Abstract
Amfepramone (AFP) is an appetite-suppressant drug used in the treatment of obesity. Nonetheless, studies on interindividual pharmacokinetic variability and its association with genetic variants are limited. We employed a pharmacokinetic and pharmacogenetic approach to determine possible metabolic phenotypes of AFP and identify genetic markers that could affect the pharmacokinetic variability in a Mexican population. A controlled, randomized, crossover, single-blind, two-treatment, two-period, and two sequence clinical study of AFP (a single 75 mg dose) was conducted in 36 healthy Mexican volunteers who fulfilled the study requirements. Amfepramone plasma levels were measured using high-performance liquid chromatography mass spectrometry. Genotyping was performed using real-time PCR with TaqMan probes. Four AFP metabolizer phenotypes were found in our population: slow, normal, intermediate, and fast. Additionally, two gene polymorphisms, ABCB1-rs1045642 and CYP3A4-rs2242480, had a significant effect on AFP pharmacokinetics (P < 0.05) and were the predictor factors in a log-linear regression model. The ABCB1 and CYP3A4 gene polymorphisms were associated with a fast metabolizer phenotype. These results suggest that metabolism of AFP in the Mexican population is variable. In addition, the genetic variants ABCB1-rs1045642 and CYP3A4-rs2242480 may partially explain the AFP pharmacokinetic variability.
Subject terms: Genetic testing, Predictive markers
Introduction
Currently, obesity is a major health problem worldwide. According to the organization for economic co-operation and development (OECD), in 2017, Mexico was among the countries with the highest obesity rates worldwide1. Today, 36.3% of adolescents aged 12–19 years, and 72.5% of adults are overweight or obese2. Strictly, obesity is abnormal fat accumulation that can compromise health and be associated with other clinical conditions such as cardiovascular diseases, diabetes, musculoskeletal disorders, and cancer3.
Obesity is mainly caused by an energy surplus between calories consumed and calories expended3. Therefore, most treatments are focused on calorie-restricted diets and an increase in physical activity. Only when aforementioned treatments yield unsatisfactory results, pharmacological treatment is recommended4,5. Anti-obesity pharmacotherapy is limited to a few options that either suppress appetite or increase energy expenditure4. At present, the Food and Drug Administration (FDA) has approved the short-term use of appetite suppressants such as amfepramone (AFP) and phentermine (PHE) to treat obesity5–7; however, adverse drug reactions have been reported for some of them4,8,9.
The drug AFP, is classified as a schedule IV controlled substance in the United States and Canada because it acts on the central nervous system10, where it not only induces the release of the neurotransmitters noradrenaline (NA) and dopamine (DA) but also suppresses their reuptake. This mechanism of action results in less appetite11,12. However, owing to the psychostimulant effects of AFP, cautions should be taken regarding the dose and in patients with a psychiatric or cardiovascular history13–15. Specifically, in a Mexican population, AFP has been proven to be effective and safe for long-term treatment16. Nonetheless, some mild to moderate adverse events have been associated with AFP use, such as dry mouth, insomnia, anxiety, irritability, constipation, headache, dizziness, and polydipsia12,13,16.
The wide range of pharmacological effects of drugs, from being safe and effective to the risk of adverse reactions, may involve genetic factors that control absorption, distribution, metabolism, and excretion (ADME) processes17,18 and, therefore, pharmacokinetic parameters19. For example, recent studies in a Mexican population have shown that genetic variation may explain 30–90% of the pharmacokinetic variability20,21 of Statins. Here, the most common genetic variation responsible for these pharmacokinetic discrepancies are polymorphisms in genes encoding metabolizing enzymes and transporters19–21. Nevertheless, no pharmacogenetic data is available for AFP.
Therefore, to date, although after oral administration, AFP is extensively biotransformed to secondary metabolites; until now, the responsible metabolic enzymes are unknown22–24. Two metabolic pathways seem to be involved: 1) N-dealkylation and reduction22,23 and 2) N-deethylation24. The enzymes encoded by CYP3A4 and CYP3A5 are the most important because they metabolize a significant percentage of drugs. These enzyme isoforms have similar substrate selectivity, and therefore metabolize the same compounds25. Previous studies reported that CYP3A4 performs N-dealkylation and N-deethylation26,27. Therefore, CYP3A4 and CYP3A5 may have an important role in AFP metabolism. However, other CYP isoforms25 and other enzymes of phase II metabolism, such as glutathione S-transferases (GSTs), may also be involved28.
Amfepramone and its metabolites are excreted via the kidney22–24. Transporter proteins known to be involved in drug removal, such as the ones encoded by ABCB1, ABCG2, and SLCO1B1, may affect AFP plasma levels in the Mexican population20,21.
Because variations in genes involved in the ADME processes may affect the pharmacokinetic profile and, therefore, the effectivity of a drug, the aims of this study were: (1) to classify the AFP metabolism, (2) to evaluate the impact of genetic polymorphism related to drug metabolism in the Mexican population on AFP pharmacokinetics, and (3) to assess a possible association between genotypes and metabolizer phenotypes.
Results
Study population
A total of 36 unrelated healthy volunteers, aged between 18 and 51 years, of either sex and of Mexican origin, were enrolled and treated with AFP. Demographical, clinical, and pharmacokinetic data are presented in Table 1. Despite the fact that height and weight values were significantly higher in males than in females (P ≤ 6.97 × 10−5); the Body Mass Index (BMI) was not significantly different. No other features displayed significant variability (Table 1).
Table 1.
Demographic data and pharmacokinetic descriptive statistics of volunteers.
| n | All subjects | Males | Females |
|---|---|---|---|
| 36 | 19 | 17 | |
| Age (years) | 24.25 ± 6.35 | 25.74 ± 8.25 | 22.59 ± 2.48 |
| BMI (kg/m2) | 23.81 ± 2.16 | 24.19 ± 1.96 | 23.39 ± 2.36 |
| Height (m) | 1.67 ± 0.10 | 1.74 ± 0.08a | 1.59 ± 0.06 |
| Weight (kg) | 66.57 ± 11.23 | 73.12 ± 9.58b | 59.25 ± 8.05 |
| Cmax (ng/mL) | 7.04 ± 2.86 | 7.40 ± 3.46 | 6.64 ± 2.01 |
| AUC0-t (ng/mL/h) | 29.87 ± 12.40 | 30.91 ± 15.57 | 28.72 ± 7.79 |
| AUC0-∞ (ng/mL/h) | 36.59 ± 13.66 | 38.38 ± 17.40 | 34.55 ± 7.71 |
| Ke | 0.1923 ± 0.0539 | 0.2056 ± 0.0637 | 0.1774 ± 0.0365 |
| T1/2 (h) | 3.95 ± 1.45 | 3.84 ± 1.83 | 4.07 ± 0.91 |
| Cl (L/h/kg) | 2.32 ± 0.90 | 2.37 ± 1.16 | 2.27 ± 0.51 |
| Vd (L/kg) | 12.92 ± 5.47 | 12.47 ± 6.38 | 13.42 ± 4.38 |
Data shown as mean ± standard deviation. aP = 5.24 × 10−6; bP = 6.97 × 10−5. BMI, body mass index; Cmax, maximum plasma concentration; AUC, area under the plasma concentration-time curve; AUC0-t, AUC from time 0 to the time of last measurement; AUC0-∞, AUC from time 0 extrapolated to infinity; Ke, elimination rate constant in the terminal drug phase; T1/2, half-life drug; Vd, volume of distribution; Cl, total drug clearance.
Amfepramone pharmacokinetics
The mean ± SD values for the pharmacokinetic parameters for all subjects were as follows: maximum plasma concentration (Cmax) = 7.04 ± 2.86 ng/mL, area under the plasma concentration-time curve from 0 to the time of last measurement (AUC0-t) = 29.87 ± 12.39 ng/mL/h, AUC from time 0 extrapolated to infinity (AUC0-∞) = 36.59 ± 13.66 ng/mL/h, elimination rate constant in the terminal drug phase (Ke) = 0.1923 ± 0.0539, half-life (T1/2) = 7.04 ± 2.86 h, total drug clearance (Cl) = 2.32 ± 0.90 L/h/kg, volume of distribution (Vd) = 7.04 ± 2.86 L/kg. No sex differences were found in pharmacokinetic parameters (Table 1). Despite the lack of significant differences in the demographic and clinical features (Table 1), there was a remarkable pharmacokinetic variability (Table 2).
Table 2.
Pharmacokinetic parameters according to metabolizer phenotype.
| Pharmacokinetic parameters | Metabolizer phenotypes for all subjects | |||
|---|---|---|---|---|
| Slow | Intermediate | Normal | Fast | |
| N | 7 | 9 | 12 | 8 |
| Cmax (ng/mL) | 11.07 ± 3.49a | 7.21 ± 0.91a | 6.51 ± 1.16a | 4.12 ± 0.89a |
| AUC0-t (ng/mL/h) | 45.84 ± 14.69b | 34.98 ± 2.13b | 26.44 ± 2.87b | 15.30 ± 3.74b |
| AUC0-∞ (ng/mL/h) | 51.61 ± 15.59c | 43.17 ± 10.58c | 32.87 ± 2.96c | 21.62 ± 4.67c |
| Ke | 0.2013 ± 0.0431 | 0.2063 ± 0.0708 | 0.1794 ± 0.0452 | 0.1880 ± 0.0577 |
| T1/2 (h) | 3.58 ± 0.80 | 4.01 ± 1.15 | 4.11 ± 1.15 | 3.96 ± 1.08 |
| Cl (L/h/kg) | 1.53 ± 0.32d | 1.80 ± 0.31d | 2.30 ± 0.20d | 3.64 ± 0.91d |
| Vd (L/kg) | 7.89 ± 2.13e | 9.59 ± 2.87e | 13.68 ± 4.08e | 19.90 ± 3.75e |
Data shown as mean ± standard deviation. aP ≤ 9.37 × 10−3; bP ≤ 0.001; cP ≤ 0.023; dP ≤ 0.026; eP ≤ 0.002; cP ≤ 9.79 × 10−6. Cmax, maximum plasma concentration; AUC, area under the plasma concentration-time curve; AUC0-t, AUC from time 0 to the time of last measurement; AUC0-∞, AUC from time 0 extrapolated to infinity; Ke, elimination rate constant in the terminal drug phase; T1/2, half-life drug; Vd, volume of distribution; Cl, total drug clearance.
Metabolic phenotype classification
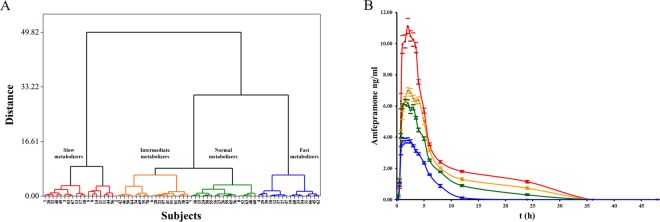
Based on the multivariate analysis20,29–31 of the parameters Cmax and AUC0-t, we identified four AFP phenotypes–slow metabolizers (n = 7), intermediate metabolizers (n = 9), normal metabolizers (n = 12), and fast metabolizers (n = 8)–as shown in Fig. 1A. The means of most pharmacokinetic parameters were significantly different among the four metabolizer phenotypes (P ≤ 0.026, Table 2). We used AUC0-t values to validate the pharmacokinetic variability. The regression analysis suggests that the four metabolizer phenotypes offer a better prediction of the variability of AUC0-t (R = 0.888, R2 = 0.788, adjusted R = 0.781, P = 5.50 × 10−13; AICC = 144.02; and ASE = 7.15). There was a > 6-fold difference in AFP pharmacokinetic parameters between the slowest metabolizer (Cmax = 17.80 ng/mL and AUC0-t = 78.85 ng/mL/h) and the fastest metabolizer (Cmax = 2.94 ng/mL and AUC0-t = 10.31 ng/mL/h). The pharmacokinetic profiles of the different metabolizer phenotypes are presented in Fig. 1B.
Figure 1.
Classification of amfepramone (AFP) metabolic phenotypes. (A) Dendrogram generated with Manhattan distance and Ward’s linkage method. (B) Pharmacokinetic profiles of different metabolic phenotypes. Mean peak plasma AFP concentration-time curves after single 75 mg dose of AFP. Data shown are mean ± standard error (SE) concentrations. For both (A,B): slow metabolizers (red), intermediate metabolizers (orange), normal metabolizers (green), and fast metabolizers (blue).
Pharmacogenetic tests
All gene polymorphisms were in Hardy Weinberg Equilibrium (HWE) (P > 0.05) and passed the quality control tests of genotyping. Genotype frequencies are presented in Table 3.
Table 3.
Polymorphisms with significant effect on AFP pharmacokinetics.
| Genotypes | n | Pharmacokinetics parameters | ||||||
|---|---|---|---|---|---|---|---|---|
| Cmax (ng/mL) | AUC0-t (ng/mL/h) | AUC0-∞ (ng/mL/h) | Ke | T1/2 (h) | Cl (L/h/kg) | Vd (L/kg) | ||
| ABCB1-rs1045642 | ||||||||
| C/C | 10 | 5.42 ± 1.88a | 23.29 ± 9.57 | 29.02 ± 9.74 | 0.2193 ± 0.0533b | 3.35 ± 0.90 | 2.94 ± 1.22 | 13.85 ± 5.13 |
| C/T | 13 | 8.15 ± 3.63 | 34.36 ± 15.42 | 42.10 ± 17.64 | 0.1901 ± 0.0628 | 4.20 ± 2.05 | 2.01 ± 0.63 | 11.85 ± 5.67 |
| T/T | 13 | 7.18 ± 2.11 | 30.45 ± 9.13 | 36.91 ± 9.01 | 0.1738 ± 0.0375 | 4.16 ± 0.95 | 2.17 ± 0.62 | 13.26 ± 5.78 |
| C/T + T/T | 26 | 7.66 ± 2.95c | 32.40 ± 12.57d | 39.50 ± 13.97e | 0.1819 ± 0.0513 | 4.18 ± 1.57 | 2.09 ± 0.62 | 12.56 ± 5.65 |
| ABCG2-rs2231142 | ||||||||
| C/C | 25 | 6.77 ± 2.33 | 28.05 ± 9.10 | 35.04 ± 11.26 | 0.1928 ± 0.0577 | 4.01 ± 1.67 | 2.39 ± 0.94 | 13.26 ± 5.43 |
| C/A | 11 | 7.66 ± 3.87 | 34.01 ± 17.66 | 40.11 ± 18.14 | 0.1911 ± 0.0462 | 3.79 ± 0.84 | 2.17 ± 0.83 | 12.14 ± 5.75 |
| SLCO1B1-rs4149056 | ||||||||
| T/T | 32 | 7.09 ± 2.95 | 30.26 ± 12.84 | 37.01 ± 14.30 | 0.1959 ± 0.0553 | 3.90 ± 1.51 | 2.32 ± 0.95 | 12.68 ± 5.58 |
| C/T | 4 | 6.65 ± 2.26 | 26.75 ± 8.62 | 33.26 ± 6.84 | 0.1638 ± 0.0323 | 4.35 ± 0.84 | 2.32 ± 0.41 | 14.79 ± 4.75 |
| DRD3-rs6280 | ||||||||
| C/C | 8 | 7.10 ± 1.71 | 29.77 ± 8.09 | 35.48 ± 9.10 | 0.1994 ± 0.0521 | 3.69 ± 0.98 | 2.22 ± 0.51 | 11.73 ± 3.66 |
| C/T | 19 | 7.11 ± 3.38 | 30.47 ± 15.03 | 36.49 ± 14.91 | 0.1959 ± 0.0499 | 3.76 ± 1.00 | 2.37 ± 1.01 | 13.01 ± 6.42 |
| T/T | 9 | 6.85 ± 2.73 | 28.71 ± 10.20 | 37.78 ± 15.51 | 0.1784 ± 0.0665 | 4.58 ± 2.36 | 2.31 ± 1.01 | 13.76 ± 4.94 |
| CYP3A4-rs2242480 | ||||||||
| C/C | 13 | 8.49 ± 3.86 | 34.77 ± 15.64 f | 40.38 ± 16.27 | 0.1993 ± 0.0454 | 3.63 ± 0.76 | 2.10 ± 0.74 | 11.12 ± 4.88 |
| C/T | 14 | 5.52 ± 1.62 g | 24.60 ± 9.68 | 32.42 ± 13.77 | 0.1924 ± 0.0688 | 4.21 ± 2.07 | 2.70 ± 1.15 | 15.05 ± 5.90 |
| T/T | 9 | 7.32 ± 1.25 | 30.99 ± 8.00 | 37.60 ± 7.50 | 0.1821 ± 0.0412 | 4.00 ± 1.06 | 2.06 ± 0.39 | 12.20 ± 4.99 |
| C/C + T/T | 22 | 8.01 ± 3.08 h | 33.23 ± 12.95i | 39.24 ± 36.59 | 0.1923 ± 0.0436 | 3.78 ± 0.89 | 2.08 ± 0.61 | 11.56 ± 4.84 |
| CYP3A5-rs776746 | ||||||||
| A/A | 1 | 6.30 ± NA | 27.00 ± NA | 34.56 ± NA | 0.1500 ± NA | 4.62 ± NA | 2.17 ± NA | 14.47 ± NA |
| A/G | 13 | 6.44 ± 2.05 | 28.21 ± 11.55 | 36.18 ± 15.16 | 0.1913 ± 0.0605 | 4.16 ± 2.06 | 2.51 ± 1.27 | 13.73 ± 6.34 |
| G/G | 22 | 7.43 ± 3.29 | 30.99 ± 13.28 | 36.92 ± 13.40 | 0.1948 ± 0.0514 | 3.79 ± 1.01 | 2.22 ± 0.63 | 12.37 ± 5.11 |
| CYP2B6-rs3745274 | ||||||||
| G/G | 14 | 7.19 ± 3.66 | 32.17 ± 16.50 | 38.09 ± 16.16 | 0.2029 ± 0.0560 | 3.69 ± 1.14 | 2.26 ± 0.90 | 12.28 ± 6.57 |
| G/T | 18 | 6.84 ± 2.46 | 27.21 ± 8.88 | 34.69 ± 12.80 | 0.1845 ± 0.0545 | 4.18 ± 1.75 | 2.46 ± 0.98 | 13.88 ± 4.79 |
| T/T | 4 | 7.43 ± 1.57 | 33.80 ± 8.84 | 39.90 ± 8.30 | 0.1905 ± 0.0504 | 3.82 ± 0.98 | 2.32 ± 0.90 | 10.83 ± 4.35 |
| GSTM3-rs1799735 | ||||||||
| *A/*A | 29 | 7.18 ± 3.12 | 29.62 ± 13.59 | 36.66 ± 15.03 | 0.1922 ± 0.0572 | 4.00 ± 1.58 | 2.37 ± 0.98 | 13.26 ± 5.80 |
| *A/*B | 7 | 6.46 ± 1.28 | 30.93 ± 5.71 | 36.28 ± 5.78 | 0.1929 ± 0.0409 | 3.72 ± 0.75 | 2.12 ± 0.40 | 11.48 ± 3.83 |
Data shown as mean ± standard deviation. aP = 0.042 (C/C vs. T/T); bP = 0.046 (C/C vs. T/T); cP = 0.031 (C/C vs. C/T + T/T); dP = 0.041 (C/C vs. C/T + T/T); eP = 0.037 (C/C vs. C/T + T/T); fP = 0.033 (C/C vs. C/T); gP = 0.021 (C/T vs. T/T); hP = 0.005 (C/T vs. C/C + T/T); iP = 0.038 (C/T vs. C/C + C/T). Cmax, maximum plasma concentration; AUC, area under the plasma concentration-time curve; AUC0-t, AUC from time 0 to the time of last measurement; AUC0-∞, AUC from time 0 extrapolated to infinity; Ke, elimination rate constant in the terminal drug phase; T1/2, half-life drug; Vd, volume of distribution; Cl, total drug clearance.
Association between genotypes and Amfepramone pharmacokinetics
Two of the analysed polymorphisms affected AFP pharmacokinetics. Cmax, AUC0-t, AUC0-∞, and Ke were affected by ABCB1-rs1045642 and CYP3A4-rs2242480 under the co-dominant, dominant, and over-dominant models (Tables 3 and 4). For ABCB1-rs1045642, the values of Cmax, AUC0-t, and AUC0-∞ in carriers of genotype C/C were significantly lower than those in carriers of the variant allele (T/T or C/T + T/T, P ≤ 0.042; Table 3), but the values of Ke (P = 0.046) in carriers of genotype C/C were significantly higher than those in carriers of the homozygous variant allele (T/T). The regression analysis confirmed our findings. Regarding CYP3A4-rs2242480, AUC0-t was significantly lower in carriers of the heterozygous genotype (C/T) than in subjects with the C/C genotype (P = 0.033). Moreover, the C/T genotype was associated with lower Cmax values than the T/T genotype (P = 0.021). In addition, the Cmax and AUC0-t values of the C/T genotype were significantly lower than those in the combined homozygous subjects (C/C + T/T; P ≤ 0.038; Table 3).
Table 4.
Analysis of predictor models for the pharmacokinetic parameters of amfepramone.
| Parameter | Predictors | Genetic model | R square | Adjusted R square | P-Value |
|---|---|---|---|---|---|
| Ke | rs1045642 | Co-dominant | 0.113 | 0.087 | 0.045* |
| Cmax | rs1045642 | Dominant | 0.173 | 0.149 | 0.012* |
| AUC0-t | rs1045642 | Dominant | 0.143 | 0.117 | 0.023* |
| AUC0-∞ | rs1045642 | Dominant | 0.163 | 0.139 | 0.037* |
| Cl | rs1045642 | Dominant | 0.185 | 0.161 | 0.009* |
| Cmax | rs224280 | Over-dominant | 0.223 | 0.200 | 0.004* |
| AUC0-t | rs224280 | Over-dominant | 0.146 | 0.121 | 0.021* |
| Cmax | rs224280, rs1045642 | Over-dominant, dominant | 0.248 | 0.202 | 0.009* |
| AUC0-t | rs224280, rs1045642 | Over-dominant, dominant | 0.181 | 0.132 | 0.037* |
| Cl | rs224280, rs1045642 | Over-dominant, dominant | 0.240 | 0.194 | 0.011* |
*P-Value supported by automatic linear modeling and log-linear regression analysis and method. Cmax, maximum plasma concentration; AUC, area under the plasma concentration-time curve; AUC0-t, AUC from time 0 to the time of last measurement; AUC0-∞, AUC from time 0 extrapolated to infinity; Ke, elimination rate constant in the terminal drug phase; Cl, total drug clearance.
The importance of aforementioned variability in AFP pharmacokinetics was confirmed by a regression analysis. However, after this analysis, the effect of ABCB1-rs1045642 polymorphism on Cl was revealed using a dominant model (Table 4). The final prediction model included ABCB1-rs1045642 and CYP3A4-rs2242480 (P ≤ 0.037; Table 4).
Association between genotypes and metabolizer phenotypes
The association between genotype and phenotype was evaluated under the various genetic models and combinations of genotypes. We found two polymorphisms associated with the fast metabolizer phenotype (Table 5). The C/C genotype of ABCB1-rs1045642 under the co-dominant and dominant model and the C/T genotype of CYP3A4-rs2242480 under the over-dominant model were significantly associated with the fast metabolizer phenotype (Table 5) after Bonferroni’s correction (P ≤ 0.049).
Table 5.
Association between genotypes and metabolizer phenotypes
| Gene | Polymorphism | Model | OR (95% CI) | P-Value | Pc-Value |
|---|---|---|---|---|---|
| ABCB1 | rs1045642 | Dominant (C/C vs. C/T + T/T) | C/C: Fast metabolizers | 0.013 | 0.041* |
| 0.29 (0.11–0.75) | |||||
| C/T + T/T: Slow/intermediate/normal metabolizers | |||||
| 2.19 (0.88–5.45) | |||||
| CYP3A4 | rs224280 | Co-dominant | C/T: Fast metabolizers | 0.033 | |
| CYP3A4 | rs224280 | Over-dominant (C/T vs. C/C + T/T) | C/C + T/T: slow metabolizers | 0.019 | 0.055 |
| 1.93 (1.36–2.75) | |||||
| CYP3A4 | rs224280 | Over-dominant (C/T vs. C/C + T/T) | C/T: Fast metabolizers | 0.018 | 0.049* |
| 0.38 (0.19–0.77) | |||||
| C/C + T/T: Slow/intermediate/normal metabolizers | |||||
| 2.86 (0.84–9.71) | |||||
| Combination | rs1045642, rs224280 | Dominant (C/C vs. C/T + T/T), Over-dominant (C/T vs. C/C + T/T) | C/C-C/T: Fast metabolizers | 0.004 | 0.014* |
| 0.14 (0.03–0.64) | |||||
| Combination | rs1045642, rs224280 | Dominant (C/C vs. C/T + T/T), Over-dominant (C/T vs. C/C + T/T) | C/T + T/T-C/C + T/T: Slow metabolizers | 0.003 | 0.012* |
| 2.64 (1.66–4.20) |
*P-Value supported by logistic regression analysis. OR, odds ratio; CI, confidence interval; Pc, Bonferroni-corrected P-values.
Discussion
In Mexico, the issue of obesity is alarming because of its high prevalence1; however, only a small fraction of the population goes to professional health services for diagnosis and treatment32. Pérez-Salgado et al. suggest that this could be because the government institutions in Mexico focus mainly on raising awareness among the population about diseases that derive from obesity32 and, therefore, research focused on improving the effectiveness of treatments is needed.
In order to contribute to health needs, the pharmacokinetic variability of one of the drugs used in the treatment of obesity was tested in a Mexican population. In this study, AFP pharmacokinetic profiles of 36 healthy Mexican volunteers were assessed under controlled conditions. In our sample, the groups of men and women had similar age range and BMI and the same ethnic group. However, they were significantly different in terms of weight and height (Table 1). These differences that exist between men and women could affect the drug disposition33 and, therefore, influence the pharmacokinetic variability34. Nevertheless, this variability could be eliminated if normalization by weight is performed33. Accordingly, in our findings, no sex differences were observed in pharmacokinetic parameters among males and females (Table 1). To our knowledge, no prior studies have reported the effect of sex on AFP pharmacokinetics.
To verify whether there exists variation in AFP pharmacokinetic profiles and investigate a possible association with polymorphisms in genes related to drug metabolism, we used a simple approach to classify the pharmacokinetic profiles using the parameters Cmax and AUC0-t. Four major metabolizer phenotypes were distinguished (slow, normal, intermediate, and fast) with significant differences in pharmacokinetic parameters among the metabolizer phenotype groups (Table 2, Fig. 1A,B). Although our metabolic classification should be validated by another phenotyping method such as pharmacometabolomics35, the pharmacokinetic variability among metabolic phenotypes is evident. This variability is confirmed by a >6-fold difference in the pharmacokinetic parameters between the slowest and the fastest metabolizers. To the best of our knowledge, this is the first report on different AFP metabolizer types. In 1973, Testa and Beckett reported minor intersubject variations under standardized conditions of AFP metabolism; however, their trials were performed on only four subjects36. During the clinical trial and at the end, no volunteers showed any adverse effects.
Although several pharmacogenetics and pharmacogenomics studies have been published, this is the first study evaluating AFP. In this study, two polymorphisms had a significant impact on AFP pharmacokinetics (P ≤ 0.046). One polymorphic variant involved a gene that encodes a transporter protein (ABCB1-rs1045642), and the other located polymorphism involved genes encoding drug-metabolizing enzymes (CYP3A4-rs2242480). It is known that ABCB1 interacts with a multitude of structurally and biochemically unrelated substrates37. Therefore, ABCB1 affects plasma concentrations of and, consequently, the pharmacological response to several drugs, with inconsistent results. The latter depends on the population as well as the substrate analyzed20,37–44, for example, in a Chinese study population, the T/T genotype was associated with lower plasma concentrations of clopidogrel and its active metabolites45. However, in Korean subjects, the T/T genotype was associated with higher Cmax and AUC values of fexofenadine hydrochloride46. These discrepancies may be due to the fact that polymorphisms with silent effects can affect the time of protein translation and, therefore, its folding, causing changes in substrate specificity47. Regarding anti-obesity drugs, Lloret et al. conducted a study on the role of ABCB1-rs1045642 gene variant on oral morphine pharmacokinetics; however, they did not find an association between the two48. Nevertheless, there is no information on the effect of ABCB1 on AFP pharmacokinetics. For the Mexican population, the C/C genotype of the rs1045642 polymorphism has been associated with the atorvastatin fast metabolizer phenotype because of a significant effect on Cmax and AUC0-t values among 60 healthy volunteers20. The results of the present study are consistent with the previous report; carriers of the C/C genotype showed significantly lower Cmax, AUC0-t, and AUC0-∞ values (Tables 3 and 4), significant variability in Ke and Cl values, and were associated with the AFP fast metabolizer phenotype after Bonferroni’s correction (Table 5). These results suggest that the effect of rs1045642 could be related to abnormal intestinal absorption and clearance of AFP. Here, the C/C genotype carriers showed higher Ke and Cl values but lower T1/2. However, only Ke was significantly different (Table 3). These findings are similar to those reported by Gonzalez-Vacarezza et al. in quetiapine pharmacokinetics49. To our knowledge, the present study is the first report on the effect of rs1045642 on AFP pharmacokinetics.
CYP3A4 encodes one of the main drug metabolizing-enzymes25. Although the CYP3A4*1B polymorphic variant is the most studied, there is no report of an association between drug metabolism and response in the Mexican population50–52. Therefore, other polymorphic variants in non-coding regions have been explored, for example, CYP3A4-rs2242480 in intron 1050. Interestingly, we found that CYP3A4-rs2242480 affected several parameters of AFP pharmacokinetics (Tables 3 and 4). Individuals with the heterozygous genotype (C/T) showed a significant effect on Cmax and AUC0-t and a significant association with the fast metabolizer phenotype (Table 5), i.e. there is an increased capacity for plasma clearance. Nonetheless, our results suggest that the C/T genotype is associated with an increased biotransformation of AFP because rs2242480 significantly affects only Cmax and AUC0-t values but not parameters such as Ke and Cl (Tables 3 and 4). These results are consistent with the previous report in a Mexican population. The rs2242480 polymorphism was the most important variant of the prediction model of the variation of AUC values of atorvastatin21 and for the increased R-warfarin clearance in subjects with a heterozygous genotype in a population from the United Kingdom53. However, the present results differ from those reported by Danielak et al. in a Polish population where no effect of rs2242480 was observed on clopidogrel pharmacokinetics50. The T allele frequency (0.44) found in our study population was different from those reported in populations from the United Kingdom (0.09)53, Poland (0.08)50, and Asia (0.29)54.
In summary, AFP pharmacokinetics differs among Mexicans, and we found two genetic polymorphisms with a significant effect on AFP pharmacokinetics, which were used in a log-linear regression analysis. Nevertheless, the interaction of these polymorphisms could be considered to produce a small or moderate effect because only 13–20% of the variability on Cmax, AUC0-t, and Cl can be predicted (Table 4). It is possible that other gene polymorphisms not evaluated in this study may contribute to AFP pharmacokinetic variability.
Furthermore, we did not measure the concentration of secondary metabolites. Such information might have provided an additional phenotyping method to support our results. Another study limitation was the small population size. A follow-up study with a larger population is required to validate our results.
Conclusion
This pilot study revealed variability in AFP pharmacokinetics in the Mexican population. Additionally, ABCB1-rs1045642 and CYP3A4-rs2242480 were found to significantly affect AFP plasma levels. These results suggest that genetic variants may be useful in studies predicting pharmacokinetic variability and those that evaluate the response to AFP treatment. Further studies in a larger population are required to validate our findings.
Material and Methods
Design
A controlled, randomized, crossover, single-blind, two-treatment, two-period, and two-sequence clinical study was conducted in healthy volunteers of Mexican origin to assess the bioequivalence of a single oral dose (75 mg) of AFP (extended-release capsule; Medix Products, S. A., Mexico City, CDMX, MEX) at Ipharma S.A. The study complies with the guidelines of the Declaration of Helsinki, the Good Clinical Practice Standards of Tokyo, and the Mexican regulations for studies of bioavailability and bioequivalence. The clinical protocol was approved by the Research and Ethics Committee of the Clinical and Experimental Pharmacology Center, Ipharma S. A. (Monterrey, NL, MEX), and the pharmacogenetic procedure was approved by the Ethics, Research, and Biosecurity Committees of the University of Monterrey (San Pedro Garza Garcia, NL, MEX; registry number 042014-CIE). The study was registered in the National Registry of Clinical Trials (RNEC; registry number /A394-16) at the Federal Commission for Protection Against Health Risks (COFEPRIS) and Australian New Zealand Clinical Trials Registry (ACTRN12619000391178; registration date: 12/03/2019). Written informed consent was obtained from all subjects or their parents/legal tutors.
Study population
From February 2017 to March 2017, 36 healthy volunteers were enrolled and randomized using R statistical software version 2.15.2. Inclusion criteria for this study were as follows: healthy males and non-pregnant, non-breastfeeding females, aged 18–55 years, with a BMI in the range 18–27 kg/m2, non-smokers, and willing to use adequate methods of contraception throughout the study. Exclusion criteria included electrocardiographic, clinical, biochemical, haematological, coagulation, or urinalysis test abnormalities; a positive test for drug or alcohol abuse, human immunodeficiency virus, hepatitis B and C viruses, and rapid plasma reagin; a medical history of serious adverse reaction or serious hypersensitivity to the test drug or to chemically related drugs; glaucoma, hypertension, anorexia and thyroid problems or existence of concurrent disease; a history of smoking, alcohol or drug abuse; the use of prescription or over-the-counter medication 3 weeks before enrolment; and participation in a clinical research study in the previous 3 months.
Randomization
The volunteers who fulfilled the requirements were selected by the clinical research coordinator of Ipharma, S.A. Before the start of the clinical study, the statistical division of Ipharma S.A. performed randomization for the allocation of drugs (R or P, file code, and subject code). The randomization was done in two blocks in a balanced design using the R statistical software version 2.15.2. and the Mersenne Twister algorithm. The participants were blinded to treatment allocation.
Sampling
A single dose of 75 mg of AFP extended-release capsule (Medix Products, S. A., Mexico City, CDMX, MEX) was administered orally after overnight fasting. The volunteers were under medical supervision throughout the protocol. Blood samples (4 mL) were collected in BD K2EDTA-coated Vacutainers (BD Diagnostics, Franklin Lakes, NJ, USA) at pre-dose (time 0) and seventeen times after drug administration (0.25, 0.5, 0.75, 1, 1.25, 1.50, 1.75, 2, 2.5, 3, 4, 6, 8, 10, 12, 24, and 48 h). Plasma was separated by centrifugation (10 min at 1600 × g at 4 °C) and stored at −65 ± 15 °C for subsequent analysis.
Determination of plasma Amfepramone concentration
Plasma AFP concentration was measured using a validated method developed by Ipharma S.A., according to Mexican regulations (NOM-177-SSA1-2013)55 and guidelines of European Medicines Agency (EMA) regarding bioanalytical method validation56.
In brief, the plasma proteins were eliminated by acetonitrile precipitation. Then, 300 µL acetonitrile was added to a 50 µL plasma sample, vortexed (60 rpm, 4 min), and centrifuged (9600 × g, 10 min, 10 °C). The supernatant (200 µL) was injected into a high-performance liquid chromatography (HPLC) system coupled to a tandem mass spectrometer 6410B (MS/MS) with a triple quadrupole detector (Agilent Technologies, Santa Clara, CA, USA). The chromatographic separation was performed at 45 °C with a Gemini C18 pre-column (Phenomenex, Torrance, CA, USA) and a Zorbax Eclipse XDB C-18 column (3.5 µm, 80 Å, 4.6 × 150 mm) (Agilent Technologies, Santa Clara, CA, USA). The injection sample volume was 5 µL. The mobile phase had a flow rate of 0.6 mL/min and consisted of 5.0 mM ammonium formate/0.1% formic acid and acetronitrile (15:85). The detection system used an ESI MS/MS precursor ion (+) 206.2 m/z and a (+) product ion 105.0 m/z. The interday linearity was assessed with calibration curves (curve range, 1 to 100 ng/mL: 1, 2.5, 5, 10, 25, 50 and 100 ng/mL). The intraday quality control was evaluated by control samples of 1.7, 7.5, 35, and 75 ng/mL each. The validated method had a coefficient of variation (CV) ≤5% for the precision and an error ≤5% for the accuracy.
Pharmacokinetic analysis
The pharmacokinetic parameters were calculated by non-compartmental methods. The maximum plasma concentration (Cmax) and time to reach Cmax (Tmax) were obtained from the concentration-time data of the plasma. The area under the plasma AFP concentration-time curve (AUC) from time 0 to the time of last measurement (AUC0-t), AUC from time 0 extrapolated to infinity (AUC0-∞) were calculated by log-linear trapezoidal rule. Ke was estimated by log-linear regression from the terminal portion of the log-transformed concentration-time plots. T1/2 was estimated by dividing 0.693 by Ke. The total drug clearance was calculated by dividing dose by AUC0-∞ and adjusting for weight. Volume of distribution was calculated as Cl divided by Ke. The AUC and Cmax values were adjusted for dose and weight (AUC/dW and Cmax/dW). The pharmacokinetic analysis was performed using R statistical software version 2.15.2.
Classification of Amfepramone metabolism
The metabolizer phenotypes were determined according to a multivariate analysis of the combined pharmacokinetic parameters Cmax and AUC0-t using a hierarchical cluster analysis (HCA)20,29. Since the clustering algorithms are dependent on the data size, we used a modified method to estimate the ideal number of clusters30,31. First, we introduced the data set of the unadjusted Cmax and AUC0-t values together with the adjusted data set. Second, Cmax and AUC0-t were standardized to minimize the effect of scale differences, and a distance matrix was made from the combined Cmax and AUC0-t values. Third, hierarchical cluster analysis (HCA) using the Ward linkage method was performed on individual Cmax and AUC0-t values. Finally, the interindividual Manhattan distances were computed. Minitab 16 software (Minitab Inc., State College, PA, USA) was used for standardization and HCA. We identified the subjects of each cluster and calculated the means of all adjusted pharmacokinetic parameters by cluster. According to the mean values of the pharmacokinetic parameters of the clusters, they were classified into metabolizer phenotypes20.
Pharmacogenetic tests
The Wizard Genomic DNA Purification kit (Promega Corp., Madison, WI, USA) was used to isolate DNA according to manufacturer’s instructions. Genomic DNA was quantified by UV absorbance using Nanodrop (Thermo Fisher Scientific Inc., Wilmington, MA, USA). The DNA purity was evaluated with the A260/280 and A260/230 ratios, and the samples were stored at −20 °C until use. The DNA samples were subjected to genotyping for the polymorphisms ABCB1-rs1045642, ABCG2-rs2231142, SLCO1B1-rs4149056, DRD3-rs6280, CYP3A4-rs2242480, CYP3A5-rs776746, CYP2B6-rs3745274, and GSTM3-rs179973557 using real-time polymerase chain reaction and Taqman probes (Applied Biosystems; Thermo Fisher Scientific Inc., Wilmington, MA, USA) according to the manufacturer’s protocol. Three quality control thresholds were applied: a genotype call rate equal to 1, an HWE test with P > 0.05, and minor allele frequency >0.01.
Statistical analysis
For sample size calculation, the intrasubject CV obtained in a previous pilot study (Amfepramone/A359-16P) was considered. It was assumed that CV was 25.77% for both Cmax and AUC. Considering a confidence level of 95%, a significance level of 5%, and a minimum power of 80%, a sample size of 30 would suffice. One-way analysis of variance (ANOVA); Kruskal Wallis test; automatic linear modelling using forward stepwise method, Akaike Information Criterion (AICC) and Overfit Prevention Criterion (ASE); and linear and logistic regression analysis model were applied to validate the phenotyping model. The HWE was determined by comparing the genotype frequencies with the expected values using the maximum likelihood method58. Differences between males and females regarding genotype frequencies were determined using a corrected χ2 test. To assess the effects of polymorphisms on the AFP pharmacokinetic parameters, comparisons between two and three groups were made. The Student’s t-test and one-way analysis of variance were used for parametric distributions, whereas Mann-Whitney U and Kruskal-Wallis H tests were used for nonparametric distributions. Post-hoc tests (Bonferroni correction and Tamhane’s T2 test) were used for pairwise comparisons. To confirm the contribution of genetic factors to the variability of pharmacokinetic parameters, automatic linear modelling using forward stepwise method, AICC and ASE as well as linear regression analysis using enter, stepwise, remove, backward, and forward methods were performed. Possible associations of genotypes or combinations of genotypes with phenotypes were evaluated using χ2 and Fisher’s exact tests and validated by logistic regression analysis. The evaluation effects of polymorphisms and associations were assessed under three different models (co-dominant, dominant, over-dominant, and recessive). The odds ratio (OR) was estimated with a 95% confidence interval (95% CI). All P values were two-tailed. Corrected P values (Pc) were obtained using the Bonferroni correction for exclusion of spurious associations. P < 0.05 was interpreted as statistically significant. The statistical analyses were performed with SPSS for Windows, V.20 (IBM Corp., NY, USA).
Trial registration
Australian New Zealand Clinical Trials Registry: ACTRN12619000391178, date registered: 12/03/2019.
Acknowledgements
The authors would like to thank QBP Marcelino Aguirre Garza, Dr. Lizeth Alejandra Martínez Jacobo from University of Monterrey and technical staff of Ipharma, S.A. for facilities, advice and technical support; and Dr. Irene Meester for reviewing and improving the manuscript. Clinical trial Pharmaceutical Research S.A.,/A394-16. Pharmacogenetic study University of Monterrey, UIN19516.
Author contributions
M.G.-S., A.L.-G. and R.B.R.L.-C. designed research and conception. E.P.-G. and R.V.-Z. collected samples and acquired clinical data of volunteers. M.G.-S., E.P.-G. and R.V.Z. conducted analytical determination M.B.L. performed genetic studies of the study subjects. M.E.G.-P. carried out the pharmacokinetic analysis data. R.B.R.L.-C. and M.E.G.-P. analyzed data. R.B.R.L.-C., M.B.L. and A.L. data interpretation. M.G.-S., A.L.-G. R.B.R.L.-C. and A.L. wrote and critically reviewed the manuscript. All authors read and approved the final manuscript.
Data availability
All data generated and analysed during this study are included in this published article, but if necessary, some additional information is available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Organisation for Economic Co-operation and Development. Obesity Update 2017, https://www.oecd.org/els/health-systems/Obesity-Update-2017.pdf (2017).
- 2.Rivera-Dommarco, J. et al. (Instituto Nacional de Salud Publica (MX), Cuernavaca, México, 2016).
- 3.World Health Organization. Obesity and overweight, http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (2017).
- 4.Bray GA, Greenway FL. Pharmacological treatment of the overweight patient. Pharmacological reviews. 2007;59:151–184. doi: 10.1124/pr.59.2.2. [DOI] [PubMed] [Google Scholar]
- 5.Wilbert B, Mohundro BL, Shaw V, Andres A. Appetite suppressants as adjuncts for weight loss. American family physician. 2011;83:1–2. [PubMed] [Google Scholar]
- 6.Manning S, Pucci A, Finer N. Pharmacotherapy for obesity: novel agents and paradigms. Therapeutic advances in chronic disease. 2014;5:135–148. doi: 10.1177/2040622314522848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang JG, Park CY. Anti-Obesity Drugs: A Review about Their Effects and Safety. Diabetes & metabolism journal. 2012;36:13–25. doi: 10.4093/dmj.2012.36.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas EA, et al. Greater hunger and less restraint predict weight loss success with phentermine treatment. Obesity. 2016;24:37–43. doi: 10.1002/oby.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray GA. Lifestyle and pharmacological approaches to weight loss: efficacy and safety. The Journal of clinical endocrinology and metabolism. 2008;93:S81–88. doi: 10.1210/jc.2008-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendricks EJ. Off-label drugs for weight management. Diabetes, metabolic syndrome and obesity: targets and therapy. 2017;10:223–234. doi: 10.2147/DMSO.S95299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arias HR, Santamaria A, Ali SF. Pharmacological and neurotoxicological actions mediated by bupropion and diethylpropion. International review of neurobiology. 2009;88:223–255. doi: 10.1016/S0074-7742(09)88009-4. [DOI] [PubMed] [Google Scholar]
- 12.Suplicy H, et al. A comparative study of five centrally acting drugs on the pharmacological treatment of obesity. International journal of obesity. 2014;38:1097–1103. doi: 10.1038/ijo.2013.225. [DOI] [PubMed] [Google Scholar]
- 13.Cercato C, et al. A randomized double-blind placebo-controlled study of the long-term efficacy and safety of diethylpropion in the treatment of obese subjects. International journal of obesity. 2009;33:857–865. doi: 10.1038/ijo.2009.124. [DOI] [PubMed] [Google Scholar]
- 14.Kalyanasundar B, et al. The efficacy of the appetite suppressant, diethylpropion, is dependent on both when it is given (day vs. night) and under conditions of high fat dietary restriction. Appetite. 2016;100:152–161. doi: 10.1016/j.appet.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Mijares M, Bernardes AM, Silva MT. Diethylpropion produces psychostimulant and reward effects. Pharmacology, biochemistry, and behavior. 2009;91:621–628. doi: 10.1016/j.pbb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Soto-Molina H, et al. Six-month efficacy and safety of amfepramone in obese Mexican patients: a double-blinded, randomized, controlled trial. International journal of clinical pharmacology and therapeutics. 2015;53:541–549. doi: 10.5414/CP202135. [DOI] [PubMed] [Google Scholar]
- 17.Leon-Cachon RB, Ascacio-Martinez JA, Barrera-Saldana HA. Individual response to drug therapy: bases and study approaches. Revista de investigacion clinica; organo del Hospital de Enfermedades de la Nutricion. 2012;64:364–376. [PubMed] [Google Scholar]
- 18.Herrera-Gonzalez S, et al. Effect of AGTR1 and BDKRB2 gene polymorphisms on atorvastatin metabolism in a Mexican population. Biomedical reports. 2017;7:579–584. doi: 10.3892/br.2017.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leon-Cachon RB, et al. Application of Genomic Technologies in Clinical Pharmacology Research. Revista de investigacion clinica; organo del. Hospital de Enfermedades de la Nutricion. 2015;67:212–218. [PubMed] [Google Scholar]
- 20.Leon-Cachon RBR, et al. A pharmacogenetic pilot study reveals MTHFR, DRD3, and MDR1 polymorphisms as biomarker candidates for slow atorvastatin metabolizers. BMC cancer. 2016;16:74. doi: 10.1186/s12885-016-2062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cruz-Correa OF, Leon-Cachon RB, Barrera-Saldana HA, Soberon X. Prediction of atorvastatin plasmatic concentrations in healthy volunteers using integrated pharmacogenetics sequencing. Pharmacogenomics. 2017;18:121–131. doi: 10.2217/pgs-2016-0072. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic acids research. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckett AH, Stanojcic M. Re-evaluation of the metabolism and excretion of diethylpropion in non-sustained and sustained release formulations. The Journal of pharmacy and pharmacology. 1987;39:409–415. doi: 10.1111/j.2042-7158.1987.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 25.Klein K, Zanger UM. Pharmacogenomics of Cytochrome P450 3A4: Recent Progress Toward the “Missing Heritability” Problem. Frontiers in genetics. 2013;4:12. doi: 10.3389/fgene.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi K, et al. Human buprenorphine N-dealkylation is catalyzed by cytochrome P450 3A4. Drug metabolism and disposition: the biological fate of chemicals. 1998;26:818–821. [PubMed] [Google Scholar]
- 27.Wang JS, Backman JT, Taavitsainen P, Neuvonen PJ, Kivisto KT. Involvement of CYP1A2 and CYP3A4 in lidocaine N-deethylation and 3-hydroxylation in humans. Drug metabolism and disposition: the biological fate of chemicals. 2000;28:959–965. [PubMed] [Google Scholar]
- 28.Jancova P, Anzenbacher P, Anzenbacherova E. Phase II drug metabolizing enzymes. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia. 2010;154:103–116. doi: 10.5507/bp.2010.017. [DOI] [PubMed] [Google Scholar]
- 29.Lindon JC, Nicholson JK. The emergent role of metabolic phenotyping in dynamic patient stratification. Expert opinion on drug metabolism & toxicology. 2014;10:915–919. doi: 10.1517/17425255.2014.922954. [DOI] [PubMed] [Google Scholar]
- 30.Green MA, et al. Who are the obese? A cluster analysis exploring subgroups of the obese. Journal of public health. 2016;38:258–264. doi: 10.1093/pubmed/fdv040. [DOI] [PubMed] [Google Scholar]
- 31.Hu CW, Kornblau SM, Slater JH, Qutub AA. Progeny Clustering: A Method to Identify Biological Phenotypes. Scientific reports. 2015;5:12894. doi: 10.1038/srep12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Salgado D, Valdes Flores J, Janssen I, Ortiz-Hernandez L. Diagnosis and treatment of obesity among Mexican adults. Obesity facts. 2012;5:937–946. doi: 10.1159/000346325. [DOI] [PubMed] [Google Scholar]
- 33.Soldin OP, Chung SH, Mattison DR. Sex differences in drug disposition. Journal of biomedicine & biotechnology. 2011;2011:187103. doi: 10.1155/2011/187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandhi M, Aweeka F, Greenblatt RM, Blaschke TF. Sex differences in pharmacokinetics and pharmacodynamics. Annual review of pharmacology and toxicology. 2004;44:499–523. doi: 10.1146/annurev.pharmtox.44.101802.121453. [DOI] [PubMed] [Google Scholar]
- 35.Huang Q, et al. A Pharmacometabonomic Approach To Predicting Metabolic Phenotypes and Pharmacokinetic Parameters of Atorvastatin in Healthy Volunteers. Journal of proteome research. 2015;14:3970–3981. doi: 10.1021/acs.jproteome.5b00440. [DOI] [PubMed] [Google Scholar]
- 36.Testa B, Beckett AH. Metabolism and excretion of diethylpropion in man under acidic urine conditions. The Journal of pharmacy and pharmacology. 1973;25:119–124. doi: 10.1111/j.2042-7158.1973.tb10604.x. [DOI] [PubMed] [Google Scholar]
- 37.Hodges LM, et al. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein) Pharmacogenetics and genomics. 2011;21:152–161. doi: 10.1097/FPC.0b013e3283385a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dessilly G, Panin N, Elens L, Haufroid V, Demoulin JB. Impact of ABCB1 1236C> T-2677G> T-3435C> T polymorphisms on the anti-proliferative activity of imatinib, nilotinib, dasatinib and ponatinib. Scientific reports. 2016;6:29559. doi: 10.1038/srep29559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregers J, et al. Polymorphisms in the ABCB1 gene and effect on outcome and toxicity in childhood acute lymphoblastic leukemia. The pharmacogenomics journal. 2015;15:372–379. doi: 10.1038/tpj.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miura M, et al. Influence of ABCB1 C3435T polymorphism on the pharmacokinetics of lansoprazole and gastroesophageal symptoms in Japanese renal transplant recipients classified as CYP2C19 extensive metabolizers and treated with tacrolimus. International journal of clinical pharmacology and therapeutics. 2006;44:605–613. doi: 10.5414/CPP44605. [DOI] [PubMed] [Google Scholar]
- 41.Suthandiram S, et al. Effect of polymorphisms within methotrexate pathway genes on methotrexate toxicity and plasma levels in adults with hematological malignancies. Pharmacogenomics. 2014;15:1479–1494. doi: 10.2217/pgs.14.97. [DOI] [PubMed] [Google Scholar]
- 42.Saiz-Rodriguez M, et al. Effect of Polymorphisms on the Pharmacokinetics, Pharmacodynamics and Safety of Sertraline in Healthy Volunteers. Basic & clinical pharmacology &. toxicology. 2018;122:501–511. doi: 10.1111/bcpt.12938. [DOI] [PubMed] [Google Scholar]
- 43.Su J, et al. ABCB1 C3435T polymorphism and response to clopidogrel treatment in coronary artery disease (CAD) patients: a meta-analysis. PloS one. 2012;7:e46366. doi: 10.1371/journal.pone.0046366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calderon-Cruz B, et al. C3435T polymorphism of the ABCB1 gene is associated with poor clopidogrel responsiveness in a Mexican population undergoing percutaneous coronary intervention. Thrombosis research. 2015;136:894–898. doi: 10.1016/j.thromres.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 45.Wang XQ, et al. Genetic polymorphisms of CYP2C19 2 and ABCB1 C3435T affect the pharmacokinetic and pharmacodynamic responses to clopidogrel in 401 patients with acute coronary syndrome. Gene. 2015;558:200–207. doi: 10.1016/j.gene.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 46.Yi SY, et al. A variant 2677A allele of the MDR1 gene affects fexofenadine disposition. Clinical pharmacology and therapeutics. 2004;76:418–427. doi: 10.1016/j.clpt.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 48.Lloret-Linares C, et al. Oral Morphine Pharmacokinetic in Obesity: The Role of P-Glycoprotein, MRP2, MRP3, UGT2B7, and CYP3A4 Jejunal Contents and Obesity-Associated Biomarkers. Molecular pharmaceutics. 2016;13:766–773. doi: 10.1021/acs.molpharmaceut.5b00656. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Vacarezza N, et al. MDR-1 genotypes and quetiapine pharmacokinetics in healthy volunteers. Drug metabolism and drug interactions. 2013;28:163–166. doi: 10.1515/dmdi-2013-0008. [DOI] [PubMed] [Google Scholar]
- 50.Danielak D, et al. Impact of CYP3A4*1G Allele on Clinical Pharmacokinetics and Pharmacodynamics of Clopidogrel. European journal of drug metabolism and pharmacokinetics. 2017;42:99–107. doi: 10.1007/s13318-016-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi WL, Tang HL, Zhai SD. Effects of the CYP3A4*1B Genetic Polymorphism on the Pharmacokinetics of Tacrolimus in Adult Renal Transplant Recipients: A Meta-Analysis. PloS one. 2015;10:e0127995. doi: 10.1371/journal.pone.0127995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zochowska D, Wyzgal J, Paczek L. Impact of CYP3A4*1B and CYP3A5*3 polymorphisms on the pharmacokinetics of cyclosporine and sirolimus in renal transplant recipients. Annals of transplantation. 2012;17:36–44. doi: 10.12659/AOT.883456. [DOI] [PubMed] [Google Scholar]
- 53.Lane S, et al. The population pharmacokinetics of R- and S-warfarin: effect of genetic and clinical factors. British journal of clinical pharmacology. 2012;73:66–76. doi: 10.1111/j.1365-2125.2011.04051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He BX, et al. A functional polymorphism in the CYP3A4 gene is associated with increased risk of coronary heart disease in the Chinese Han population. Basic & clinical pharmacology &. toxicology. 2011;108:208–213. doi: 10.1111/j.1742-7843.2010.00657.x. [DOI] [PubMed] [Google Scholar]
- 55.Secretaría de Salud & Comisión Federal para la Protección Contra Riesgos Sanitarios. (Secretaría de Gobernación, México, DF, 2013).
- 56.European Medicines Agency. Science Medicines Health. (European Medicines Agency, London, UK, 2011).
- 57.Martinez-Trevino DA, et al. Rapid Detection of the GSTM3 A/B Polymorphism Using Real-time PCR with TaqMan((R)) Probes. Archives of medical research. 2016;47:142–145. doi: 10.1016/j.arcmed.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Reed TE, Schull WJ. A general maximum likelihood estimation program. American journal of human genetics. 1968;20:579–580. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and analysed during this study are included in this published article, but if necessary, some additional information is available from the corresponding author upon reasonable request.