Supplemental Digital Content is Available in the Text.
Keywords: Spinal cord stimulation, Chemotherapy-induced peripheral neuropathy, Neuropathic pain, RNA-sequencing, Rat
Abstract
Introduction:
Paclitaxel-induced peripheral neuropathy (PIPN) is a common dose-limiting side effect of this cancer treatment drug. Spinal cord stimulation (SCS) has demonstrated efficacy for attenuating some neuropathic pain conditions.
Objective:
We aim to examine the inhibitory effect of SCS on the development of PIPN pain and changes of gene expression in the spinal cord in male rats after SCS.
Methods:
We examined whether traditional SCS (50 Hz, 6–8 h/session daily for 14 consecutive days) administered during paclitaxel treatment (1.5 mg/kg, i.p.) attenuates PIPN-related pain behavior. After SCS treatment, we performed RNA-seq of the lumbar spinal cord to examine which genes are differentially expressed after PIPN with and without SCS.
Results:
Compared to rats treated with paclitaxel alone (n = 7) or sham SCS (n = 6), SCS treatment (n = 11) significantly inhibited the development of paclitaxel-induced mechanical and cold hypersensitivity, without altering open-field exploratory behavior. RNA-seq showed that SCS induced upregulation of 836 genes and downregulation of 230 genes in the spinal cord of paclitaxel-treated rats (n = 3) as compared to sham SCS (n = 5). Spinal cord stimulation upregulated immune responses in paclitaxel-treated rats, including transcription of astrocyte- and microglial-related genes, but repressed transcription of multiple gene networks associated with synapse transmission, neuron projection development, γ-aminobutyric acid reuptake, and neuronal plasticity.
Conclusion:
Our findings suggest that traditional SCS may attenuate the development of pain-related behaviors in PIPN rats, possibly by causing aggregate inhibition of synaptic plasticity through upregulation and downregulation of gene networks in the spinal cord.
1. Introduction
Chemotherapy-induced peripheral neuropathy (CIPN) is a dose-limiting neurotoxic effect of many chemotherapeutic agents.21 Chemotherapy-induced peripheral neuropathy presents in patients as stocking-and-glove distribution paresthesia and debilitating pain that often leads to dose reduction during treatment. Advances in treatments have contributed to a 23% decrease in cancer mortality, but thereby also increased the number of cancer survivors who may experience long-lasting side effects.49 Chemotherapy-induced peripheral neuropathy affects approximately 30% of cancer patients at 6 months after treatment cessation.48 Owing to a lack of objective diagnostic criteria for CIPN, the prevalence of CIPN is likely much underestimated.
Although the impact of opioids on cancer progression is contentious, their frequent use for cancer-related pain may increase cancer recurrence and severe adverse events, abuse, and accidental death from overdose.8 A number of experimental modalities (acupuncture and exercise) and agents have demonstrated very limited efficacy for CIPN pain.38 Therefore, alternative treatments for CIPN pain are urgently needed. In particular, no available treatment can prevent incapacitating CIPN pain.21 One potential option is spinal cord stimulation (SCS), which has shown efficacy for treating refractory neuropathic pain conditions.41 Traditional SCS primarily targets the dorsal columns and activates both spinal and supraspinal neurophysiologic mechanisms to induce pain inhibition.33,51 Intriguingly, recent case reports suggested that SCS might be beneficial for alleviating established CIPN pain.5,43 Nevertheless, little is known about the efficacy or mechanisms of SCS for CIPN pain, especially whether it can be used for prevention.
When neuropathic pain has established, prolonged central sensitization may correlate with poor response to SCS.52 However, SCS applied early after injury may be more effective than later treatment, as shown in animal models and in the clinic.28,59 Furthermore, traditional SCS can acutely inhibit both short-form neuronal sensitization (wind-up) and long-term potentiation (LTP) in dorsal horn neurons, important spinal neurophysiologic mechanisms that may underlie chronic pain development.17,60,61 Accordingly, we postulate that SCS introduced early during the initiation of chemotherapy may attenuate the development of CIPN pain.
Paclitaxel is a chemotherapeutic agent commonly used to treat solid tumors (eg, ovarian, breast, and lung cancer). Its major dose-limiting toxicity is debilitating painful peripheral neuropathy, but the underlying mechanisms are not fully known.48 Previous studies focused on single nucleotide polymorphisms associated with paclitaxel-induced peripheral neuropathy (PIPN).3,29 Yet, no study has evaluated broad changes in spinal gene expression with any form of CIPN. In addition to attenuating neuronal hyperexcitability, recent studies unraveled that SCS may induce broad changes in gene expression in the spinal cord after nerve injury.55,57 To develop new therapies for CIPN pain, it is important to identify gene networks and molecular pathways that are altered by CIPN, as well as those modulated by SCS. Accordingly, we examined PIPN-related pain behavior and conducted RNA sequencing (RNA-seq) in male rats that received repetitive SCS during the development of PIPN. We hypothesized that SCS during chemotherapy treatment would broadly alter long-term gene expression in the spinal cord and attenuate PIPN pain development.
2. Materials and methods
2.1. Animals
Adult male rats (n = 34; starting weight 350–400 g; Envigo, Indianapolis, IN) were allowed to acclimate for a minimum of 48 hours before any experimental procedure. The rats were housed separately after SCS electrode implantation and were given access to food and water ad libitum. All procedures involving animals were reviewed and approved by the Johns Hopkins Animal Care and Use Committee and were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
2.2. Behavioral testing
2.2.1. Mechanical sensitivity test
All behavioral assays were conducted by an observer who was blinded to drug or SCS condition.
Animals were placed in individual Plexiglas cages with a wire mesh floor and allowed to acclimate for 1 hour. Mechanical hypersensitivity was measured with the up-down method by applying von Frey monofilaments to the midplantar surface of the hind paws as previously described.50 A series of von Frey monofilaments (0.38, 0.57, 1.23, 1.83, 3.66, 5.93, 9.13, and 13.1 g) was applied between the footpads on the plantar surface of each hind paw for 4 to 6 seconds. Monofilaments with increasing force were applied until a positive response was observed (eg, abrupt paw withdrawal, shaking, and licking). When a positive response was observed, the monofilament with the next lower force was applied. If a negative response was observed, the next higher force was used. The test continued until (1) 5 filament applications had been completed after a positive test was observed, or (2) the upper or lower end of the von Frey monofilament set was reached. The paw withdrawal threshold (PWT) was determined according to the formula provided by Dixon.12
2.2.2. Cold sensitivity test
The cold plantar assay was used to evaluate noxious cold sensitivity.4 Briefly, animals were placed individually into clear acrylic containers separated by white opaque dividers on a 3/16″ glass plate and allowed to acclimate for 20 minutes before being tested. Powdered dry ice in a cutoff 10-mL syringe was held against the glass beneath the hind paw until the paw was withdrawn. Applications were repeated at 5-minute intervals, alternating paws, for a total of 4 trials. The mean paw withdrawal latency (PWL) was calculated. To avoid potential tissue damage, the cutoff time was set to 45 seconds.
2.2.3. Open field test
The open-field test was used to assess the effect of CIPN and SCS on spontaneous exploration and locomotor activity as described in a previous study.58 Rats were placed in an open-field chamber (73 × 45-cm rectangular plastic box with a wall height of 33 cm) for 10 minutes. We analyzed the parameters of total distance traveled, number of center crossings, mean travel speed, and number of entries at the border and internal periphery in video recordings using SMART 3 software (Panlab Harvard Apparatus, Barcelona, Spain).
2.3. Drug treatment
Paclitaxel is usually administered at a dose of 1 to 2 mg/kg on alternate days for inducing the CIPN model.7,22 At these doses, animal health is not debilitated and normal weight gain is maintained; however, animals display robust hypersensitivity to mechanical and cold stimuli.7,22 After habituation to the test environments and baseline measurements of pain sensitivity, all rats were injected intraperitoneally (i.p.) on days 1, 3, 5, and 7 with either vehicle (dimethyl sulfoxide, 70% ethanol, and 0.9% saline) or paclitaxel (1.5 mg/kg). The injection concentration was 1.5 mg/mL in a mixture containing dimethyl sulfoxide, 70% ethanol, and 0.9% saline (T7402; Sigma-Aldrich, St. Louis, MO, Fig. 1A). The cumulative paclitaxel dose was 6 mg/kg. When an injection was to be given on the same day as behavioral testing, all rats were injected after the measurements were taken, but before SCS treatment for the day.
Figure 1.
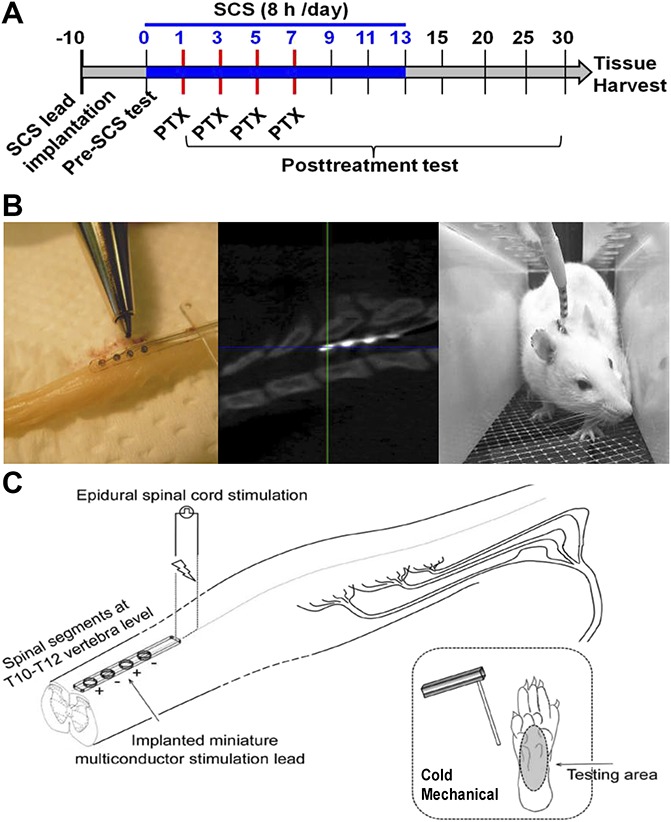
Experimental protocol and placement of SCS. (A) Experimental protocol. Rats in the SCS + paclitaxel (PTX) group (n = 11) received the same SCS (blue bar, 50 Hz, 80% motor threshold [MoT], 0.2 ms, constant current, 6 to 8 hours/session) from day 0 to day 13. Motor thresholds were measured at 4-Hz stimulation (0.2 ms). Paclitaxel (1.5 mg/kg) was administered on days 1, 3, 5, and 7 to the SCS + paclitaxel (n = 11) and paclitaxel (n = 13) rats. (B) Left: the miniature SCS lead (Medtronic); Middle: CT scan of a rat shows an implanted quadripolar lead at T10-T12 vertebral level (∼T13-L1 spinal cord). Right: the other end of the lead is connected to an external stimulator. (C) Schematic diagram of the experimental setup for SCS in vivo. CIPN, chemotherapy-induced peripheral neuropathy; SCS, spinal cord stimulation.
2.4. Electrode placement and spinal cord stimulation treatment
A sterile, quadripolar SCS electrode (Medtronic, Inc, Minneapolis, MN) that mimics clinical SCS was placed at the dorsal spinal cord of each rat (Fig. 1B, C), as described in previous studies and was validated in rats.50,62 Briefly, with rats under isoflurane anesthesia, we performed a laminectomy at the T13 vertebral level and inserted the electrode epidurally in the rostral direction. The position of the electrode was adjusted so that the contacts were at the T13-L1 spinal cord level, which corresponds to the lower thoracic-upper lumbar region (Fig. 1B, C). The paddle electrode was sutured to muscle to secure its position, and the proximal end was tunneled subcutaneously until it exited the animal at the top of its head for later connection to an external neurostimulator (Model 2100; A-M Systems, Sequim, WA). Rats that exhibited signs of spinal cord injury, poor lead placement, or damaged electrodes were euthanized and excluded from subsequent studies. In twin-pairs SCS (Fig. 1C), the first and third contacts of the lead from the rostral direction were set as anodes (+), and the second and fourth were set as cathodes (−). Traditional SCS (50 Hz, 0.2 ms, constant current, 6–8 h/session, daily) was applied at an intensity that activated low-threshold A fibers (80% motor threshold [MoT]), as described in previous studies.50,62 Before SCS each day, the MoT for each animal was determined by slowly increasing the current amplitude from zero, until muscle contraction in the midlower trunk or hind limbs was observed in response to 4-Hz stimulation at 0.2-ms pulse width.
2.5. RNA-seq
2.5.1. RNA isolation
Frozen spinal cord tissues were submerged in Trizol and homogenized in a Dounce homogenizer. Total RNA was extracted from the aqueous phase with the Quick-RNA Plus kit (Zymo, Irvine, CA) according to manufacturer instructions with on-column DNase I digestion. RNA quantity was measured by the Qubit RNA BR Assay Kit (ThermoScientific, Waltham, MA), and RNA integrity was assessed by the Bioanalyzer RNA Nano Eukaryote kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
2.5.2. RNA-seq library construction and sequencing
We used 1 μg of total RNA per sample to construct sequencing libraries (n = 1 rat/sample) as in our previous study.55 Strand-specific RNA libraries were prepared by using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) after poly(A) selection by the NEBNext poly(A) mRNA Isolation Module (New England Biolabs) according to the manufacturer's instructions. Samples were barcoded by using the recommended NEBNext Multiplex Oligos (New England Biolabs). Size range and quality of libraries were verified on the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA-seq libraries were quantified by quantitative real-time polymerase chain reaction (qPCR) with the NEBNext Library Quant Kit for Illumina (New England Biolabs). Each library was normalized to 2 nM and pooled in equimolar concentrations. Paired-end × 100 sequencing was performed on an Illumina HiSeq 4000 (Illumina, San Diego, CA). Libraries were pooled and sequenced in 3 lanes of one HiSeq 4000 flow cell to an average depth of 36.7 million reads per sample.
2.6. Validation of RNA-seq by quantitative real-time polymerase chain reaction
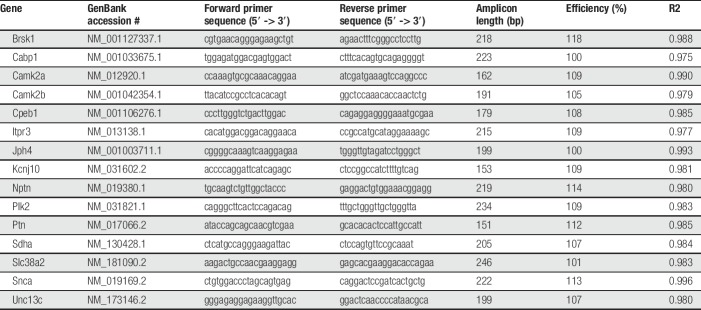
We used qPCR to confirm the relationship of gene expression trends in selected genes of paclitaxel + Sham SCS and paclitaxel + SCS groups. The first-strand cDNA synthesis from 2 µg total RNA in a 20-μL reaction was performed using random hexamer primers and the SuperScript IV Reverse Transcriptase (ThermoFisher Scientific) according to manufacturer's instructions. cDNA was diluted 1:10 with nuclease-free water and stored at −20°C. mRNA sequence for each gene was retrieved from NCBI. Forward and reverse primers for each gene were designed using the PrimerQuest Tool (IDT, Coralville, Iowa) to span one or more introns. Primers were obtained through Integrated DNA Technologies (IDT) and sequences are provided in Table 1. Each 20-μL qPCR reaction consisted of 10ul 2X Power SYBR Green Master Mix (Thermo Fisher Scientific), 200 nM each forward and reverse primer, and 2-μL diluted cDNA. Polymerase chain reaction of each target was performed using the 7900HT Fast Real-Time PCR system (Applied Biosystems, Waltham, MA) with the following thermocycling conditions: initial denaturation at 95°C for 10 minutes followed by 40 cycles of 95°C for 10 seconds and 60°C for 60 seconds. Each sample was run in triplicate for each target gene. Nuclease-free water was included in each plate as a no-template control. Polymerase chain reaction efficiencies of each primer set were determined using the slope of standard curves constructed with Cq values obtained from 5-fold serial dilutions of pooled cDNA from tissue of each group (eg, paclitaxel + Sham SCS, and paclitaxel + SCS). The efficiency was calculated using the formula: E = 10 − 1/slope. Dissociation curve analysis was used to identify amplification of nonspecific products including primer dimers. Sdha was selected as the endogenous control for relative gene expression calculations of each target gene.
Table 1.
List of primers used for qPCR validation.
2.7. Experimental design
Rats were randomized to 1 of 4 groups: (1) no treatment (Naive, n = 6), (2) paclitaxel only (paclitaxel; n = 7), (3) paclitaxel with electrode placement (Sham SCS + paclitaxel; n = 6), and (4) paclitaxel with active SCS (SCS + paclitaxel; n = 11). Pain behaviors were assessed in all animals before any experimental procedure (day 0). All behavioral assays were conducted by an observer who was blinded to treatment condition. Rats randomized to the Sham SCS + paclitaxel and SCS + paclitaxel groups were implanted with an SCS electrode on Day −10, and the SCS + paclitaxel group received SCS (50 Hz, 80% MoT, 0.2 ms, constant current, 6–8 h/session per day for 14 consecutive days on days 0–13 (Fig. 1A). Sham SCS occurred in the same environment with the leads connected to the stimulator without transmission of any electric power. Seventeen days after the last SCS treatment (ie, day 30), all animals were euthanized by overdose of isoflurane. The lumbar spinal cord (L3–L6) was harvested and flash frozen on dry ice until RNA was extracted. As RNA-seq and qPCR are exquisitely sensitive tools for measuring gene expression and variations may be caused by nonuniform separation of the dorsal and ventral spinal cord, we chose to examine the whole spinal cord segment in these experiments.
2.8. Statistical analysis
We analyzed animal behavior data using GraphPad Prism 7.0 software and compared data from mechanical allodynia and cold allodynia tests using two-way mixed-model analysis of variance (ANOVA) followed by Bonferroni post hoc test. Area under the curve was calculated by the trapezoid rule and compared with a two-way mixed-model ANOVA followed by Bonferroni's post hoc test. The data for the open-field test assays, total distance traveled, and number of center crossings were analyzed using Kruskal–Wallis one-way ANOVA followed by Dunn post hoc test. Body weight data were compared by using a two-way mixed-model ANOVA followed by Bonferroni post hoc test. The criterion level for determination of significance was set at P < 0.05. No outlier data were removed. Animal behavioral data are presented as mean ± SD.
Sequencing reads were aligned to annotated RefSeq genes of the rat reference genome (rn6) using HISAT2, filtered to remove ribosomal RNA, and visualized using the Integrative Genomics Viewer.26,45 A gene count matrix that contained raw transcript counts for uniquely mapped reads to each annotated gene was generated by using the featureCounts function of Subread.32 This count matrix was then filtered for no count genes so that only those genes with >0 reads in at least one sample were retained. To identify genes that were differentially regulated after nerve injury, we normalized and log2 transformed transcript counts using the default normalization procedures in DESeq2.37 This analysis identified differentially expressed genes between (1) Sham SCS + paclitaxel and SCS + paclitaxel, and between (2) Sham SCS + paclitaxel and Naive groups. All downstream analyses of RNA-seq data were performed on data obtained from DESeq2, which implements the procedures of Benjamini and Hochberg to adjust the P-value for multiple comparisons.32 Unless otherwise stated, an adjusted P-value (ie, false discovery rate [FDR]) < 0.05 was used to define differential expression in pairwise comparisons. We then included genes differentially expressed between the Sham SCS + paclitaxel and SCS + paclitaxel groups in gene ontology (GO) analysis to infer their functional roles and relationships. Gene ontology analysis for enriched GO biological processes in each set of differentially enriched genes identified by DESeq2 was performed with ToppGeneSuite (https://toppgene.cchmc.org).6 The International Union of Basic and Clinical Pharmacology (IUPHAR) database (http://www.guidetopharmacology.org) was used to assign categories to gene products.18 Raw and processed sequencing data are available in the NCBI GEO database under accession #GSE132491.
For qPCR, default settings were used to define quantification cycle (Cq) values using SDS software version 2.4.1 (Applied Biosystems). The Cq values were averaged over 3 replicates. If the SD of this average was >0.20, the outlying replicate was removed and the Cq was averaged over the 2 remaining technical replicates. The 2−ΔΔCT method was used to convert Cq values into relative gene expression for each gene.36
3. Results
3.1. Overall health status of the animals
All rats survived until the end of the study with the exception of 3 rats from the SCS + paclitaxel group and one rat from the Sham SCS + paclitaxel group. These rats were all noted to have had spinal cord injury, poor lead placement, or damaged electrodes, and were euthanized before the start of the experiments; thus, there are no data from these animals. All rats used in analysis survived until the end of the study and seemed healthy.
3.2. Spinal cord stimulation attenuated the development of mechanical hypersensitivity associated with paclitaxel-induced peripheral neuropathy
All rats in the SCS + paclitaxel (n = 11), Sham SCS + paclitaxel (n = 6), and paclitaxel only (n = 7) groups developed mechanical hypersensitivity by 1 week after the first paclitaxel dose, as indicated by the significant decrease in PWT from baseline (Fig. 2A). The Sham SCS + paclitaxel (n = 6) and the paclitaxel only (n = 7) groups showed no statistical differences at any time point in behavioral assays (days 0, 1, 3, 5, 7, 9, 11, 13, 15, 29, and 25; P > 0.05). Therefore, behavior data from these 2 groups were combined to form a larger composite single paclitaxel group (n = 13) for the analysis.
Figure 2.
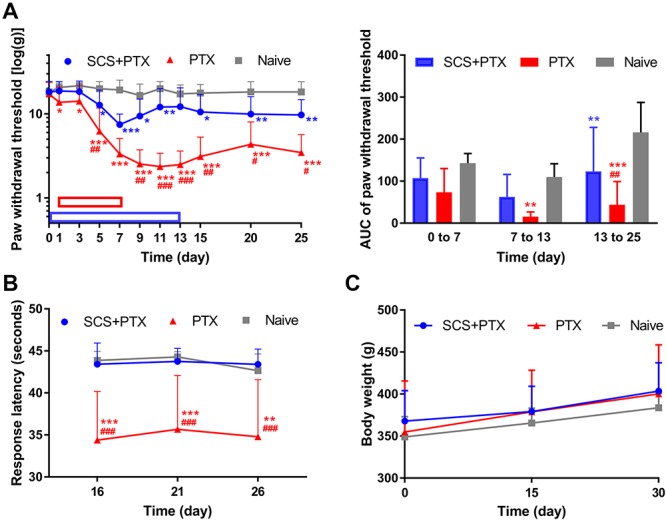
Effects of preemptive SCS on animal pain behavior after paclitaxel treatment. (A) Changes in the paw withdrawal threshold to mechanical stimuli from pre-SCS to day 25 and area under the curve (AUC) for 3 periods (early SCS: days 0–7, late SCS: days 7–13, post SCS: days 13–25). Paclitaxel (PTX; 1.5 mg/kg, i.p.) was injected on days 1, 3, 5, and 7 (red bar; SCS + PTX, n = 11; PTX, n = 13). The SCS + paclitaxel group received paclitaxel treatment and 8 hours of stimulation (50 Hz, 80% MoT)/day from day 0 to day 13 (blue bar). Naive rats (n = 6) received vehicle. (B) Paw withdrawal latency to cold stimulation (dry ice) applied to the plantar side of the hind paw. (C) Body weight. Two-way mixed-model ANOVA followed by Bonferroni post hoc test to compare specific data points. Data are expressed as mean + SD. *P < 0.05, **P < 0.01, **P < 0.001 vs Naive; #P < 0.05, ##P < 0.01; ###P < 0.001 paclitaxel vs SCS + paclitaxel. ANOVA, analysis of variance; MoT, motor threshold; SCS, spinal cord stimulation.
Compared to the naive group, rats that received only paclitaxel showed significant and lasting mechanical hypersensitivity that peaked at day 11 and lasted at least till day 25 (P < 0.001, Fig. 2A). Area under the curve revealed a significant decrease in PWT of rats both during and after paclitaxel treatment, as compared to that in the naive group (days 7–13: P = 0.004; days 13–25: P < 0.001; Fig. 2A). Importantly, SCS treatment significantly inhibited the development of paclitaxel-induced mechanical hypersensitivity (paclitaxel vs SCS + paclitaxel: day 11, P < 0.001; day 13, P < 0.001; day 15, P = 0.002; day 20, P = 0.03; day 25, P = 0.01). Area under the curve also showed that SCS treatment partially prevented the decrease in PWT after paclitaxel treatment (paclitaxel vs SCS + paclitaxel: days 13–25, P = 0.004). Spinal cord stimulation treatment did not completely block PIPN-induced mechanical hypersensitivity because PWTs in the SCS + paclitaxel group remained significantly lower than those of naive rats (day 11, P = 0.009; day 15, P = 0.02; day 20, P = 0.005; day 25, P = 0.004).
3.3. Spinal cord stimulation prevented the development of cold hypersensitivity after paclitaxel-induced peripheral neuropathy
Rats that received paclitaxel exhibited significantly shorter PWL to cold stimulation than did naive rats (day 16, P < 0.001; day 21, P < 0.001; day 26, P = 0.002; Fig. 2B). However, rats that received SCS during paclitaxel treatment did not display cold hypersensitivity, an effect that was sustained for at least 13 days after SCS had ceased (paclitaxel vs SCS + paclitaxel: day 16, P < 0.001; day 21, P < 0.001; day 26, P < 0.001). Strikingly, PWLs did not differ significantly between SCS + paclitaxel and naive rats (day 16, P > 0.99; day 21, P > 0.99; day 26, P > 0.99; Fig. 2B), suggesting a complete block of PIPN-associated cold hypersensitivity by SCS.
Body weight was measured to monitor whether the paclitaxel dosing, lead implantation, or stimulation interfered with normal growth and overall health status. Body weight averaged 367.9 ± 36.1 g (n = 11) for the SCS + paclitaxel group, 354.9 ± 60.8 g (n = 13) for the paclitaxel group, and 349.0 ± 24.1 g (n = 6) for the Naive group at baseline. Animals in all groups continued to gain weight normally, and there was no significant difference in the average weight or the amount of weight gain between groups during the 30-day testing period (days 0, 15, 30: P > 0.05, Fig. 2C).
3.4. Spinal cord stimulation and paclitaxel did not impair locomotor function
In open-field testing, we observed no significant differences in the total distance traveled (Naive vs paclitaxel: P = 0.54; paclitaxel vs SCS + paclitaxel: P > 0.99; Naive vs SCS + paclitaxel: P > 0.99; Fig. 3A, B), number of center crossings (Naive vs paclitaxel: P > 0.99; paclitaxel vs SCS + paclitaxel: P = 0.14; Naive vs SCS + paclitaxel: P > 0.99; Fig. 3C), mean travel speed, or number of entries at the border and internal periphery (data not shown) among the 3 groups. These findings suggest that neither paclitaxel nor SCS significantly affects normal activity level, gross locomotion, or exploration habits compared to those in naive rats.
Figure 3.
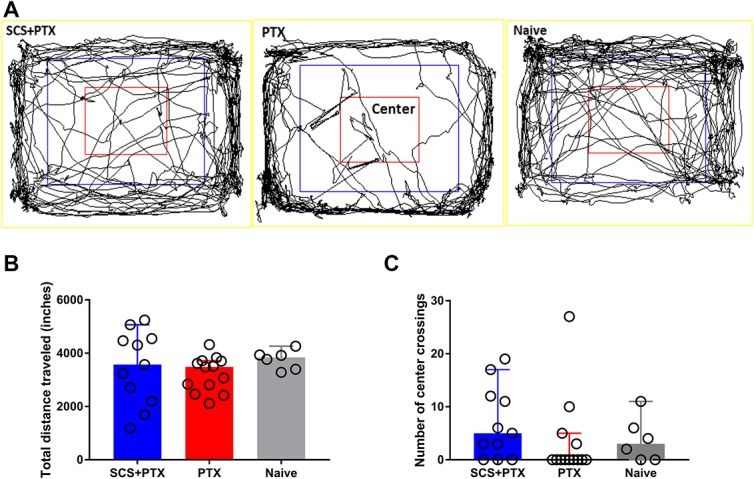
Open-field exploration. (A) Examples of rat exploration activity (10 minutes) in the open-field test at 2 weeks after paclitaxel (PTX) or SCS + paclitaxel. (B and C) Paclitaxel and SCS did not significantly change the total distance traveled (B) or the number of center crossings (C). SCS + paclitaxel, n = 11; paclitaxel, n = 13; Naive, n = 6. Kruskal–Wallis one-way ANOVA followed by Dunn post hoc test. Data are expressed as median + 95% confidence interval. Differences between groups were not statistically significant (P > 0.05). ANOVA, analysis of variance; SCS, spinal cord stimulation.
3.5. Comparisons of gene expression profiles in the spinal cord of naive, sham spinal cord stimulation + paclitaxel, and spinal cord stimulation + paclitaxel rats
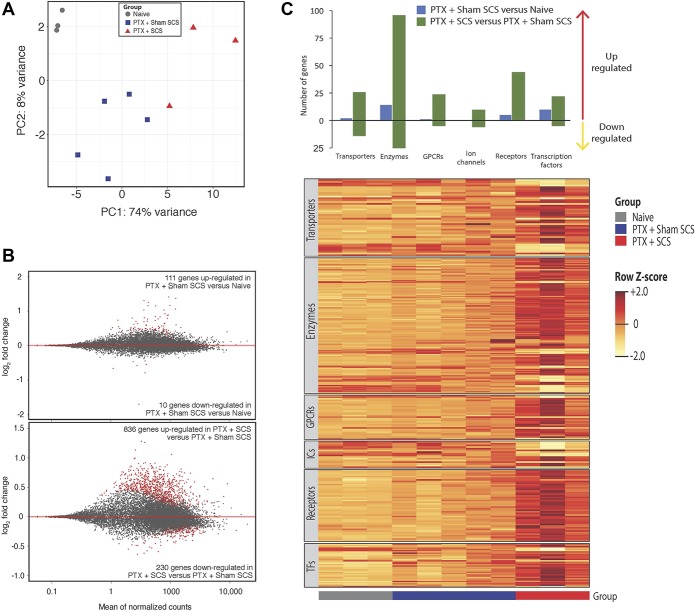
Because spinal segmental mechanisms are important for pain inhibition by SCS, we conducted RNA-seq to examine differential gene expression in the lumbar spinal cord from naive (n = 3), Sham SCS + paclitaxel (n = 5), and SCS + paclitaxel (n = 3) rats. Because our primary goal was to elucidate gene expression changes that may drive pain phenotype differentiation between Sham SCS + paclitaxel and SCS + paclitaxel rats, we did not include the paclitaxel only group in RNA-seq analysis. Because SCS electrodes were placed several weeks before tissue harvest and previous work demonstrated no major difference between naive and naive + Sham SCS group in RNA-seq analysis,57 naive + Sham SCS group was not included in this study. To examine the long-lasting changes in gene expression induced by SCS, spinal cord tissues were obtained from rats on day 30 (23 days after the last dose of paclitaxel and 17 days after the last SCS treatment) for RNA-seq. Principal component analysis showed separation of the transcriptomes from rats among these 3 groups (Fig. 4A). In comparison with naive rats, the lumbar spinal cord from Sham SCS + paclitaxel rats differentially expressed 121 (0.8%) genes (FDR < 0.05; Fig. 4B, upper). Of these 121 differentially expressed genes, 111 (91.7%) were upregulated and 10 (8.3%) were downregulated with Sham SCS + paclitaxel. In addition, 57 of the 121 genes could be classified into different gene classes (ie, transporters, enzymes, G-protein-coupled receptors, ion channels, catalytic receptors, and transcription factors) as defined by IUPHAR (Fig. 4C). Mean normalized counts, relative fold change of specific genes, and the composition of these gene classes are shown in supplemental figure 1 (available at http://links.lww.com/PR9/A52).
Figure 4.
Changes in gene expression in the lumbar spinal cord after paclitaxel and SCS. (A) Principal component analysis of libraries sequenced for RNA-seq. (B) MA-plots showing normalized mean counts (ie, average of normalized read counts across all samples) and log2 fold changes from RNA-seq data of L3-L6 spinal cord in paclitaxel (PTX)-treated rats with sham SCS vs Naive rats (TOP), and PTX-treated rats with SCS vs sham SCS (BOTTOM). DEGs are designated in red and are defined as differentially expressed genes with a false discovery rate <0.05. (C) Top: bar plot showing the numbers of differentially expressed genes upregulated and downregulated by gene class as defined by the IUPHAR. Bottom: relative expression levels for each rat are shown for each gene class represented in the bar plot. Upregulated and downregulated genes are colored in yellow and orange, respectively. Horizontal bars indicate group assignment for each rat. GPCRs, G-protein-coupled receptors; IUPHAR, International Union of Basic and Clinical Pharmacology; ICs, ion channels; SCS, spinal cord stimulation; TFs, transcription factors.
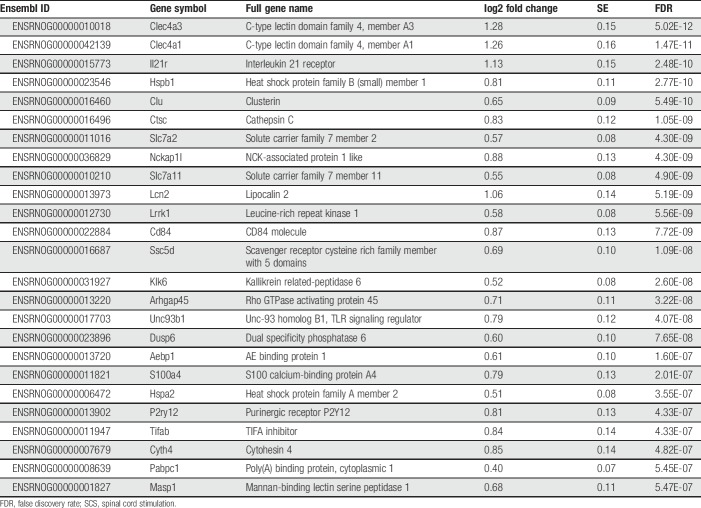
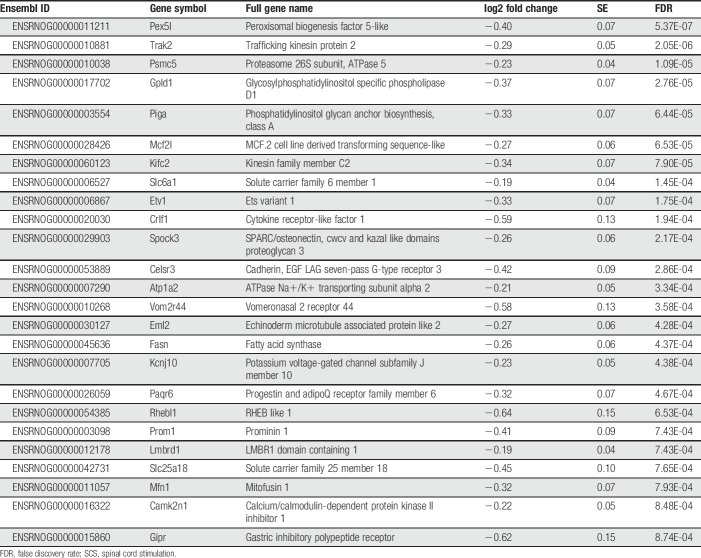
In comparison with the Sham SCS + paclitaxel group, the SCS + paclitaxel rats differentially expressed 1,066 (7.4%) genes (FDR < 0.05, Fig. 4B, lower), of which 836 (78.4%) were upregulated and 230 (21.6%) were downregulated with SCS. The most significantly upregulated and downregulated genes in SCS + paclitaxel-treated rats are listed in Tables 2 and 3, respectively. Of the 1,066 differentially expressed genes, 294 could be classified into several gene classes (ie, transporters, enzymes, G-protein-coupled receptors, ion channels, catalytic receptors, and transcription factors) as defined by IUPHAR (Fig. 4C). Mean normalized counts and relative fold change of specific genes that compose each of these gene classes are shown in supplemental figure 2 (available at http://links.lww.com/PR9/A52).
Table 2.
Top 25 upregulated genes in SCS + PTX rats by FDR (within the 11th percentile for all significantly upregulated genes).
Table 3.
Top 25 downregulated genes in SCS + PTX rats by FDR (within the 11th percentile for all significantly downregulated genes).
We conducted a technical validation of 11 genes related to regulation of synaptic plasticity (GO Biologic Process: 0048167) by qPCR. We evaluated several housekeeping genes and identified Sdha as an appropriate endogenous control gene because it showed stable gene expression among Sham SCS + paclitaxel and SCS + paclitaxel rats. These 11 genes were downregulated on RNA sequencing and this trend was confirmed with qPCR (supplemental figure 3, available at http://links.lww.com/PR9/A52).
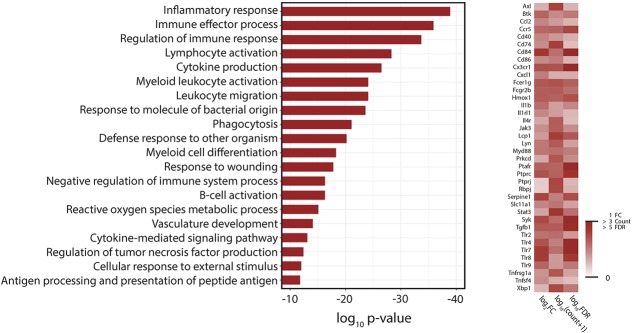
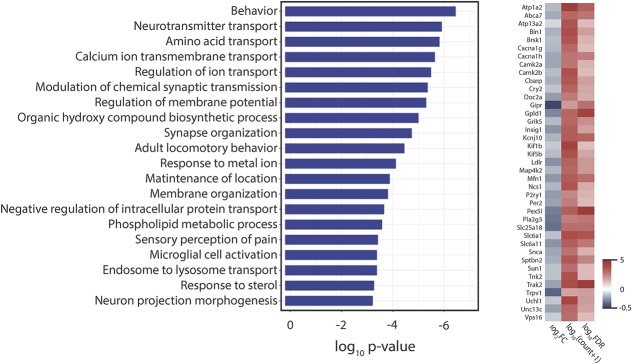
Gene ontology analysis of the genes upregulated in the SCS + paclitaxel vs those in the Sham SCS + paclitaxel group showed significant enrichment among a variety of immune-related biological processes (Fig. 5; supplemental tables 1–3, available at http://links.lww.com/PR9/A51). Intriguingly, GO analysis of the downregulated transcripts in the same group comparison showed significant enrichment among genes involved in neurotransmitter transport, synaptic transmission, synapse organization, calcium ion transmembrane transport, and microglial cell activation (Fig. 6).
Figure 5.
Gene ontology (GO) analysis of differentially expressed genes that are upregulated with SCS. Left: the top 20 GO biological processes associated with genes upregulated in the SCS + paclitaxel group as compared to levels in the Sham SCS + paclitaxel group (false discovery rate [FDR] < 0.05) as ranked by P-value. Right: heatmap of selected upregulated genes associated with multiple overrepresented GO biological processes. Data shown are relative expression (ie, log2FC), mean normalized transcript abundance across all samples (ie, log10(count + 1)), and statistical significance level (ie, −log10P-value). SCS, spinal cord stimulation.
Figure 6.
Gene ontology (GO) analysis of differentially expressed genes that are downregulated with SCS. Left: The top 20 GO biological processes associated with genes downregulated in SCS + paclitaxel group as compared to levels in the Sham SCS + paclitaxel group (false discovery rate [FDR] < 0.05) as ranked by P-value. Right: heatmap of selected downregulated genes associated with the first overrepresented GO biological process. Data shown are relative expression (ie, log2FC), mean normalized transcript abundance across all samples (ie, log10(count + 1)), and statistical significance level (ie, −log10P-value)). SCS, spinal cord stimulation.
4. Discussion
Traditional SCS induces pain inhibition by applying tonic electrical stimulation (40–80 Hz) that activates spinal cord dorsal columns. By using parameters and electrodes to mimic clinical application of SCS,50,62 we showed for the first time that SCS inhibited the development of PIPN-pain related behaviors in male rats. Furthermore, RNA-seq revealed broad changes in spinal gene expression after paclitaxel and SCS treatment.
Mechanical and cold hypersensitivities are consistently reported by CIPN patients, for which there is no effective treatment.16 Paclitaxel-induced peripheral neuropathy rats that underwent electrode placement developed both mechanical and cold hypersensitivities similar to rats that received only paclitaxel, and to those in previous reports.22,44 Importantly, preemptive SCS significantly attenuated the development of PIPN-related pain behaviors. It has been shown that SCS provides only short-lived relief of established cold allodynia.2,53 However, applying SCS before and during paclitaxel administration completely prevented the development of cold hypersensitivity in the current study. The reduction in mechanical hypersensitivity also persisted for at least 2 weeks after SCS. These findings differ from the short-term effects reported when SCS was examined in rats with established neuropathic pain.9 Similarly, human trials of SCS with a crossover design that compare various waveforms often use short washout periods of days or less in duration.27 Intriguingly, preemptive SCS may also exert therapeutic effects on ischemia-related conditions.14,34,54 Nevertheless, PIPN rats that received SCS still showed mechanical hypersensitivity from day 5 to 25. Thus, optimizing the SCS protocol (eg, 24 h/d SCS throughout the course of paclitaxel administration), alternating waveforms (eg, burst or “high dose” SCS), or adjuvant pharmacologic treatments may be tested to improve its effectiveness.10,33 We did not measure heat hypersensitivity because this is not a symptom consistently reported by CIPN patients.21,49 The equivalence in total distance traveled among the 3 groups in the open-field test indicates that paclitaxel and SCS did not severely impair the animals' locomotor function or exploration behavior.
The mechanisms by which preemptive SCS provides extended reduction of PIPN pain remain to be determined. To identify the genes and pathways that may be involved in PIPN and those related to SCS-induced pain inhibition, we performed RNA-seq of the spinal cord, which is the primary site of action for SCS. Mechanosensation is particularly relevant in peripheral neuropathies such as CIPN.13 Compared to naive rats, we observed upregulation of multiple genes related to mechanosensory function in Sham SCS + paclitaxel rats (eg, Egr1, Jun, Junb, Ptgs2, Mmp9, Prkcd, Mpo, Fos, Ptprc, Atf3, Sik1, Adamts1, Slc4a1, Tnfrsf11b, Zfp36, Sgk1, and Bcl3; supplemental figure 1, available at http://links.lww.com/PR9/A52), which may contribute to the decreased PWTs after paclitaxel. Neuroimmune responses are associated with neuropathic pain due to diabetic neuropathy and mononeuropathy.25,46 Neuroimmune activity and gliosis in the spinal cord may also be linked to the accompanying CIPN pain.39 In the dorsal root ganglion (DRG), infiltration of macrophages and proinflammatory T cells was observed with PTX administration.39,64 Hyperexcitability of DRG neurons in PIPN has been linked to monocyte chemoattractant protein-1 (MCP-1/CCL2), its receptor CCR2, and toll-like receptor 4 (TLR-4) signaling.30,31,39,63 Despite these findings, immune responses may also function to mediate regeneration and repair and enhance neuroprotection.1,15,24,55 Therefore, the broad impact of changes of immune-related genes in the spinal cord and DRG after SCS in CIPN rats necessitates further inquiry.
In line with previous findings, Sham SCS + paclitaxel rats showed upregulation of astrocyte-related genes (eg, S100a8, Mt2a) and one microglia-related gene (Mmp8), as compared to naive rats. Concurrent SCS during paclitaxel administration further enhanced immune-related genes, as indicated by increases in astrocyte-related (Gpr183, Hexb, Manf, and Mt2a) and microglia-related genes (Csf1, Cx3cr1, Itgam, and Nrros, P2rx4), when compared with levels in the sham SCS group. These findings are consistent with recent observations in peripheral nerve-injury models.55,57 However, these previous studies examined SCS for established mononeuropathies as opposed to preemptive SCS for polyneuropathy prevention in current study. To the best of our knowledge, this is the first report of transcriptome-wide assessment of spinal gene expression after CIPN with SCS. The implications of enhanced spinal glial activity and upregulation of immune-related genes after SCS of PIPN rats are unclear and warrant further investigation.
P2ry12 is a receptor for adenosine diphosphate, which mediates inhibition of adenylyl cyclase. Repetitive SCS may enhance adenosine-related activity, as indicated by increased expression of P2ry12 in paclitaxel-treated rats after SCS. This finding is in line with previous observations that activation of adenosinergic signaling enhanced SCS-induced pain inhibition,10 and A3 adenosine receptor agonist attenuated the neuroinflammatory response and inhibit PIPN.23 Thus, whereas increases in immune-related genes commonly suggest heightening of neuropathic pain, the interaction among components within the neuroimmune system during CIPN remains uncertain, and counteracting changes in expression and function of other gene families may outweigh upregulation. Although we did not observe similar changes in the spinal cord after PIPN, recent work to characterize the nociceptor translatome in a mouse PIPN model has demonstrated broad changes in the regulation of mTORC1 and MNK-eIF4E signaling networks in the DRG.40 It remains to be examined whether SCS alters gene expression and translatomes in the DRG under CIPN condition.
Spinal cord stimulation has been associated with inhibitory neurotransmission that involves γ-aminobutyric acid (GABA), serotonin, norepinephrine, acetylcholine, adenosine, and endocannabinoids.33,62 We observed downregulations of GABA reuptake-related genes (eg, Slc6a1 and Slc6a11), P2ry1, Agrn, and Grik5. Slc6a11 encodes GAT3, a GABA transporter expressed on glial cells that mediates GABA reuptake. These findings are consistent with a previous observation (eg, downregulation of spinal Slc6a11 after SCS) in rats after sciatic nerve injury.55 GABAergic mechanisms were shown to mediate SCS-induced inhibition of excitatory amino acid release in the dorsal horn of nerve-injured rats.11 Accordingly, we postulate that downregulation of GAT3 by SCS may increase GABAergic signaling that inhibits neurotransmission in PIPN rats (supplemental figure 4, available at http://links.lww.com/PR9/A52). GABAergic inhibition of excitatory amino acid release may involve suppression of calcium influx into presynaptic terminals.11 Indeed, numerous genes associated with calcium ion regulation (eg, Grik5, P2ry1, Camk2a, Sptbn2, Cacna1g, Doc2a, Ncs1, Cacna1h, Cbarp, and Unc13c) were also downregulated in paclitaxel-treated rats after SCS, which may enhance GABAergic inhibition.
P2ry1 is a subunit of P2Y receptors, which are slow G-protein-coupled ATP receptors that can increase signal duration.47 Agrn encodes agrin, which induces aggregation of acetylcholine receptors in clusters at the neuromuscular junction.42 Grik5 encodes the glutamate receptor subunit KA2,56 and its downregulation may result in destabilization of glutamate receptors on the postsynaptic membrane and a reduction in excitatory neurotransmission from peripheral afferents. Our RNA-seq study further showed that preemptive SCS in PIPN rats is associated with downregulations of genes involved in LTP, which is associated with the development of chronic pain.17,61 Spinal cord stimulation suppresses LTP in spinal wide-dynamic-range neurons.60 Camk2a, Cam2kb, and Cacng2 are linked by their association with glutamate binding, activation of AMPA receptors, and subsequent enhancement of synaptic plasticity (supplemental figure 4, available at http://links.lww.com/PR9/A52).20,35 Downregulation of AMPA receptor subunits impairs spinal synaptic plasticity and attenuates inflammatory pain.19 Thus, downregulation of key genes linked to LTP may decrease glutamatergic neurotransmission and lead to prolonged inhibition of PIPN pain by SCS. In addition, preemptive SCS-induced downregulation of genes (Camk2b, Hprt1, Camk2a, Agrn, Uchl1, L1cam, Celsr3, Cntnap1, Ptch1, Trak2, Mfn1, Ctnna2, Sptbn4, Kirrel3, Tbce, Etv1, Atl1, Brsk1, and Pdzd7) associated with neuron projection development may further inhibit the synaptic plasticity required for the transition from acute to chronic pain state.61 However, future studies are needed to explore the functional implications of changes in spinal gene expression after SCS in PIPN rats, especially the impact of these transcriptional changes on molecular function, cellular excitability, and pain behavior after PIPN.
Our study has some limitations. Because male rats have been typically studied in the existing SCS literature and CIPN literature, we chose to examine males only in this original investigation. Future investigations should examine the effects of SCS in models that include females and other chemotherapeutic agents to generalize its utility for CIPN pain prevention. New SCS waveforms to optimize the amount of electric charge transmitted from the leads to the spinal cord should also be tested in CIPN pain.33 In addition, the effect of SCS on tumor growth should be examined in the future to enhance a possible clinical applicability.
In summary, our findings suggest that traditional SCS during paclitaxel administration may prevent the development of mechanical and cold hypersensitivity in male rats. RNA-seq analysis revealed a complex interplay of upregulated and downregulated genes in the lumbar spinal cord with SCS. Technological innovation is rapidly ongoing in the field of neuromodulation, and less invasive SCS may open the horizon for new applications. Identifying specific transcriptional pathways and targets for pain prevention may enable optimization of waveforms and reasonable selection of drugs to increase therapeutic effects of SCS. Our study represents a small first step towards molecular understanding of SCS in CIPN, which may spur the clinical development of interventions to prevent CIPN pain in the future.
Disclosures
Y. Guan and S.N. Raja received research grant support from Medtronic, Inc. B. Linderoth is a consultant for Medtronic, Inc, St Jude Medical/Abbott, Boston Scientific, and Elekta AB. The remaining authors have no conflicts of interest to declare.
This study was conducted at the Johns Hopkins University and supported by a grant from National Institutes of Health (Bethesda, Maryland, USA) NS110598 (Y.G.), a grant from the Neurosurgery Pain Research Institute at the Johns Hopkins University (Y.G.), a seed grant for Stimulating and Advancing ACCM Research (StAAR) from the Department of Anesthesiology and Critical Care Medicine at the Johns Hopkins University (E.S.), a seed grant from the Blaustein Pain Research Endowment at the Johns Hopkins University (E.S.), a grant from the American Society of Regional Anesthesia and Pain Medicine (ASRA) (E.S.), and a grant from the Thompson Family Foundation Initiative-Columbia Neurology (Y.G. and E.S.). This study was also subsidized by grants from the National Institutes of Health (Bethesda, Maryland, USA): NS070814 (Y.G.), NS026363 (S.N.R.), and T32GM075774 (E.S.). Funders had no role in study design, data collection, or data interpretation, or in the decision to submit the work for publication.
Acknowledgments
The authors thank Claire F. Levine, MS, ELS (Scientific Editor, Department of Anesthesiology and Critical Care Medicine, the Johns Hopkins University), for editing the manuscript, and Medtronic, Inc. (Minneapolis, MN) for generously providing the electrodes for spinal cord stimulation. The authors also thank Rakel Tryggvadóttir and Colin Callahan for their technical assistance.
Author contributions: E. Sivanesan and Y. Guan designed the experiments; E. Sivanesan K.E. Stephens, Z. Chen, S.-Q. He, Q. Huang, N.C. Ford, and W. Duan performed the experiments; E. Sivanesan, K.E. Stephens, and Y. Guan were involved with data analysis; E. Sivanesan, K.E. Stephens, X. Gao, B. Linderoth, S.N. Raja, and Y. Guan were involved in discussion and interpretation of results; E. Sivanesan wrote the first manuscript draft; E. Sivanesan, K.E. Stephens, and Y. Guan wrote the final manuscript. All authors read and approved the final manuscript.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PR9/A52 and http://links.lww.com/PR9/A51.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painrpts.com).
References
- [1].Austin PJ, Moalem-Taylor G. The neuro-immune balance in neuropathic pain: involvement of inflammatory immune cells, immune-like glial cells and cytokines. J Neuroimmunol 2010;229:26–50. [DOI] [PubMed] [Google Scholar]
- [2].Barchini J, Tchachaghian S, Shamaa F, Jabbur S, Meyerson B, Song Z, Linderoth B, Saade N. Spinal segmental and supraspinal mechanisms underlying the pain-relieving effects of spinal cord stimulation: an experimental study in a rat model of neuropathy. Neuroscience 2012;215:196–208. [DOI] [PubMed] [Google Scholar]
- [3].Boora GK, Kanwar R, Kulkarni AA, Abyzov A, Sloan J, Ruddy KJ, Banck MS, Loprinzi CL, Beutler AS. Testing of candidate single nucleotide variants associated with paclitaxel neuropathy in the trial NCCTG N08C1 (Alliance). Cancer Med 2016;5:631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brenner DS, Golden JP, Gereau RW., IV A novel behavioral assay for measuring cold sensation in mice. PLoS One 2012;7:e39765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cata JP, Cordella JV, Burton AW, Hassenbusch SJ, Weng HR, Dougherty PM. Spinal cord stimulation relieves chemotherapy-induced pain: a clinical case report. J Pain Symptom Manage 2004;27:72–8. [DOI] [PubMed] [Google Scholar]
- [6].Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37:W305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Y, Yang C, Wang Z. Proteinase-activated receptor 2 sensitizes transient receptor potential vanilloid 1, transient receptor potential vanilloid 4, and transient receptor potential ankyrin 1 in paclitaxel-induced neuropathic pain. Neuroscience 2011;193:440–51. [DOI] [PubMed] [Google Scholar]
- [8].Cronin‐Fenton DP, Heide‐Jørgensen U, Ahern TP, Lash TL, Christiansen PM, Ejlertsen B, Sjøgren P, Kehlet H, Sørensen HT. Opioids and breast cancer recurrence: a Danish population‐based cohort study. Cancer 2015;121:3507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cui JG, Linderoth B, Meyerson BA. Effects of spinal cord stimulation on touch-evoked allodynia involve GABAergic mechanisms: an experimental study in the mononeuropathic rat. PAIN 1996;66:287–95. [DOI] [PubMed] [Google Scholar]
- [10].Cui JG, Sollevi A, Linderoth B, Meyerson BA. Adenosine receptor activation suppresses tactile hypersensitivity and potentiates spinal cord stimulation in mononeuropathic rats. Neurosci Lett 1997;223:173–6. [DOI] [PubMed] [Google Scholar]
- [11].Cui JG, O'Connor WT, Ungerstedt U, Linderoth B, Meyerson BA. Spinal cord stimulation attenuates augmented dorsal horn release of excitatory amino acids in mononeuropathy via a GABAergic mechanism. PAIN 1997;73:87–95. [DOI] [PubMed] [Google Scholar]
- [12].Dixon WJ. Efficient analysis of experimental observations. Ann Rev Pharmacol Toxicol 1980;20:441–62. [DOI] [PubMed] [Google Scholar]
- [13].Ferrier J, Pereira V, Busserolles J, Authier N, Balayssac D. Emerging trends in understanding chemotherapy-induced peripheral neuropathy. Curr Pain Headache Rep 2013;17:364. [DOI] [PubMed] [Google Scholar]
- [14].Gherardini G, Lundeberg T, Cui J-G, Eriksson SV, Trubek S, Linderoth B. Spinal cord stimulation improves survival in ischemic skin flaps: an experimental study of the possible mediation by calcitonin gene-related peptide. Plast Reconstr Surg 1999;103:1221–8. [DOI] [PubMed] [Google Scholar]
- [15].Goldstein EZ, Church JS, Hesp ZC, Popovich PG, McTigue DM. A silver lining of neuroinflammation: beneficial effects on myelination. Exp Neurol 2016;283(pt B):550–9. [DOI] [PubMed] [Google Scholar]
- [16].Griffith KA, Couture DJ, Zhu S, Pandya N, Johantgen ME, Cavaletti G, Davenport JM, Tanguay LJ, Choflet A, Milliron T. Evaluation of chemotherapy-induced peripheral neuropathy using current perception threshold and clinical evaluations. Support Care Cancer 2014;22:1161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guan Y, Wacnik PW, Yang F, Carteret AF, Chung C-Y, Meyer RA, Raja SN. Spinal cord stimulation-induced analgesia: electrical stimulation of dorsal column and dorsal roots attenuates dorsal horn neuronal excitability in neuropathic rats. Anesthesiology 2010;113:1392–405. [DOI] [PubMed] [Google Scholar]
- [18].Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, Catterall WA, Davenport AP, Delagrange P, Dollery CT, Foord SM, Gutman GA, Laudet V, Neubig RR, Ohlstein EH, Olsen RW, Peters J, Pin JP, Ruffolo RR, Searls DB, Wright MW, Spedding M. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res 2009;37:D680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hartmann B, Ahmadi S, Heppenstall PA, Lewin GR, Schott C, Borchardt T, Seeburg PH, Zeilhofer HU, Sprengel R, Kuner R. The AMPA receptor subunits GluR-A and GluR-B reciprocally modulate spinal synaptic plasticity and inflammatory pain. Neuron 2004;44:637–50. [DOI] [PubMed] [Google Scholar]
- [20].Henley JM, Wilkinson KA. Synaptic AMPA receptor composition in development, plasticity and disease. Nat Rev Neurosci 2016;17:337. [DOI] [PubMed] [Google Scholar]
- [21].Hershman DL, Lacchetti C, Loprinzi CL. Prevention and management of chemotherapy-induced peripheral neuropathy in survivors of adult cancers: American Society of Clinical Oncology clinical practice guideline summary. J Oncol Pract 2014;10:e421–4. [DOI] [PubMed] [Google Scholar]
- [22].Hopkins HL, Duggett NA, Flatters SJ. Chemotherapy-induced painful neuropathy: pain-like behaviours in rodent models and their response to commonly-used analgesics. Curr Opin Support Palliat Care 2016;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Janes K, Esposito E, Doyle T, Cuzzocrea S, Tosh DK, Jacobson KA, Salvemini D. A3 adenosine receptor agonist prevents the development of paclitaxel-induced neuropathic pain by modulating spinal glial-restricted redox-dependent signaling pathways. PAIN 2014;155:2560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jin X, Yamashita T. Microglia in central nervous system repair after injury. J Biochem 2016;159:491–6. [DOI] [PubMed] [Google Scholar]
- [25].Kim CF, Moalem-Taylor G. Detailed characterization of neuro-immune responses following neuropathic injury in mice. Brain Res 2011;1405:95–108. [DOI] [PubMed] [Google Scholar]
- [26].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kriek N, Groeneweg J, Stronks D, de Ridder D, Huygen F. Preferred frequencies and waveforms for spinal cord stimulation in patients with complex regional pain syndrome: a multicentre, double‐blind, randomized and placebo‐controlled crossover trial. Eur J Pain 2017;21:507–19. [DOI] [PubMed] [Google Scholar]
- [28].Kumar K, Rizvi S, Nguyen R, Abbas M, Bishop S, Murthy V. Impact of wait times on spinal cord stimulation therapy outcomes. Pain Pract 2014;14:709–20. [DOI] [PubMed] [Google Scholar]
- [29].Leandro-García LJ, Inglada-Pérez L, Pita G, Hjerpe E, Leskelä S, Jara C, Mielgo X, González-Neira A, Robledo M, Åvall-Lundqvist E. Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. J Med Genet 2013;50:599–605. [DOI] [PubMed] [Google Scholar]
- [30].Li Y, Zhang H, Kosturakis AK, Cassidy RM, Zhang H, Kennamer-Chapman RM, Jawad AB, Colomand CM, Harrison DS, Dougherty PM. MAPK signaling downstream to TLR4 contributes to paclitaxel-induced peripheral neuropathy. Brain Behav Immun 2015;49:255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li Y, Zhang H, Zhang H, Kosturakis AK, Jawad AB, Dougherty PM. Toll-like receptor 4 signaling contributes to Paclitaxel-induced peripheral neuropathy. J Pain 2014;15:712–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- [33].Linderoth B, Foreman RD. Conventional and novel spinal stimulation algorithms: hypothetical mechanisms of action and comments on outcomes. Neuromodulation 2017;20:525–33. [DOI] [PubMed] [Google Scholar]
- [34].Linderoth B, Gherardini G, Ren B, Lundeberg T. Preemptive spinal cord stimulation reduces ischemia in an animal model of vasospasm. Neurosurgery 1995;37:266–72. [DOI] [PubMed] [Google Scholar]
- [35].Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci 2012;13:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [37].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Majithia N, Loprinzi CL, Smith TJ. New practical approaches to chemotherapy-induced neuropathic pain: prevention, assessment, and treatment. Oncology 2016;30:1020–29. [PubMed] [Google Scholar]
- [39].Makker PG, Duffy SS, Lees JG, Perera CJ, Tonkin RS, Butovsky O, Park SB, Goldstein D, Moalem-Taylor G. Characterisation of immune and neuroinflammatory changes associated with chemotherapy-induced peripheral neuropathy. PLoS One 2017;12:e0170814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Megat S, Ray PR, Moy JK, Lou T-F, Barragan-Iglesias P, Li Y, Pradhan G, Wanghzou A, Ahmad A, Burton MD. Nociceptor translational profiling reveals the Ragulator-Rag GTPase complex as a critical generator of neuropathic pain. J Neurosci 2019;39:393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mekhail NA, Mathews M, Nageeb F, Guirguis M, Mekhail MN, Cheng J. Retrospective review of 707 cases of spinal cord stimulation: indications and complications. Pain Pract 2011;11:148–53. [DOI] [PubMed] [Google Scholar]
- [42].Mohseni P, Sung HK, Murphy AJ, Laliberte CL, Pallari H-M, Henkelman M, Georgiou J, Xie G, Quaggin SE, Thorner PS. Nestin is not essential for development of the CNS but required for dispersion of acetylcholine receptor clusters at the area of neuromuscular junctions. J Neurosci 2011;31:11547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Phan P, Khodavirdi A. Successful treatment of chemotherapy-induced peripheral neuropathy (CIPN) with spinal cord stimulation (SCS): case reports. Cancer Res 2007;67(9 supplement):35. [Google Scholar]
- [44].Polomano RC, Mannes AJ, Clark US, Bennett GJ. A painful peripheral neuropathy in the rat produced by the chemotherapeutic drug, paclitaxel. PAIN 2001;94:293–304. [DOI] [PubMed] [Google Scholar]
- [45].Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sandireddy R, Yerra VG, Areti A, Komirishetty P, Kumar A. Neuroinflammation and oxidative stress in diabetic neuropathy: futuristic strategies based on these targets. Int J Endocrinol 2014;2014:674987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Scholz J, Woolf CJ. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci 2007;10:1361. [DOI] [PubMed] [Google Scholar]
- [48].Seretny M, Currie GL, Sena ES, Ramnarine S, Grant R, MacLeod MR, Colvin LA, Fallon M. Incidence, prevalence, and predictors of chemotherapy-induced peripheral neuropathy: a systematic review and meta-analysis. PAIN 2014;155:2461–70. [DOI] [PubMed] [Google Scholar]
- [49].Shah A, Hoffman EM, Mauermann ML, Loprinzi CL, Windebank AJ, Klein CJ, Staff NP. Incidence and disease burden of chemotherapy-induced peripheral neuropathy in a population-based cohort. J Neurol Neurosurg Psychiatry 2018;89:636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shechter R, Yang F, Xu Q, Cheong Y-K, He S-Q, Sdrulla A, Carteret AF, Wacnik PW, Dong X, Meyer RA. Conventional and kilohertz-frequency spinal cord stimulation produces intensity-and frequency-dependent inhibition of mechanical hypersensitivity in a rat model of neuropathic pain. Anesthesiology 2013;119:422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sivanesan E, Maher DP, Raja SN, Linderoth B, Guan Y. Supraspinal mechanisms of spinal cord stimulation for modulation of pain: five decades of research and prospects for the future. Anesthesiology 2018;130:651–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Smits H, Ultenius C, Deumens R, Koopmans GC, Honig W, Van Kleef M, Linderoth B, Joosten EA. Effect of spinal cord stimulation in an animal model of neuropathic pain relates to degree of tactile “allodynia”. Neuroscience 2006;143:541–6. [DOI] [PubMed] [Google Scholar]
- [53].Song Z, Meyerson BA, Linderoth B. Spinal 5-HT receptors that contribute to the pain-relieving effects of spinal cord stimulation in a rat model of neuropathy. PAIN 2011;152:1666–73. [DOI] [PubMed] [Google Scholar]
- [54].Southerland EM, Milhorn DM, Foreman RD, Linderoth B, DeJongste MJ, Armour JA, Subramanian V, Singh K, Singh M, Ardell JL. Pre-emptive, but not reactive, spinal cord stimulation mitigates transient ischemia induced myocardial infarction via cardiac adrenergic neurons. Am J Physiol Heart Circ Physiol 2007;292:H311–7. [DOI] [PubMed] [Google Scholar]
- [55].Stephens K, Chen Z, Sivanesan E, Raja S, Linderoth B, Tavernn SD, Guan Y. RNA-seq of spinal cord in nerve-injured rats after spinal cord stimulation. Mol Pain 2018;14:1744806918817429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Szpirer C, Molné M, Antonacci R, Jenkins NA, Finelli P, Szpirer J, Riviere M, Rocchi M, Gilbert DJ, Copeland NG. The genes encoding the glutamate receptor subunits KA1 and KA2 (GRIK4 and GRIK5) are located on separate chromosomes in human, mouse, and rat. Proc Natl Acad Sci 1994;91:11849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tilley DM, Cedeno DL, Kelley CA, Benyamin R, Vallejo R. Spinal cord stimulation modulates gene expression in the spinal cord of an animal model of peripheral nerve injury. Reg Anesth Pain Med 2016;41:750–6. [DOI] [PubMed] [Google Scholar]
- [58].Tiwari V, Anderson M, Yang F, Tiwari V, Zheng Q, He S-Q, Zhang T, Shu B, Chen X, Grenald SA. Peripherally acting μ-opioid receptor agonists attenuate ongoing pain-associated behavior and spontaneous neuronal activity after nerve injury in rats. Anesthesiology 2018;128:1220–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Truin M, Kleef M, Linderoth B, Smits H, Janssen SP, Joosten EA. Increased efficacy of early spinal cord stimulation in an animal model of neuropathic pain. Eur J Pain 2011;15:111–17. [DOI] [PubMed] [Google Scholar]
- [60].Wallin J, Fiskå A, Tjølsen A, Linderoth B, Hole K. Spinal cord stimulation inhibits long-term potentiation of spinal wide dynamic range neurons. Brain Res 2003;973:39–43. [DOI] [PubMed] [Google Scholar]
- [61].Woolf CJ, Salter MW. Neuronal plasticity: increasing the gain in pain. Science 2000;288:1765–8. [DOI] [PubMed] [Google Scholar]
- [62].Yang F, Xu Q, Shu B, Tiwari V, He SQ, Vera-Portocarrero LP, Dong X, Linderoth B, Raja SN, Wang Y, Guan Y. Activation of cannabinoid CB1 receptor contributes to suppression of spinal nociceptive transmission and inhibition of mechanical hypersensitivity by Aβ-fiber stimulation. PAIN 2016;157:2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang H, Boyette-Davis JA, Kosturakis AK, Li Y, Yoon S-Y, Walters ET, Dougherty PM. Induction of monocyte chemoattractant protein-1 (MCP-1) and its receptor CCR2 in primary sensory neurons contributes to paclitaxel-induced peripheral neuropathy. J Pain 2013;14:1031–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhang H, Li Y, de Carvalho-Barbosa M, Kavelaars A, Heijnen CJ, Albrecht PJ, Dougherty PM. Dorsal root ganglion infiltration by macrophages contributes to paclitaxel chemotherapy-induced peripheral neuropathy. J Pain 2016;17:775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]