Abstract
Inositol phosphate synthase (IPS) is a rate-limiting enzyme in myo-inositol biosynthesis, which can regulate stress responses in plants and animals. However, there are few studies on the function of IPS in insects, especially in Apis cerana cerana. In this study, the inositol-3-phosphate synthase 1-B gene (AccIPS1-B) was isolated from Apis cerana cerana, and its connection to antioxidant defence was investigated. The open reading frame of AccIPS1-B was 1542 bp, encoding a 513 amino acid polypeptide. Quantitative real-time PCR analysis revealed that the expression level of AccIPS1-B was highest in pupae of Apis cerana cerana, and it was expressed at higher levels in the thorax than in other tissues tested. Moreover, the expression of AccIPS1-B was significantly upregulated by abiotic stresses. The recombinant AccIPS1-B also displayed significant tolerance to cumene hydroperoxide and HgCl2. In addition, knockdown of AccIPS1-B significantly suppressed the expression of most of the antioxidant genes and decreased the antioxidant enzymatic activities of SOD, POD, and GST. Taken together, these findings indicate that AccIPS1-B may be involved in the response to antioxidant defence and development in Apis cerana cerana.
Keywords: Apis cerana cerana, AccIPS1-B, Quantitative real-time PCR, Abiotic stress, Antioxidant defence
Introduction
Chinese honeybees, as pollinators, play important roles in ecological stability and agricultural development. However, the number of Chinese honeybees has decreased sharply because honeybees encounter environmental pollution and environmental changes in the process of honey gathering, flying, and drinking. Therefore, it is particularly important to the survival of Chinese honeybees to identify specific genes and proteins involved in the response to abiotic stress.
Inositol is the precursor for many inositol-containing compounds, such as signalling molecules, and plays important roles in many essential processes, including growth regulation, hormonal regulation, membrane trafficking, and signal transduction (Kaur et al. 2013; Tan et al. 2013). Therefore, regulation of inositol biosynthesis is crucial for the regulation of second messenger signalling. The de novo synthesis of inositol is catalysed by a series of enzymes in organisms. Among them, myo-inositol-1-phosphate synthase (MIPS or ino-1, EC 5.5.1.4) is the first and rate-limiting step enzyme that catalyses glucose-6-phosphate to myo-inositol-phosphate l (Frej et al. 2016; Smith and Major 2011). The first gene encoding MIPS to be identified was the yeast INO1 (Culbertson et al. 1976; Donahue and Henry 1981). To date, many MIPS genes have been identified from many living organisms, including green algae, bacteria, higher plants, and animals. MIPS genes are highly conserved among eukaryotes (Majumder et al. 2003). The typical MIPS proteins are characterized by four highly conserved regions, including NAD+-binding domains, putative catalytic domains, conserved region 1, and conserved region 2 (Frej et al. 2016).
It was reported that MIPS genes play a critical role in the responses to abiotic stress in organisms. For example, overexpression of the MIPS gene in plants could enhance various metabolites responsible for protecting plants from abiotic stress (Kusuda et al. 2015). In Larimichthys crocea, MIPS was involved in the response to cold stress (Li et al. 2018). In the gram-positive Actinobacteria Corynebacterium glutamicum, ino-1 played an important role in oxidative stress resistance (Chen et al. 2018). Moreover, deletion of the ino-1 gene led to a decrease in cell viability, an increase in ROS production, and the elevation of protein carbonylation levels under various stress conditions.
Given the important role of inositol in cellular functioning, surprisingly, little is known about MIPS in honeybees, particularly in Apis cerana cerana. In this study, AccIPS1-B was characterized from Apis cerana cerana, and the expression profiles of this gene were investigated. AccIPS1-B knockdown causes inositol depletion, which results in the downregulation of some ROS-related genes and the decrease in ROS-scavenging enzyme activities in response to oxidative stress. To our knowledge, this is the first study to examine the function of AccIPS1-B in response to abiotic stresses in honeybees.
Materials and methods
Insects and treatments
The Chinese honeybees, Apis cerana cerana, used in the following experiment were maintained at Shandong Agricultural University (Taian, China). Honeybees were generally divided into different developmental stages (larvae, pupae, and adults) according to age, eye colour, and shape (Michelette and Soares 1993). The whole bodies of the first to seventh larval instars (L1–L7), pupae (PP, prepupae; Pw, white eyes; Pp, pink eyes; Pb, brown eyes; Pd, dark eyes), and adults (A1, 1-day post-emergence workers; A15, 15-day post-emergence workers) were collected from the hive randomly and were kept in an incubator at a constant temperature (34 °C) with 70% relative humidity under a 24-h dark regimen. Then, the honeybees (A15) were divided into nine groups receiving different treatments, and each group consisted of 48 individuals. The honeybees in group 1 were injected with H2O2 (0.5 μL, final concentration of 2 mM) and treated for 0, 0.5, 1, 2, 3, and 4 h. Groups 2–4 were exposed to cold (4 °C), hot (44 °C), and UV radiation (30 mJ/cm2 UV), respectively, for 0, 0.5, 1, 2, 3, and 4 h. The honeybees in groups 5 and 6 were fed CdCl2 (3 mg/mL added to food) and HgCl2 (0.5 mg/L added to food), respectively, for 0, 0.5, 1, 1.5, 2, and 3 h. Moreover, the honeybees in groups 7–9 were exposed to different pesticides (lambda cyhalothrin and emamectin benzoate) for 0, 1, 2, 3, 4, and 5 h. For the tissue-specific expression analysis, the honeybees (A15) were dissected into different tissue parts, including the epidermis (Ep), wing (Wi), honey sac (Hs), muscle (Ms), leg (Le), abdomen (Ab), thorax (Th), sting needle (Sn), and midgut (Mi). All samples were collected at a specified point in time and stored at − 80 °C for RNA and protein extraction. Each treatment was repeated at least three times.
RNA extraction and quantitative real-time PCR
Total RNA was extracted with RNAiso Plus (TaKaRa, Dalian, China), and cDNA was synthesized by purified RNA samples using the PrimeScript™ RT reagent kit with gDNA Eraser (Vazyme, Nanjing, China). Quantitative real-time PCR (qRT-PCR) was performed using the SYBR® PrimeScript™ RT-PCR Kit (TaKaRa, Dalian, China). According to the supplier’s instructions, the PCR mixture was as follows: 1.0 μL cDNA, 6.5 μL SYBR Premix Ex Taq, 0.3 μL of each primer (10 mM each), and 5.9 μL RNase-free water in a total volume of 15 μL. The qRT-PCR reaction conditions were as follows: initial denaturation at 95 °C for 30 s, 40 cycles (95 °C for 5 s, 55 °C for 15 s, and 72 °C for 15 s), and a single melt cycle. β-Actin (XM-640276) was used as the internal reference. The relative level of gene expression was automatically calculated by CFX Manager software (version 1.1) according to the 2–ΔΔCT method. All expression levels were tested with three biological replicates, and each reaction was repeated three times. All primers used in this study are listed in Table 1.
Table 1.
The primers used in this study
| Primer name | Primer sequence (5′-3′) | Description |
|---|---|---|
| IZ1 | ATGGCTTTGAAAATTCGCGTTC | cDNA primer of AccIPS1-B, forward |
| IZ2 | TCATAATTGATTTTTGAAATTAACTTTG | cDNA primer of AccIPS1-B, reverse |
| β-s | GTTTTCCCATCTATCGTCGG | Standard control primer of qPCR, forward |
| β-x | TTTTCTCCATATCATCCCAG | Standard control primer of qPCR, reverse |
| BD1 | GGATCCATGGCTTTGAAAATTCGCGTT BamHI | Primers of constructing expression vector, forward |
| BD2 | GAGCTCTAATTGATTTTTGAAATTAACT SacI | Primers of constructing expression vector, reverse |
| IQ1 | ACGGTGACACCTGTATCAAC | qPCR primer of AccIPS1-B, forward |
| IQ2 | CTTTGTTTCCTTTGCCTAAT | qPCR primer of AccIPS1-B, reverse |
| IA1 | TAATACGACTCACTATAGGGCGAGATTGTATTGAAGCACAGTACGA | RNAi primer of AccIPS1-B, forward |
| IA2 | TAATACGACTCACTATAGGGCGATTGCACGTTGCATAGCATCAGCTA | RNAi primer of AccIPS1-B, reverse |
| GA1 | TAATACGACTCACTATAGGGCGAGTCAGCGCTGGAGGGAGTT | RNAi primer of GFP, forward |
| GA2 | TAATACGACTCACTATAGGGCGAGATCTCCGCATCTTCTAA | RNAi primer of GFP, reverse |
| AccSOD1-F | AAACTATTCAACTTCAAGGACC | qPCR primer of AccSOD1, forward |
| AccSOD1-R | CACAAGCAAGACGAGCACC | qPCR primer of AccSOD1, reverse |
| AccSOD2-F | TTGCCATTCAAGGTTCTGGTT | qPCR primer of AccSOD2, forward |
| AccSOD2-R | GCATGTTCCCAAACATCAATACC | qPCR primer of AccSOD2, reverse |
| AccGSTD-F | CGAAGGAGAAAACTATGTGGGCAG | qPCR primer of AccGSTD, forward |
| AccGSTD-R | CGTAATCCACCACCCACCTCTATCG | qPCR primer of AccGSTD, reverse |
| AccGSTO1-F | CCAGAAGTAAAAGGACAAGTTCGT | qPCR primer of AccGSTO1, forward |
| AccGSTO1-R | CCATTAACATCAACAAGTGCTGGT | qPCR primer of AccGSTO1, reverse |
| AccTpx3-F | CCTGCACCTGAATTTTCCGG | qPCR primer of AccTpx3, forward |
| AccTpx3-R | CTCGGTGTATTAGTCCATGC | qPCR primer of AccTpx3, reverse |
| AccP450-F | CGCAAAGAGAATGGGAAGG | qPCR primer of AccP450, forward |
| AccP450-R | CTTTTGTGTGACGGAGGTGC | qPCR primer of AccP450, reverse |
Protein expression
To obtain the AccIPS1-B recombinant protein, the open reading frame (ORF) of AccIPS1-B was amplified using the primers BD1/BD2 and then inserted into the expression vector pET-30a (+). Furthermore, pET-30a (+)-AccIPS1-B was transformed into Escherichia coli strain BL21 (DE3) for protein expression. The recombinant AccIPS1-B and pET-30a (+) E. coli cells were cultured at 28 °C with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). A 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used for the separation of the AccIPS1-B recombinant protein, which was then stained using Coomassie brilliant blue.
Disc diffusion assay
To evaluate the tolerance of the recombinant AccIPS1-B protein under stress, disc diffusion assays were performed as described by Yan et al. (2013). pET-30a (+) was used as a control, and recombinant AccIPS1-B E. coli cells were plated on LB agar plates containing kanamycin and then incubated at 37 °C for 1 h. Next, filter discs (8-mm-diameter) soaked in different concentrations of cumene hydroperoxide (0, 10, 20, 50, or 100 mM) and HgCl2 (0, 10, 20, 40, and 100 mM) were placed on the surface of the culture medium (Yan et al. 2013). Finally, the bacteria were cultivated at 37 °C for 24 h, and then the killing zones around the filter discs were measured three times from different angles. Every treatment was performed in triplicate.
Protein extraction and western blot
Total protein from different treatments was extracted from honeybees using a Tissue Protein Extraction Kit (Comwin Biotech, Beijing, China) and quantified using the BCA Protein Assay Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). As described by Yan et al. (2013), AccIPS1-B antibody was obtained by injecting the AccIPS1-B recombinant protein into white mice at least four times, as described by Chen et al. (2015). Anti-α-tubulin antibody (Beyotime Biotechnology, Shanghai, China) was used as the control, and western blot was performed as previously described (Chen et al. 2015). All experiments were carried out three times.
RNA interference AccIPS1-B
Partial sequence of the coding region of AccIPS1-B was amplified by PCR with primers GA1/GA2, then was used for synthesized massively double-strand RNA AccIPS1B (dsRNA-AccIPS1-B) through RiboMAX T7 large-scale RNA production systems (Promega, Madison, WI, USA), and injected into honeybees. GFP (GenBank accession number: U87974) was used as a control because none of the genes in Apis cerana cerana share homology with GFP. The 15-day post-emergence adult workers were selected for RNA interference (RNAi) experiments and divided into three groups with 40 individuals in each group. Then, 8 μg dsRNA-AccIPS1-B or dsRNA-GFP was injected into each adult between the first and second abdominal segments using a microsyringe. The last group was not injected and was used as a control. qRT-PCR and western blotting were used to examine whether AccIPS1-B was silenced. Then, the silenced honeybees were collected at different time points (12, 24, 36, 48 h) and stored at − 80 °C until use.
Antioxidant enzyme activity analysis
Total protein of the silenced honeybees was extracted in 1 mL normal saline, and concentrations were quantified using a BCA Protein Assay Kit (Nanjing Jiancheng Bioengineering Institute). Next, the activities of the antioxidant enzymes superoxide dismutase (SOD), POD, and glutathione S-transferase (GST) were determined using SOD, POD, and GST kits (Nanjing Jiancheng Bioengineering Institute), respectively, following the instructions. All experiments were repeated three times.
Bioinformatic analysis
Many IPS genes and proteins were analysed using the BLAST search program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The physical and chemical properties of AccIPS1-B were predicted by the ProtParam tool (http://www.expasy.ch/tools/protparam.html). An amino acid multiple alignment was conducted by DNAMAN (version 5.2.2). A phylogenetic tree was generated with the neighbour-joining method using MEGA 4.0 software. The protein tertiary structure was built and analysed with the SWISSMODEL software (https://swissmodel.expasy.org/). The putative cis-acting elements in the AccIPS1-B promoter region were predicted using TFBIND (http://tfbind.hgc.jp/). Statistical significance was analysed using Duncan’s multiple range tests with analysis of variance (ANOVA), and calculations were performed with SPSS Statistics. Significance was set at P < 0.01.
Results
Characterization of AccIPS-1B
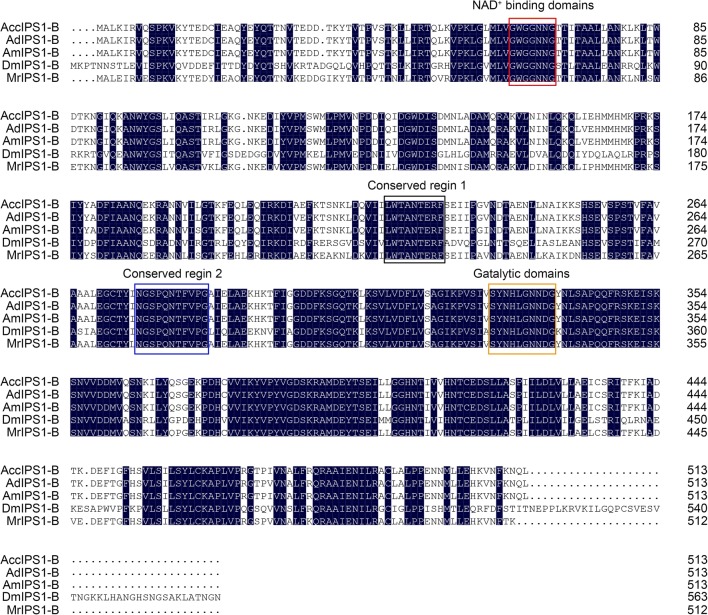
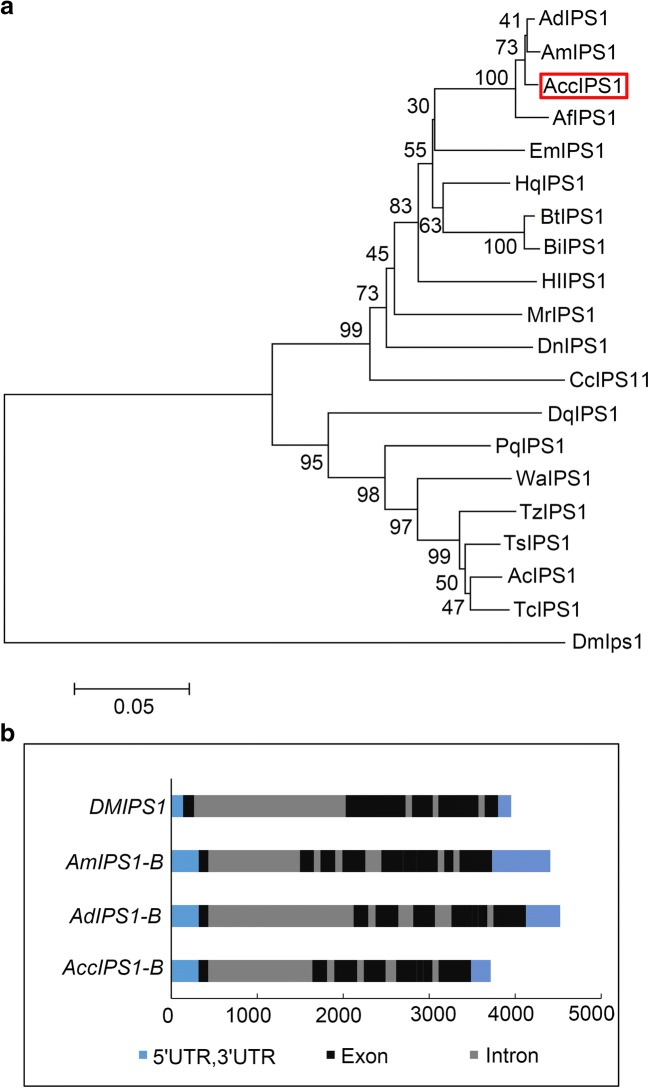
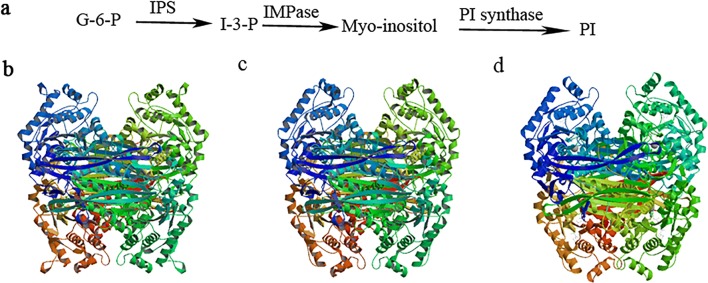
The inositol-3-phosphate synthase 1-B gene was cloned by the primers IZ1/IZ2 from Apis cerana cerana and named AccIPS1-B. The full-length cDNA sequence of AccIPS1-B (GenBank Accession no: XM017065093.1) contains a 317-bp 5′UTR, a 228-bp 3′UTR, and a 1542-bp open reading frame (ORF). The ORF of AccIPS1-B encodes a 513 amino acid polypeptide with a predicted molecular mass of 57.14 kDa and a theoretical isoelectric point (pI) of 7.81. The AccIPS1-B protein contains four highly conserved regions, including NAD+-binding domains (GWGGNNG), putative catalytic domains (SYNHLGNNDG), conserved region 1 (LWTANTERF), and conserved region 2 (NGSPQNTFVPG) (Fig. 1). The phylogenetic tree revealed that AccIPS1-B shares higher homology with AmIPS1-B and AfIPS1-B than with the other proteins (Fig. 2a). As shown in Fig. 3, the tertiary structure of the AccIPS1-B protein is similar to that of IPS in yeast and Drosophila melanogaster. To further analyse the gene structure of AccIPS1-B, a 3715-bp full-length sequence of DNA (GenBank accession number LOC108003032) was obtained from NCBI, which contained 5 introns and 6 exons. Mature mRNA of AccIPS1-B was spliced from six exons (Fig. 2b).
Fig. 1.
Multiple alignment of AccIPS1-B with other known IPS1 genes. All species abbreviations and sequence IDs are as follows: AccIPS1-B (Apis cerana cerana, XP_016920582.1), AdIPS1-B (Apis dorsata, XP_006623674.1), AmIPS1-B (Apis mellifera, XP_623377.1), DmIPS1 (Drosophila melanogaster, NP_477405.1), MrIPS1-B (Megachile rotundata, XP_003706882.1). Identical regions are shaded in black markers, and the four conserved regions are marked with boxes, including NAD+-binding domains, catalytic domains, conserved region 1, and conserved region 2
Fig. 2.
Analysis of the biological information of IPS1. a A phylogenetic tree of IPS1 from different species. All species abbreviations and sequence IDs are as follows: AccIPS1-B (Apis cerana cerana, XP_016920582.1), AdIPS1-B (Apis dorsata, XP_006623674.1), AmIPS1-B (Apis mellifera, XP_623377.1), AfIPS1-B (Apis florea, XP_003692777.1), HqIPS1-B (Melipona quadrifasciata, KOX68055.1), BtIPS1-B (Bombus terrestris, XP_003403362.1), BiIPS1-B (Bombus impatiens, XP_003484397.1), DmIPS1 (Drosophila melanogaster, NP_477405.1), HlIPS1-A (Habropoda laboriosa, XP_017791627.1), MrIPS1-B (Megachile rotundata, XP_003706882.1), DnIPS1-B (Dufourea novaeangliae, XP_015431343.1), CcIPS1-A (Ceratina calcarata, XP_017885676.1), EmIPS1-A (Eufriesea mexicana, XP_017756426.1), PgIPS1-A (Pseudomyrmex gracilis, XP_020281034.1), WaIPS1-B (Wasmannia auropunctata, XP_011702940.1), TsIPS1-A (Trachymyrmex septentrionalis, XP_018347046.1), AcIPS1-A (Atta cephalotes, XP_012062538.1), TcIPS1-A (Trachymyrmex cornetzi, XP_018374519.1), TzIPS1-A (Trachymyrmex zeteki, XP_018314715.1), DqIPS1-B (Dinoponera quadriceps, XP_014474412.1). b Genomic structure of some IPS1 genes
Fig. 3.
a Three-dimensional structure and catalysis of IPS. De novo synthesis of inositol and PI. The abbreviations and full names of the substances and enzymes in the inositol synthesis are as follows: G-6-P (glucose-6-phosphate), IPS (inositol-3-phosphate synthase), IMPase (inositol monophosphatase), PI (inositol monophosphate). b–d The tertiary structure of the IPS1 protein in yeast, honeybee, and Drosophila melanogaster, respectively. The amino acid sequences of IPS in Saccharomyces cerevisiae, Apis cerana cerana, and Drosophila melanogaster were downloaded from the NCBI database, and their GenBank accession numbers are NP_012382.2, XM017065093.1, and NP_477405.1, respectively
Identification and analysis of the promoter region
The 821-bp promoter sequence was obtained from NCBI (gene symbol LOC108003032) to examine the transcription regulatory region of AccIPS1-B using the online software program TFBIND (http://tfbind.hgc.jp/). Many important transcription factor binding sites (TFBS) were identified (Fig. 4). Some transcription factors play important roles in environmental stress and the immune response, including heat-shock factors (HSFs), activating protein-1 (AP1), and cAMP response element binding protein (CREB). In addition, several transcription factors involved in tissue development, such as the caudal-related homeobox (CdxA) protein, cell factor 2-II (CF2-II), Oct-1, broad complex (BR-C), and Sox-5, were also detected in the promoter region of AccIPS1-B.
Fig. 4.
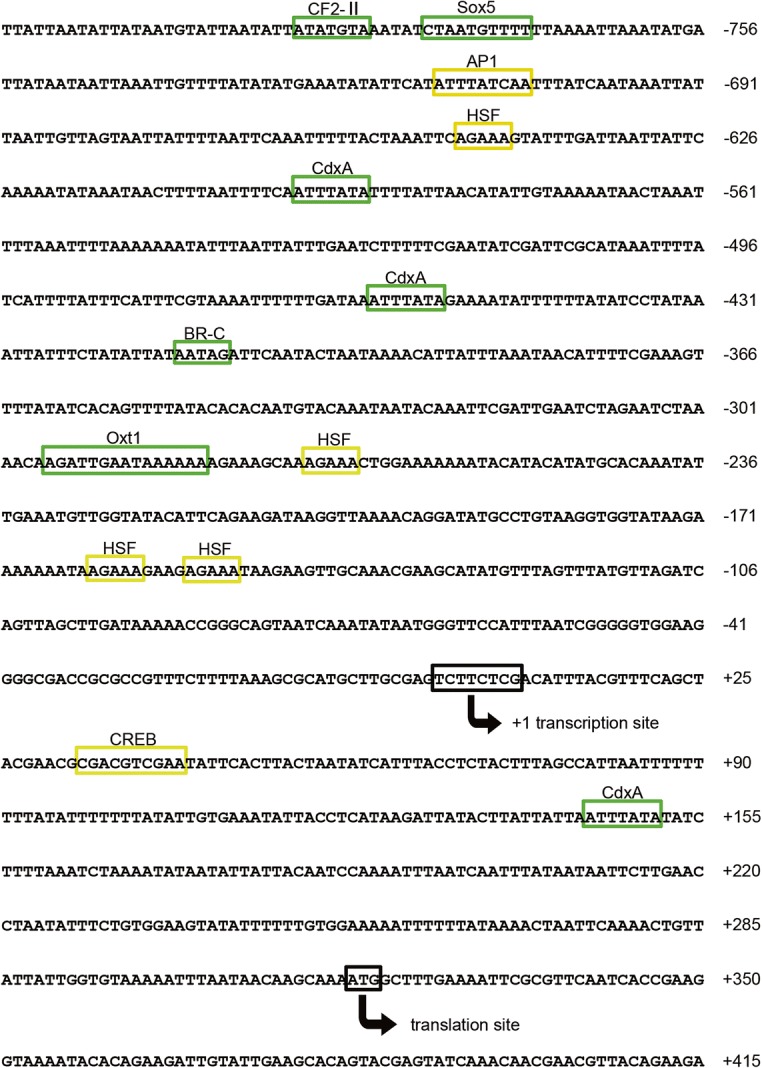
Nucleotide sequence of the 5′-flanking region of AccIPS1-B. The translation and transcription start sites are marked with arrows, and the cis-acting elements are boxed. The cis-acting elements of environmental stress and immune response are marked with yellow boxes, and the cis-acting elements involved in tissue development and early growth stages are marked with green boxes
Temporal and spatial expression of AccIPS1-B
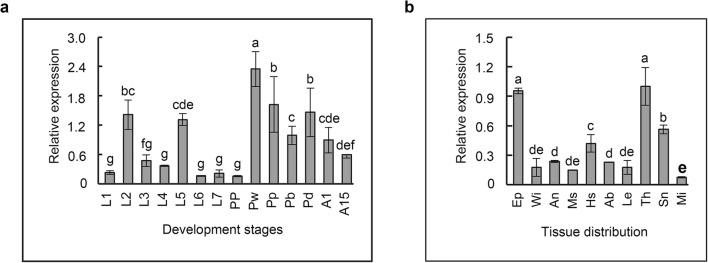
qRT-PCR was used to determine the expression profiles of AccIPS1-B during different developmental stages and in different tissues. As shown in Fig. 5a, AccIPS1-B was expressed at all developmental stages of Apis cerana cerana, and the highest expression level was found at the white eyes stage (Pw). Among the selected tissues, the highest expression of AccIPS1-B was displayed in the thorax (Th), followed by the epidermis (Ep) (Fig. 5b). The above results indicated that AccIPS1-B might play an important role in the development of Apis cerana cerana.
Fig. 5.
The expression profile of AccIPS1-B at different developmental stages and in different tissues. a The expression levels of AccIPS1-B at different developmental stages. The honeybees were larvae (L1–L7, the first to seventh instars), pupae (PP, prepupae; Pw, white eyes; Pp, pink eyes; Pb, brown eyes; and Pd, dark eyes), and adults (A1, 1 day post-emergence; A15, 15 days post-emergence). b The expression levels of AccIPS1-B in different tissues, including epidermis (Ep), wing (Wi), honey sac (Hs), muscle (Ms), leg (Le), abdomen (Ab), thorax (Th), sting needle (Sn), and midgut (Mi). Vertical bars represent the means ± SEM (n = 3). Letters above the bars designate significant differences (P < 0.01)
Expression of AccIPS1-B under abiotic stresses
To elucidate the impact of AccIPS1-B on different abiotic stress responses, its expression profile was evaluated by qRT-PCR. In the present study, the test honeybees were exposed to a variety of adverse environmental stresses, such as cold (4 °C), heat (44 °C), heavy metals (HgCl2 and CdCl2), UV radiation, and pesticides, all of which may cause the formation of ROS. As shown in Fig. 6a, the expression of AccIPS1-B was significantly upregulated under H2O2 treatment and peaked after 2 h. The AccIPS1-B transcription dramatically increased after cold and hot treatment and reached a peak after 1 and 4 h, respectively (Fig. 6b, c). In addition, AccIPS1-B transcription increased significantly under UV treatment, and the expression peak was observed at 3 h (Fig. 6d). Following the HgCl2 treatments, AccIPS1-B expression was significantly upregulated, with the highest levels observed at 1.5 h (Fig. 6e). As shown in Fig. 6f, AccIPS1-B expression under CdCl2 treatment reached a peak after 1 h. Moreover, different pesticides also significantly increased AccIPS1-B expression levels (Fig. 6g–i). All of the above results suggest that AccIPS1-B might be involved in responses to various abiotic stresses.
Fig. 6.
The transcriptional expression profile of AccIPS1-B under different abiotic stresses. Total RNA was extracted from adult bees (15 days) at the specific time points and treated with H2O2 (a), 4 °C (b), 44 °C (c), UV (d), HgCl2 (e), CdCl2 (f), and different pesticides (g–i). β-Actin (GenBank accession number: HM640276) was used as an internal control. Vertical bars represent the means ± SEM (n = 3). Letters above the bars designate significant differences (P < 0.01)
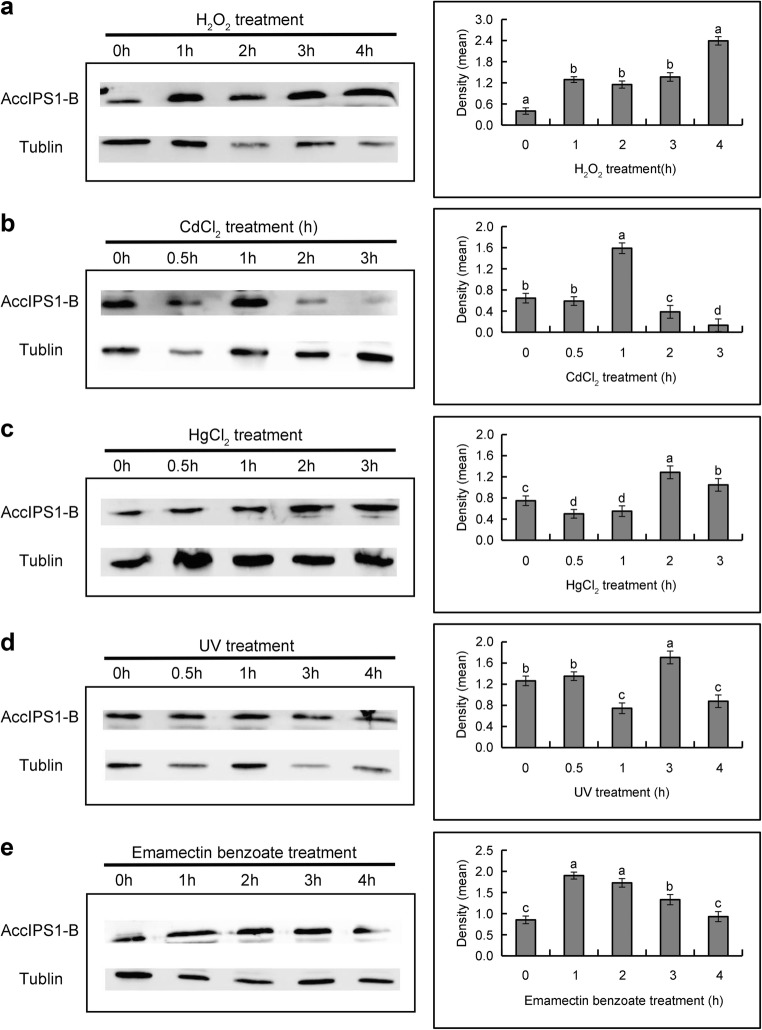
To further examine AccIPS1-B expression under different types of abiotic stresses, western blot analysis was performed as previously described (Chen et al. 2015). Following exposure of bees to UV, H2O2, CdCl2, HgCl2, emamectin benzoate, and lambda cyhalothrin, samples were collected to detect AccIPS1-B expression using anti-AccIPS1-B. The results showed that AccIPS1-B expression was significantly induced under all of the abovementioned treatments (Fig. 7a–e), which indicates that the expression levels of the AccIPS1-B protein and mRNA were consistent.
Fig. 7.
The protein profile of AccIPS1-B under different abiotic stresses was obtained using western blots. Honeybees were exposed to the following treatments: H2O2 (a), CdCl2 (b), HgCl2 (c), UV (d), and emamectin benzoate (e). Anti-α-tubulin antibody was used as an internal control
Characterization of recombinant AccIPS1-B protein
AccIPS1-B was overexpressed in E. coli with several histidine tags after IPTG induction. The recombinant AccIPS1-B protein was separated by SDS-PAGE and showed an apparent molecular mass of 64.17 kDa (containing His-tag) (Fig. 8), which is consistent with the predicted result from DNAMAN.
Fig. 8.
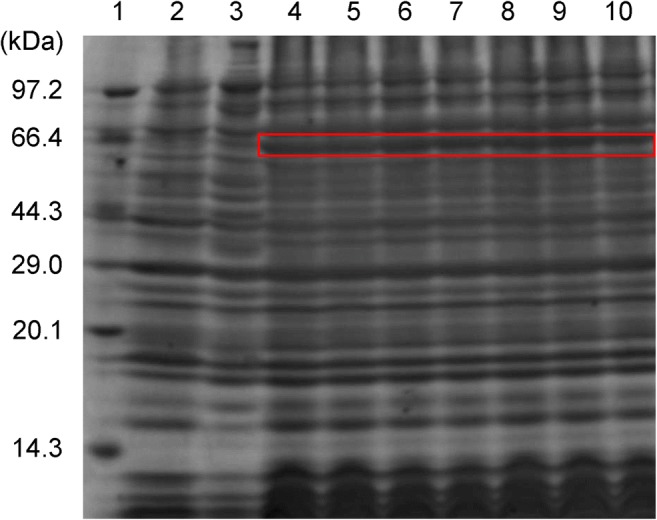
The expression of the recombinant AccIPS1-B protein in E. coli. Recombinant AccIPS1-B was separated by SDS-PAGE. Lane 1, protein molecular weight marker; lane 2, induced expression of pET30a(+) in E. coli BL21; lane 3, uninduced expression of recombinant AccIPS1-B protein; lanes 4–6, expression of recombinant AccIPS1-B protein induced at 37 °C; lanes 7–10, expression of recombinant AccIPS1-B protein induced at 28 °C. The site of recombinant AccIPS1-B is marked with a red box
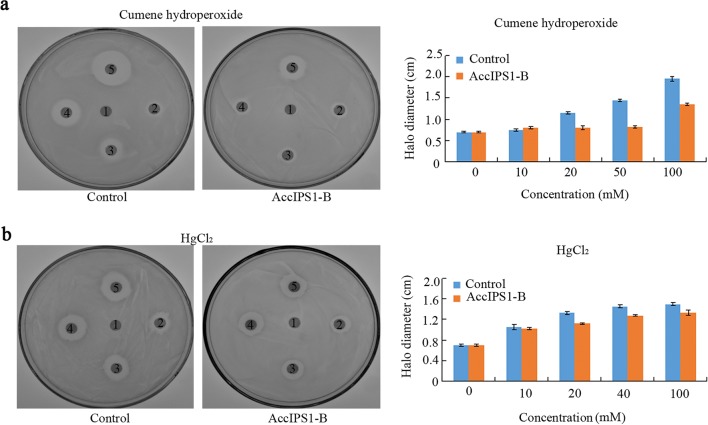
To assess the ability of AccIPS1-B to resist oxidative stress, a disc diffusion test was performed with methods similar to those in a previous report (Jia et al. 2017). Compared with plates containing control bacteria, the halo diameters of the death zones on the plates containing E. coli overexpressing AccIPS1-B were significantly smaller after exposure to different concentrations of cumene hydroperoxide and HgCl2 (Fig. 9). This result indicated that overexpression of AccIPS1-B can improve the tolerance of E. coli to oxidative stress.
Fig. 9.
Resistance of E. coli cells overexpressing AccIPS1-B to cumene hydroperoxide and HgCl2. LB agar plates were inoculated with 5 × 108 cells. E. coli cells were transfected with a plasmid to overexpress AccIPS1-B; bacteria transfected with empty pET-30a (+) vector were used as the negative controls. a The diameter (1, 2, 3, 4, 5) after soaking in different concentrations of cumene hydroperoxide (0, 10, 20, 50, or 100 mM). b The concentrations of HgCl2 were 0, 10, 20, 40, and 100 mM, respectively. After overnight exposure, the diameters of the death zones surrounding the drug-soaked filters were measured. The data were presented in three independent experiments
Knockdown of AccIPS1-B
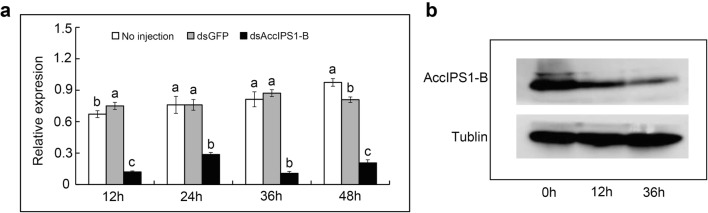
To further confirm the function of AccIPS1-B, AccIPS1-B was knocked down using the RNAi technique. Fifteen-day post-emergence adult workers were microinjected with dsRNA of GFP or AccIPS1-B. The qRT-PCR results showed that the transcript levels of AccIPS1-B after dsAccIPS1-B injection significantly decreased compared to those after dsGFP injection or those with no injection, specifically at 12 h and 36 h (Fig. 10a). Then, honeybees microinjected with dsAccIPS1-B at 12 h and 36 h were selected for protein extraction and western blot analysis. As shown in Fig. 10b, the protein expression levels of AccIPS1-B at 12 and 36 h after injection were clearly lower than those of the control groups. Thus, the results suggest that AccIPS1-B was successfully silenced.
Fig. 10.
Effects of AccIPS1-B RNAi in A. cerana cerana adults. a The transcription levels of AccIPS1-B after RNA interference in adult honeybees (15-day post-emergence workers). b Western blot analysis of the AccIPS1-B protein in control and AccIPS1-B-silenced honeybees. The data presented are from three independent experiments. Vertical bars represent the means ± SEM (n = 3). Letters above the bars designate significant differences (P < 0.01)
Response of AccIPS1-B-silenced honeybees to oxidative stress
To further explore the impact of AccIPS1-B silencing, the transcript levels of various antioxidant genes, including AccSOD1, AccSOD2, AccGSTD, AccGSTO1, AccTpx3, and AccP450, were determined using qRT-PCR at 12 and 36 h after dsAccIPS1-B injection. In addition, the activities of antioxidant enzymes (including SOD, POD, and GST) were measured. As shown in Fig. 11, the transcript levels of various antioxidant genes were significantly lower than those of the control groups. Moreover, the SOD, POD, and GST activities were significantly decreased in AccIPS1-B-silenced honeybees (Fig. 12). The results clearly suggested that AccIPS1-B may help to prevent oxidative damage to proteins and membranes that is commonly caused by abiotic stresses.
Fig. 11.
Expression patterns of antioxidant genes after AccIPS1-B knockdown. aAccSOD1 (GenBank ID: JN700517). bAccSOD2 (GenBank ID: JN637476). cAccGSTD (GenBank ID: JF798572). dAccGSTO1 (GenBank ID: KF496073). eAccP450 (GenBank ID: KC243984). fAccTPX3 (GenBank ID: JX456217). Vertical bars represent the means ± SEM (n = 3). Letters above the bars designate significant differences (P < 0.01)
Fig. 12.
The antioxidant enzymatic activities of SOD (a), POD (b), and GST (c) in AccIPS1-B-silenced honeybees compared to the GFP control. The data presented are from three independent experiments. Letters above the bars designate significant differences (P < 0.01)
Discussion
IPS catalyses the rate-limiting step in the de novo synthesis of inositol, which plays a critical role in the growth and survival of organisms (Frej et al. 2016; Seelan et al. 2011; Li et al. 2016). However, few studies of IPS have been systematically conducted in insects, especially in Apis cerana cerana. In the present study, AccIPS1-B was isolated from Apis cerana cerana, and its response to different oxidative stresses was investigated. In addition, we also examined the expression levels of some antioxidant genes and the activities of several antioxidant enzymes in AccIPS1-B-silenced honeybees. According to the results, we speculate that AccIPS1-B might play important roles in the oxidative stress response in Chinese honeybees.
IPS1 proteins of different species contain four highly conserved regions, including NAD+-binding domains (GWGGNNG), putative catalytic domains (SYNHLGNNDG), conserved region 1 (LWTANTERF), and conserved region 2 (NGSPQNTFVPG) (Frej et al. 2016). In this study, sequence alignment analysis revealed that the AccIPS1-B protein also harboured the four typically conserved domains (Fig. 1a), which suggested that they may have the same functions. In addition, our data showed that AccIPS1-B has the same Ser residue as IPS of other species, including S277 and S355 in NGSPQN and SKSNV, which may relate to the regulation of inositol synthesis (Parthasarathy et al. 2013; Zhai et al. 2016; Deranieh et al. 2013). The phylogenetic tree revealed that AccIPS1-B shared higher homology with AmIPS1-B and AfIPS1-B than with the other proteins (Fig. 2a).
It has previously been reported that IPS is involved in growth and has tissue-specific expression (Kuwano et al. 2010; Luo et al. 2011). In the present study, some predicted transcription factor binding sites related to growth and development, such as CdxA, CF2-II, Oct-1, BR-C, and Sox-5, were found in the 5′ flanking region of AccIPS1-B. Tissue-specific expression analysis indicated that AccIPS1-B was comparatively highly expressed in the epidermis and thorax (Fig. 5b). Previous studies suggested that the epidermis is the first place to resist the adverse external environment and that the thorax is the major movement centre and is required for collecting pollen and clearing combs (Meng et al. 2014; Von et al. 1994). Thus, the specific expression pattern of AccIPS1-B in different tissues may reflect the unique demands of the specific tissue. In addition, the expression of AccIPS1-B was higher in the pupae than in larvae and adults during all development stages, which indicates that AccIPS1-B might play an important role in the early developmental stages.
Honeybees usually suffer from different environmental stresses in their lifetime, including temperature, ultraviolet radiation, heavy metals, and pesticides, all of which may induce the formation of reactive oxygen species (Jia et al. 2014; Lushchak 2010). A previous study indicated that MIPS1 plays important roles in resistance to different environmental stresses (Butzin et al. 2013; Huang and Hernick 2015; Ju et al. 2004). However, the function of AccIPS1-B in response to environmental stresses remains unclear in Apis cerana cerana. In this study, some transcription factors involved in environmental stress and the immune response, including HSFs, AP1, and CREB, were identified from the promoter region of AccIPS1-B, which suggested that AccIPS1-B may have the potential to respond to oxidative stress. Furthermore, qRT-PCR and western blot results showed that the expression of AccIPS1-B could be induced by cold, heat, heavy metals, ultraviolet radiation, and pesticides (Fig. 6). In addition, analysis of the bacteriostatic experiment showed that overexpression of AccIPS1-B in E. coli protected these cells from oxidative stressors, such as HgCl2 and H2O2 (Fig. 9). These results suggest that AccIPS1-B may be involved in resisting oxidative stress.
High ROS concentrations in cells can lead to oxidative damage to nucleic acids, proteins, and lipids (Cadet et al. 2005; Kottuparambil et al. 2012; Ravanat et al. 2001). Cells possess a variety of antioxidant enzymes, including superoxide dismutases (SODs), glutathione S-transferases (GSTs), catalase (CAT), and thioredoxin peroxidase (Tpx), to scavenge ROS (Jia et al. 2014; Tang et al. 2012; Yu et al. 2011). A previous study reported that deletion of the ino-1 gene in the gram-positive Actinobacteria Corynebacterium glutamicum resulted in a decrease in cell viability and an increase in ROS production under various abiotic stresses (Chen et al. 2018), which indicates that IPS may take part in the resistance to ROS damage. In this study, we found that the transcript levels of various antioxidant genes and the enzyme activities of SOD, POD, and GST were significantly decreased when AccIPS1-B was knocked down (Fig. 12). Thus, we speculate that AccIPS1-B may play a critical role in scavenging ROS when Apis cerana cerana encounters different environmental stresses.
In conclusion, we have identified AccIPS1-B from Apis cerana cerana, which possesses the conserved domains of IPS1. The expression pattern of AccIPS1-B under different environmental stresses indicated that AccIPS1-B might participate in the response to antioxidant defence in honeybees. The recombinant AccIPS1-B also enhanced the tolerance to environmental stresses. Finally, the knockdown of AccIPS1-B significantly suppressed the expression of most of the antioxidant genes and decreased the antioxidant enzymatic activities. These findings unequivocally indicated that AccIPS1-B may be involved in the antioxidant defence response.
Funding information
This work was financially supported by the Shandong Province Modern Agricultural Technology System Innovation Team Special Fund (SDAIT-24-04).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xingqi Guo, Phone: +86-538-8242656, Email: xqguo@sdau.edu.cn.
Qinghua Sun, Phone: +86-538-8242656, Email: qhsun@sdau.edu.cn.
References
- Butzin NC, Lapierre P, Green AG, Swithers KS, Gogarten JP, Noll KM. Reconstructed ancestral myo-inositol-3-phosphate synthases indicate that ancestors of the thermococcales and thermotoga species were more thermophilic than their descendants. PLoS One. 2013;8:e84300. doi: 10.1371/journal.pone.0084300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Sage E, Thierry D. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Chen X, Yao P, Chu X, Hao L, Guo X, Xu B. Isolation of arginine kinase from Apis cerana cerana and its possible involvement in response to adverse stress. Cell Stress Chaperones. 2015;20:169–183. doi: 10.1007/s12192-014-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen K, Su T, Zhang B, Li G, Pan J, Si M. Myo-inositol-1-phosphate synthase (Ino-1) functions as a protection mechanism in Corynebacterium glutamicum under oxidative stress. Microbiologyopen. 2018;1:e721. doi: 10.1002/mbo3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Donahue TF, Henry SA. Control of inositol biosynthesis in Saccharomyces cerevisiae: inositol-phosphate synthetase mutants. J Bacteriol. 1976;126(1):243–250. doi: 10.1128/jb.126.1.243-250.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deranieh RM, He Q, Caruso JA, Greenberg ML. Phosphorylation regulates myo-inositol-3-phosphate synthase: a novel regulatory mechanism of inositol biosynthesis. J Bio Chem. 2013;288:26822–26833. doi: 10.1074/jbc.M113.479121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue TF, Henry SA. Inositol mutants of saccharomyces cerevisiae: mapping the ino1 locus and characterizing alleles of the ino1, ino2 and ino4 loci. Genetics. 1981;98(3):491–503. doi: 10.1093/genetics/98.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frej AD, Clark J, Le Roy CI, Lilla S, Thomason PA, Otto GP, ChurchillG IRH, Claus SP, Hawkins P, Stephens L, Williams RS. The inositol-3-phosphate synthase biosynthetic enzyme has distinct catalytic and metabolic roles. Mol Cell Bio. 2016;36:1464–1479. doi: 10.1128/MCB.00039-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hernick M. Recombinant expression of a functional myo-inositol-1-phosphate synthase (MIPS) in Mycobacterium smegmatis. Protein J. 2015;34:380–390. doi: 10.1007/s10930-015-9632-z. [DOI] [PubMed] [Google Scholar]
- Jia H, Sun R, Shi W, Yan Y, Li H, Guo X, Xu B. Characterization of a mitochondrial manganese superoxide dismutase gene from Apis cerana cerana and its role in oxidative stress. J Insect Physio. 2014;60:68–79. doi: 10.1016/j.jinsphys.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Jia H, Ma M, Zhai N, Liu Z, Wang H, Guo X, Xu B. Roles of a mitochondrial AccSCO2 gene from Apis cerana cerana in oxidative stress responses. J Inorg Biochem. 2017;175:9–15. doi: 10.1016/j.jinorgbio.2017.06.015. [DOI] [PubMed] [Google Scholar]
- Ju S, Shaltiel G, Shamir A, Agam G, Greenberg ML. Human 1-D-myo-inositol-3-phosphate synthase is functional in yeast. J Bio Chem. 2004;279:21759–21765. doi: 10.1074/jbc.M312078200. [DOI] [PubMed] [Google Scholar]
- Kaur H, Verma P, Petla BP, Rao V, Saxena SC, Majee M. Ectopic expression of the ABA-inducible dehydration-responsive chickpeal-myo-inositol 1-phosphate synthase 2 (CaMIPS2) in Arabidopsis enhances tolerance to salinity and dehydration stress. Planta. 2013;237:321–335. doi: 10.1007/s00425-012-1781-0. [DOI] [PubMed] [Google Scholar]
- Kottuparambil S, Shin W, Brown MT, Han T. UV-B affects photosynthesis, ROS production and motility of the freshwater flagellate, Euglena agilis Carter. Aquat Toxicol. 2012;122-123:206–213. doi: 10.1016/j.aquatox.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Kusuda H, Koga W, Kusano M, Oikawa A, Saito K, Hirai MY, Yoshida KT. Ectopic expression of myo-inositol 3-phosphate synthase induces a wide range of metabolic changes and confers salt tolerance in rice. Plant Sci. 2015;232:49–56. doi: 10.1016/j.plantsci.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Kuwano M, Mimura T, Takaiwa F, Yoshida KT. Generation of stable ‘low phytic acid’ transgenic rice through antisense repression of the 1D-myo-inositol 3-phosphate synthase gene (RINO1) using the 18-kDa oleosin promoter. Plant Biotechnol J. 2010;7:96–105. doi: 10.1111/j.1467-7652.2008.00375.x. [DOI] [PubMed] [Google Scholar]
- Li T, Sun Y, Ruan Y, Xu L, Hu Y, Hao Z, Zhang X, Li H, Wang Y, Yang L, Chen B. Potential role of D-myo-inositol-3-phosphate synthase and 14-3-3 genes in the crosstalk between Zea mays and Rhizophagus intraradices under drought stress. Mycorrhiza. 2016;26:879–893. doi: 10.1007/s00572-016-0723-2. [DOI] [PubMed] [Google Scholar]
- Li M, Miao L, Chen Y, Hou H, Chen J. Cloning of MIPS gene in large yellow croaker (Larimichthys crocea) and the expression analysis under cold treatments. J Agric Biotechnol. 2018;26:292–301. [Google Scholar]
- Luo Y, Qin G, Zhang J, Liang Y, Song Y, Zhao M, Tsuge T, Aoyama T, Liu J, Gu H, Qu LJ. D-myo-inositol-3-phosphate affects phosphatidylinositol mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell. 2011;23:1352–1372. doi: 10.1105/tpc.111.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak VI. Environmentally induced oxidative stress in aquatic animals. Aquat Toxicol. 2010;101:13–30. doi: 10.1016/j.aquatox.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Majumder AL, Chatterjee A, Ghosh DK, Majee M. Diversification and evolution of L-myo-inositol 1-phosphate synthase. FEBS Lett. 2003;553(1):3–10. doi: 10.1016/S0014-5793(03)00974-8. [DOI] [PubMed] [Google Scholar]
- Meng F, Zhang Y, Liu F, Guo X, Xu B. Characterization and mutational analysis of omega-class GST (GSTO1) from Apis cerana cerana, a gene involved in response to oxidative stress. PLoS One. 2014;9(3):e93100. doi: 10.1371/journal.pone.0093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelette ER, Soares AEE. Characterization of preimaginal developmental stages in Africanized honey bee workers (Apis mellifera L) Apidologie. 1993;24:431–440. doi: 10.1051/apido:19930410. [DOI] [Google Scholar]
- Parthasarathy RN, Lakshmanan J, Thangavel M, Seelan RS, Stagner JI, Janckila AJ, Vadna RE, Casanova MF, Parthasarathy LK. Rat brain myo-inositol 3-phosphate synthase is a phosphoprotein. Mol Cell Biochem. 2013;378:83–89. doi: 10.1007/s11010-013-1597-7. [DOI] [PubMed] [Google Scholar]
- Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B Biol. 2001;63:88–102. doi: 10.1016/S1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- Seelan RS, Pisano MM, Greene RM, Casanova MF, Parthasarathy RN. Differential methylation of the gene encoding myo-inositol 3-phosphate synthase (Isyna1) in rat tissues. Epigenomics. 2011;3:111–124. doi: 10.2217/epi.10.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TK, Major LL. Screening the maybridge rule of 3 fragment library for compounds that interact with the Trypanosoma brucei myo-inositol-3-phosphate synthase and/or show trypanocidal activity. Mol Bio Int. 2011;5:389–364. doi: 10.4061/2011/389364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Wang C, Xiang B, Han R, Guo Z. Hydrogen peroxide and nitric oxide mediated cold-induced and dehydration-induced myo-inositol phosphate synthase that confers multiple resistances to abiotic stresses. Plant Cell Environ. 2013;36:288–299. doi: 10.1111/j.1365-3040.2012.02573.x. [DOI] [PubMed] [Google Scholar]
- Tang T, Huang D, Zhou C, Li X, Xie Q, Liu F. Molecular cloning and expression patterns of copper/zinc superoxide dismutase and manganese superoxide dismutase in Musca domestica. Gene. 2012;505:211–220. doi: 10.1016/j.gene.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Von KL, Crossgrove K, Von SD, Guild GM, Beckendorf SK. The broad-complex directly controls a tissue-specific response to the steroid hormone ecdysone at the onset of Drosophila metamorphosis. EMBO J. 1994;13:3505–3516. doi: 10.1002/j.1460-2075.1994.tb06657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Jia H, Gao H, Guo X, Xu B. Identification, genomic organization, and oxidative stress response of a sigma class glutathione S-transferase gene (AccGSTS1) in the honey bee, Apis cerana cerana. Cell Stress Chaperones. 2013;18:415–426. doi: 10.1007/s12192-012-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Kang M, Meng F, Guo X, Xu B. Molecular cloning and characterization of a thioredoxin peroxidase gene from Apis cerana cerana. Insect Mol Bio. 2011;20:367–378. doi: 10.1111/j.1365-2583.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- Zhai H, Wang F, Si Z, Huo J, Xing L, An Y, He S, Liu Q. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol J. 2016;14:592–602. doi: 10.1111/pbi.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]