Abstract
Membrane integral pyrophosphatases (mPPases) are responsible for the hydrolysis of pyrophosphate. This enzymatic mechanism is coupled to the pumping of H+ or Na+ across membranes in a process that can be K+ dependent or independent. Understanding the movements and dynamics throughout the mPPase catalytic cycle is important, as this knowledge is essential for improving or impeding protein function. mPPases have been shown to play a crucial role in plant maturation and abiotic stress tolerance, and so have the potential to be engineered to improve plant survival, with implications for global food security. mPPases are also selectively toxic drug targets, which could be pharmacologically modulated to reduce the virulence of common human pathogens. The last few years have seen the publication of many new insights into the function and structure of mPPases. In particular, there is a new body of evidence that the catalytic cycle is more complex than originally proposed. There are structural and functional data supporting a mechanism involving half-of-the-sites reactivity, inter-subunit communication, and exit channel motions. A more advanced and in-depth understanding of mPPases has begun to be uncovered, leaving the field of research with multiple interesting avenues for further exploration and investigation.
Keywords: membrane-integral pyrophosphatases, human pathogens, plants, structural biology, molecular mechanism, membrane proteins, hydrolysis, ion pumping
Introduction
Pyrophosphatases (PPases) are enzymes responsible for the reversible hydrolysis of the phosphoanhydride bond in pyrophosphate (PPi) to two inorganic phosphate molecules (Kajander et al., 2013). PPases are subcategorized into three separate protein families; Family I, Family II, and membrane-integral pyrophosphatases (mPPases). Families I and II are evolutionarily unrelated soluble proteins. Despite their common enzymatic activity, mPPases are vastly different to the other PPase families. Firstly, the architecture and sequence of these protein families are unrelated. Ignoring the oligomeric structure, a single subunit of both Family I and mPPases consists of a single domain, while Family II PPases have two domains per subunit. mPPases are the only family that is embedded in the membrane: they have 15-17 transmembrane helices (TMH) per 70-81 kDa subunit (Kellosalo et al., 2012; Lin et al., 2012). Secondly, mPPases have only been reported as homodimers, whereas Family I PPases can form other oligomeric arrangements (Kankare et al., 1996; Kellosalo et al., 2012; Lin et al., 2012; Li et al., 2016; Tsai et al., 2019; Vidilaseris et al., 2019). Family I and II PPases are only responsible for removing excess waste PPi from the cytoplasm, while mPPases are primary ion pumps: they couple hydrolysis to movement of H+ and/or Na+ across a membrane (Kajander et al., 2013) and thus generate a membrane potential, which contributes to a number of cellular functions, such as acidocalcisome and vacuole regulation and energization (Shah et al., 2016). The catalytic activities of these protein families differ by orders of magnitude. The mPPases are the slowest as they can only hydrolyze ≈ 10 PPi molecules per second, followed by Family I, which can hydrolyze ≈ 200 molecules per second, and Family II which can hydrolyze ≈ 2,000 per second (Kajander et al., 2013).
mPPases are less abundant than soluble PPases but still occur in all kingdoms of life except fungi and multicellular animals (Kajander et al., 2013). In eukaryotes, they are localized in the membranes of organelles, such as the Golgi apparatus (Mitsuda et al., 2001), plant vacuoles (Gaxiola et al., 2007), or the acidocalcisomes of protists (Moreno and Docampo, 2009). They are also present in the inner cellular membrane of bacteria such as Bacteroides vulgatus (Luoto et al., 2013a). mPPases have been found to play a role in stress tolerance and plant maturation. Finally, mPPases promote energy efficiency and survival in numerous human pathogens, making them clinically relevant as potential drug targets (Luoto et al., 2011; Shah et al., 2016).
This evolutionarily ancient family evolved through a gene triplication; they thus consist of three structurally-conserved splayed 4-helix bundles made up of TMH 3-6, 9-12, and 13-16 (Au et al., 2006; Kajander et al., 2013) arranged with ~3-fold symmetry perpendicular to the membrane plane. The bundles are structurally highly similar with RMSD/Cα values of 2.1–2.9 Å between them (Kellosalo et al., 2012) and also have 23.6–26.1% sequence identity (Kellosalo et al., 2012). Phylogenic analysis suggests that hydrolysis of PPi to provide energy may have occurred prior to the adoption of ATP as the universal energy currency (Baltscheffsky et al., 1999). Therefore, mPPases may have been the first enzymes to couple phosphoanhydride bond formation/hydrolysis to chemiosmotic potential. Since the initial discovery of mPPase in Rhodospirillum rubrum, seven different mPPase subfamilies have been discovered (Table 1) (Luoto et al., 2011, 2015; Tsai et al., 2014). In brief, the different mPPases are subdivided into two main groups: (i) K+ independent, which pump protons (H+-PPases) and can be regulated by Na+, and (ii) K+ dependent, which can function in the absence of K+, but require K+ for maximal activity. Of these, there are H+-PPases, Na+ pumping (Na+-PPase), and dual Na+/H+ pumping (Na+/H+-PPase) PPases (Kajander et al., 2013). Sequence analysis suggests that the first mPPases were Na+-PPases, and that H+-PPases evolved from these four independent times (Baykov et al., 2013). Additionally, it is likely that the evolution of Na+/H+-PPases occurred separately to the H+-PPases (Luoto et al., 2013b).
Table 1.
mPPase subfamily classification.
| Cation Pumping Specificity | Monovalent Cation Dependence | Semi-Conserved Glutamate Location | Regulation | Hypothesized Sodium Binding Sites | Example | References |
|---|---|---|---|---|---|---|
| H+ | K+ | 6.57 | – | – | Vigna radiata | Tsai et al., 2014 |
| 6.53 | – | – | Carboxydothermus hydrogenoformans | Tsai et al., 2014 | ||
| 5.43 | – | – | Flavobacterium johnsoniae | Tsai et al., 2014 | ||
| - | 6.53 | Na+ and K+ | Inhibitory | Chlorobium limicola | Luoto et al., 2015 | |
| – | – | Pyrobaculum aerophilum | Luoto et al., 2011 | |||
| Na+ | K+ and Na+ | 6.53 | – | Activating and Inhibitory | Thermotoga maritima | Tsai et al., 2014 |
| Na+ and H+ | K+ and Na+ | 6.53 | – | Activating | Bacteroides vulgatus | Tsai et al., 2014 |
This review encompasses our current knowledge of mPPase function, including their evolution, role in whole organisms, and their structure and mechanism on the molecular level. We also suggest avenues for future exploration.
Function and Relevance
Plants
Under physiological conditions in plants, H+-PPases are predominantly localized to the tonoplast membrane surrounding the vacuole (Segami et al., 2014) and make up 10% of its protein components (Segami et al., 2018a). The vacuole possesses multiple functions that require the large-scale movement of molecules across its membrane. The required membrane potential is generated by the vacuolar ATPase complex (V-ATPase) in combination with H+-PPases (Kriegel et al., 2015). There is some controversy over the delineation of the roles of these proton pumps, as a V-ATPase knock-out strain was able to properly maintain acidification of the vacuole and normal function (Krebs et al., 2010). However, a more recent study suggested that lack of V-ATPase could not be compensated for by increased mPPase activity (Kriegel et al., 2015). The general consensus is that the role of H+-PPase in stress tolerance is to replace V-ATPase activity when ATP levels are low (Maeshima, 2000), but this does not fully address the roles of the proton pumps under normal conditions.
H+-PPases are important in plant maturation (Li et al., 2005) because they remove PPi from the cytoplasm (Ferjani et al., 2011; Asaoka et al., 2016). PPi is the by-product of many different cellular processes, including the biosynthesis of protein, RNA and, importantly for plants, cellulose (Maeshima, 2000). Removing the excess PPi following these reactions is critical for driving these processes. Additionally, PPi has a modulatory role as a biochemical intermediate of a number of enzymes (Heinonen, 2001), so tight control of its cytoplasmic availability is essential for normal cellular function. mPPase over-expression in Arabidopsis thaliana resulted in increased cell division and hyperplasia of different organs, in particular the leaves. In contrast, knock-out mutants, and RNA interference studies showed severely disrupted root and shoot development. Each of these were linked to increased or decreased trafficking of the phytohormone auxin, which is known to mediate organogenesis (Li et al., 2005), suggesting a role for H+-PPases in auxin regulation.
This role in auxin regulation was further highlighted in studies of transgenic plants over-expressing H+-PPase genes. Multiple studies have shown that increased polar auxin transport upon mPPase over-expression is closely related to improved root development under stress conditions (Li et al., 2005; Park et al., 2005; Pasapula et al., 2011; Zhang et al., 2011). This plays a role in drought resistance, as the larger root system provides enhanced water absorption (Zhang et al., 2011). In addition to the increased root biomass mechanism, the effect of H+-PPase over-expression on vacuolar function improved tolerance of drought and salinity. The increased electrochemical gradient may drive uptake of ions into the vacuole, producing an increase in osmotic potential and stimulating water uptake (Park et al., 2005; Brini et al., 2006; Zhao et al., 2006; Pasapula et al., 2011; Zhang et al., 2011). Further evidence of the potential of mPPases to improve crop tolerance to suboptimal conditions were the reports of increased chlorophyll content, photosynthesis, leaf water content and fiber yield, with decreased cell membrane damage in transgenic cotton plants, as compared to wild-type under low water and high salt conditions (Lv et al., 2009; Pasapula et al., 2011). Interestingly, these effects may not be predominantly due to the proton pumping activity of the vacuolar mPPases. One study saw a Vr-PPase mutant lacking proton pumping activity, but retaining hydrolysis, was sufficient to rescue the stunted knock-out phenotypes (Asaoka et al., 2016). In addition to this, an A. thaliana plant H+-PPase knockout saw no heterotrophic growth following germination, but this phenotype was rescued by soluble PPase expression, suggesting that effective PPi clearance is the primary function of H+-PPases during postgerminative growth in Planta (Ferjani et al., 2011). It is not clear why this is the case, as all plant cells express soluble Family I PPases at concentrations that should be sufficient to clear the pyrophosphate generated. An explanation could be that the soluble and mPPases function cooperatively (Segami et al., 2018b). In this model, the H+-PPase functions as the major cytosolic PPi-hydrolysis enzyme and the soluble PPases contribute to preventing accumulation to toxic levels, which would explain how soluble PPase expression was able to somewhat compensate for mPPase loss in the aforementioned study (Ferjani et al., 2011).
Human Pathogens
Protozoan Pathogens
A number of major human diseases are caused by protozoan parasites, for example, malaria (Plasmodium ssp.), toxoplasmosis (Toxoplasma gondii), trypanosomiasis (Trypanosome spp.), and leishmaniasis (Leishmania spp.) (Shah et al., 2016). These diseases each have a high prevalence and risk of fatality (Büscher et al., 2017; World Health Organization, 2018) or association with other diseases. For example, toxoplasmosis has been suggested to be associated with a number of conditions, such as psychiatric, neurological, and neoplastic disorders (Torgerson and Mastroiacovo, 2013; Flegr et al., 2014). In addition to this, several protozoan strains responsible for malaria and trypanosomiasis have emerged that are resistant to most of the current treatment regimes (Büscher et al., 2017; World Health Organization, 2018). Therefore, there is a demand for novel therapeutics for these tropical diseases.
The protozoan parasite life cycle typically involves transitions between vectors and hosts and intracellular to extracellular environments, which means the protozoan cell must survive and adapt to several different conditions (Crompton et al., 2014). In terms of mPPase function, the most relevant change to overcome is the difference in osmotic pressure the cells experience in these different environments. The main protozoan mechanism for adjusting internal osmotic pressure involves the acidocalcisome (Docampo et al., 2013), where mPPases are localized in protozoa (Scott et al., 1998; Marchesini et al., 2000). This is a small acidic compartment where numerous ions are stored, including polyphosphate, which the parasite hydrolyses or synthesizes in response to osmotic stress and to release energy (Ruiz et al., 2001). The low pH of the acidocalcisome is crucial for its function, as loss of acidity can lead to a 10-fold decrease in stored polyphosphate levels (Lemercier et al., 2002), resulting in reduced capability to respond to osmotic changes. In addition to the effects on polyphosphate storage, there are detrimental effects on intracellular pH regulation, growth rate and final cell density (Lemercier et al., 2002), suggesting that the loss of mPPases has more widespread effects than just reduced osmotic regulation.
As mPPase function in protozoa is highly important, mPPases are a validated target for pharmacological intervention. Knock-down and knock-out studies in Trypanosoma brucei (Lemercier et al., 2002) T. gondii (Liu et al., 2014), and P. falciparum (Zhang et al., 2018) have demonstrated that mPPases are required for maintaining acidocalcisome acidification, parasitic virulence and in vitro asexual bloodstage growth. Additionally, mPPase-inhibiting bisphosphonate derivatives retarded intracellular proliferation of T. gondii with no effect on host cells (Rodrigues et al., 2000).
Bacterial Pathogens
The most relevant mPPase-expressing bacterial genus to human health are Bacteroides spp., especially B. vulgatus and Bacteroides fragilis (Luoto et al., 2013a), which form a mutualistic relationship with healthy individuals as part of the gastrointestinal microflora (Wexler, 2007). However, outside this environment, they can cause bacteremia (presence of bacteria in the blood), intra-abdominal sepsis, appendicitis, gynecological, and skin infections, endocarditis, septic arthritis, and abscesses in tissues including the brain and female urogenital tract (Wexler, 2007). This is a major threat to human health, as Bacteroides reportedly have the highest resistance rates of all anaerobic pathogens (Wexler, 2007), and an associated mortality rate of over 19%, rising to 60% in untreated cases (Goldstein, 1996).
Bacterial mPPases are primarily found in species that exist under conditions of low-energy stress, such as the obligate anaerobes Bacteroides and deep-sea organisms (Thermotoga maritima and Pyrobaculum aerophilum) (Luoto et al., 2011). In these situations, mPPases may be essential to increasing energy efficiency and promoting survival. mPPases have also been shown to confer greater resistance to other stress conditions. One study revealed that expression of transgenic plant mPPases in Escherichia coli and Saccharomyces cerevisiae resulted in improved tolerance to heat, hydrogen peroxide and high salinity (Yoon et al., 2013). Therefore, we speculate that inhibiting mPPase activity could have effects on bacterial viability in response to stressors, similar to the effects seen in protozoan parasites and plants.
Structure and Mechanism
Structural Overview
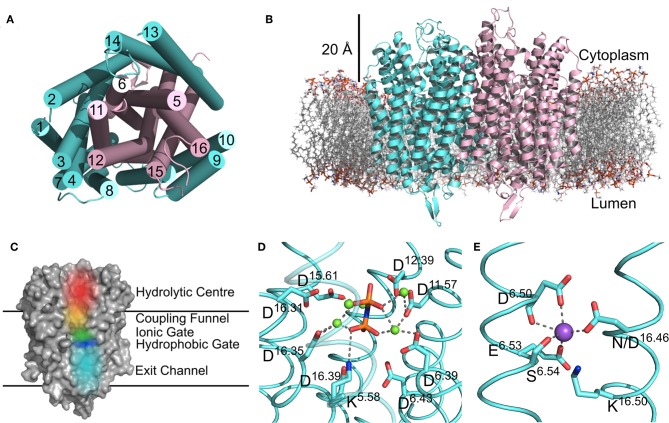
There are now several structures of mPPases (Table 2). In the current structures, each subunit of the homodimeric mPPase is formed by 16 TMH, which form two concentric rings (Figure 1A) surrounding the four catalytic regions: the hydrolytic center, the coupling funnel, the ion gate and the exit channel (Figure 1C) (Kellosalo et al., 2012; Lin et al., 2012; Li et al., 2016). The protein subunit-subunit interface is maintained by hydrophobic interactions and hydrogen bonds between residues on TMH 10 (I/R10.33,N10.42, S10.45, K10.49), 13 (L13.19, L/V13.21, N13.22, M/V13.23, I13.33, Y13.40, S13.48, G13.54, E13.59, R13.62), and 15 (S15.45, Q15.48) (Shah et al., 2016). Here, we use the Ballesteros and Weinstein numbering system, first introduced for GPCRs, so that all functional residues have the same index (Residuehelixnumber.offset) (Ballesteros and Weinstein, 1995), and which we adopted for mPPases (Tsai et al., 2014). In this system, each residue is assigned two numbers, one indicating the TMH and the other to describe its offset from a well-conserved residue close to the middle of the TMH (assigned position 50), as shown in Table 3. For example, K415 from Tm-PPase, is on TMH10 and is one before the conserved S416 and is thus designated K10.49.
Table 2.
Published mPPase structures.
| Species | Liganda | Ionsa | Conformation | Mutation | PDB | Resolution (Å) | Noteworthy | References |
|---|---|---|---|---|---|---|---|---|
| Vigna radiata | IDP | 5 Mg2+ K+ | Symmetrical IDP-Bound | – | 4A01 | 2.35 | First mPPase Structure | Lin et al., 2012 |
| PO4 | 2 Mg2+ | Relaxed Product Bound | – | 5GPJ | 3.5 | Li et al., 2016 | ||
| 2 PO4 | 5 Mg2+ K+ | Product Bound | – | 6AFS | 2.3 | Demonstrated Exit Channel Width Affects Function | Tsai et al., 2019 | |
| E6.57Q | 6AFT | 2.5 | ||||||
| L12.64M | 6AFU | 2.8 | ||||||
| L12.64K | 6AFV | 2.7 | ||||||
| T5.36D | 6AFW | 2.2 | ||||||
| E5.33A | 6AFX | 2.3 | ||||||
| E5.33S | 6AFY | 2.4 | ||||||
| E5.33H | 6AFZ | 2.5 | ||||||
| Thermotoga maritima | - | Mg2+ Ca2+ | Resting | – | 4AV3 | 2.6 | Kellosalo et al., 2012 | |
| 2 PO4 | 4 Mg2+ K+ | Product Bound | – | 4AV6 | 4.0 | Kellosalo et al., 2012 | ||
| IDP | 5 Mg2+ Na+ | Symmetrical IDP-Bound | – | 5LZQ | 3.5 | Li et al., 2016 | ||
| WO4 | 2 Mg2+ | Relaxed Product Bound | – | 5LZR | 4.0 | Li et al., 2016 | ||
| 2 ATCb IDP | 5 Mg2+ Na+ | Locked | – | 6QXA | 3.7 | Asymmetrical Structure Allosteric Inhibitor |
Vidilaseris et al., 2019 |
The number of ligands or ions per subunit.
Only bound to one of the subunits.
IDP, imidodiphosphate; ATC, N-[(2-amino-6-benzthiazolyl)methlyl]-1H-indole-2-carboxamide.
Figure 1.
Structural Features of mPPases. (A) The concentric ring arrangement of the 16 helices in one mPPase subunit, as viewed from the cytoplasmic side. (B) The dimeric protein and its positioning in the lipid bilayer viewed from the membrane plane. The mPPase subunits are colored cyan and pink. (C) A surface view of one mPPase subunit with each of the catalytic regions colored (red, hydrolytic center; yellow, coupling funnel; green, ionic gate; dark blue, hydrophobic gate; light blue, exit channel). The coordination of (D) IDP and Mg2+ (green spheres) in the hydrolytic center and (E) of Na+ (purple sphere) at the ionic gate. Figures generated using PyMol (DeLano, 2002) and the IDP-bound Tm-PPase structure (PBD: 5LZQ) (Li et al., 2016).
Table 3.
Ballesteros and weinstein numbering for mPPases.
| TMH | Signifier | % Sequence Identity | Residue in Tm-PPase | Residue in Vr-PPase | Residue in Pa-PPase |
|---|---|---|---|---|---|
| 1 | 1.50 | 38.1 | F17 | F25 | Y20 |
| 2 | 2.50 | 27.6 | K55 | K94 | R58 |
| 3 | 3.50 | 99.5 | S87 | S153 | S96 |
| 4 | 4.50 | 92.9 | G130 | G194 | G138 |
| 5 | 5.50 | 86.4 | R191 | R242 | R182 |
| 5-6 Bordera | 5.77 | 99.9 | D218 | D269 | D210 |
| 5-6 Borderb | 6.26 | 96.2 | D219 | D270 | D211 |
| 6 | 6.50 | 99.7 | D243 | D294 | D235 |
| 7 | 7.50 | 46.3 | G297 | G334 | A271 |
| 8 | 8.50 | 75.8 | L321 | L361 | L296 |
| 9 | 9.50 | 91.1 | G369 | G411 | G333 |
| 10 | 10.50 | 84.3 | S416 | S458 | S380 |
| 11 | 11.50 | 98.1 | D458 | D500 | D439 |
| 12 | 12.50 | 99.7 | K499 | K541 | K480 |
| 13 | 13.50 | 99.4 | V566 | V597 | V554 |
| 14 | 14.50 | 79 | M611 | M642 | F599 |
| 15 | 15.50 | 94 | A649 | A680 | S637 |
| 16 | 16.50 | 94.5 | K707 | K742 | K691 |
The last TMH5 residue.
The first TMH6 residue.
The inner ring is composed of six TMH (Baltscheffsky et al., 1999; Moreno and Docampo, 2009; Luoto et al., 2013a; Tsai et al., 2014, 2019; Vidilaseris et al., 2019) and the outer ring is composed of the remaining 10 (Figure 1A) (Kellosalo et al., 2012; Lin et al., 2012; Li et al., 2016). A large hydrophilic region extends about 20 Å into the cytoplasm (Figure 1B). This region has a hydrolytic center, which is lined with highly conserved negative (D5.61, D5.65, E5.76, D5.77, D6.35, D6.39, D6.43, D11.57, D15.61, D16.31, D16.35, D16.39), positive (K5.58, K15.64, K16.38) and polar (N12.53) residues that are involved in substrate binding through both direct interactions and via coordination of Mg2+ and water (Figure 1D). The hydrolytic center is followed by the coupling funnel, which is comprised of an ionic network between TMH 5-6 (R5.50, K5.58, D6.39, D6.50), 11-12 (D11.50, K12.50) and 16 (K16.38, D16.39). Below this is the ionic gate, which is situated in the center of the membrane and where the cation is bound prior to pumping (Figure 1E). This region is made up of charged and hydrophilic residues (S5.43, D6.50, E/S6.53, S6.54, E6.57, D16.46, K16.50). The hydrophobic gate located near the ionic gate and is formed by semi-conserved non-polar residues on TMH 5, 6, 12, and 16. Mutations of residues near the ionic and hydrophobic gates (I12.54, L12.64, D16.46, V16.54, L16.57) to alanine decouple the enzyme so that mPPase can hydrolyze PPi but does not pump, suggesting these regions are also essential for the coupling of hydrolysis to pumping (Asaoka et al., 2014). Finally, the residues of the exit channel are not highly conserved between mPPases. However, allosteric inhibitor binding to this region in one Tm-PPase subunit is nonetheless associated with a “locked” conformation (Vidilaseris et al., 2019) and exit channel width has been implicated in coupling hydrolysis and pumping (Tsai et al., 2019), thereby illustrating the importance of conformational changes in this region during the catalytic cycle.
Structural Differences Between Subfamilies
Cation Specificity
Mutagenesis and structural studies showed that the residues E6.53/6.57, S6.54, and D/N16.46 are involved in cation binding and pumping specificity (Asaoka et al., 2014; Li et al., 2016). The main residue regulating cation specificity is the semi-conserved glutamate on TMH6. Movement of this glutamate down one helical turn from E6.53 to E6.57 converts a Na+-PPase to a H+-PPase by destroying the Na+ binding site in the substrate bound state (Li et al., 2016). However, the reverse mutation does not yield a Na+-PPase, indicating that the factors dictating cation specificity are more complex than solely the location of a single residue.
K+ Dependence and Independence
Mutation of A12.46 to lysine in the active site converts a K+-dependent enzyme to an independent enzyme (Belogurov and Lahti, 2002). Modeling this mutation suggested that the group of the lysine sidechain functionally replaces the K+ ion at physiological PPi concentrations (Kellosalo et al., 2012; Lin et al., 2012). A systematic analysis of the potassium ion binding site or active site lysine (K+/K12.46 catalytic center) across all mPPase subfamilies revealed that the lysine contributed to, but was not essential, for the activity of K+ independent mPPases, as the activity was restored at high K+ concentrations when mutated (Artukka et al., 2018). This suggests that K12.46 masks a K+ binding site, but there appear to be larger-scale differences between the two classes than just the identity of A/K12.46 in the active site (see Inter-subunit Communication).
Na+ Regulatory Sites
The difference between Na+- or H+-PPases and Na+/H+-PPases was unclear prior to the discovery of Na+ and K+ regulated H+-PPases (Luoto et al., 2015). These are a subfamily of K+ independent H+-PPases, in which low K+ concentrations enhance H+ transport but higher Na+ and K+ concentrations inhibit ion pumping and hydrolysis. This led to the hypothesis that there are two possible Na+ binding sites; one in the ion conductance channel associated with activation of the mPPase, and the other located away from the conductance channel conferring inhibition of H+ conductance, to which K+ can bind with low affinity. This fits with the reports of H+ translocation by Na+-PPases in sub-physiological sodium concentrations (Luoto et al., 2013a), suggesting that loss of the second inhibitory site led to the evolution of the Na+/H+-PPases.
Mechanism of Pumping and Hydrolysis
Three mechanisms have been proposed for mPPase pumping and hydrolysis: (i) pumping occurs upon binding of substrate and prior to hydrolysis in a “binding change” type mechanism (Kellosalo et al., 2012), (ii) the proton released upon hydrolysis triggers release of the ions at the ion gate via a Grotthus type mechanism (Lin et al., 2012), and (iii) the hydrolysis-generated proton is directly transported in a “direct-coupling” mechanism (Baykov et al., 2013). This first mechanism assumes that the nucleophilic proton may not be involved in pumping, whereas the second and third propose that the hydrolysis-generated proton is the one pumped, thereby assuming hydrolysis precedes cation-pumping. However, neither the second nor the third mechanisms account for Na+ transport (Kajander et al., 2013), except for potentially via a “billiard-type” model in the third, where the Na+ is pushed into the exit channel by the nucleophilic water proton (Baykov et al., 2013). In our opinion, models in which hydrolysis precedes cation pumping do not conform with the studies of non-hydrolysable substrate analogs [IDP and methylene diphosphonate (MEDP)] binding to Vr-PPase (Li et al., 2016; Shah et al., 2017) resulting in a single ion turnover event.
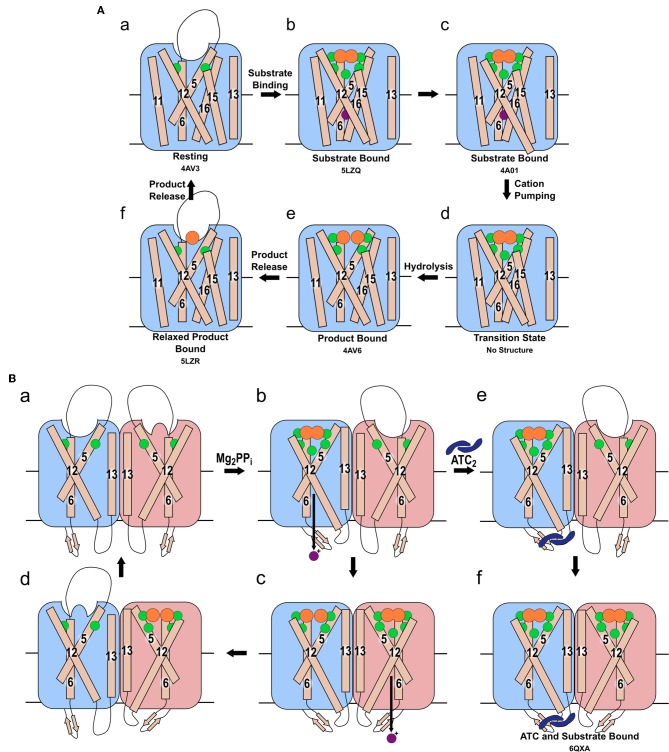
The “binding change” mechanism unifies both sodium and proton pumping, and is supported by studies indicating that charge transfer—and so presumably ion pumping—does not require hydrolysis (Li et al., 2016; Shah et al., 2017) and so presumably precedes it in the full catalytic cycle. In this model (Figure 2), the substrate binds to the hydrolytic center, which is then closed to the cytoplasm by ordering of the 5-6 loop and movement of TMHs 11-12 and 15-16 toward the center of the coupling funnel (Figure 2Aa,b). During these conformational changes, a cation is pumped out from the ionic gate (Figure 2Ac,d). The increase in overall negative charge in this region leads to downwards movement of TMH 12 by 2 Å and an associated bend of TMH11, as observed structurally (Figure 2Ab,c) (Kellosalo et al., 2012; Lin et al., 2012; Li et al., 2016). This causes deprotonation of the aspartate pair D6.43 and D16.39, which activates the water nucleophile leading to hydrolysis (Figure 2Ad,e) (Kajander et al., 2013). This process has been demonstrated to be independent of cation pumping in exit channel mutants (Asaoka et al., 2014). These mutations were initially proposed to alter the position and angle of their helices and thereby uncouple the reactions (Asaoka et al., 2014). However, more recent structural characterization of exit channel and hydrophobic gate mutants indicate that this uncoupling appear to be due to widening of the exit channel (Tsai et al., 2019), potentially allowing ion back-flow.
Figure 2.
Mechanism of pumping and hydrolysis. (A) “Binding change” mechanism. In the resting state (a), a cation may be present, but its position has not been revealed structurally. When substrate binds, the hydrolytic center is occluded by movement of the 5-6 loop and of TMH 11-12 and 15-16, and a cation localizes to the ionic gate (b). The cation is then pumped (c) and TMH 12 resumes its original position (d). Hydrolysis then occurs (e). Prior to the sequential release of the product Pi, the 5-6 loop, TMH 11-12 and 15-16 relax to their resting positions (f). (B) The proposed asymmetric cycle of the mPPase dimer. One site is capable of binding PPi, while the other is distorted (a). Upon binding of one PPi molecule and ion pumping, the other site undergoes a conformational change, triggered by the movement of TMH13, to increase its affinity for PPi (b). The second PPi molecule binds and the second cation is pumped, while the first PPi is hydrolyzed (c). Upon release of the resulting Pi, this active site is distorted to reduce affinity for PPi whilst the second bound PPi undergoes hydrolysis (d). States (e,f): the proposed result of allosteric inhibitor ATC binding to the mPPase. The dimer is trapped in a state unable to hydrolyze PPi, but capable of binding one (e) or two (f) PPi. (Green spheres: Mg2+, purple spheres: cation for pumping, orange spheres: PPi or PO4, blue dimeric shape: ATC). PDB IDs for structurally defined states are listed beneath the relevant image.
Asymmetry and Inter-subunit Communication
The events occurring in each mPPase subunit during pumping and hydrolysis are relatively well-understood (see above), but recent studies have provided evidence of asymmetry and subunit interdependence. Firstly, the role of the K+/K12.46 catalytic center in inter-subunit communication, as inferred from the effects of excess substrate was published (Artukka et al., 2018) (see Inter-subunit Communication). Shortly after this, the first structural evidence of asymmetry, further functional evidence, and a putative mechanism involving both subunits were reported (Vidilaseris et al., 2019). The earliest reports of subunit interdependence were in the 1990s in a series of studies utilizing radiation-induced damage to identify the functional unit size of mPPases (Wu et al., 1991; Sarafian et al., 1992; Tzeng et al., 1996). Several of these showed that one impaired subunit conferred compromised function to the unaffected subunit (see Inter-subunit Communication).
Functional Asymmetry
Vidilaseris et al. (2019) reported that binding of the first substrate increased the affinity of the second subunit to PPi. They also observed that the binding of first substrate molecule potentiated binding of the second. This suggests positive co-operativity for substrate binding, and thus they proposed the following mechanism (Figure 2Ba): the first substrate binds to the first subunit leading to cation translocation in this subunit (Figure 2Bb), this causes conformational changes in the second subunit to optimize PPi binding. Following the binding of the second substrate molecule, hydrolysis occurs in the first subunit and ion pumping in the second (Figure 2Bc). This then allows hydrolysis in the second subunit and product release (Figure 2Bd). However, the same study (Vidilaseris et al., 2019) also reported a 20-fold decrease in the maximal rate of hydrolysis when both subunits bind substrate, which does not appear to fit with their proposed asymmetric mechanism.
In contrast, Artukka et al. (2018) reported a decrease in affinity for PPi at the second subunit following substrate binding to the first. The binding of fluorescein 5'-isothiocyanate (FITC) to a lysine believed to be in the catalytic center (K12.50) also decreases the affinity for FITC in the second subunit (Yang et al., 2000), providing further support for an allosteric mechanism. Binding at the first site thus presumably distorts the active site in the second subunit, reducing its affinity for PPi/FITC. Negative co-operativity for substrate binding is highly relevant, because excess substrate inhibits hydrolysis in all wild-type mesophilic mPPases studied so far (Artukka et al., 2018), so excess substrate may play a role in proton pumping inhibition (Luoto et al., 2015). As there is no evidence of an alternative PPi binding site, and the hydrolytic center and coupling funnel could not accommodate further PPi molecules, the inhibition must be a result of substrate binding to the second subunit. This would only be possible in excess substrate concentrations, due to the decreased affinity at the second site. Similar to Vidilaseris et al. (2019), this study also saw a 3–16-fold decrease in maximal velocity of hydrolysis in both subunits upon binding of the second substrate. This again supports the model in which excess substrate inhibits hydrolysis through binding at this second site, thus suggesting that at moderate PPi concentrations only one subunit operates at any given time. The direct implication of this is that the symmetrical IDP bound structures are potentially not mechanistically relevant at typical PPi concentrations, but a possible artifact of the 4 mM minimum IDP concentration during crystallization (Lin et al., 2012; Li et al., 2016). These papers by Artukka et al. (2018) and Vidilaseris et al. (2019) are the first two definitive studies of possible asymmetry: further data are needed to understand fully the mechanism involving both subunits.
What could explain the inconsistencies between these two studies? Firstly, one study investigated thermophilic protein in an assay at 71°C (Vidilaseris et al., 2019), whereas, the other used mesophilic protein at 30°C (Artukka et al., 2018). The behavior could indicate different properties between thermo- and mesophilic proteins, or different activities at different temperatures. Secondly, one study was performed with purified protein in detergent micelles (Vidilaseris et al., 2019) and the other used unpurified protein in inner membrane vesicles (Artukka et al., 2018). This could suggest that the lipid environment of the enzyme could be involved in regulating the dimeric interface, a concept for which there is increasing evidence (Gupta et al., 2017; Pyle et al., 2018).
Structural Asymmetry
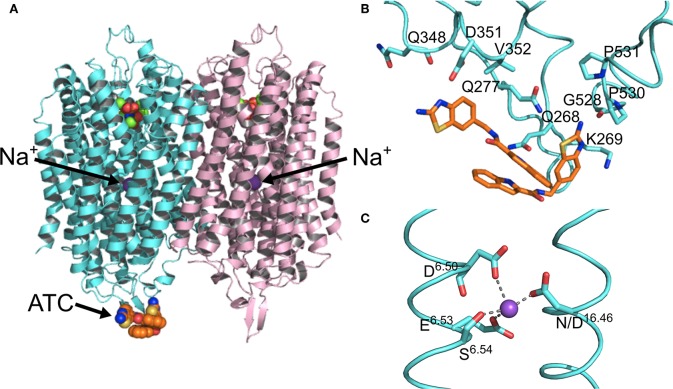
Kinetic and structural studies showed that the non-phosphorous allosteric inhibitor N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) binds as a dimer to one of the subunits in Tm-PPase (Vidilaseris et al., 2019) (Figure 3A) thereby trapping an asymmetric structure, with RMSD/Cα values reaching 1.6 Å for some of the loops vs. a highest RMSD/Cα of 0.5 Å for the equivalent loops in the symmetrical IDP-bound structure (PDB: 5LZQ). The ATC dimer bound via the β1-2 loop (Q268, K269, Q277), loop 8-9 (Q348, D351, V352), and loop 12-13 (G528, P530, P531) (Tm-PPase numbering) (Figure 3B). The chief difference between the 5LZQ and 6QXA structures is the presence of ATC in the crystals. Due to the ATC, the β1-2 loop moves closer to loop 12-13 and away from the dimer interface to an angle similar to that seen in the resting and product bound structures, indicating closure of the exit channel. Loop 12-13 also experienced some slight movements, but TMH12 still moves downwards by about 2 Å compared to the resting state. In addition, the coordination of Na+ at the ionic gate changes from pentacoordination to tetracoordination and the ion is displaced by about 1.2 Å from the IDP-only structure (Figure 3C). Thus, the ATC-bound subunit appears to be trapped in a closed exit channel state and cannot perform hydrolysis, and the unbound subunit cannot undergo its catalytic cycle, due to the ATC-bound subunit restricting its motions (Figure 2Be,f).
Figure 3.
Structural asymmetry. (A) The location of the ATC molecule (orange spheres) and the Na+ ion (purple sphere) in the mPPase dimer. (B) The binding of the two ATC molecules (orange sticks) to the β1-2 loop, loop 8-9, and loop 12-13, with the interacting residues shown at sticks (Tm-PPase numbering). (C) The tetracoordination of Na+ (purple sphere) at the ionic gate. (PDB: 6QXA) (Vidilaseris et al., 2019).
Inter-subunit Communication
Artukka et al. (2018) demonstrated that the wild-type K+/K12.46 center was important for inter-subunit communication. Substrate inhibition only occurred in K+ independent mPPases when the native K12.46 residue was present. In the K12.46A substituted enzymes there was no inhibition. Similarly, K+ dependent mPPases were not inhibited by excess PPi unless K+ was present, even in A12.46K substituted enzymes. Consequently, this suggests the lysine and potassium ion are unable to perfectly replace each other, despite the findings of previous modeling studies (Kellosalo et al., 2012). This is further evidence that inter-subunit communication in response to excess PPi is modulated by the native K+/K12.46 center, as its presence is required for excess substrate inhibition.
The recent asymmetrical ATC structure (see above) demonstrated direct linkage of TMH 12 to the dimer interface (Vidilaseris et al., 2019). Vidilaseris et al. suggest that, in the presence of K+, the movement of TMH 12 upon substrate binding to one subunit induces conformational changes in the dimer interface, which are then propagated to the other subunit. Moreover, when residues close to A12.46 were mutated to alanine, proton pumping and hydrolysis were uncoupled (Asaoka et al., 2014). This may be because this region is implicated in inter-subunit communication for cation translocation at normal PPi levels, and thus when it is mutated, cation pumping ceased. This is further supported by the fact that mPPases are dimers, even though all the required catalytic domains are contained within a single monomer.
Early studies of the functional unit size of mPPases demonstrated that mPPases had to be functional dimers for cation pumping (Sarafian et al., 1992) but a single catalytic subunit was sufficient for hydrolysis (Sarafian et al., 1992; Tzeng et al., 1996). Although these studies were limited by the utilization of radiation-induced destruction, a technique which damages the entire molecule, not just specific sites (Kempner, 1993), they are still relevant when interpreting more recent evidence of subunit interdependence.
Conclusion and Future Outlook
mPPases represent an important area of research, due to their essential functions in plants and human parasites. These functions make the enzymes both clinically and agriculturally relevant targets for modification. They are validated drug targets against a number of human diseases, and there is a wealth of evidence that manipulation of these proteins in plants can improve their tolerance to environmental stressors. An essential part of developing these modulation strategies is understanding the mPPase catalytic and cation pumping mechanism on a molecular level. Despite the recent structural and functional studies (Artukka et al., 2018; Vidilaseris et al., 2019), the mechanism involving both protein subunits remains elusive. To elucidate their full mechanism, further studies that couple time-resolved experimental techniques with molecular dynamics simulations are required.
Author Contributions
AH, AG, and AK: conceptualization, writing-review, and editing. AG and AK: funding acquisition and supervision. AH: investigation and writing-original draft preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Steven Harborne for reading an early version of this manuscript, and the members of the Goldman group for sharing unpublished results.
Footnotes
Funding. This work was supported by a BBSRC DTP fellowship (BB/M011151/1) (to AH) and grants from the BBSRC (BB/M023281/1) and the Academy of Finland (1286429) (to AG). AK was supported by a Springboard Award from the Academy of Medical Sciences (GB) and the Wellcome Trust (SBF002\1031).
References
- Artukka E., Luoto H. H., Baykov A. A., Lahti R., Malinen A. M. (2018). Role of the potassium/lysine cationic center in catalysis and functional asymmetry in membrane-bound pyrophosphatases. Biochem. J. 475, 1141–1158. 10.1042/BCJ20180071 [DOI] [PubMed] [Google Scholar]
- Asaoka M., Segami S., Maeshima M. (2014). Identification of the critical residues for the function of vacuolar H+-pyrophosphatase by mutational analysis based on the 3D structure. J. Biochem. 156, 333–344. 10.1093/jb/mvu046 [DOI] [PubMed] [Google Scholar]
- Asaoka M. M., Segami S., Ferjani A., Maeshima M. (2016). Contribution of PPi-hydrolyzing function of vacuolar H+-pyrophosphatase in vegetative growth of arabidopsis: evidenced by expression of uncoupling mutated enzymes. Front. Plant Sci. 7, 1–12. 10.3389/fpls.2016.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. M., Barabote R. D., Hu K. Y., Saier M. H. (2006). Evolutionary appearance of H+-translocating pyrophosphatases. Microbiology 152, 1243–1247. 10.1099/mic.0.28581-0 [DOI] [PubMed] [Google Scholar]
- Ballesteros J. A., Weinstein H. (1995). Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in g protein-coupled receptors. Methods Neurosci. 25, 366–428. 10.1016/S1043-9471(05)80049-7 [DOI] [Google Scholar]
- Baltscheffsky M., Schultz A., Baltscheffsky H. (1999). H+-proton-pumping inorganic pyrophosphatase: a tightly membrane-bound family. FEBS Lett. 452, 121–127. 10.1016/S0014-5793(99)00617-1 [DOI] [PubMed] [Google Scholar]
- Baykov A. A., Malinen A. M., Luoto H. H., Lahti R. (2013). Pyrophosphate-fueled Na+ and H+ transport in prokaryotes. Microbiol. Mol. Biol. Rev. 77, 267–276. 10.1128/MMBR.00003-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov G. A., Lahti R. (2002). A lysine substitute for K+: A460K mutation eliminates K+ dependence in H+-pyrophosphatase of Carboxydothermus hydrogenoformans. J. Biol. Chem. 277, 49651–49654. 10.1074/jbc.M210341200 [DOI] [PubMed] [Google Scholar]
- Brini F., Hanin M., Mezghani I., Berkowitz G. A., Masmoudi K. (2006). Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress Tolerance in Arabidopsis thaliana plants. J. Exp. Bot. 8, 301–308. 10.1093/jxb/erl251 [DOI] [PubMed] [Google Scholar]
- Büscher P., Cecchi G., Jamonneau V., Priotto G. (2017). Human African trypanosomiasis. Lancet 390, 2397–2409. 10.1016/S0140-6736(17)31510-6 [DOI] [PubMed] [Google Scholar]
- Crompton P. D., Moebius J., Portugal S., Waisberg M., Hart G., Garver L. S., et al. (2014). Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Annu. Rev. Immunol. 32, 157–187. 10.1146/annurev-immunol-032713-120220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. (2002). Pymol: an open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 40. [Google Scholar]
- Docampo R., Jimenez V., Lander N., Li Z. H., Niyogi S. (2013). New insights into roles of acidocalcisomes and contractile vacuole complex in osmoregulation in protists. Int. Rev. Cell Mol. Biol. 305, 69–113. 10.1016/B978-0-12-407695-2.00002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani A., Segami S., Horiguchi G., Muto Y., Maeshima M., Tsukaya H. (2011). Keep an eye on PPi: the vacuolar-Type H+-pyrophosphatase regulates postgerminative development in Arabidopsis. Plant Cell. 23, 2895–2908. 10.1105/tpc.111.085415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegr J., Prandota J., Sovičková M., Israili Z. H. (2014). Toxoplasmosis – a global threat. correlation of latent toxoplasmosis with specific disease burden in a set of 88 countries. PLoS ONE 9:e90203. 10.1371/journal.pone.0090203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola R. A., Palmgren M. G., Schumacher K. (2007). Plant proton pumps. FEBS Lett. 581, 2204–2214. 10.1016/j.febslet.2007.03.050 [DOI] [PubMed] [Google Scholar]
- Goldstein E. J. (1996). Anaerobic bacteremia. Clin. Infect. Dis. 23, S97–101. 10.1093/clinids/23.Supplement_1.S97 [DOI] [PubMed] [Google Scholar]
- Gupta K., Donlan J. A. C., Hopper J. T. S., Uzdavinys P., Landreh M., Struwe W. B., et al. (2017). Robinson. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424. 10.1038/nature20820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen J. K. (2001). Biological Role of Inorganic Pyrophosphate. Boston, MA: Springer US; 10.1007/978-1-4615-1433-6 [DOI] [Google Scholar]
- Kajander T., Kellosalo J., Goldman A. (2013). Inorganic pyrophosphatases: one substrate, three mechanisms. FEBS Lett. 587, 1863–1869. 10.1016/j.febslet.2013.05.003 [DOI] [PubMed] [Google Scholar]
- Kankare J., Salminen T., Lahti R., Cooperman B. S., Baykov A. A., Goldman A. (1996). Sructure of Escherichia coli inorganic pyrophosphatase at 2.2 Å resolution. Acta Crystallogr Sect D Biol Crystallogr. 52, 551–563. 10.1107/S0907444996000376 [DOI] [PubMed] [Google Scholar]
- Kellosalo J., Kajander T., Kogan K., Pokharel K., Goldman A. (2012). The structure and catalytic cycle of a sodium-pumping pyrophosphatase. Science 337, 473–476. 10.1126/science.1222505 [DOI] [PubMed] [Google Scholar]
- Kempner E. S. (1993). Damage to proteins due to the direct action of ionizing radiation. Q. Rev. Biophys. 26, 27–48. 10.1017/S0033583500003954 [DOI] [PubMed] [Google Scholar]
- Krebs M., Beyhl D., Gorlich E., Al-Rasheid K. A. S., Marten I., Stierhof Y. D., et al. (2010). Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 107, 3251–3256. 10.1073/pnas.0913035107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel A., Andrés Z., Medzihradszky A., Krüger F., Scholl S., Delang S., et al. (2015). Job sharing in the endomembrane system: vacuolar acidification requires the combined activity of V-ATPase and V-PPase. Plant Cell 27, 3383–3396. 10.1105/tpc.15.00733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier G., Dutoya S., Luo S., Ruiz F. A., Rodrigues C. O., Baltz T., et al. (2002). A vacuolar-type H+-pyrophosphatase governs maintenance of functional acidocalcisomes and growth of the insect and mammalian forms of Trypanosoma brucei. J. Biol. Chem. 277, 37369–37376. 10.1074/jbc.M204744200 [DOI] [PubMed] [Google Scholar]
- Li J., Yang H., Peer W. A., Richter G., Blakeslee J., Bandyopadhyay A., et al. (2005). Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310, 121–125. 10.1126/science.1115711 [DOI] [PubMed] [Google Scholar]
- Li K. M., Wilkinson C., Kellosalo J., Tsai J. Y., Kajander T., Jeuken L. J., et al. (2016). Membrane pyrophosphatases from Thermotoga maritima and Vigna radiata suggest a conserved coupling mechanism. Nat. Commun. 7:13596. 10.1038/ncomms13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. M., Tsai J. Y., Hsiao C. D., Huang Y. T., Chiu C. L., Liu M. H., et al. (2012). Crystal structure of a membrane-embedded H+-translocating pyrophosphatase. Nature 484, 399–403. 10.1038/nature10963 [DOI] [PubMed] [Google Scholar]
- Liu J., Pace D., Dou Z., King T. P., Guidot D., Li Z. H., et al. (2014). A vacuolar-H+-pyrophosphatase (TgVP1) is required for microneme secretion, host cell invasion, and extracellular survival of Toxoplasma gondii. Mol. Microbiol. 93, 698–712. 10.1111/mmi.12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoto H. H., Baykov A. A., Lahti R., Malinen A. M. (2013b). Membrane-integral pyrophosphatase subfamily capable of translocating both Na+ and H+. Proc. Natl. Acad. Sci. U.S.A. 110, 1255–1260. 10.1073/pnas.1217816110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoto H. H., Belogurov G. A., Baykov A. A., Lahti R., Malinen A. M. (2011). Na+-translocating membrane pyrophosphatases are widespread in the microbial world and evolutionarily precede H+-translocating pyrophosphatases. J. Biol. Chem. 286, 21633–21642. 10.1074/jbc.M111.244483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoto H. H., Nordbo E., Baykov A. A., Lahti R., Malinen A. M. (2013a). Membrane Na+-pyrophosphatases can transport protons at low sodium concentrations. J. Biol. Chem. 288, 35126–35137. 10.1074/jbc.M113.510909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoto H. H., Nordbo E., Malinen A. M., Baykov A. A., Lahti R. (2015). Evolutionarily divergent, Na+-regulated H+-transporting membrane-bound pyrophosphatases. Biochem. J. 467, 281–291. 10.1042/BJ20141434 [DOI] [PubMed] [Google Scholar]
- Lv S. L., Lian L. J., Tao P. L., Li Z. X., Zhang K. W., Zhang J. R. (2009). Overexpression of Thellungiella halophila H+-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta 229, 899–910. 10.1007/s00425-008-0880-4 [DOI] [PubMed] [Google Scholar]
- Maeshima M. (2000). Vacuolar H+ pyrophosphatase. Biochim. Biophys. Acta 1465, 37–51. 10.1016/S0005-2736(00)00130-9 [DOI] [PubMed] [Google Scholar]
- Marchesini N., Luo S., Rodrigues C. O., Moreno S. N., Docampo R. (2000). Acidocalcisomes and a vacuolar H+-pyrophosphatase in malaria parasites. Biochem. J. 347, 243–253. 10.1042/bj3470243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Enami K., Nakata M., Takeyasu K., Sato M. H. (2001). Novel type Arabidopsis thaliana H+-PPase is localized to the golgi apparatus. FEBS Lett. 488, 29–33. 10.1016/S0014-5793(00)02400-5 [DOI] [PubMed] [Google Scholar]
- Moreno S. N., Docampo R. (2009). The role of acidocalcisomes in parasitic protists. J. Eukaryot. Microbiol. 56, 208–213. 10.1111/j.1550-7408.2009.00404.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Li J., Pittman J. K., Berkowitz G. A., Yang H., Undurraga S., et al. (2005). Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc. Natl. Acad. Sci. U.S.A. 102, 18830–18835. 10.1073/pnas.0509512102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapula V., Shen G., Kuppu S., Paez-Valencia J., Mendoza M., Hou P., et al. (2011). Expression of an arabidopsis vacuolar H+-pyrophosphatase Gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol. J. 9, 88–99. 10.1111/j.1467-7652.2010.00535.x [DOI] [PubMed] [Google Scholar]
- Pyle E., Kalli A. C., Amillis S., Hall Z., Lau A. M., Hanyaloglu A. C., et al. (2018). Structural lipids enable the formation of functional oligomers of the eukaryotic purine symporter UapA. Cell Chem. Biol. 25, 840–848. 10.1016/j.chembiol.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C. O., Scott D. A., Bailey B. N., De Souza W., Benchimol M., Moreno B., et al. (2000). Vacuolar proton pyrophosphatase activity and pyrophosphate (PPi) in Toxoplasma gondii as possible chemotherapeutic targets. Biochem. J. 349, 737–745. 10.1042/bj3490737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F. A., Rodrigues C. O., Docampo R. (2001). Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 276, 26114–26121. 10.1074/jbc.M102402200 [DOI] [PubMed] [Google Scholar]
- Sarafian V., Potier M., Poole R. J. (1992). Radiation-inactivation analysis of vacuolar H+-ATPase and H+-pyrophosphatase from Beta-Vulgaris L.: functional sizes for substrate hydrolysis and for H+ transport. Biochem. J. 283, 493–497. 10.1042/bj2830493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. A., de Souza W., Benchimol M., Zhong L., Lu H. G., Moreno S. N., et al. (1998). Presence of a plant-like proton-pumping pyrophosphatase in acidocalcisomes of Trypanosoma cruzi. J. Biol. Chem. 273, 22151–22158. 10.1074/jbc.273.34.22151 [DOI] [PubMed] [Google Scholar]
- Segami S., Asaoka M., Kinoshita S., Fukuda M., Nakanishi Y., Maeshima M. (2018a). Biochemical, structural and physiological characteristics of vacuolar H+-pyrophosphatase. Plant Cell Physiol. 59, 1300–1308. 10.1093/pcp/pcy054 [DOI] [PubMed] [Google Scholar]
- Segami S., Makino S., Miyake A., Asaoka M., Maeshima M. (2014). Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in arabidopsis: artifactual bulbs and native intravacuolar spherical structures. Plant Cell 26, 3416–3434. 10.1105/tpc.114.127571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S., Tomoyama T., Sakamoto S., Gunji S., Fukuda M., Kinoshita S., et al. (2018b). Vacuolar H+-pyrophosphatase and cytosolic soluble pyrophosphatases cooperatively regulate pyrophosphate levels in Arabidopsis thaliana. Plant Cell 30, 1040–1061. 10.1105/tpc.17.00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N., Vidilaseris K., Xhaard H., Goldman A. (2016). Integral membrane pyrophosphatases: a novel drug target for human pathogens? AIMS Biophys. 3, 171–194. 10.3934/biophy.2016.1.171 [DOI] [Google Scholar]
- Shah N. R., Wilkinson C., Harborne S. P., Turku A., Li K. M., Sun Y. J., et al. (2017). Insights into the mechanism of membrane pyrophosphatases by combining experiment and computer simulation. Struct. Dyn. 4, 1–12. 10.1063/1.4978038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson P. R., Mastroiacovo P. (2013). The global burden of congenital toxoplasmosis: a systematic review. Bull. World Health Organ. 91, 501–508. 10.2471/BLT.12.111732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. Y., Kellosalo J., Sun Y. J., Goldman A. (2014). Proton/sodium pumping pyrophosphatases: the last of the primary ion pumps. Curr. Opin. Struct. Biol. 27, 38–47. 10.1016/j.sbi.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Tsai J. Y., Tang K. Z., Li K. M., Hsu B. L., Chiang Y. W., Goldman A., et al. (2019). Roles of the hydrophobic gate and exit channel in Vigna radiata pyrophosphatase ion translocation. J. Mol. Biol. 431, 1619–1632. 10.1016/j.jmb.2019.03.009 [DOI] [PubMed] [Google Scholar]
- Tzeng C. M., Yang C. Y., Yang S. J., Jiang S. S., Kuo S. Y., Hung S. H., et al. (1996). Subunit structure of vacuolar proton-pyrophosphatase as determined by radiation inactivation. Biochem. J. 316, 143–147. 10.1042/bj3160143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidilaseris K., Kiriazis A., Turku A., Khattab A., Johansson N. G., Leino T. O., et al. (2019). Asymmetry in catalysis by Thermotoga maritima membrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor. Sci. Adv. 5:eaav7574. 10.1126/sciadv.aav7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. 10.1128/CMR.00008-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2018). Global Malaria Programme. World Malaria Report 2017. [Google Scholar]
- Wu J. J., Ma J. T., Pan R. L. (1991). Functional size analysis of pyrophosphatase from Rhodospirillum rubrum determined by radiation inactivation. FEBS Lett. 283, 57–60. 10.1016/0014-5793(91)80552-E [DOI] [PubMed] [Google Scholar]
- Yang S. J., Jiang S. S., Van R. C., Hsiao Y. Y., Pan R. (2000). A lysine residue involved in the inhibition of vacuolar H+-pyrophosphatase by fluorescein 5'-isothiocyanate. Biochim. Biophys. Acta 1460, 375–383. 10.1016/S0005-2728(00)00203-6 [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Kim S. Y., Kim I. S. (2013). Stress response of plant H+-PPase-expressing transgenic Escherichia coli and Saccharomyces cerevisiae: a potentially useful mechanism for the development of stress-tolerant organisms. J. Appl. Genet. 54, 129–133. 10.1007/s13353-012-0117-x [DOI] [PubMed] [Google Scholar]
- Zhang H., Shen G., Kuppu S., Gaxiola R., Payton P. (2011). Creating drought- and salt-tolerant cotton by overexpressing a vacuolar pyrophosphatase gene. Plant Signal. Behav. 6, 861–863. 10.4161/psb.6.6.15223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wang C., Otto T. D., Oberstaller J., Liao X., Adapa S. R., et al. (2018). Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360:eaap7847. 10.1126/science.aap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F. Y., Zhang X. J., Li P. H., Zhao Y. X., Zhang H. (2006). Co-expression of the Suaeda salsa SsNHX1 and arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single SsNHX1. Mol. Breed. 17, 341–353. 10.1007/s11032-006-9005-6 [DOI] [Google Scholar]