Abstract
Chronic wasting disease (CWD) is a highly infectious prion disease of cervids. Accumulation of prions, the disease-specific structural conformers of the cellular prion protein (PrPC), in the central nervous system, is the key pathological event of the disorder. The analysis of cervid PrPC sequences revealed the existence of polymorphism at position 226, in which deer PrP contains glutamine (Q), whereas elk PrP contains glutamate (E). The effects of this polymorphism on CWD are still unknown. We determined the high-resolution nuclear magnetic resonance structure of the mule deer prion protein that was compared to previously published PrP structures of elk and white-tailed deer. We found that the polymorphism Q226E could influence the long-range intramolecular interactions and packing of the β2−α2 loop and the C-terminus of the α3 helix of cervid PrP structures. This solvent-accessible epitope is believed to be involved in prion conversion. Additional differences were observed at the beginning of the well-defined C-terminus domain, in the α2−α3 region, and in its interactions with the α1 helix. Here, we highlight the importance of the PrP structure in prion susceptibility and how single amino acid differences might influence the overall protein folding.
Introduction
Chronic wasting disease (CWD) is an infectious prion disease of free-ranging cervids. It has been reported in both captive and wild cervid species, including elk (Cervus canadensis), mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), red deer (Cervus elaphus), reindeer (Rangifer tarandus), and moose (Alces alces).1−7 The disease has now been reported in 26 states of the United States, three provinces of Canada, South Korea, Norway, Finland, and Sweden.7−12
Prion diseases, also known as transmissible spongiform encephalopathies (TSEs), are rapid, progressive, and devastating neurodegenerative disorders, caused by misfolding events of the main α-helical cellular prion protein (PrPC) to a β-sheet-enriched, partially protease-resistant, and infectious isoform (PrPSc or prion).13,14 Although there is a lack of high-resolution three-dimensional (3D) structural data for PrPSc (mostly because of its insolubility and propensity to aggregate),15 the PrPC structure has been solved by solution-state nuclear magnetic resonance (NMR) and crystallographic methods. PrPC consists of a highly flexible N-terminal segment (residues 23–124) and a folded C-terminus domain (residues 125–231).15,16 The C-terminus domain contains three α-helices, spanning residues 144–154 (α1), 173–194 (α2), and 200–228 (α3), and two short antiparallel β-strands comprising residues 128–131 (β1) and 161–164 (β2). The PrP expressed by mammalian species exhibits a similar fold, with the local sequence and structure variations most prominently localized at the interface of the β2−α2 loop and in the C-terminus part of the α3 helix.17
Among the mammalian prion diseases, CWD is the most infectious form. Free-ranging cervids are at the highest risk of exposure to CWD prions through direct horizontal transmission via infectious agents such as saliva, urine, and feces or through an indirect transmission occurring by environmental exposure to contaminated and infectious material.18,19 The ability of PrPSc to selectively infect some mammalian species rather than others is known as species barriers.20,21 The primary structural identity between PrPC and PrPSc facilitates prion transmission, thus influencing resistance or susceptibility to prion conversion.22,23 Naturally occurring PrP polymorphisms that alter prion disease susceptibility have been documented in many species.24 In humans, the polymorphic residue at codon 129 (M129V) influences the susceptibility to prion diseases,25 where the presence of valine induces the formation of unstable intermolecular β-sheets, conflicting with the spatially adjacent residues.26,27
Polymorphisms and few allelic variations in the well-conserved Prnp gene within the family Cervidae may influence the different susceptibility of CWD progression and PrPSc infection.28,29 Polymorphisms M132L and S225F in elk and mule deer are related to increased resistance to CWD.28,30,31 Additionally, a single difference in primary structure exists between elk and deer PrP; elk PrP contains glutamic acid (E) at position 226, whereas deer PrP contains glutamine (Q) at this position28,32 (Figure 1). Polymorphism Q226E is related to the identification of biologically distinct prion strains on the basis of different disease progressions in deer and elk.33,34 Recently, it was shown that amino acid variation at residue 226 of deer and elk PrP controls the disease onset and conformational features of the resulting prions, thus confirming the presence of different cervid strains.35 Moreover, replacement of the coding sequence of mouse PrP with the deer or elk sequence renders the mice highly susceptible to CWD prions. Therefore, the analysis of structural features of PrP is of outstanding importance for a better understanding of the pathogenesis and transmission of TSEs.
Figure 1.
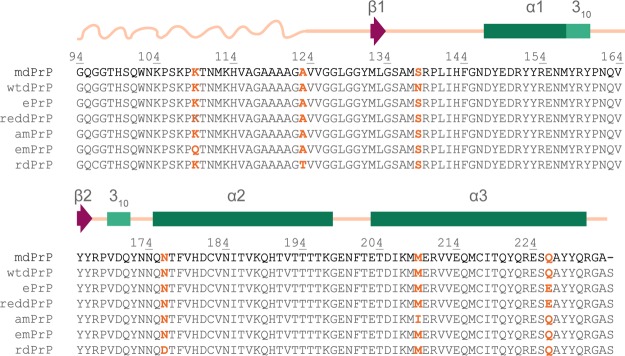
Sequence alignment of PrPs of cervid subspecies with confirmed CWD. Amino acid variants are marked with orange color. Residue numbering is based on the mdPrP amino acid sequence. Secondary structural elements are summarized based on the mdPrP structural model presented in this article, with the α-helices of mdPrP denoted by green rectangles, 310-helices by light green rectangles, β-strands by magenta arrows, flexible N-terminal tail by a curved line, and linkers between the secondary structure elements by straight lines, both lines colored champagne pink.
In the current study, we have determined a high-resolution structure of the truncated recombinant mule deer PrP (from residues 94 to 233, hereafter indicated as mdPrP) with the use of NMR spectroscopy. A comparison to previously determined PrP structures from the white-tailed deer and Rocky Mountain elk36,37 provides insights that may contribute to our understanding of how the single polymorphism Q226E between deer and elk can alter the structure and help to explain the substantial differences in biochemical properties, pathogenesis, and formation of different strains of CWD prions among cervids.38 We hypothesized that the presence of polymorphism Q226E, as the most critical for CWD among the six identified differences in amino acid sequences, could influence the long-range intramolecular interactions including the packing of the β2−α2 loop and the C-terminus of the α3 helix. This solvent-accessible epitope has been studied greatly in view of its role in prion conversion.39,40 Additionally, the changes from the neutral to negatively charged side chain at position 226 will influence the electrostatic surface potential in this region, which is of great relevance for the intermolecular interactions between PrPC and PrPSc among cervids.
Results and Discussion
Amino Acid Alignment and mdPrP Construct
The amino acid sequences of PrPs from various cervid subspecies related to CWD are highly evolutionary-conserved. The alignment of amino acid sequences of mdPrP, white-tailed deer (wtdPrP), elk (ePrP), red deer PrP (reddPrP), American moose PrP (amPrP), Eurasian moose PrP (emPrP), and reindeer PrP (rdPrP) showed differences in the amino acid residues at positions 109, 123, 138, 176, 209, and 226 (Figure 1; numbering is based on the amino acid sequence of the mdPrP construct used herein for structure determination). A simple perusal of the differences shows that the three of them are positioned within the well-defined secondary structural elements. Truncated recombinant mdPrP from residues 94 to 233 with serine at position 138 and glutamine at position 226 was used for structural characterization and comparison with previously resolved wtdPrP and ePrP structures.
Resonance Assignment and Structure Calculation
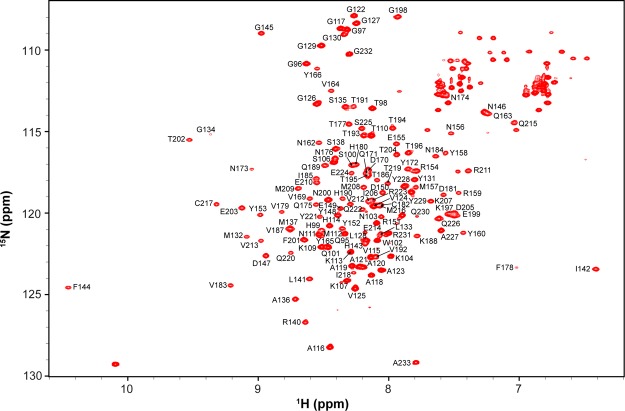
The 15N-heteronuclear single quantum coherence (HSQC) spectrum of 13C, 15N doubly labeled mdPrP presents a favorable dispersion of cross-peaks, indicating a high potential for in-depth structural determination (Figure 2). Standard two-dimensional and 3D NMR experiments were used for the assignment of backbone and side-chain resonances of mdPrP. In short, the sequence-specific assignment of the backbone 1H, 15N, 13Cα, 13Cβ, and 13CO resonances for mdPrP was obtained using the 15N-HSQC spectrum and triple-resonance HNCO, HN(CO)CA, HNCA, CBCA(CO)NH, and HNCACB experiments.41 The 1H and 13C resonances of aliphatic and aromatic side chains were assigned using 13C-HSQC in combination with HAHB(CO)NH, CC(CO)NH, (H)CCH- total correlation spectroscopy (TOCSY), and 13C-edited nuclear Overhauser enhancement spectroscopy (NOESY)-HSQC experiments.42 NOE contacts were determined in 3D 15N and 13C-edited NOESY-HSQC experiments. The overall completeness of chemical shift assignment was 99.1%. Noteworthily, the side-chain resonances including CHε of His99, His114, and Tyr152 and CHζ of Phe178 and Phe201 could not be unambiguously assigned; however, this agrees very well with the final structure and properties of the studied protein. We have considered the use of residual dipolar couplings for structure improvement, especially interhelical orientations. However, the use of aligning media was showed to induce sample precipitation of PrPs.
Figure 2.
15N-HSQC spectrum of mdPrP with the amino acid assignment. Cross-peaks of the side chains of Asn, Gln, and Trp are not marked.
The high-resolution structure of mdPrP was calculated using 545 intraresidual, 618 sequential, 482 medium-range, and 559 long-range distance restraints complemented with 168 backbone torsion angle restraints (Table 1). The calculated structure of mdPrP (PDB ID: 6FNV) is composed of two distinct domains. The highly disordered N-terminal domain consists of residues from Gly94 to Gly122, whereas the well-defined C-terminus domain is composed of residues from Ala123 to Ala233 and exhibits a backbone root-mean-square deviation (rmsd) of 0.42 Å (Figure 3A and Table 1).
Table 1. NMR Restraints and Structural Statistics for an Ensemble of 20 Lowest Energy Structures of mdPrP.
| NOE upper distance limitsa | |
|---|---|
| total | 2204 |
| intraresidue (|i – j| = 0) | 545 |
| sequential (|i – j| = 1) | 618 |
| medium-range (1 < |i – j| < 5) | 482 |
| long-range (|i – j| ≥ 5) | 559 |
| torsion angle restraintsa | |
|---|---|
| backbone (φ/ψ) | 168 |
| rmsd to the mean coordinates (Å) | |
|---|---|
| ordered backbone atoms (123–231) | 0.42 ± 0.08 |
| ordered heavy atoms (123–231) | 0.78 ± 0.09 |
| Ramachandran plot (123–231)b | |
|---|---|
| residues in most favored regions (%) | 95.0 |
| residues in additional allowed regions (%) | 5.0 |
| structure Z scoresb | |
|---|---|
| first generation packing quality | 0.763 ± 0.513 |
| second generation packing quality | 5.102 ± 1.464 |
| Ramachandran plot appearance | –0.453 ± 0.282 |
| chi-1/chi-2 rotamer normality | –3.717 ± 0.483 |
| backbone conformation | –0.541 ± 0.234 |
| rms Z scoresb | |
|---|---|
| bond lengths | 1.143 ± 0.003 |
| bond angles | 0.465 ± 0.011 |
| omega angle restraints | 0.481 ± 0.032 |
| side-chain planarity | 0.358 ± 0.029 |
| improper dihedral distribution | 0.571 ± 0.017 |
| inside/outside distribution | 1.030 ± 0.011 |
Figure 3.
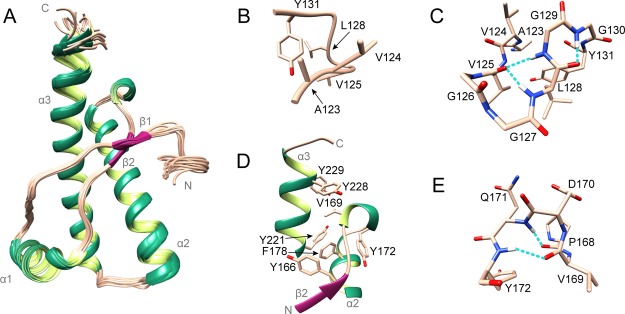
Structure of mdPrP. (A) Ensemble of 20 lowest energy structures of mdPrP (residues form Ala123 to Ala233). α-Helices and 310-helix are colored green, β-sheets are colored magenta, and loops are colored champagne pink. (B) Well-defined region between residues Ala123 and Tyr131. (C) Residues from Ala123 to Tyr131 involved in the formation of α-helical turn (Val125–Leu128) and γ-turn (Leu128–Gly130). (D) Hydrophobic pocket in the proximity of the β2−α2 loop and the C-terminus of the α3 helix. (E) 310-Helix from residues Pro168 to Tyr172 inside the β2−α2 loop. Residues are presented as sticks in champagne pink and the hydrogen bonds in panels (C,E) are shown as dashed lines in cyan.
The C-terminus domain of mdPrP is characterized by a compact set of three α-helices and a short antiparallel β-sheet. The α1 helix is composed of residues from Asp147 to Asn156 and is followed by the 310-helix turn from Met157 to Arg159. The geometry of α1 helix is classified as kinked according to the HELANAL web server.46,47 The α2 and α3 helices are longer than the α1 helix and are composed of residues from Gln175 to Lys197 and from Glu203 to Gln230, respectively. The geometries of α2 and α3 helices are linear and of curved type, respectively. The helices α2 and α3 form a twisted V-shaped skeleton that serves as a platform for anchoring the α1 helix and β-sheet. The antiparallel β-sheet is formed at the beginning of the C-terminus domain and consists of two β strands, β1 and β2, that are composed of residues from Met132 to Leu133 and from Tyr165 to Tyr166, respectively. The structure of mdPrP is stabilized by a disulfide bond between Cys182 and Cys217 that is located in the middle of α2 and α3 helices.
Unique Structural Features of the mdPrP Protein
We observed the structuring of the region at the beginning of the C-terminus domain of mdPrP. This region consists of nine residues from Ala123 to Tyr131 and adopts a well-defined structure with the backbone rmsd of 0.22 Å (Figure 3B). It is characterized by an α-helical turn and a γ-turn that are stabilized by three hydrogen bonds (Figure 3C). The carbonyl group of Val125 is involved in a bifurcated hydrogen bond with the amide protons of Leu128 and Gly129 in the α-helical turn. In addition, a hydrogen bond is formed between the carbonyl group of Leu128 and the amide proton of Gly130 in the γ-turn. An hydrophobic pocket in this region is formed by Val125, Leu128, and Tyr131 (Figure 3B,C).
Additionally, the β2−α2 loop of the mdPrP structure is well defined with one turn of 310-helix from the residues Val169 to Gln171 (Figure 3D) held together by the Gln171HN–Pro168O and Tyr172HN–Val169O hydrogen bonds (Figure 3E). The β2−α2 loop is further stabilized by hydrophobic and aromatic interactions with the nearby amino acid residues at the C-terminus of the α3 helix. The hydrophobic pocket defined by the β2−α2 loop and the C-terminus of the α3 helix is composed of residues Tyr166, Val169, Tyr172, Phe178, Tyr221, and Tyr228 (Figure 3D).
The above structuring of the region before the well-defined C-terminus domain and the β2−α2 loop is supported further with the study of backbone dynamics. We analyzed 118 resolved amide resonances of mdPrP on a fast (picoseconds to nanoseconds) timescale with the use of 15N relaxation time measurements at two magnetic fields (14.1 and 18.8 T). The resulting 15N longitudinal (R1), transverse (R2), and rotating frame (R1ρ) relaxation rates combined with {1H}–15N heteronuclear NOE (hNOE) conform to the flexible N-terminal tail and a well-structured C-terminus domain of the mdPrP structure (Figure 4).
Figure 4.
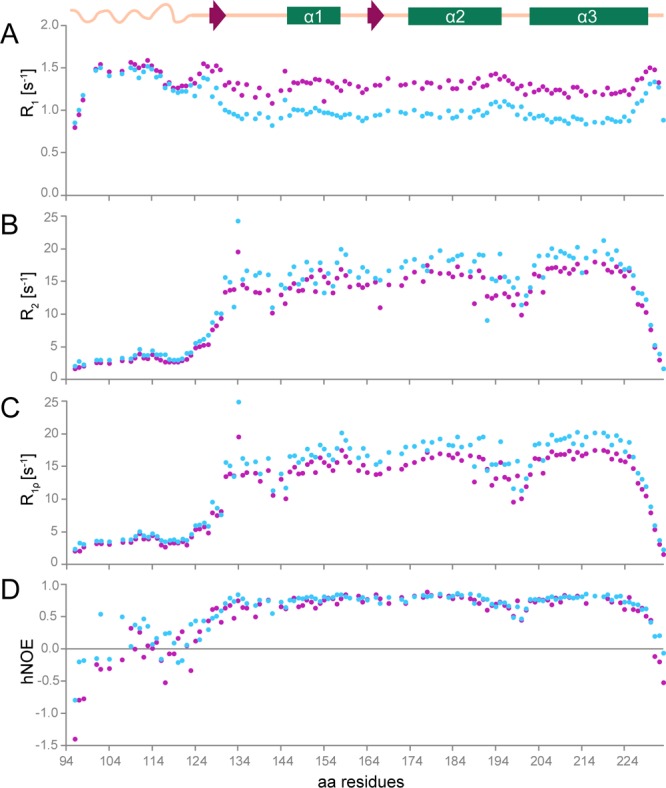
15N amide backbone relaxation rates and hNOE of mdPrP. (A) 15N longitudinal (R1 = 1/T1), (B) transverse (R2 = 1/T2), (C) spin–lattice relaxation rates in the rotation frame (R1ρ = 1/T1ρ), and (D) hNOE at 298 K at a magnetic field of 14.1 (magenta) and 18.8 T (blue). A schematic presentation of the secondary structure elements of mdPrP is at the top of the figure. For clarity, error bars are not shown here as they are within the size of the data points in the above graphics but are presented in the Supporting Information in Figure S1.
Furthermore, 15N relaxation data indicate structuring for residues from Ala123 to Tyr131 at the beginning of the C-terminus domain and for the β2−α2 loop. On the other hand, 15N relaxation data show an increased mobility for residues from Lys197 to Phe201 that connect α2 and α3 helices and for residues from Tyr229 to Ala233 at the C-terminus of the mdPrP structure. However, the relative lower values of R2 and R1ρ relaxation rates for residues around Ile142 and Gln189 are not indicative of a well-defined secondary structure and suggest more complex dynamics coupled to their intricate tertiary interaction. Few amino acid residues could not be analyzed because of the cross-peak overlap (for details, see Methods).
Comparison of Structures of mdPrP and Other Cervids
Cervid prion proteins exhibit a well-conserved amino acid sequence, which may suggest similarity of their 3D structures. We compared our mdPrP structure with the structures of previously determined PrPs from white-tailed deer and Rocky mountain elk and observed several differences. For easier comparison of cervids’ PrP structures, we unified the residue numbering based on the mdPrP amino acid sequence. Here, we have to mention that the wtdPrP structure was determined in complex with an antibody fragment POM1 at pH 6.8; however, the PDB entry for wtdPrP includes residues from 128 to 228 (PDB ID: 4YXH).36 The ePrP structure consists of residues from 124 to 234 (PDB ID: 1XYW) and was determined at pH 4.5.37 We used pH of 5.5 for structure determination because the lower pH prevents aggregation and enables longevity of the prion protein samples that is necessary for structure determination by NMR spectroscopy. Additionally, it has been suggested that misfolding of PrPs in prion disease occurs in endosomes that exhibit a low pH (pH ≈ 5).48 In general, the fold of mdPrP is grossly similar to wtdPrP and ePrP structures (Figure 5A), even though the structures were determined under different sample conditions.
Figure 5.
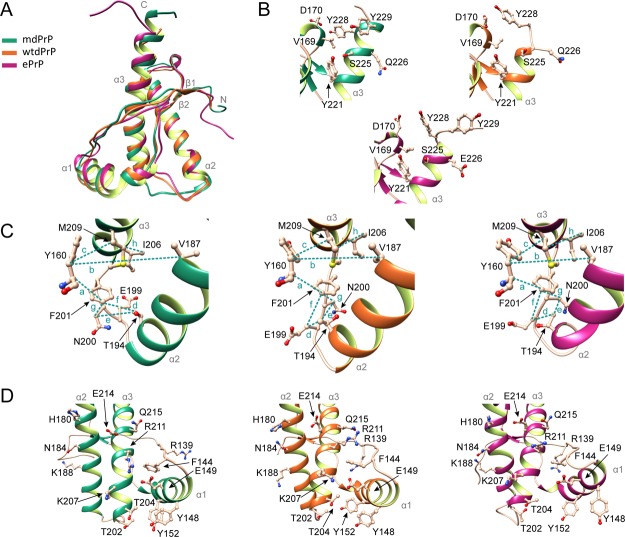
Comparison of mdPrP, wtdPrP, and ePrP structures. (A) Superposition of well-defined C-terminus domains from amino acids Ala123–Ala233 of mdPrP (green), wtdPrP (orange), and ePrP (magenta). The selected residues are presented as ball-and-stick and colored in champagne pink with marked heteroatoms. (B) Structural diversity at the end of the α3 helix and the β2−α2 loop. (C) Spatial orientation of residues in the proximity of the α2−α3 loop with marked distances. Selected distances among residues are indicated with dashed lines and small letters (see Table 2 for distance information). (D) Structural differences in orientations at the α1 helix with respect to the α2−α3 V-shaped skeleton.
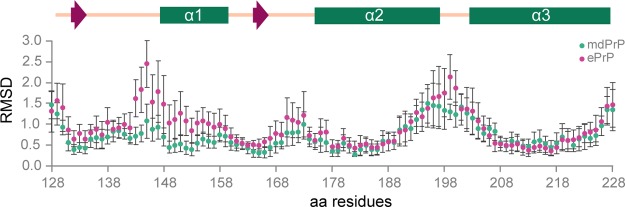
We compared the chemical shifts (δ) of amide proton (HN), Cα, and Cβ atoms of amino acids from 124 to 233 between the mdPrP and ePrP structures determined by NMR spectroscopy (Figure S2). The chemical shifts of HN, Cα, and Cβ atoms of mdPrP and ePrP showed good agreement along the sequence. Slight, if not negligible, differences have been observed for δ(HN) and δ(Cα) for the amino acid residues in the α2−α3 loop, which could indicate different long-range interactions among amino acids in this region. The calculated rmsd for the protein backbone of the three compared structures (residues 128–228) is 1.2 Å. The local backbone rmsd values per residue are in good agreement with the observed differences among the examined structures (Table S1, Figures 5 and 6). The main differences in backbone rmsd values between the mdPrP and ePrP structures have been detected at the N-terminal of the α1 helix and at the α2−α3 loop (Figure 6).
Figure 6.
Local rmsd values for backbone atoms per residue (from 128 to 228) of mdPrP (green) and ePrP (magenta) with respect to the wtdPrP structure that was determined by X-ray. Standard deviations are reported for the ensemble of 20 lowest energy structures of mdPrP and ePrP.
However, despite a very high level of numerical similarity, structural differences are observed at the beginning of the C-terminus domains, β2−α2 loops and their interactions with α3 helices, at the beginning of α1 helices and their interactions with α2 and α3 helices, and α2−α3 loops (Figures 5 and S3).
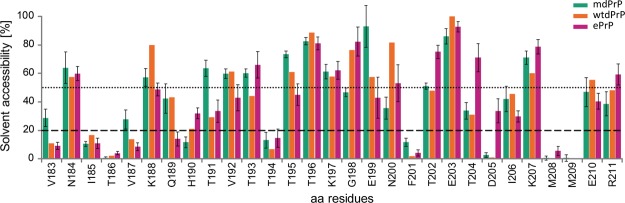
Residues from Val124 to Gly130 in ePrP exhibit an extended backbone conformation. Similarly, the residues from Leu128 to Gly130 in the wtdPrP structure have no observed structuring. Both regions of ePrP and wtdPrP structures exhibit no hydrogen bonds in contrast to mdPrP, where we observed three hydrogen bonds (Figures 3C, 5A and S3). No hydrophobic pocket was observed in this region of ePrP or wtdPrP structures, whereas mdPrP exhibits a well-defined hydrophobic pocket composed by the residues Val125, Leu128, Tyr131, and Ile185. The backbone structures of the three proteins exhibit similar conformations after the residue Gly130. Interestingly, within the examined structures, Tyr131 exhibits a diverse side-chain orientation and distinct stacking with Tyr166 and Ile185. The relative position of Tyr131 side chain and its interactions with the residues in proximity might have an impact on the formation of the α-helical turn and γ-turn in mdPrP in contrast to wtdPrP and ePrP. Solvent accessibility analysis with the GETAREA program49,50 showed that the Tyr131 residue is protected from exchange with solvents in all three structures (Figure S4). However, the distance between Tyr131 and Ile185 in the α2 helix is 2 times longer in mdPrP and ePrP structures in comparison to the wtdPrP structure (Table S2). In contrast, the distance between Leu128 and Ile185 is shorter in the mdPrP structure with respect to the distances in ePrP and wtdPrP structures (Table S2). These observations indicate differences in interactions between the β1−α1−β2 and α2−α3 subdomains of the compared structures. It was previously proposed that different side-chain orientations of Tyr131 play an important role in the interactions between these subdomains and furthermore could also affect the flexibility of the β2−α2 loop region.36,55 In this way, the region from the residues Ala123 to Tyr131 could additionally stabilize the mdPrP structure through its interactions with the antiparallel β-sheet and α2 helix and in this way prevent PrPC-to-PrPSc conversion.
The β2−α2 loop of mdPrP comprising residues 168–178 is well-defined, composed of 310-helix, and stabilized with aromatic and hydrophobic interactions with the nearby residues. The backbone orientations of β2−α2 loops of the three examined structures are similar, whereas small differences are notable in the side-chain orientations of Val169 and Asp170 (Figure 5B). Moreover, different orientations of glutamine and asparagine side chains in the β2−α2 loops (residues Gln171, Asn173, Asn174, and Asn176) are observed and might be related to the long-range interactions and orientations of the aromatic moiety of Tyr131 in the three compared structures (Figure S3). The structural features of β2−α2 loop have been extensively discussed in the literature.22,56−60 The presence of Asn/Gln residues in the β2−α2 loop can be a strong determinant for prion conversion that overrides the differences in the sequence and has influence on the appearance of prions according to the zipper model between the cervid and human PrP.22 Moreover, insertion of additional Gln residues into the β2−α2 loop of mouse PrP promotes prion protein conversion,61−63 whereas several substitutions (at positions 169, 171, 173, and 177; residues numbering based on the mdPrP sequence) in the β2−α2 loop of PrPC are believed to prevent the spontaneous prion formation by influencing the structural stability of the β2−α2 loop.56−59,64
Additionally, the structures and interactions of side chains in the β2−α2 loops are influenced by the orientations of side chains in the C-terminus of the α3 helix including the residue at position 226. The α3 helix of wtdPrP is shorter and ends with Ser225, possibly because of the shorter amino acid sequence36 with respect to the mdPrP structure. The C-terminus of ePrP protein is unstructured after Tyr228 (Figure 5B). Tyr228 and Tyr229 have different side-chain orientations in mdPrP in comparison to the ePrP structure, as a result of their distinct relative orientation, that lead to hydrophobic interactions with the residues Val169, Asp170, and Ser225 and additionally stabilize the end of the C-terminus part in mdPrP (Figure 5B and Table S2). Tyr228 of the wtdPrP structure is involved in stacking interactions with Asp170 in the β2−α2 loop. However, the interactions between the residues at the end of the α3 helix and Gln226 are not observed in wtdPrP, resulting in higher solvent accessibility of Ser225 and Gln226 in comparison to mdPrP and ePrP (Figures 5B and S3). Importantly, beside the polymorphism Q226E, mule deer exhibits serine-to-asparagine polymorphism at position 138, which is processed as a pseudogene,69,70 and serine-to-phenylalanine polymorphism at codon 225.30 Allele Phe225 in mule deer could contribute to CWD resistance in view of the reported prolonged incubation period with respect to the Ser225 mule deer homozygote.30 Interestingly, it has been shown that polymorphisms at residues 225 and 226 affect the interactions between the β2−α2 loop and α3 helix and therefore prion propagation within deer and elk.40,71 Our results showed that Ser225 is protected from solvents as it is involved in the interaction with Tyr228 in mdPrP. Additionally, Tyr228 in mdPrP is protected from solvents by the stacking interaction with Val169. These data contribute to the understanding at the molecular level and are in agreement with the structural and molecular dynamics studies of inter- and intraspecies PrP transmission related to cervids that pointed out a critical role of residues 225 and 226 in PrPC-to-PrPSc conversion and strain propagation.40
In the three structures, α2 and α3 helices form a V-shaped skeleton that slightly differs in the spatial orientation of the helices. The interhelical angle between the α2 and α3 helices of mdPrP is 44.5°, whereas its value in wtdPrP and ePrP is 49.8 and 52.2°, respectively. The hydrophobic and aromatic residues of α2 and α3 helices have preserved architectures that are stabilized by a disulfide bond in the three structures. However, significant differences are observed for the side-chain orientations of His190 and Thr194 in the α2 helix and Ile206, Met209, and Glu210 in the α3 helix (Figures 5C and S1). These residues are spatially close to the loop that connects α2 and α3 helices. Surprisingly, the α2−α3 loop of mdPrP exhibits a unique backbone conformation with different orientations of the side chains of Glu199, Asn200, and Phe201 with respect to wtdPrP and ePrP. However, the hydrophobic interactions of Phe201 and Tyr160 are preserved in the three structures. Major differences are observed for distances Tyr160Cα–Val201Cβ and Glu199Cβ–Phe201Cζ that are up to 2 Å longer in mdPrP with respect to wtdPrP and ePrP (Table 2). The opposite is observed for distances Thr194Cγ2–Asn200Cβ, Asn200Cβ–Phe201Cζ, and Ile206Cγ2–Met209Cγ that are shorter in mdPrP with respect to the other two cervid structures (Figure 5C and Table 2). The residue Glu199 is more exposed to the solvents, whereas residue Asn200 is less solvent-exposed in mdPrP in comparison to wtdPrP and ePrP (Figure 7). Different side-chain orientations in the α2−α3 loop could influence the interactions of residues in helices that are spatially close to this region. In early events of oligomerization, it is believed that the α1 helix moves away from the α2−α3 V-shaped skeleton. This is responsible for the increased local structural dynamics that is reflected in greater exposure of the amide hydrogen atoms in the α1 helix of mouse PrP.65−68 Importantly, the stabilization of interactions or covalent linkage by a disulfide bond between the subdomains β1−α1−β2 and α2−α3 is supposed to prevent the oligomerization.65
Table 2. Distances between C Atoms of Selected Amino Acid Residues in Proximity of the α2−α3 Loop in mdPrP, wtdPrP, and ePrP Structuresa.
| markb | distance | mdPrP (Å) | wtdPrP (Å) | ePrP (Å) |
|---|---|---|---|---|
| A | Tyr160Cα–Phe201Cζ | 6.5 ± 0.3 | 5.3 | 5.5 ± 0.4 |
| B | Tyr160Cβ–Val187Cβ | 11.5 ± 0.4 | 9.1 | 8.9 ± 0.3 |
| C | Tyr160Cβ–Met209Cβ | 5.2 ± 0.1 | 4.7 | 5.1 ± 0.3 |
| D | Thr194Cγ2–Glu199Cβ | 4.8 ± 0.4 | 5.7 | 4.2 ± 0.6 |
| E | Thr194Cγ2–Asn200Cβ | 5.7 ± 0.5 | 8.3 | 6.4 ± 1.3 |
| F | Glu199Cβ–Phe201Cζ | 7.3 ± 0.5 | 6.8 | 6.5 ± 0.7 |
| G | Asn200Cβ–Phe201Cζ | 4.0 ± 0.2 | 7.5 | 7.5 ± 0.4 |
| H | Ile206Cγ2–Met209Cγ | 4.8 ± 0.1 | 6.5 | 6.8 ± 0.2 |
Reported distances are average values obtained from the coordinates of the structural ensemble for mdPrP (PDB id 6FNV) and ePrP (PDB id 1XYW) that were determined by NMR and for wtdPrP (PDB id 4YXH) determined by X-ray crystallography. Standard deviations are reported for the ensemble of 20 lowest energy structures for mdPrP and ePrP.
Figure 7.
Solvent accessibility of selected residues that belong to the α2 and α3 helices. Hatched and dotted lines at 20 and 50% indicate the limits of amino acid residue accessibility to solvents (>50%) or burial in solvent-inaccessible regions (<20%). Standard deviations are reported for the ensemble of 20 lowest energy structures of mdPrP and ePrP that have been determined by NMR.
Structure variations were also observed in the orientations of the three helices and at the end of the α1 helix and its interactions with the α2−α3 loop. The aromatic interactions between Tyr148 and Tyr152 differ among the three structures (Figure 5D). Furthermore, 2 times shorter distances of Tyr148 with Thr202 and Thr204 are observed in mdPrP and ePrP structures compared to wtdPrP (Table S2). A similar trend in distances was observed between Tyr152 and Thr202. In contrast, the distances between Tyr152 and Thr204 are similar for all the three structures. Thr202 and Thr204 are more solvent-exposed in the ePrP structure with respect to mdPrP and wtdPrP structures (Figure 7). We observed a closer anchoring of the α1 helix to α2 and α3 helices in the mdPrP structure with respect to ePrP and wtdPrP.
Effect of Polymorphism Q226E on Electrostatic Surface Potential
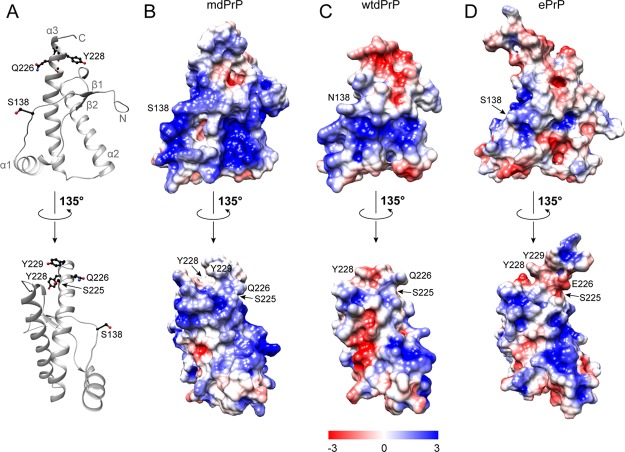
MdPrP, wtdPrP, and ePrP are known for their polymorphisms at positions 138 and 226. S138N and Q226E polymorphisms have a major impact on the electrostatic surface potential of the examined structures (Figure 8).
Figure 8.
Electrostatic surface potential of the three cervid PrPs. (A) Ribbon presentation of the mdPrP backbone orientation used in panels (B–D). Residues Ser138, Ser225, Gln226, Tyr228, and Tyr229 are presented as ball-and-stick and colored black. Electrostatic surface potentials of (B) mdPrP, (C) wtdPrP, and (D) ePrP. Regions of positive and negative charges are depicted from blue to red according to the presented charge legend. Orientation of structures is preserved in all panels. The lower set of structures is rotated by 135°.
Our results show that variations in the electrostatic surface potential among the three proteins are mostly clustered at the β1−α1 loop, at the beginning of the α2 helix, at the V-shaped skeleton where the antiparallel β sheet is in proximity to the α2 and α3 helices, and at the C-terminus of the α3 helix (Figure 8).
In contrast to the wtdPrP and ePrP structures, a large contiguous area of positive electrostatic potential is observed on the surface of the mdPrP structure. The region around residue 138 is positively charged in mdPrP, whereas the corresponding region in wtdPrP and ePrP proteins is neutral. Additional variations of positive charge in the mdPrP and wtdPrP structures to a predominantly neutral state in the ePrP structure are observed in the middle of α2 and α3 helices. Polymorphism Q226E is reflected in the charge of preferentially positive surface areas in the β2−α2 loop and the C-terminus of the α3-helix in mdPrP and wtdPrP in comparison to the negative electrostatic potential in ePrP. However, the C-terminus of wtdPrP is negatively charged to mdPrP and ePrP. Residues 225 and 226 are located in a distal region of the α3 helix that participates in interactions with the β2−α2 loop to form a solvent-accessible contiguous epitope.72 Our data suggest that different distributions of electrostatic potential between mdPrP and ePrP proteins may facilitate intramolecular interactions between two allelic variants in deer subspecies in case of S225F and Q226E polymorphisms and influence the early stages of prion conversion and neuropathology of CWD among cervids.
Conclusions
The spread of CWD in North America and the most recent cases of CWD-infected cervid subspecies in Europe have raised concerns for public health and perceived risks for possible CWD transmission to humans through the consumption of CWD-infected venison.8,51−53 The possible spontaneous spread of the disease among cervid subspecies in Eurasia represents a global threat.54 Structural studies on mdPrP at the molecular level are important for understanding the still unknown reasons for the appearance of the detected and confirmed cases of CWD in captive mule deer3 and the progressive spread and identification of the disease in other cervids.
Previous findings suggest that the primary structural differences at residue 226 identify biologically distinct prion strains on the basis of different disease progressions in deer and elk33 and have a role in dictating the selection of different CWD prion strains in gene-targeted mice.35 These findings suggest that the observed differences are related to an altered structure of PrPC caused by the Q226E polymorphism, highlighting the importance of amino acid sequence variations affecting the local changes of 3D structures, whereas the globular fold remains similar. A detailed comparative structural analysis of the examined PrP of cervids could provide insights into pathogenesis suggesting that the structures of deer and elk prion proteins may determine prion strain mutation in these cervids.
Our comparative analysis uncovered the structural determinants of mdPrP that are manifested in diverse structural rearrangements and distinct electrostatic surface potentials with respect to the wtdPrP and ePrP structures. The region at the beginning of the C-terminus domain could protect the β-sheet from solvents, force the closer packing of β1−α1−β2 to α2−α3 subdomains and raise the structural stability of mdPrP. These structural features could have a major effect on the prion conversion. In our previous studies, we have found that amino acid substitution at position 226 has dramatic effects on CWD prion replication, pathogenesis, and biochemical properties.73 Tg(DeerPrP) has a longer incubation time compared with Tg(ElkPrP) mice after inoculation with CWD prions. On the other hand, Tg(DeerPrP) mice were susceptible to SSBP/1,74 whereas Tg(ElkPrP) mice were completely resistant.71 In addition, Q226 CWD prions display more resistance to guanidine denaturation than the E226 CWD prions.35 Our current findings suggest that the long-range interactions in the mdPrP protein might stabilize the overall structure, thus impacting the PrPC-to-PrPSc conversion. The distribution of electrostatic surface potential on the mdPrP protein may lead to different intermolecular interactions between PrPC and PrPSc and in this way may represent a step toward understanding the underlining mechanism of CWD prion transmission.
Methods
Plasmid Construction for NMR Sample Preparation
The recombinant dePrP (94–233) was obtained using the QuikChange kit (Stratagene) utilizing primers 5′-CAGAGAGAATCCCAGGCTTATTACCAAAGA-3′ and 5′-TCTTTGGTAATAAGCCTGGGATTCTCTCTG-3′ and ePrP(94–234) as templates. The DNA product was then inserted into pProExHTa (Invitrogen), containing the cleavage site between the His6 tag and the protein fragment. The cloned DNA sequences were verified by sequencing.
Prion Protein Expression and Purification
A freshly transformed overnight culture of E. coli BL21 (DE3) cells (Stratagene) was added at 37 °C to 2 L of the minimal medium plus ampicillin (100 mg/mL). For isotope labeling, 4 g/L [13C6] glucose and 1 g/L [15N] ammonium chloride were added. At 0.8 OD600, expression was induced with isopropyl β-d-galactopyranoside to a final concentration of 0.8 mM. Cells were grown in a Biostat B plus 2 L vessel (Sartorius) and harvested 18 h after inoculation. The bacterial paste was resuspended in 25 mM Tris-HCl, 0.8% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride, pH 8.0, and lysed by a Panda homogenizer. A crude extract was loaded onto a 5 mL HisTrap column (GE Healthcare) equilibrated in a binding buffer [2 M GndHCl, 500 mM NaCl, 20 mM Tris-HCl, and 20 mM imidazole (pH 8.0)] and eluted with 500 mM imidazole. The purified protein was lyophilized and dissolved in 8 M GndHCl. The protein was diluted to a final concentration of 0.5 mg/mL in a tobacco etch virus (TEV) reaction buffer (50 mM Tris base, 1 mM ethylenediaminetetraacetic acid, and 5 mM dithiothreitol), and TEV protease was added to the final concentration of 75 μg/mL. The reaction was incubated at 22 °C overnight. The cleaved sample was loaded onto a 5 mL HisTrap column (GE Healthcare) equilibrated with a binding buffer [500 mM NaCl and 20 mM Tris-HCl (pH 8.0)], and the flow-through containing only the cleaved protein was collected. The purified protein was lyophilized and redissolved in 8 M GndHCl. Refolding was performed by dialysis against a refolding buffer [20 mM sodium acetate and 0.005% NaN3 (pH 5.5)] using a Spectra/Por membrane (molecular weight, 3000). The purified protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis under reducing conditions, western blot, and electrospray mass spectrometry. The purification and expression of TEV protease were obtained as described earlier.75
NMR Spectroscopy and Structure Calculation
All NMR experiments used for structure determination were performed on a 13C, 15N isotopically labeled mdPrP sample on a Varian VNMRS 800 MHz spectrometer equipped with a triple 1H/13C/15N resonance cryogenic probe head operating at 25 K with inverse detection. The sample temperature was calibrated using the methanol-d3 standard sample to ensure consistent sample temperature. The sample temperature for all experiments was 298 K. The sample contained 0.48 mM of mdPrP in a 20 mM sodium phosphate buffer, pH 5.5. NMR experiments for NH and HC detection were performed in 90%/10% H2O/D2O and in a 100% deuterated buffer, respectively. The sequence-specific assignment of the backbone 1H, 15N, 13Cα, 13Cβ, and 13CO resonances for mdPrP was obtained using the 15N-HSQC spectrum and triple-resonance NMR experiments HNCO, HN(CO)CA, HNCA, CBCA(CO)NH, and HNCACB.41 The 1H and 13C resonances of aliphatic and aromatic side chains were assigned using 13C-HSQC in combination with HAHB(CO)NH, CC(CO)NH, (H)CCH-TOCSY, and 13C-edited NOESY-HSQC experiments.42 NOE contacts were determined in 3D 15N and 13C-edited NOESY-HSQC experiments. Structure modeling of mdPrP was performed using the program CYANA 3.1.76 Structure refinement using the explicit solvent model was performed by the YASARA program.77 An ensemble of 20 lowest energy structures of mdPrP was validated by the web server software ICING44 and PSVS.45
Backbone amide relaxation measurements including 15N longitudinal (R1), transverse (R2), rotating frame (R1ρ) relaxation rates, and {1H}–15N heteronuclear NOE were obtained at two different magnetic fields (14.1 and 18.8 T) at 298 K.78 Residues Gln95, Thr98, Ser100, Asn103, Ser106, Met137, His143, Tyr165, Asp170, Gln171, Asn174, Asn176, His180, Cys182, Val187, Met216, and Ile218 could not be analyzed because of the cross-peak overlap.
All recorded spectra were processed with NMRPipe software79 and analyzed with CARA80 and SPARKY software.81 The prediction of backbone dihedral angles was made by the TALOS+ program.82 Alignment was prepared using ClustalO.83 An analysis was performed by the web server GETAREA.49,50 The potentials were calculated at an experimental pH of 5.5 of mdPrP using PDB2PQR server,84 APBS,85 and PROPKA.86,87
Glossary
Abbreviations
- δ
chemical shift
- amPrP
America moose prion protein
- CWD
chronic wasting disease
- ePrP
elk prion protein
- emPrP
Eurasian moose prion protein
- HSQC
heteronuclear single quantum coherence
- hNOE
{1H}–15N heteronuclear nuclear Overhauser enhancement
- mdPrP
mule deer prion protein
- PrP
prion protein
- PrPC
cellular isoform of prion protein
- PrPSc
prion–protease-resistant and infectious isoform
- R1
longitudinal relaxation rate (1/T1)
- R2
transverse relaxation rate (1/T2)
- R1δ
relaxation rate in rotating frame (1/T1δ)
- reddPrP
red deer prion protein
- rdPrP
reindeer prion protein
- TOCSY
total correlation spectroscopy
- TSE
transmissible spongiform encephalopathy
- wtdPrP
white-tailed deer prion protein
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.9b02824.
15N amide backbone relaxation rates and hNOE of mdPrP; chemical shifts of HN, Cα, and Cβ atoms of mdPrP and ePrP structures; structural diversity of mdPrP, wtdPrP, and ePrP; and solvent accessibility of the selected amino acid residues of mdPrP, wtdPrP, and ePrP (PDF)
Author Contributions
U.S. and G.S. contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding was provided by the Slovenian NMR center through the Slovenian Research Agency grant no. P1-0242 and the CERIC-ERIC Consortium for access to experimental facilities. This work was also supported by a fellowship from the HEaD “Higher Education and Development” project (funded by the Friuli Venezia Giulia autonomous Region (Italy) through the Operational Program of the European Social Fund 2014/2020; grant no. FP1619889004) (to G.S.).
The authors declare no competing financial interest.
Notes
The atomic coordinates and structure factors of mdPrP (UniProtKB—P47852, PRIO_ODOHE) have been deposited in the Protein Data Bank (PDB ID: 6FNV) and Biological Magnetic Resonance Bank (BMRB ID: 34236).
Supplementary Material
References
- Belay E. D.; Maddox R. A.; Williams E. S.; Miller M. W.; Gambetti P.; Schonberger L. B. Chronic wasting disease and potential transmission to humans. Emerging Infect. Dis. 2004, 10, 977. 10.3201/eid1006.031082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley N. J.; Hoover E. A. Chronic wasting disease of cervids: current knowledge and future perspectives. Annu. Rev. Anim. Biosci. 2015, 3, 305–325. 10.1146/annurev-animal-022114-111001. [DOI] [PubMed] [Google Scholar]
- Williams E. S.; Young S. Chronic wasting disease of captive mule deer: A spongiform encephalopathy 1. J. Wildl. Dis. 1980, 16, 89–98. 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- Williams E. S.; Young S. Spongiform encephalopathy of rocky mountain elk 1. J. Wildl. Dis. 1982, 18, 465–471. 10.7589/0090-3558-18.4.465. [DOI] [PubMed] [Google Scholar]
- Spraker T. R.; Miller M. W.; Williams E. S.; Getzy D. M.; Adrian W. J.; Schoonveld G. G.; Spowart R. A.; O’Rourke K. I.; Miller J. M.; Merz P. A. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J. Wildl. Dis. 1997, 33, 1–6. 10.7589/0090-3558-33.1.1. [DOI] [PubMed] [Google Scholar]
- Baeten L. A.; Powers B. E.; Jewell J. E.; Spraker T. R.; Miller M. W. A natural case of chronic wasting disease in a free-ranging moose (Alces alces shirasi). J. Wildl. Dis. 2007, 43, 309–314. 10.7589/0090-3558-43.2.309. [DOI] [PubMed] [Google Scholar]
- Benestad S. L.; Mitchell G.; Simmons M.; Ytrehus B.; Vikøren T. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet. Res. 2016, 47, 88. 10.1186/s13567-016-0375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benestad S. L.; Telling G. C. Chronic wasting disease: an evolving prion disease of cervids. Handb. Clin. Neurol. 2018, 153, 135–151. 10.1016/b978-0-444-63945-5.00008-8. [DOI] [PubMed] [Google Scholar]
- Sohn H.-J.; Jae-Hoon K.; Jin-Ju N.; Yi-Seok J.; Young-Hwa J.; Soo-Whan A.; Ok-Kyung K.; Dae-Yong K.; BALACHANDRAN A. A case of chronic wasting disease in an elk imported to Korea from Canada. J. Vet. Med. Sci. 2002, 64, 855–858. 10.1292/jvms.64.855. [DOI] [PubMed] [Google Scholar]
- Stokstad E. Norway seeks to stamp out prion disease. Science 2017, 356, 12. 10.1126/science.356.6333.12. [DOI] [PubMed] [Google Scholar]
- Gale P.; Roberts H.. Update on Chronic Wasting Disease in Europe, 2018.
- Pirisinu L.; Tran L.; Chiappini B.; Vanni I.; Di Bari M. A.; Vaccari G.; Vikøren T.; Madslien K. I.; Våge J.; Spraker T.; Mitchell G.; Balachandran A.; Baron T.; Casalone C.; Rolandsen C. M.; Røed K. H.; Agrimi U.; Nonno R.; Benestad S. L. Novel Type of Chronic Wasting Disease Detected in Moose (Alces alces), Norway. Emerging Infect. Dis. 2018, 24, 2210–2218. 10.3201/eid2412.180702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran M.; Mahmood S. An overview of animal prion diseases. Virol. J. 2011, 8, 493. 10.1186/1743-422x-8-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby D. W.; Prusiner S. B. Prions. Cold Spring Harbor Perspect. Biol. 2011, 3, a006833. 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surewicz W. K.; Apostol M. I.. Prion protein and its conformational conversion: a structural perspective. Prion Proteins; Springer, 2011; pp 135–167. [DOI] [PubMed] [Google Scholar]
- Riek R.; Hornemann S.; Wider G.; Billeter M.; Glockshuber R.; Wüthrich K. NMR structure of the mouse prion protein domain PrP (121–231). Nature 1996, 382, 180. 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- Biljan I.; Ilc G.; Plavec J.. Analysis of Prion Protein Structure Using Nuclear Magnetic Resonance Spectroscopy. Prions Methods in Molecular Biology; Springer, 2017; pp 35–49. [DOI] [PubMed] [Google Scholar]
- Williams E. S. Chronic wasting disease. Vet. Pathol. 2005, 42, 530–549. 10.1354/vp.42-5-530. [DOI] [PubMed] [Google Scholar]
- Miller M. W.; Williams E. S. Horizontal prion transmission in mule deer. Nature 2003, 425, 35–36. 10.1038/425035a. [DOI] [PubMed] [Google Scholar]
- Hill A. F.; Collinge J.. Prion strains and species barriers. Prions; Karger Publishers, 2004; Vol. 11, pp 33–49. [DOI] [PubMed] [Google Scholar]
- Moore R.-A.; Vorberg I.; Priola S.-A.. Species barriers in prion diseases—brief review. Infectious Diseases from Nature: Mechanisms of Viral Emergence and Persistence; Springer, 2005; pp 187–202. [DOI] [PubMed] [Google Scholar]
- Kurt T. D.; Jiang L.; Fernández-Borges N.; Bett C.; Liu J.; Yang T.; Spraker T. R.; Castilla J.; Eisenberg D.; Kong Q.; Sigurdson C. J. Human prion protein sequence elements impede cross-species chronic wasting disease transmission. J. Clin. Invest. 2015, 125, 1485–1496. 10.1172/jci79408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeting B.; Khan M. Q.; Chakrabartty A.; Pai E. F. Structural factors underlying the species barrier and susceptibility to infection in prion disease. Biochem. Cell Biol. 2010, 88, 195–202. 10.1139/o09-172. [DOI] [PubMed] [Google Scholar]
- Goldmann W.; Hunter N.; Smith G.; Foster J.; Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 1994, 75, 989–995. 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- Apetri A. C.; Vanik D. L.; Surewicz W. K. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90-231. Biochemistry 2005, 44, 15880–15888. 10.1021/bi051455+. [DOI] [PubMed] [Google Scholar]
- Palmer M. S.; Dryden A. J.; Hughes J. T.; Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature 1991, 352, 340–342. 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- Fernández-Borges N.; Espinosa J. C.; Marín-Moreno A.; Aguilar-Calvo P.; Asante E. A.; Kitamoto T.; Mohri S.; Andréoletti O.; Torres J. M. Protective Effect of Val129-PrP against Bovine Spongiform Encephalopathy but not Variant Creutzfeldt-Jakob Disease. Emerging Infect. Dis. 2017, 23, 1522–1530. 10.3201/eid2309.161948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. J.; Samuel M. D.; O’Rourke K. I.; Johnson C. J. The role of genetics in chronic wasting disease of North American cervids. Prion 2012, 6, 153–162. 10.4161/pri.19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. C.; Mateus-Pinilla N. E.; Diffendorfer J.; Jewell E.; Ruiz M. O.; Killefer J.; Shelton P.; Beissel T.; Novakofski J. Prion sequence polymorphisms and chronic wasting disease resistance in Illinois white-tailed deer (Odocoileus virginianus). Prion 2008, 2, 28–36. 10.4161/pri.2.1.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J. E.; Conner M. M.; Wolfe L. L.; Miller M. W.; Williams E. S. Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J. Gen. Virol. 2005, 86, 2127–2134. 10.1099/vir.0.81077-0. [DOI] [PubMed] [Google Scholar]
- O’Rourke K. I.; Spraker T. R.; Zhuang D.; Greenlee J. J.; Gidlewski T. E.; Hamir A. N. Elk with a long incubation prion disease phenotype have a unique PrPd profile. NeuroReport 2007, 18, 1935–1938. 10.1097/WNR.0b013e3282f1ca2f. [DOI] [PubMed] [Google Scholar]
- Cervenáková L.; Rohwer R.; Williams E. S.; Brown P.; Gajdusek D. C. High sequence homology of the PrP gene in mule deer and Rocky Mountain elk. Lancet 1997, 350, 219–220. 10.1016/s0140-6736(05)62387-2. [DOI] [PubMed] [Google Scholar]
- Perrott M. R.; Sigurdson C. J.; Mason G. L.; Hoover E. A. Evidence for distinct chronic wasting disease (CWD) strains in experimental CWD in ferrets. J. Gen. Virol. 2012, 93, 212–221. 10.1099/vir.0.035006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. S.; Young S. Neuropathology of chronic wasting disease of mule deer (Odocoileus hemionus) and elk (Cervus elaphus nelsoni). Vet. Pathol. 1993, 30, 36–45. 10.1177/030098589303000105. [DOI] [PubMed] [Google Scholar]
- Bian J.; Christiansen J. R.; Moreno J. A.; Kane S. J.; Khaychuk V.; Gallegos J.; Kim S.; Telling G. C. Primary structural differences at residue 226 of deer and elk PrP dictate selection of distinct CWD prion strains in gene-targeted mice. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 12478. 10.1073/pnas.1903947116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral P. K.; Swayampakula M.; Aguzzi A.; James M. N. G. X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins. J. Struct. Biol. 2015, 192, 37–47. 10.1016/j.jsb.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Gossert A. D.; Bonjour S.; Lysek D. A.; Fiorito F.; Wuthrich K. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 646–650. 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers R. C.; Kang H.-E.; Napier D.; Browning S.; Seward T.; Mathiason C.; Balachandran A.; McKenzie D.; Castilla J.; Soto C.; Jewell J.; Graham C.; Hoover E. A.; Telling G. C. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 2010, 328, 1154–1158. 10.1126/science.1187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B.; Hornemann S.; Damberger F. F.; Wüthrich K. Prion protein NMR structure from tammar wallaby (Macropus eugenii) shows that the β2−α2 loop is modulated by long-range sequence effects. J. Mol. Biol. 2009, 389, 833–845. 10.1016/j.jmb.2009.04.040. [DOI] [PubMed] [Google Scholar]
- Angers R.; Christiansen J.; Nalls A. V.; Kang H.-E.; Hunter N.; Hoover E.; Mathiason C. K.; Sheetz M.; Telling G. C. Structural effects of PrP polymorphisms on intra-and interspecies prion transmission. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 11169–11174. 10.1073/pnas.1404739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L. E.; Ikura M.; Tschudin R.; Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson. 1990, 89, 496–514. 10.1016/0022-2364(90)90333-5. [DOI] [PubMed] [Google Scholar]
- Bax A.; Grzesiek S.. Methodological advances in protein NMR. NMR of Proteins; Springer, 1993; pp 33–52. [Google Scholar]
- Laskowski R. A.; Rullmann J. A. C.; MacArthur M. W.; Kaptein R.; Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. 10.1007/bf00228148. [DOI] [PubMed] [Google Scholar]
- Doreleijers J. F.; Sousa da Silva A. W.; Krieger E.; Nabuurs S. B.; Spronk C. A. E. M.; Stevens T. J.; Vranken W. F.; Vriend G.; Vuister G. W. CING: an integrated residue-based structure validation program suite. J. Biomol. NMR 2012, 54, 267–283. 10.1007/s10858-012-9669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A.; Tejero R.; Montelione G. T. Evaluating protein structures determined by structural genomics consortia. Proteins: Struct., Funct., Bioinf. 2007, 66, 778–795. 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Bansal M. Geometrical and sequence characteristics of α-helices in globular proteins. Biophys. J. 1998, 75, 1935–1944. 10.1016/s0006-3495(98)77634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M.; Kumart S.; Velavan R. HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 2000, 17, 811–819. 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- Van der Kamp M. W.; Daggett V. Influence of pH on the human prion protein: insights into the early steps of misfolding. Biophys. J. 2010, 99, 2289–2298. 10.1016/j.bpj.2010.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraczkiewicz R.; Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998, 19, 319–333. . [DOI] [Google Scholar]
- Sridharan S.; Nicholls A.; Sharp K. A. A rapid method for calculating derivatives of solvent accessible surface areas of molecules. J. Comput. Chem. 1995, 16, 1038–1044. 10.1002/jcc.540160810. [DOI] [Google Scholar]
- Vaske J. J.; Miller C. A.; Ashbrook A. L.; Needham M. D. Proximity to chronic wasting disease, perceived risk, and social trust in the managing agency. Hum. Dimens. Wildl. 2018, 23, 115–128. 10.1080/10871209.2018.1399317. [DOI] [Google Scholar]
- Hannaoui S.; Schatzl H. M.; Gilch S. Chronic wasting disease: Emerging prions and their potential risk. PLoS Pathog. 2017, 13, e1006619 10.1371/journal.ppat.1006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell L.; Greig J.; Mascarenhas M.; Otten A.; Corrin T.; Hierlihy K. Current evidence on the transmissibility of chronic wasting disease prions to humans—a systematic review. Transboundary Emerging Dis. 2018, 65, 37–49. 10.1111/tbed.12612. [DOI] [PubMed] [Google Scholar]
- Sutherland W. J.; Butchart S. H. M.; Connor B.; Culshaw C.; Dicks L. V.; Dinsdale J.; Doran H.; Entwistle A. C.; Fleishman E.; Gibbons D. W.; Jiang Z.; Keim B.; Roux X. L.; Lickorish F. A.; Markillie P.; Monk K. A.; Mortimer D.; Pearce-Higgins J. W.; Peck L. S.; Pretty J.; Seymour C. L.; Spalding M. D.; Tonneijck F. H.; Gleave R. A. A 2018 Horizon Scan of Emerging Issues for Global Conservation and Biological Diversity. Trends Ecol. Evol. 2018, 33, 47–58. 10.1016/j.tree.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Paramithiotis E.; Pinard M.; Lawton T.; LaBoissiere S.; Leathers V. L.; Zou W.-Q.; Estey L. A.; Lamontagne J.; Lehto M. T.; Kondejewski L. H.; Francoeur G. P.; Papadopoulos M.; Haghighat A.; Spatz S. J.; Head M.; Will R.; Ironside J.; O’Rourke K.; Tonelli Q.; Ledebur H. C.; Chakrabartty A.; Cashman N. R. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 2003, 9, 893. 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- Bett C.; Fernández-Borges N.; Kurt T. D.; Lucero M.; Nilsson K. P. R.; Castilla J.; Sigurdson C. J. Structure of the β2-α2 loop and interspecies prion transmission. FASEB J. 2012, 26, 2868–2876. 10.1096/fj.11-200923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt T. D.; Bett C.; Fernández-Borges N.; Joshi-Barr S.; Hornemann S.; Rülicke T.; Castilla J.; Wüthrich K.; Aguzzi A.; Sigurdson C. J. Prion transmission prevented by modifying the β2-α2 loop structure of host PrPC. J. Neurosci. 2014, 34, 1022–1027. 10.1523/jneurosci.4636-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson C. J.; Nilsson K. P. R.; Hornemann S.; Manco G.; Fernández-Borges N.; Schwarz P.; Castilla J.; Wüthrich K.; Aguzzi A. A molecular switch controls interspecies prion disease transmission in mice. J. Clin. Invest. 2010, 120, 2590. 10.1172/jci42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt T. D.; Aguilar-Calvo P.; Jiang L.; Rodriguez J. A.; Alderson N.; Eisenberg D. S.; Sigurdson C. J. Asparagine and glutamine ladders promote cross-species prion conversion. J. Biol. Chem. 2017, 292, 19076–19086. 10.1074/jbc.m117.794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldarulo E.; Barducci A.; Wüthrich K.; Parrinello M. Prion protein β2−α2 loop conformational landscape. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 9617–9622. 10.1073/pnas.1712155114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piening N.; Nonno R.; Di Bari M.; Walter S.; Windl O.; Agrimi U.; Kretzschmar H. A.; Bertsch U. Conversion efficiency of bank vole prion protein in vitro is determined by residues 155 and 170, but does not correlate with the high susceptibility of bank voles to sheep scrapie in vivo. J. Biol. Chem. 2006, 281, 9373–9384. 10.1074/jbc.m512239200. [DOI] [PubMed] [Google Scholar]
- Scott M.; Groth D.; Foster D.; Torchia M.; Yang S.-L.; DeArmond S. J.; Prusiner S. B. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell 1993, 73, 979–988. 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- Avbelj M.; Hafner-Bratkovič I.; Jerala R. Introduction of glutamines into the B2–H2 loop promotes prion protein conversion. Biochem. Biophys. Res. Commun. 2011, 413, 521–526. 10.1016/j.bbrc.2011.08.125. [DOI] [PubMed] [Google Scholar]
- Kurt T. D.; Jiang L.; Bett C.; Eisenberg D.; Sigurdson C. J. A proposed mechanism for the promotion of prion conversion involving a strictly conserved tyrosine residue in the β2-α2 loop of PrPC. J. Biol. Chem. 2014, 289, 10660–10667. 10.1074/jbc.m114.549030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghiaian F.; Daubenfeld T.; Quenet Y.; Van Audenhaege M.; Bouin A.-P.; Van Der Rest G.; Grosclaude J.; Rezaei H. Diversity in prion protein oligomerization pathways results from domain expansion as revealed by hydrogen/deuterium exchange and disulfide linkage. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 7414–7419. 10.1073/pnas.0607745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J.; Udgaonkar J. B. Structural effects of multiple pathogenic mutations suggest a model for the initiation of misfolding of the prion protein. Angew. Chem., Int. Ed. Engl. 2015, 54, 7529–7533. 10.1002/anie.201501011. [DOI] [PubMed] [Google Scholar]
- Miller M. B.; Wang D. W.; Wang F.; Noble G. P.; Ma J.; Woods V. L. Jr; Li S.; Supattapone S. Cofactor molecules induce structural transformation during infectious prion formation. Structure 2013, 21, 2061–2068. 10.1016/j.str.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J.; Kumar H.; Sabareesan A. T.; Udgaonkar J. B. Rational stabilization of helix 2 of the prion protein prevents its misfolding and oligomerization. J. Am. Chem. Soc. 2014, 136, 16704–16707. 10.1021/ja510964t. [DOI] [PubMed] [Google Scholar]
- Brayton K. A.; O’Rourke K. I.; Lyda A. K.; Miller M. W.; Knowles D. P. A processed pseudogene contributes to apparent mule deer prion gene heterogeneity. Gene 2004, 326, 167–173. 10.1016/j.gene.2003.10.022. [DOI] [PubMed] [Google Scholar]
- O’Rourke K. I.; Spraker T. R.; Hamburg L. K.; Besser T. E.; Brayton K. A.; Knowles D. P. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J. Gen. Virol. 2004, 85, 1339–1346. 10.1099/vir.0.79785-0. [DOI] [PubMed] [Google Scholar]
- Angers R. C.; Seward T. S.; Napier D.; Green M.; Hoover E.; Spraker T.; O’Rourke K.; Balachandran A.; Telling G. C. Chronic wasting disease prions in elk antler velvet. Emerging Infect. Dis. 2009, 15, 696. 10.3201/eid1505.081458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez D. R.; Damberger F. F.; Wüthrich K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J. Mol. Biol. 2010, 400, 121–128. 10.1016/j.jmb.2010.04.066. [DOI] [PubMed] [Google Scholar]
- Moreno J. A.; Telling G. C.. Insights into mechanisms of transmission and pathogenesis from transgenic mouse models of prion diseases. Prions; Springer, 2017; pp 219–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. M.; Browning S. R.; Seward T. S.; Jewell J. E.; Ross D. L.; Green M. A.; Williams E. S.; Hoover E. A.; Telling G. C. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. J. Gen. Virol. 2008, 89, 598–608. 10.1099/vir.0.83168-0. [DOI] [PubMed] [Google Scholar]
- Tropea J. E.; Cherry S.; Waugh D. S.. Expression and purification of soluble His 6-tagged TEV protease. High throughput Protein Expression and Purification; Springer, 2009; pp 297–307. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Krieger E.; Koraimann G.; Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins: Struct., Funct., Bioinf. 2002, 47, 393–402. 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Kay L. E.; Torchia D. A.; Bax A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. 10.1007/bf00197809. [DOI] [PubMed] [Google Scholar]
- Keller R. L. J.The Computer Aided Resonance Assignment Tutorial; CANTINA Verlag, 2004. [Google Scholar]
- Goddard K. D.SPARKY 3; University of California: San Francisco, 2008. [Google Scholar]
- Shen Y.; Delaglio F.; Cornilescu G.; Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T. J.; Nielsen J. E.; McCammon J. A.; Baker N. A. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. A.; Sept D.; Joseph S.; Holst M. J.; McCammon J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 10037–10041. 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søndergaard C. R.; Olsson M. H.; Rostkowski M.; Jensen J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of p K a values. J. Chem. Theory Comput. 2011, 7, 2284–2295. 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Olsson M. H. M.; Søndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: consistent treatment of internal and surface residues in empirical p K a predictions. J. Chem. Theory Comput. 2011, 7, 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.