,Walsh EM, et al. 2014. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 4:928–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alekseyenko AA,Walsh EM,Zee BM, et al. 2017. Ectopic protein interactions within BRD4-chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad SciUSA. 114:E4184–E4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonescu CR,Owosho AA,Zhang L, et al. 2017. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: a clinicopathologic and molecular study of 115 cases. Am J Surg Pathol. 41:941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Italiano A,Sung YS,Zhang L, et al. 2012. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 51:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Specht K,Sung YS,Zhang L,Richter GH,Fletcher CD,Antonescu CR. 2014. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosomes Cancer. 53:622–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bettegowda C,Agrawal N, Jiao Y, et al. 2011. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 333:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang S-C,Zhang L,Sung Y-S, et al. 2016. Recurrent CIC gene abnormalities in angiosarcomas: a molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 gene alterations. Am J Surg Pathol. 40:645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson S,Perrin V,Guillemot D, et al. 2018. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 245: 29–40. [DOI] [PubMed] [Google Scholar]
- 13.Sturm D,Orr BA,Toprak UH, et al. 2016. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 164: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson BC,Sung YS,Rosenblum MK, et al. 2018. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abo RP,Ducar M,Garcia EP, et al. 2015. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 43:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagle N,Berger MF,Davis MJ, et al. 2012. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haack H,Johnson LA,Fry CJ, et al. 2009. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol. 33:984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French CA. 2012. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 7:247–265. [DOI] [PubMed] [Google Scholar]
- 19.Reynoird N,Schwartz BE,Delvecchio M, et al. 2010. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 29:2943–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alekseyenko AA,Walsh EM,Wang X, et al. 2015. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 29:1507–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filippakopoulos P,Qi J,Picaud S, et al. 2010. Selective inhibition of BET bromodomains. Nature. 468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz BE,Hofer MD,Lemieux ME, et al. 2011. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 71:2686–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stathis A,Zucca E,Bekradda M, et al. 2016. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 6:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hung YP,Fletcher CD,Hornick JL. 2016. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histologic mimics. Mod Pathol. 29:1324–1334. [DOI] [PubMed] [Google Scholar]
- 25.Mertens F,Wiebe T,Adlercreutz C,Mandahl N,French CA. 2007. Successful treatment of a child with t(15;19)-positive tumor. Pediatr Blood Cancer. 49:1015–1017. [DOI] [PubMed] [Google Scholar]
- 26.Stelow EB,Bellizzi AM,Taneja K, et al. 2008. NUT rearrangement in undifferentiated carcinomas of the upper aerodigestive tract. Am J Surg Pathol. 32:828–834. [DOI] [PubMed] [Google Scholar]
- 27.Tilson MP,Bishop JA. 2014. Utility of p40 in the differential diagnosis of small round blue cell tumors of the sinonasal tract. Head Neck Pathol. 8:141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.den Bakker MA,Beverloo BH,van den Heuvel-Eibrink MM, et al. 2009. NUT midline carcinoma of the parotid gland with mesenchymal differentiation. Am J Surg Pathol. 33:1253–1258. [DOI] [PubMed] [Google Scholar]
- 29.Antonescu CR,Dal Cin P. 2014. Promiscuous genes involved in recurrent chromosomal translocations in soft tissue tumours. Pathology. 46: 105–112. [DOI] [PubMed] [Google Scholar]
- 30.Mertens F,Antonescu CR,Mitelman F. 2016. Gene fusions in soft tissue tumors: recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer. 55:291–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangray SKD,Le Guellec S,Fridman E, et al. 2018. USCAP 2018 abstracts: genitourinary pathology (894–1126). Mod Pathol. 31: 323–403.29551799 [Google Scholar]