Abstract
Environmentally-responsive genes can affect fruit red colour via the activation of MYB transcription factors. The apple B-box (BBX) gene, BBX33/CONSTANS-like 11 (COL11) has been reported to influence apple red-skin colour in a light- and temperature-dependent manner. To further understand the role of apple BBX genes, other members of the BBX family were examined for effects on colour regulation. Expression of 23 BBX genes in apple skin was analysed during fruit development. We investigated the diurnal rhythm of expression of the BBX genes, the anthocyanin biosynthetic genes and a MYB activator, MYB10. Transactivation assays on the MYB10 promoter, showed that BBX proteins 1, 17, 15, 35, 51, and 54 were able to directly function as activators. Using truncated versions of the MYB10 promoter, a key region was identified for activation by BBX1. BBX1 enhanced the activation of MYB10 and MdbHLH3 on the promoter of the anthocyanin biosynthetic gene DFR. In transformed apple lines, over-expression of BBX1 reduced internal ethylene content and altered both cyanidin concentration and associated gene expression. We propose that, along with environmental signals, the control of MYB10 expression by BBXs in ‘Royal Gala’ fruit involves the integration of the expression of multiple BBXs to regulate fruit colour.
Subject terms: Light responses, Secondary metabolism
Introduction
Apple is an important fruit crop, both commercially and from a dietary nutrition perspective. Compounds found in apple have been shown to have positive effects on human health1,2. These compounds are derived from secondary metabolites and comprise a diverse set of bioactive phytochemicals including phenylpropanoids, which contain the flavonoid subclass that includes anthocyanins, the pigments responsible for the red colour of apple. Potential health benefits include anti-inflammatory, anti-carcinogenic, and antioxidant properties3,4. Fruit colour is also a strong market driver, with consumer preferences varying between countries and regions5,6. In general, consumer appeal tends towards red colour, with red apples being preferred to yellow or green cultivars5–7. Apple colour and patterning varies naturally within and between cultivars, and in many apple cultivars this is influenced by environmental conditions, modulating the concentrations of chlorophyll, carotenoid, and anthocyanin8,9. The red colour of apple skin is composed almost exclusively of the anthocyanin derivative, cyanidin-3-O-galactoside.
Anthocyanin is important for plants as it protects against photo-oxidative and heat damage caused by sunlight, as well as influencing pollination and seed dispersal10–12. The accumulation of anthocyanins in plants appears to be linked to biotic and abiotic stress, with anthocyanin concentrations being increased in response to herbivory, fungal and viral pathogens, wounding, temperature, high light and UV, mineral imbalance, drought, salinity, anoxia, ozone, herbicides13. Research on the regulation of anthocyanin production has identified both the biosynthetic pathways genes14–16 and the major regulating transcription factors (TFs), comprising the MYB-bHLH-WD40 (MBW) complex17. This complex binds promoter regions of anthocyanin biosynthetic genes and MYB10 and increases transcription, as shown for Arabidopsis (Arabidopsis thaliana)17, nectarine18, petunia (Petunia hybrida)15, strawberry19, legumes20, Norway spruce21, and apple22, and reviewed by Li23.
The MYBs that regulate anthocyanin accumulation in apple have been well studied22,24–26. Alleles MYBA and MYB1 are identical and share 98% sequence homology with MYB10, which differs by three amino acids in the open reading frame25. The R2R3 binding region of these and anthocyanin-related MYBs in other plant species is highly conserved25. In apple, MYB10, bHLH3/33 and a WD40 protein TTG1 have been shown to form the MBW complex seen in other species to regulate anthocyanin concentrations26. Patterning of apple fruit skin includes striped and blushed, and this variation can be explained by the differential expression of MYB10 and degree of methylation in the promoter27.
Repressors also play an important role in the regulation of anthocyanin production. The genes involved and the mechanisms by which they operate are beginning to be resolved. For example, two repressors have been reported in petunia, MYB27 and MYBx15,28. MYB27 is a R2R3 type MYB, which can interfere with the MBW complex by preventing its formation or being incorporated, and converting the complex into a repression complex. MYBx is a R3 type MYB that also appears to compete for interaction with the bHLH component of the MBW complex. MYB27 contains an EAR (ethylene-responsive element binding factor associated amphiphilic repression domain) and TLLLFR motif, both of which appear to confer the ability to repress anthocyanin15. MYBL2, an R3-MYB, has been identified in Arabidopsis as a repressor of anthocyanin and also contains the TLLLFR motif29,30. Similar repression mechanisms have also been identified in strawberry31. In apple, a family of MYB repressors of anthocyanin were identified after heat treatment and were characterised by the EAR motif32.
Repression and activation of the anthocyanin pathway is responsive to environmental signals. Lin-Wang et al.32 have shown that heat reduces apple skin MYB10 expression, while Xie et al.33 showed that cold induces bHLH3 to bind with MYB1 and upregulate the promoters of anthocyanin biosynthetic genes. Appropriate light conditions are also required for anthocyanin accumulation in many apple cultivars34,35 and anthocyanin has been shown to increase in response to ultraviolet-B (UV-B) irradiation and to absorb UV-B7,36. Long-term changes, enduring after six weeks of postharvest storage, were found in apples grown on trees covered by a canopy of UV-reducing film37. The reduction of UV affected a wide range of fruit qualities, including a delay in ripening, reduction in fruit size, and decrease of anthocyanin and flavonol concentrations. Many of the transcription factors responsible for these environmentally induced anthocyanin changes have been identified. As a consequence of these genes being responsive to environmental signals they can exhibit expression rhythms that follow the stimulus. These rhythms can be detected by measuring transcription. In Arabidopsis, ELONGATED HYPOCOTYL 5 (HY5), a basic leucine zipper (bZIP) transcription factor, receives signals from photoreceptors and regulates anthocyanin by directly binding promotors of PAP1 (AtMYB75) and anthocyanin biosynthetic genes38. HY5 is also involved in low temperature-induced anthocyanin accumulation39. LIGHT-REGULATED ZINC FINGER PROTEIN 1 (LZF1, or B-BOX DOMAIN PROTEIN 22) has been found to act downstream of HY5 to inhibit anthocyanin40. The photoreceptor UV RESISTANCE LOCUS 8 (UVR8) provides light-signalling control of various pathways, including regulating HY5 and interacting with CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)41,42. COP1 is a RING finger type ubiquitin E3 ligase involved in the targeted degradation of various anthocyanin influencing factors, including MYB1 in apple43–45.
Although many transcription factors have been identified in the regulation of anthocyanin biosynthesis, a full understanding of how light regulates this process is still being developed. The role BBXs play is an example. Studies focusing on specific members of the BBX gene family in apple have shown that UV-B light in particular upregulates MdCOL11/BBX33 expression in a temperature-dependent manner7. MdBBX33 is a close homolog of AtBBX22/LZF1, also known as STH3 (Salt Tolerance Homolog 3) and DBB3 (Double B-Box zinc finger 3)7,40,46. Bai et al.7 generated transgenic lines of Arabidopsis overexpressing apple BBX33, which had increased anthocyanin accumulation in seedlings and showed also that the expression profiles of BBX33 and MYB10 are related during different temperature and light regimes. Using dual luciferase assays it was shown that BBX33 upregulates the MYB10 promoter. These findings suggest that BBX33 functions downstream of HY5 and upstream of MYB10 as a component in the environmental sensing pathway, which leads to anthocyanin production in apple.
Discovery of the first BBX, in Arabidopsis, came from characterising the flowering pathway. CONSTANS (CO/BBX1) was shown to be important for regulating transcription levels of floral integrators where it is a key factor controlling photoperiodic flowering response in Arabidopsis47–51. CO/BBX1 was subsequently identified as a member of the larger BBX gene family. This is a family characterised by the proteins they encode possessing two highly conserved domains, an N-terminal zinc finger BBX52,53 and a C-terminal CCT (CO, CO-like, TIMING OF CAB EXPPRESSION 1) domain54–56. Both domains are involved with mediating transcriptional regulation and protein-protein interaction. Arabidopsis has 32 BBX genes (1–32), which fall into five groups (I-V) based on common variations in the conserved BBX and CCT domains53. Twenty-one of the 32 AtBBX gene sequences contain two BBXs in tandem, the remaining 11 contain one BBX55,57. This classification of BBX genes using number of conserved motifs has been reiterated by Huang et al.58 in rice. Further analysis, which included comparison of BBX sequences across a diverse range of species, has been conducted by Crocco and Botto53 where 214 BBX proteins were identified and compared, providing evolutionary insight into these genes.
More recently, Liu et al.59 reported a comprehensive list of the apple BBX gene family. A total of 64 MdBBXs were identified. The phylogeny and motif structure were reported, along with chromosomal locations. Although naming these genes as ‘COLs’ (for CONSTANS-LIKE) has been reported56,60–64, Liu et al.59 and others53,55 selected ‘BBX’ as the convention so as to include genes that do not contain the 3’ CCT motif, and to align with the naming convention reported by Khanna et al.57 in Arabidopsis.
Here we examined members of the BBX gene family in apple, using the naming convention from Liu et al.59, with an emphasis on potential roles in anthocyanin regulation. The expression of 23 BBXs, representing the five sub-clades in the gene family, in Malus x domestica ‘Royal Gala’ fruit skin during fruit development was assessed. Given that many BBX genes, including Arabidopsis CO, COL1 and COL2, show diurnal patterns of expression64,65, a detailed examination of diurnal expression patterns in both candidate apple BBX genes and anthocyanin structural genes was performed. Expression profiles of anthocyanin biosynthetic genes and the MYB regulator were tested for expression correlations. A subset of the BBX proteins were screened for their ability to activate the promoter of MYB1/10, which was also tested for any dependent BBX-related motifs. Furthermore, the activation of the promoter of the anthocyanin biosynthetic gene DIHYDROFLAVONOL 4-REDUCASE (DFR) by MYB10 and bHLH3 was assessed in the presence of BBX1. Finally, we generated and characterized five transgenic lines of apple trees overexpressing BBX1 and measured postharvest metrics and metabolite content differences compared to wildtype.
Materials and Methods
Plant material
Apple fruit tissue used for the development series was sampled from ‘Royal Gala’ trees grown in standard conditions in a glasshouse in 2012/2013 at seven time points during development: 35, 65, 85, 110, 120, 130 and 140 days after full bloom (DAFB) at 1300 h. Full bloom occurred on 28 September 2012 and at each time point five fruit were harvested and pooled. Apple fruit tissue used for diurnal sampling were collected from ‘Royal Gala’ apple trees in the Plant and Food Research orchard, Motueka, Nelson, New Zealand, on 8–10 February 2011 (season one) and from the Plant and Food Research orchard, Mt Albert, Auckland, New Zealand, on 27 February - 2 March 2012 (season two). Apple leaf tissue was harvested at the same time as fruit using five pooled leaves per time point. Fruit sampling began at 120 DAFB. Due to length of time course, number of fruit harvested per time point had to be limited to three fruit at each time point, which occurred at 1300 h, and then pooled. To help with this, two seasons were sampled. Apple trees over-expressing BBX1 were grown in the Plant and Food Research glasshouse in Auckland. Three pooled fruit were used for each replicate and run in quadruplicate for gene expression analysis and the average reading for five fruit is presented for ethylene, weight, firmness, and soluble solids content (measured as °Brix). High performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LCMS) data are also presented as the average reading from five fruit. For each fruit all of the skin was removed using a scalpel and immediately frozen in liquid nitrogen along with leaf samples, ground to powder without thawing, then stored at −80 °C until RNA extraction for RT-qPCR analysis.
Promoter and gene cloning
Full-length coding sequences of MYB10, MYB8, bHLH3, bHLH15 and BBX1, 9, 10, 15, 17, 33, 35, 37, 42, 43/44, 47, 51, 54, 57 were isolated from ‘Royal Gala’ cDNA and inserted into the pGEM-T Easy vector (Promega, Madison, USA) before being inserted into the pHEX2 binary vector via restriction site cloning66. Promoter sequences for the R1 allele of MYB10, DFR, BBX1, and BBX15 were isolated from ‘Royal Gala’ genomic DNA and inserted into the cloning site of pGreenII08005’-LUC as previously described by Espley et al.67. Truncated versions of the promotor of MYB10 (∆a and ∆b) were generated by designing primers to amplify 834 bp and 405 bp fragments upstream of the MYB10 start site. Primer sequences are given in Supplementary Table 1.
Dual luciferase assay of transiently transformed Nicotiana benthamiana leaves
Nicotiana benthamiana plants were used for transient transformation assays. These plants were grown at Plant and Food Research, Auckland, according to standard glasshouse management practices. Agrobacterium tumefaciens GV3101 was transformed by electroporation to introduce constructs. N. benthamiana leaves were transiently transformed for the dual luciferase assay as previously reported by Lin-Wang et al.68.
Metabolite analysis
Anthocyanin content was measured by HPLC. For the more comprehensive analysis of the BBX1 over-expressing lines, HPLC was used to detect anthocyanins and polyphenols. Ground apple skin tissue (100 mg) was freeze-dried for 12 hours. Acidified (0.5% HCL) methanol (5x volume) was added to the tissue and placed in the dark for 3 hours to extract. Samples were centrifuged at 16,000 rpm for 4 minutes, then supernatant transferred to a new tube for vacuum evaporation. Extract was then re-suspended in 500 µL of 20% methanol, filtered into a new tube and vacuum evaporated. Extract was then finally re-suspended in 20 µL methanol for analysis. Extract was analysed on a Dionex Ultimate 3000 RS system as previously described by Dare et al.69.
Gene expression analysis by RT-qPCR
RNA was isolated using Spectrum™ RNA extraction kit (Sigma-Aldrich) according to the manufacturer’s instructions. RNA quantification was performed using a NanoDrop™ ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Thermo Scientific, USA) and visualized by gel electrophoresis. First strand cDNA synthesis of 2 µg total RNA was performed using Qiagen QuantiTect® Reverse Transcription Kit (Qiagen). Expression analysis was performed using four technical replicates on a Roche LightCycler® 480 platform and LightCycler FastStart SYBR Green I Master Mix as described previously22. Data was analysed using the Target/Reference ratio calculated with the LightCycler 480® software version 1.5.0.39 (Roche) and primer efficiency corrections added. CP values for housekeeper, Actin, did not vary by more the 1.72 cycles during the 48 h and 52 h of sampling for season one and two respectively. Primers were designed to optimise efficiency as previously described32. Primer sequences are given in Supplementary Table 1.
Internal ethylene assessment
Internal ethylene concentration of apple fruit was determined using a needle and syringe to extract gas from the internal core cavity of attached fruit during season 2 of sampling at 130 DAFB at 1300 h. BBX1 over-expressing lines samples were taken at 125 DAFB at 1300 h. Core cavity gas (1 mL) was injected into a gas chromatograph (Hewlett Packard, 5890 series II) and run according to the method of Johnston et al.70.
Statistical analysis
To test correlations between genes, the gene expression data were used to generate correlation coefficient values using two-sided Pearson’s product-moment correlation in R version 3.2.471. Differences between averages were analysed using Analysis of Variance (ANOVA) in GenStat version 17.1.0.14713. Post hoc pairwise differences were determined using Fisher’s LSD test at the 5% significance level72.
Stable transformation of apple
The cDNA sequence of apple BBX1 was cloned into the binary vector pSAK27722 driven by the 35 S promoter and transferred into Agrobacterium strain LBA4404. Transgenic lines of ‘Royal Gala’ plants were generated by Agrobacterium-mediated transformation of leaf pieces using a method previously reported by Yao et al.73. Transgenic plants were rooted and grown in a containment glasshouse at The New Zealand Institute for Plant and Food Research Limited (PFR), Auckland, New Zealand, with flowering and fruiting occurring three years after planting.
Results
Anthocyanin concentration and gene expression of the BBX family in fruit skin during development
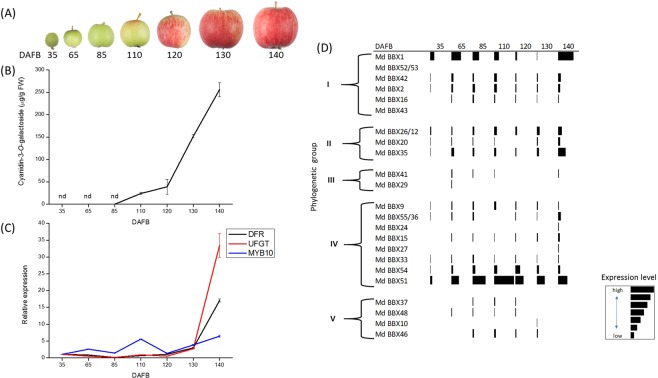
Apple fruit were assessed at seven time points during development: 35, 65, 85, 110, 120, 130, and 140 DAFB (Fig. 1A). At each time point, concentrations of cyanidin-3-O-galactoside, the expression of apple anthocyanin biosynthetic genes DFR and URIDINE DIPHOSPHATE (UDP)-GLUCOSE FLAVONOID 3-O-GLYCOSYLTRANSFERASE (UFGT), and the expression of anthocyanin-regulating transcription factor MYB10, as well as that of 23 genes from the BBX family were measured. Cyanidin-3-O-galactoside content was detectable at 110 DAFB, increasing up to 140 DAFB (Fig. 1B). Expression of DFR, UFGT, and MYB10 increased during development, with the highest expression for all three genes seen at 140 DAFB (Fig. 1C). Transcripts of a subset of the BBX family genes, representing all five sub clades59, were also examined in apple skin during fruit development (Fig. 1D). This identified BBXs with expression patterns that coincided with anthocyanin accumulation in developing fruit. The highest and lowest expression for the genes varied over the six time points measured within and between the different groups of BBXs. Transcript abundance of many of the BBXs started low and then increased during fruit development (data presented at higher resolution in Supplementary Fig. 1). BBX51 and BBX1 were the two most highly expressed BBXs examined. Expression of BBX51 was highest at 110 DAFB, while BBX1 had an expression peak early in development (65 DAFB) and then again at 140 DAFB. Some genes, such as BBX15, were relatively lowly expressed but still displayed increasing expression during fruit development (Supplementary Fig. 1). Overall, BBXs from groups I, II and III had variable patterns of gene expression in the skin during fruit development, with the highest expression occurring early, middle, or late in the sampling. Group IV BBXs generally displayed their lowest expression at 35 DAFB, with the exception of BBX27 (Supplementary Fig. 1). Most group V BBXs started with low expression and peaked at different times around the mid-range of the sampling period.
Figure 1.
Assessment of skin anthocyanin accumulation during apple fruit development compared with BBX gene expression. Apple fruit were assessed at seven time points; 35, 65, 85, 110, 120, 130, and 140 DAFB. (A) Photographs of representative apple fruit at each stage of development assessed showing accumulation of red colour. (B) Cyanidin-3-O-galactoside accumulation in apple fruit skin at seven time points during development. Values are the average concentration of cyanidin as determined by HPLC from two pooled fruit per time point. Time points where no cyanidin was detected are indicated by ‘nd’. Error bars show SE of four technical replicates. (C) Relative expression of DFR, UFGT, and MYB10 in apple fruit skin. Expression is relative to Actin normalized to 35 DAFB. Error bars show SE of four technical replicates. (D) Table showing relative expression level of selected BBXs in apple fruit skin at seven time points during development. Phylogenetic groups reported by Liu et al.59 indicated. Black bar at each DAFB indicates level of expression relative to the other BBX genes. Key shows that the length of the black bars indicates level of BBX expression relative to Actin and normalised to highest expressed BBX (BBX51 at 110 DAFB).
Measuring diurnal patterns of transcription in apple leaves and fruit
Having demonstrated that the transcription of certain BBX genes increases during fruit development in a similar pattern to the concentration of anthocyanin and key anthocyanin-related genes, we then tested the transcript abundance of selected genes during a diurnal time series at 120 DAFB. This is the stage at which anthocyanin is rapidly accumulating. To confirm that variation in expression could be detected in apple diurnally, we tested transcription of apple GIGANTEA (GI) in leaf and fruit tissue (Fig. 2). GI was selected as a positive control because of its role in the circadian pathway and its known pattern of expression74,75. GI was found to be expressed in a highly regular pattern (high in the evening and low in the morning) in accordance with findings in Arabidopsis74,75. The expression of BBX1 showed a distinct pattern of expression in the apple leaf, with the highest expression levels around 0400 h and lowest in the evening. Other genes, such as BBX51, DFR, HY5, and COP1 also exhibited daily transcript fluctuations in apple leaves (Supplementary Fig. 2).
Figure 2.
Diurnal gene expression levels at four hour intervals over two seasons. Gene expression for apple Gigantea (GI) and BBX1 in apple leaf tissue and fruit skin tissue labelled, all other genes are fruit skin results. Other genes tested were light-associated COP1, HY5, and UVR8, temperature-linked genes CBF2 and HSFA2a and BBX genes 15, 33, and 51 from BBX group IV over two seasons. Expression results were normalised to Actin and error bars are SE of four technical replicates. Daylight hours were from 0640 h to 2040 h for season 1 and from 0700 h to 2000 h for season 2, rounded to the nearest 10 min. Black bars indicate night.
Diurnal rhythms of gene expression were assessed in fruit skin over two seasons and two locations, to control for environmental and seasonal variation (Fig. 2). GI expression in skin showed a diurnal expression rhythm with the highest expression in the evening at 2000 h and lowest at 0800 h for both seasons and sites, as was seen in leaves. BBX1 peaked at 0400 h and was lowest in the evening, as was seen in leaves. A selection of light and temperature genes were also measured in apple skin to ascertain if detection of daily rhythms for genes reported to be involved in light and temperature perception was also possible in apple skin. These included COP1, HY5, and UVR8 involved in light response (Fig. 2), and CBF2 and HSFA2a involved in temperature-regulated expression (Fig. 2)76,77. A further six genes were tested (Supplementary Fig. 3). COP1 and HY5 expression patterns were similar, displaying peaks during daylight. These findings for apple COP1 support work in Arabidopsis where COP1 expression is high in the afternoon43. Peaks in the expression of UVR8 coincided with light periods except for the first night of season 2. As expected, CBF2 expression was elevated at night and HSFA2a was highest during the day (Fig. 2).
BBXs 51, 33, and 15 were also selected for assessment (Fig. 2). BBX51 was chosen as it showed the highest expression during development (Fig. 1D), BBX33 because of its involvement in anthocyanin accumulation7, and BBX15 as it showed increased transcription over fruit development (Fig. 1D). Expression of BBX51 was highest in the morning and lowest in the evening. Expression of BBX33 was not found to be as rhythmic as the other BBX genes tested for season 1, and in season 2 it showed highest expression in the evening and lowest in the morning. BBX15 was also highest in the evening and lowest in the morning, for both seasons tested.
Correlation analysis of MYB10 and anthocyanin pathway genes
To further investigate diurnal gene expression in apple fruit skin tissue, we analysed the expression of anthocyanin structural genes PHENYLALANINE AMMONIA LYASE (PAL), CHALCONE SYNTHASE (CHS), CHALCONE ISOMERASE (CHI), FLAVANONE 3-HYDROXYLASE (F3H), DFR, LEUCOANTHOCYANIDIN DIOXYGENASE/ANTHOCYANIDIN SYNTHASE (LDOX/ANS), UFGT, and the MYB10 TF (Fig. 3A). All the biosynthetic genes have a similar diurnal expression pattern to MYB10, with correlation analysis confirming a positive linear relationship, with four of the seven genes tested being statistically significant (p < 0.05) (Fig. 3B). However, no direct diurnal correlation with MYB10 and BBX transcripts was found (Supplementary Table 2), even when taking into account a possible delay in the correlation by analysing for the effect on MYB10 with a four- or eight-hour delay (data not shown).
Figure 3.

(A) Diurnal gene expression at four-hour intervals for MYB10 and seven anthocyanin biosynthetic genes (PAL, CHS, CHI, F3H, DFR, LDOX, and UFGT). Expression was normalised to Actin and error bars are SE of four technical replicates. Daylight hours were 0640 h to 2040 h, rounded to the nearest 10 min. Black bars indicate night. (B) Table of Pearson’s product-moment correlation coefficient values for seven anthocyanin biosynthetic genes compared to MYB10 (p-value < 0.05*, <0.01**, <0.001***).
Activation of the MYB10 promoter by BBX genes
Members of the apple BBX gene family were selected for promoter activation assays based on their expression during development (Fig. 1D and Supplementary Fig. 1). These 14 BBX genes were screened for activation of the MYB10 promoter using the dual luciferase system in N. benthamiana. The MYB10 promoter was infiltrated alone, or co-infiltrated with MYB10 + /- MdbHLH3 (Fig. 4). Compared with the controls, BBX1 alone activated the MYB10 promoter and showed an additive affect when co-infiltrated with MYB10, but no further activation was found when bHLH3 was added. BBX51 and BBX54 both activated the promoter when co-infiltrated with MYB10, but this effect was largely lost after the addition of bHLH3. BBX15 did not activate the promoter alone; however, the promoter was activated by the addition of MYB10 and MYB10 plus bHLH3. The highest activation of the promotor was when BBX35 was co-infiltrated with MYB10 and bHLH3. From these data, we identified BBX1, 15, 35, 51 and 54 as BBXs capable of activating the promoter of MYB10. Furthermore, the addition of the bHLH co-factor gene displayed all three possible effects, depending on the BBX: no change, or decreased or increased activation.
Figure 4.

Transient expression assay of apple BBXs effect on the promoter of apple MYB10 in tobacco. pR1 denotes the MYB10 promoter to distinguish it from the MYB10 protein. Each BBX was infiltrated with pR1 (light grey bars), with pR1 and MYB10 (medium grey), and with pR1, MYB10, and bHLH3 (dark grey). Phylogenetic groups as reported by Liu, et al.59 are indicated (groups I, II, IV, and V) and error bars show SE of four biological replicates.
Deletions in the MYB10 promotor reveal specific sites required for BBX1 activation
To further characterise the activation of the MYB10 promoter by BBX genes, we generated two truncated versions of the 1704 bp promoter sequence of MYB10 (Fig. 5A), MYB10∆a and MYB10∆b at 834 bp and 405 bp, respectively, upstream of the ATG start site. We assessed the promoter sequence for cis-elements which might facilitate interaction with BBXs and found CCAAT motifs present. In yeast these motifs are bound by HEME ACTIVATOR PROTEINS (HAP), also known as NUCLEAR FACTOR Y (NF-Y) proteins, facilitated by domains similar to the CCT domain of many plant BBXs in plants, including BBX78,79. Due to some BBXs containing the CCT domain it is possible they will bind the same CCAAT motif as the HAP proteins. The full MYB10 promotor sequence contains three CCAAT motifs, MYB10∆a contains one, while MYB10∆b contains none. The infiltration of BBX1 alone resulted in a slight activation of MYB10 and MYB10∆a compared with MYB10 or bHLH3 controls (Fig. 5B). Most notably, the ability for the promoter to be activated by MYB10 and BBX1 dropped significantly for the Δb promotor sequence (Fig. 5B). This also applied if bHLH3 was present. Apple MYB8 was included as a control to show specificity of the MYB10 infiltration co-factor to BBX1 activation of the promoter22.
Figure 5.
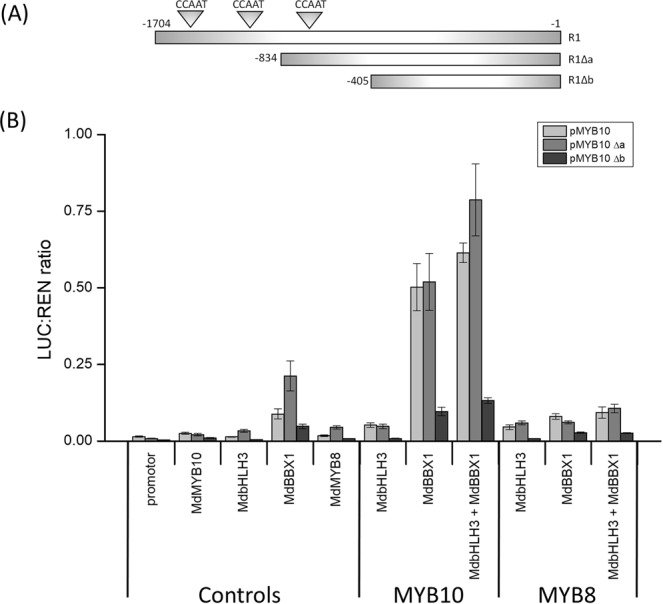
Identification of the region of the MYB10 promoter activated by BBX1. (A) Schematic of the full length and two truncated versions (Δa and Δb) of the MYB10 promoter used to identify the region sensitive to the presence of BBX1. Position of three CCAAT motifs indicated. (B) Transient expression assay using the full length MYB10 promoter and the two truncated versions. Full length and truncated promoters were run alone, with MYB10, and with MYB8 to test activation capability of BBX1 with and without a bHLH cofactor. Error bars show SE of four biological replicates.
The apple DFR promoter is activated by BBX1
Having demonstrated that some BBXs activated the MYB10 promoter (Fig. 4), the promoter of a gene encoding an anthocyanin biosynthetic enzyme, DFR, was selected to test for activation by BBX1. The promoter of DFR was selected as it displayed the most significant correlation with the expression of MYB10 (Fig. 3) and is a key step in the anthocyanin pathway, being one of the late biosynthetic enzymes. BBX1 was chosen because of its gene expression profile, both in the fruit development and diurnal rhythm experiments as well as its ability to activate the MYB10 promoter. Infiltrations were performed in N. benthamiana using the dual luciferase assay (Fig. 6). Infiltration of BBX1 alone did not activate the DFR promoter. Co-infiltration of both MYB10 and bHLH3 resulted in strong activation and this was further strengthened by the addition of BBX1. We checked the DFR promoter sequence for CCAAT motifs, finding two, one at −425 and the other at −659 upstream of the start site.
Figure 6.
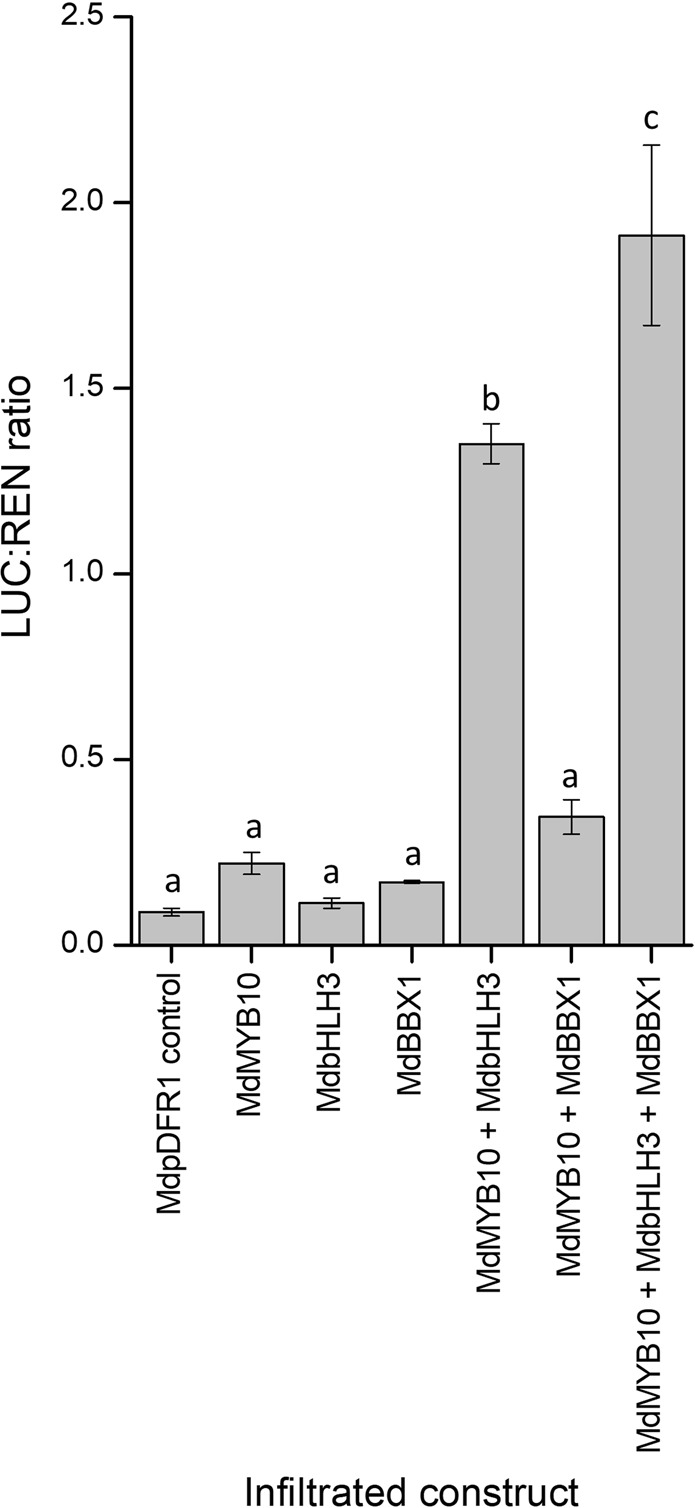
Transient expression assay of the activation of the promoter of the anthocyanin biosynthetic gene DFR by BBX1 in the presence of MYB10 and bHLH3. Error bars show SE of four technical replicates. Differences between averages were statistically analysed using ANOVA and pairwise comparison performed using Fisher’s LSD test at the 5% significance level. Error bars show SE of four biological replicates.
Over-expression of BBX1 in transgenic ‘Royal Gala’ apple
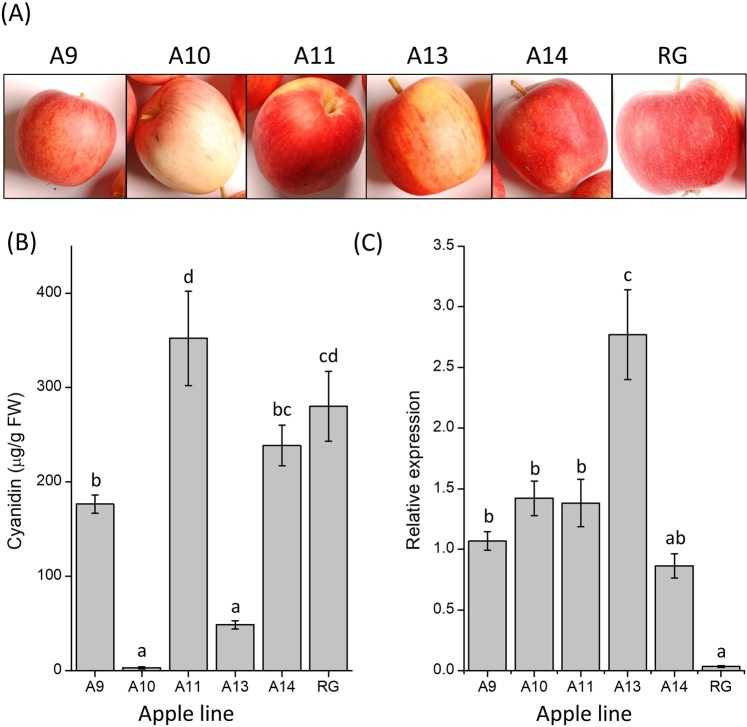
The apple tree cultivar ‘Royal Gala’ was transformed with the BBX1 cDNA driven by the CaMV35S promoter. Trees were grown in the glasshouse until sufficiently mature to flower. Apple fruit from five independent transgenic lines were harvested and phenotypically assessed 125 DAFB (Fig. 7A). HPLC analysis confirmed that the concentration of cyanidin-3-O-galctoside varied in the different lines, but mainly showed a reduction compared with the control (Fig. 7B). This suggests that other partners of this transcription factor are required for activation. The over-expression of BBX1 in transgenic fruit was confirmed by RT-qPCR and was shown to be highly elevated in all the lines tested compared with the control (Fig. 7C). However, overexpression of BBX1 did not result in more anthocyanin accumulation in the fruit skin of transgenic apples. Fruit weight, firmness and soluble solids content, expressed in °Brix values were also recorded, showing a significant increase of firmness and soluble solids content in the over-expression lines compared with those in the ‘Royal Gala’ controls (Supplementary Fig. 4). Ethylene concentration and transcript abundance for MYB10, DFR, and ethylene pathway genes ACS and ACS were also assessed (Supplementary Fig. 5). Other polyphenolics in the over-expression lines were generally similar to those found in ‘Royal Gala’, although some differences were observed, such as an increase in chlorogenic acid (Supplementary Table 3).
Figure 7.
Characterisation of 35S:BBX1 transgenic lines. (A) Images of representative apples from each of the lines. (B) Cyanidin concentration in fruit skin for the transgenic lines compared with ‘Royal Gala’ control (RG). Cyanidin concentrations reported are the sums of cyanidin 3-O-galactoside, cyanidin 3-O-glucoside, and cyanidin 3-O-arabinoside in µg/g FW. Cyanidin 3-O-galactoside accounted for over 90% in every sample tested. (C) Gene expression of BBX1 in the 35S:BBX1 transgenic lines compared with that in ‘Royal Gala’ control fruit (RG). Results are average expression of three biological replicates and four technical replicates relative to Actin. Error bars show SE of four biological replicates. Differences between averages were statistically analysed using ANOVA and pairwise comparison performed using Fisher’s LSD test at the 5% significance level.
Discussion
In this study the putative role of BBX genes on anthocyanin accumulation in apple was assessed. This family of transcription factors control diverse processes in plants with at least one, BBX33, having a direct role in anthocyanin accumulation in apple fruit skin7. To study the BBX family we tested gene expression through fruit development and examined the possible role of a number of BBX genes in directly activating anthocyanin biosynthesis. Diurnal rhythms of candidate BBX genes were measured and BBX1 was chosen to advance into stable transgenic work after showing the ability to activate MYB10 and DFR.
BBX transcriptional levels vary during fruit apple development
Anthocyanin accumulation in apple skin generally occurs late in fruit development and is a result of the anthocyanin-regulating MYB induction of the genes encoding anthocyanin biosynthetic enzymes. The expression patterns of a subset of BBX genes was assessed during fruit development and compared with both the anthocyanin content and the expression of DFR, UFGT and MYB10. This showed that BBX genes have a range of expression patterns, some of which increased with anthocyanin accumulation in fruit skin, similar to banana BBX expression during fruit development80. A number of these genes showed an expression patterns correlating with anthocyanin induction, including in particular BBX1, 15, 33, 51 and 54. BBX1 showed the highest expression outside group IV BBXs. BBX15, 51 and 54 showed increasing expression during development, peaking during the period of highest anthocyanin accumulation, and these are all in group IV, which contains BBX33, already shown to activate the anthocyanin pathway in apple7. In Arabidopsis, group IV BBXs, in particular, have been identified as linked to light response and so were of interest to assess for any diurnal rhythms they might possess81.
The expression pattern of BBX genes demonstrate a diurnal rhythm
BBX genes have been shown to be diurnally regulated in banana and potato80,82. In apple, expression analysis over a 48-hour period late in fruit development was analysed (Fig. 2). Strong patterns in expression were seen for GI, light, and temperature-related genes in fruit skin, and these daily rhythms appeared stable in the orchard-grown fruit. BBX1, 15, 33 and 51 were chosen based on their expression patterns during fruit development. All four BBXs displayed diurnal patterns of expression for both seasons, except for BBX33, which displayed no obvious pattern for season 1. BBX33 influences anthocyanin in apple7 and appears to have been influenced by an unknown factor in season 1 that did not affect BBX1, BBX15 or BBX51. The diurnal variation in BBX transcription recorded here signifies the importance of sampling times for BBXs, and indeed all environmentally regulated genes, when considering experimental design. For example, results varied as much as six-fold for BBX1 between sampling at 0800 h or 2000 h.
The expression of anthocyanin-related genes demonstrates a clear diurnal rhythm, co-ordinated by MYB10
The diurnal rhythm demonstrated by a number of the BBX genes suggests that diurnal rhythms may also be important in anthocyanin regulation. We tested key genes in the anthocyanin pathway, and showed that transcript accumulation of these anthocyanin biosynthetic genes followed a very similar pattern over the 48-hour sampling period (Fig. 3). This was also evident for the MYB regulator, MYB10, confirmed by statistical analysis. This suggests that the pathway genes are modulated in a highly controlled manner, although further work is necessary to examine the interplay of the transcriptional control and environmental factors determining this phenomenon. We did not find any correlation between expression patterns of MYB10 and BBX1, 15, 33 or 51 (Supplementary Table 2A). Interestingly, BBX1 did have a significant correlation with temperature (Supplementary Fig. 2B), a relationship supported by previous findings7,32. Temperature readings during the sampling period are provided in Supplementary Fig. 6. Furthermore, BBX33 transcription had a significant correlation with HY5 transcription for season 1, supporting the model that BBX33 influences anthocyanin via HY5 proposed by Bai et al.7 (Supplementary Table 2C). This correlation, however, was not seen for season 2 and so appears to vary seasonally and geographically.
BBX genes can directly activate the apple anthocyanin-regulating MYB
To further determine the role of BBXs, we tested different sub-groups of the BBX family for transactivation of the promoter of the anthocyanin regulator MYB10 using luciferase assays. Activation capability was not specific to particular groups. As BBX33 has been published by Bai et al.7 and group IV has been reported as important in light signalling81, we focused on this group by testing more of its members. There was substantial variation in the effect of addition of MYB10 and bHLH3; for example, BBX15 had little activation without MYB10 and bHLH3, whereas BBX17 had the same degree of activation regardless of the presence of MYB10 or bHLH3. The highest activating BBXs were BBX1, 35, 15, 51 and 54, to which the closest predicted amino acid (AA) BLAST hits in Arabidopsis were AtBBX5 (AtCOL4), AtBBX12, AtBBX21, and AtBBX24, respectively (with apple BBX51 and 54 both being most similar to AtBBX24). In Arabidopsis these BBXs have been reported to be involved with photosynthesis, photomorphogenesis, and various stress responses57,83. AtBBX21 and AtBBX24 specifically have been linked to the regulation of HY584, which is supported by results here that showed developmental transcription, diurnal rhythm, and MYB10 activation for the apple homologues BBX15 and BBX51. Compared with the highest activating BBXs, BBX33 did not highly activate MYB10, which, being involved in anthocyanin accumulation, suggests a low threshold is required for BBX activation of the anthocyanin pathway. This ability of BBXs to activate MYB10 suggests the presence of cis elements in the MYB10 promoter that might be bound by BBX proteins.
A BBX-related cis element is necessary for BBX transactivation of the MYB10 promoter
Data from the promoter deletion study (Fig. 5) showed that BBX activation of the MYB10 promoter is partly dependent on the presence of upstream sequences. The shortest promoter fragment, ∆b, was not able to activate the reporter to the same extent as either the full promoter tested or ∆a. This suggests that BBX1 requires a motif that is absent in the most truncated version. Previous research on CONSTANS binding with the FLOWERING LOCUS T (FT) promoter has identified two possible motifs in Arabidopsis. A CCAAT box binding domain was identified by Wenkel et al.78, and this relatively common eukaryotic cis-element binds with the NF-Y/HAP complex to drive transcription. The full-length MYB10 promoter showed a high level of transactivation in the presence of BBX1 (Fig. 5B). There are three CCAAT cis-elements in the 5’ region of this promoter sequence (Fig. 5A). There was no significant reduction when the same assay was performed using the truncated ∆a promoter fragment. This contains just one CCAAT domain. However, when ∆b containing no CCAAT motifs was used, there was a dramatic reduction in activation. This suggests the region between MYB10∆a and MYB10∆b is crucial for the effects of BBX1 and indicates that the CCAAT motif may be essential for BBX binding. However, there was still some activation, indicating that another element may be present. Bai et al.7 reported activation of MYB10 also being influenced by ACGT-containing elements and Polymorphic G-boxes, and found activation occurring down to a −314 bp promotor fragment, suggesting a possible binding site between −405bp and −314bp. PLACE (Plant cis-acting regulatory DNA elements) analysis for this region identified many potential sites, but no CCAAT, ACGT-containing elements, or G-boxes. A second, more specific element has been described for CO binding of the FT promoter85. This sequence was identified as TGTG(N2-3)ATG, but was not found in the MYB10 promoter. As well as the MYB10 promotor, light-induced factors could also be upregulating the anthocyanin biosynthetic genes directly by binding their promotors.
BBX1 in combination with MYB10 and bHLH3 activates the DFR promotor
Previous results indicated that light responsive regulators of anthocyanin could be acting on the structural genes in the pathway directly38,86. Transient expression assays were performed on the promotor of DFR to test this for BBX1. BBX1 did not display a significant activation of the DFR promotor when infiltrated alone (Fig. 6). As expected, a strong activation was seen when the DFR promoter was infiltrated with MYB10 and bHLH3, which was enhanced with the co-infiltration of BBX1, suggesting some combinatorial effect of members of the MBW complex and BBX1. BBX33 has previously been shown to be regulated by HY5 and to regulate MYB107.
BBX1 overexpression in apple may affect fruit maturation, but did not increase anthocyanin accumulation
BBX1 was over-expressed in apple trees. Fruit weight was not significantly different between the transgenic lines and the ‘Royal Gala’ control. There was, however, a significant difference in fruit firmness and soluble solids content (Supplementary Fig. 4). Ethylene was also significantly lower in the transgenic fruit (Supplementary Fig. 5A), which alongside the difference in firmness and soluble solids content, suggests an effect on fruit maturity. Transcript abundance for ethylene genes did not show the same pattern as ethylene measurements, which were all reduced in the over-expressing lines, suggesting the control of other factors (Supplementary Fig. 5B). BBXs have been linked to regulating shade avoidance, which is controlled by hormones that influence plant growth, such as ethylene87. The anthocyanin content of the transgenic fruit was either similar to control fruit, as shown in the independent lines A11 and A14, or reduced as shown in A9, A10, A13 (Fig. 6B). Although the expression of BBX1 was high compared with that in the ‘Royal Gala’ control (Fig. 6C), the expression levels of the other genes tested were more similar to MYB10 than to BBX1 (Supplementary Fig. 5B), indicating that the over-expression did not overcome the control MYB10 has over the anthocyanin pathway, particularly as the anthocyanin content correlated to the transcript abundance of MYB10. Other environmental conditions, such as temperature, are likely to contribute to the results of this study. It is possible that thresholds of temperature sensitive expression of BBX1 were not reached during this work, and therefore anthocyanin accumulation was not affected. Future experiments could test this by manipulating temperature and measuring BBX1 expression along with anthocyanin concentration.
Conclusion
BBX genes play a diverse set of roles in plants. The few reports to date have classified the family and focused on individual BBXs7,59. Here, a wide range of BBXs were scrutinised to determine their potential as activators of the anthocyanin pathway, identifying candidates worth pursuing. The expression of a number of BBXs correlated with cyanidin-3-O-galactoside during fruit development. BBX1 activated the promoters of MYB10 and DFR; however, when BBX1 was over-expressed in apple, there was no apparent increase in anthocyanin. Taken together, this evidence suggests that there may be an element of control of the anthocyanin pathway by BBXs with other partner transcription factors. Identifying transcriptional partners that influence colour in apple alongside the previously demonstrated effect of BBX337 will aid the development of apple varieties with predictable colour development.
Supplementary information
Acknowledgements
We are grateful to Monica Dragulescu and glasshouse staff for management of plants and to Geeta Chhiba for providing media and buffers. We also thank Tony McGhie for help with HPLC. KM was funded by the Finnish horticultural foundation Maiju ja Yrjö Rikalan Puutarhasäätiö.
Author contributions
B.P., R.E., J.P. and A.A. contributed to project planning; A.D. conducted HPLC analysis; R.E., R.H., K.M. and B.P. performed RT-PCR; A.F. and R.D. collected plant material; B.P. conducted statistical analysis; K.M., S.H. and B.P. performed transient assays; S.T. generated transgenic apple lines; B.P., R.E., J.P. and A.A. wrote the manuscript; All authors contributed to editing the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54166-2.
References
- 1.Boyer J, Liu RH. Apple phytochemicals and their health benefits. Nutrition Journal. 2004;3:1–15. doi: 10.1186/1475-2891-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Espley, R. & Martens, S. In Bioactives in Fruit 81–100 (John Wiley & Sons, Ltd, 2013).
- 3.He J, Giusti MM. Anthocyanins: Natural Colorants with Health-Promoting Properties. Annual Review of Food Science and Technology. 2010;1:163–187. doi: 10.1146/annurev.food.080708.100754. [DOI] [PubMed] [Google Scholar]
- 4.Scalbert, A. & Williamson, G. Dietary Intake and Bioavailability of Polyphenols. The Journal of Nutrition130 (2000). [DOI] [PubMed]
- 5.Cliff M, Sanford K, Wismer W, Hampson C. Use of Digital Images for Evaluation of Factors Responsible for Visual Preference of Apples by Consumers. HortScience. 2002;37:1127–1131. doi: 10.21273/HORTSCI.37.7.1127. [DOI] [Google Scholar]
- 6.Marjorie C, Margaret C. Development of a model for prediction of consumer liking from visual attributes of new and established apple cultivars. Journal of American Pomological Society. 2002;56:223. [Google Scholar]
- 7.Bai S, et al. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta. 2014;240:1051–1062. doi: 10.1007/s00425-014-2129-8. [DOI] [PubMed] [Google Scholar]
- 8.Delgado-Pelayo R, Gallardo-Guerrero L, Hornero-Méndez D. Chlorophyll and carotenoid pigments in the peel and flesh of commercial apple fruit varieties. Food Research International. 2014;65, Part B:272–281. doi: 10.1016/j.foodres.2014.03.025. [DOI] [Google Scholar]
- 9.Lancaster JE, Grant JE, Lister CE, Taylor MC. Skin Color in Apples - Influence of Copigmentation and Plastid Pigments on Shade and Darkness of Red Color in Five Genotypes. Journal of the American Society for Horticultural Science. 1994;119:63–69. doi: 10.21273/JASHS.119.1.63. [DOI] [Google Scholar]
- 10.Li P, Cheng L. The elevated anthocyanin level in the shaded peel of ‘Anjou’ pear enhances its tolerance to high temperature under high light. Plant Science. 2009;177:418–426. doi: 10.1016/j.plantsci.2009.07.005. [DOI] [Google Scholar]
- 11.Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytologist. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- 12.Felicetti DA, Schrader LE. Changes in pigment concentrations associated with the degree of sunburn browning of ‘Fuji’ apple. Journal of the American Society for Horticultural Science. 2008;133:27–34. doi: 10.21273/JASHS.133.1.27. [DOI] [Google Scholar]
- 13.Gould, K. & Lister, C. In Flavonoids 397–441 (CRC Press, 2005).
- 14.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 15.Albert, N. et al. A Conserved Network of Transcriptional Activators and Repressors Regulates Anthocyanin Pigmentation in Eudicots. Vol. 26 (2014). [DOI] [PMC free article] [PubMed]
- 16.Henry-Kirk RA, McGhie TK, Andre CM, Hellens RP, Allan AC. Transcriptional analysis of apple fruit proanthocyanidin biosynthesis. Journal of experimental botany. 2012;63:5437–5450. doi: 10.1093/jxb/ers193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baudry A, et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. The Plant Journal. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 18.Ravaglia D, et al. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biology. 2013;13:68. doi: 10.1186/1471-2229-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaart JG, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytologist. 2013;197:454–467. doi: 10.1111/nph.12017. [DOI] [PubMed] [Google Scholar]
- 20.Albert, N. W. Subspecialization of R2R3-MYB Repressors for Anthocyanin and Proanthocyanidin Regulation in Forage Legumes. Frontiers in Plant Science6 (2015). [DOI] [PMC free article] [PubMed]
- 21.Nemesio-Gorriz M, et al. Identification of Norway Spruce MYB-bHLH-WDR Transcription Factor Complex Members Linked to Regulation of the Flavonoid Pathway. Frontiers in plant science. 2017;8:305. doi: 10.3389/fpls.2017.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espley R, et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signaling & Behavior. 2014;9:e27522. doi: 10.4161/psb.27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takos A, et al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ban Y, et al. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- 26.Allan A, Hellens R, Laing W. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008;13:99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Telias A, et al. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biology. 2011;11:93. doi: 10.1186/1471-2229-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albert NW, et al. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 29.Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. The Plant Journal. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 30.Dubos C, et al. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. The Plant Journal. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- 31.Aharoni A, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal. 2001;28:319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 32.Lin-Wang, K. U. I. et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant, Cell & Environment, 1–15 (2011). [DOI] [PubMed]
- 33.Xie X-B, et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant, Cell &. Environment. 2012;35:1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 34.Dong Y-H, et al. Expression of pigmentation genes and photo-regulation of anthocyanin biosynthesis in developing Royal Gala apple flowers. Functional Plant Biology. 1998;25:245–252. doi: 10.1071/PP97108. [DOI] [Google Scholar]
- 35.Jakopic J, Stampar F, Veberic R. The influence of exposure to light on the phenolic content of ‘Fuji’ apple. Scientia Horticulturae. 2009;123:234–239. doi: 10.1016/j.scienta.2009.09.004. [DOI] [Google Scholar]
- 36.Ban Y, Honda C, Bessho H, Pang X-M, Moriguchi T. Suppression subtractive hybridization identifies genes induced in response to UV-B irradiation in apple skin: isolation of a putative UDP-glucose 4-epimerase. Journal of Experimental Botany. 2007;58:1825–1834. doi: 10.1093/jxb/erm045. [DOI] [PubMed] [Google Scholar]
- 37.Henry-Kirk RA, et al. Solar UV light regulates flavonoid metabolism in apple (Malus x domestica) Plant, Cell & Environment. 2018;41:675–688. doi: 10.1111/pce.13125. [DOI] [PubMed] [Google Scholar]
- 38.Shin DH, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Letters. 2013;587:1543–1547. doi: 10.1016/j.febslet.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Zheng S, Liu Z, Wang L, Bi Y. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. Journal of Plant Physiology. 2011;168:367–374. doi: 10.1016/j.jplph.2010.07.025. [DOI] [PubMed] [Google Scholar]
- 40.Chang C-sJ, et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. The Plant Journal. 2008;54:205–219. doi: 10.1111/j.1365-313X.2008.03401.x. [DOI] [PubMed] [Google Scholar]
- 41.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. The EMBO Journal. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Gregan S, Winefield C, Jordan B. From UVR8 to flavonol synthase: UV-B-induced gene expression in Sauvignon blanc grape berry. Plant, Cell & Environment. 2015;38:905–919. doi: 10.1111/pce.12349. [DOI] [PubMed] [Google Scholar]
- 43.Deng X-W, Caspar T, Quail P. H. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes & Development. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- 44.Li Y-Y, et al. MdCOP1 Ubiquitin E3 Ligases Interact with MdMYB1 to Regulate Light-Induced Anthocyanin Biosynthesis and Red Fruit Coloration in Apple. Plant Physiology. 2012;160:1011–1022. doi: 10.1104/pp.112.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier A, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. The Plant Journal. 2013;74:638–651. doi: 10.1111/tpj.12153. [DOI] [PubMed] [Google Scholar]
- 46.Datta S, et al. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box Protein Involved in Light-Dependent Development and Gene Expression, Undergoes COP1-Mediated Ubiquitination. The Plant Cell. 2008;20:2324–2338. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayama R, Coupland G. The Molecular Basis of Diversity in the Photoperiodic Flowering Responses of Arabidopsis and Rice. Plant Physiology. 2004;135:677–684. doi: 10.1104/pp.104.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 49.Valverde F, et al. Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science. 2004;303:1003. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 50.Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G. Mutagenesis of Plants Overexpressing CONSTANS Demonstrates Novel Interactions among Arabidopsis Flowering-Time Genes. The Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samach A, et al. Distinct Roles of CONSTANS Target Genes in Reproductive Development of Arabidopsis. Science. 2000;288:1613. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 52.Borden KLB. RING domains: master builders of molecular scaffolds?1. Journal of Molecular Biology. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 53.Crocco CD, Botto JF. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene. 2013;531:44–52. doi: 10.1016/j.gene.2013.08.037. [DOI] [PubMed] [Google Scholar]
- 54.Strayer C, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 55.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends in Plant Science. 2014;19:460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 56.Robson F, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. The Plant Journal. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- 57.Khanna R, et al. The Arabidopsis B-Box Zinc Finger Family. The Plant Cell. 2009;21:3416–3420. doi: 10.1105/tpc.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J, Zhao X, Weng X, Wang L, Xie W. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLoS ONE. 2012;7:e48242. doi: 10.1371/journal.pone.0048242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu, X., Li, R., Dai, Y., Chen, X. & Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Molecular Genetics and Genomics (2017). [DOI] [PubMed]
- 60.Jeong D-H, Sung S-K, An G. Molecular cloning and characterization of constans-like cDNA clones of the fuji apple. Journal of Plant Biology. 1999;42:23–31. doi: 10.1007/BF03031143. [DOI] [Google Scholar]
- 61.Lagercrantz U, Axelsson T. Rapid Evolution of the Family of CONSTANS LIKE Genes in Plants. Molecular Biology and Evolution. 2000;17:1499–1507. doi: 10.1093/oxfordjournals.molbev.a026249. [DOI] [PubMed] [Google Scholar]
- 62.Jin J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon S, Rühl M, de Montaigu A, Wötzel S, Coupland G. Evolution of CONSTANS Regulation and Function after Gene Duplication Produced a Photoperiodic Flowering Switch in the Brassicaceae. Molecular Biology and Evolution. 2015;32:2284–2301. doi: 10.1093/molbev/msv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. The Plant Journal. 2001;26:15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- 65.Suarez-Lopez P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 66.Hellens R, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Espley R, et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin-Wang K, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biology. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dare AP, et al. Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. The Plant Journal. 2017;91:237–250. doi: 10.1111/tpj.13559. [DOI] [PubMed] [Google Scholar]
- 70.Johnston JW, Gunaseelan K, Pidakala P, Wang M, Schaffer RJ. Co-ordination of early and late ripening events in apples is regulated through differential sensitivities to ethylene. Journal of Experimental Botany. 2009;60:2689–2699. doi: 10.1093/jxb/erp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL, https://www.R-project.org/ (2016).
- 72.Hayter AJ. The Maximum Familywise Error Rate of Fisher’s Least Significant Difference Test. Journal of the American Statistical Association. 1986;81:1000–1004. doi: 10.1080/01621459.1986.10478364. [DOI] [Google Scholar]
- 73.Yao J-L, Cohen D, Atkinson R, Richardson K, Morris B. Regeneration of transgenic plants from the commercial apple cultivar’Royal Gala’. Plant Cell Reports. 1995;14:407–412. doi: 10.1007/BF00234044. [DOI] [PubMed] [Google Scholar]
- 74.Fowler S, et al. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO Journal. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller TA, Muslin EH, Dorweiler JE. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta. 2008;227:1377–1388. doi: 10.1007/s00425-008-0709-1. [DOI] [PubMed] [Google Scholar]
- 76.Dong MA, Farré EM, Thomashow MF. Circadian Clock-Associated 1 And Late Elongated Hypocotyl Regulate Expression Of The C-Repeat Binding Factor (CBF) pathway in Arabidopsis. Proceedings of the National Academy of Sciences. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giorno F, Guerriero G, Baric S, Mariani C. Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics. 2012;13:1–13. doi: 10.1186/1471-2164-13-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wenkel S, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell Online. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gnesutta N, et al. CONSTANS imparts DNA sequence-specificity to the histone-fold NF-YB/NF-YC dimer. The Plant Cell. 2017;29:1516–1532. doi: 10.1105/tpc.16.00864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaurasia AK, et al. Molecular characterization of CONSTANS-Like (COL) genes in banana. Physiology and Molecular Biology of Plants. 2016;22:1–15. doi: 10.1007/s12298-016-0345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarmiento Felipe. The BBX subfamily IV. Plant Signaling & Behavior. 2013;8(4):e23831. doi: 10.4161/psb.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Talar U, Kiełbowicz-Matuk A, Czarnecka J, Rorat T. Genome-wide survey of B-box proteins in potato (Solanum tuberosum)—Identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS One. 2017;12:e0177471. doi: 10.1371/journal.pone.0177471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ji-Hee M, Jung-Sung C, Kyeong-Hwan L, Soo KC. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. Journal of Integrative Plant Biology. 2015;57:313–324. doi: 10.1111/jipb.12246. [DOI] [PubMed] [Google Scholar]
- 84.Yadukrishnan, P., Job, N., Johansson, H. & Datta, S. Opposite Roles of Group IV BBX Proteins: Exploring Missing Links between Structural and Functional Diversity. Plant Signaling & Behavior, 1–13 (2018). [DOI] [PMC free article] [PubMed]
- 85.Tiwari SB, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytologist. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 86.Chattopadhyay S, Ang L-H, Puente P, Deng X-W, Wei N. Arabidopsis bZIP Protein HY5 Directly Interacts with Light-Responsive Promoters in Mediating Light Control of Gene Expression. The Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crocco CD, et al. The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nature Communications. 2015;6:6202. doi: 10.1038/ncomms7202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.