Abstract
The RTN4 gene plays a role in the development and progression of cancer. This case–control study aimed to investigate the association between the RTN4 gene polymorphism and its plasma level with the risk of nasopharyngeal carcinoma (NPC) in a Chinese population.
RTN4 gene polymorphisms (rs2920891, rs17046583, rs117465650, rs10496040, and rs2588519) in 220 patients with NPC and 300 healthy controls were analyzed using Snapshot single-nucleotide polymorphism genotyping assays. The plasma level of RTN4 was measured using the enzyme-linked immunosorbent assay.
The allele frequencies of RTN4 gene polymorphisms showed no significant difference between the patients and controls (P > .05). Nevertheless, the rs2920891 polymorphism in a dominant model (A/C+C/C) and codominant model (A/C) was significantly associated with the susceptibility to NPC (P = .017, odds ratio [OR] = 1.54, 95% confidence interval [CI] = 1.08–2.21 and P = .034, OR = 1.64, 95% CI = 1.13–2.38, respectively). The plasma level of RTN4 was significantly higher in patients with NPC in comparison with the controls (P < .001). Furthermore, we observed that patients with NPC carrying the rs2920891 A/C+C/C genotype had a higher RTN4 level than those carrying the A/A genotype (P < .001).
Our findings indicated that the rs2920891 polymorphism may be associated with increased susceptibility to NPC, possibly by increasing plasma RTN4.
Keywords: RTN4, gene polymorphism, nasopharyngeal carcinoma, susceptibility
1. Introduction
Nasopharyngeal carcinoma (NPC) is a malignancy of the head and neck and is characterized by the invasion of adjacent regions and metastasis to regional lymph nodes or distant organs. This distinct malignancy commonly occurs in Southeast Asia, North Africa, and especially southern China.[1] Interestingly, the incidence rate in southern China is 100 times higher than that in Western countries, which is around 15 to 50 per 100,000 people each year.[1] However, the exact pathogenesis of NPC is not completely understood.
Various risk factors have been proposed to be associated with the pathogenesis of NPC, including environmental exposure, Epstein–Barr virus infection, and genetic factors.[2–4] Previously, several candidate genes were found to affect the susceptibility of individuals to NPC, such as interleukin-1, interleukin-10, and osteopontin.[2–4] Identification of the relationship between susceptibility genes and NPC may help elucidate the disease mechanisms and provide effective treatment and prevention measures.
The reticulon-4 (RTN4) gene is located on the chromosome 2p12-14 and produces 3 isoforms (Nogo-A, Nogo-B, and Nogo-C) through alternative promoter usage and splicing.[5,6] Interestingly, these 3 different isoforms share a C-terminal domain, known as the highly conserved reticulon homology domain, which is composed of 188 amino acid residues.[5,6] In addition, they have a wide range of tissue expression patterns and multiple regulatory functions. Nogo-A (largest isoform) is a neurite outgrowth inhibitor and is mainly secreted in the central nervous system.[7] Nogo-B is expressed in various tissues, including vessel walls, smooth muscles, skeletal muscles, and endothelium, and it regulates the migration of olfactory ensheathing cells,[8] tumor cell migration and invasion,[8] and vascular homeostasis and remodeling.[9–11] Nogo-C, a shorter protein of the Nogo family, is secreted in the liver, vascular smooth muscles, skeletal muscles, heart, and neurons, and it regulates cardiomyocyte/hepatocellular carcinoma cell apoptosis.[9–11]
Few studies have considered the important role of Nogo isoforms. Nogo-B is a mediator of vascular homeostasis and remodeling.[11] Kritz et al[11] reported that the lack of Nogo-B could enhance the apoptosis of hepatic stellate cells, and the higher expression of Nogo-B could inhibit apoptosis. However, Tagami et al[12] found that Nogo-B could interact with Bcl-XL/-2, promoting localization on the endoplasmic reticulum (ER) and decreasing anti-apoptotic activity. Nevertheless, various studies have indicated that the overexpression of Nogo-B could induce the apoptosis of cells via ER stress and ER-specific signaling pathways.[13] A previous study reported the tumor-inhibiting activity of Nogo-A, which has been observed to attenuate the malignancy of oligodendroglial tumors.[13] In addition, Nogo-C expression in HEK293 cells was found to promote apoptosis by inducing the activation of caspase-3 and p53 through the JNK-c-Jun-dependent pathway.[14] The studies suggest that Nogo proteins may play a critical role in cellular apoptosis, especially in cancer cells.
It has been shown that single-nucleotide polymorphisms (SNPs) in RTN4 may affect the susceptibility of individuals to a variety of human diseases.[15–17] Nevertheless, no studies to date have investigated the association of RTN4 gene polymorphism and its plasma level between the risk of NPC in Chinese. Therefore, we evaluated the association of RTN4 gene polymorphisms (rs2920891, rs17046583, rs117465650, rs10496040, and rs2588519) with the susceptibility to NPC and further investigated the effect of the SNPs on RTN4 plasma level in patients with NPC.
2. Materials and methods
2.1. Participants
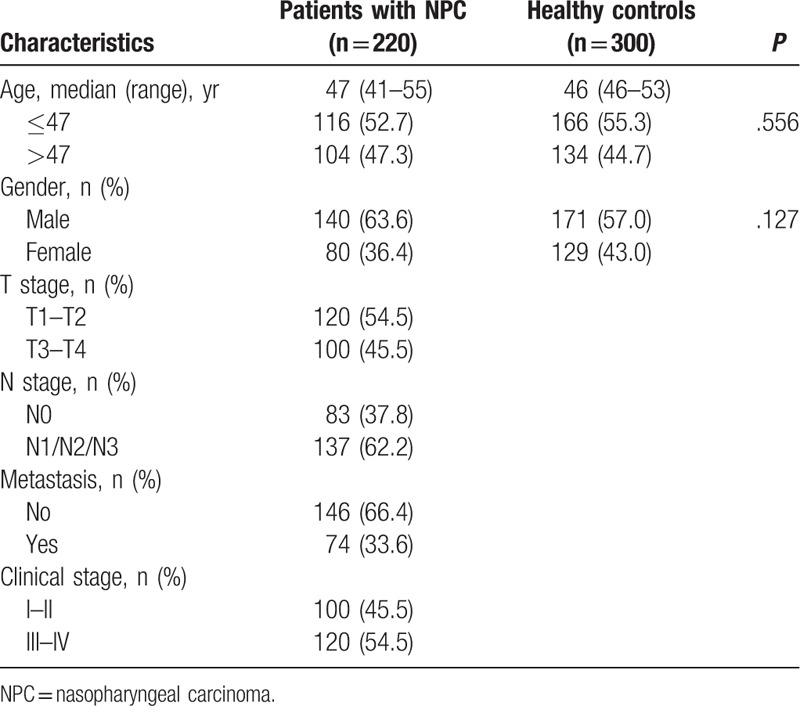
Our study was approved by the ethics committee of the Youjiang Medical University for Nationalities Affiliated Hospital (no: yyfyll2016005), and all participants provided written informed consent. The study population included 220 patients with NPC and 300 healthy controls selected between May 2014 and February 2017 at the Affiliated Hospital of Youjiang Medical University for Nationalities (Guangxi, China). The patients were included if they were pathologically diagnosed with NPC and had not received any chemotherapy or chemotherapy. All clinical data were extracted from medical records, including gender, age, tumor location, and metastasis. Patients with a history of familial cancer were excluded. Control subjects were selected randomly from routine health examination in the same hospital and matched to each case by age and gender. The controls were eligible if they had no history of cancer, chronic rhinitis, nasosinusitis, and allergic rhinitis. Detailed characteristics are shown in Table 1. The study protocol adhered to the ethical principles for medical research of the Helsinki Declaration.
Table 1.
Characteristics of patients with NPC and healthy controls.
2.2. Determination of the RTN4 genotype
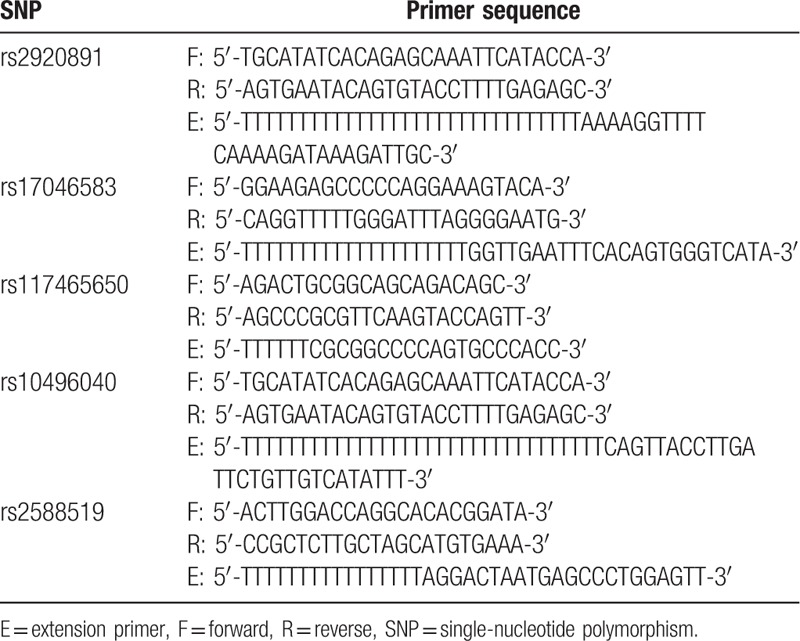
Genomic DNA was extracted from the peripheral blood using a whole-blood genomic DNA isolation kit (Tiangen Inc, Beijing, China). RTN4 polymorphisms were genotyped using the multiple single-nucleotide primer extension technique. The primers were designed according to the GenBank sequences (Table 2). To confirm the genotyping results, 10% of the samples were randomly selected and analyzed repeatedly, and the results were 100% consistent.
Table 2.
Primer sequences for genotyping RTN4 SNPs.
2.3. Plasma RTN4 quantification
Blood samples were collected from the participants and centrifuged at 1000g for 15 minutes, and plasma samples were kept at −80°C until analysis. The level of plasma RTN4 was measured using enzyme-linked immunosorbent assay kits (LifeSpan BioSciences, Inc, Seattle, WA). The detection range was 0.156 to 10 ng/mL.
2.4. Statistical analysis
Demographic and clinical data were analyzed using the Pearson Chi-squared test. The Hardy–Weinberg equilibrium (HWE) was assessed by Chi-squared test. The genotype and allele frequencies of RTN4 gene polymorphisms were compared between patients with NPC and controls by Chi-squared test or Fisher exact test. Genotypic association and haplotype analyses were conducted using SNPstats (Sole et al, 2006). After logistic regression analyses, the association of RTN4 polymorphisms with NPC risk was assessed using odds ratio (OR) and 95% confidence interval (CI). All statistical analyses were performed using SPSS 17.0 statistical software (SPSS Inc, Chicago, IL). A P-value <.05 was considered statistically significant.
3. Results
3.1. Participant general information
The general information of participants is shown in Table 1. The median age of the controls was 46.0 years (range, 46.0–53.0 years). The median age of the patients was 47.0 years (range, 41.0–55.0 years). The patients and controls showed no statistical differences in age (P = .556) and gender (P = .127).
3.2. Allele and genotype frequencies of the RTN4 gene
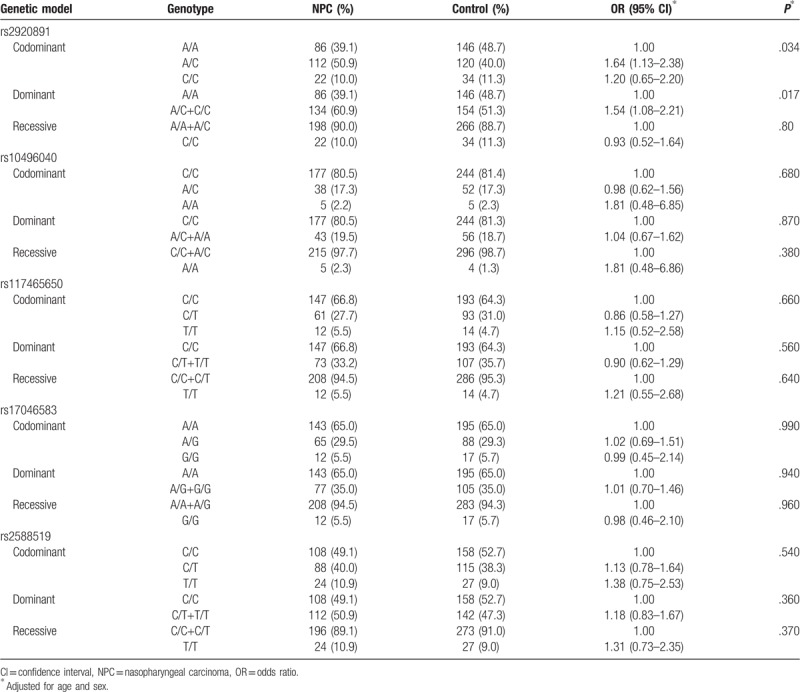
Five SNPs were successfully genotyped in 220 patients with NPC and 300 healthy controls. The allele and genotype frequencies of RTN4 gene polymorphisms in the patients and controls are shown in Tables 3 and 4. The genotype frequencies of the 5 polymorphisms in the patients and controls were in HWE (P > .05). The allele frequencies of the polymorphisms were not significantly different between the patients and controls (P > .05). For the rs2920891 polymorphism, a significantly higher NPC risk was associated with the A/C+C/C genotype compared with the A/A genotype in a dominant model (P = .017, OR = 1.54, 95% CI = 1.08–2.21). In addition, a considerably higher NPC risk was associated with the A/C genotype in a codominant model (P = .034, OR = 1.64, 95% CI = 1.13–2.38). Nevertheless, no associations were found between NPC risk and the rs10496040, rs117465650, rs17046583, and rs2588519 polymorphisms in genotypic association analyses (P > .05).
Table 3.
Allele frequencies of 5 RTN4 SNPs in patients with NPC and healthy controls.
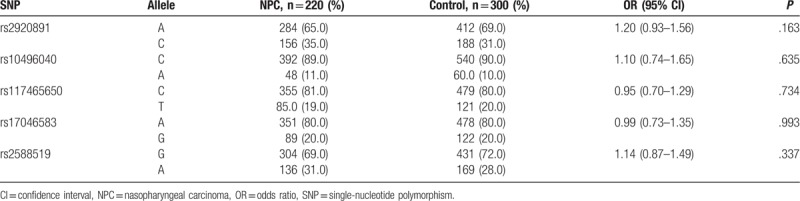
Table 4.
Genotype frequencies of 5 RTN4 SNPs in patients with NPC and healthy controls.
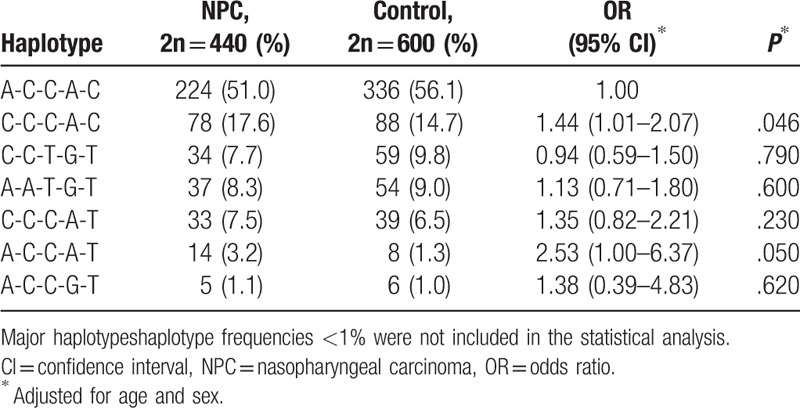
3.3. Haplotype analysis of the RTN4 gene
Haplotype analysis of RTN4 polymorphisms was performed using SNPstats software. As shown in Table 5, the major haplotype (A-C-C-A-G) accounted for 51.0% and 56.1% of the distribution in patients with NPC and healthy controls, respectively. Adjusted for age and sex, the C-C-C-A-G haplotype was associated with an increased risk of NPC in the patients compared with the controls (P = .046, OR = 1.44, 95% CI = 1.01–2.07).
Table 5.
Haplotype distribution in patients with NPC and healthy controls.
3.4. Association between clinical characteristics and RTN4 polymorphisms
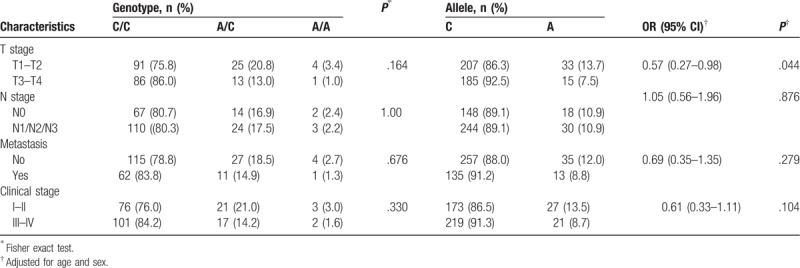
We further performed stratified analysis to determine the genotype and allele distribution of RTN4 polymorphisms in patients with different stages of NPC (T stage, N stage, metastasis, and clinical stage) (Tables 6 and 7). The results showed that the rs2920891 and rs10496040 polymorphisms were associated with clinical stage (P = .027) and T stage (P = .044), respectively. However, other polymorphisms were not associated with clinical characteristics (data not shown).
Table 6.
Relationship between patient characteristics and the genotype and allele distribution of rs2920891.
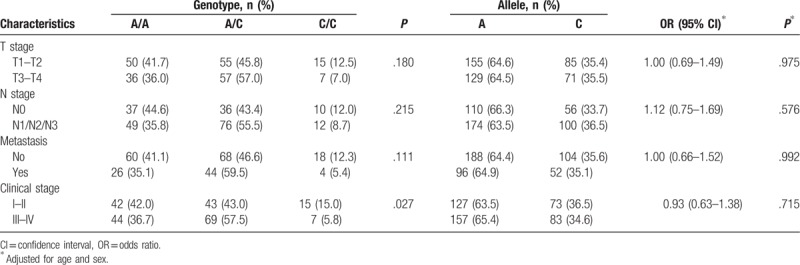
Table 7.
Relationship between patient characteristics and the genotype and allele distribution of rs10496040.
3.5. Association between RTN4 gene polymorphisms and plasma RTN4 levels
The plasma RTN4 levels of patients with NPC and healthy controls were measured (Fig. 1A). The plasma level of RTN4 was significantly higher in patients with NPC than in the controls (P < .001), and the median plasma RTN4 level was 2.05 ng/mL (range 1.54–2.24 ng/mL) in patients with NPC (n = 220) and 0.83 ng/mL (range 0.49–1.17 ng/mL) in the controls (n = 300). We also investigated the correlation between RTN4 gene polymorphisms and plasma RTN4 levels. Notably, the rs2920891 polymorphism was significantly associated with plasma RTN4 level in patients with NPC. Patients with NPC carrying the rs2920891 A/C+C/C genotype had a higher RTN4 level than those carrying the A/A genotype (Fig. 1B; P < .001). However, there was no relationship between plasma RTN4 and rs17046583, rs117465650, rs10496040, and rs2588519 (P > .05, data not shown).
Figure 1.
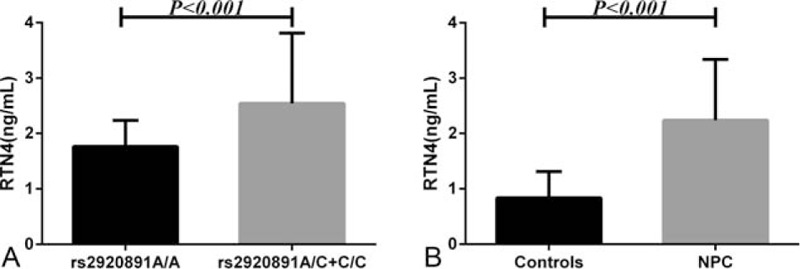
Enzyme-linked immunosorbent assay analysis of RTN4 expression. (A) RTN4 plasma level in patients with NPC (n = 220) and healthy controls (n = 300). (B) RTN4 plasma level in patients carrying the rs2920891 A/A genotype (n = 86) and patients carrying the A/C+C/C genotype (n = 134).
4. Discussion
This is the 1st study to investigate the relationship between RTN4 gene polymorphism and individual susceptibility to NPC in a Chinese population and the association of RTN4 gene polymorphism with the plasma level of RTN4 and clinical features. We demonstrated that the rs2920891 polymorphism was associated with an increased risk of NPC. The increased risk was observed in a dominant model (A/C+C/C genotype, P = .017, OR = 1.54, 95% CI = 1.08–2.21) and a codominant model (A/C genotype, P = .034, OR = 1.64, 95% CI = 1.13–2.38) and associated with the C-C-C-A-G haplotype (P = .046, OR = 1.44, 95% CI = 1.01–2.07). Notably, the rs2920891 and rs10496040 polymorphisms were associated with clinical stage (P = .027) and T stage (P = .044), respectively. Moreover, patients with NPC carrying the rs2920891 A/C+C/C genotype had a higher level of RTN4. These findings indicated that the rs2920891 polymorphism may contribute to the susceptibility to NPC.
Several studies have demonstrated the association of RTN4 gene polymorphisms with the risk of various tumors. Chen et al[14] observed that the rs34917480 polymorphism in the RTN4 gene could contribute to the risk of nonsmall-cell lung cancer in a Chinese population. Furthermore, sex, age, and environmental exposure could influence the carcinogenic effects of rs34917480. A similar result was observed in a study of uterine leiomyoma (UL), suggesting a high UL risk among homozygous carriers of rs34917480.[18] Moreover, Zhang et al[18] demonstrated statistically significant differences in the TATC and CAA (insertion/deletion) polymorphisms in the RTN4 gene between patients with cervical squamous cell carcinoma (CSCC) and control subjects, and the polymorphisms may be associated with the advanced clinical stage of CSCC. These studies investigated the association of SNPs with several cancers, and the results were consistent. Here, we reported for the 1st time that the rs2920891 polymorphism in RTN4 contributed to an elevated risk of NPC in Chinese patients and was related to the abnormal expression of RTN4. We believe that rs2920891, located in intron 1 of the RTN4 gene, may inhibit transcriptional activity. Moreover, recent studies have established that introns can serve as important gene regulatory structures with various functional effects, such as RNA editing, noncoding RNA, and transacting elements.[19,20] We do not know of studies that have investigated the effect of RTN4 gene polymorphisms on the risk of NPC; hence, we could not compare this study with other similar studies.
There were some potential limitations in our study. First, the sample size of this study was not large enough. Second, selection bias in the hospital-based case–control study was inevitable. However, deviation from the HWE was not detected in all SNPs, indicating minimal probability. Nevertheless, our findings should be interpreted with caution. Further studies with larger samples and ethnically different populations could help elucidate the relationship between these polymorphisms and the susceptibility to NPC.
5. Conclusion
Our results demonstrated that the rs2920891 polymorphism may contribute to the susceptibility to NPC. To our knowledge, this is the 1st study to investigate the effect of RTN4 SNPs on the risk of NPC in a Chinese population and evaluate the difference in RTN4 expression between patients with NPC and healthy controls. Nevertheless, to determine the exact mechanism of RTN4 in NPC, further studies with larger samples are needed.
Author contributions
Conceptualization: Junli Wang, Renguang Tang.
Data curation: Fenglian Yang, Shixian Yang, Jin Liu, Xiaoxia Pang, Feng Shi, Haimei Qin.
Formal analysis: Shixian Yang.
Investigation: Fenglian Yang, Shixian Yang, Jin Liu, Xiaoxia Pang, Feng Shi, Haimei Qin.
Supervision: Junli Wang.
Writing – original draft: Fenglian Yang.
Writing – review & editing: Junli Wang, Renguang Tang.
Junli Wang orcid: 0000-0002-2340-3297.
Footnotes
Abbreviations: CSCC = cervical squamous cell carcinoma, CI = confidence interval, ER = endoplasmic reticulum, HWE = Hardy–Weinberg equilibrium, NPC = nasopharyngeal carcinoma, OR = odds ratio, RTN4 = reticulon-4, SNP = single-nucleotide polymorphism, UL = uterine leiomyoma.
How to cite this article: Yang F, Yang S, Liu J, Pang X, Shi F, Qin H, Wang J, Tang R. Impact of RTN4 gene polymorphism and its plasma level on susceptibility to nasopharyngeal carcinoma: a case–control study. Medicine. 2019;98:47(e17831).
FY and SY contributed equally to the work.
This study was approved by the ethics committee of the Youjiang Medical University for Nationalities Affiliated Hospital (no: yyfyll2016005). All participants provided written informed consent. All participants gave consent for publication.
This study was supported by grants from the National Natural Science Foundation of China (no: 81560461), the medical high-level talent training plan and thousands of young- and middle-aged backbone teachers cultivation plan of Guangxi Province, China, and the 2018 to 2020 professional and experimental practice teaching base construction projects of Guangxi Province, China.
The authors have no conflicts of interest to disclose.
References
- [1].Chua MLK, Wee JTS, Hui EP, et al. Nasopharyngeal carcinoma. Lancet 2016;387:1012–24. [DOI] [PubMed] [Google Scholar]
- [2].Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma--review of the molecular mechanisms of tumorigenesis. Head Neck 2008;30:946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Turunen A, Rautava J, Grenman R, et al. Epstein-Barr virus (EBV)-encoded small RNAs (EBERs) associated with poor prognosis of head and neck carcinomas. Oncotarget 2017;8:27328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2006;15:1765–77. [DOI] [PubMed] [Google Scholar]
- [5].Yang J, Yu L, Bi AD, et al. Assignment of the human reticulon 4 gene (RTN4) to chromosome 2p14-->2p13 by radiation hybrid mapping. Cytogenet Cell Genet 2000;88:101–2. [DOI] [PubMed] [Google Scholar]
- [6].Oertle T, Huber C, van der Putten H, et al. Genomic structure and functional characterisation of the promoters of human and mouse nogo/rtn4. J Mol Biol 2003;325:299–323. [DOI] [PubMed] [Google Scholar]
- [7].Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol 2007;8:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Su Z, Cao L, Zhu Y, et al. Nogo enhances the adhesion of olfactory ensheathing cells and inhibits their migration. J Cell Sci 2007;120(Pt 11):1877–87. [DOI] [PubMed] [Google Scholar]
- [9].Miao RQ, Gao Y, Harrison KD, et al. Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc Natl Acad Sci U S A 2006;103:10997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kondo Y, Jadlowiec CC, Muto A, et al. The Nogo-B-PirB axis controls macrophage-mediated vascular remodeling. PLoS One 2013;8:e81019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kritz AB, Yu J, Wright PL, et al. In vivo modulation of Nogo-B attenuates neointima formation. Mol Ther 2008;16:1798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tagami S, Eguchi Y, Kinoshita M, et al. A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene 2000;19:5736–46. [DOI] [PubMed] [Google Scholar]
- [13].Kuang E, Wan Q, Li X, et al. ER stress triggers apoptosis induced by Nogo-B/ASY overexpression. Exp Cell Res 2006;312:1983–8. [DOI] [PubMed] [Google Scholar]
- [14].Chen Y, Tang X, Cao X, et al. Human Nogo-C overexpression induces HEK293 cell apoptosis via a mechanism that involves JNK-c-Jun pathway. Biochem Biophys Res Commun 2006;348:923–8. [DOI] [PubMed] [Google Scholar]
- [15].Novak G, Kim D, Seeman P, et al. Schizophrenia and Nogo: elevated mRNA in cortex, and high prevalence of a homozygous CAA insert. Brain Res Mol Brain Res 2002;107:183–9. [DOI] [PubMed] [Google Scholar]
- [16].Chen Y, Zhou B, Li H, et al. Analysis of RTN4 3’UTR insertion/deletion polymorphisms in ventricular septal defect in a Chinese Han population. DNA Cell Biol 2011;30:323–7. [DOI] [PubMed] [Google Scholar]
- [17].Shi S, Zhou B, Wang Y, et al. Genetic variation in RTN4 3’-UTR and susceptibility to cervical squamous cell carcinoma. DNA Cell Biol 2012;31:1088–94. [DOI] [PubMed] [Google Scholar]
- [18].Zhang K, Bai P, Shi S, et al. Association of genetic variations in RTN4 3’-UTR with risk of uterine leiomyomas. Pathol Oncol Res 2013;19:475–9. [DOI] [PubMed] [Google Scholar]
- [19].Jacquier A. Group II introns: elaborate ribozymes. Biochimie 1996;78:474–87. [DOI] [PubMed] [Google Scholar]
- [20].Qaddourah RH, Magdoud K, Saldanha FL, et al. IL-10 gene promoter and intron polymorphisms and changes in IL-10 secretion in women with idiopathic recurrent miscarriage. Hum Reprod 2014;29:1025–34. [DOI] [PubMed] [Google Scholar]


