Abstract
Oral diseases are the complex host responses composed of a broad array of inflammatory cells, and cytokines, chemokines, and mediators derived from the cells resident in the gingival tissues, as well as from the emigrating inflammatory cells. A chronic polymicrobial challenge to the local host tissues triggers this response, which under certain circumstances, and in a subset of the population, leads to the progressing soft and hard tissue destruction that characterizes periodontitis. The red complex has been proposed as a pathogenic consortium, consisting of P. gingivalis, T. denticola, and T. forsythia. This review has attempted to examine the virulence potential and determinants of these commensal opportunists.
Keywords: Biofilm, periodontitis, red complex, virulence factor
Introduction
Periodontal diseases are multifactorial infections elicited by a complex of bacterial species that interact with host tissues and cells causing the release of a broad array of inflammatory cytokines, chemokines, and mediators, some of which lead to the destruction of the periodontal structures, including the tooth-supporting tissues, alveolar bone, and periodontal ligament.[1]
Bacteria associated with periodontal disease trigger inflammatory responses in the immune cells, which in later stages of the disease, causes loss of both soft and hard tissue structures supporting the teeth. Until now, only a handful of bacteria have been characterized as infectious agents of periodontal disease.[2]
The search for the etiologic agents of periodontal diseases has been in progress for over a century. The search started in the “golden age of microbiology” (approx. 1880–1920), when the etiologic agents of many medically important infections were determined, parallel investigations of the etiology of periodontal diseases were initiated in this era.[3]
Periodontitis is a dysbiotic disease resulting from deviation in subgingival gram-positive bacteria to gram-negative bacteria. The development of periodontal dysbiosis occurs over a broadened timeframe, which slowly turns the symbiotic association of the host and the microbe to pathogenic form. Among microbial complexes, the first, such complex that has been related to disease is the orange complex consisting of anaerobic gram-negative species such as Prevotella intermedia, Prevotella nigrescens, Prevotella micros, and Fusobacterium nucleatum, which on disease progression shifts toward red-complex consisting of Tannerella forsythia, Tannerella denticola, and Porphyromonas gingivalis.[4]
Over 13,000 subgingival plaque samples from 185 adult subjects used cluster analysis and community ordination techniques to demonstrate the presence of specific microbial groups within dental plaque. Six closely associated groups of bacterial species were recognized which included the Actinomyces, a yellow complex consisting of members of the genus Streptococcus, a green complex consisting of Capnocytophaga species, A. actinomycetemcomitans serotype a, E. corrodens and Campylobacter and a purple complex consisting of V. parvula and Actinomyces odontolyticus. These groups of species are early colonizers of the tooth surface whose growth usually precedes the multiplication of the predominantly gram-negative orange and red complexes.[5]
The orange complex consists of Campylobacter gracilis, C. rectus, C. showae, E. nodatum, F. nucleatum subspecies, F. periodonticum, P. micros, P. intermedia, P. nigrescens, and S. constellatus, while the red complex consists of B. forsythus, P. gingivalis, and T. denticola. These two complexes are comprised of the species thought to be the major etiologic agents of periodontal diseases. The red complex bacteria are known to occur together in plaque samples often adjacent to the epithelial lining of the periodontal pocket in deeper areas. This is mainly due to interspecies interaction, co-aggregation, and metabolic interdependency among these three bacterial species.[6]
The red complex, which appears later during biofilm development, comprises species that are considered periodontal pathogens, namely, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia (previous names Bacteroides forsythus, or Tannerella forsythensis).[5] The red complex presents a portion of the climax community in the biofilms at sites expressing progressing periodontitis.
Members of the red complex are rarely found in the absence of members of the orange complex. With increasing colonization by the orange complex, more sites were colonized by increasing numbers of the red complex [Figures 1 and 2].
Figure 1.
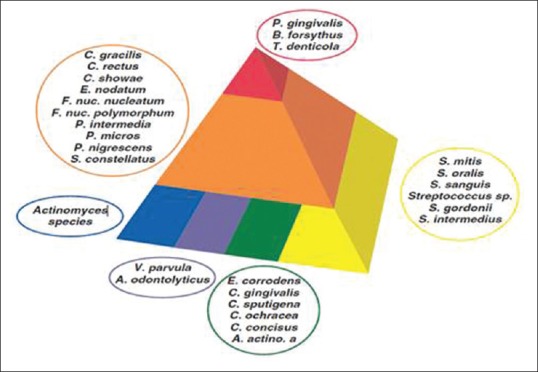
Diagram showing association among subgingival species. The base of the pyramid is comprised of species thought to colonize the tooth surface and proliferate at an early stage
Figure 2.
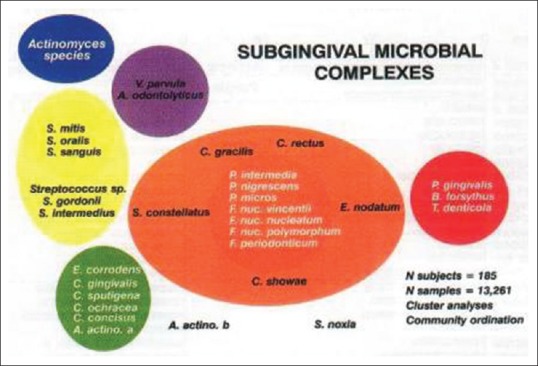
Different subgingival microbial complexes
Species in the red complex exhibited a very strong relationship with pocket depth and bleeding on probing. For example, B. forsythus and P. gingivalis (and T. denticola, data not shown) increased in prevalence and numbers with increasing pocket depth. Using statistical analysis, Socransky et al. clustered frequently occurring bacterial species into complexes that were color-coded to facilitate discussion [Table 1].
Table 1.
Bacterial clusters described by Socransky et al. (1998)
| Complex | Bacterial cluster |
|---|---|
| Red | Treponema denticola, Porphyromonas gingivalis, Tannerella forsythensis |
| Orange |
Fusobacterium nucleatum, Prevotella intermedia, Prevotella nigrescens, Peptostreptococcus micros (Associated species included) Eubacterium nodatum, Campylobacter rectus, Campylobacter showae, Streptococcus constellatus, Campylobacter gracilis) |
| Yellow |
Streptococcus sanguis, Streptococcus oralis, Streptococcus mitis, Streptococcus gordonii, Streptococcus intermedius |
| Green |
Carpnocytophaga, Camplobacter concisus, Eikenella corrodens, Actinobacillus actinomycemcomitans (serotype a) |
| Purple |
Veillonella parvula, Actinomyces odontolyticus, Actinobacillus actinomycemcomitans (serotype b), Selenomonus noxia, Actinomyces naesludii |
The virulence potential and determinants of these commensal opportunists (P. gingivalis, T. denticola, T. forsythia) including requirements for adherence, sustenance (i.e. accessing required nutritional materials for growth and replication), and evasion of host responses are discussed.[1]
Porphyromonas gingivalis
P. gingivalis has long been considered an important member of periodontopathic microbiota involved in periodontal disease progression and bone and tissue destruction.[7] Darveau et al. classified this small, gram-negative, black-pigmented anaerobe as a bonafide periodontal pathogen.[8] Interactions between P. gingivalis and other members of the oral microbiota including Streptococcus spp. and F. nucleatum resulted in specific co-aggregation, which contributes to the ability of the microorganism to effectively colonize the subgingival sulcus. The initial event in the pathogenicity of P. gingivalis is its interaction (adherence) in the oral cavity.[9] To accomplish this, P. gingivalis employs several bacterial components: fimbriae, proteases, hemagglutinins, and lipopolysaccharide.[10,11]
Morphological characteristics
Members of the P. gingivalis species are gram-negative, non-motile, asacchrolytic, obligatory anaerobic coccobacilli of approximately 0.5–0.8 to 1.0–3.5 m diameter.[12] They exhibit smooth, round colonies. When grown on a blood agar surface, the colonies initially are white to cream-colored. With time (4–8 days) these colonies darken from their edge towards the center and a deep red to black color, which correlates with the concentration of proto heme is observed.
Virulence factors
These include
Capsule
Outer membrane and its associated LPS
Fimbriae
Proteinases
Selected enzymes.
Capsule
The presence of a capsule in P. gingivalis has been considered an important antiphagocytic virulence factor. Capsule ensures increased resistance to phagocytosis, serum resistance, and decreased chemotaxis of PMN's.
Bacterial fimbriae
They are cellular appendages which are of two types, those that are involved in the interaction with other bacteria and mammalian cells (adhesions) and referred to as type-specific fimbriae, and those that involved in bacterial conjugation referred to as sex pili. Type-specific fimbriae apart from adhesion produce and delivers selected toxins and is also involved in motility. Each fimbria is approximately 3–25 nm in diameter and 3–25 m long and are arranged in a peritrichious fashion.
An important characteristic of the P. gingivalis fimbriae is their chemotactic ability. This ability to sense host stimuli could have a significant effect on the formation of an inflammatory lesion as well as the progression of periodontal tissue and bone destruction.[13]
Outer membrane proteins
P. gingivalis contains about 20 major proteins, ranging in size from approximately 20 to 90 KDa. Many in vitro studies have concentrated on the effects of “major outer sheath membrane proteins” on epithelial cells, fibroblasts, and a variety of bone cells. These structures, which are released from the outer membrane during growth, are referred to as outer membrane vesicles.[14]
Mihara and Holt[15,16,17] purified a 24-kDa protein from the outer membrane vesicles of P. gingivalis strain W50 and observed the purified protein to be capable of stimulating thymidine incorporated human gingival fibroblasts. Due of its significant fibroblast-stimulating ability, these authors named this 24-kDa protein as a “fibroblast activating factor”.
Proteinases
One of the potentially significant virulence characteristics of P. gingivalis is the large number of hydrolytic, proteolytic, and lipolytic enzymes that are produced by essentially all of the known strains. Important P. gingivalis associated proteases (proteinases capable of hydrolyzing peptide bonds) are: trypsin, thiol, caseinolytic proteinases, and two peptidases.
The proteinases cleave polypeptides after arginine or lysine-specific proteinases. At least 40 different proteinases have been described as being produced by P. gingivalis. The Arg- and Lys-proteinases are cysteine proteinases and have been given the common name, gingipains. The P. gingivalis collagenase has been classified as a proteinase with a hydrolytic predilection for collagen.[18] Genes coding for collagenase, a protease-hemagglutinin gene, a broad spectrum protease, an endothelin converting like enzyme, a dipeptidyl peptidase, and a reported protease called periodontain have all been isolated.[19,20]
Treponema denticola
Spirochetes were first observed in the oral cavity in humans by Van Leeuwenhoek[21] and have been detected in the periodontal lesion. Shinn, in 1962, accidentally discovered that patients receiving metronidazole for Trichomonal Vaginitis reduced signs and symptoms of ANUG. This study in those days clearly indicated that specific groups of organisms; mainly Spirochetes were involved in specific disease processes.
Morphological characteristics
Spirochetes are long, thin, corkscrew-like gram-negative anaerobic bacteria whose characteristic motility and morphology can readily be discerned by darkfield and phase contrast microscopic examination. The spiral-shaped cells of T. denticola are covered with an outer sheath consisting of a fragile envelope-like structure.[22] Periplasmic flagella are located on the cytoplasmic membrane and are covered with the outer sheath. T. denticola typically produces four flagella, which are intertwined around the cytoplasmic cylinder. The major outer sheath protein (Msp) is the predominant protein in the outer sheath [Figure 3].[23]
Figure 3.
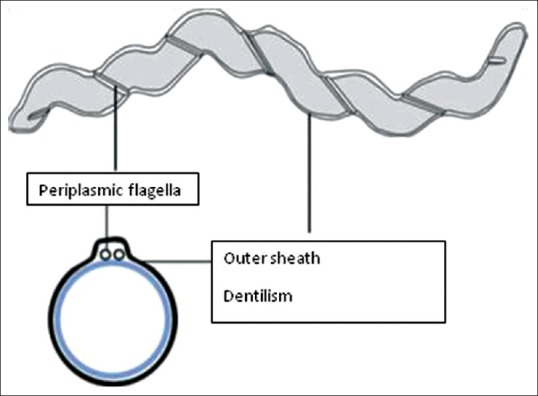
Structure of Treponema denticola
Spirochetes and periodontal disease
The role of Spirochetes in other forms of periodontal disease is less clear. The organisms have been considered as possible periodontal pathogens since the late 1800s. Over two decades ago, Armitage et al. was one of the first to establish a positive relationship between the percentage of spirochetes in periodontal pockets and clinical measures of inflammation, increased dental plaque, increasing gingival exudate, bleeding on probing, periodontal pocket depth, and the loss of connective tissue attachment.[24] The major reason for the interest in this group of organisms has been their increased numbers in sites with increased pocket depths. Healthy sites rarely exhibit the presence of microorganisms like spirochetes however these microorganisms are present in low to moderate number in many deep pockets around the teeth.
Bacterial co-aggregation and bacteria-host interactions
T. denticola interacts with other oral bacterial species, notably P. gingivalis and F. nucleatum. This co -aggregation likely plays a role in the progression of periodontal disease (i.e. biofilm formation), since Kigure et al. reported that these two members of the red complex occurred physically related in the formation of biofilm.[25] The protein, LrrA binds human epithelial cells to T. forsythia, another member of the red complex.[26] P. gingivalis fimbriae mediate attachment to host cells, as well as participate in the interaction (coaggregation) between several different plaque-forming bacterial species (i.e. A. naeslundii, S. gordonii, and S. oralis).
Virulence factors
Mainly produces hydrolytic enzymes like
Hyalouronidase
Collagenase
Protease
Phospholipase
Phosphatases
Others are-
Major outer sheath protein (Msp)
OppA (ortholog)
FH- like binding protein
Dentilism
Leucine-rich repeat A
Tannerella forsythia
Formerly, many of bacteroides were referred to as the “Black Pigmented bacteroides” or the “BPB” group of organisms, since the bacterial colonies often produce a black or beige pigment especially when cultured on blood media.
It is an anaerobic gram-negative member of the Cytophaga- Bacteroides family and was initially named Bacteroides forsythus. T. forsythia is associated, more frequently and or at higher levels, with various forms of periodontal disease, including gingivitis, chronic, and aggressive periodontitis, than with healthy individuals. T. forsythia infection is more likely to cause periodontitis in overweight women than in normal-weight women. Overweight or obese individuals have an overgrowth of T. forsythia, thus subjecting these individuals to a higher risk of developing periodontal disease.[27]
Virulence factors
So far, only a few putative virulence factors have been identified in T. forsythia, including:- Trypsin-like and PrtH proteases, Sialidases SiaH and NanH, leucine-rich repeat (Lrr) cell surface-associated and secreted protein BspA, α-D-glucosidase, and N-acetyl-b-glucosaminidase Hemagglutinin
Biofilm activity
T. forsythia has been shown to form poor monospecies biofilms in vitro.[28] However, when an operon involved in exopolysaccharide synthesis in T. forsythia was disrupted, a significant increase in biofilm formation was observed. The study suggested that surface exopolysaccharides in T. forsythia play inhibitory roles in biofilm development by T. forsythia. Interestingly, it was found that T. forsythia also forms mixed synergistic biofilms with F. nucleatum. F. nucleatum is considered to be a bridge bacterium due to its ability to co-aggregate with both early- and late-colonizing bacteria, thereby facilitating dental plaque formation.
Polymicrobial Nature of Chronic Periodontitis
It is widely accepted that T. denticola, Porphyromonas gingivalis, and Tannerella forsythia form a bacterial consortium, often referred to as the “red complex”, that is strongly associated with the clinical progression of chronic periodontitis.[29,30]
The unifying features of the Red complex bacteria are their extracellular proteolytic activity, their complex anaerobic fermentation of amino acids, production of toxic metabolites, and outer membrane vesicles.
Of the three species, only the Treponeme is motile and able to respond chemotactically to environmental stimuli. The in vivo interactions of these species are still poorly characterized but some studies have indicated that P. gingivalis may be needed for T. denticola colonization and presence in subgingival plaque.[31,32]
A recent study of the bacterial composition of subgingival plaque in individuals with chronic periodontitis showed that P. gingivalis, T. denticola, and Tannerella forsythia were routinely found together in subgingival plaque.[33] Interestingly, P. gingivalis or T. denticola were rarely found in subgingival plaque without T. forsythia. Mineoka et al. have recently found a similar relationship between T. forsythia and P. gingivalis. T. forsythia has also been found to be more prevalent than P. gingivalis in subgingival plaque. This may suggest that T. forsythia colonizes plaque before P. gingivalis and T. denticola.
Synergism between Different Species of Red Complex
The complex was considered the most significant complex in periodontal disease progression because of the members of this consortium increase in numbers and prevalence with increasing clinical parameters of periodontal disease.
P. gingivalis, T. forsythia, and T. denticola are found together in plaque samples often adjacent to the epithelial lining of the periodontal pocket of the gingival sulcus. P. gingivalis benefits from what appears to be a nutritional interdependence with T. denticola.
Both P. gingivalis and T. denticola have several characteristics that make them prime candidates as pathogens involved in the clinical destruction of periodontal tissues:
They occur concomitantly with the clinical signs of periodontal destruction.
They appear closely linked topologically in the developing biofilm.
In vitro studies demonstrate their ability to produce a number of outer membrane-associated proteinases.
Gmur et al. showed a strong relationship between T. forsythia and P. gingivalis in the subgingival plaques taken at different pocket depths. P. gingivalis was never detected in the absence of T. forsythia.
Members of the red complex were found in high numbers in adult periodontitis lesions and in sites with deeper pockets or more advanced lesions. In 4 to 6 mm pockets, T. denticola was detected at the surface layer of the plaque while P. gingivalis cells were detected in the layer beneath. In deeper pockets, the species coexisted in large numbers. This trio has been shown to produce proteolytic enzymes such as those sought in the “BANA” test and in the proposed SK013 peptidase test.
A new member of the Bacteriodetes phylum Candidatus Bacteroides periocalifornicus (CBP) bacterium has been identified recently and is present in dental plaque, as well as shows strong association with the well-known pathogenic “red complex” that resides in deep periodontal pockets. CBP is orally ubiquitous, existing in both healthy and diseased individuals, but not present in gut or skin samples. CBP also increases with increased pocket depth, coexists with both F. nucleatum, T. denticola, and P. gingivalis. Its abundance is strongly correlated with members of the red complex, but not healthy commensals, all of which suggest that CBP is a novel candidate member of the symbiotic and pathogenic red complex.[34]
Detection of Red Complex Species
Enzyme tests
Porphyromonas gingivalis, Tannerella forsythensis, previously called Bacteroides forsythus and Treponema denticola are among the few subgingival microflorae associated with periodontal disease which possess a trypsin-like enzyme capable of hydrolyzing the synthetic peptide N-benzoyl-DL-arginine-B-naphthylamide, commonly referred to as BANA.
The commercially available BANA test (Oral B, Perioscan TM) is a modification of the BANA hydrolysis test adapted from the trypsin-like enzyme of the API-ZYM Kit. BANA is a rapid and effective diagnostic aid and has shown to correlate well with the clinical indices used to diagnose periodontal disease. BANA is able to detect between 103–105 targets/samples which is within the same range as immunoassays and nucleic acid probes.
SK-013 (carbobenzoxy-glycyl-arginine-3, 5-dibromo-4-hydroxy- arginine) has also been used for the detection of peptidase activity of T. denticola, P. gingivalis and T. forsythensis in plaque samples. However, BANA appears to be more frequently employed.
Diagnostic tests such as BANA-hydrolysis, have confirmed the role of Treponema denticola, Porphyromonas gingivalis and Tannerella forsythensis as indicators of infection and their strong association with severe periodontal disease, led to the designation of this group of bacteria as the red complex.[35,36]
Implications for clinical practice
The primary care physician is the first contact of a patient for the consultation of illness. Early diagnosis and a multidisciplinary approach are key components for managing complex diseases. Increased awareness of oral health and the identification of various disease-causing pathogens help to rule out the risk factors involved in oral diseases. Certain therapies have shown promising results that further needs evaluation in prospective clinical trials. Elimination of these pathogens from the site of infection remains a perplexing task, which demands the use of antibiotics. The emergence of drug-resistant forms has spurred interest in identifying novel therapeutic targets against these pathogens. Acetaminophen (APAP) and ibuprofen (IB) are drugs commonly used to reduce pain due to their anti-inflammatory, antipyretic, and analgesic effect. These drugs have antimicrobial potential against red-complex pathogens, namely, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. These two drugs were found to target vital proteins involved in the cellular process, metabolism, and virulence of red complex pathogens.[37]
Reserpine is another known potent drug having antibacterial activity against red-complex pathogens. Reserpine targets the vital protein transporters such as ABC transporter and efflux pumps which are known to play a crucial role in the survival of bacterial cells. Hence, it destroys the bacterial cells that have the potential for the disease.[38]
Conclusion
This review has provided data supporting the production of these species by a number of biomolecules that have homologs in other pathogens and that have been identified to endow these microorganisms with an enhanced resistance to host innate and adaptive immune responses. There exists little concrete information concerning whether the red complex consortium represents a footnote of environmental changes occurring with the periodontal disease process or synergistically contributes to the initiation and progression of tissue destructive events. The future research in this area should direct some effort into moving from cataloging the biofilm consortia towards defining the crucial components that underpin these purported in vivo pathogenic properties.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The red complex, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 2.Larsen T, Fiehn N-E. Dental biofilm infections–An update. APMIS. 2017;125:376–84. doi: 10.1111/apm.12688. [DOI] [PubMed] [Google Scholar]
- 3.Dumitrescu AL. Norway: Elsevier; 2006. Etiology and Pathogenesis of Periodontal Disease; p. 39. [Google Scholar]
- 4.Shaikh HFM, Patil SH, Pangam TS, Rathod KV. Polymicrobial synergy and dysbiosis: An overview. J Indian Soc Periodontol. 2018;22:101–6. doi: 10.4103/jisp.jisp_385_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carranza FA, Newman MG, Takei HH, Klokkevold PR. 10th ed. Missouri: WB Saunders; 2006. Clinical Periodontology; p. 143. [Google Scholar]
- 6.Nayak A, Bhat K, Shivanaikar S, Pushpa P, Kugaji M, Kumbar V. Detection of red complex organisms in chronic periodontitis by multiplex polymerase chain reaction. J Adv Clin Res Insights. 2018;5:139–44. [Google Scholar]
- 7.Kheur S, Hazarey VK, Kapley A, Purohit H. Tracking of Actinobacillus actinomycetemcomitans in subgingival plaque of aggressive periodontitis patients. J Int Clin Dent Res Organ. 2010;2:64–9. [Google Scholar]
- 8.Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 9.Kuramitsu HK, Ellen RP. Oral Bacterial Ecology: The Molecular Basis. Norfolk, UK: Horizon Scientific Press; 2000. [Google Scholar]
- 10.Hamada S, Amano A, Kimura S, Nakagawa I, Kawabata S, Morisaki I. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129–38. doi: 10.1111/j.1399-302x.1998.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 11.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors in Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 12.Slots J, Lisgarten MA. Bacteroides gingivalis, Bacteroides intermedians and Aggregatibacter actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988;15:85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 13.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 14.Mayrand D, Grenier D. Biological activities of outer membrane vesicles. Can J Microbiol. 1989;35:607–13. doi: 10.1139/m89-097. [DOI] [PubMed] [Google Scholar]
- 15.Mihara J, Holt SC. Purification and characterization of fibroblast-activating factor isolated from Porhyromonas gingiualis W50. Infect Immun. 1993;61:588–95. doi: 10.1128/iai.61.2.588-595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihara J, Miyazawa Y, Holt SC. Modulation of growth and function of human gingival fibroblasts by fibroblast-activating factor derived from Porphyrornonas gingiualis W50. Infect Immun. 1993;61:596–601. doi: 10.1128/iai.61.2.596-601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mihara J, Yoneda T, Holt SC. Role of Porphyrornonas gingiualis- derived fibroblast-activating factor in bone resorption. Infect Immun. 1993;61:3562–4. doi: 10.1128/iai.61.8.3562-3564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourgeau G, Lapointe H, Peloquin P, Mayrand D. Cloning, expression, and sequencing of a protease gene (tpr) from Porpkyromonas gingivalis W83 in Escherichia coli. Infect Immun. 1992;60:3186–92. doi: 10.1128/iai.60.8.3186-3192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato T, Takahashi N, Kuramitsu HK. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene which expresses novel collagenase activity. J Bacteriol. 1992;174:3889–95. doi: 10.1128/jb.174.12.3889-3895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson D, Potempa J, Kordula T, Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human a1-proteinase inhibitor. J Biol Chem. 1999;274:12245–51. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 21.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by Tannerella forsythia. Infect Immun. 2006;74:5023–8. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, et al. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishihara K. Virulence factors of Treponema denticola. Periodontology 2000. 2010;54:117–35. doi: 10.1111/j.1600-0757.2009.00345.x. [DOI] [PubMed] [Google Scholar]
- 24.Armitage GC, Dickinson WR, Jenderseck RS, Levine SM, Chambers DW. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J Periodontol. 1982;53:550–6. doi: 10.1902/jop.1982.53.9.550. [DOI] [PubMed] [Google Scholar]
- 25.Kigure T, Saito A, Seida K, Yamada S, Ishihara K, Okuda K. Distribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different periodontal pocket depths examined by immunohistochemical methods. J Periodontal Res. 1995;30:332–41. doi: 10.1111/j.1600-0765.1995.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 26.Ikegami A, Honma K, Sharma A, Kuramitsu HK. Multiple functions of the leucine-rich repeat protein Lrra of Treponema denticola. Infect Immun. 2004;72:4619–27. doi: 10.1128/IAI.72.8.4619-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanner AC, Izard J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000. 2006;42:88–113. doi: 10.1111/j.1600-0757.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 28.Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 2007;42:156–66. doi: 10.1016/j.micpath.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Lamont RJ, Jenkinson HF. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–63. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 31.Simonson LG, McMohan KT, Childers DW, Morton HE. Bacterial synergy of Treponema denticola and Porphyromonas gingivalis in a multinational population. Oral Microbiol Immunol. 1992;7:111–2. doi: 10.1111/j.1399-302x.1992.tb00519.x. [DOI] [PubMed] [Google Scholar]
- 32.Sela M. Role of Treponema denticola in periodontal diseases. Crit Rev Oral Biol Med. 2001;12:399–413. doi: 10.1177/10454411010120050301. [DOI] [PubMed] [Google Scholar]
- 33.Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 2009;24:469–77. doi: 10.1111/j.1399-302X.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 34.Pedro J, Torres, John Thompson, Jeffrey S. McLean, Scott T. Kelley, Anna Edlund. Discovery of a novel periodontal disease-associated bacterium. Microb Ecol. 2019;77:267–76. doi: 10.1007/s00248-018-1200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WJ Loesche, DE Lopatin, Giordano J, Alcoforado G, Hujoel P. Comparison of the benzoyl-DL-arginine-naphthylamide (BANA) test, DNA probes, and immunological reagents for ability to detect anaerobic periodontal infections due to Porphyromonas gingivalis, Treponema denticola, and Bacteroides forsythus. J Clin Microbiol. 1992;30:427–33. doi: 10.1128/jcm.30.2.427-433.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laughon BE, Syed SA, Loesche WJ. API ZYM system for identification of Bacteroides spp., Capnocytophaga spp., and spirochetes of oral origin. J Clin Microbiol. 1982;15:97–102. doi: 10.1128/jcm.15.1.97-102.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vijayashree Priyadharsini J. In silico validation of non-antibiotic drugs, acetaminophen, and ibuprofen as antibacterial agents against red complex pathogens. J Periodontol. 2019 doi: 10.1002/JPER.18-0673. doi: 10.1002/JPER.18-0673. [DOI] [PubMed] [Google Scholar]
- 38.Ushanthika T, Smiline Girija AS, Paramasivam A, Priyadharsini JV. An in silico approach towards identification of virulence factors in red complex pathogens targeted by reserpine. Nat Prod Res. 2019;17:1–6. doi: 10.1080/14786419.2019.1641811. [DOI] [PubMed] [Google Scholar]


