Supplemental Digital Content is available in the text.
Keywords: atrial fibrillation, biomarkers, cardiovascular diseases, genome-wide association study, heart rate, retina
Objective:
The retina may provide readily accessible imaging biomarkers of global cardiovascular health. Increasing evidence suggests variation in retinal vascular traits is highly heritable. This study aimed to identify the genetic determinants of retinal vascular traits.
Approach and Results:
We conducted a meta-analysis of genome-wide association studies for quantitative retinal vascular traits derived using semi-automatic image analysis of digital retinal photographs from the GoDARTS (Genetics of Diabetes Audit and Research in Tayside; N=1736) and ORCADES (Orkney Complex Disease Study; N=1358) cohorts. We identified a novel genome-wide significant locus at 19q13 (ACTN4/CAPN12) for retinal venular tortuosity (TortV), and one at 13q34 (COL4A2) for retinal arteriolar tortuosity (TortA); these 2 loci were subsequently confirmed in 3 independent cohorts (Ntotal=1413). In the combined analysis of discovery and replication cohorts, the lead single-nucleotide polymorphism in ACTN4/CAPN12 was rs1808382 (βs.d.=-0.109; SE=0.015; P=2.39×10-13) and in COL4A2 was rs7991229 (βs.d.=0.103; SE=0.015; P=4.66×10-12). Notably, the ACTN4/CAPN12 locus associated with TortV is also associated with coronary artery disease, heart rate, and atrial fibrillation.
Conclusions:
Genetic determinants of retinal vascular tortuosity are also linked to cardiovascular health. These findings provide a molecular pathophysiological foundation for the use of retinal vascular traits as biomarkers for cardiovascular diseases.
Highlights.
Emerging evidence indicates that retinal tortuosity traits are associated with vascular health and highly heritable. However, the genetic architecture of retinal vascular tortuosity has not been investigated.
By using a meta-analysis of genome-wide association studies, we found a novel association at 19q13 (ACTN4/CAPN12) for retinal venular tortuosity (TortV), and one at 13q34 (COL4A2) for retinal arteriolar tortuosity (TortA) at discovery stage and validated in 3 independent cohorts.
In-silico look-ups indicate that the significant associations between lead single-nucleotide polymorphisms at 19q13 and coronary artery disease, cardiovascular vascular risk factors, atrial fibrillation, and heart rate. Colocalization eQTL study (expression quantitative trait loci) found CAPN12 as most likely to be a causal gene for TortV in heart and blood vessel tissues.
Our findings highlight genetic impacts on TortV, and their association with cardiovascular disease and may provide a molecular pathophysiological foundation for the use of retinal vascular traits as biomarkers for cardiovascular diseases.
Retinal vascular traits can be readily measured noninvasively from fundus images, and changes in these traits have been linked to a number of clinical conditions associated with vascular health, including cardiovascular disease,1,2 stroke,3 hypertension,4 and neurodegenerative disease.5 The association between retinal vascular calibers and cardiovascular disease has been reported in numerous studies, and structural variation in retinal vasculature could predict cardiovascular risk.6–8 More recently, deep learning applied to retinal images has been successfully used to predict cardiovascular risk factors and outcomes.9 Whereas this potentially powerful approach indicated that vascular regions of the retina appeared important, it cannot provide a molecular pathoetiological basis for the link between retinal vasculature and cardiovascular disease.
Population-based studies have demonstrated a significant genetic component to variation in retinal blood vessel width.10 Evidence suggests that retinal vascular tortuosity, a potentially important vascular parameter, is also associated with a range of cardiovascular risk factors.11,12 Heritability estimates for retinal arterial tortuosity range from 50% to 82% and 21% for retinal venular tortuosity,10,13 indicating a substantial genetic contribution to the variation in these parameters. Recent genome-wide association studies (GWAS) found a number of loci for the more widely investigated retinal traits, central retinal vein equivalent (CRVE)14–16 and central retinal arteriolar equivalent (CRAE)14–16 as well as retinal image-derived optic disk morphology parameters.17
To our knowledge, no studies have performed a genome-wide scan on retinal vascular tortuosity traits. Understanding the molecular genetic architecture of retinal tortuosity traits would provide a molecular pathophysiological basis linking retinal microvascular features with systemic vascular pathology. We, therefore, performed a sufficiently powered GWAS to identify genetic variants influencing the retinal vascular tortuosity traits; arteriolar tortuosity (TortA), maximum TortA (TortAmax), venular tortuosity (TortV), and maximum TortV (TortVmax). We also examined other previously investigated retinal parameters, including CRAE, CRVE, arteriole-to-venule ratio (AVR), as well as the nonvascular optic disc radius (ODradius). We have investigated whether any of the significant variants associated with tortuosity traits are also associated with cardiovascular-related outcomes in the previously published GWAS results. There is ample evidence indicating that increased resting heart rate is linked to various cardiovascular events and associated with the increased risk of plaque rupture in coronary atherosclerosis patients.18,19 Therefore, we also checked whether there is any association between TortV-associated variants with heart rate in independent samples from United Kingdom Biobank.
Subjects and Methods
Data analysis, methods, and all other supporting materials are available in the online-only Data Supplement document. The data that support the findings of this study are available to the researchers from the corresponding author upon request.
Study Participants
Participants in the discovery phase of this study were obtained from 2 independent cohorts, the GoDARTS (Genetics of Diabetes Audit and Research in Tayside20) and the ORCADES (Orkney Complex Disease Study).21 Three independent cohorts of individuals of European ancestry were used at the replication stage, including the Lothian Birth Cohort 1936 (LBC1936),22 the Croatia-Korčula, and Croatia-Split study. Detailed descriptions of each cohort are presented in the online-only Data Supplement note.
Retinal Vascular Parameter Measurement
Standard digital retinal photographs used for diabetes mellitus retinal screening were obtained from the clinical record of patients with type 2 diabetes mellitus in GoDARTS. A total of 1744 images (661 images from the GoDARTS data set 1 and 1083 images from GoDARTS data set 2) were selected for analysis after quality control (QC). Similarly, 1595 individual’s retinal images from ORCADES were used for this study after QC. The Vascular Assessment and Measurement Platform for Images of Retina 3.1, semi-automatic software, was used to measure retinal vascular traits in fundus images (Figure I in the online-only Data Supplement) from both GoDARTS and ORCADES. Standard protocols were followed to measure the retinal vessel parameters,23 including CRAE, CRVE, AVR, ODradius, TortA, TortAmax, TortV, and TortVmax (online-only Data Supplement). Tortuosity mean values were normalized by natural log transformation for association analysis (Figure II in the online-only Data Supplement). Smaller values indicate straighter vessels. After quality assessment and processing, a total of 644, 387, and 382 individuals’ retinal fundus images from the LBC1936, Croatia- Korčula, and Croatia-Split cohorts, respectively, were selected for the analysis. In these cohorts, retinal tortuosity traits were quantified using SIVA v3.124 (Singapore I Vessels Assessment), semi-automated software, and normalized by natural log transformation for association analysis (online-only Data Supplement).
Genotyping, QC, and Imputation
GoDARTS participants were genotyped using the Affymetrix 6.0 (n=927) and Illumina Human Omni Express (n=809) platforms. ORCADES samples were genotyped with either the Illumina HumanHap300 bead chip (n=890) or the Illumina Omni1M (n=304) or Illumina Omni Express bead chips (n=1073). Genotype data quality was assessed and imputed on the basis of 1000 Genome Projects reference panel. Imputed genotypes for 658, 1078, 1358 individuals from the GoDARTS data set 1, GoDARTS data set 2, and ORCADES cohorts, respectively, were used for the three independent GWAS analysis.
LBC1936 samples were genotyped using the Illumina Human 610Quad BeadChip. A total of 1398 participants from the 2 independent Croatian replication cohorts were available for the analysis, and subjects were genotyped on different genotyping platforms including Illumina CNV370v1 and CNV370-Quadv3 for Croatia-Korčula (n=378), and Illumina CNV370-Quadv3 and IlluminaOmniExpressExome-8v1_A for Croatia-Split (n=376). More details on QC, imputation, and processing can be found in the online-only Data Supplement.
Statistical Analyses
We performed association analyses with each data set from GoDARTS separately for each of the 8 retinal traits using SNPTEST V2.525 linear regression assuming an additive genetic model, adjusting for 3 ancestry principal components, age at eye examination, and gender. Association analysis in ORCADES was performed using linear mixed modeling to account for relatedness and assuming an additive genetic model, adjusting for 3 ancestry principal components, age and gender, using the mmscore function in ProbABEL.26 Then, we performed the meta-analysis using a fixed-effects model in GWAMA27 with the QC filtered GWAS summary results (imputation quality score >0.4 and minor allele frequency >0.03) from the GoDARTS and ORCADES. Manhattan plots, Quantile-Quantile plots, and forest plots were generated using in-built R scripts, and metafor—R package. Regional plots were generated using the Locus Zoom tool.28
Conditional analyses were performed in SNPTEST v2.5 using the genome-wide significant loci in the COL4A2 region, conditioned on the lead single-nucleotide polymorphism (SNP) (rs56399312). In addition, this newly discovered locus was conditioned on previously reported genome-wide significant SNPs (rs4773144,29 rs11617955,30 and rs951520331) associated with coronary artery disease (CAD). We performed an association test with an additive model adjusted for age, gender, and the first 3 principal components ancestry in the diabetes mellitus cohort (GoDARTS) using 759 samples without any cardiovascular events before the retinal screening date. We investigated the in-silico functional effects of the sentinel variants for each retinal vascular traits using various bioinformatics databases.32–37 Details can be found in the online-only Data Supplement.
The top 3 SNPs (P≤1.07×10−07) near ACTN4, TMEM132D, and COL4A2 from the discovery stage for the tortuosity traits were taken forward for examination in 3 replication cohorts of European ancestry. In the LBC1936 cohort, association analyses were performed for arterial and venular tortuosity traits using linear regression model adjusting for age, sex, and 3 ancestry principal components, using mach2qtl. Similarly, in the Croatia—Split, and Korčula cohorts, association analyses were performed for each trait separately using linear mixed models implemented in the hglm R package, accounting for kinships derived using the gkin function of the GenABEL package.38
Summary association statistics for lead SNPs associated with TortA and TortV from the 2 discovery and 3 replication cohorts were combined and effect estimates from each cohort were presented in the forest plots using metafor—R package. Because of the variability in the β values and SE between the discovery and replication studies arising from different measurement algorithms, we standardized the effect estimates (using Cohen d) from each of the individual study cohorts. Cohen d is a scale-free interpretation which provides a standardized effect size measurement between the studies.39 In this study, Cohen d was calculated by the effect estimate divided by the SD.
In-Silico Look-Ups of the Novel Variants for Clinical Outcomes
To investigate the association of the lead SNPs for TortA, and TortV with cardiovascular outcomes, we performed in-silico look-ups using summary association results from the Coronary Artery Disease Genome-wide Replication and meta-analysis plus C4D consortium,29 Global Lipid Genetics Consortium analysis40, and International consortium for blood pressure GWAS analysis.41 A recent study reported the association of ACTN4 locus with heart rate.42 To examine whether the lead SNPs associated with TortV in ACTN4 were also associated with heart rate, we checked the linkage disequilibrium (r2>0.8) between our SNPs and the index SNP (rs11083475) for heart rate in that study. Furthermore, we investigated the association of these SNPs with pulse rate in UK Biobank data.43 Additionally, we explored the relationship between the TortV-associated variants with atrial fibrillation (AF) using the summary statistics data from a recent large-scale meta-analysis of GWAS of AF.44 To identify the potential causal/target gene for TortV-associated variants, colocalization analysis was performed eCAVIAR.45 Details of these studies and online in-silico functional annotation resources have been described in the online-only Data Supplement on methods.
Results
Meta-Analysis of Discovery GWAS
The characteristics of the discovery study cohorts and overall study design can be found in Table I in the online-only Data Supplement and Figure 1. We combined the summary GWAS results from the GoDARTS and ORCADES cohorts for each trait using a fixed-effect meta-analysis, which overall showed no evidence of excessive amount of false-positive associations (genomic inflation factor of 0.99). Table II in the online-only Data Supplement presents the results from the meta-analysis and independent cohort GWAS analysis. Manhattan plots, Quantile-Quantile plots, and regional plots are shown in Figures 2 and 3 and Figures III through V in the online-only Data Supplement, respectively.
Figure 1.
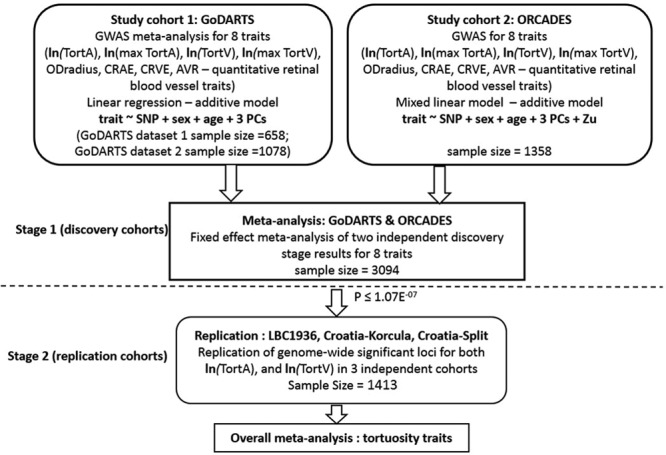
Study Design. Natural log-transformed data—TortA, TortAmax, TortV, TortVmax, u is the genetic value for each subject under a random-effects model, covariance amongst subjects assumed to be proportionate to the genomic relationship matrix. All Croatia indicates Croatia island of Korcula, Croatia-Split; AVR, arteriole-to-venule ratio; CRAE, Central Retinal Arteriolar Equivalent; CRVE, Central Retinal Venular Equivalent; GoDARTS, Genetics of Diabetes Audit and Research in Tayside; LBC1936, Lothian Birth Cohorts 1936; ODradius, Optic Disc Radius; ORCADES, Orkney Complex Disease Study; PC, principal components; TortA, retinal arteriolar tortuosity; TortAmax, maximum retinal arteriolar tortuosity; TortV, retinal venular tortuosity; and TortVmax, maximum retinal arteriolar tortuosity.
Figure 2.
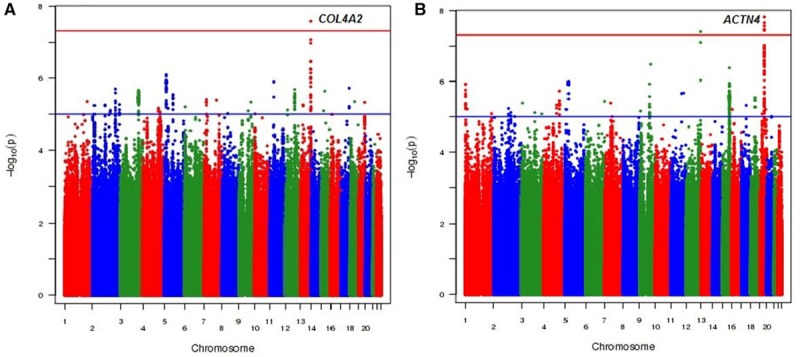
Manhattan plots for meta-analysis of genome-wide association results from 2 independent discovery cohorts. A, The results for the arteriolar tortuosity (TortA) and (B) represents the results for the venular tortuosity trait (TortV). The blue and red horizontal lines indicate the suggestive and genome-wide significance threshold (P<5×10−8), respectively.
Figure 3.
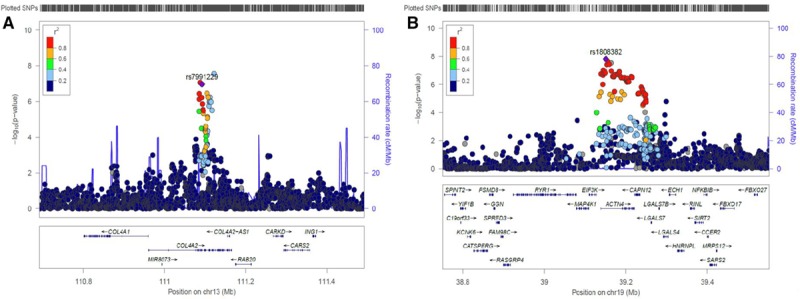
Regional association and recombination plots of variants that reached P value <5×10-7 in the meta-analysis of the 2 discovery study cohorts (GoDARTS and ORCADES). A, Lead SNP for TortA; B, lead SNP for TortV. Each plot was created using LocusZoom for the lead single-nucleotide polymorphism (SNP) in genomic region 400 kb in either side of the significant signal. Blue spikes represent the estimated recombination rates. Color scale (high to low r2) circles depicts the pairwise correlation (r2) between lead SNP and other SNPs in the loci. The lead SNP in that region is indicated by purple color solid diamond, and gene annotations in this region are shown in the bottom. GoDARTS indicates Genetics of Diabetes Audit and Research in Tayside; and ORCADES, Orkney Complex Disease Study.
This analysis revealed one genome-wide significant (P<5×10−8) SNP, rs56399312, associated with TortA at 13q34, in COL4A2 with moderate heterogeneity (I2=0.50; β=0.182, SE=0.032, P=2.70×10−8), and another SNP rs9515212 near COL4A2 that was just below the threshold for genome-wide significance (β=0.151, SE=0.028, P=8.59×10−8). Conditional analysis on the lead SNP indicated that these are not independent signals (Table III in the online-only Data Supplement). Two genome-wide significant SNPs were associated with TortV, at 19q13 in ACTN4 (lead SNP rs1808382; β=−0.123, SE=0.022, P=1.55×10−8; no heterogeneity, I2=0.00), and at 12q24.33 near TMEM132D (lead SNP rs73157566; β=−0.294, SE=0.054, P=4.07×10−8; low heterogeneity, I2=0.10); these associations at both these loci have not been reported previously with any retinal vascular parameters.
Moreover, we replicated 3 loci out of 8 previously reported loci for CRVE14,16 but did not replicate any of the previously reported SNPs associated with CRAE.15 Finally, we replicated a previously reported genome-wide significant locus for ODradius at 10q21.3 near PBLD (lead SNP rs61854835; β=−3.840, SE=0.575, P=4.06×10−11) and confirmed a number of other loci for this trait17,46–48 (Table IV in the online-only Data Supplement).
Replication of Novel Associations With Vessel Tortuosity in Independent Cohorts
As candidates to carry forward for replication, we selected 3 lead SNPs from both loci (total 6 SNPs) (ACTN4/CAPN12 andTMEM132D) for TortV and COL4A2 for TortA that reached significance P≤1.07×10−07 and had similar effect size and direction across the discovery cohorts. The characteristics of the replication cohorts are presented in Table V in the online-only Data Supplement.
Two TortA-associated SNPs, rs7991229, and rs9515212 in COL4A2 reached nominal significance (P<0.05) in the LBC1936, Croatian cohorts. P values were significant after multiple testing in stage 2 and combined analyses (based on 2 independent tests). The lead SNP (rs56399312) from discovery stage did not reach significant P value in the LBC1936 but had similar effect size and direction. Two TortV-associated SNPs, rs1808382 and rs3786835 in ACTN4/CAPN12 reached suggestive significance (P<1×10−04) in the combined analysis of replication cohorts whereas rs73157566 near TMEM132D did not replicate. Table contains the summary statistics from replication cohorts and meta-analysis of these cohorts.
Table 1.
Results of Discovery, Replication, and Overall Meta-Analysis for Tortuosity Traits
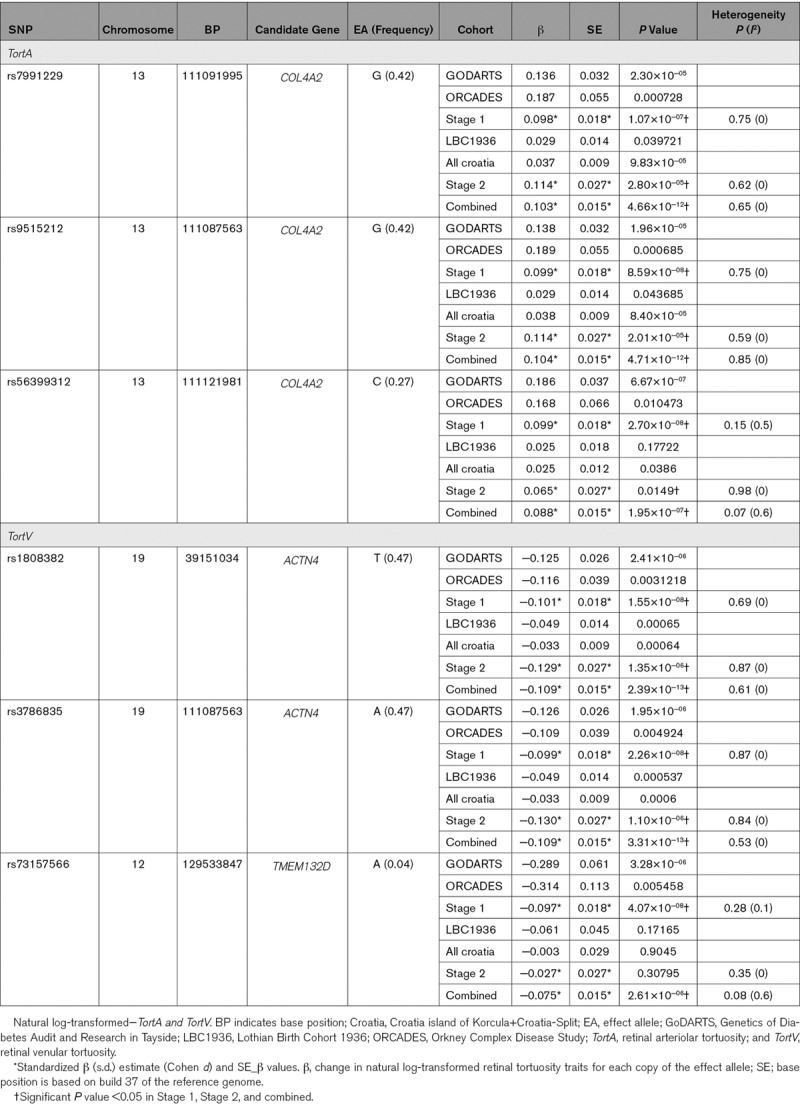
Meta-Analysis of Discovery and Replication Cohorts
In the overall meta-analysis, SNPs at 13q34, COL4A2 (TortA) and 19q13, ACTN4 (TortV) display genome-wide significant associations. Although TortA-associated SNPs, rs9515212 and rs7991229, were not genome-wide significant in the discovery meta-analysis, they reached genome-wide significance in the overall meta-analysis, and no heterogeneity (I2=0.00) was observed across different cohorts; Poverall=4.66×10−12 and Poverall=4.71×10−12, respectively. The lead SNP in COL4A2 for TortA (rs56399312) at the discovery stage did not reach genome-wide significance in the overall meta-analysis (Poverall=1.95×10−07), suggesting it is not likely to be the causal association at this locus that warrants further study. For TortV the lead SNPs, rs1808382 (Poverall=2.39×10−13) and rs3786835 (Poverall=3.31×10−13) near ACTN4/CAPN12, increased genome-wide significance and show no heterogeneity of effect sizes across studies (I2=0.00; Table). These SNPs are in tight linkage disequilibrium and, therefore, do not represent independent signals. The effect estimates from each of the individual study cohorts were standardized using Cohen d. Forest plots for lead SNPs in the combined analysis are shown in Figure 4.
Figure 4.
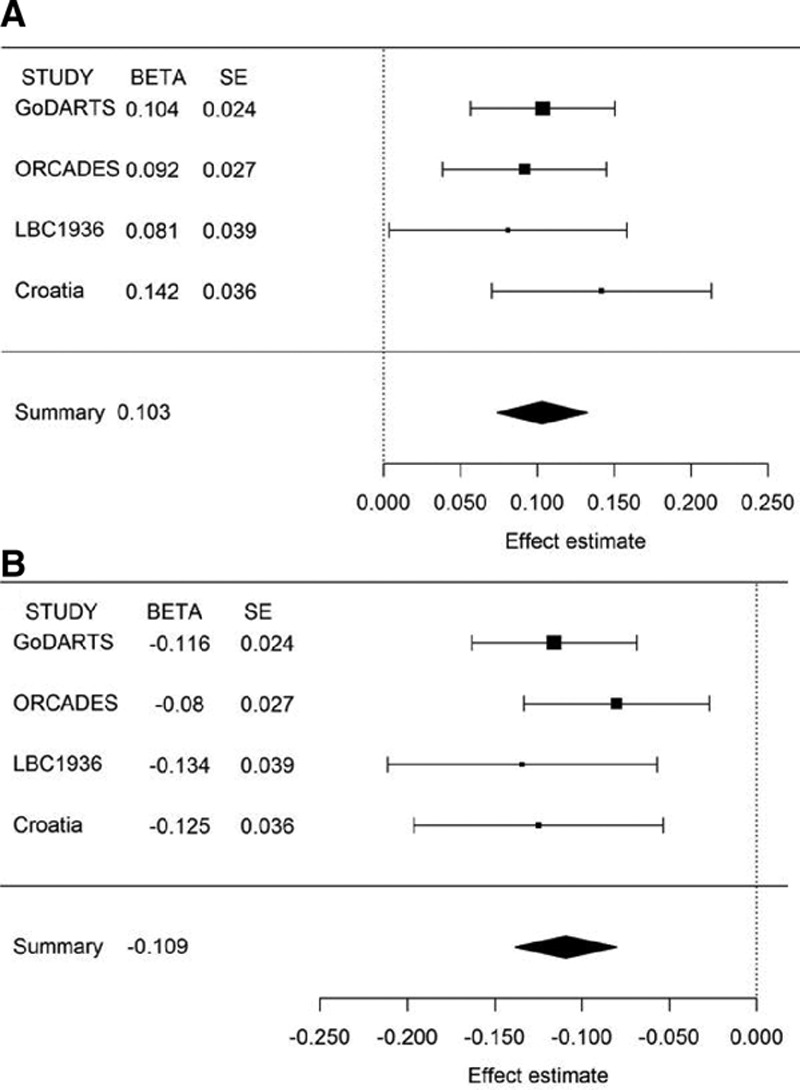
Forest plots for the genome-wide significant hits in overall meta-analysis. A, rs7991229 for TortA; B, rs1808382 for TortV. The plots represent standardized β (βsd) and SE from GoDARTS (Genetics of Diabetes Audit and Research in Tayside), ORCADES (Orkney Complex Disease Study), LBC1936 (Lothian Birth Cohorts 1936), Croatia-Korcula Split, and meta-analysis study. Standardized β estimate: change in natural log-transformed retinal tortuosity traits for each copy of the effect allele.
Functional Annotation: TortA-Associated Variants
COL4A2 encodes collagen alpha-2(IV) (collagen type IV α 2), one of the 6 subunits of type IV collagens which are major structural components of basement membranes, forming a thin sheet of fibers under the endothelium controlling passage of vasoactive substances. These are conserved across species and C-terminal noncollagenous domains play a role in angiogenesis.49 Recent GWAS report that common variants around COL4A2 and COL4A1 (a paralogue immediately proximal to COL4A2, with which it shares a promoter and is co-expressed), are associated with coronary artery calcification,50 arterial stiffness,51 and CAD.29–31,52 Gene expression data from GeneAtlas,53 a human protein-coding transcriptome study validated the high expression of COL4A2 in retinal microvessel endothelial cells (Figure VI in the online-only Data Supplement) whereas COL4A1 is weakly expressed in retina indicating a specific role of COL4A2 in the retinal vasculature. TortA-associated variants near COL4A2 significantly alter transcription factor binding motifs and have putative effects on transcription as annotated by ENCODE (Table VI in the online-only Data Supplement). Additionally, expression data from the GTEx database34 confirmed that these significant SNPs are associated with the expression of COL4A2 in heart left ventricle and artery aorta, shown in Table VII and Figure VII in the online-only Data Supplement, and these SNPs are in linkage disequilibrium (rs9515212 and rs7991229; r2=0.99, D′=1).
Association of TortA-Associated Variants With Cardiovascular Risk Factors
Lead SNPs associated with TortA remained significant after conditioning on the previously reported cardiovascular risk variants in COL4A2 (rs11617955,29 rs4773144,30 and rs951520331; Figure VIII and Table VIII in the online-only Data Supplement). Conversely, the lead SNPs for TortA were not associated with CAD and myocardial infarction risk in the Coronary Artery Disease Genome-wide Replication and meta-analysis plus C4D consortium meta-analysis29 (Table IX in the online-only Data Supplement). Finally, the CAD-associated variants specifically in COL4A1 from Coronary Artery Disease Genome-wide Replication and meta-analysis plus C4D were not associated with TortA, whereas CAD-associated COL4A2 variants are only weakly associated with TortA (Table X in the online-only Data Supplement). Retinal arteriolar tortuosity traits have been previously associated with blood pressure,10,12 which may, therefore, link these variants with CAD; however, we found no evidence for an association between these lead variants and blood pressure in the International consortium for blood pressure GWAS analysis41 (Table XI in the online-only Data Supplement).
Functional Annotation: TortV-Associated Variants
ACTN4 encodes α-actinin 4 (alpha-actinin-4), a cross-linking protein belonging to the spectrin superfamily and mutations in this gene cause focal segmental glomerulosclerosis in humans. ACTN2, a homolog of ACTN4, interacts with ACTN4 and missense mutations in ACTN2 are linked to a range of cardiac diseases.54 Annotation by ENCODE33 indicates that the 2 genome-wide significant variants (rs1808382 and rs3786835) associated with TortV near ACTN4 may have direct regulatory effects as they are located within a DNase I hypersensitivity site and in genomic regions enriched for promoter/enhancer histone marks in heart tissues (Table VI in the online-only Data Supplement). ACTN4 and CAPN12 (calcium-activated neural proteases 12) overlap by 339 bases at their 3′ ends and multitissue expression quantitative trait loci (eQTL) analysis confirms that these SNPs in ACTN4 are associated with mRNA expression of both ACTN4 and CAPN12 in aorta, tibial artery, atrial appendage, and left ventricle of the heart (Table VII and Figure IX in the online-only Data Supplement). Additionally, this analysis indicates that the T allele at rs1808382 is correlated with lower ACTN4 (artery aorta; P=2.1×10−03) and this correlation is even stronger with CAPN12 (artery aorta; P=2.0×10−07). Whereas gene expression data using Genevestigator validated the high expression of ACTN4 in arterial tissue, the highest expression of CAPN12 appears to be in the hematopoietic system.
Colocalization Analysis of SNPs in ACTN4/CAPN12
We further conducted eQTL/trait colocalization analyses with eCAVIAR45 and estimated the colocalization posterior probability for the lead variants that are associated with TortV and eQTLs. We found that the CAPN12 has substantially higher colocalization posterior probability in heart left ventricle and artery tibial tissues than the ACNT4 which indicates moderate colocalization between CAPN12 eQTL and TortV (Table XIII in the online-only Data Supplement). The heart left ventricle is considered as the most relevant tissue (colocalization posterior probability; 0.38) for the TortV-associated lead variant (rs1808382)-CAPN12 eQTL whereas for the other 3 tissues (heart atrial appendage; 0.22, artery aorta; 0.27, and artery tibial; 0.37) are considered as the relevant tissues for the second top variant (rs3786835) associated with TortV-CAPN12 eQTL (Table XIII in the online-only Data Supplement).
Association of TortV-Associated Variants With Cardiovascular Risk Factors
Lead SNPs in ACTN4 were significantly associated with CAD in the Coronary Artery Disease Genome-wide Replication and meta-analysis plus C4D consortium meta-analysis29 (Table IX in the online-only Data Supplement) and were associated with CAD risk factors include HDL (high-density lipoprotein) cholesterol and triglycerides in the Global Lipid Genetics Consortium analysis40 but not associated with LDL (low-density lipoprotein) cholesterol in the Global Lipid Genetics Consortium analysis or blood pressure in the International Consortium for blood pressure GWAS analysis41 (Table XI in the online-only Data Supplement). Furthermore, we have confirmed the association between TortV and top variants in ACTN4/CAPN12 in a sensitivity analysis that only included GoDARTS samples without any cardiovascular events before the retinal screening date (Table XII in the online-only Data Supplement). Moreover, a recent meta-analysis of 35 GWAS studies reported the association of SNP (rs11083475) in the ACTN4 locus with increased resting heart rate,42 which may increase cardiovascular disease risk. This signal appears the same as that for TortV with strong linkage disequilibrium being observed between the lead SNPs for TortV and the index SNP for heart rate. Furthermore, we found that these SNPs were associated with heart rate in UK Biobank43 (Table XIV and Figure X in the online-only Data Supplement). Interestingly, TortV-associated SNPs also associated with AF in the recent AF GWAS analysis44 (Table XI in the online-only Data Supplement).
Discussion
In this first GWAS meta-analysis for quantitative retinal vascular tortuosity traits, we identify a novel locus for retinal arteriolar tortuosity (COL4A2) and for retinal venular tortuosity (ACTN4/CAPN12), which were firmly established by replication in 3 independent cohorts. Power calculations indicated that a sample size of 4507 (stage 1 and stage 2), and 3094 (stage 1) and the effect size of 0.10 using Bonferroni correction (P<5×10−8) was adequate to provide 80% statistical power to detect the associations. Notably, we also identified in the discovery studies a genome-wide significant signal at a previously reported locus in/near ATOH7/PBLD for the optic disc radius and replicated previously identified variants for CRVE which validate our retinal traits measurement methods. However, we did not replicate the previously reported SNPs for CRAE, but the direction of the effect was consistent. This may due to differences in the phenotyping, smaller effect sizes for certain SNPs compared to the previous studies and need for more power to replicate this association (Table XV in the online-only Data Supplement).15,16
There are few limitations in this study. The meta-analysis of the lead SNPs for TortA and TortV in the replication cohorts (stage 2) lacks genome-wide significant P values which may due to the limited sample sizes. Other possible limitation of this study is that the retinal vascular traits were measured using different software in the discovery (eg, Vascular Assessment and Measurement Platform for Images of Retina) and replication cohorts (eg, SIVA). In spite of this limitation, our findings show consistent, homogeneous effects on tortuosity across 5 fairly diverse cohorts of European ancestry comprising individuals with and without diabetes. This highlights the robust nature of these genetic effects on retinal vascular topology. Also, these aspects strongly support the robustness our study design and findings.
Previous studies have reported association between COL4A2 and CAD but the TortA-associated variants in COL4A2 in the present study are not associated with cardiovascular disease and similarly COL4A2 variants that are associated with CAD do not appear to be associated with arteriolar tortuosity suggesting that variants in this gene complex may be involved differentially in the pathophysiology of microvascular and macrovascular diseases. However, more work has to be done to determine the distinct role of genetic variants in COL4A2/COL4A1 in different clinical conditions. In contrast, we found that retinal venular tortuosity-associated variants near ACTN4/CAPN12 were associated with CAD, heart rate, and AF. Furthermore, the lead variant influences the expression of the ACTN4/CAPN12 genes in the heart and blood vessel tissues. But our colocalization eQTL analysis found CAPN12 as the most likely causal gene at the chromosome 19 locus. Our sensitivity analyses including samples without CAD before the date of acquisition of the measured retinal image indicates that the relationship between genetic predictors of retinal venular tortuosity and cardiovascular diseases is not due to reverse causation and demonstrate the robustness of our findings.
Notably, the TortV-associated variants have shared genetic architecture with other cardiovascular-related traits including HDL cholesterol and AF. A recent study reported the relationship between retinal venular tortuosity and lower HDL cholesterol in the Asian-based cross-sectional cohorts.12 Our in-silico look-ups indicate that the genetic determinants for TortV near ACTN4 also associated with HDLC in the Global Lipid Genetics Consortium analysis GWAS but not associated with the LDLC which is consistent with the Asian-based study.12 A study from the ORCADES and Croatia-Korcula cohorts reported a weak association between retinal arteriolar tortuosity and systolic blood pressure and no significant association between retinal venular tortuosity trait and blood pressure.10 A recent larger epidemiological study in European-based prospective cohort reported an association with systolic blood pressure for TortA, and a weaker association for TortV.55 In this regard, neither the TortA nor TortV-associated variants were associated with systolic and diastolic blood pressure in the International Consortium for blood pressure GWAS analysis; therefore, it seems unlikely that observed associations with CAD or related traits are mediated through blood pressure.
In summary, this first GWAS for retinal arteriolar and venular tortuosity reveals associated SNPs of strong effects influencing the expression of COL4A2 and ACTN4/CAPN12, respectively. Our results demonstrate that the TortA-associated variants in COL4A2 are independent of CAD, myocardial infarction, and blood pressure, and point to a selective role of COL4A2 rather than COL4A1 in the retinal vessels. Strikingly, we found TortV-associated ACTN4/CAPN12 SNPs are associated with CAD, HDL cholesterol, AF, and heart rate but not associated with blood pressure. Our findings appear to indicate CAPN12 as the causal gene for TortV through colocalization analysis. However, detailed investigation and functional validation of this new finding are essential to elucidate the causal role of this locus and the relative contribution of ACTN4 and CAPN12 in the observed cardiovascular pathophysiology. These findings highlight the potential genetic impacts of retinal vasculature to provide new insights into cardiovascular disease.
Acknowledgments
The study was designed by C.N.A. Palmer, A.S.F. Doney, and E. Trucco for the GoDARTS (Genetics of Diabetes Audit and Research in Tayside) cohort, J.F. Wilson for the ORCADES (Orkney Complex Disease Study) cohort, I.J. Deary for the LBC1936 (Lothian Birth Cohort 1936) cohort, O. Polasek for the Croatia-Split, and Croatia-Korcula cohorts. The Vascular Assessment and Measurement Platform for Images of Retina (VAMPIRE) software was developed by E. Trucco, T. MacGillivray, D. Relan, E. Brown, and B. Dhillon. Retinal images were collected, and analysis was performed by E.Trucco, T. MacGillivray, J.F. Wilson, L. Ballerini, M. Kirin, D. Relan, V. Vitart, S.S. Vaidya, and H. Campbell. Genotype data processing and statistical analysis were conducted by A. Veluchamy, K.E. Schraut, P.K. Joshi, L. Ballerini, M. Kirin, S. Harris, V. Vitart, C. Hayward, and K. Zhou. Bioinformatics analysis was performed by A. Veluchamy. The article was drafted by A. Veluchamy, C.N.A. Palmer, and A.S.F. Doney and revised by E. Trucco, J.F. Wilson, T. MacGillivray, I.J. Deary, S. Harris, E.R. Pearson, V. Vitart, C. Hayward, and K. Zhou. All the authors reviewed the article and approved the final version. We are grateful to all the participants in the GoDARTS study, the general practitioners, the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The study complies with the Declaration of Helsinki. We acknowledge The National Institute for Health Research (NIHR) global health research unit on global diabetes outcomes research at the University of Dundee (INSPIRED project) Award number 16/136/102. We acknowledge the support of the Health Informatics Centre, University of Dundee for managing and supplying the anonymized data and NHS Tayside, the original data owner. We acknowledge the invaluable contributions of the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney. We thank the Lothian Birth Cohort 1936 (LBC1936) participants and team members who contributed to these studies. We acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to the University of Split and Zagreb Medical Schools and Croatian Institute for Public Health. Support from NHS Lothian R&D, and Edinburgh Imaging and the Edinburgh Clinical Research Facility at the University of Edinburgh is gratefully acknowledged.
Sources of Funding
The Wellcome Trust United Kingdom Type 2 Diabetes Case-Control Collection (GoDARTS [Genetics of Diabetes Audit and Research in Tayside]) was funded by The Wellcome Trust (072960/Z/03/Z, 084726/Z/08/Z, 084727/Z/08/Z, 085475/Z/08/Z, and 085475/B/08/Z) and as part of the EU IMI-SUMMIT program. ORCADES (Orkney Complex Disease Study) was supported by the Chief Scientist Office of the Scottish Government (CZB/4/276 and CZB/4/710), the Royal Society, the MRC Human Genetics Unit, Arthritis Research UK and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Wellcome Trust Clinical Research Facility in Edinburgh. Phenotype collection for LBC1936 was supported by Age UK (The Disconnected Mind project). Genotyping for LBC1936 was funded by the BBSRC (BB/F019394/1). The LBC1936 work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1); funding from the BBSRC and Medical Research Council (MRC) is gratefully acknowledged. The Croatia-Korčula and Croatia-Split study were funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (Contract No. LSHG-CT-2006-018947), European Commission Framework 7 project BBMRI-LPC (FP7 313010), the Republic of Croatia Ministry of Science, Education and Sports research grant (216-1080315-0302) and the Croatian Science Foundation (grant 8875). The SNP genotyping for the Korčula cohort was performed in Helmholtz Zentrum München, Neuherberg, Germany. Vascular Assessment and Measurement Platform for Images of Retina (VAMPIRE) team: Parts of the VAMPIRE software and its use for measuring the image set described here was funded by the Leverhulme Trust project RPG-419 Discovery of retinal biomarkers for genetics with large cross-linked data sets. VAMPIRE 3.1 has been further developed under funding from EPSRC (EPSRC EP/M005976/1) and the EU (REVAMMAD FP7-PEOPLE ITN, grant agreement 316990). For the analysis of the association of the identified genetic variants with heart rate, this research has been conducted using the UK Biobank Resource under Application Number 20405.
Disclosures
I.J. Deary reports grants from Age UK, grants from Biotechnology and Biological Sciences Research Council and grants from the Medical Research Council during the conduct of the study. H. Campbell has received funding from EU IMI, Sanofi for work on pneumococcal disease, CRUK for cancer research, WHO and Gates foundation for research on child health during the conduct of the study. The other authors report no conflicts.
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AF
- atrial fibrillation
- AVR
- arteriole-to-venule ratio
- CAD
- coronary artery disease
- CRAE
- retinal arteriolar equivalent
- CRVE
- central retinal vein equivalent
- eQTL
- expression quantitative trait loci
- GoDARTS
- Genetics of Diabetes Audit and Research in Tayside
- GWAS
- Genome-wide association studies
- HDL
- high-density lipoprotein
- LBC1936
- Lothian Birth Cohort 1936
- LDL
- low-density lipoprotein
- ODradius
- optic disc radius
- ORCADES
- Orkney Complex Disease Study
- SNP
- single-nucleotide polymorphism
- QC
- quality control
- TortA
- arteriolar tortuosity
- TortV
- venular tortuosity
These authors contributed equally to this article.
For Sources of Funding and Disclosures, see page 2551.
The online-only Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.119.312552.
References
- 1.Flammer J, Konieczka K, Bruno RM, Virdis A, Flammer AJ, Taddei S. The eye and the heart. Eur Heart J. 2013;34:1270–1278. doi: 10.1093/eurheartj/eht023. doi: 10.1093/eurheartj/eht023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liew G, Mitchell P, Rochtchina E, Wong TY, Hsu W, Lee ML, Wainwright A, Wang JJ. Fractal analysis of retinal microvasculature and coronary heart disease mortality. Eur Heart J. 2011;32:422–429. doi: 10.1093/eurheartj/ehq431. doi: 10.1093/eurheartj/ehq431. [DOI] [PubMed] [Google Scholar]
- 3.Baker ML, Hand PJ, Wang JJ, Wong TY. Retinal signs and stroke: revisiting the link between the eye and brain. Stroke. 2008;39:1371–1379. doi: 10.1161/STROKEAHA.107.496091. doi: 10.1161/STROKEAHA.107.496091. [DOI] [PubMed] [Google Scholar]
- 4.Ding J, Wai KL, McGeechan K, Ikram MK, Kawasaki R, Xie J, Klein R, Klein BB, Cotch MF, Wang JJ, et al. Meta-Eye Study Group. Retinal vascular caliber and the development of hypertension: a meta-analysis of individual participant data. J Hypertens. 2014;32:207–215. doi: 10.1097/HJH.0b013e32836586f4. doi: 10.1097/HJH.0b013e32836586f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacCormick IJ, Czanner G, Faragher B. Developing retinal biomarkers of neurological disease: an analytical perspective. Biomark Med. 2015;9:691–701. doi: 10.2217/bmm.15.17. doi: 10.2217/bmm.15.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nägele MP, Barthelmes J, Ludovici V, Cantatore S, von Eckardstein A, Enseleit F, Lüscher TF, Ruschitzka F, Sudano I, Flammer AJ. Retinal microvascular dysfunction in heart failure. Eur Heart J. 2018;39:47–56. doi: 10.1093/eurheartj/ehx565. doi: 10.1093/eurheartj/ehx565. [DOI] [PubMed] [Google Scholar]
- 7.Wang JJ, Liew G, Klein R, Rochtchina E, Knudtson MD, Klein BE, Wong TY, Burlutsky G, Mitchell P. Retinal vessel diameter and cardiovascular mortality: Pooled data analysis from two older populations. Eur Heart J. 2007;28:1984–1992. doi: 10.1093/eurheartj/ehm221. doi:10.1093/eurheartj/ehm221. [DOI] [PubMed] [Google Scholar]
- 8.Wang SB, Mitchell P, Liew G, Wong TY, Phan K, Thiagalingam A, Joachim N, Burlutsky G, Gopinath B. A spectrum of retinal vasculature measures and coronary artery disease. Atherosclerosis. 2018;268:215–224. doi: 10.1016/j.atherosclerosis.2017.10.008. doi:10.1016/j.atherosclerosis.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, Peng L, Webster DR. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2:158–164. doi: 10.1038/s41551-018-0195-0. doi:10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 10.Kirin M, Nagy R, MacGillivray TJ, Polašek O, Hayward C, Rudan I, Campbell H, Wild S, Wright AF, Wilson JF, et al. Determinants of retinal microvascular features and their relationships in two European populations. J Hypertens. 2017;35:16468–1659. doi: 10.1097/HJH.0000000000001408. doi:10.1097/HJH.0000000000001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owen CG, Rudnicka AR, Nightingale CM, Mullen R, Barman SA, Sattar N, Cook DG, Whincup PH. Retinal arteriolar tortuosity and cardiovascular risk factors in a multi-ethnic population study of 10-year-old children; the Child Heart and Health Study in England (CHASE). Arterioscler Thromb Vasc Biol. 2011;31:1933–1938. doi: 10.1161/ATVBAHA.111.225219. doi: 10.1161/ATVBAHA.111.225219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung CY, Zheng Y, Hsu W, Lee ML, Lau QP, Mitchell P, Wang JJ, Klein R, Wong TY. Retinal vascular tortuosity, blood pressure, and cardiovascular risk factors. Ophthalmology. 2011;118:812–818. doi: 10.1016/j.ophtha.2010.08.045. doi: 10.1016/j.ophtha.2010.08.045. [DOI] [PubMed] [Google Scholar]
- 13.Taarnhøj NC, Munch IC, Sander B, Kessel L, Hougaard JL, Kyvik K, Sørensen TI, Larsen M. Straight versus tortuous retinal arteries in relation to blood pressure and genetics. Br J Ophthalmol. 2008;92:1055–1060. doi: 10.1136/bjo.2007.134593. doi: 10.1136/bjo.2007.134593. [DOI] [PubMed] [Google Scholar]
- 14.Kamran Ikram M, Xueling S, Jensen RA, et al. Four novel loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation In vivo. PLoS Genet. 2010;6:18–12. doi: 10.1371/journal.pgen.1001184. doi:10.1371/journal.pgen.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sim X, Jensen RA, Ikram MK, Cotch MF, Li X, MacGregor S, Xie J, Smith AV, Boerwinkle E, Mitchell P, et al. Wellcome Trust Case Control Consortium 2; Global BPGen Consortium. Genetic loci for retinal arteriolar microcirculation. PLoS One. 2013;8:e65804. doi: 10.1371/journal.pone.0065804. doi: 10.1371/journal.pone.0065804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen RA, Sim X, Smith AV, Li X, Jakobsdóttir J, Cheng CY, Brody JA, Cotch MF, Mcknight B, Klein R, et al. CHARGE Exome Chip Blood Pressure Consortium. Novel genetic loci associated with retinal microvascular diameter. Circ Cardiovasc Genet. 2016;9:45–54. doi: 10.1161/CIRCGENETICS.115.001142. doi: 10.1161/CIRCGENETICS.115.001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macgregor S, Hewitt AW, Hysi PG, Ruddle JB, Medland SE, Henders AK, Gordon SD, Andrew T, McEvoy B, Sanfilippo PG, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010;19:2716–2724. doi: 10.1093/hmg/ddq144. doi: 10.1093/hmg/ddq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aune D, Sen A, ó’Hartaigh B, Janszky I, Romundstad PR, Tonstad S, Vatten LJ. Resting heart rate and the risk of cardiovascular disease, total cancer, and all-cause mortality - A systematic review and dose-response meta-analysis of prospective studies. Nutr Metab Cardiovasc Dis. 2017;27:504–517. doi: 10.1016/j.numecd.2017.04.004. doi: 10.1016/j.numecd.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhang D, Shen X, Qi X. Resting heart rate and all-cause and cardiovascular mortality in the general population: a meta-analysis. CMAJ. 2016;188:E53–E63. doi: 10.1503/cmaj.150535. doi:10.1503/cmaj.150535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hébert HL, Shepherd B, Milburn K, et al. Cohort profile: Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS). Int J Epidemiol. 2017:1–12. doi: 10.1093/ije/dyx140. doi:10.1093/ije/dyx140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deary IJ, Gow AJ, Pattie A, Starr JM. Cohort profile: the lothian birth cohorts of 1921 and 1936. Int J Epidemiol. 2012;41:1576–1584. doi: 10.1093/ije/dyr197. doi:10.1093/ije/dyr197. [DOI] [PubMed] [Google Scholar]
- 23.MacGillivray TJ, Cameron JR, Zhang Q, El-Medany A, Mulholland C, Sheng Z, Dhillon B, Doubal FN, Foster PJ, Trucco E, et al. UK Biobank Eye and Vision Consortium. Suitability of UK biobank retinal images for automatic analysis of morphometric properties of the vasculature. PLoS One. 2015;10:e0127914. doi: 10.1371/journal.pone.0127914. doi: 10.1371/journal.pone.0127914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh V, Cheung CY, Zheng Y, Wong TY, Wong W, Aung T. Relationship of retinal vascular tortuosity with the neuroretinal rim: the singapore malay eye study. Invest Ophthalmol Vis Sci. 2010;51:3736–3741. doi: 10.1167/iovs.09-5008. doi: 10.1167/iovs.09-5008. [DOI] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 26.Aulchenko SY, Struchalin VM, van Duijn MC. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2011;27:2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Ng FL, Chan K, Pu X, Poston RN, Ren M, An W, Zhang R, Wu J, Yan S, et al. Coronary-heart-disease-associated genetic variant at the COL4A1/COL4A2 locus affects COL4A1/COL4A2 expression, vascular cell survival, atherosclerotic plaque stability and risk of myocardial infarction. PLoS Genet. 2016;12:e1006127. doi: 10.1371/journal.pgen.1006127. doi: 10.1371/journal.pgen.1006127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. doi:10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speir ML, Zweig AS, Rosenbloom KR, Raney BJ, Paten B, Nejad P, Lee BT, Learned K, Karolchik D, Hinrichs AS, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44(D1):D717–D725. doi: 10.1093/nar/gkv1275. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ENCODE Project Consortium. A user’s guide to the Encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. doi:10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. doi:10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinlan AR. BEDTools: the swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–11.1234. doi: 10.1002/0471250953.bi1112s47. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol Rev Camb Philos Soc. 2007;82:591–605. doi: 10.1111/j.1469-185X.2007.00027.x. doi: 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 40.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. doi:10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, Nolte IM, Segrè AV, Holm H, Handsaker RE, et al. Global BPgen Consortium; CARDIoGRAM Consortium; PR GWAS Consortium; QRS GWAS Consortium; QT-IGC Consortium; CHARGE-AF Consortium. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621–631. doi: 10.1038/ng.2610. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carolina R, Mark D. C, Lu-Chen W, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hormozdiari F, van de Bunt M, Segrè AV, Li X, Joo JWJ, Bilow M, Sul JH, Sankararaman S, Pasaniuc B, Eskin E. Colocalization of GWAS and eQTL signals detects target genes. Am J Hum Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramdas WD, van Koolwijk LM, Ikram MK, Jansonius NM, de Jong PT, Bergen AA, Isaacs A, Amin N, Aulchenko YS, Wolfs RC, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khor CC, Ramdas WD, Vithana EN, Cornes BK, Sim X, Tay WT, Saw SM, Zheng Y, Lavanya R, Wu R, et al. Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum Mol Genet. 2011;20:1864–1872. doi: 10.1093/hmg/ddr060. doi: 10.1093/hmg/ddr060. [DOI] [PubMed] [Google Scholar]
- 48.Springelkamp H, Mishra A, Hysi PG, Gharahkhani P, Höhn R, Khor CC, Cooke Bailey JN, Luo X, Ramdas WD, Vithana E, et al. NEIGHBORHOOD Consortium. Meta-analysis of genome-wide association studies identifies novel loci associated with optic disc morphology. Genet Epidemiol. 2015;39:207–216. doi: 10.1002/gepi.21886. doi: 10.1002/gepi.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo DS, Labelle-Dumais C, Gould DB. COL4A1 and COL4A2 mutations and disease: insights into pathogenic mechanisms and potential therapeutic targets. Hum Mol Genet. 2012;21(R1):R97–R110. doi: 10.1093/hmg/dds346. doi: 10.1093/hmg/dds346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, Sun YV, Province MA, Aspelund T, Dehghan A, et al. CARDIoGRAM Consortium. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarasov KV, Sanna S, Scuteri A, Strait JB, Orrù M, Parsa A, Lin PI, Maschio A, Lai S, Piras MG, et al. COL4A1 is associated with arterial stiffness by genome-wide association scan. Circ Cardiovasc Genet. 2009;2:151–158. doi: 10.1161/CIRCGENETICS.108.823245. doi: 10.1161/CIRCGENETICS.108.823245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Cardiogenics; CARDIoGRAM Consortium. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu C, Bagnall RD, Ingles J, Yeates L, Kennerson M, Donald JA, Jormakka M, Lind JM, Semsarian C. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 2010;55:1127–1135. doi: 10.1016/j.jacc.2009.11.016. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Owen CG, Rudnicka AR, Welikala RA, Fraz MM, Barman SA, Luben R, Hayat SA, Khaw KT, Strachan DP, Whincup PH, et al. Retinal vasculometry associations with cardiometabolic risk factors in the European Prospective Investigation of Cancer-Norfolk Study. Ophthalmology. 2019;126:96–106. doi: 10.1016/j.ophtha.2018.07.022. doi: 10.1016/j.ophtha.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


