Abstract
The epithelial-to-mesenchymal transition (EMT) is an essential developmental process which can be hijacked by cancer cells, leading to enhanced metastasis and chemoresistance in experimental models. Recent studies have linked gene expression of EMT-associated gene signatures to increased inflammatory immune response in multiple cancer types. However, these studies did not account for the potential confounding effects of gene expression by tumor-infiltrating mesenchymal stromal cells. In this study, we comprehensively dissect the associations between multiple EMT transcription factors and EMT markers with stromal and immune tumor infiltration. We find that EMT-related genes are highly correlated with intratumoral stromal cell abundance and identify a specific relationship between stroma-corrected ZEB1 expression and decreased immune activity in multiple cancer types. We derive a stroma-corrected ZEB1-activated transcriptional signature and demonstrate that this signature includes several known inhibitors of inflammation, including BMPR2. Finally, multivariate survival analysis reveals that ZEB1 and its expression signature are significantly associated with reduced overall survival in breast cancer patients. In conclusion, this study identifies a novel association between stroma-adjusted ZEB1 expression and tumor immune activity and addresses the critical issue of confounding between EMT-associated genes and tumor stromal content.
Subject terms: Cancer genomics, Data mining, Gene regulatory networks
Introduction
The epithelial-to-mesenchymal transition (EMT) is a critical process in early development by which embryonic cells migrate to properly form new tissues1. Accumulated experimental evidence has suggested that EMT is pathologically re-activated in epithelial cancers, leading to an increased propensity for chemotherapeutic resistance, tumor recurrence, and distal metastatic progression1,2. Breast cancer has served as one of the primary models for studying the role of EMT in cancer progression. It has been demonstrated both in vitro and in vivo that breast cancer cells that undergo EMT gain stemlike features, increased chemoresistance, and an enhanced ability to invade local tissues and colonize distant organs3. However, the role of EMT in patient outcomes remains controversial.
An EMT can be provoked by several signaling pathways which converge on the activation of a group of EMT-promoting transcription factors (EMT-TFs), including members of the SNAI, TWIST, ZEB and FOX transcription factor families4. This diverse group of transcription factors can directly repress the expression of critical epithelial adhesion proteins, like E-cadherin (CDH1), and upregulate mesenchymal cytoskeletal and extracellular matrix components1,4. Mechanistically, ectopic expression of any individual EMT-TF is sufficient to induce an EMT. Despite this commonality, the relationship between different EMT-TFs is complex and not fully understood. Developmental studies have demonstrated EMT-TFs have distinct roles during embryogenesis5. In parallel, recent studies have demonstrated that EMT-TFs have different proclivities for promoting cancer chemoresistance and metastasis6,7. Despite these differences, few studies have examined multiple EMT-TFs simultaneously at either the functional or genomic level5.
Experimentally, EMT has been shown to suppress the immune response in multiple cancer types8,9. Interestingly, EMT can be induced by inflammatory signals10. These post-EMT cells can then act to suppress inflammation within tumors10. Several recent studies of gene expression data from patient-derived tumor samples have attempted to evaluate the relationship between EMT and antitumor immune activity, with conflicting results. Multiple groups have linked the expression of EMT-associated genes to increased immune cell infiltration and increased intratumoral inflammation11,12. Conversely, a study in lung squamous and adenocarcinoma found that an EMT gene signature was associated with decreased T-cell infiltration and increased expression of immunosuppressive cytokines13.
Other studies have attempted to use similar EMT-associated gene signatures to predict survival outcomes in patients14. However, these prior inquiries did not account for potential confounding by stromal cell infiltration into the tumor sample15. The patient-derived tumor sequencing data used in these studies were generated by bulk tumor sequencing, which generates an expression measurement for each gene averaged across all of the cells present within the sample. Consequently, the presence of both mesenchymal stromal cells and immune cells can contribute to the overall gene expression signal and confound evaluations of the contribution of a gene or gene signature to both tumor biology and clinical outcomes. As EMT-associated genes are highly expressed by mesenchymal cell types, the potential for confounding is substantial. For example, a recent study demonstrated that EMT and stromal cell signatures can confound one another and in general are associated with an altered response to immunotherapy in urothelial carcinoma16.
An example of this potential confounding effect is shown in Supplemental Fig. 1. Gene X is a gene that is associated with the mesenchymal phenotype, being expressed at a higher level in fibroblasts and EMT-like cancer cells than in epithelial cancer cells (Supplemental Fig. 1A). Two hypothetical tumor samples, sample A and sample B, have an equal proportion of fully epithelial cancer cells to cancer cells undergoing EMT (5:1). However, sample B contains a higher number of infiltrating mesenchymal stromal cells. As a result, the estimated gene expression of the mesenchymal-associated gene Gene X will be higher in sample B than sample A, even after normalization (Supplemental Fig. 1B). If the effect of stromal cell abundance is not considered, a researcher may make spurious conclusions about the associations between the expression of Gene X and both tumor biology and patient outcomes. However, gene signature-based deconvolution methods can be used to estimate the abundance of stromal cells present within the tumor and then correct the gene expression levels to represent the cancer-specific expression of Gene X17. Thus, it is critical to account for tumor purity in any study that attempts to associate tumor gene expression to clinical or pathologic characteristics.
In this study, we used multiple gene signature-based methods to evaluate the association between the expression of EMT-TFs and EMT marker genes with both stromal and immune cell content in tumor samples. These analyses revealed critical differences between different EMT-TFs and markers, highlighting the complexity of EMT-associated processes. Our results demonstrated that the stroma-adjusted expression of one EMT-TF, ZEB1, has a unique inverse association with immune cell abundance and activity in multiple cancer types. As well-characterized EMT markers did not recapitulate this relationship, we derived a stroma-corrected, ZEB1-regulated gene signature with an integrative transcriptomic approach. The ZEB1 gene signature showed similar associations with both stromal and immune cell abundance as ZEB1 and contained several known repressors of the immune response, including the receptor BMPR2. Finally, a multivariate survival analysis demonstrated that ZEB1 and members of the ZEB1 gene signature are significant independent predictors of overall survival in breast cancer. Our results confirm the importance of considering the complicated cellular composition of tumors when performing gene association analyses. Importantly, this study demonstrates that stroma-corrected ZEB1 transcriptional activity is associated with decreased abundance of tumor-infiltrating immune cells and decreased intratumoral inflammation, providing new insight into the role of ZEB1 as a suppressor of antitumor immune activity.
Results
EMT-TFs exhibit distinct correlative relationships with stromal and immune cell abundance in primary tumor samples
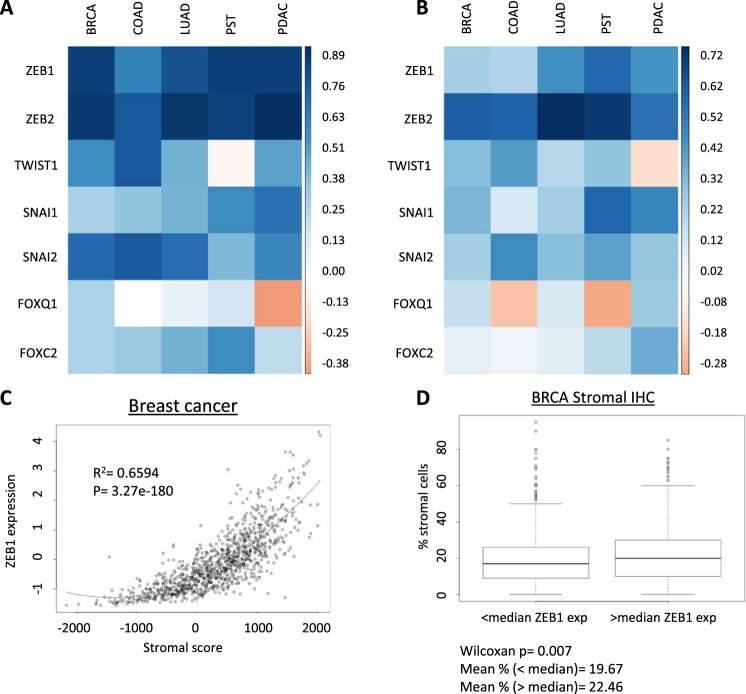
As tumor-infiltrating stromal cells express high levels of mesenchymal-associated genes, we examined whether the expression of a set of well-characterized EMT-driving transcription factors was correlated with stromal cell infiltration in multiple cancer types. Normalized mRNA expression data from TCGA studies of breast, prostate, lung, colorectal and pancreatic adenocarcinomas was downloaded for ZEB1, ZEB2, SNAI1, SNAI2, TWIST1, FOXQ1 and FOXC218,19. These data were combined with stromal and immune cell abundance scores generated by the ESTIMATE approach, then evaluated for the Spearman correlation coefficient between both scores and the expression of each EMT-TF20. ZEB1, ZEB2, SNAI1, SNAI2 and FOXC2 exhibited consistent positive correlations with the ESTIMATE stromal score in all five cancer types (Fig. 1A, Supplemental Table 1). Of these, ZEB1 (r = 0.60 to 0.84) and ZEB2 (0.75 to 0.89) exhibited the strongest overall correlations. However, ZEB2 is one of the 141 genes used to generate the ESTIMATE stromal score. Interestingly, TWIST1 was not significantly correlated with stromal score in prostate cancer samples, while FOXQ1 was not significantly correlated with stromal cell abundance in colorectal and lung adenocarcinoma. Also, FOXQ1 was significantly inversely correlated with stromal score in pancreatic ductal adenocarcinoma. The distinct patterns of association of each EMT-TF demonstrate the unique expression profile of different EMT-TFs, highlighting the potential functional differences between these factors.
Figure 1.
EMT-TFs are significantly positively correlated with stromal and immune cell abundance in multiple cancer types. (A) Heatmap illustrating Spearman correlation coefficients for selected EMT-TFs and ESTIMATE stromal score across five cancer types (breast (BRCA), colorectal (COAD), lung (LUAD), prostate (PST) and pancreatic (PDAC) adenocarcinoma. (B) Heatmap illustrating Spearman correlation coefficients for selected EMT-TFs and ESTIMATE immune score across same five cancer types. (C) Scatterplot illustrating correlation between ZEB1 expression and stromal score in BRCA dataset. Table shows correlation coefficients for both ER+ and ER− subsets. (D) Comparison of immunohistochemistry-estimated percentage of stromal cells between patients with less or greater than median ZEB1 (z score = −0.3026) mRNA expression (n = 1078 for all BRCA analyses).
Next, this approach was repeated to determine the correlation between EMT-TFs and immune cell abundance. ZEB1, ZEB2, SNAI1, and SNAI2 were significantly and positively correlated with the ESTIMATE immune score in all five cancer types examined (Fig. 1B). However, FOXQ1, TWIST1 and FOXC2 were either not significantly correlated or were modestly inversely correlated with immune score (Fig. 1B).
After removing ZEB2 from consideration, ZEB1 was most significantly correlated with stromal infiltration in breast cancer (r2 = 0.65, p = 3.99 × 10−180) (Fig. 1C). To confirm this observation, the relationship between ZEB1 mRNA expression and an immunohistochemistry-based estimation of the percent of stromal cell abundance was evaluated. As immunohistochemistry is a semi-quantitative approach for estimating cell content, samples were split into two groups by median ZEB1 expression. ZEB1 was significantly associated with stromal cell abundance (Wilcoxon p = 0.007), despite previous observations that IHC-based estimates of tumor stromal content are less sensitive than genomics-based approaches (Fig. 1D)21.
From these analyses, we concluded that several EMT-TFs are correlated with stromal cell abundance within tumors and that ZEB1 has a particularly strong and reproducible association with tumor stromal content in breast cancer. Similarly, multiple EMT-TFs are positively associated with immune cell content before considering the potential effect of stromal cell abundance. This relationship may explain the prior reports of a positive correlation between EMT and immune activity.
Stroma-corrected ZEB1 expression is inversely correlated with total immune cell abundance in primary tumor samples
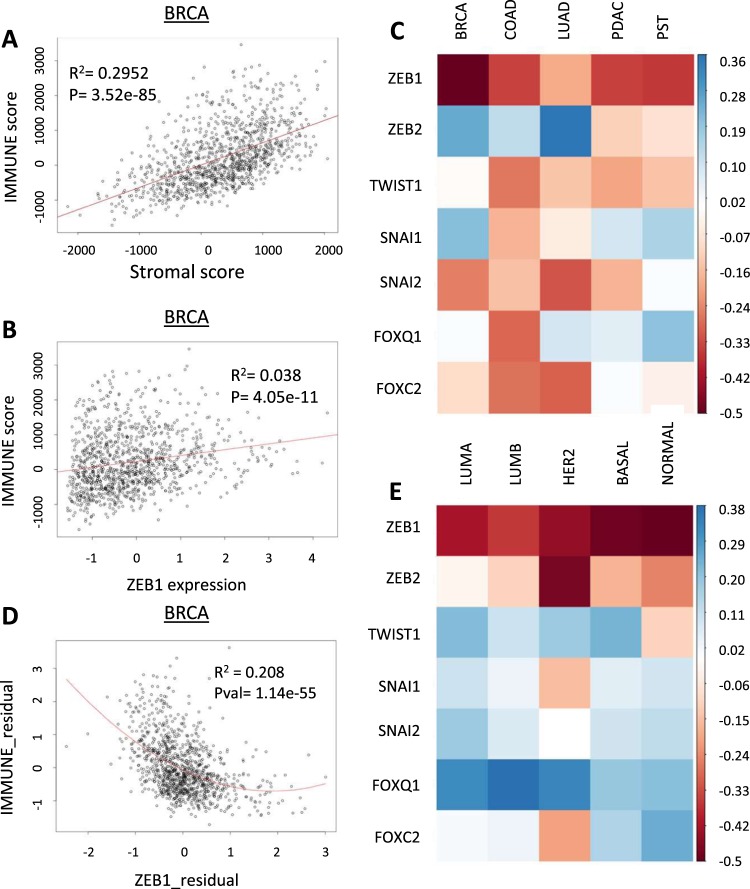
To reproduce prior reports, we evaluated the association between immune and stromal cell infiltration. We found that stromal cell and immune cell abundance were highly correlated (Fig. 2A)21. We noted a discrepancy between the correlation values between EMT-TFs and stromal cell abundance when compared against immune cell abundance: In general, the correlation coefficients were much lower between EMT-TFs and immune cell content. For example, the coefficient of determination for the relationship between ZEB1 and immune score in breast cancer was 0.038, much lower than the correlation between stromal and immune scores (Fig. 2B).
Figure 2.
Stroma-corrected ZEB1 expression is inversely correlated with total immune cell abundance in tumor samples. (A) Correlation between ESTIMATE stromal and immune scores in BRCA dataset. (B) Correlation between ZEB1 expression and immune scores in BRCA. (C) Heatmap illustrating partial Spearman correlation coefficients of EMT-TFs with ESTIMATE immune score in five cancer types. (D) Residual plot of ZEB1 expression and ESTIMATE immune score, adjusted by stromal score. (E) Partial correlation coefficients for EMT-TFs and immune score by PAM50 subtypes of breast cancer.
To correct for possible stromal cell confounding, the partial correlation coefficient was calculated for each EMT-TF with immune score. The partial correlation approach considers the correlations between two variables which are each highly correlated with a third variable, then corrects for the possible confounding effect of that third variable22. This analysis was repeated for each EMT-TF in each tumor type and plotted as a heatmap (Fig. 2C, Supplemental Table 2). As expected, correcting for stromal cell abundance significantly altered the relationships between EMT-TF expression and immune cell content. Uniquely, stroma-corrected ZEB1 expression was significantly inversely correlated with total immune cell abundance in all five tumor types (Fig. 2C). TWIST1, SNAI2 and FOXC2 also showed moderate inverse correlations with immune cell content in four of five cancer types (Fig. 2C). The inverse relationship between ZEB1 and immune cell content was strongest in breast cancer (partial r = −0.5, r2 = 0.208) (Fig. 2C,D). As breast cancer is a heterogenous disease comprised of multiple molecular subtypes, we re-evaluated this association by PAM50 subtypes. The PAM50 approach uses gene expression data to cluster breast cancer samples into five subtypes: Luminal A and B, Her2, Basal, and Normal23. PAM50 scores for the TCGA breast cancer cohort were obtained for a recent study24. Despite the differences between PAM50 subtypes, stroma-corrected ZEB1 expression was consistently inversely correlated with immune cell abundance in all subtypes (Fig. 2E, Supplemental Table 3). However, ZEB1 exhibited the strongest inverse associations in the basal and normal PAM50 samples (partial r = −0.492 and −0.504, respectively).
Stroma-corrected ZEB1 expression is inversely associated with the abundance of multiple immune cell types and with immune cell recruitment to breast tumors
Our results demonstrated that the positive associations identified between EMT-TF expression and immune cell content were due to confounding by stromal cell abundance. By adjusting for stromal cell abundance, partial correlation analysis indicates a general trend towards an inverse association between EMT-TF expression and immune cell abundance in tumors.
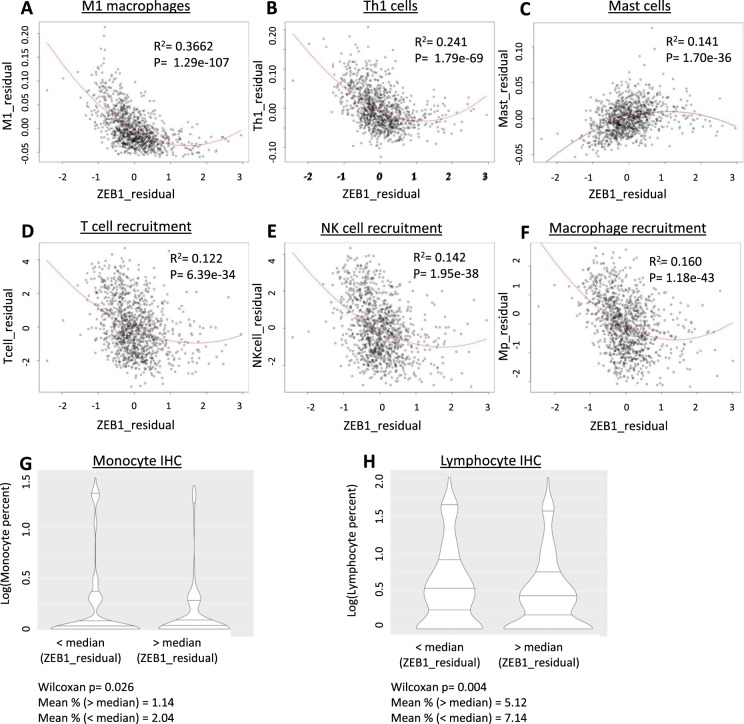
Interestingly, stroma-corrected ZEB1 expression showed a strong and reproducible inverse relationship with immune cell abundance, particularly in breast cancer samples. To investigate this association in more detail, the xCELL algorithm was used to generate estimations of the abundance of different cell types within breast tumor samples25. The partial correlation coefficient for each cell type with ZEB1, corrected for stromal cell abundance, was then generated (Supplemental Table 4). We observed that ZEB1 was inversely correlated with multiple immune cell types, including M1 macrophages and Th1 cells (Fig. 3A,B). Both M1 macrophages and Th1 cells are known to promote inflammation and the antitumor immune response26. Th1 cells are known activators of antitumor macrophage activity, lending further credence to our results26,27. ZEB1 was moderately positively associated with the infiltration of T-regulatory cells, well-characterized suppressors of the antitumor immune response (partial r = 0.177)28. Unexpectedly, ZEB1 was also positively associated with mast cell abundance (partial r = 0.40, r2 = 0.141) (Fig. 3C). Mast cells are typically considered immunosuppressive within the tumor microenvironment, and the presence of mast cells has been associated with worse prognosis29. When we generated a correlation matrix for all 64 cell types in the breast cancer dataset, we found that mast cells were only modestly positively correlated with other immune cell types and were negatively correlated with Th1 cell abundance (Supplemental Fig. 2).
Figure 3.
Stroma-corrected ZEB1 expression is inversely correlated with the abundance of multiple immune cell types and with immune cell recruitment to breast tumors. (A–C) Residual plot of ZEB1 expression and xCELL macrophage (A), Th1 cell (B) and mast cell (C) abundance corrected for stromal cell content. (D–F) Residual plot of ZEB1 expression and TIP-based estimation of T cell (D), natural killer cell (E) and macrophage (F) recruitment. (G) Immunohistochemistry measurement of monocyte percentage in tumor samples compared by samples with greater or less than median stroma-adjusted ZEB1 expression (median percentage 0%). (H) Immunohistochemistry measurement of lympocyte percentage in tumor samples compared by samples with greater or less than median stroma-adjusted ZEB1 expression (median percentage 1%).
As several of the immune cell types which were inversely correlated with ZEB1 are known to activate the immune response, we dissected the immune activity profile of the breast cancer tumor samples using the Tumor ImmunoPhenotype server algorithm (TIP)30. Like xCELL, TIP uses gene sets to evaluate immune activity in bulk tumor samples. However, instead of evaluating by cell type, TIP estimates the relative activity of different stages of the immune response, from tumor-antigen presentation to tumor cell killing by immune cells30. TIP scores were downloaded and compared with ZEB1 expression by partial correlation analysis. Stroma-corrected ZEB1 expression was significantly inversely correlated with five out of seven “steps” of the immune cycle: Antigen release, immune cell priming and activation, immune cell recruitment to the tumor, tumor infiltration, and cell killing (Table 1). ZEB1 exhibited a modest positive correlation with cancer cell antigen presentation and was not correlated with cancer cell recognition by immune cells (Table 1). Of the immune cycle steps, stroma-corrected ZEB1 expression was most significantly associated with immune cell activation and recruitment (Steps 3 and 4) (Table 1). Step 4 was then subset by recruitment of different immune cell types. By cell type, stroma-adjusted ZEB1 expression was most significantly inversely associated with macrophage, T cell, and NK cell recruitment (Fig. 3D–F).
Table 1.
Stroma-corrected ZEB1 expression is inversely associated with immune cell infiltration and activity in breast tumors.
| Step of immune response | Description | Partial corr. | p.value |
|---|---|---|---|
| Step 1 | Cancer Cell Ag Release | −0.1373 | 5.26E-06 |
| Step 2 | Cancer Ag Presentation | 0.1469 | 1.08E-06 |
| Step 3 | Priming and Activation | −0.2222 | 1.10E-13 |
| Step 4: Recruitment | T cell.recruiting | −0.3592 | 1.36E-34 |
| CD4 T cell.recruiting | −0.2060 | 6.22E-12 | |
| CD8 T cell.recruiting | −0.3026 | 1.45E-24 | |
| Th1 cell.recruiting | −0.2746 | 2.40E-20 | |
| Dendritic cell.recruiting | −0.2695 | 1.27E-19 | |
| Th22 cell.recruiting | −0.2518 | 2.95E-17 | |
| Macrophage.recruiting | −0.4693 | 6.69E-61 | |
| Monocyte.recruiting | −0.1986 | 3.57E-11 | |
| Neutrophil.recruiting | −0.2568 | 6.67E-18 | |
| NK cell.recruiting | −0.3840 | 1.11E-39 | |
| Eosinophil.recruiting | −0.4318 | 8.16E-51 | |
| Basophil.recruiting | −0.3382 | 1.26E-30 | |
| Th17 cell.recruiting | −0.1572 | 1.78E-07 | |
| B cell.recruiting | −0.1088 | 0.000317 | |
| Th2 cell.recruiting | −0.3104 | 8.07E-26 | |
| Treg cell.recruiting | −0.1810 | 1.71E-09 | |
| MDSC.recruiting | −0.2294 | 1.67E-14 | |
| Step 5 | Tumor infiltration | −0.1302 | 1.59E-05 |
| Step 6 | Recognition | −0.0379 | 0.210775 |
| Step 7 | Cell killing | −0.1947 | 8.56E-11 |
The results generated by TIP reproduced the results from the xCELL-based approach, increasing our confidence that stroma-corrected ZEB1 expression is indeed associated with decreased immune cell abundance and antitumor immune activity in breast cancer. However, both methods are rely on GSEA-based approaches. As a confirmation that ZEB1 expression was inversely associated with inflammatory activity in breast tumors, the partial correlations between stroma-adjusted ZEB1 expression and the expression of nine pro-inflammatory cytokines (CCL2, CCL4, IFNG, IL6, IL18, IL1A, IL1B, and MIF, TNF) were calculated. When corrected for stromal abundance, ZEB1 was significantly inversely correlated with all the cytokines examined (Supplemental Fig. 3A). ZEB1 was most significantly correlated with decreasing expression of IL-18 (partial r = −0.403), MIF (partial r = −0.394), CCL2 (partial r = −0.367) and CCL4 (partial r = −0.396). As these cytokines have been implicated in macrophage activation and polarization to the M1 phenotype, these data further support a role for ZEB1 in the suppression of antitumor macrophage activation31,32.
As an alternative to transcriptomic data, we investigated the association between stroma-corrected ZEB1 expression with immunohistochemistry-based measures of immune cell infiltration. When samples were split by median stroma-adjusted ZEB1 expression, ZEB1 was significantly associated with a lower percentage of monocytes (p = 0.026) and lymphocytes (p = 0.004) in primary tumor samples (Fig. 3G,H). Finally, we validated that the association between stroma-adjusted ZEB1 expression and immune infiltration was not specific to the TCGA BRCA cohort by repeating these analyses in the METABRIC breast cancer expression dataset33. ESTIMATE stromal and immune scores were generated with the ESTIMATE R package and the prior analyses were repeated (Supplemental Data 1)20. Unadjusted ZEB1 expression was significantly correlated with stromal score (r = 0.54) (Supplemental Fig. 3B) When ZEB1 expression was adjusted by stromal score, ZEB1 was significantly inversely correlated with immune score (partial r = −0.28) (Supplemental Fig. 3C).
EMT marker genes exhibit distinct patterns of association with tumor stromal and immune cell abundance
Together, our results strongly indicated that stroma-corrected ZEB1 expression is associated with a reduced antitumor immune response in breast cancer. Specifically, ZEB1 is inversely correlated with both the abundance of pro-inflammatory and immune-activating cells and with measures of immune activation and immune cell recruitment. This finding suggested two possible explanatory hypotheses: (1) That ZEB1 is a marker for EMT, and that EMT is driving the inverse association, or (2) that genes specifically under the regulation of ZEB1 are responsible for this relationship.
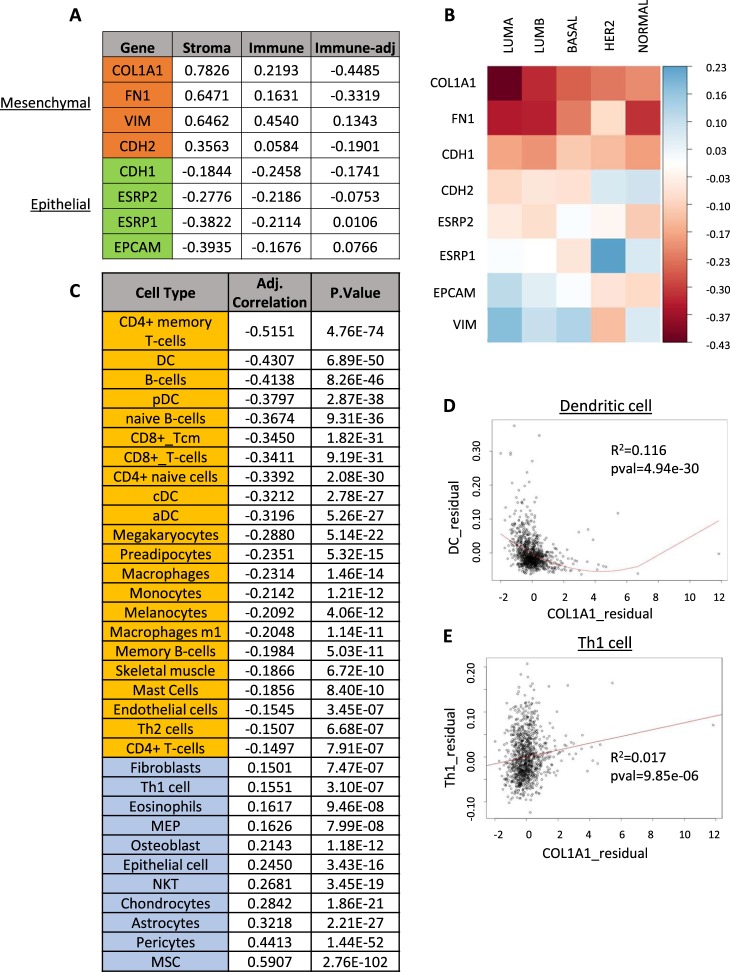
To investigate the possibility that the association we observed between stroma-corrected ZEB1 expression and anti-tumor immune activity were due to its role as a marker for EMT, we examined the relationship of a representative sample of well-characterized EMT maker genes with both stromal and immune cell abundance. Based on a literature review, the genes collagen 1a1 (COL1A1), fibronectin (FN1), vimentin (VIM) and N-cadherin (CDH2) were selected as mesenchymal markers, while E-cadherin (CDH1), epithelial splicing regulatory protein 1 and 2 (ESRP1,2), and epithelial cell adhesion marker (EPCAM) were selected as epithelial markers34. As expected, the mesenchymal marker genes were all positively correlated with stromal cell abundance, while the epithelial markers were inversely correlated (Fig. 4A). The same pattern was observed for the association between gene expression and immune cell content (Fig. 4A). A more complicated picture emerged when the partial correlation between each gene and immune cell abundance, adjusted for stromal cell abundance, was calculated. COL1A1 and FN1 exhibited strong inverse correlations with immune cell abundance, as expected (Fig. 4A). However, CDH2 exhibited a much lower inverse correlation, while VIM was positively correlated with immune cell abundance. Stroma-adjusted epithelial genes also showed a more complex relationship to immune cell content: EPCAM was modestly positively correlated with immune cells, ESRP1 was not significantly correlated, and both ESRP2 and CDH1 were moderately inversely correlated (Fig. 4A). We further profiled these associations by breaking the analysis up by PAM50 subtype. COL1A1, FN1 and CDH1 were all consistently negatively correlated with immune cell content, while the other markers did not exhibit consistent directional associations (Fig. 4B, Supplemental Table 5).
Figure 4.
Association of EMT markers with stromal and immune cell abundance in breast tumor samples. (A) Correlation of selected EMT marker genes with stromal, immune, and stroma-adjusted immune abundance. Mesenchymal markers are highlighted in orange, and epithelial genes are highlighted in green. (B) Heatmap representing partial correlation coefficients for EMT marker genes and immune cell abundance, adjusted for stromal cell content. (C) Partial correlation coefficients for COL1A1 and xCELL cell types adjusted for stromal content. (D,E) Residual plot of adjusted COL1A1 expression and dendritic cell (D) and Th1 cell (E) content.
These results suggested that a simple relationship between EMT and immune cell content was unlikely. However, the specific association of stroma-corrected COL1A1 expression with immune cell content was consistent with that observed for ZEB1. To further evaluate this correspondence, we calculated the partial correlation coefficients for COL1A1 with xCELL estimates of cell types (Fig. 4C, Supplemental Table 6). While COL1A1 was inversely associated with immune cell abundance, the cell types which showed the strongest associations were generally different from those associated with ZEB1. Both ZEB1 and COL1A1 exhibited a consistent inverse association with dendritic cell abundance (Fig. 4D, Supplemental Tables 4 and 6). However, COL1A1 was strongly inversely correlated with CD4+ memory T-cells, a relationship ZEB1 did not share. Similarly, ZEB1 was significantly inversely correlated with Th1 cell and macrophage abundance, while COL1A1 exhibited a much weaker association with both cell types (Fig. 4C,E). As a result, we concluded that the relationships of ZEB1 and COL1A1 expression to antitumor immunity are at least partially independent.
Derivation of a high confidence stroma-corrected ZEB1 expression signature
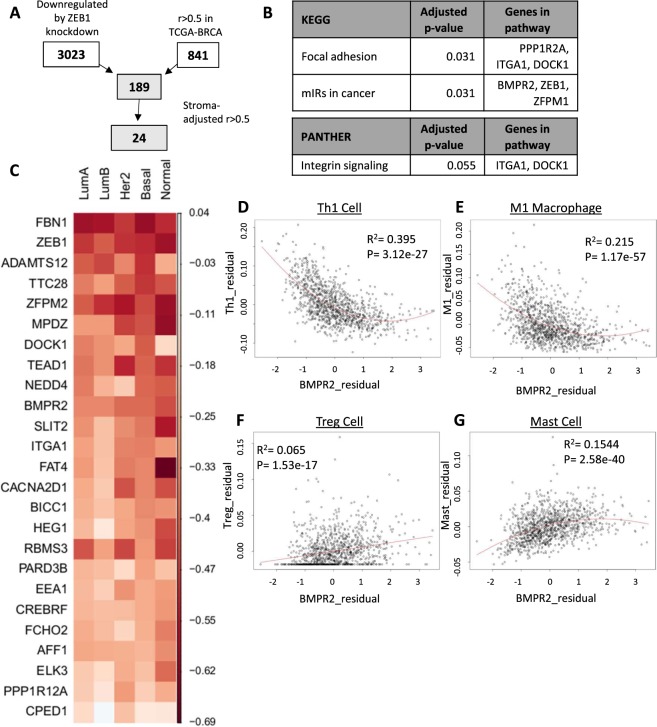
Since our results indicated that a general association between EMT-related gene expression and reduced immune cell activity was unlikely, we hypothesized that ZEB1 may specifically regulate the immune response by activating the expression of immune-modulating genes. To identify a set of high-confidence ZEB1-regulated genes, an integrative transcriptomic pipeline was used, as illustrated in Fig. 5A: First, a previous study in which ZEB1 was knocked down with shRNA in the MDA-MB-231 cell line and analyzed for differential gene expression was identified35. MDA-MB-231 is a mesenchymal breast cancer cell with high expression of ZEB1, providing an excellent model for studying ZEB1 transcriptional activity in a breast cancer model35. This data was reanalyzed to identify a set of 3023 genes which were significantly (FDR < 0.05, log(fold change) <−2) downregulated by knockdown of ZEB1. Next, the set of genes which were highly correlated with ZEB1 in the breast cancer TCGA dataset was curated with cBioPortal (Spearman r >0.5, 823 genes)18,19. A union of these two gene sets identified 186 genes which were both highly associated with ZEB1 expression and differentially expressed upon ZEB1 knockdown. This overlap was highly significant by the hypergeometric test, confirming that ZEB1-correlated genes are likely to be regulated by ZEB1 transcriptional activity (1.5-fold enrichment, p = 4.20 × 10−09). The correlation between each gene and ZEB1 was then adjusted by stromal score using partial correlation analysis. This approach identified 24 genes which remained significantly positively correlated with ZEB1 (partial r > 0.5) (Supplemental Fig. 4). Pathway analysis of this gene set identified significant enrichment for focal adhesion and integrin signaling, as well as for genes targeted by cancer-associated microRNAs (Fig. 5B).
Figure 5.
Stroma-adjusted ZEB1 transcriptional program is associated with decreased immune cell abundance in breast tumors. (A) Flowchart describing method for deriving ZEB1 transcriptional program in breast cancer. (B) Pathway analysis for 24-gene ZEB1 program. (C) Heatmap illustrating the partial correlation coefficients of ZEB1 program genes and immune content, adjusted for stromal content. (D–G) Residual plots for stroma-adjusted BMPR2 expression and Th1 (D), M1 macrophage (E), T-regulatory (F), and mast (G) cell content.
After deriving the ZEB1 transcription program, we evaluated the association between the individual genes within the signature and stromal cell infiltration. As expected, all genes exhibited a significant positive association with stromal infiltration (Table 2). We then adjusted the expression of each gene by stromal score and examined the association between measures of immune activity. We found that all 24 genes were significantly inversely correlated with ESTIMATE immune score after adjustment for stromal score (average r = −0.21, range −0.09 to −0.55) (Table 2). As before, this association was then evaluated in the context of PAM50 subtypes. All stroma-adjusted genes except CPED1 showed a consistent inverse association with immune cell abundance, although not all the partial correlation coefficients reached significance (Fig. 5C).
Table 2.
The ZEB1 transcriptional program is inversely associated with immune activity in breast tumors.
| Gene | Stromal score correlation | p.Value | Adj. Correlation | p.value |
|---|---|---|---|---|
| HEG1 | 0.842 | 5.81e-296 | −0.238 | 1.22E-15 |
| RBMS3 | 0.744 | 1.99e-193 | −0.381 | 4.40E-39 |
| FBN1 | 0.843 | 3.83e-296 | −0.584 | 4.10E-101 |
| ZFPM2 | 0.803 | 3.83e-296 | −0.462 | 5.42E-59 |
| FAT4 | 0.735 | 2.07e-186 | −0.318 | 3.35E-27 |
| SLIT2 | 0.697 | 3.22e-160 | −0.325 | 2.04E-28 |
| ELK3 | 0.666 | 2.20e-141 | −0.208 | 3.14E-12 |
| BICC1 | 0.736 | 4.43e-187 | −0.266 | 3.57E-19 |
| CPED1 | 0.715 | 7.40e-172 | −0.098 | 0.001126 |
| ADAMTS12 | 0.624 | 4.08e-119 | −0.439 | 9.89E-53 |
| ITGA1 | 0.616 | 2.24e-115 | −0.292 | 5.28E-23 |
| CACNA2D1 | 0.522 | 1.90e-77 | −0.286 | 4.38E-22 |
| BMPR2 | 0.453 | 1.83e-56 | −0.366 | 5.18E-36 |
| TTC28 | 0.495 | 7.43e-69 | −0.373 | 1.49E-37 |
| NEDD4 | 0.415 | 6.21e-47 | −0.338 | 9.59E-31 |
| TEAD1 | 0.359 | 1.15e-34 | −0.385 | 5.11E-40 |
| EEA1 | 0.322 | 8.14e-28 | −0.235 | 2.78E-15 |
| CREBRF | 0.319 | 2.48e-27 | −0.241 | 5.42E-16 |
| PARD3B | 0.342 | 1.57e-31 | −0.230 | 1.20E-14 |
| FCHO2 | 0.325 | 1.87e-28 | −0.246 | 1.36E-16 |
| AFF1 | 0.269 | 1.13e-19 | −0.265 | 4.45E-19 |
| DOCK1 | 0.315 | 1.20e-26 | −0.343 | 1.36E-31 |
| PPP1R12A | 0.269 | 1.37e-19 | −0.182 | 1.37E-09 |
| MPDZ | 0.269 | 1.19e-19 | −0.354 | 1.05E-33 |
As we hypothesized that genes within the ZEB1 transcriptional signature might be responsible for the observed inverse relationship to immune activity, a literature review was performed to identify known modulators of the immune response within the gene set. Of the 24 genes, bone morphogenic protein receptor 2 (BMPR2) was most consistently associated with immune activity in the literature. Inactivation or decreased expression of BMPR2 has a well-characterized association with familial pulmonary arterial hypertension, an effect which may be due to the role of BMPR2 in suppressing the response to TNF-mediated GM-CSF release36,37. BMPR2 inactivation has also been shown to increase intratumoral inflammation in mouse models of breast cancer38.
Since BMPR2 has a well-demonstrated association with reducing inflammation both physiologically and in models of breast cancer, we examined the specific associations between BMPR2 expression and the xCELL-determined abundance of cell types. Like ZEB1, BMPR2 was strongly negatively correlated with both Th1 cells and M1 macrophages, supporting an association with reduced pro-inflammatory cells within breast tumors (Fig. 5D,E, Supplemental Table 7). BMPR2 exhibited a more robust positive association with T-regulatory cell abundance than ZEB1 (Fig. 5F). Like ZEB1, BMPR2 was also positively correlated with mast cell infiltration (Fig. 5G). These findings confirmed that the ZEB1 transcriptional signature shares an overall inverse association with antitumor immunity and suggests that genes contained within this signature may be functionally responsible for this association.
ZEB1 expression and ZEB1 transcription activity is associated with worse overall survival in breast cancer
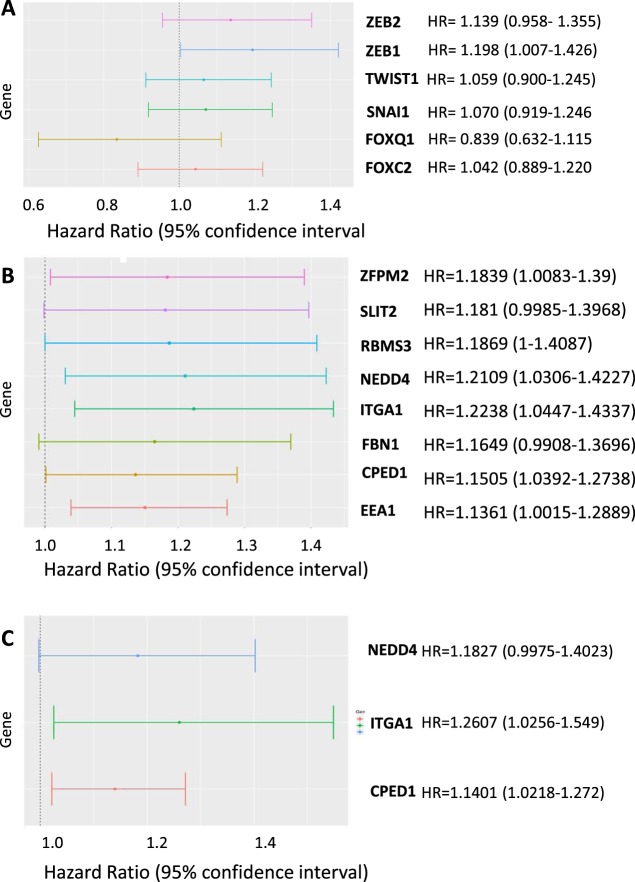
Finally, we evaluated the associations of ZEB1, other EMT-TFs and the ZEB1 program genes with overall survival in breast cancer patients. We observed that no EMT-TF was significantly associated with survival when included in a univariate Cox regression model. However, when the known prognostic factors of pathologic stage, the age of the patient at diagnosis and estrogen receptor status were included in the model, only ZEB1 was a significant independent predictor of overall survival (HR 1.21, 95% CI 1.00–1.42, p = 0.045) (Fig. 6A). Next, we included each ZEB1 program gene independently in the same multivariate model. We found that increased expression of 8 of 24 genes were significantly associated with decreased survival time (Fig. 6B).
Figure 6.
ZEB1 and the ZEB1 transcriptional program are associated with worse overall survival in breast cancer. (A) Forest plot of hazard ratios for multiple EMT-TFs in breast cancer, individually included in a multivariate Cox proportional hazards model. (B) Forest plot of 8 significantly prognostic genes within the ZEB1 transcriptional signature individually included in the multivariate Cox model. (C) Three genes within signature which remain significantly prognostic after adjusting for stromal score.
Finally, we examined how adjusting gene expression by stromal score affected the association with overall survival. Stromal score itself was not a significant independent prognostic factor (data not shown). When adjusted for stromal score, three genes (ITGA1, NEDD4, CPED1) remained significant independent prognostic factors, although all eight genes still exhibited a general trend towards an association with worse overall survival (Fig. 6C). This decrease in the number of genes reaching statistical significance was due to widening of the confidence interval for the residuals, rather than a directional effect. Together, these data indicate that expression of the ZEB1 transcriptional program is associated with reduced overall survival in breast cancer.
Discussion
This study demonstrates that stromal cells within tumors can significantly affect the statistical evaluation of associations between gene expression levels, tumor immune activity, and patient outcomes. By correcting for stromal cell abundance, our analyses demonstrated that previous attempts to study EMT using bulk tumor sequencing data were likely confounded. Our results also suggest that EMT-associated transcription factors and marker genes cannot be treated as interchangeable representatives of a common program. In fact, these results show a diverse range of associations between EMT-associated genes and both tumor stromal and immune cell content, indicated that each gene may have a unique functional association with the tumor microenvironment.
Previous studies have reported a significant association between EMT and immune activity in several cancers without accounting for the gene expression contribution of the stromal compartment11,13. We observed that the association between several EMT markers and immune cell infiltration were significantly influenced by stromal content. This suggests that previous findings linking EMT to altered immune activity may have been confounded by stroma, a possibility that has been suggested by other groups39. It was also shown that only some EMT markers have strong inverse associations with immune cell abundance, indicating that prior attempts to generalize an association between EMT-affiliated gene expression with the tumor microenvironment were misguided.
When adjusted for stromal cell abundance, ZEB1 gene expression is significantly inversely correlated with multiple measures of the immune response. This effect was strongest with ZEB1 among the several EMT-TFs examined. These results suggest that ZEB1 transcriptional activity in tumor cells may modulate the antitumor immune response. However, as this study relied on retrospective and correlative data, it is not yet possible to determine the causal relationship implied by this association.
Importantly, this study identified a novel inverse association between the transcription factor ZEB1 and immune activity in multiple cancer types. By deriving a set of ZEB1 activated and correlated genes, we demonstrated that ZEB1 transcriptional activity is also correlated with decreased tumor immune activity and provided a set of potential functional regulators of the tumor immune response for further investigation.
In addition to BMPR2, several genes within the stroma-adjusted ZEB1 transcriptional program have been characterized as modulators of immune activity. Fibrillins, including the ZEB1 program gene fibrillin 1 (FBN1), are critical regulators of immune activity40. Inactivating mutations in the cell-matrix interaction domain of FBN1 have been demonstrated to lead to significantly increased pro-inflammatory cell infiltration and fibrosis in the skin41. Another member of the ZEB1 transcriptional program, the ROBO receptor ligand SLIT2, has been demonstrated to inhibit the response of antigen-presenting cells to allergic skin reactions42. SLIT-ROBO signaling has also been implicated in the inhibition of leukocyte chemotaxis43. Beyond the set of genes identified by this study, ZEB1 has also been shown to directly regulate the expression of IL-6 and other cytokines by breast cancer cells to promote accumulation of myeloid-derived suppressor cells in a mouse model of breast cancer44. As IL-6 has also been shown to recruit mast cells to the tumor microenvironment, ZEB1-mediated IL-6 expression may explain the significant positive association we observed between ZEB1 expression and the abundance of intratumoral mast cells29. ZEB1 has also been shown experimentally to regulate PD-L1 expression in breast tumors, although our analysis did not identify a significant correlation between ZEB1 and PD-L1 expression45. Inhibiting ZEB1-mediated immune suppression, either directly or by inhibiting ZEB1-regulated immune modulators like BMPR2, may be a therapeutic approach to promote antitumor immune activity in breast cancer.
Methods
Expression and clinical data
Primary tumor RNAseqV2 expression and clinicopathologic data for the TCGA breast, lung, prostate, pancreatic and colon adenocarcinoma cohorts was downloaded from the cBioPortal in the form of z-scores19. Data was combined by comparison of sample ID and filtered to include primary tumor samples only. For the METABRIC cohort, microarray expression data in the form of normalized log intensity level was downloaded from the cBioPortal33. Microarray data from Lehmann et al. (E-MTAB-3482) for MDA-MB-231 with knockdown of either GFP (control) or ZEB1 were downloaded from https://www.ebi.ac.uk/arrayexpress 35.
Measures of tumor purity and immune cell content
ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data), which estimates stromal and immune cell content in tumors by using ssGSEA to rank samples on the expression of two 141-gene sets, was used to determine tumor purity. ESTIMATE stromal and immune scores for all TCGA datasets were obtained from the ESTIMATE online platform and matched to gene expression and clinical data from cBioPortal using sample ID codes46. For METABRIC data, ESTIMATE scores were generated using the estimate R package20. Immune activity scores were obtained from the TIP server30. Stromal and immune cell IHC percent estimates were obtained for the TCGA breast cancer dataset using Firebrowse and formatted with the psichomics R package47. Patients were split into two groups by median ZEB1 expression and evaluated by Wilcoxon rank-sum test to determine if the two groups exhibited significant differences in IHC-measured purity.
For cell-type specific analyses, the xCELL algorithm was used to generate estimates for the relative proportions of 64 cell types25. xCELL uses a deconvolution-based GSEA approach to determine the proportion of each cell type. xCELL scores for the TCGA dataset were downloaded from the xCELL server (http://xcell.ucsf.edu/). For immune activity profiling, the Tumor ImmunoPhenotype (TIP) pipeline was used30. This approach uses a similar, GSEA-based approach to evaluate the relative activity of the seven “steps” of the immune cycle within bulk tumor samples. TIP scores for the BRCA TCGA subset were downloaded from the online TIP server (http://biocc.hrbmu.edu.cn/TIP/).
Regression analyses
The association between gene expression, measures of tumor purity and clinical characteristics was performed using both linear models and partial correlations. Univariable analysis of variance (ANOVA) was used to test the significance each predictor variable. Multivariable ANOVA was used to identify a parsimonious set of predictor variables The ppcor R package was used to generate Spearman’s partial correlation coefficients22. The partial correlation coefficients were illustrated by plotting the residuals of the final parsimonious model excluding the variable of interest versus the residuals of a model with the variable of interest as the response versus the same parsimonious set of predictors (excluding the variable of interest). Linear models were used to generate the coefficient of determination for each association. As many of the relationships are quadratic in nature, the r2 value on plots may not precisely correspond to r values determined by partial correlation analysis.
Derivation of a ZEB1 transcriptional signature
First, microarray data from Lehmann et al. were processed and analyzed for differential expression using the Transcriptome Analysis Console (TAC) from ThermoFisher. The set of significantly downregulated genes (FDR < 0.05, log(Fold change) <−2) was obtained. Next, genes which were significantly (Spearman r >0.5) correlated with ZEB1 in the breast cancer TCGA dataset were identified and downloaded. The two gene sets were compared for intersection, with 186 genes contained in both sets. Finally, the BRCA-TCGA expression data for these genes in the breast cancer dataset was downloaded. The partial correlation for each gene with ZEB1 was computed, adjusting for the stromal cell content using the ppcor package. The overlap between ZEB1-regulated and -correlated genes was tested for significance using the hypergeometric test.
Survival analyses
Survival analysis was conducted with the survival package in R48. Univariate and multivariate Cox proportional hazards models were generated. The multivariate model included patient stage, age at diagnosis and estrogen receptor status, as these were all significant independent clinical predictors of overall survival. Plots representing the hazard ratio and 95% confidence interval were generated with ggplot2 in R49.
Supplementary information
Acknowledgements
This study was supported by T32-CA009531 (CJB.) and internal grant support from the Molecular Therapeutics program at Karmanos Cancer Institute (GW).
Author contributions
Study was conceived by C.J.B. and G.W. and designed by C.J.B. Analyses were performed by C.J.B. and I.J.C. and interpreted by C.J.B. G.D., D.W. and M.R. provided critical advice and feedback during the execution of the study. Funding was obtained by C.J.B. and G.W. Manuscript was written by C.J.B. with input and consent of all authors.
Data availability
All data from the TCGA and METABRIC was downloaded by either cBioPortal or Firehose. Microarray data from Lehmann et al. was obtained from GEO. All final outputs from our analyses are available as csv files upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54282-z.
References
- 1.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nistico P., Bissell M. J., Radisky D. C. Epithelial-Mesenchymal Transition: General Principles and Pathological Relevance with Special Emphasis on the Role of Matrix Metalloproteinases. Cold Spring Harbor Perspectives in Biology. 2012;4(2):a011908–a011908. doi: 10.1101/cshperspect.a011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat Cell Biol. 2019;21:102–112. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- 6.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 7.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dongre A, et al. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 2017;77:3982–3989. doi: 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Ricciardi M, et al. Epithelial-to-mesenchymal transition (EMT) induced by inflammatory priming elicits mesenchymal stromal cell-like immune-modulatory properties in cancer cells. Br J Cancer. 2015;112:1067–1075. doi: 10.1038/bjc.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mak MP, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lou Y, et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chae YK, et al. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC) Sci Rep. 2018;8:2918. doi: 10.1038/s41598-018-21061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George JT, Jolly MK, Xu S, Somarelli JA, Levine H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017;77:6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chockley PJ, Keshamouni VG. Immunological Consequences of Epithelial-Mesenchymal Transition in Tumor Progression. J Immunol. 2016;197:691–698. doi: 10.4049/jimmunol.1600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, et al. EMT- and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat Commun. 2018;9:3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris JS, Kopetz S. Tumor Microenvironment in Gene Signatures: Critical Biology or Confounding Noise? Clin Cancer Res. 2016;22:3989–3991. doi: 10.1158/1078-0432.CCR-16-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosuke Yoshihara, H. K. a. R. G. V. & estimate: Estimate of Stromal and Immune Cells in Malignant Tumor Tissues from Expression Data. R package version 1.0.13/r21, https://R-Forge.R-project.org/projects/estimate/ (2016).
- 21.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S. ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients. Commun Stat Appl. Methods. 2015;22:665–674. doi: 10.5351/CSAM.2015.22.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger AC, et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 2018;33:690–705 e699. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 27.Heusinkveld M, et al. M2 macrophages induced by prostaglandin E2 and IL-6 from cervical carcinoma are switched to activated M1 macrophages by CD4+ Th1 cells. J Immunol. 2011;187:1157–1165. doi: 10.4049/jimmunol.1100889. [DOI] [PubMed] [Google Scholar]
- 28.Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol. 2019;16:356–371. doi: 10.1038/s41571-019-0175-7. [DOI] [PubMed] [Google Scholar]
- 29.Dalton DK, Noelle RJ. The roles of mast cells in anticancer immunity. Cancer Immunol Immunother. 2012;61:1511–1520. doi: 10.1007/s00262-012-1246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L, et al. TIP: A Web Server for Resolving Tumor Immunophenotype Profiling. Cancer Res. 2018;78:6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 31.Sierra-Filardi E, et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. J Immunol. 2014;192:3858–3867. doi: 10.4049/jimmunol.1302821. [DOI] [PubMed] [Google Scholar]
- 32.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann W, et al. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun. 2016;7:10498. doi: 10.1038/ncomms10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, et al. Inflammation, endothelial injury, and persistent pulmonary hypertension in heterozygous BMPR2-mutant mice. Am J Physiol Heart Circ Physiol. 2008;295:H677–690. doi: 10.1152/ajpheart.91519.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawada H, et al. Reduced BMPR2 expression induces GM-CSF translation and macrophage recruitment in humans and mice to exacerbate pulmonary hypertension. J Exp Med. 2014;211:263–280. doi: 10.1084/jem.20111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owens P, et al. Disruption of bone morphogenetic protein receptor 2 (BMPR2) in mammary tumors promotes metastases through cell autonomous and paracrine mediators. Proc Natl Acad Sci USA. 2012;109:2814–2819. doi: 10.1073/pnas.1101139108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terry S, et al. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11:824–846. doi: 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeyer KA, Reinhardt DP. Fibrillin-containing microfibrils are key signal relay stations for cell function. J Cell Commun Signal. 2015;9:309–325. doi: 10.1007/s12079-015-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerber EE, et al. Integrin-modulating therapy prevents fibrosis and autoimmunity in mouse models of scleroderma. Nature. 2013;503:126–130. doi: 10.1038/nature12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan H, et al. Neuronal repellent Slit2 inhibits dendritic cell migration and the development of immune responses. J Immunol. 2003;171:6519–6526. doi: 10.4049/jimmunol.171.12.6519. [DOI] [PubMed] [Google Scholar]
- 43.Wu JY, et al. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katsura A, et al. ZEB1-regulated inflammatory phenotype in breast cancer cells. Mol Oncol. 2017;11:1241–1262. doi: 10.1002/1878-0261.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saraiva-Agostinho Nuno, Barbosa-Morais Nuno L. psichomics: graphical application for alternative splicing quantification and analysis. Nucleic Acids Research. 2018;47(2):e7–e7. doi: 10.1093/nar/gky888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T, T. A Package for Survival Analysis in S. version 2.38, https://CRAN.R-project.org/package=survival. (2015).
- 49.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from the TCGA and METABRIC was downloaded by either cBioPortal or Firehose. Microarray data from Lehmann et al. was obtained from GEO. All final outputs from our analyses are available as csv files upon request.