Abstract
Branched amphiphilic peptide capsules (BAPCs) are an efficient transport system that can deliver nucleic acids, small proteins, and solutes. The ability of BAPCs to break down is essential to their adoption as a delivery vehicle for human and agricultural applications. Until now, however, BAPCs were shown to be inert to mammalian degradation systems. Here, we demonstrate, using BAPCs encapsulating the toxic urea analogue thiourea, that the common soil fungus Aspergillus nidulans can degrade BAPCs. We provide evidence that this degradation is extracellular through the action of secreted proteases. Our data indicate that BAPCs are likely biodegradable in the environment.
Introduction
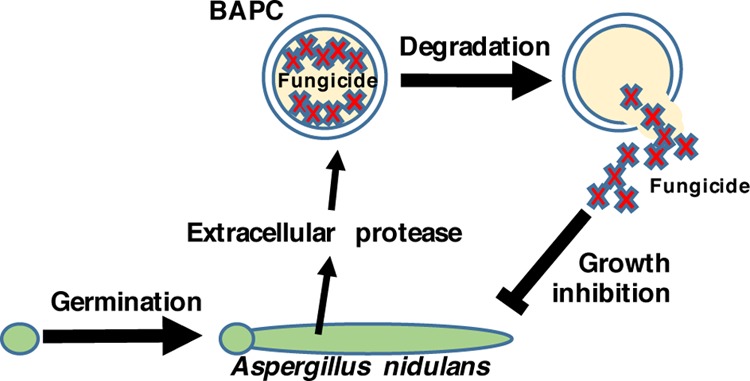
An efficient amino acid-derived nanoscale delivery method that facilitates oral delivery and cellular uptake of biopesticides that is biodegradable could greatly reduce the environmental load of active ingredients currently employed in agriculture. The Tomich lab has developed such a delivery agent. It is a self-assembling cationic nanocapsule composed of branched amphiphilic peptides (BAPs). As the peptides anneal, they form peptide bilayer-delimited capsules capable of encapsulating solutes in an aqueous environment. These BAP capsules (BAPCs) are prepared from two branched peptides—bis(Ac-FLIVI)-K-KKKK-CO-NH2 (CAS RN 1492039-67-4) and bis(Ac-FLIVIGSII)-K-KKKK-CO-NH2 (CAS RN 1492039-68-5).1 The two peptides can assemble either singly or together using different assembly temperatures to alter the sizes of the nanocapsules.2 Fully formed BAPCs range in diameter from 10 to >500 nm depending on the annealing conditions.3 BAPCs encapsulating fluorescent dyes are taken up by several epithelial cell lines, including HeLa (CCL-2), HEK-293 (CRL-11268), CasKi (CRL-1550), and N/N 1003A rabbit lens epithelial cells, through the endocytic pathway and then become localized in the perinuclear space.4,5 They show minimal cytotoxicity to cultured HeLa cells.6 In all previous studies, BAPCs have proven to be resistant to cellular degradative machinery and encapsulated active ingredients were not released from the capsules.5 This lack of break down currently limits the utility of BAPCs for encapsulated drug delivery.
The cation surface of these nanoparticles can also bind and deliver a variety of nucleic acids. BAPCs were used in insects for oral delivery of dsRNAs directed to silence genes involved in the protein refolding pathway.7 If the capsules are to be deployed in the environment, they need to be biodegradable to avoid environmental accumulation.
For this study, the common soil fungus Aspergillus nidulans was chosen to demonstrate fungal biodegradation of BAPCs. The Aspergillus genus is ubiquitous and very frequently occurs in soil.8−11 A well-known characteristic of fungi is the secretion of extracellular proteases for breaking down proteins in the surrounding environment for nutrient acquisition, making them excellent candidates to possess BAPC peptide capsule degradation capability.12 Here, we show that this fungus degrades BAPCs. It can be inferred that the capsules would be broken down in the soil and, therefore, will not build up in the environment. It is important to note that proteases are conserved and therefore other species are expected to have similar BAPC degradation properties.
Results and Discussion
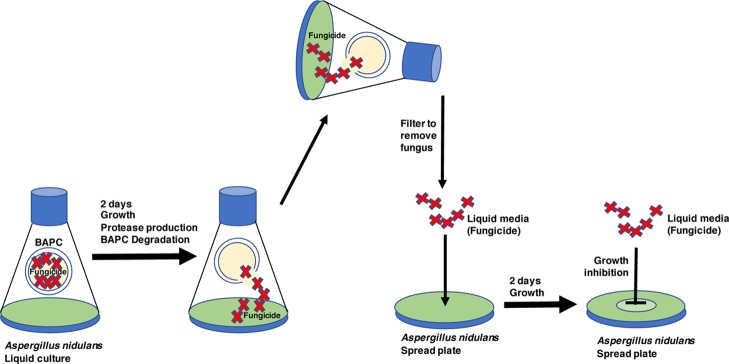
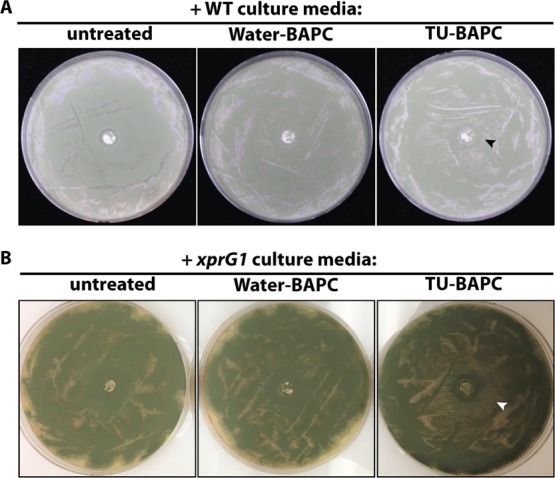
To determine whether A. nidulans can degrade the peptide capsules, water soluble thiourea (TU), an analogue of urea known to be toxic when taken up by A. nidulans,13 was encapsulated during BAPC formation and used to assess whether TU could be released from the BAPCs and cause growth inhibition on A. nidulans spread plates (Scheme 1). We reasoned that the peptide capsules are likely to be degraded by proteases, and therefore used media containing milk as a de-repressing nitrogen source known to favor protease expression.12Figure 1A shows the results of treating wild type MH1 with the TU-encapsulated BAPCs compared with water-encapsulated BAPCs. The lawn of A. nidulans growth is visible as the green color of the asexual spores (conidia) when treated with water-BAPCs, while treatment with TU-BAPCs conferred a zone of growth inhibition at the center of the plate (mean diameter ± standard error = 38.4 ± 4.36 mm) characterized by mycelium with reduced conidiation (asexual spore production) and displaying a brown pigmentation reaction encircled by a less intense green color, typical of reduced conidiation. A level of background growth occurs within the zone of TU-BAPC inhibition as the fungus must grow prior to producing the degrading enzymes that release the TU. Because the BAPCs were thoroughly washed to remove any surface-bound TU, the observed growth inhibition suggests that the contents of the BAPCs were released through the disassembly of the peptide bilayer through the action of one or more proteases.
Scheme 1. Spread Plate Assay for BAPC Degradation.
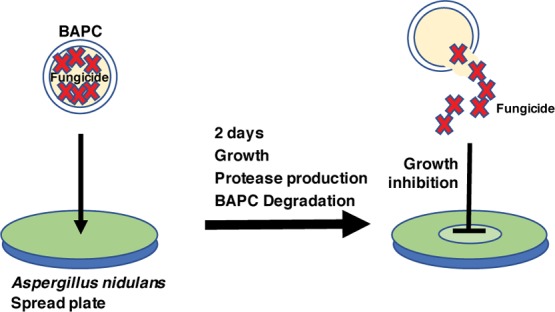
Figure 1.
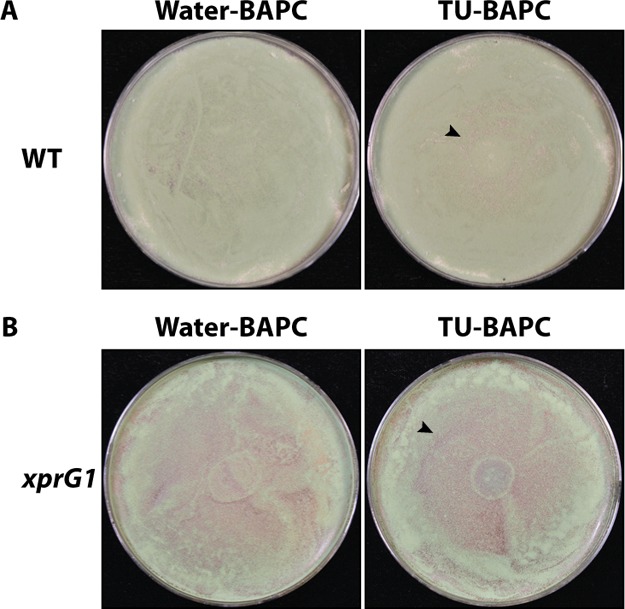
Degradation of BAPCs containing TU leads to growth inhibition on A. nidulans spread plates. (A) MH1 (wild type, WT) spread on 1% milk plates. −40 μL of BAPCs encapsulating water, left, 40 μL of BAPCs encapsulating 1.865 M TU, right, were spotted at the center of the plate. (B) MK85 (xprG1) spread on 1% milk plates—left, 40 μL of BAPCs encapsulating water, right, 40 μL of BAPCs encapsulating TU. The arrowheads indicate the zone of inhibition.
To confirm that fungal proteases are involved, cultures were established using the mutant strain, MK85 (xprG1). This strain has increased extracellular protease production because of a gain-of-function mutation in the p53-like transcription factor XprG, which regulates the expression of extracellular protease genes.14,15 The results of this experiment showed a larger and clearer zone of inhibition for MK85 (mean diameter ± standard error = 45.6 ± 3.52 mm) than for the wild type strain MH1 (Figure 1B).
Next, we developed an assay for the degradation of TU-containing BAPCs by MH1 (wild type) or MK85 (xprG1) fungal cells in liquid culture (Scheme 2). The liquid cultures were prepared, then the filtered fungus-free media was concentrated, and spotted at the center of a wild type MH1 spread plate (Figure 2A,B). No growth inhibition was evident from the filtrate from either the wild type or xprG1 water-BAPC or no BAPC treatment. In contrast, a zone of mild growth inhibition, evident as reduced green density of conidiation at the center of the plate was observed for the wild type TU-BAPC filtrate (Figure 2A). This indicates that secreted proteases were present in the media of the liquid cultures and that they had broken down TU-encapsulated BAPCs thereby releasing the fungicide. A larger zone of inhibition was observed for the TU-BAPC-treated filtrate from the MK85 (xprG1) culture (Figure 2B). This is consistent with the involvement of proteases in BAPC degradation. It is important to note the possibility of an alternative interpretation that the BAPCs could have been degraded intracellularly with the subsequent excretion of the fungicide into the media. However, because of the known elevated expression of extracellular proteases in the xprG1 gain-of-function mutant, we favor the interpretation that the degradation is performed by an extracellular protease.
Scheme 2. Liquid Culture Assay for BAPC Degradation.
Figure 2.
Concentrated media from liquid cultures grown with BAPC-containing TU inhibits A. nidulans growth on spread plates. (A) Media from MH1 (WT) 1% milk liquid cultures spotted on MH1 1% milk plates. Left to right-untreated (no BAPCs), BAPCs encapsulating water, BAPCs encapsulating TU. (B) Media from MK85 (xprG1) 1% milk liquid cultures spotted on MH1 (WT) 1% milk plates, left to right—untreated (no BAPCs), BAPCs encapsulating water, BAPCs encapsulating TU. The arrowheads indicate the zone of inhibition. Photographs in A and B were taken under different lighting; adjustments were made to brightness and contrast.
Conclusions
BAPCs are being tested for their ability to deliver nucleic acids designed to be biocides. In all animal species tested, the BAPCs remain intact and show no cytotoxicity when given at clinical dosages.7 An early concern has been the accumulation of the peptide nanoparticles in the environment, should they be used on an agricultural scale. Based on our experiments, it is clear that a wild type ubiquitous soil fungus can proteolyze these structures. Furthermore, the increased diameter of the zone of inhibition observed with the MK85 gain-of-function mutant, which was originally selected for elevated extracellular protease production, provided evidence that the fungus produced an extracellular protease capable of BAPC degradation. The candidate enzyme(s) responsible for BAPC degradation are being sought. ε-Poly-l-lysine-degrading enzyme from Streptomyces albulus (BAE53412.2) is known to break down the bond found in the branch point in BAPCs.16 Preliminary sequence database searches reveal sequence similarity to several hypothetical proteases in A. nidulans. There are many other species with potential orthologues to the assumed BAPC-degrading enzyme according to sequence similarity.
There is much work to be done to give a comprehensive molecular analysis of BAPC breakdown. BAPCs can now be considered biodegradable because of the ability of A. nidulans to degrade the capsules and release their contents. A. nidulans, like other Aspergillus species, is widely distributed in the soil in a range of biomes and latitudes.10 Studies of fungal density in cultivated agricultural soils report filamentous fungi to occur at 104 to 105 colony forming units per gram of soil.17,18 Our results, when considered with the abundance of Aspergilli in soils and longer environmental time-frames than the two-day time-point used in our studies, suggest that the accumulation of BAPCs in the environment should not present a sizable hurdle to their introduction as biocide carriers. The accumulation of nanoparticles in the soil will be avoided if BAPCs are used to deliver biocides composed of biodegradable nucleic acids.
Experimental Section
BAPC Formation
The two self-assembling peptides, bis(Ac-FLIVI)-K-KKKK-CO-NH2 and bis(Ac-FLIVIGSII)-K-KKKK-CO-NH2, were chemically synthesized as previously described.4 BAPCs were prepared by mixing equimolar concentrations (1 mM each) of the two branched peptides dissolved in trifluoroethanol. Under these conditions the peptides do not associate and remain monomeric. They were dried in vacuo and rehydrated dropwise in sterile water for controls or with water containing the active ingredient. The nascent particles were subjected to a temperature shift protocol previously described yielding 20–30 nm nanoparticles.3 Because it takes ∼1600 peptides to make one BAPC, the final concentration was 1.3 μM. BAPCs loaded with the active ingredient were dried and then resuspended in 50 μL of water at a final concentration of 20 μM BAPCs.
Fungicide Encapsulation
Water soluble TU (Sigma, St. Louis, MO) was encapsulated at a concentration of 1.865 M during BAPC assembly. The TU-encapsulated BAPCs were washed several times to remove externally bound active ingredient, as previously described.
A. nidulans Strains and Growth
A. nidulans strain MH1 (biA1 veA1) was used as wild type (M.J. Hynes, University of Melbourne; deposited in the Fungal Genetics Stock Center (FGSC), Manhattan KS, as FGSC A2291).19A. nidulans MK85 (biA1 xprG1 niiA4 veA1) is an xprG1 gain-of-function mutant de-repressed for extracellular proteases (M.E. Katz, University of New England; FGSC A1630).14,15 Fungal growth and manipulations were conducted, and growth media were prepared as previously described.20 Spore suspensions were generated, by surface scraping, at a density of one 12–14 mm diameter colony per 400 μL of 0.005% Tween-80 (Sigma), grown from point-inoculation on complete media for two days at 37 °C.
Solid media were prepared using polystyrene 100 × 15 mm Petri dishes (KORD-Valmark Labware Products) and Aspergillus nitrogen-free medium (ANM) containing 1% glucose as the carbon source, supplements to repair auxotrophies, and 1% (w/v) skim milk (Kroger Co.) as the nitrogen source.14
Plate Tests for Degradation of Fungicide-Encapsulated BAPC
A. nidulans spore suspension (50 μL) was spread on the plates using glass beads (≤160 μm) (Sigma) to obtain homogeneous lawns, as described.21 After 20 min, the TU-BAPC samples described above were spotted at the center of the plates at volumes of 10 and 40 μL. Plates were incubated at 37 °C and analyzed at 24 and 48 h. Three biological replicates were performed.
BAPC Degradation in Liquid Culture Assay
Two mL liquid cultures comprising supplemented liquid ANM with 1% milk as the nitrogen source and 50 μL of BAPC (1.3 μM) were inoculated with 50 μL of A. nidulans spore suspension. The test sample consisted of BAPCs containing a 1.865 M TU solution, and BAPC-free and water-encased BAPC controls were included. The samples, in 25 mL conical flasks, were grown with shaking at 160 oscillations per minute for two days at 37 °C in an orbital shaking incubator. After incubation, conditioned media was filtered through Miracloth (Millipore-Sigma). Filtered media (20 μL) was spotted onto a thick chromatography paper (grade 238, Ahlstrom Filtration Inc.) 5 mm diameter disk and placed in the center of a supplemented ANM-1% milk plate spread with spores 20 min prior. The remaining media was freeze-dried then rehydrated with a small volume (250 μL) to increase its concentration. Concentrated media (20 μL) were then spotted on the chromatography paper disks on milk plates. Three biological replicates were performed.
Acknowledgments
This project was supported by a Cancer Research Award from the Johnson Center for Basic Cancer Research, Kansas State University, and a K-INBRE STAR Traineeship funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103418. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. Contribution no. 19-333-J from the Kansas Agricultural Experiment Station. Publication of this article was funded in part by the Kansas State University Open Access Publishing Fund. Contribution.
Author Present Address
§ Johns Hopkins University Bloomberg School of Public Health, 1928 Fleet Street, APT B, Baltimore, Baltimore, MD, USA 21231, 6209317564.
The authors declare the following competing financial interest(s): JMT is a stockholder in the firm Phoreus Biotechnology that has licensed the IP developed by Professor Tomich at Kansas State University. All of the research for this project was carried out in a non-affiliated laboratory. EMW and RBT have no conflicts to declare.
References
- Barros S. M.; Whitaker S. K.; Sukthankar P.; Avila L. A.; Gudlur S.; Warner M.; Beltrão E. I. C.; Tomich J. M. A review of solute encapsulating nanoparticles used as delivery systems with emphasis on branched amphipathic peptide capsules. Arch. Biochem. Biophys. 2016, 596, 22–42. 10.1016/j.abb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros S. d. M.; Avila L. A.; Whitaker S. K.; Wilkinson K. E.; Sukthankar P.; Beltrão E. I. C.; Tomich J. M. Branched amphipathic peptide capsules: different ratios of the two constituent peptides direct distinct bilayer structures and sizes. Langmuir 2017, 33, 7096–7104. 10.1021/acs.langmuir.7b00912. [DOI] [PubMed] [Google Scholar]
- Sukthankar P.; Whitaker S. K.; Garcia M.; Herrera A.; Boatwright M.; Prakash O.; Tomich J. M. Thermally induced conformational transitions in branched amphiphilic peptide capsules. Langmuir 2015, 31, 2946–2955. 10.1021/la504381y. [DOI] [PubMed] [Google Scholar]
- Gudlur S.; Sukthankar P.; Gao J.; Avila L. A.; Hiromasa Y.; Chen J.; Iwamoto T.; Tomich J. M. Peptide nanovesicles formed by the self-assembly of branched amphiphilic peptides. PLoS One 2012, 7, e45374 10.1371/journal.pone.0045374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukthankar P.; Avila L. A.; Whitaker S. K.; Iwamoto T.; Morgenstern A.; Apostolidis C.; Liu K.; Hanzlik R. P.; Dadachova E.; Tomich J. M. Branched amphiphilic peptide capsules: cellular uptake and retention of encapsulated solutes. Biochim. Biophys. Acta, Biomembr. 2014, 1838, 2296–2305. 10.1016/j.bbamem.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila L. A.; Aps L. R. M. M.; Sukthankar P.; Ploscariu N.; Gudlur S.; Šimo L.; Szoszkiewicz R.; Park Y.; Lee S. Y.; Iwamoto T.; Ferreira L. C. S.; Tomich J. M. Branched Amphiphilic Cationic Oligopeptides Form Peptiplexes with DNA: A Study of Their Biophysical Properties and Transfection Efficiency. Mol. Pharm. 2015, 12, 706–715. 10.1021/mp500524s. [DOI] [PubMed] [Google Scholar]
- Avila L. A.; Chandrasekar R.; Wilkinson K. E.; Balthazor J.; Heerman M.; Bechard J.; Brown S.; Park Y.; Dhar S.; Reeck G. R.; Tomich J. M. Delivery of lethal dsRNAs in insect diets by branched amphiphilic peptide capsules. J. Controlled Release 2018, 273, 139–146. 10.1016/j.jconrel.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P.; Riley R.; Wiebenga A.; Aguilar-Osorio G.; Amillis S.; Uchima C. A.; Anderluh G.; Asadollahi M.; Askin M.; Barry K.; Battaglia E.; Bayram Ö.; Benocci T.; Braus-Stromeyer S. A.; Caldana C.; Cánovas D.; Cerqueira G. C.; Chen F.; Chen W.; Choi C.; Clum A.; Dos Santos R. A. C.; Damásio A. R. d. L.; Diallinas G.; Emri T.; Fekete E.; Flipphi M.; Freyberg S.; Gallo A.; Gournas C.; Habgood R.; Hainaut M.; Harispe M. L.; Henrissat B.; Hildén K. S.; Hope R.; Hossain A.; Karabika E.; Karaffa L.; Karányi Z.; Kraševec N.; Kuo A.; Kusch H.; LaButti K.; Lagendijk E. L.; Lapidus A.; Levasseur A.; Lindquist E.; Lipzen A.; Logrieco A. F.; MacCabe A.; Mäkelä M. R.; Malavazi I.; Melin P.; Meyer V.; Mielnichuk N.; Miskei M.; Molnár Á. P.; Mulé G.; Ngan C. Y.; Orejas M.; Orosz E.; Ouedraogo J. P.; Overkamp K. M.; Park H.-S.; Perrone G.; Piumi F.; Punt P. J.; Ram A. F. J.; Ramón A.; Rauscher S.; Record E.; Riaño-Pachón D. M.; Robert V.; Röhrig J.; Ruller R.; Salamov A.; Salih N. S.; Samson R. A.; Sándor E.; Sanguinetti M.; Schütze T.; Sepčić K.; Shelest E.; Sherlock G.; Sophianopoulou V.; Squina F. M.; Sun H.; Susca A.; Todd R. B.; Tsang A.; Unkles S. E.; van de Wiele N.; van Rossen-Uffink D.; Oliveira J. V. d. C.; Vesth T. C.; Visser J.; Yu J.-H.; Zhou M.; Andersen M. R.; Archer D. B.; Baker S. E.; Benoit I.; Brakhage A. A.; Braus G. H.; Fischer R.; Frisvad J. C.; Goldman G. H.; Houbraken J.; Oakley B.; Pócsi I.; Scazzocchio C.; Seiboth B.; vanKuyk P. A.; Wortman J.; Dyer P. S.; Grigoriev I. V. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhady A.; Giné A.; Topalovic O.; Jacquiod S.; Sørensen S. J.; Sorribas F. J.; Heuer H. Microbiomes associated with infective stages of root-knot and lesion nematodes in soil. PLoS One 2017, 12, e0177145 10.1371/journal.pone.0177145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klich M. A. Biogeography of Aspergillus species in soil and litter. Mycologia 2002, 94, 21–27. 10.2307/3761842. [DOI] [PubMed] [Google Scholar]
- Grishkan I. Thermotolerant mycobiota of Israeli soils. J. Basic Microbiol. 2018, 58, 30–40. 10.1002/jobm.201700517. [DOI] [PubMed] [Google Scholar]
- Cohen B. L. Regulation of intracellular and extracellular neutral and alkaline proteases in Aspergillus nidulans. Microbiology 1973, 79, 311–320. 10.1099/00221287-79-2-311. [DOI] [PubMed] [Google Scholar]
- Pateman J. A.; Kinghorn J. R.; Dunn E.; Forbes E. Ammonium regulation in Aspergillus nidulans. J. Bacteriol. 1973, 1, 674–675. 10.1042/bst0010674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M. E.; Flynn P. K.; vanKuyk P. A.; Cheetham B. F. Mutations affecting extracellular protease production in the filamentous fungus Aspergillus nidulans. MGG, Mol. Gen. Genet. 1996, 250, 715–724. 10.1007/s004380050125. [DOI] [PubMed] [Google Scholar]
- Katz M. E.; Gray K.-A.; Cheetham B. F. The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet. Biol. 2006, 43, 190–199. 10.1016/j.fgb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Kito M.; Takimoto R.; Yoshida T.; Nagasawa T. Purification and characterization of an epsilon-poly-L-lysine-degrading enzyme from an epsilon-poly-L-lysine-producing strain of Streptomyces albulus. Arch. Microbiol. 2002, 178, 325–330. 10.1007/s00203-002-0459-6. [DOI] [PubMed] [Google Scholar]
- Horn B. W. Relationship between soil densities of Aspergillus species and colonization of wounded peanut seeds. Can. J. Microbiol. 2006, 52, 951–960. 10.1139/w06-050. [DOI] [PubMed] [Google Scholar]
- Zablotowicz R. M.; Abbas H. K.; Locke M. A. Population ecology of Aspergillus flavus associated with Mississippi Delta soils. Food Addit. Contam. 2007, 24, 1102–1108. 10.1080/02652030701546198. [DOI] [PubMed] [Google Scholar]
- Hynes M. J.; Pateman J. A. The use of amides as nitrogen sources by Aspergillus nidulans. J. Gen. Microbiol. 1970, 63, 317–324. 10.1099/00221287-63-3-317. [DOI] [PubMed] [Google Scholar]
- Todd R. B.; Davis M. A.; Hynes M. J. Genetic manipulation of Aspergillus nidulans: meiotic progeny for genetic analysis and strain construction. Nat. Protoc. 2007, 2, 811–821. 10.1038/nprot.2007.112. [DOI] [PubMed] [Google Scholar]
- Grünbacher A.; Throm T.; Seidel C.; Gutt B.; Röhrig J.; Strunk T.; Vincze P.; Walheim S.; Schimmel T.; Wenzel W.; Fischer R. Six Hydrophobins Are Involved in Hydrophobin Rodlet Formation in Aspergillus nidulans and Contribute to Hydrophobicity of the Spore Surface. PLoS One 2014, 9, e94546 10.1371/journal.pone.0094546. [DOI] [PMC free article] [PubMed] [Google Scholar]