Abstract
PURPOSE
Mutation testing of the key genes involved in melanoma oncogenesis is now mandatory for the application of targeted therapeutics. However, knowledge of the mutational profile of melanoma remains largely unknown in Brazil.
PATIENTS AND METHODS
In this study, we assessed the mutation status of melanoma driver genes BRAF, NRAS, TERT, KIT, and PDGFRA in a cohort of 459 patients attended at Barretos Cancer Hospital between 2001 and 2012. We used polymerase chain reaction followed by Sanger sequencing to analyze the hot spot mutations of BRAF exon 15 (V600E), NRAS (codons 12/13 and 61), TERT (promoter region), KIT (exons 9, 11, 13, and 17), and PDGFRA (exons 12, 14, and 18) in tumors. The mutational profile was investigated for associations with demographic, histopathologic, and clinical features of the disease.
RESULTS
The nodular subtype was most frequent (38.9%) followed by the superficial spreading subtype (34.4%). The most frequent tumor location was in the limbs (50.0%). The mutation rates were 34.3% for TERT and 34.1% for BRAF followed by NRAS (7.9%), KIT (6.2%), and PDGFRA (2.9%). The BRAF (P = .014) and TERT (P = .006) mutations were associated with younger patients and with different anatomic locations, particularly in the trunk, for the superficial spreading and nodular subtypes, respectively (P = .0001 for both). PDGFRA mutations were associated with black skin color (P = .023) and TERT promoter mutations with an absence of ulceration (P = .037) and lower levels of lactate dehydrogenase. There was no association between patient survival rates and mutational status.
CONCLUSION
The similar mutational profile we observe in melanomas in Brazil compared with other populations will help to guide precision medicine in this country.
INTRODUCTION
Melanoma is a deadly disease with an increased global incidence in recent decades.1 GLOBOCAN estimated 287,723 new melanoma cases worldwide and 60,712 deaths in 2018.1 The INCA (Instituto Nacional de Câncer) estimate for Brazil was 6,260 new melanoma cases in 2018.2 Surgical resection of early stages is usually straightforward and uneventful, but advanced presentations are still life threatening and often require complex and costly treatments.3
In the past decade, the clinical management of patients with melanoma has changed significantly with the discovery of the high frequency of a target mutation in the mitogen-activated protein kinase (MAPK) pathway in this disease.4 Now patients are treated with anti-BRAF and anti-MEK that specifically target the pathway and inhibit its activity. The BRAF V600 mutant kinase of the MAPK pathway is present in approximately 37%-50% of all melanomas, and the use of this targeted therapy has become part of standard treatment.5,6
Other newly identified molecular alterations that characterize the genomic mutational landscape of melanoma comprise four main groups: the BRAF subtype, which is defined by the presence of BRAF hot spot mutations and is related to younger patients when BRAF occurs with MITF amplification; the RAS subtype, which is defined by the presence of NRAS hot spot mutations and is related to MAPK activation and AKT3 overexpression; the NF1 subtype, which is associated with disease in older patients and a higher mutation burden; and the triple wild type, which is a heterogenous subtype defined by a lack of ultraviolet signature with higher copy number and complex rearrangements.7 In addition to these four main molecular subtypes of melanoma, mutations in the TERT promoter region have been identified with high mutation frequency (approximately 50%), and their presence is associated with lower patient survival.8
CONTEXT
Key Objective
Do Brazilian patients share the same frequencies of hot spot mutations of essential genes in melanoma development that have been observed worldwide?
Knowledge Generated
We found that TERT promoter was the most mutated hot spot in 308 Brazilian patients with melanoma (34.3%) followed by BRAF (34.1%), NRAS (7.9%), KIT (6.2%), and PDGFRA (2.9%). The Brazilian population has similar frequencies of mutation for these 5 key genes to that observed in other countries. The small differences that we found could be the result of ethnic aspects, the proportion of primary and metastatic cases included in the studies, and/or the proportion of melanoma subtypes.
Relevance
Our study provides the relative frequencies of driver genes in melanoma development after undergoing target therapy. These findings allow for more accurate predictions of the financial impact of the implementation of these treatment options in the Brazilian health care system.
Several subtypes of melanoma arise in body parts protected from direct ultraviolet light, including uveal, mucosal, and acral melanomas.9 Of the various subtypes, acral melanoma is one of the rarest, accounting for < 2% of all melanoma cases. This melanoma can occur in the skin but exhibits unique clinical characteristics that affect only the palms of the hands, the soles of the feet, and the subungual area.10 The proportion of acral melanoma is much higher in Asian and dark-skinned individuals than in white populations.11 Those types of melanoma have been described with patterns of molecular alteration that distinguish them from the cutaneous melanomas, such as a higher frequency of KIT and PDGFRA mutations and amplifications.12 However, the blockade of these genes has so far demonstrated little clinical benefit.13
Melanoma has predominantly been investigated in white populations, and few studies have addressed its molecular characterization in an ethnically diverse and interbred population. Several studies have focused on the genetic ancestry in populations from southeastern and northern Brazil. These studies have shown that considerable admixture exists in the Brazilian population, with an ancestry proportion of approximately 60% European, 20% African, 7% Native American, and 7% East Asian.14-19 Furthermore, the EPIGEN-Brazil project studied 6,487 Brazilians from three population-based cohorts with different geographic and demographic backgrounds and showed that the ancestry proportions fluctuated in different regions.20 These findings corroborate with demographic studies that have shown that the Brazilian population is one of the most heterogeneous in the world as a result of 5 centuries of interethnic mating.15
The lack of knowledge of the frequency and diversity of these molecular changes in Brazilian patients with melanoma makes it difficult to evaluate the cost-benefit of implementing targeted therapeutics in this country. However, these treatments clearly need to be introduced into the public health care system because a significant number of patients would benefit from increases in survival rates and improved quality of life. In this context, we aimed to determine the mutational frequency of BRAF, NRAS, KIT, PDGFRA, and TERT driver genes in 459 tumors from Brazilian patients with melanoma and to evaluate the association between mutational status and clinicopathologic features of patients in the cohort.
PATIENTS AND METHODS
This retrospective study evaluated 459 patients with melanoma attended at Barretos Cancer Hospital from 2001 to 2012. The inclusion criteria were a diagnosis of melanoma, presence of formalin-fixed paraffin-embedded (FFPE) tumor tissue for analysis, and at least 1 treatment performed at Barretos Cancer Hospital. This study was previously approved by the institutional ethical board (548/2011).
Patients and Clinical Data
Demographic data (age, sex, skin color, and place of birth) and date of primary tumor and its characteristics (location, histopathologic subtype, Breslow depth, Clark level, ulceration, and mitotic rate/mm2) were collected from medical records. The retrieved clinical data included the presence or absence of regional lymph node metastases and any features of distant metastases (localization and serum lactate dehydrogenase [LDH]).
Progression-free and melanoma-specific survivals were calculated, and the patients were categorized according to validated diagnostic criteria (TNM classification). Survival tests were performed from the date of primary tumor diagnosis, regional metastasis, and distant metastasis to the last event (clinical evaluation or death).
DNA Isolation From FFPE Tissue
DNA was isolated from FFPE tissue sections representative of the tumor lesions as previously described but with some modifications. Briefly, serial adjacent 10-μm-thick unstained sections cut from paraffin blocks and 1 representative hematoxylin and eosin section were analyzed by a qualified pathologist for identification and selection of the tumor area, which was macrodissected into a microfuge tube using a sterile needle (Neolus 25G × 0.5 mm; Terumo Medical Corporation, Tokyo, Japan). The macrodissected tissue was deparaffinized by sequential washing with xylol and ethanol (100%-70%-50%) and allowed to air dry. DNA was extracted using a QIAamp DNA Micro Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions and then quantified using the NanoDrop 2000 system (Thermo Fisher Scientific, Waltham, MA). DNA samples were stored at −20°C until required for genetic analysis. Because the melanin protein present in melanoma tumor cells can be copurified during DNA extraction, and this contamination may inhibit subsequent polymerase chain reaction (PCR) methods, we used a protocol established by our group to remove melanin from DNA of pigmented samples.21
Mutation Analysis of TERT, KIT, PDGFRA, BRAF, and NRAS Hot Spot Regions
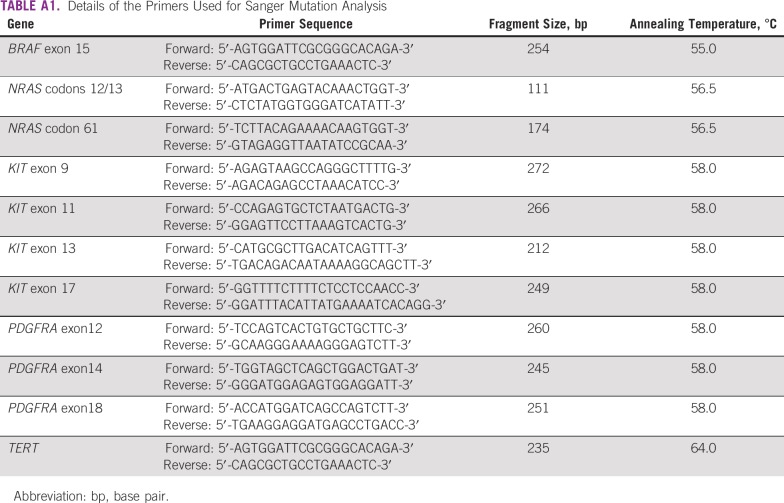
The analysis of hot spot mutations of TERT promoter regions (containing the sites c.-124:C>T and c.-146:C>T), KIT (exons 9, 11, 13, and 17), PDGFRA (12, 14, and 18), BRAF (exon 15), and NRAS (codons 12/13 and 61) was performed by PCR followed by direct Sanger sequencing, as described previously by our group.22,23 Briefly, using specific pairs of primers (Appendix Table A1), the target regions were amplified by PCR using a Veriti PCR thermal cycler (Applied Biosystems, Foster City, CA). We used an initial denaturation at 95°C for 15 minutes followed by 40 cycles of 95°C denaturation for 45 seconds; specific annealing temperature was for 90 seconds and the 72°C elongation phase for 45 seconds followed by a 72°C final elongation for 7 minutes. Amplification of PCR products was confirmed by gel electrophoresis.
Sequencing PCR was performed using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and a 3500xL Genetic Analyzer (Applied Biosystems). Confirmation of all mutations was performed by repeating the PCR and sequencing the altered regions.
Statistical Analysis
To identify associations among demographic, clinical, histopathologic, and molecular characteristics, the Fisher’s exact test or χ2 test was used. Associations of variables to cancer-specific survival were tested using the Kaplan-Meier method. The log-rank test was applied to evaluate the differences between the curves. Patients with melanoma in situ were excluded from the survival analyses. For survival tests, patients with multiple tumor samples were considered mutated when at least 1 sample presented the alteration. Multivariable analyses were performed with the Cox regression method for variables that were significant in the univariable analysis or considered to fit in the multivariable model. The significance level considered for all tests was 95% (P ≤ .05).
RESULTS
Patients and Tumor Features
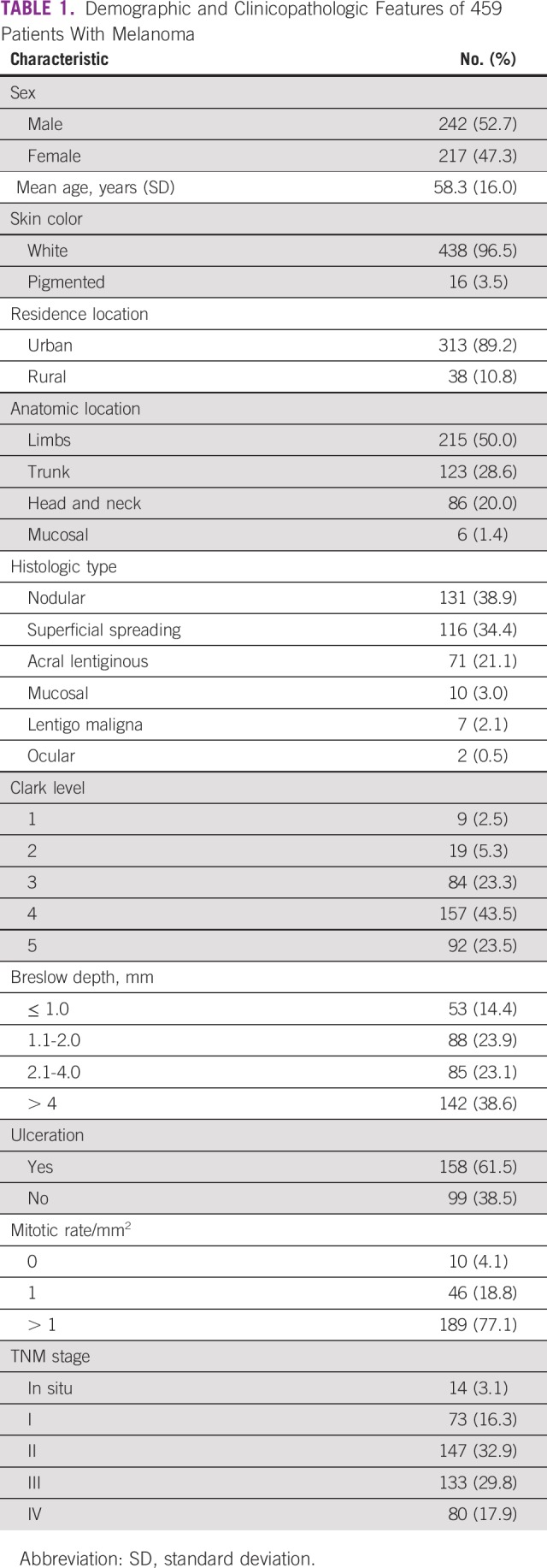
A summary of the clinicopathologic features of all patients in the cohort is listed in Table 1. The mean age of the patients was 58 years, and the majority (96.5%) were self-described as white. The most frequent anatomic location for the primary tumor was in the limbs (50%). Histologically, the predominant subtype was nodular melanoma (38.9%) followed by superficial spreading (34.4%) and acral lentiginous subtype (21.1%). The majority of patients had TNM stage II or III disease, and the most relevant clinical information, such as Clark level, Breslow depth, ulceration, and mitotic index, was related to local advanced tumors.
TABLE 1.
Demographic and Clinicopathologic Features of 459 Patients With Melanoma
Mutation Profile
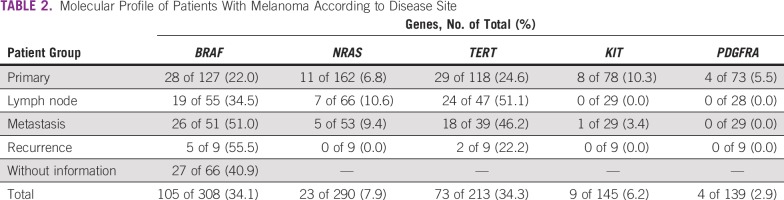
The quality of samples used in this study was excellent, with sequence data being unavailable in only 32.9% (151 of 459 patients) because of poor quality and/or DNA recovery issues. Overall, we observed that 308 (67.1%) of 459 patients had conclusive results for BRAF mutation, 290 (63.1%) for NRAS, 213 (46.4%) for TERT promoter, 145 (31.6) for KIT, and 139 (30.2%) for PDGFRA (Table 2).
TABLE 2.
Molecular Profile of Patients With Melanoma According to Disease Site
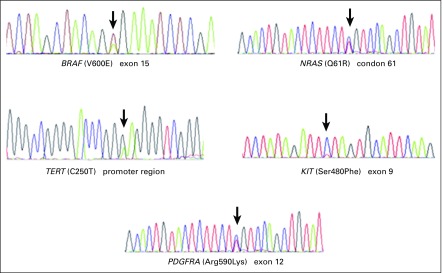
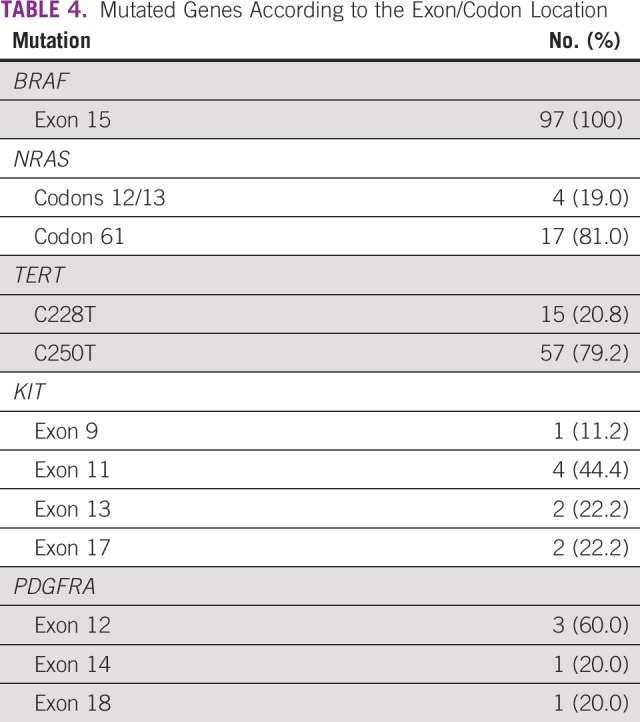
Appendix Figure A1 and Figure A2 show examples of the sequencing profiles used to identify the different somatic mutations studied in the cohort. We found that TERT promoter mutations were the most frequently observed alteration, with 34.3% of the samples (73 of 213) having this change. We found that 20.8% had TERT promoter mutation in the c.-124:C>T region and the remaining 79.2% in the c.-146:C>T region (Tables 2 and 3). The second most common mutated gene was BRAF, with 34.1% of the samples (105 of 308) comprising 89.7% V600E, 8.3% V600K, 1.0% V600M, and 1% T599I (Tables 2 and 3). NRAS mutations were found in approximately 8% of samples (23 of 290), with 19.0% at codons 12/13 and 81.0% at codon 61 (Tables 2-4). Mutations in the KIT gene were found in 6% of melanomas (9 of 145), with 11.2% at exon 9, 44.4% at exon 11, 22.2% at exon 13, and 22.2% at exon 17 (Tables 2-4). The least frequently mutated gene was PDGFRA, with only 3% of tumors (4 of 139) having this mutation and 60.0% being located at exon 12, 20.0% at exon 14, and 20.0% at exon 18 (Tables 2-4).
TABLE 3.
Characterization of Mutations in the Samples According to the Target Genes
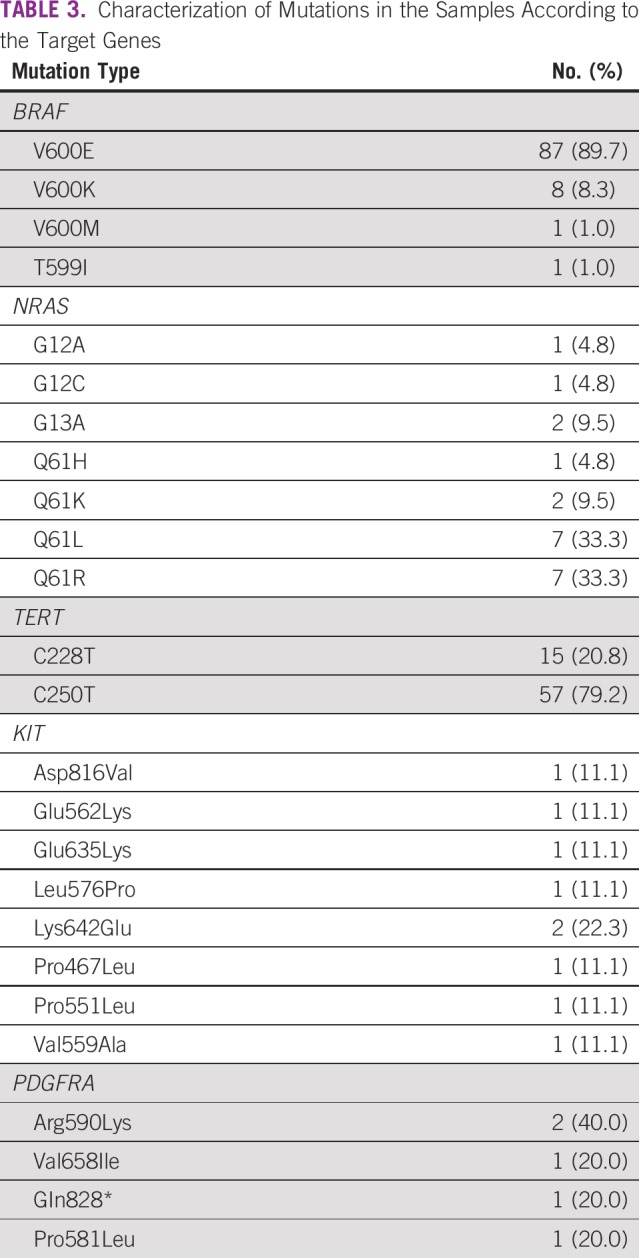
TABLE 4.
Mutated Genes According to the Exon/Codon Location
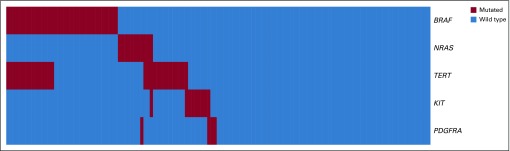
BRAF mutations were mutually exclusive with mutations in NRAS. Mutations in TERT occurred with a higher frequency together with BRAF mutations compared with the much lower rate of co-occurrence of NRAS and TERT mutations. Mutations in KIT and PDGFRA were rare in the cohort, and only 1 tumor had mutations of both genes (Fig 1).
FIG 1.
Mutational profile of 133 patients with melanoma.
Appendix Table A2 lists the comparisons between frequencies of mutation across the 5 genes in our study and that in studies with other populations. The observed differences can be the result of several reasons, including ethnic aspects; the proportion of primary and metastatic cases included in the studies; and/or the proportion of melanoma subtypes, such as the acral and mucosal groups.
Association of Mutation Profile and Patient Clinicopathologic Features
We observed that patients < 50 years of age presented with a higher frequency of mutations that affected BRAF (P = .014) and TERT (P = .006; Table 5). BRAF and TERT mutations are associated with different anatomic locations, especially the trunk region and the superficial spreading and nodular tumor types, respectively (P = .0001 for all; Table 6). In addition, mutations in TERT were associated with the absence of ulceration (P = .037) and LDH ≤ 400 U/L (Table 6). We found that PDGFRA mutations occurred more frequently in patients with black skin color (P = .023; Table 5).
TABLE 5.
Demographic Data of 459 Patients With Melanoma According to Mutational Profile
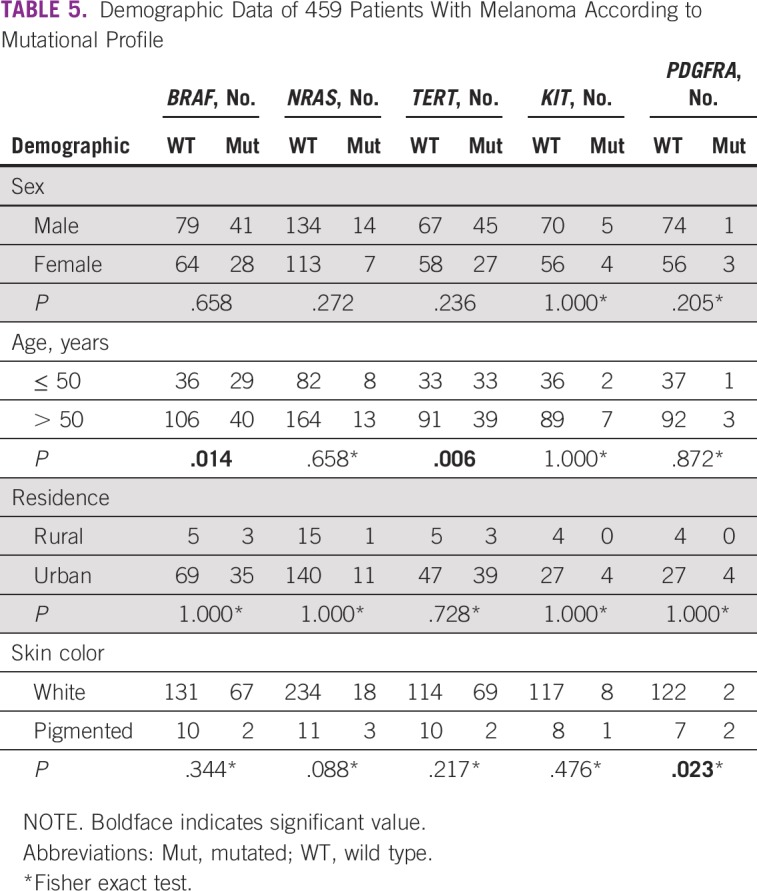
TABLE 6.
Clinical and Tumor Characteristics of 459 Patients With Melanoma According to Mutational Profile
Specific-cancer survival was estimated in all 459 patients, with a mean of 88.8 months (standard deviation, 3.8 months), median of 68.2 months, and rate of 51.1% at 5 years. Characteristics such as ulceration (P = .101), age (P = .078), mitotic index (P = .542), and LDH value (P = .377) were not related to different survival rates. However, TNM staging (P = .001), sex (P = .0001), anatomic location (P = .0001), histologic subtype (P = .0009), Clark level (P = .0001), and Breslow depth (P = .0001) were associated with different survival rates.
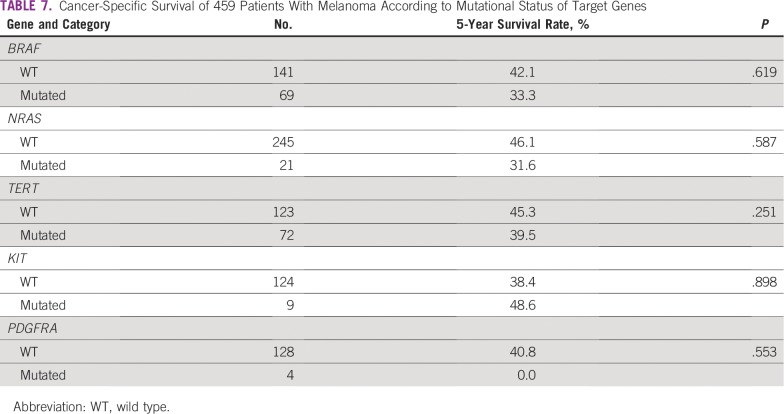
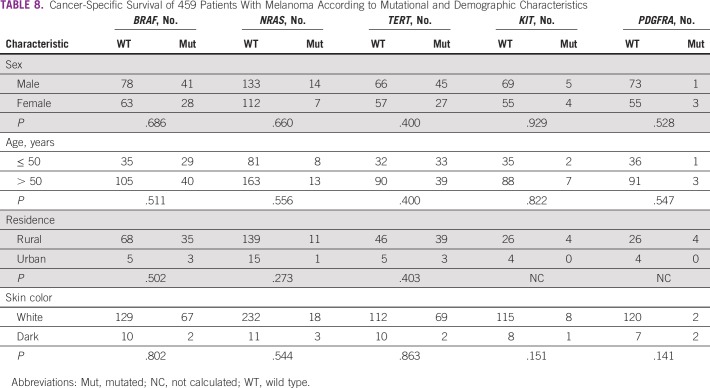
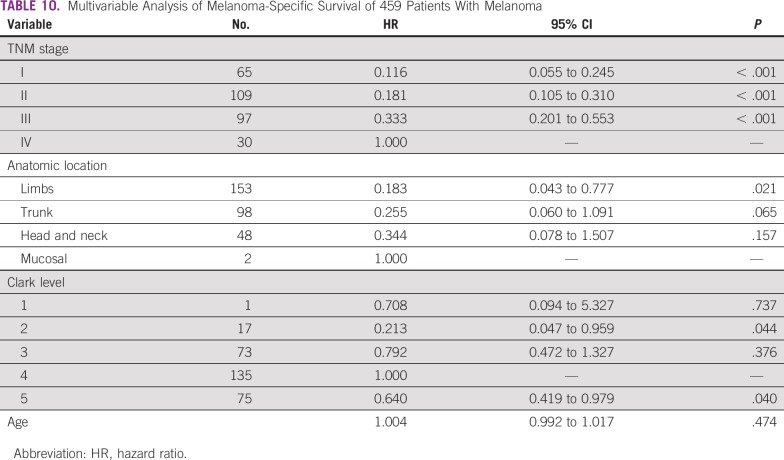
In addition, we observed that the mutation status of the 5 genes did not influence 5-year survival rates (Table 7). Similarly, the results showed no difference in cancer-specific survival according to mutations in target genes and the patient/tumor characteristics (Tables 8 and 9). Multivariable analysis demonstrated that TNM stage IV, anatomic location, and Clark level were independently associated with different survival rates (Table 10).
TABLE 7.
Cancer-Specific Survival of 459 Patients With Melanoma According to Mutational Status of Target Genes
TABLE 8.
Cancer-Specific Survival of 459 Patients With Melanoma According to Mutational and Demographic Characteristics
TABLE 9.
Cancer-Specific Survival of 459 Patients With Melanoma According to Mutational Status of Target Genes and Clinical Characteristics
TABLE 10.
Multivariable Analysis of Melanoma-Specific Survival of 459 Patients With Melanoma
DISCUSSION
This study is important because it was previously unknown whether the frequencies of established somatic mutations, known to be involved in melanoma development worldwide, are similar in Brazilian patients. Our findings are particularly relevant because the Brazilian population has a strong ethnic mixture from Africa, Europe, Brazil (indigenous), and Asia.16 In the current study, the frequency of BRAF and TERT promoter mutations was approximately 34% of the samples, with NRAS, KIT, and PDGFRA being approximately 8%, 6%, and 3% of samples, respectively.
Most studies conducted in other populations found higher frequencies of BRAF and TERT promoter mutations compared with our study (Appendix Table A2). Specifically, in the case of The Cancer Genome Atlas analysis, this is probably due to the majority of the samples being derived from metastatic cases that are known to have higher frequencies of mutations affecting these genes. In addition, another source of bias in the The Cancer Genome Atlas cohort is that the acral lentiginous melanoma subtype with lower frequencies of BRAF mutations was not included. NRAS, KIT, and PDGFRA mutations also have lower frequencies in our cohort, and these mutational alterations are usually more frequent in the acral subtype, as described previously.19,24,25
Many studies have shown the predominance of BRAF mutations in young patients, which were also confirmed in our study.7,26 In addition, we found an association between BRAF mutations and the superficial spreading subtype tumors that occurred at an anatomic location in the trunk. This result agrees with that of Egberts et al,25 who found an association between mutations in this gene and tumor localization in the trunk. However, these authors also reported an association of BRAF mutations and thin melanomas and absence of ulceration, which was not observed in our study. Thomas et al26 showed that in 812 primary melanoma tumors, BRAF mutations were associated with the superficial and nodular disseminated subtype as well as demonstrated associations of this mutation with mitotic index, which was not detected in our study. In agreement with both studies, the presence of BRAF mutations in tumors was not associated with cancer-specific survival.24,25
We found an association between mutations in TERT gene in patients < 50 years of age. In younger patients, the tumors were often located in the trunk, whereas this association was demonstrated elsewhere in older patients, with locations such as the head and neck or extremities being more common.24 Macerola et al27 showed no relationship between mutations and the presence of ulceration, but they did report an association between mitotic index and mutation, which we did not observe in our study.
The BRAF and TERT mutations co-occurrence was present in some of our patients. As pointed out by Shain et al,28 the distinct evolutionary progression of BRAF nevi and early selection of TERT promoter mutations may explain this observation.
A substantial increase in effective therapies for patients with advanced melanoma in the past several years has been significantly improving patient outcomes. On the other hand, the costs associated with these new systemic treatments are an important consideration, especially in terms of future public health spending. Several studies have shown the high costs of targeted therapy for melanoma treatment in different countries and the subsequent impact on the budget of the health care system.29-34 In a recent study, Corrêa et al29 estimated the incremental budget impact of targeted therapy for first-line treatment of advanced nonsurgical and metastatic melanoma compared with dacarbazine treatment in the Brazilian population between 2018 and 2020. The costs were estimated to be more than Brazilian real $700 million over the 3 years, which represents the annual cost for using targeted therapy that is 18-30 times more than the cost of traditional chemotherapy. The prevalence of the BRAF V600 mutation in this study was estimated on the basis of data from the scientific literature because of the absence of Brazilian information. Our study provides the frequencies of BRAF mutation according to cancer staging, which provides a better prediction for a cost-effective analysis of the budget impact of targeted therapy using national frequencies.
During this research development, we faced challenges in PCR amplification, including the presence of melanin, which copurifies during DNA extraction and is an inhibitor of DNA polymerase. Because no protocol for melanin removal was available, we have developed a method to solve this issue.21 We recovered many samples using our protocol, but 151 samples could not be tested probably because of tissue fixation issues and the long years the samples have been stored.
Overall, Brazilian patients with melanoma have similar frequencies of BRAF, NRAS, TERT, KIT, and PDGFRA mutations observed in other populations, which indicates that the high rate of miscegenation in Brazil is not a relevant factor for melanoma development in our patients. Moreover, Brazil can benefit from all currently used targeted therapies, and we are well prepared for emerging therapeutics directed at these more frequent mutational groupings.
ACKNOWLEDGMENT
We thank Jeremy Squire, PhD, for English evaluation and for critical review of this manuscript.
APPENDIX
FIG A1.
Sequencing electropherograms. Hot spot mutations are exemplified.
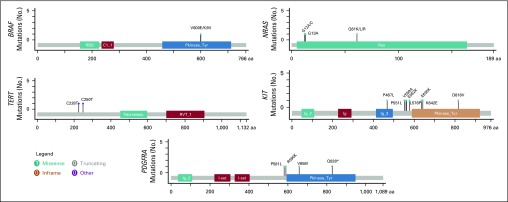
FIG A2.
Lollipop plots of driver mutations identified in BRAF, NRAS, TERT, KIT, and PDGFRA. aa, amino acids.
TABLE A1.
Details of the Primers Used for Sanger Mutation Analysis
TABLE A2.
Comparison of Molecular Change Frequencies of Melanomas Among Countries
Footnotes
Supported by the São Paulo Research Foundation (grant #2012/04194-1, V.L.V.) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for the master’s fellowship grant (A.L.S.A.V).
AUTHOR CONTRIBUTIONS
Conception and design: Cristovam Scapulatempo-Neto, Rui M. Reis, Vinicius L. Vazquez
Financial support: Rui M. Reis, Vinicius L. Vasquez
Provision of study material or patients: Anna Luiza S.A. Vicente, Cristovam Scapulatempo-Neto
Collection and assembly of data: Anna Luiza S.A. Vicente, Camila S. Crovador, Graziela Macedo, Cristovam Scapulatempo-Neto
Data analysis and interpretation: Anna Luiza S.A. Vicente, Rui M. Reis
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jgo/site/misc/authors.html.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Cristovam Scapulatempo-Neto
Speakers’ Bureau: AstraZeneca, Merck Sharp & Dohme
Research Funding: AstraZeneca (Inst)
Patents, Royalties, Other Intellectual Property: Patent BR 10 2014 003033 6, a molecular test microRNA for determination of primary sites of carcinoma of unknown primary (Inst)
Rui M. Reis
Research Funding: MSD Oncology
No other potential conflicts of interest were reported.
REFERENCES
- 1.Rastrelli M, Tropea S, Rossi CR, et al. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005–1011. [PubMed] [Google Scholar]
- 2. Instituto Nacional de Câncer: Estimativa 2018: Incidência de Câncer no Brasil, 2017. https://www.inca.gov.br/sites/ufu.sti.inca.local/files//media/document//estimativa-incidencia-de-cancer-no-brasil-2018.pdf.
- 3.Domingues B, Lopes JM, Soares P, et al. Melanoma treatment in review. ImmunoTargets Ther. 2018;7:35–49. doi: 10.2147/ITT.S134842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long GV, Hauschild A, Santinami M, et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med. 2017;377:1813–1823. doi: 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 9.Clark WH, Jr, From L, Bernardino EA, et al. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Res. 1969;29:705–727. [PubMed] [Google Scholar]
- 10.Bradford PT, Goldstein AM, McMaster ML, et al. Acral lentiginous melanoma: Incidence and survival patterns in the United States, 1986-2005. Arch Dermatol. 2009;145:427–434. doi: 10.1001/archdermatol.2008.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchetti MA, Chung E, Halpern AC. Screening for Acral lentiginous melanoma in dark-skinned individuals. JAMA Dermatol. 2015;151:1055–1056. doi: 10.1001/jamadermatol.2015.1347. [DOI] [PubMed] [Google Scholar]
- 12.Carrera C, Puig-Butille JA. Clinical, epidemiological, and molecular heterogeneity in acral melanoma. J Invest Dermatol. 2018;138:254–255. doi: 10.1016/j.jid.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 13.Hodi FS, Corless CL, Giobbie-Hurder A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol. 2013;31:3182–3190. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campanella NC, Berardinelli GN, Scapulatempo-Neto C, et al. Optimization of a pentaplex panel for MSI analysis without control DNA in a Brazilian population: Correlation with ancestry markers. Eur J Hum Genet. 2014;22:875–880. doi: 10.1038/ejhg.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manta FS, Pereira R, Caiafa A, et al. Analysis of genetic ancestry in the admixed Brazilian population from Rio de Janeiro using 46 autosomal ancestry-informative indel markers. Ann Hum Biol. 2013;40:94–98. doi: 10.3109/03014460.2012.742138. [DOI] [PubMed] [Google Scholar]
- 16.Pena SD, Di Pietro G, Fuchshuber-Moraes M, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lins TC, Vieira RG, Abreu BS, et al. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am J Hum Biol. 2010;22:187–192. doi: 10.1002/ajhb.20976. [DOI] [PubMed] [Google Scholar]
- 18.Bakos L, Masiero NC, Bakos RM, et al. European ancestry and cutaneous melanoma in Southern Brazil. J Eur Acad Dermatol Venereol. 2009;23:304–307. doi: 10.1111/j.1468-3083.2008.03027.x. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez VL, Vicente AL, Carloni A, et al. Molecular profiling, including TERT promoter mutations, of acral lentiginous melanomas. Melanoma Res. 2016;26:93–99. doi: 10.1097/CMR.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 20.Kehdy FS, Gouveia MH, Machado M, et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A. 2015;112:8696–8701. doi: 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vicente ALSA, Bianchini RA, Laus AC, et al. Comparison of protocols for removal of melanin from genomic DNA to optimize PCR amplification of DNA purified from highly pigmented lesions. Histol Histopathol. 2019;34:1089–1096. doi: 10.14670/HH-18-112. [DOI] [PubMed] [Google Scholar]
- 22.Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 23.Terada T. Low incidence of KIT gene mutations and no PDGFRA gene mutations in primary cutaneous melanoma: An immunohistochemical and molecular genetic study of Japanese cases. Int J Clin Oncol. 2010;15:453–456. doi: 10.1007/s10147-010-0087-0. [DOI] [PubMed] [Google Scholar]
- 24.Mazurenko NN, Tsyganova IV, Lushnikova AA, et al. Spectrum of oncogene mutations is different in melanoma subtypes [in Russian] Mol Biol (Mosk) 2015;49:1022–1029. doi: 10.7868/S0026898415060166. [DOI] [PubMed] [Google Scholar]
- 25.Egberts F, Bohne AS, Krüger S, et al. Varying Mutational alterations in multiple primary melanomas. J Mol Diagn. 2016;18:75–83. doi: 10.1016/j.jmoldx.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Thomas NE, Edmiston SN, Alexander A, et al. Association between NRAS and BRAF mutational status and melanoma-specific survival among patients with higher-risk primary melanoma. JAMA Oncol. 2015;1:359–368. doi: 10.1001/jamaoncol.2015.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macerola E, Loggini B, Giannini R, et al. Coexistence of TERT promoter and BRAF mutations in cutaneous melanoma is associated with more clinicopathological features of aggressiveness. Virchows Arch. 2015;467:177–184. doi: 10.1007/s00428-015-1784-x. [DOI] [PubMed] [Google Scholar]
- 28. doi: 10.1056/NEJMoa1502583. Shain A.H., Yeh I, Kovalyshyn I, et al: The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med 373:1926-1936, 2015 . [DOI] [PubMed] [Google Scholar]
- 29.Corrêa FM, Guerra RL, Fernandes RRA, et al. Target therapy versus dacarbazine in first-line treatment of advanced non-surgical and metastatic melanoma: Budget impact analysis from the perspective of the Brazilian National Health System, 2018-2020. Epidemiol Serv Saude. 2019;28:e2018325. doi: 10.5123/S1679-49742019000200013. [DOI] [PubMed] [Google Scholar]
- 30. doi: 10.1007/s40258-017-0347-5. Elliott TM, Whiteman DC, Olsen CM, et al: Estimated healthcare costs of melanoma in Australia over 3 years post-diagnosis. Appl Health Econ Health Policy 15:805-816, 2017 [Erratum: Appl Health Econ Health Policy 15:817-818, 2017] [DOI] [PubMed] [Google Scholar]
- 31.Gordon LG, Rowell D. Health system costs of skin cancer and cost-effectiveness of skin cancer prevention and screening: A systematic review. Eur J Cancer Prev. 2015;24:141–149. doi: 10.1097/CEJ.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 32. Pike E, Torkilseng EB, Saeterdal I, et al: A Health Technology Assessment of the New Drugs for Inoperable or Metastatic Malignant Melanoma Patients. Oslo, Norway, Knowledge Centre for the Health Services at The Norwegian Institute of Public Health, 2015. [PubMed] [Google Scholar]
- 33.Vouk K, Benter U, Amonkar MM, et al. Cost and economic burden of adverse events associated with metastatic melanoma treatments in five countries. J Med Econ. 2016;19:900–912. doi: 10.1080/13696998.2016.1184155. [DOI] [PubMed] [Google Scholar]
- 34.Lyth J, Carstensen J, Synnerstad I, et al. Stage-specific direct health care costs in patients with cutaneous malignant melanoma. J Eur Acad Dermatol Venereol. 2016;30:789–793. doi: 10.1111/jdv.13110. [DOI] [PubMed] [Google Scholar]